Abstract
The aryl hydrocarbon receptor (AhR), a soluble cytosolic protein, mediates many of the toxic effects of TCDD and related chemicals. The toxic effects are largely cell, tissue, and promoter context dependent. Although many details of the overall dioxin signal transduction have been elucidated, the transcriptional regulation of dioxin-induced genes like cyp1A1 is not yet completely understood. Previously, we have shown that the co-regulator RIP140 is a potential AhR coactivator. In this report, the role of coactivator, SRC-1, in AhR-mediated transcriptional regulation was examined. SRC-1 increased AhR-mediated, TCDD-dependent reporter gene activity threefold in Hepa-1 and COS-1 cells. In in vitro interaction assays, SRC-1 was found to interact with AhR but not with ARNT. SRC-1 interacted weakly with AhR in the absence of TCDD and the addition of ligand further increased SRC-1 binding to AhR. Deletional mapping studies of the AhR revealed that SRC-1 binds to the AhR transactivation domain. Finer mapping of the SRC-1-interacting subdomains in the AhR transactivation domain suggested that the Q-rich subdomain was necessary and sufficient for interaction, similar to that seen with RIP140. Using GFP-tagged constructs, SRC-1 was shown to interact with AhR in cells. Unlike RIP140, LXXLL motifs in SRC-1 were necessary for interaction with AhR in vitro and for coactivation in Hepa-1 cells. The recruitment of certain coactivators by a variety of receptors suggests possible common coactivator pools and competition among receptors for limiting coactivators. Examination of the role of SRC-1 in AhR/ARNT transactivation in ARNT-deficient mutant Hepa-1 c4 cells demonstrates that the AhR transactivation domain is sufficient for enhanced coactivation mediated by SRC-1 in the presence of a transactivation domain deleted ARNT protein.
Keywords: Ary hydrocarbon receptor (AhR), Dioxin, Coactivator, SRC-1
THE aryl hydrocarbon receptor (AhR) is known to mediate the toxic and adaptive responses to TCDD and other environmental contaminants (45). However, the biological role of the AhR is poorly understood. AhR null mice exhibit phenotypes that include an altered liver pathology associated with accelerated rates of apoptosis resulting from an aberrant accumulation of hepatic retinoic acid (3,63). Thus, the AhR may, directly or indirectly, control levels of a cytochrome P450 that is responsible for catabolizing retinoic acid (3,63). Although normal at birth, AhR-null mice displayed a slightly slower growth rate than wild-type mice for the first few weeks of life (33). Other reports indicate that AhR-deficient mice exhibit immune system impairment and hepatic defects at maturity, in addition to slow early growth, suggesting a role for the AhR in normal liver growth and development (53). A myriad of reproductive defects in AhR-deficient female mice, including female mortality, small litter size, and death of −/− pups after weaning, were also reported (1).
The AhR is found in the cytosol in a heterotetrameric complex with two Hsp90 molecules and one X-associated protein 2 (XAP2) molecule (5,29,32). Upon ligand binding, the complex translocates to the nucleus where the AhR heterodimerizes with Ah receptor nuclear translocator (ARNT) (49) after Hsp90 dissociation (13,43). The AhR/ARNT complex binds to dioxin response elements (DREs) upstream of certain genes, including cyp1A1, cyp1B1, and cyp1A2, leading to their enhanced transcription (36,51,57). Although much is known about the overall TCDD-signal transduction pathway, the mechanism of the AhR-mediated transactivation of TCDD-responsive genes is yet to be completely understood. Using the cyp1A1 gene as a model, the binding of the AhR/ARNT to DREs was shown to promote disruption of chromatin in the enhancer and promoter-proximal regions (22,23,40). Basal transcription factors, TFIIB (55), TBP, and TFIIF (50), have been suggested to interact with the AhR. Recently, coactivator, ERAP140, and co-repressor, SMRT, were shown to interact with the AhR and ARNT and alter the AhR-mediated transactivation in MCF-7 cells (37). Another coactivator, CBP, was shown to interact with ARNT preferentially (24), although whether CBP is involved in AhR/ARNT transcriptional complexes in vivo remains to be determined. Previously, we characterized the interaction between co-regulator, RIP140, and the AhR (25). Unlike steroid receptors, the AhR transactivation domain (TAD) interaction with RIP140 was not mediated by LXXLL motifs in vitro. In addition, the Q-rich subdomain of the AhR TAD was necessary and sufficient for in vitro interaction with RIP140 (25). Collectively, these studies suggest that the AhR is capable of recruiting a number of coactivators that were originally identified as steroid receptor coactivators.
Nuclear receptor coactivator, SRC-1, has been shown to interact with a number of type I and type II steroid receptors (8,42) both in vitro and in vivo, and enhance transcriptional activation potential in reporter gene assays in cells. In cell-free transcription assays with PRE chromatin templates, SRC-1 potentiated transcription by ligand-activated progesterone receptor (PR) (28). Several other transcription factors, including AP-1 (27) and cyclin D1 (65), have been reported to interact with SRC-1. Although considered a positive modulator of PR and glucocorticoid receptor (GR), SRC-1, a truncated form of full-length SRC-1 (F-SRC-1), has been observed to repress AR-mediated transactivation of an ARE-driven reporter gene. Furthermore, the interaction between the N- and C-terminus TADs of AR appeared to be disrupted by SRC-1 (17). In the case of PPARα, SRC-1 was not found to be required for PPARα-regulated gene expression in SRC-1 −/− mice (47). SRC-1 is a modular coactivator possessing intrinsic transcriptional activity with two activation domains: AD-1 and AD-2 (41). In addition, SRC-1 possesses his-tone acetyltransferase activity and is known to specifically acetylate histones H3 and H4 (54). Basal transcription factors, TFIIB and TBP, have also been shown to interact with SRC-1, suggesting its possible role as an adaptor molecule (16). SRC-1, along with other coactivators, possesses short signature motifs (LXXLL), which are necessary and sufficient for interaction with many nuclear receptors (8,12,30,38,52,56,59,64). Whether these motifs are required for interaction with other enhancer binding transcription factors will require further study. SRC-1 has been observed to exist in distinct stable complexes along with TIF2 and PR in vivo, suggesting that the assembly of transcriptional complexes involves recruitment of distinct subclasses of preformed co-regulator complexes (31).
SRC-1 is expressed as several isoforms, including splice variants, SRC-la and SRC-le. SRC-le is expressed more abundantly than SRC-la and appears to potentiate ER transactivation to a higher degree than SRC-la (11,20). The expression of SRC-1 mRNA in vivo is known to be regulated by hormones T3 and E2 in the anterior pituitary. A distinct tissue-specific pattern of expression of SRC-1 mRNA was also observed (34,35). SRC-1-null mice were viable and fertile, although the uterus, prostate, testis, and mammary glands exhibited decreased growth and development in response to steroid hormones. Interestingly, TIF2 mRNA expression was increased in these mice, suggesting a compensatory mechanism may have been induced (62).
In this report, the role of SRC-1 in AhR transactivation is examined in Hepa-1, COS-1, and ARNT-deficient mutant Hepa-1 cell lines. SRC-1 potentiated the AhR-mediated transactivation in Hepa-1 and COS-1 cells in a TCDD-dependent and SRC-1 dose-dependent manner. The TAD of the AhR, but not of ARNT, was shown to interact with SRC-1, specifically via the Q-rich subdomain of the AhR. Using GFP-tagged constructs, the interaction of SRC-1 with the AhR TAD was demonstrated in intact cells. Unlike RIP140, SRC-1 interaction was found to be mediated via LXXLL motifs in vitro. In addition, these motifs were required for SRC-1 coactivation in intact cells. Thus, SRC-1 appears to be another potential coactivator for the AhR/ARNT system.
MATERIAL AND METHODS
Plasmids
The plasmid pSG5/SRC-1a expressing SRC-1a, and pSG5/SRC-la mut1234 expressing SRC-1a mut1234 were gifts of Malcolm Parker (20). β-Globin/mAhR (32), used in COS-1 transfections, mAhR/Flag, mAhRΔTAD/Flag, NΔ315 mAhR/Gal4/Flag, pcDNA3/mARNT-Flag , and pcDNA3/mARNT-474-Flag (ARNTΔTAD) (25) used in in vitro translations were described previously. pGEX-mAhR/TAD, pGEX-mARNT/TAD, pGEX-hAhR600-800, pGEX-hAhR500-713, pGEX-hAhR600-713, pGEX-hAhR500-600, and pGEX-hAhR713-848, expressing GST fusions of AhR TAD deletion mutants, were also previously described (25). pGFP-AhR/TAD and pGFP-ARNT/TAD used in GFP colocalization experiments were also previously described (25). Escherichia coli strain DH5α (Life Technologies Inc., Baltimore, MD) was used for all plasmid preparations and E. coli BL21 (DE3) (Novagen, Madison, WI) was used in the expression of GST fusion proteins.
Generation of GST Fusion Proteins
GST and GST fusion proteins were expressed in E. coli BL21 (DE3) and purified using glutathione-agarose following manufacturer’s (Amersham-Pharmacia Biotech, Arlington Heights, IL) recommendations.
In Vitro Binding Assay for Interaction of SRC-1 With AhR
mAhR/Flag, mARNT/Flag, and other FLAG-tagged cDNA constructs were in vitro transcribed and translated separately using the TNT Coupled Reticulocyte Lysate system (Promega, Madison, WI) and mixed with [35S]methionine-labeled in vitro transcribed/translated SRC-1 and incubated at 4°C for 1.5 h. The complexes were immunoprecipitated with anti-FLAG M2 affinity gel in CSB buffer [150 mM NaCl, 15 mM CHAPS, 18.75% glycerol (v/v), 0.125% Nonidet-P40 (v/v), 20 mM Tris (pH 8.0), and 2 mg/ml bovine serum albumin] for 2 h, followed by several washes with CSB buffer without bovine serum albumin. The immunoprecipitated complexes were eluted with 50 μg of FLAG peptide in MENG buffer containing 150 mM NaCl plus 3 mg/ml soybean trypsin inhibitor. The elution was repeated and eluted proteins were separated on an 8% polyacryamide gel. The gel was treated with Amplify (Amersham-Pharmacia Biotech) and the radioisotope was detected by fluorography.
For GST pull-down assays, GST or GST fusion proteins immobilized on 30 μl of glutathione agarose (Sigma, St. Louis, MO) were incubated with [35S]methionine-labeled, in vitro translated SRC-1 in HEMG [40 mM HEPES (pH 7.8), 0.1 M NaCl, 0.2 mM EDTA, 5 mM MgCl2, 0.1% (v/v) Triton X-100, 10% glycerol] buffer. Following a 1.5-h incubation at 4°C, the agarose was washed four times with HEMG buffer. The immobilized proteins were eluted with 2× tricine sample buffer, separated by SDS-PAGE, and analyzed by fluorography.
Cell Culture and Transient Transfection Experiments
Hepa-1, COS-1, and Hepa c4 mutant cells (10) were routinely cultured in α-modified Eagle’s medium containing 10% fetal bovine serum (v/v). For reporter gene assays, Hepa-1 cells were transfected with a total of 1 μg of DNA using LipofectAMINE reagent (Life Technologies, Inc.) when performed in 24-well plates. Cells were cotransfected with 100 ng of reporter plasmid, pGUDLUC 6.1, and the β-galactosidase internal control plasmid, pCMV-βgal, along with variable amounts of SRC-1-expressing plasmid, pSG5/SRC-1a, or control plasmid.
COS-1 cells were transfected in six-well plates with a total of 2.5 μg of DNA using LipofectAMINE reagent. The transfected DNA included 100 ng of pc-DNA3/βmAhR, 200 ng of reporter plasmid, pGUDLUC 6.1, and the β-galactosidase internal control plasmid, pDJM-Δgal, along with variable amounts of pSG5/SRC-la, or control plasmid.
For Hepa-1 mutant c4 cell transfections, cells were grown to 90% confluency in six-well plates before transfection. Cells were transfected for a period of 12 h with a total of 2.5 μg of DNA using LipofectAMINE reagent. The transfected DNA included 25 ng of pcDNA3/mARNT-Flag or pcDNA3/mARNT474-Flag (ARNTΔTAD), 200 ng each of reporter plasmid, pGUDLUC 6.1, and the β-galactosidase internal control plasmid, pDJM-βgal, along with 2 μg of pSG5/SRC-1a, or control plasmid.
In each case, the LipofectAMINE complexes were removed and replaced with α-modified Eagle’s medium containing 10% fetal bovine serum. Twenty-four hours posttransfection the cells were treated with 10 nM TCDD or DMSO for 12 h, harvested, and extracts were assayed for luciferase activity. Luciferase activity was normalized to the observed β-galactosidase activity and expressed as relative luciferase units (RLU). All transfections were performed in triplicate.
For expression of GFP and its fusion proteins, COS-1 cells were seeded onto 2-mm2 coverslips in 60-mm dishes and transfected with a total of 400 ng of GFP or GFP fusion plasmid along with pSG5/SRC-1a or control plasmid. The cells were washed with phosphate-buffered saline and visualized using fluorescence microscopy 24 h after transfection.
Statistical Analysis
Experiments were done in triplicate and statistical analysis was performed using the Standard t-test in the SigmaStat program (SSSP Inc., Chicago, IL). Sample comparisons with p < 0.05 were considered to be significantly different from each other.
RESULTS
SRC-1 Modulates the AhR/ARNT-Mediated Transactivation of Reporter Genes
Previously, we have shown that the nuclear receptor co-regulator, RIP140, is a potential coactivator for the AhR/ARNT system and is able to modulate the AhR-mediated transactivation in a ligand-dependent manner (25). SRC-1 has been shown to upregulate the transactivation potential of a myriad of nuclear receptors, including ER, PR, GR, TR, and RXR (8,42). In an effort to understand the mechanism of the AhR/ARNT transactivation of target genes and determine the role of other potential coactivators, we examined the effect of ectopic expression of SRC-1 on DRE-driven reporter gene activity in two cell lines: Hepa-1 and COS-1. pGUDLUC 6.1, the reporter gene used in these assays, contains a luciferase gene driven by four DREs. Hepa-1 cells, which express substantial amounts of mAhR, when transfected with increasing amounts of pSG5/SRC-la led to a dose-dependent increase in reporter gene activity (Fig. 1A). The reporter gene activity observed was TCDD dependent. A threefold increase in reporter gene activity was seen with 800 ng of cotransfected pSG5-SRC-la. Unlike RIP140, where a biphasic response was seen in Hepa-1 cells (25), SRC-1 titration led to increased activity, even at higher amounts of cotransfected SRC-1, without a substantial increase in basal activity.
FIG. 1.

Effect of SRC-1 overexpression on AhR-mediated transactivation in COS-1 and Hepa 1c1c7 cells. Hepa 1c1c7 cells (A) and COS-1 (B) were cotransfected with increasing amounts of pSG5-SRC-1a, 100 ng of DRE-driven luciferase gene containing plasmid, pGUDLUC 6.1, and pCMV/LacZ (100 ng), and empty plasmid. COS-1 cells were also cotransfected with 100 ng pcDNA3/βmAhR where indicated. Luciferase activity was measured following a 12-h induction with 10 nM TCDD or carrier solvent and normalized to β-galactosidase activity. Each transfection was performed in triplicate. For statistical analysis, the values from the SRC-1 transfected samples were compared with those of SRC-1 nontransfected samples within each cell line. Asterisks indicate that statistically significant differences were observed (p < 0.05).
The effect of SRC-1 on DRE-driven reporter gene activity was also examined in COS-1 cells, which contain negligible amounts of the AhR. This provides an experimental system where the importance of SRC-1 can be examined in the presence or absence of exogenous AhR. In the absence of transfected AhR, a fourfold increase in reporter gene activity was detected upon SRC-1 cotransfection, which may be due to the low amounts of endogenous AhR present in COS-1 cells. However, no significant increase in basal transcriptional activity was observed. Upon transfection of 50 ng of AhR, a TCDD-dependent 14-fold increase in reporter gene activity is seen (Fig. 1B). Interestingly, cotransfection of small amounts (10 ng to 1.5 μg) of SRC-1 did not have any significant effect on the AhR-mediated transcriptional activation of the reporter gene in this cell line. A significant increase in activity was observed only when transfected with higher amounts of SRC-1 and then it increased the AhR-mediated transcriptional activation threefold in a TCDD-dependent manner. Unlike RIP140 (25), SRC-1 did not induce a biphasic response in reporter gene activity in COS-1 cells.
In Vitro Interaction of SRC-1 With the AhR
The interaction of SRC-1 with the AhR and ARNT was investigated using coimmunoprecipitation assays with in vitro transcribed/translated SRC-1 and FLAG-tagged AhR or ARNT constructs. Radiolabeled SRC-1 interacted weakly with full-length AhR in the absence of TCDD (Fig. 2). This is in contrast to the ability of AhR to interact strongly with RIP140 even in the absence of TCDD (25). The deletion of the AhR TAD essentially abolished any AhR-SRC-1 interaction. AhR-NΔ315, which lacks the bHLH and parts of the PAS domains, was competent in recruiting SRC-1. On the other hand, ARNT failed to bind SRC-1. As a control, in vitro translations carried out in the absence of any plasmid were incubated with SRC-1 and immunoprecipitated using α-FLAG antibodies. No SRC-1 interaction was detected in this case, indicating that the AhR-SRC-1 interaction was specific and not due to background binding to the M2 resin. AhR-SRC-1 interaction was also examined in the presence of TCDD. In contrast to AhR-RIP140 interaction, the addition of TCDD led to a substantial increase in SRC-1 binding to the AhR. This is in agreement with reports of ligand-dependent SRC-1 interaction with nuclear receptors.
FIG. 2.

In vitro interaction of SRC-1 with AhR/ARNT and mapping of the SRC-1 binding sites on AhR. FLAG-tagged full-length AhR (AhR), AhR without the transactivation domain (AhRΔTAD), N-terminal deletion of AhR (AhRNΔ315), full-length ARNT (ARNT), and ARNTΔTAD cDNAs were in vitro transcribed/translated and were incubated with [35S]methionine-labeled in vitro translated SRC-1 and immunoprecipitated with anti-FLAG M2 affinity gel. The bound proteins were eluted with FLAG peptide, analyzed by SDS-PAGE, and visualized by fhiorography. 1/10 input represents l/10th of the amount of [35S]methionine-labeled SRC-1 used in the binding reactions.
Finer Mapping of the SRC-1 Binding Sites on the AhR TAD
The hAhR TAD can be further divided into three subdomains: the acidic, Q rich, and P/S/T rich. In order to map the SRC-1 binding sites on the AhR TAD, GST fusions of deletion mutants (containing various combinations of the different subdomains) of the hAhR TAD were used in GST pull-down assays with [35S]methionine-labeled SRC-1. The interaction of SRC-1 with GST fusions of mAhR and hAhR was compared and found to be similar, although SRC-1 bound slightly better to the hAhR than to the mAhR (Fig. 3). The acidic subdomain deletion mutant was still capable of weakly binding SRC-1. Deletion of the P/S/T subdomain also led to a decrease in binding to SRC-1, although some binding was clearly retained. When the individual subdomains were tested for interaction with SRC-1, the Q-rich subdomain was found to be sufficient for binding. However, the acidic or the P/S/T subdomains bound SRC-1 very weakly, suggesting that the Q-rich subdomain was necessary and key for an efficient level of SRC-1 interaction. However, it is important to note that the acidic and P/S/T subdomains may participate in the overall interaction with SRC-1. As additional controls, the interaction of SRC-1 with GST-Spl, with a predominantly Q-rich TAD, and GST-VP16, with an acidic TAD, was examined. While GST and GST-VP16 failed to significantly bind SRC-1, weak binding of SRC-1 to GST-Spl was observed. These results are similar to those obtained for AhR-RIP140 interaction, thereby implying a dominant role for the Q-rich subdomain of the AhR in coactivator recruitment and the possibility of a common interface for coactivator interaction.
FIG. 3.

Identification of SRC-1 binding subdomain of the AhR TAD. GST, GST-Sp1, GST-VP16, or GST fusion proteins of truncations of human AhR transactivation domain were immobilized on 30 μl of glutathione agarose. [35S]Methionine-labeled in vitro translated full-length SRC-1 was incubated with the immobilized GST or GST fusion proteins for 1.5 h in CSB buffer. The agarose was washed several times with CSB buffer and the proteins were eluted with 2× TSB, subjected to SDS-PAGE and fluorography.
Interaction of SRC-1 With AhR in Intact Cells
To test for interaction of AhR and SRC-1 in intact cells, COS-1 cells were transfected with plasmids expressing GFP, GFP-AhR/TAD, or GFP-ARNT/TAD along with pSG5/SRC-la or control vector. GFP, GFP-AhR/TAD, and GFP-ARNT/TAD when expressed by themselves are expected to be localized to the cytosol of cells, because they lack a putative nuclear localization signal. Interaction of the fusion proteins with SRC-1, a nuclear protein, would be expected to lead to cotranslocation of the fusion proteins to the nucleus. GFP alone, when expressed in COS-1 cells, was localized predominantly to the cytosol, but to some extent was also found in the nucleus (Fig. 4), which has also been observed by the manufacturer (Clontech, Palo Alto, CA). Upon coexpression of SRC-1, the distribution of GFP remained the same, suggesting a lack of interaction between GFP and SRC-1. GFP-AhR/TAD, when cotransfected with a control plasmid, displayed predominantly cytosolic distribution and, to some extent, to the nucleus. However, upon coexpression of SRC-1, GFP-AhR/TAD localized extensively in the nucleus in discreet foci, suggesting that SRC-1 interacts with AhR in intact cells. Previous reports indicate that the AhR is uniformly distributed and does not appear to be associated with nuclear pores, membranes, or nucleoli (46). Thus, our results may suggest a possible shift in the nuclear localization pattern upon interaction with coactivators. A similar shift in localization pattern was observed in the case of RIP140 (25) and has also been seen in reports with GFP-GR (15,39) and GFP-TR (26). Localization of GFP-ARNT/TAD, on the other hand, remained unchanged in the absence or presence of ectopically expressed SRC-1, indicating that the SRC-1 interacts with the AhR predominantly.
FIG. 4.

Interaction of SRC-1 with AhR in intact cells. (A–F) In vivo interaction using GFP-tagged proteins. COS-1 cells were transfected with GFP or GFP fusion constructs and pSG5-SRC-la or control expression vector and visualized by microscopy at 100× magnification 24 h after transfection. (A) GFP, (B) GFP plus SRC-1a, (C) GFP-AhR, (D) GFP-AhR plus SRC-1a, (E) GFP-ARNT, (F) GFP-ARNT plus SRC-1a.
Role of LXXLL Motifs in AhR–SRC-1 Interaction
Previously, we have shown that the interaction of co-regulator RIP140 with the AhR involves mechanisms that do not require the LXXLL motifs, at least in vitro (25). In this report, the role of LXXLL motifs in AhR-SRC-1 interaction was examined in GST pull-down assays. SRC-1a wt or SRC-1a mut, with all the LXXLL motifs mutated, were in vitro translated in the presence of [35S]methionine and incubated with either GST, GST-ER-HBD, or GST-AhR-TAD. No binding of SRC-1 to GST-ER-HBD was observed in the absence of ligand, E2 (Fig. 5). The addition of E2 led to strong recruitment of SRC-1 to GST-ER-HBD, as previously seen in other reports (8,42). As expected, SRC-1a mut did not interact with GST-ER-HBD either in the presence or absence of E2, indicating that the LXXLL motifs are required for interaction. Next, the interaction of wt and mutant SRC-1 with GST-AhR-TAD was examined. SRC-1a wt interacted with the AhR TAD, although the interaction was weaker than that of ER and SRC-1. SRC-1 mut displayed greatly reduced binding to AhR, indicating that the LXXLL motifs may play a role in AhR-SRC-1 interaction. These finding are in contrast with that of RIP140, where the LXXLL motifs did not appear to play a role in in vitro binding to AhR, suggesting a possible diversity in the mechanism of recruitment of coactivators to the AhR.
FIG. 5.

Role of LXXLL motif in the binding of SRC-1 to AhR and ER transactivation domains. SRC-1a or SRC-1a mut, in which all the LXXLL motifs were mutated, were in vitro transcribed and translated in the presence of [35S]methionine and incubated in the presence of CSB buffer with either GST, GST-mAhRTAD or GST-ER-HBD immobilized on 30 μl of glutathione agarose. Following a 1.5-h incubation, the agarose was washed with CHAPS buffer. The bound proteins were eluted with 2× TSB and subjected to SDS-PAGE and visualized by fluorography. 1/10 input represents 1/10th of the amount of [35S]methionine-labeled SRC-1 used in the binding reactions.
Role of LXXLL Motifs in Potentiating AhR Transactivation in Intact Cells
The role of LXXLL motifs was examined in intact cells in DRE-driven reporter gene assays. Hepa-1 cells were transfected with increasing amounts of either SRC-la wt or SRC-la mut and a plasmid containing a DRE-driven luciferase gene. As expected, SRC-1 wt was able to potentiate AhR transcriptional activation by about threefold over background (Fig. 6). However, mutated SRC-1 was unable to positively modulate the AhR transactivation in intact cells, suggesting that the LXXLL motifs are required for SRC-1 coactivation of AhR. Neither the wt nor the mutant SRC-1 significantly influenced the basal activity. This observation further substantiates the requirement of LXXLL motifs for SRC-1 functionality. This ligand- and LXXLL motif-dependent interaction of SRC-1 with AhR thus appears to be similar to that of its interaction with nuclear/steroid receptors.
FIG. 6.
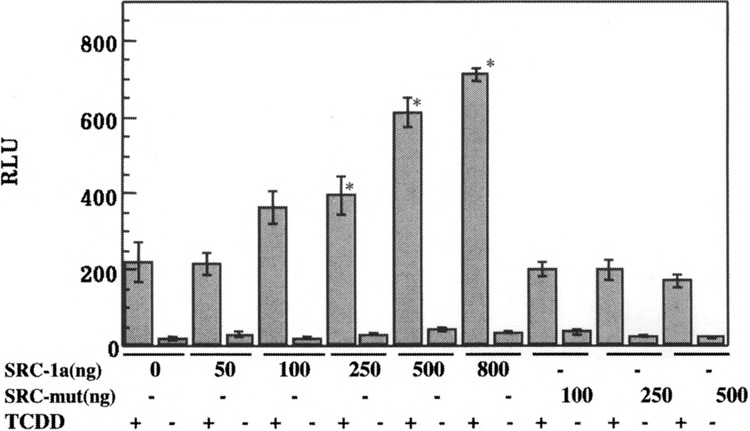
Role of LXXLL motifs in modulation of AhR transactivation by SRC-1. Hepa-1 cells were transfected with increasing amounts of pSG5-SRC-1a, expressing wt SRC-1 or pSG5-SRC-1a mut, expressing a SRC-1 LXXLL mutant along with 100 ng each of pGUDLUC6.1 and an internal control vector, pCMV-β-gal. The cells were treated with TCDD or DMSO 24 h after transfection for a period of 12 h and harvested. The extracts were assayed for luciferase and β-gal activity. Each transfection was performed in triplicate. For statistical analysis, the values from the SRC-1 transfected samples were compared with those of SRC-1 nontransfected samples within each cell line. Asterisks indicate that statistically significant differences were observed (p < 0.05).
Role of ARNT in the SRC-1 Coactivation
To further understand recruitment of SRC-1 to AhR/ARNT complex in intact cells, Hepa c4 mutant cells, which do not express ARNT, were employed. This experimental system allows dissection of the role of the transactivation domains of each of the AhR/ARNT heterodimer partners in vivo by transfecting either the full-length ARNT or a truncated ARNT lacking the TAD. Nontransfected c4 cells fail to respond to TCDD and show negligible background activity of the reporter gene in the presence or absence of TCDD (Fig. 7), indicating that ARNT is required for AhR transactivation. Cotransfection of pSG5-SRC-la, alone, does not significantly increase reporter gene activity over background, implying that SRC-1 fails to coactivate by itself in the absence of the AhR/ARNT complex, thus demonstrating that SRC-1 does not stimulate basal transcription. Transfection of full-length ARNT resulted in a 13-fold increase in transcriptional activity in the presence of TCDD. The cotransfection of pSG5-SRC-la increases reporter gene activity by 2.4-fold (Fig. 7, Table 1), similar to that observed in the case of COS-1 and Hepa-1 cells. On the other hand, cotransfection of ARNTΔTAD increases reporter gene activity fourfold over background, suggesting that AhR/ARNTΔTAD complex can still potentiate transcription of target genes, albeit at a lower efficiency. A similar effect has also been previously observed with Hepa c4 mutant cells (48). Interestingly, SRC-1 cotransfection leads to a 3.1-fold increase in reporter gene activity, clearly indicating that the ARNT TAD is not required for SRC-1 coactivation and providing indirect, in vivo evidence that SRC-1 potentially interacts, directly or indirectly, via the AhR.
FIG. 7.
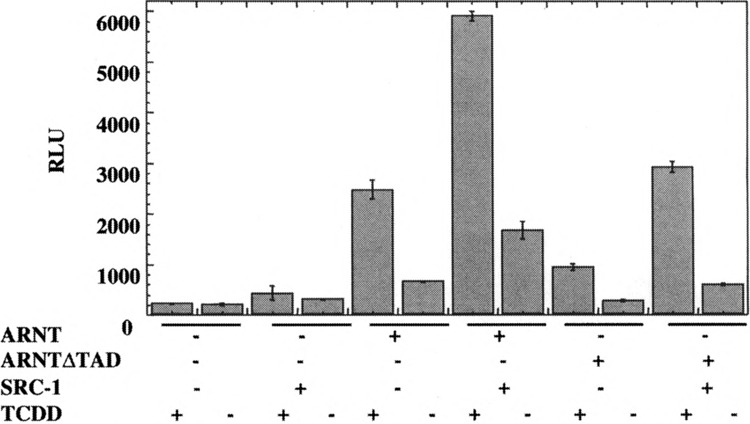
Role of ARNT-TAD in the SRC-1 modulation of transactivation of DRE-driven reporter gene in mutant c4 Hepa-1 cells. Mutant Hepa-1 c4 cells transfected in six-well plates for 12 h. The transfected DNA included 25 ng of pcDNA3/mARNT-Flag or pcDNA3/mARNT-474-Flag, 200 ng each of reporter plasmid, pGUDLUC 6.1, and the β-galactosidase internal control plasmid, pDJM-βgal, along with 2 μg of pSG5/SRC-1a, or control plasmid. The cells were treated with 10 nM TCDD or DMSO for 12 h and harvested. The extracts were assayed for luciferase activity that was normalized to β-gal activity and expressed as RLU. Each transfection was performed in triplicate. For statistical analysis, the values from the SRC-1 transfected samples were compared with those of SRC-1 nontransfected samples within each cell line.
TABLE 1.
EFFECT OF ECTOPIC EXPRESSION OF SRC-1 AND ARNT-FL AND ARNTΔTAD ON DRE-DRIVEN REPORTER GENE ACTIVITY IN MUTANT HEPA-1 c4 CELLS
| Transfected Plasmid | TCDD | RLU ± SE | Fold Induction |
|---|---|---|---|
| ARNT-FL | + | 2486.3 ± 181.8 | 1 |
| ARNT-FL + SRC-1 | + | 5912.3 ± 102.6 | 2.4 |
| ARNTΔTAD | + | 947.8 ± 69.2 | 1 |
| ARNTΔTAD + SRC-1 | + | 2930.6 ± 105.2 | 3.1 |
DISCUSSION
Exposure to TCDD results in a myriad of species-, cell-, and tissue-specific responses, including hydronephrosis, cleft palate formation, and induction of thymic hypoplasia (44,45). It has been suggested that structural differences between mammalian and fish AhRs may account for differences in relative potencies of the mono-ortho PCBs between mammals and fish: this may imply one possible mechanism for species-specific responses (2), however the precise mechanism(s) behind tissue- and cell-specific responses still need to be elucidated. At the molecular level, the AhR-regulated human NAD(P)H:quinone oxidoreductase2 (NQO2) gene is expressed in human heart, brain, liver, and skeletal muscle, but not in the placenta. In addition, large variations in levels of expression of the NQO2 and NQO1 genes are seen among various tissues (18). The expression and in-ducibility of the cytochrome P-4501A1 activity also exhibit tissue-specific differences (58). In TCDD nonresponsive fibroblasts, the chromatin structure of cyp1A1 gene lacked open regions, while in the two responsive cell types, keratinocytes and HepG2, several constitutive hypersensitive sites, as well as AhR ligand-induced alterations in the chromatin structure, were detected (9). In the nonresponsive fibroblast nuclear extracts, two novel constitutive protein-XRE complexes were detected. The fibroblast factor(s) were immunochemically distinct from the receptor but exhibited indistinguishable DNA binding specificity, which may be due to putative repressors(s) (9). Alternatively, it is possible coactivators may be able to form transcriptionally competent complexes in nonfibroblast cells, but not in fibroblast cells. Thus, coactivators and corepressors may play a role in the tissue-specific responses to TCDD.
The expression of co-regulators also appears to be tissue specific. For example, the expression of coactivator SRC-1 mRNA was found to be highest in the olfactory epithelium, suggesting that it may be involved in the development and function of the olfactory system (34). ElA-associated 300-kDa proteins, and co-repressors, SMRT, and N-CoR gene expression also exhibit distinct tissue-specific patterns. In addition, hormones T3 and E2 appeared to regulate the expression of SRC-1 mRNA in certain tissues (35). These observations suggest a role for coactivators and corepressors in tissue specificity of various responses. We are attempting to understand the role of coactivators in the transcriptional regulation of TCDD-responsive genes. To this end, we had previously characterized the role of RIP 140 in the AhR transactivation. In this report the role of SRC-1 in AhR transactivation is investigated and the results may further indicate that the AhR is capable of recruiting a number of coactivators that bind steroid receptors.
SRC-1 increased the AhR transcriptional activation of the DRE-driven reporter gene in an AhR- and TCDD-dependent manner in Hepa-1 and COS-1 cells. It is important to note that SRC-1 did not appear to increase basal activity in either cell line significantly. Interestingly, the response to SRC-1 ectopic expression appeared to be cell line dependent. In Hepa-1 cells, ectopic expression of increasing amounts of SRC-1 led to a gradual increase in reporter gene activity. However, in COS-1 cells SRC-1 failed to potentiate AhR transactivation when relatively small amounts of SRC-1 expression plasmid were cotransfected. A steep increase in reporter activity was noticed when cotransfected SRC-1 DNA was increased from 1.5 to 2 μg. This steep curve may indicate higher levels of an existing pool of SRC-1 or related coactivators in COS-1 cells, and hence a high level of ectopic SRC-1 expression is required for potentiating AhR transactivation. These results, in two different cell lines, nonetheless indicate that SRC-1 is a potential AhR coactivator. Thus, SRC-1 appears to be a common coactivator, which can modulate transactivation of several receptor types.
In vitro interaction assays using FLAG-tagged AhR and ARNT constructs argue that SRC-1 is recruited to the AhR TAD, but not to ARNT. Interestingly, in vitro, AhR without the bHLH and parts of the PAS domain interacts with SRC-1 strongly. This was also seen in the case of RIP140 (25). This would suggest that the N-terminal domain may sterically hinder coactivator recruitment by the AhR TAD. Alternatively, it has been suggested that the N-terminus may harbor a repressor domain that, when deleted, leads to higher levels of transactivation (60). SRC-1 interaction was increased in the presence of TCDD, similar to that seen with several nuclear/steroid receptors. The binding of ligand to nuclear/steroid receptors induces a conformational change such that the AF-2 amphipathic α-helix (helix 12) is aligned over the LBD, unlike the case of the unliganded receptor, where helix 12 protrudes away from the LBD. This realignment is believed to lead to the formation of a new interaction surface for coactivators (4). However, this is in contrast to our previous observations with the RIP140–AhR interaction where the addition of ligand failed to influence interaction between RIP140 and AhR (25). It is important to note that the AhR functional domains are arranged differently compared to the steroid receptors. In particular, the TAD of the AhR is distinct and separated from the ligand binding domain. This, however, does not rule out the possibility of a conformational change in AhR TAD in vivo upon ligand binding. It is possible that RIP140 may interact with AhR in the nucleus prior to the actual formation of AhR-ARNT heterodimer. It may also modulate the heterodimerization event itself. In contrast, SRC-1 and other potential coactivators/accessory protein complexes may be recruited to the AhR, predominantly after heterodimerization and DNA binding.
Finer mapping of SRC-1 binding sites on AhR TAD using GST fusion proteins in pull-down assays suggests that the Q-rich subdomain is necessary and sufficient for SRC-1–AhR interaction. Spl, which has a Q-rich TAD, also seemed to recruit SRC-1, albeit weakly. While RIP140 could interact with the Q-rich subdomain of AhR TAD, it failed to interact with Spl (25). Collectively, these results hint at the diversity and complexity of mechanisms for recruiting different coactivators. Nonetheless, there appears to be a common dominant interaction surface for coactivators on the AhR. Analysis of the predicted secondary structure indicates that there may be two helices in the AhR Q-rich subdomain between amino acids 612 and 650. α-Helices have been implicated in forming interaction surfaces on TADs of several nuclear/steroid receptors (7,14,59). The fact that the acidic and P/S/T subdomains do not appear to be required is interesting. The acidic subdomain may be primarily involved in interaction with basal transcription factors and not coactivator complexes. Alternatively, the acidic- and P/S/T-rich subdomains may be functional in a tissue- or promoter-specific fashion. The complex AhR TAD may thus be involved in imparting cell and tissue specificity. Certain coactivators may be recruited to specific subdomains, depending on the cell type and promoter context.
The LXXLL motifs of SRC-1 were found to be required for in vitro interaction and for potentiating AhR transactivation in intact cells. The LXXLL motifs of coactivators have been suggested to make contacts via the hydrophobic residues to the amphipathic helix in nuclear receptors, including TRβ (7) and PPARγ (30). In contrast, we have previously noted that the LXXLL motifs in co-regulator RIP140 did not appear to play a crucial role in the in vitro interaction with AhR (25). This also lends credence to the idea that the mechanism of RIP140 interaction with AhR may be atypical compared to recruitment of other coactivators and may thus be involved in AhR transactivation in a different capacity than SRC-1 or other related coactivators.
Examination of the role of SRC-1 in the case of ARNT-deficient Hepa c4 cells indicated that SRC-1 stimulated AhR transactivation, even in the absence of ARNT TAD, which suggests that the SRC-1 primarily functions via the AhR. This is further reinforcement of the idea that the AhR is the dominant partner in regulating TCDD-responsive genes (22). ARNT, however is required for AhR transactivation as indicated by the fact that SRC-1 fails to coactivate in the absence of any ARNT ectopic expression in c4 cells. ARNT TAD is also required for optimal AhR/ARNT transcriptional activity because the AhR/ARNTΔTAD leads to only partial transcriptional activity in the absence of ectopic SRC-1 expression. It is crucial to note that SRC-1 did not increase reporter gene activity in the absence of TCDD in the mutant cell line, indicating it is not involved in enhancing basal transcription from the reporter construct. It is possible that ARNT plays a role in recruiting different partners to different genes in response to diverse signals. For example, while ARNT recruits AhR to DREs in response to treatment with TCDD, ARNT also enlists HIF1-α to HREs in response to hypoxia (61). So, ARNT may be a central integrator for several signals/stimuli, which suggests that its heterodimer partners may play a more active role in recruiting coactivator complexes.
Collectively, these results indicate that certain steroid receptor coactivators appear to be involved in AhR/ARNT transcriptional activation. In addition, there appears to be a common coactivator or coactivator complexes, which are recruited to several receptor types in response to appropriate signals/stimuli. Several reports provide evidence for the possibility of an interplay between the AhR and other receptors, including the ER (21), AR (19), and HIF-lα (6). Competition for limited coactivators or pools of coactivators may provide a partial explanation for this interplay. Finally, whether coactivators are, in part, responsible for tissue- and cell-specific differences in TCDD responses remains to be established.
ACKNOWLEDGMENTS
We thank Malcolm Parker for SRC-la constructs, Michael Denison for pGUDLUC6.1, Ronald Hines for pRNH235, Steve Safe for TCDD, Oliver Hankinson for pcDNA3/βmAhR and Hepa-1 c4 mutant cell-lines, Christopher Bradfield for NΔ315 mAhR/ GAL4/pGEM-7Zf, Tom Gilmore for GST-Spl, and Christopher Glass for pGEX-GST-ER(HBD3) constructs. We thank Marcia Perdew for critically reviewing the manuscript. This work was supported by a grant from NIEHS (ES 04869).
REFERENCES
- 1. Abbott B. D.; Schmid J. E.; Pitt J. A.; Buckalew A. R.; Wood C. R.; Held G. A.; Diliberto J. J. Adverse reproductive outcomes in the transgenic Ah receptor-deficient mouse. Toxicol. Appl. Pharmacol. 155:62–70; 1999. [DOI] [PubMed] [Google Scholar]
- 2. Abnet C. C.; Tanguay R. L.; Heideman W.; Peterson R. E. Transactivation activity of human, zebrafish, and rainbow trout aryl hydrocarbon receptors expressed in COS-7 cells: Greater insight into species differences in toxic potency of polychlorinated dibenzo-p-dioxin, dibenzofuran, and biphenyl congeners. Toxicol. Appl. Pharmacol. 159:41–51; 1999. [DOI] [PubMed] [Google Scholar]
- 3. Andreola F.; Fernandez-Salguero P. M.; Chiantore M. V.; Petkovich M. P.; Gonzalez F. J.; De Luca L. M. Aryl hydrocarbon receptor knockout mice (AHR−/−) exhibit liver retinoid accumulation and reduced retinoic acid metabolism. Cancer Res. 57:2835–2838; 1997. [PubMed] [Google Scholar]
- 4. Bourguet W.; Ruff M.; Chambon P.; Gronemeyer H.; Moras D. Crystal structure of the ligand-binding domain of the human nuclear receptor RXR-alpha. Nature 375:377–382; 1995. [DOI] [PubMed] [Google Scholar]
- 5. Carver L. A.; Bradfield C. A. Ligand-dependent interaction of the aryl hydrocarbon receptor with a novel immunophilin homolog in vivo. J. Biol. Chem. 272:11452–11456; 1997. [DOI] [PubMed] [Google Scholar]
- 6. Chan W. K.; Yao G.; Gu Y. Z.; Bradfield C. A. Cross-talk between the aryl hydrocarbon receptor and hypoxia inducible factor signaling pathways. Demonstration of competition and compensation. J. Biol. Chem. 274:12115–12123; 1999. [DOI] [PubMed] [Google Scholar]
- 7. Darimont B. D.; Wagner R. L.; Apriletti J. W.; Stallcup M. R.; Kushner P. J.; Baxter J. D.; Fletterick R. J.; Yamamoto K. R. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 12:3343–3356; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ding X. F.; Anderson C. M.; Ma H.; Hong H.; Uht R. M.; Kushner P. J.; Stallcup M. R. Nuclear receptor-binding sites of coactivators glucocorticoid receptor interacting protein 1 (GRIP1) and steroid receptor coactivator 1 (SRC-1): Multiple motifs with different binding specificities. Mol. Endocrinol. 12:302–313; 1998. [DOI] [PubMed] [Google Scholar]
- 9. Gradin K.; Wilhelmsson A.; Poellinger L.; Berghard A. Nonresponsiveness of normal human fibroblasts to dioxin correlates with the presence of a constitutive xenobiotic response element-binding factor. J. Biol. Chem. 268:4061–4068; 1993. [PubMed] [Google Scholar]
- 10. Hankinson O.; Andersen R. D.; Birren B. W.; Sander F.; Negishi M.; Nebert D. W. Mutations affecting the regulation of transcription of the cytochrome PI-450 gene in the mouse Hepa-1 cell line. J. Biol. Chem. 260:1790–1795; 1985. [PubMed] [Google Scholar]
- 11. Hayashi Y.; Ohmori S.; Ito T.; Seo H. A splicing variant of steroid receptor coactivator-1 (SRC-IE): The major isoform of SRC-1 to mediate thyroid hormone action. Biochem. Biophys. Res. Commun. 236:83–87; 1997. [DOI] [PubMed] [Google Scholar]
- 12. Heery D. M.; Kalkhoven E.; Hoare S.; Parker M. G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387:733–736; 1997. [DOI] [PubMed] [Google Scholar]
- 13. Henry E. C.; Gasiewicz T. A. Transformation of the aryl hydrocarbon receptor to a DNA-binding form is accompanied by release of the 90 kDa heat-shock protein and increased affinity for 2,3,7,8-tetrachlorodibenzo-p-dioxin. Biochem. J. 294:95–101; 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Henttu P. M.; Kalkhoven E.; Parker M. G. AF-2 activity and recruitment of steroid receptor coactivator 1 to the estrogen receptor depend on a lysine residue conserved in nuclear receptors. Mol. Cell. Biol. 17:1832–1839; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Htun H.; Barsony J.; Renyi I.; Gould D. L.; Hager G. L. Visualization of glucocorttcoid receptor translocation and intranuclear organization in living cells with a green fluorescent protein chimera. Proc. Natl. Acad. Sci. USA 93:4845–4850; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ikeda M.; Kawaguchi A.; Takeshita A.; Chin W. W.; Endo T.; Onaya T. CBP-dependent and independent enhancing activity of steroid receptor coactivator-1 in thyroid hormone receptor-mediated transactivation. Mol. Cell. Endocrinol. 147:103–112; 1999. [DOI] [PubMed] [Google Scholar]
- 17. Ikonen T.; Palvimo J. J.; Janne O. A. Interaction between the amino- and carboxyl-terminal regions of the rat androgen receptor modulates transcriptional activity and is influenced by nuclear receptor coactivators. J. Biol. Chem. 272:29821–2988; 1997. [DOI] [PubMed] [Google Scholar]
- 18. Jaiswal A. K. Human NAD(P)H:quinone oxidoreductase2. Gene structure, activity, and tissue-specific expression. J. Biol. Chem. 269:14502–14508; 1994. [PubMed] [Google Scholar]
- 19. Jana N. R.; Sarkar S.; Ishizuka M.; Yonemoto J.; Tohyama C.; Sone H. Cross-talk between 2,3,7,8-tetrachlorodibenzo-p-dioxin and testosterone signal transduction pathways in LNCaP prostate cancer cells. Biochem. Biophys. Res. Commun. 256:462–468; 1999. [DOI] [PubMed] [Google Scholar]
- 20. Kalkhoven E.; Valentine J. E.; Heery D. M.; Parker M. G. Isoforms of steroid receptor co-activator 1 differ in their ability to potentiate transcription by the oestrogen receptor. EMBO J. 17:232–243; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kharat I.; Saatcioglu F. Antiestrogenic effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin are mediated by direct transcriptional interference with the liganded estrogen receptor. Cross-talk between aryl hydrocarbon-and estrogen-mediated signaling. J. Biol. Chem. 271:10533–10537; 1996. [DOI] [PubMed] [Google Scholar]
- 22. Ko H. P.; Okino S. T.; Ma Q.; Whitlock J. P. Jr. Dioxin-induced CYP1A1 transcription in vivo: The aromatic hydrocarbon receptor mediates transactivation, enhancer-promoter communication, and changes in chromatin structure. Mol. Cell. Biol. 16:430–436; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ko H. P.; Okino S. T.; Ma Q.; Whitlock J. P. Jr. Transactivation domains facilitate promoter occupancy for the dioxin-inducible CYP1A1 gene in vivo. Mol. Cell. Biol. 17:3497–3507; 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kobayashi A.; Numayama-Tsuruta K.; Sogawa K.; Fujii-Kuriyama Y. CBP/p300 functions as a possible transcriptional coactivator of Ah receptor nuclear translocator (Arnt). J. Biochem. (Tokyo) 122:703–710; 1997. [DOI] [PubMed] [Google Scholar]
- 25. Kumar M. B.; Tarpey R. W.; Perdew G. H. Differential recruitment of coactivator RIP 140 by Ah and estrogen receptors. Absence of a role for LXXLL motifs. J. Biol. Chem. 274:22155–22164; 1999. [DOI] [PubMed] [Google Scholar]
- 26. Lee C. H.; Chinpaisal C.; Wei L. N. Cloning and characterization of mouse RIP140, a corepressor for nuclear orphan receptor TR2. Mol. Cell. Biol. 18:6745–6755; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee S. K.; Kim H. J.; Na S. Y.; Kim T. S.; Choi H. S.; Im S. Y.; Lee J. W. Steroid receptor coactivator-1 coactivates activating protein-1-mediated transactivations through interaction with the c-Jun and c-Fos subunits. J. Biol. Chem. 273:16651–16654; 1998. [DOI] [PubMed] [Google Scholar]
- 28. Liu Z.; Wong J.; Tsai S. Y.; Tsai M. J.; O’Malley B. W. Steroid receptor coactivator-1 (SRC-1) enhances ligand-dependent and receptor-dependent cell-free transcription of chromatin. Proc. Natl. Acad. Sci. USA 96:9485–9490; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ma Q.; Whitlock J. P. Jr. A novel cytoplasmic protein that interacts with the Ah receptor, contains tetra-tricopeptide repeat motifs, and augments the transcriptional response to 2,3,7,8-tetrachlorodibenzo-p-dioxin. J. Biol. Chem. 272:8878–8884; 1997. [PubMed] [Google Scholar]
- 30. Mclnerney E. M.; Rose D. W.; Flynn S. E.; Westin S.; Mullen T. M.; Krones A.; Inostroza J.; Torchia J.; Nolte R. T.; Assa-Munt N.; Milburn M. V.; Glass C. K.; Rosenfeld M. G. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 12:3357–3368; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McKenna N. J.; Nawaz Z.; Tsai S. Y.; Tsai M. J.; O’Malley B. W. Distinct steady-state nuclear receptor coregulator complexes exist in vivo. Proc. Natl. Acad. Sci. USA 95:11697–11702; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Meyer B. K.; Pray-Grant M. G.; Vanden Heuvel J. P.; Perdew G. H. Hepatitis B vims X-associated protein 2 is a subunit of the unliganded aryl hydrocarbon receptor core complex and exhibits transcriptional enhancer activity. Mol. Cell. Biol. 18:978–988; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mimura J.; Yamashita K.; Nakamura K.; Morita M.; Takagi T. N.; Nakao K.; Ema M.; Sogawa K.; Yasuda M.; Katsuki M.; Fujii-Kuriyama Y. Loss of teratogenic response to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in mice lacking the Ah (dioxin) receptor. Genes Cells 2:645–654; 1997. [DOI] [PubMed] [Google Scholar]
- 34. Misiti S.; Koibuchi N.; Bei M.; Farsetti A.; Chin W. W. Expression of steroid receptor coactivator-1 mRNA in the developing mouse embryo: A possible role in olfactory epithelium development. Endocrinology 140:1957–1960; 1999. [DOI] [PubMed] [Google Scholar]
- 35. Misiti S.; Schomburg L.; Yen P. M.; Chin W. W. Expression and hormonal regulation of coactivator and corepressor genes. Endocrinology 139:2493–2500; 1998. [DOI] [PubMed] [Google Scholar]
- 36. Nebert D. W.; Negishi M.; Chen Y. T.; Tukey R. H. Cloning genes that encode inducible forms of P-450. Biochem. Soc. Trans. 12:99–101; 1984. [DOI] [PubMed] [Google Scholar]
- 37. Nguyen T. A.; Hoivik D.; Lee J. E.; Safe S. Interactions of nuclear receptor coactivator/corepressor proteins with the aryl hydrocarbon receptor complex. Arch. Biochem. Biophys. 367:250–257; 1999. [DOI] [PubMed] [Google Scholar]
- 38. Nolte R. T.; Wisely G. B.; Westin S.; Cobb J. E.; Lambert M. H.; Kurokawa R.; Rosenfeld M. G.; Willson T. M.; Glass C. K.; Milburn M. V. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature 395:137–143; 1998. [DOI] [PubMed] [Google Scholar]
- 39. Ogawa H.; Inouye S.; Tsuji F. I.; Yasuda K.; Umesono K. Localization, trafficking, and temperature-dependence of the Aequorea green fluorescent protein in cultured vertebrate cells. Proc. Natl. Acad. Sci. USA 92:11899–903; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Okino S. T.; Whitlock J. P. Jr. Dioxin induces localized, graded changes in chromatin structure: Implications for CyplAl gene transcription. Mol. Cell. Biol. 15:3714–3721; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Onate S. A.; Boonyaratanakornkit V.; Spencer T. E.; Tsai S. Y.; Tsai M. J.; Edwards D. P.; O’Malley B. W. The steroid receptor coactivator-1 contains multiple receptor interacting and activation domains that cooperatively enhance the activation function 1 (AF1) and AF2 domains of steroid receptors. J. Biol. Chem. 273:12101–12108; 1998. [DOI] [PubMed] [Google Scholar]
- 42. Onate S. A.; Tsai S. Y.; Tsai M. J.; O'Malley B. W. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science 270:1354–1357; 1995. [DOI] [PubMed] [Google Scholar]
- 43. Perdew G. H. Chemical cross-linking of the cytosolic and nuclear forms of the Ah receptor in hepatoma cell line 1c1c7. Biochem. Biophys. Res. Commun. 182:55–62; 1992. [DOI] [PubMed] [Google Scholar]
- 44. Peters J. M.; Narotsky M. G.; Elizondo G.; Fernandez-Salguero P. M.; Gonzalez F. J.; Abbott B. D. Amelioration of TCDD-induced teratogenesis in aryl hydrocarbon receptor (AhR)-null mice. Toxicol. Sci. 47:86–92; 1999. [DOI] [PubMed] [Google Scholar]
- 45. Poland A.; Knutson J. C. 2,3,7,8-Tetrachlorodibenzo-p-dioxin and related halogenated aromatic hydrocarbons: Examination of the mechanism of toxicity. Annu. Rev. Pharmacol. Toxcol. 22:517–554; 1982. [DOI] [PubMed] [Google Scholar]
- 46. Pollenz R. S.; Sattler C. A.; Poland A. The aryl hydrocarbon receptor and aryl hydrocarbon receptor nuclear translocator protein show distinct subcellular localizations in Hepa 1c1c7 cells by immunofluorescence microscopy. Mol. Pharmacol. 45:428–438; 1994. [PubMed] [Google Scholar]
- 47. Qi C.; Zhu Y.; Pan J.; Yeldandi A. V.; Rao M. S.; Maeda N.; Subbarao V.; Pulikuri S.; Hashimoto T.; Reddy J. K. Mouse steroid receptor coactivator-1 is not essential for peroxisome proliferator-activated receptor alpha-regulated gene expression. Proc. Natl. Acad. Sci. USA 96:1585–1590; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Reisz-Porszasz S.; Probst M. R.; Fukunaga B. N.; Hankinson O. Identification of functional domains of the aryl hydrocarbon receptor nuclear translocator protein (ARNT). Mol. Cell. Biol. 14:6075–6086; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Reyes H.; Reisz-Porszasz S.; Hankinson O. Identification of the Ah receptor nuclear translocator protein (Arnt) as a component of the DNA binding form of the Ah receptor. Science 256:1193–1195; 1992. [DOI] [PubMed] [Google Scholar]
- 50. Rowlands J. C.; McEwan I. J.; Gustafsson J. A. Trans-activation by the human aryl hydrocarbon receptor and aryl hydrocarbon receptor nuclear translocator proteins: Direct interactions with basal transcription factors. Mol. Pharmacol. 50:538–548; 1996. [PubMed] [Google Scholar]
- 51. Rushmore T. H.; King R. G.; Paulson K. E.; Pickett C. B. Regulation of glutathione S-transferase Ya sub-unit gene expression: Identification of a unique xenobiotic-responsive element controlling inducible expression by planar aromatic compounds. Proc. Natl. Acad. Sci. USA 87:3826–3830; 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schaufele F. Regulation of estrogen receptor activation of the prolactin enhancer/promoter by antagonistic activation function-2-interacting proteins. Mol. Endocrinol. 13:935–945; 1999. [DOI] [PubMed] [Google Scholar]
- 53. Schmidt J. V.; Su G. H.; Reddy J. K.; Simon M. C.; Bradfield C. A. Characterization of a murine Ahr null allele: Involvement of the Ah receptor in hepatic growth and development. Proc. Natl. Acad. Sci. USA 93:6731–6736; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Spencer T. E.; Jenster G.; Burcin M. M.; Allis C. D.; Zhou J.; Mizzen C. A.; McKenna N. J.; Onate S. A.; Tsai S. Y.; Tsai M. J.; O’Malley B. W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature 389:194–198; 1997. [DOI] [PubMed] [Google Scholar]
- 55. Swanson H. I.; Yang J. H. The aryl hydrocarbon receptor interacts with transcription factor IIB. Mol. Pharmacol. 54:671–677; 1998. [PubMed] [Google Scholar]
- 56. Takeshita A.; Yen P. M.; Ikeda M.; Cardona G. R.; Liu Y.; Koibuchi N.; Norwitz E. R.; Chin W. W. Thyroid hormone response elements differentially modulate the interactions of thyroid hormone receptors with two receptor binding domains in the steroid receptor coactivator–1. J. Biol. Chem. 273:21554–21562; 1998. [DOI] [PubMed] [Google Scholar]
- 57. Tukey R. H.; Nebert D. W. Regulation of mouse cytochrome P3-450 by the Ah receptor. Studies with a P3-450 cDNA clone. Biochemistry 23:6003–6008; 1984. [DOI] [PubMed] [Google Scholar]
- 58. Tuteja N.; Gonzalez F. J.; Nebert D. W. Developmental and tissue-specific differential regulation of the mouse dioxin-inducible PI-450 and P3-450 genes. Dev. Biol. 112:177–184; 1985. [DOI] [PubMed] [Google Scholar]
- 59. Westin S.; Kurokawa R.; Nolte R. T.; Wisely G. B.; Mclnerney E. M.; Rose D. W.; Milburn M. V.; Rosenfeld M. G.; Glass C. K. Interactions controlling the assembly of nuclear-receptor heterodimers and co-activators. Nature 395:199–202; 1998. [DOI] [PubMed] [Google Scholar]
- 60. Whitelaw M. L.; Gustafsson J. A.; Poellinger L. Identification of transactivation and repression functions of the dioxin receptor and its basic helix-loop-helix/PAS partner factor Arnt: Inducible versus constitutive modes of regulation. Mol. Cell. Biol. 14:8343–8355; 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wood S. M.; Gleadle J. M.; Pugh C. W.; Hankinson O.; Ratcliffe P. J. The role of the aryl hydrocarbon receptor nuclear translocator (ARNT) in hypoxic induction of gene expression. Studies in ARNT-deficient cells. J. Biol. Chem. 271:15117–15123; 1996. [DOI] [PubMed] [Google Scholar]
- 62. Xu J.; Qiu Y.; DeMayo F. J.; Tsai S. Y.; Tsai M. J.; O’Malley B. M. Partial hormone resistance in mice with disruption of the steroid receptor coactivator-1 (SRC-1) gene. Science 279:1922–1925; 1998. [DOI] [PubMed] [Google Scholar]
- 63. Zaher H.; Fernandez-Salguero P. M.; Letterio J.; Sheikh M. S.; Fornace A. J. Jr.; Roberts A. B.; Gonzalez F. J. The involvement of aryl hydrocarbon receptor in the activation of transforming growth factor-beta and apoptosis. Mol. Pharmacol. 54:313–321; 1998. [DOI] [PubMed] [Google Scholar]
- 64. Zhu Y.; Qi C.; Jain S.; Rao M. S.; Reddy J. K. Isolation and characterization of PBP, a protein that interacts with peroxisome proliferator-activated receptor. J. Biol. Chem. 272:25500–25506; 1997. [DOI] [PubMed] [Google Scholar]
- 65. Zwijsen R. M.; Buckle R. S.; Hijmans E. M.; Loomans C. J.; Bernards R. Ligand-independent recruitment of steroid receptor coactivators to estrogen receptor by cyclin Dl. Genes Dev. 12:3488–3498; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]


