Abstract
The Bomanins (Boms) are a family of a dozen secreted peptides that mediate the innate immune response governed by the Drosophila Toll receptor. We recently showed that deleting a cluster of 10 Bom genes blocks Toll-mediated defenses against a range of fungi and gram-positive bacteria. Here, we characterize the activity of individual Bom family members. We provide evidence that the Boms overlap in function and that a single Bom gene encoding a mature peptide of just 16 amino acids can act largely or entirely independent of other family members to provide phenotypic rescue in vivo. We further demonstrate that the Boms function in Drosophila humoral immunity, mediating the killing of the fungal pathogen Candida glabrata in an in vitro assay of cell-free hemolymph. In addition, we find that the level of antifungal activity both in vivo and in vitro is linked to the level of Bom gene expression. Although Toll dictates expression of the antimicrobial peptides (AMPs) drosomycin and metchnikowin, we find no evidence that Boms act by modifying the expression of the mature forms of these antifungal AMPs.
Keywords: Antifungal activity, Antimicrobial peptides, Bomanins, Drosophila, Effector, Host defense, NF-κB, Toll receptor
Introduction
The innate immune system provides essential defenses against pathogens in both plants and animals, and is the sole basis for defense in all nonvertebrates. Substantial insight into this system grew out of the discovery of the Toll signaling pathway and its translation from insects to humans [1, 2]. We now have a fairly comprehensive picture of how pathogen invasion of body tissues is recognized and how an ensuing cascade of signaling triggers immune defense activation [3, 4]. In many organisms, these defenses are demonstrably essential: infection of wild-type hosts by a range of bacteria, fungi, or viruses results in minimal morbidity or mortality, whereas hosts with compromised innate immune functions succumb, often rapidly. Nevertheless, a fundamental question remains: what effector activities mediate survival in the absence of a lymphocyte-directed adaptive response?
The study of innate antimicrobial effectors began in the 1920s with the discovery of the enzyme lysozyme, which Fleming [5] found can lyse bacterial cell walls. In the 1970s, studies on insects identified activities in hemolymph that kill bacteria or fungi and increase in response to the activation of innate defenses [6, 7]. This culminated in the identification and isolation of the first 3 antimicrobial peptides (AMPs) in 1980 [8]. There was a flurry of interest in AMPs as potential therapeutic agents, and a general assumption that they represent the primary effector mechanism. However, very few mutations have been reported that link the inactivation of particular AMP genes to an immune deficiency. Moreover, in Drosophila, such a link was established, not for an AMP, but rather for a set of novel effectors, the Bomanin, or Bom, gene family.
Bomanin genes encode 3 forms of mature peptide, short, tailed, and bicipital, that are secreted into the hemolymph upon Toll pathway activation [9, 10]. Ten of the twelve defined Bomanin genes are tandemly arrayed in a cluster at position 55C on the Drosophila melanogaster second chromosome. We have demonstrated that excising this cluster disrupts immune defenses to the same extent and with the same specificity as blocking the entire Toll pathway [10]. This finding raises a number of questions regarding Bom peptides as immune effectors. Are they humoral effectors or do they promote cellular immunity? Do they complex with one another or do they act as individual peptides? Is each Bomanin specific for a different pathogen?
In this paper, we explore the effector role of Boms via a two-pronged approach. We begin by using transgenic studies to define the activity of particular Bom genes. We then use microbiological and biophysical assays to define the nature of Bom activity in vitro. Together, these 2 approaches reveal that the expression of individual Boms in hemolymph can provide both resistance to infection in vivo and pathogen killing in vitro.
Materials and Methods
Flies and Transgenic Strains
Flies were raised at 25°C on standard corn-meal molasses food. The w1118 strain was used as the wild type. Bom∆55C flies were described previously [10].
All transgenic constructs were based on a genomic fragment (hereafter referred to as “55C-Right”) that encodes the four 55C Bom genes (Bom3, Bom836, Bom065, and Bom068) remaining in the Bom∆left second chromosome deficiency [10]. Specifically, genomic DNA encompassing region 2R: 18,387,209 to 18,391,496 was amplified and cloned into attB-pNot-CaSpeR [11]. Next, individual Bom ORFs were deleted using PCR SOEing [12], in such a way as to generate 4 constructs, each deleted for 3 of the 4 Bom genes in 55C-Right.
To generate transgenic constructs expressing a single Bom gene under the control of the Bom3 promoter (pBOM3), the starting point was the 55C-Right construct with Bom3 intact and the remaining Bom ORFs deleted. The Bom3 ORF was then swapped out for another ORF using PCR SOEing, placing the introduced ORF directly downstream of pBOM3.
All constructs were introduced into the Drosophila genome by ΦC31-mediated transgenesis [13] at an attP landing site located at 86Fb (BDSC stock #24749). Stable lines established for each construct carry homozygous transgenes on the third chromosome in a Bom∆55C background.
CRISPR Deletion of 55C Region
Cloning and injections were performed using established protocols [14]. A pair of gRNAs designed to delete the region 2R: 18,380,931 to 18,391,053 were cloned into pU6-BbsI-chiRNA (Add gene plasmid # 45946). Homology arms (766 bp left and 840 bp right) were cloned into pDsRed-attP (Addgene plasmid #51019). pBS-Hsp70-Cas9 (Addgene plasmid #46294) was used as the Cas9 source. Constructs were injected into w1118 flies.
Microbial Culture
Micrococcus luteus was cultured overnight in LB media at 37°C, heat-killed by autoclaving for 20 min at 121°C, and then concentrated to OD600 = 300 in 20% glycerol. For infection experiments, the Candida glabrata strain CBS 138 [ATCC 2001] was cultured overnight in YPD media at 37°C and concentrated to OD600 = 100 in fresh PBS containing 0.01% Tween. For antimicrobial assay experiments, C. glabrata was grown to mid-log phase in YPD and then diluted 1: 1,000 in YPD.
Drosophila Infection, Survival Assays, and Statistical Analyses
Septic wounding, survival assays, and statistical analyses using GraphPad Prism were conducted, essentially as previously described [10].
Bomanin Peptide Preparations
The 16-amino acid mature forms of Bom1 (GNVIINGDCRVCNVHG) and Bom3 (GNVIINGDCRVCNVRA) were synthesized by Lifetein and Biomatik, respectively, using a solid support resin with Fmoc and Boc chemistry. Peptides were delivered at 95% purity, confirmed by HPLC as well as mass spectrometry. Based on previously published mass spectrometry analysis [9], an intramolecular disulfide bond was introduced into both synthetic peptides and Bom1 was amidated at the carboxyl-terminus. Bom1 was dissolved in sterile water, and Bom3 was dissolved in 50% DMSO. Peptide concentrations were determined using the Qubit protein assay (Life Technologies), and were measured to be 43 μM (Bom1) and 56 μM (Bom3). These peptide solutions were introduced neat into the in vitro antimicrobial assay.
Hemolymph Preparation and Antimicrobial Assays
Toll pathway activation was achieved by wounding male flies (aged 2–7 days) with heat-killed M. luteus. After incubating the flies at 29°C for 24 h, cell-free hemolymph was collected from groups of 70 flies by a modification of the standard method [15]. Flies were loaded into a Zymo-Spin IC column (Zymo Research) previously washed twice with 200 μL sterile water. The flies were covered with 2-mm glass beads (Walter Stern) and centrifuged at 13,500 rpm atop a collection tube, for 20 min at 4°C, yielding approximately 4 μL of cell-free hemolymph.
Antimicrobial assays with synthetic peptide or hemolymph were carried out as prescribed for standard antimicrobial peptide assays [16]. Two microliters of peptide, hemolymph, or a combination of the two was mixed with an equal volume of log-phase C. glabrata suspended at 1: 1,000 in YPD. Samples were incubated either at room temperature for 1 h or on ice for 24 h. Following incubation, samples were diluted to 100 μL with YPD, spread onto YPD plates, and grown overnight at 37°C. Yeast colonies were counted the next day.
Hemolymph Extraction and MALDI-TOF Analysis
After Toll activation as above, hemolymph samples for MALDI-TOF were extracted from male flies (aged 2–7 days) using glass capillaries, and transferred with a Narishige IM-300 microinjector into 2 µL 0.1% trifluoroacetic acid (TFA)/50% acetonitrile (ACN). The samples were then mixed 1: 1 with α-cyano-4-hydroxycinnam ic acid (HCCA) matrix (Agilent: 2037A), and 2 μL was spotted onto a MALDI plate where sample and matrix cocrystallized in situ under a flow of warm air.
MALDI mass spectra were acquired using a Bruker Biflex IV MALDI-TOF mass spectrometer operated in either linear or reflectron mode with positive polarization. Desorption and ionization were achieved using a 337-nm laser. As an external mass calibration for linear mode spectra, equal parts of peptide calibration standard II (Bruker: 8222570) and protein calibration standard I (Bruker: 8206355) were mixed with HCCA matrix and spotted onto the same sample plate. For the calibration of reflectron mode spectra, peptide calibration standard II was mixed with HCCA matrix. For each genotype and condition, spectra from at least 5 individual flies were collected, yielding reproducible results. To identify particular peptides, the m/z values of peaks in our spectra were matched to those of the corresponding peaks in a prior study [9]. Spectra, for which representative examples are shown, were visualized using R 3.3.2, RStudio 1.0.136 and ggplot2 2.2.1.
Results
A Transgenic Assay Reveals the Effector Activity of a Single Bom Peptide
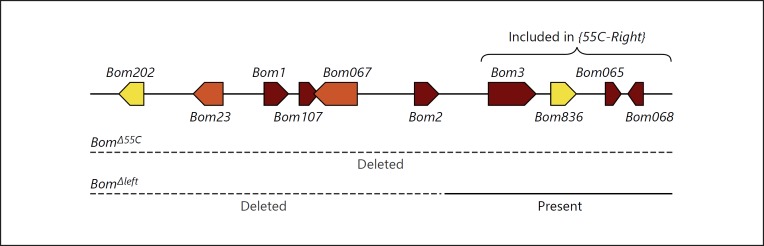
Ten Bomanin genes are clustered on the D. melanogaster second chromosome (Fig. 1). Our prior studies [10] defined 2 deletions affecting this cluster, designated 55C for its position on the polytene map. Bom∆55C flies, which lack the entire 55C Bom cluster, fail to mount a successful immune response against pathogens normally counteracted by the Toll pathway. In contrast, flies carrying the smaller Bom∆left deletion are susceptible to a number of pathogens, but have wild-type resistance to a relatively weak pathogen, the yeast C. glabrata. Because Bom∆left lacks the 6 genes on the left (proximal) side of the Bom cluster, the 4 remaining Bom genes provided a useful starting point for defining the functional unit of Bom activity.
Fig. 1.
Organization of the Bom family cluster at cytogenetic map position 55C. This figure reflects a renaming of the Bom family members, with the previously used CG or IM prefixes replaced with the Bom prefix, while retaining the gene number for both DIMs (1 or 2 digits) and CGs (last 3 digits) for continuity with prior literature [9, 17]. Accordingly, the 2 remaining Bom genes, located outside the 55C region, are now Bom778 and Bom791. For the 10 genes in the drawing, arrowhead direction denotes gene orientation. The color indicates the form of the encoded Bom peptide: short (red), tailed (yellow), or bicipital (orange). The bracket at the top encompasses the Bom genes in the 55C-Right genomic construct and the dotted lines below illustrate the extent of the chromosomal deletions in Bom∆55C and Bom∆left.
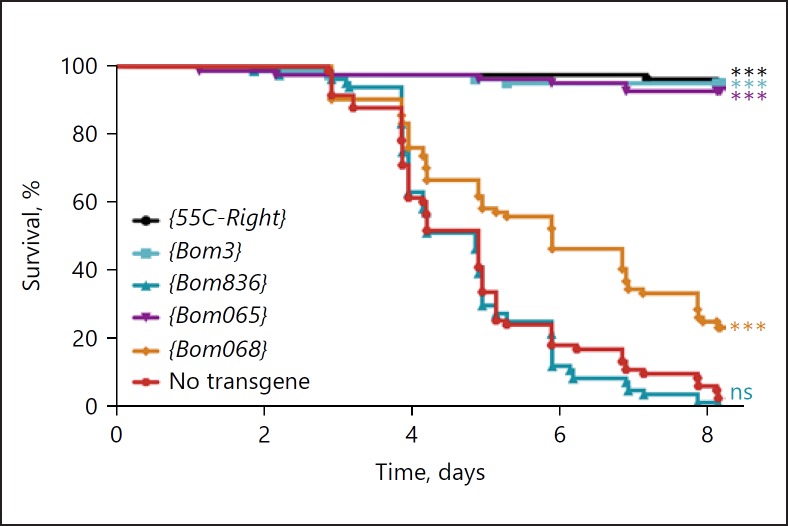
We generated a transgenic construct, 55C-Right, spanning the 4 intact Bom genes to the right of the deletion in Bom∆left. By generating flies carrying the 55C-Right transgene on each third chromosome and the Bom∆55C deletion on each second chromosome, we could recapitulate the genome of Bom∆left. Using 55C-Right as a template, we next generated 4 related constructs, each expressing only a single ORF from the 4 Bom genes originally present. We then introduced 2 copies of the wild-type or single-gene 55C-Right constructs into the Bom∆55C background and assayed survival after challenge with C. glabrata (Fig. 2).
Fig. 2.
A single Bom gene rescues the Bom∆55C immunodeficiency toward C. glabrata. The graph illustrates the survival of transgenic adults at indicated intervals after infection with C. glabrata. Each transgene encodes either the 4 Bom peptides of 55C-Right or a single one of these peptides, as indicated. All transgenes were present in 2 copies in a Bom∆55C background. Each curve represents the pooled results of 3 independent experiments involving ≥25 flies per genotype. Survival curves were compared using the Gehan-Breslow-Wilcoxon test. Significance is shown relative to the no transgene control (Bom∆55C) and adjusted for multiple comparisons (*** p < 0.0001; ns, not significant, p > 0.05).
The data in Figure 2 demonstrate that a single Bom gene, either Bom3 or Bom065, can restore resistance to C. glabrata in Bom∆55C flies. We noted that both Bom3 and Bom065 encode short-form Bom peptides, defined as having a mature length of just 16 amino acids [10]. A third short-form Bom gene, Bom068, provided lesser but still significant rescue. Intriguingly, the resistance strength appeared to correlate with the Bom transcript level, as measured in Toll-induced wild-type adults (Table 1). Thus, resistance was higher for Bom3 and Bom065 and lower for Bom068.
Table 1.
Survival after infection with C. glabrata correlated with total Bom transcript level
Robust Expression of Short-Form Boms Conveys Resistance to C. glabrata
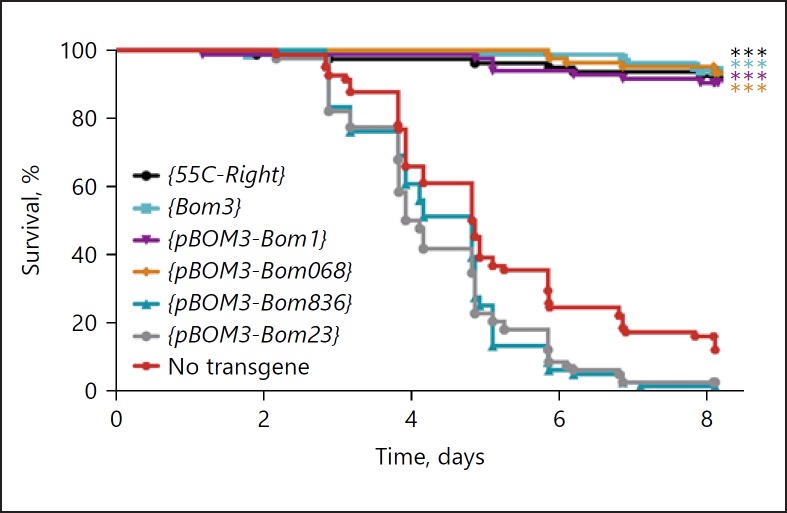
By further modifying the 55C-Right construct, we set out to compare the activity of different Bom forms when expressed at comparable levels. Specifically, we replaced the Bom3 coding region with that of other Bom genes, i.e., we fused different Bom ORFs to the strongly Toll-responsive Bom3 promoter (pBOM3). In this fashion, we generated 55C-Right constructs that should express a single, short, tailed, or bicipital Bom at the high levels normally observed for Bom3 upon Toll pathway activation. We then assayed survival after challenge with C. glabrata (Fig. 3).
Fig. 3.
Short-form Bom peptides mediate survival after infection with C. glabrata. The graph plots the survival of adults at indicated intervals after infection with C. glabrata. Bom transgenes were expressed under control of either their endogenous promoter or pBOM3. All transgenes were present in 2 copies in a Bom∆55C background. Data were captured and analyzed as in Figure 2.
All 3 short-form Bom genes tested provided full resistance to C. glabrata when expressed under the control of pBOM3. This included Bom068, which provided considerably weaker resistance when driven by its own promoter (Fig. 2 vs. Fig. 3). These data support the hypothesis that it is the level of expression, rather than the sequence composition, of short-form Boms that determines their contribution to survival. Furthermore, we found no evidence that the short-form Bom peptides vary intrinsically in specificity for C. glabrata.
We turned next to representative examples of tailed and bicipital Bomanins. A tailed form, Bom836, provided no rescue of Bom∆55C when controlled by its endogenous promoter. Analysis of the endogenous transcript levels following Toll activation revealed far less transcript accumulation for Bom836 than for either Bom3 or Bom065 (Fig. 2; Table 1). However, the Bom836 ORF failed to generate detectable rescue even when fused to pBOM3 (Fig. 3). Likewise, we did not observe rescue with pBOM3-driven expression of a bicipital form, Bom23 (Fig. 3). Thus, neither the tailed nor the bicipital Bom exhibited activity against C. glabrata. For subsequent studies, we therefore focused on the short-form Boms, for which our survival assay provides a robust readout in vivo for an individual gene product.
Short-Form Boms Mediate Cell-Free Killing of C. glabrata
Having found that expression of a single short-form Bom could rescue adults otherwise immunodeficient against C. glabrata infection, we turned our attention to in vitro studies. In initial experiments, we assayed synthetic, mature short-form Bom peptides under conditions permissive for antimicrobial activity against C. glabrata. No such activity was detected with the synthetic forms of mature Bom1 or Bom3 (data not shown).
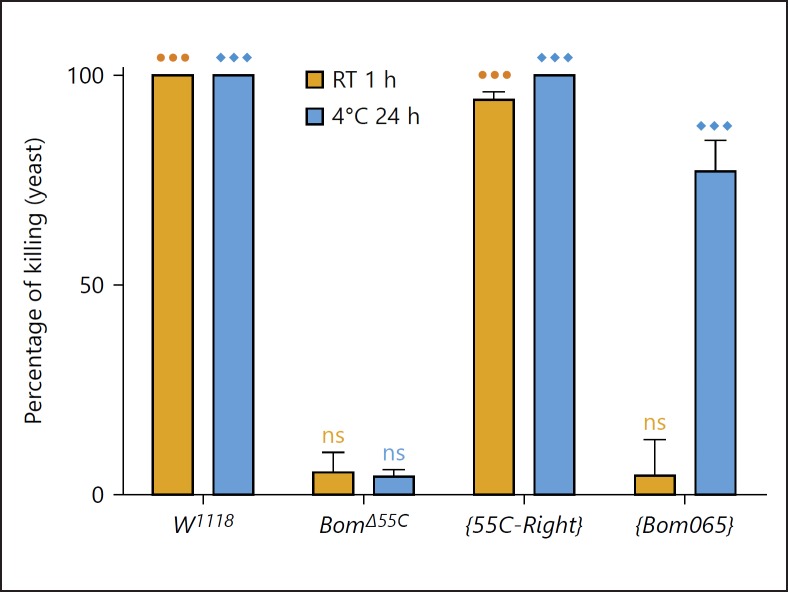
In parallel, we set out to explore the in vitro activity of hemolymph, in which many of the Bom peptides are abundantly expressed upon infection. We stimulated the Toll pathway, and then collected cell-free hemolymph after 24 h and mixed it with an equal volume of mid-log C. glabrata. After incubation, the mixture was plated and the yeast colonies were counted the next day. Two alternative conditions were used for incubation. The first, a 1-h incubation at room temperature, was designed to mimic conditions under which infection would normally occur. The second, a 24-h incubation at 4°C, was designed to increase assay sensitivity. Wild-type hemolymph was fungicidal under either condition, with no detectable C. glabrata colonies formed (Fig. 4).
Fig. 4.
Fungicidal activity of hemolymph is Bom-dependent. Hemolymph activity was assayed for 4 sets of flies: w1118 (wild-type), Bom∆55C, and Bom∆55C carrying either a 55C-Right (four-gene) or Bom065 (single-gene) construct. All flies had been pricked 1 day earlier with heat-killed M. luteus to induce Toll-mediated gene expression. Fungicidal activity was assayed by incubating cell-free hemolymph with C. glabrata for 1 h at room temperature (RT) or for 24 h at 4°C before spreading the mixture onto a yeast (YPD) plate. Percent killing was calculated by comparing the colony count for each sample to that for yeast mixed with a w1118 uninduced hemolymph control. Experiments were performed in triplicate; error bars represent standard error of the mean. One-way ANOVA followed by Tukey's test was performed for each incubation condition. Significance is shown relative to the w1118 uninduced control (⚫⚫⚫ and ◆◆◆ = p < 0.0001; ns, not significant, p > 0.05).
When we assayed hemolymph from Bom∆55C adults, there was a clear and dramatic difference from the wild type; deleting the 55C Bom genes eliminated the fungicidal activity of hemolymph. The Bom genes thus mediate a humoral antimicrobial activity against C. glabrata. Furthermore, the Bom-dependent activity of hemolymph in vitro parallels the Bom-dependent resistance observed in vivo against the same pathogen.
Hemolymph Activity Correlates with Bom Gene Dosage
To assay the effect of gene dosage on the level of Bom-dependent hemolymph activity, we took advantage of the genetic backgrounds representing different overall levels of Bom expression. We first assayed hemolymph from Bom∆55C; {55C-Right} flies. Antimicrobial activity was comparable to the wild type under both conditions (Fig. 4). We next assayed hemolymph from Bom∆55C; {Bom065} flies, in which expression of a single Bom peptide (Bom065) confers C. glabrata resistance in the Bom∆55C background (Fig. 2). In this case we found no fungicidal activity by the Bom∆55C; {Bom065} hemolymph at room temperature, but readily detectable and significant killing at 4°C overnight (Fig. 4). Thus, the strength of antimicrobial activity in vitro, like resistance in vivo, appears to correlate with the overall level of short-form Bom expression.
The Levels and Composition of Antifungal AMPs Appear Bom-Independent
Although the Bomanins mediate antimicrobial activity, they could act upstream of known AMPs, e.g., to bring about accumulation or modification of AMP gene products. In the case of antifungal defenses, there are 2 AMPs that are robustly induced upon Toll activation and display strong antifungal activity in vitro: drosomycin (Drs) and metchnikowin (Mtk) [17, 18, 19] (a third AMP, cecropin, is active against a wide range of pathogens [20], but is inactive against C. glabrata [21]). We have previously shown that induction of mRNA expression by Toll does not require Bom function [10]. To determine if Boms act on these AMPs at the protein level, we turned to mass spectrometry.
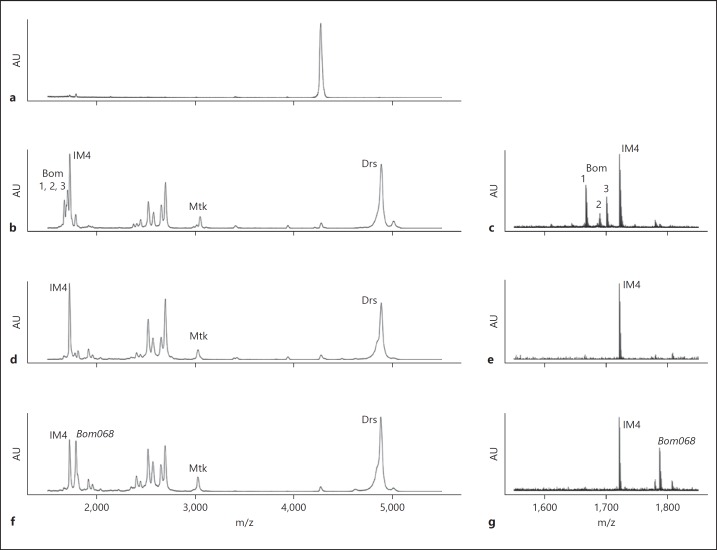
Uttenweiler-Joseph et al. [9] reported an approach for characterizing the peptide composition of hemolymph from a single Drosophila adult. Using matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS), they identified singly charged ions for a set of 24 immune-induced molecules (IMs). Adopting their approach, we obtained near identical spectrograms (Fig. 5a, b, compare with Fig. 3 in [9]), enabling us to identify individual peptides by their m/z ratio. Upon inducing innate immune signaling, the IMs were readily detectable, including Drs (IM19), Mtk (IM17), and 3 Boms (1, 2 and 3) in the 55C cluster (Fig. 5b, c). The m/z for an additional 55C Bom peptide, Bom23, falls outside the mass range examined.
Fig. 5.
Analysis of hemolymph from adult Drosophila by MALDI-TOF MS. Hemolymph (50 nL) was collected from a single fly from each genotype and analyzed by MALDI-TOF MS using the linear (a, b, d, f) or reflectron mode (c, e, g) (see Materials and Methods). Representative MALDI spectra from ≥5 such samples are shown for hemolymph from a w1118 fly either uninduced (control) (a), or induced 24 h earlier by Toll pathway stimulation with heat-killed M. luteus (b, c), and induced flies of the following genotypes: Bom∆55C (d, e) and Bom∆55C; {pBOM3-Bom068} (f, g). AU, arbitrary units. The y axis was scaled to the tallest peak within the window examined. Numbering and naming of induced peptides corresponds to published conventions [9]. As observed previously [9], a prominent peak appears in control spectra that becomes insignificant in the context of Toll induction. Drs, drosomycin; Mtk, metchnikowin.
Next, we carried out parallel experiments with hemolymph from Bom∆55C flies (Fig. 5d, e). As expected, no peaks were found with an m/z ratio matching that of Bom1, Bom2, or Bom3. Peaks were present, however, for both Drs and Mtk. In the case of Drs, the signal remained robust and with the same signal maximum. We noted, however, that Drs has a relatively high molecular weight, precluding detection of a minor change in m/z in our study.
For Mtk, we did observe a significant difference in peak maxima, with a shift from 3,044 m/z in the w1118 control to 3,022 m/z in Bom∆55C. This shift, however, is not Bom-dependent. Rather, it reflects a difference in Mtk isoform between the 2 genetic backgrounds. There are 2 known isoforms of Mtk, encoding either a His or Arg residue at position 3 in the mature peptide. Both isoforms are active [19]. The m/z values corresponding to the 2 species exactly match those we detected here.
To verify that the Bom phenotypes do not reflect a difference in Mtk isoform, we first sequenced the Mtk gene in the w1118 and Bom∆55C backgrounds. This analysis confirmed the isoform identification by mass spectrometry: MtkR3 in the w1118 background and MtkH3 in the Bom∆55C background. Next, we used CRISPR-Cas9 to generate a Bom 55C deletion in the w1118 background, eliminating the polymorphism in Mtk between the control and experimental samples. Using the newly derived Bom∆55C-2 flies, we obtained survival curves after infection identical to those of the previously published TALEN-generated Bom∆55C flies (online suppl. Fig. 1; see www.karger.com/doi/10.1159/000489831 for all online suppl. material).
Taken together, these studies reveal that deleting the Bom 55C cluster has no readily detectable effect on accumulation or posttranslational modification of 2 major Toll-induced antifungal peptides in hemolymph.
Refined Understanding of the Bom Repertoire
Six of the 55C Boms are not detected even in wild-type hemolymph. Why? They are unlikely to represent pseudogenes, since they are well conserved at the amino acid level and lack any obvious nonsense or missense mutations. Instead, we favor 2 explanations. First, for a Bom with robust expression at the mRNA level, the peptide is likely present but masked. For example, we noted that the calculated mass of the amidated form of Bom065 is 1,723.8 m/z. This peptide is likely to fall in the shoulder of the robust peak for IM4 (calculated mass 1,721.9 m/z) and therefore not be detected. Second, a number of Boms may have been missed simply because their level of immune-induced expression is relatively low. Here, Bom068 provides a useful example. Compared to other short-form Boms, Bom068 mRNA accumulates to only modest levels upon Toll activation (Table 1). Consistent with this weak expression, we observed modest resistance to C. glabrata infection in flies expressing Bom068 from its endogenous promoter in a Bom∆55C background, and were unable to detect the peptide by MALDI-TOF. However, when the Bom068 ORF was heterologously expressed under control of the strongly Toll-responsive pBOM3 promoter, we observed wild-type resistance to C. glabrata infection and, furthermore, ready detection of a peak at 1,787 m/z, which matches the predicted value on MALDI-TOF for Bom068 that has undergone proteolytic processing but not amidation (Fig. 5f, g).
Discussion
Individual 55C Bom Genes Mediate Antimicrobial Defense
Transgenic analyses revealed that a single, short-form 55C Bom gene provides resistance to C. glabrata in the absence of the remaining 9 genes from the cluster. Furthermore, every short-form Bom gene tested displayed such activity. The highly similar structure of the short-form Boms thus appears to reflect a shared activity found in each. These data make it unlikely that all 10 genes of the cluster form a supramolecular complex or are all part of a common activity cascade.
We did not find transgenic activity for tailed or bicipital Boms. Two explanations appear very reasonable. First, these Bom forms might have specificity for particular pathogens other than C. glabrata. Bom activity has been observed not just for yeast, but also for representatives of hyphal fungi and gram-positive bacteria [10]. Second, the tailed and bicipital Boms might be active only in combination with other Bom family members.
Boms exist in a cluster across the Drosophila genus. Why did such a cluster arise if individual genes are active and interchangeable, at least in certain circumstances? Our results indicate that, for particularly virulent pathogens, such as Enterococcus faecalis, successful immune defense requires very high levels of Bomanins [10]. We hypothesize that a single transcription unit is insufficient to provide enough Bom peptide in the time frame required. According to this model, duplication provided multiple, near-identical genes that could act as a battery for defense. Several lines of evidence support this idea. First, there are multiple, strongly Toll-responsive Bomanin genes, each immediately downstream of a canonical TATA start site, and, almost always, a perfect or near-perfect Toll-responsive NF-κB element [22]. Second, the 55C-Right construct, inserted at 86Fb and lacking more than half the 55C cluster, expressed each of the 4 remaining Bom genes at the same level as in the full-length cluster, suggesting an absence of control elements shared across the 10 genes (online suppl. Table 1).
If duplication is driven by a need for a high overall level of expression, what should we make of the variation we observed in the sequence of Bom family members? It could provide sequence specificity for Bom action. It could also provide protection against pathogen countermeasures, varying the residues targeted by microbial enzymes or inhibitors. Lastly, variation could also be a simple consequence of duplication and drift.
It is important to note that removing the 55C Bom cluster drastically reduces, but does not eliminate, Bom expression. Two additional Bomanins, Bom778 (tailed) and Bom791 (bicipital), remain intact, as do 2 additional genes (IM4, IM14) that are immune-induced and encode peptides that bear sequence similarity to the Bomanins. For this reason, caution is required when drawing conclusions with regard to the complete repertoire of the Bomanin family.
Bom Peptides Are Required for the Toll-Directed Humoral Defense
Bomanins are clearly required for the humoral response, as a hemolymph preparation free from intact cells provides Bomanin-dependent fungicidal activity in vitro. Furthermore, this activity acts rapidly, killing C. glabrata within an hour under normal growth conditions. An involvement in humoral immunity makes good sense, as the Bomanins are secreted from the fat body into the hemolymph [9]. Indeed, they are loaded into hemolymph at concentrations sufficiently high as to suggest a stoichiometric role in defense rather than a role as cytokines.
To date, synthetic Boms have not been active in our in vitro assays. We have no easy explanation for this. The problem is not one of concentration, since MALDI-TOF revealed that synthetic peptide concentrations were well-matched to those of the endogenously expressed forms. Furthermore, the measured mass-to-charge (m/z) values were also identical. Nor is the problem one of the exogenous peptides requiring a partner protein or other factor, since the synthetic Bomanins are also inactive when mixed with induced Bom∆55C hemolymph (unpubl. observ.). This failure to observe complementation in vitro suggests that the synthetic forms lack a conformation or stable interaction required for activity and which is normally provided in vivo prior to release into the hemolymph.
Loss-of-function mutations in multiple family members provide a powerful tool for dissecting effector function. For example, in this study, we were able to define an essential role for the Bom family, assess the significance of gene duplication and divergence, and define effector specificity both in vivo and in vitro. Whereas small genes of overlapping function are a poor target for traditional mutagenesis, generating mutations in a number of such genes is now both straightforward and feasible. More insights and more surprises lie ahead.
Disclosure Statement
The authors declare no competing financial interests.
Funding Sources
This work was supported by National Institutes of Health (NIH. gov) grant R01 GM050545 (to S.A.W.) and NIH predoctoral training grant T32 GM008666 (to S.J.H.L.). The funders had no role in study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
Supplementary Material
Supplementary data
Acknowledgements
The authors would like to thank Yongxuan Su and the Molecular Mass Spectrometry Facility at UCSD for guidance in preparation and analysis of samples for mass spectrometry. We would like to acknowledge the technical assistance of Yangyang Xu and Shelief Juarez in development of the hemolymph assay. We appreciate the contributions of materials and advice from Toto Olivera and Joanna Gajewiak, and access to equipment provided by Ethan Bier and Amy Pasquinelli. We also thank Russell Doolittle, Eric Bennett, Bill McGinnis, and Scott Rifkin for helpful discussions and advice, and Emily Troemel, Matt Daugherty, Alisa Huffaker, Lianne Cohen, and Alexa Clemmons for helpful comments on the manuscript. Lastly, we express our gratitude for the enthusiasm and assistance provided by Bo Zhou, who is greatly missed.
References
- 1.Anderson KV, Nüsslein-Volhard C. Information for the dorsal-ventral pattern of the Drosophila embryo is stored as maternal mRNA. Nature. 1984;311:223–227. doi: 10.1038/311223a0. [DOI] [PubMed] [Google Scholar]
- 2.Hoffmann JA, Kafatos FC, Janeway CA, Ezekowitz RAB. Phylogenetic perspectives in innate immunity. Science. 1999;284:1313–1318. doi: 10.1126/science.284.5418.1313. [DOI] [PubMed] [Google Scholar]
- 3.Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 4.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 5.Fleming A. On a remarkable bacteriolytic element found in tissues and secretions. Proc R Soc B Biol Sci. 1922;93:306–317. [Google Scholar]
- 6.Boman HG, Nilsson I, Rasmuson B. Inducible antibacterial defence system in Drosophila. Nature. 1972;237:232–235. doi: 10.1038/237232a0. [DOI] [PubMed] [Google Scholar]
- 7.Boman HG, Nilsson-Faye I, Paul K, Rasmuson T. Characteristics of an inducible cell-free antibacterial reaction in hemolymph of Samia cynthia pupae. Infect Immun. 1974;10:136–145. doi: 10.1128/iai.10.1.136-145.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hultmark D, Steiner H, Rasmuson T, Boman HG. Insect immunity. Purification and properties of three inducible bactericidal proteins from hemolymph of immunized pupae of Hyalophora cecropia. Eur J Biochem. 1980;106:7–16. doi: 10.1111/j.1432-1033.1980.tb05991.x. [DOI] [PubMed] [Google Scholar]
- 9.Uttenweiler-Joseph S, Moniatte M, Lagueux M, Van Dorsselaer A, Hoffmann JA, Bulet P. Differential display of peptides induced during the immune response of Drosophila: a matrix-assisted laser desorption ionization time-of-flight mass spectrometry study. Proc Natl Acad Sci. 1998;95:11342–11347. doi: 10.1073/pnas.95.19.11342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clemmons AW, Lindsay SA, Wasserman SA. An effector peptide family required for Drosophila Toll-mediated immunity. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1004876. e1004876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu F, Posakony JW. An enhancer composed of interlocking submodules controls transcriptional autoregulation of suppressor of hairless. Dev Cell. 2014;29:88–101. doi: 10.1016/j.devcel.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 13.Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM, et al. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics. 2014;196:961–971. doi: 10.1534/genetics.113.160713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neyen C, Bretscher AJ, Binggeli O, Lemaitre B. Methods to study Drosophila immunity. Methods. 2014;68:116–128. doi: 10.1016/j.ymeth.2014.02.023. [DOI] [PubMed] [Google Scholar]
- 16.Wiegand I, Hilpert K, Hancock RE. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc. 2008;3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 17.De Gregorio E, Spellman PT, Rubin GM, Lemaitre B. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc Natl Acad Sci USA. 2001;98:12590–12595. doi: 10.1073/pnas.221458698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fehlbaum P, Bulet P, Michaut L, Lagueux M, Broekaert WF, Hetru C, et al. Septic injury of Drosophila induces the synthesis of a potent antifungal peptide with sequence homology to plant antifungal peptides. J Biol Chem. 1994;269:33159–33163. [PubMed] [Google Scholar]
- 19.Levashina EA, Ohresser S, Bulet P, Reichhart J, Hetru C, Hoffmann JA. Metchnikowin, a novel immune-inducible proline-rich peptide from Drosophila with antibacterial and antifungal properties. Eur J Biochem. 1995;233:694–700. doi: 10.1111/j.1432-1033.1995.694_2.x. [DOI] [PubMed] [Google Scholar]
- 20.Ekengren S, Hultmark D. Drosophila cecropin as an antifungal agent. Insect Biochem Mol Biol. 1999;29:965–972. doi: 10.1016/s0965-1748(99)00071-5. [DOI] [PubMed] [Google Scholar]
- 21.Lowenberger C, Charlet M, Vizioli J, Kamal S, Richman A, Christensen BM, et al. Antimicrobial activity spectrum, cDNA cloning, and mRNA expression of a newly isolated member of the cecropin family from the mosquito vector Aedes aegypti. J Biol Chem. 1999;274:20092–20097. doi: 10.1074/jbc.274.29.20092. [DOI] [PubMed] [Google Scholar]
- 22.Busse MS, Arnold CP, Towb P, Katrivesis J, Wasserman SA. A κB sequence code for pathway-specific innate immune responses. EMBO J. 2007;26:3826–3835. doi: 10.1038/sj.emboj.7601798. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data