Abstract
Purpose
Non-small-cell lung cancer (NSCLC) is the one of the most common malignancies worldwide, and occurs at a higher frequency in male individuals. Little is known about the role of the long intergenic noncoding RNA for kinase activation (LINK-A) in NSCLC, so in the present study we assessed its potential role on cell proliferation in NSCLC.
Methods
Expression levels of LINK-A in NSCLC tissues and cell lines were detected by quantitative reverse-transcription polymerase chain reaction. LINK-A was knocked down and overexpressed separately in A549 cells and NCI-H1299 cells. The effect of LINK-A expression on cell proliferation was determined by MTT assay. The correlation between LINK-A and hexokinase II (HKII) expression was investigated by Western blot and HKII Activity Assay. Glucose consumption and lactate production assay were used to investigate the aerobic glycolysis in NSCLC cells. The effect of LINK-A in vivo was determined by xenograft assay.
Results
LINK-A expression levels were increased in NSCLC tissues compared with normal tissues. Moreover, LINK-A expression was positively correlated with NSCLC clinicopathological characteristics and survival rate, while knockdown of LINK-A reduced NSCLC cell proliferation. LINK-A expression was also positively correlated with HKII, and NSCLC cells with low LINK-A expression were found to have significantly reduced HKII protein expression, accompanied by a reduction in enzyme activity levels. Both in vitro and in vivo experiments showed that LINK-A expression affected glucose consumption and lactate production through regulation of HKII expression.
Conclusion
These data suggest that the functions of LINK-A in NSCLC might play a key role in tumor progression and that LINK-A could be a promising predictive biomarker and potential therapeutic target for NSCLC.
Keywords: non-small-cell lung cancer, LINK-A, proliferation, hexokinase II, aerobic glycolysis, prognosis
Introduction
Lung cancer is the leading cause of cancer-related deaths worldwide, accounting for 28% of cancer deaths in males and 26% in females.1 Indeed, data from the American Cancer Society in 2013 showed that lung and bronchial cancer were the top cause of cancer death in men over 40 years old and women above 60 years old in the United States.2 Non-small-cell lung cancer (NSCLC) is the main pathological type of lung cancer with a poor 5-year survival rate.3–5 To facilitate the improvement of new diagnoses and therapeutic strategies, it is crucial to understand the molecular mechanisms underlying the development and progression of NSCLC.
Approximately 5%–10% of the human genome is stably transcribed, of which most sequences are non-protein-coding genes.6 Long noncoding RNA (lncRNA) is a functional RNA molecule >200 nucleotides in length transcribed from non-protein-coding DNA sequences located between protein-coding genes. It has recently been found to be closely associated with tumor biology.7 Thousands of human lncRNAs have been identified, although only 1% have a clearly defined and proven function.8 A growing number of studies suggest that lncRNAs are involved in gene regulation through participating in transcriptional and epigenetic regulation in eukaryotes, thereby affecting the biological functions of cells.9,10 While many lncRNAs are closely related to disease, including tumors, most of their biological functions and mechanisms remain unclear.11–15
In tumor cells, the expression of tissue-specific lncRNA may serve as a diagnostic marker. For example, Yao et al16 observed low CAMD1-AS1 expression in renal clear cell carcinoma, and reported a worse prognosis for patients with lower expression levels, indicating a possible role for CADM1-AS1 as a tumor suppressor gene. In NSCLC, MALAT1 was shown to promote the growth and colony formation of NSCLC cells, while interference with MALAT1 expression markedly reduced the migration ability of pulmonary adenocarcinoma.17,18
Abnormal changes in glucose, lipid, and protein metabolism may exist in tumor cells, particularly an abnormal glucose metabolism. Even in the presence of oxygen, glucose metabolism in the mitochondria of tumor cells is transformed from oxidative phosphorylation to glycolysis, which consumes large amounts of glucose and generates lactic acid; this phenomenon is known as the Warburg effect.19,20 A significantly elevated expression of hexokinase (HK) in tumor cells compared with normal tissues has been reported in various malignant tumors, including malignant pleural mesothelioma, myeloma, colon cancer, and pancreatic cancer, with HKII elevation being the most evident.21–23 HKII is the major isozyme that is overexpressed in tumors. It contributes to aerobic glycolysis, so is thought to be a pivotal player in the Warburg effect and is proposed as a metabolic target for cancer therapeutic development.24,25
Long intergenic noncoding RNA for kinase activation (LINK-A) is a newly discovered lncRNA that appears to correlate with tumor progression. LINK-A and LINK-A-dependent signaling pathway activation were recently reported to correlate with malignant tumors, including triple-negative breast cancer and glioma.26,27 However, little is known about the role of LINK-A in NSCLC. In the present study, we assessed the potential role of LINK-A in NSCLC cell proliferation, and discovered that LINK-A expression is increased in NSCLC tissues and positively correlates with clinical outcomes.
Materials and methods
Patients, tissue specimens, and cell lines
One hundred and thirteen pairs of NSCLC tissues and corresponding noncancerous adjacent tissues were obtained from patients diagnosed with NSCLC at Department of Thoracic Surgery of The First Affiliated Hospital of Chengdu Medical College. All involved patients were informed and consent was written and collected. The study was approved by the Clinical Research Ethics Committees of The First Affiliated Hospital of Chengdu Medical College (No CDM107632). Clinical data were collected from all patients at the same time.
Human NSCLC cell lines (A549, H1703, SK-MES-1, and NCI-H1299) were purchased from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, People’s Republic of China). Cells were cultured in Roswell Park Memorial Institute-1640 medium (Thermo Fisher Scientific, Waltham, MA, USA) containing 10% fetal bovine serum (Thermo Fisher Scientific) and maintained at 37°C with 5% CO2.
RNA extraction and real-time PCR analysis
Total RNA was extracted with Trizol® (Thermo Fisher Scientific, Carlsbad, CA, USA) and used as a template for cDNA synthesis (TaKaRa Biotechnology Co. Ltd, Dalian, People’s Republic of China). LINK-A expression levels were detected by quantitative reverse-transcription polymerase chain reaction (qRT-PCR) using the SYBR Green PCR master mix (TaKaRa Biotechnology Co. Ltd) in 20 μL reactions on the cobas z480 analyzer (Roche Molecular Systems, Inc., Pleasanton, CA, USA). PCR conditions were as follows: 95°C for 30 seconds, then 40 cycles of 95°C for 5 seconds, and 60°C for 30 seconds. The comparative Ct method was used to quantify transcripts. LINK-A primers were as follows: LINK-A forward 5′-TTCCCCCATTTTTCCTTTTC-3′ and reverse 5′-CTCTGGTTGGGTGACTGGTT-3′; β-actin was used as an internal control and was amplified using primers 5′-AGCGAGCATCCCCCAAAGTT-3′ and 5′-GGGCACGAAGGCTCATCATT-3′.
Construction of the LINK-A expression plasmid and infection
Full-length LINK-A cDNA fragments were obtained by RT-PCR and cloned into the pGEM-T-Easy vector (Promega, Madison, WI, USA). Sequences from LINK-A-positive clones were then subcloned into the pCDNA3.1 (+) vector (Invitrogen). The LINK-A expression plasmid pCDNA3.1-LINK-A was infected into NCI-H1299 cells using Lipofectamine 2000 (Thermo Fisher Scientific).
RNA interference
To obtain a stable LINK-A-knockdown cell line, a lentivirus carrying LINK-A-shRNA (System Biosciences, Palo Alto, CA, USA) was infected into A549 cells using polybrene (4 μg/mL, Sigma-Aldrich Co., St Louis, MO, USA). At 48 hours post viral infection, the medium was changed to Roswell Park Memorial Institue-1640 containing 10% fetal bovine serum, followed by selection with 1 μg/mL of puromycin (Sigma) for 14 days. The shRNA targeting LINK-A sequence was 5′-TGTCTAAGGTGGAGATTAC-3′. The target sequence for HKII was 5′-CGGACAGAACACGGAGAGdTdT-3′ (Thermo Fisher Scientific). The siRNA was transfected into NSCLC cells separately using Lipofectamine 2000 reagent (Thermo Fisher Scientific) in accordance with the manufacturer’s instructions.
Western blot analysis
Total protein from cultured cells was isolated with lysis buffer (Pierce, Appleton, WI, USA) and quantified using the Bradford method. Twenty microgram of protein was resolved and separated by 10% SDS-PAGE and then transferred onto a PVDF membrane (Millipore, Billerica, MA, USA). Antibodies against HKII (1:1,000, Cell Signaling Technology, Danvers, MA, USA) and β-actin (1:2,000, Santa Cruz Biotechnology Inc., Dallas, TX, USA) were used to incubate the membrane overnight at 4°C. The expression of β-actin was quantified as a loading control. The secondary antibody (1:2,000, Cell Signaling Technology) was then added and incubated for 2 hours at room temperature. The results of protein expression were visualized using ECL (Tanon, Shanghai, People’s Republic of China) and detected using BioImaging Systems (Tanon).
Cell proliferation assay
LINK-A-overexpressing NCI-H1299 cells and LINK-A-knockdown A549 cells were seeded separately at a density of 3×103 cells/well in 96-well plates in 100 μL medium. Then, 10 μL of MTT was added to each well and cells were incubated at 37°C for 4 hours in the dark. The medium was removed and 100 μL of dimethyl sulfoxide was added to each well. The absorption value at 490 nm was then recorded.
Glucose consumption and lactate production assay
NCI-H1299 cells infected with the pCDNA3.1-LINK-A plasmid were seeded in 35 mm dishes and incubated for 36 hours. Stable LINK-A-knockdown A549 cells and corresponding control cells were incubated for 34 hours. The supernatants of the 2 cell lines were collected by centrifugation at 800 rpm 5 minutes to remove the cells. Glucose consumption was measured using the Glucose Assay Kit (Sigma, GAGO-20). Lactate production levels in the culture media were measured using the Lactate Assay kit (BioVision, K607-100, Milpitas, CA, USA) according to the manufacturer’s instructions.
HK II activity assay
HKII activity was measured using the PicoProbe™ Hexokinase Activity Assay Kit (BioVision). Relative HKII activities in treated and untreated cells were expressed as percentages of the HKII activity in control cells, which was arbitrarily set as 100. 3-Bromopyruvic acid (3-BrPA; BioVision) was used as a HK inhibitor to treat NCI-H1299 cells overexpressing LINK-A.
In vivo tumor xenograft assay
All animal experiments were approved by the Animal Care and Use Committee of Chengdu Medical College. LINK-A-knockdown A549 cells and control cells were suspended in 40% Matrigel matrix (BD Biosciences, Franklin Lakes, NJ, USA) in PBS at a density of 1×107 cells/mL. Six-week-old male athymic mice were kept in a specific pathogen-free facility. Cells were injected subcutaneously into the flank region of nude mice using a 1–3 cm1 tuberculin syringe. Tumor size was measured every 7 days with a caliper, and tumor volume was calculated using the following equation: tumor volume (mm3) = L×W2/2, where L = length, and W = width. When the biggest tumor was nearly 400 mm3, which is the ethical endpoint to nude mice, the tumors were removed, photographed, and weighed.
Immunohistochemistry (IHC) analysis
Expression level of HK II was evaluated by IHC analysis. Formalin-fixed, paraffin-embedded tumor sections were deparaffinized using a graded ethanol series, then incubated in 3% hydrogen peroxide for 10 minutes to block endogenous peroxidase. Primary antibody for HK II (1:150 dilutions; Cell Signaling Technology) was applied overnight at 4°C. Finally, immunoreactivity was scored as the nuclear staining intensity (on a scale of 0–3: negative =0, weak =1, moderate =2, strong =3) × the percentage of cells stained (positively recorded on an ordered categorical scale: 0= zero, 1=1%–25%, 2=26%–50%, 3=51%–100%), which resulted in a scale of 0–9. Immunoreactivity was evaluated by separately evaluated by 2 senior pathologists. Any discrepancy between the 2 evaluators was resolved by reevaluation and careful discussion until agreement was reached.
Statistical analyses
All data are presented as mean ± SD. χ2 test was used to evaluate the relationship between LINK-A expression and clinicopathologic features. The comparison of multiple groups was analyzed using analysis of variance with Holm–Sidak’s or Dunnett’s multiple comparisons test (GraphPad Prism 6.0, GraphPad Software, La Jolla, CA, USA). P<0.05 was considered statistically significant. All data analysis was performed with SPSS16.0 software (SPSS Inc., Chicago, IL).
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the Clinical Research Ethics Committees of The First Affiliated Hospital of Chengdu Medical College and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. All procedures performed in studies involving animals were in accordance with the ethical standards of the Institutional Review Board of Chengdu Medical College.
Results
LINK-A is upregulated in NSCLC and correlates with patient outcome
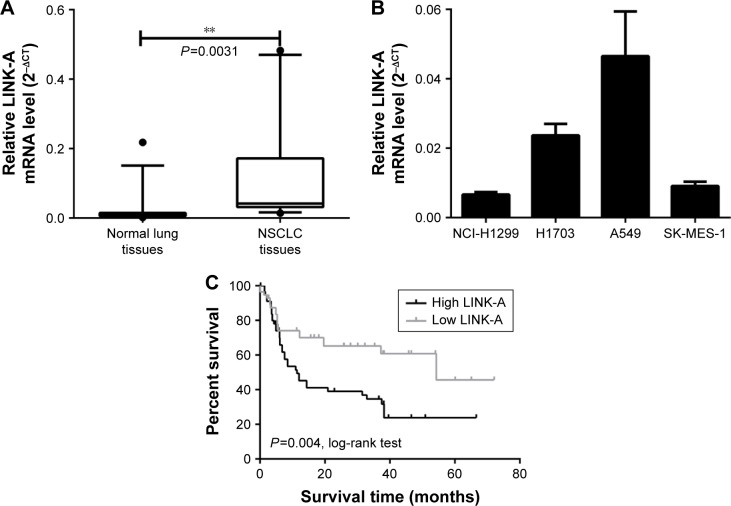
In this study, we used qRT-PCR to evaluate lncRNA LINK-A expression in 113 pairs of NSCLC and noncancerous adjacent tissues. As shown in Figure 1A, LINK-A mRNA expression levels in NSCLC tissues were significantly higher than in corresponding noncancerous adjacent tissues (P=0.0031).
Figure 1.
LINK-A is overexpressed in NSCLC and correlates with patient outcome.
Notes: (A) qRT-PCR evaluation of LINK-A expression in NSCLC samples compared with adjacent normal tissues. (B) Expression levels of LINK-A expression in different NSCLC cell lines, including A549, H1703, SK-MES-1, and NCI-H1299 cells. (C) Overall survival Kaplan–Meier analysis of NSCLC patients. **P<0.01.
Abbreviations: LINK-A, long intergenic noncoding RNA for kinase activation; NSCLC, non-small-cell lung cancer; qRT-PCR, quantitative reverse-transcription polymerase chain reaction.
LINK-A expression was then measured by qRT-PCR in NSCLC cell lines, including NCI-H1299, H1703, SK-MES-1, and A549. LINK-A showed the highest expression in A549 cells among the 4 cell lines and the lowest in NCI-H1299 cells (Figure 1B). Therefore, we used A549 cells and NCI-H1299 cells for further experiments.
To identify the potential association between LINK-A expression and tumor clinicopathological features in NSCLC, the patients were classified into low-LINK-A or high- LINK-A groups according to LINK-A mRNA expression levels (Table 1). LINK-A expression levels significantly correlated with tumor size (P=0.019), tumor grade (P=0.032), and recurrence status (P=0.002). The log-rank test showed that low LINK-A expression patients had significantly higher survival rates than those with high levels of LINK-A expression (P=0.004, Figure 1C).
Table 1.
LINK-A mRNA expression and clinicopathological factors in NSCLC patients
| Characteristic | Number of patients | LINK-A mRNA expression
|
||
|---|---|---|---|---|
| Low | High | P-value | ||
| Gender | 0.159 | |||
| Male | 90 | 35 | 55 | |
| Female | 23 | 15 | 8 | |
| Age | 0.106 | |||
| <65 | 66 | 25 | 41 | |
| ≥65 | 47 | 25 | 22 | |
| Size of tumor | 0.019a | |||
| ≤3 cm | 58 | 34 | 24 | |
| >3 cm | 55 | 16 | 39 | |
| Tumor grade | 0.0012a | |||
| I | 32 | 20 | 12 | |
| II | 30 | 17 | 13 | |
| III | 51 | 13 | 38 | |
| Recurrence status | 0.0023a | |||
| Positive | 61 | 35 | 26 | |
| Negative | 52 | 15 | 37 | |
Note:
Statistically significant difference (P<0.05).
Abbreviations: LINK-A, long intergenic noncoding RNA for kinase activation; NSCLC, non-small-cell lung cancer.
Overexpression of LINK-A promotes NSCLC cell proliferation
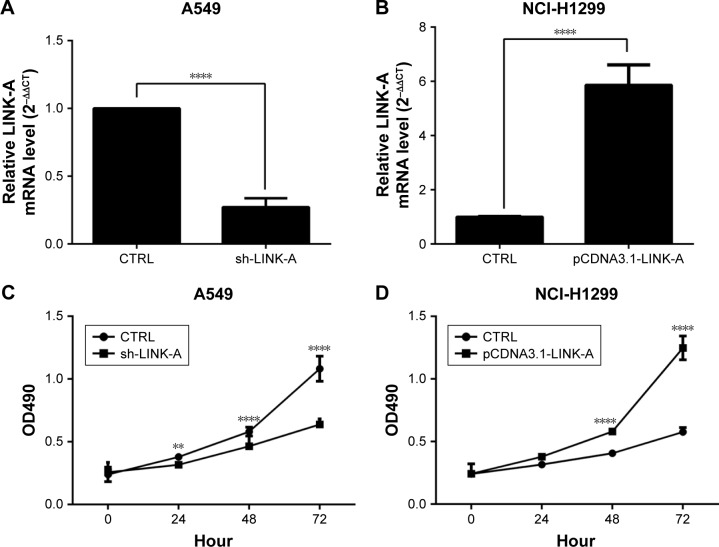
To determine the biological function of LINK-A in NSCLC cells, we knocked down LINK-A expression in A549 cells by lentivirus-mediated RNA interference, and overexpressed it in NCI-H1299 cells by infecting plasmid pCDNA3.1-LINK-A (Figure 2A and B). The MTT assay showed that LINK-A silencing inhibited the proliferation of A549 cells (Figure 2C). In contrast, NCI-H1299 cells showed a significant increase in growth compared with the control group (Figure 2D).
Figure 2.
Overexpression of LINK-A promotes NSCLC cell proliferation.
Notes: qRT-PCR evaluation of LINK-A expression in LINK-A-knockdown A549 (A) and LINK-A-overexpressing NCI-H1299 cells (B). Cell proliferation assay was performed in LINK-A-knockdown A549 (C) and LINK-A-overexpressing NCI-H1299 cells (D). **P<0.01; ****P<0.001.
Abbreviations: LINK-A, long intergenic noncoding RNA for kinase activation; NSCLC, non-small cell lung cancer; qRT-PCR, quantitative reverse-transcription polymerase chain reaction; CTRL, control.
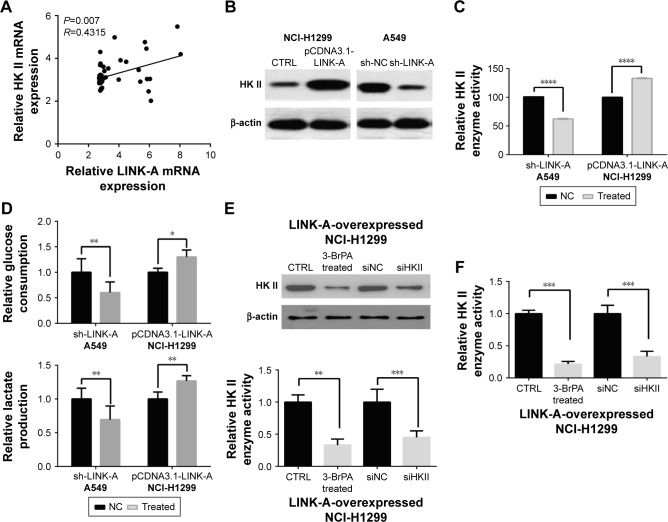
LINK-A affects aerobic glycolysis and promotes cell proliferation through regulating HKII expression in NSCLC cells
To investigate the role of LINK-A expression in aerobic glycolysis, we analyzed the expression of glycolysis genes in GEO profile data (GDS3627). We found that the expression level of LINK-A was positively correlated with HKII (Figure 3A, P=0.007). This was verified by Western blot analysis revealing lower HKII levels in A549 cells following LINK-A knockdown, and upregulated HKII levels in LINK-A-overexpressing NCI-H1299 cells (Figure 3B). The corresponding HKII enzyme activity showed similar results, being reduced by 35.8% in A549 cells and increased by 32.9% in NCI-H1299 cells (Figure 3C). Next, we detected lactate production and glucose consumption in infected cells to investigate the aerobic glycolysis in NSCLC cells (Figure 3D). Knockdown of LINK-A inhibited glucose consumption by 39.7% and lactate levels by 30.6% in A549 cells. In NCI-H1299 cells, glucose consumption and lactate production were increased by 30.1% and 26.7%, respectively.
Figure 3.
Expression of LINK-A affects the aerobic glycolysis of NSCLC cells.
Notes: (A) Analysis of the expression correlation between LINK-A and HK II in the GEO profile database (GDS1479). (B) Expression of HK II in LINK-A-knockdown A549 and LINK-A-overexpressing NCI-H1299 cells. (C) HKII enzyme activity changes were determined in LINK-A-knockdown A549 and LINK-A-overexpressing NCI-H1299 cells. (D) Lactate production and glucose consumption were measured in LINK-A-knockdown A549 and LINK-A-overexpressing NCI-H1299 cell media. (E) Test of HK II enzyme activity in LINK-A-overexpressed-NCI-H1299 cells after 3-BrPA and HK II specific siRNA treatment. (F) Treatment with 3-BrPA and siRNA inhibit NSCLC cancer cell proliferation in cells overexpressing LINK-A. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001.
Abbreviations: LINK-A, long intergenic noncoding RNA for kinase activation; NSCLC, non-small-cell lung cancer; HK II, hexokinase II.
To further determine whether LINK-A plays a role in cancer cell proliferation through the regulation of HKII expression in aerobic glycolysis, 3-BrPA (50 μM, an HK inhibitor) and specific siRNA were used to reduce HKII expression and activity in NCI-H1299 cells overexpressing LINK-A (Figure 3E). Treatment with 3-BrPA inhibited cancer cell proliferation by 31.27%, and siRNA by 38.44% in cells overexpressing LINK-A (Figure 3F).
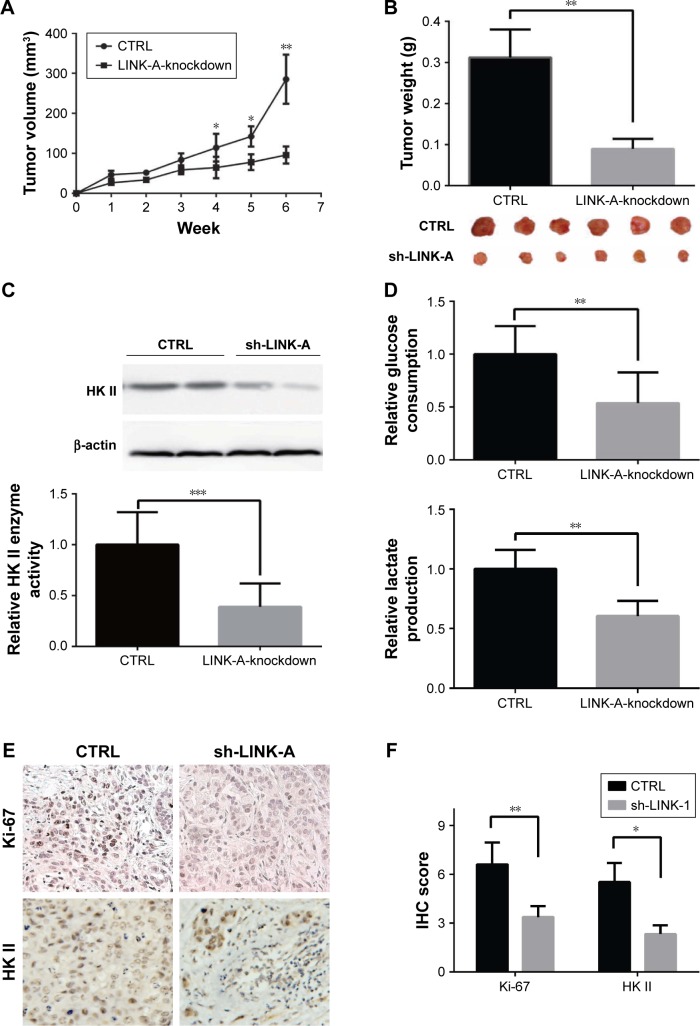
Expression of LINK-A affects NSCLC tumorigenicity in vivo
To assess the role of LINK-A in vivo, we performed the nude mouse xenograft assay for further study. The results showed that tumor volume of LINK-A-knockdown xenografts were significantly decreased compared with the control group (Figure 4A). The average tumor weight of xenografts from LINK-A-knockdown cell-injected mice was significantly lower than that of the control group (P=0.0023; Figure 4B). HKII protein levels and enzyme activity in LINK-A-knockdown xenografts were significantly decreased compared with the control group (Figure 4C). Similarly, glucose consumption and lactate production were reduced by 46.3% and 39.6%, respectively, compared with the control group (Figure 4D). Moreover, tumor growth related protein, Ki-67, and HK II protein levels were decreased in LINK-A-knockdown xenografts (Figure 4E and F). These data suggest LINK-A promotes NSCLC tumorigenesis through aerobic glycolysis in vivo.
Figure 4.
Expression of LINK-A affects NSCLC tumorigenicity in vivo.
Notes: (A) In vivo tumor growth curve of LINK-A-knockdown and control xenografts over 4 weeks. (B) Photograph and average tumor weight of control and LINK-A-knockdown xenografts at the endpoint. (C) HKII protein levels and enzyme activity in LINK-A-knockdown xenografts. (D) Glucose consumption and lactate production in LINK-A-knockdown xenografts. (E) IHC detected Ki-67 and HK II expression in xenograft groups. (F) Evaluation of Ki-67 and HK II protein level by IHC data. *P<0.05; **P<0.01; ***P<0.001.
Abbreviations: LINK-A, long intergenic noncoding RNA for kinase activation; NSCLC, non-small cell lung cancer; HK II, hexokinase II; IHC, Immunohistochemistry; CTRL, control.
Discussion
Despite the rapid progress of lung cancer-related clinical medicine and experimental oncology, the treatment and prognosis of NSCLC remain unsatisfactory. The 5-year overall survival rate is reported to be about 11%, and nearly half of all patients are diagnosed at an advanced stage after metastasis has occurred in distant organs.28,29 Therefore, it is of great importance to elucidate the underlying mechanisms of NSCLC and to search for effective molecular targets to improve disease therapy and prognosis. Previous studies have suggested that lncRNA expression is abnormal in chronic lymphocytic leukemia, colorectal carcinoma, and hepatocellular carcinoma, with most of these lncRNAs being located near fragile sites or oncogenes.30 The role of some lncRNAs in oncogenesis has been addressed, such as that of HOTAIR, located within HOXC on chromosome 12q13.13, in breast cancer.31 In the present study, we investigated the expression level of a recently discovered lncRNA, LINK-A, in human NSCLC tissues. qRT-PCR showed for the first time that LINK-A mRNA expression was significantly higher in human NSCLC tissues than in noncancerous adjacent tissues. More importantly, we found that NSCLC patients with high LINK-A expression levels had significantly lower survival rates compared with those with low LINK-A, and that LINK-A expression correlated with NSCLC clinical pathological factors.
Many reported lncRNAs affect tumor biological progression through regulating important signaling pathways. For example, HOTAIR binds to PRC2, while some silent genes within the HOXD locus induce the trimethylation of histone H3 lysine 27, and then remodel the gene expression pattern in mammary epithelial cells.32,33 The ZXF1 sequence (7,291 bp), located between bases at loci 90692441 and 90699731 on human chromosome 10, interferes with the progression of pulmonary adenocarcinoma by regulating the transforming growth factor-β signaling pathway and the α-smooth muscle actin gene.34 Moreover, Qiu et al35 observed that the overexpression of lncRNA colon cancer-associated transcript 2 was specifically correlated with the development of pulmonary adenocarcinoma and could promote the invasion of carcinoma cells.
In triple-negative breast cancer, LINK-A expression and activation were reported to promote breast cancer glycolysis reprogramming and tumorigenesis through HIF1α signaling pathway.26 Similarly, LINK-A was found to correlate with glioma cell growth and invasion via LDHA.27 To identify the biological effect of LINK-A on NSCLC progression, we constructed 2 specific cell lines: LINK-A-overexpressing NCI-H1299 and LINK-A-knockdown A549 cells. A cell proliferation assay indicated that LINK-A silencing inhibited the proliferation of LINK-A-overexpressing NSCLC cells, while an in vivo study in nude mice confirmed that the knockdown of LINK-A expression significantly reduced tumor weight and volume.
HKII is overexpressed in most cancer cells, including breast cancer, malignant pleural mesothelioma, myeloma, colon cancer, pancreatic cancer, and glioblastoma (GBM).36–42 A recent study conducted by Wolf et al43 demonstrated that HKII may play an important role in GBM aerobic glycolysis. HKI was mainly expressed in normal brain tissues and low-grade GBM, whereas HKII expression was found mainly in high-grade GBM. Other research has also shown that the proliferating rate of the hepatocellular carcinoma cell line SNU-449 is accelerated by 1.5–2.0-fold following the exogenous infection of HKII.44 Therefore, the upregulation of HKII may influence the biological behavior of carcinomas through promoting efficient glycolysis in tumor cells.
Although LINK-A and HKII were both found to be correlated with cancer cells invasion or migration,27,45 here we detect migration and invasion abilities in LINK-A-overexpressed and -knockdown NSCLC cells. The data shows that neither overexpression nor knockdown of LINK-A has any effects on NSCLC cell migration and invasion compared with parental cells (data not shown). This result hints LINK-A may has other target gene(s) in NSCLC cells.
In the present study, we found that LINK-A affects NSCLC cell glycolysis. NSCLC cells with high levels of LINK-A expression had significantly increased HKII protein levels accompanied by increased enzyme activity, whereas the opposite was observed in A549 cells with low LINK-A expression. In GEO profile data and our collected NSCLC tumors, the mRNA level of HKII has positive correlation to LINK-A; however, we found no correlation between mRNA level of LINK-A and the protein level of HKII when assessed by IHC. Moreover, the mRNA level of HKII also showed no positive correlation with its protein level in NSCLC tumors (data not shown). We propose that LINK-A has directed or indirect regulation on HKII transcription, so we can find a positive correlation between LINK-A and HKII using their mRNA levels. However, in tumors, the protein level of HKII has many influencing factors,46,47 such as hypoxia and posttranscriptional modification; these influencing factors disturbed the protein level which is in accordance to our finding.
We also showed that LINK-A expression affects glucose consumption and lactate production both in vitro and in vivo. Tumor cell metabolism involves multiple processes that are regulated by the abnormal expression of coding and noncoding genes. Glucose consumption may be as much as 10-fold higher than that of normal cells; consequently, the production of the glycolysis product lactic acid is also increased, which is favorable for the growth and proliferation of tumor cells. Some researchers argue that these characteristics of tumor cells help them escape apoptosis, which is associated with poor prognosis in tumor patients.25,48 The pharmacologic inhibition (3-BrPA) and siRNA of HKII were previously found to suppress LINK-A overexpression-induced cell proliferation (Figure 3E and F), suggesting that LINK-A promotes cell proliferation and aerobic glycolysis in NSCLC cells via the regulation of HKII. Combined with cell proliferation results, this indicates that LINK-A affects tumor progression through glycolysis.
Conclusion
This study reports for the first time that LINK-A is upregulated in NSCLC tissue. The expression of LINK-A positively correlates with clinicopathological characteristics and survival rate in NSCLC. Moreover, LINK-A affected the proliferation and aerobic glycolysis of NSCLC cells through regulating HKII. These data suggest that the functions of LINK-A in NSCLC might play a key role in tumor progression and could be a promising predictive biomarker and potential therapeutic target for NSCLC.
Informed consent
All the details relating to individual participants are included in the manuscript. All listed authors meet the authorship criteria, and all authors are in agreement with the content of the manuscript and have obtained written informed consent to publish.
Acknowledgments
This work was supported by the Natural Science Foundation of China (81602636), the Fundamental Research Funds for the Central Universities (2242015K40034), Nanjing Medical Science and Technology Development Project (ZKX15049) and Jiangsu Postdoctoral Research Grant (1601182B).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.van Zandwijk N, Fong KM. Update in lung cancer: prologue to a modern review series. Respirology. 2015;20(2):183–184. doi: 10.1111/resp.12457. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 3.Stinchcombe TE. Recent advances in the treatment of non-small cell and small cell lung cancer. F1000Prime Rep. 2014;6:117. doi: 10.12703/P6-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 5.Sève P, Reiman T, Dumontet C. The role of betaIII tubulin in predicting chemoresistance in non-small cell lung cancer. Lung Cancer. 2010;67(2):136–143. doi: 10.1016/j.lungcan.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 6.Bertone P, Stolc V, Royce TE, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306(5705):2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- 7.Kapranov P, Cheng J, Dike S, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316(5830):1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 8.Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145(2):178–181. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 9.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21(6):354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Kotake Y, Nakagawa T, Kitagawa K, et al. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30(16):1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Printz C. Missing links: new survey shows need for improvements in cancer diagnosis. Cancer. 2013;119(12):2203–2204. doi: 10.1002/cncr.28189. [DOI] [PubMed] [Google Scholar]
- 12.Polyak K, Garber J. Targeting the missing links for cancer therapy. Nat Med. 2011;17(3):283–284. doi: 10.1038/nm0311-283. [DOI] [PubMed] [Google Scholar]
- 13.Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer? Hum Mol Genet. 2010;19(R2):R152–R161. doi: 10.1093/hmg/ddq353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naemura M, Murasaki C, Inoue Y, Okamoto H, Kotake Y. Long Noncoding RNA ANRIL Regulates Proliferation of Non-small Cell Lung Cancer and Cervical Cancer Cells. Anticancer Res. 2015;35(10):5377–5382. [PubMed] [Google Scholar]
- 15.Dang Y, Lan F, Ouyang X, et al. Expression and clinical significance of long non-coding RNA HNF1A-AS1 in human gastric cancer. World J Surg Oncol. 2015;13:302. doi: 10.1186/s12957-015-0706-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yao J, Chen Y, Wang Y, et al. Decreased expression of a novel lncRNA CADM1-AS1 is associated with poor prognosis in patients with clear cell renal cell carcinomas. Int J Clin Exp Pathol. 2014;7(6):2758–2767. [PMC free article] [PubMed] [Google Scholar]
- 17.Ji P, Diederichs S, Wang W, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22(39):8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt LH, Spieker T, Koschmieder S, et al. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thorac Oncol. 2011;6(12):1984–1992. doi: 10.1097/JTO.0b013e3182307eac. [DOI] [PubMed] [Google Scholar]
- 19.Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11(5):325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 20.Warburg O. Origin of cancer cells. Oncologia. 1956;9(2):75–83. [PubMed] [Google Scholar]
- 21.Kaira K, Serizawa M, Koh Y, et al. Relationship between 18F-FDG uptake on positron emission tomography and molecular biology in malignant pleural mesothelioma. Eur J Cancer. 2012;48(8):1244–1254. doi: 10.1016/j.ejca.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 22.Nakano A, Miki H, Nakamura S, et al. Up-regulation of hexokinaseII in myeloma cells: targeting myeloma cells with 3-bromopyruvate. J Bioenerg Biomembr. 2012;44(1):31–38. doi: 10.1007/s10863-012-9412-9. [DOI] [PubMed] [Google Scholar]
- 23.Izuishi K, Yamamoto Y, Sano T, et al. Molecular mechanism underlying the detection of colorectal cancer by 18F-2-fluoro-2-deoxy-D-glucose positron emission tomography. J Gastrointest Surg. 2012;16(2):394–400. doi: 10.1007/s11605-011-1727-z. [DOI] [PubMed] [Google Scholar]
- 24.Mathupala SP, Ko YH, Pedersen PL. Hexokinase-2 bound to mitochondria: cancer’s stygian link to the “Warburg Effect” and a pivotal target for effective therapy. Semin Cancer Biol. 2009;19(1):17–24. doi: 10.1016/j.semcancer.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vander Heiden MG. Targeting cancer metabolism: a therapeutic window opens. Nat Rev Drug Discov. 2011;10(9):671–684. doi: 10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- 26.Lin A, Li C, Xing Z, et al. The LINK-A lncRNA activates normoxic HIF1α signalling in triple-negative breast cancer. Nat Cell Biol. 2016;18(2):213–224. doi: 10.1038/ncb3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu D, Zhao B, Cao X, Wan J. Long non-coding RNA LINK-A promotes glioma cell growth and invasion via lactate dehydrogenase A. Oncol Rep. 2017;38(3):1525–1532. doi: 10.3892/or.2017.5806. [DOI] [PubMed] [Google Scholar]
- 28.Yang Y, Li H, Hou S, Hu B, Liu J, Wang J. The noncoding RNA expression profile and the effect of lncRNA AK126698 on cisplatin resistance in non-small-cell lung cancer cell. PLoS One. 2013;8(5):e65309. doi: 10.1371/journal.pone.0065309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verdecchia A, Francisci S, Brenner H, et al. Recent cancer survival in Europe: a 2000-02 period analysis of EUROCARE-4 data. Lancet Oncol. 2007;8(9):784–796. doi: 10.1016/S1470-2045(07)70246-2. [DOI] [PubMed] [Google Scholar]
- 30.Calin GA, Liu CG, Ferracin M, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12(3):215–229. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 31.Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Enfield KS, Pikor LA, Martinez VD, Lam WL. Mechanistic Roles of Noncoding RNAs in Lung Cancer Biology and Their Clinical Implications. Genet Res Int. 2012;2012:737416–16. doi: 10.1155/2012/737416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang L, Zhou XF, Pan GF, Zhao JP. Enhanced expression of long non-coding RNA ZXF1 promoted the invasion and metastasis in lung adenocarcinoma. Biomed Pharmacother. 2014;68(4):401–407. doi: 10.1016/j.biopha.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 35.Qiu M, Xu Y, Yang X, et al. CCAT2 is a lung adenocarcinoma-specific long non-coding RNA and promotes invasion of non-small cell lung cancer. Tumour Biol. 2014;35(6):5375–5380. doi: 10.1007/s13277-014-1700-z. [DOI] [PubMed] [Google Scholar]
- 36.Tong SY, Lee JM, Ki KD, et al. Correlation between FDG uptake by PET/CT and the expressions of glucose transporter type 1 and hexokinase II in cervical cancer. Int J Gynecol Cancer. 2012;22(4):654–658. doi: 10.1097/IGC.0b013e31824864e6. [DOI] [PubMed] [Google Scholar]
- 37.Gregersen LH, Jacobsen A, Frankel LB, Wen J, Krogh A, Lund AH. MicroRNA-143 down-regulates Hexokinase 2 in colon cancer cells. BMC Cancer. 2012;12:232. doi: 10.1186/1471-2407-12-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang L, Wang J, Xiong H, et al. Co-targeting hexokinase 2-mediated Warburg effect and ULK1-dependent autophagy suppresses tumor growth of PTEN- and TP53-deficiency-driven castration-resistant prostate cancer. EBioMedicine. 2016;7:50–61. doi: 10.1016/j.ebiom.2016.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katagiri M, Karasawa H, Takagi K, et al. Hexokinase 2 in colorectal cancer: a potent prognostic factor associated with glycolysis, proliferation and migration. Histol Histopathol. 2017;32(4):351–360. doi: 10.14670/HH-11-799. [DOI] [PubMed] [Google Scholar]
- 40.Ho N, Morrison J, Silva A, Coomber BL. The effect of 3-bromopyruvate on human colorectal cancer cells is dependent on glucose concentration but not hexokinase II expression. Biosci Rep. 2016;36(1):e00299. doi: 10.1042/BSR20150267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deng Y, Lu J. Targeting hexokinase 2 in castration-resistant prostate cancer. Mol Cell Oncol. 2015;2(3):e974465. doi: 10.4161/23723556.2014.974465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderson M, Marayati R, Moffitt R, Yeh JJ. Hexokinase 2 promotes tumor growth and metastasis by regulating lactate production in pancreatic cancer. Oncotarget. 2017;8(34):56081–56094. doi: 10.18632/oncotarget.9760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolf A, Agnihotri S, Micallef J, et al. Hexokinase 2 is a key mediator of aerobic glycolysis and promotes tumor growth in human glioblastoma multiforme. J Exp Med. 2011;208(2):313–326. doi: 10.1084/jem.20101470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ahn KJ, Hwang HS, Park JH, et al. Evaluation of the role of hexokinase type II in cellular proliferation and apoptosis using human hepatocellular carcinoma cell lines. J Nucl Med. 2009;50(9):1525–1532. doi: 10.2967/jnumed.108.060780. [DOI] [PubMed] [Google Scholar]
- 45.Giatromanolaki A, Sivridis E, Arelaki S, Koukourakis MI. Expression of enzymes related to glucose metabolism in non-small cell lung cancer and prognosis. Exp Lung Res. 2017;43(4–5):167–174. doi: 10.1080/01902148.2017.1328714. [DOI] [PubMed] [Google Scholar]
- 46.Sebastian S, Kenkare UW. Expression of two type II-like tumor hexokinase RNA transcripts in cancer cell lines. Tumour Biol. 1998;19(4):253–260. doi: 10.1159/000030015. [DOI] [PubMed] [Google Scholar]
- 47.Zhao W, Chang C, Cui Y, et al. Steroid receptor coactivator-3 regulates glucose metabolism in bladder cancer cells through coactivation of hypoxia inducible factor 1α. J Biol Chem. 2014;289(16):11219–11229. doi: 10.1074/jbc.M113.535989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]