Abstract
Surface-enhanced Raman scattering (SERS) has a broad application prospect in the field of tumor detection owing to its ultrahigh detective sensitivity. However, SERS analysis of serum remain a challenge in terms of repeatability and stability due to the maldistribution of the silver nanoparticles (Ag-NPs)-serum. With the aim to make up for this shortcoming, we report a new method for obtaining stable serum Raman signals utilizing the ordered arrays of pyramidal silicon (PSi) and Ag-NPs. We prove the practicability of this method by detecting the samples of serum from 50 lung cancer patients and 50 normal healthy people. Principal component analysis (PCA) of the serum SERS spectra shows that the spectral data of the two sample groups can form obvious and completely separated clusters. The receiver operating characteristic curve provides the sensitivity (100%) and specificity (90%) from the PCA-LDA method. This research indicates that a stable and label-free analysis technique of serum SERS based on Ag-NPs/PSi and PCA-LDA is promising for noninvasive lung cancer diagnoses.
OCIS codes: (240.6695) Surface-enhanced Raman scattering, (170.1470) Blood or tissue constituent monitoring, (170.4580) Optical diagnostics for medicine
1. Introduction
Despite the rapid development of medical science, the detection of lung cancer is still quite challenging. The actual detection methods of lung cancer, such as computed tomography (CT) and positron emission tomography (PET), have considerable limitations, which affect their diagnostic effectiveness. For instance, CT and PET scans expose people to a relatively high dose of radiation in comparison with other types of diagnostic tests and are likely to cause new cancers in some patients [1,2]. As an ultrasensitive and specific detection technique for biological and chemical sensing, surface-enhanced Raman scattering (SERS) is burgeoning in molecular detection [3–5]. Compared with conventional detection methods based on X-ray mammography, fluorescence, radio-immunoassay, and enzyme-linked immunosorbent assay (ELISA), SERS analysis has many advantages in disease diagnosis [6,7]. In recent years, SERS technology, providing rich fingerprints information of biomolecular structures and conformations, has received widespread attention owing to its prospect in biomedical applications [8–14]. In particular, SERS is widely used in the analysis of complex biomolecules due to its non-destructive, ultra sensitive and fast detection ability for samples, ranging from tissue [9], cell [10], and saliva [11] to DNA [12] and RNA [8].
Generally, the enhancement of SERS is mainly achieved by nano-scale metals (gold, silver, etc.) [15–17]. The experimental results show that silver nanoparticles (Ag-NPs) had stronger SERS enhanced performance compared to gold nanoparticles (Au-NPs) [17]. Furthermore, SERS substrates based on Ag have the virtue of good stability and high sensitivity [18,19]. Besides the metal species, the enhancement of SERS signal is also affected by the microstructure of the SERS substrate [20]. In SERS detections, surface morphologies of the metal would play key roles in the effect and the sensitivity [21,22].
Blood analysis is minimally invasive and relatively simple, and blood is the preferred material for non-invasive diagnosis of cancer patients [23]. In contrast with normal people, the biomolecular composition and content in the blood of cancer patients will produce many subtle changes, such as nucleic acid [24], ribonucleic acid [25], proteins [26], and lipids [13], and their changes will be better reflected by Raman scattering signals. Currently, the main objectives for blood SERS analysis include blood cells [27], blood plasma [13], and serum [28]. Compared with SERS immunoassay approach based on the principle of immunoaffinity, label-free SERS may be cheaper and easier to operate [29]. In addition, common anticoagulant agents, such as the ethylene diamine tetraacetic acid (EDTA) and citrate, also produce weak interference to SERS spectra. Consequently, SERS analysis based on serum samples is of great value in the diagnosis of cancer patients. In recent years, many researchers have applied the SERS to perform label-free and non-invasive detection of serum including nasopharyngeal [13], colorectal [24], prostate [30], liver [31], and so forth. Most of these studies used a common method that the serum was mixed with the noble-metal nanoparticles (NMNPs). However, this method accompanies problem of relatively poor stability due to the random distribution of NMNPs.
In this paper, we proposed a novel strategy for serum SERS analysis in lung cancer diagnosis by combining Ag-NPs with pyramidal silicon (PSi) as a substrate. Ag-NPs/PSi substrate can provide larger surface area and capture more biomolecular information [32]. The regular array of the NMNPs may help to improve the repeatability and uniformity of measurements. The measured serum SERS spectra acquired from patients with lung cancer and normal human volunteers were analyzed by principal component-linear discriminate analysis (PCA-LDA). The accuracy of PCA-LDA diagnosis algorithm was evaluated and confirmed using the receiver operating characteristic (ROC) curve. The new method demonstrated that serum SERS based on Ag-NPs/PSi-substrates has an extensive prospect in the detection and analysis of tumor.
2. Experimental section
2.1. Subjects and protocol
In this prospective study, the experimental serums were obtained from 50 lung cancer patients and 50 healthy volunteers in Shandong Cancer Hospital & Institute, from January to April this year. According to histopathological diagnosis, we divided the experimental samples into cancer and healthy groups. The samples of cancer group were taken from lung cancer patients of stage III/IV, while the samples of healthy control group were obtained from healthy subjects. The age ranges for the lung cancer patients and healthy volunteers were 39–90 and 34–75 years, respectively. Table 1 shows the simple information about these people. All patients have confirmed by clinical and histopathological diagnoses. The research was authorized by an institutional review board of the Shandong Cancer Hospital & Institute. According to the code of ethics published by council for international organizations of medical sciences (CIOMS), all blood sample providers have signed written informed consents [33].
Table 1. Clinical information of lung cancer patients and normal human volunteers.
| Lung cancer patients (n = 50) | Normal human volunteers (n = 50) | |
|---|---|---|
| Age | ||
| Mean | 61 | 58 |
| SD | 11 | 12 |
| Gender | ||
| Male | 30 | 28 |
| Female | 20 | 22 |
| TNM stage | ||
| III | 23 | N/A |
| IV | 27 | N/A |
2.2 Preparation of pyramidal silicon, silver colloids, and human blood serum samples
PSi was prepared based on the modified method from Zhang [32], and was manufactured using the anisotropic etching of single crystal silicon with the assist of sodium hydroxide (NaOH). According to the method described by Zhang et al. [34], Ag colloids were synthesized by employing the polyvinylpyrrolidone (PVP, Mw = 55000) to decompose silver nitrate (AgNO3). Briefly, a total of 0.5 g PVP and 0.1 g AgNO3 were added to 40 mL ethylene glycol solution (C2H6O2, 99.0%) with continuously stirring. Then, acetone (CH3COCH3, 99.5%) was added to the mixture, and the resulting mixture was rinsed using demonized water (18.25 MΩ·cm−1) until the mixture becomes stable milky gray silver colloid.
All subjects had to fast for 12 hours before blood samples were collected. Between 7 A.M. and 8 A.M., three milliliters of peripheral blood samples were collected on each subject. The cells in the blood samples were removed using a centrifugal machine (3000 rpm, 10 min), and the supernatant (serum) was obtained. Before measuring SERS, 20 µl of Ag sol was uniformly mixed with 20 µl of serum using a pipette tip, and then the mixture was incubated in the refrigerator (4, 1.5 h). Finally, the mixture was coated on the surface of PSi substrates using a spinner with 2000 rpm for 60 seconds rotating speed for SERS measurement.
2.3. SERS measurements
The SERS spectra of 600-1800 cm−1 were acquired by a Raman system (Horiba JY HR evolution, France) with a 532 nm laser excitation. Studies have shown that excitation light sources of 532 nm wavelength can be used for Raman detection of biological samples (such as tissue cells and plasma) [35,36]. The laser excitation with a power of approximately 0.48 mW was focused on the sample surface through a × 50 objective lens (NA = 0.5). The spectral resolution was 2 cm−1. The acquisition time of 8 s and accumulations of 2 times were set for all experiments. To obtain the mean SERS signal of each sample, 12 spectral data were collected from each serum sample.
3. Results and discussion
3.1 Validation of measuring SERS spectra using Ag-NPs/PSi
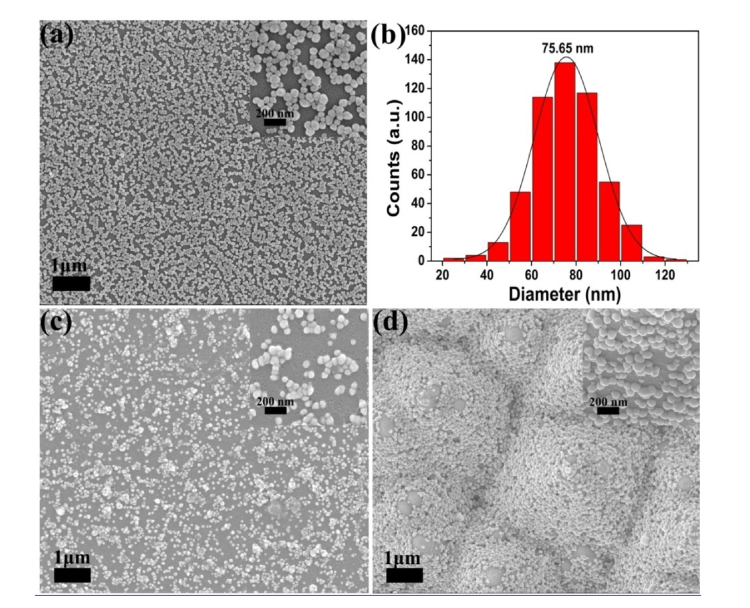
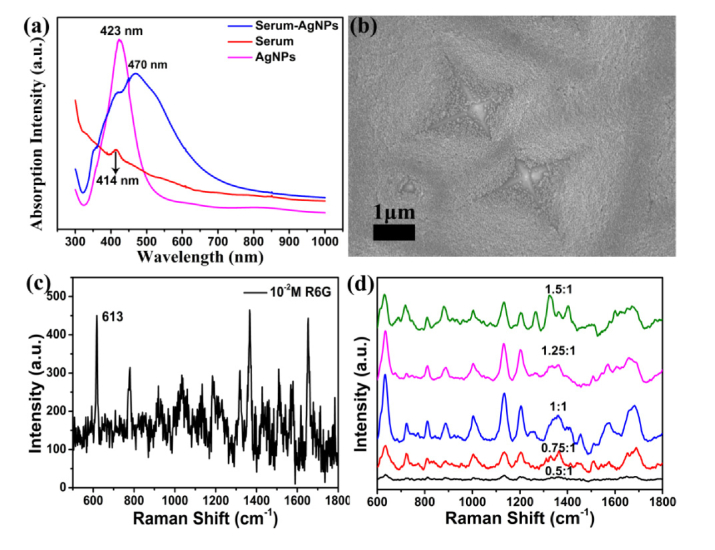
The scanning electron microscopy (SEM) image of Ag-NPs on a silicon wafer was shown in Fig. 1(a). The histograms (Fig. 1(b)) exhibit the diameter distributions calculated by the Nano Measurer software according to the statistics of 500 Ag-NPs. The average diameter of Ag-NPs is 75.65 nm, and the standard deviation is 10.3 nm. SEM images were further carried out to investigate the surface morphology of the mixture of serum-Ag-NPs on the silicon wafer (see Fig. 1(c)) and PSi substrate (see Fig. 1(d)), separately. The illustrations are magnified SEM images of Ag-NPs on the silicon wafer, the mixture of serum-Ag-NPs on the silicon wafer, and the mixture of serum-Ag-NPs on the PSi substrate, respectively. As shown in Fig. 1(d), the mixtures of serum-Ag-NPs on the PSi substrate have better regular array, and the adsorption of silver particles to biochemical substances in serum leads to the aggregation of silver particles. Fig. 8(a) in the Appendix shows the UV-Vis absorption spectra of the Ag-NPs, serum, and serum-Ag NPs mixture. The maximum absorption of Ag-NPs is 423 nm, shifting to 470 nm for the serum-Ag-NPs mixture. The band shift demonstrates that aggregation of serum-Ag-NPs occurs to some extent.
Fig. 1.
(a) Scanning electron microscopy (SEM) image of Ag-NPs on the silicon wafer. (b) The size distribution of 500 Ag-NPs counted from (a). SEM was carried out to investigate the surface morphology of the mixture of serum-Ag-NPs on the silicon wafer (c) and PSi substrate (d), respectively.
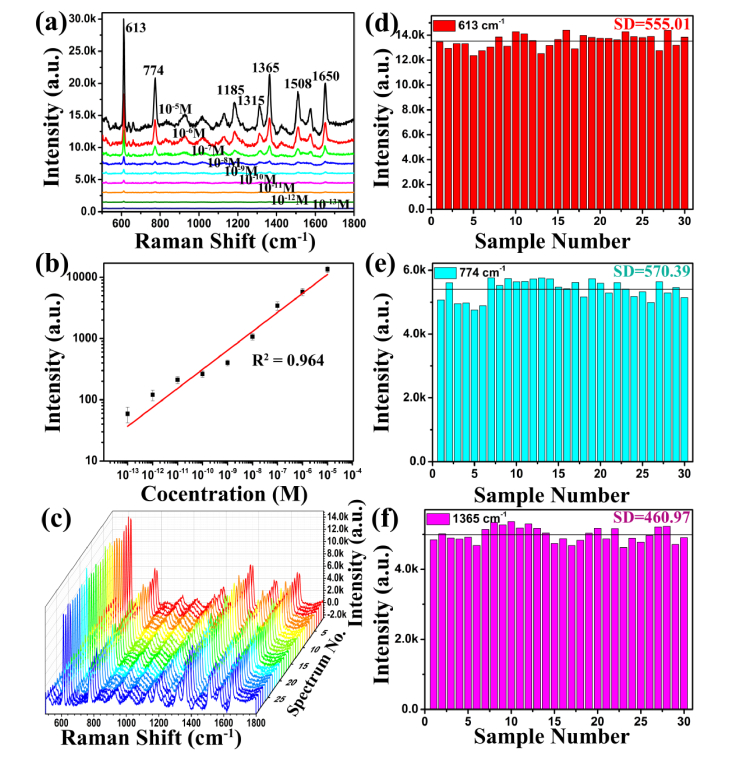
To further insight into the sensibility of Ag-NPs/PSi substrate, the Ag-NPs/PSi substrate was obtained by depositing Ag-NPs on the pyramidal silicon with a dip-coating method (see Fig. 8(b)). 2 μL rhodamine 6G (R6G) water solution at concentrations ranging from 10−5 M to 10−13 M were dropped on the Ag-NPs/PSi substrates, and Raman detection was performed after they were naturally dried, and the results are given in Fig. 2(a). Meanwhile, the value of R2 is 0.964 on the Ag-NPs/PSi substrates, as shown in Fig. 2(b). Fig. 8(c) shows the ordinary Raman spectrum of R6G at a concentration of 10−2 M under the same test conditions. It can be found that the R6G spectral signal measured on the Ag-NPs/PSi substrate is greatly improved. To further investigate the homogeneity and stability of SERS substrate, R6G aqueous solution with a concentration of 10−5 M was used to investigate the performance of the SERS substrate, and the SERS spectra of R6G water solutions (10−5 M) from 30 positions in a same Ag-NPs/PSi substrate are given in Fig. 2(c). The 30 Raman spectra obtained under the same experimental conditions have no obvious difference, which means that SERS substrate has higher homogeneity. Randomly select 613 cm−1 (red), 774 cm−1 (pale blue), and 1365 cm−1 (lavender) peaks to further study the stability of the Raman spectra collected with Ag-NPs/PSi substrates, and the results are shown in Fig. 2(d)-2(f), respectively. According to the above results, it is found that Ag-NPs/PSi substrates have excellent performance including high sensitivity and stability.
Fig. 2.
(a) SERS spectra of R6G with the different concentrations from 10−5 M to 10−13 M detected on the Ag-NPs/PSi substrate. (b) The intensity of peaks at 613 cm−1 for R6G as a function of the molecular concentration. (c) SERS spectra of 30 random spots from R6G molecules (10−5 M) on Ag-NPs/PSi substrates. (d)-(f) Box plots of average and median concentrations of the peaks at 613, 774, and 1365 cm−1 from R6G (10−5 M) molecules based on Ag-NPs/PSi substrates.
3.2 SERS spectra of clinical serum samples
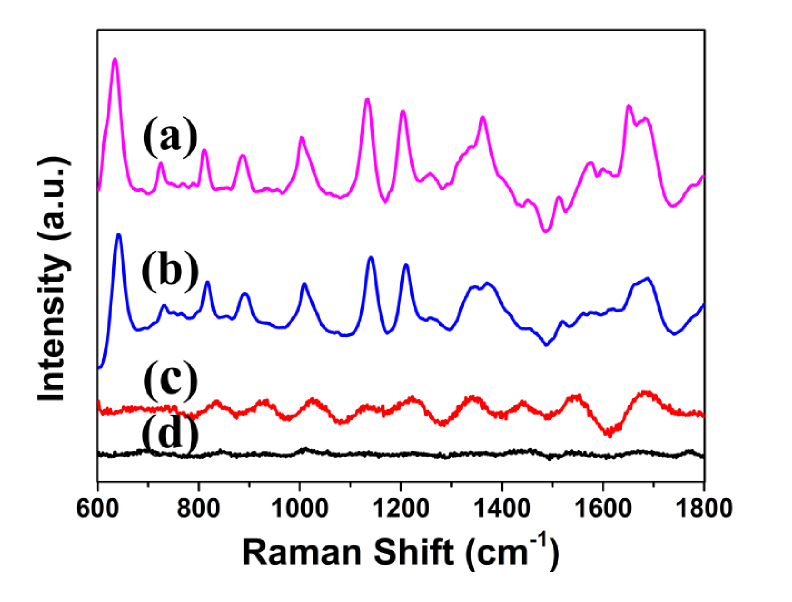
Figure 3 shows four spectra of serum samples from the same patient. Figure 3(a) and 3(b) shows the SERS spectra of serum with silver colloids on the PSi substrate and silicon substrate, respectively. It can be clearly seen that the Raman scattering signal is significantly enhanced. Moreover, the SERS signal is more pronounced in Fig. 3(a) than in Fig. 3(b) due to the change in the structure of substrate. The structure of PSi substrate can increase the specific surface area and capture more biomolecules [32]. The conventional Raman spectrum of serum samples without Ag sol is shown in Fig. 3(c). As a result of the existence of strong fluorescence background, normal Raman scattering signal is covered, and only a few of Raman peaks are observed in Fig. 3(c). Comparing Fig. 3(b) with Fig. 3(c), it can be clearly observed that the Raman scattering signal is considerably enhanced, which means that there is strong interaction between serum and Ag sol, so that the biomolecules in serum are adsorbed on the surface of Ag particles. The Raman spectrum of Ag sol is shown in Fig. 3(d). It is worth noting that there is no distinct signal in the range of fingerprint spectrum, showing that the Ag sol cannot generate disturbance to the SERS spectra in our measurement. In addition, we also studied the spectral signals of Ag sol and serum mixed in different proportions (Fig. 8(d)).
Fig. 3.
The SERS spectrum of serum with added silver colloid on the PSi substrate (a) and silicon substrate (b). The Raman spectrum of serum sample without Ag sol (c). The Raman spectrum of Ag sol without serum sample (d).
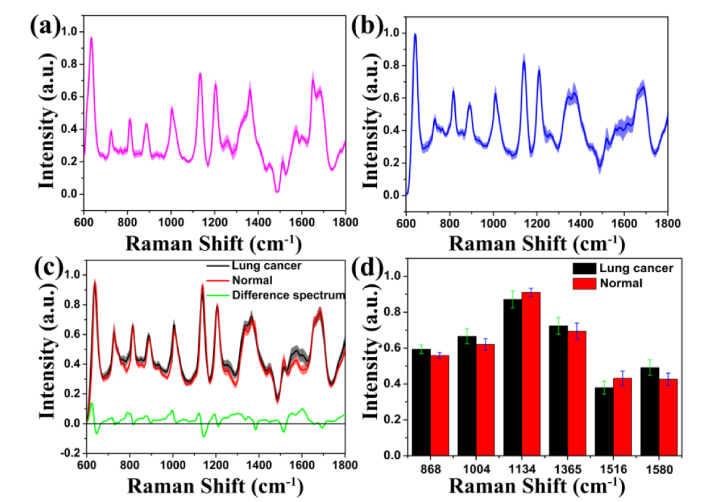
We removed the fluorescence background from the original Raman spectrum data by applying the method of Zhao et al. [37]. To reduce the influence of the spectral intensity between different samples on the shape of the spectra, normalization (integral area under the curve) was performed on all SERS spectra data. Fig. 4(a) and 4(b) show the normalized mean (mean ± SD) SERS spectra of the same serum-NPs sample on the PSi substrate and silicon substrate, respectively. The shaded part represents the standard deviations of the mean value, which can reflect the fluctuation range of the spectral data. It can be found that the standard deviation of the SERS spectra obtained from the same sample on the PSi substrate is smaller than that on the silicon substrate. Figure 4(c) represents the normalized average (mean ± SD) SERS spectra of serum from the two sample groups. The distinct SERS bands at 638, 725, 813, 850, 886, 932, 1004, 1134, 1207, 1365, 1447, 1516, 1573, 1580, and 1683 cm−1 can be steadily and clearly detected in healthy and cancerous serum samples. By contrast, serum SERS spectra of lung cancer patients display lower intensities at 638, 813, 1134, 1207, 1516, and 1683 cm−1, while are higher at 725, 886, 932, 1004, 1365, 1447, 1573, and 1580 cm−1. The difference in normalized spectral intensity (bottom in Fig. 4(c)) can more clearly discriminate the cancerous and healthy serum. The Raman scattering signals with significant differences between lung cancer serum and healthy serum are located at around 886, 1004, 1134, 1363, 1516 and 1580 cm−1 (see Fig. 4(d)).
Fig. 4.
Normalized mean (mean ± SD) SERS spectra of a same serum sample on the PSi substrate (a) and silicon substrate(b), respectively. Normalized mean (mean ± SD) SERS spectra of lung cancer and normal serum samples on the PSi substrate (c). Difference spectrum calculated from the mean SERS spectra of normal and cancer serum samples (bottom in (c)). The shaded part represents the standard deviations of the mean value. Plot shows the intensity value of the selected peaks with the most distinguishable differences between normal human serum and lung cancer serum (d).
In routine clinical diagnosis, blood samples can reflect some important subtle changes in tumor metabolism, such as amino acid metabolism and biomarker production [38,39]. Compared with normal lung serum, the contents and conformations of biomolecular components in cancerous serum (e.g. DNA/RNA, proteins, and lipids) may generate subtle changes. To better analyze SERS characteristics of biomolecular in lung serum, we tentatively assign the SERS spectra data obtained from the experiment, as listed in Table 2 [13,24,26,36,40–42]. The intensities of SERS bands of cancerous and healthy serum at 638 and 725 cm−1 occur obvious difference, this peaks showed diagnostic significance in cancer diagnosis [43]. Compared with normal serums, the SERS signals of tyrosine (638 cm−1), l-Serine (813 cm−1), and l-Tryptophan (1207 cm−1) in serums of lung cancer patients show a lower intensity (see Fig. 4(d)). On the whole, the content of amino acid is decreased in the serum biological molecules for lung cancer patients. The Raman peak at 725 cm−1 belongs to the C-H bending of the DNA/RNA. This study shows that the DNA/RNA bases content in lung cancer serum is higher than that in normal serum. This result is consistent with the study of blood plasma SERS in patients with nasopharyngeal carcinoma or colorectal cancer [13,24]. Furthermore, the intensity of the SERS peak at 1573 cm−1 related to the DNA/RNA band is also relatively increased. In contrast with studies of hepatocellular cancer [44], the intensity of band at 886 cm−1 in the serum SERS of lung was increased, this may be due to the differences in protein metabolism between different cancers. The peak at 1004 cm−1 belongs to the C–C stretching mode of phenylalanine. The strong band at 1134 cm−1 corresponds to the C–N stretching mode of mannose. The SERS band at 1445 cm−1 belongs to the CH2 bending mode of collagen and phospholipids, which was lower in normal serum than in cancerous serum. This peak has diagnostic significance in medical research [45]. The Raman line at 1516 cm−1 corresponds to the C = C stretch vibrations of the conjugated backbone [46], and can also been found in serum SERS of dengue virus [36]. The SERS band at 1580 cm−1 corresponds to the C = C bending vibrations of the phenylalanine [47]. The SERS signal of cancer patient at 1580 cm−1 is considerably increased, which has been reported in the Raman spectroscopy of malignant lung tissues [48]. Compared with healthy volunteers, the intensity of most Raman peaks in the serum of lung cancer patients was increased, which may be due to the abnormal proliferation of cancer cells. In summary, serum SERS based on Ag-NPs/PSi substrate is a powerful analytical tool for the fast prediction of lung diseases and it has capable of revealing biomolecular changes of the serum in lung cancer patients.
Table 2. SERS peak positions and tentative vibrational mode assignment.
| Peak position (cm−1) | Vibrational mode | Major assignments |
|---|---|---|
| 638 | ν(C–S) | L-Tyrosine, lactose |
| 725 | δ(C–H) | Adenine, coenzyme A, DNA/RNA |
| 813 | ν(C–C–O) | L-Serine, glutathione |
| 850 | Tyrosine | |
| 886 | δ(C–O–H) or ring bending | Tryptophan, glutathione, tryptophan, D-(C)-galactosamine |
| 932 | C-C stretching | Proteins |
| 1004 | νs(C–C) | Phenylalanine |
| 1134 | ν(C–N) | D-Mannose |
| 1207 | Ring vibration | L -Tryptophan, phenylalanine |
| 1257 | Amide III | |
| 1365 | Tryptophan | |
| 1445 | δ(CH2) | Collagen, phospholipids |
| 1516 | ν(C = C) | Carotenoids |
| 1573 | DNA/RNA bases | |
| 1580 | δ(C = C) | Phenylalanine, acetoacetate, riboflavin |
| 1683 | Amide I |
Abbreviations: ν, stretching vibration; δ, bending vibration; νs, symmetric stretch.
3.3 Statistical analysis
The spectral intensity of normal and cancer serums varied significantly and partially overlapped between different subjects. To further verify the ability of the novel SERS strategy to distinguish between normal and lung cancer patients, the PCA-LDA-based diagnostic algorithm was implemented on the SERS spectra of lung serums. The PCA-LDA method has been applied to the detection and assortment of cancer serum and plasma [13,24,26]. In this paper, the normalized SERS spectra data were analyzed using PCA and LDA methods with SPSS software package (SPSS Inc., Chicago).
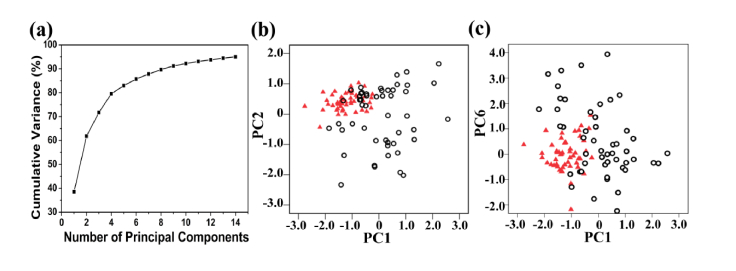
PCA can compress complex SERS spectral data into simple principal components (PCs), which cannot only better explain the difference between spectral data, but also reduce the number of variables used in LDA. A broken line diagram of the cumulative variance of the varying number of PCs is shown in Fig. 5(a). It is interesting that the accumulation of variance is mainly concentrated in the first 4 PCs, while the remaining PCs contribute less to the total variance. Three PCs (PC1, PC2, and PC6) accounting for 64.6% of the total variance were found to be the most diagnostically significant (P < 0.05) for healthy and cancerous serum by an independent-sample T test on all PC scores. A scatter plot of PC1 and PC2 for the healthy and cancerous serum samples is shown in Fig. 5(b). Figure 5(c) also presents a similar scatter plot of the PC1 and PC6. Scatter diagrams show that cancer patients and healthy subjects have separate data distributions except for a small amount of data overlap, suggesting that PCA can distinguish between normal and cancer serum SERS spectra. In addition, normal serum samples show a more concentrated regional distribution, while cancer serum samples are more widely distributed. Blood contains a variety of biological components (proteins, lipids, sugars, etc.), which can transport nutrients, metabolic waste and defense against harmful substances. The metabolism of the healthy human body is relatively stable, so that the biological components in the serum are in a relatively stable state for a long time. However, there are significant differences in serum components associated with cancer among patients due to the heterogeneity of cancer progression. Thus, cancer serum samples show more extensive regional distribution on the PCA plot compared to healthy serum samples.
Fig. 5.
(a) Cumulative variance as a function of the number of PCs for PCA. (b) The plot of the first two principal component (PC1, PC2) for the normal group (red triangles) vs. the lung cancer group (black circles). (c) A plot of the two principal component (PC1, PC6) for the normal group (red triangles) vs. the lung cancer group (black circles).
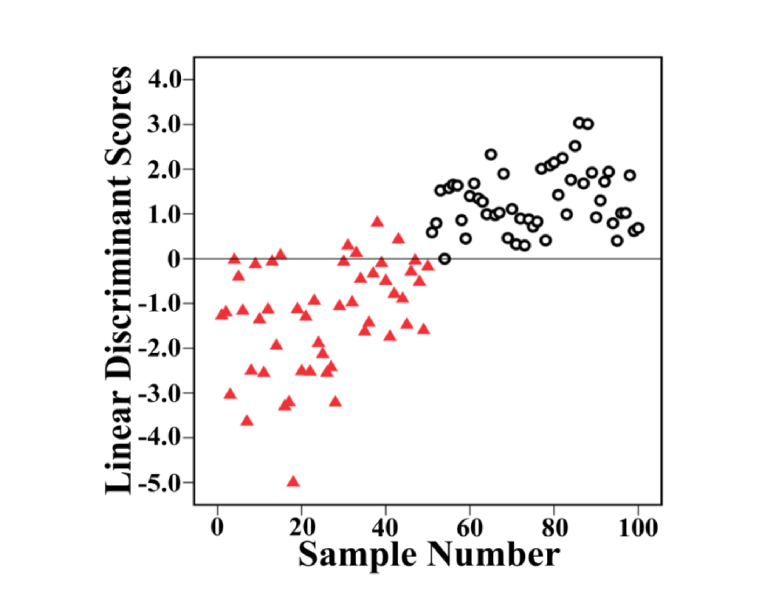
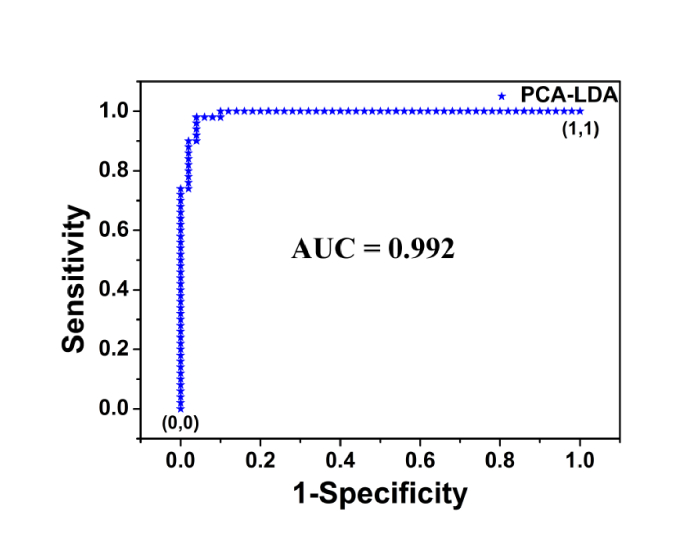
The first 14 PCs, explaining the 95% of the spectral variance, were used to build LDA diagnostic classifier. Figure 6 shows the posterior probabilities of cancerous and healthy volunteer samples in the LDA model. Using a discrimination threshold of 0, 50 cancer and 50 normal samples were discriminated correctly based on the PCA-LDA diagnostic algorithm. To further confirm the performance of diagnostic models developed by multivariate statistical methods, receiver operating characteristic (ROC) curve of PCA-LDA diagnosis algorithms was calculated and shown in Fig. 7. There is a clear dividing line between serum SERS spectra of normal and lung cancer samples, and only 5 normal samples are misjudged as cancer samples. The integration area under the ROC (AUC) curve is a quantitative indicator to represent classifier performances, and is positively correlated with the diagnostic result [49]. According to the Youden’s index, 100% of the diagnostic sensitivity and 90% of the specificity are achieved in the ROC curve. The AUC is 0.992 for PCA-LDA-based diagnostic algorithms, which indicates the efficient performance of the new diagnostic method. The above results show that the combination of the PCA-LDA algorithm and serum SERS data in fingerprint region has high diagnostic performance. In summary, this new method has great potential for steady, label-free, and noninvasive cancer detection and screening.
Fig. 6.
Scatter plots of the linear discriminant scores belonging to the normal (red triangles) and lung cancer samples (black circles), calculated from the data sets of serum SERS.
Fig. 7.
The ROC curve of the discrimination result for the PCA-LDA-based SERS spectral classification. The AUC for PCA-LDA is 0.992.
4. Conclusions
In the current work, we not only measured the serum SERS spectra of normal people and lung cancer patients applying a novel strategy, but also verified the results with PCA-LDA multivariate analysis. The results show that the substrate has high stability and sensitivity in the study of cancer serum SERS, and there are significant differences in SERS spectra between normal and cancer lung serum, and high diagnostic sensitivity (100%) and specificity (90%) can be achieved using the PCA-LDA diagnostic algorithm. The change of Raman signal of biomolecules in serum of cancer patients may be caused by malignant proliferation of cancer cells. This preliminary study implies that the serum SERS based on Ag-NPs/PSi substrate combined with PCA-LDA algorithm may have giant potential in the preoperative diagnosis and screening of lung tumors. Our next step will be to conduct more detailed investigations to completely evaluate the efficacy of this new cancer screening technique.
Appendix
Fig. 8.
(a) The UV/visible absorption spectra of the Ag-NPs, serum, and serum-Ag-NPs mixture. (b) SEM image of the surface morphology of Ag-NPs on the PSi substrate. (c) Raman spectrum of R6G (10−2 M) on the silicon wafer. (d) Spectral images of Ag sol mixed with serum in different ratios.
Funding
National Natural Science Foundation of China (11774208, 11474187, 11747072, 11674199, 11604040); Shandong Province Natural Science Foundation (ZR2018MA040, ZR2017BA004, ZR2016AM19); A Project of Shandong Province Higher Educational Science and Technology Program (J18KZ011); China Postdoctoral Science Foundation (2016M602716); Undergraduate research fund project of Shandong Normal University (2017BKSKY43).
Disclosures
The authors declare that there are no conflicts of interest related to this article.
References and links
- 1.Bird R. E., Wallace T. W., Yankaskas B. C., “Analysis of cancers missed at screening mammography,” Radiology 184(3), 613–617 (1992). 10.1148/radiology.184.3.1509041 [DOI] [PubMed] [Google Scholar]
- 2.Berrington de Gonzalez A., Berg C. D., Visvanathan K., Robson M., “Estimated risk of radiation-induced breast cancer from mammographic screening for young BRCA mutation carriers,” J. Natl. Cancer Inst. 101(3), 205–209 (2009). 10.1093/jnci/djn440 [DOI] [PubMed] [Google Scholar]
- 3.Barhoumi A., Halas N. J., “Label-free detection of DNA hybridization using surface enhanced Raman spectroscopy,” J. Am. Chem. Soc. 132(37), 12792–12793 (2010). 10.1021/ja105678z [DOI] [PubMed] [Google Scholar]
- 4.Eom G., Kim H., Hwang A., Son H. Y., Choi Y., Moon J., Kim D., Lee M., Lim E. K., Jeong J., “Nanogap‐Rich Au Nanowire SERS Sensor for Ultrasensitive Telomerase Activity Detection: Application to Gastric and Breast Cancer Tissues Diagnosis,” Adv. Funct. Mater. 27(37), (2017). [Google Scholar]
- 5.Li M., Cushing S. K., Zhang J., Suri S., Evans R., Petros W. P., Gibson L. F., Ma D., Liu Y., Wu N., “Three-dimensional hierarchical plasmonic nano-architecture enhanced surface-enhanced Raman scattering immunosensor for cancer biomarker detection in blood plasma,” ACS Nano 7(6), 4967–4976 (2013). 10.1021/nn4018284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li M., Cushing S. K., Zhang J., Lankford J., Aguilar Z. P., Ma D., Wu N., “Shape-dependent surface-enhanced Raman scattering in gold-Raman probe-silica sandwiched nanoparticles for biocompatible applications,” Nanotechnology 23(11), 115501 (2012). 10.1088/0957-4484/23/11/115501 [DOI] [PubMed] [Google Scholar]
- 7.He S., Liu K.-K., Su S., Yan J., Mao X., Wang D., He Y., Li L.-J., Song S., Fan C., “Graphene-based high-efficiency surface-enhanced Raman scattering-active platform for sensitive and multiplex DNA detection,” Anal. Chem. 84(10), 4622–4627 (2012). 10.1021/ac300577d [DOI] [PubMed] [Google Scholar]
- 8.Ma D., Huang C., Zheng J., Tang J., Li J., Yang J., Yang R., “Quantitative detection of exosomal microRNA extracted from human blood based on surface-enhanced Raman scattering,” Biosens. Bioelectron. 101, 167–173 (2018). 10.1016/j.bios.2017.08.062 [DOI] [PubMed] [Google Scholar]
- 9.Wang Y. W., Doerksen J. D., Kang S., Walsh D., Yang Q., Hong D., Liu J. T., “Multiplexed Molecular Imaging of Fresh Tissue Surfaces Enabled by Convection-Enhanced Topical Staining with SERS-Coded Nanoparticles,” Small 12(40), 5612–5621 (2016). 10.1002/smll.201601829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X.-P., Walkenfort B., König M., König L., Kasimir-Bauer S., Schlücker S., “Fast and reproducible iSERS microscopy of single HER2-positive breast cancer cells using gold nanostars as SERS nanotags,” Faraday Discuss. 205, 377–386 (2017). 10.1039/C7FD00135E [DOI] [PubMed] [Google Scholar]
- 11.Hernández-Arteaga A., Nava J. J. Z., Kolosovas-Machuca E. S., Velázquez-Salazar J. J., Vinogradova E., José-Yacamán M., Navarro-Contreras H. R., “Diagnosis of breast cancer by analysis of sialic acid concentrations in human saliva by surface-enhanced Raman spectroscopy of silver nanoparticles,” Nano Res. 10(11), 3662–3670 (2017). 10.1007/s12274-017-1576-5 [DOI] [Google Scholar]
- 12.Qi J., Zeng J., Zhao F., Lin S. H., Raja B., Strych U., Willson R. C., Shih W.-C., “Label-free, in situ SERS monitoring of individual DNA hybridization in microfluidics,” Nanoscale 6(15), 8521–8526 (2014). 10.1039/C4NR01951B [DOI] [PubMed] [Google Scholar]
- 13.Feng S., Chen R., Lin J., Pan J., Chen G., Li Y., Cheng M., Huang Z., Chen J., Zeng H., “Nasopharyngeal cancer detection based on blood plasma surface-enhanced Raman spectroscopy and multivariate analysis,” Biosens. Bioelectron. 25(11), 2414–2419 (2010). 10.1016/j.bios.2010.03.033 [DOI] [PubMed] [Google Scholar]
- 14.Jiang T., Wang X., Zhou J., Jin H., “The construction of silver aggregate with inbuilt Raman molecule and gold nanowire forest in SERS-based immunoassay for cancer biomarker detection,” Sens. Actuators B Chem. 258, 105–114 (2018). 10.1016/j.snb.2017.11.084 [DOI] [Google Scholar]
- 15.Yang Y., Liu J., Fu Z.-W., Qin D., “Galvanic replacement-free deposition of Au on Ag for core-shell nanocubes with enhanced chemical stability and SERS activity,” J. Am. Chem. Soc. 136(23), 8153–8156 (2014). 10.1021/ja502472x [DOI] [PubMed] [Google Scholar]
- 16.Bai T., Sun J., Che R., Xu L., Yin C., Guo Z., Gu N., “Controllable preparation of core-shell Au-Ag nanoshuttles with improved refractive index sensitivity and SERS activity,” ACS Appl. Mater. Interfaces 6(5), 3331–3340 (2014). 10.1021/am405357v [DOI] [PubMed] [Google Scholar]
- 17.Tang L., Li S., Han F., Liu L., Xu L., Ma W., Kuang H., Li A., Wang L., Xu C., “SERS-active Au@Ag nanorod dimers for ultrasensitive dopamine detection,” Biosens. Bioelectron. 71, 7–12 (2015). 10.1016/j.bios.2015.04.013 [DOI] [PubMed] [Google Scholar]
- 18.Lu W., Luo Y., Chang G., Sun X., “Synthesis of functional SiO2-coated graphene oxide nanosheets decorated with Ag nanoparticles for H2O2 and glucose detection,” Biosens. Bioelectron. 26(12), 4791–4797 (2011). 10.1016/j.bios.2011.06.008 [DOI] [PubMed] [Google Scholar]
- 19.Jiang T., Zhang L., Zhou J., “Silver nanocube-mediated sensitive immunoassay based on surface-enhanced Raman scattering assisted by etched silicon nanowire arrays,” Analyst (Lond.) 139(22), 5893–5900 (2014). 10.1039/C4AN01293C [DOI] [PubMed] [Google Scholar]
- 20.Zhang C., Jiang S. Z., Huo Y. Y., Liu A. H., Xu S. C., Liu X. Y., Sun Z. C., Xu Y. Y., Li Z., Man B. Y., “SERS detection of R6G based on a novel graphene oxide/silver nanoparticles/silicon pyramid arrays structure,” Opt. Express 23(19), 24811–24821 (2015). 10.1364/OE.23.024811 [DOI] [PubMed] [Google Scholar]
- 21.Kneipp K., Kneipp H., Itzkan I., Dasari R. R., Feld M. S., “Ultrasensitive chemical analysis by Raman spectroscopy,” Chem. Rev. 99(10), 2957–2976 (1999). 10.1021/cr980133r [DOI] [PubMed] [Google Scholar]
- 22.Jayanthi V. S. P. K. S. A., Das A. B., Saxena U., “Recent advances in biosensor development for the detection of cancer biomarkers,” Biosens. Bioelectron. 91, 15–23 (2017). 10.1016/j.bios.2016.12.014 [DOI] [PubMed] [Google Scholar]
- 23.Premasiri W. R., Lee J. C., Ziegler L. D., “Surface-enhanced Raman scattering of whole human blood, blood plasma, and red blood cells: cellular processes and bioanalytical sensing,” J. Phys. Chem. B 116(31), 9376–9386 (2012). 10.1021/jp304932g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin D., Feng S., Pan J., Chen Y., Lin J., Chen G., Xie S., Zeng H., Chen R., “Colorectal cancer detection by gold nanoparticle based surface-enhanced Raman spectroscopy of blood serum and statistical analysis,” Opt. Express 19(14), 13565–13577 (2011). 10.1364/OE.19.013565 [DOI] [PubMed] [Google Scholar]
- 25.Chen Y., Chen G., Feng S., Pan J., Zheng X., Su Y., Chen Y., Huang Z., Lin X., Lan F., Chen R., Zeng H., “Label-free serum ribonucleic acid analysis for colorectal cancer detection by surface-enhanced Raman spectroscopy and multivariate analysis,” J. Biomed. Opt. 17(6), 067003 (2012). 10.1117/1.JBO.17.6.067003 [DOI] [PubMed] [Google Scholar]
- 26.Lin J., Chen R., Feng S., Pan J., Li Y., Chen G., Cheng M., Huang Z., Yu Y., Zeng H., “A novel blood plasma analysis technique combining membrane electrophoresis with silver nanoparticle-based SERS spectroscopy for potential applications in noninvasive cancer detection,” Nanomedicine (Lond.) 7(5), 655–663 (2011). 10.1016/j.nano.2011.01.012 [DOI] [PubMed] [Google Scholar]
- 27.Wang X., Qian X., Beitler J. J., Chen Z. G., Khuri F. R., Lewis M. M., Shin H. J. C., Nie S., Shin D. M., “Detection of circulating tumor cells in human peripheral blood using surface-enhanced Raman scattering nanoparticles,” Cancer Res. 71(5), 1526–1532 (2011). 10.1158/0008-5472.CAN-10-3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu R., Zi X., Kang Y., Si M., Wu Y., “Surface‐enhanced Raman scattering study of human serum on PVA: Ag nanofilm prepared by using electrostatic self‐assembly,” J. Raman Spectrosc. 42(2), 137–144 (2011). 10.1002/jrs.2665 [DOI] [Google Scholar]
- 29.Neng J., Harpster M. H., Zhang H., Mecham J. O., Wilson W. C., Johnson P. A., “A versatile SERS-based immunoassay for immunoglobulin detection using antigen-coated gold nanoparticles and malachite green-conjugated protein A/G,” Biosens. Bioelectron. 26(3), 1009–1015 (2010). 10.1016/j.bios.2010.08.015 [DOI] [PubMed] [Google Scholar]
- 30.Li S., Zhang Y., Xu J., Li L., Zeng Q., Lin L., Guo Z., Liu Z., Xiong H., Liu S., “Noninvasive prostate cancer screening based on serum surface-enhanced Raman spectroscopy and support vector machine,” Appl. Phys. Lett. 105(9), 091104 (2014). 10.1063/1.4892667 [DOI] [Google Scholar]
- 31.Xiao R., Zhang X., Rong Z., Xiu B., Yang X., Wang C., Hao W., Zhang Q., Liu Z., Duan C., Zhao K., Guo X., Fan Y., Zhao Y., Johnson H., Huang Y., Feng X., Xu X., Zhang H., Wang S., “Non-invasive detection of hepatocellular carcinoma serum metabolic profile through surface-enhanced Raman spectroscopy,” Nanomedicine (Lond.) 12(8), 2475–2484 (2016). 10.1016/j.nano.2016.07.014 [DOI] [PubMed] [Google Scholar]
- 32.Zhang C., Man B., Jiang S., Yang C., Liu M., Chen C., Xu S., Qiu H., Li Z., “SERS detection of low-concentration adenosine by silver nanoparticles on silicon nanoporous pyramid arrays structure,” Appl. Surf. Sci. 347, 668–672 (2015). 10.1016/j.apsusc.2015.04.170 [DOI] [Google Scholar]
- 33.Sciences C. I. O. M., “International ethical guidelines for biomedical research involving human subjects,” Bull. Med. Ethics 17(182), 17–23 (2002). [PubMed] [Google Scholar]
- 34.Zhang F., Braun G. B., Shi Y., Zhang Y., Sun X., Reich N. O., Zhao D., Stucky G., “Fabrication of Ag@SiO(2)@Y(2)O(3):Er nanostructures for bioimaging: tuning of the upconversion fluorescence with silver nanoparticles,” J. Am. Chem. Soc. 132(9), 2850–2851 (2010). 10.1021/ja909108x [DOI] [PubMed] [Google Scholar]
- 35.Liu W., Wang H., Du J., Jing C., “Raman microspectroscopy of nucleus and cytoplasm for human colon cancer diagnosis,” Biosens. Bioelectron. 97, 70–74 (2017). 10.1016/j.bios.2017.05.045 [DOI] [PubMed] [Google Scholar]
- 36.Saleem M., Bilal M., Anwar S., Rehman A., Ahmed M., “Optical diagnosis of dengue virus infection in human blood serum using Raman spectroscopy,” Laser Phys. Lett. 10(3), 035602 (2013). 10.1088/1612-2011/10/3/035602 [DOI] [Google Scholar]
- 37.Zhao J., Lui H., McLean D. I., Zeng H., “Automated autofluorescence background subtraction algorithm for biomedical Raman spectroscopy,” Appl. Spectrosc. 61(11), 1225–1232 (2007). 10.1366/000370207782597003 [DOI] [PubMed] [Google Scholar]
- 38.Hocker J. R., Peyton M. D., Lerner M. R., Mitchell S. L., Lightfoot S. A., Lander T. J., Bates-Albers L. M., Vu N. T., Hanas R. J., Kupiec T. C., Brackett D. J., Hanas J. S., “Serum discrimination of early-stage lung cancer patients using electrospray-ionization mass spectrometry,” Lung Cancer 74(2), 206–211 (2011). 10.1016/j.lungcan.2011.03.014 [DOI] [PubMed] [Google Scholar]
- 39.Lai H.-S., Lee J.-C., Lee P.-H., Wang S.-T., Chen W.-J., “Plasma free amino acid profile in cancer patients,” Semin. Cancer Biol. 15(4), 267–276 (2005). 10.1016/j.semcancer.2005.04.003 [DOI] [PubMed] [Google Scholar]
- 40.Lin D., Pan J., Huang H., Chen G., Qiu S., Shi H., Chen W., Yu Y., Feng S., Chen R., “Label-free blood plasma test based on surface-enhanced Raman scattering for tumor stages detection in nasopharyngeal cancer,” Sci. Rep. 4(1), 4751 (2015). 10.1038/srep04751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang J., Lin D., Lin J., Yu Y., Huang Z., Chen Y., Lin J., Feng S., Li B., Liu N., Chen R., “Label-free detection of serum proteins using surface-enhanced Raman spectroscopy for colorectal cancer screening,” J. Biomed. Opt. 19(8), 087003 (2014). 10.1117/1.JBO.19.8.087003 [DOI] [PubMed] [Google Scholar]
- 42.Shen X. W., Huang C. Z., Li Y. F., “Localized surface plasmon resonance sensing detection of glucose in the serum samples of diabetes sufferers based on the redox reaction of chlorauric acid,” Talanta 72(4), 1432–1437 (2007). 10.1016/j.talanta.2007.01.066 [DOI] [PubMed] [Google Scholar]
- 43.Han H., Yan X., Dong R., Ban G., Li K., “Analysis of serum from type II diabetes mellitus and diabetic complication using surface-enhanced Raman spectra (SERS),” Appl. Phys. B 94(4), 667–672 (2009). 10.1007/s00340-008-3299-5 [DOI] [Google Scholar]
- 44.Wang J., Feng S., Lin J., Zeng Y., Li L., Huang Z., Li B., Zeng H., Chen R., “Serum albumin and globulin analysis for hepatocellular carcinoma detection avoiding false-negative results from alpha-fetoprotein test negative subjects,” Appl. Phys. Lett. 103(20), 204106 (2013). 10.1063/1.4830047 [DOI] [Google Scholar]
- 45.Liu C.-H., Das B. B., Sha Glassman W. L., Tang G. C., Yoo K. M., Zhu H. R., Akins D. L., Lubicz S. S., Cleary J., Prudente R., et al., “Raman, fluorescence, and time-resolved light scattering as optical diagnostic techniques to separate diseased and normal biomedical media,” J. Photochem. Photobiol. B 16(2), 187–209 (1992). 10.1016/1011-1344(92)80008-J [DOI] [PubMed] [Google Scholar]
- 46.Movasaghi Z., Rehman S., Rehman I. U., “Raman spectroscopy of biological tissues,” Appl. Spectrosc. Rev. 42(5), 493–541 (2007). 10.1080/05704920701551530 [DOI] [Google Scholar]
- 47.Stone N., Stavroulaki P., Kendall C., Birchall M., Barr H., “Raman spectroscopy for early detection of laryngeal malignancy: preliminary results,” Laryngoscope 110(10), 1756–1763 (2000). 10.1097/00005537-200010000-00037 [DOI] [PubMed] [Google Scholar]
- 48.Huang Z., McWilliams A., Lui H., McLean D. I., Lam S., Zeng H., “Near-infrared Raman spectroscopy for optical diagnosis of lung cancer,” Int. J. Cancer 107(6), 1047–1052 (2003). 10.1002/ijc.11500 [DOI] [PubMed] [Google Scholar]
- 49.Obuchowski N. A., Lieber M. L., Wians F. H., Jr., “ROC curves in clinical chemistry: uses, misuses, and possible solutions,” Clin. Chem. 50(7), 1118–1125 (2004). 10.1373/clinchem.2004.031823 [DOI] [PubMed] [Google Scholar]