Abstract
GATA4 is a transcription factor that is responsible for tissue-specific gene regulation in many tissues, and more recent studies showed that it is necessary for osteoblast differentiation. Previously, we showed that in vivo deletion of Gata4 using Cre-recombinase under the control of the Col1a1 2.3 kb promoter, showed significantly reduced trabecular bone properties. To understand the role of GATA4 in more differentiated cells, GATA4fl/fl mice were crossed with mice expressing Cre-recombinase under the control of the osteocalcin promoter. MicroCT analysis of trabecular bone properties of the femur and tibia from 14-week-old female osteocalcin-Cre/ GATA4fl/fl (OCN-cKO) mice showed a significant reduction in percentage bone volume, a decrease in trabecular number and an increase in trabecular spacing. In vivo, histomorphometric analysis revealed a decrease in the number of osteoblasts and an increase in the number of osteoclasts in the tibiae of OCN-cKO mice. In vivo and in vitro systems correlated a decrease in Gata4 mRNA with increased RANKL gene expression. To determine if RANKL is a direct target of GATA4, chromatin immunoprecipitation (ChIP)-sequencing was performed, and it demonstrated that GATA4 is recruited to seven enhancers near RANKL. Furthermore, when Gata4 is knocked down, the chromatin at the RANKL region is further opened, as detected by a reduction in histone 3 lysine 27 trimethylation (H3K27me3) and an increase in histone 3 lysine 4 dimethylation (H3K4me2) in the RANKL locus. In vitro, TRAP staining of cells from bone marrow cultures from Gata4 knockout cells show that the increased levels of RANKL are sufficient for osteoclast formation. Together, the data suggest that GATA4 directly represses RANKL expression via seven cis-regulatory regions and plays an important role in maintaining proper bone development and osteoclast formation.
Keywords: GATA4, Bone, Osteoblast, Osteoclast, RANKL
1. Introduction
Skeletal remodeling is due to the coupled effects of both osteoblasts and osteoclasts. At homeostasis bone resorption by osteoclasts is followed by an equal amount of bone formation by osteoblasts. Osteoclast differentiation is activated by RANKL (Receptor Activator of NFκB Ligand, TNFSF11) binding to the RANK receptor (TNFRSF11a), and subsequent activation of the NFκB pathway and osteoclast gene transcription. RANKL is expressed in osteoblasts, T-cells and B-cells to initiate osteoclastogenesis [1]. Cytokines, vitamin D and PTH have each been shown to increase RANKL expression [2]. RANKL is also regulated post-transcriptionally by the decoy receptor osteoprotegerin (OPG, TNFRSF11b), which binds to RANKL protein and inhibits RANK activation. RANKL can also be cleaved from the cell membrane by MMPs, leading to a less potent activator [3]. Increased RANKL can lead to bone loss, and the RANKL neutralizing antibody Denosumab is clinically approved to treat osteoporosis [4]. Thus, understanding RANKL regulation is key to preventing and treating osteoporosis.
Several transcription factors, including CREB [5], ATF4 [6] and NFATC1 [7], have been shown to bind to the promoter of RANKL and regulate its expression. More recently, ChIP-sequencing has revealed long distance enhancers regulating RANKL expression. The vitamin D receptor (VDR) binds to osteoblast- and T-cell-specific enhancers that are up to 150 kilobases (kb) upstream of the RANKL promoter [8]. However, RANKL repression is not well understood.
GATA4 is a key transcription factor that regulates RUNX2, BMP, TGFβ, Fas Ligand, and other key proteins in osteoblast differentiation and function [9–11]. GATA4 is necessary for proper bone development and estrogen signaling in bone [9,10]. Global knockout of Gata4 in the mouse results in early embryonic lethality [12,13]. Conditional Gata4 knockout mice with Cre-recombinase expression driven by the 2.3-kb promoter of Col1a1 showed reduced trabecular bone qualities in both young and adult mice [9,11]. To determine if GATA4 plays a role in later stages of osteoblastogenesis, osteocalcin-Cre mice [14] were crossed with GATA4-floxed mice. We show here that these mice had reduced percentage bone volume, a decrease in the trabecular number and an increase in trabecular separation. These changes were due in part to increased expression of RANKL and an increase in osteoclasto-genesis in the absence of Gata4.
2. Materials and methods
2.1. Animals
All animal experiments comply with the ARRIVE guidelines. Animal experiments were approved by the Institutional Animal Care and Use Committee at the University of Tennessee Health Science Center. Animals were maintained in a specific pathogen free environment at 20–26 °C with a relative humidity of 30–70% and a 12h light/dark cycle. Commercial rodent chow (LM-485, Teklad, Madison, WI) and drinking water were available ad libitum. GATA4fl/fl mice were purchased from Jackson Laboratory (Gata4tm1.1Sad/J, JAX stock #008194). The floxed homozygous mice (GATA4fl/fl) were backcrossed for 10 generations to FVB background and then to C57BL/6 for 10 generations. The C57BL/6 GATA4fl/fl mice were crossed to B6N.FVB-Tg (BGLAP-cre)1Clem/J (C57BL/6NJ) from Jackson Labs (JAX stock #019509 [14]) to produce GATA4fl/fl/Cre+ (OCN cKO) and GATA4fl/fl/ Cre− littermate control mice in the C57BL/6NJ background. No gross abnormalities were observed at birth in GATA4 cKO animals. 14-week-old adult (10 wildtype (WT) and 10 knockouts (cKO)) were euthanized and the right hind limbs cleaned and stored in phosphate-buffered saline (PBS) at −20 °C for μCT until measurements were made. The left femur was transferred to formalin and the left tibia was transferred to ethanol and stored at 4 °C for histology.
GATA4-Flag-biotin mice (Flag-bio, Gt(ROSA)26Sortm1(birA)Mejr Gata4tm3.1Wtp/J) [15] were obtained from Jackson Labs.
2.2. Micro-computed tomography (μCT)
Tibiae and femurs from 14-week-old mice were stored in PBS and then scanned using a Scanco μCT 40 (Brüttisellen, Switzerland) set at 55 kVp/109 μA. The entire femur and tibia were scanned in the sample holder with 12.3-mm diameter at medium resolution. These tubes were filled with PBS and the top of the tube was covered with Parafilm to prevent dehydration. A scout view of each bone was taken and the sample height was adjusted to ensure the bone was within the field of view. The images were obtained at 6 μm resolution. The integration time and Gaussian filter used for these samples was 300 ms and 1, respectively. Solid three-dimensional models were reconstructed from these images automatically after completion of each cone-beam image stack with the built-in software. The trabecular parameters were calculated on 200 slices of trabecular bone from a region 50 slices below the growth plate as described in [16] with a threshold of 250 and quoted using American Society of Bone and Mineral Research nomenclature. μCT was also used to measure the cortical properties of each diaphysis on 100 slices of cortical bone from the region closest to the center of the shaft of the bone.
2.3. Bone histomorphometric analysis and serum markers of bone resorption/formation
Histomorphometric analyses were performed on the left tibiae. The bones were embedded in methylmethacrylate. 5 μm sections of the diaphysis of the proximal tibia, were mounted to glass slides with Haupt’s solution, and stained with toluidine blue and osteoclasts were visualized by TRAP staining. Histomorphometric analysis was performed using a Nikon Eclipse E400 microscope equipped with fluorescent light, an Olympus DP73 camera and OsteoMeasure7 v4.3.0.0 software (OsteoMetrics, Inc.). The cancellous bone of the secondary spongiosa was analyzed for enumeration of osteoblast number (N.Ob), osteoclast number (N.Oc), and osteoclast surface (Oc.S) on the TRAP stained slides. For the measurement of the bone formation rate, the mice were injected i.p. with calcein (Sigma catalog #C0875) 10 and 3 days before sacrifice and unstained sections were used to record fluorescent double label analysis to determine mineral apposition rate (MAR) and bone formation rate (BFR). The results are reported using the abbreviations recommended by the American Society for Bone and Mineral Research Histomorphometric Nomenclature Committee [17].
Rat/mouse N-terminal propeptide of type I procollagen (P1NP) Enzyme Immunoassay (EIA) Kit and RatLaps (C-terminal telopeptides of type I collagen, CTX-I) EIA were purchased from Immunodiagnostic Systems Inc., and the assays were performed using mouse serum, as per the manufacturer’s protocol.
2.4. TRAP staining and RANKL immunohistochemistry (IHC)
Paraffin-embedded sections of the left femurs were hydrated and then analyzed for TRAP (TRACP & ALP Double-Stain Kit, Takara) and the nuclei were stained with methyl green. For immunohistochemistry, paraffin-embedded sections of the left femurs were hydrated and then probed with an antibody to RANKL (Abcam, Cambridge, MA, catalog #ab45039). Nuclei were co-stained with hematoxylin.
2.5. Isolation and culture of osteoblasts
Mouse calvarial osteoblasts were isolated from 2-day-old CD1 or Flag-bio mice by sequential collagenase digestion [18]. Experiments were performed in α-MEM media with 10% fetal bovine serum (Omega Scientific, Tarzana, CA, USA). The cells were incubated for 40min in α-MEM-1.0 mg/mL collagenase P-1.25% trypsin at 37 °C. The cells were then washed in α-MEM and then incubated in α-MEM-1.0 mg/mL collagenase P-1.25% trypsin for 1 h at 37 °C. Collagenase digestion was stopped by addition of complete α-MEM media containing 10% FBS. The cells from second digest were obtained and allowed to proliferate in α-MEM media containing 10% FBS. Where indicated, the cells were treated for 14 days with differentiation media (α-MEM media supplemented with 50 μg/mL L-ascorbic acid 2-phosphate and 5 mM β-glycerophosphate) and the media was changed after every 3 days.
Mouse bone marrow cells were isolated from long bones of 3-month-old WT and GATA4 cKO mice. Each hind limb was cleaned off by scraping the diaphysis and pulling the tissue towards the end of the bones. The ends of the femur and tibia just below the end of marrow cavity were chopped with a sharp rongeur. The bone marrow cells were then flushed and seeded in a 6-well plate at a seeding density of 2 × 106 cells/well and left undisturbed for 5 days. Bone marrow cells were treated for 14 days with differentiation media (α-MEM media supplemented with 50 μg/mL L-ascorbic acid 2-phosphate and 5 mM β-glycerophosphate) and the media was changed after every 3 days. Adenovirus expressing Cre-recombinase (catalog # 1700) (or GFP (as a control, catalog #1060) was purchased from Vector Biolabs (Malvern, PA) and used at an MOI of 10.
2.6. Short hairpin knockdown of Gata4
Lentivirus shC (a short hairpin that does not recognize any mammalian DNA) and shGATA4 were purchased from Sigma Aldrich (St. Louis, MO, USA). Two different shRNA from The RNAi Consortium (TRC) in pLKO vector were used to knockdown mouse Gata4 (TRCN0000095215: CCGGCCCAATCTCGATATGTTTGATCTCGAGATC AAACATATCGAGATTGGGTTTTTG and TRCN0000095217: CCGGCAT CTCCTGTCACTCAGACATCTCGAGATGTCTGAGTGACAGGAGATGTTT TTG). The calvarial cells were plated in six-well plates at a seeding density of 2 × 105 cells per well. After 24 h, the cells were infected with lentivirus at an MOI of 50 in α-MEM with 8 μg/mL polybrene per well.The plates were centrifuged at 1400 ×g at 30 °C for 45 min and left undisturbed for 24 h after which the cells were washed with PBS and mineralization media was added. The knockdown of Gata4 was con-firmed by qPCR.
2.7. RNA and qPCR
Total RNA was isolated from mouse cells differentiated in osteogenic media for 14 days using TRIzol (Invitrogen, Carlsbad, CA, USA). cDNA was prepared using Superscript III First Strand Synthesis Kit according to manufactures guideline and then quantified using TaqMan® Universal Master Mix II (Applied Biosystems; Foster City, CA, USA) or SYBR Green (Sigma-Aldrich. St. Louis, MO, USA). The qPCR cycling conditions for TaqMan were initial denaturation at 50 °C for 2 min; 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s; 60 °C for 1 min. The SYBR Green conditions used were 95 °C for 10 min; followed by 40 cycles of 95 °C for 15 s; 60 °C for 1 min. Gata4 was quantified by TaqMan using primers from ThermoScientific (Assay Mm00484689_m1 and normalized to β-actin (Mm00607939_s1)). The oligonucleotide specific primers for SYBR Green assays are listed in Supplemental Tables 1 and 2. For analysis of the data, the cDNA values were normalized to β-actin.
2.8. Chromatin immunoprecipitation
For chromatin immunoprecipitation, the calvarial cells were plated at a seeding density of 2 × 105 cells and left undisturbed for 24 h prior to silencing using lentivirus directed towards shC or shGATA4 as described above. 24 h after silencing, the cells were washed with PBS and complete MEM media was added to each well and left for additional three days, after which ChIP was performed using truChIP™ Ultra Low Cell Chromatin Shearing Kit (Covaris, Inc., Woburn, Massachusetts). The histone 3 lysine 4 dimethylation (H3K4me2, catalog #07–030) and histone 3 lysine 27 trimethylation (H3k27me3, catalog #17–622) antibodies were purchased from Millipore (Burlington, Massachusetts). The antibodies were incubated with protein A magnetic beads (Dynabeads, ThermoFisher Scientific catalog # 10002D) and then added to chromatin.
The calvarial cells from 2-day-old Flag-bio pups were grown in α-MEM media with 10% fetal bovine serum. Once confluent, the cells were fixed with 16% formaldehyde for 5 min, and the excess formaldehyde was quenched with 10× glycine for an additional 5 min. 100 μL of streptavidin-coupled Dynabeads (ThermoFisher Scientific, catalog # 65–601) were used for each ChIP reaction. The sheared chromatin was pre-cleared for one hour with 100 μL of protein A magnetic beads (Dynabeads, ThermoFisher Scientific catalog # 10002D). The precleared chromatin was then transferred to the streptavidin beads and incubated overnight at 4 °C. After the incubation, the samples were sequentially washed with cold SDS wash buffer, high salt buffer, LiCl buffer, and TE buffer [19]. To reverse the cross-links the IP beads and the input were resuspended in elution buffer and placed on water bath at 70 °C overnight followed but incubation on heat block for 2 h at 55 °C and an additional 1 h at 37 °C with proteinase K and RNAse, after which the DNA was purified using QIAquick PCR purification kit (Qiagen, Valencia, CA, USA) and was validated by qPCR with primers listed in Supplemental Table 1. Each ChIP was performed in biological triplicates. ChIP DNA was sequenced on an Illumina NextSeq 500. The sequences were aligned to the mouse genome (mm10) using BWA [20] and peaks were called using HOMER [21] with a P value of 0.001.
2.9. Mouse osteoclast generation
The femur and tibia from WT and OCN cKO mice were excised and osteoclasts were cultured as previously described [22]. The cells were plated at a seeding density of 1 × 105 cells/well in a 96 well plate in osteoblast differentiation media and M-CSF (Sigma), with or without RANKL (Sigma). The media was changed every three days and the cells were fixed using the TRAP staining kit (Takara) as per the manufacturer’s protocol on day 8.
2.10. RANKL-promoter luciferase
U2OS osteosarcoma cells were plated in 24-well tissue culture dishes and transfected with RANKL-promoter-luciferase (a kind gift from Charles O’Brien [23], pcDNA3-GATA4 (a kind gift from Dr. Michael Parmacek [24]), and pRL-SV40 (Promega, Madison, WI, USA) as a transfection control. Each transfection was done in triplicate and the entire assay was repeated three times. 24 h after transfection, cells were assayed according to the manufacturer’s protocol (Promega, Dual-Luciferase Reporter Assay System). To obtain “relative luciferase” all values were normalized as follows: the raw firefly luciferase value was divided by the internal Renilla luciferase value (pRL-SV40) and then divided by the vector control.
2.11. Statistical analysis
The data shown are the mean ± standard deviation. Statistical analyses were made using Sigma Plot (version 11.0 Systat Software Inc.). Data were analyzed using two-tailed unpaired Students t-test and a P value < 0.05 was considered statistically significant.
3. Results
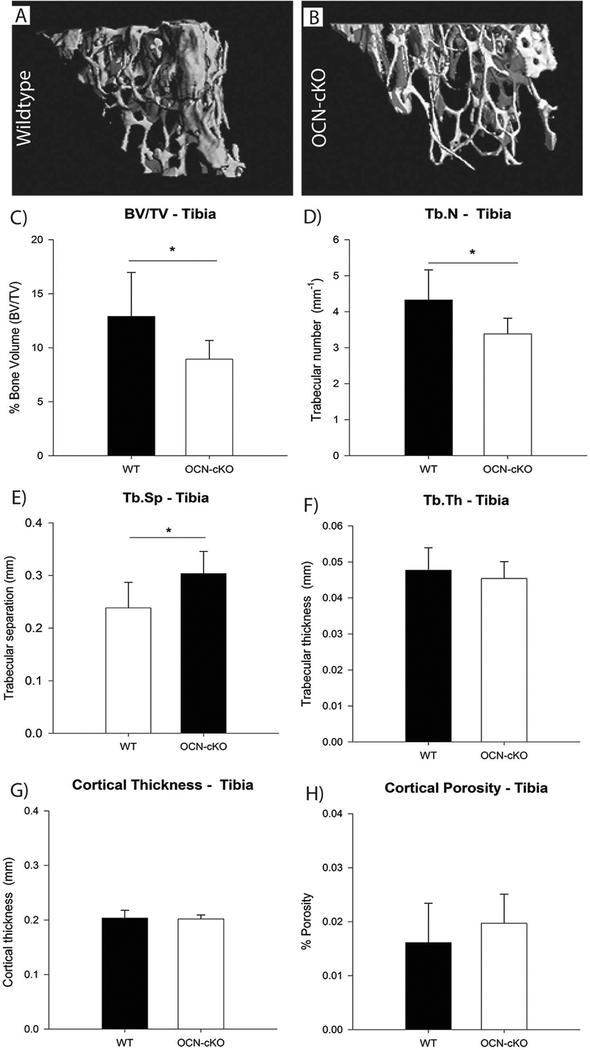
We have shown that GATA4 regulates genes involved in early osteoblastogenesis, including RUNX2 and TGFβ [9,11]. To determine if there is also a role for GATA4 in adult bone osteoblast differentiation, GATA4 floxed mice were crossed to mice expressing Cre-recombinase controlled by the osteocalcin promoter [14]. At 14 weeks of age the bones of these mice, here on called OCN-cKO, were analyzed by μCT, and representative images are shown (Fig. 1A–B). The percentage bone volume [bone volume (BV)/ tissue volume (TV)] was significantly reduced in tibiae of female OCN-cKO mice compared with wildtype (WT) littermates (Fig. 1C). In addition, there was a significant decrease in the trabecular number and a significant increase in trabecular separation (Fig. 1D–E), while no change was seen in trabecular thickness (Fig. 1F). Furthermore, no significant changes in cortical bone thickness nor porosity were detected between OCN-cKO mice and their littermate controls (Fig. 1G–H).
Fig. 1.
Trabecular bone volume is decreased in GATA4 OCN-cKO mice. (A, B) μCT reconstruction of trabecular bone in the distal tibia metaphysis of 14-week-old female mice. (C) Mean percentage bone volume (BV/TV), (D) trabecular number (Tb.N), (E) trabecular separation (Tb.Sp), (F) trabecular thickness (Tb.Th), (G) cortical thickness and (H) cortical porosity of WT and OCN-cKO mice. Black bars indicate wildtype (WT) mice; white bars indicate GATA4 OCN-cKO mice. Data are mean ± standard deviation from 12 WT and 12 cKO mice. Student’s t-test: *P < 0.05; **P < 0.01 compared with WT.
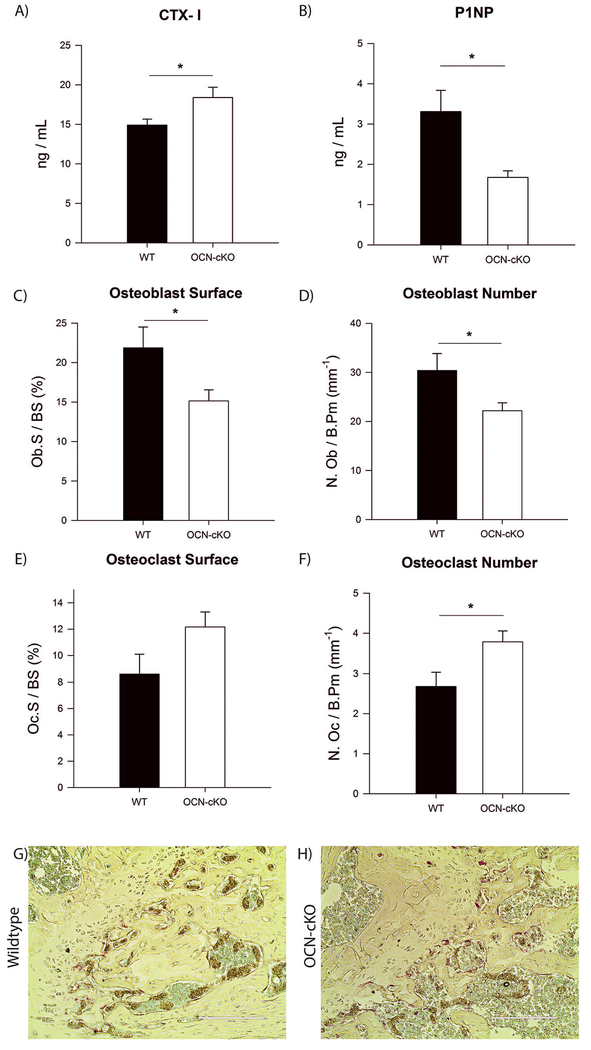
The serum of WT and OCN-cKO mice were analyzed for the bone resorption marker C-terminal telopeptides of type I collagen (CTX-I) and the bone formation marker N-terminal propeptide of type I pro-collagen (P1NP) by enzyme immunoassay (EIA). OCN-cKO mice had an increase in CTX-1 and a decrease in P1NP (Fig. 2A–B). The bones of the WT and OCN-cKO mice were further analyzed by histomorphometric analysis. OCN-cKO mice had a decrease in the osteoblast surface (Fig. 2C) and osteoblast number (Fig. 2D). OCN-cKO mice had a slight increase in osteoclast surface that was not statistically significant (Fig. 2E), but did have a significant increase in the osteoclast number (Fig. 2F). To directly visualize the increase in osteoclast number, the femurs of the OCN-cKO mice were analyzed by TRAP staining for the number of osteoclasts compared to Cre-negative littermates. The trabecular bone from OCN-cKO mice contained 3–4 times as many TRAP-positive cells as those from wildtype mice (Fig. 2G–H). The trabecular bone from Col1a1-cKO mice also had more TRAP-positive cells than those from wildtype mice (Supplemental Fig. 2). Together, these data identify a role for GATA4 in mature osteoblasts to regulate both bone formation and resorption.
Fig. 2.
Bone formation is decreased and resorption is increased due to a reduced number of osteoblasts and an increased number of osteoclasts, respectively. (A) Serum from WT and OCN-cKO mice was assayed using RatLaps (C-terminal telopeptides of type I collagen, CTX-I) EIA. (B) Serum from WT and OCN-cKO mice was assayed using P1NP EIA. (C) Bone histomorphometric analysis was performed on WT and OCN-cKO tibiae. Osteoblast surface, (D) osteo-blast number, (E) osteoclast surface and (F) osteoclast number. (G–H) Paraffin-embedded WT and OCN-cKO femurs were stained for TRAP (pink) and counter-stained with methyl green to identify nuclei. Student’s t-test: *P < 0.05. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
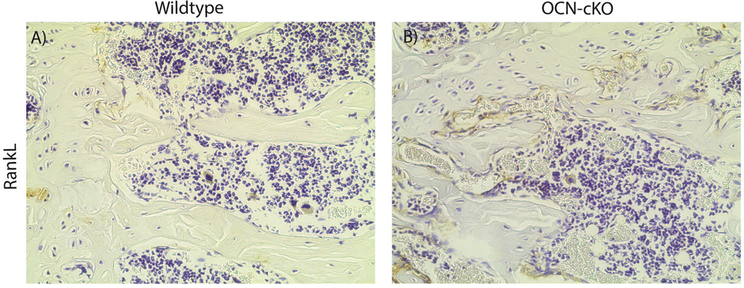
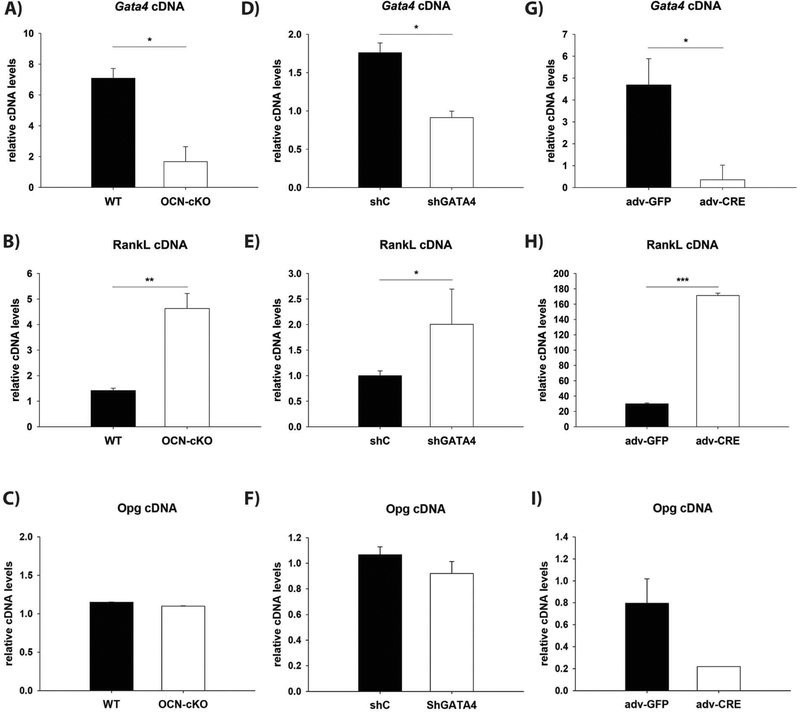
RANKL is a necessary activator of osteoclastogenesis [25]. Therefore, RANKL protein was analyzed in the femurs of the OCN-cKO mice by immunohistochemistry. RANKL was detected at an enhanced level in trabecular osteoblasts of the knockout mice (Fig. 3A and B). The expression of RANKL was subsequently examined in three parallel systems with reduced GATA4 expression (Fig. 4). First, bone marrow was obtained from OCN-cKO mice and Cre-negative littermates and differentiated for two weeks to osteoblasts, in vitro, with β-glycerophosphate and ascorbic acid (Fig. 4A). The second system utilized calvarial osteoblasts that had GATA4 silenced with lentiviral shRNA or a control shRNA (Fig. 4D). The calvarial osteoblasts were differentiated for two weeks in vitro. The third used bone marrow from GATA4fl/fl mice and Cre-recombinase was added using adenovirus, in vitro (Fig. 4G). In all three systems, a reduction in GATA4 (Fig. 4A, D and G) correlated with an increase in RANKL mRNA (Fig. 4B, E and H). The expression of the RANKL decoy Osteoprotegerin (OPG) did not change in the osteoblasts from OCN-cKO mice nor in the calvarial osteoblasts that had GATA4 silenced with lentiviral shRNA (Fig. 4C, F). There was, however, a decrease in OPG in the adenovirus system (Fig. 4I). As previously reported, GATA4 regulates also RUNX2, BMP4 and BMP6 (Supplemental Fig. 3). The results strongly argue that GATA4 protein expression mediates silencing of RANKL mRNA and protein.
Fig. 3.
GATA4 OCN-cKO mice have an increased expression of RANKL. (A, B). Immunohistochemistry for RANKL in WT (A) and OCN-cKO (B) mice.
Fig. 4.
GATA4 expression is inversely correlated with RANKL mRNA. (A, B, C) Bone marrow from WT and OCN-cKO mice was differentiated for two weeks with β-glycerophosphate and ascorbic acid and then mRNA was obtained. (D, E, F) Calvarial osteoblasts were isolated and the silenced with lentivirus to Gata4 (shG4) or control (shC) lentivirus. The cells were differentiated for two weeks with β-glycerophosphate and ascorbic acid, in vitro. (G, H, I) GATA4fl/fl bone marrow treated with Adenovirus GFP (adv-GFP) or Adenovirus Cre recombinase (Adv-CRE). qPCR was performed with primers to Gata4 (A, D, G), RANKL (B, E, H) and osteoprotegerin (OPG: C, F, I). qPCR data was normalized to β-actin (Actb) mRNA. Student’s t-test: *P < 0.05; **P < 0.01, ***P < 0.001 compared with WT.
The best way to determine if GATA4 directly regulates RANKL transcription by binding to the promoter and/or enhancers of the RANKL gene, is by conducting chromatin immunoprecipitation (ChIP) assays. Based on the fact that GATA4 rarely binds to promoters of genes, it is necessary to perform ChIP-sequencing (ChIP-seq) to identify GATA4 binding sites in the RANKL locus and surrounding regions in an unbiased manner.
Unfortunately, commercially available antibodies to GATA4 do not perform well in ChIP assays. To overcome this limitation, we used a mouse with GATA4 tagged with the Flag-epitope and a biotinylation site at the endogenous locus (herein called Flag-bio mice). This system was used previously to successfully demonstrate that GATA4 binds to the Runx2 promoter in osteoblasts [11]. Others have used the Flag-bio mice for ChIP-sequencing experiments in other organs [26–28]. Calvaria from these mice were obtained and ChIP was performed with streptavidin beads. As a control, wildtype mice were also used to demonstrate specificity. ChIP and ChIP-sequencing were performed on these samples. HOMER was used to identify peaks and, genome-wide, 5983 sites were identified for GATA4 at a P-value of 0.001. GATA4 was enriched in introns (52.1%), at “distal intergenic” regions (30.1%) and at the promoter of genes (7%) (Supplemental Fig. 4). The GATA4 motif WGATAA (Supplemental Fig. 4) was found to be significantly enriched in the ChIP peaks with a p-value of 2.4 × 10−6.
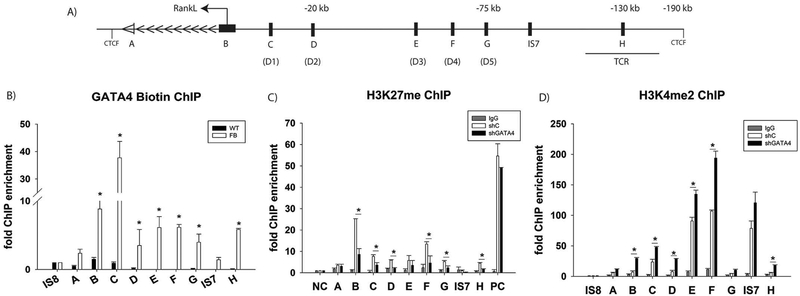
As detected by ChIP-sequencing, the RANKL locus contains eight GATA4 binding sites (Regions A–H (Fig. 5A)). Each of these sites was analyzed by the PATCH transcription factor motif software (http://gene-regualtion.com) for the GATA family motif. All eight regions contained one or more GATA motifs (Supplemental Fig. 5). Directed ChIP-qPCR demonstrated that GATA4 is recruited to seven of these sites (Fig. 5B). Region A was only marginally increased over background DNA. Region B is the promoter of RANKL and GATA4 is significantly enriched there (> 8-fold). Regions C–G have been shown to be vitamin D Receptor (VDR) binding sites and demonstrate significant GATA4 enrichment [29]. Region H is 150 kb upstream of the RANKL proximal promoter and is a novel potential RANKL enhancer. Two intergenic sequences (IS7 and IS8) were not shown to be enriched for GATA4 by directed ChIP or ChIP sequencing. The GATA4 binding sites near RANKL are all within previously published CTCF insulator regions [8]. Together, the data identify several potential GATA4 regulatory cis-elements within the RANKL locus in primary calvarial osteoblast cells.
Fig. 5.
GATA4 regulates RANKL at eight different loci. Schematic diagram of the RANKL genomic locus. The arrows indicate the direction of gene transcription. The GATA4 binding sites are denoted A-H. VDR sites are denoted D1-D5. The TCR (T cell control region) and CTCF binding sites are indicated [8]. The distances from the transcriptional start site are marked. (B) ChIP was performed using streptavidin-coated beads with wildtype or Flag-biotin-GATA4 calvarial osteoblasts. qPCR was performed with primers to the indicated regions. (C) Calvarial osteoblasts were transduced with either shC or shGATA4 lentivirus. ChIP was performed with an antibody to H3K27me3 or normal IgG. qPCR was performed to the indicated regions. Each PCR was normalized to input and represented as fold enrichment over Actb. Intergenic sequence 7 (IS7) is a negative control. An additional negative control (NC) and positive control (PC) region were used. N = 3, Student’s t-test: * = P < 0.05. (D) Calvarial osteoblasts were transduced with either shC or shGATA4 lentivirus. ChIP was performed with an antibody to H3K4me2 or normal IgG. qPCR was performed to the indicated regions. Each PCR was normalized to input and represented as fold enrichment over a negative intergenic sequence (IS8). Intergenic sequence 7 (IS7) is a negative control. N = 3, Student’s t-test: *P < 0.05; **P < 0.01, ***P < 0.001.
To further determine if GATA4 regulates RANKL by binding to the RANKL promoter, a luciferase assay was performed using a 704 base pair promoter region [23]. U2OS cells were transiently transfected with the RANKL-promoter luciferase reporter plasmid, along with pcDNA3-GATA4 or control pcDNA3 plasmid. GATA4 increased the luciferase expression by over 3-fold (Supplementary Fig. 6).
We, and others, have demonstrated that GATA4 regulates gene transcription by controlling the chromatin state [9,10,30]. We have shown that there is “open” chromatin, as marked by histone 3 lysine 4 dimethylation (H3K4me2), where GATA4 binds when promoting osteoblast specific genes (i.e. Runx2) and in its absence there is closed or “poised” chromatin, as marked by histone 3 lysine 27 trimethylation (H3K27me3) at the same enhancer region [11]. Therefore, we hypothesized that the RANKL locus would be “poised” for transcription, as marked H3K27me3, and that a decrease in GATA4 expression would change the histone modifications to an “open” conformation (H3K4me2). Publicly available H3K27me3 ChIP-seq datasets show the highest levels of H3K27me3 at the promoter of RANKL in non-bone tissues where RANKL is not expressed (Supplemental Fig. 7). In calvarial osteoblasts expressing GATA4, the highest level of H3K27me3 was also at the promoter of RANKL. A site on chromosome 2 that was found to be enriched for H3K27me3 in many publicly available datasets (positive control, PC) showed a high level of H3K27me3 that did not change after silencing GATA4 (Fig. 5C). Each region A–H near RANKL is significantly enriched for H3K27me3, when compared to the ACTB (β-actin) promoter, which is constitutively active, or to an immunoprecipitation with normal rabbit IgG (Fig. 5C). Silencing of Gata4 in calvarial osteoblasts from wildtype CD1 cells led to a reduction in H3K27me3 at regions B, C, D, F, G and H.
Next, we hypothesized that silencing of GATA4 would relieve repression of RANKL and lead to an “open” chromatin capable of transcriptional activation and that this would be accompanied by an increase of H3K4me2 in these regions that were identified by ChIP-Seq. To this end, we isolated calvaria from wildtype CD1 mice as previously described and conducted ChIP for H3K4me2 in the presence (shC) or absence of GATA4 expression (shGATA4, Fig. 5D). Regions A–D, G and H are not enriched for H3K4me2 in shC osteoblasts. Regions E, F and IS7 were enriched for H3Kme2 in shC cells. When GATA4 was silenced with lentiviral shRNA there is a dramatic increase in H3K4me2 at six of the RANKL enhancer sites (regions B–F and H), correlating with an increase in mRNA levels for RANKL, previously shown (Fig. 4). Together, the ChIP experiments demonstrate that GATA4 directly binds to the DNA and represses RANKL transcription by regulating the chromatin compaction.
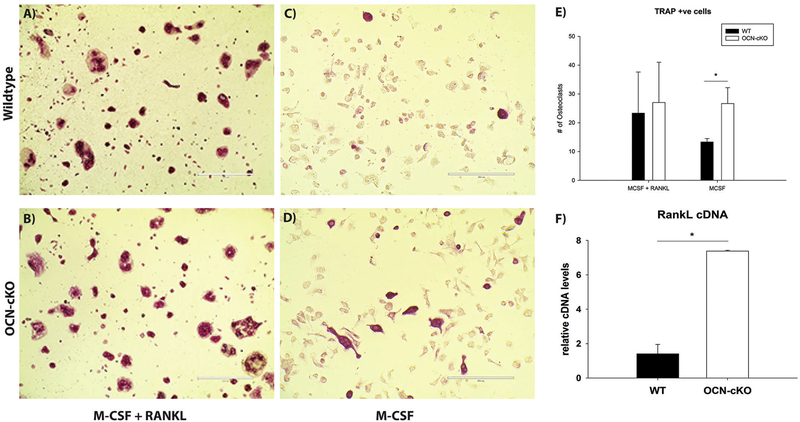
Bone marrow-derived monocytes/macrophages require both macrophage colony stimulating factor (M-CSF) and RANKL to differentiate into osteoclasts, in vitro. The osteoclast cells can then be identified by TRAP staining. To determine if the increase in RANKL mRNA levels are sufficient for enhanced osteoclastogenesis, bone marrow cells from WT and OCN-cKO mice were cultured in vitro. The cells were then differentiated for seven days with either M-CSF plus RANKL or M-CSF alone to generate osteoclasts (Fig. 6). TRAP-positive multinuclear cells could be observed after one week with M-CSF and RANKL, in both the WT and OCN-KO cells (Fig. 6A–B). The TRAP-positive cells were quantified with no difference noted in the M-CSF plus RANKL treatment (Fig. 6E), in either wildtype or OCN-cKO cells. In contrast, when M-CSF-alone was added to the culture media very few osteoclasts were formed in the wildtype cells, but over 7-fold more osteoclasts were differentiated in cells from the OCN-cKO mice (Fig. 6B and D). The cells from the OCN-cKO mice were confirmed to express higher mRNA levels of RANKL (Fig. 6F). This experiment confirms that GATA4 down-regulates RANKL in wildtype osteoblasts and that the level of RANKL in OCN-cKO osteoblasts is both functional and sufficient for osteoclastogenesis.
Fig. 6.
Loss of GATA4 leads to a sufficient level of RANKL for osteoclastogenesis. Bone marrow from wildtype or OCN-cKO was differentiated in the osteogenic differentiation media with M-CSF and RANKL (A, B) or M-CSF without RANKL (C, D). The number of TRAP-positive cells from five wells of A–D was enumerated (E). RANKL cDNA was quantified by qPCR (F) from cells treated identically to those in A–D. *P < 0.05.
4. Discussion
Here we investigated the role of GATA4 in adult osteoblasts in vivo and in vitro and we showed that GATA4 regulates bone mineral density in OCN-expressing osteoblasts. GATA4 represses osteoclastogenesis by regulating RANKL expression by binding to seven regions in the RANKL locus to repress its transcription.
We have previously shown that GATA4 regulates RUNX2 expression and other early osteoblast genes, including TGF-β [9]. GATA4 is highly expressed early in osteoblastogenesis and its expression decreases with differentiation. It is surprising to find that GATA4 is also necessary in osteocalcin-expressing cells, which are later in the differentiation pathway. Knockout of Gata4 with OCN-Cre results in a decrease in BV/ TV and trabecular number, along with an increase in trabecular spacing. The trabecular region also contained an increase in osteoclast number, as determined by TRAP staining, to at least partially explain the changes in BV/TV, trabecular number and trabecular spacing.
From an unbiased approach (ChIP-seq), we discovered that GATA4 overlaps with previously published VDR enhancers. GATA4 sites C-G are also vitamin D receptor binding sites [29]. The GATA4 site “G” that is located 75 kb away from the RANKL promoter has been extensively studied. This site is also known as the VDR site 5 (mRL-D5) and the vitamin D receptor, RUNX2, c-fos, STAT3/5 and CREB bind at this site [23,31]. This enhancer has been deleted in mice, resulting in decreased RANKL expression and increased bone mass [32], confirming that this region regulates RANKL transcription. Deletion of this enhancer also delays the progression of atherosclerotic plaque development and plaque calcification in hypercholesterolemic mice [33]. These long-range enhancers are key to RANKL expression and function.
RANKL mRNA increases with a loss of Gata4, suggesting that GATA4 is located at the RANKL enhancers to repress transcription. GATA4 is located almost equally near up-regulated genes (48%) and down-regulated genes (52%) in the ilium [34]. GATA4 has been shown to open chromatin, specifically by recruiting p300 to activate transcription [27]. However, the mechanism of GATA4 repression of target genes remains to be determined.
In summary, we show here that GATA4 directly represses RANKL expression via seven cis-regulatory regions. Furthermore, in osteoblasts, GATA4 is necessary for proper bone mineral density and regulation of osteoclast differentiation.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Award Number AR-064354–01 to SAK. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Disclosure statement
The authors have nothing to disclose.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bone.2018.07.014.
References
- [1].Rao S, et al. , RANKL and RANK: from mammalian physiology to cancer treatment, Trends Cell Biol. 28 (3) (2017) 213–223. [DOI] [PubMed] [Google Scholar]
- [2].O’Brien CA, Control of RANKL gene expression, Bone 46 (4) (2010) 911–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Martin A, et al. , Estrogens antagonize RUNX2-mediated osteoblast-driven osteoclastogenesis through regulating RANKL membrane association, Bone 75 (2015) 96–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Scott LJ, Denosumab: a review of its use in postmenopausal women with osteoporosis, Drugs Aging 31 (7) (2014) 555–576. [DOI] [PubMed] [Google Scholar]
- [5].Park HJ, et al. , The cooperation of CREB and NFAT is required for PTHrP-induced RANKL expression in mouse osteoblastic cells, J. Cell. Physiol 230 (3) (2015) 667–679. [DOI] [PubMed] [Google Scholar]
- [6].Baek K, et al. , Isoproterenol increases RANKL expression in a ATF4/NFATc1-dependent manner in mouse osteoblastic cells, Int. J. Mol. Sci 18 (10) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Park HJ, et al. , TNFalpha increases RANKL expression via PGE(2)-induced activation of NFATc1, Int. J. Mol. Sci 18 (3) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bishop KA, et al. , Mouse Rankl expression is regulated in T cells by c-Fos through a cluster of distal regulatory enhancers designated the T cell control region, J. Biol. Chem 286 (23) (2011) 20880–20891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Guemes M, et al. , GATA4 is essential for bone mineralization via ERalpha and TGFbeta/BMP pathways, J. Bone Miner. Res 29 (1) (2014) 2676–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Miranda-Carboni GA, et al. , GATA4 regulates estrogen receptor-alpha-mediated osteoblast transcription, Mol. Endocrinol 25 (7) (2011) 1126–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Khalid AB, et al. , GATA4 directly regulates Runx2 expression and osteoblast differentiation, JBMR Plus 2 (2) (2018) 81–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Molkentin JD, et al. , Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis, Genes Dev. 11 (8) (1997) 1061–1072. [DOI] [PubMed] [Google Scholar]
- [13].Kuo CT, et al. , GATA4 transcription factor is required for ventral morphogenesis and heart tube formation, Genes Dev. 11 (8) (1997) 1048–1060. [DOI] [PubMed] [Google Scholar]
- [14].Zhang M, et al. , Osteoblast-specific knockout of the insulin-like growth factor (IGF) receptor gene reveals an essential role of IGF signaling in bone matrix mineralization, J. Biol. Chem 277 (46) (2002) 44005–44012. [DOI] [PubMed] [Google Scholar]
- [15].He A, et al. , PRC2 directly methylates GATA4 and represses its transcriptional activity, Genes Dev. 26 (1) (2012) 37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Khalid AB, et al. , Mechanical and material properties of cortical and trabecular bone from cannabinoid receptor-1-null (Cnr1−/−) mice, Med. Eng. Phys 38 (10) (2016) 1044–1054. [DOI] [PubMed] [Google Scholar]
- [17].Dempster DW, et al. , Standardized nomenclature, symbols, and units for bone histomorphometry: a 2012 update of the report of the ASBMR Histomorphometry Nomenclature Committee, J. Bone Miner. Res 28 (1) (2013) 2–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Krum SA, et al. , Unique ER{alpha} cistromes control cell type-specific gene regulation, Mol. Endocrinol 22 (11) (2008) 3121–3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].He A, Pu WT, Genome-wide location analysis by pull down of in vivo biotinylated transcription factors, Curr. Protoc. Mol. Biol (2010) 21.20.1–21.20.15 Chapter 21:p. Unit 21.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Li H, Durbin R, Fast and accurate short read alignment with Burrows-Wheeler transform, Bioinformatics 25 (14) (2009) 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Heinz S, et al. , Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities, Mol. Cell 38 (4) (2010) 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Krum SA, et al. , Estrogen protects bone by inducing Fas ligand in osteoblasts to regulate osteoclast survival, EMBO J. 27 (3) (2008) 535–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Fu Q, Manolagas SC, O’Brien CA, Parathyroid hormone controls receptor activator of NF-kappaB ligand gene expression via a distant transcriptional enhancer, Mol. Cell. Biol 26 (17) (2006) 6453–6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Morrisey EE, et al. , GATA-6: a zinc finger transcription factor that is expressed in multiple cell lineages derived from lateral mesoderm, Dev. Biol 177 (1) (1996) 309–322. [DOI] [PubMed] [Google Scholar]
- [25].Boyce BF, Xing L, Biology of RANK, RANKL, and osteoprotegerin, Arthritis Res. Ther 9 (Suppl. 1) (2007) S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].He A, et al. , Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart, Proc. Natl. Acad. Sci. U. S. A 108 (14) (2011) 5632–5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].He A, et al. , Dynamic GATA4 enhancers shape the chromatin landscape central to heart development and disease, Nat. Commun 5 (2014) 4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zheng R, et al. , Function of GATA factors in the adult mouse liver, PLoS One 8 (12) (2013) e83723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kim S, et al. , Activation of receptor activator of NF-kappaB ligand gene expression by 1,25-dihydroxyvitamin D3 is mediated through multiple long-range enhancers, Mol. Cell. Biol 26 (17) (2006) 6469–6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Cirillo LA, et al. , Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4, Mol. Cell 9 (2) (2002) 279–289. [DOI] [PubMed] [Google Scholar]
- [31].Onal M, et al. , Unique distal enhancers linked to the mouse Tnfsf11 gene direct tissue-specific and inflammation-induced expression of RANKL, Endocrinology 157(2) (2016) 482–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Galli C, et al. , Targeted deletion of a distant transcriptional enhancer of the receptor activator of nuclear factor-kappaB ligand gene reduces bone remodeling and increases bone mass, Endocrinology 149 (1) (2008) 146–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Shamsuzzaman S, et al. , Deletion of a distal RANKL gene enhancer delays progression of atherosclerotic plaque calcification in hypercholesterolemic mice, J. Cell. Biochem 118 (12) (2017) 4240–4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Aronson BE, et al. , GATA4 represses an ileal program of gene expression in the proximal small intestine by inhibiting the acetylation of histone H3, lysine 27, Biochim. Biophys. Acta 1839 (11) (2014) 1273–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.