Abstract
Objective
We sought to identify AMPK-regulated genes via bioinformatic analysis of microarray data generated from skeletal muscle of animal models with genetically altered AMPK activity. We hypothesized that such genes would play a role in metabolism. Ganglioside-induced differentiation-associated protein 1 (GDAP1), a gene which plays a role in mitochondrial fission and peroxisomal function in neuronal cells but whose function in skeletal muscle is undescribed, was identified and further validated. AMPK activation reduced GDAP1 expression in skeletal muscle. GDAP1 expression was elevated in skeletal muscle from type 2 diabetic patients but decreased after acute exercise.
Methods
The metabolic impact of GDAP1 silencing was determined in primary skeletal muscle cells via siRNA-transfections. Confocal microscopy was used to visualize whether silencing GDAP1 impacted mitochondrial network morphology and membrane potential.
Results
GDAP1 silencing increased mitochondrial protein abundance, decreased palmitate oxidation, and decreased non-mitochondrial respiration. Mitochondrial morphology was unaltered by GDAP1 silencing. GDAP1 silencing and treatment of cells with AMPK agonists altered several genes in the core molecular clock machinery.
Conclusion
We describe a role for GDAP1 in regulating mitochondrial proteins, circadian genes, and metabolic flux in skeletal muscle. Collectively, our results implicate GDAP1 in the circadian control of metabolism.
Keywords: AMPK, Skeletal muscle, GDAP1, Diabetes, Circadian, Mitochondria
Highlights
-
•
Transcriptomic studies reveal GDAP1 mRNA is inversely associated with AMPK activity.
-
•
GDAP1 silencing increases mitochondrial protein abundance in skeletal muscle.
-
•
GDAP1 silencing influences expression of core molecular clock machinery.
-
•
GDAP1 is a AMPK target involved in metabolism and circadian gene expression.
Skeletal muscle is a major regulator of whole-body metabolic homeostasis. AMP-activated protein kinase (AMPK) is of particular importance for metabolic regulation in skeletal muscle given its role in glucose transporter type 4 (GLUT4)-mediated glucose uptake, inhibition of fatty-acid synthesis, and activation of mitochondrial biogenesis [1]. Importantly, skeletal muscle of type 2 diabetic patients retains the ability to activate AMPK during exercise, despite exhibiting several metabolic deficiencies, including impaired insulin-stimulated glucose uptake [2]. This is of clinical relevance given that prescriptions of lifestyle interventions, which include regular exercise training, have been reported to be as good as, or superior to, pharmacological treatment to preventing the development of type 2 diabetes in at-risk individuals [3], [4]. To gain insight into mechanisms by which AMPK modifies metabolic health, we have developed several genetically-modified animal models, which exhibit enhanced or diminished AMPK-mediated phosphorylation [5].
Skeletal muscle of mice with chronically activated AMPK activity exhibit a differential gene expression profile, which opposes that of mice with chronically impaired AMPK activity [6]. Pharmacological treatment of mice by oral gavage for six days with the AMPK agonist 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) increases the expression of oxidative genes in gastrocnemius muscle [7], implicating AMPK as a direct regulator of gene expression. Supporting evidence for AMPK as a transcriptional regulator comes from the findings that its catalytic subunit contains a nuclear export sequence, and its phosphorylation targets include cytoplasmic and nuclear proteins [8], [9]. Although microarray studies demonstrate that AMPK modulates the expression of specific candidate genes, these putative AMPK-dependent genes have not been independently validated for a role in metabolic homeostasis.
Transcriptomic studies reveal that ganglioside-induced differentiation-associated protein 1 (GDAP1) mRNA is inversely associated with AMPK activity [6], [7]. GDAP1 encodes for ganglioside-induced differentiation-associated protein 1 (GDAP1) [6], [7]. Mutations in GDAP1 lead to an incurable neurological disorder, Charcot-Marie Tooth disease, which is characterized by altered mitochondrial morphology [10], [11]. GDAP1 may be autoinhibitory, playing a role in mitochondrial fission in its active conformation [12]. GDAP1 is linked to insulin signaling and altered substrate handling of carbohydrates and lipids in a Drosophila model of GDAP1 function [13]. Given that the form and function of mitochondria are subject to disruption by improper GDAP1 expression, and that GDAP1 expression is under the control of AMPK, we hypothesized that GDAP1 plays a role in skeletal muscle metabolism.
In this study, we first utilized publically-available transcriptomics data from rodent models of altered AMPK activation in skeletal muscle to identify novel AMPK-regulated gene candidates for further validation in the context of metabolic homeostasis. Specifically, we compared the skeletal muscle transcriptomic profiles of AMPKγ3R225Q transgenic mice, AMPKγ3−/− knockout mice, and AICAR-treated wild-type mice. We have previously reported evidence for AMPKγ3-dependent transcriptional regulation in skeletal muscle [6]. Additionally, we have shown that phosphorylation of the AMPK target ACC is increased in skeletal muscle of AMPKγ3R225Q transgenic mice under basal conditions and in response to either AICAR-stimulated or exercise compared with wild-type mice, and unaltered between AMPKγ3−/− knockout and wild-type mice [5], [14]. These genetically modified mouse models have been useful to identify AMPKγ3-dependent gene transcription and metabolic regulation. In terms of the AMPKγ3−/− mouse model, despite a relatively low contribution to global AMPK activity, the γ3 unit may have very specific and important influences on the outcomes of AMPK activation. Thereafter, we validated the role of one such candidate gene, GDAP1, in skeletal muscle metabolism. Our approach identifies GDAP1 as a novel AMPK target that plays a role in metabolic processes and circadian gene expression in skeletal muscle.
1. Methods
1.1. Bioinformatic analysis
To identify candidate genes regulated by AMPK activity in skeletal muscle, microarray data were downloaded from the Gene Expression Omnibus (GEO) corresponding to models wherein AMPK activity was experimentally increased (GSE11804 and GSE4065) or decreased (GSE4063). Data from GSE11804 compared mice injected for six days with AICAR to saline-injected control mice [6]. Data from GSE4065 and GSE4063 compared wild-type littermates to either AMPKγ3R225Q or AMPKγ3−/− mice, representing gain or loss of AMPK function respectively [7]. All data were preprocessed by the robust multi-array average method [15] then non-annotated genes were filtered out, and unpaired t-tests were used with α = 0.05 to determine which transcripts were differentially expressed in each of the three cohorts. To identify AMPK-regulated genes, gene sets from each of the cohorts were further filtered to include only genes which were either 1) upregulated in both models of AMPK activation and downregulated in a model of AMPK inhibition, or 2) downregulated in both models of AMPK activation and upregulated in a model of AMPK inhibition.
1.2. AMPKγ3−/− and AMPKγ3R225Q mice
Skeletal muscle was harvested from male AMPKγ3−/− or AMPKγ3R225Q mice (both generated on a C57BL/6 background) and wild-type littermates, generated as described [5]. Animals were housed with same sex littermates in groups of 2–4 mice in cages with wood-chip bedding, clumps of tissue paper, and a cardboard shelter. The researchers handling the animals were blinded to the genotype. Mice were either starved for 12 h or had ad libitum access to food before being anesthetized with tribromoethanol. Thereafter, tissues were collected and frozen with clamps pre-cooled in liquid nitrogen. mRNA was extracted and converted to cDNA from gastrocnemius muscle as described [16]. Animal research was approved under license number N263/12 by the local ethical committee, the Stockholm North Animal Research Ethics Committee.
1.3. Study participants
Skeletal muscle cDNA from a previously published cohort of type 2 diabetes patients and normal glucose tolerant age-matched controls was used in this study [17]. The clinical characteristics of the study participants have been previously reported [17]. These participants donated skeletal muscle biopsies at three timepoints: at rest, immediately after 30 min of cycle ergometry at 85% of maximum heart rate, and 3 h after the exercise bout. Muscle biopsies were homogenized, RNA isolated, and converted to cDNA in a reverse transcription reaction, as described [17]. Muscle biopsies were also obtained from fasted normal glucose tolerant volunteers and primary cultures were prepared as described [16], [18], [19]. Informed consent was received from all study participants. The experimental procedures were conducted according to the Declaration of Helsinki and approved under the license number 2005/1080-31/4 by the local ethical committee for the participants who served as donors for the primary human skeletal muscle cells and under the license number 2013/647-31/3 for the participants who donated skeletal muscle biopsies.
1.4. Growth, differentiation, and siRNA-transfection in primary human skeletal muscle cells
Cells were grown and differentiated as previously described [16], [18], [19]. In one experiment, cells were differentiated then treated with 0.5 mM AICAR for 24 h prior to being harvested to interrogate gene expression. For one circadian experiment, skeletal muscle myotubes were differentiated into myotubes and then incubated with differentiation medium with 50% horse serum for 2 h before replacement with normal media to induce circadian gene oscillations, as previously described [20]. In all other experiments, cells were studied after being transfected to silence GDAP1 gene expression. In an initial experiment to interrogate if transfection with siRNA against GDAP1 would reduce protein abundance of GDAP1, cells were harvested 48 h after a single transfection, or after two transfections separated by 48 h. After determining that protein abundance had the greatest reduction after two transfections separated by 48 h (thus, 96 h after the first transfection), the same transfection timing was used for all other experiments. The transfection methods employed have been described earlier [19], using 5 μM of either silencer select Negative control No.2 (no. 4390847) or validated silencer select siRNA s28923 to target GDAP1 for two separate 5-h transfection periods separated by ∼48 h (Thermo Fisher Scientific, Waltham, MA). Two days after the final transfection, primary human skeletal muscle cells used to evaluate gene expression, protein signaling, metabolic function, or mitochondrial morphology.
1.5. GDAP1 silencing and AMPK-agonist treatment in primary human skeletal muscle cells
In the experiments performed using siRNA-transfections, the cells were treated for 48 h with either of the AMPK-activating compounds, 0.5 mM AICAR or 0.125 mM phenformin. Cells were subsequently harvested for gene expression analysis via RT-qPCR or protein signaling by western blot analysis as described [16].
1.6. Metabolic phenotyping of cells after GDAP1 silencing
Palmitate oxidation in primary human skeletal muscle cells was analyzed by exposing cells to 0–5 g glucose/L media for 6 h in the presence of radiolabeled palmitate (9, 10-[3H]palmitate, NET043005MC; PerkinElmer, Waltham, MA) as described [16]. The supernatant was subsequently processed and subjected to scintillation counting, and data were normalized to the protein content (Protein Assay Dye Reagent #5000006; Bio-Rad, Hercules, CA).
Glucose oxidation and glycogen synthesis were assayed as described [16], [19]. Cells were subjected to a 5-h serum starvation prior to exposure to insulin (10 or 120 nM) and radiolabeled glucose (D-[U-14C]glucose, NEC042B005MC; PerkinElmer). In the glycogen synthesis assay, cells were exposed to insulin and radiolabeled glucose for 2 h, then harvested, processed, and subjected to scintillation counting. In the glucose oxidation assay, cells were exposed to insulin and radiolabeled glucose for 4 h in air-tight cell-culture wells prior to an addition of 150 μL 2 M HCl; for 1 h. The CO2 released from this reaction was collected into small cups in the cell-culture wells containing 300 μL of 2 M NaOH prior to being subjected to scintillation counting. In both assays, radioactive counts were normalized to protein content, quantified by a Bradford colorometric assay (Bio-Rad).
Glucose uptake in primary human skeletal muscle cells was assessed by exposing cells to 2-[1,2-3H]deoxy-d-glucose, as described [19]. Cells were processed and subjected to scintillation counting. To analyze lactate production, media was collected from the same cells prior to exposure to the radiolabeled glucose. Lactate was measured in a colorimetric assay using N-methylphenazonium methyl sulphate, iodonitrotetrazolium chloride, lactic acid, β-nicotinamide adenine dinucleotide hydrate, and lactate dehydrogenase [21] (Sigma–Aldrich, St. Louis, MO). Radioactive counts corresponding to glucose uptake and lactate measured by the colorimetric assay were normalized to protein quantified by a Bradford colorometric assay (Bio-Rad).
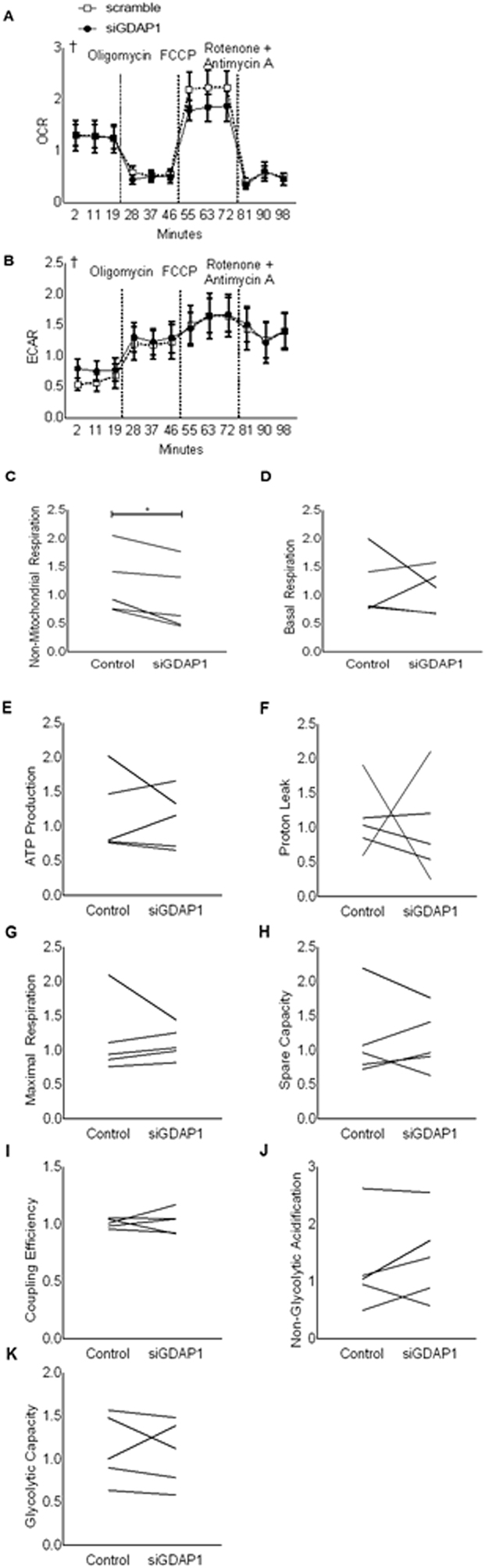
To assess the mitochondrial function of primary human skeletal muscle cells after GDAP1 silencing, cells were subjected to a Seahorse XF Mito Stress Test using the manufacturer's instructions for timing (Agilent, Santa Clara, CA). Oxygen consumption rates (OCR) and extracellular acidification rates (ECAR) were measured at three timepoints under unstimulated conditions, then after treatment with 1 μM oligomycin, 2 μM FCCP, and 0.75 μM rotenone + antimycin A. OCR and ECAR were normalized to protein as quantified by a Pierce BCA protein assay kit (Thermo Fisher Scientific).
1.7. Mitochondrial morphology with confocal microscopy
Mitochondrial morphology was examined using confocal microscopy (LSM880, Carl Zeiss AG, Jena, Germany) after staining cells with ViaFluor SE (Biotium Inc., Fremont, CA), Hoechst dye, and MitoTracker Deep Red (Thermo Fisher Scientific) according to manufacturer's instructions. Images from elongated multinucleated cells were captured using a 40x-objective on a confocal microscope. Mitochondrial networks were qualitatively assessed by researchers who were blinded to the treatment conditions. Additionally, images were quantified via Mitochondrial Network Analysis (MiNA) toolset and ImageJ [22].
1.8. Gene expression analysis
cDNA was prepared from human skeletal muscle, AMPKγ3−/− mice, and primary human skeletal muscle cells. Gene expression was determined by using Fast SYBR Green Master Mix (Thermo Fisher Scientific) and self-designed oligonucleotides (oligonucleotide sequences provided in Table 1).
Table 1.
Oligonucleotides used in the qPCR analysis.
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| Mm_B2m | TGCTATCCAGAAAACCCCTCA | GGGTGAATTCAGTGTGAGCC |
| Mm_Gdap1 | GGTCTTGGATCAGGTGGAAA | GATGCAATGTGACAGCCAGT |
| Hs_B2M | TGTCTTTCAGCAAGGACTGG | AGCAAGCAAGCAGAATTTGG |
| Hs_TBP | AGTTCTGGGATTGTACCGCA | TATATTCGGCGTTTCGGGCA |
| Hs_TFRC | CTGCAAAATCCGGTGTAGGC | TGAAGTTGCTGGTACCAAGAAC |
| Hs_RPLP0 | AGATTCGGGATATGCTGTTGGC | TCGGGTCCTAGACCAGTGTTC |
| Hs_GDAP1 | TGGAGAAAGTCTTGGATCAGGT | CTGCCAGGGTGAAGGATTC |
| Hs_DES | GGCTACCAGGACAACATTGC | GATTGATCCGGCTCTCCTCT |
| Hs_PDK4 | CACAGACAGGAAACCCAAGC | GCCCGCATTGCATTCTTAAA |
| Hs_DNM1L | TGGTGAAGCGGCAAATCAAA | TGCCACTAAGTTATGGACCATT |
| Hs_FIS1 | CAGTTTGAGTACGCCTGGTG | TCTCGTATTCCTTGAGCCGG |
| Hs_MFN2 | GAACTGTCTGGGACCTTTGC | CCAACCGGCTTTATTCCTGAG |
| Hs_MARCH5 | GTGACAGTGATGCAGGTTGT | TTCGAGTATTTGCGCCACAG |
| Hs_NR1D2 | TGGAGTTCATGCTTGCGAAG | ACCAAACCGAACAGCATCTC |
| Hs_PER3 | AGCCTTACAAGCTGGTTTGC | GTAGCATCCCTGGCTGTCTC |
| Hs_NPAS2 | TGAATCTGACCACACCTGCT | CTCTGGGCGTACTTGACTTG |
| Hs_DBP | CCCAGCTGATCTTGCCCTAT | GGCTCCAGTATTTCTCATCCTTC |
1.9. Protein abundance in primary human skeletal muscle cells
Cells were scraped into lysis buffer, supernatants were centrifuged, and the protein concentration was quantified by a Pierce protein assay kit (Thermo Fisher Scientific). Samples were diluted in Laemmli buffer to equal protein concentration prior to being subjected to western blot analysis as described [16]. Commercially available antibodies including p-ACC2S222 (#3661) and ACC (#3676) from Cell Signaling Technology (Beverly, MA), MYH1/2 (#sc-53088) from Santa Cruz Biotechnology (Dallas, TX), Desmin (#ab15200) and Total OXPHOS (#110411) from Abcam (Cambridge, UK), GDAP1 (#H00054332-A01) from Abnova (Taipei, Taiwan), and β-actin (#A5441) from Sigma–Aldrich were used for western-blot analysis. GDAP1 (#H00054332-A01) from Abnova (Taipei, Taiwan) was optimized for human skeletal muscle cells, but we were unable to optimize this antibody in mouse skeletal muscle.
1.10. Experimental outcomes
Gene expression of GDAP1 was the primary experimental outcome to validate the bioinformatics analysis. After validating that GDAP1 mRNA is negatively regulated by AMPK activity, secondary experimental outcomes included assessing the effects of GDAP1 silencing on the expression of metabolic genes, abundance of mitochondrial proteins, and metabolic phenotyping via assays of palmitate oxidation, lactate production, glucose uptake, glycogen synthesis, glucose oxidation, and Seahorse readings. Tertiary experimental outcomes were assessments of mitochondrial function via the Seahorse assays and morphology as indicated by confocal microscopy.
1.11. Statistical analysis
When the underlying assumptions were not violated, parametric tests were utilized to make statistical inferences. When assumptions were violated, nonparametric tests were used. The omnibus tests applied to interrogate that data are indicated in respective figure legends. The Benjamini-Hochberg false-discovery rate correction method was used in all post hoc pairwise-comparisons, except in the analysis of the microarray data for identification of candidate genes. The threshold for significance (α) was set to 0.05. R base v 3.4.3 and open-source packages were used for inferential statistics, while Graphpad Prism v7.04 (La Jolla, CA) was used for generation of figures. All data points are shown in figures.
2. Results
2.1. Identification and validation of GDAP1 as an AMPK-regulated gene
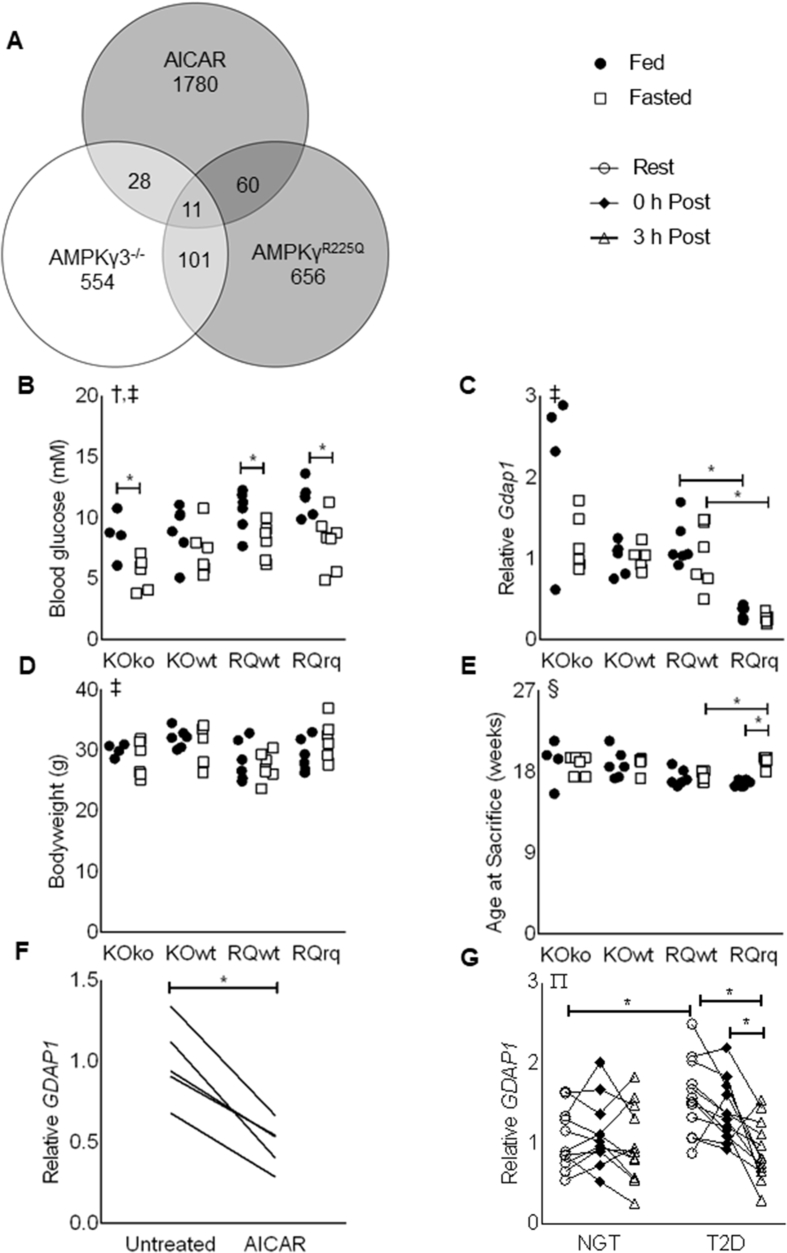
By overlaying the significantly regulated genes from the three GSE datasets, we identified 11 candidate genes which were either upregulated (Lpin1, Ugp2, Gpx3, Phlda3) or downregulated (Hhatl, Fam20c, Kansl1l, Dpp8, Gdap1, Aamdc, Asph) respectively, due to AMPK activity (Figure 1A and Table 2). Of these candidate genes, Gdap1 was selected for further validation. Gdap1 is a known regulator of mitochondrial dynamics in neuronal tissue, and mutations of the gene lead to Charcot-Marie Tooth diseases, a motor and sensory neuropathy affecting 1/2500 people [23].
Figure 1.
Identification of GDAP1 as an AMPK-regulated gene in skeletal muscle. (A) Venn diagram illustrating the number of significantly regulated genes due to increased AMPK activation (shaded circles) or decreased AMPK activation (empty circle). (B) Blood glucose, (C) mRNA of Gdap1 normalized to B2m in gastrocnemius muscle, (D) bodyweight, and (E) age of AMPKγ3 KO mice (KOko), wild-type littermates (KOwt), AMPKR225Q mice (RQrq), and wild-type littermates (RQwt) under fed (full circles) or fasted (empty boxes) conditions. All data are plotted from n = 4–7 mice per group. (F) mRNA of GDAP1 in primary human skeletal muscle cells. Expression was normalized to the geometric mean of B2M, TBP, and RPLP0. All data are plotted from n = 4 matched samples. (G) mRNA of GDAP1 in vastus lateralis muscle from individuals with normal glucose tolerance (NGT) or type-2 diabetes (T2D) at rest (open circles), 0-h post exercise (filled diamonds), or 3-h post exercise (empty triangles). Expression was normalized to B2M. All data are plotted from n = 11–12 per group. ∗Significant pairwise difference between subsets of data indicated by brackets as detected by paired-t test (panel C only) or pairwise post hoc tests after FDR-correction, †Significant effect of treatment as detected by 2-way Independent ANOVA. ‡Significant effect of genotype as detected by 2-way Independent ANOVA. §Significant difference between the groups as detected by Kruskal–Wallis test (assumptions for 2-way ANOVA were not met). ∏Significant condition-by-time interaction as detected by 2-way mixed-model ANOVA.
Table 2.
Candidate genes identified by bioinformatics analysis.
| Gene ID |
GSE4063 AMPKγ3−/− |
GSE4065 AMPKγ3R225Q |
GSE11804 AICAR-treated |
|||
|---|---|---|---|---|---|---|
| Fold-change | p-value | Fold-change | p-value | Fold-change | p-value | |
| Hhatl | 1.23 | 0.017 | −1.25 | 0.034 | −1.11 | 0.043 |
| Fam20c | 1.20 | 0.006 | −1.13 | 0.001 | −1.35 | 0.009 |
| Kansl1l | 1.19 | 0.046 | −1.37 | <0.001 | −1.09 | 0.001 |
| Dpp8 | 1.17 | 0.021 | −1.15 | 0.048 | −1.23 | 0.014 |
| Gdap1 | 1.17 | 0.020 | −1.29 | 0.002 | −1.29 | 0.046 |
| Aamdc | 1.13 | 0.012 | −1.31 | 0.019 | −1.11 | 0.039 |
| Asph | 1.10 | 0.027 | −1.15 | 0.005 | −1.08 | 0.027 |
| Lpin1 | −1.19 | 0.006 | 1.16 | 0.014 | 1.33 | <0.001 |
| Ugp2 | −1.20 | 0.003 | 1.32 | <0.001 | 1.16 | 0.004 |
| Gpx3 | −1.26 | 0.017 | 2.24 | <0.001 | 1.21 | 0.015 |
| Phlda3 | −1.36 | 0.016 | 1.19 | 0.005 | 1.33 | 0.010 |
Data were collected from a public repository and processed as described in the methods. The eleven genes are presented with their gene ID, fold-change as compared to the control in the respective dataset, and p-value after unpaired t-tests were performed.
To validate the findings from the bioinformatic analysis, Gdap1 expression was assessed in gastrocnemius muscle of AMPKγ3−/− and AMPKR225Q mice. Blood glucose was reduced in mice subjected to a 12-h fast (Figure 1B), and GDAP1 expression was inversely related to AMPK activity (Figure 1C). Gdap1 expression was reduced in skeletal muscle of mice with constitutively active AMPKγ3 (AMPKR225Q mice) as compared to wild-type littermates. Two minor caveats to this finding are that bodyweight was different due to genotype, but no pairwise comparisons were significantly different (Figure 1D), and the fasted AMPKR225Q mice were older than fed counterparts and fasted wild-type controls (Figure 1E).
To investigate if GDAP1 expression was directly reduced by AMPK activation, differentiated primary human skeletal muscle cells were treated with 0.5 mM AICAR, for 24 h. According to MIT assays, cell viability was unaffected. GDAP1 expression was reduced in cells treated with AICAR as compared to untreated control cells (Figure 1F). Furthermore, GDAP1 expression was elevated at baseline in vastus lateralis from type 2 diabetic patients but decreased in response to an acute exercise bout (Figure 1G).
2.2. Effects of GDAP1 silencing on gene expression and protein abundance
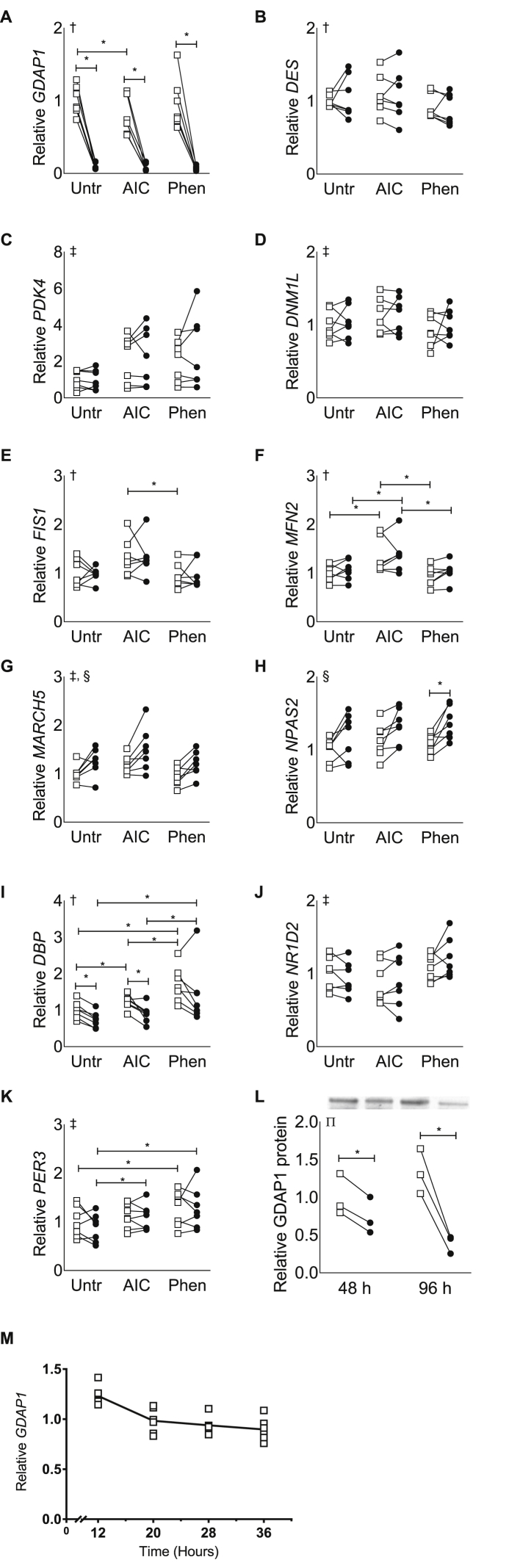
After validating that GDAP1 is expressed in primary human skeletal muscle cells and directly regulated by AMPK activity, we performed siRNA-mediated gene silencing of GDAP1 to interrogate the effects on gene expression and protein abundance. In conjunction with the gene-silencing experiments, cells were treated with or without the AMPK activators AICAR (0.5 mM) or phenformin (0.125 mM). The transfection procedure achieved consistent reductions of GDAP1 expression (Figure 2A,L). We confirmed our initial finding that AICAR-treatment reduces GDAP1 expression, although phenformin had no effect. DES expression was not altered due to GDAP1 silencing in any of the treatments, indicating that differentiation was unaltered due to GDAP1 silencing (Figure 2B). AMPK agonists increased PDK4 expression, although no pairwise comparisons were significant, and there was no effect of GDAP1 silencing (Figure 2C). Several genes which encode proteins responsible for maintenance of mitochondrial morphology were increased due to AICAR-treatment including DNM1L, FIS1, and MFN2, but neither phenformin treatment nor GDAP1 silencing altered the expression of these genes (Figure 2D–F). MARCH5, another gene involved in mitochondrial morphology, was increased in response to AICAR treatment and GDAP1 silencing (Figure 2G). The circadian gene NPAS2 was also increased due to GDAP1 silencing, but unaffected by AMPK agonists (Figure 2H). In contrast, the circadian gene DBP was reduced by GDAP1 silencing and increased due to phenformin treatment (Figure 2I). Two other circadian genes, NR1D2 and PER3, showed altered expression in response to AMPK agonists, but were unaffected by GDAP1 silencing (Figure 2J–K). Given these effects on circadian genes, we assessed whether GDAP1 had circadian oscillations in cultured skeletal muscle cells after serum shock (this induces core-clock oscillation as previously described [20]) (Figure 2M). GDAP1 gene expression had values of 0 for circadian oscillatory amplitude and period length after serum shock, when assessed by JTK_CYCLE, a non-parametric algorithm for detecting rhythmic components [24].
Figure 2.
GDAP1 silencing in primary human skeletal muscle cells affects MARCH5, NPAS2, and DBP mRNA expression. Primary human skeletal muscle cells were used for qPCR analysis of gene expression after normalization to the geometric mean of B2M and TFRC. Cells were transfected with a non-targeting scrambled control RNA (open squares) or siRNA targeted against GDAP1 (black circles) and treated with regular media (Unt), AICAR (AIC), or phenformin (Phen). Individual graphs report results for (A) GDAP1, (B) DES, (C) PDK4, (D) DNM1L, (E) FIS1, (F) MFN2, (G) MARCH5, (H) NPAS2, (I) DBP, (J) NR1D2, and (K) DBP expression. (L) GDAP1 protein 48 h after a single transfection (48 h) or after two transfections separated by 48 h (96 h). (M) GDAP1 expression after serum shock to induce circadian oscillations in primary human skeletal muscle cells. All data are plotted from n = 7 matched samples (A–K) or n = 3 matched samples (L). ∗Significant pairwise difference between subsets of data indicated by brackets as detected by pairwise post hoc tests after FDR-correction. †Significant differences among groups as detected by Friedman's test (assumptions for 2-way-RM ANOVA were not met). ‡Significant effect of pharmacological treatment as detected by 2-way-RM ANOVA. §Significant effect of GDAP1 silencing as detected by 2-way-RM ANOVA. ∏Significant treatment-by-time interaction as detected by 2-way-RM ANOVA.
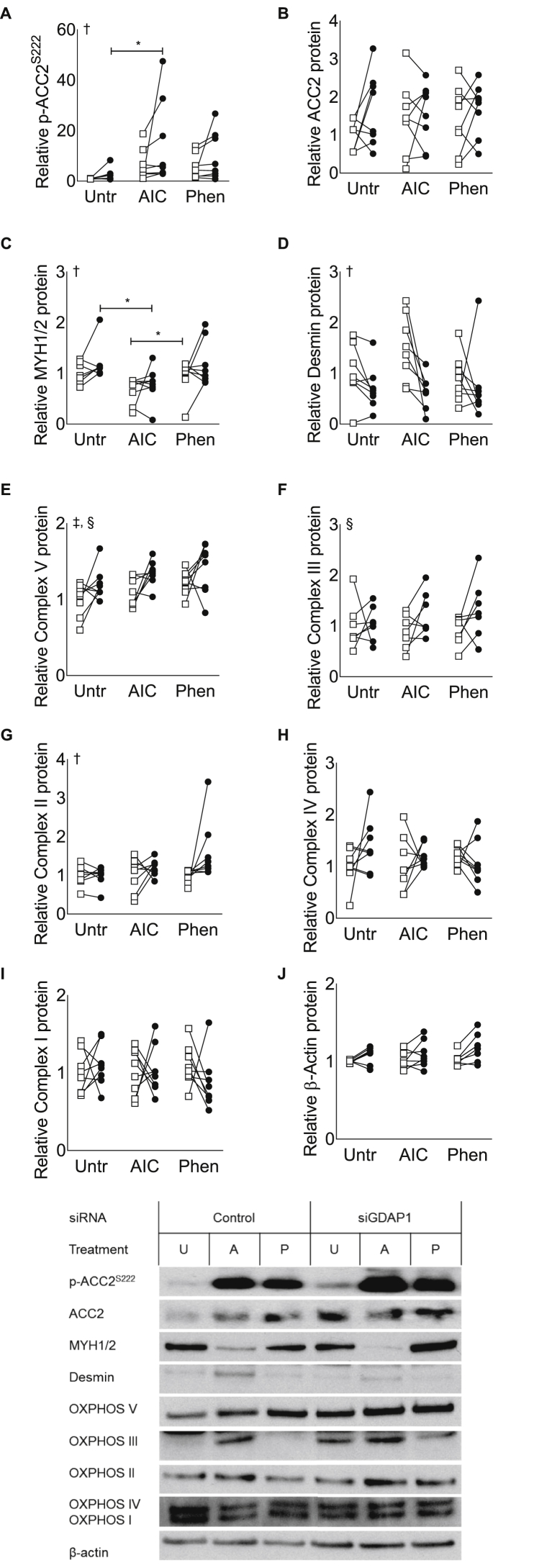
We next investigated whether AMPK activation and GDAP1 silencing altered the abundance of various proteins involved in metabolism. Treatment of cells with AICAR or phenformin increased phosphorylated acetyl CoA carboxylase (ACC), p-ACC2S222, without altering total ACC2 (Figure 3A–B). Myosin heavy-chain was reduced in AICAR-treated cells (Figure 3C). There was a tendency for GDAP1 silencing to reduce desmin protein abundance, but post hoc pairwise comparisons did not reveal a significant difference (Figure 3D). The abundance of mitochondrial complexes V and III were increased by GDAP1 silencing (Figure 3E–F). Mitochondrial complex II abundance was altered due to treatment AMPK-agonist treatment, but no pairwise post hoc comparisons were significant (Figure 3G). Mitochondrial complexes IV and I, as well as abundance of the reference protein β-actin, were unaltered by GDAP1 silencing or AMPK agonists, (Figure 3H–J).
Figure 3.
GDAP1 silencing increases mitochondrial proteins for Complex V and II. Primary human skeletal muscle cells were used for western blot analysis of protein abundance. Cells were transfected with a non-targeting scrambled control RNA (open squares) or siRNA targeted against GDAP1 (black circles) and treated with regular media (Unt), AICAR (AIC), or phenformin (Phen). (A) p-ACC2S222. (B) ACC2. (C) MYH1/2. (D) Desmin. (E) OXPHOS Complex V. (F) OXPHOS Complex III. (G) OXPHOS Complex II. (H) OXPHOS Complex IV. (I) OXPHOS Complex I. (J) β-Actin. All data are plotted from n = 7–8 matched samples. ∗Significant pairwise difference between subsets of data indicated by brackets as detected by pairwise post hoc tests after FDR-correction, †Significant differences among groups as detected by Friedman's test (assumptions for 2-way-RM ANOVA were not met), ‡Significant effect of pharmacological treatment as detected by 2-way-RM ANOVA, §Significant effect of GDAP1 silencing as detected by 2-way-RM ANOVA.
2.3. Metabolic phenotype of GDAP1-deficient human skeletal muscle cells
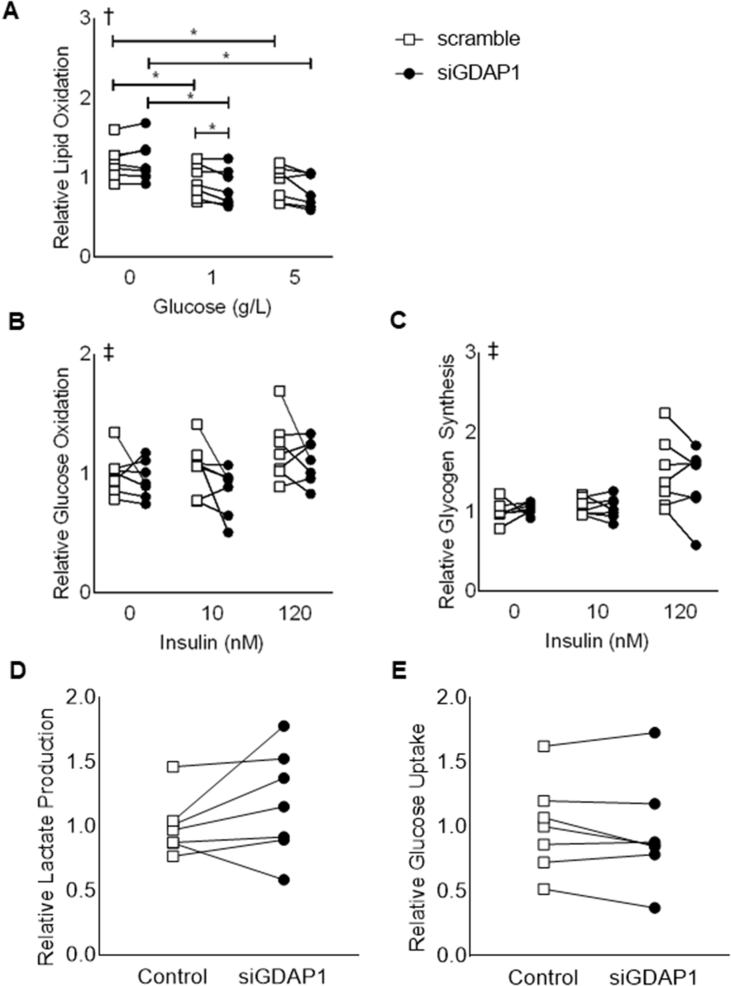
We next determined the influence of GDAP1 on glucose and lipid metabolism in cultured human skeletal muscle cells. Following GDAP1 silencing, cells were incubated in 0–5 g/L glucose. GDAP1 silencing reduced lipid oxidation of skeletal muscle cells, specifically in the presence of 1 g/L glucose (Figure 4A). However, we were unable to detect differences in glucose oxidation or glycogen synthesis in the presence or absence of insulin in GDAP1-silenced cells (Figure 4B–C). Furthermore, lactate production and glucose uptake was unaltered by GDAP1 silencing (Figure 4D–E).
Figure 4.
GDAP1 silencing reduces lipid oxidation without affecting glucose metabolism. Primary human skeletal muscle cells were used to assess lipid oxidation and glucose metabolism. Cells were transfected with a non-targeting scrambled control RNA (open squares) or siRNA targeted against GDAP1 (black circles). (A) Lipid oxidation in the presence of 0–5 g/L glucose. (B) Glucose oxidation in the absence or presence of 10 or 120 nM insulin. (C) Glycogen synthesis in the absence or presence of 10 or 120 nM insulin. (D) Lactate production. (E) Glucose uptake. In all panels, data were normalized to protein content, and all data are plotted for n = 7 matched samples. ∗Significant pairwise difference between subsets of data indicated by brackets as detected by pairwise post hoc tests after FDR-correction. †Significant interaction between treatment dose and GDAP1 silencing as detected by 2-way-RM ANOVA. ‡Significant effect of treatment dose as detected by 2-way-RM ANOVA.
To further investigate the metabolic role of GDAP1, cells were subjected to a Seahorse XF Mito Stress Test. While the sequential addition of oligomycin, FCCP, and rotenone + antimycin A altered OCR and ECAR, GDAP1 silencing was without effect (Figure 5A–B). A benefit of using the Seahorse Mito Stress test to investigate OCR and ECAR is that it permits the calculation of various metabolic parameters by comparing the responses of cells at different timepoints and under different pharmacological stimuli. We found that non-mitochondrial respiration was decreased by GDAP1 silencing (Figure 5C). However, other metabolic parameters including basal respiration, ATP production, proton leak, maximal respiration, oxidative spare capacity, coupling efficiency, non-glycolytic acidification, and glycolytic capacity were unaffected by GDAP1 silencing (Figure 5D–K).
Figure 5.
GDAP1 silencing reduces non-mitochondrial respiration. Primary human skeletal muscle cells were used in a Mito Stress Test after being transfected with a non-targeting scrambled control RNA (open squares) or siRNA targeted against GDAP1 (black circles). (A) Relative oxygen consumption rate are shown, and vertical dashed lines represent sequential injections of oligomycin, then FCCP, and finally rotenone + antimycin A. (B) Relative extracellular consumption rate (C) Non-mitochondrial respiration. (D) Basal respiration. (E) ATP production. (F) Proton leak. (G) Maximal respiration. (H) Spare capacity. (I) Coupling efficiency. (J) Non-glycolytic acidification. (K) Glycolytic capacity. All data were normalized to protein content. Mean ± SEM (A–B) or all individual responses (C–H) are plotted as relative rates from n = 5 matched samples. ∗Significant pairwise difference between subsets of data indicated by brackets as detected by paired-t test. †Significant differences among groups as detected by Friedman's test (assumptions for 2-way-RM ANOVA were not met).
2.4. Effect of GDAP1 silencing on mitochondrial morphology in primary human myotubes
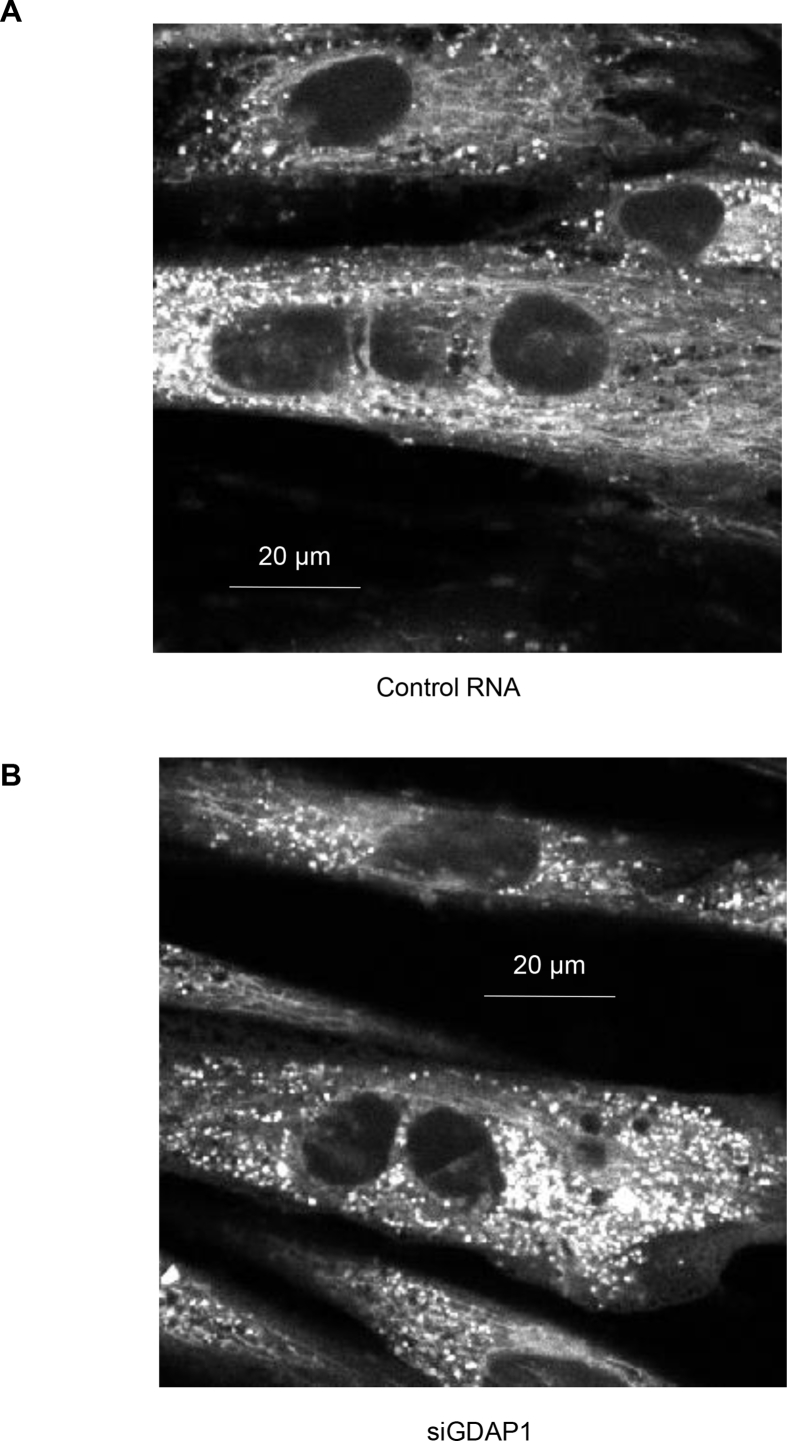
Morphology of mitochondrial networks in primary human myotubes was unaltered by GDAP1 silencing (Figure 6A–B). Although individual images of single myotubes under any given condition may have appeared to have more punctated mitochondrial or more robust networks, these observations were not consistent between the treatment groups. Additionally, quantitative analysis via MiNA toolset [22] identified no statistical differences in mitochondrial network size, amount or branch length (Table 3).
Figure 6.
Mitochondrial networks in primary skeletal muscle are unaltered by GDAP1 silencing. Primary human skeletal muscle cells were used for visualizing the mitochondrial networks. Sample images are shown where the white indicates the MitoTracker Deep Red dye. (A) Cells transfected with control RNA. (B) Cells transfected with siRNA against GDAP1.
Table 3.
Mitochondrial morphology.
| Mitochondrial Analysis | Treatment | Relative Value (AU) | SEM |
|---|---|---|---|
| Mean Branch Length | SCR | 10.99 | 0.7718 |
| Mean Branch Length | siGDAP1 | 11.64 | 0.3837 |
| Mean Branch Length | NT | 11.45 | 0.995 |
| Median Branch Length | SCR | 8.938 | 0.4427 |
| Median Branch Length | siGDAP1 | 9.453 | 0.1862 |
| Median Branch Length | NT | 9.214 | 0.638 |
| Mean Network Size (Branches) | SCR | 52.58 | 7.206 |
| Mean Network Size (Branches) | siGDAP1 | 50.4 | 3.164 |
| Mean Network Size (Branches) | NT | 50.76 | 1.472 |
| Median Network Size (Branches) | SCR | 4.417 | 0.4167 |
| Median Network Size (Branches) | siGDAP1 | 5.042 | 0.2083 |
| Median Network Size (Branches) | NT | 3.75 | 0.25 |
| Networks | SCR | 60 | 18.33 |
| Networks | siGDAP1 | 42.29 | 0.9583 |
| Networks | NT | 61 | 7.333 |
Mitochondrial morphology was examined using confocal microscopy (LSM880. Carl Zeiss AG. Jena, Germany) after staining cells with ViaFluor SE (Biotium Inc., Fremont. CA), Hoechst dye, and MitoTracker Deep Red (Thermo Fisher Scientific), according to manufacturer's instructions. Images from elongated multinucleated cells were captured using a 40x-objective on a confocal microscope. Images were analyzed via Mitochondrial Network Analysis (MiNA) toolset and ImageJ [22]. There were no quantitative statistical differences between mitochondrial networks. SCR = Scramble. NT = Non-transfected, AU = Arbitrary units (relative), SEM = Standard Error of Mean. Results are mean ± SEM for N = 3 with 3 images per subject per condition.
3. Discussion
AMPK plays an indirect role in regulating the gene expression profile of skeletal muscle via interactions with p53, Class IIA histone deacetylases (HDACs), peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α), cAMP-response-element binding protein (CREB) family proteins, forkhead box protein O (FoxO) family proteins, and transcriptional regulators [25]. Our research adds to the understanding of the role of AMPK in metabolic regulation by overlaying results from microarray studies to identify novel AMPK-dependent candidate genes in skeletal muscle. Several of these AMPK-regulated genes warrant further investigation including adipogenesis associated Mth938 domain-containing protein (AAMDC), aspartate beta-hyroxylase (ASPH), dipeptidyl peptidase 8 (DPP8), KAT8 regulatory NSL complex subunit 1 like (KANSL1L), and lipin 1 (LPIN1). Since DPP8 is closely related to DPP4, a pharmacological target for the treatment of type 2 diabetes, and LPIN1 has been linked to AMPK activity in rodent liver's subjected to metabolic stress [26], these targets warrant special attention for future studies.
Here we interrogated the role of GDAP1 in the regulation of metabolism in skeletal muscle. Using AMPKγ3 subunit transgenic and knockout mouse models and primary human muscle cells, we demonstrate that AMPK activity is inversely coupled to GDAP1 gene expression, indicative of AMPK regulation. We also provide evidence that GDAP1 expression is elevated in vastus lateralis muscle of type 2 diabetic patients and reduced 3-h after an acute exercise bout. Our findings couple AMPK activity to the level of GDAP1 gene expression, and provide novel evidence for a role of this gene in skeletal muscle metabolism. The precise link between AMPK activity and GDAP1 regulation is unknown. YY1 may transcriptionally regulate GDAP1 via a functional binding site in GDAP1's core promoter [27]. In turn, YY1 is regulated by PGC-1α [28], this may be the indirect mechanism by which AMPK regulates GDAP1 [29]. GDAP1 has mainly been studied in neuronal development and dysfunction. Given that GDAP1−/− mice have enlarged neuronal mitochondria, GDAP1 protein has been implicated in mitochondrial division [30], [31]. The muscular dysfunction and eventual atrophy exhibited in GDAP1−/− mice and patients with Charcot-Marie Tooth disease have classically been attributed to a secondary functional loss at the neuromuscular junctions as a consequence of disruption of the axonal arms in peripheral nerves.
We next assessed the effect of GDAP1 knockdown on mitochondrial form or function in skeletal muscle. Despite an increase in mitochondrial complex proteins, GDAP1 silencing reduced lipid oxidation in primary human muscle. GDAP1 silencing reduced non-mitochondrial respiration, suggesting alterations in peroxisomal function. Effective peroxisomal processing of acyl chains alleviates some of the metabolic burden caused by obesity and insulin resistance [32]. Moreover, impaired mitochondrial fatty-acid oxidation can be compensated for by increased peroxisomal oxidation [33]. Although GDAP1 is localized to the mitochondria, GDAP1 knockdown reduced peroxisomal fragmentation [34]. Our data may be explained by a potential cell-specific role of GDAP1, wherein it primarily regulates mitochondrial fission in nerve cells, and is delegated to peroxisomal regulation in skeletal muscle. Thus, it is perhaps unsurprising that we failed to detect robust alterations in mitochondrial morphology or mitochondrial function due to GDAP1 silencing. A “peroxisome-centric” role of GDAP1 in skeletal muscle may also explain why GDAP1 silencing did not alter the other metabolic parameters investigated herein.
Disruptions in GDAP1 may perturb whole-body metabolism. Genome-wide association studies (GWAS) compiled in the new GWAS catalog link GDAP1 to various symptoms of metabolic disorder [35]. Specifically, single-nucleotide polymorphisms (SNPs) in GDAP1 associate with waist circumference [36], [37], [38], [39], [40], [41], [42], [43], cholesterol, body-fat distribution, BMI, triglycerides, and blood pressure [44], and obesity-related traits such as BMI, fatty acids, metabolic rate, and sleep [36], [45], [46], [47]. Our findings coincide with the GWAS data in that GDAP1 knockdown reduced palmitate oxidation in skeletal muscle, which may lead to an increased risk of metabolic derangement through the accumulation of intramuscular lipid species, including diacylglycerols and ceramides [48], [49].
Given that GDAP1 mutations cause neurological disorder and SNPs in the gene associate with aspects of the metabolic syndrome, the teleological conundrum remains as to why AMPK activation reduces GDAP1 expression. We initiated our research by identifying a relationship between AMPK activation and GDAP1 expression. However, we extended our findings to several models including mice with differing levels of AMPK activity, primary human skeletal muscle cells treated with AICAR and studies in patients with type 2 diabetes. Furthermore, using the gene expression omnibus (GEO) [50], we observed that GDAP1 expression in gastrocnemius muscle from mice is responsive to a 12 h fast [51] or lifelong caloric restriction [52]. Collectively, these data support our hypothesis that downregulation of GDAP1 expression by AMPK activation may be an adaptive response to metabolic stress given that AMPK activation is an acute event that prioritizes cell energy utilization towards rapid ATP production. Although chronic disruption of GDAP1 has negative consequences for mitochondrial and peroxisomal function, a temporary impairment of translation of the GDAP1 gene by AMPK activity is unlikely to produce long-term negative outcomes, and in type 2 diabetes, it may even be beneficial.
An unexpected discovery is that both GDAP1 silencing and AMPK agonists alter the expression of several genes which are canonically associated with the circadian rhythm. Primary human skeletal muscle cells can be synchronized via serum-shock or serum-starvation protocols [53] and are therefore a useful model to study circadian biology. The effects of GDAP1 silencing on the circadian machinery are apparently robust, since we observed consistent changes in NPAS2 and DBP, despite the fact that the cells used in our studies were not synchronized. The link between GDAP1 and circadian gene expression is unknown. Because NPAS2 and DBP are antiphase to one another [54], our finding that these genes are regulated in opposing directions due to GDAP1 silencing is not surprising. Since DBP expression is elevated in liver of mice subjected to diet-induced obesity [55], the decrease due to GDAP1 silencing may be beneficial for metabolic regulation. NPAS2 can substitute for the CLOCK gene to code for a protein capable of pairing with BMAL1 to induce transcription of other genes [56]. Moreover, NPAS2 has a greater amplitude in skeletal muscle circadian rhythm as compared to CLOCK [56]. Our findings of increased NPAS2 expression in GDAP1-silenced cells strengthen the case for further study of this AMPK-regulated target. We have previously reported an interaction between AMPK activity and other molecular components of the circadian clock [57], but this is the first evidence that AMPK agonists impact DBP, NR1D2, and PER3. It appears that in cultured primary human skeletal muscle cells, GDAP1 is not circadian per se. However, the circadian clock is has multiple feedback loops, both positive and negative, ranging from an intracellular, to a systemic level. Our discoveries regarding the ability of AMPK agonists and GDAP1 silencing to alter core components of the circadian clock has major implications for the role of these genes in preserving metabolic health.
In conclusion, using publically available microarray data, we identified several novel AMPK-regulated genes including GDAP1. Other AMPK-regulated candidate genes –AAMDC, ASPH, DPP8, KANSL1L, and LPIN1– warrant future research. We reveal a role for GDAP1 in modulating mitochondrial protein abundance, lipid oxidation, and non-mitochondrial respiration. In the absence of direct evidence for a role of GDAP1 in modulating mitochondrial function, we suggest that it plays a role in maintaining peroxisomal integrity in skeletal muscle. We also report GDAP1 silencing and AMPK agonists interact in a previously undescribed manner to alter the expression of key molecular components of the core circadian clock machinery. Collectively, our results highlight a role for GDAP1 in the regulation of metabolic processes and circadian gene expression in skeletal muscle.
Author contributions
DGL, RJOS, AK, and JRZ conceived the study or parts of the study. DGL was responsible for statistical design and the analysis plan. DGL, RJOS, and BG generated the data. DGL, BG, AK, and JRZ analyzed and the interpreted the data. All authors participated in preparation of the manuscript and approved the final version for publication. The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request. J.R.Z. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Acknowledgements
The Strategic Research Program in Diabetes at Karolinska Institutet (2009-1068), European Research Council Ideas Program (ICEBERG, ERC-2008-AdG23285), Swedish Research Council (2011-3550), Swedish Diabetes Foundation (DIA2012-082; DIA2012-047), Swedish Foundation for Strategic Research (SRL10-0027), Diabetes Wellness Sweden (2949/2014SW), Novo Nordisk Foundation and Stockholm County Council (NNF14OC0011493 and 20150326) supported this research. The Novo Nordisk Foundation Center for Basic Metabolic Research is an independent Research Center at the University of Copenhagen partially funded by an unrestricted donation from the Foundation. Håkan Karlsson and Petter Alm assisted with the initial collection and analysis of biopsies from our human participants. Nicolas Pillon and Jonathon Smith assisted with assay development and data collection, respectively and are affiliated with Department of Physiology and Pharmacology, Integrative Physiology, Karolinska Institutet, Stockholm, Sweden. Florian Salomons assisted with confocal microscopy and is affiliated with the Department of Cell and Molecular Biology, Karolinska Institutet, Stockholm, Sweden.
Conflicts of interest
All authors approved the final version of the manuscript. None of the authors have a potential conflict of interest to report regarding this article. J.R.Z. is the guarantor of this work. Thus, she has full access to all the data of the study, and takes responsibility for the integrity of the data and the accuracy of the data analysis.
References
- 1.Kjobsted R., Hingst J.R., Fentz J., Foretz M., Sanz M.N., Pehmoller C. AMPK in skeletal muscle function and metabolism. The FASEB Journal. 2018;32:1741–1777. doi: 10.1096/fj.201700442R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Musi N., Fujii N., Hirshman M.F., Ekberg I., Froberg S., Ljungqvist O. AMP-activated protein kinase (AMPK) is activated in muscle of subjects with type 2 diabetes during exercise. Diabetes. 2001;50:921–927. doi: 10.2337/diabetes.50.5.921. [DOI] [PubMed] [Google Scholar]
- 3.Lindstrom J., Louheranta A., Mannelin M., Rastas M., Salminen V., Eriksson J., The Finnish Diabetes Prevention Study (DPS) Lifestyle intervention and 3-year results on diet and physical activity. Diabetes Care. 2003;26:3230–3236. doi: 10.2337/diacare.26.12.3230. [DOI] [PubMed] [Google Scholar]
- 4.Knowler W.C., Barrett-Connor E., Fowler S.E., Hamman R.F., Lachin J.M., Walker E.A. Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. New England Journal of Medicine. 2002;346:393–403. doi: 10.1056/NEJMoa012512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnes B.R., Marklund S., Steiler T.L., Walter M., Hjalm G., Amarger V. The 5'-AMP-activated protein kinase gamma3 isoform has a key role in carbohydrate and lipid metabolism in glycolytic skeletal muscle. Journal of Biological Chemistry. 2004;279:38441–38447. doi: 10.1074/jbc.M405533200. [DOI] [PubMed] [Google Scholar]
- 6.Nilsson E.C., Long Y.C., Martinsson S., Glund S., Garcia-Roves P., Svensson L.T. Opposite transcriptional regulation in skeletal muscle of AMP-activated protein kinase gamma3 R225Q transgenic versus knock-out mice. Journal of Biological Chemistry. 2006;281:7244–7252. doi: 10.1074/jbc.M510461200. [DOI] [PubMed] [Google Scholar]
- 7.Narkar V.A., Downes M., Yu R.T., Embler E., Wang Y.X., Banayo E. AMPK and PPARdelta agonists are exercise mimetics. Cell. 2008;134:405–415. doi: 10.1016/j.cell.2008.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salt I., Celler J.W., Hawley S.A., Prescott A., Woods A., Carling D. AMP-activated protein kinase: greater AMP dependence, and preferential nuclear localization, of complexes containing the alpha2 isoform. Biochemical Journal. 1998;334(Pt 1):177–187. doi: 10.1042/bj3340177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kazgan N., Williams T., Forsberg L.J., Brenman J.E. Identification of a nuclear export signal in the catalytic subunit of AMP-activated protein kinase. Molecular Biology of the Cell. 2010;21:3433–3442. doi: 10.1091/mbc.E10-04-0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pakhrin P.S., Xie Y., Hu Z., Li X., Liu L., Huang S. Genotype-phenotype correlation and frequency of distribution in a cohort of Chinese Charcot-Marie-Tooth patients associated with GDAP1 mutations. Journal of Neurology. 2018;265:637–646. doi: 10.1007/s00415-018-8743-9. [DOI] [PubMed] [Google Scholar]
- 11.Baloh R.H., Schmidt R.E., Pestronk A., Milbrandt J. Altered axonal mitochondrial transport in the pathogenesis of Charcot-Marie-Tooth disease from mitofusin 2 mutations. Journal of Neuroscience. 2007;27:422–430. doi: 10.1523/JNEUROSCI.4798-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huber N., Bieniossek C., Wagner K.M., Elsasser H.P., Suter U., Berger I. Glutathione-conjugating and membrane-remodeling activity of GDAP1 relies on amphipathic C-terminal domain. Scientific Reports. 2016;6:36930. doi: 10.1038/srep36930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lopez Del Amo V., Palomino-Schatzlein M., Seco-Cervera M., Garcia-Gimenez J.L., Pallardo F.V., Pineda-Lucena A. A Drosophila model of GDAP1 function reveals the involvement of insulin signalling in the mitochondria-dependent neuromuscular degeneration. Biochimica et Biophysica Acta. 2017;1863:801–809. doi: 10.1016/j.bbadis.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 14.Barnes B.R., Long Y.C., Steiler T.L., Leng Y., Galuska D., Wojtaszewski J.F. Changes in exercise-induced gene expression in 5'-AMP-activated protein kinase gamma3-null and gamma3 R225Q transgenic mice. Diabetes. 2005;54:3484–3489. doi: 10.2337/diabetes.54.12.3484. [DOI] [PubMed] [Google Scholar]
- 15.Eijssen L.M., Jaillard M., Adriaens M.E., Gaj S., de Groot P.J., Muller M. User-friendly solutions for microarray quality control and pre-processing on ArrayAnalysis.org. Nucleic Acids Research. 2013;41:W71–W76. doi: 10.1093/nar/gkt293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lassiter D.G., Nylen C., Sjogren R.J.O., Chibalin A.V., Wallberg-Henriksson H., Naslund E. FAK tyrosine phosphorylation is regulated by AMPK and controls metabolism in human skeletal muscle. Diabetologia. 2018;61:424–432. doi: 10.1007/s00125-017-4451-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mudry J.M., Alm P.S., Erhardt S., Goiny M., Fritz T., Caidahl K. Direct effects of exercise on kynurenine metabolism in people with normal glucose tolerance or type 2 diabetes. Diabetes Metabolism Research Reviews. 2016;32:754–761. doi: 10.1002/dmrr.2798. [DOI] [PubMed] [Google Scholar]
- 18.Mudry J.M., Lassiter D.G., Nylen C., Garcia-Calzon S., Naslund E., Krook A. Insulin and glucose alter death-associated protein kinase 3 (DAPK3) DNA methylation in human skeletal muscle. Diabetes. 2017;66:651–662. doi: 10.2337/db16-0882. [DOI] [PubMed] [Google Scholar]
- 19.Massart J., Sjogren R.J.O., Lundell L.S., Mudry J.M., Franck N., O'Gorman D.J. Altered miR-29 expression in type 2 diabetes influences glucose and lipid metabolism in skeletal muscle. Diabetes. 2017;66:1807–1818. doi: 10.2337/db17-0141. [DOI] [PubMed] [Google Scholar]
- 20.Balsalobre A., Damiola F., Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 21.Babson A.L., Phillips G.E. A rapid colorimetric assay for serum lactic dehydrogenase. Clinica Chimica Acta. 1965;12:210–215. doi: 10.1016/0009-8981(65)90032-x. [DOI] [PubMed] [Google Scholar]
- 22.Valente A.J., Maddalena L.A., Robb E.L., Moradi F., Stuart J.A. A simple ImageJ macro tool for analyzing mitochondrial network morphology in mammalian cell culture. Acta Histochemica. 2017;119:315–326. doi: 10.1016/j.acthis.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 23.Bertholet A.M., Delerue T., Millet A.M., Moulis M.F., David C., Daloyau M. Mitochondrial fusion/fission dynamics in neurodegeneration and neuronal plasticity. Neurobiology of Disease. 2016;90:3–19. doi: 10.1016/j.nbd.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 24.Hughes M.E., Hogenesch J.B., Kornacker K. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. Journal of Biological Rhythms. 2010;25:372–380. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McGee S.L., Hargreaves M. AMPK-mediated regulation of transcription in skeletal muscle. Clinical Science (London) 2010;118:507–518. doi: 10.1042/CS20090533. [DOI] [PubMed] [Google Scholar]
- 26.Hu M., Wang F., Li X., Rogers C.Q., Liang X., Finck B.N. Regulation of hepatic lipin-1 by ethanol: role of AMP-activated protein kinase/sterol regulatory element-binding protein 1 signaling in mice. Hepatology. 2012;55:437–446. doi: 10.1002/hep.24708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ratajewski M., Pulaski L. YY1-dependent transcriptional regulation of the human GDAP1 gene. Genomics. 2009;94:407–413. doi: 10.1016/j.ygeno.2009.08.014. [DOI] [PubMed] [Google Scholar]
- 28.Cunningham J.T., Rodgers J.T., Arlow D.H., Vazquez F., Mootha V.K., Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450:736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- 29.Canto C., Auwerx J. PGC-1alpha, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Current Opinion in Lipidology. 2009;20:98–105. doi: 10.1097/MOL.0b013e328328d0a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niemann A., Huber N., Wagner K.M., Somandin C., Horn M., Lebrun-Julien F. The Gdap1 knockout mouse mechanistically links redox control to Charcot-Marie-Tooth disease. Brain. 2014;137:668–682. doi: 10.1093/brain/awt371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barneo-Munoz M., Juarez P., Civera-Tregon A., Yndriago L., Pla-Martin D., Zenker J. Lack of GDAP1 induces neuronal calcium and mitochondrial defects in a knockout mouse model of charcot-marie-tooth neuropathy. PLoS Genetics. 2015;11 doi: 10.1371/journal.pgen.1005115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noland R.C., Woodlief T.L., Whitfield B.R., Manning S.M., Evans J.R., Dudek R.W. Peroxisomal-mitochondrial oxidation in a rodent model of obesity-associated insulin resistance. American Journal of Physiology-Endocrinology and Metabolism. 2007;293:E986–E1001. doi: 10.1152/ajpendo.00399.2006. [DOI] [PubMed] [Google Scholar]
- 33.Wicks S.E., Vandanmagsar B., Haynie K.R., Fuller S.E., Warfel J.D., Stephens J.M. Impaired mitochondrial fat oxidation induces adaptive remodeling of muscle metabolism. Proceedings of the National Academy of Sciences of the USA. 2015;112:E3300–E3309. doi: 10.1073/pnas.1418560112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huber N., Guimaraes S., Schrader M., Suter U., Niemann A. Charcot-Marie-Tooth disease-associated mutants of GDAP1 dissociate its roles in peroxisomal and mitochondrial fission. EMBO Reports. 2013;14:545–552. doi: 10.1038/embor.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.MacArthur J., Bowler E., Cerezo M., Gil L., Hall P., Hastings E. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog) Nucleic Acids Research. 2017;45:D896–D901. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Comuzzie A.G., Cole S.A., Laston S.L., Voruganti V.S., Haack K., Gibbs R.A. Novel genetic loci identified for the pathophysiology of childhood obesity in the Hispanic population. PLoS One. 2012;7 doi: 10.1371/journal.pone.0051954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heard-Costa N.L., Zillikens M.C., Monda K.L., Johansson A., Harris T.B., Fu M. NRXN3 is a novel locus for waist circumference: a genome-wide association study from the CHARGE Consortium. PLoS Genetics. 2009;5 doi: 10.1371/journal.pgen.1000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu C.T., Monda K.L., Taylor K.C., Lange L., Demerath E.W., Palmas W. Genome-wide association of body fat distribution in African ancestry populations suggests new loci. PLoS Genetics. 2013;9 doi: 10.1371/journal.pgen.1003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shungin D., Winkler T.W., Croteau-Chonka D.C., Ferreira T., Locke A.E., Magi R. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187–196. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Croteau-Chonka D.C., Marvelle A.F., Lange E.M., Lee N.R., Adair L.S., Lange L.A. Genome-wide association study of anthropometric traits and evidence of interactions with age and study year in Filipino women. Obesity (Silver Spring) 2011;19:1019–1027. doi: 10.1038/oby.2010.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wen W., Kato N., Hwang J.Y., Guo X., Tabara Y., Li H. Genome-wide association studies in East Asians identify new loci for waist-hip ratio and waist circumference. Scientific Reports. 2016;6:17958. doi: 10.1038/srep17958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Southam L., Gilly A., Suveges D., Farmaki A.E., Schwartzentruber J., Tachmazidou I. Whole genome sequencing and imputation in isolated populations identify genetic associations with medically-relevant complex traits. Nature Communications. 2017;8:15606. doi: 10.1038/ncomms15606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fox C.S., Liu Y., White C.C., Feitosa M., Smith A.V., Heard-Costa N. Genome-wide association for abdominal subcutaneous and visceral adipose reveals a novel locus for visceral fat in women. PLoS Genetics. 2012;8 doi: 10.1371/journal.pgen.1002695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lowe J.K., Maller J.B., Pe'er I., Neale B.M., Salit J., Kenny E.E. Genome-wide association studies in an isolated founder population from the Pacific Island of Kosrae. PLoS Genetics. 2009;5 doi: 10.1371/journal.pgen.1000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Melka M.G., Bernard M., Mahboubi A., Abrahamowicz M., Paterson A.D., Syme C. Genome-wide scan for loci of adolescent obesity and their relationship with blood pressure. Journal of Clinical Endocrinology & Metabolism. 2012;97:E145–E150. doi: 10.1210/jc.2011-1801. [DOI] [PubMed] [Google Scholar]
- 46.Norris J.M., Langefeld C.D., Talbert M.E., Wing M.R., Haritunians T., Fingerlin T.E. Genome-wide association study and follow-up analysis of adiposity traits in Hispanic Americans: the IRAS Family Study. Obesity (Silver Spring) 2009;17:1932–1941. doi: 10.1038/oby.2009.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scuteri A., Sanna S., Chen W.M., Uda M., Albai G., Strait J. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genetics. 2007;3:e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brons C., Grunnet L.G. Mechanisms in endocrinology: skeletal muscle lipotoxicity in insulin resistance and type 2 diabetes: a causal mechanism or an innocent bystander? European Journal of Endocrinology. 2017;176:R67–R78. doi: 10.1530/EJE-16-0488. [DOI] [PubMed] [Google Scholar]
- 49.Gemmink A., Goodpaster B.H., Schrauwen P., Hesselink M.K.C. Intramyocellular lipid droplets and insulin sensitivity, the human perspective. Biochimica et Biophysica Acta. 2017;1862:1242–1249. doi: 10.1016/j.bbalip.2017.07.010. [DOI] [PubMed] [Google Scholar]
- 50.Edgar R., Domrachev M., Lash A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Research. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hakvoort T.B., Moerland P.D., Frijters R., Sokolovic A., Labruyere W.T., Vermeulen J.L. Interorgan coordination of the murine adaptive response to fasting. Journal of Biological Chemistry. 2011;286:16332–16343. doi: 10.1074/jbc.M110.216986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Edwards M.G., Anderson R.M., Yuan M., Kendziorski C.M., Weindruch R., Prolla T.A. Gene expression profiling of aging reveals activation of a p53-mediated transcriptional program. BMC Genomics. 2007;8:80. doi: 10.1186/1471-2164-8-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hansen J., Timmers S., Moonen-Kornips E., Duez H., Staels B., Hesselink M.K. Synchronized human skeletal myotubes of lean, obese and type 2 diabetic patients maintain circadian oscillation of clock genes. Scientific Reports. 2016;6:35047. doi: 10.1038/srep35047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Noshiro M., Furukawa M., Honma S., Kawamoto T., Hamada T., Honma K. Tissue-specific disruption of rhythmic expression of Dec1 and Dec2 in clock mutant mice. Journal of Biological Rhythms. 2005;20:404–418. doi: 10.1177/0748730405280195. [DOI] [PubMed] [Google Scholar]
- 55.Hsieh M.C., Yang S.C., Tseng H.L., Hwang L.L., Chen C.T., Shieh K.R. Abnormal expressions of circadian-clock and circadian clock-controlled genes in the livers and kidneys of long-term, high-fat-diet-treated mice. International Journal of Obesity (London) 2010;34:227–239. doi: 10.1038/ijo.2009.228. [DOI] [PubMed] [Google Scholar]
- 56.Dyar K.A., Ciciliot S., Wright L.E., Bienso R.S., Tagliazucchi G.M., Patel V.R. Muscle insulin sensitivity and glucose metabolism are controlled by the intrinsic muscle clock. Molecular Metabolism. 2014;3:29–41. doi: 10.1016/j.molmet.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vieira E., Nilsson E.C., Nerstedt A., Ormestad M., Long Y.C., Garcia-Roves P.M. Relationship between AMPK and the transcriptional balance of clock-related genes in skeletal muscle. American Journal of Physiology-Endocrinology and Metabolism. 2008;295:E1032–E1037. doi: 10.1152/ajpendo.90510.2008. [DOI] [PubMed] [Google Scholar]