Abstract
The estimation of clinical prognosis for diffuse large B-cell lymphoma (DLBCL) with a quick, cost-efficient method is necessary because of the clinical heterogeneity of this disease, which leads to death, relapsed or refractory disease in approximately 40% of patients. We analyzed 320 cases diagnosed from 2007 to 2013 treated with R-CHOP therapy at Tokai University Hospital and associated institutions. DLBCL was classified according to the cell-of-origin using the Hans algorithm [germinal center B-cell-like (GCB) vs non-GCB subtypes], and into 6 subgroups derived from combinations of CD10, BCL6 and MUM1 markers. The percentage of GCB and non-GCB (NGCB) subtypes was 35% and 65%, respectively. GCB-DLBCL was characterized by lower BCL2 immunohistochemical expression, extranodal sites <1, better therapeutic response, and favorable overall survival (OS) and progression free survival (PFS) (P<0.01). The most frequent subgroup was NGCB-1 (CD10-BCL6+MUM1+, 51%) followed by GCB-1 (CD10+BCL6+or-MUM1+, 21%), NGCB-2 (CD10-BCL6-MUM1+, 13%), GCB-2 (CD10+BCL6+or-MUM1-, 10%), GCB-3 (CD10-BCL6+MUM1-, 4%) and NGCB-3 (CD10-BCL6-MUM1-, 2%). In comparison with GCB-2 and GCB-3 (both MUM1-), the GCB-1 (MUM1+) was characterized by favorable PFS (5-year PFS 84% vs 65%, OR 0.368, P<0.05), independent of high LDH (associated with unfavorable PFS, OR 7.04, P<0.01) in the multivariate analysis. This predictive value of MUM1 was independent of CD10. Interestingly, triple-negative NGCB-3 tended to have a more favorable prognosis than the other NGCB subgroups.
In conclusion, the Hans classifier is a valid method to evaluate the prognosis of DLBCL NOS. In the GCB subtypes, GCB subtypes, MUM1-positivity is associated with a more favorable outcome (PFS).
Keywords: diffuse large B cell lymphoma, Hans classifier, cell-of-origin, clinicopathological characteristics, prognosis
INTRODUCTION
Diffuse large B-cell lymphoma (DLBCL) is the most common type of non-Hodgkin lymphoma (NHL), and accounts for 30%–40% of all adult NHL.1,2 The diagnostic category of DLBCL is heterogeneous, and the clinicopathological and molecular genetic diversity of DLBCL was reflected in the 2008 WHO classification of lymphomas. Furthermore, the updated revision in 20163 described many subgroups and entities based on distinct morphological, immunophenotypical, molecular and clinical parameters. Although DLBCL is a potentially curable disease with current therapy, particularly for those who achieve complete remission with first-line treatment, approximately 40% of patients will die of relapsed or refractory disease.2,4 Factors that contribute to the outcome include age, socioeconomic conditions, comorbid conditions, performance status and several clinical features.5,6 Therefore, it is necessary to stratify patients according to their prognostic risk in a suitable and cost-effective manner.
Prognostic factors in DLBCL include standard and well-established factors such as the International Prognostic Index (IPI; with its variants as the original, R-IPI, age-adjusted IPI and NCCN-IPI), immunohistochemistry for cell-of-origin (e.g. Hans, Choi and Tally algorithms), imaging by positron emission tomography (PET)/CT scans, fluorescence in situ hybridization (FISH) for MYC and BCL2 rearrangements, and absolute lymphocyte and monocyte count. In addition, non-routine techniques include gene expression profiling (GEP, frozen tissue-derived or new Lymph2Cx assays that quantify RNA transcript levels in formalin-fixed paraffin-embedded tissue sections), and serum immunoglobulin-free light chain, vitamin D3 and cytokine/chemokine (sIL2R, IL6 and TNFA) levels.2,7-12
GEP of DLBCL reported by Alizadeh et al. in 2000 found the molecular subtypes of germinal center B-cells (GCB) and activated B-cells (ABC) to have different overall survival (OS) curves with CHOP therapy.13 This established a putative cell-of-origin (COO) for DLBCL. Subsequently, the Hans classifier (i.e. algorithm) was used to immunohistochemically categorize DLBCL into two major classes: the germinal center B-cell-like (GCB) DLBCL and the non-GCB DLBCL for ABC, based on three markers, CD10, BCL6 and MUM1. Considering the cDNA microarray classification as the gold standard, the sensitivity of TMA was 71% for the GCB group and 88% for non-GCB group. Although the concordance rate of the Hans classifier and GEP was approximately 80%, the algorithm stratified the patients according to prognosis in a comparable way with that reported using the cDNA microarray: the 5-year OS for the GCB group was 76% compared with only 34% for the non-GCB group (P<0.001).7 It is important to note that the three groups, GCB, ABC and non-classifiable, were separated based on gene expression, and the prognosis of the non-classifiable group was similar with that of the ABC group.13
In the age of new generation sequencing (NGS) technologies and new therapies, it is important to assess whether the use of the Hans classifier is still a simple, cost-efficient and reliable tool to stratify DLBCL patients according to their prognosis. The Hans algorithm only identifies 2 groups: GCB and non-GCB (NGCB), but based on three markers, 8 possible combinations or 6 simplified subgroups may be created, and to our knowledge further combinations have not been fully analyzed.
In this paper, 320 cases of DLBCL with standard RCHOP-based therapy were assessed, and the Hans classifier and the 6 subgroups were evaluated and found to be correlated with several clinical variables, including overall and progression-free survival. We found that within the Hans GCB subtype, MUM1 positivity was associated with the best outcome.
PATIENTS AND METHODS
Patients
Of the 370 patients diagnosed with DLBCL between January 2007 and December 2013 at Tokai University Hospital, 345 patients were treated at our hospital or at the related institutions of Atsugi Satoh Hospital, Isehara Kyodo Hospital, Ebina General Hospital, Ozawa Hospital and Japanese Red Cross Hadano Hospital. From these initial 345 patients, 25 were excluded due to insufficient immunohistochemical data for the Hans classification because of unavailable paraffin embedded tissue and/or complete clinical information. Therefore, the final dataset comprised 320 patients. CD5-positive cases represented 12% of the cases.
Immunohistochemistry
Following deparaffinization, heat-induced antigen retrieval techniques were used for each antibody. Mouse monoclonal antibodies against human CD3 (non-glycosylated epsilon chain, clone LN10, 1:200 dilution), CD5 (4C7, 1:400), CD10 (56C6, 1:100), CD20 (L26, 1:200), BCL-2 (bcl-2/100/D5, 1:400), BCL-6 (LN22, 1:100) and MUM-1 (EAU32, 1:100) (Novocastra, Leica Microsystems K.K., Tokyo, Japan) were used as primary antibodies. Detection of signals was carried out using the Leica Bond-Max fully automatic IHC system, Bond Polymer Refine detection kit and Bond Epitope Retrieval Solution 2 (EDTA based pH 9.0) for 20 min for antigen retrieval according to the manufacturer’s instructions (DS9800 and AR9640, Leica Microsystems). CD20 staining was performed to assess tumor involvement and the evaluation for the percentage of stained tumor cells was performed by visual estimation under a microscope (BX51, Olympus K.K., Tokyo Japan). Cases were considered positive if 30% or more of the tumor cells were stained with an antibody, the same criteria as described by Hans CP et al.7 The DLBCL subtypes of GCB or non-GCB were categorized using CD10, BCL6 and MUM1 according to the Hans algorithm.7 Detection of latent Epstein-Barr virus (EBV) infection was performed by means of in situ hybridization for EBV encoded mRNA using the EBER Probe, Anti-Fluorescein Antibody and Bond Polymer Refine Detection (Bond ISH EBER Probe, #BP0589; Bond Ready-to-Use primary antibody, anti-fluorescein antibody, #AR0833; Leica Microsystems).
Six subgroups with the Hans classifier
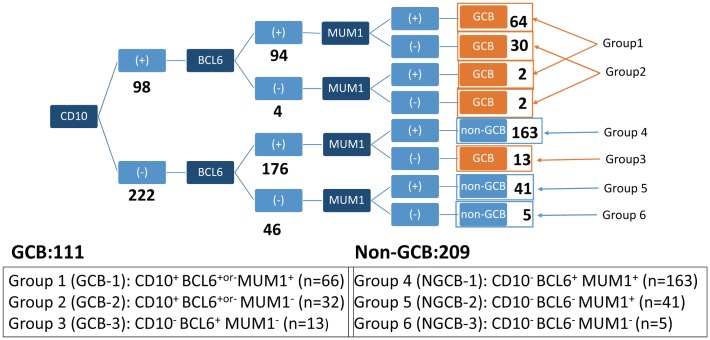
Based on the three markers of the Hans classifier, 8 possible combinations were possible. As the subgroups of CD10+ BCL6− MUM1+ and CD10+ BCL6− MUM1− included only a few cases, they were merged with others (Fig. 1). In this paper, the subgroups according to the 6 combinations were as follows: GCB-1 (CD10+ BCL6+or- MUM1+), GCB-2 (CD10+ BCL6+or- MUM1−), GCB-3 (CD10− BCL6+ MUM1−), NGCB-1 (CD10− BCL6+ MUM1+), NGCB-2 (CD10− BCL6− MUM1+) and NGCB-3 (CD10− BCL6− MUM1−) (Fig. 1).
Fig. 1.
Subgroup classification based on the combination of CD10, BCL6 and MUM1 of the Hans classifier.
The most frequent subgroups were the NGCB-1 (group 4) and GCB-1 (group 1) at 51% and 21%, respectively.
The BCL6+or- cases were defined as samples with immunohistochemical expression of BCL6 above or less than 30%. For example, group 1 of GCB-1 was characterized by being CD10-positive, MUM1-positive and either positive or negative for BCL6.
Clinical information
Staging procedures were standard, and included patient history, physical examination, performance status according to the Eastern Cooperative Oncology Group (ECOG) scale, presence of B symptoms, blood cell counts, serum biochemistry, including lactate dehydrogenase (LDH) and soluble IL-2 receptor, PET/CT scan and bone marrow biopsy. Both IPI and R-IPI (the revised version of the IPI for patients treated with immunochemotherapy)14 were calculated. Tumor responses were evaluated with PET/CT scans and the patients were classified by the best tumor response according to the response criteria for malignant lymphoma. OS was defined as the duration from the date of diagnosis of DLBCL to the date of death of any cause or the last follow-up. Progression-free survival was calculated as the period from the date of treatment initiation to the date of recurrence, exacerbation or death. The date of recurrence or exacerbation refers to the day when recurrence or exacerbation was diagnosed through blood testing, imaging or biopsy. The chemotherapy treatment was either R-CHOP or R-CHOP-like regimens. This study was approved by the institutional review board (IRB 14R-080) and conducted in accordance with the Helsinki Declaration of 1975 as revised in 2008.
Statistical analysis
Fisher’s exact test was used to compare proportions between groups. Survival curves were built using the Kaplan–Meier method and the comparison of the survival distributions using the log-rank test as well as multivariate analysis. A P-value less than 0.05 was used as the cutoff for significance. EZR (Easy R) software for medical statistics was used for all analyses.15
RESULTS
Correlation with the immunohistochemical characteristics
From the total of 320 cases, 111 (35%) had the GCB DLBCL phenotype and 209 (65%) had the non-GCB DLBCL phenotype. The CD10− BCL6+ MUM1+ phenotype (NGCB-1) was the most common at 51% (163 cases), followed by the CD10+ BCL6+or- MUM1+ phenotype [GCB-1 (21%, 66 cases)]. The distribution of the remaining cases according to the 6 combinations was as follows: the CD10+ BCL6+or- MUM1− phenotype [GCB-2 (10%, 32 cases)], the CD10− BCL6+ MUM1− phenotype [GCB-3 (4%, 13 cases), the CD10− BCL6− MUM1+ phenotype [NGCB-2 (13%, 41 cases)] and the CD10− BCL6− MUM1− phenotype [NGCB-3 (2%, 5 cases) (Fig. 1).
Immunohistochemical results other than CD10, BCL6 and MUM1 are shown in Table 1. No significant differences were seen between the GCB and non-GCB subtype in terms of the prevalence of CD5 or EBV-ISH positivity. The GCB subtype was characterized by lower BCL2 expression than the non-GCB subtype, 73.4% vs 89.9% (P<0.01).
Table 1. Immunostaining and FISH results for the GCB and non-GCB groups.
| Marker | GCB (n=111) | non-GCB (n=209) | Total (n=320) | P- value | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | |||||||
| CD10+ | 98 | 88.3 | 0 | 0 | 98 | 30.6 | - | |||||
| BCL6+ | 107 | 96.4 | 163 | 78 | 270 | 84.4 | - | |||||
| MUM1+ | 66 | 59.5 | 204 | 97.6 | 270 | 84.4 | - | |||||
| BCL2+ | 80 (n=109) | 73.4 | 186 (n=207) | 89.9 | 266 (n=316) | 84.2 | 0.0003 | |||||
| CD5+ | 10 (n=110) | 9.1 | 29 (n=208) | 13.9 | 39 (n=318) | 12.3 | 0.281 | |||||
| EBV-ISH+ | 3 (n=111) | 2.7 | 16 (n=204) | 7.8 | 19 (n=315) | 6 | 0.083 | |||||
Correlation with clinical characteristics
No differences were found between GCB and non-GCB subtypes for most of the clinical variables, except for extranodal sites and achievement of complete remission. No differences were found regarding the age at the time of diagnosis, percentage of elderly patients (≥60 years), sex, prevalence of B symptoms, bone marrow involvement, elevated LDH (≥220 IU/L) at the first visit, clinical stage, or high-intermediate or high IPI. However, a higher prevalence of extranodal sites >1 was observed in the non-GCB group, 72.3% vs 57.3% (P=0.008) (Table 2). The patients received similar R-CHOP-like regimens as the initial treatment. However, the rates of complete remission were higher for the GCB subtype, 77.5% vs 52.6%, respectively, indicating a significantly better therapeutic response (P<0.0001) (Table 3).
Table 2. Clinical characteristics of the GCB and non-GCB patients.
| Variable | GCB (n=111) | non-GCB (n=209) | Total (n=320) | P- value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | |||||
| Age (years; mean ± STD) | 64.66 ± 13.53 | - | 67.56 ± 12.63 | - | - | - | 0.06 | |||
| Age >60 | 76 | 68.5 | 164 | 78.5 | 240 | 75 | 0.06 | |||
| Male | 68 | 61.3 | 104 | 49.8 | 172 | 53.8 | 0.06 | |||
| B symptoms | 24 (n=110) | 21.8 | 58 (n=207) | 28 | 82 (n=317) | 25.9 | 0.28 | |||
| Extranodal | 63 (n=110) | 57.3 | 151 (n=209) | 72.3 | 214 (n=319) | 67.1 | 0.008 | |||
| BM invasion | 11 (n=106) | 10.4 | 32 (n=208) | 15.4 | 43 (n=314) | 13.7 | 0.3 | |||
| High LDH (>220 U/L) | 65 (n=110) | 59.1 | 134 (n=209) | 64.1 | 199 (n=319) | 62.4 | 0.4 | |||
| Stage III/IV | 54 (n=110) | 49.1 | 117 (n=208) | 56.2 | 171 (n=318) | 53.8 | 0.24 | |||
| IPI H-I/H | 44 (n=109) | 40.4 | 96 (n=208) | 46.2 | 140 (n=317) | 44.2 | 0.34 | |||
Table 3. Initial treatment.
| 1st line therapy | GCB (n=111) | non-GCB (n=209) | Total (n=320) | P-value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | |||||
| R-CHOP-like*1 | 96 | 86.5 | 195 | 93.3 | 291 | 90.9 | ||||
| Other regimens*2 | 6 | 5.4 | 5 | 2.4 | 11 | 3.4 | - | |||
| Operation | 1 | 0.9 | 1 | 0.5 | 2 | 0.6 | - | |||
| Radiation | 0 | 0 | 1 | 0.5 | 1 | 0.3 | - | |||
| Not available*3 | 5 | 4.5 | 6 | 2.9 | 11 | 3.4 | - | |||
| Unknown | 3 | 2.7 | 1 | 0.5 | 4 | 1.3 | - | |||
| Achieve CR1 | 86 | 77.5 | 110 | 52.6 | 196 | 61.3 | 1.28E-05 | |||
1* R-CHOP, R-THP-COP, Rw-CHOP, R-COP, R-CHP, R-CHO.
2* CHOP, R, DA-EPOCH-R, HDMA, Obinutuzumab-CHOP.
3* PSL.
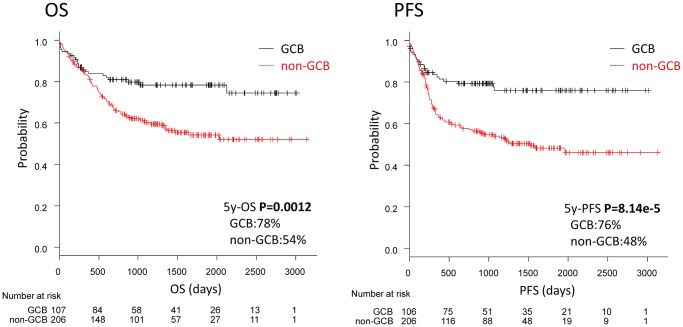
The non-GCB subtype was associated with an unfavorable prognosis. Five-year OS rates for the GCB and non-GCB groups were 78% and 54%, respectively, indicating significantly higher survival among GCB patients (P=0.0012). The five-year PFS rate of 76% among GCB patients was also significantly greater than the 48% rate among non-GCB patients (P<0.001) (Fig. 2).
Fig. 2.
Survival according to the Hans classifier (GCB vs non-GCB).
GCB patients were characterized by a favorable outcome, with higher OS and PFS than the non-GCB patients (P<0.01).
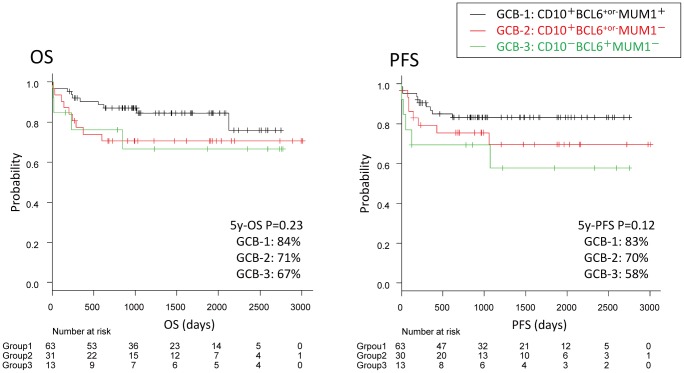
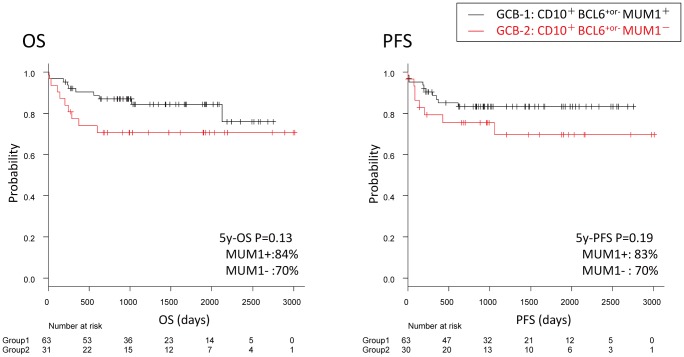
No differences were found regarding the OS between the GCB groups 1, 2 and 3 (P=0.23); the five-year OS rates were 84%, 71%, and 67%, respectively. The five-year PFS rates were 83%, 70%, and 58%, respectively, indicating that PFS rates tended to be poor for the MUM1-negative groups (Groups 2 and 3), although the differences were not significant (P=0.12; differences across groups) (Fig. 3).
Fig. 3.
Survival in the GCB group.
No differences were observed between GCB groups 1, 2 and 3 for OS (P=N.S.). Regarding PFS, the MUM1+ GCB-1 group tended to have a more favorable PFS than the MUM1- GCB-2+3 group, although not significantly (P=0.12).
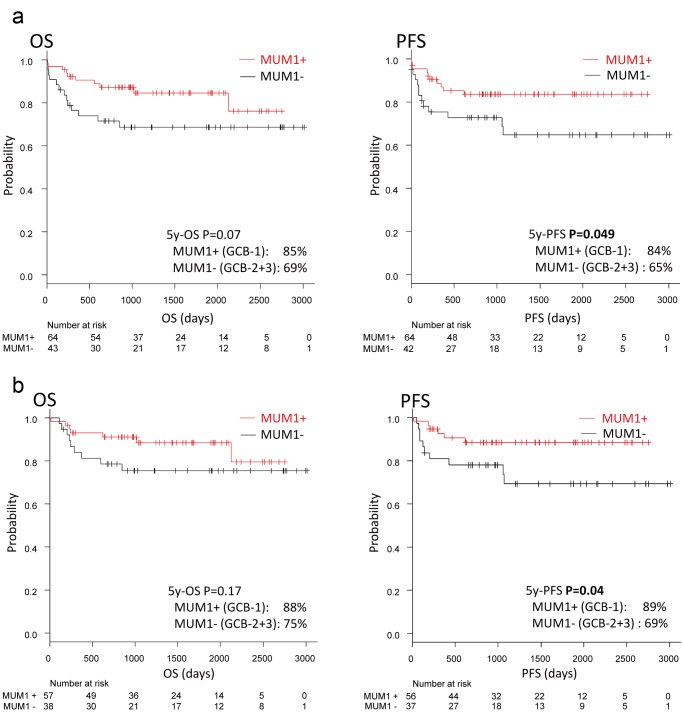
When the whole GCB subtype was stratified according to MUM1 expression (i.e. GCB-1 vs GCB-2+3), MUM1-positivity was associated with a significantly higher PFS (five-year PFS, 65% vs 84%, P=0.049; OS, not significant, P=0.07) (Fig. 4a). Further multivariate analysis demonstrated MUM1 and LDH to be significant predictors correlated with PFS: MUM1 positivity as protective [OR: 0.368, 95% confidence interval (CI): 0.137–0.989; P=0.048] and high LDH (>220 IU/L) as a risk variable (OR: 7.04, 95%CI: 1.91–25.9; P=0.003) (Table 4). The relationship between MUM1 positivity with OS and PFS was maintained in the GCB subtype when only those receiving R-CHOP-like therapy were examined. Of note, in this series, rituximab was used in 297 of 320 cases (93%) and R-CHOP-like therapy corresponded to 291 cases (91%). MUM1-positive cases had a higher PFS than the MUM1-negative cases (five-year PFS, 89% vs 69%, P=0.04) (Fig. 4b).
Fig. 4.
Survival between MUM1+ vs MUM1- in the GCB group.
Fig. 4a. MUM1+ vs MUM1− in GCB
(a) When the whole GCB subtype was stratified according to MUM1 expression, GCB-1 vs GCB2+3, the subgroup with MUM1-positivity was characterized by a significant PFS (P=0.049).
Fig. 4b. MUM1+ vs MUM1− in GCB with R-CHOP like regimens
Survival between MUM1+ vs MUM1- in the GCB group.
(b) The same results were observed when selecting only the patients receiving R-CHOP-like regimens.
Table 4. Univariate and multivariate analyses of PFS event in the GCB group.
| Variable | PFS event- (n=84) | PFS event+ (n=24) | P-value | Odds Ratio | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | Univariate | Multivariate | (95% CI) | ||||
| Gender (Male) | 50 | 59.5 | 16 | 24 | 0.637 | |||||
| Age >60 | 54 | 64.3 | 19 | 79.2 | 0.22 | |||||
| B symptom | 18 | 21.4 | 5 | 20.8 | 1 | |||||
| Stage III/IV | 37 | 44 | 15 | 62.5 | 0.164 | |||||
| IPI HI-H | 29 | 34.5 | 14 | 58.3 | 0.057 | - | ||||
| Extranodal | 44 | 52.4 | 17 | 70.8 | 0.161 | |||||
| BM invasion (n=105) | 5 | 6.2 | 6 | 25 | 0.0164 | - | ||||
| High LDH | 42 | 50 | 21 | 87.5 | 0.0009 | 0.003 | 7.04 (1.91-25.9) | |||
| CD10 | 76 | 90.5 | 19 | 79.2 | 0.158 | |||||
| BCL6 | 82 | 97.6 | 22 | 91.7 | 0.213 | |||||
| MUM1 | 55 | 65.5 | 10 | 41.7 | 0.057 | 0.048 | 0.368 (0.137-0.989) | |||
| CD5 | 7 | 8.4 | 3 | 12.5 | 0.69 | |||||
| BCL2 (n=106) | 60 | 72.3 | 17 | 73.9 | 1 | |||||
| EBV-ISH | 3 | 3.6 | 0 | 0 | 1 | |||||
Univariate analysis identified bone marrow invasion and LDH as significant predictors. However, MUM1 positivity was not identified. The multivariate analysis was tested by logistic regression (stepwise backward elimination using P-values), and variables that had a P-value ≤0.1 in the univariate analysis were included. LDH and MUM1 positivity were identified as significant predictors. High LDH was >220 IU/L.
There are some reports that document CD10 positivity as a predictor for favorable prognosis. We compared the survival curves of the MUM1-positive and -negative subgroups restricted to CD10-positive GCB patients, i.e., Groups 1 and 2, and found that prognosis tended to be better in Group 1 (the MUM1-positive subgroup) in terms of both OS and PFS, although the differences were not significant (Fig. 5). There were no significant differences between the two groups in terms of clinicopathological characteristics (Table 5). In the multivariate analysis of PFS in the CD10-positive GCB patients, high LDH was identified as a significant risk predictor (OR: 8.96, 95%CI = 1.94–41.5; P= 0.005) (Table 6).
Fig. 5.
Survival between MUM1+ vs MUM- in the CD10+GCB patients.
In the GCB subtype with positive CD10 expression, the expression of MUM1 was not correlated with the prognosis of the patients (P>0.05).
Table 5. Clinicopathological characteristics of MUM1-positive and -negative subgroups of the CD10+GCB patients.
| Variable | MUM1+ (n=66) | MUM1- (n=32) | P-value | ||||
|---|---|---|---|---|---|---|---|
| n | % | n | % | ||||
| Age >60 | 45 | 68.2 | 20 | 62.5 | 0.65 | ||
| Male | 42 | 63.6 | 19 | 59.4 | 0.82 | ||
| B symptoms | 16 | 24.2 | 6 | 19.4 | 0.8 | ||
| Extranodal | 35 | 53 | 18 (n=31) | 58.1 | 0.67 | ||
| BM invasion | 5 (n=64) | 7.8 | 5 (n=29) | 17.2 | 0.28 | ||
| High LDH | 36 | 54.5 | 20 (n=31) | 64.5 | 0.39 | ||
| Stage III-IV | 33 | 50 | 17 (n=31) | 54.8 | 0.67 | ||
| IPI H-I/H | 25 (n=65) | 38.5 | 14 (n=31) | 45.2 | 0.66 | ||
| CD5 | 7 (n=65) | 10.8 | 1 | 3.1 | 0.44 | ||
| EBV-ISH | 1 | 1.5 | 2 | 6.2 | 0.25 | ||
Table 6. Univariate and multivariate analyses of PFS events in the CD10+GCB group.
| Variable | PFS event- (n=76) | PFS event+ (n=19) | P-value | Odds Ratio | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | Univariate | Multivariate | (95% CI) | ||||
| Age >60 | 47 | 61.8 | 15 | 78.9 | 0.19 | |||||
| Male | 45 | 59.2 | 14 | 73.7 | 0.3 | |||||
| B symptom | 17 | 22.4 | 4 | 21.1 | 0.93 | |||||
| Stage III/IV | 36 | 47.4 | 12 | 63.2 | 0.31 | |||||
| IPI HI-H | 27 | 35.5 | 11 | 57.9 | 0.12 | - | ||||
| Extranodal | 38 | 50 | 13 | 68.4 | 0.2 | - | ||||
| BM invasion | 5 | 6.8 | 5 | 26.3 | 0.03 | - | ||||
| High LDH | 37 | 48.7 | 17 | 89.5 | 0.0015 | 0.005 | 8.96 (1.94-41.5) | |||
| MUM1 | 55 | 72.4 | 10 | 52.6 | 0.11 | - | ||||
| CD5 | 7 (n=75) | 9.3 | 1 | 5.3 | 1 | |||||
| BCL2 | 55 (n=75) | 73.3 | 13 | 68.1 | 0.78 | |||||
| EBV-ISH | 3 | 3.9 | 0 | 0 | 1 | |||||
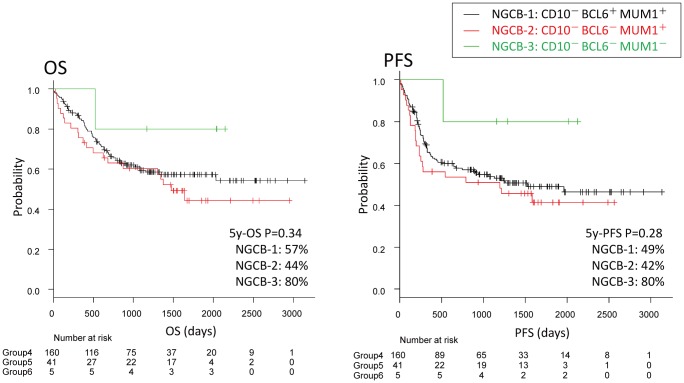
No significant differences in the OS or PFS survival curves were observed between the three subgroups of the non-GCB group. However, the CD10, BCL6 and MUM1 triple-negative subgroup 6 (NGCB-3) tended to have a better prognosis than the other two subgroups (Fig. 6).
Fig. 6.
Survival in the non-GCB group.
No significant differences were found between the non-GCB subgroups (P>0.05). However, the triple-negative NGCB-3 (CD10- BCL6- MUM1-) tended to be associated with a better prognosis.
DISCUSSION
In this paper, we report that the Hans classifier is still a useful algorithm to predict the clinical outcome of DLBCL in the rituximab era, with the GCB subtype associated with a favorable prognosis (OS and PFS). Moreover, we evaluated the usefulness of subdividing DLBCL into 6 subgroups and we found that in the GCB subtype, the expression of MUM1 was associated with a favorable PFS.
DLBCL is a heterogeneous disease both clinically and morphologically. Despite improvements in clinical responses due to the introduction of rituximab, it is still important to identify patients who may benefit from more aggressive or experimental therapies at diagnosis. The IPI reflects a mixture of underlying biological and genetic differences, and is able to stratify the patients according to their risk, but elucidation of these underlying factors remains incomplete. Advances using cDNA microarray and immunohistochemistry with tissue microarray7,13 were able to divide DLBCL into different subgroups with different prognoses as GCB, ABC and type 3, and GCB vs non-GCB. For the Hans classifier, by using three immunohistochemical markers, CD10, BCL6 and MUM1, which identify molecules whose mRNA expression was highly associated with the cDNA microarrays, frozen tissue was unneeded. However, these studies were from the pre-rituximab era, and it is necessary to evaluate if the Hans algorithm is still the most rapid and cost-effective tool for Western populations and Asian countries such as Japan. In our series of 320 cases, the GCB and non-GCB subtypes accounted for 35% and 65% respectively, and the GCB subtype was associated with a more favorable outcome. The classifier, however, did not correlate with most of the clinical variables, similar with the initial report by Hans CP et al.7 On the other hand, the GCB subtype was associated with lower prevalence of extranodal sites and higher clinical response to treatment. Interestingly, we also found that the GCB subtype had lower BCL2 expression.
As three simple markers can predict the prognosis of a heterogeneous entity, such as DLBCL, the markers likely reflect the pathogenic mechanisms. CD10 (membrane metalloendopeptidase, MME, located at 3q25.2) is a membrane-associated, neural peptidase that is expressed in a variety of human tissues, but is restricted to the germinal center cells of reactive lymphoid tissues16-18 and present on leukemic cells of the pre-B phenotype, which represents 85% of cases of human acute lymphocytic leukemia.19 CD10 is thought to be expressed during the first stages of heavy chain gene rearrangement, and in an immunological context, it is thought that the enzyme modulates the enkephalin-mediated inflammatory response.20 BCL6 (B-cell CLL/lymphoma 6, 3q27.3) is a zinc-finger protein that acts as a transcriptional repressor.21 BCL6 is expressed in germinal center B cells and a subset of CD4+T cells, and controls germinal center formation as well as Th2-type inflammation.22-26 MUM1 (interferon regulatory factor 4, IRF4, 6p25.3) is a lymphoid-specific member of the interferon regulatory factor family of transcription factors.27 MUM1 is normally expressed in plasma cells and in a minor subset of germinal center cells; it has been reported that MUM1 expression in DLBCL ranges from 50% to 77%.28-30 As a transcriptional activator, MUM1 binds to the interferon-stimulated response element (ISRE) of the MHC class I promoter and binds the immunoglobulin lambda chain enhancer, together with PU.1. MUM1 regulates germinal center cell formation,31 class-switch recombination and plasma cell differentiation.32,33 Although MUM1 expression was associated with poor survival in the pre-rituximab era,34 recent studies have failed to find a correlation.35 Interestingly, translocations activating MUM1 (IRF4) comprise a subtype of germinal center-derived B-cell lymphoma affecting predominantly children and young adults.36 In addition to the original Hans classifier, we permutated the 3 markers and identified 8 combinations. In our series, the most frequent phenotype was non-GCB CD10− BCL6+ MUM1+ (51%; NGCB-1) followed by GCB CD10+ BCL6+or- MUM1+ (21%; GCB-1) and non-GCB CD10- BCL6- MUM1+ (13%; NGCB-2). In terms of OS, we did not find differences between the GCB groups 1, 2 and 3, but we found a trend regarding the PFS; MUM1-negative groups had poorer PFS. Furthermore, when the GCB subtype was stratified according to MUM1 expression, MUM1-positive cases tended to have favorable OS and significant PFS. In a multivariate analysis for PFS, MUM1 was an independent protective variable, independent from the risk variable of LDH. This protective effect may be related with the function of MUM1 as regulator of germinal center formation.31 Translocations activating MUM1 were common in GCB lymphoma of children and young adults. Unfortunately, we did not perform FISH in our series and were unable to test the rearrangement status. We also examined if the prognostic relevance of MUM1 was related with CD10 expression, but we found no correlation. Finally, although not significant, we found that the triple-negative non-GCB (CD10- BCL6- MUM1-; NGCB-3) tended to be correlated with improved outcomes, but the reason for this observation is unknown.
Lenz et al. studied the molecular characteristics of the different cell-of-origin subtypes of DLBCL,37 and data from NGS recently became available from the TCGA Research Network with information about CD10, BCL6 and MUM1 markers. However, to date, no correlation with the Hans classifier is available for this NGS data and its prognostic relevance remains untested. A group claimed that new technologies, such as the Lymph2Cx assay, may be more robust, reliable methods for predicting outcomes of DLBCL patients treated with R-CHOP chemotherapy than the Hans algorithm in a series of 82 cases.38 The Lymphoma/Leukemia Molecular Profiling Project’s Lymph2Cx assay project had >95% concordance with the original cell-of-origin assignment based on GEP in 119 cases.11 NGS and Lymph2Cx assays may be attractive for clinical trials in the future considering that the relationship between cell-of-origin by immunohistochemistry and prognosis is controversial.39 Our research based on 320 cases demonstrated that the Hans classifier is still a valid and inexpensive method to estimate the prognosis of DLBCL patients.
In conclusion, GCB DLBCL is associated with a favorable outcome. Within the GCB group, the MUM1-positive subgroup (GCB-1) had a significantly better PFS than the negative subgroup, and MUM1 positivity was also identified as a predictor in the multivariate analysis. We did not examine IG/IRF4 translocation in the present study, but doing so may be necessary in the future.
Footnotes
CONFLICTS OF INTEREST: The authors declare no conflicts of interest.
REFERENCES
- 1.Coiffier B, Lepage E, Briere J, Herbrecht R, Tilly H, et al. : CHOP chemotherapy plus rituximab compared with CHOP alone in elderly patients with diffuse large-B-cell lymphoma. N Engl J Med 346: 235-242, 2002. 10.1056/NEJMoa011795 [DOI] [PubMed] [Google Scholar]
- 2.Vaidya R, Witzig TE: Prognostic factors for diffuse large B-cell lymphoma in the R(X)CHOP era. Ann Oncol 25: 2124-2133, 2014. 10.1093/annonc/mdu109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swerdlow SH, Campo E, Pileri SA, Harris NL, Stein H, et al. : The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 127: 2375-2390, 2016. 10.1182/blood-2016-01-643569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Habermann TM, Weller EA, Morrison VA, Gascoyne RD, Cassileth PA, et al. : Rituximab-CHOP versus CHOP alone or with maintenance rituximab in older patients with diffuse large B-cell lymphoma. J Clin Oncol 24: 3121-3127, 2006. 10.1200/JCO.2005.05.1003 [DOI] [PubMed] [Google Scholar]
- 5.Smith A, Crouch S, Howell D, Burton C, Patmore R, et al. : Impact of age and socioeconomic status on treatment and survival from aggressive lymphoma: a UK population-based study of diffuse large B-cell lymphoma. Cancer Epidemiol 39: 1103-1112, 2015. 10.1016/j.canep.2015.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kallam A, Armitage JO, Chen B, Vose JM, Bhatt VR: Center Effect and Socioeconomic Determinants of Overall Survival (OS) of Diffuse Large B Cell Lymphoma (DLBCL). Blood 128: 5949, 2016 [Google Scholar]
- 7.Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, et al. : Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood 103: 275-282, 2004. 10.1182/blood-2003-05-1545 [DOI] [PubMed] [Google Scholar]
- 8.Choi WW, Weisenburger DD, Greiner TC, Piris MA, Banham AH, et al. : A new immunostaining algorithm classifies diffuse large B-cell lymphoma into molecular subtypes with high accuracy. Clin Cancer Res 15: 5494-5502, 2009. 10.1158/1078-0432.CCR-09-0113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scott DW: Cell-of-Origin in Diffuse Large B-Cell Lymphoma: Are the Assays Ready for the Clinic? Am Soc Clin Oncol Educ Book 458-e466, 2015. 10.14694/EdBook_AM.2015.35.e458 [DOI] [PubMed] [Google Scholar]
- 10.Meyer PN, Fu K, Greiner TC, Smith LM, Delabie J, et al. : Immunohistochemical methods for predicting cell of origin and survival in patients with diffuse large B-cell lymphoma treated with rituximab. J Clin Oncol 29: 200-207, 2011. 10.1200/JCO.2010.30.0368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scott DW, Wright GW, Williams PM, Lih CJ, Walsh W, et al. : Determining cell-of-origin subtypes of diffuse large B-cell lymphoma using gene expression in formalin-fixed paraffin-embedded tissue. Blood 123: 1214-1217, 2014. 10.1182/blood-2013-11-536433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dlouhy I, Filella X, Rovira J, Magnano L, Rivas-Delgado A, et al. : High serum levels of soluble interleukin-2 receptor (sIL2-R), interleukin-6 (IL-6) and tumor necrosis factor alpha (TNF) are associated with adverse clinical features and predict poor outcome in diffuse large B-cell lymphoma. Leuk Res 59: 20-25, 2017. 10.1016/j.leukres.2017.05.014 [DOI] [PubMed] [Google Scholar]
- 13.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, et al.: Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature Feb 3;403:503-511, 2000 [DOI] [PubMed]
- 14.Sehn LH, Berry B, Chhanabhai M, Fitzgerald C, Gill K, et al. : The revised International Prognostic Index (R-IPI) is a better predictor of outcome than the standard IPI for patients with diffuse large B-cell lymphoma treated with R-CHOP. Blood 109: 1857-1861, 2007. 10.1182/blood-2006-08-038257 [DOI] [PubMed] [Google Scholar]
- 15.Kanda Y: Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transplant 48: 452-458, 2013. 10.1038/bmt.2012.244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dogan A, Bagdi E, Munson P, Isaacson PG: CD10 and BCL-6 expression in paraffin sections of normal lymphoid tissue and B-cell lymphomas. Am J Surg Pathol 24: 846-852, 2000. 10.1097/00000478-200006000-00010 [DOI] [PubMed] [Google Scholar]
- 17.Dyhdalo KS, Lanigan C, Tubbs RR, Cook JR: Immunoarchitectural patterns of germinal center antigens including LMO2 assist in the differential diagnosis of marginal zone lymphoma vs follicular lymphoma. Am J Clin Pathol 140: 149-154, 2013. 10.1309/AJCPHPVH4M7MTWUN [DOI] [PubMed] [Google Scholar]
- 18.Ohshima K, Kawasaki C, Muta H, Muta K, Deyev V, et al. : CD10 and Bcl10 expression in diffuse large B-cell lymphoma: CD10 is a marker of improved prognosis. Histopathology 39: 156-162, 2001. 10.1046/j.1365-2559.2001.01196.x [DOI] [PubMed] [Google Scholar]
- 19.Uckun FM, Sather H, Gaynon P, Arthur D, Nachman J, et al. : Prognostic significance of the CD10+CD19+CD34+ B-progenitor immunophenotype in children with acute lymphoblastic leukemia: a report from the Children’s Cancer Group. Leuk Lymphoma 27: 445-457, 1997. 10.3109/10428199709058311 [DOI] [PubMed] [Google Scholar]
- 20.Shipp MA, Stefano GB, Switzer SN, Griffin JD, Reinherz EL: CD10 (CALLA)/neutral endopeptidase 24.11 modulates inflammatory peptide-induced changes in neutrophil morphology, migration, and adhesion proteins and is itself regulated by neutrophil activation. Blood 78: 1834-1841, 1991 [PubMed] [Google Scholar]
- 21.Chang CC, Ye BH, Chaganti RS, Dalla-Favera R: BCL-6, a POZ/zinc-finger protein, is a sequence-specific transcriptional repressor. Proc Natl Acad Sci USA 93: 6947-6952, 1996. 10.1073/pnas.93.14.6947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flenghi L, Bigerna B, Fizzotti M, Venturi S, Pasqualucci L, et al. : Monoclonal antibodies PG-B6a and PG-B6p recognize, respectively, a highly conserved and a formol-resistant epitope on the human BCL-6 protein amino-terminal region. Am J Pathol 148: 1543-1555, 1996 [PMC free article] [PubMed] [Google Scholar]
- 23.Falini B, Mason DY: Proteins encoded by genes involved in chromosomal alterations in lymphoma and leukemia: clinical value of their detection by immunocytochemistry. Blood 99: 409-426, 2002. 10.1182/blood.V99.2.409 [DOI] [PubMed] [Google Scholar]
- 24.Falini B, Fizzotti M, Pileri S, Liso A, Pasqualucci L, et al. : Bcl-6 protein expression in normal and neoplastic lymphoid tissues. Ann Oncol 8(suppl 2): 101-104, 1997. 10.1093/annonc/8.suppl_2.S101 [DOI] [PubMed] [Google Scholar]
- 25.Cattoretti G, Chang CC, Cechova K, Zhang J, Ye BH, et al. : BCL-6 protein is expressed in germinal-center B cells. Blood 86: 45-53, 1995 [PubMed] [Google Scholar]
- 26.Ye BH, Cattoretti G, Shen Q, Zhang J, Hawe N, et al. : The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat Genet 16: 161-170, 1997. 10.1038/ng0697-161 [DOI] [PubMed] [Google Scholar]
- 27.Mamane Y, Heylbroeck C, Génin P, Algarté M, Servant MJ, et al. : Interferon regulatory factors: the next generation. Gene 237: 1-14, 1999. 10.1016/S0378-1119(99)00262-0 [DOI] [PubMed] [Google Scholar]
- 28.Natkunam Y, Warnke RA, Montgomery K, Falini B, van De Rijn M: Analysis of MUM1/IRF4 protein expression using tissue microarrays and immunohistochemistry. Mod Pathol 14: 686-694, 2001. 10.1038/modpathol.3880373 [DOI] [PubMed] [Google Scholar]
- 29.Falini B, Fizzotti M, Pucciarini A, Bigerna B, Marafioti T, et al. : A monoclonal antibody (MUM1p) detects expression of the MUM1/IRF4 protein in a subset of germinal center B cells, plasma cells, and activated T cells. Blood 95: 2084-2092, 2000 [PubMed] [Google Scholar]
- 30.Tsuboi K, Iida S, Inagaki H, Kato M, Hayami Y, et al. : MUM1/IRF4 expression as a frequent event in mature lymphoid malignancies. Leukemia 14: 449-456, 2000. 10.1038/sj.leu.2401696 [DOI] [PubMed] [Google Scholar]
- 31.Willis SN, Good-Jacobson KL, Curtis J, Light A, Tellier J, et al. : Transcription factor IRF4 regulates germinal center cell formation through a B cell-intrinsic mechanism. J Immunol 192: 3200-3206, 2014. 10.4049/jimmunol.1303216 [DOI] [PubMed] [Google Scholar]
- 32.Klein U, Casola S, Cattoretti G, Shen Q, Lia M, et al. : Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol 7: 773-782, 2006. 10.1038/ni1357 [DOI] [PubMed] [Google Scholar]
- 33.Nutt SL, Hodgkin PD, Tarlinton DM, Corcoran LM: The generation of antibody-secreting plasma cells. Nat Rev Immunol 15: 160-171, 2015. 10.1038/nri3795 [DOI] [PubMed] [Google Scholar]
- 34.Chang CC, McClintock S, Cleveland RP, Trzpuc T, Vesole DH, et al. : Immunohistochemical expression patterns of germinal center and activation B-cell markers correlate with prognosis in diffuse large B-cell lymphoma. Am J Surg Pathol 28: 464-470, 2004. 10.1097/00000478-200404000-00005 [DOI] [PubMed] [Google Scholar]
- 35.Horn H, Ziepert M, Wartenberg M, Staiger AM, Barth TF, et al. : Different biological risk factors in young poor-prognosis and elderly patients with diffuse large B-cell lymphoma. Leukemia 29: 1564-1570, 2015. 10.1038/leu.2015.43 [DOI] [PubMed] [Google Scholar]
- 36.Salaverria I, Philipp C, Oschlies I, Kohler CW, Kreuz M, et al. : Translocations activating IRF4 identify a subtype of germinal center-derived B-cell lymphoma affecting predominantly children and young adults. Blood 118: 139-147, 2011. 10.1182/blood-2011-01-330795 [DOI] [PubMed] [Google Scholar]
- 37.Lenz G, Wright GW, Emre NCT, Kohlhammer H, Dave SS, et al. : Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proc Natl Acad Sci USA 105: 13520-13525, 2008. 10.1073/pnas.0804295105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoon N, Ahn S, Yong Yoo H, Jin Kim S, Seog Kim W, et al. : Cell-of-origin of diffuse large B-cell lymphomas determined by the Lymph2Cx assay: better prognostic indicator than Hans algorithm. Oncotarget 8: 22014-22022, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Read JA, Koff JL, Nastoupil LJ, Williams JN, Cohen JB, et al. : Evaluating cell-of-origin subtype methods for predicting diffuse large B-cell lymphoma survival: a meta-analysis of gene expression profiling and immunohistochemistry algorithms. Clin Lymphoma Myeloma Leuk 14: 460-467, 2014. 10.1016/j.clml.2014.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]