Abstract
Regulatory brain cytoplasmic 200 RNA (BC200RNA) is highly expressed in human mammary carcinoma cells Here, we ask whether BC200 RNA becomes detectable in peripheral blood of patients with invasive breast cancer Using quantitative reverse-transcription PCR (qtr.-PCR) methodology, we observed that BC200 RNA blood levels were significantly elevated, in comparison with healthy subjects, in patients with invasive breast cancer prior to tumorectomy (p=0.001) and in patients with metastatic breast cancer (p=0.003) In patients with invasive breast cancer who had recently undergone tumorectomy, BC200 RNA blood levels were not distinguishable from levels in healthy subjects However, normality analysis revealed a heterogeneous distribution of patients in this group, including a subgroup of individuals with high residual BC200 RNA blood levels In blood from patients with invasive breast cancer, BC200 RNA was specifically detected in the mononuclear leukocyte fraction The qtr.-PCR approach is sensitive enough to detect as few as three BC200 RNA-expressing tumor cells Our work establishes the potential of BC200 RNA detection in blood to serve as a molecular indicator of invasive breast malignancy.
INTRODUCTION
Neuronal BC RNAs, an abundant subtype of small cytoplasmic RNAs, are translational regulators that operate in the translational control of gene expression in mammalian nerve cells1,2 BC RNAs interact with components of the eukaryotic translational machinery Specifically, primate BC200 RNA and rodent BC1 RNA target eukaryotic initiation factors (eIFs) 4A and 4B, canonical translation factors that are required for the recruitment of small ribosomal subunits to mRNAs with higher-order structure content in their 5’ untranslated regions2–6 While BC RNAs act as translational repressors in the basal default state, BC RNA control can reversibly switch from repressive to permissive on receptor activation in neurons6 Specifically, stimulation of group I metabotropic glutamate receptors causes rapid activation of protein phosphatase 2A (PP2A), which in turn triggers dephosphorylation of eIF4B at serine 406 (S406) Thus dephosphorylated, eIF4B interacts with BC RNAs eIF4B at significantly reduced affinity, as a result allowing the factor to engage the small ribosomal subunit for translation initiation6
BC RNAs are not typically expressed in somatic mammalian cell types other than neurons2,7 However, in a notable exception to this neuronal specificity, BC200 RNA was found expressed at high levels in human invasive breast carcinoma cells8,9 In contrast, BC200 RNA was not detected in normal human breast epithelial cells or in benign tumors of the breast The combined data suggested that regulatory BC200 RNA operates in invasive or preinvasive breast carcinoma cells where it causes dysregulation of cellular translational control9,10.
Despite the fact that a combination of factors, including early detection (eg, mammography) and improved therapeutic intervention (resection, adjuvant therapy), has in recent years resulted in a decrease of breast cancer mortality in the USA, breast cancer remains a leading cause of death from malignancies in women11 However, mammography has been a subject of controversial debate,12–15 and therapeutic intervention has been hampered by a lack of indicators, molecular and otherwise, that would aid physicians and patients in breast cancer treatment decisions In particular, any improvement in our ability to predict treatment outcome, and to prognosticate tumor progression and recurrences, would have potentially significant clinical use.
Here, we hypothesized that invasive breast cancers may disseminate BC200-expressing cells into the circulation Considering that BC200 RNA (1) is a small regulatory RNA with significant structural stability16 and (2) is expressed at high levels in breast cancer cells,8,9 we further hypothesized that the RNA may be persistently expressed in circulating tumor cells (CTCs) derived from invasive breast cancers The goal of the present study was to test this hypothesis, using reverse-transcription (RT)-PCR and quantitative RT-PCR (qtr.-PCR) methodology, and to ascertain the potential of an RNA-based blood test for invasive breast cancer.
MATERIALS AND METHODS
Subject recruitment
Peripheral blood was collected from the following subject groups:
Healthy subjects: This group comprised apparently healthy women without diagnosis, or signs or symptoms, of cancer (n=12).
No evidence of disease: This group comprised previous patients with breast cancer who had completed treatment, including adjuvant therapy, at least 15 years prior to blood collection These patients were under survey-lance, with no evidence of current disease, at the time of blood collection (n=10).
Primary disease I: This group comprised patients with operable breast cancer after diagnosis but before resection of the primary tumor (n=14).
Primary disease II: This group comprised patients with operable breast cancer after tumorectomy who, at the time of blood collection, were either undergoing adjuvant therapy or had recently completed adjuvant therapy (recently being defined as within 1 month before blood collection; n=12).
Metastatic disease: This group comprised patients with active metastatic disease, stage IV (n=23) Patient parameters are listed in table 1 Patients’ records and oncological assessments were reviewed by three oncologists (EQC, RLC, and GAS).
Table 1.
Classification of breast cancer
| No evidence of disease | Primary disease I (presurgery) |
Primary disease II (postsurgery) |
Metastatic disease | |
|---|---|---|---|---|
| No of patients | 10 | 14 | 12 | 23 |
| Age (mean) | 62.9 | 59.1 | 53.8 | 53.7 |
| Premenopausal | 1 | 5 | 5 | 8 |
| Postmenopausal | 9 | 9 | 7 | 15 |
| Stage 1 | 6 | 0 | 1 | 0 |
| Stage II | 3 | 5 | 7 | 0 |
| Stage III | 1 | 9 | 4 | 0 |
| Stage IV | 0 | 0 | 0 | 23 |
| Tumor size 1–1.9 cm | 6 | 0 | 2 | 1 |
| Tumor size 2–3.9 cm | 1 | 3 | 7 | 2 |
| Tumor size >4 cm | 1 | 11 | 3 | 14 |
| Unknown | 2 | 0 | 0 | 6 |
| Grade 1 or 2 | 3 | 5 | 3 | 6 |
| Grade 3 | 4 | 8 | 7 | 12 |
| Unknown | 3 | 1 | 2 | 5 |
| Lymph node 0 | 7 | 3 | 6 | 4 |
| Lymph nodes 1–3 | 3 | 11 | 6 | 15 |
| Unknown | 0 | 0 | 0 | 4 |
| Estrogen receptor + | 6 | 7 | 8 | 15 |
| Estrogen receptor − | 1 | 7 | 4 | 8 |
| Unknown | 3 | 0 | 0 | 0 |
| Progesterone receptor + | 5 | 7 | 8 | 10 |
| Progesterone receptor − | 2 | 7 | 4 | 13 |
| Unknown | 3 | 0 | 0 | 0 |
| Her2/neu + | 4 | 12 | 8 | 19 |
| Her2/neu − | 2 | 1 | 4 | 4 |
| Unknown | 4 | 1 | 0 | 0 |
Specimen characteristics
Blood samples were drawn from the median cubital vein on the anterior forearm and collected in BD Vacutainer tubes containing EDTA (BD, Franklin Lakes, NJ) Then 250 µL aliquots were quick-frozen at –80°C A 250 µL aliquot typically generated 15 µg total RNA Furthermore, 10 ng total RNA was used per reaction, that is, 30 ng for triplicate BC200 RNA amplification and 30 ng for triplicate control RNA amplification reaction, in total 60 ng per experiment.
RNA isolation, RT-PCR, and qtr.-PCR
Total RNA was extracted from 250 µL whole blood using the Ribopure Extraction Kit (Ambion, Austin, TX) RNA samples were treated with the Turbo DNA-free kit (Ambion) to minimize DNA contamination After DNase treatment, each sample was tested for residual genomic DNA by omitting the RT reaction Only samples that did not exhibit detectable DNA background were used in this work The reverse transcription step was performed as follows, using random hexamer primers and SuperScript III (Thermo Fisher, Springfield, NJ): 5 min at 25°C, 1 hour at 50°C, and 15 min at 70°C PCR amplification reactions were carried out in a final volume of 50 µL using 10 U of Platinum Taq DNA Polymerase (Invitrogen, Carlsbad, CA) PCR amplification conditions were as follows: 1 cycle of 1 min at 94°C, 45 s at 57°C, 1 min at 72°C, followed by 37 cycles of 30 s at 94°C, 45 s at 57°C, 1 min at 72°C, and a final cycle of 1 min at 94°C, 45 s at 57°C, 15 min 72°C Conditions for qPCR were as follows: 30 s at 95°C, 30 s at 58°C, 30 s at 72°C, carried out for 40 cycles qPCR was performed with the IQ SYBR Green Supermix dye kit (Bio-Rad, Hercules, CA), using a CFX96 Real-Time PCR System (Bio-Rad) Primers for the amplification of BC200 RNA were as follows:
BC200 forward primer: 5’ CCTGGGCAATATAGCGAGAC 3’
BC200 reverse primer: 5’ GGGTTGTTGCTTTGAGGGA 3’
The predicted amplification product corresponds to nt 91–188 of the 3’ region of BC200 RNA The 97-nt-length PCR product was sequenced and verified as corresponding to the above 3’ BC200 RNA segment.
As an internal standard for qtr.-PCR, we used the mRNA encoding acidic ribosomal protein (ARP, also known as ribosomal protein P0, NM_001002), using ARP primers as described17 ARP mRNA was used to normalize the amount of total RNA in each qtr.-PCR reaction18 As an independent internal control, we used primers specific for Glyceralde-hyde 3-phosphate dehydrogenase (GAPDH, SABioscience, Frederick, MD), as described by the manufacturer qtr.-PCR data were analyzed using the cycle threshold (Ct) method19 For each sample, the Ct for BC200 RNA was normalized to the Ct for ARP mRNA, resulting in a ΔCt that reflects the relative level of BC200 RNA in that sample For a blood sample from a patient with breast cancer, the ΔCt was normalized to the mean ΔCt obtained with samples from healthy subjects, resulting in a ΔΔCt for the patient sample 2−ΔΔCt was then calculated as a measure of the fold increase of BC200 RNA in the patient’s blood sample, relative to control blood samples from healthy subjects.
Blood fractionation
To prepare blood fractions, in particular the mononu-clear leukocyte (MNL) fraction, which includes CTCs, we performed cell fractionation using the Ficoll-Paque procedure (Histopaque 1077; Sigma-Aldrich, St Louis, MO), as described20 In brief, blood samples were diluted with phosphate buffered saline (PBS) (without Ca2+ or Mg2+) and were layered on 3 mL of a Ficoll gradient After centrifugation at 300×g for 10 min, MNL, RBC/PML (red blood cell/polymorphonuclear leukocyte fraction), and plasma fractions were collected, washed with PBS, and RNA-extracted using Ribopure.
Limiting dilution experiments with MCF-7 cells
MCF-7 cells (ATCC, Manassas, VA) were used as a breast cancer cell system For limiting dilution experiments, we used cell lysates to avoid random variations in the number of cells included in the assay Lysates of known numbers of MCF-7 cells were prepared using lysis buffer (10 mM glycine, 1% Triton X-100) A given amount of MCF-7 cell lysate, representing the desired number of cells, was then added to total RNA isolated from 104 Baby Hamster Kidney (BHK) cells or to 10 ng of total RNA isolated from blood of healthy subjects BC200 RNA levels in MCF-7 cells were established against the same amount of total RNA from the blood of healthy subjects that was used in figure 1 BHK cells were obtained from ATCC Total RNA was prepared using TRIzol (Invitrogen) Samples were DNase-digested and reverse transcribed as described above.
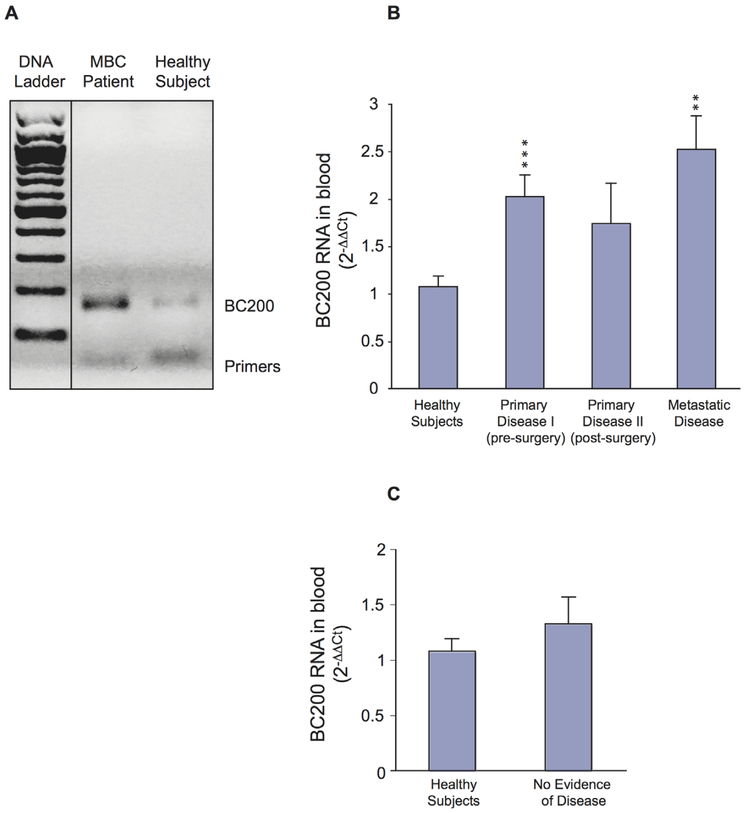
Figure 1.
BC200 RNA blood levels in patients with breast cancer (A) High levels of BC200 RNA are detected in peripheral blood from a patient with metastatic breast cancer (MBC) Total RNA was isolated from whole blood The RT-PCR product was resolved on a 12% agarose gel (bands at the bottom of the gel represent primers used in the PCR reaction) Medical history: The patient was diagnosed with breast cancer 2 years prior to BC200 RNA analysis She underwent surgery and adjuvant therapy, including six cycles of doxorubicin and cyclophosphamide followed by anastrozole (Arimidex) Two years later, she was diagnosed with bone metastases and was scheduledto undergo a new treatment cycle with paclitaxel (Taxol) and trastuzumab (Herceptin) Blood for BC200 RNA analysis was drawn after diagnosis of recurrent disease but before Taxol/Herceptin treatment It appears that in this case, that is, prior to the initiation of adjuvant therapy, BC200 RNA blood levels are higher than average BC200 RNA blood levels in metastatic disease (B) Bar diagram shows qRTPCR analysis of BC200 RNA expression levels in whole blood from healthy subjects and from three groups of patients with breast cancer (presurgery, postsurgery, metastatic disease) BC200 RNA amplification data were normalized to ARP mRNA17,18 Data are shown in the format mean±SEM, with each data point representing three experiments for each blood sample analyzed BC200 RNA blood levels are shown as fold increase in comparison with the control group (healthy subjects), calculated as 2−ΔΔCt (Materials and methods section)19 Statistical analysis: non-parametric Kruskal-Wallis analysis (p=0.005) followed by non-parametric Mann-Whitney U-test; **p=0.003, ***p=0.001 (C) BC200 RNA blood levels in former patients with breast cancer with no evidence of recurrent or residual disease are similar to those in healthy subjects Statistical analysis: Kruskal-Wallis (p=0.668).
Expression of BC200 RNA in tumor cell lines
MCF-10A, MCF-7, BHK, and HeLa cells were collected and washed with PBS Total RNA was extracted with TRIzol (Invitrogen) Reverse transcription and qPCR amplification reactions were carried out using the same reagents, primers, and amplification conditions as described above qtr.-PCR results were analyzed using the cycle threshold (Ct) method (for details, see previous section).
Statistical methods
Statistical analysis was performed on 71 blood samples in five groups To analyze the significance of BC200 RNA blood levels among groups, we used non-parametric Kruskal-Wallis one-way analysis of variance by ranks The non-parametric Mann-Whitney U-test was used for comparisons of two groups Receiver operating characteristic (ROC) analysis was used to establish discriminative power of BC200 RNA as a diagnostic and prognostic indicator In ROC analysis, sensitivity is plotted against (1−specificity)21 The area under the curve (AUC) is an index of the discriminative diagnostic power of a given test, with an ideally performing test having an AUC of 10 Statistical analysis was performed using SPSS software.
RESULTS
BC200 RNA in blood of patients with breast cancer
We initially used RT-PCR to establish whether BC200 RNA is detectable in peripheral blood of patients with invasive breast cancer RT-PCR amplification revealed that BC200 RNA levels in blood from a patient with breast cancer with metastatic disease were substantially higher than those from a healthy subject (figure 1A).
Subsequently, we collected peripheral blood from a total of 71 subjects (Materials and methods section) BC200 RNA levels were established in blood samples from these subjects by qtr.-PCR The results are shown in diagrammatic form in figure 1B,C High levels of BC200 RNA were detected in blood samples from patients with breast cancer with active disease BC200 RNA blood levels were significantly elevated in patients with either primary or metastatic disease, in comparison with healthy subjects (figure 1B) Specifically, patients with breast cancer with primary disease who had not yet undergone tumorectomy exhibited a substantial increase of BC200 RNA levels (twofold in comparison with healthy subjects; figure 1B, group Primary Disease I) This increase was highly significant (***p=0.001, Mann-Whitney U-test), suggesting high discriminating power Patients with metastatic disease (figure 1B, group Metastatic Disease) exhibited an almost two-and-a-half-fold average increase of BC200 RNA blood levels, in comparison with healthy subjects (**p=0.003, Mann-Whitney U-test).
In postsurgery patients (figure 1B, group Primary Disease II), BC200 RNA blood levels had decreased and were not significantly different from BC200 RNA blood levels in healthy subjects This result is consistent with the notion that BC200 RNA in peripheral blood of patients with invasive breast cancer (ie, group Primary Disease I) is contributed by CTCs that have been released from the primary tumor, and that further release of cells would cease after tumor resection Similar observations have been reported after excision of primary malignancies other than breast cancer22 However, BC200 RNA blood levels of the post-surgery patient group were also not significantly different from levels of the presurgery patient group The data indicate that in postsurgery patients, BC200 RNA is detected in blood at levels intermediate between presurgery patients and healthy subjects In addition, the SEM in postsurgery group is relatively high, an indication of heterogeneity in BC200 RNA blood levels in this patient group It is possible that such heterogeneity is a reflection of varying treatment success, for example, a relatively high number of residual CTCs in patients with high blood levels of BC200 RNA.
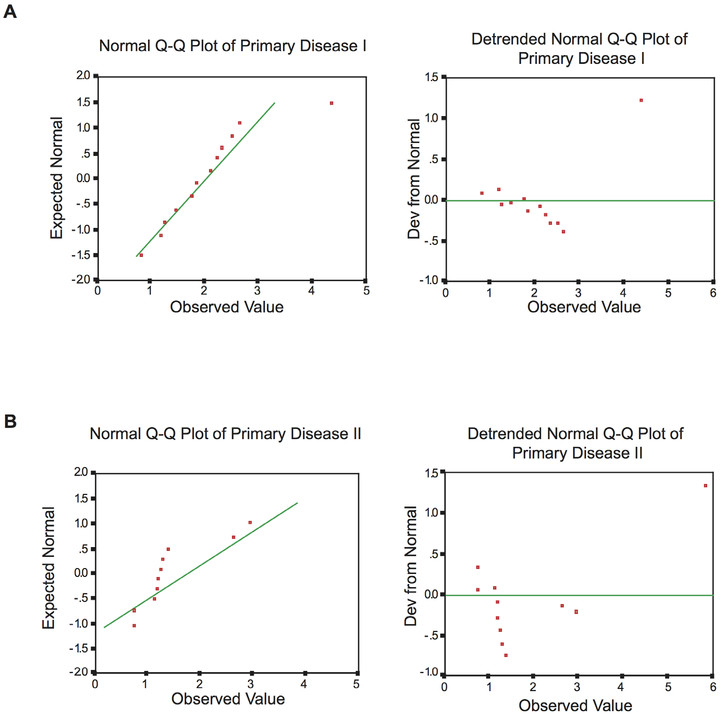
The above considerations were confirmed by normality analysis Normal Q-Q plots and detrended normal Q-Q plots show that BC200 RNA levels in the Primary Disease I group (figure 2A) conform to a normal distribution (shown in green) In contrast, BC200 RNA levels in the Primary Disease II group (postsurgery primary disease breast cancer patients) displayed a marked departure from normal distribution (figure 2B) Kolmogorov-Smirnov analysis of normality indicated a high significance of this deviation (p<0.0005), a result consistent with a heterogeneous population distribution The skewness for this patient group was 23, a likely reflection of the presence within this group of a subgroup of individuals with high residual BC200 RNA blood levels We note that some intragroup heterogeneity was observed for BC200 RNA blood levels in all patient groups, but for groups other than postsurgery primary disease patients, such deviation from normality did not reach statistical significance (Kolmogorov-Smirnov, p>0.2) The combined data indicate heterogeneity of BC200 RNA blood levels among post-surgery patients, a heterogeneity that likely reflects, within this group of patients, the presence of subgroups with high versus low residual BC200 RNA levels.
Figure 2.
Sample distribution analysis of Primary Disease I and Primary Disease II patient groups (normal and detrended normal Q-Q plots) Normality analysis of Primary Disease I (primary disease patients prior to tumorectomy) sample distribution shows no significant deviation from normal distribution (green line, A) Conversely, analysis of Primary Disease II group (postsurgery primary disease patients) sample distribution shows a significant departure from normality (green line, B) Kolmogorov-Smirnov, p<0.0005.
We also examined blood samples from former patients with breast cancer with no evidence of residual disease These patients had been undergoing surveillance for several years post-treatment and were considered disease free at the time of blood sample collection In blood samples from these patients, BC200 RNA levels were significantly lower than in samples from patients with active disease, primary or metastatic, and were not significantly different from BC200 RNA levels in samples from healthy subjects (figure 1C).
Patient tumor characteristics are listed in table 1 Characteristics are variable among patient groups, and such variability could potentially impact BC200 RNA expression levels It is noted that while previous work has established significant differences in BC200 RNA tissue levels between patients with invasive breast cancer and healthy subjects, no significant differences in such levels were observed between patients with breast cancer with different subtypes of invasive disease9.
Discriminative diagnostic power
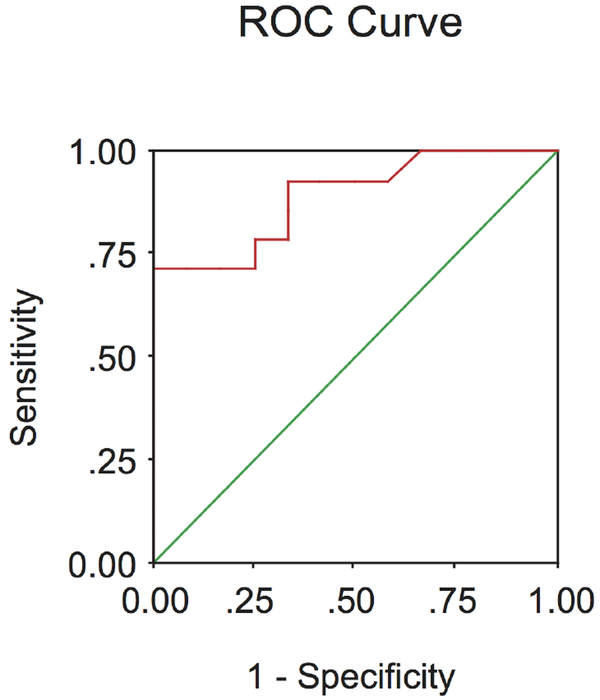
ROC analysis was performed to calculate the diagnostic efficacy of BC200 RNA blood levels as a molecular indicator of invasive breast cancer, that is, the ability of the test to correctly classify subjects with and without disease In ROC analysis, sensitivity is plotted against (1−specificity), and the AUC is used as an index of diagnostic power, varying from 05 (no diagnostic power) to 10 (perfect diagnostic power)21 Comparing the presurgery invasive breast cancer group (Primary Disease I) with the healthy subjects group, an AUC of 089 (with a 95% CI of 077 to 10) was obtained for BC200 RNA blood levels (figure 3) These data indicate high discriminative power of BC200 RNA in peripheral blood as a molecular indicator of invasive breast cancer They are in agreement with the observed lack of false positives in the healthy subject group (ie, BC200 RNA levels in all cases below mean±SEM of Primary Disease I group) Analogously, an AUC of 081 (95% CI 067 to 095) was obtained when comparing the metastatic disease group with the healthy subjects group, again indicating high discriminating diagnostic power (data not illustrated).
Figure 3.
ROC analysis of BC200 RNA expression in peripheral blood of patients with invasive breast cancer To ascertain the discriminative diagnostic power of the BC200 RNA blood test, receiver operating characteristic (ROC) analysis was performedby plotting sensitivity against (1−specificity), comparing BC200 RNA levels in blood from healthy subjects with BC200 RNA levels in presurgery patients with invasive breast cancer (group Primary Disease I) The obtained ROC curve shows an area under the curve of 089 (95% CI of 077 to 10), indicating a high discriminative efficacy of the BC200 RNA test.
It is noted that the above ROC analyses apply to the presurgery patients as BC200 RNA blood levels were not analyzed for detection of residual disease in patients following tumor resection.
BC200 RNA levels in cellular fractions from peripheral blood
The data obtained are consistent with the notion that cancer cells expressing BC200 RNA enter the circulation as CTCs If so, BC200 RNA would be expected to become detectable in the white blood cell fraction (ie, MNL) To test this hypothesis, we performed Ficoll fractionation of blood from patients with invasive breast cancer We separated three blood fractions: plasma, MNL (also called buffy coat), and RBC, a fraction containing also PML.
BC200 RNA levels in MNL fractions from patients with breast cancer were increased by more than twofold in comparison with MNL fractions from healthy donors (table 2).
Table 2.
Expression of BC200 RNA in the mononuclear cell fraction of peripheral blood of patients with breast cancer
| Blood fraction | BC200 level (2−⍰ΔCtt±SEM) | Student’s t-test |
|---|---|---|
| MNL | 2.24±.56 | p<0.05 |
| PML/RBC | nd | – |
| Plasma | nd | – |
Total RNA was isolated after cell fractionation, and qtr.-PCR was performed with three fractions: MNLs, containing CTCs, PMLs, and RBCs, and plasma BC200 RNA levels were elevated in the MNL cell fraction from patients with breast cancer, compared with the MNL cell fraction from healthy subjects In PML/RBC fractions, BC200 RNA was not reliably detectable (nd) by qtr.-PCR BC200 RNA was also not detected in plasma samples 2−ΔΔCt represents the fold increase of BC200 RNA levels in a patient’s blood sample, relative to blood samples from healthy subjects.
BC200 RNA, brain cytoplasmic 200 RNA; CTC, circulating tumor cell; MNL, mononuclear leukocyte; PML, polymorphonuclear leukocyte; RBC, red blood cell.
In PML/RBC fractions (containing mainly anucleate erythrocytes), BC200 RNA could not be reliably detected in samples from either patients with breast cancer or healthy subjects (table 2) BC200 RNA was also not detectable in plasma fractions from patients with breast cancer, a result to suggest that BC200 RNA is not released into the circulation in acellular form, that is, as ‘free RNA’ Thus, BC200 RNA is reliably detected in MNL fractions derived from peripheral blood of patients with invasive breast cancer These data suggest that BC200 RNA levels in peripheral blood are in fact reflecting the presence of CTCs that express the RNA.
Sensitivity of BC200 RNA detection in blood
To address the question of sensitivity (ie, the minimum number of cancer cells that we can reliably detect), we performed limiting dilution experiments using MCF-7 cells, a cell line derived from a human breast adenocarcinoma Total RNA from given numbers of MCF-7 cells was analyzed against a background of total RNA from 104 cultured BHK cells (non-primate BHK cells do not express human BC200 RNA)2.
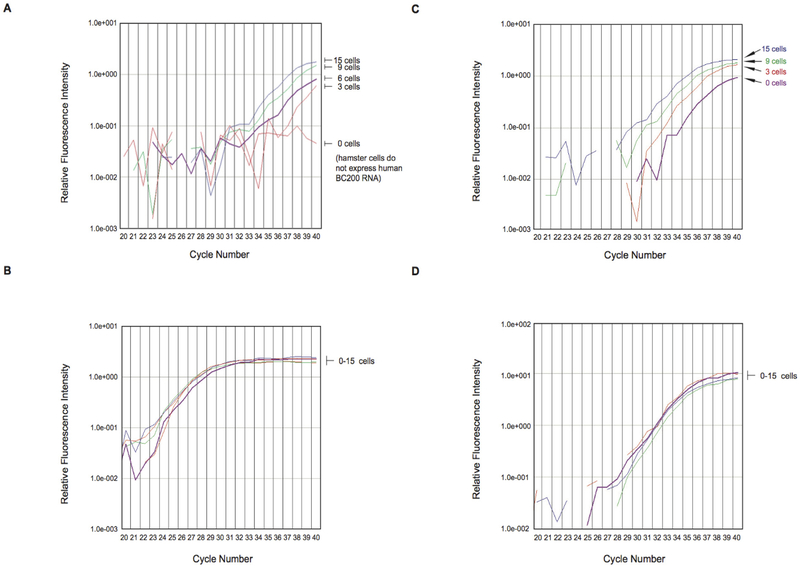
The qRT.-PCR amplification plot shows that the BC200 RNA approach can detect as few as three MCF-7 cells and, additionally, that it can detect small changes in the number of such cells (figure 4A,B) BC200 RNA is thus a highly sensitive indicator of invasive breast cancer cells in blood Conversely, the mRNA encoding the housekeeping protein ARP produced amplification plots that clustered together (figure 4A,B), that is, did not reflect the number of MCF-7 cells used This result was expected considering that the gene is constitutively expressed irrespective of cell type.
Figure 4.
(A and B) Limiting dilution experiments with MCF-7 breast cancer cell RNA in Baby Hamster Kidney (BHK) cell RNA (A) The panel shows qtr.-PCR amplification plots for BC200 RNA Total RNA isolated from MCF-7 cells, representing the respective number of cells indicated, was diluted with total RNA from 104 BHK cells and subjected to qtr.-PCR amplification The curves show that BC200 RNA amplification allowed detection of as few as three MCF-7 cells The zero MCF-7 cells sample failed to reach amplification threshold after 40 cycles, indicating that no BC200 RNA was detected (B) Control amplification experiments were performed with acidic ribosomal protein (ARP) mRNA In contrast to BC200 amplification, ARP amplification resulted in plots that were grouped together, indicating constant expression levels independent of MCF-7 cell numbers (C and D) Limiting dilution experiments: MCF-7 breast cancer cell RNA in RNA from blood of healthy subjects (C) The panel shows qtr.-PCR amplification plots for BC200 RNA Total RNA isolated from MCF-7 cells (from 3 to 15 cells) was diluted with total RNA (10 ng) from blood of healthy human subjects BC200 RNA qtr.-PCR amplification allowed detection of three MCF-7 cells (D) Control amplification experiments were performed with GAPDH mRNA As in the experiments with ARP mRNA, GAPDH amplification plots were grouped together.
Analogous limiting dilution experiments were performed in which RNA from MCF-7 breast cancer cells was analyzed against a background of RNA from whole blood of healthy subjects Using both ARP and GAPDH mRNAs as internal qtr.-PCR standards, we were able to detect BC200 RNA from three MCF-7 cells (figure 4C,D) BC200 RNA levels in samples containing three MCF-7 cell equivalents were significantly elevated, compared with samples containing 0 MCF-7 cell equivalents (Student’s t-test, p=0.0325) The strength of the qtr.-PCR BC200 signal again increased proportionally with the number of MCF-7 cell equivalents (figure 4C,D) The amplification signal in 0 MCF-7 cell samples is consistent with the baseline expression of BC200 RNA observed in blood from healthy subjects (see also figure 1A,B).
Expression of BC200 RNA in cancer cell lines
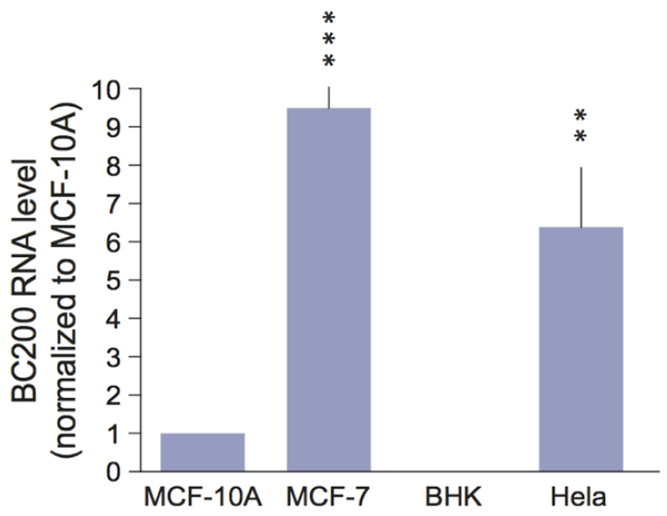
The above results with MCF-7 cells raise the question whether expression of BC200 RNA correlates with tumor cell invasiveness To address this question, we established expression levels in MCF-7 cells and HeLa cells in comparison with levels in MCF-10A cells, a cell line derived from non-tumorigenic breast epithelial cells BHK cells served as a negative control Results from these experiments (figure 5) indicate that MCF-7 cells and HeLa cells express significantly more BC200 RNA than MCF-10A cells Neither HeLa cells nor BHK cells are breast cancer cell lines The HeLa cell result is therefore in confirmation of earlier data indicating that BC200 RNA is expressed in primary cervical cancer cells, although at moderate levels8.
Figure 5.
Expression of BC200 RNA in tumor cell lines Expression levels in MCF-10A cells, MCF-7 cells, BHK cells, and HeLa cells were established by qRT-PCR BC200 RNA expression was significantly elevated in MCF-7 cells and HeLa cells, in comparison with MCF-10A cells (the latter normalized to 1) Quantitative analysis: one-way ANOVA, p=0.0003; Dunnett’s post hoc analysis, comparison of MCF-7 cells (p<0.001) and of HeLa cells (p<0.01) with MCF-10A cells BHK cells were usedas a negative control as expression of BC200 RNA is specific to primates2.
HeLa cells are considered highly invasive23 MCF-7 cells, in contrast, are normally poorly invasive; however, invasiveness and metastatic activity can be induced, for example, by VEGF-C expression24 Expression of BC200 RNA in these two cell types is thus reminiscent of expression in invasive breast cancer and in ductal carcinomas in situ (DCIS) BC200 RNAs is expressed at high levels (similar to levels in invasive ductal carcinomas) in high-grade comedo DCIS but only at background levels in non-high-grade DCIS9 These results were interpreted to indicate that expression of BC200 RNA is associated with invasive carcinomas and potentially invasive (‘preinvasive’) carcinomas in situ The data presented in figure 5 lend further support to this notion.
DISCUSSION
The significance of regulatory RNAs in eukaryotic cellular form and function is increasingly being recognized1,2 However, the role of such RNAs in tumorigenesis and tumor development, and their potential use as clinical tools, remains poorly understood and underexplored10 We have previously reported that human BC200 RNA is expressed at high levels in invasive breast cancer cells9 We now show that in patients with invasive breast cancer, BC200 RNA can be detected in peripheral blood with high specificity and sensitivity The data illustrate the potential of BC200 RNA detection in blood as a minimally invasive molecular indicator of breast malignancies.
We have developed qtr.-PCR methodology to discriminate BC200 RNA blood levels in patients with invasive breast cancer from those in healthy subjects and from those in patients with a past diagnosis of breast cancer but no evidence of recurrent disease Not included in the present work were patients with benign, non-invasive breast tumors such as fibroadenomas as tissue expression levels of BC200 RNA in such tumors have previously been shown not to be significantly different from those in healthy breast tissue9 The difference in blood levels of BC200 RNA between untreated invasive disease subjects and healthy subjects is highly significant (p=0.001), and ROC analysis confirmed high specificity and sensitivity of the technique (AUC=089) Mammography has a median sensitivity of 79%, ranging from 74% to 85%25,26
A BC200 RNA blood test would therefore be a valuable clinical complement to mammography in screening for breast cancer Of all malignancies examined, only adenocarcinomas of the breast, mucoepidermoid carcinomas of the parotid, and melanomas were found to express robust levels of BC200 RNA8 Thus, a screening result of elevated BC200 RNA blood levels would be an indication of the presence of one of these malignancies (which can easily be differentiated).
In patients with breast cancer who have undergone treatment (ie, resection and adjuvant therapy), average BC200 RNA blood levels were not significantly different from levels in healthy subjects, indicating that treatment has resulted in a significant reduction of BC200-expressing cells This result is consistent with previous observations that treatment can reduce CTC numbers in blood22 However, BC200 RNA blood levels in postsurgery primary disease breast cancer patients were also not significantly different from those in presurgery patients, and normality analysis revealed that some patients in the postsurgery group exhibited elevated BC200 RNA levels even after treatment We hypothesize that treatment efficacy may have been suboptimal in such cases and that, as a consequence, tumor cells will continue to be present in the circulation Such information may be valuable for individual breast cancer treatment decisions and outcome predictions CTCs derived from patients with breast cancer have been advanced as predictors of recurrent or metastatic disease27–29 We propose that high residual BC200 RNA blood levels after treatment can serve as an index of high relapse potential.
Tumor biomarker development has often been beset with difficulties30 Even prostate-specific antigen (PSA), widely used for prostate cancer screening, has been of debatable clinical use because of its rather poor sensitivity and specificity31 We suggest that one of the underlying reasons for the paucity of suitable molecular tumor markers is the limited stability and/or availability of tumor-relevant mRNAs or proteins This contrasts with the high stability and abundance of a regulatory RNA such as BC200 RNA, molecular features that result in superior reliability and consistency of detection Our work demonstrates the potential clinical use of BC200 RNA as a minimally invasive and highly discriminating molecular indicator of invasive breast malignancies.
Significance of this study
What is already known about this subject?
-
►
Breast cancer is a leading cause of death from malignancies in women.
-
►
Regulatory brain cytoplasmic 200 (BC200) RNA is expressed at high levels in breast cancer cells.
-
►
Circulating tumor cells (CTCs) have been advanced as predictors of recurrent or metastatic disease.
What are the new findings?
-
►
Quantitative reverse-transcription PCR methodology was used to establish the potential of a BC200 RNA-based blood test for invasive breast cancer.
-
►
In patients with invasive breast cancer, BC200 RNA can be detected in peripheral blood with high specificity and sensitivity
-
►
Detection of BC200 RNA is a CTC surrogate marker of high discriminating power.
How might these results change the focus of research or clinical practice?
-
►
BC200 RNA detection in blood isa minimally invasive and highly discriminating molecular indicator of invasive breast malignancies High residual BC200 RNA blood levels after treatment can serve as an indicator of high relapse potential.
Acknowledgements
Part of the work described here is covered by US patent 9,777,334 (HT, AI, GAS, 2017, Cancer blood test using BC200 RNA isolated from peripheral blood for diagnosis and treatment of breast cancer) We thank Dr Ellen Hsu for assistance with blood fractionation AI remembers Guglielmina Marucchi who died of breast cancer on August 25, 2008
Funding This work was supported, in part, by a Friends for an Earlier Breast Cancer Test Foundation grant (AI), a Susan G Komen for the Cure grant BCTR90106 (HT), and NIH grants DA026110 and NS046769 (HT)
Footnotes
Competing interests None declared.
Patient consent Obtained.
Ethics approval 267093–11.
Provenance and peer review Not commissioned; externally peer reviewed.
References
- 1.Iacoangeli A, Tiedge H Translational control at the synapse: role of RNA regulators. Trends Biochem Sci 2013;38:47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eom T, Muslimov IA, Iacoangeli A, et al. Dendritic targeting and regulatory RNA control of local neuronal translation In: Sossin W, ed Oxford handbook of neuronal protein synthesis New York: Oxford University Press, 2018. Published online at http://www.oxfordhandbookscom/;paper version in press [Google Scholar]
- 3.Wang H, Iacoangeli A, Lin D, et al. Dendritic BC1 RNA in translational control mechanisms J Cell Biol 2005;171:811–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin D, Pestova TV, Hellen CU, et al. Translational control by a small RNA: dendritic BC1 RNA targets the eukaryotic initiation factor 4A helicase mechanism Mol Cell Biol 2008;28:3008–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eom T, Berardi V, Zhong J, et al. Dual nature of translational control by regulatory BC RNAs. Mol Cell Biol 2011;31:4538–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eom T, Muslimov IA, Tsokas P, et al. Neuronal BC RNAs cooperate with eIF4B to mediate activity-dependent translational control J Cell Biol 2014;207:237–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin Y, Brosius J, Tiedge H Neuronal BC1 RNA: co-expression with growth-associated protein-43 messenger RNA Neuroscience 2001;103:465–79 [DOI] [PubMed] [Google Scholar]
- 8.Chen W, Böcker W, Brosius J, et al. Expression of neural BC200 RNA in human tumours J Pathol 1997;183:345–51. [DOI] [PubMed] [Google Scholar]
- 9.Iacoangeli A, Lin Y, Morley EJ, et al. BC200 RNA in invasive and preinvasive breast cancer Carcinogenesis 2004;25:2125–33. [DOI] [PubMed] [Google Scholar]
- 10.Sosińska P, Mikuła-Pietrasik J, Książek K The double-edged sword of long non-coding RNA: the role of human brain-specific BC200 RNA in translational control, neurodegenerative diseases, and cancer Mutat Res Rev Mutat Res 2015;766:58–67. [DOI] [PubMed] [Google Scholar]
- 11.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008 CA Cancer J Clin 2008;58:71–96 [DOI] [PubMed] [Google Scholar]
- 12.Huynh PT, Jarolimek AM, Daye S The false-negative mammogram Radiographics 1998;18:1137–54 [DOI] [PubMed] [Google Scholar]
- 13.Gøtzsche PC, Olsen O Is screening for breast cancer with mammography justifiable? Lancet 2000;355:129–34. [DOI] [PubMed] [Google Scholar]
- 14.Kriege M, Brekelmans CT, Boetes C, et al. Efficacy of MRI and mammography for breast-cancer screening in women with a familial or genetic predisposition N Engl J Med 2004;351:427–37. [DOI] [PubMed] [Google Scholar]
- 15.Kriege M, Brekelmans CT, Obdeijn IM, et al. Factors affecting sensitivity and specificity of screening mammography and MRI in women with an inherited risk for breast cancer Breast Cancer Res Treat 2006;100:109–19. [DOI] [PubMed] [Google Scholar]
- 16.Tiedge H, Chen W, Brosius J Primary structure, neural-specific expression, and dendritic location of human BC200 RNA J Neurosci 1993;13:2382–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iqbal J, Dai K, Seimon T, et al. IRE1beta inhibits chylomicron production by selectively degrading MTP mRNA Cell Metab 2008;7:445–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bièche I, Lerebours F, Tozlu S, et al. Molecular profiling of inflammatory breast cancer: identification of a poor-prognosis gene expression signature Clin Cancer Res 2004;10:6789–95. [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD Analysis of relative gene expression data using real-time quantitative PCR and the 2 Delta Delta C(T)) Method Methods 2001;25:402–8. [DOI] [PubMed] [Google Scholar]
- 20.Fraczek M, Sanocka D, Kamieniczna M, et al. Proinflammatory cytokines as an intermediate factor enhancing lipid sperm membrane peroxidation in in vitro conditions J Androl 2008;29:85–92. [DOI] [PubMed] [Google Scholar]
- 21.Hanley JA, McNeil BJ The meaning and use of the area under a receiver operating characteristic (ROC) curve Radiology 1982;143:29–36. [DOI] [PubMed] [Google Scholar]
- 22.Patel H, Le Marer N, Wharton RQ, et al. Clearance of circulating tumor cells after excision of primary colorectal cancer Ann Surg 2002;235:226–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramer R, Hinz B Inhibition of cancer cell invasion by cannabinoids via increased expression of tissue inhibitor of matrix metalloproteinases-1 J Natl Cancer Inst 2008;100:59–69. [DOI] [PubMed] [Google Scholar]
- 24.Comşa Ş, Cîmpean AM, Raica M The story of MCF-7 breast cancer cell line: 40 years of experience in research Anticancer Res 2015;35:3147–54. [PubMed] [Google Scholar]
- 25.Miglioretti DL, Smith-Bindman R, Abraham L, et al. Radiologist characteristics associated with interpretive performance of diagnostic mammography J Natl Cancer Inst 2007;99:1854–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taplin S, Abraham L, Barlow WE, et al. Mammography facility characteristics associated with interpretive accuracy of screening mammography J Natl Cancer Inst 2008;100:876–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boral D, Vishnoi M, Liu HN, et al. Molecular characterization of breast cancer CTCs associated with brain metastasis Nat Commun 2017;8:196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cabel L, Proudhon C, Gortais H, et al. Circulating tumor cells: clinical validity and utility Int J Clin Oncol 2017;22:421–30. [DOI] [PubMed] [Google Scholar]
- 29.Cherdyntseva NV, Litviakov NV, Denisov EV, et al. Circulating tumor cells in breast cancer: functional heterogeneity, pathogenetic and clinical aspects Exp Oncol 2017;39:2–11. [PubMed] [Google Scholar]
- 30.McShane LM, Altman DG, Sauerbrei W, et al. Reporting recommendationsfor tumor marker prognostic studies (REMARK) J Natl Cancer Inst 2005;97:1180–4. [DOI] [PubMed] [Google Scholar]
- 31.Garnick MB, Fair WR Combating prostate cancer Sci Am 1998;279:74–83. [DOI] [PubMed] [Google Scholar]