Abstract
Mitochondrial respiratory disorders are incurable progressive degenerative diseases with multi-organ system manifestations. These orphan diseases are caused by mutations in the nuclear or mitochondrial genome affecting the oxidative phosphorylation (OXPHOS) system responsible for ATP synthesis. Currently, therapeutic treatments are not available to patients, resulting in significant disability and a poor prognosis. Patients exhibit a constellation of complex neurological and multisystem phenotypic symptoms. The hallmark of these diseases is their clinical heterogeneity and high variability among patients. Consequently, establishing an accurate diagnosis remains a challenging, invasive, and time-consuming process due to the limited sensitivity, specificity and reliability of the current serum biomarkers used in clinical settings. Recent mouse model-based research combined with patient studies led to the identification of fibroblast growth factor 21 (FGF-21) as a promising serum biomarker. With its high specificity and sensitivity, FGF-21 is a promising diagnostic tool for muscle-affecting mitochondrial respiratory disorders, which might be a useful first-line diagnostic tool instead of the invasive muscle biopsy currently performed in clinical settings. Discovering additional diagnostic biomarkers is critical for establishing an accurate diagnosis given the high clinical heterogeneity of these mitochondrial respiratory diseases. Ultimately, these novel biomarkers might be instrumental to monitor the progression of these diseases and the efficacy of novel therapeutic interventions.
Keywords: Biomarkers, Creatine, Fibroblast-growth factor 21, Lactate, Mitochondrial respiratory disorders, Pyruvate, Oxidative phosphorylation
Key Concepts and Clinical Features of Mitochondrial Respiratory Disorders
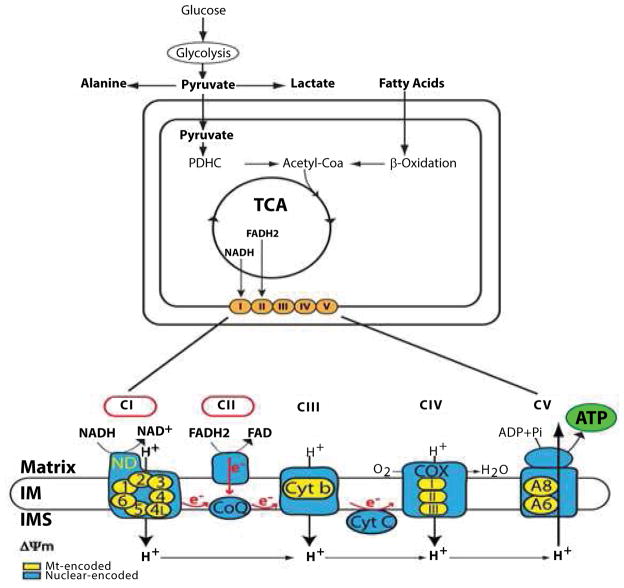
Mitochondrial Respiratory Disorders (MRDs) are a group of rare and incurable diseases defined by insufficient ATP levels due to impaired oxidative phosphorylation (OXPHOS) [1–3]. The OXPHOS system, which is embedded in the inner mitochondrial membrane, is responsible to produce ATP upon electron transfer through a series of four OXPHOS respiratory complexes, with complexes I and II being the two points of entry for electrons and ATP synthesis occurring at complex V, also called ATP synthase (Figure 1). Except for complex II that solely contains nuclear-encoded subunits, the OXPHOS complexes are composed of subunits encoded by both the nuclear and mitochondrial genomes [4, 5].
Figure 1.
The Mitochondrial OXPHOS system and ATP synthesis. Biochemical pathways relevant to oxidative phosphorylation are indicated. Glucose gives rise to cytosolic pyruvate via glycolysis, which is then transported into the mitochondrial matrix to generate mitochondrial acetyl CoA, a key substrate of the tricarboxylic acid (TCA) cycle. Beta oxidation of fatty acids also result in acetyl CoA synthesis in the mitochondrial matrix. NADH and FADH2, two reducing agents generated by the TCA cycle, provide electrons to CI and CII of the OXPHOS system (orange circles), which is embedded in the mitochondrial inner membrane. A high magnification containing the series of five multi-subunit respiratory complexes (CI to CV) is shown below. Red circles indicate the two points of entry for electrons at CI and CII, which are transferred to CIII and CIV via the electron carriers, Coenzyme Q10 (CoQ) and cytochrome c (Cyt C). ATP synthesis occurs at complex V (CV) using the proton gradient, a key component of the mitochondrial membrane potential (ΔΨm), generated during transfer of electrons. The nuclear-encoded subunits are indicated in blue, while the mitochondrial-encoded subunits are shown in yellow.
MRD patients harbor inherited mutations that map in their mitochondrial or nuclear genome thereby impairing OXPHOS activity and ATP production [6]. Thus, the retrograde signaling pathway is impaired due to an accumulation of NAD+ and AMP, a direct consequence of a defective OXPHOS system (Figure 2). This affects the activity of the key regulators, Sirt1 and AMPK, and their main substrate, PGC-1α, required to activate the anterograde signaling for a full mitobiogenic response, thereby failing to rectify the chronic ATP deficit (Figure 2). Currently, no therapeutic options are available to these patients to mitigate their ATP insufficiency, resulting in significant disability, a poor prognosis and premature death [7,8]. MRDs can manifest at any age, ranging from the neonatal phase to adulthood, with variable severity. In the case of severe defects in the OXPHOS system, organogenesis is affected at the onset since it coincides with a metabolic shift from glycolytic to oxidative respiration to ensure optimal ATP production during high-energy embryonic developmental stages [9]. Embryonic differentiation of neural stem cells into neurons, astrocytes, or oligodendrocytes consumes about 50% of cellular ATP to execute key differentiation processes [10]. Nevertheless, MRDs always worsen over time due to their progressive degenerative characteristics. Most MRD patients display heterogeneous clinical symptoms affecting several organs with high-energy demand, such as the central nervous system (CNS), peripheral nerves, skeletal and cardiac muscles, kidneys, and endocrine organs [11,12]. MRD patients exhibit symptoms with variable intensity that fall into two clinical groups: 1) central neurodegenerative phenotypes, including encephalopathy, stroke-like episodes, migraines, seizure, ataxia and dementia; and 2) peripheral neuronal and muscular phenotypes, such as myopathy, cardiomyopathy, peripheral neuropathy, sensorineural deafness, and optic atrophy [13].
Figure 2.
Integrated mitochondrial anterograde and retrograde signaling regulating ATP levels in healthy subjects (left panel) and MRD patients (right panel). Low ATP levels are accompanied by decreased NADH:NAD+ and ATP/AMP ratios, triggering a retrograde signaling that promotes PGC-1α activation via Sirt1-mediated de-acetylation and AMPK-mediated phosphorylation. PGC-1αactivation induces mitochondrial biogenesis and bioenergetics, resulting in regulation of a genetic program via anterograde signaling. Successful coordination of anterograde and retrograde signaling results in increased ATP levels. In MRD patients, chronic ATP deficit is accompanied by high NADH: NAD+ ratio and low ATP:AMP ratio, resulting in a deficient retrograde signaling that fails to activate PGC-1α via Sirt1 and AMPK and subsequent anterograde signaling. This cascade of molecular events results in chronic low ATP deficit.
Clinical heterogeneity is most acute in patients affected with a specific MRD due to mutations in the mitochondrial (mt) genome. These mutations, which are maternally inherited, alter either mitochondrial protein synthesis, when mapped in a mt-tRNA or mt-rRNA gene, or the OXPHOS system, when mapped in one the 13 mt genes encoding for a subunit of an OXPHOS complex [4]. Most pathogenic mtDNA mutations only affect a subset of the multi-copy mt genome, causing heteroplasmy, which is defined as the presence of wild-type (WT) and mutated mtDNAs in a mitochondrion [14,15]. Heteroplasmy is dictated by the ratio of WT and mutated mtDNAs in mitochondria, which results in a mixed population of healthy and diseased mitochondria within a cell. A mitochondrion is considered diseased or dysfunctional if its mutant mtDNAs surpass a certain threshold, overwhelming its WT mtDNAs, and vice versa for healthy/functional mitochondria. Therefore, a diseased phenotype occurs when mitochondrial heteroplasmic reaches a certain threshold, which ranges from 60 to 90% depending of the tissue and mutation, leading to insufficient ATP levels [14,16]. Thus, the degree of heteroplasmy influences the severity of the diseased phenotype as well as the heterogeneity of clinical symptoms. This is best exemplified by the mitochondrial respiratory disorder MELAS (Mitochondrial Encephalopathy with Lactic Acidosis and Stroke-like episodes), which is due to a maternally inherited A to G substitution at position 3243 of the mitochondrial gene for tRNALeu(UUR), known as the A3243G MELAS mutation [17,18]. This progressive neurodegenerative disease has an early onset of heterogeneous clinical symptoms that include encephalopathy, seizures, stroke-like episodes, and chronic lactic acidosis [19–21]. Among siblings, high clinical variability is often observed. Furthermore, the MELAS mutation can cause cardiomyopathy or myopathy in some families, while causing hearing loss and diabetes in others [22–24].
The prevalence of these rare MRDs at about 1 in 5000 live births in the United States is most likely underestimated based on recent epidemiological studies on newborn cord bloods that revealed 1 in 200 newborn carrying a potentially pathogenic mtDNA mutation [25,26]. With improved clinical diagnosis and advances in next-generation sequencing (NGS) technology, more patients will be accurately diagnosed, resulting in increased prevalence of these orphan diseases [27,28].
Current Metabolite Biomarkers Used in Clinical Setting for Diagnosis of a Mitochondrial Respiratory Disorder
Establishing an accurate diagnosis of MRD is often challenging, time consuming, and costly in part due to the absence of sensitive and specific biomarkers [29,30]. Due to extreme phenotypic variability among patients, the diagnostic process requires a set of clinical assessment combined with complex biochemical, histological and genetic analyses [31,32]. In the case of a single syndromic phenotype with known causative genes, such as Leber hereditary optic neuropathy (LHON), DNA sequencing easily confirms its clinical diagnosis since primary LHON mitochondrial DNA mutations are responsible for about 95% of LHON cases by directly affecting the enzymatic activities of the OXPHOS system and subsequently ATP levels [33–35].
Several conventional metabolite biomarkers, such as lactate, pyruvate, amino acids, and creatine, are routinely measured in plasma and/or cerebrospinal fluid (CSF) and provide a minimally invasive evaluation in patients suspected of having an MRD [8,30]. Due to their limited specificity, sensitivity and consistency, altered levels are only suggestive of a specific MRD [29].
Lactate levels are not consistently elevated in blood or CSF of patients affected with MRD. For example, patients diagnosed with LHON, Leigh disease, Kearns-Sayre syndrome and complex I deficiency display normal lactate levels [36], while most of the MELAS patients exhibit elevated levels of lactate in blood and CSF [18,19]. More specifically, high lactate levels are common in patients affected with neurodegenerative MRDs, such as MELAS, MERFF (Myoclonus Epilepsy and Ragged-Red Fibers) and MDS (Mitochondrial DNA Depletion Syndrome) [40]. Since increased lactate levels are also observed in unrelated pathologies, such as CNS infection, seizures, and stroke, lactate by itself is not a specific and reliable biomarker to definitively establish an MRD diagnosis [38].
Pyruvate is another plasma metabolite integrated into diagnostic chemistry profile currently performed in clinical settings for diagnosis [29]. The fact that pyruvate levels are prone to inaccurate measurement due to instability and susceptibility to inadequate specimen collection diminishes its reliability as a diagnostic biomarker. Finally, measurement of the ratio of lactate to pyruvate (L:P) in blood or CSF only provides valuable diagnostic information in MRDs affecting the CNS given the elevated lactate levels in blood and/or CSF [39, 40].
Elevated blood or CSF levels of amino acids, such as alanine, glycine, and proline, have also been reported in MRD patients as a result of a defective OXPHOS system and subsequent changes in the NADH:NAD+ redox signature (Figure 2) [29]. However, their sensitivity and specificity remain uncertain. For example, elevated alanine levels are not only found in MRD patients, but also in patients affected with pyruvate metabolism disorders characterized by an accumulation of cytosolic pyruvate converted into alanine (Figure 1) [41].
Recent studies investigated whether creatine could be used as a biomarker for MRDs given its known link with mitochondrial bioenergetics [42]. While plasma creatine levels were elevated in patients with specific MRDS, such as MELAS, MERFF and mtDNA deletion, when compared to healthy subjects [43,44], such correlation was not consistently observed among patients affected with different MRDs from another independent cohort [45]. Thus, additional studies are required to validate the specificity and reliability of creatine as a biomarker for a definitive diagnosis of MRD.
FGF-21: A promising Diagnostic Biomarker for a Group of MRDs
Since none of the metabolite biomarkers currently used in a clinical setting is reliable, specific and discriminative, there is an urgent need to discover serum biomarkers for a definitive diagnosis in patients suspected to have an MRD [8,46]. Although global metabolic profiling using plasma from patients already properly diagnosed with an MRD was used to identify promising biomarkers for MRDs, this comprehensive approach has yet to lead to the discovery of consistent and sensitive diagnostic biomarkers [43].
Recent mouse studies combined with patient studies have revealed fibroblast growth factor 21 (FGF-21) as unexpected potential biomarker [47]. FGF-21 is a circulating cytokine known to be involved in carbohydrate and lipid metabolism as well as secreted upon prolonged fasting in humans [48–51]. FGF-21 levels were significantly increased in the blood and OXPHOS-deficient muscle fibers of the “deletor” mouse model, which mimics key features of late-onset mitochondrial myopathy upon expression of a dominant patient mutation in the mitochondrial replicative helicase Twinkle [52,53]. Moreover, FGF-21 levels correlate with the severity of the OXPHOS deficit and the progression of mitochondrial myopathy. Interestingly, the “deletor” mice have skeletal muscle fibers with characteristics of a pseudo-starvation state, despite their normal nutritional state, which is congruent with FGF-21 being secreted from the liver in response to fasting [54].
The feasibility of FGF-21 as a serum diagnostic biomarker was assessed in patients genetically diagnosed with a specific MRD, patients with non-mitochondrial neurological disorders affecting muscles and healthy subjects [55]. This comprehensive multicenter study has revealed increased FGF-21 serum levels in patients with an MRD affecting skeletal muscles. However, patients with an MRD mainly affecting the nervous system, such as MIRAS (Mitochondrial Recessive Ataxia Syndrome), exhibited lower FGF-21 serum concentrations that those with mitochondrial diseases manifesting in skeletal muscle. The fact that FGF-21 levels were unaltered in patients with non-mitochondrial muscle diseases, implying that FGF-21 serum levels are a direct result of both OXPHOS deficit skeletal muscle pathology [55]. Most importantly, the sensitivity and specificity of FGF-21 was estimated at 92% for muscle-manifesting MRDs, making FGF-21 a reliable first-line diagnostic tool for these diseases instead of the more invasive muscle biopsy, which is currently the gold standard diagnostic tool [8]. In addition, FGF-21 could be a diagnostic indicator of the progression of the disease since FGF-21 levels correlate with the severity of the symptoms. One of the siblings with MELAS exhibiting limited cardiomyopathy and myopathy had lower FGF-21 levels than his sibling with severe phenotypic manifestation of myopathy [55]. These clinical observations are in agreement with results from studies using the “deletor” mice showing increased FGF-21 levels upon progression of mitochondrial myopathy [53]. Ultimately, FGF-21 may be a promising biomarker to monitor efficacy of therapeutic intervention and therefore promote the design of novel therapeutic strategies for the currently intractable MRDs.
Conclusion
The fact that the FGF-21 biomarker is specific for MRDs affecting skeletal muscles emphasizes the urgent need for large-scale clinical analysis and identification of novel diagnostic biomarkers tailored to the complexity and clinical heterogeneity of those MRDs. Such discoveries will most likely establish a shift in the diagnostic pathway used in clinical settings for differential and accurate diagnosis for patients suspected of having an MRD. Such progress in translational research will result in a more precise assessment of the prevalence of those mitochondrial diseases than currently estimated. In fact, recent epidemiological studies on newborn cord bloods have revealed that 1 in 200 newborns harbor mutations affecting the OXPHOS system and therefore are at risk of developing MRD. In sum, the use of new biomarkers combined with the advent of NGS technique will bring the field of mitochondrial medicine forward and accelerate the discovery of novel therapeutic strategies.
Acknowledgments
This work was supported by the National Institute of Health, National Institute of Neurological Disorders and Stroke (Grant Number R21NS085282 to AC).
Abbreviations
- CNS
Central nervous system
- CSF
Cerebrospinal fluid
- FGF-21
Fibroblast growth factor 21
- LHON
Leber Hereditary optic neuropathy
- MDS
Mitochondrial DNA depletion syndrome
- MELAS
Mitochondrial encephalopathy with lactic acidosis and stroke-like episodes
- MIRAS
Mitochondrial recessive ataxia syndrome
- MRD
Mitochondrial respiratory disorder
- MRRF
Myoclonus epilepsy and ragged-red fibers
- mtDNA
mitochondrial DNA
- NGS
Next-generation sequencing
- OXPHOS
oxidative phosphorylation
- WT
Wild type
Footnotes
The authors confirm that there is no conflict of interest to declare.
References
- 1.DiMauro S, Schon EA. Mitochondrial respiratory-chain diseases. New Engl J Med. 2003;348:2656–2668. doi: 10.1056/NEJMra022567. [DOI] [PubMed] [Google Scholar]
- 2.DiMauro S. Mitochondrial medicine. Biochem Biophys Acta. 2004;1658:107–114. doi: 10.1016/j.bbabio.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 3.Reineke F, Smeitink JA, van der W. gene expression and control in mitochondrial disorders. Biochem Biophysic Acta. 2009;1792:1113–1121. doi: 10.1016/j.bbadis.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 5.Wallace DC. The mitochondrial genome in human adaptive radiation and disease: on the road to therapeutics and performance enhancement. Gene. 2005;354:169–180. doi: 10.1016/j.gene.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 6.Ylikallio E, Suomalainen A. Mechanisms of mitochondrial diseases. Annals Med. 2012;44:41–59. doi: 10.3109/07853890.2011.598547. [DOI] [PubMed] [Google Scholar]
- 7.Kerr DS. Treatment of mitochondrial electron transport chain disorders: a review of clinical trials over the past decade. Mol Genet Metab. 2010;99:246–255. doi: 10.1016/j.ymgme.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 8.Suomalainen A. Therapy for mitochondrial disorders: Little proof, high research activity, some promise. Sem Fetal Neonatal Med. 2011;16:236–240. doi: 10.1016/j.siny.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 9.Uittenbogaard M, Chiaramello A. Mitochondrial biogenesis: a therapeutic target for neurodevelopmental disorders and neurodegenerative diseases. Current Pharm Des. 2014;20:5574–5593. doi: 10.2174/1381612820666140305224906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernstein BW, Bamburg JR. Actin-ATP hydrolysis is a major energy drain for neurons. J Neurosci. 2003;23:1–6. doi: 10.1523/JNEUROSCI.23-01-00002.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DiMauro S, Schon EA. Mitochondrial disorders in the nervous system. Annu Rev Neurosci. 2008;31:91–123. doi: 10.1146/annurev.neuro.30.051606.094302. [DOI] [PubMed] [Google Scholar]
- 12.DiMauro S, Schon EA. The clinical maze of mitochondrial neurology. Nat Rev Neurol. 2013;9:429–444. doi: 10.1038/nrneurol.2013.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kisler JE, Whittaker RG, McFarland R. Mitochondrial diseases in childhood: a clinical approach to investigation and management. Dev Med Child Neurol. 2010;52:422–433. doi: 10.1111/j.1469-8749.2009.03605.x. [DOI] [PubMed] [Google Scholar]
- 14.Wallace DC, Chalkia D. Mitochondrial DNA Genetics and the heteroplasmy conundrum in evolution and disease. Cold Spring Harb Perspect Biol. 2013;5:a021220. doi: 10.1101/cshperspect.a021220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stewart JB, Chinnery PF. The dynamics of mitochondrial DNA heteroplasmy: implications for human health and disease. Nature Rev Genet. 2015;16:530–542. doi: 10.1038/nrg3966. [DOI] [PubMed] [Google Scholar]
- 16.Park CB, Larsson NG. Mitochondrial DNA mutations in disease and aging. J Cell Biol. 2011;193:809–818. doi: 10.1083/jcb.201010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobayashi Y, Momoi MY, Tominaga K, Shimoizumi H, Nihei K, et al. Respiration-deficient cells are caused by a single point mutation in the mitochondrial tRNA-Leu (UUR) gene in mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes (MELAS) Am J Hum Genet. 1991;49:590–599. [PMC free article] [PubMed] [Google Scholar]
- 18.Hirano M, Pavlakis SG. Mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes (MELAS): current concepts. J Child Neurol. 1994;9:4–13. doi: 10.1177/088307389400900102. [DOI] [PubMed] [Google Scholar]
- 19.Sproule DM, Kaufmann P. Mitochondrial encephalopathy, lactic acidosis, and stroke-like episodes. Ann NY Acad Sci. 2008;1142:133–158. doi: 10.1196/annals.1444.011. [DOI] [PubMed] [Google Scholar]
- 20.Kaufmann P, Engelstad K, Wei Y, Kulikova R, Oskoui M, et al. Spectrum of the mitochondrial DNA mutations of Leber’s hereditary optic neuropathy in Koreans. J Neurol. 2003;250:278–281. doi: 10.1007/s00415-003-0985-4. [DOI] [PubMed] [Google Scholar]
- 21.El-Hattab AW, Adenisa AM, Jones J, Scaglia F. MELAS syndrome: Clinical manifestations, pathogenesis, and treatment options. Mol Genet Metab. 2015;116:4–12. doi: 10.1016/j.ymgme.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 22.Ciacci F, Silvestri G, Shanske S, Sciacco M, Hirano M, et al. A typical clinical presentations associated with the MELAS mutation at position 3243 of human mitochondrial DNA. Neuromuscul Disord. 1993;3:43–50. doi: 10.1016/0960-8966(93)90040-q. [DOI] [PubMed] [Google Scholar]
- 23.Reardon W, Ross RJ, Sweeney MG, Luxon LM, Pembrey ME, et al. Diabetes mellitus associated with a pathogenic point mutation in mitochondrial DNA. Lancet. 1992;340:1376–1379. doi: 10.1016/0140-6736(92)92560-3. [DOI] [PubMed] [Google Scholar]
- 24.Finsterer J. Genetic pathogenetic, and phenotypic implications of the mitochondrial A3243G tRNALeu(UUR) mutation. Acta Neurol Scand. 2007;116:1–14. doi: 10.1111/j.1600-0404.2007.00836.x. [DOI] [PubMed] [Google Scholar]
- 25.Skladal D, Halliday J, Thornburn DR. Minimum birth prevalence of mitochondrial respiratory chain disorders in children. Brain. 2003;126:1905–1912. doi: 10.1093/brain/awg170. [DOI] [PubMed] [Google Scholar]
- 26.Elliott HR, Samuels DC, Eden JA, Relton CL, Chinnery PF. Pathogenic mitochondrial DNA mutations are common in the general population. Amer J Ham Genet. 2008;83:254–260. doi: 10.1016/j.ajhg.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Calvo SE, Mootha VK. The mitochondrial proteome and human disease. Annu Rev Genomics Hum Genet. 2010;11:25–44. doi: 10.1146/annurev-genom-082509-141720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lieber DS, Calvo SE, Shanahan K, Slate NG, Liu S, et al. Targeted exome sequencing of suspected mitochondrial disorders. Neurology. 2013;80:1762–1770. doi: 10.1212/WNL.0b013e3182918c40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haas RH, Parikh S, Falk MJ, Saneto RP, Wolf NI, et al. The in-depth evaluation of suspected mitochondrial disease. Mol Genet Metab. 2008;94:16–37. doi: 10.1016/j.ymgme.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parikh S, Goldstein A, Koening MK, Scaglia F, Enns G, et al. Diagnosis and management of mitochondrial disease: a consensus statement from the Mitochondrial Medicine Society. Genet Med. 2015;17:689–701. doi: 10.1038/gim.2014.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wolf NI, Smeitink JA. Mitochondrial disorders: a proposal for consensus diagnostic criteria in infants and children. Neurology. 2002;59:1402–1405. doi: 10.1212/01.wnl.0000031795.91814.d8. [DOI] [PubMed] [Google Scholar]
- 32.Liang C, Ahmad K, Sue CM. The broadening spectrum of mitochondrial disease: Shifts in the diagnostic paradigm. Biochem Biophys Acta. 2014;1840:1360–1367. doi: 10.1016/j.bbagen.2013.10.040. [DOI] [PubMed] [Google Scholar]
- 33.Wallace DC, Singh G, Lott M, Hodge JA, Schurr TG, et al. Mitochondrial DNA mutation associated with Leber’s hereditary optic neuropathy. Science. 1988;242:1427–1430. doi: 10.1126/science.3201231. [DOI] [PubMed] [Google Scholar]
- 34.Kim JY, Hwang JM, Chang BL, Park SS. Spectrum of the mitochondrial DNA mutations of Leber’s hereditary optic neuropathy in Koreans. J Neurol. 2003;250:278–281. doi: 10.1007/s00415-003-0985-4. [DOI] [PubMed] [Google Scholar]
- 35.Yu-Wai-Man P, Griffiths PG, Hudson G, Chinnery PF. Inherited mitochondrial optic neuropathies. J Med Genet. 2009;46:145–158. doi: 10.1136/jmg.2007.054270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Triepels RH, Loeffen JL, Buskens CA, Smeets RJ, Rubio Gazalbo ME, et al. Leigh syndrome associated with a mutation in the NDUFS7 (PSST) nuclear encoded subunit of complex I. Ann Neurol. 1999;45:787–790. doi: 10.1002/1531-8249(199906)45:6<787::aid-ana13>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 37.El-Hattab A, Scaglia F. Mitochondrial DNA depletion syndromes: Review and updates of genetic basis, manisfestations, and therapeutic options. Neurotherapeutics. 2013;10:186–198. doi: 10.1007/s13311-013-0177-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chow SL, Rooney ZJ, Cleary MA, Clayton PT, Leonard JV. The significance of elevated CSF lactate. Arch Dis Child. 2005;90:1188–1189. doi: 10.1136/adc.2005.075317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Debray FG, Mitchell GA, Allard P, Robinson BH, Hanley JA, et al. Diagnostic accuracy of blood lactate-to-pyruvate molar ration in the differential diagnosis of congenital lactic acidosis. Clin Chem. 2007;53:916–921. doi: 10.1373/clinchem.2006.081166. [DOI] [PubMed] [Google Scholar]
- 40.Yamada K, Toribe Y, Yanagihara K, Mano T, Akagi M, et al. Diagnostic accuracy of blood and CSF lactate in identifying children with mitochondrial diseases affecting the central nervous system. Brain Dev. 2012;34:92–97. doi: 10.1016/j.braindev.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 41.Clarke C, Xiao R, Place E, Zhang Z, Sondheimer N, et al. Mitochondrial respiratory chain disease discrimination by retrospective cohort analysis of blood metabolites. Mol Genet Metab. 2013;110:145–152. doi: 10.1016/j.ymgme.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walliman T, Dolder M, Schlattner U, Eder M, Hornemann T, et al. Some new aspects of creatine kinase (CK): compartmentation, structure, function and regulation for cellular and mitochondrial bioenergetics and physiology. Biofactors. 1998;8:229–234. doi: 10.1002/biof.5520080310. [DOI] [PubMed] [Google Scholar]
- 43.Shaham O, State NG, Goldberger O, Xu Q, Ramanathan A, et al. A plasma signature of human mitochondrial disease revealed through metabolic profiling of spent media from cultured muscle cells. Proc Natl Acad Sci USA. 2010;107:1571–1575. doi: 10.1073/pnas.0906039107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boenzi S, Martinelli D, Carrozzo R, Piemonte F, DiCiomma V, et al. Plasma creatine is elevated in mitochondrial disorders: a new biomarker for the diagnosis. J Inherit Metab Dis. 2011;34:S49–S286. [Google Scholar]
- 45.Pajares S, Arias A, García-Villoria J, Briones P, Ribes A. Role of creatine as biomarker of mitochondrial diseases. Mol Genet Metab. 2013;108:119–124. doi: 10.1016/j.ymgme.2012.11.283. [DOI] [PubMed] [Google Scholar]
- 46.Mancuso M, Orsucci D, Coppedè F, Nesti C, Choub A, et al. Diagnostic approach to mitochondrial disorders: the need for a reliable biomarker. Curr Mol Med. 2009;9:1095–1107. doi: 10.2174/156652409789839099. [DOI] [PubMed] [Google Scholar]
- 47.Suomalainen A. Fibroblast growth factor 21: a novel biomarker for human muscle-manifesting mitochondrial disorders. Expert Opin Med Diagn. 2013;7:313–317. doi: 10.1517/17530059.2013.812070. [DOI] [PubMed] [Google Scholar]
- 48.Kharitonenkov A, Shiyanova TL, Koester A, Ford AM, Micanovic R, et al. FGF-21 as a novel metabolic regulator. J Clin Invest. 2005;115:1627–1635. doi: 10.1172/JCI23606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reitman ML. FGF21: a missing link in the biology of fasting. Cell Metab. 2007;5:405–407. doi: 10.1016/j.cmet.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 50.Chen WW, Li L, Yang GY, Li K, Qi XY, et al. Circulating FGF-21 levels in normal subjects and in newly diagnosed patients with type 2 diabetes mellitus. Exp Clin Endocrinol Diabetes. 2008;116:65–68. doi: 10.1055/s-2007-985148. [DOI] [PubMed] [Google Scholar]
- 51.Zhang X, Yeung DC, Karpisek M, Stejskal D, Zhou ZG, et al. Serum FGF21 levels are increased in obesity and are in independently associated with the metabolic syndrome in humans. Diabetes. 2008;57:1246–1253. doi: 10.2337/db07-1476. [DOI] [PubMed] [Google Scholar]
- 52.Tyynismaa H, Mjosund KP, Wanrooij S, Lappalainen I, Ylikallio E, et al. Mutant mitochondrial helicase Twinkle causes multiple mtDNA deletions and a late-onset mitochondrial disease in mice. Proc Natl Acad Sci USA. 2005;102:17687–17692. doi: 10.1073/pnas.0505551102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tyynismaa H, Carroll CJ, Raimundo N, Ahola-Erkkila S, et al. Mitochondrial myopathy induces a starvation-like response. Hum Mol Genet. 2010;19:3948–3958. doi: 10.1093/hmg/ddq310. [DOI] [PubMed] [Google Scholar]
- 54.Badman MK, Pissios P, Kennedy AR, Koukos G, Flier JS, et al. Hepatic fibroblast growth factor 21 is regulated by PPARalpha and is a key mediator of hepatic lipid metabolism in ketotic states. Cell Metab. 2007;5:426–437. doi: 10.1016/j.cmet.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 55.Suomalainen A, Elo JM, Pietiläinen KH, Hakonen AH, Sevastianova K, et al. FGF-21 as a biomarker for muscle-manifesting mitochondrial respiratory chain deficiencies: a diagnostic study. Lancet Neurol. 2011;10:806–818. doi: 10.1016/S1474-4422(11)70155-7. [DOI] [PMC free article] [PubMed] [Google Scholar]