Abstract
The suprachiasmatic nucleus (SCN) is the neural network that drives daily rhythms in behavior and physiology. The SCN encodes environmental changes through the phasing of cellular rhythms across its anteroposterior axis, but it remains unknown what signaling mechanisms regulate clock function along this axis. Here we demonstrate that arginine vasopressin (AVP) signaling organizes the SCN into distinct anteroposterior domains. Spatial mapping of SCN gene expression using in situ hybridization delineated anterior and posterior domains for AVP signaling components, including complementary patterns of V1a and V1b expression that suggest different roles for these two AVP receptors. Similarly, anteroposterior patterning of transcripts involved in Vasoactive Intestinal Polypeptide- and Prokineticin2 signaling was evident across the SCN. Using bioluminescence imaging, we then revealed that inhibiting V1A and V1B signaling alters period and phase differentially along the anteroposterior SCN. V1 antagonism lengthened period the most in the anterior SCN, whereas changes in phase were largest in the posterior SCN. Further, separately antagonizing V1A and V1B signaling modulated SCN function in a manner that mapped onto anteroposterior expression patterns. Lastly, V1 antagonism influenced SCN period and phase along the dorsoventral axis, complementing effects on the anteroposterior axis. Together, these results indicate that AVP signaling modulates SCN period and phase in a spatially specific manner, which is expected to influence how the master clock interacts with downstream tissues and responds to environmental changes. More generally, we reveal anteroposterior asymmetry in neuropeptide signaling as a recurrent organizational motif that likely influences neural computations in the SCN clock network.
Keywords: suprachiasmatic, spatiotemporal, anteroposterior, vasopressin, circadian, RRID: IMSR_JAX:006852, RRID: AB_2313978, RRID: AB_518682, RRID: AB_2556546, RRID: AB_2340474
Graphical Abstract
We investigated the patterning of peptide signaling in the master circadian clock of mammals. Vasopressin signaling differentially modulated cellular rhythms along the anteroposterior axis of the network, suggesting a functional compartmentalization orthogonal to classic models. These results indicate peptide signaling regulates three-dimensional timekeeping across the master clock network.
Introduction
Daily rhythms in behavior and physiology are programmed by the circadian system, a collection of endogenous biological clocks that sustain ca. 24 h period and thus allow organisms to anticipate recurring daily environmental events relevant for survival (Mohawk et al., 2012). At the molecular level, circadian timekeeping is sustained by interconnected feedback loops that regulate the expression of clock genes (Buhr & Takahashi, 2013; Mackey, 2007). In mammals, CLOCK and BMAL1 dimers activate transcription of Period (Per) and Cryptochrome (Cry) genes, whose protein products inhibit their own transcription. Over time, PER and CRY proteins are degraded, allowing their transcription to recommence the following day. This molecular oscillator operates in nearly every cell of the body to regulate cellular physiology in a tissue-specific manner (Zhang et al., 2014).
At the system level, local tissue clocks in the brain and body are coordinated by the suprachiasmatic nucleus (SCN) of the anterior hypothalamus (Evans, 2016). Serving as the master clock, the SCN sends temporal cues to downstream tissues to coordinate them with one another and the 24 h day. Like other cells, individual SCN neurons are capable of sustained circadian timekeeping. However, intercellular signaling among SCN neurons drives several emergent network properties that are essential for master clock function. Networked SCN neurons produce cellular rhythms that are more precise, higher amplitude, and more robust than those of isolated SCN neurons (Herzog et al., 2004). Further, circadian period varies among individual SCN neurons (Welsh et al., 1995), and coordination of an ensemble period is needed for the transmission of coherent daily output signals (Low-Zeddies & Takahashi, 2001; Smyllie et al., 2016). In contrast to the tendency of networked SCN neurons to adopt a common period, SCN neurons display differences in the phase of cellular activity that contribute to master clock function in key ways. For instance, the phasing of cellular rhythms across the SCN network is thought to influence the daily patterning of daily outputs (Kalsbeek & Buijs, 2002; Rohling et al., 2006; Yamaguchi et al., 2003). In addition, cellular phase relationships across the anteroposterior axis of the SCN network likely encode photoperiod to regulate seasonal changes in behavior and physiology (Hazlerigg et al., 2005; Inagaki et al., 2007; Rohling et al., 2006). Thus, interactions among SCN neurons distributed across the network are critical for coordinating cellular rhythms so that the population forms a functional circadian pacemaker. Despite its importance for master clock function, the mechanisms that regulate spatiotemporal organization across the anteroposterior axis of the SCN network remain unknown.
As noted, emergent properties of the SCN network, including coordinated circadian phase and period, derive from intercellular signaling mediated by numerous signaling molecules (Southey et al., 2014). Based on differences in neuropeptide expression, the SCN is classically segregated into two compartments known as the shell and core (Abrahamson & Moore, 2001; Antle et al., 2003). The SCN shell spans the anteroposterior extent of the network and contains a large population of neurons that express arginine vasopressin (AVP). In contrast, the SCN core is enveloped by the SCN shell and contains neurons that express vasoactive intestinal polypeptide (VIP). VIP signaling is an important modulator of SCN function required for maintaining synchrony of cellular rhythms (Vosko et al., 2007). In contrast, the role of AVP remains unclear. Historically, AVP has been considered mainly as an SCN output signal (Antle et al., 2003; Kalsbeek et al., 2010), but recent evidence suggests that AVP may also regulate the function of the SCN network itself (Ingram et al., 1998; Li et al., 2009; Yamaguchi et al., 2013). Because most previous studies have focused on shell-core circuitry along the dorsoventral extent of the SCN, the degree to which neuropeptide signaling contributes to spatiotemporal organization along the anteroposterior axis of the network remains unclear.
Given that AVP neurons span the anteroposterior extent of the network, we hypothesized that AVP signaling modulates cellular rhythms across this domain. To test this hypothesis, we first investigated the anteroposterior expression of signaling components for AVP and other neuropeptides known to interact with AVP using in situ hybridization. We found evidence for patterned expression of all these pathways in the anterior versus posterior poles of the SCN network, including differential expression of AVP receptor V1a and V1b transcripts that suggest potentially diverging functions of AVP along the SCN anteroposterior axis. We then demonstrate that this anteroposterior asymmetry in transcript expression is associated with regional differences in the effect of AVP receptor antagonism on the period and phase of cellular rhythms across the SCN network. This work reveals the functional relevance of the anteroposterior axis of the SCN and identifies regional AVP signaling as the first major mechanism that modulates cellular rhythms across this dimension. Finally, we also present in situ data showing complementary expression of ligand/receptor pairs for both Vasoactive Intestinal Polypeptide (VIP) and Prokineticin2 (PROK2) signaling systems across the anteroposterior SCN. This suggests that anteroposterior asymmetry in intercellular signaling is a general SCN property.
Materials and methods
Animal and husbandry conditions
PERIOD2::LUCIFERASE (PER2::LUC) mice (Yoo et al., 2004, RRID: IMSR_JAX:006852), generated on a C57BL/6 background, and wild type C57BL/6 mice were bred and raised under a 24 h light:dark cycle with 12 h of light and 12 h of darkness (LD12:12). Vip−/− and Vip+/+ mice generated on a C57BL/6 background were bred and raised under LD14:10. Throughout life, ambient temperature was maintained at 22 ± 2 °C, and mice had ad libitum access to water and food (Teklad Rodent Diet #8604). These studies were conducted in accordance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and approved by the Institutional Animals Care and Use Committee at Marquette University, the Perelman School of Medicine at the University of Pennsylvania, and the Johns Hopkins Medical Institute.
In situ hybridization
Gene expression for Avp and Prok2 was assessed in adult male and female 2-to-6-month old C57BL/6 Vip+/+ and Vip−/− mice, with sexes apportioned equally among groups. Gene expression for V1a, V1b, Vipr2, and Prokr2 was assessed in adult 3-month-old male C57BL/6 wild type mice. Mice were entrained to LD 12:12 and released into constant darkness (DD) for 24 hours. Brains were flash frozen on dry ice during the second day in DD and processed for chromogenic in situ hybridization (Bedont et al., 2014). SCN were sectioned at 25 μm into five series on a Leica CM3050 cyrostat and stored at −80° C. Using RNAase-free technique, slide-mounted slices were fixed in 4% PFA in 1X phosphate buffer solution (PBS), acetylated in 0.1M triethanolamine hydrochloride (pH 8, Sigma, Cat#T9534), and activated with acetic anhydride (Sigma Cat#320102), interspersed with 1X PBS washes. Slides were then blocked in hybe buffer containing 50% formamide (Sigma, Cat#F7503) in 5X Saline-Sodium Citrate buffer (SCC, Sigma, S6639), 5X Denhardt’s solution (Sigma, Cat#D2532), 250 μg/mL yeast tRNA (Sigma Cat#10109525001), and 500 μg/mL salmon sperm DNA (Sigma Cat#11467140001) at room temperature for an hour. Slides were incubated overnight at 68° C in 100 μL of hybridization buffer spiked with 1μL of digoxigenin-labeled probe under a siliconized coverslip. The following day, slides were decoverslipped in 5X SSC, washed twice in 0.2X SSC for an hour at 65° C, transitioned to B1 buffer (100 mM Tris pH 7.5, 150 mM NaCl), blocked in B2 buffer (B1 buffer + 5% heat-inactivated sheep serum) at room temperature for an hour, then incubated overnight at 4° C in B2 buffer + 1:5000 Roche anti-digoxigenin-alkaline phosphatase antibody (Sigma Cat#11093274910). Finally, slides were washed in B1 buffer, transitioned to B3 buffer (100 mM Tris pH 9.5, 100 mM NaCl, 50 mM MgCl2), and laid out to colorize in humidified chambers at room temperature in B3 buffer + 0.34 mg/mL NBT (Thermo Fisher Cat#PI-34035), 0.18 mg/mL BCIP (Sigma Cat#11585002001), and 0.24 mg/mL levamisole hydrochloride (Sigma Cat#L9756). The colorization reaction was monitored under a dissecting microscope and extinguished in stop buffer (10 mM Tris pH 7.5, 50 mM EDTA) while the reaction was still within its linear range. As a quality control to ensure linearity, the reaction was terminated for a given probe either before the signal at peak expression saturated or before the background staining increased, whichever occurred first. Slides were then coverslipped in gelvatol. Probes generated from Open Biosystems clones were used for Avp (BE945936), Vipr2 (AI840195), and Prok2 (BU938827). Other probes were generated from TOPO clones made in-house using inserts amplified with these primers (written 5′-to-3′): V1a (TGTTTTTCCCATTGCTGTGA, CACAACAACCCTGTGAGGTG), V1b (AACTGTCCCGATTCTAACCTGA, GGAGGAGGACTCACCAATACAG), and Prokr2 (GGGACAGGCTTTACCACAGA, ACAGGGATGCTGAGGGTATG).
Densitometry analysis of in situ hybridizations
Bright field images were obtained with a 1-by-2 mosaic at 20X on an Axioskop 2 Mot Plus (Zeiss Instruments) or at 10X on an EVOS FL (Thermo Fisher). Using ImageJ, rolling ball correction and subtraction of signal from adjacent cell-poor domains were used to correct for background. To quantify density of gene expression within each SCN slice, a region of interest (ROI) was drawn around the boundaries of the SCN, and average signal intensity within that region was measured, producing a measure of signal density. For Avp, signal density of the SCN shell alone was measured by drawing an ROI specifically around the Avp+ subdomain of the SCN. SCN slices from different brains were aligned on SCN morphology and size. The aligned planes of section were numbered from 1–5; Plane 1 corresponds to the anterior-most plane of the SCN, while Plane 5 corresponds to the posterior-most plane of the SCN. To calculate the average signal density of gene expression across the entire SCN for each brain (defined here as Whole SCN) in a manner that accounted for differences in SCN size across planes, the average signal density of Planes1–5 was averaged after weighting the values from each plane by SCN area (as determined from the size of the ROI used to measure gene expression in the SCN of each slice). Finally, the signal for each probe was normalized for graphing purposes by setting the highest single-plane intensity to 100. Also, Vip−/− values were normalized to peak WT values to facilitate genotype comparisons.
SCN collection and bioluminescence recording
Male and female PER2::LUC mice (5–11 months old) were anesthetized with isoflurane and sacrificed by cervical dislocation 4–6 hours before lights-off (defined as Zeitgeber Time 12, ZT12). Brains were removed and sectioned in the coronal plane with a motorized vibratome (Leica Biosystems, VT1200S) in HBSS supplemented with HEPES, NaCHO3, and 0.01% penicillin/streptomycin. Three consecutive SCN slices (150μm) were collected, trimmed by hand with a scalpel, and cultured on a membrane with 1.18mL DMEM containing 0.2mM beetle luciferin (Gold Biotechnologies), HEPES, NaCHO3, 2% B27, and 0.01% penicillin/streptomycin. SCN slices were cultured in OPC 21268 (V1A antagonist, Cat#3924, Tocris Bioscience, Minneapolis, MN, USA), SR 49059 (V1A antagonist, Cat#2310, Tocris Bioscience, Minneapolis, MN, USA), and/or SSR 149415 (V1B antagonist, Cat#1116, Axon Medchem, Reston, VA, USA) at 2μM, 10μM, 50μM, 100μM, 200μM, or 400μM. Vehicle-treated control slices were cultured with < 0.1% DMSO. PER2::LUC rhythms were monitored for at least five days inside a light-tight chamber heated to 37° C. Bioluminescence imaging was conducted using a Stanford Photonics XR Mega-10Z cooled intensified CCD camera on a Zeiss AxioObserver Z1 microscope with a 10x Fluar objective lens. Images were collected with Piper Software (Stanford Photonics) at 15 frames/sec, filtered in real-time to eliminate single-image noise events (i.e., cosmic rays), and stored as 90-sec images collected once every 10min. A 2 h moving average was then applied, images were converted to 8-bit, pixel dimensions were reduced in half, and three consecutive images were summed to produce a series of 30 min images.
SCN bioluminescence analyses
PER2::LUC rhythms were analyzed with Lumicycle and Matlab-based scripts as described previously (Evans et al., 2011). Time in vitro was normalized across samples to account for differences in the clock time of the start of recording by expressing the start time of recording relative to Zeitgeber Time (e.g., if a recording started 1 h after lights-off, the ZT start time would be ZT13). To assess whole tissue PER2::LUC rhythms from imaging, a time series was generated using a hand-drawn region encompassing each lobe (ImageJ Software) and analyzed with Lumicycle software. PER2::LUC rhythms from SCN sub-regions and cell-like ROIs were extracted and analyzed using Matlab-based scripts. For individual phase maps, a time series was generated for each 12-pixel diameter ROI on a uniform grid with 2-pixel spacing. The time series for each ROI was judged to exhibit a significant circadian rhythm if the autocorrelation coefficient with a lag of 24h was significant at α = 0.05, a local maximum occurred in the autocorrelation corresponding to a lag between 18h and 30h, and the signal-to-noise ratio of the time series was at least 0.75. The linear trend was eliminated and a Butterworth filter was applied once forward and once backward to remove high- and low-frequency interference. For composite maps illustrating the results for a whole group, a representative slice was selected for each coronal position and other slices collected at that position were aligned to that sample by minimizing the sum of squared difference of the 24h-summed bioluminescence profiles. Using composite maps, spatially segregated regions were selected for analyses of cellular ROIs. An iterative process was employed to locate and extract data from cell-like ROIs after background and local noise subtraction. Period was calculated as the average daily period over the first three cycles in vitro. Phase was calculated as the number of hours between the start of the recording and the time of peak PER2::LUC expression on the first cycle in vitro. Because time in vitro is corrected for the ZT start time of recording, this measure is referred to as “ZT Peak Time”. To examine phase relationships across the SCN network in average phase maps, regional ZT peak time was expressed relative to peak time of the whole anterior slice on the first cycle in vitro, and referred to as “Relative Peak Time”. Lastly, damping rate of PER2::LUC rhythms was recorded using Lumicycle analysis software, which calculates the number of days required for the rhythm amplitude to decrease to 1/e of the initial value (ca. 37%).
Immunohistochemistry
To assess the relationship between regional differences in PER2::LUC rhythms and neuropeptide expression, SCN slices like those used for imaging were processed for arginine vasopressin (AVP) and vasoactive intestinal polypeptide (VIP) expression, as in previous work (Evans et al., 2011). Specifically, SCN slices were sectioned at 150μm and cultured immediately with colchicine-treated medium (25μg/ml) for 24 h, fixed with 4% paraformaldehyde, and then cyprotected in sucrose solution. Free-floating slices were rinsed in PBS, blocked in PBS diluted (1:200) + 2.5% normal donkey serum + 0.3% TritonX-100 (PBS+NDS+TrX), and then incubated in primary antibodies (anti-AVP, 1:1K Cat#T-5048, RRID: AB_2313978, anti-VIP, 1:500, Cat#T-4246, RRID: AB_518682; both from Peninsula Laboratories, San Carlos, CA, USA) diluted in PBS+NDS+TrX for 48 h at 4° C. Slices were rinsed in PBS before 2 h incubation in secondary antibodies diluted in PBS+NDS+TrX (1:200; Alexa Fluor 488 Donkey Anti-Rabbit, Cat#125719, RRID: AB_2556546, Alexa Fluor 594 Donkey Anti-Guinea Pig Cat#104099, RRID: AB_2340474; both from Jackson ImmunoResearch West Grove, PA, USA). Slices were rinsed in PBS before mounting onto microscope slides with Prolong Gold AntiFade,(Invitrogen, Carlsbad, CA, USA), Fluorescence images were obtained with a Nikon 80i microscope fitted with a Retiga 2000R digital camera (QImaging, Surrey, BC, Canada) connected to a computer running NIS Elements-D software (Nikon Instruments, Melville NY, USA).
Statistical analyses
Rhythmicity of gene expression in Planes 1–5 and whole SCN was assessed using JTK-Cycle version 3.1 (Hughes et al., 2010). Other statistical analyses were conducted using JMP software. Full factorial ANCOVA was used for in situ data (main factors: Circadian Time (CT), Plane of section, CT*Plane) and PER2::LUC imaging data (main factors: Antagonist Group, SCN Slice/Region, Group*Slice/Region), with Mouse ID# included as the ANCOVA regression factor to account for multiple slices/cells being collected from the same mouse. In addition, in situ data were binned into subjective Day (CT04-CT12 for all probes except Prok2, which used CT00-CT08) and subjective Night (CT16-24 for all probes except Prok2, which used CT12-CT20) phases of the circadian cycle and submitted to one-way ANCOVA to assess overall spatial differences in gene expression. Also, one-way ANCOVA was used to test within-groups differences in PER2::LUC rhythms across slices or regions. Post hoc comparisons used Tukey’s HSD and/or least square mean (LSM) contrasts. To compare rhythmic gene expression in each plane to the Whole SCN, we conducted LSM contrasts using a separate full factorial ANCOVA for each plane (α = 0.01 to account for five models generated to analyze all five planes). All data are represented in figures and text as Mean ± SEM.
Results
Anteroposterior asymmetry in SCN expression of AVP signaling components
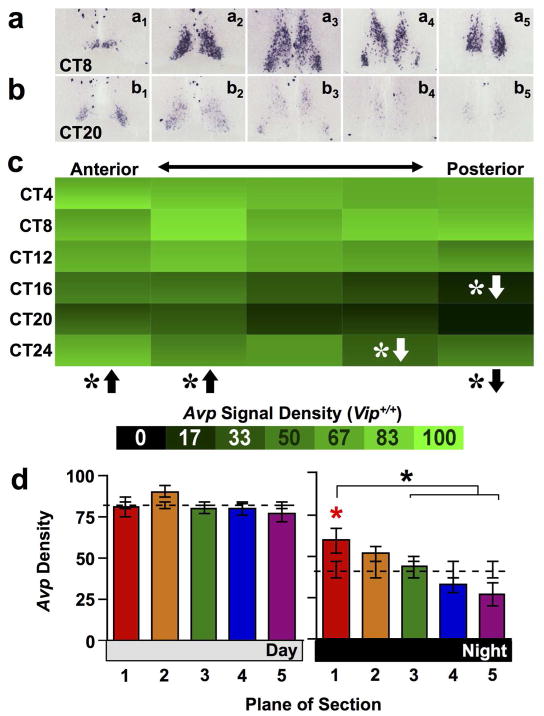
Hypothesizing that anteroposterior differences in SCN function are regulated by AVP signaling, we used in situ hybridization to assess Avp expression along five planes of the SCN shell. As expected, Avp was rhythmically expressed in each anteroposterior plane with a peak during mid-subjective day (Figure 1a–c, JTK-Cycle: Plane1 p < 0.05, Planes2–5 p < 0.0001, Whole Shell p < 0.0001). The density of Avp expression also varied spatially across the anteroposterior SCN (Figure 1a–c, full factorial ANCOVA effect of Plane: p < 0.0001). Relative to average density of expression across the Whole SCN shell, the SCN anterior shell had higher signal density and the posterior shell had lower signal density when overall Avp expression was assessed independent of circadian time (Figure 1a–c, LSM Contrasts: Planes 1,4,5 p < 0.05). This spatial gradient interacted with the circadian regulation of Avp (full factorial ANCOVA interaction: p < 0.05), reflecting higher Avp signal density in the anterior SCN shell relative to the Whole Shell specifically during subjective night (Figure 1d, Day one-way ANCOVA: p = 0.28; Night one-way ANCOVA: p < 0.01; LSM Contrasts: Plane 1 p < 0.05), coincident with the timing of the trough of the Avp rhythm. Elevated Avp expression in the anterior shell during the subjective nighttime was the primary driver of a 3-fold lower amplitude rhythm relative to that displayed by the posterior shell (JTK-Cycle: Amplitude for Planes1–5 = 6.97, 13.37, 13.22, 15.19, 20.05). Similar spatiotemporal patterns were evident when Avp expression density was measured using an ROI that encompassed the entire SCN rather than just the SCN shell (data not shown).
Figure 1. Avp expression across the anteroposterior SCN in wild type mice.
a–b) Representative slices illustrating Avp expression across the anteroposterior SCN in wild type mice. Subscript indicates plane of section from Plane 1 (most anterior) to Plane 5 (most posterior). Images for each circadian time (CT) illustrate the times of peak and trough expression. c) Heat maps illustrating temporal patterns of Avp expression for each plane of section 1–5, corresponding to plane of section in the raw images immediately above (n = 3 slices per circadian time point for each plane). For each cell, variance is encoded as a vertical color gradient depicting +/− SEM. White asterisks with directional arrows indicate that Avp expression in that plane of section was higher (up arrow) or lower (down arrow) density relative to the Whole SCN at that circadian time point. LSM Contrasts: * p < 0.01. Black asterisks with directional arrows below heat maps indicate that Avp expression in that plane of section differed from Whole SCN across the 24-hour cycle overall. LSM Contrasts: * p < 0.05. d) Normalized Avp expression (Mean ± SEM) during subjective day (CT04-CT12) and subjective night (CT16-24) across the anteroposterior SCN (n = 9 slices per time bin per plane). Dashed line indicates average density for the Whole SCN. Asterisks with brackets indicate differences among SCN planes- Tukey’s HSD, * p < 0.05. Asterisks located immediately above bars (color-coded in online version) indicate SCN planes that differ from Whole SCN- LSM Contrasts * p < 0.01.
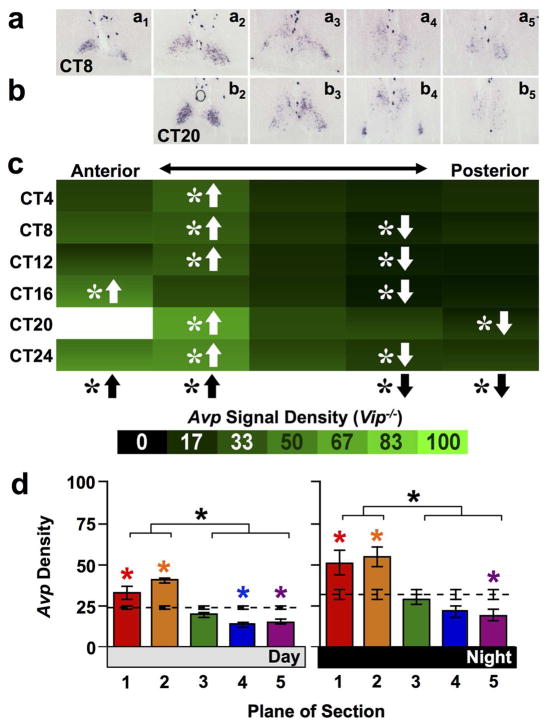
Since prior work has shown that SCN Avp transcription is reduced markedly by the absence of VIP signaling (Bedont et al., 2017; Harmar et al., 2002), we assessed Avp expression in Vip−/− mice to test how VIP signaling interacts with the anteroposterior gradient of Avp expression. As expected, Avp expression in the SCN shell of Vip−/− mice was decreased relative to wild type mice (Figure 2a–c, full factorial ANCOVA effect of Genotype: p < 0.0001). Despite displaying lower Avp expression, Vip−/− mice were similar to WT mice in that the overall density of Avp expression was highest in the anterior shell and lowest in the posterior shell (Figures 2a–c, full factorial ANCOVA effect of Plane and interaction: p < 0.0001; LSM Contrasts: Planes 1,2,4,5 p < 0.001). However in contrast to WT mice, the anterior shell displayed higher Avp signal density than the Whole Shell during both subjective day and night phases of the circadian cycle (Figure 2d, Day and Night one way ANCOVA: p < 0.0001, LSM Contrasts: Planes 1,2,5 p < 0.01, Plane 4 (Day only) p < 0.01). Surprisingly, weak Avp rhythmicity persisted in Vip−/− SCN shell (JTK-Cycle: Whole Shell p < 0.001, Plane2 p = 0.06, Plane3–5 p < 0.05), but the phase of the rhythm was inverted relative to wildtype mice (Figure 2c–d). Also, there was a trend toward greater rhythm amplitude in the anterior shell compared to the posterior shell (Amplitude for Planes2–5 = 7.81, 3.46, 3.73, 2.47), which also differed from wild type mice. Together, these data demonstrate that the density of Avp expression is elevated in the anterior SCN in a VIP-independent manner, which is obscured in wild type animals by a stronger, anti-phasic rhythm of Avp expression driven by VIP signaling.
Figure 2. Avp expression across the anteroposterior SCN in Vip−/− mice.
a–b) Representative slices illustrating Avp expression in Vip−/− mice. c) Heat maps illustrating temporal patterns of Avp expression, with density values of Vip−/− SCN normalized to peak values in wild type SCN to facilitate genotype comparisons. n = 3 slices per circadian time point (except CT20 Plane 1, n = 1 slice). d) Normalized Avp expression (Mean ± SEM) during subjective day (CT04-CT12) and subjective night (CT16-24) across the anteroposterior SCN. n = 7–9 slices per Day/Night time point for each plane. Other conventions as in Figure 1.
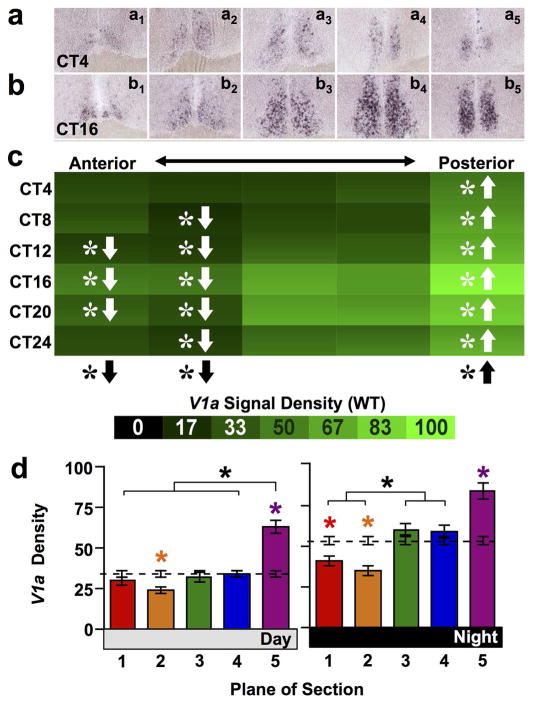
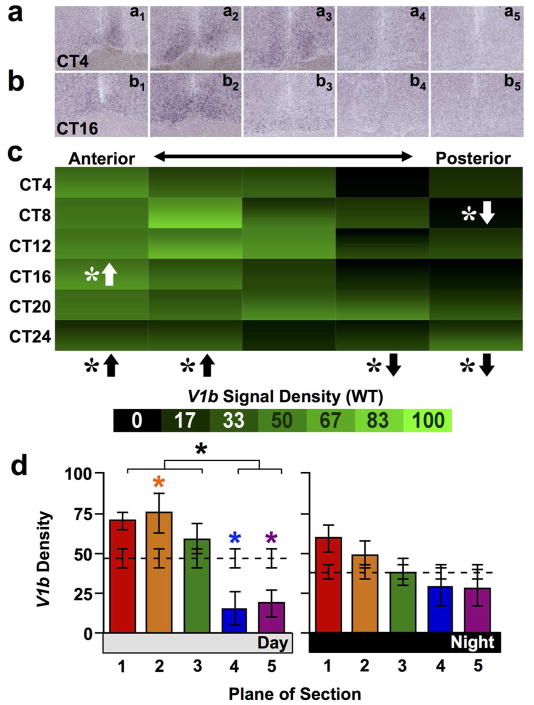
Complementing Avp, the AVP receptors V1a and V1b were asymmetrically expressed across the anteroposterior SCN in wild type mice. V1a expression was rhythmic in all but the anterior-most SCN (Figure 3a–c, JTK-Cycle: Whole SCN p < 0.0001, Plane1 p = 0.09, Plane2–5 p < 0.01) and with a greater amplitude rhythm in the posterior SCN (Amplitude for Planes1–5 = 2.22, 2.77, 9.10, 7.68, 7.32). Further, V1a was differentially expressed across the anteroposterior SCN (full factorial ANCOVA effect of Plane: p < 0.0001) in a manner that interacted with circadian time (full factorial ANCOVA interaction: p < 0.01). Relative to the Whole SCN, V1a density was higher in the posterior-most SCN and lower in the anterior SCN (Figure 3c, LSM Contrasts: Planes 1,2,5 p < 0.0001) during both subjective day and night (Figure 3d, one-way ANCOVA p < 0.0001 for both Day and Night; LSM Contrasts: Planes 2,5 p < 0.05, Plane 1 (Night only) p< 0.05). In contrast, overall V1b expression was not rhythmic (Figure 4a–c, JTK-Cycle: Whole SCN p = 0.43, Planes1–5 p > 0.1). Overall V1b signal density was higher in anterior SCN and lower in posterior SCN compared to Whole SCN (Figure 4c, full factorial ANCOVA effect of Plane: p < 0.0001, LSM Contrasts: Planes 1,2,4,5 p < 0.05), and spatial differences in V1b expression did not interact with circadian time (ANCOVA interaction: p = 0.13). Nevertheless, anteroposterior differences in V1b signal density were evident during subjective daytime (Figure 4d, Day one-way ANCOVA: p < 0.0001, LSM Contrasts: Planes 1,4,5 p < 0.05), but not subjective nighttime (Figure 4d, Night one-way ANCOVA: p = 0.14).
Figure 3. V1a expression across the anteroposterior SCN in wild type mice.
a–b) Representative slices illustrating V1a expression. c) Heat maps illustrating temporal patterns of V1a expression. n = 3–4 slices per circadian time point for each plane. d) Normalized V1a expression (Mean ± SEM) during subjective day (CT04-CT12) and subjective night (CT16-24) across the anteroposterior SCN. n = 11–12 slices per group per plane. Other conventions as in Figure 1.
Figure 4. V1b expression across the anteroposterior SCN in wild type mice.
a–b) Representative slices illustrating V1b expression. c) Heat maps illustrating temporal patterns of V1a expression. n = 3–4 slices per circadian time point for each plane. d) Normalized V1a expression (Mean ± SEM) during subjective day (CT04-CT12) and subjective night (CT16-24) across the anteroposterior SCN. n = 11–12 slices per group per plane. Other conventions as in Figure 1.
Anteroposterior gene expression as a general principle of SCN organization
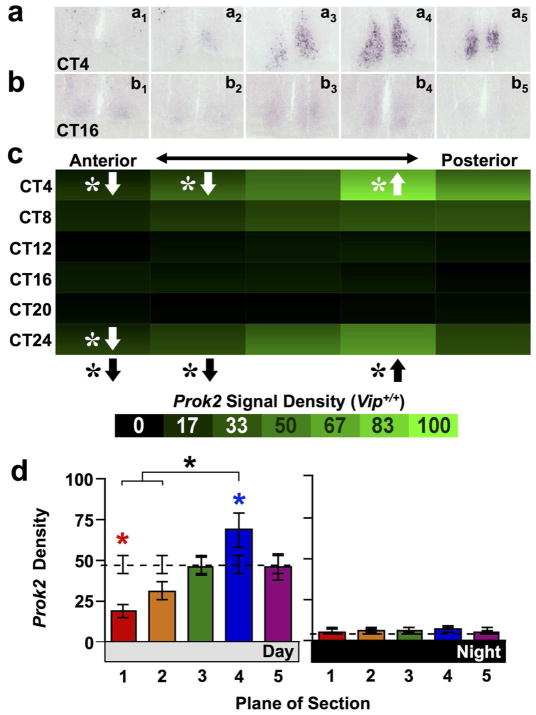
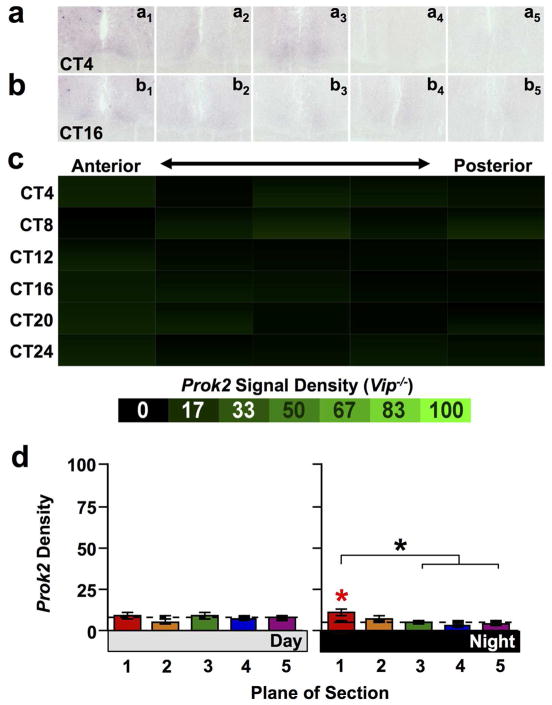
Next we tested whether asymmetry was specific to Avp and V1 expression by assessing anteroposterior differences in the expression of other SCN neuropeptide signaling systems that interact with AVP signaling. Expression of the Prok2 transcript is regulated by V1A signaling (Li et al., 2009). Thus, we hypothesized that expression of PROK2 signaling components might be asymmetrically distributed across the anteroposterior SCN. In agreement with this hypothesis, the overall density of Prok2 expression in wild type mice varied spatially across the anteroposterior SCN (Figure 5a–c, full factorial ANCOVA effect of Plane and interaction: p < 0.0001). Relative to the Whole SCN, the overall density of Prok2 expression was higher in posterocentral SCN (Plane 4) and lower in the anterior SCN (Figure 5a–c, LSM Contrasts: Planes 1,2,4 p < 0.05). Prok2 signal density was concentrated in Plane 4 specifically during subjective day (Figure 5d; Day one-way ANCOVA: p < 0.001; LSM Contrasts: Planes 1,4 p < 0.05; Night one-way ANCOVA: p = 0.95). In addition, Prok2 expression was significantly rhythmic in all slices except those collected from the anterior-most SCN (Figure 5c, JTK-Cycle: Whole SCN p < 0.001, Plane1 p > 0.1, Planes2–5 p < 0.05), with the largest amplitude rhythms observed in the posterior SCN (Amplitude for Planes1–5 = 0.77, 4.95, 8.05, 11.88, 7.24). Similar to Avp, peak Prok2 expression was reduced in Vip−/− mice relative to wild type mice (Figure 6c–d, full factorial ANCOVA effect of Genotype: p < 0.0001). However, Prok2 was not rhythmic in Vip−/− SCN (Figure 6c, JTK-Cycle: Whole SCN p = 0.87, Planes1–5 p > 0.1), and Prok2 expression in Vip−/− SCN did not differ spatially (full factorial ANCOVA effect of Plane: p = 0.14, ANCOVA interaction: p = 0.53). Interestingly, a weak enrichment of Prok2 signal density was detected in the anterior-most SCN compared to whole SCN specifically during subjective night (Figure 6d, Day one-way ANCOVA: p = 0.79; Night one-way ANCOVA: p < 0.01; LSM Contrasts: Plane 1 p < 0.01), which was opposite the spatiotemporal patterns of Prok2 expression detected in wild-type mice.
Figure 5. Prok2 expression across the anteroposterior SCN in wild type mice.
a–b) Representative slices illustrating Prok2 expression. c) Heat maps illustrating temporal patterns of V1a expression. n = 3 slices per circadian time point for each plane. d) Normalized V1a expression (Mean ± SEM) during subjective day (CT00-CT08) and subjective night (CT12-CT20) across the anteroposterior SCN. n = 9 slices per group per plane. Other conventions as in Figure 1.
Figure 6. Prok2 expression across the anteroposterior SCN in Vip−/− mice.
a–b) Representative slices illustrating Prok2 expression. c) Heat maps illustrating temporal patterns of V1a expression. n = 3 slices per circadian time point for each plane. d) Normalized V1a expression (Mean ± SEM) during subjective day (CT00-CT08) and subjective night (CT12-CT20) across the anteroposterior SCN. n = 9 slices per group per plane. Other conventions as in Figure 1.
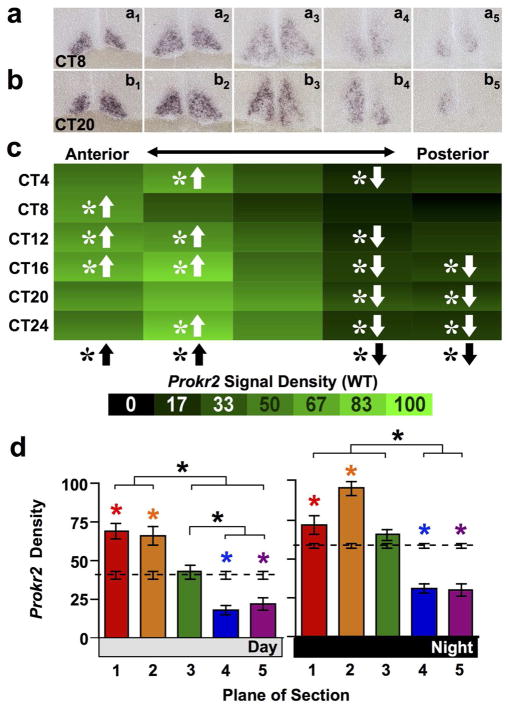
Next we tested spatiotemporal patterns in the expression of the SCN PROK2 receptor Prokr2 in wild type mice. Like rhythmically expressed V1a, Prokr2 expression displayed a spatial patterning that was complementary to its ligand. Prokr2 was only rhythmically expressed in the anterior and middle SCN (Figure 7a–c, JTK-Cycle: Whole SCN p < 0.0001, Planes2–3 p < 0.01, Plane4 p = 0.06, Planes1,5 p > 0.1). Spatially, density of Prokr2 expression varied along the anteroposterior SCN in a manner that did not interact significantly with circadian time (Figure 7c, full factorial ANCOVA effect of Plane: p < 0.0001; ANCOVA interaction: p = 0.1), reflecting consistently higher anterior and lower posterior signal density relative to average density across the whole SCN (Figure 7c, LSM Contrasts: Planes 1,2,4,5 p < 0.0001). This was true during both subjective day and night (Figure 7d, one-way ANCOVA p < 0.0001 for both Day and Night; LSM Contrasts: Planes 1,2,4,5 p < 0.05).
Figure 7. Prokr2 expression across the anteroposterior SCN in wild type mice.
a–b) Representative slices illustrating Prokr2 expression. c) Heat maps illustrating temporal patterns of V1a expression. n = 3–4 slices per circadian time point for each plane. d) Normalized V1a expression (Mean ± SEM) during subjective day (CT04-CT12) and subjective night (CT16-24) across the anteroposterior SCN. n = 11–12 slices per group per plane. Other conventions as in Figure 1.
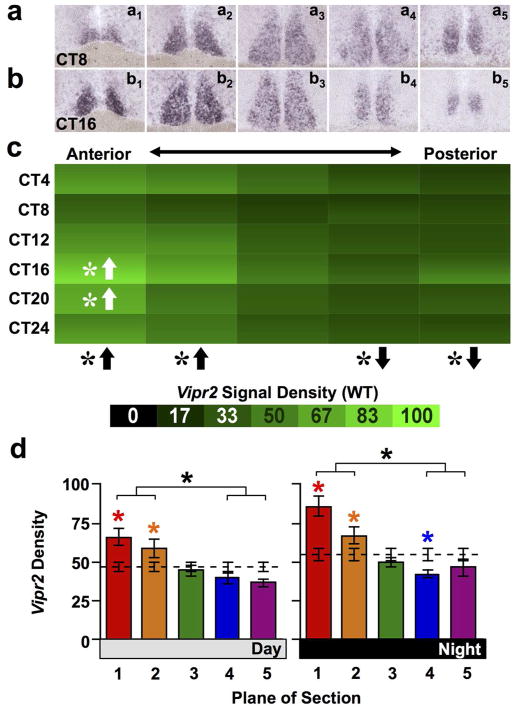
Finally, given the spatially modulated dependence of Avp expression upon VIP signaling, we tested for polarization of VIP signaling components along the anteroposterior SCN. We chose to focus on the VIP receptor Vipr2 because it is expressed across the full anteroposterior extent of the SCN and anteroposterior variations in its expression have not been examined closely. In contrast, Vip is well known to be concentrated in the core domain of the SCN (Abrahamson & Moore, 2001; An et al., 2012; Bedont et al, 2014). Only the anterior-most SCN plane displayed rhythmic Vipr2 expression (Figure 8a–c, JTK-Cycle: Plane1 p < 0.05, Whole SCN p = 0.11, Planes2–4 p > 0.1, Plane5 p = 0.08). Further, Vipr2 expression varied spatially in a manner that did not interact with circadian time (Figure 8a–c, full factorial ANCOVA effect of Plane: p < 0.0001; ANCOVA interaction: p = 0.87) due to higher density of Vipr2 signal in anterior SCN and lower density in posterior SCN compared to whole SCN (Figure 8a–c, LSM Contrasts: Planes 1,2,4,5 p < 0.05). Higher anterior Vipr2 signal density compared to whole SCN was detectable during both subjective day and night (Figure 8d, one-way ANCOVA: p < 0.0001 for both Day and Night; LSM Contrasts: Planes 1,2 p < 0.05, Plane 4 (Night only) p < 0.05). Collectively, these results reveal that signaling components for three distinct neuropeptides (i.e., Avp, Prok, Vip) are expressed asymmetrically in complementary patterns along the anteroposterior SCN, suggesting that this is a general organizational motif in the master clock.
Figure 8. Vipr2 expression across the anteroposterior SCN in wild type mice.
a–b) Representative slices illustrating Vipr2 expression. c) Heat maps illustrating temporal patterns of V1a expression. n = 3–4 slices per circadian time point for each plane. d) Normalized V1a expression (Mean ± SEM) during subjective day (CT04-CT12) and subjective night (CT16-CT24) across the anteroposterior SCN. n = 11–12 slices per group per plane. Other conventions as in Figure 1.
V1 antagonism modulates SCN period and phase in a dose-dependent manner
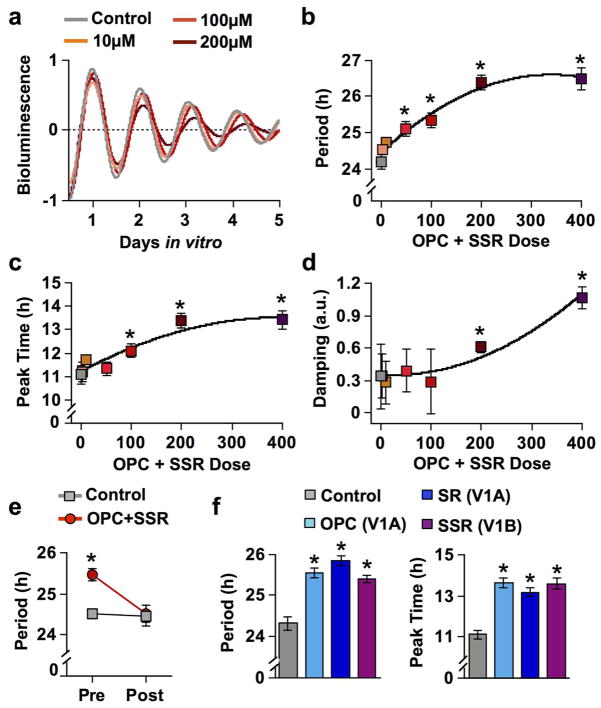
To test whether regional differences in Avp and V1 expression modulate SCN function, we performed PER2::LUC bioluminescence recording on SCN slices cultured with V1 antagonists (Figure 9a). To select an effective dose for V1 antagonism, we first used luminometry to record whole-tissue PER2::LUC rhythms from SCN slices cultured with antagonists for both V1A and V1B receptors (OPC and SSR, respectively) at one of seven doses (0μM, 2μM, 10μM, 50μM, 100μM, 200μM, or 400μM). In control SCN slices, the period of PER2::LUC rhythms was ca. 24 h (Figure 9b). V1 antagonism at doses ≥ 50μM increased the period of SCN rhythms, with the magnitude of the effect proportional to dose (Figure 9b). Consistent with its period lengthening effect, V1 antagonism delayed the time of peak PER2::LUC expression on the first cycle in vitro in a manner dependent on antagonist dose (Figure 9c). As expected, there was a positive correlation between period and phase (Spearman’s Rho: R2= 0.55, p < 0.0001). In addition, 200μM and 400μM OPC+SSR increased the damping rate of PER2::LUC rhythms, but rhythm damping was not produced by lower doses of V1 antagonism (Figure 9d). Collectively, these data indicate that AVP signaling regulates the period and phase of SCN rhythms.
Figure 9. V1 antagonism lengthens SCN period and delays phase in a dose-dependent manner.
a) PER2::LUC rhythms of SCN slices at different doses of OPC+SSR. b) Dose response curve of average period length (± SEM) of SCN slices in vitro. c) Dose response curve for average ZT peak time (± SEM) of SCN slices in vitro. d) Dose response curve for rhythm damping (± SEM) in SCN slices in vitro. e) Period length of SCN slices cultured with 100μM OPC+SSR or vehicle control before (pre) and after (post) medium exchange to washout drugs. f) Period length and peak time of SCN slices cultured with vehicle control or 100μM single receptor antagonists: OPC (V1A), SR (V1A), or SSR (V1B). LSM Contrasts *p < 0.01 different from control; n = 4–9 slices per group. Note that small error bars may be obscured by symbols.
We proceeded to test if the effects of V1 antagonism were reversible, using the 100μM dose that altered both SCN period and phase without damping PER2::LUC rhythms. Upon drug washout, SCN period returned to ca. 24 h (Figure 9e), indicating that the effect of V1 antagonism was not permanent. Next we tested the role of each V1 receptor separately by culturing SCN slices with one of two different antagonists of the V1A receptor (OPC, SR) or the one available antagonist of the V1B receptor (SSR). Similar to dual antagonism, each V1 antagonist when applied alone increased period by 1–2 h and caused a 1–2 h delay in phase (Figure 9f). The two different V1A antagonists each elicited comparable effects on SCN period and phase (Figure 9f), thus demonstrating that these changes are not due to off-target effects. V1B antagonism likewise lengthened period and delayed phase, providing complementary evidence that AVP signaling modulates SCN function by affecting the period and phase of the network.
AVP signaling modulates SCN period in a region-specific manner
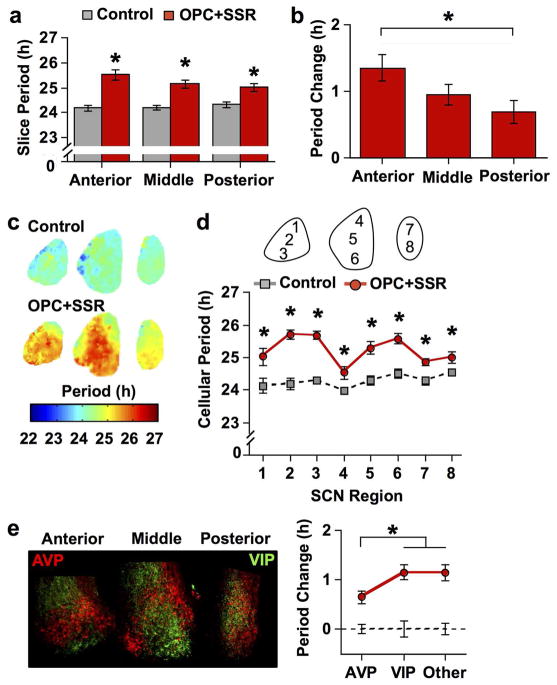
Next we investigated whether spatial patterning of V1 receptors influences the function of the SCN network along the anteroposterior axis. Three consecutive SCN slices were collected from each mouse for PER2::LUC imaging and cultured with medium containing either vehicle or 100μM OPC+SSR (i.e., the dose that altered period and phase without damping PER2::LUC rhythms, Figure 9). As expected, the period of SCN rhythms in vehicle-treated slices was ca. 24 h regardless of anteroposterior position (Figure 10a, one-way ANCOVA effect of Slice: p = 0.69), consistent with previous work using this approach (Evans et al., 2011). V1 antagonism increased SCN period by 1–2 h (Figure 10a, full factorial ANCOVA effect of Antagonist: p < 0.0001), with larger period lengthening effect in the anterior SCN than in the posterior SCN (Figure 10b, one way ANCOVA effect of Slice p < 0.05, Tukey’s HSD: p < 0.05). These results indicate that AVP signaling modulates SCN period differentially across the anteroposterior extent of the master clock network.
Figure 10. V1 antagonism lengthens SCN period in a region-dependent manner.
a) Average period length (± SEM) in vitro of SCN slices collected from three anteroposterior positions. b) Average change in cellular period (± SEM) relative to control conditions (line at 0) across anteroposterior positions. c) Period maps (color-coded in online version) illustrating regional effects of V1 antagonism. d) Average period length (± SEM) of cellular rhythms in different SCN regions. e) Magnitude of the period lengthening across neurochemically-distinct regions. Left: Representative images illustrating AVP- and VIP-immunoreactivity in cultured SCN slices (color-coded in online version). Note VIP+ neurons are located in the ventral region of the anterior and middle SCN slice, which can be distinguished from VIP+ fibers in other regions by the brighter, circular staining pattern. Right: Average change in cellular period (± SEM) relative to control conditions (dashed line at 0) across neuropeptide regions. Regions used for grouping are AVP: 1, 4, 7, 8; VIP: 3, 6; Other: 2, 5. For A–E) * p < 0.01, + p < 0.05, n = 7–8 slices per group for each plane. Note that small error bars may be obscured by symbols.
Having established that period-lengthening effects of V1 antagonism vary across the anteroposterior SCN, we next tested for regional differences in the coronal plane of each slice. Similar to our whole slice analyses (Figure 10a), SCN neurons in control slices displayed ca. 24 h rhythms (Figure 10c–d, one-way ANCOVA effect of Region p = 0.13). V1 antagonism increased cellular period in a manner that varied by region (Figure 10c–d, full factorial ANCOVA effect of Antagonist and interaction: p < 0.005). Interestingly, V1 antagonism lengthened the period of cellular rhythms to the largest extent in the central and ventral regions of the anterior and middle SCN slice (Figure 10c–d). Phase mapping analyses suggested that regional differences in the magnitude of period changes were reminiscent of the shell-core compartmentalization of the SCN network (Figure 10c), with the most affected regions lying within the SCN core. Indeed, the greatest change in cellular period was observed in non-AVP expressing regions (Figure 10e, Tukey’s HSD: p < 0.05). Together, these results indicate that AVP signaling acts to shorten the period of SCN neurons throughout the network, with the greatest effect on the period of non-AVP neurons.
AVP signaling influences phase differences among SCN neurons
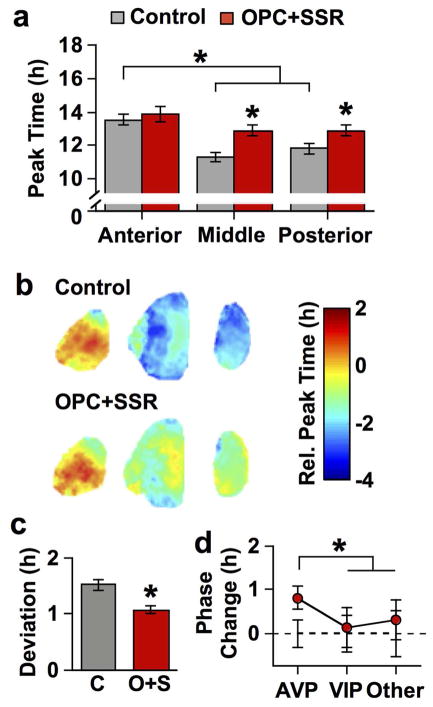
Given the effect on period length varied by SCN region, we predicted that V1 antagonism would alter phase relationships among SCN neurons. Under control conditions, the SCN network displayed gradients in the phase of cellular rhythms across its three dimensions (Figure 11a–b, one-way ANCOVA: p < 0.0001), as shown previously (Evans et al., 2011, 2013). One of the most pronounced features is a regional node within the anterior SCN that peaks 2–4 h later than regions in the middle and posterior SCN (Figure 11b). V1 antagonism reduced the magnitude of phase differences across the anteroposterior SCN (Figure 11a, one-way ANCOVA p > 0.2) by delaying the phase of the middle and posterior SCN (Figure 11a–b, full factorial ANCOVA effect of Antagonist: p < 0.001, LSM Contrasts: p < 0.05). Consistent with reduced regional phase differences, V1 antagonism also reduced deviation in the phasing of cellular rhythms across the anteroposterior SCN (Figure 11c, Student’s t test: p < 0.05). Interestingly, the largest effect of V1 antagonism was on the phase of cellular rhythms in AVP-expressing regions (Figure 11d, Tukey’s HSD: p < 0.05). Collectively, these results suggest that AVP signaling contributes to phase heterogeneity within the SCN network by influencing cellular phase, with the largest influence on SCN neurons that express AVP.
Figure 11. V1 antagonism alters SCN phase relationships by delaying phase in a region-specific manner.
a) Average peak time (± SEM) on the first day in vitro normalized to the zeitgeber time (ZT) on the day of slice collection. b) Average phase maps for control and OPC+SSR anteroposterior SCN slices (color-coded in online version). c) Standard deviation of peak time displayed by SCN neurons across all three slices collected from each mouse. d) Average change in cellular phase (± SEM) relative to control conditions (dashed line at 0) across neurochemically-distinct regions. Regions used for each neurochemical group are AVP: 1, 4, 7, 8, Other: 2, 5, VIP: 3, 6. For A–E) * p < 0.01; +, p < 0.05; n = 7–8 slices per group for each plane. Note that small error bars may be obscured by symbols.
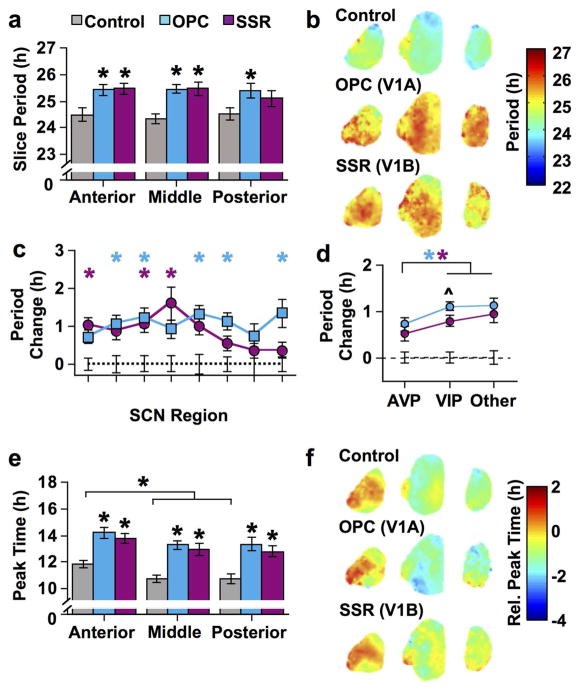
V1A and V1B receptors both contribute to period and phase modulation
To test the role of each V1 receptor in modulating the spatiotemporal function of the SCN network, we cultured SCN slices with single antagonists targeting either the V1A (OPC) or V1B (SSR) receptor. Similar to dual antagonism, each antagonist alone increased period by 1–2 h in the anterior and middle SCN (Figure 12a, full factorial ANCOVA effect of Antagonist: p < 00001, LSM Contrasts: p < 0.0005), but only V1A antagonism lengthened period in the posterior SCN (Figure 12a, LSM Contrasts: p < 0.0005). Similarly, there were regional differences in the effect of V1A versus V1B antagonism on cellular rhythms (Figure 12b–d, full factorial ANCOVA effect of Antagonist: p < 00001, LSM Contrasts: p < 0.0005). In contrast, both V1A and V1B antagonists influenced the phasing of SCN rhythms to a similar extent (Figure 12e–f, full factorial ANCOVA effect of Antagonist: p < 00001, LSM Contrasts: p < 0.0001). These results suggest that AVP signaling can influence SCN period and phase by activating either V1 receptor subtype.
Figure 12. Signaling through V1A and V1B AVP receptors influences SCN period and phase.
a) Average period (± SEM) in vitro of SCN slices. b) Average period maps for each group (color-coded in online version). c) Average change in cellular period (± SEM) relative to control conditions (dashed line at 0) in different SCN regions. d) Average change in cellular period (± SEM) relative to control conditions (dashed line at 0) across neurochemically-distinct regions. Regions used for each neurochemical group are AVP: 1, 4, 7, 8, Other: 2, 5, VIP: 3, 6. e) Average ZT peak time (± SEM) in vitro of SCN slices. f) Average phase maps for each group (color-coded in online version). For a & e) * p < 0.01; + p < 0.05. For c–d) * p < 0.01 different from control (color-coded in online version to represent specific antagonist group); ^ p < 0.01 difference between V1A and V1B antagonists; n = 7–8 slices per group for each plane. Note that small error bars may be obscured by symbols.
Discussion
A conserved principle in neuroscience is that network dynamics are dictated by cellular interactions integrated across time and space. In the SCN, cellular interactions determine collective period and phase, which are properties critical for clock function. Here we demonstrate that AVP signaling is an important mechanism that modulates SCN spatiotemporal function differentially along the anteroposterior axis of the network. Notably, we find that anteroposterior asymmetries in the expression of AVP signaling components are associated with regional differences in the magnitude of period and phase effects produced by V1 antagonism. This underscores the growing appreciation that SCN spatiotemporal organization is critical to understanding the function of the master circadian clock, although the anteroposterior spatial domain is rarely examined in studies of SCN function. Anteroposterior asymmetries are also evident in the expression of other important neuropeptides and receptors, suggesting that patterned signaling along this axis is a general organizational property of the SCN network. Collectively, these results provide new mechanistic insight into SCN spatiotemporal organization with important implications for master clock function and how it adjusts to changing environmental conditions.
AVP signaling modulates SCN period in a spatiotemporal manner
The present study provides evidence that AVP signaling directly influences the period of SCN rhythms, which has not been appreciated fully in previous work. Pharmacological inhibition of V1 receptors has been observed to lengthen period of organotypic SCN slices collected from postnatal PER2::LUC mice (Edwards et al., 2016), but changes in SCN period have not been reported in other work using lower doses (Maywood et al., 2011). We reconciled these observations by constructing a dose response curve, which demonstrates that period is lengthened by V1 antagonism in a dose-dependent manner in our slice preparation (i.e., acutely dissected SCN slices collected from adult mice). Because the antagonist dose/type and the age/nature of the preparation can vary across labs, these may be important factors to consider given evidence that the role of AVP signaling changes with age (Ono et al., 2016).
Genetic approaches examining the influence of AVP signaling on SCN function have likewise produced conflicting results. The free-running period of locomotor rhythms is lengthened in Brattleboro rats (Groblewski et al., 1981) and V1a−/− mice (Li et al., 2009), but not V1a−/−V1b−/− mice (Yamaguchi et al., 2013). Recent work focusing on the role of AVP clock neurons rather than AVP signaling per se has found that the period of behavioral activity rhythms is lengthened in Avp-Cre;Bmal1lox/lox mice, which lack a functional molecular clock in AVP neurons and show abrogated AVP expression (Jin et al., 1999; Mieda et al., 2015). However, correlational studies have not found a consistent relationship between the number of AVP neurons and circadian period in genetic models with naturally occurring variation in AVP expression (Bult et al., 1993; Scarbrough and Turek, 1996). The reason for discrepant results across different genetic models remains unclear, but may involve developmental compensation and/or differential roles for V1A and V1B receptors in regulating period. Our results suggest that AVP signaling can influence SCN function by activating either V1 receptor subtype, which is consistent with previous work indicating that V1A and V1B receptors have similar affinity for AVP and both receptor subtypes are linked to Gq signaling (Koshimizu et al., 2012; Maybauer et al., 2008). Given similarities in the structure and function of the V1A and V1B receptors, spatiotemporal differences in their regulation of SCN function likely reflect their differential expression patterns along the anteroposterior axis of the network. Interestingly, only the V1B-antagonist SSR lengthened period in the anterior SCN where V1b receptor transcripts are most concentrated. In contrast, only the V1A-antagonist OPC lengthened period in the posterior SCN where V1a transcripts are most concentrated. Previous work suggests the single antagonists used here can affect both V1 receptors at high doses when bath applied to transfected HEK293 cells (Oshikawa et al., 2004), although the interface slice preparation used here is expected to reduce the dose delivered to the tissue. Since regional effects of single antagonists were observed on SCN period length, this suggests that antagonists were at least partially selective for the different V1 receptors at the doses used here.
Overall, these results imply that AVP signaling shortens SCN period, which may be due to specific clock properties of AVP or AVP-responsive neurons and/or the unchecked influence of other SCN factors. One possible mechanism underlying this influence of AVP signaling is that AVP neurons have a shorter intrinsic period, which they communicate to the rest of the network. Consistent with this possibility, AVP release rhythms in vitro are shorter than those for VIP release (Noguchi & Watanabe, 2008; Noguchi et al., 2004), suggesting that AVP neurons in the SCN have faster cellular clocks than VIP neurons. However, cell-type specific period is not evident in isolated SCN neurons categorized post hoc into neuropeptide subgroups (Webb et al., 2009), and AVP neurons themselves display regional differences in period length (Noguchi et al., 2004). Thus, it seems more likely that network connectivity tunes the period of SCN neurons.
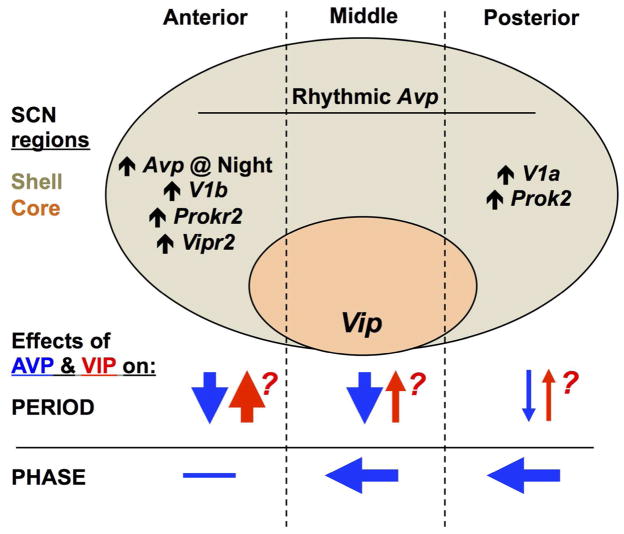
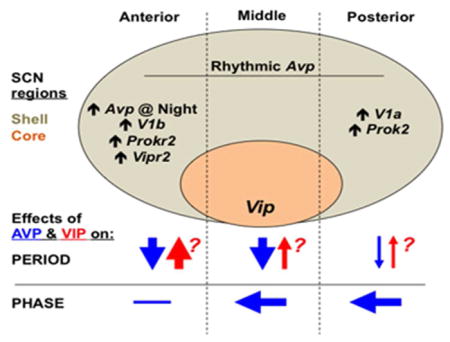
We speculate that countervailing influences of AVP and VIP work together to constrain SCN period to ca. 24 hours, which is the defining feature of the circadian clockworks (Figure 13). Thus, V1 inhibition may lengthen period by removing the AVPergic check on VIP signaling. This model is supported by our observation of stronger period-lengthening effects of V1 antagonism in VIP expressing regions (Figure 10e) and in the anterior SCN, which is enriched for Vipr2 (Figure 8a–d). Consistent with this model, SCN slices that contain fewer VIP expressing neurons display shorter periods (Noguchi & Watanabe, 2008). Further support for VIP as a competitive driver of period is provided by complementary work indicating that VIP signaling lengthens SCN period and entrains cellular rhythms of AVP neurons (Noguchi & Watanabe, 2008; Watanabe et al., 2000; Yamaguchi et al., 2003). With this in mind, it is notable that activating VIP neurons with Gq signaling lengthens SCN period (Brancaccio et al., 2013) by a magnitude similar to AVP receptor antagonism (Figure 10a, Edwards et al., 2016). Determination of SCN period may also involve additional signals that interact with VIP and AVP. One potential candidate is PROK2, which is expressed asymmetrically across the anteroposterior SCN (Figure 5) and lengthens behavioral circadian period in Lhx1 mutant mice that are deficient in a range of SCN neuropeptides (Bedont et al., 2014). Importantly, enriched expression of V1b, Vipr2, and Prokr2 together with enhanced V1-mediated period responses in the anterior SCN suggest that this site preferentially integrates multiple neuropeptide signals that modulate period in opposite directions (Figure 13).
Figure 13. Model of the regionalized role of AVP in SCN period and phase computation.
Schematic representation of the SCN network in the sagittal plane, with the location of the shell and core compartments illustrated and subdivided into anterior, middle, and posterior regions. Avp is rhythmically expressed throughout the SCN in wild type mice, but higher basal Avp expression and other region-specific patterns of gene expression for signaling transcripts are indicated where appropriate. Arrows below illustrate putative effects of AVP and VIP signaling on period and phase. Question marks alongside symbols for VIP signaling reflect the speculative nature of region-specific effects because it is unknown how VIP deficiency affects clock function across the anteroposterior SCN.
AVP signaling modulates phase relationships among SCN neurons
Our work also suggests that AVP signaling regulates SCN phase relationships to increase the magnitude of regional phase differences. SCN neurons adopt specific phase relationships (Evans et al., 2013; Inagaki et al., 2007), and the spatiotemporal organization across the network contributes to the fidelity and temporal patterning of SCN outputs to downstream tissues (Evans et al., 2015; Kalsbeek & Buijs, 2002). We find that antagonizing AVP receptors reduces SCN phase differences by preferentially delaying the phase of cellular clocks in the middle-to-posterior regions of the network. A reduction of cellular phase differences is also suggested by SCN imaging data collected from V1a−/−V1b−/− mice (Yamaguchi et al., 2013), although this was not analyzed directly in this previous report. Further, V1 antagonism reduces the magnitude of intra-SCN phase differences across the dorsoventral axis of Cry1−/−;Cry2−/− SCN when rhythms were rescued by co-culture with a wild type SCN (Ono et al., 2016). Lastly, V1a−/− mice and Avp-Cre;Bmal1lox/lox mice display a long duration of behavioral activity relative to controls (Li et al., 2009; Mieda et al., 2015). This is consistent with our in vitro results because the duration of behavioral activity and the magnitude of SCN phase dispersion are inversely related (Evans & Gorman, 2016). This overall pattern of results suggests that SCN phase dispersion may be reduced in the absence of AVP signaling in vivo. Future work should assess how AVP signaling influences SCN organization and interacts with VIP signaling across the lifespan (Duncan et al., 2001; Ono et al., 2016).
By modulating SCN phase relationships, AVP signaling may have pronounced consequences for SCN function and how the circadian system adjusts to changing environmental conditions. Phase relationships among SCN neurons dictate the waveform of circadian rhythms under different photoperiods (Hazlerigg et al., 2005; Inagaki et al., 2007). AVP signaling may modulate the phase of the posterior SCN relative to the rest of the nucleus (Figure 13) and regulate plasticity of behavioral rhythms during seasonal changes in the environment (Evans et al., 2011, 2013; Inagaki et al., 2007; Naito et al., 2008). Further, Avp expression in the SCN increases under long day photoperiods in some species (Duncan et al., 1995; Hazlerigg et al., 2005; Tournier et al., 2003), suggesting that the role of AVP signaling may depend on environmental conditions. Increased phase synchrony among SCN neurons in the absence of AVP signaling may also contribute to accelerated recovery from jet lag observed in V1a−/−V1b−/− mice (Yamaguchi et al., 2013) by aligning the circadian times at which light resets cellular clocks in the SCN (vanderLeest et al., 2009).
Considering that AVP influences phase relationships among SCN neurons, it remains curious that exogenous AVP does not induce phase shifts in vivo despite its ability to alter SCN electrical properties in vitro in a V1-dependent manner (reviewed in Ingram et al., 1998). This stands in contrast to strong effects of exogenous VIP on both phase and network synchrony (Albers et al., 1991; Maywood et al., 2011; Piggins et al., 1995). This discrepancy may reflect regional differences in signal transduction for AVP versus VIP signaling and/or the degree to which each influences the expression of other SCN neuropeptides (Figures 1–2, Figures 5–6; Bedont et al., 2017; Harmar et al., 2002; Li et al., 2009).
Anterior and posterior SCN are neurochemically and functionally distinct subdomains
The anteroposterior asymmetries in expression of both Avp and its receptors described here markedly expands on previous work focused on Avp expression along the dorsoventral axis (Hamada et al., 2004; Jin et al., 1999; Yoshikawa et al., 2015). Although RNA and protein expression can differ, the match between regional patterns of transcript expression and antagonist-driven effects suggests that spatial asymmetries are likewise evident at the post-transcriptional level. Strong effects of V1 antagonism on the phase of the posterior SCN correspond with highly rhythmic V1a expression that oscillates in anti-phase to Avp in this specific region of the network. Based on the relative phasing of these rhythms, AVP signaling would be predicted to have discrete (i.e., non-parametric) effects on the phase of cells in this specific region (Pittendrigh, 1981). In contrast, period lengthening effects of V1 antagonism were greatest in the anterior SCN, which displays constitutively higher V1b expression than the posterior SCN. This pattern of receptor expression likely contributes to stronger period effects in the anterior SCN by inducing a more tonic (i.e., parametric) response to AVP (Aschoff, 1965). Lower amplitude rhythms of V1a and Avp expression in the anterior SCN may further contribute to the tonic nature of AVP signaling in this specific domain. Furthermore, the expression of VIP and PROK2 signaling components suggests a common posterior-to-anterior directionality to their signaling, with both receptors having denser expression in the anterior SCN and both ligands having denser expression in the middle-to-posterior SCN. Collectively, these results support the view that the anterior and posterior SCN are distinct compartments (Figure 13). Together with accumulating evidence of other structural and functional differences along the anteroposterior axis of the SCN (Bedont & Blackshaw, 2015; Evans et al., 2011; Inagaki et al., 2007; Lokshin et al., 2015; Morin, 2007) this leads us to postulate the existence of discrete anterior and posterior SCN subdomains, oriented orthogonally to and overlapping the classical core/shell axis of the network.
In summary, our work identifies AVP as the first known signaling mechanism that functionally organizes molecular clock properties across all three dimensions of the SCN network. First, we find compartmentalized expression of not only AVP, but also VIP and PROK2 signaling components that suggest regionally distinct interactions between AVP and other key signaling pathways. We then identify AVP signaling as an important modulator of molecular clock properties along the anteroposterior SCN, with region-specific effects on period and phase that coincide with V1a and V1b expression patterns. Finally, we show that the SCN shell and core differ in their period and phase responses to V1 antagonism, complementing previous work documenting effects of VIP in the dorsoventral axis (Pauls et al., 2014). Together, these findings suggest a model in which AVP and VIP signaling together dictate spatiotemporal patterning of clock properties across all three dimensions of the SCN (Figure 13). In this manner, our work reinforces the view that SCN organization is mediated by multiple signaling mechanisms released by distinct cell groups interacting in a recursive manner (Evans et al., 2013; Maywood et al., 2011). The continuing challenge to understanding the SCN circuit will be to unravel how signals from different neuronal subgroups are integrated across the three dimensions of the master clock network.
Acknowledgments
We would like to thank Harshida Pancholi and Mary Bozsik for their assistance in executing these experiments. Also, we are grateful to Stanford Photonics for their equipment and technical assistance. This work was supported by an NSF graduate research fellowship (JLB), the Howard Hughes Medical Institute (AS), the Johns Hopkins Brain Science Institute (SB, SH), NIH R01NS091234 (JAE), and the Whitehall Foundation (JAE).
References
- Abrahamson EE, Moore RY. Suprachiasmatic nucleus in the mouse: Retinal innervation, intrinsic organization and efferent projections. Brain Res. 2001;916(1–2):172–191. doi: 10.1016/s0006-8993(01)02890-6. [DOI] [PubMed] [Google Scholar]
- Albers HE, Liou SY, Stopa EG, Zoeller RT. Interaction of colocalized neuropeptides: Functional significance in the circadian timing system. J Neurosci. 1991;11(3):846–851. doi: 10.1523/JNEUROSCI.11-03-00846.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An S, Tsai C, Ronecker J, Bayly A, Herzog ED. Spatiotemporal distribution of vasoactive intestinal polypeptide receptor 2 in mouse suprachiasmatic nucleus. J Comp Neurol. 2012;520(12):2730–2741. doi: 10.1002/cne.23078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antle MC, Foley DK, Foley NC, Silver R. Gates and oscillators: A network model of the brain clock. J Biol Rhythms. 2003;18(4):339–350. doi: 10.1177/0748730403253840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschoff J. Response curves in circadian periodicity. In: Aschoff J, editor. Circadian Clocks. Amsterdam: North Holland; 1965. pp. 95–111. [Google Scholar]
- Bedont JL, Blackshaw S. Constructing the suprachiasmatic nucleus: A watchmaker’s perspective on the central clockworks. Front Syst Neurosci. 2015;9:74. doi: 10.3389/fnsys.2015.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedont JL, LeGates TA, Buhr E, Bathini A, Ling JP, Bell B, Wu MN, Wong PC, Van Gelder RN, Mongrain V, Hattar S, Blackshaw S. An LHX1-regulated transcriptional network controls sleep/wake coupling and thermal resistance of the central circadian clockworks. Curr Biol. 2017;27:128–136. doi: 10.1016/j.cub.2016.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedont JL, LeGates TA, Slat EA, Byerly MS, Wang H, Hu J, Rupp AC, Qian J, Wong GW, Herzog ED, Hattar S, Blackshaw S. Lhx1 controls terminal differentiation and circadian function of the suprachiasmatic nucleus. Cell Rep. 2014;7(3):609–622. doi: 10.1016/j.celrep.2014.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brancaccio M, Maywood ES, Chesham JE, Loudon AS, Hastings MH. A Gq-Ca2+ axis controls circuit-level encoding of circadian time in the suprachiasmatic nucleus. Neuron. 2013;78(4):714–728. doi: 10.1016/j.neuron.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhr ED, Takahashi JS. Handb Exp Pharmacol. 217. 2013. Molecular components of the mammalian circadian clock; pp. 3–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan MJ, Cheng X, Heller KS. Photoperiodic exposure and time of day modulate the expression of arginine vasopressin mRNA and vasoactive intestinal peptide mRNA in the suprachiasmatic nuclei of Siberian hamsters. Brain Res Mol Brain Res. 1995;32(2):181–186. doi: 10.1016/0169-328x(95)00072-z. [DOI] [PubMed] [Google Scholar]
- Duncan MJ, Herron JM, Hill SA. Aging selectively suppresses vasoactive intestinal peptide messenger RNA expression in the suprachiasmatic nucleus of the Syrian hamster. Brain Res Mol Brain Res. 2001;87(2):196–203. doi: 10.1016/s0169-328x(01)00015-8. [DOI] [PubMed] [Google Scholar]
- Edwards MD, Brancaccio M, Chesham JE, Maywood ES, Hastings MH. Rhythmic expression of cryptochrome induces the circadian clock of arrhythmic suprachiasmatic nuclei through arginine vasopressin signaling. Proc Natl Acad Sci U S A. 2016;113(10):2732–2737. doi: 10.1073/pnas.1519044113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JA. Collective timekeeping among cells of the master circadian clock. J Endocrinol. 2016;230(1):R27–49. doi: 10.1530/JOE-16-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JA, Gorman MR. In synch but not in step: Circadian clock circuits regulating plasticity in daily rhythms. Neuroscience. 2016;320:259–280. doi: 10.1016/j.neuroscience.2016.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JA, Leise TL, Castanon-Cervantes O, Davidson AJ. Intrinsic regulation of spatiotemporal organization within the suprachiasmatic nucleus. PLoS One. 2011;6(1):e15869. doi: 10.1371/journal.pone.0015869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JA, Leise TL, Castanon-Cervantes O, Davidson AJ. Dynamic interactions mediated by nonredundant signaling mechanisms couple circadian clock neurons. Neuron. 2013;80(4):973–983. doi: 10.1016/j.neuron.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JA, Suen TC, Callif BL, Mitchell AS, Castanon-Cervantes O, Baker KM, Kloehn I, Baba K, Teubner BJ, Ehlen JC, Paul KN, Bartness TJ, Tosini G, Leise T, Davidson AJ. Shell neurons of the master circadian clock coordinate the phase of tissue clocks throughout the brain and body. BMC Biol. 2015;13:43. doi: 10.1186/s12915-015-0157-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groblewski TA, Nunez AA, Gold RM. Circadian rhythms in vasopressin deficient rats. Brain Res Bull. 1981;6(2):125–130. doi: 10.1016/s0361-9230(81)80036-6. [DOI] [PubMed] [Google Scholar]
- Hamada T, Antle MC, Silver R. Temporal and spatial expression patterns of canonical clock genes and clock-controlled genes in the suprachiasmatic nucleus. Eur J Neurosci. 2004;19(7):1741–1748. doi: 10.1111/j.1460-9568.2004.03275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmar AJ, Marston HM, Shen S, Spratt C, West KM, Sheward WJ, Morrison CF, Dorin JR, Piggins HD, Reubi JC, Kelly JS, Maywood ES, Hastings MH. The VPAC(2) receptor is essential for circadian function in the mouse suprachiasmatic nuclei. Cell. 2002;109(4):497–508. doi: 10.1016/s0092-8674(02)00736-5. [DOI] [PubMed] [Google Scholar]
- Hazlerigg DG, Ebling FJ, Johnston JD. Photoperiod differentially regulates gene expression rhythms in the rostral and caudal SCN. Curr Biol. 2005;15(12):R449–450. doi: 10.1016/j.cub.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Herzog ED, Aton SJ, Numano R, Sakaki Y, Tei H. Temporal precision in the mammalian circadian system: A reliable clock from less reliable neurons. J Biol Rhythms. 2004;19(1):35–46. doi: 10.1177/0748730403260776. [DOI] [PubMed] [Google Scholar]
- Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: An efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms. 2010;25(5):372–380. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki N, Honma S, Ono D, Tanahashi Y, Honma K. Separate oscillating cell groups in mouse suprachiasmatic nucleus couple photoperiodically to the onset and end of daily activity. Proc Natl Acad Sci U S A. 2007;104(18):7664–7669. doi: 10.1073/pnas.0607713104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram CD, Ciobanu R, Coculescu IL, Tanasescu R, Coculescu M, Mihai R. Vasopressin neurotransmission and the control of circadian rhythms in the suprachiasmatic nucleus. Prog Brain Res. 1998;119:351–364. doi: 10.1016/s0079-6123(08)61580-0. [DOI] [PubMed] [Google Scholar]
- Jin X, Shearman LP, Weaver DR, Zylka MJ, de Vries GJ, Reppert SM. A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell. 1999;96(1):57–68. doi: 10.1016/s0092-8674(00)80959-9. [DOI] [PubMed] [Google Scholar]
- Kalsbeek A, Buijs RM. Output pathways of the mammalian suprachiasmatic nucleus: Coding circadian time by transmitter selection and specific targeting. Cell Tissue Res. 2002;309(1):109–118. doi: 10.1007/s00441-002-0577-0. [DOI] [PubMed] [Google Scholar]
- Kalsbeek A, Fliers E, Hofman MA, Swaab DF, Buijs RM. Vasopressin and the output of the hypothalamic biological clock. J Neuroendocrinol. 2010;22(5):362–372. doi: 10.1111/j.1365-2826.2010.01956.x. [DOI] [PubMed] [Google Scholar]
- Koshimizu TA, Nakamura K, Egashira N, Hiroyama M, Nonoguchi H, Tanoue A. Vasopressin V1a and V1b receptors: From molecules to physiological systems. Physiol Rev. 2012;92(4):1813–1864. doi: 10.1152/physrev.00035.2011. [DOI] [PubMed] [Google Scholar]
- Li JD, Burton KJ, Zhang C, Hu SB, Zhou QY. Vasopressin receptor V1a regulates circadian rhythms of locomotor activity and expression of clock-controlled genes in the suprachiasmatic nuclei. Am J Physiol. 2009;296(3):R824–830. doi: 10.1152/ajpregu.90463.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokshin M, LeSauter J, Silver R. Selective distribution of retinal input to mouse SCN revealed in analysis of sagittal sections. J Biol Rhythms. 2015;30(3):251–257. doi: 10.1177/0748730415584058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low-Zeddies SS, Takahashi JS. Chimera analysis of the Clock mutation in mice shows that complex cellular integration determines circadian behavior. Cell. 2001;105(1):25–42. doi: 10.1016/s0092-8674(01)00294-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey SR. Biological Rhythms Workshop IA: Molecular basis of rhythms generation. Cold Spring Harb Symp Quant Biol. 2007;72:7–19. doi: 10.1101/sqb.2007.72.060. [DOI] [PubMed] [Google Scholar]
- Maybauer MO, Maybauer DM, Enkhbaatar P, Traber DL. Physiology of the vasopressin receptors. Best Pract Res Clin Anaesthesiol. 2008;22(2):253–263. doi: 10.1016/j.bpa.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Maywood ES, Chesham JE, O’Brien JA, Hastings MH. A diversity of paracrine signals sustains molecular circadian cycling in suprachiasmatic nucleus circuits. Proc Natl Acad Sci U S A. 2011;108(34):14306–14311. doi: 10.1073/pnas.1101767108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieda M, Ono D, Hasegawa E, Okamoto H, Honma K, Honma S, Sakurai T. Cellular clocks in AVP neurons of the SCN are critical for interneuronal coupling regulating circadian behavior rhythm. Neuron. 2015;85(5):1103–1116. doi: 10.1016/j.neuron.2015.02.005. [DOI] [PubMed] [Google Scholar]
- Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annu Rev Neurosci. 2012;35:445–462. doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin LP. SCN organization reconsidered. J Biol Rhythms. 2007;22(1):3–13. doi: 10.1177/0748730406296749. [DOI] [PubMed] [Google Scholar]
- Naito E, Watanabe T, Tei H, Yoshimura T, Ebihara S. Reorganization of the suprachiasmatic nucleus coding for day length. J Biol Rhythms. 2008;23(2):140–149. doi: 10.1177/0748730408314572. [DOI] [PubMed] [Google Scholar]
- Noguchi T, Watanabe K. Regional differences in circadian period within the suprachiasmatic nucleus. Brain Res. 2008;1239:119–126. doi: 10.1016/j.brainres.2008.08.082. [DOI] [PubMed] [Google Scholar]
- Noguchi T, Watanabe K, Ogura A, Yamaoka S. The clock in the dorsal suprachiasmatic nucleus runs faster than that in the ventral. Eur J Neurosci. 2004;20(11):3199–3202. doi: 10.1111/j.1460-9568.2004.03784.x. [DOI] [PubMed] [Google Scholar]
- Oshikawa S, Tanoue A, Koshimizu T, Kitagawa Y, Tsujimoto G. Vasopressin stimulates insulin release from islet cells through V1b receptors: A combined pharmachological/knockout approach. Mol Pharmacol. 2004;65(3):623–9. doi: 10.1124/mol.65.3.623. [DOI] [PubMed] [Google Scholar]
- Ono D, Honma S, Honma K. Differential roles of AVP and VIP signaling in the postnatal changes of neural networks for coherent circadian rhythms in the SCN. Sci Adv. 2016;2(9):e1600960. doi: 10.1126/sciadv.1600960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauls S, Foley NC, Foley DK, LeSauter J, Hastings MH, Maywood ES, Silver R. Differential contributions of intra-cellular and inter-cellular mechanisms to the spatial and temporal architecture of the suprachiasmatic nucleus circadian circuitry in wild-type, cryptochrome-null and vasoactive intestinal peptide receptor 2-null mutant mice. Eur J Neurosci. 2014;40(3):2528–2540. doi: 10.1111/ejn.12631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggins HD, Antle MC, Rusak B. Neuropeptides phase shift the mammalian circadian pacemaker. J Neurosci. 1995;15(8):5612–5622. doi: 10.1523/JNEUROSCI.15-08-05612.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittendrigh CS. Circadian Systems: Entrainment. In: Aschoff J, editor. Biological Rhythms. Vol. 4. New York: Plenum Press; 1981. pp. 94–124. [Google Scholar]
- Rohling J, Meijer JH, VanderLeest HT, Admiraal J. Phase differences between SCN neurons and their role in photoperiodic encoding; a simulation of ensemble patterns using recorded single unit electrical activity patterns. J Physiol Paris. 2006;100(5–6):261–270. doi: 10.1016/j.jphysparis.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Smyllie NJ, Chesham JE, Hamnett R, Maywood ES, Hastings MH. Temporally chimeric mice reveal flexibility of circadian period-setting in the suprachiasmatic nucleus. Proc Natl Acad Sci U S A. 2016;113(13):3657–3662. doi: 10.1073/pnas.1511351113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southey BR, Lee JE, Zamdborg L, Atkins N, Jr, Mitchell JW, Li M, Gillette MU, Kelleher NL, Sweedler JV. Comparing label-free quantitative peptidomics approaches to characterize diurnal variation of peptides in the rat suprachiasmatic nucleus. Anal Chem. 2014;86(1):443–452. doi: 10.1021/ac4023378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournier BB, Menet JS, Dardente H, Poirel VJ, Malan A, Masson-Pevet M, Pevet P, Vuillez P. Photoperiod differentially regulates clock gene expression in the suprachiasmatic nucleus of Syrian hamster. Neuroscience. 2003;118(2):317–322. doi: 10.1016/s0306-4522(03)00008-3. [DOI] [PubMed] [Google Scholar]
- vanderLeest HT, Rohling JH, Michel S, Meijer JH. Phase shifting capacity of the circadian pacemaker determined by the SCN neuronal network organization. PLoS One. 2009;4(3):e4976. doi: 10.1371/journal.pone.0004976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosko AM, Schroeder A, Loh DH, Colwell CS. Vasoactive intestinal peptide and the mammalian circadian system. Gen Comp Endocrinol. 2007;152(2–3):165–175. doi: 10.1016/j.ygcen.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Vanecek J, Yamaoka S. In vitro entrainment of the circadian rhythm of vasopressin-releasing cells in suprachiasmatic nucleus by vasoactive intestinal polypeptide. Brain Res. 2000;877(2):361–366. doi: 10.1016/s0006-8993(00)02724-4. [DOI] [PubMed] [Google Scholar]
- Webb AB, Angelo N, Huettner JE, Herzog ED. Intrinsic, nondeterministic circadian rhythm generation in identified mammalian neurons. Proc Natl Acad Sci U S A. 2009;106(38):16493–16498. doi: 10.1073/pnas.0902768106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh DK, Logothetis DE, Meister M, Reppert SM. Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron. 1995;14(4):697–706. doi: 10.1016/0896-6273(95)90214-7. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Isejima H, Matsuo T, Okura R, Yagita K, Kobayashi M, Okamura H. Synchronization of cellular clocks in the suprachiasmatic nucleus. Science. 2003;302(5649):1408–1412. doi: 10.1126/science.1089287. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Suzuki T, Mizoro Y, Kori H, Okada K, Chen Y, Fustin JM, Yamazaki F, Mizuguchi N, Zhang J, Dong X, Tsujimoto G, Okuno Y, Doi M, Okamura H. Mice genetically deficient in vasopressin V1a and V1b receptors are resistant to jet lag. Science. 2013;342(6154):85–90. doi: 10.1126/science.1238599. [DOI] [PubMed] [Google Scholar]
- Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, Menaker M, Takahashi JS. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A. 2004;101(15):5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa T, Nakajima Y, Yamada Y, Enoki R, Watanabe K, Yamazaki M, Sakimura K, Honma S, Honma K. Spatiotemporal profiles of arginine vasopressin transcription in cultured suprachiasmatic nucleus. Eur J Neurosci. 2015;42(9):2678–2689. doi: 10.1111/ejn.13061. [DOI] [PubMed] [Google Scholar]
- Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc Natl Acad Sci U S A. 2014;111(45):16219–16224. doi: 10.1073/pnas.1408886111. [DOI] [PMC free article] [PubMed] [Google Scholar]