Abstract
Introduction
Defects in the DNA damage response (DDR) drive the development of cancer by fostering DNA mutation but also provide cancer-specific vulnerabilities that can be exploited therapeutically. The recent approval of three different PARP inhibitors for the treatment of ovarian cancer provides the impetus for further developing targeted inhibitors of many of the kinases involved in the DDR, including inhibitors of ATR, ATM, CHEK1, CHEK2, DNAPK and WEE1.
Areas covered
We summarise the current stage of development of these novel DDR kinase inhibitors, and describe which predictive biomarkers might be exploited to direct their clinical use.
Expert opinion
Novel DDR inhibitors present promising candidates in cancer treatment and have the potential to elicit synthetic lethal effects. In order to fully exploit their potential and maximize their utility, identifying highly penetrant predictive biomarkers of single agent and combinatorial DDR inhibitor sensitivity are critical. Identifying the optimal drug combination regimens that could used with DDR inhibitors is also a key objective.
Keywords: Cancer, Cell cycle, DNA damage response (DDR), Kinase inhibitors, Replication stress
1. Introduction
The genome is constantly exposed to a series of endogenous and exogenous agents that damage and alter the normal composition of the double helix [1]. If left unrepaired, the “DNA lesions” that are the result of this damage have the potential to either subtly alter cell behaviour by causing DNA mutations, or in extreme cases, to impair the fitness of cells. For example, unrepaired DNA damage can result in mutations that impair the ability to encode a fully functional transcriptome, thus impairing the fitness of cells. Likewise, DNA lesions that prevent DNA replication can lead to cell death, as can gross changes to the number and structure of chromosomes [1]. Given the level of threat to the genome and the potentially dire consequences of not repairing DNA damage once it occurs, it is unsurprising that relatively complex molecular networks that maintain the integrity of the genome have evolved. These networks are collectively known as the DNA damage response (DDR) [2]. Whilst a reductionist view of DDR networks might be that these operate by simply detecting and repairing DNA lesions, a more holistic understanding suggests that, like most other cellular processes, the DDR does not operate in isolation and interacts with, for example, DNA replication and transcriptional machinery [3], cell cycle control molecular networks [4], molecular pathways that control energy metabolism [5], programmed cell death [5], and systems that control innate immunity [6]. These interactions with ostensibly distinct molecular processes ensure, for example, that the cell cycle is stalled to allow DNA repair [4] or that cells with excessive levels of DNA damage are removed from the population. In doing so, the integrity of the genome within a single cell is maintained, and that those cells that do divide, transmit a functional genome to daughter cells.
Understanding how the DDR functions is an intense area of study, particularly from the perspective of how defects in the DDR influence cancer development and treatment. The link between defects in the DDR and cancer pathogenesis has been established via multiple lines of evidence. These include: (i) genetic studies, where defects in tumour suppressor genes that control the DDR (e.g. BRCA1, BRCA2, PALB2, RAD51C, RAD51D, FANC-family genes, MLH1, etc. (reviewed in [2])) predispose to familial forms of cancer; (ii) cytogenetic and genomic studies, where the number and type of different DNA mutations and forms of genomic instability found in human tumours often betray the DNA repair defects that have moulded tumour genomes [7]; and (iii) functional studies, where experimental induction of specific DNA repair defects causes cancer in animal models [8]. A straightforward hypothesis is that DNA mutations that result from DDR defects can, in some cases, allow cells to acquire the characteristics, or hallmarks, of cancer (e.g. ability to evade programmed cell death, independence from inhibitory growth signals etc. [9]). Moreover, DDR defects inevitably cause genetic diversity to emerge within a cell population and thus provide a likely driver for the molecular and phenotypic heterogeneity seen within tumours as well as the ability of tumour cell populations to evolve in the face of selective pressure [10]. The ability of DDR defects to result in disordered, mutated genomes might also be enhanced by commonly occurring defects in gatekeeper tumour suppressor genes such as p53, ATM, CHK1 and CHK2 [11]. These genes normally encode proteins whose function is to induce cell cycle arrest in response to DNA damage; the partial or complete inactivation of these gatekeeper tumour suppressors often allows cells to circumvent cell cycle checkpoints and to continue to proliferate even in the face of persistent DNA damage (reviewed in [2]). Similarly, the inactivation of specific tumour suppressor proteins such as ATM (Ataxia telangiectasia mutated), allows cells to proliferate in the face of replication fork stress, i.e. the stalling or slowing of replication forks [12,13]. This replication fork stress appears to be a feature of pre-neoplastic lesions and is associated with the activation of oncogenes such as Cyclin E (CCNE1) or Myc. The hyper-activation of Cyclin E and Myc normally drives cells into a state of senescence [12,13] and whilst inactivation of ATM circumvents this event, the resultant cells divide with unresolved replication fork associated DNA damage [14]. As well as being driven by oncogene activation, replication fork stress can also arise through a variety of additional causes, including an excess of naturally occurring secondary structures within the DNA double helix, therapy induced DNA lesions that stall replication forks, nucleotide depletion, collisions between the replication and transcription machinery, or an enhanced incorporation of ribonucleotides into DNA [15]. In many cases, the replication fork stress that ensues can be tolerated so that it does not impair the fitness of cells (for example by inactivation of ATM as described above) but can lead to an increasingly disordered genome and often generates an increased reliance on DDR proteins such as ATR (Ataxia telangiectasia and Rad3-related protein) that are involved in stabilising replication forks [14].
There is also a certain duality in how the DDR influences the real-world outcome for people with cancer; whilst defects in the DDR undoubtedly drive the development of cancer, these also provide somewhat cancer-specific vulnerabilities that often form the basis of how a patient might be best treated. For example, many of the chemotherapy or radiotherapy treatment regimens commonly used in the treatment of cancer generate DNA lesions, including abnormal covalent bonds (“cross links”) within the double helix. In tumour cells with particular DDR defects, these DNA lesions are ineffectively recognised and/or repaired, which often leads to cytotoxicity; conversely most normal cells, which in principle have a better capacity to process DNA damage, are relatively unharmed. Of course, highly proliferative normal tissues, such as the epithelial lining of the gastrointestinal tract and many myeloid cell lineages, are often not spared from the cytotoxic effect of chemotherapy treatment; the result for the patient receiving such treatment is often a series of deleterious side effects that significantly impair their quality of life. Nevertheless, in some patients, DNA damaging chemotherapy and/or radiotherapy can either extend survival or be curative, demonstrating that exploiting the DDR defects in tumour cells have real therapeutic value.
The challenge of trying to develop what might be better tolerated treatment approaches for cancer that exploit DDR defects has, until now, largely been addressed by the discovery and development of targeted agents that either inhibit specific DNA repair or cell cycle checkpoint proteins. The central premise behind developing such targeted DDR inhibitors is that these might generate DNA lesions that selectively target any one of a number of characteristics recurrently seen in tumour cells, including existing DDR defects (by exploiting synthetic lethal interactions), replication fork stress, genomic instability or existing defects in cell cycle control that prevent the effective repair of DNA damage. In addition, targeted DDR inhibitors might also show some utility when combined with other treatment modalities including combinations with DNA damaging chemotherapies, radiotherapy or immunotherapy, based partially on the assumption that such combinations will also elicit DNA lesions and/or a DNA damage response that is selectively cytotoxic to tumour cells [16].
In this review article, we summarise much of the recent data describing the discovery and development of novel targeted DDR kinase inhibitors. Arguably the first class of targeted DDR inhibitors to be approved for use in the treatment of cancer are the PARP inhibitors, including rucaprib (Clovis), olaparib (AstraZeneca) and niraparib (Tesaro), each of which is approved for the treatment of ovarian cancer [17]. As PARP inhibitors have recently been reviewed elsewhere [17], we focus here on inhibitors of kinases involved in the DDR: Ataxia telangiectasia mutated (ATM), Ataxia telangiectasia and Rad3-related protein (ATR), DNA Dependent Protein Kinase (DNA-PK) (all Phosphatidyl-Inositol Kinase-like Kinase (PIKK) enzymes [18]), CHK1, CHK2 and WEE1. In each case, we will focus on a number of key issues we believe to be important to the eventual successful clinical development of these agents, namely: (i) what are the optimal predictive biomarkers that might predict favourable patient responses to each agent and what are the mechanisms of action that explain biomarker/drug sensitivity relationships (Table 1); (ii) what mechanisms of drug resistance might be important for each target; (iii) what drug combination approaches might be appropriate for each drug class?
Table 1. Candidate predictive biomarkers identified from pre-clinical studies.
| Drug | Candidate biomarker |
|---|---|
| ATM inhibitor | p53 status [26,34,35] |
| ATR deficiency [36–38] | |
| Potential synthetic lethal interactions between ATM and VHK1, SETD2, SMO, KDM6A, VHL [41] | |
| ATR inhibitor | Silencing of ATRIP, RAD17, RAD9A, RAD1, HUS1, TOPBP1, XPF, ERCC1 [58] |
| ATM deficiency [36,48,59] | |
| POLD1 deficiency [61] | |
| ARID1A deficiency [62] | |
| Alternative lengthening of telomeres [66,67] | |
| EWS-FLI1/ERG Ewing sarcoma fusion gene [64] | |
| SS18-SSX synovial sarcoma fusion gene [65] | |
| ATR inhibitor + Cisplatin combination | Silencing of 53BP1, REV3L or REV7 [56] |
| DNA-PK inhibitor | Potential synthetic lethal interactions between DNA-PK and AKT1, CDK4, CDK9, CHK1, IGFR1, mTOR, VHL1, RRM2 silencing [41] |
| ATM loss [78] | |
| CHK1 inhibitors | p53 status [87–90] |
| CHK1 inhibitors +MK2 inhibitors | KRAS and BRAF-mutant tumours[98] |
| WEE1 inhibitors | p53 status [110] |
| Increased expression of EZH2, Cyclin B1, B2 and E1[108] | |
| Low expression of PKMYT1[111] | |
| Defects in FANCC, FANCG and BRCA2 [113] | |
| H3K36Me3-deficiency [114] | |
| WEE1 inhibitors + mTOR inhibitors | NRAS and KRAS- mutations [112] |
2. ATM as a cancer drug target
2.1. ATM function
The PIKK family member Ataxia telangiectasia mutated (ATM) is activated at DNA double strand breaks (DSBs) and, amongst a number of substrates, phosphorylates the kinase CHK2 and the tumour suppressor p53 to activate the G1/S checkpoint [5] (Figure 1). ATM also activates CHK1 and CHK2 via phosphorylation to induce an intra-S or G2/M cell cycle arrest [19,20]. Additionally, ATM phosphorylates hundreds of other proteins associated with a wide variety of molecular processes including DNA repair, chromatin structure, transcription and apoptosis [5] (Figure 2). Like most kinases involved in the DDR, ATM also has “non-canonical” functions; these include the regulation of the spliceosome in the face of replication blocking lesions [21].
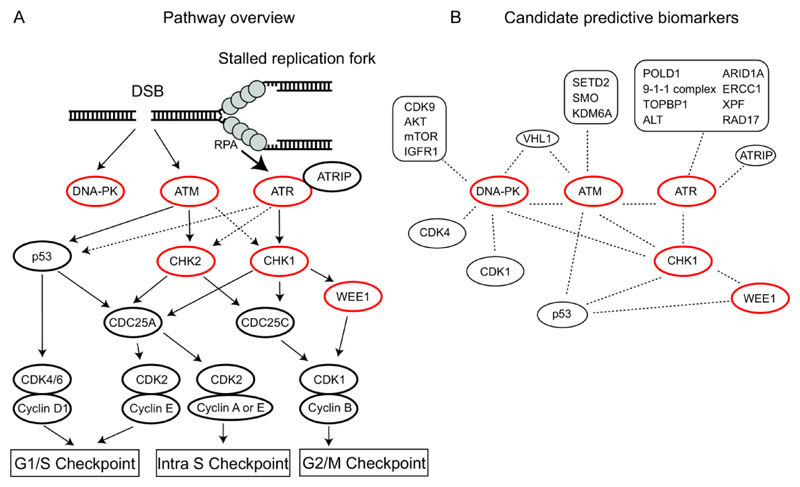
Figure 1. DDR pathway overview and candidate predictive biomarkers.
A) Ataxia-telangiectasia mutated (ATM) is recruited to and activated at DNA Double Strand Breaks (DSBs). Once activated, ATM phosphorylates p53 and CHK2, resulting in a G1/S cell cycle arrest via CDC25A and Cyclin-CDK (Cyclin-dependent kinase) complexes. Ataxia-telangiectasia and Rad3-related (ATR) is activated at persistent stretches of single strand DNA (ssDNA) coated by Replication Protein A (RPA); such ssDNA stretches occur at sites of resection or stalled replication forks. ATR primarily activates CHK1. CHK1 can signal via CDC25A to activate the intra S checkpoint or via CDC25A and WEE1 to activate the G2/M checkpoint, depending on the phase of the cell cycle the damage is detected in. DNA dependent protein kinase (DNA-PK), activated at DSBs, does not play a role in the regulation of cell cycle progression after DNA damage. Kinases discussed in this review are highlighted in red. Dashed arrows indicate indirect regulation. B) Candidate predictive biomarkers for ATM, ATR, DNA-PK, CHK1 and WEE1 discussed in this review. ALT: Alternative lengthening of telomeres.
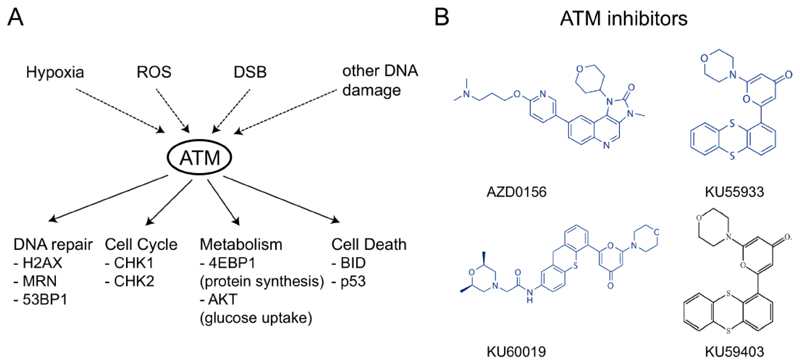
Figure 2. ATM function and ATM inhibitors.
A) ATM can be activated by hypoxia, reactive oxygen species (ROS), DNA double strand breaks (DSBs) or other types of DNA damage. Activated ATM phosphorylates multiple substrates and primary roles for ATM are illustrated, with key substrates indicated below. B) The structure of ATM inhibitors discussed in this review.
ATM is a well-recognised tumour suppressor, and deleterious mutations and/or deletions in ATM are extremely common in several solid tumour types including gastric, colorectal and prostate cancers. In addition, several B-cell lymphoma subtypes including Mantle Cell Lymphoma (MCL) and chronic lymphocytic leukaemia (CLL), are characterised by high rates of ATM alterations, (50% in MCL [22]). Germ-line mutations in ATM results in Ataxia Telangiectasia (A-T), a recessive disorder characterised by neural degeneration, ataxia and increased predisposition to cancer [5].
2.2. ATM inhibitors
The kinase activity of ATM can be inhibited using small molecule ATP analogues (Figure 2). KU55933, a potent and selective inhibitor of ATM, was developed by KuDOS pharmaceuticals (later acquired by AstraZeneca) [23]. This toolbox inhibitor does not possess all of the properties of a drug-like compound but nevertheless has utility in interrogating ATM function. Like its structural derivative KU60019 [24], the therapeutic potential of KU55933 is limited by poor aqueous solubility and in vivo bioavailability issues [25]. Another structurally related compound, KU59403, has improved bioavailability and solubility and also increased selectivity for ATM, compared to the structurally related PIKKs such as ATR and DNA-PK [26]. AstraZeneca has also recently initiated phase I clinical trials of another ATM inhibitor, AZD0156 [27], used either as a single agent or in combination with the PARP inhibitor olaparib (NCT02588105 – clinicaltrials.gov), suggesting that drug-like ATM-inhibitors can be developed.
2.3. Combination therapy with ATM inhibitors: pre-clinical evidence
In pre-clinical experiments, ATM inhibitors have been shown to be potent sensitizers to IR [23] (Table 2). In addition, ATM inhibitors have also been shown to increase the sensitivity of cells to topoisomerase II poisons (etoposide and doxorubicin) as well as to the radiomimetic bleomycin, agents which induce DNA DSBs [23,28]. ATM inhibitors also potentiate the toxicity of topoisomerase I poisons such as camptothecin, which while they do not directly induce DNA DSB, do generate a covalent protein-DNA lesion that induces DSBs during S-phase as a result of collapsed replication forks [29].
Table 2. Drug combinations associated with each DDR kinase inhibitor.
| Drug target | Inhibitor | Combination | Stage of development |
|---|---|---|---|
| ATM | AZD0156 | + olaparib | Clinical trial phase I (NCT02588105) |
| KU-55933 | + IR or radiomimetic + topoisomerase I poisons |
Pre-clinical data [23] | |
| ATR | VE-821 | + cisplatin | Pre-clinical data [37] |
| AZD6738 | + carboplatin + olaparib + anti- PD-L1 antibody (MEDI4736) |
Phase I trial (NCT02264678) | |
| AZD6738 and VX970 | + Standard chemotherapy | Phase I trials (Reviewed in [68]) | |
| VX970 | + cisplatin | Pre-clinical data [65] | |
| + olaparib | Pre-clinical data [65] | ||
| DNA-PK | VX-984 | + doxorubicin | Phase I (NCT NCT02644278) |
| CHK1 | MK8776 (SCH900776), LY2603618, CCT245737, GDC-0575 | + Standard chemotherapy | Phase I trials (Reviewed in [68]) |
| + WEE1 inhibitor | Pre-clinical data [96,97] | ||
| CHK2 | PV1019 | + topotecan + camptothecin + IR |
Pre-clinical data [101] |
| CCT241533 | + PARP inhibitors | Pre-clinical data [103] | |
| CHK1/2 | AZD7762 | + irinotecan + gemcitabine |
Pre-clinical data [104,105] |
| WEE1 | AZD1775 | + CHK1 inhibition | Pre-clinical data [105,108,115] |
| + cisplatin | Pre-clinical data [119] | ||
| + cytarabine | Pre-clinical data [118] | ||
| + panobinostat | Pre-clinical data [117] |
Abbreviations: IR = Ionizing radiation.
In response to DNA damage, ATM directly phosphorylates CtBP-interacting protein (CTIP), an endonuclease that initiates the 5’-3’ resection of DNA DSBs, a step critical for DSB repair by homologous recombination (HR) [30]. Given this, ATM-deficiency results in a mild homologous repair deficiency phenotype (HRD) [31], causing ATM-deficient cells to be sensitive to agents that target HR defect including cisplatin and PARP inhibitors (PARPi) [32,33]. This also suggests that ATM inhibitors might sensitise HR proficient cells to PARP inhibition or that ATM inhibitors could be used to potentiate the effect of PARPi.
2.4. Biomarkers to predict ATM inhibitor sensitivity: preclinical evidence
Biddlestone-Thorpe and colleagues demonstrated that exposure to KU60019 induced greater radiation sensitivity in p53-mutant xenograft models compared to p53 wild type xenografts [34]. However, this p53-selective effect was not seen when KU59403 was assessed in in vitro studies or in xenografts [26]. It remains unclear whether these differences are due to the different inhibitors used and/or the different model systems used to assess radiosensitivity. In MCL, a B-cell lymphoma subtype, p53-defective tumour cell lines have also been shown to be more sensitive to the combination of ATMi and PARPi than their p53-proficient counterparts [35]. This finding thus provides some support to the idea that a ATMi/PARPi combination should be assessed in p53-mutant MCL.
ATM inhibitors could also be utilized to target tumour cells with loss of function mutations in genes previously shown to be synthetic lethal with ATM. For example ATM and ATR have been shown to be synthetic lethal, [36–38] suggesting that ATM inhibitors could be used in tumours with partial suppression of ATR function, although it is as yet unclear how this partial inactivation of ATR might be best measured in clinical biopsies. Furthermore, it has been demonstrated that BRCA1-deficient cells that have become PARP inhibitor resistant due to loss of either of two DNA repair proteins 53BP1 or REV7, can be resensitised to PARPi via exposure to an ATMi [39,40]. We also note that recently described CRISPR-based approaches to identify synthetic lethal approaches in human cells have identified a series of additional synthetic lethal (SL) interactions involving ATM that might be assessed as predictive biomarkers of ATM inhibitor sensitivity, including SL between ATM and CHK1, SETD2, SMO, KDM6A and VHL [41]. Thus far, no mechanisms of resistance to ATM inhibitors have been reported.
3. ATR as a cancer drug target
3.1. ATR function
The PIKK Ataxia telangiectasia and Rad3-related protein (ATR) is activated by regions of single stranded (ss) DNA [42]. Such ssDNA can result from nucleolytic processing of DNA DSBs and is also found at stalled replication forks when the activity of the replicative DNA helicase (MCM complex) becomes uncoupled from the activity of the DNA polymerase machinery [43]. Following activation, ATR phosphorylates a series of substrates, triggering a wide array of responses including the instigation of cell cycle checkpoints via CHK1 and WEE1 kinases, blocking replication origin firing, the repair of damaged DNA and also apoptosis [43] (Figure 1 and 3). CHK1 activates WEE1 via phosphorylation [44] and WEE1 in turn phosphorylates CDK1 (also known as CDC2) on tyrosine 15, thereby inhibiting CDK1 activity and preventing mitotic entry [45].
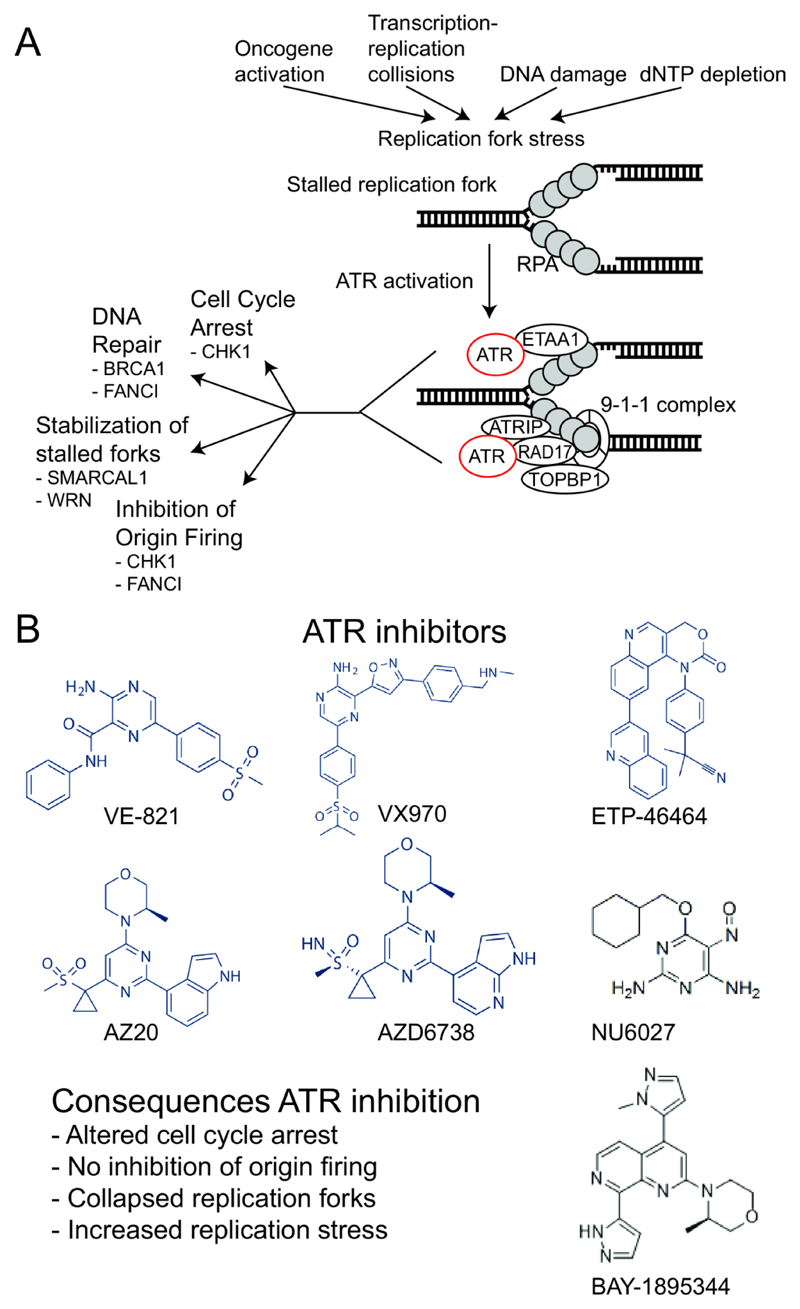
Figure 3. ATR function and ATR inhibitors.
A) Oncogene activation, collisions between the transcription and replication machinery, DNA damage and dNTP starvation are all causes of replication stress. A characteristic of replication stress is the presence of stalled replication forks. At these stalled forks the exposed ssDNA is covered by RPA. ATR interacting protein (ATRIP) binds to RPA coated ssDNA and recruits ATR to the site of ssDNA. RAD17, the Rad9–Rad1–Hus1 (9-1-1) complex and (DNA) topoisomerase II binding protein 1 (TOPBP1) are also recruited and all these proteins are required for ATR activation. ATR can also be activated via Ewing Tumour Associated Antigen 1 (ETAA1), which interacts directly with RPA coated ssDNA. Once activated, ATR can arrest the cell cycle via CHK1, initiate DNA repair, facilitate the stabilization of stalled forks and/or inhibit the firing of new origins to prevent further fork stalling. Below each function of ATR key substrates are shown. B) ATR inhibitors described in the main text are shown and the main consequences of ATR inhibition are listed.
3.2. ATR inhibitors
Several specific, potent ATR small molecule inhibitors have been reported from both academic research groups as well as pharmaceutical companies including AstraZeneca, Bayer and Vertex Pharmaceuticals. The toolbox ATR inhibitor VE-821 (Vertex) is a competitive ATP analogue inhibitor of ATR which blocks in vitro kinase activity with a Ki of 6 nM and a greater than 600 fold selectivity for ATR compared to ATM and DNA-PK [46]. The clinical ATR inhibitor VX-970 (previously VE-822, now M6620 since acquisition by Merck KGaA) is a structural analogue of VE-821 with superior potency, selectivity, and pharmacodynamic properties and as such was selected for clinical trial assessment [47]. Like the Vertex ATR inhibitors, the AstraZeneca toolbox ATR inhibitor AZ20 is a competitive ATP analogue inhibitor of ATR, which has an in vitro IC50 of 5 nM (50 nM in cell based assays) [48]. AZD6738 is an orally available analogue of AZ20 with superior solubility and pharmacodynamic properties [49], which is being used in clinical trials (including NCT01955668). In separate work, EPT-46464, developed by Toledo et al., was identified as a potent and selective inhibitor of ATR by screening a library of small molecule inhibitors using a cell based assay for the ATR-dependent phosphorylation of histone H2AX [14]. NU6027, developed by Peasland et al. was originally developed as a CDK2 inhibitor. However this compound was subsequently discovered to potently inhibit ATR, with an IC50 of 6.7 μM [50]. Another ATR inhibitor that has recently entered the first stages of clinical testing as a monotherapy for the treatment of cancer is BAY1895344 (Bayer, see NCT03188965).
3.3. Combination therapy with ATR inhibitors: pre-clinical evidence
ATR inhibitors (ATRi) potently sensitize multiple tumour cell models to platinum-based cross-linking agents (e.g. cisplatin and oxalipaltin), PARP inhibitors, IR, UV light exposure, the chemotherapeutic gemcitabine and poisons of either topoisomerase I or II [47,50–54] (Table 2). Interestingly, VE-821 also potentiates the toxicity of CHK1 inhibitors [55]. Importantly the synergy between VE-821 and cisplatin was much more pronounced in p53/ATM-deficient cancer cell lines compared to non-tumour cell lines, suggesting a potential tumour-specific biomarker for this drug combination [37]. An siRNA screen also found that silencing of the trans-lesion synthesis (TLS) polymerase ζ, consisting of REV3L and REV7, and the DNA DSB repair factor 53BP1, resulted in increased sensitivity to the VE-821/cisplatin combination [56]. Recurrent REV3L mutations have been identified in cisplatin-resistant squamous cell carcinoma of head and neck [57], making this a tumour subtype which might respond to the ATRi/cisplatin combination.
3.4. Biomarkers to predict ATR inhibitor sensitivity: preclinical evidence
Several groups have identified candidate biomarkers of ATR inhibitor sensitivity. Using an RNA interference (RNAi)-based chemosensitisation screen, Mohni and colleagues found that the silencing of ATR itself, as well as many of the proteins required for ATR activation (ATRIP, RAD17, RAD9A, RAD1, HUS1, TOPBP1), profoundly sensitized cells to VE-821 [58]. In addition, loss of ATM or the nucleotide excision repair proteins ERCC1 and ERCC4 (XPF) sensitized cells to VE-821 [58]. The sensitivity of ATM-deficient cells to ATR inhibition has also been observed with the ATRi AZD6738 [48,59] and ATM-deficient MCL models were shown to be sensitive to ATR inhibitors [60].
In independent synthetic lethal screens, VE-821 was also found to selectively target cells deficient in the replicative polymerase component protein POLD1 [61]. Although not mechanistically addressed, it seems likely that loss of POLD1 might cause increased replication stress resulting in a greater dependence on ATR activity. Similarly, a third synthetic lethal screen identified silencing of the chromatin-remodelling tumour suppressor gene ARID1A, to cause ATR inhibitor sensitivity, an effect later reproduced in multiple in vitro and in vivo models of ARID1A defective cancer [62]. Mechanistically, loss of ARID1A function results in a defect in the recruitment of the topoisomerase, TOP2A, to DNA; this TOP2A defect likely causes an inability to effectively decatenate DNA after replication and thus a reliance upon ATR [62]. As ARID1A is commonly mutated in a large number of tumours, including uterine, stomach, bladder and ovarian clear cell carcinomas [63], detecting tumour specific ARID1A mutations could provide a straightforward way to stratify patients for ATR inhibitor therapy.
Recent pre-clinical studies in Ewing sarcoma (ES) and synovial sarcoma (SS) have also suggested that the pathognomonic EWS-FLI1/ERG and SS18-SSX gene fusions found in ES and SS, respectively, might also cause sensitivity to ATR inhibitors [64,65]. These cancer driver fusions, which are caused by chromosomal translocations, cause replication fork stress and a reliance upon ATR function; as a result ES and SS tumour cells and xenografts are profoundly sensitive to drug like ATR inhibitors [64,65]. Finally, cells that utilize the alternative form of telomere lengthening (ALT) also display increased sensitivity to VE-821 [66], suggesting that surrogate biomarkers of ALT could direct the use of ATRi. However, the generality of this observation was recently questioned by a study by Deeg and colleagues, who failed to observe ALT-specific ATRi sensitivities in a distinct set of tumour cell models [67], compared to those used to originally identify the ALT/ATRi phenotype [66]. This might suggest that whilst an ATRi-sensitivity phenotype might be associated with ALT, this effect might not be fully penetrant.
3.5. Clinical trials
VX-970 and AZD6738 are currently being evaluated in several on-going clinical trials; these include trials where ATR inhibitors are used either as single agents or in combination with standard of care chemotherapies (reviewed in [68]). Consistent with much of the preclinical development of AZD6738, the on-going clinical trials have largely focused on using ATM-deficiency as a biomarker for patient selection (eg. NCT02264678). This Phase I/Ib trial (NCT02264678) is also determining the safety of combining AZD6738 with carboplatin, the PARPi olaparib or the anti-PD-L1 antibody MEDI4736 in solid tumours. Treating cancers with PARPi might increase the number of stalled replication forks, making cells more reliant on ATR to maintain genomic integrity [69]. Treatment with ATR inhibitors could potentially make tumours more immunogenic by increasing their mutational load, potentially enhancing the effect of immune checkpoint inhibitors such as MEDI4736 [70].
3.6. Resistance to ATR inhibitors
A recent in vitro CRISPR (Clustered regularly interspaced short palindromic repeats)–Cas9 mutagenesis genetic perturbation screen demonstrated that loss of the cell-cycle control phosphatase, CDC25A, results in ATR inhibitor resistance [71] (see also the later section “CHK1 function“). In this particular case, loss of CDC25A elicits cell cycle arrest in ATRi exposed cells, prior to mitosis, reducing the DSB load that ATR inhibitors might otherwise generate [71]. This resistance-causing effect can be reversed by forcing mitotic entry using a WEE1 inhibitor [71]. It remains yet to be established whether loss of CDC25A will represent a clinically relevant mechanism of ATRi resistance, but if this this indeed the case, a combination of ATRi plus WEE1 inhibitor might provide a mechanism based approach to dealing with such an event.
4. DNA-PK as a cancer drug target
4.1. DNA-PK function
DNA-PK is a PIKK that consists of the DNA Dependent Protein Kinase catalytic subunit (DNA-PKcs) and the Ku70/Ku80 heterodimer. DNA-PKcs is an essential component of the canonical Non-Homologous End Joining (NHEJ) pathway [72]. NHEJ is required for the repair of DSBs as well as during the generation of antibody diversity in mature B-cells (class switch recombination (CSR)). During CSR, programmed DSBs are formed, which are repaired via NHEJ. The NHEJ defect caused by DNA-PKcs mutations likely contributes to the radiosensitivity and immunodeficiency observed in DNA-PKcs mutant patients [73]. Conversely, elevated expression of DNA-PK is associated with radio-resistance in cervical and prostate cancer [74,75]. Apart from a role in NHEJ, DNA-PKcs has also been linked to telomere maintenance, transcription and several other functions [72] (Figure 4).
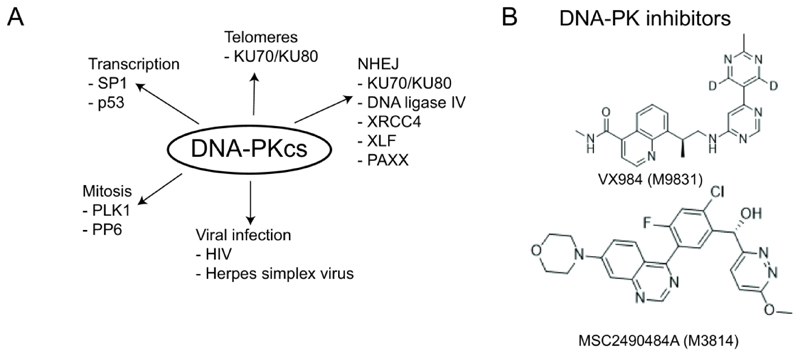
Figure 4. DNA-PKcs function and DNA-PK inhibitors.
A) DNA-PKcs primarily plays a role in non-homologous end joining (NHEJ). DNA-PKcs has also been shown to play a role in transcription via p53 and SP1. DNA-PKcs interacts with Polo-like kinase 1 (PLK1) and protein phosphatase 6 (PP6), both of which play a role in mitosis. Upon infection with Herpes Simplex virus, DNA-PKcs is degraded, while upon HIV infection, DNA-PKcs is activated to mediate p53-dependent apoptosis. DNA-PKcs is also required for telomere protection, together with KU70 and KU80. B) The structure of DNA-PK inhibitors discussed in the main text.
4.2. DNA-PK inhibitors
Currently there are two relatively specific DNA-PK inhibitors (DNA-PKi) in phase I clinical trials. MSC2490484A (M3814), developed by Merck KGaA, is being tested as a single agent in advanced solid tumours and CLL (NCT02316197) and in combination with fractionated palliative radiotherapy in solid tumours (NCT02516813). The reason for specific inclusion of CLL patients in the first clinical trial is because these patients often show deletion of the ATM gene [76], which is likely to be synthetically lethal with DNA-PK. VX-984 (M9831, developed by Vertex Pharmaceuticals and now licenced to Merck KGaA), is being tested as a single agent but also in combination with pegylated liposomal doxorubicin in patients with solid tumours (NCT02644278).
4.3. Biomarkers to predict DNA-PK inhibitor sensitivity: preclinical evidence
Several synthetic lethal interactions have been described for DNA-PKcs. In an shRNA screen, Zhou et al. found that DNA-PKcs depletion is lethal in MYC-dependent human cancers [77], primarily as DNA-PKcs inhibition leads to a reduction in MYC mRNA and protein expression, thus targeting MYC addicted tumour cells [77]. Defects in ATM and DNA-PKcs also form a synthetic lethal interaction, observed both in vitro as well as in vivo using a toolbox DNA-PK inhibitor [78]. A synthetic lethal interaction was also identified between DNA-PKcs and the mismatch repair (MMR) protein MSH3. Dietlein et al. profiled mutations in 67 different cancer cell lines and found that mutations in MSH3, BRCA1, BRCA2 and other HR-associated genes correlated with DNA-PKcs addiction [79]. Exposure of MSH3-deficient cancer cells to the toolbox DNA-PK inhibitor, KU60648 (KuDOS, AZ) lead to apoptosis. MSH3-deficient cells also displayed a reduction in HR, which might explain the synthetic lethality between MSH3 and DNA-PKcs. Additionally, a recent CRISPR screen for genetic interactions identified potential synthetic lethal interactions between DNA-PKcs (PRKDC) and: AKT1, CDK4, CDK9, CHK1, IGFR1, mTOR, VHL and RRM2 [41]; whether these synthetic lethal interactions can be replicated with small molecule DNAPK inhibitors remains to be seen.
Since DNA-PKcs is involved in NHEJ, a mechanism used to repair IR induced DSBs, a DNA-PK inhibitor could in principle be used to enhance the effect of radiotherapy. The maximum dose of radiotherapy applied is normally limited by normal tissue toxicity and some tumour cells are inherently more radio-resistant. Therefore the addition of agents that enhance the effect of radiotherapy could improve tumour response. However, currently most improvements in radiotherapy are made by technological advances that allow more precise targeting of the radiation to the location containing the tumour based on anatomical information [80]. Still, for some tumours in difficult to reach areas, treatment with a DNA-PK inhibitor might improve treatment outcome. A potential disadvantage of such an approach is that the DNA-PK inhibitor, delivered systemically, might increase tumour radio-sensitivity but also the deleterious side effects of radiation in normal, surrounding, tissue. Finally, as far as we are aware, mechanisms of resistance to DNA-PK inhibitors, either used a single agents or in combination, have not as yet been identified in pre-clinical studies.
4.4. Combination therapy with DNA-PK inhibitors: pre-clinical evidence
DNA-PK inhibitors could be used to enhance agents that cause DSBs that require NHEJ for their repair, including topoisomerase II inhibitors etoposide or doxorubicin, or IR (Table 2). Combination of the toolbox DNA-PKi KU60648 with etoposide resulted in increased survival of mice with Eμ:Myc;Arf–/–-driven lymphomas expressing an shRNA targeting Atm, providing proof of principle that such an approach might be feasible [78].
DNA-PKi could potentially limit mutational processes, especially in HR-defective cancers. In these cancers, DSBs are repaired via NHEJ and other non-conservative pathways; this leads to deletions, translocations and other chromosomal rearrangements [81]. One hypothesis is that these alterations fuel cancer development, metastasis and resistance. Inhibition of DNA-PK could prevent NHEJ and therefore potentially limit the mutational burden. This might raise the possibility of using DNA-PK inhibitors to prevent tumorigenesis in patients at high risk of developing HR-defective cancers, such as those with BRCA1 or BRCA2 mutations. Such an approach might be relevant if prophylactic use of a DNA-PK inhibitor could be shown to be non-toxic.
5. CHK1 as a cancer drug target
5.1. CHK1 function
ATR, but also ATM, phosphorylates and activates the kinase CHK1 in response to DNA damage [82] (Figure 1). CHK1 regulates the intra-S checkpoint via phosphorylation of CDC25A. This phosphorylation results in degradation of CDC25A and a subsequent reduction in CDK2 activity in S-phase [83]. CHK1 also phosphorylates CDC25C and WEE1, events which control mitotic entry and thereby the G2/M checkpoint [84].
5.2. CHK1 inhibitors
The most specific CHK1 inhibitors that have reached the stage of clinical testing (used either as single agents or in combination with antimetabolites) include LY2603618 (Eli Lilly), SCH900776 (MK-8776; Merck KGaA), GDC-0575 (Genentech), and CCT245737 (SRA373, Sierra Oncology) (reviewed in [4,68]). Tumour types included in these phase I/II clinical trials include non-small cell lung cancer (NSCLC) (NCT01139775), Acute Myeloid Leukaemia (AML) (NCT01870596), lymphoma (NCT01564251) as well as a range of other types of tumours (NCT02797977, NCT02797964). Although some of these studies are still on-going, recent results suggest early evidence of clinical efficacy of CHK1 inhibitors in subsets of patients and hint at certain histological subtypes that may be more sensitive to CHK1 inhibitors than others, such as NSCLC [85] and AML [86].
5.3. Biomarkers to predict CHK1 inhibitor sensitivity: preclinical evidence
Although results from clinical trials show the clinical potential of CHK1 inhibitors, particularly in combination with antimetabolites, they also underline the need for patient stratifying biomarkers. Various in vitro studies have shown that CHK1 inhibition allows selective targeting of p53-mutant cells, and CHK1 inhibition also potentiates the cytotoxicity of topoisomerase inhibitors and IR in p53-deficient, but not in p53-proficient cells [87–89]. However, other recent studies, including clinical studies, report no clear correlation between p53-deficiency and response to CHK1 inhibitors [90].
γH2AX and CHK1 phosphorylation have been used as predictive pharmacodynamic biomarkers of CHK1 inhibitor-chemotherapy combination sensitivity, including combinations of CHK1 inhibitors with gemcitabine or camptothecin [91]. pCHK1 (S296) was identified as a predictive biomarker of CHK1 inhibitor sensitivity in ovarian and ER/PR/HER2-negative (triple-negative) breast cancers and γH2AX was predictive in another breast cancer subtype, luminal breast cancer [92]. In addition to CHK1 and γH2AX phosphorylation, Cyclin B1 levels were found to be an efficacy-predicting biomarker for CHK1 inhibitors [93], and a subset of cancer cell lines showed acute sensitivity to the CHK1 inhibitor MK-8776 as mono-therapy due to CDK2 activation in S-phase [94].
5.4. Combination therapy with CHK1 inhibitors: pre-clinical evidence
In addition to combinations with antimetabolites or other classes of cytotoxic agents (e.g. gemcitabine and topoisomerase inhibitors that synergise with CHK1 inhibitors [87,95]), a number of other CHK1 inhibitor combination approaches have been identified in pre-clinical models (Table 2). CHK1 inhibition together with WEE1 inhibition has shown promising and strong synergistic activity in AML-derived leukemic cells [96] and in malignant melanoma tumour cell lines and xenografts [97]. In addition, a cancer-specific synthetic lethality between ATR and CHK1 kinase activities has been reported, suggesting tumours with reduced ATR activity could be sensitive to CHK1 inhibitors [55]. In combination with MK2 inhibitors, CHK1 inhibitors also target KRAS- or BRAF-mutant tumour cells [98].
6. CHK2 as a cancer drug target
6.1. CHK2 function
The CHK2 kinase is primarily activated by ATM. After phosphorylation by ATM, CHK2 homo-dimerises [99]. Similar to CHK1, CHK2 also regulates the CDC25 family of phosphatases and affects the intra-S as well as the G2/M checkpoint [20]. Additionally CHK2 also affects the G1/S checkpoint by phosphorylating CDC25C [100] (Figure 1).
6.2. CHK2 inhibitors
At the moment two relatively specific toolbox CHK2 inhibitors are used in pre-clinical studies: PV1019 (NIH) [101] and CCT241533 (ICR) [102]. PV1019 is an ATP-competitive inhibitor that is reported to be selective for CHK2 and that synergises with topotecan, camptothecin, and IR in human tumour cell lines [101]. CCT241533 also binds the ATP-binding pocket in CHK2 and prevents CHK2 autophosphorylation at Serine 516 [103]. This CHK2 inhibitor does not synergise with gemcitabine, etoposide, or mitomycin C, but does enhance the effect of the PARP inhibitors rucaparib and olaparib [103].
The kinase domains in CHK1 and CHK2 are highly conserved and AZD7762 (AstraZeneca) targets CHK1 and CHK2 with similar potency [104]. In vivo, AZD7762 enhanced the effect of the topoisomerase I inhibitor irinotecan in a colon cancer xenograft model [104]. In a pancreatic tumour model, AZD7762, when used with gemcitabine, sensitized tumour cell lines and patient derived xenografts (PDX) to IR [105]. So far, three clinical trials with AZD7762 have been initiated, one of which has been completed and two have been terminated (NCT00937664, NCT00413686, NCT00473616). No further development of AZD7762 appears to be planned, possibly due to the cardio-toxicity associated with this inhibitor [106]. Whether this cardio-toxicity is an on-target effect of AZD7762 and a potentially a common characteristic of CHK1 inhibitors is not clear from current data and remains to be investigated.
7. WEE1 as a cancer drug target
7.1. WEE1 function
CHK1 actives the kinase WEE1 via phosphorylation [44] and WEE1 in turn phosphorylates CDK1 (also known as CDC2) on Tyrosine 15 (Y15), thereby inhibiting CDK1 activity and preventing mitotic entry [45] (Figure 1). To allow mitosis to occur, WEE1 is phosphorylated by Polo-like Kinase 1 (PLK1), which triggers WEE1 degradation [107].
At present there is only one WEE1 inhibitor being tested in phase I and II clinical trials. AZD1775 (MK1775, Merck KGaA, now AstraZeneca) inhibits the tyrosine 15 phosphorylation on CDK1 and the G2/M checkpoint. WEE1 inhibition forces cells into mitosis before replication has been completed, resulting in abnormal mitoses being formed and in some cases, apopotosis [108]. In head and neck cancer, patients with p53 mutations often respond poorly to cisplatin treatment. Osman et al. showed that platinum-resistant cell lines and xenografts could be sensitized to cisplatin by AZD1775 [109].
7.2. Biomarkers to predict WEE1 inhibitor sensitivity: preclinical evidence
Several potential biomarkers for AZD1775 sensitivity have been proposed. In in vitro studies, mutations in p53 sensitize tumour cells to WEE1 inhibition [110]. Loss of p53 function abrogates the G1 checkpoint and one hypothesis suggests that the greater reliance upon the G2/M cell cycle checkpoint can be targeted in p53 defective cells with WEE1 inhibitors. In p53-mutant cells, elevated expression of Histone-lysine N-methyltransferase Enhancer of zeste homolog 2 (EZH2) and the mitotic cyclins (Cyclin B1, B2 and E1) also results in an increased reliance on WEE1 to prevent pre-mature mitotic entry [108]. Lower expression of PKMYT1, a kinase that regulates mitotic entry, also sensitizes cells to WEE1 inhibition [111]. Additionally, Weisberg et al. found that AZD1775 potentiates mTOR inhibition in NRAS- and KRAS-mutant positive AML cell lines and primary patient samples [112]. Pancreatic cancer cell lines with defects in DDR genes such as FANCC, FANCG and BRCA2 are also more sensitive to AZD1775 [113]. Lastly, it has been shown that AZD1775 can inhibit H3K36me3-deficient tumour cells and in vivo tumours [114]. H3K36me3 is required to facilitate expression of Ribonucleoside-diphosphate reductase subunit M2 (RRM2). RRM2 catalyses the synthesis of deoxyribonucleotides from ribonucleotides, which are required for DNA synthesis. WEE1 inhibition results in the degradation of RRM2 and in H3K36me3-deficient cells, the degradation of the already low level of RRM2 results in dNTP (deoxyribonucleotide triphosphate) starvation and ultimately cell death.
7.3. Combination therapy with WEE1 inhibitors: pre-clinical evidence
WEE1 inhibition has been shown to synergise with CHK1 inhibition independently of p53 status [115] (Table 2). The combination has been shown to be effective in MCL cell lines [116] and in breast cancer cells [108]. Combinations with other DNA damaging or targeted agents, such as the HDAC inhibitor panobinostat, which down-regulates CHK1 [117], cytarabine, which interferes with DNA synthesis [118] or cisplatin [119], have been investigated as well.
While tumours with mutations in BRCA1 and BRCA2 are highly sensitive to PARP inhibitors, secondary “reversion” mutations in these genes restore some DNA repair function and cause PARP inhibitor resistance, in both pre-clinical models and in the clinic [120,121]. Dréan et al. recently found that whilst tumour cells with reversion mutations are profoundly resistant to multiple PARP inhibitors, they still retain the AZD1775 sensitivity seen in tumour cells without reversions [122]. In mice bearing xenografts consisting of both BRCA2 revertant and non-revertant tumour cells, the PARP inhibitor olaparib had little therapeutic effect as it targeted non-revertant cells but not revertant tumour cells which eventually dominated the tumour cell population [122]. Conversely, treatment with AZD1775 extended the survival of mice as it effectively targeted both revertant and non-revertant tumour cells [122], raising the clinically testable hypothesis that WEE1 inhibitors could serve some utility in either delaying the onset of PARPi resistance or targeting resistant disease once it emerges.
7.4. Clinical trials using WEE1 inhibitors
Currently twenty-eight clinical trials with MK1775/AZD1775 are registered (reviewed in [68]) and at this point in time, the results of two phase I trials and one phase II trial have been reported. So far, two partial responses to AZD1775 as a single agent have been observed in patients with either BRCA1 or BRCA2 mutations [123]. In a follow-up phase II study, one patient showed a prolonged complete response to AZD1775 [124]. This patient, diagnosed with serous ovarian cancer, had mutations in p53, BRCA1, MYC and Cyclin E.
7.5. Mechanisms of WEE1 inhibitor resistance
In vitro, one mantle cell leukemia (MCL)-derived cell line has been described that is resistant to both the CHK1 inhibitor PF-00477736 and AZD1775 [125]. MCL is characterized by Cyclin D1 overexpression and resistant cells exhibited a decrease in Cyclin D1 expression, as well as an upregulation of pro-survival pathways [125]. Another possible mechanism of resistance could be the restoration of the G1 checkpoint via restoration of p53 function. Li et al. found that inhibition of MDM2 with nutlin, resulting in activation of p53, reduced the cytotoxic effects of AZD1775 in p53-proficient cell lines [126]. Reactivation of the G1 checkpoint could allow tumour cells to stall the cell cycle to repair DNA damage, thus reducing the cytotoxic effects of AZD1775.
8. Conclusions
As described above, the identification of predictive biomarkers, optimal drug combinations and mechanisms of drug resistance are as critical a part of the drug discovery and development process for DDR kinase inhibitors, as they should be for all new cancer drugs. It seems reasonable to think that the efforts to dissect each of these areas for the DDR kinase inhibitors that are already being assessed in clinical trials and those yet to enter first in human studies, will have a positive effect on streamlining and optimising the route towards eventual drug approval.
9. Expert Opinion
Predictive biomarkers of sensitivity to DDR inhibitors
As for any anti-cancer treatment, it seems reasonable to assume that identifying predictive biomarkers of sensitivity to DDR inhibitors will be critical to the successful use of these agents. As we have described above, some of the first steps have been taken to define such predictive biomarkers. In some cases, such as in the case of ATR inhibitors, pre-clinical work has identified candidate predictive biomarkers (e.g. ATM loss, ARID1A mutation, Ewings sarcoma translocation etc.) that could now be assessed in clinical trials. Nevertheless, there are still a number of areas clearly worthy of further investigation. These include:
-
(i)
more pre-clinical research is required to identify predictive biomarkers not only of single agent DDR inhibitor sensitivity but also for combination therapy involving DDR inhibitors. The assumption that factors that cause single agent DDRi sensitivity will also predict sensitivity to combination therapy, and visa versa, might not be wholly true and so an increased focus on identifying predictive biomarkers of DDR combination therapy response is required;
-
(ii)
at present, much of the focus on identifying predictive DDRi biomarkers has used a “forward translation” where candidate biomarkers are identified using pre-clinical approaches, informing the design or analysis of clinical trials. As the number of clinical trials involving DDRi increases, taking “reverse translation” approaches, where observations made in clinical trials are then validated in pre-clinical experiments, should become an integral part of identifying and understanding predictive biomarkers;
-
(iii)
in pre-clinical experimental systems, replication fork stress is often associated with DDRi sensitivity. Replication fork stress (i.e. the slowing and stalling of replication forks), has many causes and likely encapsulates a broad range of molecularly diverse phenotypes, each of which might result in a different DDRi sensitivity effect. With this in mind, additional focus is required to define clinically measurable predictive biomarkers that are related to replication fork stress but which also define a specific DDRi (or other drug) sensitivity.
Mechanisms of resistance
Based on what is understood about mechanisms of resistance to DNA damaging chemotherapies, other kinase inhibitors and the approved DDRi class, PARP inhibitors, it might be interesting to predict which mechanisms might cause resistance to novel DDR kinase inhibitors. Firstly, mutations in the catalytic domains of DDR kinases that allow catalysis in the presence of small molecule inhibitors might very well cause drug resistance (analogous to EGFR catalytic domain mutations that cause gefitinib and erlotinib resistance [127]). Secondary “revertant” mutations in synthetic lethal partners of DDRi kinase inhibitor targets might also cause drug resistance, in much the same way that revertant mutations in BRCA1 or BRCA2 cause PARP inhibitor resistance [120,121]. One might predict, for example, that resistance to ATM/ATR inhibitor synthetic lethality in ATM mutant cancers might be driven by revertant mutations in ATM that restore ATM function. Of course, this mechanism would only operate if the continued fitness of tumour cells was not impaired by restoring ATM function. Analogous to this concept, additional alterations in proteins that restore DDR pathway function might also drive resistance to DDR kinase inhibitors, similar to 53BP1 or REV7 defects restoring DSB resection and PARP inhibitor resistance in BRCA1 defective cells [17]. Again, taking the ATM/ATR inhibitor synthetic lethal effect as an example, one might expect restoration of ATM pathway function via elevated function of downstream ATM signalling components to cause ATRi resistance. Finally, alterations in cell cycle control proteins are likely candidates for mediating resistance, as illustrated by CDC25A loss of function causing ATR inhibitor resistance [71]. These predictions of course are predicated on prior knowledge, and we would still argue that taking relatively unbiased approaches (such as genetic perturbation screens) to uncover mechanisms of resistance are also likely to be informative as taking hypotheses-testing approaches.
Novel drug combinations: combining DDR inhibitors with immunotherapy
In addition to the potential of using DDR inhibitors in combination regimens with other DDR inhibitors or with standard-of-care chemotherapy or radiotherapy treatments, some consideration might be given to identifying optimised approaches to using DDR inhibitors alongside immunotherapy agents including immune checkpoint inhibitors [68,70,128]. Transformed or malignant cells can be recognised and in some cases eliminated by the adaptive and innate immune response [129]. In many cases, however, tumours acquire the ability to suppress these anti-cancer immune responses. For example, tumour cells inactivate T cells by expressing Programmed Death-Ligand 1 (PD-L1); interaction between PD-L1 on tumour cells and Programmed Death Receptor 1 (PD-1) on the surface of T cells drives T cell inactivation. This inactivation can be reversed by the use of anti-PD-L1 therapeutic antibodies such as atezolizumab (Roche) or duvalumab (AstraZeneca), eliciting an anti-tumour cell immune response. Although there is a relative paucity of pre-clinical data describing the effectiveness of DDR inhibitor/immunotherapy combinations (largely, we presume because of the relative complexity of effectively modelling immunotherapy responses in pre-clinical models), such an approach might offer several advantages. For example, if DDR inhibitors and immunotherapy agents demonstrate non-overlapping mechanisms of action and therefore are less likely to display overlapping mechanisms of resistance, it might make sense to consider DDR inhibitor/immunotherapy combinations that do not necessarily elicit synergistic/supra-additive effects on tumour cells, but nevertheless extend survival. Secondly, DDR inhibitors might themselves either stimulate or enhance anti-tumour cell immune responses to the extent that DDR inhibitor/immunotherapy combinations are effective. For example, DNA damage is known to enhance signalling pathways that activate the innate immune response, including the the cGAS-STING pathway that monitors cytoplasmic DNA [130]; using DDR inhibitors to exacerbate forms of DNA damage that activate cGAS-STING signalling in a cancer-specific fashion might very well enhance the effectiveness of immune modulating therapies. Thirdly, DDR inhibitors might be used to enhance the mutational burden of tumours; this in turn might elevate the neo-antigen load, a likely determinant of anti-tumour immune responses [131]. However, there is a potentially limiting factor that could also limit the potential of DDR inhibitor/immunotherapy combinations; it is possible that DDR inhibitors might increase the formation of mutations and neoepitopes in a relatively heterogeneous fashion, generating multiple subclones, each with a distinct neoepitope profile. In such a scenario, the absence of a relatively clonal neoepitope might result in an anti-cancer immune response that only removes a minority of tumour cells.
Article highlights box.
Inhibitors of the DDR kinases ATM, ATR, DNA-PK, CHK1, CHK2 and WEE1 are now being assessed in phase I and phase II clinical trials for the treatment of cancer.
These novel DDR inhibitors (DDRi) have the potential to exploit synthetic lethal interactions that operate in tumour cells, as well as exploiting other commonly found hallmarks of cancer, including genomic instability and replication fork stress
Pre-clinical studies have identified a series of candidate predictive biomarkers of sensitivity to DDRi, some of which are suitable for assessment in clinical trials.
Thus far, few mechanisms of DDRi resistance have been identified, although such effects seem likely
Drug combination strategies might be used to enhance the overall effectiveness of DDRi, but as for single agent DDRi use, predictive biomarkers are required to direct the use of combination approaches
Acknowledgements
We thank Breast Cancer Now and Cancer Research UK for funding the work in our laboratory and their continued support. EDGF is the recipient of a Rubicon Fellowship.
List of abbreviations
- 53BP1
TP53-binding protein 1
- AKT1
RAC-alpha serine/threonine-protein kinase
- ALT
Alternative Lengthening of Telomeres
- AML
Acute Myeloid Leukaemia
- Arf
Tumor suppressor ARF
- ARID1A
AT-rich interactive domain-containing protein 1A
- ATM
Ataxia telangiectasia mutated
- ATMi
ATM inhibitor
- ATP
Adenosine Triphosphate
- ATR
Ataxia telangiectasia and Rad3-related protein
- ATRi
ATR inhibitor
- ATRIP
ATR Interacting Protein
- BRCA1
Breast cancer type 1 susceptibility protein
- BRCA2
Breast cancer type 2 susceptibility protein
- CCNE1
Cyclin E1
- CD8
T-cell surface glycoprotein CD8
- CDC2
CDK1
- CDC25A
M-phase inducer phosphatase 1
- CDC25C
M-phase inducer phosphatase 3
- CDK1
Cyclin-dependent Kinase 1
- CDK2
Cyclin-dependent Kinase 2
- CDK4
Cyclin-dependent Kinase 4
- CDK9
Cyclin-dependent Kinase 9
- CHK1
Checkpoint kinase-1
- CHK1i
CHK1 inhibitor
- CHK2
Checkpoint kinase-2
- CHK2i
CHK2 inhibitor
- CLL
Chronic Lymphocytic leukaemia
- CRISPR
Clustered regularly interspaced short palindromic repeats
- CSR
Class Switch Recombination
- CTIP
CtBP-interacting protein
- CTLA-4
Cytotoxic T-lymphocyte protein 4
- DDR
DNA Damage Response
- DNA
Deoxyribonucleic Acid
- DNA-PK
DNA-dependent Protein Kinase
- DNA-PKcs
DNA-dependent Protein Kinase catalytic subunit
- DNA-PKi
DNA-PK inhibitor
- dNTP
Deoxyribonucleotide triphosphate
- DSB
Double Strand Break
- dsDNA
Double stranded DNA
- EGFR
Epidermal growth factor receptor
- ERCC1
Excision Repair Cross-Complementation Group 1
- ERCC4
Excision Repair Cross-Complementation Group 4
- EZH2
Enhancer of zeste homolog 2
- FANC
Fanconi anemia
- FANCC
Fanconi anemia group C protein
- FANCG
Fanconi anemia group G protein
- H2AX
Histone H2A.X
- HDAC
Histone Deacetylase
- HR
Homologous Recombination
- HRD
Homologous Recombination Deficiency
- IGFR1
Insulin-like growth factor I
- IR
Ionising Radiation
- KDM6A
Lysine-specific demethylase 6A
- KRAS
Kirsten rat sarcoma viral oncogene
- MCL
Mantle Cell Lymphoma
- MDM2
Double minute 2 protein
- MHC
Major histocompatibility complex
- MK2
MAP kinase-activated protein kinase 2
- MLH1
MutL protein homolog 1
- MMC
Mitomycin C
- MMR
Mismatch repair
- MSH3
MutS Homolog 3
- mTOR
Mechanistic target of rapamycin
- NHEJ
Non-homologous end joining
- NKG2D
Natural Killer Group 2, member D
- NKT
Natural Killer T-cell
- NSCLC
Non-Small Cell Lung Cancer
- PARP
Poly ADP ribose polymerase
- PARPi
PARP inhibitor
- PD1
Programmed cell death protein 1
- PDL1
Programmed death-ligand 1
- PIKK
Phosphatidyl inositol 3’ kinase-related kinases
- PKMYT1
Protein Kinase, membrane associated tyrosine/threonine 1
- PLK1
Polo-Like Kinase 1
- POLD1
Polymerase D1
- RAD1
RAD1 checkpoint DNA exonuclease
- RAD17
RAD17 checkpoint Clamp Loader component
- RAD9A
RAD9 checkpoint Clamp component A
- RAD51C
DNA repair protein RAD51 homolog 3
- RAD51D
DNA repair protein RAD51 homolog 4
- REV3L
REV3 like, DNA directed polymerase zeta catalytic subunit
- REV7
MAD2L2: Mitotic Arrest Deficient 2 like 2
- RNA
Ribonucleic Acid
- RRM2
Ribonucleoside-diphosphate reductase subunit M2
- SETD2
SET domain-containing protein 2
- SMO
Smoothened homolog
- ssDNA
Single Stranded DNA
- TLS
Trans Lesion Synthesis
- TOP2A
Topoisomerase IIA
- TOPBP1
DNA topoisomerase 2-binding protein 1
- UV
Ultraviolet (light)
- VHL
Von Hippel-Lindau disease tumor suppressor
- WEE1
WEE1 G2 checkpoint kinase
- WEE1i
WEE1 inhibitor
- XPF
Xeroderma Pigmentosum, complementation group F
- γH2AX
H2AX phosphorylated at S139
Footnotes
Disclosure statement CJL and CW are inventors on patents describing the use of DDR inhibitors in cancer and stand to gain for their use as part of the ICR “Rewards to Inventors” scheme. CJL receives/has received research funding from Merck KGaA, Vertex and Astra Zeneca.
References
•= of interest, ••= of considerable interest
- [1].Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- [2].Lord CJ, Ashworth A. The DNA damage response and cancer therapy. Nature. 2012;481:287–294. doi: 10.1038/nature10760. [DOI] [PubMed] [Google Scholar]
- [3].Rundle S, Bradbury A, Drew Y, et al. Targeting the ATR-CHK1 Axis in Cancer Therapy. Cancers (Basel) 2017;9:41. doi: 10.3390/cancers9050041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Otto T, Sicinski P. Cell cycle proteins as promising targets in cancer therapy. Nat Rev Cancer. 2017;17:93–115. doi: 10.1038/nrc.2016.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol. 2013;14:197–210. [PubMed] [Google Scholar]
- [6].Bhattacharya S, Srinivasan K, Abdisalaam S, et al. RAD51 interconnects between DNA replication, DNA repair and immunity. Nucleic Acids Res. 2017;45:4590–4605. doi: 10.1093/nar/gkx126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kersten K, de Visser KE, van Miltenburg MH, et al. Genetically engineered mouse models in oncology research and cancer medicine. EMBO Mol Med. 2016;9:137–153. doi: 10.15252/emmm.201606857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- [10].McGranahan N, Swanton C. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell. 2017;168:613–628. doi: 10.1016/j.cell.2017.01.018. [DOI] [PubMed] [Google Scholar]
- [11].Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Karakaidos P, Zacharatos P, Kotsinas A, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005:907–913. doi: 10.1038/nature03485. [•• Describes the activation of the DDR in response to oncogene activation] [DOI] [PubMed] [Google Scholar]
- [13].Bartkova J, Rezaei N, Liontos M, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [•• Describes the activation of the DDR in response to oncogene activation] [DOI] [PubMed] [Google Scholar]
- [14].Toledo LI, Murga M, Zur R, et al. A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nat Struct Mol Biol. 2011;18:721–727. doi: 10.1038/nsmb.2076. [• Describes the identification of ATR inhibitors and synthtic lethal interactions with Cyclin E1 overexpression and/or p53 loss] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat Cell Biol. 2014;16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Drean A, Lord CJ, Ashworth A. PARP inhibitor combination therapy. Crit Rev Oncol. 2016;108:73–85. doi: 10.1016/j.critrevonc.2016.10.010. [DOI] [PubMed] [Google Scholar]
- [17].Lord CJ, Ashworth A. PARP inhibitors: Synthetic Lethality in the clinic. Science (80-. ) 2017;355:1152–1158. doi: 10.1126/science.aam7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lavin MF, Khanna KK, Beamish H, Spring K, Watters D, Shiloh Y. Relationship of the ataxia-telangiectasia protein ATM to phosphoinositide 3-kinase. Trends Biochem Sci. 1995;20:382–383. doi: 10.1016/s0968-0004(00)89083-0. [DOI] [PubMed] [Google Scholar]
- [19].Dai Y, Grant S. New insights into checkpoint kinase 1 in the DNA damage response signaling network. Clin Cancer Res. 2010;16:376–383. doi: 10.1158/1078-0432.CCR-09-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zannini L, Delia D, Buscemi G. CHK2 kinase in the DNA damage response and beyond. J Mol Cell Biol. 2014;6:442–457. doi: 10.1093/jmcb/mju045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Tresini M, Warmerdam DO, Kolovos P, et al. The core spliceosome as target and effector of non-canonical ATM signalling. Nature. 2015;523:53–58. doi: 10.1038/nature14512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Beà S, Valdés-Mas R, Navarro A, et al. Landscape of somatic mutations and clonal evolution in mantle cell lymphoma. Proc Natl Acad Sci U S A. 2013;110:18250–18255. doi: 10.1073/pnas.1314608110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hickson I, Zhao Y, Richardson CJ, et al. Identification and Characterization of a Novel and Specific Inhibitor of the Ataxia-Telangiectasia Mutated Kinase ATM Identification and Characterization of a Novel and Specific Inhibitor of the Ataxia-Telangiectasia Mutated Kinase ATM. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- [24].Golding SE, Rosenberg E, Valerie N, et al. Improved ATM kinase inhibitor KU-60019 radiosensitizes glioma cells, compromises insulin, AKT and ERK prosurvival signaling, and inhibits migration and invasion. Mol Cancer Ther. 2009;8:2894–2902. doi: 10.1158/1535-7163.MCT-09-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Weber AM, Ryan AJ. ATM and ATR as therapeutic targets in cancer. Pharmacol Ther. 2015:124–138. doi: 10.1016/j.pharmthera.2014.12.001. [DOI] [PubMed] [Google Scholar]
- [26].Batey MA, Zhao Y, Kyle S, et al. Preclinical evaluation of a novel ATM inhibitor, KU59403, in vitro and in vivo in p53 functional and dysfunctional models of human cancer. Mol Cancer Ther. 2013;12:959–967. doi: 10.1158/1535-7163.MCT-12-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Pike KG. Identifying high quality, potent and selective inhibtors of ATM kinase: Discovery of AZD0156. Proceeding 107th Annu Meet if Am Assoc Cancer Res. 2016 [Google Scholar]
- [28].Nakayama Y, Igarashi A, Kikuchi I, et al. Bleomycin-induced over-replication involves sustained inhibition of mitotic entry through the ATM/ATR pathway. Exp Cell Res. 2009;315:2515–2528. doi: 10.1016/j.yexcr.2009.06.007. [DOI] [PubMed] [Google Scholar]
- [29].Hsiang YH, Lihou MG, Liu LF. Arrest of Replication Forks by Drug-stabilized Topoisomerase I-DNA Cleavable Complexes as a Mechanism of Cell Killing by Camptothecin. Cancer Res. 1989;49:5077–5082. [PubMed] [Google Scholar]
- [30].Sartori AA, Lukas C, Coates J, et al. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Bakr A, Oing C, Köcher S, et al. Involvement of ATM in homologous recombination after end resection and RAD51 nucleofilament formation. Nucleic Acids Res. 2015;43:3154–3166. doi: 10.1093/nar/gkv160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Williamson C, Muzik H, Turhan A. ATM deficiency sensitizes mantle cell lymphoma cells to poly (ADP-ribose) polymerase-1 inhibitors. Mol Cancer Ther. 2010;9:347–357. doi: 10.1158/1535-7163.MCT-09-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].McCabe N, Turner NC, Lord CJ, et al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 2006;66:8109–8115. doi: 10.1158/0008-5472.CAN-06-0140. [DOI] [PubMed] [Google Scholar]
- [34].Biddlestone-Thorpe L, Sajjad M, Rosenberg E, et al. ATM kinase inhibition preferentially sensitizes p53-mutant glioma to ionizing radiation. Clin Cancer Res. 2013;19:3189–3200. doi: 10.1158/1078-0432.CCR-12-3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Williamson CT, Kubota E, Hamill JD, et al. Enhanced cytotoxicity of PARP inhibition in mantle cell lymphoma harbouring mutations in both ATM and p53. EMBO Mol Med. 2012;4:515–527. doi: 10.1002/emmm.201200229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Menezes DL, Holt J, Tang Y, et al. A Synthetic Lethal Screen Reveals Enhanced Sensitivity to ATR Inhibitor Treatment in Mantle Cell Lymphoma with ATM Loss-of-function. Mol Cancer Res. 2014;13 doi: 10.1158/1541-7786.MCR-14-0240. pii: molcanres.0240.2014. [DOI] [PubMed] [Google Scholar]
- [37].Reaper PM, Griffiths MR, Long JM, et al. Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nat Chem Biol. 2011;7:428–430. doi: 10.1038/nchembio.573. [• Describes the synergy between small molecule ATR inhibitors and platinum salts in pre-clinical models of p53 mutant cancer] [DOI] [PubMed] [Google Scholar]
- [38].Cui Y, Palii SS, Innes CL, et al. Depletion of atr selectively sensitizes atmdeficient human mammary epithelial cells to ionizing radiation and dna-damaging agents. Cell Cycle. 2014;13:3541–3550. doi: 10.4161/15384101.2014.960729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Bunting SF, Callén E, Wong N, et al. 53BP1 inhibits homologous recombination in brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Xu G, Chapman JR, Brandsma I, et al. REV7 counteracts DNA double-strand break resection and affects PARP inhibition. Nature. 2015;521:541–544. doi: 10.1038/nature14328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Shen JP, Zhao D, Sasik R, et al. Combinatorial CRISPR–Cas9 screens for de novo mapping of genetic interactions. Nat Methods. 2017;14 doi: 10.1038/nmeth.4225. [•• CRISPR-Cas9 genetic screen that exploits a "dual-guide RNA" approach which identifies synthetic lethal interactions involving DDR and tumour suppressor genes.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lecona E, Fernández-Capetillo O. Replication stress and cancer: It takes two to tango. Exp Cell Res. 2014:26–34. doi: 10.1016/j.yexcr.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Yazinski SA, Zou L. Functions, Regulation, and Therapeutic Implications of the ATR Checkpoint Pathway. Annu Rev Genet. 2015:1–19. doi: 10.1146/annurev-genet-121415-121658. [DOI] [PubMed] [Google Scholar]
- [44].Lee J, Kumagai a, Dunphy WG. Positive regulation of Wee1 by Chk1 and 14-3-3 proteins. Mol Biol Cell. 2001;12:551–563. doi: 10.1091/mbc.12.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Parker LL, Piwnica-Worms H. Inactivation of the p34cdc2-Cyclin B Complex by the Human WEE1 Tyrosine Kinase. Science (80-. ) 1992;257:1955–1957. doi: 10.1126/science.1384126. [DOI] [PubMed] [Google Scholar]
- [46].Charrier JD, Durrant SJ, Golec JMC, et al. Discovery of Potent and Selective Inhibitors of Ataxia Telangiectasia Mutated and Rad3 Related (ATR) Protein Kinase as Potential Anticancer Agents. J Med Chem. 2011;54:2320–2330. doi: 10.1021/jm101488z. [DOI] [PubMed] [Google Scholar]
- [47].Fokas E, Prevo R, Pollard JR, et al. Targeting ATR in vivo using the novel inhibitor VE-822 results in selective sensitization of pancreatic tumors to radiation. Cell Death Dis. 2012;3:e441. doi: 10.1038/cddis.2012.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Foote KM, Blades K, Cronin A, et al. Discovery of 4-{4-[(3R)- 3-Methylmorpholin-4-yl]-6-[1- (methylsulfonyl)cyclopropyl]pyrimidin-2-yl}-1H-indole (AZ20): A Potent and Selective Inhibitor of ATR Protein Kinase with Monotherapy in Vivo Antitumor Activity. J Med Chem. 2013;56:2125–2138. doi: 10.1021/jm301859s. [DOI] [PubMed] [Google Scholar]
- [49].Karnitz LM, Zou L. Molecular pathways: Targeting ATR in cancer therapy. Clin Cancer Res. 2015;21:4780–4785. doi: 10.1158/1078-0432.CCR-15-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Peasland A, Wang L-Z, Rowling E, et al. Identification and evaluation of a potent novel ATR inhibitor, NU6027, in breast and ovarian cancer cell lines. Br J Cancer. 2011;105:372–381. doi: 10.1038/bjc.2011.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Prevo R, Fokas E, Reaper PM, et al. The novel ATR inhibitor VE-821 increases sensitivity of pancreatic cancer cells to radiation and chemotherapy. Cancer Biol Ther. 2012;13:1072–1081. doi: 10.4161/cbt.21093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Huntoon CJ, Flatten KS, Wahner Hendrickson AE, et al. ATR inhibition broadly sensitizes ovarian cancer cells to chemotherapy independent of BRCA status. Cancer Res. 2013;73:3683–3691. doi: 10.1158/0008-5472.CAN-13-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Jossé R, Martin SE, Guha R, et al. ATR inhibitors VE-821 and VX-970 sensitize cancer cells to topoisomerase I inhibitors by disabling DNA replication initiation and fork elongation responses. Cancer Res. 2014;74:6968–6978. doi: 10.1158/0008-5472.CAN-13-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Biskup E, Naym DG, Gniadecki R. Small-molecule inhibitors of Ataxia Telangiectasia and Rad3 related kinase (ATR) sensitize lymphoma cells to UVA radiation. J Dermatol Sci. 2016;3 doi: 10.1016/j.jdermsci.2016.09.010. [DOI] [PubMed] [Google Scholar]
- [55].Sanjiv K, Hagenkort A, Calderón-Montaño JM, et al. Cancer-Specific Synthetic Lethality between ATR and CHK1 Kinase Activities. Cell Rep. 2016;14:298–309. doi: 10.1016/j.celrep.2015.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Mohni KN, Thompson PS, Luzwick JW, et al. A synthetic lethal screen identifies DNA repair pathways that sensitize cancer cells to combined ATR inhibition and cisplatin treatments. PLoS One. 2015;10 doi: 10.1371/journal.pone.0125482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Huang KK, Jang KW, Kim S, et al. Exome sequencing reveals recurrent REV3L mutations in cisplatin-resistant squamous cell carcinoma of head and neck. Sci Rep. 2016;6 doi: 10.1038/srep19552. 19552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Mohni KN, Kavanaugh GM, Cortez D. ATR pathway inhibition is synthetically lethal in cancer cells with ercc1 deficiency. Cancer Res. 2014;74:2835–2845. doi: 10.1158/0008-5472.CAN-13-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Kwok M, Davies N, Agathanggelou A, et al. ATR inhibition induces synthetic lethality and overcomes chemoresistance in TP53 or ATM defective chronic lymphocytic leukemia cells. Blood. 2016;127:582–595. doi: 10.1182/blood-2015-05-644872. [DOI] [PubMed] [Google Scholar]
- [60].Menezes DL, Holt J, Tang Y, et al. A Synthetic Lethal Screen Reveals Enhanced Sensitivity to ATR Inhibitor Treatment in Mantle Cell Lymphoma with ATM Loss-of-function. Mol Cancer Res. 2014 doi: 10.1158/1541-7786.MCR-14-0240. [DOI] [PubMed] [Google Scholar]
- [61].Hocke S, Guo Y, Job A, et al. A synthetic lethal screen identifies ATR-inhibition as a novel therapeutic approach for POLD1-deficient cancers. Oncotarget. 2016;7:7080–7095. doi: 10.18632/oncotarget.6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Williamson CT, Miller R, Pemberton HN, et al. ATR inhibitors as a synthetic lethal therapy for tumours deficient in ARID1A. Nat Commun. 2016;7 doi: 10.1038/ncomms13837. 13837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Kadoch C, Hargreaves DC, Hodges C, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Nieto-Soler M, Morgado-Palacin I, Lafarga V, et al. Efficacy of ATR inhibitors as single agents in Ewing sarcoma. Oncotarget. 2016;7:58759–58767. doi: 10.18632/oncotarget.11643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Jones S, Fleuren E, Frankum J. ATR is a therapeutic target in synovial sarcoma. Cancer Res. doi: 10.1158/0008-5472.CAN-17-2056. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Flynn RL, Cox KE, Jeitany M, et al. Alternative lengthening of telomeres renders cancer cells hypersensitive to ATR inhibitors. Science (80-. ) 2015;347:273–277. doi: 10.1126/science.1257216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Deeg KI, Chung I, Bauer C, et al. Cancer cells with alternative lengthening of telomeres do not display a general hypersensitivity to ATR inhibition. Front Oncol. 2016;6:0–13. doi: 10.3389/fonc.2016.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Brown JS, OCarrigan B, Jackson SP, et al. Targeting DNA Repair in Cancer: Beyond PARP Inhibitors. Cancer Discov. 2016:20–38. doi: 10.1158/2159-8290.CD-16-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Colicchia V, Petroni M, Guarguaglini G, et al. PARP inhibitors enhance replication stress and cause mitotic catastrophe in MYCN-dependent neuroblastoma. Oncogene. 2017:1–10. doi: 10.1038/onc.2017.40. [DOI] [PubMed] [Google Scholar]
- [70].Mouw KW, Goldberg MS, Konstantinopoulos PA, et al. DNA Damage and Repair Biomarkers of Immunotherapy Response. Cancer Discov. 2017:617–632. doi: 10.1158/2159-8290.CD-17-0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Ruiz S, Mayor-Ruiz C, Lafarga V, et al. A Genome-wide CRISPR Screen Identifies CDC25A as a Determinant of Sensitivity to ATR Inhibitors. Mol Cell. 2016;62:307–313. doi: 10.1016/j.molcel.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Jette N, Lees-Miller SP. The DNA-dependent protein kinase: A multifunctional protein kinase with roles in DNA double strand break repair and mitosis. Prog Biophys Mol Biol. 2015;117:194–205. doi: 10.1016/j.pbiomolbio.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].van der Burg M, van Dongen JJM, van Gent DC. DNA-PKcs deficiency in human: long predicted, finally found. Curr Opin Allergy Clin Immunol. 2009;9:503–509. doi: 10.1097/ACI.0b013e3283327e41. [DOI] [PubMed] [Google Scholar]
- [74].Beskow C, Skikuniene J, Holgersson a, et al. Radioresistant cervical cancer shows upregulation of the NHEJ proteins DNA-PKcs, Ku70 and Ku86. Br J Cancer. 2009;101:816–821. doi: 10.1038/sj.bjc.6605201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Bouchaert P, Guerif S, Debiais C, et al. DNA-PKcs expression predicts response to radiotherapy in prostate cancer. Int J Radiat Oncol Biol Phys. 2012;84:1179–1185. doi: 10.1016/j.ijrobp.2012.02.014. [DOI] [PubMed] [Google Scholar]
- [76].Austen B, Powell JE, Alvi A, et al. Mutations in the ATM gene lead to impaired overall and treatment-free survival that is independent of IGVH mutation status in patients with B-CLL. Blood. 2005;106:3175–3182. doi: 10.1182/blood-2004-11-4516. [DOI] [PubMed] [Google Scholar]
- [77].Zhou Z, Patel M, Ng N, et al. Identification of synthetic lethality of PRKDC in MYC-dependent human cancers by pooled shRNA screening. BMC Cancer. 2014;14:944. doi: 10.1186/1471-2407-14-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Riabinska A, Daheim M, Herter-Sprie GS, et al. Therapeutic Targeting of a Robust Non-Oncogene Addiction to PRKDC in ATM-Defective Tumors. Sci Transl Med. 2013;5:189ra78. doi: 10.1126/scitranslmed.3005814. [DOI] [PubMed] [Google Scholar]
- [79].Dietlein F, Thelen L, Jokic M, et al. A functional cancer genomics screen identifies a druggable synthetic lethal interaction between MSH3 and PRKDC. Cancer Discov. 2014;4:592–605. doi: 10.1158/2159-8290.CD-13-0907. [DOI] [PubMed] [Google Scholar]
- [80].Baumann M, Krause M, Overgaard J, et al. Radiation oncology in the era of precision medicine. Nat Rev Cancer. 2016;16:234–249. doi: 10.1038/nrc.2016.18. [DOI] [PubMed] [Google Scholar]
- [81].Patel AG, Sarkaria JN, Kaufmann SH. Nonhomologous end joining drives poly(ADP-ribose) polymerase (PARP) inhibitor lethality in homologous recombination-deficient cells. Proc Natl Acad Sci U S A. 2011;108:3406–3411. doi: 10.1073/pnas.1013715108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Gatei M, Sloper K, Sörensen C, et al. Ataxia-telangiectasia-mutated (ATM) and NBS1-dependent phosphorylation of Chk1 on Ser-317 in response to ionizing radiation. J Biol Chem. 2003;278:14806–14811. doi: 10.1074/jbc.M210862200. [DOI] [PubMed] [Google Scholar]
- [83].Busino L, Donzelli M, Chiesa M, et al. Degradation of Cdc25A by beta-TrCP during S phase and in response to DNA damage. Nature. 2003;426:87–91. doi: 10.1038/nature02082. [DOI] [PubMed] [Google Scholar]
- [84].Sanchez Y, Wong C, Thoma RS, et al. Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through Cdc25. Science. 1997;277:1497–1501. doi: 10.1126/science.277.5331.1497. [DOI] [PubMed] [Google Scholar]
- [85].Calvo E, Chen V, Marshall M, et al. Preclinical analyses and phase I evaluation of LY2603618 administered in combination with pemetrexed and cisplatin in patients with advanced cancer. Invest New Drugs. 2014;32:955–968. doi: 10.1007/s10637-014-0114-5. [DOI] [PubMed] [Google Scholar]
- [86].Karp JE, Thomas BM, Greer JM, et al. Phase I and pharmacologic trial of cytosine arabinoside with the selective checkpoint 1 inhibitor Sch 900776 in refractory acute leukemias. Clin Cancer Res. 2012;18:6723–6731. doi: 10.1158/1078-0432.CCR-12-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Chen Z, Xiao Z, Gu WZ, et al. Selective Chk1 inhibitors differentially sensitize p53-deficient cancer cells to cancer therapeutics. Int J Cancer. 2006;119:2784–2794. doi: 10.1002/ijc.22198. [DOI] [PubMed] [Google Scholar]
- [88].Bridges KA, Chen X, Liu H, et al. MK-8776, a novel chk1 kinase inhibitor, radiosensitizes p53-defective human tumor cells. Oncotarget. 2016;15 doi: 10.18632/oncotarget.12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Zhang Y, Hunter T. Roles of Chk1 in cell biology and cancer therapy. Int J Cancer. 2014;134:1013–1023. doi: 10.1002/ijc.28226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Scagliotti G, Kang J, Smith D, et al. Phase II evaluation of LY2603618, a first-generation CHK1 inhibitor, in combination with pemetrexed in patients with advanced or metastatic non-small cell lung cancer. Invest New Drugs. 2016;34:625–635. doi: 10.1007/s10637-016-0368-1. [•• Phase II clinical trial data for the CHK1 inhibitor LY2603618] [DOI] [PubMed] [Google Scholar]
- [91].Rawlinson R, Massey AJ. γH2AX and Chk1 phosphorylation as predictive pharmacodynamic biomarkers of Chk1 inhibitor-chemotherapy combination treatments. BMC Cancer. 2014;14:483. doi: 10.1186/1471-2407-14-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Bryant C, Rawlinson R, Massey AJ. Chk1 Inhibition as a novel therapeutic strategy for treating triple-negative breast and ovarian cancers. BMC Cancer. 2014;14 doi: 10.1186/1471-2407-14-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Xiao Z, Xue J, Gu W-Z, et al. Cyclin B1 is an efficacy-predicting biomarker for Chk1 inhibitors. Biomarkers. 2008;13:579–596. doi: 10.1080/13547500802063240. [DOI] [PubMed] [Google Scholar]
- [94].Sakurikar N, Thompson R, Montano R, et al. A subset of cancer cell lines is acutely sensitive to the Chk1 inhibitor MK-8776 as monotherapy due to CDK2 activation in S phase. Oncotarget. 2016;7:1380–1394. doi: 10.18632/oncotarget.6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Matthews DJ, Yakes FM, Chen J, et al. Pharmacological abrogation of S-phase checkpoint enhances the anti-tumor activity of gemcitabine in vivo. Cell Cycle. 2007;6:104–110. doi: 10.4161/cc.6.1.3699. [DOI] [PubMed] [Google Scholar]
- [96].Chaudhuri L, Vincelette ND, Koh BD, et al. CHK1 and WEE1 inhibition combine synergistically to enhance therapeutic efficacy in acute myeloid leukemia ex vivo. Haematologica. 2014;99:688–696. doi: 10.3324/haematol.2013.093187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Magnussen GI, Emilsen E, Giller Fleten K, et al. Combined inhibition of the cell cycle related proteins Wee1 and Chk1/2 induces synergistic anti-cancer effect in melanoma. BMC Cancer. 2015;15:462. doi: 10.1186/s12885-015-1474-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Dietlein F, Kalb B, Jokic M, et al. A Synergistic Interaction between Chk1- and MK2 Inhibitors in KRAS-Mutant Cancer. Cell. 2015;162:146–159. doi: 10.1016/j.cell.2015.05.053. [•• select useful drug combinations and the authors define a biomarker for CHK1 and MK2] [DOI] [PubMed] [Google Scholar]
- [99].Ahn JY, Li X, Davis HL, et al. Phosphorylation of threonine 68 promotes oligomerization and autophosphorylation of the Chk2 protein kinase via the forkhead-associated domain. J Biol Chem. 2002;277:19389–19395. doi: 10.1074/jbc.M200822200. [DOI] [PubMed] [Google Scholar]
- [100].Bartek J, Falck J, Lukas J. CHK2 kinase--a busy messenger. Nat Rev Mol Cell Biol. 2001;2:877–886. doi: 10.1038/35103059. [DOI] [PubMed] [Google Scholar]
- [101].Jobson AG, Lountos GT, Lorenzi PL, et al. Cellular inhibition of checkpoint kinase 2 (Chk2) and potentiation of camptothecins and radiation by the novel Chk2 inhibitor PV1019 [7-nitro-1H-indole-2-carboxylic acid {4-[1-(guanidinohydrazone)-ethyl]-phenyl}-amide] J Pharmacol Exp Ther. 2009;331:816–826. doi: 10.1124/jpet.109.154997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Caldwell JJ, Welsh EJ, Matijssen C, et al. Structure-based design of potent and selective 2-(quinazolin-2-yl)phenol inhibitors of checkpoint kinase 2. J Med Chem. 2011;54:580–590. doi: 10.1021/jm101150b. [DOI] [PubMed] [Google Scholar]
- [103].Anderson VE, Walton MI, Eve PD, et al. CCT241533 is a potent and selective inhibitor of CHK2 that potentiates the cytotoxicity of PARP inhibitors. Cancer Res. 2011;71:463–472. doi: 10.1158/0008-5472.CAN-10-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Zabludoff SD, Deng C, Grondine MR, et al. AZD7762, a novel checkpoint kinase inhibitor, drives checkpoint abrogation and potentiates DNA-targeted therapies. Mol Cancer Ther. 2008;7:2955–2966. doi: 10.1158/1535-7163.MCT-08-0492. [DOI] [PubMed] [Google Scholar]
- [105].Morgan MA, Parsels LA, Zhao L, et al. Mechanism of radiosensitization by the Chk1/2 inhibitor AZD7762 involves abrogation of the G2 checkpoint and inhibition of homologous recombinational DNA repair. Cancer Res. 2010;70:4972–4981. doi: 10.1158/0008-5472.CAN-09-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Sausville E, LoRusso P, Carducci M, et al. Phase I dose-escalation study of AZD7762, a checkpoint kinase inhibitor, in combination with gemcitabine in US patients with advanced solid tumors. Cancer Chemother Pharmacol. 2014;73:539–549. doi: 10.1007/s00280-014-2380-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Watanabe N, Arai H, Nishihara Y, et al. M-phase kinases induce phospho-dependent ubiquitination of somatic Wee1 by SCFbeta-TrCP. Proc Natl Acad Sci U S A. 2004;101:4419–4424. doi: 10.1073/pnas.0307700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Aarts M, Sharpe R, Garcia-Murillas I, et al. Forced mitotic entry of S-phase cells as a therapeutic strategy induced by inhibition of WEE1. Cancer Discov. 2012;2:524–539. doi: 10.1158/2159-8290.CD-11-0320. [DOI] [PubMed] [Google Scholar]
- [109].Osman AA, Monroe MM, Ortega Alves MV, et al. Wee-1 kinase inhibition overcomes cisplatin resistance associated with high-risk TP53 mutations in head and neck cancer through mitotic arrest followed by senescence. Mol Cancer Ther. 2015;14:608–619. doi: 10.1158/1535-7163.MCT-14-0735-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Hirai H, Iwasawa Y, Okada M, et al. Small-molecule inhibition of Wee1 kinase by MK-1775 selectively sensitizes p53-deficient tumor cells to DNA-damaging agents. Mol Cancer Ther. 2009;8:2992–3000. doi: 10.1158/1535-7163.MCT-09-0463. [DOI] [PubMed] [Google Scholar]
- [111].Guertin AD, Li J, Liu Y, et al. Preclinical evaluation of the WEE1 inhibitor MK-1775 as single-agent anticancer therapy. Mol Cancer Ther. 2013;12:1442–1452. doi: 10.1158/1535-7163.MCT-13-0025. [DOI] [PubMed] [Google Scholar]
- [112].Weisberg E, Nonami A, Chen Z, et al. Identification of Wee1 as a novel therapeutic target for mutant RAS-driven acute leukemia and other malignancies. Leukemia. 2014;29:27–37. doi: 10.1038/leu.2014.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Lal S, Zarei M, Chand SN, et al. WEE1 inhibition in pancreatic cancer cells is dependent on DNA repair status in a context dependent manner. Sci Rep. 2016;6 doi: 10.1038/srep33323. 33323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Pfister SX, Markkanen E, Jiang Y, et al. Inhibiting WEE1 Selectively Kills Histone H3K36me3-Deficient Cancers by dNTP Starvation. Cancer Cell. 2015;28:557–568. doi: 10.1016/j.ccell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Carrassa L, Chilà R, Lupi M, et al. Combined inhibition of Chk1 and Wee1: In vitro synergistic effect translates to tumor growth inhibition in vivo. Cell Cycle. 2012;11:2507–2517. doi: 10.4161/cc.20899. [DOI] [PubMed] [Google Scholar]
- [116].Chila R, Basana A, Lupi M, et al. Combined inhibition of Chk1 and Wee1 as a new therapeutic strategy for mantle cell lymphoma. Oncotarget. 2015;6:3394–3408. doi: 10.18632/oncotarget.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Wang G, Niu X, Zhang W, et al. Synergistic antitumor interactions between MK-1775 and panobinostat in preclinical models of pancreatic cancer. Cancer Lett. 2015;356:656–668. doi: 10.1016/j.canlet.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Ford JB, Baturin D, Burleson TM, et al. AZD1775 sensitizes T cell acute lymphoblastic leukemia cells to cytarabine by promoting apoptosis over DNA repair. Oncotarget. 2015;2 doi: 10.18632/oncotarget.4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Jhuraney A, Woods NT, Wright G, et al. PAXIP1 potentiates the combination of WEE1 inhibitor AZD1775 and platinum agents in lung cancer. Mol Cancer Ther. 2016:1669–1681. doi: 10.1158/1535-7163.MCT-15-0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Edwards SL, Brough R, Lord CJ, et al. Resistance to therapy caused by intragenic deletion in BRCA2. Nature. 2008;451:1111–1115. doi: 10.1038/nature06548. [DOI] [PubMed] [Google Scholar]
- [121].Sakai W, Swisher EM, Karlan BY, et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. 2008;451:1116–1120. doi: 10.1038/nature06633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Drean A, Williamson CT, Brough R, et al. Modelling therapy resistance in BRCA1/2 mutant cancers. Mol Cancer Ther. 2017 doi: 10.1158/1535-7163.MCT-17-0098. molcanther.0098.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Do K, Wilsker D, Ji J, et al. Phase I study of single-agent AZD1775 (MK-1775), a wee1 kinase inhibitor, in patients with refractory solid tumors. J Clin Oncol. 2015;33:3409–3415. doi: 10.1200/JCO.2014.60.4009. [•• Phase I clinical trial data for the WEE1 inhibitor AZD1775.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Leijen S, van Geel RMJM, Sonke GS, et al. Phase II Study of WEE1 Inhibitor AZD1775 Plus Carboplatin in Patients With TP53-Mutated Ovarian Cancer Refractory or Resistant to First-Line Therapy Within 3 Months. J Clin Oncol. 2016 doi: 10.1200/JCO.2016.67.5942. JCO.2016.67.5942. [•• Phase II clinical trial data for the WEE1 inhibitor AZD1775] [DOI] [PubMed] [Google Scholar]
- [125].Restelli V, Chilà R, Lupi M, et al. Characterization of a mantle cell lymphoma cell line resistant to the Chk1 inhibitor PF-00477736. Oncotarget. 2015;6:37229–37240. doi: 10.18632/oncotarget.5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Li Y, Saini P, Sriraman A, et al. Mdm2 inhibition confers protection of p53-proficient cells from the cytotoxic effects of Wee1 inhibitors. Oncotarget. 2015;6:32339–32352. doi: 10.18632/oncotarget.5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Kwak EL, Sordella R, Bell DW, et al. Irreversible Inhibitors of the EGF Receptor May Circumvent Acquired Resistance to Irreversible inhibitors of the EGF receptor may circumvent acquired resistance to gefitinib. Source Proc Natl Acad Sci United States Am. 2005;102:7665–7670. doi: 10.1073/pnas.0502860102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Nakad R, Schumacher B. DNA Damage Response and Immune Defense: Links and Mechanisms. Front Genet. 2016;7:147. doi: 10.3389/fgene.2016.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Kyi C, Postow MA. Checkpoint blocking antibodies in cancer immunotherapy. FEBS Lett. 2014:368–376. doi: 10.1016/j.febslet.2013.10.015. [DOI] [PubMed] [Google Scholar]
- [130].Parkes EE, Walker SM, Taggart LE, et al. Activation of STING-dependent innate immune signaling by s-phase-specific DNA damage in breast cancer. J Natl Cancer Inst. 2017;109 doi: 10.1093/jnci/djw199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Lennerz V, Fatho M, Gentilini C, et al. The response of autologous T cells to a human melanoma is dominated by mutated neoantigens. Proc Natl Acad Sci U S A. 2005;102:16013–16018. doi: 10.1073/pnas.0500090102. [DOI] [PMC free article] [PubMed] [Google Scholar]