Abstract
142 protein structure models were submitted to second Cryo-EM model challenge (2015–2016). Accuracy of the models was evaluated with 54 evaluation scores. Results of the descriptive statistical analysis of the scores are provided in this article.
Specifications table
| Subject area | Structural Biology |
| More specific subject area | Cryo-EM Models |
| Type of data | Figures |
| How data was acquired | Computational analysis |
| Data format | Analyzed |
| Experimental factors | None |
| Experimental features | None |
| Data source location | Rutgers University |
| Data accessibility | http://model-compare.emdatabank.org/data/scores |
| http://model-compare.emdatabank.org/em_score_boxplots.cgi |
Value of the data
-
•
The data reveal ranges of evaluation scores for models submitted to cryo-EM model challenge and provide descriptive statistics.
-
•
The data show differences in accuracy of cryo-EM models generated with different modeling techniques.
-
•
The data can serve as a benchmark for future cryo-EM modeling challenges.
-
•
The data can be compared with the data in other structure modeling experiments (e.g., CASP [1]).
1. Data
142 protein structure models were submitted on eight modeling targets of the second Cryo-EM model challenge (2015–2016) [2]. A computational system was developed to estimate the accuracy of the models [3]. Each model was evaluated using a suite of 15 software packages and, as a result, 54 accuracy scores were generated per model. All scores are presented in tables and graphs of the dedicated web infrastructure (http://model-compare.emdatabank.org). Some scores were analyzed in the accompanying paper [3]. This article provides results of the descriptive statistical analysis of the complete set of evaluation scores.
2. Materials and methods
2.1. Model types
Each model submitted to the second cryo-EM model challenge was accompanied by basic information about the modeling technique used. This information indicated whether model was built ab initio by fitting coordinates to density maps or by optimizing already available related structures. Based on this information, models were binned into two categories: ab initio or optimized. Fig. 1, Fig. 2, Fig. 3 in this paper show distributions of the evaluation scores separately for ab initio and optimized models, while Figs. 4 and 5 show the distributions for all models in one dataset (see description of the data presented in the figures in Section 4.4 below).
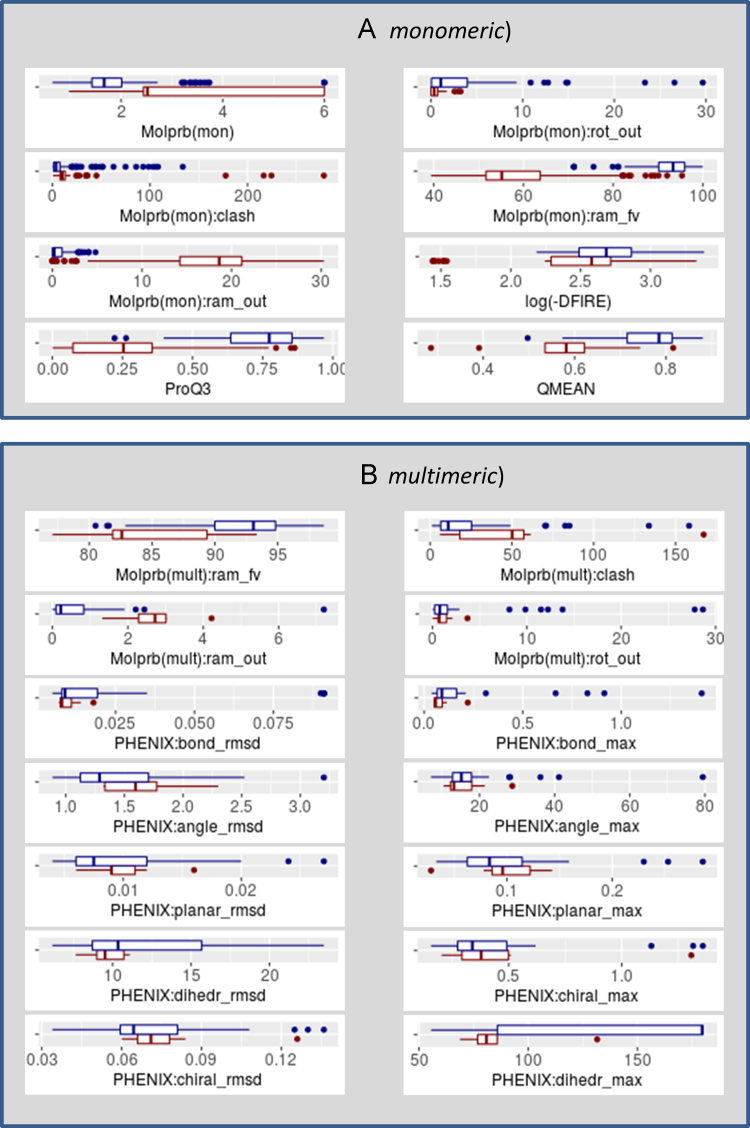
Fig. 1.
Distribution of the evaluation scores calculated exclusively from the model coordinates for different types of models. For each measure (specified in the x-axis title), a blue boxplot shows the score distribution for models built starting from reference structure, while a red boxplot – for models built ab initio. Panel (A) shows evaluation scores for models’ subunits (monomeric evaluation mode), while panel (B) for whole multimeric models.
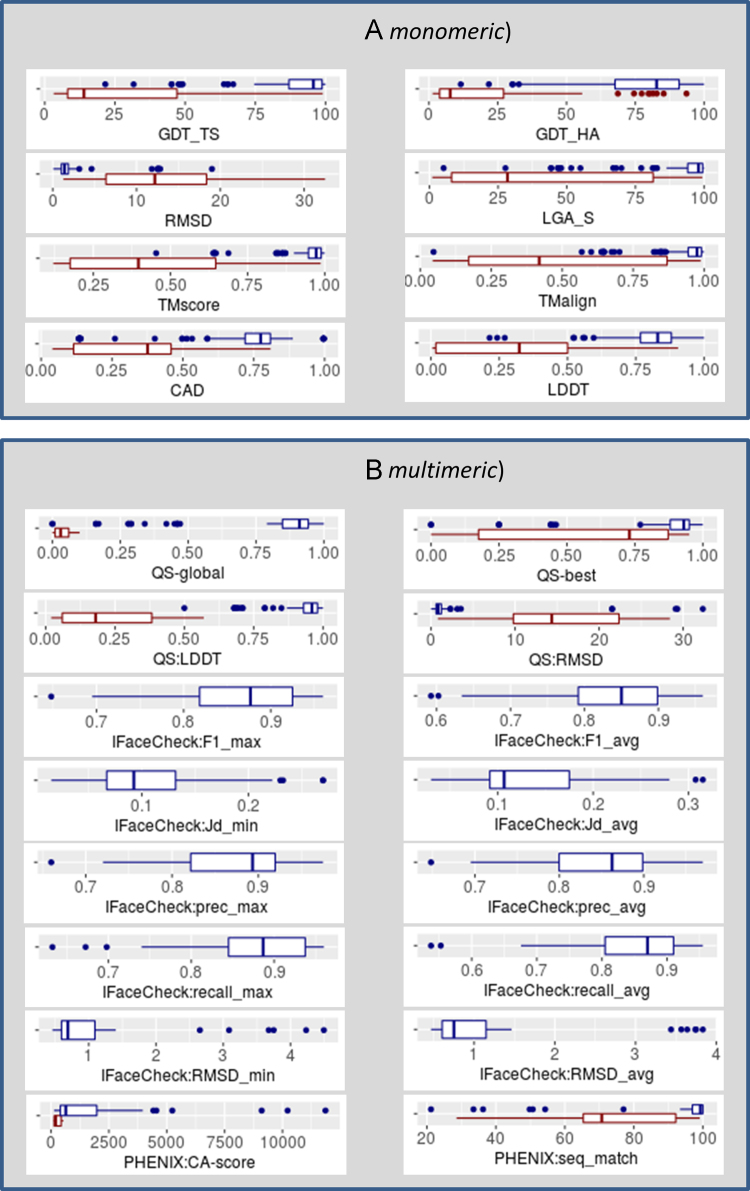
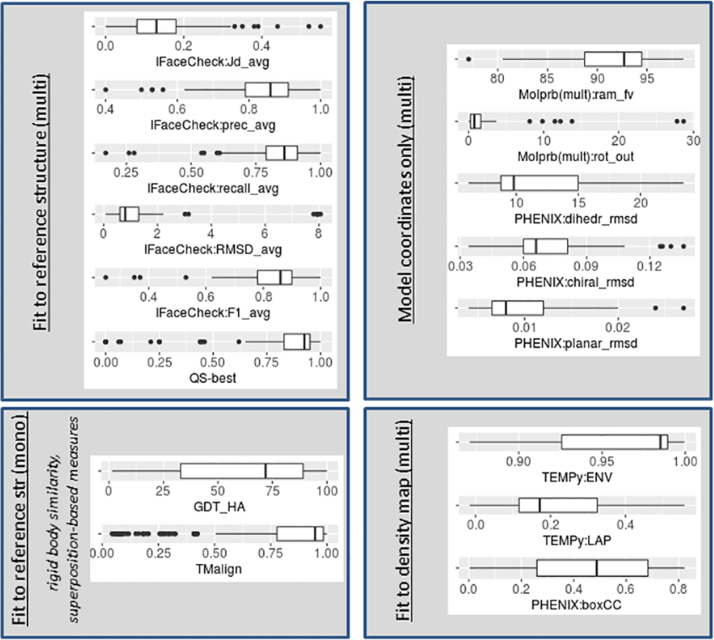
Fig. 2.
Distribution of the evaluation scores calculated by comparing models with reference structures. For each measure (specified in the x-axis title), a blue boxplot shows the score distribution for models built starting from reference structure, while a red boxplot – for models built ab initio. Panel (A) shows evaluation scores for models’ subunits (monomeric evaluation mode), while panel (B) for whole multimeric models.
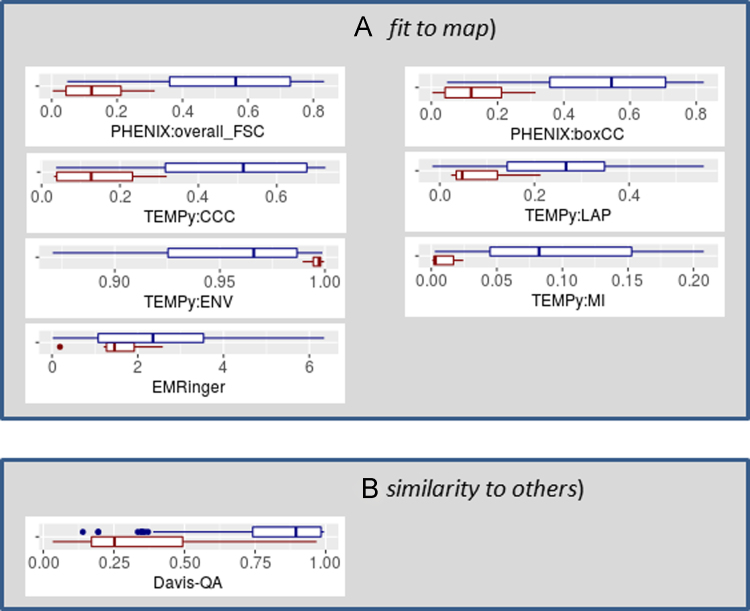
Fig. 3.
Distribution of the evaluation scores estimating fit of models to density maps (panel A) and similarity of models to other submitted models (panel B). For each measure (specified in the x-axis title), a blue boxplot shows the score distribution for models built starting from reference structure, while a red boxplot – for models built ab initio.
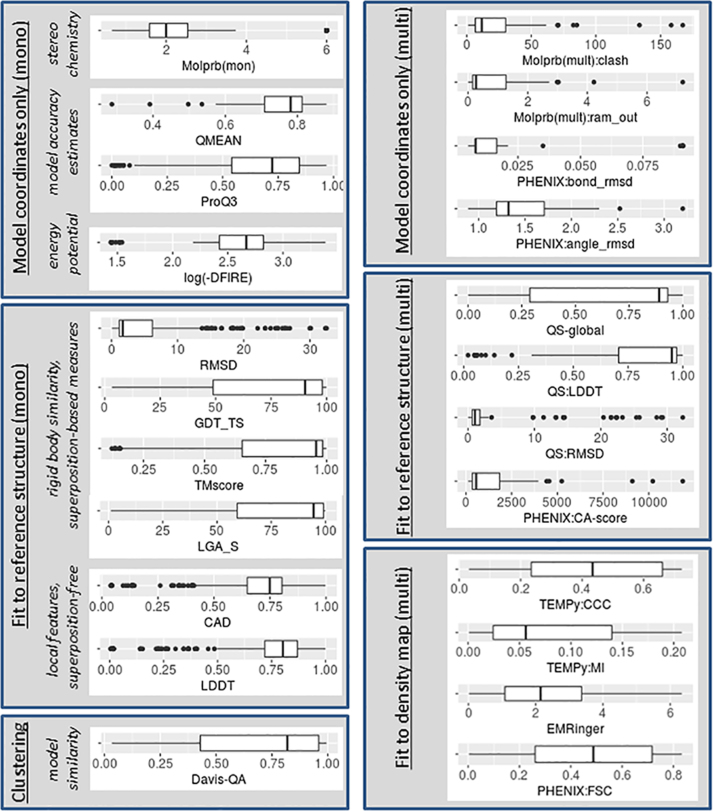
Fig. 4.
Distribution of the evaluation scores shown in Fig. 8 of Ref. [3] for all submitted models (optimization and ab initio models pulled together). Left set of boxplots shows scores from multimeric evaluations, right set – from monomeric ones.
Fig. 5.
Distribution of the evaluation scores not included in Fig. 4 for all submitted models.
2.2. Box plots
Data in the Fig. 1, Fig. 2, Fig. 3, Fig. 4, Fig. 5 are presented as box plots. Box boundaries correspond to the Q1 = 25th (bottom) and Q3 = 75th (top) percentiles in the data; the horizontal line inside the box corresponds to the median (Q2). The height of the box defines the interquartile range (IQR = Q3-Q1). The height of the whiskers shows the range of the values outside the interquartile range, but within 1.5 IQR. The dots correspond to outliers, i.e. values outside the 1.5 IQR range.
2.3. Evaluation tracks and packages
Submitted models were evaluated in four evaluation tracks:
-
(1)
directly from the model coordinates, i.e. without referring to density maps or other available structures (software packages used: MolProbity 4.4 [4], phenix.model_vs_map module (from PHENIX 1.11.1-2575 distribution [5]), DFIRE [6], ProQ3 [7], QMEAN [8]);
-
(2)
comparing the model coordinates to those of reference structures (software packages used: LGA [9], TM [10], [11], LDDT [12], CAD [13], QS-score [14], IFaceCheck [15], phenix.chain_comparison module [5]);
-
(3)
checking fit of the model coordinates to the experimental 3DEM density maps (software packages used: TEMPy 1.0 [16], [17], EMRinger [18], phenix.model_vs_map module [5]);
-
(4)
comparing coordinates of each model to those of other submitted models (software packages used: Davis-QAconsensus [19]).
2.4. Score distributions
Detailed explanation of the evaluation scores is provided in the accompanying paper [3].
Fig. 1 illustrates distributions of evaluation scores calculated directly from model coordinates (evaluation track 2.3.1). Panel (A) shows scores calculated on representative model subunits, while panel (B) – on whole multimeric structures. The figure includes box plots for the following scores:
Panel (A):
-
•
Molprb(mon) – MolProbity׳s combined MPscore;
-
•
Molprb(mon):rot_out – MolProbity׳s rotamer outlier score;
-
•
Molprb(mon):clash – MolProbity׳s clash score;
-
•
Molprb(mon):ram_fv – MolProbity׳s Ramachandran favored score;
-
•
Molprb(mon):ram_out – MolProbity׳s Ramachandran outlier score;
-
•
Log(-DFIRE) – logarithm of negative DFIRE energy score;
-
•
ProQ3 – score from the ProQ3 program ran with default parameters;
-
•
QMEAN – score from the QMEAN program ran with default parameters.
Panel (B):
-
•
[Molprb(mult):ram_fv /Molprb(mult):clash /Molprb(mult):ram_out /Molprb(mult):rot_out] – MolProbity׳s [Ramachandran favored score /clash score /Ramachandran outlier score /rotamer outlier] score;
-
•
[PHENIX:bond_rmsd /PHENIX:angle_rmsd /PHENIX:planar_rmsd /PHENIX:chiral_rmsd /PHENIX:dihedr_rmsd] – the RMSD on [bond /angle /planarity /chirality /dihedral angle] deviations calculated with the phenix.model_vs_map module;
-
•
PHENIX:bond_max – the maximum deviation of bond distances from ideal values (in Å);
-
•
PHENIX:angle_max – the maximum deviation of angles from ideal values (in deg.);
-
•
PHENIX:planar_max – the maximum deviation of peptide bond planarity from ideal values (in deg.);
-
•
PHENIX:chiral_max – the maximum deviation of chirality score from ideal values;
-
•
PHENIX:dihedr_max – the maximum deviation of dihedral angles from ideal values (in deg.).
Fig. 2 illustrates distributions of evaluation scores calculated by comparing models with reference structures (evaluation track 2.3.2). Panel (A) shows scores calculated on representative model subunits, while panel (B) – on whole multimeric structures. The figure includes box plots for the following scores:
Panel (A):
-
•
GDT_TS, GDT_HA – scores from the LGA package ran with parameters: ‘-3 -sda -d:4’);
-
•
LGA_S – score from the LGA package ran with parameters: ‘-4 -sia -d:4’);
-
•
RMSD – root mean square deviation on Cα atoms of the representative chain (as reported by the LGA package);
-
•
TMscore, TMalign – scores from the TM package ran with default parameters;
-
•
CAD – CAD_aa variant of the CAD score calculated on all atoms;
-
•
LDDT – a score from the LDDT package run with 15 Å inclusion radius.
Panel (B):
-
•
[QS-global /QS-best] – Quaternary Structure score calculated on [all interfaces /best interface] with the QS-score package;
-
•
[QS:LDDT /QS:RMSD] – [LDDT and /Cα RMSD] scores calculated on all chains by the QS-score package;
-
•
IFaceCheck:F1_max – maximum F1 statistics from among those calculated on all interfaces calculated with the IFaceCheck package;
-
•
IFaceCheck:Jd_min – the minimum Jaccard distance from among those calculated on all interfaces;
-
•
IFaceCheck:prec_max – maximum precision from among those calculated on all interfaces;
-
•
IFaceCheck:recall_max – maximum recall from among those calculated on all interfaces;
-
•
IFaceCheck:RMSD_min – minimum RMSD on target interface atoms from among those calculated on all interfaces;
-
•
[IFaceCheck:F1_avg /IFaceCheck:Jd_avg /IFaceCheck:prec_avg /IFaceCheck:recall_avg /IFaceCheck:RMSD_avg] – the [F1 /Jaccard distance /precision /recall /interface RMSD] scores averaged over all interfaces;
-
•
[PHENIX:CA-score /PHENIX:seq_match] –scores generated with the phenix.chain_comparison module.
Fig. 3 illustrates distributions of evaluation scores estimating fit of models to density maps (panel A, evaluation track 2.3.3) and similarity of models to other submitted models (panel B, evaluation track 2.3.4). The figure includes box plots for the following scores:
Panel (A):
-
•
[PHENIX:overall_FSC /PHENIX:boxCC] – the [overall Fourier Shell Correlation in reciprocal Fourier space /per-chain box cross-correlation] calculated with the phenix.model_vs_map module;
-
•
[TEMPy:CCC /TEMPy:LAP /TEMPy:ENV /TEMPy:MI] – TEMPY׳s [cross-correlation coefficient /Laplacian-filtered cross-correlation /envelope /mutual information] scores;
-
•
EMRinger – EMRinger score calculated using the phenix:emringer module.
Panel (B):
-
•
Davis-QA – a model consensus score calculated by averaging the GDT_TS scores from pairwise comparisons of the model to all others.
Figs. 4 and 5 illustrate distributions of evaluation scores presented in Fig. 1, Fig. 2, Fig. 3 when all models (optimization and ab initio) are grouped in one dataset. Score names are as described above for Fig. 1, Fig. 2, Fig. 3.
Acknowledgements
Authors acknowledge support of the National Institute of General Medical Sciences, USA: Grant R01GM079429 to WC, and Grant P01GM063210 to PDA.
Footnotes
Transparency data associated with this article can be found in the online version at 10.1016/j.dib.2018.08.214.
Transparency document. Supplementary material
Supplementary material
References
- 1.Moult J., Fidelis K., Kryshtafovych A., Schwede T., Tramontano A. Critical assessment of methods of protein structure prediction (CASP)—round XII. Proteins Struct. Funct. Bioinforma. 2018;86:7–15. doi: 10.1002/prot.25415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lawson C., Kryshtafovych A., Chiu W., Adams P., Brünger A., Kleywegt G., Patwardhan A., Read R., Schwede T., Topf M., Afonine P., Avaylon J., Baker M., Braun T., Cao W., Chittori S., Croll T., DiMaio F., Frenz B., Grudinin S., Hoffmann A., Hryc C., Joseph A.P., Kawabata T., Kihara D., Mao B., Matthies D., McGreevy R., Nakamura H., Nakamura S., Nguyen L., Schroeder G., Shekhar M., Shimizu K., Singharoy A., Sobolev O., Tajkhorshid E., Teo I., Terashi G., Terwilliger T., Wang K., Yu I., Zhou H., Sala R. CryoEM models and associated data submitted to the 2015/2016 EMDataBank model challenge. Zotero. 2018 [Google Scholar]
- 3.Kryshtafovych A., Adams P.D., Lawson C.L., Chiu W. Evaluation system and web infrastructure for the second cryo‐EM model challenge. J. Struct. Biol. 2018;204(1):96–108. doi: 10.1016/j.jsb.2018.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen V.B., Arendall W.B., 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. (pii) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., McCoy A.J., Moriarty N.W., Oeffner R., Read R.J., Richardson D.C., Richardson J.S., Terwilliger T.C., Zwart P.H. PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. (pii) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou H., Zhou Y. Distance-scaled, finite ideal-gas reference state improves structure-derived potentials of mean force for structure selection and stability prediction. Protein Sci. 2002;11:2714–2726. doi: 10.1110/ps.0217002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uziela K., Shu N., Wallner B., Elofsson A. ProQ3: improved model quality assessments using Rosetta energy terms. Sci. Rep. 2016;6:33509. doi: 10.1038/srep33509. (pii) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benkert P., Kunzli M., Schwede T. QMEAN server for protein model quality estimation. Nucleic Acids Res. 2009;37:W510–W514. doi: 10.1093/nar/gkp322. (doi:gkp322 [pii]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zemla A. LGA: a method for finding 3D similarities in protein structures. Nucleic Acids Res. 2003;31:3370–3374. doi: 10.1093/nar/gkg571. 〈http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12824330〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y., Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. (doi:33/7/2302 [pii]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y., Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins. 2004;57:702–710. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]
- 12.Mariani V., Biasini M., Barbato A., Schwede T. lDDT: a local superposition-free score for comparing protein structures and models using distance difference tests. Bioinformatics. 2013;29:2722–2728. doi: 10.1093/bioinformatics/btt473. (pii) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olechnovic K., Kulberkyte E., Venclovas C. CAD-score: a new contact area difference-based function for evaluation of protein structural models. Proteins. 2013;81:149–162. doi: 10.1002/prot.24172. [DOI] [PubMed] [Google Scholar]
- 14.Bertoni M., Kiefer F., Biasini M., Bordoli L., Schwede T. Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology. Sci. Rep. 2017;7:10480. doi: 10.1038/s41598-017-09654-8. (pii) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lafita A., Bliven S., Kryshtafovych A., Bertoni M., Monastyrskyy B., Duarte J.M., Schwede T., Capitani G. Assessment of protein assembly prediction in CASP12. Proteins Struct. Funct. Bioinforma. 2018;86:247–256. doi: 10.1002/prot.25408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vasishtan D., Topf M. Scoring functions for cryoEM density fitting. J. Struct. Biol. 2011;174:333–343. doi: 10.1016/j.jsb.2011.01.012. (pii) [DOI] [PubMed] [Google Scholar]
- 17.Farabella I., Vasishtan D., Joseph A.P., Pandurangan A.P., Sahota H., Topf M. a Python library for assessment of three-dimensional electron microscopy density fits. J. Appl. Crystallogr. 2015;48:1314–1323. doi: 10.1107/S1600576715010092. (pii) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barad B.A., Echols N., Wang R.Y., Cheng Y., DiMaio F., Adams P.D., Fraser J.S. EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat. Methods. 2015;12:943–946. doi: 10.1038/nmeth.3541. (pii) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kryshtafovych A., Monastyrskyy B., Fidelis K. CASP prediction center infrastructure and evaluation measures in CASP10 and CASP ROLL. Proteins Struct. Funct. Bioinforma. 2014;82:7–13. doi: 10.1002/prot.24399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material