Abstract
The stage- and tissue-specific expression of many eukaryotic genes is regulated by cis-regulatory elements, some of which are located in proximity to the start site of transcription whereas others have been identified at considerable distances. In previous studies we have identified far upstream DNase I-hypersensitive sites in the murine α1(I) collagen (Col1a1) gene, which may play a role in the regulation of this abundantly expressed gene. Here we have cloned several of these sites into reporter gene constructs containing the Col1a1 promoter driving the green fluorescent protein (GFP) reporter gene and tested their possible functions in transfection experiments and transgenic mice. In transient and stable transfections none of the hypersensitive sites had a significant effect on Col1a1 promoter activity, indicating that they do not contain a classical transcriptional enhancer. In transgenic animals one element located at −18 to −19.5 kb enhanced the position-independent activity of the linked Col1a1 promoter and may be part of a locus control region. Another element located at −7 to −8 kb specifically enhanced reporter gene expression in the uteri of transgenic mice, suggesting that it contains a novel transcriptional enhancer that may be involved in the regulation of type I collagen expression in tissue remodeling in the uterus during the estrous cycle. Our studies also demonstrate the versatility of the GFP reporter gene for use in transgenic animals because it can be analyzed in live animals, whole mount embryos, histological thin sections, or primary cell cultures, and it can be quantified very sensitively in tissue or cell extracts using a fluorometer.
Keywords: α1(1) collagen, Green fluorescent protein, Transgenic mice, Distal regulatory elements, Enhancer, Uterus
THE collagen superfamily contains at least 19 different collageneous proteins, and several more proteins are known that contain collagen-like domains (22). Type I collagen, a fibrillar collagen, is the most abundant of the collagens and probably the most abundant protein in vertebrates. It has diverse biological functions: it provides tensile strength to connective tissues such as bone, skin, tendons, and ligaments; it forms supporting frameworks of connective tissue in all major internal organs such as liver, spleen, heart, and the vascular system; it promotes cell migration and differentiation during embryonic development; and it is the major substance produced during wound healing and in tissue repair processes. Thus, the Col1a1 and Col1a2 genes, which code for the α1 and α2 subunits of type I collagen, respectively, are expressed during embryonic development, in a variety of cell types, and under various physiological conditions, and their regulation is accordingly complex (4,7,33,38). Moreover, various human disorders are associated with under- or overexpression of type I collagen (22). An elucidation of the complex molecular mechanisms involved in the stage- and tissue-specific regulation of type I collagen expression is therefore not only essential for understanding normal mammalian development and morphogenesis, but also human disease processes.
Numerous studies have addressed the molecular mechanisms involved in the regulation of type I collagen genes in various species, including human, rat, mouse, and chicken. However, despite considerable efforts, many details of type I collagen gene regulation remain elusive. It has generally been reported that the α1 and α2 promoters confer basic tissue-specific expression to reporter gene or minigene constructs in transfection experiments and transgenic animals (25,26,29,33,36). In addition, cis-regulatory elements have been identified that mediate the effect of various modulators of type I collagen expression (17,28,34) or are required for high levels of expression in different collagen-producing cell types (2,29,30).
Several lines of evidence indicate that, in addition to regulatory elements in the proximal promoter, more distal regulatory elements in both 5′- and 3′-flanking region may contribute to the correct stage-and tissue-specific expression of the type I collagen genes. A cluster of DNase I-hypersensitive sites in the distal 5′-flanking sequence of the murine Col1a2 gene has been shown to have transcriptional enhancer activity in transgenic mice (5). A chromatin structure analysis of the murine Col1a1 gene has revealed the presence of several distal 5′ and 3′ DNase I-hypersensitive sites at positions very similar to hypersensitive sites in the homologous human COL1a1 gene (1,31). An E-box in the 3′-flanking region of the Col1a1 gene binds transcription factors USF-1 and USF-2 and stimulates Col1a1 gene transcription (27). The fact that several of the distal 5′-hypersensitive sites are present in collagen-producing but not in nonproducing cells (31) suggests that they function in Col1a1 gene regulation. In a first attempt to elucidate their function we have used reporter gene constructs containing the Col1a1 promoter driving the firefly luciferase or green fluorescent protein (GFP) reporter genes in transient and stable transfection experiments and to generate transgenic mouse lines. We report here that a novel regulatory element located 7–8 kb upstream of the start site of transcription specifically enhances reporter gene expression in the uterus of transgenic animals and that another element located 18–19.5 kb upstream of the start site of transcription enhances position-independent expression of the reporter gene. Moreover, our studies demonstrate the versatility of the GFP reporter gene for use in transgenic animals.
MATERIALS AND METHODS
Reporter Gene Constructs
The construction of the reporter genes used in this study required multiple cloning steps, the details of which have been described (19) and/or will be made available upon request. Briefly, two different sets of reporter genes were used. The first set contained the Col1a1 promoter from −1622 (PstI) to +111 (XbaI), the luciferase reporter gene, and various combinations of upstream DNase-hypersensitive sites. These constructs were made by replacing the SV40 promoter in the plasmid pGL2 (Promega) with the Col1a1 promoter. A unique NotI site was then inserted upstream of the promoter into which NotI cassettes containing the upstream hypersensitive sites were cloned. The second set of reporter genes was constructed by cloning the Col1a1 promoter from −3122 (KpnI) to +111 (XbaI) and the EGFP reporter gene from the plasmid pEGFP-1 (Clontech) into pPCR-Script (Stratagene). This plasmid (pCol9GFP) was then further modified to allow insertion of the NotI cassettes containing the upstream hypersensitive sites into a unique NotI site, and the subsequent removal of all vector sequences using two newly introduced SrfI sites to prepare DNA constructs for the generation of transgenic mice. Various DNA fragments containing the previously described Col1a1 upstream hypersensitive sites (31) were cloned into pBluescript (Stratagene). Second NotI sites were then inserted into the plasmids so that the inserts were flanked by two NotI sites and could be recovered and cloned into the reporter genes as NotI cassettes. Orientation of all inserts and identity of the constructs were verified by multiple restriction enzyme digests and DNA sequence analysis when necessary. The plasmid pSVCol9GFP was made by cloning the Col1a1 promoter from −3122 (KpnI) to +111 (XbaI) into the plasmid pEGFP-1 (Clontech), which contains the SV40 enhancer. A schematic representation of the GFP reporter constructs is shown in Fig. 1. The luciferase reporter constructs were used only in transient transfection assays, the results of which are not shown (see the text), and they are therefore not included in Fig. 1.
FIG. 1.
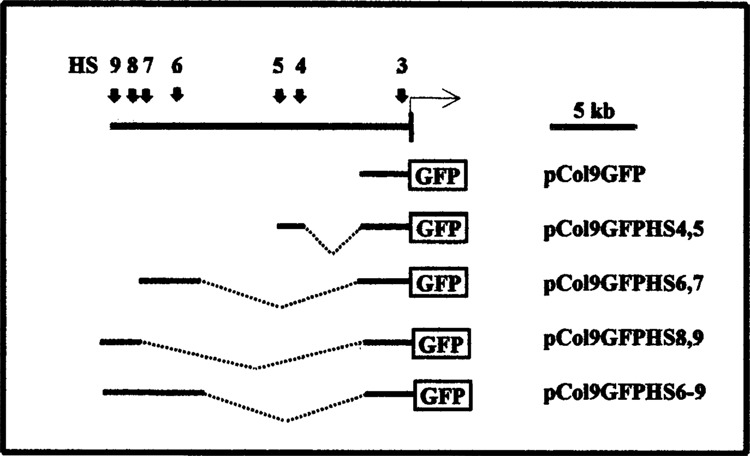
Reporter gene constructs. The vertical arrows in the uppermost line show the location of previously mapped DNase I-hypersensitive sites 3–9 in the 5′-flanking region of the murine Col1a1 gene (6,30). The vertical bar shows part of the first exon and the horizontal arrow the start of transcription. Below the reporter gene constructs used in this study are shown schematically. pCol9GFP contains 3.2 kb of the Col1a1 promoter linked to the EGF reporter gene. The other constructs contain the various hypersensitive sites as indicated.
Transient and Stable Transfections and Reporter Gene Assays
Transient and stable transfections and luciferase reporter gene assays were performed as previously described (24,25). GFP reporter gene expression in transfected cells was analyzed by visual inspection using an Olympus inverted fluorescent microscope, by determining fluorescence in cell extracts using a TD700 fluorometer, or by fluorescence-activated cell sorting using a FACScan (Becton-Dickinson). Cell extracts for fluorescence measurements were prepared as described below for tissue extracts. For fluorescence-activated cell sorting the cells were trypsinized and resuspended in PBS.
Generation of Transgenic Mice
Transgenic mice were generated at the UNC Transgenic Facility. DNA sequences containing the various reporter gene constructs were isolated from vector sequences by gel purification and injected into single-cell mouse embryos. The embryos were then reimplanted into pseudopregnant C3H × C57B1 foster mothers. At approximately 3 weeks of age tail clippings were obtained from the pups and analyzed for GFP expression using a fluorescent microscope. Subsequently, DNA was extracted from the tail clippings and presence of the transgene was determined using PCR and GFP-specific primers. Transgene copy numbers were determined by Southern blot analysis of liver DNA using standard procedures.
GFP Expression in Whole Mount Embryos, Tissues, and Primary Fibroblast Cultures From Transgenic Mice
Embryos were collected at different stages of development, day 1 being the day after the vaginal plug. The embryos were fixed in PBS containing 4% paraformaldehyde and 0.2% Tween 20 and stored at 4°C. Fluorescence was stable under these conditions for many months. For GFP analysis in tissue thin sections of the tissues were fixed for 24 h at 4°C in PBS containing 4% paraformaldehyde. Blocks of tissues were mounted in O.C.T. compound (Lab-Tek) on specimen holders, frozen in liquid nitrogen, and cut into thin sections using a cryotome. Sections were mounted on Superfrost/Plus microscope slides (Fisher), stained with DAPI for 30 s, covered with FluorSave reagent (Calbiochem), and analyzed under a fluorescent microscope using DAPI and GFP-specific filter sets. To analyze transgene expression in tissues animals were sacrificed at between 4 and 20 weeks of age and tissues harvested and homogenized with a polytron homogenizer in 1 ml PBS containing 0.5% Triton X100, 10 μg/ml leupeptin, 20 μg/ml aprotinin, and 0.1 mM PMSF. Homogenates were microcentrifuged for 20 min and the supernatants removed to determine protein concentrations using a BCA protein assay reagent (Pierce) and fluorescence using a TD700 fluorometer. To prepare primary fibroblast cultures small pieces of skin were obtained from the ears of mice, sterilized in 70% ethanol, washed in PBS, and minced into small pieces with a scalpel. The pieces were then incubated in 200 U/ml collagenase at 37°C for 24 h and subsequently in trypsin at 37°C for 30 min, and cells were collected by centrifugation and grown in minimal essential medium containing 10% fetal calf serum. In later experiments cells from ear biopsies were prepared using collagenase/dispase (Boehringer) as recommended by the supplier.
RESULTS
The Col1a1 Upstream DNase-Hypersensitive Sites Have No Significant Effect on Col1a1 Promoter Activity in Transient or Stable Transfections
We have recently identified a series of distal DNase-hypersensitive sites in the 5′-flanking region of the murine Col1a1 gene (31) (Fig. 1). The fact that most of these sites were found in collagen-producing 3T3 fibroblasts and osteoblasts but not in nonproducing WEHI 3B myelomonocytic leukemia cells strongly suggests that they function in the regulation of Col1a1 gene expression. To analyze their possible functions we have constructed a series of reporter genes containing the Col1a1 promoter from −1622 to +111, the luciferase reporter gene, and the individual DNase-hypersensitive sites 4, 5, 6, 7, and 8 in both orientations as well as DNase-hypersensitive sites 4+5 and 6+7 in both orientations. The reporter genes were transiently transfected into NIH 3T3 fibroblasts and luciferase activity was determined after 48 h, normalized to transfection efficiency using a cotransfected β-gal reporter plasmid as described (24), and compared to luciferase activity of a reporter gene containing the Col1a1 promoter only. None of the DNase-hypersensitive sites tested in this series of experiments had a significant effect on Col1a1 promoter activity; HS 6 showed an approximately twofold reduction and HS 8 an approximately twofold stimulation of reporter gene expression (data not shown).
Some regulatory elements exert their effect through an alteration of chromatin structure and can therefore only be detected in stable transfections after integration into the host cell DNA. Other elements regulate gene expression in very specialized tissues or cell types and cannot be detected in transfection experiments at all, but require introduction into transgenic animals. We therefore wished to analyze the Col1a1 upstream hypersensitive sites in stable transfections and transgenic animals. However, after the construction of the luciferase reporter genes used in the experiments described in the previous paragraph, new regulatory elements important for expression in some tissues in transgenic animals were discovered in the Col1a1 promoter between −900 and −3200 (29). We therefore decided to construct a new series of reporter genes that included these additional promoter sequences for use in stable transfections and transgenic mice. These elements have no effect on Col1a1 promoter activity in skin fibroblasts (29) and therefore should not have affected the results of the transient transfection experiments, which were performed in fibroblasts. We also took advantage of the availability of the EGFP reporter gene, which is optimized for expression in mammalian cells. We thus constructed the reporter gene pCol9GFP, which contains the Col1a1 promoter from −3122 to +111 and the EGFP reporter gene and several derivatives containing the various upstream DNase-hypersensitive sites as shown in Fig. 1, or the SV40 enhancer, and performed transient and stable transfection experiments. As for the luciferase reporter constructs described in the previous paragraph, none of the upstream DNase-hypersensitive sites showed a significant effect on Col1a1 promoter activity in transient transfections (data not shown). When the constructs were stably transfected into NIH 3T3 cells and GFP expression analyzed by FACS analysis we found that the SV40 enhancer had a slightly stimulating effect on Col1a1 promoter activity whereas the various upstream DNase-hypersensitive sites showed a slightly inhibitory effect on the level of GFP expression and little or no effect on the number of GFP-expressing cells (Fig. 2).
FIG. 2.
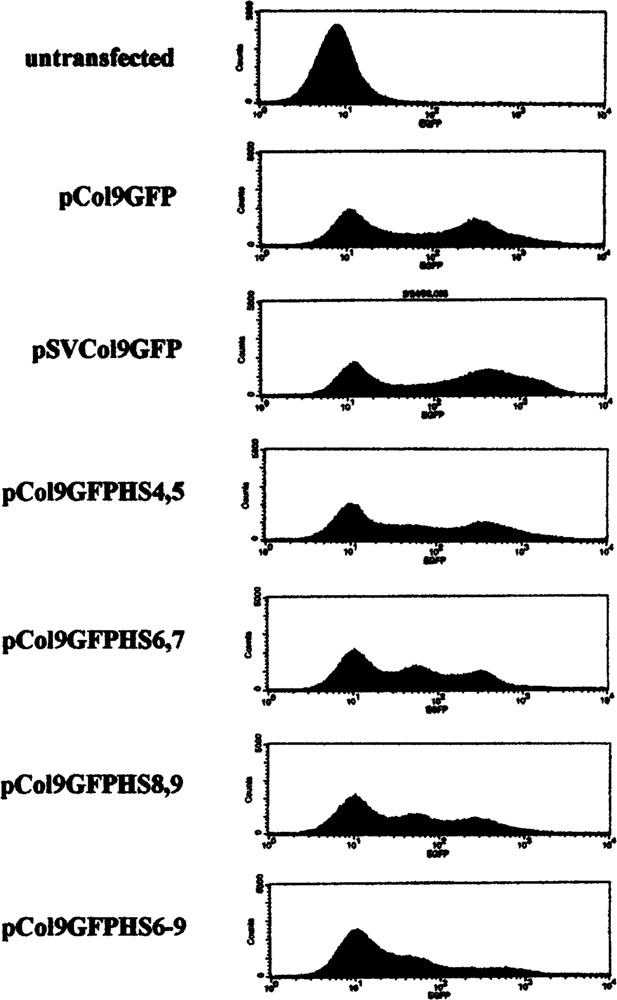
Upstream hypersensitive sites show marginal effect on Col1a1 promoter activity in stable transfections. The indicated reporter constructs were stably transfected into NIH 3T43 fibroblasts and GFP expression analyzed by FACS and compared to untransfected cells as described in Materials and Methods.
Generation of Transgenic Mice and Analysis of Transgene-Expressing Animals
We chose three of the reporter constructs for initial introduction into transgenic mice: 1) the promoter-only construct pCol9GFP as a control; 2) pCol9GFPHS-4,5 because we had preliminary evidence that DNase-hypersensitive sites 4 and/or 5 may contain a transcriptional enhancer; and 3) pCol9GFP-HS8,9 because HS 8 contains a topoisomerase II cleavage site and HS 9 a nuclear matrix attachment region (MAR), and these sites therefore resemble chromatin domain border elements or locus control regions (LCRs; a manuscript describing these findings is in preparation). Multiple transgenic mouse lines were established for each of the constructs and GFP expression was analyzed during embryonic development and in adult animals. Transgene-positive founder animals were identified by PCR amplification of GFP sequences in tail DNA, and transgene-expressing founders and offspring were identified by inspection of GFP expression in tail clippings, toe clippings, or whole live animals under a fluorescent stereomicroscope (in some very highly expressing animals GFP was detectable as a greenish stain with the naked eye without fluorescent light). Expression in the tail was higher than in any other tissue in all founders with all three constructs and in all offspring analyzed (except in the uteri of some animals containing pCol9GFPHS-4,5; see below). Moreover, an analysis of a large number of PCR-positive but tail expression-negative animals revealed no indication of aberrant transgene expression in any of the lines (i.e., no animal lacking microscopically detectable fluorescence in the tail expressed the transgenes in other tissues when assayed by the very sensitive fluorimetric analysis of tissue extracts). Thus, fluorescence in the tail was a very reliable indicator of transgene expression for all three constructs.
Transgene copy numbers in expressing lines were determined by Southern blot analysis of liver DNA. Different lines harbored between 1 and >100 transgene copies (Table 1), and in most lines the Southern blot hybridization patterns were consistent with multiple transgene copies arranged in a head-to-tail fashion at a single integration site (data not shown).
TABLE 1.
COPY NUMBER AND GFP EXPRESSION IN TRANSGENIC MOUSE LINES HARBORING DIFFERENT REPORTER CONSTRUCTS
| Mouse Line | rel GFP* | % Cells† | Copy No. | GFP/Copy |
|---|---|---|---|---|
| pCol9GFP | ||||
| 2111 | 5.1 | 12 | 4 | 1.3 |
| 2116 | 10.7 | 17 | 2 | 5.3 |
| 2118 | 28.6 | 14 | 10 | 2.9 |
| 2125 | 11.5 | 35 | 2 | 5.8 |
| 2131 | 9.0 | 15 | 2 | 4.5 |
| 2132 | 4.2 | 3 | 6 | 7.0 |
| pCol9GFP-HS4.5 | ||||
| 2018 | 83.9 | 73 | 7 | 12.0 |
| 2023 | 11.7 | 8 | ∼35 | 0.3 |
| 2024 | 8.2 | 60 | 1 | 8.2 |
| 2025 | 18.3 | 35 | 6 | 3.1 |
| 2027 | 17.7 | 37 | <100 | 0.2 |
| 2031 | 3.4 | 15 | 20 | 0.2 |
| 2033 | 100 | 90 | 50 | 2.0 |
| 2036 | 93.3 | 77 | 50 | 1.9 |
| 2048 | 31.4 | 37 | 10 | 3.1 |
| 2052 | 3.0 | 0 | 4 | 0.8 |
| pCol9GFP-HS8.9 | ||||
| 2063 | 14.0 | 52 | 1 | 14.0 |
| 2606 | 11.0 | 30 | 14 | 0.8 |
| 2608 | 6.7 | 0 | 2 | 3.4 |
| 2610 | 40.6 | 31 | 10 | 4.1 |
| 2617 | 50.0 | 50 | 5 | 10.0 |
| 2623 | 23.3 | 43 | 3 | 7.8 |
| 2624 | 0.9 | 0 | 6 | 0.2 |
GFP expression was measured in tail extracts from each individual founder animal and normalized to expression in the highest expression 2033 (100%).
Percent of GFP-expressing cells in primary dermal fibroblast cultures from each of the founders.
Upstream Hypersensitive Sites Enhance Position-Independent Activity of the Col1a1 Promoter
We compared GFP expression in the tail of all founders at similar ages and found that expression in the different lines varied ∼100-fold (Table 1). The relative level of expression in a given line was stable (i.e., was very similar in several generations of offspring) (data not shown). We found that the level of GFP expression showed a good correlation to the proportion of cells in primary dermal fibroblasts that express the transgene (Table 1, see below). There was no classical enhancement of expression by HS4,5 or HS8,9 (Table 1) and GFP expression per copy number varied greatly in the different lines with each of the constructs (i.e., there was no copy number dependence of expression). There was, however, a significant increase in the position-independent transgene expression in mouse lines harboring the construct pCol9GFP-HS8,9 (Fig. 3). While construct pCol9GFP was expressed in 16.7% (6/36) and pCol9GFP-HS4,5 in 31.3% (10/31) of the PCR-positive founders, construct pCol9GFP-HS8,9 was expressed in 66.7% (8/12) of the PCR-positive founders.
FIG. 3.
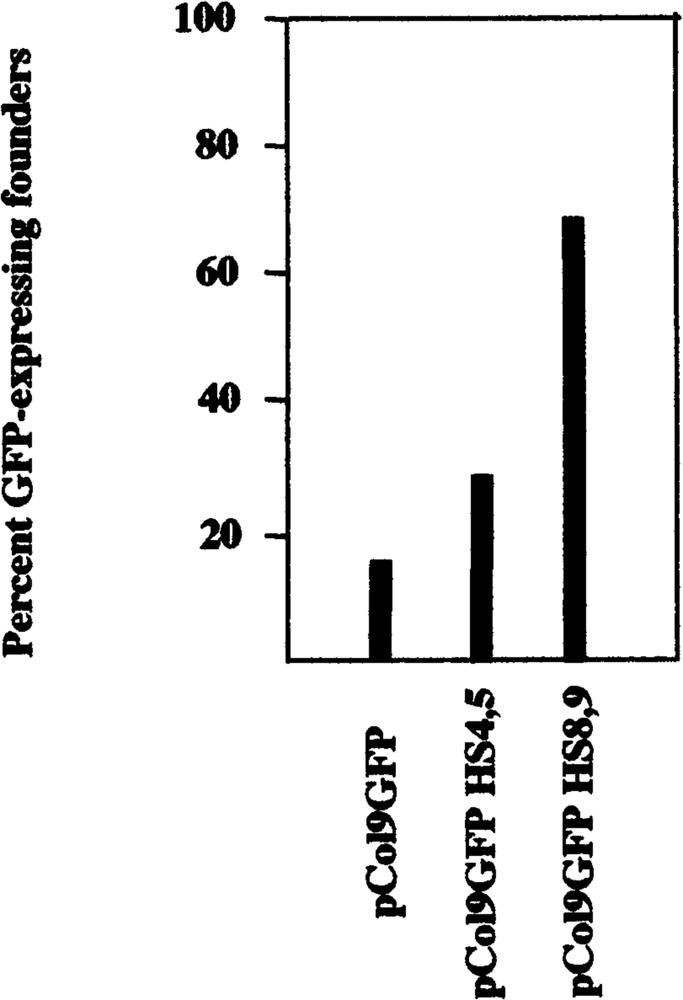
Upstream hypersensitive site HS8,9 significantly enhanced position-independent transgene expression. Transgene-containing animals were identified by PCR amplification of GFP sequences in tail DNA and transgene-expressing animals by fluorescence analysis in tail and other tissues as described in the text.
The Col1a1 Promoter Directs Basal Stage-and Tissue-Specific Expression of the GFP Reporter Gene
A typical example of a transgenic embryo and a negative littermate at day 18 of embryonic development is shown in Fig. 4. High levels of expression can be seen in tendon (tail), bone (calvarium, ossification centers in the digits), and skin. Expression patterns at this level of analysis were undistinguishable in embryos containing the different constructs (compare the animal in Fig. 4 containing pCol9GFPHS-4,5 with those in Fig. 5D containing pCol9GFP and Fig. 5E containing pCol9GFPHS-8,9). Transgene expression at different stages of development is shown in Fig. 5. Expression was detectable throughout embryos at days 12 and 14 of development (Fig. 5A, B). At day 16 the highest expression was seen in bone (Fig. 5C) and at days 17 and 18 in tail, skin, and bone (Fig. 5D, E). Because several previous studies have shown that reporter genes driven by type I collagen promoters are expressed in a correct stage- and tissue-specific manner (3,25,29,30,35,36), and the results shown here confirm these earlier reports, we have so far not analyzed expression of our constructs during embryonic development in any more details than shown here.
FIG. 4.
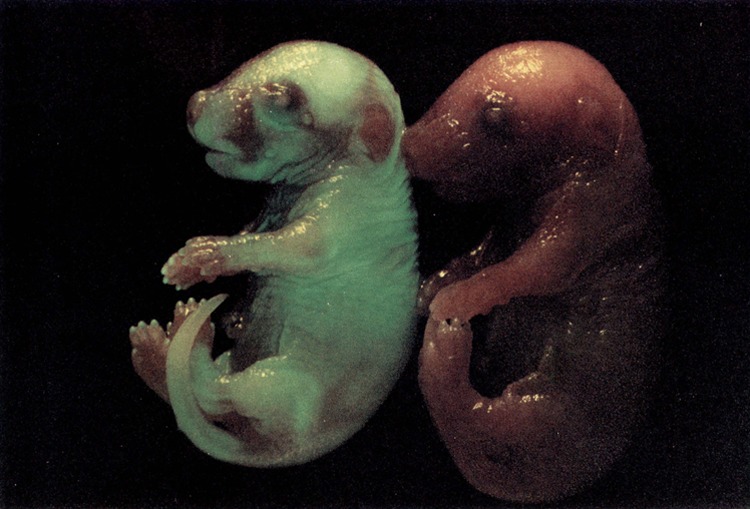
GFP expression in a whole mount embryo containing the reporter gene pCol9GFP-HS4,5 and a negative littermate at day 18 of embryonic development.
FIG. 5.
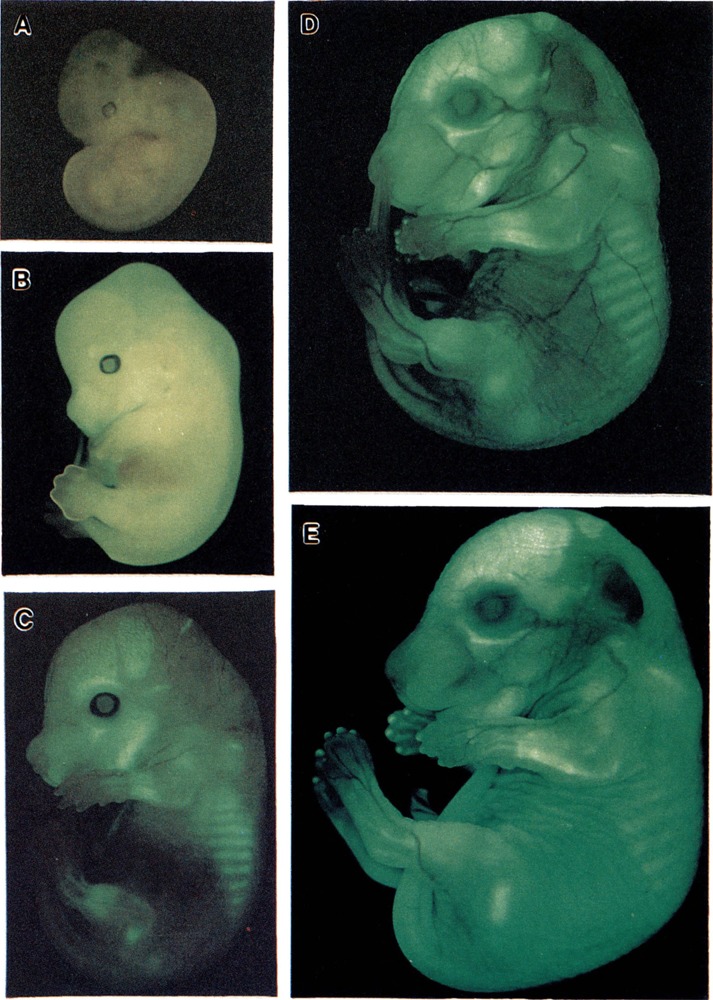
GFP expression in whole mount embryos containing the different reporter genes (see the text) at various stages of embryonic development. (A) day 12, (B) day 14, (C) day 16, (D) day 17, (E) day 18 (p.c).
Expression of the three reporter constructs was also analyzed in adult mice, and our results (Table 2) confirm previous reports that the Col1a1 promoter is sufficient for correct tissue-specific activity in adult animals (29,30,35,36). Highest expression of all constructs was seen in the tail, followed by skin and bone; all major internal organs expressed at much lower, although clearly detectable, levels. Again, there were no significant differences in the relative levels of expression of the three different constructs in different tissues except for the significant higher levels of GFP in the uteri of mice containing the construct pCol9GFPHS-4,5 (see next paragraph).
TABLE 2.
GFP EXPRESSION IN TISSUE EXTRACTS OF ADULT TRANSGENIC MICE HARBORING DIFFERENT REPORTER CONSTRUCTS
| Construct | pCol9GFP | pCol9GFP-HS4,5 | PCol9GFP-HS8.9 |
|---|---|---|---|
| Tail | 100 | 100 | 100 |
| Bone | 16.9 (12.8) | 23.5 (7.7) | 21.7 (7.3) |
| Skin | 10.5 (7.8) | 41.8(15.2) | 33.8 (14.1) |
| Muscle | 0.7 (0.3) | 1.7 (0.8) | 1.3 0.4) |
| Heart | 0.3 (0.2) | 0.7 (0.5) | 0.8 (1.4) |
| Spleen | 0.3 (0.1) | 0.7 (0.3) | 0.2 (0.2) |
| Kidney | 0.3 (0.6) | 1.1 (0.9) | 1.8 (1.6) |
| Liver | 0.3 (0.2) | 0.2 (0.1) | 0.1 (0.0) |
| Lung | 0.1 (0.1) | 1.3 (0.8) | 0.2 (0.2) |
| Brain | 0.3 (0.0) | 0.6 (0.4) | 0.4 (0.2) |
| Thymus | n.d. | n.d. | 0.3 (0.1) |
| Uterus | 9.6 (5.1) | 91.0(41.3) | 13.0(19.8) |
GFP expression was measured in tissue extracts from one or several offspring of each of the founder animals and normalized to expression in the tail (100%). For each tissue the mean relative expression is shown with the SD in parentheses, n.d. = not determined.
Upstream DNase-Hypersensitive Sites 4,5 Specifically Enhance Col1a1 Promoter Activity in the Uteri of Transgenic Mice
Female mice containing the construct pCol9GFP-HS4,5 showed a significant enhancement of GFP expression in the uterus relative to GFP expression in the tail (91.0%) compared to mice containing the promoter-only construct pCol9GFP (9.6%; Table 2, Fig. 6). The level of expression in uterus in these mice exceeded that in skin and bone of the same mice and was seen in offspring of all the different founders (i.e., was not position dependent). Moreover, no enhancement of uterus-specific expression was seen in mice containing construct pCol9GFP-HS-8,9 (13.0%, Table 1, Fig. 6), and the enhancement was thus specific for pCol9GFP-HS4,5. These results suggest that DNase-hypersensitive sites 4 and/or 5 contain a novel transcriptional enhancer that may be involved in the regulation of type I collagen expression in tissue remodeling in the uterus during the estrous cycle.
FIG. 6.
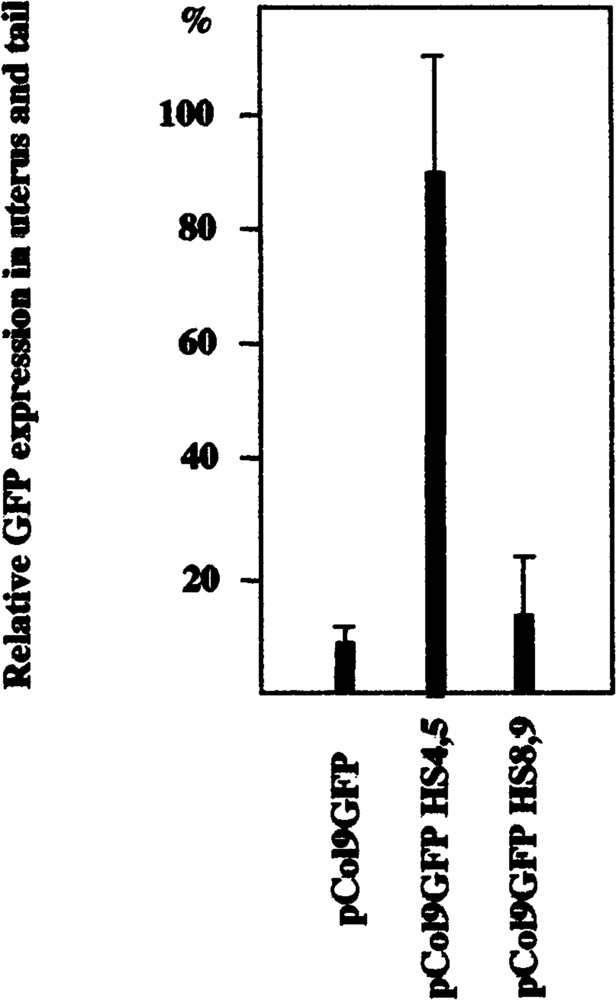
HS4,5 enhanced GFP expression in uterus of transgenic mice. Extracts were prepared from tails and uteri of transgenic animals and GFP expression determined by fluorometry as described in Materials and Methods. The data are derived from comparing GFP expression in age-matched offspring from all six founders containing construct pCol9GFP with offspring from seven founders containing pCol9GFP-HS4,5 and offspring from all seven founders containing pCol9GFP-HS8,9 and are shown as expression in uterus in percent of expression in tail of the same animal. The enhanced uterus-specific expression in animals containing HS4,5 is statistically highly significant (p < 0.001).
To determine which uterine cells expressed the GFP reporter gene we prepared thin sections from the uteri of sexually mature female transgenic mice and analyzed them by fluorescent microscopy. A typical example of GFP expression in the uterus of a mouse containing pCol9GFP-HS4,5 is shown in Fig. 7. High levels of GFP were detectable in endometrial stromal cells and the myometrium, but not in luminal epithelial cells or uterine glands. Similar expression patterns were observed in the uteri of transgenic mice containing the other constructs, although expression was less intense (not shown). We do not know at what stage of the estrous cycle the mice were at the time of analysis and how GFP expression might change throughout the estrous cycle. We are currently performing more detailed analyses of the effect of female sex steroid hormones on the expression of the different reporter constructs.
FIG. 7.
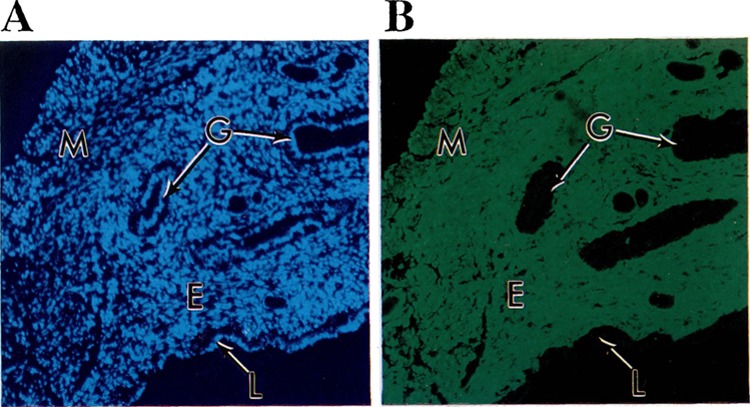
GFP expression in the uterus of a transgenic animal. Thin sections of the uterus of an offspring of founder 2033 containing the construct pCol9GFP-HS4,5 were prepared as described in Materials and Methods and analyzed under the fluorescent microscope with filters specific for DAPI stain (A) or GFP (B). Strong fluorescence can be seen in the endometrium (E) and myometrium (M), but not in the epithelial cells lining the uterine lumen (L) or uterine glands (G).
Different Levels of Transgene Expression Result From Different Degrees of Variegation
Different levels of transgene expression in different cell or mouse lines can result either from different levels of expression in every cell of a given population or from different proportions of cells within a given population expressing the transgene (variegated expression). Several cis-regulatory elements have been described that prevent variegated expression (12,37,39). In order to find out what determines the different expression levels of our constructs in the different lines (which was not related to transgene copy number, Table 1) and to examine whether the DNase-hypersensitive sites analyzed here had any effect on variegated expression of the linked Col1a1 promoter, we prepared primary fibroblast cultures from each of the founders and quantified the proportions of GFP-expressing cells for each line. The results are shown in Table 1 and Fig. 8. With some exceptions, there was a good correlation between the relative fluorescence in tail extracts and the number of GFP-positive cells, indicating that the level of transgene expression in most of the lines is determined primarily by variegation. Moreover, this correlation was seen with all three constructs used (i.e., none of the DNase-hypersensitive sites analyzed affected variegated expression of the linked transgene).
FIG. 8.
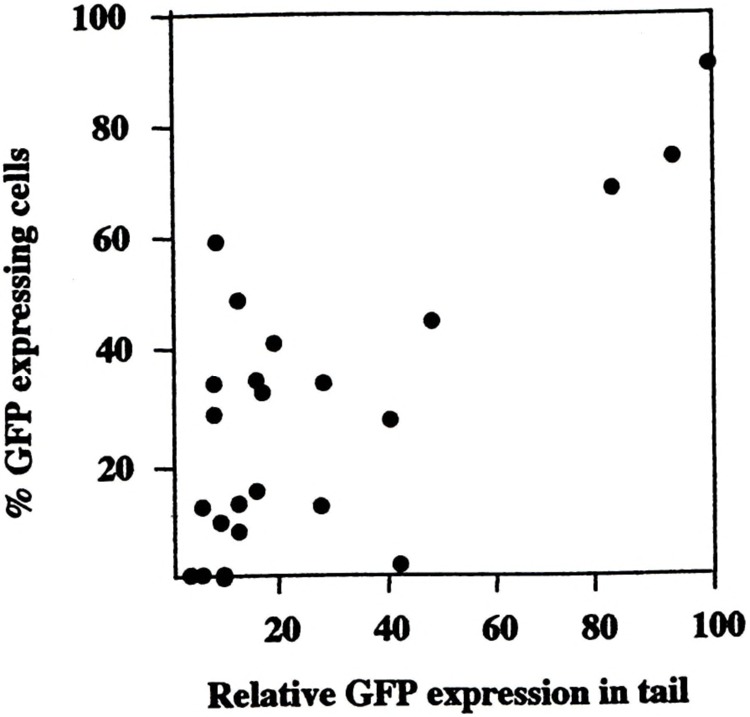
Correlation between relative GFP expression in the tails of transgenic animals and percentage of GFP-expressing cells in primary dermal fibroblast cultures. Primary dermal fibroblast cultures were established from each of the founders as described in Materials and Methods and the number of expressing cells determined by microscopy and plotted against the relative level of GFP expression in the tails of each animal.
DISCUSSION
The expression of eukaryotic genes is regulated by the concerted actions of proximal and distal cis-regulatory elements. While the proximal elements interact directly with the basic transcription machinery and trans-activating factors, distal elements are thought to exert their effects by establishing and maintaining a transcription-competent chromatin environment. A complete understanding of the stage- and tissue-specific regulation of a particular gene requires knowledge of the functions, modes of action of, and interaction between both types of elements. In the work described here we have analyzed the role of far upstream DNA sequences in the regulation of the murine Col1a1 gene expression. These sequences are candidates for cis-regulatory elements because they were identified as DNase-hypersensitive sites in chromatin from collagen-expressing but not from nonexpressing cells [except HS 8 (31)]. We have now inserted several of these upstream hypersensitive sites individually or in combinations into reporter gene constructs and have performed transient and stable transfections and generated transgenic mice. In a series of transient transfection experiments using the luciferase reporter gene driven by ∼1.6 kb of the Col1a1 promoter we found no significant effect of these elements on Col1a1 promoter activity (data not shown). We then inserted several combinations of hypersensitive sites into construct pCol9GFP, which contains 3.2 kb of Col1a1 promoter sequence (Fig. 1) and thus all previously identified Col1a1 promoter regulatory elements (29). In stable transfections most of the constructs showed somewhat reduced levels of expression compared to pCol9GFP, but the numbers of expressing cells remained similar (Fig. 2). This indicates that none of the upstream hypersensitive sites tested shows classical enhancer activity, which would have either increased levels of Col1a1 promoter activity or the number of transfected cells expressing the constructs. Similarly, the hypersensitive site-containing constructs analyzed so far in transgenic mice (HS4,5 and HS8,9) are on the average not expressed at higher levels than the construct containing the promoter only (i.e., they show no enhancer activity) (Table 1). The Col1a1 gene thus may differ from the Col1a2 gene, which has recently been shown to have a potent far upstream enhancer located between 13.5 and 19.5 kb 5′ of the Col1a1 start site of transcription (5) (i.e., at a similar location as the hypersensitive sites 6–9 tested here). However, the Col1a2 enhancer was tested in transgenic animals and it was not reported whether it is active in stable transfection assays. Therefore, it remains possible, although unlikely considering the results of the transfection experiments (Fig. 2), that hypersensitive sites 6 and/or 7 or a combination of sites 6 through 9 contain enhancer activity in transgenic mice. We are currently generating additional strains of transgenic mice to test this. Other possibilities are that a Col1a1 enhancer is located elsewhere (further upstream or in the 3′-flanking region) or that the Col1a1 gene does not contain a classical transcriptional enhancer.
We did, however, observe a significant increase in the proportion of transgene-positive animals expressing the construct containing hypersensitive sites 8 and 9 (Fig. 3). This indicates that in vivo these sites at least in part shield the linked Col1a1 promoter from position effects. We have recently found that these sites contain an in vivo topoisomerase II cleavage site and a nuclear matrix attachment region and could therefore constitute the locus control region (LCR) of the Col1a1 domain (manuscript in preparation). LCRs are modular cis-regulatory elements, which are thought to be necessary for a position-independent, copy number-dependent, stage- and tissue-specific, and high-level expression of a linked transgene (14,18). However, a recent report that the much studied β-globin LCR is not necessary for an open chromatin structure or developmentally regulated transcription (11) raises new questions about the exact nature and function of LCRs. In many cases LCRs are composed of multiple DNase-hypersensititive sites with different functions (enhancers, insulaters, chromatin opening elements, nuclear matrix attachment regions) that act in a synergistic manner. Because position independence of pCol9GFP-HS8,9 was significantly enhanced, although not to 100%, and the construct was not expressed in a copy number-dependent fashion (Table 1), the most likely interpretation is that hypersensitive sites 8 and 9 are part of an LCR and need additional sequences for full activity. While HS 6 and 7 are good candidates for being such additionally required sequences, their presence in reporter constructs did not have the expected effect in stable transfections (Fig. 2). This may suggest that other, so far unidentified, sequences located further upstream may be required for full LCR function. We are currently pursuing these questions.
Expression of a gene or transgene is said to be variegated when it is expressed only in a certain proportion of cells in a population of a given cell type, and the degree of variegation is position dependent and is thought to spread from regions of heterochromatin. In several instances cis-regulatory elements (enhancers, LCRs) have been shown to function by overcoming such heterochromatin-mediated position effect variegation (12,37,39). With some exceptions we have observed a good correlation between the relative intensity of GFP expression in tail and other tissues and the number of GFP-expressing cells in primary fibroblast cultures (Table 1, Fig. 8), indicating that the overall level of transgene expression in most strains is determined by different degrees of variegation. The Col1a1 upstream regulatory elements studied here, notably HS8,9, which may be part of a Col1a1 LCR (see above), were not able to prevent variegation. This confirms that they do not constitute an enhancer of the type that can overcome variegated expression (37,39) or a fully functional LCR. Moreover, variegation showed a tendency to increase with age of the animals (data not shown), an observation also reported by others (37). The phenomenon of variegated expression is of interest because it is possible that the regulation not only of transgenes, but also of endogenous tissue-specific genes, occurs through this mode (i.e., the number of cells in a given tissue modulating expression of a gene in response to physiological stimuli may vary rather than the level at which the gene is expressed in every cell). Similarly, it has been proposed that variegation may be involved in the acquisition and maintenance of particular cellular phenotypes (lineage commitment) during differentiation (12,18). The transgenic mice described in this article allow us to address these interesting questions.
The most intriguing result of this study was the finding that the construct pCol9GFP-HS4,5 was expressed at approximately 10-fold higher levels in the uteri of transgenic mice than the other constructs (Table 2, Fig. 6). This suggests that hypersensitive sites 4 and/or 5 contain a tissue-specific enhancer that may be involved in the steroid hormone-dependent stromal cell proliferation and extracellular collagen deposition during the estrous cycle in the uterus and possibly in other organs or tissues. During the proestrous phase of the estrous cycle the ovarian hormones estrogen and progesterone stimulate growth and proliferation of the endometrium, which decreases in size in the postestrous phase and is sloughed unless implantation occurs. Thus, there is a constant remodeling of connective tissue, blood vessels, glands, and other parts of the uterus throughout the estrous cycle. The transgenic mice analyzed in this study presumably were in various stages of the estrous cycle and were synthesizing connective tissue at widely varying rates at the times of analysis. That would explain the large variation in GFP expression observed in the uteri (Table 2). Estrogen is known to affect type I collagen synthesis in the uterus (10,21), osteoblasts and osteosarcoma cells (20,23), and other tissues (8,10,13). Estrogen also induces expression of the progesterone receptor, and progesterone plays a role in the proliferation and differentiation of the endometrial stroma (15). It is not known whether this effect of estrogen or progesterone on collagen synthesis is direct or requires other mediators. The DNA sequences requirements for estrogen receptor binding to estrogen response elements have been very well defined (9), while progesterone response elements are less well characterized (15). A sequence analysis of 1.3 kb of DNA containing hypersensitive sites 4 and 5 has not revealed the presence of estrogen or progesterone response elements (unpublished sequence data). We are currently performing experiments to more precisely identify the cis-regulatory elements within HS4,5 and the trans-acting factors involved in mediating the effect of steroid hormones on Col1a1 gene expression.
The use of the GFP gene as a reporter gene in transgenic mice offers several advantages over the use of other reporter genes such as the CAT, β-galactosidase, or luciferase genes. Because fluorescence analysis requires no fixation or staining it is readily detectable using a fluorescent microscope (or a handheld lamp with appropriate filters) in live newborn or adult animals. It can be microscopically analyzed in unfixed or fixed whole mount embryos (Figs. 4, 5), in whole organs or tissues such as tail, toes, uterus, skin, and bone, or (in highly expressing animals) kidney, heart, lung, and muscle. In fixed embryos and frozen organs or tissues fluorescence remains detectable for many months. GFP can also be easily detected in histological analyses of thin sections of different organs (Fig. 7) or quantified very sensitively in tissue or cell extracts using a fluorometer (Tables 1 and 2). Finally, GFP expression can be observed in live transfected cells or primary cell cultures such as dermal fibroblasts or marrow stromal cells, and the effect of modulators of (in this instance) Col1a1 promoter activity can be monitored over long periods of time in such cultures. The transgenic mouse lines expressing the GFP gene driven by the Col1a1 promoter described here should prove useful tools to study many aspects of the regulation of type I collagen and other extracellular matrix components.
ACKNOWLEDGMENTS
This work was supported by NIH grants AR41909 (M.B.), AA10459 (R.A.R.), GB41804 (D.A.B.), and P50-AA11605 (R.A.R. and D.A.B).
REFERENCES
- 1. Barsh G. S.; Roush C. L.; Gelinas R. E. DNA and chromatin structure of the human alpha1 (I) collagen gene. J. Biol. Chem. 259:14906–14913; 1984. [PubMed] [Google Scholar]
- 2. Bedalov A.; Salvatori R.; Dodig M.; Kronenberg M. S.; Kapural B.; Bogdanovic Z.; Kream B. E.; Woody C. O.; Clark S. H.; Mack K.; Rowe D. W.; Lichtler A. L. Regulation of COL1A1 gene expression in type I collagen producing tissues: Identification of a 49 base pair region which is required for transgene expression in bone of transgenic mice. J. Bone Miner. Res. 10:1443–1452; 1995. [DOI] [PubMed] [Google Scholar]
- 3. Bogdanovic Z.; Bedalov A.; Krebsbach P. H.; Pavlin D.; Woody C. O.; Clark S. H.; Thomas H. F.; Rowe D. W.; Kream B. E.; Lichtler A. C. Upstream regulatory elements necessary for expression of the rat COL1A1 promoter in transgenic mice. J. Bone Miner. Res. 9:285–292; 1994. [DOI] [PubMed] [Google Scholar]
- 4. Bornstein P. Regulation of expression of the al (I) collagen gene: A critical appraisal of the role of the first intron. Matrix Biol. 15:3–10; 1996. [DOI] [PubMed] [Google Scholar]
- 5. Bou-Gharios G.; Garrett L. A.; Rossert J.; Niederreiter K.; Eberspacher H.; Smith C.; Black C.; de-Crombrugghe B.. A potent far-upstream enhancer in the mouse pro α1 (I) collagen gene regulates expression of reporter genes in transgenic mice. J. Cell Biol. 134:1333–11344; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Breindl M.; Harbers K.; Jaenisch R. Retrovirus-induced lethal mutation in collagen I gene of mice is associated with an altered chromatin structure. Cell 38:9–16; 1984. [DOI] [PubMed] [Google Scholar]
- 7. Brenner D. A.; Rippe R. A.; Rhodes K.; Trotter J. R.; Breindl M. Fibrogenesis and type I collagen gene regulation. J. Lab. Clin. Med. 124:755–760; 1994. [PubMed] [Google Scholar]
- 8. Chang W. Y.; Wilson M. J.; Birch L.; Prins G. S. Neonatal estrogen stimulates proliferation of periductal fibroblasts and alters the extracellular matrix composition in the rat prostate. Endocrinology 140:405–415; 1999. [DOI] [PubMed] [Google Scholar]
- 9. Driscoll M. D.; Sathya G.; Muyan M.; Klinge C. M.; Hilf R.; Bambara R. A. Sequence requirements for estrogen receptor binding to estrogen response elements. J. Biol. Chem. 273:29321–29330; 1998. [DOI] [PubMed] [Google Scholar]
- 10. Dyer R. F.; Sodek J.; Heersche J. M. N. The effect of 17β-estradiol on collagen and noncollagenous protein synthesis in the uterus and some periodontal tissues. Endocrinology 107:1014–1022; 1980. [DOI] [PubMed] [Google Scholar]
- 11. Epner E.; Reik A.; Cimbora D.; Telling A.; Bender M. A.; Fiering S.; Enver T.; Martin D. I. K.; Keller G.; Groudine M. The β-globin LCR is not necessary for an open chromatin structure or developmentally regulated transcription of the native mouse β-globin locus. Mol. Cell 2:447–455; 1998. [DOI] [PubMed] [Google Scholar]
- 12. Festenstein R.; Tolaini M.; Corbella P.; Mamalaki C. ; Parrington J.; Fox M.; Miliou A.; Jones M.; Ki-oussis D. Locus control region function and heterochromatin-induced position effect variegation. Science 271:1123–1125; 1996. [DOI] [PubMed] [Google Scholar]
- 13. Fischer G. M. Comparison of collagen dynamics in different tissues under the influence of estradiol. Endocrinology 93:1216–1224; 1973. [DOI] [PubMed] [Google Scholar]
- 14. Fraser P.; Grosveld F. Locus control regions, chromatin activation and transcription. Curr. Opin. Genet. Dev. 10:361–365; 1998. [DOI] [PubMed] [Google Scholar]
- 15. Graham J. D.; Clarke C. L. Physiological action of progesterone in target tissues. Endocr. Rev. 18:502–519; 1997. [DOI] [PubMed] [Google Scholar]
- 16. Higgs D. R. Do LCRs open chromatin domains? Cell 95:299–302; 1998. [DOI] [PubMed] [Google Scholar]
- 17. Inagaki Y.; Truter S.; Ramirez F. Transforming growth factor-beta stimulates alpha 2 (I) collagen gene expression through a cis-acting element that contains an Sp1-binding site. J. Biol. Chem. 296:14828–14834; 1994. [PubMed] [Google Scholar]
- 18. Kioussis D.; Festenstein R. Locus control regions: Overcoming heterochromatin-induced gene inactivation in mammals. Curr. Opin. Genet. Dev. 7:614–619; 1997. [DOI] [PubMed] [Google Scholar]
- 19. Krempen K. Analysis of upstream regulatory elements of the murine αl(I) collagen gene. Master’s thesis, San Diego State University; 1998. [Google Scholar]
- 20. Mahonen A.; Jukkola A.; Risteli L.; Risteli J.; Mäenpää P. Type I procollagen synthesis is regulated by steroids and related hormones in human osteosarcoma cells. J. Cell. Biochem. 68:151–163; 1998. [PubMed] [Google Scholar]
- 21. Mariotti A.; Soderholm K. J.; Johnson S. The in vivo effects of bisGMA on murine uterine weight, nucleic acids and collagen. Eur. J. Oral Sci. 106:1022–1027; 1998. [DOI] [PubMed] [Google Scholar]
- 22. Prockop D. J.; Kivirikko K. I. Collagens: Molecular biology, diseases, and potentials for therapy. Annu. Rev. Biochem. 64:403–434; 1995. [DOI] [PubMed] [Google Scholar]
- 23. Qu Q.; Perala-Heape M.; Kapanen A.; Dahllund J.; Salo J.; Vaananen H. K.; Harkonen P. Estrogen enhances differentiation of osteoblasts in mouse bone marrow culture. Bone 22:201–209; 1998. [DOI] [PubMed] [Google Scholar]
- 24. Rhodes K.; Rippe R. A.; Umezawa A.; Nehls M.; Brenner D. A.; Breindl M. DNA methylation represses murine α1(I) collagen promoter by an indirect mechanism. Mol. Cell. Biol. 14:5950–5960; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rhodes K.; Hall K.; Lee K. E.; Razzaghi H.; Breindl M. Correct cell and differentiation-specific expression of a murine α1 (I) collagen minigene in in vitro differentiating embryonal carcinoma cells. Gene Expr. 6:35–44; 1996. [PMC free article] [PubMed] [Google Scholar]
- 26. Rippe R. A.; Lorenzen S.-I.; Brenner D. A.; Breindl M. Regulatory elements in the 5′-flanking region and the first intron contribute to transcriptional control of the mouse alpha 1 type I collagen gene. Mol. Cell. Biol. 9:2224–2227; 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rippe R. A.; Umezawa A.; Kimball J. P.; Breindl M.; Brenner D. A. Binding of upstream stimulatory factor to an E-box in the 3′-flanking sequence stimulates α1 (I) collagen gene transcription. J. Biol. Chem. 272:1753–1760; 1997. [DOI] [PubMed] [Google Scholar]
- 28. Ritzenthaler J. D.; Goldstein R. H.; Fine A.; Lichtler A.; Rowe D. W.; Smith B. D. Transforming-growth-factor-β activation elements in the distal promoter regions of the rat αl type I collagen gene. Biochem. J. 280:157–162; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rossert J. A.; Eberspaecher H.; deCrombrugghe B. Separate cis-acting DNA elements of the mouse pro-α1 (I) collagen promoter direct expression of reporter genes to different type I collagen-producing cells in transgenic mice. J. Cell Biol. 129:1421–1432; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rossert J. A.; Chen S. S.; Eberspaecher H.; Smith C. D. ; deCrombrugghe B. Identification of a minimal sequence of the mouse α1 (I) collagen promoter that confers high-level osteoblast expression in transgenic mice and that binds a protein selectively present in osteoblasts. Proc. Natl. Acad. Sci. USA 93:1027–1031; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Salimi-Tari P.; Cheung M.; Safar C. A.; Tracy J. T.; Tran I.; Harbers K.; Breindl M. Molecular cloning and chromatin structure analysis of the murine α1 (I) collagen gene domain. Gene 198:61–72; 1997. [DOI] [PubMed] [Google Scholar]
- 32. Schmidt A.; Rossi P.; deCrombrugghe B. Transcriptional control of the mouse α2 (I) collagen gene: Functional deletion analysis of the promoter and evidence for cell-specific expression. Mol. Cell. Biol. 6:347–354; 1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Slack J. D.; Liska D. J.; Bornstein P. Regulation of expression of the type I collagen genes. Am. J. Med. Genet. 45:140–151; 1993. [DOI] [PubMed] [Google Scholar]
- 34. Slack J. L.; Parker M. I.; Bornstein P. Transcriptional repression of the α1 (I) collagen gene by ras is mediated in part by an intronic API site. J. Cell. Biochem. 58:380–392; 1995. [DOI] [PubMed] [Google Scholar]
- 35. Sokolov B. P.; Mays P. K.; Khillan J. S.; Prockop D. J. Tissue- and development-specific expression in transgenic mice of a type I procollagen (COL1A1) minigene construct with 2.3 kb of the promoter region and 2 kb of 3′-flanking region. Specificity is independent of the putative regulatory sequences in the first intron. Biochemistry 32:9242–9249; 1993. [DOI] [PubMed] [Google Scholar]
- 36. Sokolov B. P.; Ala-Koko L.; Dhulipala R.; Arita M.; Khillan J. S.; Prockop D. J. Tissue-specific expression of the gene for type I procollagen (COL1A1) in transgenic mice. J. Biol. Chem. 270:9622–9629; 1995. [DOI] [PubMed] [Google Scholar]
- 37. Sutherland H. G. E.; Martin D. I. K.; Whitelaw E. A globin enhancer acts by increasing the proportion of erythrocytes expressing a linked transgene. Mol. Cell. Biol. 17:1607–1614; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vuorio E.; de Crombrugghe B. The family of collagen genes. Annu. Rev. Biochem. 59:837–872; 1990. [DOI] [PubMed] [Google Scholar]
- 39. Walters M. C.; Magis W.; Fiering S.; Scalzo D.; Groudine M.; Martin D. I. K. Transcriptional enhancers act in cis to suppress position-effect variegation. Genes Dev. 10:185–195; 1996. [DOI] [PubMed] [Google Scholar]


