Abstract
The intestinal mucosal development of piglets (Sus scrofa) during the weaning stage is important to their disease susceptibility and later growth. Quantitative real-time PCR (RT-qPCR) is commonly used to screen for differentially expressed genes and, for accurate results, proper reference housekeeping genes are essential. Here we assessed the mRNA expression of 18 well-known candidate reference genes at different parts of the gastrointestinal tract (GIT) of piglets during the weaning process by RT-qPCR assay. GeNorm analysis revealed that B2M/HMBS/HPRT1 were the three most stable reference genes and GAPDH was the least stable gene in the duodenum, jejunum, ileum, colon, and whole GIT. BestKeeper analysis found that B2M/HMBS/PGK11, HMBS/B2M/HPRT1, B2M/HMBS/HSPCB, B2M/HPRT1/HMBS, and B2M/HMBS/HPRT1 were the most stable genes in the duodenum, jejunum, ileum, colon, and whole GIT, respectively, whereas GAPDH, B-actin, and 18S rRNA were the least stable genes at different parts of the GIT. To confirm the crucial role of appropriate housekeeping genes in obtaining reliable results, we analyzed the expression of ALP using each of the 18 reference genes to normalize the RT-qPCR data. We found that the expression levels of ALP normalized using the most stable reference genes (B2M/HMBS/HPRT1) differed greatly from the expression levels obtained when the data were normalized using the least stable genes (GAPDH, B-actin, and 18S). We concluded that B2M/HMBS/HPRT1 were the optimal reference genes for gene expression analysis by RT-qPCR in the intestinal mucosal development stages of piglets at weaning. Our findings provide a set of porcine housekeeping reference genes for studies of mRNA expression in different parts of the pig intestine.
Introduction
The weaning process is a widespread health concern in the swine industry [1,2] because the rapid dietary shift from sow milk to solid diet contributes to dramatic shifts in the intestinal structure and function and can lead to post-weaning crypt elongation and villous atrophy [3]. The developing intestinal mucosa, which is the first line of defense against a hostile environment within the intestinal lumen [4], is particularly vulnerable to disruptions during the transformation from lactation to weaning and post-weaning that can have long-term impacts on disease susceptibility of mammals [1]. Therefore, understanding the underlying mechanisms associated with intestinal mucosal development is of great importance to later growth [5].
Gene expression analysis, especially by Quantitative real-time PCR (RT-qPCR), has already provided insights into the potential mechanisms involved in the intestinal mucosal development in pig [6,7]. Unlike other methods used in gene expression studies, RT-qPCR data are generally normalized against a set of housekeeping reference genes to minimize the differences among starting materials [8]. Of particular note is the evidence from Huggett et al. (2005), which indicated inappropriate normalization of RT-qPCR data resulted in incorrect conclusions [9]. Thus, the identification of an appropriate set of housekeeping genes as endogenous reference genes is crucial to normalize RT-qPCR data [10,11]. Ideal housekeeping genes are expected to be expressed at a constant level in different tissues at all developmental stages, be unaffected by experimental treatments, and should pass through the same steps involved in gene analysis for qualification [9]. However, several commonly used housekeeping genes, such as B-actin (beta-actin), GAPDH (glyceraldehyde-3-phosphate dehydrogenase), and 18S (18S ribosomal RNA), are context-dependent because their expression levels may vary significantly with the experimental conditions being investigated [12], and no set of housekeeping genes appear to be generally applicable for all circumstances. Consequently, the validity of candidate housekeeping genes should be well assessed to circumvent problems for data normalization [11]. To date, information about constant housekeeping genes in the intestinal mucosa of piglets in early life is largely lacking.
In this study, we analyzed the mRNA expression of 18 commonly used housekeeping genes in the duodenum, jejunum, ileum, and colon of piglets at 0, 7, 14, and 21 days post-weaning (dpw) to identify suitable reference genes for RT-qPCR assays. The 18 reference genes were 5S (5S ribosomal RNA), RPL4 (ribosomal protein L4), CANX (calnexin), ALDOA (aldolase A), PPARGC1A (PPARG coactivator 1 alpha), HSPCB (heat shock 90 kD protein 1, beta), TBP (TATA box binding protein), B2M (beta-2-microglobulin), HMBS (hydroxymethylbilane synthase), HPRT1 (hypoxanthine phosphoribosyltransferase 1), B-actin (beta-actin), 18S (18S ribosomal RNA), YWHAZ (3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta), RPL32 (ribosomal protein L32), PGK1 (phosphoglycerate kinase 1), PPIA (peptidylprolyl isomerase A), TOP2B (topoisomerase II beta), and GAPDH (glyceraldehyde-3-phosphate dehydrogenase).
Materials and methods
Animal experiments
The animal procedures used in this study were according to the guidelines of the China Animal Protection Association, and all of the work was approved by the Shenzhen Animal Care Committee.
Piglets and feeding
A total of 32 conventionally-raised Landrace-Duroc piglets (Sus scrofa) weaned at day 26 at a 300-sow batch-farrowing facility (Shenzhen Premix Inve Nutrition Co., Ltd, Shenzhen, China) were used in this study. The randomly selected piglets had an average initial body weight of 6.38 kg and were healthy with no clinical signs of disease. The piglets were assigned randomly to four pens of eight piglets and fed a diet, formulated as a powder, without any in-feed antibiotics based on the guidelines of the National Research Council [13]. All piglets had ad libitum access to feed and water. The given and remaining feed was weighted daily at 0800 h.
Sample collection and processing
One piglet (close to the average body weight in each pen) was chosen from each pen at each time point (i.e., 0, 7, 14, and 21 dpw) and sacrificed with sodium pentobarbital (150.00 mg/kg body weight) by intravenous injection. Then, the whole intestine encompassing duodenum, jejunum, ileum, and colon were dissected out. After slitting the intestine lengthwise and gently rinsing with ice-cold 1×PBS, the mucosa was scraped separately from the duodenum, jejunum, ileum, and colon with a glass slide and the samples were immediately placed in liquid nitrogen for further analysis.
RNA preparation and RT-qPCR
The RNA preparation and RT-qPCR protocols were as described previously [11]. Briefly, total RNA was extracted from all the samples with Trizol reagent (Invitrogen, Carlsbad, CA, USA). The concentration of the extracted RNA was measured in a NanoDrop ND-1000 spectrophotometer (Agilent Technologies, Palo Alto, CA, USA). The purity of the RNA was measured based on the A260:A230 and A260:A280 ratios, and the quality of RNA was analyzed by 2% agarose gel electrophoresis.
The A260:A230 and A260:A280 ratios ranged from 2.00 to 2.10 and 1.90 to 2.05, respectively, suggesting that all the RNA samples were of high quality. To synthesize the cDNA, 1.00 μg of total RNA from each sample was reverse transcribed using a PrimeScript™ RT Reagent Kit with a gDNA Eraser (Takara Bio Inc., Otsu, Japan), according to the manufacturer’s protocol.
RT-qPCR assays
The RT-qPCR reactions were carried out in a 96-well plate using a Bio-Rad CFX96TM Real-time PCR Detection System (Bio-Rad, California, USA). The RT-qPCR reaction protocol was as described previously [7]. Briefly, the RT-qPCR reactions were performed in a final volume of 15.0 μl, containing 7.5 μl of 2×SYBR Premix EX Taq II, 5.5 μl of DNase (deoxyribonuclease)/RNase (ribonuclease)-free water, 1 μl of cDNA, and 500 nM of each primer. The PCR cycling program was 95°C (5 min), followed by 40 cycles of 95°C (10 s), 60°C (20 s), and 72°C (20 s). A melting curve analysis was conducted by heating the samples from 65°C to 95°C along continuous fluorescent acquisition. All the RT-qPCR reactions were carried out in triplicate for each sample.
Selection of reference genes, a target gene, and primer design
We selected 18 well-known candidate reference genes and one target gene for the RT-qPCR assay. The porcine reference genes were obtained from a literature review and the sequences of the selected genes were obtained from the GenBank database [7,14–16]. The primer pairs were designed using Primer Premier 5 (http://www.premierbiosoft.com/primerdesign/), and details of the primer sequences are provided in Table 1.
Table 1. Primers and relative information of the reference and target genes.
| Gene symbol | Primer sequence (5'-3') | Amplicon length (bp) | Tm(°C) | Source |
|---|---|---|---|---|
| YWHAZ | F: ATGCAACCAACACATCCTATC | 178 | 62 | Erkens et al. (2006) |
| R: GCATTATTAGCGTGCTGTCTT | ||||
| UBC | F: TTGAGGGGAGGTTTCTAA | 82 | 58.4 | M18159 |
| R: GAATGCAACAACTTTATTGA | ||||
| TBP | F: GATGGACGTTCGGTTTAGG | 124 | 55.9 | Erkens et al. (2006) |
| R: AGCAGCACAGTACGAGCAA | ||||
| RPL32 | F: AGCCCAAGATCGTCAAAAAG | 165 | 55 | NM_001001636 |
| R: TGTTGCTCCCATAACCAATG | ||||
| RPL19 | F: GCTTGCCTCCAGTGTCC | 82 | 62 | AF435591 |
| R: GTTGGCGTTGGCGATTT | ||||
| PPIA | F: GGGAGAAAGGATTTGGTTAT | 175 | 62 | NM_214353 |
| R: ATGGACAAGATGCCAGGAC | ||||
| PPARGC1A | F: CCTGCATGAGTGTGTGCTCT | 107 | 59 | Muráni et al. (2007) |
| R: CTCAGAGTCCTGGTTGCACA | ||||
| PGK1 | F: GGGCTAAGCAGATTGTATG | 180 | 52.8 | NM_001099932 |
| R: TGACTTTATCCTCCGTGTT | ||||
| HSPCB | F: GGCAGAAGACAAGGAGAAC | 131 | 55.9 | AF288819 |
| R: CAGACTGGGAGGTATGGTAG | ||||
| CANX | F: CAATGATGGATGGGGTCTGAA | 135 | 60 | Muráni et al. (2007) |
| R: AACACAGGTAATGCCACAGTCAA | ||||
| ALDOA | F: GAACCAACGGCGAGACAA | 142 | 55.6 | AY359812 |
| R: ATGATGGCGAGGGAGGAG | ||||
| 5S | F: GCCCGATCTCGTCTGATCT | 93 | 54 | AF329851 |
| R: AGCCTACAGCACCCGGTATT | ||||
| 18S | F: CCCACGGAATCGAGAAAGAG | 125 | 53 | Wang et al.(2016) |
| R: TTGACGGAAGGGCACCA | ||||
| B2M | F: CAAGATAGTTAAGTGGGATCG | 161 | 60 | Wang et al.(2016) |
| R: TGGTAACATCAATACGATTTC | ||||
| B-actin | F: GGATGCAGAAGGAGATCACG | 134 | 51 | Wang et al.(2016) |
| R: ATCTGCTGGAAGGTGGACAG | ||||
| GAPDH | F: CCTTCCGTGTCCCTACTGC | 195 | 55.9 | AF017079 |
| R: CATCAAAGGTAGAAGAGTGAGTGTC | ||||
| HMBS | F: AGGATGGGCAACTCTACCTG | 83 | 59.5 | Wang et al.(2016) |
| R: GATGGTGGCCTGCATAGTCT | ||||
| HPRT | F: GGACTTGAATCATGTTTGTG | 91 | 59.5 | Wang et al.(2016) |
| R: CAGATGTTTCCAAACTCAAC | ||||
| ALP | F: CTAAAGGGGCAGATGAATGG | 105 | 55.9 | Lackeyram et al.(2010) |
| R: CACCTGTCTGTCCACGTTGT | ||||
Prior to the RT-qPCR assay, we performed agarose-gel-electrophoresis and conventional PCRs (S1 Fig) to verify the amplified products and assess the gene-specific primers. All the RT-qPCR products were sequenced and aligned against the pig genome using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to validate their identity. Primer specificity was further assessed by melting curve analysis. Only a single band of the expected size should be observed for each of the primers and the sequences should exactly match the GenBank sequences. Further, the dissociation step of the melting curve analysis should produce a single unique peak, indicating the primers are suitable for use in RT-qPCR assays.
Data analysis
We separately pooled the RNA samples collected from the four parts of the intestine at the different growth stages, and then pooled together the RNA samples from all four parts of the intestine at the different growth stages to assess the whole gastrointestinal tract (GIT). Subsequently, the threshold cycle (CT) values of the 18 references genes were analyzed by two-way ANOVA using the GLM procedure in the SAS software (SAS Institute Inc., Cary, NC, USA). The model included the main effects of the different parts of the GIT, growth stages, and their interaction.
We also used two classic software programs, GeNorm (Version 3.4) [17] and BestKeeper (Version 1) [18], to evaluate the 18 candidate reference genes. The GeNorm algorithm defines the standard deviation (SD) of the mRNA expression ratio of reference genes as a pairwise variation under the assumption that the ratio of the mRNA expression is identical across all chosen samples and the tested genes are not co-regulated [11]. A gene-stability measure (M value) is then calculated as the average pairwise variation of a specific reference gene compared with other reference genes. The reference gene with the highest M value is considered the least stable gene. Then, all candidate reference genes are ranked by the stepwise elimination of reference genes with the highest M values. GeNorm can also be used to assess the number of reference genes that are required for dependable normalization.
Unlike GeNorm, BestKeeper uses raw data instead of the relative mRNA expression levels [11]. BestKeeper estimates reference gene stability according to the SD values of each gene and generates an index that is analyzed based on the geometric mean of the CT values. The most stable reference gene is the one that has the highest Pearson coefficients of correlation with the BestKeeper index as well as the lowest SD values.
To further assess the efficacy of the selected reference genes for normalization, the relative mRNA expression of a selected target gene (ALP) was calculated during the piglet weaning stages using a 2−ΔΔCT method where ΔΔCT = [(CTALP−CTReference gene)Post weaning-day x−(CTALP−CTReference gene)Post weaning-day 7] [19].
Normalization was performed with the 18 references genes and the relative mRNA expression of ALP at each part of the GIT was analyzed using the one-factor ANOVA program with the different growth stages as the one factor. The significant results obtained by one-way ANOVA were used in a Duncan’s multiple-range test for multiple-comparisons. The data were expressed as mean ± standard error of the mean (SEM), and differences were considered significant at P < 0.05.
Results
Primer validation
The amplification efficiency of all the primer pairs varied from 90% to 103% and the coefficient of determination (R2) ranged between 0.998 and 1.000 (S2 Fig). Single peaks were observed for the products of all the primer pairs according to the melting curve analysis, and the sequences of the amplified DNA fragments (S1 Table) matched the sequences of the reference and target genes in GenBank (Table 2).
Table 2. Sequencing results of genes using BLASTN from NCBI against nucleotide collection.
| Gene Name | Best hit in NCBI | Identity |
|---|---|---|
| YWHAZ | Sus scrofa 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta (YWHAZ), transcript variant X1, mRNA | 100% |
| UBC | Sus scrofa ubiquitin C (UBC), transcript variant X3, mRNA | 98% |
| TBP | PREDICTED: Sus scrofa TATA box binding protein (TBP), mRNA | 95% |
| RPL32 | Sus scrofa ribosomal protein L32 (RPL32), mRNA | 97% |
| RPL19 | Sus scrofa ribosomal protein L19 (RPL19), mRNA | 96% |
| PPIA | PREDICTED: Sus scrofa peptidylprolyl isomerase A (cyclophilin A) (PPIA), transcript variant X2, mRNA | 99% |
| PPARGC1A | Sus scrofa PPARG coactivator 1 alpha (PPARGC1A), mRNA | 97% |
| PGK1 | Phosphoglycerate kinase 1 [Sus scrofa] | 100% |
| HSPCB | Sus scrofa heat shock 90kD protein 1, beta (HSPCB) Mrna | 96% |
| CANX | Sus scrofa calnexin (CANX), mRNA | 98% |
| ALDOA | Fructose-bisphosphate aldolase A [Sus scrofa] | 96% |
| 5S | Human 5S ribosomal RNA gene | 98% |
| 18S | Sus scrofa 18S ribosomal RNA (RN18S), ribosomal RNA | 99% |
| B2M | Sus scrofa beta-2-microglobulin (B2M), mRNA | 99% |
| B-actin | Sus scrofa actin, beta (ACTB), mRNA | 98% |
| GAPDH | Sus scrofa glyceraldehyde-3-phosphate dehydrogenase (GAPDH), mRNA | 97% |
| HMBS | Sus scrofa hydroxymethylbilane synthase (HMBS), mRNA | 96% |
| HPRT1 | Sus scrofa hypoxanthine phosphoribosyltransferase 1 (HPRT1), mRNA | 100% |
| ALP | Sus scrofa alkaline phosphatase, intestinal (ALPI), transcript variant 2, mRNA | 99% |
Expression profiling of the 18 candidate reference genes
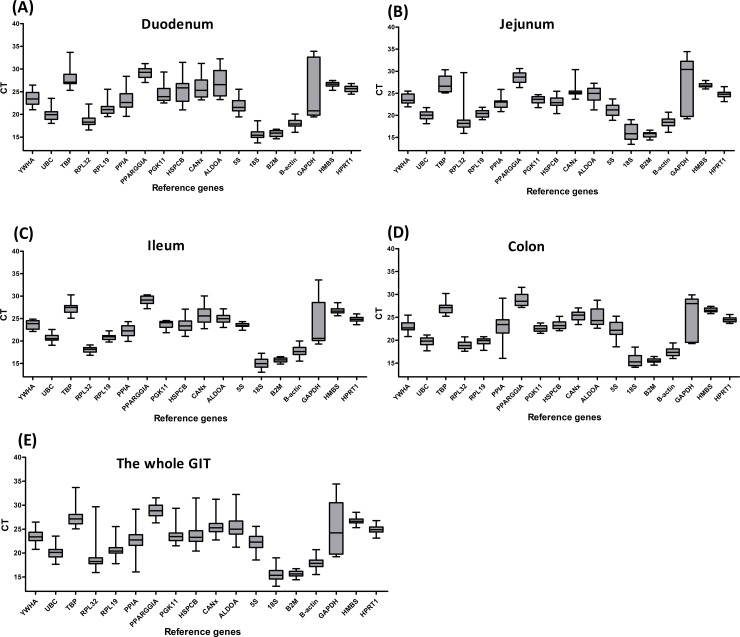
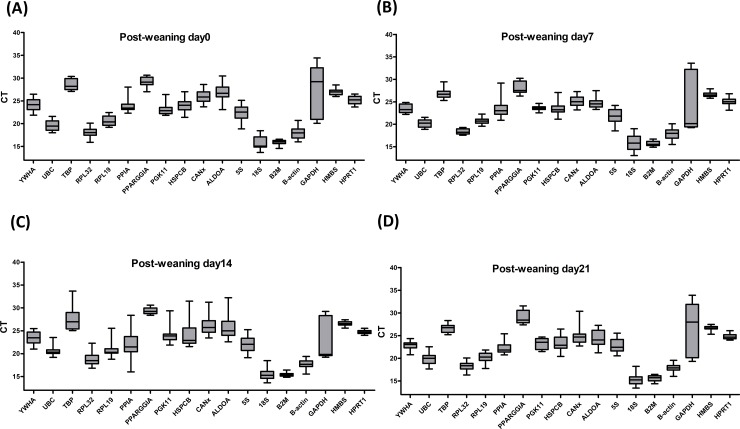
The mRNA expression of all the candidate reference genes was assayed in all the mucosa samples from different parts of the GIT. As was shown in Fig 1, the average CT values ranged from 13.70 to 33.90 in the duodenum, 18.48 to 34.25 in the jejunum, 14.19 to 31.55 in the ileum, and 13.62 to 30.25 in the colon. At different growth stages (Fig 2), the average CT values ranged from 13.70 to 34.44 at 0 dpw, 13.05 to 33.60 at 7 dpw, 13.64 to 34.63 at 14 dpw, and 13.44 to 34.01 at 21 dpw. Although the average CT values varied significantly, the overall distribution of the CT values was similar at the four different parts of the GIT. A similar trend in the distribution of the CT values was observed at the different growth stages (0, 7, 14, and 21 dpw).
Fig 1.
Distribution of the CT values of the reference genes in the duodenum (A), jejunum (B), ileum (C), colon (D) and whole gastrointestinal tract (E) of piglets. The boxes encompass the 25th to the 75th percentiles. The middle line in a box marks the median. Whisker caps indicate the minimum and maximum values.
Fig 2. Distribution of the CT values of the reference genes at different growth stages of the gastrointestinal tract.
The boxes encompass the 25th to the 75th percentiles. The middle line in a box marks the median. Whisker caps indicate the minimum and maximum values.
The CT values of reference genes HMBS (P >0.05) and B2M (P >0.05) were not significantly altered at the different parts of the GIT or at the different growth stages, whereas the other 16 reference genes (P <0.05) were significantly changed at the different parts of GIT and at different growth stages (Table 3). These results showed that HMBS and B2M had the most stable CT values compared with the CT values of the other reference genes, implying that both HMBS and B2M are optimal reference genes.
Table 3. A comparison of CT values of the 18 reference genes at different GITs and growth stages.
| Gene | GIT | Age (Post-weaning) | SEM | P-value | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Duodenum | Jejunum | Ileum | Colon | Day 0 | Day 7 | Day 14 | Day 21 | GIT | Age | GIT*Age | GIT | Age | GIT*Age | |
| YWHA | 23.61 | 23.62 | 23.03 | 23.61 | 24.09a | 23.44b | 23.47b | 22.89c | 0.185 | 0.185 | 0.369 | 0.065 | <0.001 | <0.001 |
| UBC | 20.07b | 20.01b | 19.69b | 20.61a | 19.62c | 20.26ab | 20.63a | 19.86bc | 0.189 | 0.189 | 0.379 | 0.010 | 0.002 | <0.001 |
| TBP | 28.09a | 27.07b | 27.01b | 27.49ab | 28.57a | 26.84bc | 27.60b | 26.65c | 0.292 | 0.292 | 0.585 | 0.041 | <0.001 | 0.003 |
| RPL32 | 18.73 | 18.46 | 18.87 | 18.05 | 18.11b | 18.33b | 19.29a | 18.38b | 0.302 | 0.302 | 0.605 | 0.237 | 0.037 | 0.062 |
| RPL19 | 21.39a | 20.41b | 19.68c | 20.91ab | 20.61ab | 20.69ab | 20.93a | 20.17b | 0.185 | 0.185 | 0.371 | <0.001 | 0.039 | <0.001 |
| PPIA | 23.27 | 22.76 | 22.75 | 22.29 | 23.73a | 23.29a | 21.84b | 22.20b | 0.332 | 0.332 | 0.664 | 0.232 | <0.001 | <0.001 |
| PPARGGIA | 29.15 | 28.59 | 28.77 | 29.06 | 29.18a | 28.15b | 29.32a | 28.92a | 0.207 | 0.207 | 0.415 | 0.209 | 0.001 | <0.001 |
| PGK11 | 24.57a | 23.48b | 22.53c | 23.69b | 23.19b | 23.63ab | 24.21a | 23.23b | 0.217 | 0.217 | 0.435 | <0.001 | 0.004 | 0.001 |
| HSPCB | 25.16a | 23.06b | 23.29b | 23.58b | 24.12a | 23.63ab | 24.11a | 23.23b | 0.28 | 0.28 | 0.561 | <0.001 | 0.081 | <0.001 |
| CANx | 25.93 | 25.46 | 25.3 | 25.57 | 25.90a | 25.15b | 26.28a | 24.93b | 0.259 | 0.259 | 0.518 | 0.361 | 0.001 | <0.001 |
| ALDOA | 26.87a | 24.78b | 25.09b | 25.08b | 26.84a | 24.75c | 25.88b | 24.34c | 0.29 | 0.29 | 0.579 | <0.001 | <0.001 | <0.001 |
| 5S | 22.09b | 21.12c | 22.19b | 23.52a | 22.41ab | 21.68b | 22.01b | 22.83a | 0.252 | 0.252 | 0.503 | <0.001 | 0.012 | <0.001 |
| 18S | 15.72a | 16.09a | 15.74a | 15.07b | 15.76ab | 16.03a | 15.49ab | 15.35b | 0.223 | 0.223 | 0.446 | 0.015 | 0.141 | <0.001 |
| B2M | 15.78 | 15.67 | 15.53 | 15.74 | 15.92 | 15.72 | 15.47 | 15.61 | 0.092 | 0.064 | 0.103 | 0.333 | 0.102 | 0.081 |
| B-actin | 17.99ab | 18.47a | 17.40b | 17.77b | 18.07 | 17.92 | 17.71 | 17.93 | 0.199 | 0.199 | 0.398 | 0.003 | 0.654 | 0.042 |
| GaPDH | 24.96b | 27.23a | 25.22b | 23.82b | 27.33a | 24.49b | 22.95b | 26.46a | 0.648 | 0.648 | 1.296 | 0.004 | <0.001 | <0.001 |
| HMBS | 26.65 | 26.80 | 26.61 | 26.74 | 26.91 | 26.64 | 26.58 | 26.67 | 0.099 | 0.099 | 0.197 | 0.508 | 0.094 | 0.052 |
| HPRT1 | 25.61a | 24.73b | 24.53b | 24.83b | 25.22a | 24.98ab | 24.71b | 24.79b | 0.11 | 0.11 | 0.22 | <0.001 | 0.007 | 0.001 |
a,b,c Mean values within a row with unlike superscript letters were significantly different (P< 0·05).
GeNorm analysis
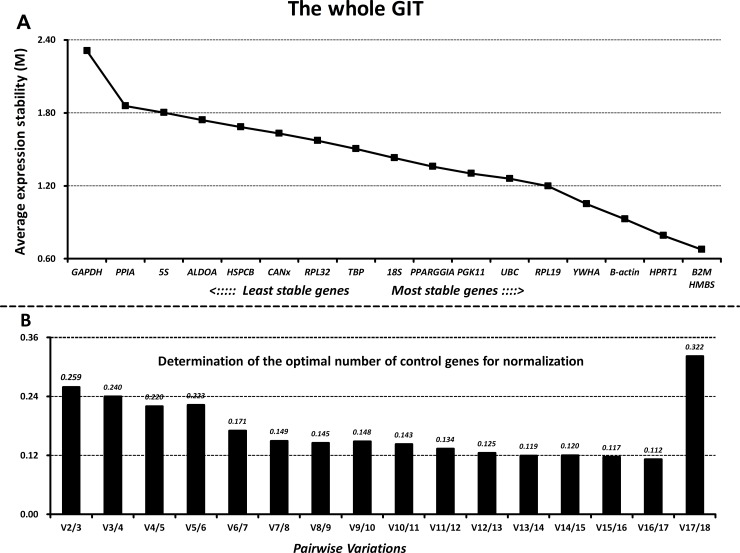
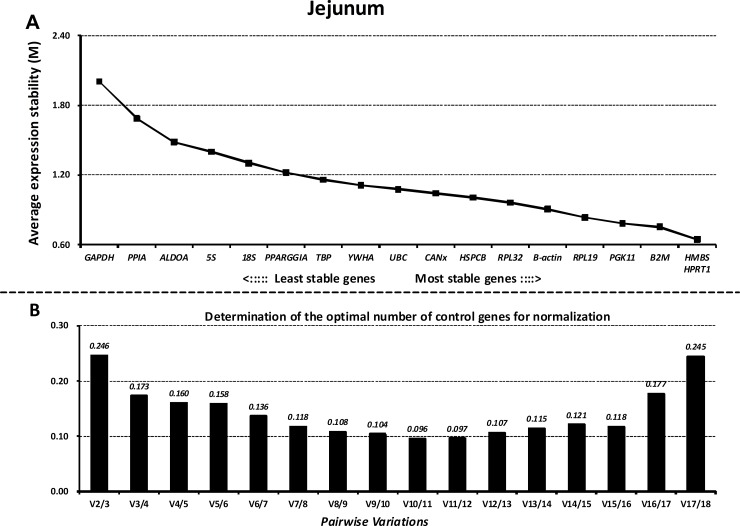
Next, we ranked all the reference genes based on their stability using GeNorm. The M values of all the reference genes at different parts of the GIT and the whole GIT are shown in Figs 3A–7A. A greater value of M means higher variation in gene expression. At different parts of the GIT as well as the whole GIT, HMBS, B2M, and HPRT1 had the lowest M values and the most stable mRNA expression, whereas GAPDH exhibited significant variations and was the least stable gene. Moreover, the candidate genes HPRT1/HMBS/B2M, HMBS/RPL32/PGK11, B2M/ HMBS/HPRT1, and HPRT1/YWHA/B2M had the lowest M values at 0, 7, 14, and 21 dpw, respectively, whereas GAPDH again exhibited significant variations and was the least stable gene (S3A–S6A Figs).
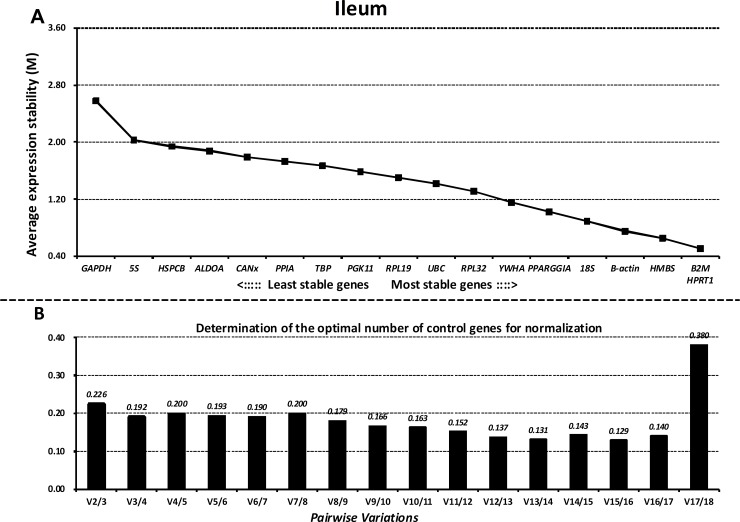
Fig 3. Gene expression stability and ranking of the reference genes in the duodenum as calculated by GeNorm.
A) Rank order of gene expression stability based on average expression stability values (M) for the reference genes from least stable (left) to most stable (right). B) Pairwise variation analysis (V) to determine the optimal number of reference genes for RT-qPCR data normalization.
Fig 7. Gene expression stability and rankings of the reference genes in the whole gastrointestinal tract as calculated by GeNorm.
A) Rank order of gene expression stability based on average expression stability values (M) for the reference genes from least stable (left) to most stable (right). B) Pairwise variation analysis (V) to determine the optimal number of reference genes for RT-qPCR data normalization.
Fig 4. Gene expression stability and rankings of the reference genes in the jejunum as calculated by GeNorm.
A) Rank order of gene expression stability based on average expression stability values (M) for the reference genes from least stable (left) to most stable (right). B) Pairwise variation analysis (V) to determine the optimal number of reference genes for RT-qPCR data normalization.
Fig 5. Gene expression stability and rankings of the reference genes in the ileum as calculated by GeNorm.
A) Rank order of gene expression stability based on average expression stability values (M) for the reference genes from least stable (left) to most stable (right). B) Pairwise variation analysis (V) to determine the optimal number of reference genes for RT-qPCR data normalization.
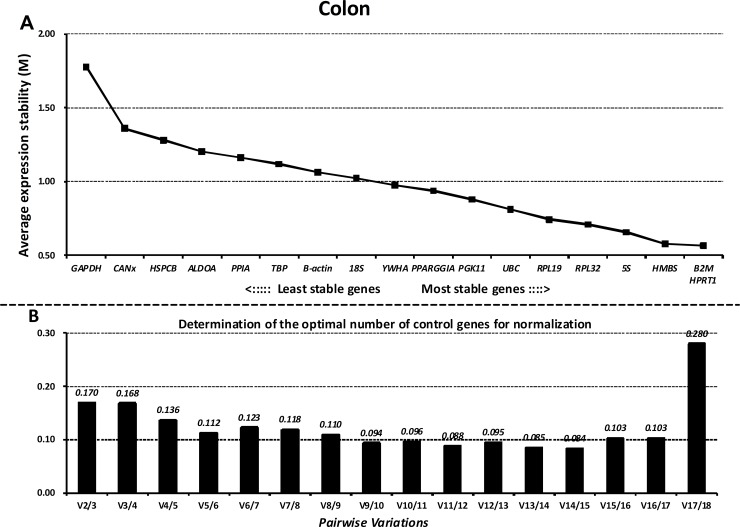
Fig 6. Gene expression stability and rankings of the reference genes in the colon as calculated by GeNorm.
A) Rank order of gene expression stability based on average expression stability values (M) for the reference genes from least stable (left) to most stable (right). B) Pairwise variation analysis (V) to determine the optimal number of reference genes for RT-qPCR data normalization.
Additionally, GeNorm determined the optimal number of reference genes needed to normalize RT-qPCR data on the basis of the average value of pairwise variations (Vn/n+1). In this study, when the cut-off M value was < 0.24, the inclusion of an additional reference gene did not further improve the normalization. On the basis of this threshold, the values of V3/4 values fell below 0.24 for different parts of the GIT (Figs 3B–7B) and different growth stages (S3B–S6B Figs), suggesting that the three selected reference genes (B2M/HMBS/HPRT) would be sufficient to normalize the data across all the samples.
BestKeeper analysis
The correlation of repeated pairwise variations was calculated to assess the optimal reference genes using BestKeeper. BestKeeper requires that the SD values of selected reference genes should not exceed 1.00, and the most stable reference gene also should have a Pearson coefficient of correlation (R) close to 1.00. Table 4 showed the R values of all the reference genes at different parts of the GIT ranked according to the BestKeeper analysis. In the duodenum, B2M, HMBS, and PGK11 were the most stable reference genes with R values of 0.97, 0.92, and 0.89 respectively. In the jejunum, HMBS, B2M, and HPRT1 were the most stable with R values of 0.87, 0.81, and 0.62 respectively. In the ileum, B2M, HMBS, and HSPCB were the most stable with R values of 0.82, 0.82, and 0.77 respectively. In the colon, B2M, HPRT1, and HMBS were the most stable with R values of 0.79, 0.67, and 0.61 respectively. In the whole GIT, B2M, HMBS, and HPRT1 were the most stable with R values of 0.78, 0.70, and 0.59 respectively.
Table 4. Reference genes stability calculated by BestKeeper based on the CT.
| a) Duodenum | b) Jejunum | |||||
| Gene | Coeff.of corr.[R] | Std dev[± CT] | Gene | Coeff.of corr.[R] | Std dev[± CT] | |
| B2M | 0.97 | 0.56 | HMBS | 0.87 | 0.59 | |
| HMBS | 0.92 | 0.83 | B2M | 0.81 | 0.32 | |
| PGK11 | 0.89 | 0.81 | HPRT1 | 0.62 | 0.74 | |
| HPRT1 | 0.87 | 0.55 | RPL19 | 0.62 | 0.68 | |
| UBC | 0.84 | 0.74 | B-actin | 0.52 | 0.99 | |
| RPL32 | 0.59 | 0.83 | PGK11 | 0.48 | 0.47 | |
| TBP | 0.57 | 0.47 | UBC | 0.46 | 0.99 | |
| RPL19 | 0.3 | 0.93 | YWHA | 0.1 | 0.77 | |
| B-actin | 0.16 | 0.56 | ||||
| c) Ileum | d) Colon | |||||
| Gene | Coeff.of corr.[R] | Std dev[± CT] | Gene | Coeff.of corr.[R] | Std dev[± CT] | |
| B2M | 0.82 | 0.52 | B2M | 0.79 | 0.52 | |
| HMBS | 0.82 | 0.59 | HPRT1 | 0.67 | 0.47 | |
| HSPCB | 0.77 | 0.55 | HMBS | 0.61 | 0.64 | |
| HPRT1 | 0.67 | 0.37 | RPL19 | 0.44 | 0.62 | |
| YWHA | 0.53 | 0.92 | PPARGGIA | 0.41 | 0.88 | |
| PGK11 | 0.24 | 0.61 | YWHA | 0.36 | 0.9 | |
| RPL19 | 0.23 | 0.82 | RPL32 | 0.32 | 0.59 | |
| 5S | 0.05 | 0.98 | ||||
| e) The whole GIT | ||||||
| Gene | coeff. of corr. [r] | std dev [± CP] | ||||
| B2M | 0.78 | 0.50 | ||||
| HMBS | 0.70 | 0.43 | ||||
| HPRT1 | 0.59 | 0.59 | ||||
| UBC | 0.52 | 0.89 | ||||
| B-actin | 0.43 | 0.85 | ||||
| RPL32 | 0.43 | 0.94 | ||||
| YWHA | 0.40 | 0.98 | ||||
| RPL19 | 0.17 | 0.86 | ||||
Coeff. of corr. [R]: Pearson coefficient of correlation; Std dev [± CT]: the standard deviation of the CT.
At different growth stages (S2 Table), PGK11/HPRT1/B2M (R values 0.89, 0.79, and 0.76), HPRT1/HMBS/TBP (R values 0.74, 0.68, and 0.66), HPRT1/PGK11/B2M (R values 0.95, 0.91, and 0.91), and B2M/HMBS/HPRT1 (R values 0.66, 0.63, and 0.51) were the most stable reference genes at 0, 7, 14, and 21 dpw, respectively.
Reference gene validation
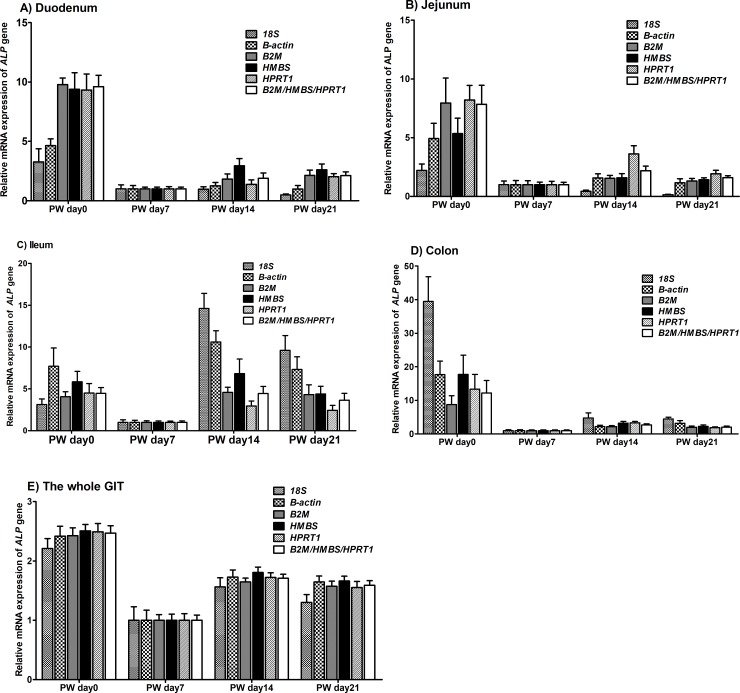
According to the results of GeNorm and BestKeeper, the optimal number of reference genes in this study are three (B2M, HMBS and HPRT1), and we then used ALP as a target gene to assess the reference genes by normalizing its expression against all the reference genes as well as the geometric mean of B2M/HMBS/HPRT1 (S3–S7 Tables). Generally, the mRNA expression pattern of ALP in the duodenum, jejunum, colon and the whole GIT was similar when normalized using HMBS, B2M, HPRT1 compared with the geometric mean of B2M/HMBS/HPRT1 (Fig 8). However, in the ileum, the ALP expression obtained by normalized against the geometric mean of B2M/HMBS/HPRT1 significantly changed when compared with its expression normalized against HMBS, B2M, HPRT1 at 0, 14, 21 dpw. Therefore, it suggested that the three selected reference genes (B2M/HMBS/HPRT) instead of one individual reference gene (e.g., HMBS, B2M, HPRT1) would be more sufficient to normalize the data across all the samples.
Fig 8.
Effect of normalization on ALP expression values in the duodenum (A), jejunum (B), ileum (C), colon (D) and the whole gastrointestinal tract (E) of piglets. The expression of ALP was normalized to 18S, B-actin, HMBS, B2M and HPRT1 and the geometric mean of B2M/HMBS/HPRT1 expression and is shown relative to its expression at different post-weaning stages. Error bars indicate mean ± standard error of the mean (SEM).
Furthermore, we also assessed the relative mRNA expression of ALP normalized against some commonly used reference genes (e.g., B-actin and 18S) and the geometric mean of B2M/HMBS/HPRT1 (Fig 8). In the duodenum and jejunum, compared with normalization against B-actin and 18S, normalization against the geometric mean of B2M/HMBS/HPRT1 produced a much higher value (about 2.0-fold) for ALP expression at the peak of relative expression at 0 dpw. In the ileum, compared with normalization against B-actin and 18S, the normalization against the geometric mean of B2M/HMBS/HPRT1, gave a much lower value for ALP expression at 14 and 21 dpw. In the colon, the ALP expression obtained by normalized against the geometric mean of B2M/HMBS/HPRT1 and B-actin was significantly lower than its expression normalized against 18S at 0 dpw. In the whole GIT, the peak of the ALP expression normalized against 18S and B-actin did not significantly change when compared with normalization against the geometric mean of B2M/HMBS/HPRT1. Thus, at different parts of the GIT, normalization against some commonly used reference genes produced dramatic differences in ALP expression that were not seen when normalized against the geometric mean of B2M/HMBS/HPRT1.
Discussion
Because gene expression analysis by RT-qPCR assay has high specificity, accuracy, and is cost-effective, it is commonly used to identify genes associated with novel metabolic or signaling pathways involved in multiple biological processes in pig, which is a good model for human health. It is therefore critical to select appropriate housekeeping reference genes for RT-qPCR data normalization in gene expression analysis [20]. A total of 665 published papers (up to 31 December 2017) involving Sus scrofa (i.e., swine, pig, boar, sow, or boar) that were related to mRNA gene expression analysis using miscellaneous technologies were available in NCBI’s PubMed database (S8 Table). In these publications, the three most commonly used housekeeping genes were B-actin (158 papers, 23.72%), GAPDH (140 papers, 21.02%), and 18S (67 papers, 10.67%). Less than 25% of the publications used more than one endogenous control gene to assess gene expression, although normalization with a single housekeeping gene can produce false results [21]. Therefore, a set of constant housekeeping genes for pig is important for determining mRNA abundance at different developmental stages, in different tissues, and in environment-specific samples [22]. In this study, we systematically evaluated the reliability of 18 commonly used housekeeping genes across the whole GIT and developed a new set of constant housekeeping genes in pig at the weaning stage. We identified B2M, HMBS, and HPRT1 as the optimal housekeeping genes for normalization of RT-qPCR expression data of genes involved in the intestinal development of piglets because of their high expression abundance and high stability in different parts of the GIT (e.g., duodenum, jejunum, ileum, and colon).
B2M, is ubiquitously expressed in nucleated cells and is present in most biological fluids, such as urine, synovial fluid, and serum [23,24]. Besides coding the light chain of major histocompatibility complex (MHC) class I molecules, B2M also plays multiple roles in the immune system of pigs [24]. In studies that use pig as an animal model, B2M was a commonly used reference gene in longissimus thoracis et lumborum muscle [25] and duodenum [7], but it was also identified as the least stable reference gene in other tissues, such as oocytes and preimplantation embryos [26], left ventricle [27], ovary [28], and skeletal muscle [29]. HPRT1 is conserved across prokaryotes and eukaryotes and plays a vital role in the generation of purine nucleotides via the purine salvage pathway [30]. HPRT1 is considered to be a somatic cell genetic marker with clinical significance in human disease, and is therefore one of the best characterized genes in human [31]. HPRT1 is a frequently used housekeeping gene in different pig tissues, such as diaphragm, heart, lung [32], skeletal muscle [8], right and left atrium, left ventricle [27], and duodenum [7]. However, in a study that assessed a set of reference genes for different tissues (liver, lung, kidney, spleen, stomach, small intestine, and large intestine) of different pig breeds (Berkshire, Landrace, Duroc, and Yorkshire), HPRT1 was the least stable housekeeping gene due to its inconsistent expression patterns [6]. The HMBS gene sequence is about 10-kb long, has 15 exons, and encodes a major protein involved in the heme biosynthetic pathway that regulates autosomal dominant disorders, such as acute intermittent porphyria [33]. HMBS also stimulates transferase activity that catalyzes the conversion of guanine to guanosine monophosphate and hypoxanthine to inosine monophosphate via the transfer of the 5-phosphoribosyl group from 5-phosphoribosyl 1-pyrophosphate [33]. HMBS was identified as the most stable housekeeping gene in the duodenum of piglets [7], but was found to be the least stable housekeeping gene in adipose tissue [22] and uterus [28].
The most frequently used housekeeping genes, GAPDH, B-actin, and 18S [9], were not identified as ideal endogenous control genes in this study, agreeing with the findings of Nygard et al. (2007) [34], Cinar et al. (2012) [35], Zhang et al. (2012) [20], Gessner et al. (2013) [36], Li et al. (2016) [37], and Wang et al. (2016) [7], but contradicting the findings of Erkens et al. (2006) [14], Kuijk et al. (2007) [26], and Li et al. (2011) [38]. Because RT-qPCR has dynamic range and increased sensitivity compared with traditional quantitation techniques, the well-known housekeeping genes, especially GAPDH, B-actin, and 18S, have been shown to be significantly affected by biological processes, different experimental treatments, and different cell types and tissues [39]. The optimal housekeeping genes identified in this study were different from those chosen in previous studies because the samples that were used varied in cell types [40], tissues [6,32,34], developmental stages [20,29], and breeds [6,15]. Reference genes used as ideal endogenous control genes are therefore those that are commonly assumed to be constant and highly expressed under specific circumstances or in specific tissues [8].
To validate the stability of the housekeeping genes we used GeNorm and BestKeeper. According to GeNorm, B2M/HMBS/HPRT1 were the optimal housekeeping genes in the duodenum, jejunum, ileum, and colon as well as the whole GIT. According to BestKeeper, B2M/HMBS/PGK11, HMBS/B2M/HPRT1, B2M/HMBS/HSPCB, B2M/HPRT1/HMBS, and B2M/HMBS/HPRT1 had the highest stability in the duodenum, jejunum, ileum, colon and the whole GIT, respectively. In addition to the GeNorm and BestKeeper analyses, we assessed the impact of normalization on the expression pattern of a target gene (ALP) using all the reference genes and the geometric mean. ALP is an important regulator involved in the primary digestive and absorptive functions of the small intestine and is expressed mainly in the intestinal mucosa of pig [41]. ALP is involved in the hydrolysis of monophosphate esters and transcellular solute transport, and participates in the absorption of fat, which is crucial to gut mucosal structures and functions in weaned-piglets [16]. Because of its critical role in intestinal development [7], we selected ALP as the target gene to validate the reference genes. Intriguingly, the mRNA expression of ALP at different parts of the GIT, normalized against B2M, HMBS, HPRT1, and the geometric mean of all the reference genes, was similar at all post-weaning growth stages tested. However, compared with the results obtained by normalized against B2M, HMBS, HPRT1, and the geometric mean of all the reference genes, underestimation or overestimation of ALP expression occurred when the ALP RT-qPCR expression data were normalized to other commonly used reference genes, including GAPDH, B-actin, or 18S. Thus, substantial differences in the relative expression levels of ALP were obtained when different reference genes were chosen for normalization, indicating the need for accurate validation of reference genes.
Conclusion
The results of the GeNorm and BestKeeper analyses indicated that B2M/HMBS/HPRT1 were the optimal reference genes in duodenum, jejunum, ileum, and colon, as well as the whole GIT of pig, whereas other commonly used reference genes (e.g., GAPDH, B-actin, and 18S) were the least stable. Our results provide a set of optimal endogenous control genes for normalization of RT-qPCR expression data for intestinal studies of pig during the weaning stage.
Supporting information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Agarose gel (2%) electrophoresis shows a band at the expected size for each reference gene. M: DNA Marker I (MD101) (100–600 bp), 1: HMBS, 2: ALP, 3: 18S, 4: TBP, 5: RPL19, 6: PPARGC1A, 7: 5S, 8: CANX, 9: HSPCB, 10: B-actin, 11: UBC, 12: B2M, 13: HPRT1, 14: PGK1, 15: YWHAZ, 16: GAPDH, 17: PPIA, 18: RPL32, and 19: ALDOA.
(DOCX)
(DOCX)
A) Rank order of gene expression stability based on average expression stability values (M) for the reference genes from least stable (left) to most stable (right). B) Pairwise variation analysis (V) to determine the optimal number of reference genes for RT-qPCR data normalization.
(DOCX)
A) Rank order of gene expression stability based on average expression stability values (M) for the reference genes from least stable (left) to most stable (right). B) Pairwise variation analysis (V) to determine the optimal number of reference genes for RT-qPCR data normalization.
(DOCX)
A) Rank order of gene expression stability based on average expression stability values (M) for the reference genes from least stable (left) to most stable (right). B) Pairwise variation analysis (V) to determine the optimal number of reference genes for RT-qPCR data normalization.
(DOCX)
A) Rank order of gene expression stability based on average expression stability values (M) for the reference genes from least stable (left) to most stable (right). B) Pairwise variation analysis (V) to determine the optimal number of reference genes for RT-qPCR data normalization.
(DOCX)
Acknowledgments
We thank Margaret Biswas, PhD, from Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The author(s) received no specific funding for this work.
References
- 1.Lallès J-P, Bosi P, Smidt H, Stokes CR. Weaning-a challenge to gut physiologists. Livest Sci. 2007;108:82–93. 10.1016/j.livsci.2007.01.091 [DOI] [Google Scholar]
- 2.Wang S, Guo C, Zhou L, Zhang Z, Huang Y, Yang J, et al. Comparison of the biological activities of Saccharomyces cerevisiae-expressed intracellular EGF, extracellular EGF, and tagged EGF in early-weaned pigs. Appl Microbiol Biotechnol. 2015;99:7125–7135. 10.1007/s00253-015-6468-6 [DOI] [PubMed] [Google Scholar]
- 3.Boudry G, Péron V, Le Huërou-Luron I, Lallès JP, Sève B. Weaning induces both transient and long-lasting modifications of absorptive, secretory, and barrier properties of piglet intestine. J Nutr.2004;134:2256–2262. 10.1093/jn/134.9.2256 [DOI] [PubMed] [Google Scholar]
- 4.Barbara G. Mucosal barrier defects in irritable bowel syndrome. Who left the door open? Am J Gastroenterol. 2006;101:1295–1298. 10.1111/j.1572-0241.2006.00667.x [DOI] [PubMed] [Google Scholar]
- 5.Lallès J-P, Bosi P, Smidt H, Stokes CR. Nutritional management of gut health in pigs around weaning. Proc Nutr Soc. 200766:260–268. 10.1017/S0029665107005484 [DOI] [PubMed] [Google Scholar]
- 6.Park S-J, Kwon SG, Hwang JH, Park DH, Kim TW, Kim CW, et al. Selection of appropriate reference genes for RT-qPCR analysis in Berkshire, Duroc, Landrace, and Yorkshire pigs. Gene.2015; 558:152–158. 10.1016/j.gene.2014.12.052 [DOI] [PubMed] [Google Scholar]
- 7.Wang S, Guo C, Zhou L, Zhong Z, Zhu W, Huang Y, et al. Effects of dietary supplementation with epidermal growth factor-expressing Saccharomyces cerevisiae on duodenal development in weaned piglets. Br J Nutr. 2016;115:1509–1520. 10.1017/S0007114516000738 [DOI] [PubMed] [Google Scholar]
- 8.Feng X, Xiong Y, Qian H, Lei M, Xu D, Ren Z. Selection of reference genes for gene expression studies in porcine skeletal muscle using SYBR green qPCR. J Biotechnol. 2010;150: 288–293. 10.1016/j.jbiotec.2010.09.949 [DOI] [PubMed] [Google Scholar]
- 9.Huggett J, Dheda K, Bustin S, Zumla A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005;6:279–284. 10.1038/sj.gene.6364190 [DOI] [PubMed] [Google Scholar]
- 10.Dheda K, Huggett J, Chang J, Kim L, Bustin S, Johnson MA, et al. The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Anal Biochem. 2005;344:141–143. 10.1016/j.ab.2005.05.022 [DOI] [PubMed] [Google Scholar]
- 11.Zhu W, Lin Y, Liao H, Wang Y. Selection of reference genes for gene expression studies related to intramuscular fat deposition in Capra hircus skeletal muscle. PloS One. 2015;10:e0121280 10.1371/journal.pone.0121280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bruge F, Venditti E, Tiano L, Littarru G, Damiani E. Reference gene validation for qPCR on normoxia-and hypoxia-cultured human dermal fibroblasts exposed to UVA: is β-actin a reliable normalizer for photoaging studies? J Biotechnol. 2011;156:153–162. 10.1016/j.jbiotec.2011.09.018 [DOI] [PubMed] [Google Scholar]
- 13.Cunha T. Swine feeding and nutrition. Elsevier; 2012.
- 14.Erkens T, Van Poucke M, Vandesompele J, Goossens K, Van Zeveren A, Peelman LJ. Development of a new set of reference genes for normalization of real-time RT-PCR data of porcine backfat and longissimus dorsi muscle, and evaluation with PPARGC1A. BMC Biotechnol. 2006;6:41 10.1186/1472-6750-6-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muráni E, Schellander K, Wimmers K, Murániová M, Ponsuksili S. Identification of genes differentially expressed during prenatal development of skeletal muscle in two pig breeds differing in muscularity. BMC Dev Biol. 2007;7:109 10.1186/1471-213X-7-109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lackeyram D, Yang C, Archbold T, Swanson KC, Fan MZ. Early weaning reduces small intestinal alkaline phosphatase expression in pigs. J Nutr. 2010;140: 461–468. 10.3945/jn.109.117267 [DOI] [PubMed] [Google Scholar]
- 17.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:research0034. 0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26:509–515. [DOI] [PubMed] [Google Scholar]
- 19.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3:1101–1108. [DOI] [PubMed] [Google Scholar]
- 20.Zhang J, Tang Z, Wang N, Long L, Li K. Evaluating a set of reference genes for expression normalization in multiple tissues and skeletal muscle at different development stages in pigs using quantitative real-time polymerase chain reaction. DNA Cell Biol. 2012;31:106–113. 10.1089/dna.2011.1249 [DOI] [PubMed] [Google Scholar]
- 21.Wong ML, Medrano JF. Real-time PCR for mRNA quantitation. Biotechniques. 2005;39:75–88. 10.2144/05391RV01 [DOI] [PubMed] [Google Scholar]
- 22.Gu Y, Li M, Zhang K, Chen L, Jiang A, Wang JY, et al. Evaluation of endogenous control genes for gene expression studies across multiple tissues and in the specific sets of fat‐and muscle‐type samples of the pig. J Anim Breed Genet. 2011;128:319–325. 10.1111/j.1439-0388.2011.00920.x [DOI] [PubMed] [Google Scholar]
- 23.Drüeke TB, Massy ZA. Progress in uremic toxin research: beta2‐microglobulin. Wiley Online Library; 2009.
- 24.Le TM, Le QVC, Truong DM, Lee H-J, Choi M-K, Cho H, et al. β2-microglobulin gene duplication in cetartiodactyla remains intact only in pigs and possibly confers selective advantage to the species. PloS One. 2017;12:e0182322 10.1371/journal.pone.0182322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McBryan J, Hamill RM, Davey G, Lawlor P, Mullen AM. Identification of suitable reference genes for gene expression analysis of pork meat quality and analysis of candidate genes associated with the trait drip loss. Meat Sci. 2010;86:436–439. 10.1016/j.meatsci.2010.05.030 [DOI] [PubMed] [Google Scholar]
- 26.Kuijk EW, du Puy L, van Tol HT, Haagsman HP, Colenbrander B, Roelen BA. Validation of reference genes for quantitative RT-PCR studies in porcine oocytes and preimplantation embryos. BMC Dev Biol. 2007;7:58 10.1186/1471-213X-7-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martino A, Cabiati M, Campan M, Prescimone T, Minocci D, Caselli C, et al. Selection of reference genes for normalization of real-time PCR data in minipig heart failure model and evaluation of TNF-alpha mRNA expression. J Biotechnol. 2011;153:92–99. 10.1016/j.jbiotec.2011.04.002 [DOI] [PubMed] [Google Scholar]
- 28.Martinez-Giner M, Noguera JL, Balcells I, Fernandez-Rodriguez A, Pena RN. Selection of internal control genes for real-time quantitative PCR in ovary and uterus of sows across pregnancy. PLoS One. 2013;8:e66023 10.1371/journal.pone.0066023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Zhao Y, Li J, Liu H, Ernst CW, Liu X, et al. Evaluation of housekeeping genes for normalizing real-time quantitative PCR assays in pig skeletal muscle at multiple developmental stages. Gene. 2015; 565:235–241. 10.1016/j.gene.2015.04.016 [DOI] [PubMed] [Google Scholar]
- 30.Guibinga G-H, Hsu S, Friedmann T. Deficiency of the housekeeping gene hypoxanthine–guanine phosphoribosyltransferase (hprt) dysregulates neurogenesis. Mol Ther. 2010;18:54–62. 10.1038/mt.2009.178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keebaugh AC, Sullivan RT, Thomas JW, Program NCS. Gene duplication and inactivation in the HPRT gene family. Genomics. 2007;89:134–142. 10.1016/j.ygeno.2006.07.003 [DOI] [PubMed] [Google Scholar]
- 32.Svobodová K, Bílek K, Knoll A. Verification of reference genes for relative quantification of gene expression by real-time reverse transcription PCR in the pig. J Appl Genet. 2008;49:263–265. 10.1007/BF03195623 [DOI] [PubMed] [Google Scholar]
- 33.Yang CC, Kuo HC, You HL, Wang J, Huang CC, Liu CY, et al. HMBS mutations in Chinese patients with acute intermittent porphyria. Ann Hum Genet. 2008;72:683–686. 10.1111/j.1469-1809.2008.00463.x [DOI] [PubMed] [Google Scholar]
- 34.Nygard AB, Jorgensen CB, Cirera S, Fredholm M. Selection of reference genes for gene expression studies in pig tissues using SYBR green qPCR. BMC Mol Biol. 2007;8:67 10.1186/1471-2199-8-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cinar MU, Islam MA, Uddin MJ, Tholen E, Tesfaye D, Looft C, et al. Evaluation of suitable reference genes for gene expression studies in porcine alveolar macrophages in response to LPS and LTA. BMC Res Notes. 2012;5:107 10.1186/1756-0500-5-107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gessner DK, Fiesel A, Most E, Dinges J, Wen G, Ringseis R, et al. Supplementation of a grape seed and grape marc meal extract decreases activities of the oxidative stress-responsive transcription factors NF-κB and Nrf2 in the duodenal mucosa of pigs. Acta Vet Scand. 2013;55:18 10.1186/1751-0147-55-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li X, Huang K, Chen F, Li W, Sun S, Shi XE, et al. Verification of suitable and reliable reference genes for quantitative real-time PCR during adipogenic differentiation in porcine intramuscular stromal-vascular cells. Animal. 2016;10:947–952. 10.1017/S1751731115002748 [DOI] [PubMed] [Google Scholar]
- 38.Li Q, Domig KJ, Ettle T, Windisch W, Mair C, Schedle K. Evaluation of potential reference genes for relative quantification by RT-qPCR in different porcine tissues derived from feeding studies. Int J Mol Sci. 2011;12:1727–1734. 10.3390/ijms12031727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol. 2000;25:169–193. [DOI] [PubMed] [Google Scholar]
- 40.Facci MR, Auray G, Meurens F, Buchanan R, van Kessel J, Gerdts V. Stability of expression of reference genes in porcine peripheral blood mononuclear and dendritic cells. Vet Immunol Immunopathol. 2011;141:11–15. 10.1016/j.vetimm.2011.01.005 [DOI] [PubMed] [Google Scholar]
- 41.Jian B, Liu B, Bi Y, Hou W, Wu C, Han T. Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Mol Biol. 2008;9:59 10.1186/1471-2199-9-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Agarose gel (2%) electrophoresis shows a band at the expected size for each reference gene. M: DNA Marker I (MD101) (100–600 bp), 1: HMBS, 2: ALP, 3: 18S, 4: TBP, 5: RPL19, 6: PPARGC1A, 7: 5S, 8: CANX, 9: HSPCB, 10: B-actin, 11: UBC, 12: B2M, 13: HPRT1, 14: PGK1, 15: YWHAZ, 16: GAPDH, 17: PPIA, 18: RPL32, and 19: ALDOA.
(DOCX)
(DOCX)
A) Rank order of gene expression stability based on average expression stability values (M) for the reference genes from least stable (left) to most stable (right). B) Pairwise variation analysis (V) to determine the optimal number of reference genes for RT-qPCR data normalization.
(DOCX)
A) Rank order of gene expression stability based on average expression stability values (M) for the reference genes from least stable (left) to most stable (right). B) Pairwise variation analysis (V) to determine the optimal number of reference genes for RT-qPCR data normalization.
(DOCX)
A) Rank order of gene expression stability based on average expression stability values (M) for the reference genes from least stable (left) to most stable (right). B) Pairwise variation analysis (V) to determine the optimal number of reference genes for RT-qPCR data normalization.
(DOCX)
A) Rank order of gene expression stability based on average expression stability values (M) for the reference genes from least stable (left) to most stable (right). B) Pairwise variation analysis (V) to determine the optimal number of reference genes for RT-qPCR data normalization.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.