Abstract
Rhizosphere bacterial communities of kodo millet plant was analyzed from a large metagenome sequence dataset. Plant rhizosphere samples of kodo millet was collected in replicates and the metagenomic sequence data were obtained through shotgun sequencing. Overall sequences in the dataset were 476,649 comprising total read length of 179,349,372 base pairs. Taxonomic data analysis led to characterize α-diversity of 107 species. Dominance of actinobacteria followed by unclassified sequences (derived from Bacteria) was recorded. Raw data along with the analysis result is publicly available from the MG-RAST server with ID mgm4761530.3.
Specification table
| Subject area | Biology |
| More specific subject area | Metagenomics |
| Type of data | DNA sequences |
| How data was acquired | Shotgun DNA sequencing using Illumina HiSeq |
| Data format | Analyzed data |
| Experimental factors | Collection of rhizosphere in replicates, extraction of metagenomic DNA from the rhizosphere of 2 months old kodo plants |
| Experimental features | Shotgun sequencing of the metagenomic DNA followed by bioinformatics analysis for microbial community composition |
| Data source location | Jagdalpur, Chhattisgarh, India (latitude: 19.07 and longitude: 81.96) |
| Data accessibility | Data is available from MG-RAST server (ID: mgm4761530.3) (http://metagenomics.anl.gov/mgmain.html?mgpage=overview&metagenome=mgm4761530.3). |
| Related research article | None |
Value of the data
-
•
The data highlights rhizosphere bacterial diversity of kodo millet plants grown under low-fertility soils and drought-prone conditions.
-
•
Analysis reveals dominance of actinobacteria in the rhizosphere of kodo plant.
-
•
The dataset shows diversity of plant growth promoting bacteria (PGPB).
-
•
The data enhances our understanding on dominant microbial inhabitants of millet rhizosphere that may further be exploited for growing crops under harsh abiotic conditions and low-fertility soil status.
1. Data
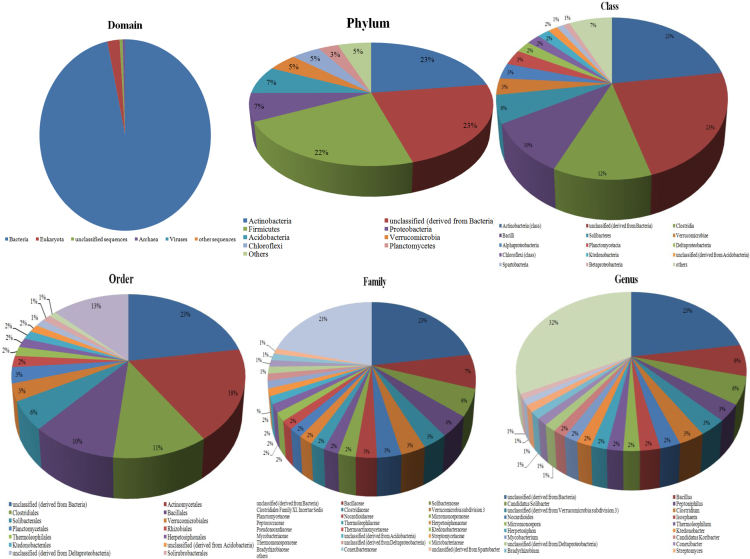
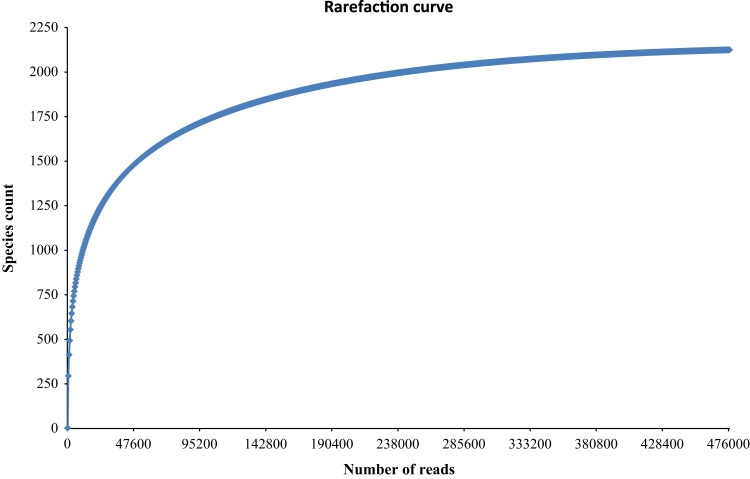
The rhizosphere metagenomic shotgun sequencing data was obtained. Total number of sequences were 476,649 with total read length of 179,349,372 base pairs (Table 1). Bacterial community structure in the kodo rhizosphere is reflected in Fig. 1, species richness in Fig. 2 and the α-diversity of 107 species is shown in Fig. 3.
Table 1.
Details about the raw and processed sequences of the kodo rhizosphere metagenome.
| Information about uploaded data | |
| Number of basepair | 179,349,372 bp |
| Number of sequences | 476,649 |
| Mean sequence length | 376 ± 76 bp |
| Mean GC percent | 57 ± 3% |
| Information after quality control analysis | |
| bp count | 22,138,479 bp |
| Sequences count | 98,133 |
| Mean sequence length | 226 ± 124 bp |
| Mean GC percent | 57 ± 3% |
| About processed sequences | |
| Predicted protein features | 679 |
| Predicted rRNA features | 34,247 |
Fig. 1.
Abundance of bacterial communities at different taxonomic units (Domain, Phylum, Class, Order, Family and Genus). Groups occupying less than 1% of the distribution were clubbed together and was designated as ׳Others׳.
Fig. 2.
Rarefaction curve of species richness.
Fig. 3.
α-diversity of the data set. The min, max, and mean values along with the standard deviation ranges (σ and 2σ) in varying shades are shown. The α-diversity of this metagenome dataset is shown in red. Alpha diversity sum up the diversity of organisms in a particular sample by a single number.
Out of total reads, 96.66% were assigned to bacteria (Supplementary Table 1). Actinobacteria was the most dominant phylum (22.76%) followed by unclassified bacteria (22.64%) and Firmicutes (22.2%) (Supplementary Table 2). Dominance of actinobacteria was also evident at the class level (Supplementary Table 3). At the order level, unclassified bacteria and Actinomycetales were the most dominating communities (Supplementary Table 4). Unclassified bacteria were also observed at family (Supplementary Table 5) and genus (Supplementary Table 6) level, though families of actinobacteria also exhibited significant proportion.
2. Experimental design, materials, and methods
2.1. Sample collection
Rhizosphere samples of kodo plants was obtained from the field of the College of Agriculture, Jagdalpur, Chhattisgarh, India (19.07N;81.96E) in April 2017.
2.2. DNA extraction
Total DNA was isolated through the FastDNA™ SPIN Kit following manufacturer instructions. Community DNA was purified and characterized through agarose-gel electrophoresis and NanoDrop spectrophotometer.
2.3. Metagenome sequencing
For the isolated DNA, amplicon sequencing was carried out with Illumina HiSeq sequencing system.
2.4. Initial pre-processing and QC check
The paired end fastq read files of the rhizosphere metagenomic dataset was processed through the standard pipeline of MG-RAST server [1] with default parameters.
2.5. Taxonomic analysis
For the taxonomic assignments, dataset was processed via MG-RAST server [1] by aligning the reads against the RefSeq protein database which provides search against various sequence databases at the same time [1]. Parameters taken were maximum E-value: 1 × 10−5, minimum percentage identity: 60%, and minimum alignment length: 15.
Acknowledgments
Ratna Prabha is thankful to Science and Engineering Research Board, Department of Science and Technology, Ministry of Science and Technology, Government of India for financial support in the form of SERB National PostDoctoral Fellowship (Grant: PDF/2016/000714).
Footnotes
Transparency data associated with this article can be found in the online version at https://doi.org/10.1016/j.dib.2018.09.006.
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.dib.2018.09.006.
Contributor Information
Ratna Prabha, Email: ratnasinghbiotech30@gmail.com.
Mukesh K. Verma, Email: vc@csvtu.ac.in.
Transparency document. Supplementary material
Supplementary material
Appendix A. Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
References
- 1.Meyer F., Paarmann D., D׳Souza M., Olson R., Glass E.M., Kubal M., Paczian T., Rodriguez A., Stevens R., Wilke A., Wilkening J., Edwards R.A. The metagenomics RAST server – a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinform. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material