Chemical gradients shape the cystic fibrosis lung microbiome, with marked effects on the outcome of antimicrobial treatments.
Abstract
Environmental microbial communities are stratified by chemical gradients that shape the structure and function of these systems. Similar chemical gradients exist in the human body, but how they influence these microbial systems is more poorly understood. Understanding these effects can be particularly important for dysbiotic shifts in microbiome structure that are often associated with disease. We show that pH and oxygen strongly partition the microbial community from a diseased human lung into two mutually exclusive communities of pathogens and anaerobes. Antimicrobial treatment disrupted this chemical partitioning, causing complex death, survival, and resistance outcomes that were highly dependent on the individual microorganism and on community stratification. These effects were mathematically modeled, enabling a predictive understanding of this complex polymicrobial system. Harnessing the power of these chemical gradients could be a drug-free method of shaping microbial communities in the human body from undesirable dysbiotic states.
INTRODUCTION
Since the time of Sergei Winogradsky (c. 1880s), environmental microbiologists have shown that chemical parameters, such as pH, oxygen, and redox state, shape the structure and function of microbial communities (1, 2). Chemical gradients create unique niche spaces that can be colonized by particular microbes, resulting in a stratified microbial ecosystem that responds uniquely to environmental change (3, 4). Microbial communities that cause chronic infections of the human airway, skin, gut, and other organs or surfaces are not too dissimilar in their responses to these factors from those found in the environment, but how chemical gradients shape the niche partitioning and physiology in an infectious context has not been extensively investigated. Instead, research has focused on identifying virulence mechanisms of individual bacterial pathogens within the community. For example, although the cystic fibrosis (CF) lung microbiome has been extensively characterized (5) and virulence mechanisms of its principle pathogens have been studied for decades (6, 7), little is known about what drives community interactions associated with disease (5, 8).
The importance of chemical gradients in shaping human microbial communities has become particularly pertinent in CF lung disease. Microenvironments are known to exist in different regions of the lung, resulting in microbial and chemical heterogeneity (9), and various chemical gradients have been measured in vivo (10, 11). Recently, a study of the microbial ecology of the CF microbiome showed that a shift in community physiology to anaerobic fermentation occurred during acute flares of CF disease called pulmonary exacerbations (CFPEs) (12). CFPEs are a detrimental event in the lives of CF patients contributing significantly to their morbidity and mortality, but they have a poorly understood etiology (13, 14). The Climax and Attack model of CF microbial ecology proposes that there are two distinct microbial communities in the CF lung that are dynamically changing during CFPEs (15, 16). The subsequent observation of microbial physiology changes associated with these events led to the hypothesis that chemical gradients may play a role in the dynamics of the lung microbiome. Therefore, in this study, we explore competition outcomes between constituents of the CF microbiome in chemical gradients using an experimental microcosm of a mucus-plugged bronchiole called the WinCF system (12). WinCF is based on the Winogradsky column [a century-old method for studying microbial communities in soil and sediment (2, 17)]. Principles of the Winogradsky column are maintained, including the ability to obtain visual and quantitative output of microbial behavior through chemical gradients, but WinCF is unique in that it contains media mimicking lung mucus in a thin cylindrical tube that is similar in diameter to lung bronchioles (18). Here, we show that pH and oxygen gradients strongly partition the CF microbiome into two communities favoring opposing niche space. These gradients govern competition outcomes between community constituents, have effects on death and resistance to antimicrobial therapy, and can be harnessed to predictably manipulate the community with support from mathematical modeling.
RESULTS
Microbial community changes in WinCF pH gradients
To test the effects of pH on the CF microbiome, sputum samples from patients (n = 18; table S1) were grown in a WinCF pH gradient from 5.0 to 8.5 (fig. S1). The artificial sputum medium (ASM) used for the WinCF media was buffered to a gradient of pH, and sputum was mixed with the media before inoculation via capillary action into the WinCF columns. Phenol red and bromocresol purple dyes were added to monitor the initial pH gradient and any changes due to growth. Growth of the sputum microbiome resulted in the production of gas bubbles and changing of the medium pH from its initial starting point (Fig. 1, A and B). Gas production, an indicator of microbial fermentation in WinCF (12), occurred in all columns and was most abundant at pH 6.5 (Fig. 1A) but was reduced at elevated pH [P < 0.05, analysis of variance (ANOVA); table S2 and Fig. 1A]. For the CF samples, the gas bubble production at pH 8.0 and 8.5 was significantly lower than at pH 5.5, 6.0, and 6.5 (one-way ANOVA, P < 0.05; Fig. 1 and table S2). Quantitation of changes in the WinCF pH indicator dyes enabled an assessment of the influence of the initial pH gradient on subsequent culture pH after growth. At the two highest pH starting points, the WinCF pH indicator dye did not change, demonstrating that, when the CF microbiome was grown in high pH, it maintained that pH, which is a trend not evident in control samples (Fig. 1B and table S2). Therefore, as described above, low gas production was associated with the maintenance of a high-pH environment (Fig. 1, A and B, and table S2).
Fig. 1. WinCF microbial physiology and diversity changes in response to pH and oxygen gradients.
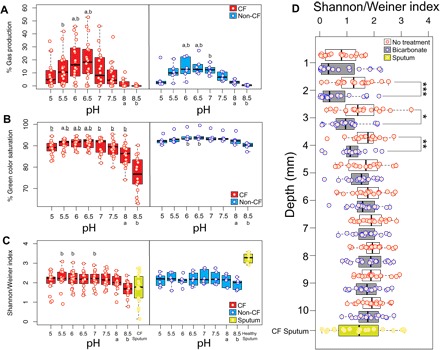
Boxplots of the amount of gas produced (A), change in green color saturation (B), and Shannon-Weiner diversity index (C) in the WinCF capillary columns through the pH gradient in both CF and non-CF sputum samples. Letters above the boxplot denote significant differences between the 8 and 8.5 pH samples. (D) Boxplot of the Shannon-Weiner index at each millimeter depth through the WinCF capillary columns with and without bicarbonate treatment. Sputum samples are shown as a reference when applicable. ***P < 0.001, **P < 0.01, *P < 0.05 from Mann-Whitney U test.
Microbial community sequencing of the WinCF tubes using 16S rRNA gene amplification allowed for an assessment of the taxa associated with these chemically induced changes. The high-pH and low–gas production community had a less diverse microbiome (Fig. 1C), and accordingly, there was a positive correlation between microbial diversity and gas production (Pearson’s r = 0.475, P < 0.001). Microbes associated with high gas production included Veillonella, Streptococcus, Prevotella, Atopobium, and Bulledia, a consortium previously shown to co-occur in vivo (16). A random forests (RF) linear model was used to determine whether the microbiome data were reflecting the pH of the initial starting culture. This model was able to accurately predict the starting pH on the basis of the microbiome data, thus indicating that the pH gradient had a highly significant effect on the community structure (Fig. 2A and fig. S2). This finding was verified by the balance trees approach to niche partitioning in microbiome data, which accounts for challenges with data compositionality (fig. S3, A to C) (19). The most diverse community was present at pH 6.5, whereas the pH extremes were occupied by fewer taxa, particularly higher pH (fig. S3, B and C). To determine the effects of pH on particular bacterial genera, we applied a generalized linear mixed model (GLMM) to test the distribution of the most abundant microbes through the pH gradient. Pseudomonas (P = 0.02), Haemophilus (P < 0.0001), Achromobacter (P = 0.0001), and the anaerobe Fusobacterium (P < 0.0001) were positively correlated with increasing pH, whereas the anaerobic genera Veillonella (P < 0.0001), Prevotella (P < 0.0001), Actinomyces (P < 0.0001), and Streptococcus (P < 0.0001) were strongly negatively correlated (Fig. 2B). Thus, the diverse low-pH community was primarily composed of fermentative anaerobes commonly observed in microbiome profiles from CF sputum. In contrast, the high-pH group comprised facultative anaerobes and organisms more commonly considered pathogenic. RF linear models and GLMM were also used to identify microbes associated with elevated gas production. Again, the community was partitioned into anaerobes associated with high gas production and pathogens with low gas production (Supplementary Materials). Thus, the pH gradient markedly altered the physiology and diversity of the CF microbiome, revealing the presence of two physiologically different communities in the sputum samples: one, a low-diversity community of microorganisms that when placed in a high-pH environment maintained that high pH and did not ferment, and the other, a high-diversity community of bacteria that lowered the pH of their environment and produced abundant gas from fermentation.
Fig. 2. Effect of pH and oxygen gradients on microbial community in WinCF system.
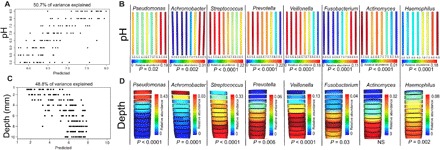
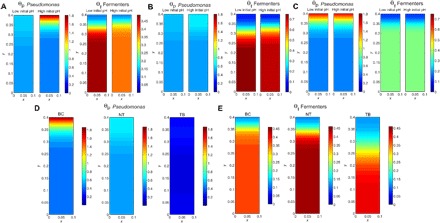
RF linear models (A and C) and bacterial genus distributions (B and D) in the WinCF pH and oxygen experiments. An RF linear model was run on the (A) pH and (C) depth of each 16S rDNA microbiome profile from the WinCF experiments. The model predicts the pH or depth of a sample on the basis of the microbiome data, providing a measure of the strength of the overall microbial changes through the gradients. The percent variance of the data in the model is also shown. The distributions of bacterial genera of interest are visualized for pH (B) and oxygen (D) using a visual depiction of the WinCF system experiments generated with the ‘ili software (45). The color scale for each WinCF tube represents the relative abundance of that particular operational taxonomic unit (OTU) and is shown for each panel, as they are not all at the same scale. In the oxygen experiment, each layer represents 1 mm into the WinCF media, and in the pH experiment, each column represents the WinCF columns grown in the pH gradient with 0.5 units from 5 to 8.5. The normalized abundance of bacterial genera of interest are shown as a heat map, with red being the most abundant and blue being the least abundant. The P value from the LMM for each genus in each gradient is shown.
Microbial community changes in WinCF oxygen gradients
An oxygen microelectrode showed that the WinCF columns contained steep oxygen gradients similar to those measured in the lung (10) and sputum (fig. S4) (11). This enabled analysis of the CF microbiome through an oxygen gradient similar to the pH experiments, by first gelling the media and sectioning the WinCF cultures into 1-mm sections from 1- to 10-mm depth after the growth of the sputum samples (n = 19; figs. S1 and S5). Although an RF linear model showed that the microbial community was strongly shaped by depth, there was little change after 6 mm (Fig. 2C). Microbial community diversity increased with increasing vertical depth (Pearson’s r = 0.472; Fig. 1D), mirroring the results of the pH experiments, where the chemical gradient partitioned the community into low diversity and high diversity. The CF community separated into microbes with a niche in the upper layers of the mucus (1 to 4 mm), the middle layers of the mucus (4 to 7 mm), and the deepest layers of the mucus (7 to 10 mm; fig. S3, E and F). The genera were normally distributed around a depth of 4.5 mm, with few bacteria exclusively preferring the aerobic layers (1 to 2 mm). Pseudomonas was especially affected by the oxygen gradient, being strongly negatively correlated with depth (P < 0.0001; Fig. 2D). Many anaerobes increased their relative abundance with depth including Streptococcus (P < 0.0001), Prevotella (P = 0.006), Veillonella (P < 0.0001), Fusobacterium (P = 0.03), and the facultative anaerobe Haemophilus (P = 0.002; Fig. 2D).
To assess the combined effect of pH and oxygen on the CF microbiome, we added bicarbonate to the aerobic interface of the WinCF columns. HCO3− was added to the top of the media to mimic the inhaled therapy of this ion, which is being developed as a treatment for CF to alleviate its decreased transport in the lung epithelium (20). Bicarbonate further decreased the community diversity, but only in the 2-, 3-, and 4-mm depths (P < 0.05, Mann-Whitney U test; Fig. 1D). The heightened effect of the pH and oxygen in combination demonstrates the potential to manipulate a polymicrobial infection with simple chemistry.
Metabolite production changes in chemical gradients
The chemical gradients also affected community physiology as determined by tandem liquid chromatography–mass spectrometry (LC-MS/MS) and gas chromatography–mass spectrometry (GC-MS) metabolomics. The metabolomes were highly affected by the pH, gas production, and oxygen variables (fig. S2). One of the most strongly affected metabolites was a by-product of microbial tryptophan metabolism: indoleacetate. This molecule was positively correlated to gas production (LMM, P < 0.0001; table S3) and associated with a Streptococcus OTU (Spearman’s ρ = 0.775), Bulledia moorei (ρ = 0.694), and Prevotella melaninogenica (ρ = 0.581). We detected the short-chain fatty acids (SCFAs) acetic acid, propionic acid, and butyric acid in both the WinCF and sputum samples, indicating that the culture system induced similar metabolic activity to that in vivo. Propionic acid was positively correlated with gas production (LMM, P < 0.0001) and anaerobes (Supplementary Materials), but negatively correlated with pH (P < 0.0001) and Pseudomonas (Pearson’s r = −0.416, P < 0.001). Butyric acid was positively correlated with pH (P = 0.005). Acetic acid was not correlated with either variable.
Pseudomonas aeruginosa produces a variety of specialized metabolites, many of which act as antimicrobials and virulence factors, including quinolones, phenazines, rhamnolipids, and the siderophore pyochelin (Fig. 3, fig. S6, and table S4). Production of these molecules was also influenced by the chemical gradients. Phenazine-1-carboxylic acid (PCA) was positively correlated with pH (P = 0.0005) and pyocyanin (PYO; P = 0.01), and the rhamnolipid Rha-Rha-C10-C10 (P = 0.01) were negatively correlated with gas production. The oxygen gradient markedly altered the production of these virulence factors, as they all decreased in abundance through the WinCF column depth, but some were more strongly affected than others (Fig. 3 and table S4). An LMM showed that the most strongly affected metabolites were 2-nonyl-4-hydroxyquinolone (NHQ; P < 0.0001), PYO (P < 0.0001), PCA (P = 0.0001), pyochelin (P < 0.0001), Rha-Rha-C10-C10 (P < 0.0001), 2-heptyl-4-hydroxyquinolone-N-oxide (P = 0.0003), 2-heptyl-4-hydroxyquinolone (HHQ; P = 0.0003), and 2-heptyl-3-hydroxy-4-quinolone (PQS; P = 0.0003; Fig. 3). PQS was most abundant in the first two oxic millimeters and dropped in abundance drastically with depth, reflecting its association with molecular oxygen (21). Phenazines, which are redox-active antibiotics produced by P. aeruginosa and many other bacteria (22), were depleted in the anoxic layers, although PCA was less affected than PYO. PYO was also less abundant when gas production was highest in the pH experiments.
Fig. 3. Molecular networks of P. aeruginosa virulence factors and their abundance through depth in the WinCF columns.
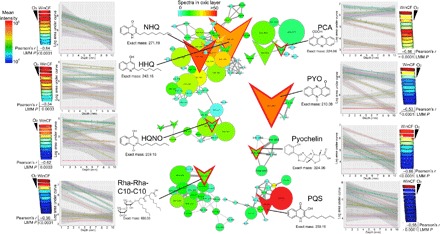
The data are mapped onto a model of the WinCF oxygen experiment using the ‘ili software (45). The log mean intensity of each P. aeruginosa virulence metabolite through the 1- to 10-mm-depth sections of the WinCF media is shown as a heat map with the oxygen penetration shown for reference. The Pearson’s r value of the correlation with depth is shown as well as the P value from the LMM. Plots of the log area under the curve compared to depth in the WinCF media are shown colored by each patient for each metabolite of interest. The molecular networks show the metabolites of interest and their related molecules. Each node represents a unique MS/MS spectrum from the molecular networking algorithm, and edges between the nodes represent a cosine similarity between them above 0.7. Colors of the nodes are scaled to their abundance in the oxic layer of the WinCF columns (1 mm) in spectral counts, and the node size is scaled to the overall abundance of the metabolite. Structures and mass in daltons of each virulence metabolite are shown. HQNO, 2-heptyl-4-hydroxyquinolone-N-oxide.
Effects of inhaled therapies on a chemically stratified microbial community
Because of the strong impact of chemical gradients on the CF microbiome, we attempted to determine whether the stratification would affect the results of inhaled CF treatments. Sodium bicarbonate and the anti-Pseudomonas antibiotic tobramycin were added to the top of the culture in separate WinCF columns (n = 19) before growth to simulate inhalation into an airway with mucus-plugged bronchioles. Bicarbonate increased Pseudomonas abundance in the aerobic and microaerophillic layers and decreased the abundance of Veillonella (Fig. 4 and fig. S7). Tobramycin induced drastic changes in the microbial community structure through the oxygen gradient. The antibiotic killed Pseudomonas and Veillonella in all regions of the column. Streptococcus was killed in the oxic layers but survived below a depth of 5 mm, while Prevotella bloomed below this depth. Stenotrophomonas thrived in the presence of tobramycin, and reads mapping to the mitochondrion of the fungus Aspergillus were also detected in the oxic layers (Fig. 4). Thus, tobramycin inhibited the growth of many bacteria normally found in the upper layers of the mucus and left niche space for resistant organisms to flourish. Many anaerobes survived in the deeper anoxic layers of the mucus because the antibiotic existed in a gradient of concentration unable to reach them at depth (verified in LC-MS/MS data; fig. S8). This shows that the inherent community stratification due to the effects of oxygen had a profound effect on the tobramycin treatment (fig. S7). This phenomenon creates the potential for the development of resistance due to lower drug concentrations deeper in lung mucus.
Fig. 4. The 3D models of the WinCF oxygen experiments with treatment visualized using the ‘ili software (45).
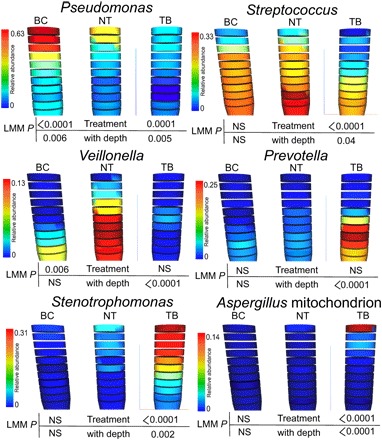
Each column represents the WinCF medium that is sectioned in 1-mm depths. The sections are colored according to the normalized abundance of bacterial genera of interest. The untreated samples (NT), bicarbonate (BC)–treated samples, and tobramycin (TB)–treated samples are shown, as well as the LMM P values with and without the depth as a mixed effect. NS, not significant.
Unexpectedly, tobramycin was of very low abundance after community growth compared to controls and undetectable in 15 of the 19 patients’ WinCF columns. Evidence of propionylation in the metabolomics data indicates that this scarcity was due to aminoglycoside modifying enzymes from CF bacteria changing the molecule (23). Although various acetylations have been reported on tobramycin as a mechanism of inactivation (23), in our data, we only detected the uncommon propionylation of the middle saccharide of tobramycin (fig. S9) (24). Thus, tobramycin that was directly added to the WinCF columns was being altered by the CF microbiome through aminoglycoside-modifying enzymes. Other modifications were likely occurring because the compound was scarce in the LC-MS data, but the specific mechanisms of this metabolism remain unknown. When intact tobramycin was detected and in high abundance, it corresponded to samples that contained Aspergillus in the oxic layers (r = 0.577, P < 0.001; Fig. 5, A and B), and in these same samples, metabolites from Aspergillus fumigatus were detected (Fig. 5C and fig. S10). The abundance of these metabolites through the WinCF gradient matched the relative abundance of Aspergillus mitochondrion reads. Collectively, these experiments show the complexity of the microbial response to antibiotic treatments, which include resistance mechanisms and complex death and survival responses for individual microbes.
Fig. 5. Profiles of Aspergillus mitochondrion reads, tobramycin abundance, and Aspergillus specialized metabolite abundance through the WinCF oxygen gradients in the 19 patients tested in this study.
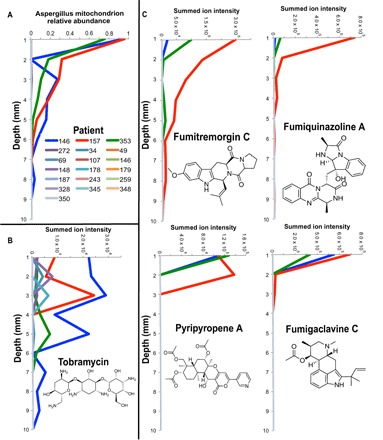
(A) Relative abundance of 16S rDNA gene reads in each patient mapping to the Aspergillus mitochondrion in tobramycin-treated WinCF tubes through the 10-mm depth. (B) Summed ion intensity of tobramycin in each patient through the WinCF 10-mm depth treated with tobramycin. (C) Spectral counts of A. fumigatus specialized metabolites in same samples.
Predictive mathematical modeling of CF microbiome in chemical gradients
The predictability of the anaerobe and pathogen response in the WinCF columns enabled mathematical modeling of their behavior through pH and oxygen gradients and a determination of its most significant drivers. Modeling allows testing, in a simplified setting without confounding factors, of hypotheses relating chemical and community structures that are not easily assayed in the laboratory. The model includes two microbial communities, P. aeruginosa and fermentative anaerobes, with their populations denoted by θp and θf, respectively. Physiological parameters in the model are sourced from literature describing P. aeruginosa’s behavior in the CF lung (see Supplementary Materials). The pathogen is facultatively anaerobic, raises the pH of its environment, and produces antimicrobial metabolites. Fermenters consume sugar, produce acid, and are killed by oxygen (12, 25). In accordance with our experimental results, the contours of θf and θp, when all quantities reached steady state, show that the growth of fermenters was restricted to the anaerobic region and was better in the low–initial pH case (Fig. 6A). P. aeruginosa had limited growth in low pH but thrived in the aerobic region in high pH. Thus, at different mucus pH, the carrying capacity of a mucus-plugged bronchiole for either P. aeruginosa or fermentative anaerobes is starkly different. This matched the experimental results and supports the mutually exclusive dynamic between anaerobes and P. aeruginosa observed in other studies (12, 16). Removal of P. aeruginosa’s pH dependence or production of antimicrobial factors from the model revealed that these parameters were crucial for the results observed (Fig. 6, B and C). Thus, the strong niche partitioning from the pH experiments may be due to a favored pH range for each organism. Each bacterium alone can grow in a wider range of pH, but when in competition with others, the community strongly partitioned. Incorporating bicarbonate and tobramycin treatments into the model mirrored our experimental results. Bicarbonate raises the pH of the environment, causing P. aeruginosa to bloom, with little effect on the anaerobes (Fig. 6, D and E). Tobramycin kills P. aeruginosa and slightly inhibits the anaerobes (fig. S11).
Fig. 6. Results of mathematical model simulations on the CF microbiome response to pH.
(A) Contours of P. aeruginosa θp and the fermentative anaerobe θf growth in the WinCF capillary column environment when started at low or high pH. The aerobic portion of the column is at the top of the figure, and the anaerobic portions are at the bottom. Depth, width, and intensity of growth parameters are on unitless scales. (B) Contours of θp and θf given by the simulation with the effect of inhibition chemical I turned off (set β1 = 0 in the model); therefore, in this case, P. aeruginosa has no effect on the growth of fermenters. (C) Contours of θp and θf with the effect pH on the growth of P. aeruginosa turned off (set β2 = 0 in the model). In this case, there is no difference between the low and high initial pH as expected. Contours of (D) θp and (E) θf resulting from the simulated treatment of bicarbonate (BC) and tobramycin (TB) from the top portion of the media are compared to the untreated case (NT). These results demonstrate the predictability of the community response to inhaled CF treatments.
DISCUSSION
Antibiotic treatment regimens against polymicrobial infections target pathogens cultured from clinical samples. This approach is naïve to the fact that these pathogens are only a single member of a complex community, and treatment outcomes are complicated by species interactions and the chemical conditions that shape them. Here, we demonstrate that a pathogenic community from the human airway is structured in a laboratory microcosm by pH and oxygen gradients, which are chemical conditions commonly encountered in chronic infections. These gradients affect community metabolism, virulence factor production, treatment, and resistance. Similar chemical gradients are believed to structure the human gut microbiome during health and disease (26), suggesting that the influence of this chemistry is centrally relevant in other organ systems. This finding mirrors the basic dogma of environmental microbiology that chemical gradients drive the ecology and physiology of microbial communities in soil, sediment, and other microbial ecosystems.
pH and oxygen gradients partitioned the CF microbiome into two mutually exclusive and fundamentally different microbial communities: a diverse community of fermentative anaerobes preferring lower pH and anoxia and a nonfermentative group of classic pathogens preferring high pH and high oxygen. This partitioning was consistent across individuals despite each patient having a unique microbiome. These two communities represent those of the Climax and Attack model of CF microbial ecology proposed to be important to pulmonary exacerbations (15, 16). Recent studies have identified signatures of microbial fermentation and the expansion of anaerobes during CFPEs (12, 27, 28). This study shows that the lung microbiome can be shifted to favor a fermentative community through lowering pH and decreasing oxygen. The dominance of anaerobic bacteria in a low-oxygen mucus environment was somewhat surprising because P. aeruginosa is known to flourish in anaerobic environments by nitrate reduction in vitro and in vivo (29, 30). In the WinCF tubes, P. aeruginosa growth by denitrification was outcompeted by anaerobes, even when nitrate was available in the WinCF medium (31). Thus, a chemically driven dysbiosis that favors anaerobic fermentation may explain the dominance of these organisms during exacerbations and the sometimes extremely low pH values measured in CF sputum (11, 12, 28). Although the causes of CFPE remain mostly unknown, they continue to be a serious clinical problem facing physicians. Chemical shifts that favor fermentation and acid production could result in a positive feedback loop, further favoring an anaerobe-dominated community and a further drop in pH. Strong community shifts may be the underlying cause of CFPEs, but further research is needed to confirm this possibility and to determine whether microbial community changes are a consequence of a developing CFPE or its fundamental cause. An important context for this study is the fact that, although the WinCF system is meant to mimic a CF bronchiole, it does not contain host cells or components of the inflammatory response that also shape the makeup and physiology of the CF microbiome. Further research into the role that lower lung pH has on both the microbial community and the host response (32) will be crucial to understand the causes of CFPEs.
The chemical gradients also altered aspects of community metabolism through the production of indoles and organic acids. In the case of the latter, lowering lung mucus pH favored anaerobic fermentation and the production of propionic acid. In the lung, this could contribute to a further decrease in the airway surface liquid pH and the low pH of sputum observed in other studies (11). P. aeruginosa has been shown to feed on propionic acid produced by anaerobes in vitro and express genes for its metabolism in vivo (25). The bacterium’s negative correlation with this SCFA may therefore be due to its metabolism of this compound during cross-feeding with anaerobes (25). Cross-feeding in such a system may further connect the Climax and Attack communities and potentially worsen lung infections if P. aeruginosa is stimulated by the by-products of anaerobic fermentation (27).
Small-molecule virulence factors from P. aeruginosa were also altered by the chemical gradients. Phenazines, quinolones, rhamnolipids, and the siderophore pyochelin all decreased with depth in the WinCF tubes. While this effect is not unexpected considering the decreased abundance of P. aeruginosa in the anoxic layers, some metabolites were more strongly affected than others. The P. aeruginosa quorum-sensing metabolite PQS was highly abundant in the oxic layers (1 to 2 mm) and then quickly depleted. This pattern likely reflects the requirement of molecular oxygen for its synthesis (21) and explains its absence in LC-MS/MS profiles of CF sputum (33). HHQ, NHQ, and PCA, however, reached far into the anoxic mucus layers, where P. aeruginosa itself was not detected. This finding demonstrates that some virulence metabolites and quorum-sensing molecules from the bacterium can readily diffuse through a mucus-filled environment, while others are more static. PYO was depleted in the anoxic layers and less abundant when gas production was highest in the pH experiments. This indicates that PYO production, believed to be important for P. aeruginosa’s competitive advantage in the CF lung, is diminished or inhibited when grown in competition with anaerobic bacteria. The altered production of virulence metabolites through oxygen and pH gradients shows that the action of these molecules, including their inhibitory effect on host and microbial cells (34, 35), may be overwhelmed by simple changes in oxygen and pH and through competition with other bacteria in the airway.
Particularly relevant to clinical treatments against CF infections, chemical gradients altered antibiotic treatment outcomes against pathogens in the WinCF lung-bronchiole model system. Simulated tobramycin treatment drastically altered the community structure. The drug killed aerobically growing Pseudomonas in the upper layers, a physiological susceptibility that has been previously described (36), but surprisingly, tobramycin also induced a bloom of Stenotrophomonas and the fungus Aspergillus in its place. Stenotrophomonas maltophilia is highly resistant to tobramycin (37) and many other antibiotics (38). These experiments show that inhalation of tobramycin may induce a bloom of resistant S. maltophilia and possibly contribute to the increasing prevalence of this pathogen in CF patients (38). The effect of tobramycin on the growth of Aspergillus, a common aerobic fungal pathogen (39), was particularly remarkable. It demonstrates that antibiotic treatment aimed at killing bacterial pathogens may have unwanted consequences, including the growth of other more resistant pathogens and fungi. Furthermore, the inherent stratification of the CF microbiome induced by oxygen had a direct effect on tobramycin treatment. Many anaerobes, including Prevotella and Streptococcus, survived, but only in the deeper anoxic layers of the mucus because the antibiotic existed in a gradient of concentration unable to reach them at depth. This phenomenon also creates the potential for the development of resistance as the limited drug penetration results in lower concentrations deeper in the mucus. Further work on the interaction between tobramycin, Aspergillus spp., and CF bacteria (40) is needed to better understand the dynamics of this inhaled treatment in CF lungs. Collectively, these results demonstrate that, because of the oxygen stratification of the community, antibiotic treatment against bacterial pathogens can have complex and potentially undesirable consequences. These consequences include the growth of more resistant bacteria, the survival of species with anaerobic niche space, the development of unique mechanisms of resistance, and the growth of fungi that replace the niche previously occupied by the targeted pathogen, the latter being a phenomenon observed in vivo (41).
Because of the strong effects of pH and oxygen gradients observed in this study, we attempted to manipulate the CF microbiome in a predictable manner by simulating inhaled bicarbonate treatment that has a direct effect on lung pH. In clinical trials to treat consequences of cystic fibrosis transmembrane conductance regulator (CFTR) dysfunction, bicarbonate lowered community diversity by inducing a P. aeruginosa bloom. This mimicked the effect of our initial pH experiment that showed that the pathogen favors a high-pH niche space. This effect only occurred in the microaerophillic regions of the WinCF tubes, indicating that inherent effects of oxygen stratification also impart an effect on pH manipulation. This led to our attempt to mathematically model the CF microbiome competitive interactions in this environment and how they are affected by these treatments. Our model validated the hypothesis that P. aeruginosa and fermentative anaerobes partition into separate communities driven by the initial mucus pH, oxygen, and products of their separate metabolisms. The model showed that at extremes of mucus pH the carrying capacity of a mucus-plugged bronchiole for either P. aeruginosa or fermentative anaerobes is starkly different. At high initial pH, P. aeruginosa dominates the aerobic portion of the tubes, but at low initial pH, fermentative anaerobes thrive, but only in the anaerobic regions. This finding was mirrored in our experimental results and supports the mutually exclusive dynamic between anaerobes and P. aeruginosa observed in other studies (12, 16). Mechanisms within the model were then iteratively removed to determine which were the most responsible for the community partitioning. P. aeruginosa’s inherent preference for a higher pH and its production of antimicrobial factors were crucial for the experimental results observed. Thus, these two factors should be further explored when investigating the dynamics surrounding this pathogen and anaerobic bacteria in CF lungs, especially as it relates to exacerbations. Collectively, the results of our bicarbonate experiments and mathematical modeling show that care should be taken when developing this compound as an inhaled therapy for CF due to the potential for P. aeruginosa stimulation. However, inhaled bicarbonate in combination with antibiotics such as tobramycin may be an advantageous means of selecting for the growth of P. aeruginosa, enabling the antibiotic to more effectively kill it. Combinatorial therapies that pair the manipulative power of basic chemistry, such as pH and oxygen, with targeted antimicrobials may be a novel approach to antimicrobial therapy allowing for more targeted killing on the basis of a fundamental understanding of the microbial ecology of a system.
In sum, these experiments show that the same chemical influences long known to govern microbial community structure in environmental systems also strongly shape microbial communities in the human body. The effect of pH and oxygen gradients on microbial community structure has direct implications for competition outcomes, virulence factor production, antibiotic treatment efficacy, and resistance. It may be possible to take advantage of the overwhelming influence of these simple chemical parameters to manipulate a pathogenic or dysbiotic microbiome to an appropriate state that enables more targeted therapy or the maintenance of a healthier microbial community composition.
MATERIALS AND METHODS
Sample collection
Samples were collected from the University of California San Diego (UCSD) adult CF clinic during routine visits from patients. Sputum samples were expectorated into a sputum cup and transported back to the laboratory within 3 hours of collection. All samples were collected via the UCSD institutional review board–approved protocol for human subject research (project #160078). A total of 18 sputum samples were collected from CF patients for the pH experiments and another 19 for the oxygen experiments (table S1). In addition, six sputum samples were collected from healthy volunteers after saline mouth rinse and expectoration of airway mucus as a comparison for the pH experiments. Samples were split into four aliquots: one fresh for WinCF culturing and three aliquots frozen at −80°C for 16S ribosomal RNA (rRNA) gene sequencing and LC-MS/MS and GC-MS metabolomics.
WinCF chemistry experiments
For the pH experiments, ASM was prepared according to (12, 42) and then split into eight different stocks that were buffered to a pH gradient from 5 to 8.5 (0.5 pH unit intervals) with a phosphate buffer concentration of 50 mM. The desired pH for each interval was achieved by adding the appropriate amount of monosodium phosphate and disodium phosphate and then bringing the media to the appropriate pH using HCl or NaOH. The pH dyes phenol red and bromocresol purple were added to the media to aid in pH gradient visualization (fig. S1A). After homogenization, 50 μl of each sputum sample was inoculated into 250 μl of ASM media at eight different pH intervals within 6 hours of sample collection. This mixture was vortexed and inoculated into capillary columns (nonheparinized Fisherbrand and Microhematocrit capillary columns, Fisher Scientific) in triplicate, as described in (12). All WinCF columns were incubated at 37°C for 48 hours, and then the medium from each triplicate capillary column was removed and pooled. Three separate 50-μl aliquots of this pooled mixture were then prepared for 16S rRNA gene sequencing and metabolomics analysis of nonvolatile compounds using LC-MS/MS and volatiles using GC-MS. The oxygen experiments were done using the same medium formulation, but a 0.5% agar solution was mixed with the medium before inoculation (fig. S1B). The WinCF columns were created by drawing the warm molten media and agar into a 1000-μl pipette tip, sealing the pointed end while leaving the wide end open to the air and allowing it to cool. Three columns were created for each of the 19 sputum samples acquired, the first left untreated, the second treated with 20 μl of tobramycin (60 mg/ml), and the third being treated with 20 μl of 7% sodium bicarbonate. The treatments were added to the top open portion of the columns before incubation. Control columns with no sputum were generated and analyzed in the same manner. The columns were incubated as described above. After incubation, they were frozen at −80°C and sectioned with a sterile razorblade in 1-mm sections to analyze the microbial and chemical difference through a 10-mm depth using a sterile razorblade. The sections were stored in microcentrifuge columns. For 16S rDNA sequencing, 200 μl of 1× phosphate-buffered saline was added to each column. Extractions for metabolomics were done directly on the sectioned media. Further details for both procedures are available in Supplementary Methods.
16S rRNA gene sequencing
The microbiome of both the sputum samples and the media from the WinCF columns after incubation in the pH and oxygen experiments were analyzed using 16S rRNA gene amplicon sequencing. For the sputum samples, an aliquot of the sample was frozen at −80°C before sequencing in batch with all other samples. DNA extraction, 16S rRNA gene variable region 4 (V4) polymerase chain reaction, and amplicon preparation for sequencing were performed according to protocols benchmarked for the Earth Microbiome Project found here http://www.earthmicrobiome.org/protocols-and-standards/ (see Supplementary Methods).
Metabolomics
For LC-MS/MS analysis, metabolites from both the sputum and WinCF media from the pH and oxygen experiments were extracted using a successive ethyl acetate and methanol extraction procedure as outlined in (33). The metabolite extracts were then diluted fourfold in methanol containing 2 μM glycocholic acid or ampicillin (Fisher Scientific) used as an MS internal standard. The chromatographic separation was conducted on a ThermoScientific UltiMate 3000 Dionex UPLC system (Fisher Scientific) with eluent further analyzed on a Bruker Daltonics MaXis qTOF mass spectrometer (Bruker). Metabolites were separated using a Kinetex 2.6-μm C18 (30 × 2.10 mm) UPLC column. Ratios of water and acetonitrile for mobile phases A and B (98:2 and 2:98, respectively) containing 0.1% formic acid and a linear gradient from 0 to 100% for a total run time of 840 s at a flow rate of 0.5 ml min−1 were used. For further details see Supplementary Methods.
The GC-MS analysis for the pH samples was conducted via headspace sampling with a polydimethylsiloxane/divinylbenzene (PDMS/DVB) df 65-μm solid-phase microextraction (SPME) fiber. A Thermo Scientific TRACE 1310 GC [TG-5ms 5; length, 30 m; ID (inner diameter) , 0.25 mm; film thickness column, 0.25 μm] and a TSQ 8000 EVO mass spectrometer (Thermo Fisher Scientific), equipped with electron ionization (EI) source, were used for the analysis. The frozen samples were transferred into 1.5-ml borosilicate glass vials, capped with silicon septum cap, placed onto tray of the GC-MS instrument, and allowed to thaw. The PDMS/DVB SPME (Supelco) tip was inserted by the sampling robot into the vial, and the sample was heated to 160°C and agitated for 10 min. Upon sample extraction, the SPME was inserted into the GC inlet maintained at 250°C, and the extracted compounds were desorbed for 1 min, cryofocused at −10°C on the head of the column, and then analyzed with a gas chromatographic protocol outlined in Supplementary Methods.
Data processing
Sequencing data were prepared and analyzed using the online tool Qiita (https://qiita.ucsd.edu/). The data were processed with the Deblur algorithm to identify unique taxa in the 16S rDNA amplicons (43). The taxonomic information was collapsed to the genus level for all statistical analyses except regressions between metabolites and bacterial abundance that were analyzed at the deblurred OTU level.
The Bruker LC-MS/MS.d data files were converted to .mzXML format and uploaded to the Global Natural Products Social Molecular Networking database for molecular networking and spectral annotation [GNPS; (http://gnps.ucsd.edu) (44)]. Molecular network were generated using the following parameters: parent and fragment tolerance of 0.2 Da (fragment tolerance of 0.5 Da for oxygen experiment), four minimum matched fragment ions to create a cluster node, minimum cosine score of 0.7, and a minimum cluster size of 2. The GNPS libraries were searched using a minimum cosine of 0.7 and minimum matched fragment ions of 4. Molecular networks were visualized using the Cytoscape software v3.3.0. Annotations from GNPS were shaped as arrowheads in the network to distinguish them from other nodes. The sizes of the nodes were scaled to the total ion count of the cluster. Edges in the network were created using the cosine score between two nodes, and the thickness of the edge was scaled to that value. Links to the pH network and oxygen networks are found here: pH (http://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=1974c72bc4bf414faa1f5a3330e648ab) and oxygen (http://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=34d825dbf4e9466e81d809faf814995b). The MS1-based feature detection for metabolite abundance estimation in the pH experiments was performed using the Knime OpenMS workflow using Optimus software (https://github.com/MolecularCartography/Optimus) (see Supplementary Methods for details). Metabolite annotations based on MS/MS library matches GNPS were linked to the MS1 features using the mass/charge ratio and retention time. Column blank samples and internal standard blanks were included in the Optimus run to remove background contaminant peaks from the data.
The GC-MS data were analyzed using the Thermo Scientific TraceFinder software (Thermo Fisher Scientific). The samples with the most diverse peak count were screened for metabolites of interest. Peaks of these metabolites were found and integrated using ICIS integration algorithm with peak threshold of 1% of largest peak. The National Institute of Standards and Technology (NIST) 2014 EI library was used to perform spectral matches with forward and reverse search. Known background contaminants (such as siloxanes and phthalates) were removed from targeted list of molecules, and 37 compounds of interest were selected. Batch mode with the developed method was used to find and integrate all targeted compounds. In addition, the abundances of specific metabolites of interest (acetic acid, propionic acid, and butyric acid) were manually calculated using the area under the curve feature of Thermo Xcalibur QualBrowser software (Thermo Fisher Scientific) to validate quantification of these targeted metabolites.
Statistical analysis
The microbiome OTU table for all samples was used to calculate the Shannon diversity index. Samples were rarefied to 1000 reads per sample before statistical analysis. Diversity calculations were done using the vegan package in the R statistical software. Regressions between diversity measures and WinCF variables were carried out using Pearson’s r. The RF algorithm was used to develop both a classification and linear model of both the metabolome and microbiome data based on the linear variables of the capillary columns and by sample type. RF analyses were carried out using the randomForest package in R with 5000 trees. Variable importance plots were used to identify OTUs and metabolites that were most correlated to the linear variables.
A generalized linear mixed-effects model (GLMM) was applied to model the relationship between bacterial sequence counts (at the genus level) as the dependent variable and pH and depth as the independent variables. The GLMM model accounts for non-normal distribution of data and to assign a random effect to individual (patient), as data were gathered over time on the same individuals with differing microbial sputum profiles. To model metabolite intensity as the dependent variable and pH and depth variables as independent, a standard linear mixed-effects model (LMM) was used. The R packages MASS and nlme were used to perform the modeling.
Balance trees were used to assess the effect of the gradients and gas production on the WinCF model microbiome at the taxonomic level of genera. Balances represent the log ratios of groups of microbes mitigating many of the issues associated with compositionally (19). The 16S rRNA gene sequencing data from the pH experiment were used to help develop the balance tree method and have been previously published (19). Here, we have built two trees, one to characterize the pH gradient, where microbes are split according to where they are most abundant along the pH gradient and another to characterize effects of the oxygen gradient. Balances were computed from these trees via the isometric log-ratio transformation using Gneiss (19). Two different LMMs were run on these balances to test for differences in pH and depth while accounting for the patient-specific differences.
Data visualization
The ‘ili software (http://ili.embl.de/) was used to visualize microbial and metabolite changes through the pH and oxygen gradients. 3D.stl models of the WinCF columns and its 1-mm sections for the oxygen experiment and the eight capillary columns from the pH experiment were created using the GeoMagic Wrap software. The location of each sample in the.stl file was recorded using the x, y, and z coordinates from GeoMagic Wrap. The normalized abundance of OTUs or metabolites from the multiomics data was mapped on to the appropriate sample locations in the WinCF columns using the corresponding x, y, and z coordinates from the.stl file in the ‘ili software to help visualize the microbial and chemical changes in the context of the experimental design. Additional information on materials and methods is available in the Supplementary Materials.
Supplementary Material
Acknowledgments
We would like to thank N. Abualkheir for assistance with Aspergillus culturing. Funding: The authors would like to acknowledge Vertex Pharmaceuticals and the Cystic Fibrosis Research Innovation Award for funding R.A.Q., NSF grant number DGE-1144086 awarded to J.T.M., the NIH/National Institute of Allergy and Infectious Diseases for funding grant number 1 U01 AI124316-01 (a systems biology approach to treatment of multidrug resistant pathogens), the NSF award numbers 1516951 and 1517100 awarded to T.Z. and I.K., respectively, and the Bruker Corporation and the NIH grant number GM S10RR029121 for funding instrumentation. Author contributions: The project was designed by R.A.Q. and P.C.D. R.A.Q., A.T., and W.C. completed experiments. Data were generated by R.A.Q., A.A., D.W., A.V.M., and G.A. Data were analyzed by R.A.Q., J.T.M., and R.d.S. Funding acquisition and project management was provided by P.C.D., R.K., and D.C. The mathematical model was generated by T.Z. and I.K. J.T.M., R.d.S., and L.-F.N. analyzed data. R.A.Q., W.C., and D.C. contributed to the sample collection. Competing interests: All authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. The sequencing data are available under EBI submission numbers ERP109668 and ERP109670 and are also available in Qiita (https://qiita.ucsd.edu) under IDs: 10511 (pH), 10863 (oxygen). LC-MS/MS data are available at the MassIVE data repository of GNPS (http://gnps.ucsd.edu) under MassIVE IDs: MSV000080397 and MSV000079677. The GC-MS data are available at MassIVE under ID MSV000081455. Code for the statistical analysis using the LMMs is available at https://github.com/DorresteinLaboratory/Mixed_Models, and the mathematical model is available at https://github.com/zhangzhongxun/WinCF_model_Code/tree/7c683dfba0fd9c0bbec11d31295bcf71934ea0ae.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/9/eaau1908/DC1
Fig. S1. Schematic of the experimental design with the WinCF system modified for the pH experiments and the oxygen experiments.
Fig. S2. Actual measurement versus predicted value from RF machine learning algorithm on the microbiome and metabolome data through pH, gas production, and depth variables.
Fig. S3. Niche partitioning of CF lung microbiota in the pH and oxygen experiments.
Fig. S4. O2 microenvironment (% air saturation) through the WinCF vertical depth gradient after incubation with sputum from two patients compared to a noninoculated control.
Fig. S5. Microbiome profiles of individual patients in the pH and oxygen experiments.
Fig. S6. Molecular network of rhamnolipids and quinolones detected in the LC-MS/MS data.
Fig. S7. GLMM results for different bacterial genera on a per patient basis.
Fig. S8. Mean abundance of tobramycin by ion count in the WinCF columns after incubation.
Fig. S9. Tobramycin and N-propionyl tobramycin identification from polar LC-MS/MS data.
Fig. S10. A. fumigatus metabolites in tobramycin-treated WinCF columns.
Fig. S11. Mean abundance of pooled anaerobes in the WinCF columns after the different treatments.
Fig. S12. WinCF model equations.
Fig. S13. Principle co-ordinate analysis (PCoA) plots of metabolome and microbiome data from all samples.
Table S1. Patient samples and information collected in this study.
Table S2. ANOVA of qualitative and quantitative variables measured during the WinCF pH gradient experiments.
Table S3. Metabolites that most changed with the WinCF gas production gradient according to an RF variable importance plot from the untreated samples.
Table S4. Mean abundance through the depth gradient (1 to 10 mm) of P. aeruginosa virulence factor metabolites detected in the WinCF depth experiments and the corresponding Pearson’s correlation (r).
Table S5. Deblurred OTUs and their sequences that most changed with the WinCF depth gradient according to an RF variable importance plot from the untreated samples.
Table S6. Results of the assessment of bias in WinCF system.
Table S7. Confusion matrix and out-of-bag error from an RF classification of the pH experiment metabolomics data based on patient source.
Supplementary Methods
REFERENCES AND NOTES
- 1.Winogradsky S. N., O roli mikrobov v obshchem krugovorote zhizni (On the role of microbes in the cycle of life). Arkhiv Biol. Nauk. 7, 1–27 (1897). [Google Scholar]
- 2.Dworkin M., Gutnick D., Sergei Winogradsky: A founder of modern microbiology and the first microbial ecologist. FEMS Microbiol. Rev. 36, 364–379 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Fenchel T., Finlay B., Oxygen and the spatial structure of microbial communities. Biol. Rev. Camb. Philos. Soc. 83, 553–569 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Lauber C. L., Hamady M., Knight R., Fierer N., Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 75, 5111–5120 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caverly L. J., Zhao J., LiPuma J. J., Cystic fibrosis lung microbiome: Opportunities to reconsider management of airway infection. Pediatr. Pulmonol. 50, S31–S38 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Véron M., Berche P., Virulence and antigens of Pseudomonas aeruginosa. Bull. Inst. Pasteur. 74, 295–337 (1976). [Google Scholar]
- 7.Veesenmeyer J. L., Hauser A. R., Lisboa T., Rello J., Pseudomonas aeruginosa virulence and therapy: Evolving translational strategies. Crit. Care Med. 37, 1777–1786 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lynch S. V, Bruce K. D., The cystic fibrosis airway microbiome. Cold Spring Harb. Perspect. Med. 3, a009738 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garg N., Wang M., Hyde E., da Silva R. R., Melnik A. V., Protsyuk I., Bouslimani A., Lim Y. W., Wong R., Humphrey G., Ackermann G., Spivey T., Brouha S. S., Bandeira N., Lin G. Y., Rohwer F., Conrad D. J., Alexandrov T., Knight R., Dorrestein P. C., Three-dimensional microbiome and metabolome cartography of a diseased human lung. Cell Host Microbe. 22, 705–716.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Worlitzsch D., Tarran R., Ulrich M., Schwab U., Cekici A., Meyer K. C., Birrer P., Bellon G., Berger J., Weiss T., Botzenhart K., Yankaskas J. R., Randell S., Boucher R. C., Döring G., Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Invest. 109, 317–325 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cowley E. S., Kopf S. H., LaRiviere A., Ziebis W., Newman D. K., Pediatric cystic fibrosis sputum can be chemically dynamic, anoxic, and extremely reduced due to hydrogen sulfide formation. MBio 6, e00767 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quinn R. A., Whiteson K., Lim Y.-W., Salamon P., Bailey B., Mienardi S., Sanchez S. E., Blake D., Conrad D., Rohwer F., A Winogradsky-based culture system shows an association between microbial fermentation and cystic fibrosis exacerbation. ISME J. 9, 1024–1038 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rogers G. B., Hoffman L. R., Johnson M. W., Mayer-Hamblett N., Schwarze J., Carroll M. P., Bruce K. D., Using bacterial biomarkers to identify early indicators of cystic fibrosis pulmonary exacerbation onset. Expert. Rev. Mol. Diagn. 11, 197–206 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Boer K., Vandemheen K. L., Tullis E., Doucette S., Fergusson D., Freitag A., Paterson N., Jackson M., Lougheed M. D., Kumar V., Aaron S. D., Exacerbation frequency and clinical outcomes in adult patients with cystic fibrosis. Thorax 66, 680–685 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Conrad D., Haynes M., Salamon P., Rainey P. B., Youle M., Rohwer F., Cystic fibrosis therapy: A community ecology perspective. Am. J. Respir. Cell Mol. Biol. 48, 150–156 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quinn R. A., Whiteson K., Wei Lim Y., Zhao J., Conrad D., LiPuma J. J., Rohwer F., Widder S., Ecological networking of cystic fibrosis lung infections. NPJ Biofilms Microbiomes 2, 4 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winogradsky S., On the nitrifying organisms. Compt. Rend. l’Acad. Sci. 110, 1013–1016 (1890). [Google Scholar]
- 18.Sriramulu D. D., Lünsdorf H., Lam J. S., Römling U., Microcolony formation: A novel biofilm model of Pseudomonas aeruginosa for the cystic fibrosis lung. J. Med. Microbiol. 54, 667–676 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Morton J. T., Sanders J., Quinn R. A., McDonald D., Gonzalez A., Vázquez-Baeza Y., Navas-Molina J. A., Song S. J., Metcalf J. L., Hyde E. R., Lladser M., Dorrestein P. C., Knight R., Balance trees reveal microbial niche differentiation. mSystems 2, e00162-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rowe S., Clancy J. P., Wilschanski M., Nasal potential difference measurements to assess CFTR ion channel activity. Methods Mol. Biol. 741, 69–86 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schertzer J. W., Brown S. A, Whiteley M., Oxygen levels rapidly modulate Pseudomonas aeruginosa social behaviours via substrate limitation of PqsH. Mol. Microbiol. 77, 1527–1538 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mavrodi D. V, Parejko J. A., Mavrodi O. V., Kwak Y.-S., Weller D. M., Blankenfeldt W., Thomashow L. S., Recent insights into the diversity, frequency and ecological roles of phenazines in fluorescent Pseudomonas spp. Environ. Microbiol. 15, 675–686 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Mingeot-Leclercq M.-P., Glupczynski Y., Tulkens P. M., Aminoglycosides: Activity and resistance. Antimicrob. Agents Chemother. 43, 727–737 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen W., Green K. D., Garneau-Tsodikova S., Cosubstrate tolerance of the aminoglycoside resistance enzyme Eis from Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 56, 5831–5838 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flynn J. M., Niccum D., Dunitz J. M., Hunter R. C., Evidence and role for bacterial mucin degradation in cystic fibrosis airway disease. PLOS Pathog. 12, e1005846 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rigottier-Gois L., Dysbiosis in inflammatory bowel diseases: The oxygen hypothesis. ISME J. 7, 1256–1261 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whiteson K. L., Meinardi S., Lim Y. W., Schmieder R., Maughan H., Quinn R., Blake D. R., Conrad D., Rohwer F., Breath gas metabolites and bacterial metagenomes from cystic fibrosis airways indicate active pH neutral 2,3-butanedione fermentation. ISME J. 8, 1247–1258 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carmody L. A., Caverly L. J., Foster B. K., Rogers M. A. M., Kalikin L. M., Simon R. H., VanDevanter D. R., LiPuma J. J., Fluctuations in airway bacterial communities associated with clinical states and disease stages in cystic fibrosis. PLOS ONE 13, e0194060 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Line L., Alhede M., Kolpen M., Kühl M., Ciofu O., Bjarnsholt T., Moser C., Toyofuku M., Nomura N., Høiby N., Jensen P. Ø., Physiological levels of nitrate support anoxic growth by denitrification of Pseudomonas aeruginosa at growth rates reported in cystic fibrosis lungs and sputum. Front. Microbiol. 5, 554 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schobert M., Jahn D., Anaerobic physiology of Pseudomonas aeruginosa in the cystic fibrosis lung. Int. J. Med. Microbiol. 300, 549–556 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Quinn R. A., Lima Y. W., Maughanb H., Conradc D., Rohwera F., Whiteson K. L., Biogeochemical forces shape the composition and physiology of polymicrobial communities in the cystic fibrosis lung. MBio. 5, e00956-13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pezzulo A. A., Tang X. X., Hoegger M. J., Alaiwa M. H. A., Ramachandran S., Moninger T. O., Karp P. H., Wohlford-Lenane C. L., Haagsman H. P., van Eijk M., Bánfi B., Horswill A. R., Stoltz D. A., McCray P. B., Welsh M. J., Zabner J., Reduced airway surface pH impairs bacterial killing in the porcine cystic fibrosis lung. Nature 487, 109–113 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quinn R. A., Phelan V. V., Whiteson K. L., Garg N., Bailey B. A., Lim Y. W., Conrad D. J., Dorrestein P. C., Rohwer F. L., Microbial, host and xenobiotic diversity in the cystic fibrosis sputum metabolome. ISME J. 10, 1483–1498 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zulianello L., Canard C., Köhler T., Caille D., Lacroix J.-S., Meda P., Rhamnolipids are virulence factors that promote early infiltration of primary human airway epithelia by Pseudomonas aeruginosa. Infect. Immun. 74, 3134–3147 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Allen L., Dockrell D. H., Pattery T., Lee D. G., Cornelis P., Hellewell P. G., Whyte M. K. B., Pyocyanin production by Pseudomonas aeruginosa induces neutrophil apoptosis and impairs neutrophil-mediated host defenses in vivo. J. Immunol. 174, 3643–3649 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Hill D., Rose B., Pajkos A., Robinson M., Bye P., Bell S., Elkins M., Thompson B., MacLeod C., Aaron S. D., Harbour C., Antibiotic susceptabilities of Pseudomonas aeruginosa isolates derived from patients with cystic fibrosis under aerobic, anaerobic, and biofilm conditions. J. Clin. Microbiol. 43, 5085–5090 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mooney L., Kerr K. G., Denton M., Survival of Stenotrophomonas maltophilia following exposure to concentrations of tobramycin used in aerosolized therapy for cystic fibrosis patients. Int. J. Antimicrob. Agents 17, 63–66 (2001). [DOI] [PubMed] [Google Scholar]
- 38.Sánchez M. B., Antibiotic resistance in the opportunistic pathogen Stenotrophomonas maltophilia. Front. Microbiol. 6, 658 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.King J., Brunel S. F., Warris A., Aspergillus infections in cystic fibrosis. J. Infect. 72, S50–S55 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Moree W. J., Phelan V. V., Wu C.-H., Bandeira N., Cornett D. S., Duggan B. M., Dorrestein P. C., Interkingdom metabolic transformations captured by microbial imaging mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 109, 13811–13816 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bargon J., Dauletbaev N., Köhler B., Wolf M., Posselt H.-G., Wagner T. O. F., Prophylactic antibiotic therapy is associated with an increased prevalence of Aspergillus colonization in adult cystic fibrosis patients. Respir. Med. 93, 835–838 (1999). [DOI] [PubMed] [Google Scholar]
- 42.Comstock W. J., Huh E., Weekes R., Watson C., Xu T., Dorrestein P. C., Quinn R. A., The WinCF model—An inexpensive and tractable microcosm of a mucus plugged bronchiole to study the microbiology of lung infections. J. Vis. Exp. e55532 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Amir A., McDonald D., Navas-Molina J. A., Kopylova E., Morton J. T., Zech Xu Z., Kightley E. P., Thompson L. R., Hyde E. R., Gonzalez A., Knight R., Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems 2, e00191-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang M., Carver J. J., Phelan V. V., Sanchez L. M., Garg N., Peng Y., Nguyen D. D., Watrous J., Kapono C. A., Luzzatto-Knaan T., Porto C., Bouslimani A., Melnik A. V., Meehan M. J., Liu W.-T., Crüsemann M., Boudreau P. D., Esquenazi E., Sandoval-Calderón M., Kersten R. D., Pace L. A., Quinn R. A., Duncan K. R., Hsu C.-C., Floros D. J., Gavilan R. G., Kleigrewe K., Northen T., Dutton R. J., Parrot D., Carlson E. E., Aigle B., Michelsen C. F., Jelsbak L., Sohlenkamp C., Pevzner P., Edlund A., McLean J., Piel J., Murphy B. T., Gerwick L., Liaw C.-C., Yang Y.-L., Humpf H.-U., Maansson M., Keyzers R. A., Sims A. C., Johnson A. R., Sidebottom A. M., Sedio B. E., Klitgaard A., Larson C. B., P C. A. B., Torres-Mendoza D., Gonzalez D. J., Silva D. B., Marques L. M., Demarque D. P., Pociute E., O’Neill E. C., Briand E., Helfrich E. J. N., Granatosky E. A., Glukhov E., Ryffel F., Houson H., Mohimani H., Kharbush J. J., Zeng Y., Vorholt J. A., Kurita K. L., Charusanti P., McPhail K. L., Nielsen K. F., Vuong L., Elfeki M., Traxler M. F., Engene N., Koyama N., Vining O. B., Baric R., Silva R. R., Mascuch S. J., Tomasi S., Jenkins S., Macherla V., Hoffman T., Agarwal V., Williams P. G., Dai J., Neupane R., Gurr J., Rodríguez A. M. C., Lamsa A., Zhang C., Dorrestein K., Duggan B. M., Almaliti J., Allard P.-M., Phapale P., Nothias L.-F., Alexandrov T., Litaudon M., Wolfender J.-L., Kyle J. E., Metz T. O., Peryea T., Nguyen D.-T., VanLeer D., Shinn P., Jadhav A., Müller R., Waters K. M., Shi W., Liu X., Zhang L., Knight R., Jensen P. R., Palsson B. Ø., Pogliano K., Linington R. G., Gutiérrez M., Lopes N. P., Gerwick W. H., Moore B. S., Dorrestein P. C., Bandeira N., Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 34, 828–837 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Protsyuk I., Melnik A. V., Nothias L.-F., Rappez L., Phapale P., Aksenov A. A., Bouslimani A., Ryazanov S., Dorrestein P. C., Alexandrov T., 3D molecular cartography using LC–MS facilitated by Optimus and ’ili software. Nat. Protoc. 13, 134–154 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Weiss S., Van Treuren W., Lozupone C., Faust K., Friedman J., Deng Y., Xia L. C., Xu Z. Z., Ursell L., Alm E. J., Birmingham A., Cram J. A., Fuhrman J. A., Raes J., Sun F., Zhou J., Knight R., Correlation detection strategies in microbial data sets vary widely in sensitivity and precision. ISME J. 10, 1669–1681 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edlund A., Yang Y., Yooseph S., Hall A. P., Nguyen D. D., Dorrestein P. C., Nelson K. E., He X., Lux R., Shi W., McLean J. S., Meta-omics uncover temporal regulation of pathways across oral microbiome genera during in vitro sugar metabolism. ISME J. 9, 2605–2619 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brodersen K. E., Nielsen D. A., Ralph P. J., Kühl M., A split flow chamber with artificial sediment to examine the below-ground microenvironment of aquatic macrophytes. Mar. Biol. 161, 2921–2930 (2014). [Google Scholar]
- 49.Sumner L. W., Amberg A., Barrett D., Beale M. H., Beger R., Daykin C. A., Fan T. W.-M., Fiehn O., Goodacre R., Griffin J. L., Hankemeier T., Hardy N., Harnly J., Higashi R., Kopka J., Lane A. N., Lindon J. C., Marriott P., Nicholls A. W., Reily M. D., Thaden J. J., Viant M. R., Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 3, 211–221 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Palmer K. L., Mashburn L. M., Singh P. K., Whiteley M., Cystic fibrosis sputum supports growth and cues key aspects of Pseudomonas aeruginosa physiology. J. Bacteriol. 187, 5267–5277 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pluskal T., Castillo S., Villar-Briones A., Orešič M., MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics 11, 395 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/9/eaau1908/DC1
Fig. S1. Schematic of the experimental design with the WinCF system modified for the pH experiments and the oxygen experiments.
Fig. S2. Actual measurement versus predicted value from RF machine learning algorithm on the microbiome and metabolome data through pH, gas production, and depth variables.
Fig. S3. Niche partitioning of CF lung microbiota in the pH and oxygen experiments.
Fig. S4. O2 microenvironment (% air saturation) through the WinCF vertical depth gradient after incubation with sputum from two patients compared to a noninoculated control.
Fig. S5. Microbiome profiles of individual patients in the pH and oxygen experiments.
Fig. S6. Molecular network of rhamnolipids and quinolones detected in the LC-MS/MS data.
Fig. S7. GLMM results for different bacterial genera on a per patient basis.
Fig. S8. Mean abundance of tobramycin by ion count in the WinCF columns after incubation.
Fig. S9. Tobramycin and N-propionyl tobramycin identification from polar LC-MS/MS data.
Fig. S10. A. fumigatus metabolites in tobramycin-treated WinCF columns.
Fig. S11. Mean abundance of pooled anaerobes in the WinCF columns after the different treatments.
Fig. S12. WinCF model equations.
Fig. S13. Principle co-ordinate analysis (PCoA) plots of metabolome and microbiome data from all samples.
Table S1. Patient samples and information collected in this study.
Table S2. ANOVA of qualitative and quantitative variables measured during the WinCF pH gradient experiments.
Table S3. Metabolites that most changed with the WinCF gas production gradient according to an RF variable importance plot from the untreated samples.
Table S4. Mean abundance through the depth gradient (1 to 10 mm) of P. aeruginosa virulence factor metabolites detected in the WinCF depth experiments and the corresponding Pearson’s correlation (r).
Table S5. Deblurred OTUs and their sequences that most changed with the WinCF depth gradient according to an RF variable importance plot from the untreated samples.
Table S6. Results of the assessment of bias in WinCF system.
Table S7. Confusion matrix and out-of-bag error from an RF classification of the pH experiment metabolomics data based on patient source.
Supplementary Methods


