Summary
There are 180 currently recognised species of RNA virus that can infect humans and, on average, 2 new species are added every year. RNA viruses are routinely exchanged between humans and other hosts (particularly other mammals and sometimes birds) over both epidemiological and evolutionary time: 89% of human-infective species are considered zoonotic and many of the remainder have zoonotic origins. Some viruses that have crossed the species barrier into humans have persisted, and become human-adapted viruses, as exemplified by the emergence of HIV-1. Most, however, have remained as zoonoses, and a substantial number have apparently disappeared again. We still know relatively little about what determines whether a virus is able to infect, transmit from and cause disease in humans, but there is evidence that factors such as host range, cell receptor usage, tissue tropisms and transmission route all play a role. Although systematic surveillance for potential new human viruses in non-human hosts would be enormously challenging, we can reasonably aspire much better knowledge of the diversity of mammalian and avian RNA viruses than exists at present.
Introduction
Viruses account for only a small fraction of the 1400 or more different species of pathogen that plague humans – the great majority are bacteria, fungi or helminths [1]. However, as both the continuing toll of childhood infections such as measles and recent experience of AIDS and influenza pandemics illustrate, viruses are rightly high on the list of global public health concerns [2]. Moreover, the great majority of newly recognised human pathogens over the past few decades have been viruses [3] and a large fraction of emerging infectious disease “events” have involved viruses [4].
There are two kinds of virus: RNA viruses and DNA viruses. The latter largely consist, with the exception of a handful of pox and herpes viruses, of viruses which have probably been present in and coevolved with humans for long periods of time. RNA viruses are very different. The majority of RNA viruses that infect humans are zoonotic, meaning that they can infect vertebrate hosts other than humans. Many of those that are not regarded as zoonotic are believed to have had recent (in evolutionary terms) zoonotic origins. So it is the RNA viruses that are of greatest interest in the context of One Health.
In this chapter we review current knowledge of how RNA viruses in humans and other vertebrates are related, in terms of both of their evolution and their ecology, with the intention of trying to understand where human RNA viruses have come from in the past and where new ones might emerge from in the future. Until recently, research on these topics was essentially a series of case studies. There has been extraordinary work done detailing events such as the historical emergence of HIV-1 in central Africa [5] and the more recent emergence of Nipah virus in south-east Asia [6]. But while every emergence event is a fascinating story in its own right, our aim here is to look beyond the specifics and to try to identify any underlying generalities that tell us something useful about the emergence of RNA viruses as a biological process.
We begin by comparing the RNA viruses reported to infect humans with RNA virus diversity as a whole and explore the overlap between viruses in humans and viruses in other kinds of host. Next, we refine the analysis by distinguishing between viruses according to their ability not just to infect humans but also to transmit from one human to another, which is a pre-requisite for a virus being able to cause major epidemics and/or become an established, endemic human pathogen. We then consider in more detail the subset of human RNA viruses that can persist in human populations without the need for a non-human reservoir. Next, we attempt to identify characteristics of RNA viruses that allow them to cross the species barrier and those that predispose them to cause severe disease, as such viruses are of particular public health concern. We go on to discuss how new human RNA viruses arise (sometimes to subsequently disappear again). From the information assembled we construct a conceptual model of the relationship between RNA viruses in humans and other hosts. We consider how this model might be of practical value, concentrating on risk assessments for newly discovered viruses and also the much-discussed topic of the design of surveillance programmes for emerging infectious diseases.
Diversity of Human RNA Viruses
The diversity of human RNA viruses was recently surveyed using a formal methodology [3] and we update that information here. All RNA viruses known to infect humans were included, with the exception of those only known to do so as the result of deliberate laboratory exposures.
In this chapter, we use virus species as designated by the 9th report of the International Committee for the Taxonomy of Viruses (ICTV) [7] (noting that this differs from earlier ICTV reports used in previous work and that it will doubtless change again in the not-too-distant future). ICTV designations may not always accurately reflect the biological meaning of a “species”, i.e. reproductive isolation. The operational criteria used for RNA viruses may include any or all of: phylogenetic relatedness based on sequence data, serological cross-reactivity, host range and transmission route. It is also important to note that any analysis at the level of a virus species implicitly ignores a great deal of biomedically relevant diversity. This point is best illustrated by the influenza A viruses: the epidemiology and public health importance of seasonal influenza A and the H5N1 or H7N9 ‘bird flu’ variants are very different, but all are included within a single “species”. Less variable virus species than influenza A may still contain multiple serotypes and other functionally distinct subtypes. Despite these limitations, the “species” remains the most useful unit for studying virus diversity currently available.
Updating the earlier survey [3] with new taxonomic information [7] reveals 180 recognised species of RNA virus that have been reported to infect humans. These viruses represent 50 genera and 17 families (with one genus, the deltaviruses, currently unassigned to a family). It is not immediately obvious what we should make of this. Is 180 a large number or a small one? Should we be surprised that it is not much higher, or that it is not much lower? We consider such questions further below. We can, at least, be sure that 180 is an underestimate. New human RNA virus species are still being discovered or recognised at a rate of approximately 2 per year, although recent work [8] has suggested that the pool of undiscovered species could be much smaller than previously proposed [3]. Even if we still have very little idea of the number of species “out there”, it is, as we will consider in detail later on, possible to say something about where “out there” is.
The possibility of large numbers of as yet unrecognised viruses also raises the spectre of ascertainment bias. Certain kinds of RNA virus may be under-represented, perhaps dramatically so, among those currently recognised. These might be viruses from particular taxonomic groups, those associated with less severe disease or certain kinds of symptoms, or simply those that are rare and/or occur in less-studied regions of the world. While this is clearly an issue, it is worth pointing out that both the rates and kinds of RNA viruses being discovered or recognised have been remarkably consistent for the past half century, despite massive changes in the technologies for virus detection and identification and considerable variability in the effort put into virus discovery in different places and at different times [3].
RNA Viruses of Humans and Non-Humans
One striking observation is that 160 species of human infective RNA virus species (89% of the total) are regarded as zoonotic, i.e. they can also infect other kinds of vertebrate hosts. (The definition of zoonotic ignores arthropod vectors; these are regarded as specialised transmission routes rather than alternative host species). The non-human hosts usually (over 90% of all zoonotic RNA virus species) include other mammals and less commonly (under 40%) birds. Humans rarely, if ever, share their RNA viruses with anything else. Although the bias towards sharing viruses with other mammals is obvious, it is less clear whether we preferentially share viruses with particular kinds of mammal. Many human viruses (both RNA and DNA) are shared with ungulates, carnivores, rodents, primates or bats [3], but our knowledge of the host range of most viruses is too incomplete to be confident about any underlying patterns. The remaining 20 RNA viruses are not known to naturally infect non-human hosts. However, most of these have close relatives that can infect other mammals. The only exceptions are hepatitis C, hepatitis delta and rubella virus.
The overlap between the ability to infect humans and the ability to infect other mammals can be illustrated in other ways too. Of the 62 recognised RNA virus genera containing species that can infect at least one kind of mammal there are 50 (81%) that contain species that can infect humans. And of the 19 recognised RNA virus families that contain species reported to infect mammals all but 2 include species found in humans. The exceptions are the Nodaviridae – which are essentially insect viruses – and the Arteriviridae – which contains species infecting a range of different mammals, notably including simian haemorrhagic fever virus (SHFV).
The fact that human infective species are distributed so widely among the RNA viruses of mammals strongly suggests that, in evolutionary terms, the ability to infect humans is very easily acquired by these viruses. It also implies that many, perhaps most, human RNA viruses need not have arisen by evolving from other human RNA viruses. This idea is supported by a recent analysis of the relationship between phylogeny and host range for three RNA virus families – Paramyxo-, Calici- and Rhabdoviridae – and two genera – Alpha- and Flaviviruses – which concluded that the majority of speciation events were associated with host species jumps [9]. Note that this pattern contrasts markedly with the human DNA viruses, among which taxa such as the Papillomaviridae and the Anelloviridae appear to have undergone extensive diversification within humans.
The Pathogen Pyramid
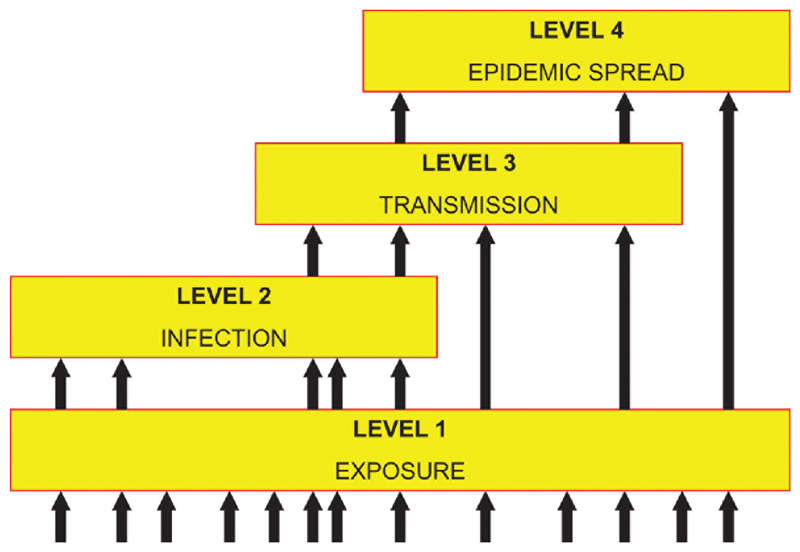
The categorization of viruses based simply on their ability to infect humans fails to distinguish between a vast range of epidemiologies, from occasional very mild cases of Newcastle disease virus infection to pandemics of influenza A or HIV-1. A useful conceptual framework for thinking about this issue is the pathogen pyramid [10]. The version of pyramid used here has four levels (Figure 1).
Figure 1.
A representation of the pathogen pyramid. Each level of the pyramid represents a different degree of interaction between a virus and a human host. Level 1 corresponds to exposure of humans, level 2 to the ability to infect humans, level 3 to the ability to transmit from one human to another, and level 4 to the ability to cause epidemics or persist as an endemic infection. Arrows indicate pathways that viruses may take to reach each level. For example, a level 4 virus may arrive at that state directly, simply by exposure to the virus from a nonhuman reservoir. This is known as an “off-the-shelf” virus. Alternatively, it may initially enter the population as a level 2 or 3 virus—not capable of sustained transmission—but evolve the ability to transmit between humans at a sufficiently high rate to persist within a human population. This is known as a “tailor-made” virus. Adapted from reference 25. doi:10.1128/microbiolspec.OH-0001-2012.f1
Level 1 corresponds to human exposure, whether via ingestion, inhalation, the bite of an arthropod vector or any other route. As discussed in the previous section, the most important sources of exposure are other mammals and, to a lesser degree, birds. There are no good estimates of the total diversity of mammal and bird viruses but it seems likely that the human population is exposed to hundreds, perhaps thousands, of species on a regular basis. The major determinants of the rate of exposure to new viruses will be the ecology and behaviour of humans, the non-human virus reservoir(s) and (in some cases) arthropod vectors.
Level 2 corresponds to human infection, which we take to mean the ability to enter and replicate in human cells in vivo. For all (known) RNA viruses there are associated host responses, although not all infections necessarily lead to clinical symptoms of disease. Key determinants of the ability to infect humans include the route of entry (e.g. needle sharing has created a new entry route for blood-borne viruses) and the molecular biology of the human virus interaction (discussed in more detail below). Of the 180 recognised species of RNA virus that can infect humans, almost 60% (107 species) are restricted to Level 2 (see Figure 2).
Figure 2.
All currently recognized human-infective RNA viruses categorized with respect to their ability to infect and transmit from humans (levels 2, 3, and 4 of the virus pyramid—see Fig. 1) and distinguished in terms of transmission route (green for vector-borne transmission, blue for other routes) and nature of diagnostic evidence (the viruses not in boldface type have only been reported in humans using serology-based methods). doi:10.1128/microbiolspec.OH-0001-2012.f2
Level 3 corresponds to the ability both to infect humans and to transmit from one human to another. The ability to transmit refers all kinds of transmission route, including vectors. Less than half of human-infective RNA viruses (73 species in all) are able to transmit between humans. A minority of these (26 species) are restricted to Level 3 (see Figure 2).
Level 4 corresponds to the ability to transmit sufficiently well that the virus can invade human populations, causing epidemics and/or establishing itself as an endemic human pathogen. In epidemiological parlance this corresponds to the condition that R0 is greater than one within the human population, where R0 is the basic reproduction number, defined as the number of secondary cases generated by a single primary case introduced into a large population of naïve hosts. In contrast, Level 3 viruses have an R0 of less than one in humans, which implies that although self-limiting outbreaks are possible, the infection cannot “take off” and cause a major epidemic. Although R0 is partly determined by the transmissibility of the virus, it is also a function of the behaviour and demography of the human host population; for example, changes in living conditions, travel patterns, sexual behaviour (for sexually-transmitted viruses) can all greatly influence R0. This argument is reflected in the term “crowd diseases”, which implies that certain human viruses (and other pathogens) could only become established once critical host population densities had been reached [10]. Our best estimate is that there are 47 Level 4 RNA virus species in humans (see Figure 2).
A useful exercise is to consider what kinds of viruses are found at Levels 2, 3 and 4 in the pyramid. There appear to be three major determinants of this: taxonomy (at the level of both family and genus); transmission route (especially the distinction between vector-borne transmission and other routes); and host range (expressed here as the ability to infect different mammalian orders). These three factors are not independent [1]; in particular, there are very few vector-borne viruses with narrow host ranges [11].
Nonetheless, several patterns can be identified. First, only two vector-borne viruses are found at the top of the pyramid (Level 4): yellow fever and dengue (see Figure 2). It is not immediately apparent why this should be so; we will consider this point further later on. Second, viruses with a host range that is, as far as we know, restricted to primates are rarely found lower down the pyramid (Levels 2 and 3), with a few exceptions such as the simian foamy viruses. The obvious implication is that if a virus is capable of infecting and transmitting from our closest relatives then it is very likely to have the same capabilities in us. Patterns are also apparent in the taxonomy of human infective viruses: for example, the Bunyaviridae, Rhabdoviridae, Arenaviridae and Togaviridae (with the exception of rubella, which is atypical of that group) are not represented at Level 4 at all. This reflects the fact that these four families are made up of viruses that are vector-borne and/or are not primate specialists.
Finally, it is worth noting that the ‘shape’ of the pathogen pyramid for RNA viruses differs from that for non-viral human pathogens. Most strikingly, much smaller fractions of recognised species of bacteria, fungi, protozoa or helminths are capable of extensive spread in human populations (i.e. are found at Level 4). On the other hand, human DNA viruses are even more concentrated at the top of the pyramid with almost 90% species at Level 4. These patterns could simply be an artefact of our incomplete knowledge of virus diversity at lower levels of the pyramid, but they could also reflect real biological differences between viruses and other kinds of pathogen: viruses (especially DNA viruses) may be more likely to speciate within humans; or viruses (especially RNA viruses) that jump the species barrier into humans may be more capable of spreading in human populations or of rapidly evolving that capability (see below).
Human-Adapted RNA Viruses
There is a semantic argument that only those viruses which are capable of persisting in human populations in the absence of a non-human reservoir should be described as “human” viruses. In our terminology these are, by definition, the Level 4 viruses, comprising 47 species, 20 of which are not known to have any natural hosts other than humans. These 47 viruses – referred to here as “human-adapted” – represent 12 different families and 29 different genera. Their most striking common characteristic is that almost all of them are transmitted by ingestion, inhalation or direct contact; just two are transmitted by vectors.
There are several possible routes for a virus to reach Level 4 on the pathogen pyramid (indicated by the arrows in Figure 1). One possibility is that humans are exposed to a virus that is already capable of effective transmission between humans, i.e. the virus is pre-adapted to humans (noting that this does not preclude further adaptation once the virus has entered the human population). These have been termed “off-the-shelf” viruses. Such viruses may be rare, perhaps extremely rare, variants of the population in the non-human reservoir, in which case the main determinants of the rate at which such viruses enter the human population will be the amount of genetic variability within the reservoir and the rate at which humans are exposed to the pre-adapted variants.
Another possibility is that the virus first enters the human population with limited ability to transmit between humans (i.e. Level 3) but that it is able to evolve that ability before the otherwise self-limiting chain of infections dies out [12]. These have been termed “tailor-made” viruses. Key determinants of the rate at which such viruses invade the human population are the frequency of primary infections and the virus mutation rate. We note that for a Level 2 virus to evolve human transmissibility, this would have to happen during the course of a primary infection. Such infections presumably give evolution relatively little material to work with and it may be that Level 2 viruses are “dead ends” in an evolutionary sense as well as an epidemiological sense. For example, rabies infections are relatively common in humans and are likely to have been so for thousands of years, but human transmissible variants have failed to materialise (with the proviso that rabies is technically a Level 3 pathogen because of rare instances of human-to-human transmission via organ transplants).
The origins of the human-adapted RNA viruses are of considerable interest, not least as a possible pointer to the likely sources of future viral threats to human health. It has previously been noted [10] that we have information on the origins of only a small minority of human pathogens, including RNA viruses. However, as stated above, it seems likely that many of them arose by species jumps from other mammals or (less often) birds, perhaps followed by some diversification within humans (e.g. human enteroviruses or parainfluenza viruses). The direct transmission routes used by most of these viruses are consistent with their being crowd diseases; that is, in contrast to vector-borne viruses, the basic reproduction number is a function of human population density.
Mechanisms
As explained above, whether a virus is found at Level 2, 3 or 4 of the pyramid reflects its ability to transmit from one human to another. Human demography and behaviour play a key role in this but, of course, intrinsic properties of the virus are also crucial.
The first consideration is the ability of the virus to infect humans at all. Given the importance of this topic we know surprisingly little about it. In effect, the question comes down to factors which restrict host range. Empirically, it does seem that the species barriers between different mammals, including humans, are very leaky: the majority of known mammal RNA viruses are capable of infecting multiple species. Only two studies [3, 13], however, have looked systematically at mechanisms, showing that use of a phylogenetically conserved receptor to gain entry to host cells is a necessary but not sufficient condition for a virus to be able to infect both humans and non-primates. This result appears robust, but the data are incomplete because the cell receptor has yet to be identified for the majority of human viruses.
Gaining entry to host cells is only the first step in initiating an infection. The virus must also be capable of replication in host cells, release from host cells, evading the innate immune response and perhaps becoming systemic. All of these processes depend on the specifics of the molecular interplay between virus and host, and all can contribute to the species barrier and host range restriction [14]. The species barrier may be quantitative rather than qualitative, perhaps expressed by the need for a higher infective dose. In one of very few experimental studies of the species barrier [15] it was found that the LD50 for rabies virus obtained from foxes was up to a million times lower for foxes than it was for cats and dogs. Similarly, there is evidence that human influenza A viruses can replicate in chimpanzees, but do so at a much lower rate [14].
The ability to get into (i.e. infect) a host does not equate with the ability to get out of (i.e. transmit from) that host. A key determinant of the ability to transmit is the virus’s capacity to invade and replicate in cells of particular tissues, notably the lower gastrointestinal tract, the upper respiratory tract, the urogenital tract, or possibly the blood or skin. In a few cases, the determinants of tissue tropisms are well understood. For example, H5N1 influenza A transmits well from ducks and poultry but not from humans. This is because it utilises a variant of the sialic acid receptor in the host cell membrane that occurs in the upper respiratory tract of ducks and poultry but is confined to the lower respiratory tract of humans [14].
Tissue tropisms inevitably play a key role in determining the route of virus transmission (e.g. respiratory, faecal-oral, arthropod vector). It has been suggested that altering tissue tropism is harder for a virus to achieve than switching host species [9]. This idea is borne out by the observation that transmission route tends to be a relatively deep-rooted trait in virus phylogenies, often to the level of family; in marked contrast to host range, which tends to be far more labile.
These few mechanistic and ecological insights fall well short of a proper understanding of why some kinds of viruses tend to occur at higher or lower levels of the pathogen pyramid. Host relatedness seems to play a role; hence viruses from other primates do seem more likely to be transmissible in humans than those acquired from non-primates, an idea supported by other studies of host relatedness and pathogen transmissibility [16]. But not all highly transmissible human viruses have been acquired from other primates. Transmission route is also important; vector-borne viruses in particular seem to be relatively good at infecting humans but relatively poor at being transmitted by humans [17]. It is possible that although humans are frequently exposed to vector-borne viruses, some of which are capable of setting up an infection, these viruses are not easily able to adapt to a new host (perhaps because any adaptation to a new vertebrate host must not compromise their interaction with the invertebrate vector [14]). Those that have adapted to humans – dengue and yellow fever – are ones that probably originated in other primates.
Virulence
In public health terms the ability of a virus to spread through human populations is, of course, only part of the story: human RNA viruses also vary enormously in the degree of harm they cause, a characteristic referred to as virulence. In the context of human infections we generally regard a pathogen as virulent if it has a high case-fatality ratio, or if infection routinely results in severe clinical disease. On this basis, HIV-1, SARS coronavirus or rabies would be regarded as virulent whereas most enteroviruses (excepting polio) or rhinoviruses would not.
Pathogen virulence is a very complex phenomenon, reflecting properties of the pathogen, the host and the interaction between them. It has been variously proposed that virulence will be influenced by transmission route, host range, level of the pathogen pyramid and the time that the pathogen and the host have had to coevolve (see [18] for an introduction to a large body of literature). These characteristics are not independent so hypothesis testing is not straightforward, although some theories do look promising. For example, the only two recent instances of newly emerging Level 4 pathogens – HIV-1 and SARS coronavirus – are/were both spectacularly virulent, in line with ideas that the virulence of novel host-pathogen combinations need not be near any evolutionary optimum. The only two Level 4 RNA viruses which are vector borne – dengue and yellow fever – are also relatively virulent, in line with ideas that vector-borne diseases can be more virulent as an ambulant host is not needed for transmission. There are also good examples of very virulent RNA viruses, such as rabies, for which humans are effectively dead-end hosts, in line with ideas that such infections are not subject to any evolutionary constraints as they do not contribute to the next generation of infections. On the other hand, many Level 2 viruses, such as Newcastle disease, Sindbis and others, result in only mild infections, so rabies may just lie at one end of a broad spectrum.
Another idea is that viruses acquired from particular kinds of reservoir, primates versus non-primates or mammals versus birds, might be especially virulent. The evidence, however, is inconsistent in this regard. It is true that some highly virulent human viruses, such as HIV-1 and dengue, were acquired from or are shared with other primates, our closest relatives. On the other hand, some highly virulent viruses are ultimately acquired from hosts much more distantly related to humans, such as H5N1 influenza A from birds or SARS and Nipah viruses from bats.
This important topic would clearly benefit from a systematic survey of the virulence of human RNA viruses (none has been published to date) which could be used to construct formal tests of the various hypotheses about pathogen virulence to be found in the evolutionary biology literature.
Emergence and the Changing Cast of RNA Viruses
New RNA virus species continue to be discovered, identified or recognised in humans. Recent examples include Nelson Bay orthoreovirus, Irkut virus, primate T-lymphotropic virus 3, human coronavirus HKU1, and human rhinovirus C. Moreover, there is usually a backlog of reports of new human viruses which have yet to be formally recognised as species. Not all of these viruses will have recently invaded human populations; many will turn out to be long-standing human pathogens which have only recently been recognised, or accepted as ‘species’.
It is therefore important to recognise that the continued accumulation of recognised human RNA virus species may reflect less the possibility that genuinely new viruses are continually emerging, most likely acquired from non-human reservoirs, than the fact that we are still getting to grips with the taxonomic diversity of viruses that have been with us for some time. This distinction between viruses that we have only just discovered and viruses that have only just discovered us is, of course, crucial in the context of emerging infectious diseases. If most of the so-called “new” viruses are not new at all then this implies that events such as the advent of HIV/AIDS in the early 1900s or the curtailed SARS epidemic in 2003 may be just unusual, one-off occurrences with their own specific causes. If, on the other hand, genuinely new viruses are appearing all the time then the HIVs and SARS coronavirus are more accurately regarded as just the highly visible tip of a much larger iceberg. Without a much more detailed and thorough understanding of the phylogenies and origins of all human viruses, not just those with high public health profiles, we cannot resolve this question.
Perhaps the most striking feature of recently discovered RNA viruses is that they tend to be much like the RNA viruses that we already knew about. They are members of the same virus families, have the same transmission routes, and share the same kinds of non-human host. If these newly recognised viruses are indeed emerging then it seems as though here is nothing special about emergence, at least from a biologist’s perspective. Even if this is correct, it is still often suggested that the rate, if not the biology, of pathogen emergence is higher in the early 21st century than it has been in the past. This reflects the notion that a variety of so-called drivers of emergence, ranging from human population growth to changes in farming methods, are combining to create a ‘perfect storm’. This idea is difficult to evaluate critically. Arguably there has been only a handful global emergence events in the past century, notably those involving HIV-1, influenza A and SARS coronavirus. This is not a strikingly large number given that many of the other 40 or so human-adapted RNA viruses may have emerged only in the past few millennia. Of course, it could be argued that less dramatic events such as the geographical spread of West Nile virus or outbreaks of Ebola are more frequent now than they have been in the past, but that claim is even harder to test with any rigour.
Another side to this issue is rarely discussed. One recent study [8] reports that while the number of virus species accumulates, at the same time many of those recognised in past years or decades seem to have disappeared, these making up about one-third of the total. There is, of course, one well-known example of the eradication of a human RNA virus through human intervention, SARS coronavirus, accompanying the even more impressive story of the eradication of smallpox, a DNA virus. However, there are many more examples of viruses which seem to have disappeared of their own accord, an unexpected observation worthy of careful consideration. There are several possibilities. First of all, rare infections, especially those with mild or common clinical presentations, may simply have been missed or no-one has bothered to report them. Another possibility is that reports from earlier times are unreliable; for example, it is striking that no human cases of foot-and-mouth disease have been noticed since a handful of reports in the mid-1960s. But it seems likely that many of the missing viruses have indeed disappeared, at least temporarily, from humans, even if they are still present in non-human reservoirs. Some, of course, could re-appear in humans at some point in the future: this has happened for the bat lyssaviruses, for example, and is a worrying possibility for SARS coronavirus.
The implication of these missing viruses is that the extant diversity of human RNA viruses is perhaps closer to 100 species than the figure of 180 given earlier. The number of missing species corresponds, very roughly, to an average loss rate of 1 per year [8]. Another way of expressing this is that there would have to be one new or re-discovered species of human RNA virus reported every year just to maintain the level of diversity that we are aware of at present.
A Conceptual Model
All of the above is consistent with the following conceptualisation of the relationship between RNA viruses which can infect humans and those found in other kinds of host, particularly other mammals. Rather than being distinct groups, viruses of humans and viruses of other mammals are readily interchanged over evolutionary time. Some of the viruses that cross the species barrier into humans persist and may become human-adapted viruses, though this seems to be a relatively rare occurrence. Many of the others remain as zoonoses, and yet others disappear again. The repertoire of human viruses is therefore not fixed but is dynamic, over time scales measured in decades [8]. However, this process is far from random. Although humans share their RNA viruses with many different mammalian taxa, those from other primates appear most likely to be capable of spreading through human populations. Similarly, although almost every family of viruses found in mammals contains species found in humans, some virus families seem to be capable of, at best, limited spread in human populations. This conceptual model is illustrated diagrammatically in Figure 3.
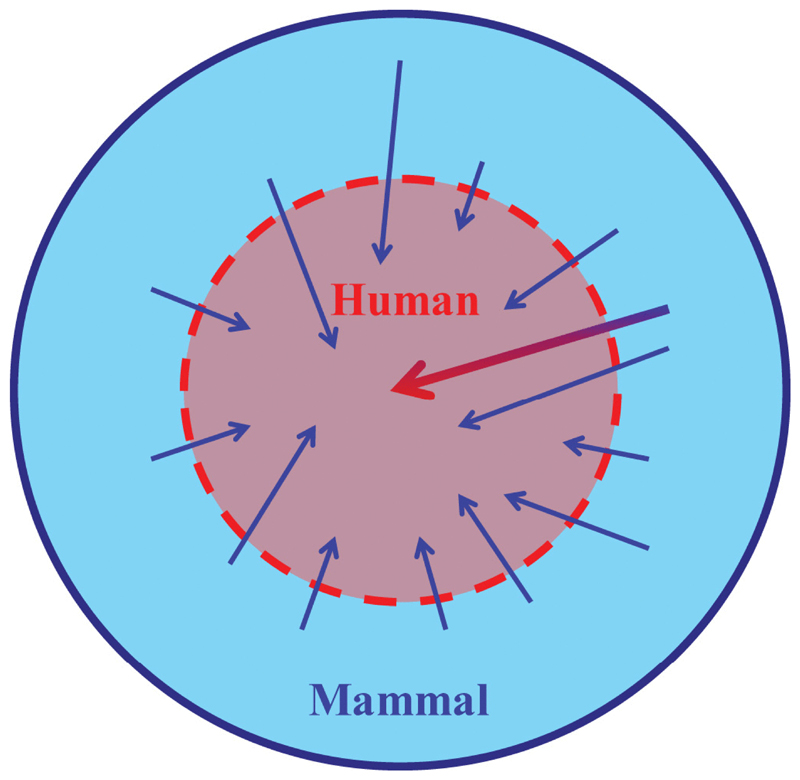
Figure 3.
A schematic representation of the relationship between human viruses and viruses from other mammals. Human viruses are depicted as a subset of mammal viruses, only partially protected by a species barrier. There are frequent minor incursions of zoonotic viruses (small arrows), and many of these may not persist in human populations. Occasionally there may be a much more significant event (large arrow) whereby a mammal virus proves capable of einvolving adaptation to infect and transmit from humans. doi:10.1128/microbiolspec.OH-0001-2012.f3
Surveillance and Risk Assessment
Our conceptual model has practical implications for both disease surveillance and risk assessment, especially in the context of newly emerging infectious diseases.
The importance of early detection of potential epidemics or pandemics cannot be over-stressed, a point made by several major studies [2]. The early detection through clinical surveillance of SARS, coupled with effective intervention based on case isolation and quarantine, prevented a potentially catastrophic pandemic [19]. A matter of some debate is whether or not surveillance should be extended into the non-human reservoirs of infection from which novel human pathogens are most likely to emerge – a concept sometimes referred to as ‘getting ahead of the curve’.
It helps, of course, if we know what we are looking for and where best to look for it [20]. We currently have only the beginnings of answers to these questions. Viruses, especially respiratory viruses, are often picked out as the most obvious threat to global public health [2]. New viruses are very likely to have a zoonotic origin, almost certainly acquired from mammals or birds. Emergence events are most likely to occur in regions – so-called ‘hotspots’ – which combine high human population densities with high densities of domestic animals and/or a high diversity of wildlife [4]. All of this information is useful, but falls well short of a recipe for designing a feasible global surveillance system [20].
One strategy to increase the likelihood of early detection is to implement sentinel surveillance in people in close, high risk contact with animal populations, such as bush meat hunters or slaughterhouse workers. In tandem with recent advances in the technologies available for virus detection – especially those based on high throughput nucleic acid sequencing – such programmes should at least improve our knowledge of the diversity of viruses there is “out there” that humans are exposed to, a process sometimes referred to as “chatter” [10]. Pathogen discovery programmes, particularly in under-studied taxa such as wild rodents and bats [21], should also add greatly to our knowledge of potential threats to human health.
Once a novel or previously unknown virus is identified it is obviously important to assess any potential risk to public health. Initial assessments are generally based on the kinds of comparative biology approach discussed in this Chapter. A recent example of this is Schmallenberg virus, a novel virus first detected in sheep and cattle in northern Europe in 2011. Schmallenberg is an orthobunyavirus, a diverse genus of vector-borne bunyaviruses that are found in a variety of hosts but especially in ungulates. Given these characteristics, and despite the fact that some distantly related orthobunyaviruses – notably Oropouche – do cause disease in and may even be transmitted by humans, Schmallenberg was provisionally designated low risk to humans and no human cases have yet been found [22]. The even more recently reported MERS coronavirus [23] has rightly caused much more concern.
Concluding Remarks
Emerging diseases caused by RNA viruses are a One Health issue. There is a continual interchange, over both epidemiological and evolutionary time scales, between viruses in humans and viruses in other animals that we cannot ignore. RNA viruses that pose serious threats to global public health have arisen repeatedly by jumping into humans from other animals. This has been going on for millennia and it continues today, as fast as ever and perhaps even faster. We have to anticipate that new viral threats will emerge in coming years or decades and we need to be prepared to rise to these new challenges as they appear.
It is worth pointing out that viruses were discovered in non-human animals (foot-and-mouth disease virus at the very end of the 19th century) before they were identified in humans. The same is true [24] for several important kinds of viruses, such as retroviruses (and lentiviruses specifically), rotaviruses, papillomaviruses and coronaviruses. A corollary of this is that veterinary rather than medical expertise may, at least initially, be our best source of knowledge about newly discovered viruses.
We have discussed the need for more effective surveillance for novel viruses but concluded that although attempts to characterise the kinds of viruses most likely to emerge are useful precise prediction is not a realistic objective, for now at least. On the other hand, there could be considerable benefit from a better understanding of RNA virus diversity in the most important host species. At present we do not even have a complete inventory of the viruses in humans, and whilst we have some knowledge of the viruses in major livestock species we know very little about the viruses present in wild mammals or birds. These gaps can and should be filled: we need to know what is out there now, and what might be waiting around the corner.
Acknowledgements
We thank Conor O’Halloran for assistance with data collation. We are grateful to past and present members of Epigroup and numerous collaborators for many fruitful discussions. This work is part of the VIZIONS project funded by a Wellcome Trust Strategic Award.
References
- 1.Taylor LH, Latham SM, Woolhouse MEJ. Risk factors for human disease emergence. Phil Trans R Soc Lond B. 2001;356:983–989. doi: 10.1098/rstb.2001.0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.King DA, Peckham C, Waage JK, Brownlie J, Woolhouse MEJ. Infectious diseases: preparing for the future. Science. 2006;313:1392–1393. doi: 10.1126/science.1129134. [DOI] [PubMed] [Google Scholar]
- 3.Woolhouse MEJ, Scott FA, Hudson Z, Howey R, Chase-Topping M. Human viruses: discovery and emergence. Phil Trans R Soc Lond B. 2012;367:2864–2871. doi: 10.1098/rstb.2011.0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, Daszak P. Global trends in emerging infectious diseases. Nature. 2008;451:990–994. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharp PM, Hahn BH. The evolution of HIV-1 and the origin of AIDS. Phil Trans R Soc Lond B. 2010;365:2487–2494. doi: 10.1098/rstb.2010.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Epstein JH, Field HE, Luby S, Pulliam JRC, Daszak P. Nipah virus: impact, origins and causes of emergence. Curr Inf Dis Rep. 2006;8:59–63. doi: 10.1007/s11908-006-0036-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy. Elsevier; Amsterdam, The Netherlands: 2012. p. 1327. [Google Scholar]
- 8.Woolhouse MEJ, Adair K. The diversity of human RNA viruses. Future Virology. 2013;8:159–171. doi: 10.2217/fvl.12.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kitchen A, Shackelton LA, Holmes EC. Family level phylogenies reveal modes of macroevolution in RNA viruses. Proc Natl Acad Sci USA. 2011;108:238–243. doi: 10.1073/pnas.1011090108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolfe ND, Dunavan CP, Diamond J. Origins of major human infectious diseases. Nature. 2007;447:279–283. doi: 10.1038/nature05775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woolhouse MEJ, Taylor LH, Haydon DT. Population biology of multi-host pathogens. Science. 2001;292:1109–1112. doi: 10.1126/science.1059026. [DOI] [PubMed] [Google Scholar]
- 12.Antia R, Regoes RR, Koella JC, Bergstrom CT. The role of evolution in the emergence of infectious diseases. Nature. 2003;426:658–661. doi: 10.1038/nature02104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bae S-E, Son HS. Classification of viral zoonosis through receptor pattern analysis. BMC Bioinf. 2011;12:96. doi: 10.1186/1471-2105-12-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuiken T, Holmes EC, McCauley J, Rimmelzwaan GF, Williams CS, Grenfell BT. Host species barriers to influenza virus infections. Science. 2006;312:394–397. doi: 10.1126/science.1122818. [DOI] [PubMed] [Google Scholar]
- 15.Blancou J, Aubert MFA. Transmission du virus de la rage: importance del la barrière d’espèce. Bull Acad Natle Méd. 1997;181:301–312. [PubMed] [Google Scholar]
- 16.Streicker DG, Turmelle AS, Vonhof MJ, Kuzmin IV, McCracken GF, Rupprecht CE. Host phylogeny constrains cross-species emergence and establishment of rabies virus in bats. Science. 2010;329:676–679. doi: 10.1126/science.1188836. [DOI] [PubMed] [Google Scholar]
- 17.Woolhouse MEJ, Adair K. Ecological and taxonomic variation among human RNA viruses. Journal of Clinical Virology. 2013 doi: 10.1016/j.jcv.2013.02.019. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ebert D, Bull J. The evolution and expression of virulence, Chapter 12. In: Stearns SC, Koella JC, editors. Evolution in Health and Disease. 2nd ed. Oxford University Press; Oxford, UK: 2008. pp. 153–167. [Google Scholar]
- 19.Stöhr K. A multicentre collaboration to investigate the cause of severe acute respiratory syndrome. Lancet. 2003;361:1730–1733. doi: 10.1016/S0140-6736(03)13376-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morse SS, Mazet JAK, Woolhouse M, Parrish CR, Carroll D, Karesh WB, Zambrana-Torrelio C, Lipkin WI, Daszak P. Predicting and preventing the next pandemic zoonosis. Lancet. 2012;380:1956–1965. doi: 10.1016/S0140-6736(12)61684-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drexler JF, et al. Bats host major mammalian paramyxoviruses. Nat Comm. 2012;3:796. doi: 10.1038/ncomms1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ducomble T, Wilking H, Stark K, Takla A, Askar M, Schaade L, Nitsche A, Kurth A. Lack of evidence for Schmallenberg virus infection in highly exposed persons, Germany, 2012. Emerg Inf Dis. 2012;18:1333–1335. doi: 10.3201/eid1808.120533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cotten M, et al. Full-genome deep sequencing and phylogenetic analysis of novel human betacoronavirus. Emerg Inf Dis. 2013;19:736–742. doi: 10.3201/eid1905.130057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palmarini M. A veterinary twist on pathogen biology. PLoS Path. 2007;3:e12. doi: 10.1371/journal.ppat.0030012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woolhouse M, Antia R. Emergence of new infectious diseases. In: Stearns SC, Koella JK, editors. Evolution in Health and Disease. 2nd edn. Oxford: Oxford University Press; 2008. pp. 215–228. [Google Scholar]