Summary
The majority of commensal oral streptococci are able to generate hydrogen peroxide (H2O2) during aerobic growth, which can diffuse through the cell membrane and inhibit competing species in close proximity. Competing H2O2 production is mainly dependent upon the pyruvate oxidase, SpxB and to a lesser extent the lactate oxidase LctO, both of which are important for energy generation in aerobic environments. Several studies point to a broad impact of H2O2 production in the oral environment, including a potential role in biofilm homeostasis, signaling, and interspecies interactions. Here, we summarize the current research regarding oral streptococcal H2O2 generation, resistance mechanisms, and the ecological impact of H2O2 production. We also discuss the potential therapeutic utility of H2O2 for the prevention/treatment of dysbiotic diseases as well as its potential role as a biomarker of oral health.
Introduction
The human host is closely associated with complex microbial communities consisting primarily of bacteria, but also of viruses, fungi, archaea, and certain protozoa1. The colonization of oral mucosal and hard tissue surfaces by microorganisms is accompanied by the vigorous intra- and interspecies exchange of information among diverse microbial communities as well as cross-kingdom communication between the microbes and host 2,3. The ideal outcome of microbial colonization is to create a host-protective environment characterized by a consortium of commensal and mutualistic flora species. The protection afforded by the resident commensal flora can be viewed both temporally as well as spatially: temporally, since the initial colonization after birth can contribute to a lifetime of proper host immune system function 4 and spatially, since community interactions amongst the flora as well as flora-host interactions both yield mostly local effects 5. To receive these protective benefits from the flora, it is critical to maintain microbial symbiosis 6. There are various recognized mechanisms that promote a symbiotic relationship with the flora, including the production of hydrogen peroxide (H2O2) by the commensal oral streptococci.
The oral cavity has several surfaces covered with microbes dwelling in biofilms and the composition of the biofilm community is determined by the respective environment 7–9. For example, the process of dental biofilm formation has been the center of several in vivo oral microbial ecology studies 10–13. Distinctive patterns of colonization sequences have been described, which can develop into a mature biofilm community with enormous complexity and structure. Initial colonization of the saliva bathed tooth surface in the first 4 to 8 hours is dominated by oral streptococci and Corynebacterium ssp. as recently shown 12,14. This dominance extended up to 16 hours into biofilm development and Streptococcus accounted for about 20% of the taxa 14. Incidentally, oral streptococci are also the main source for ecologically relevant H2O2 production 15. The growth of initial colonizers during biofilm development in the first 16 hours seemed to be exponential, suggesting ideal conditions for facultative anaerobic and aerotolerant species. Further biofilm development favors the integration of Gram-negative anaerobic species including Fusobacterium, Prevotella and Porphyromonas, but growth of the biofilm population seemed to slow down after 16 hours 14.
Initial biofilm development requires the formation of saliva-derived macromolecular complexes on the tooth surface leading to the formation of the acquired enamel pellicle. This process takes place within seconds after a clean enamel surface is exposed to saliva 16. Mechanistically, the abundance of streptococci as initial colonizers can be explained by surface adhesins specifically recognizing salivary proteins that are part of the acquired enamel pellicle. Integration of other species, including later colonizers, occurs via specific cell-surface receptors also displayed by streptococci 17, thus streptococci together with a few other species are able to determine the spatial and temporal development of oral biofilms and are important for biofilm homeostasis. Several synergistic and antagonistic mechanisms have been described that contribute to oral biofilm dynamics 18, and H2O2 production seems to play a significant role 15.
In the time since our previous review about the role of H2O2 in oral biofilm ecology 15, there have been important advances in our understanding of oral microbial ecology and species composition in health and disease. One of the key outcomes of those studies is that polymicrobial diseases such as caries and periodontal disease are the ecological consequences of dysbiosis. For example, caries is triggered by overgrowth of a subset of the oral flora that are inherently aciduric and acidogenic 19. We have previously suggested that the conserved ability of commensal oral streptococci to produce H2O2 is important for oral health 20. Here we review the latest evidence detailing our current knowledge of the biology of streptococcal H2O2 production as well as the role of H2O2 as key component of oral ecology.
Relevant Chemistry of hydrogen peroxide and reactive oxygen species (ROS) production
i). Oxygen redox pathways generate H2O2 as an intermediate
The generation of H2O2 during cellular redox reactions occurs due to the sequential univalent reduction of molecular oxygen 21. H2O2 is the second intermediate formed by the addition of one electron and two protons to the highly unstable superoxide anion. H2O2 is relatively stable and unreactive in an abiotic environment 22,23. Pure solutions of macromolecules like nucleic acids, proteins (metal-free), lipids, and polysaccharides are typically resistant to H2O2 oxidation. However, this is not the case in a biological environment, which is replete with transition metals such as Fe(ii) or Cu2+ that act as reducing agents for H2O2 22,24. The further reduction of H2O2 results in the addition of an electron and a proton to generate H2O and the extremely reactive and short-lived hydroxyl radical, which is the major reactive oxygen species (ROS) reacting with biomolecules 21. The half-life of the hydroxyl radical in a biological system is only about 1 ns. Its reaction with biomolecules is non-selective and is only limited by diffusion 25.
ii). H2O2 involvement in the production of hydroxyl radicals via Fenton chemistry
In a cellular context, the generation of hydroxyl radicals is primarily catalyzed by Fenton chemistry via the transition metal ion pool inside the cell, especially soluble Fe(II) 26. Fe(II) donates one electron to one H2O2 molecule to generate a hydroxyl radical, a hydroxide ion, and oxidized Fe(III). The hydroxyl radical will subsequently react rapidly with any organic cellular compound in the immediate vicinity 27
iii). Cytotoxicity of H2O2
Much of the cytotoxicity of H2O2 is the result of oxidative DNA damage induced by hydroxyl radicals generated through Fenton chemistry 27,28. Hydroxyl radicals can trigger direct strand breakage through the oxidation of the ribose moieties of the DNA backbone 27,29,30. In addition, the nucleobases can be oxidized, with guanine being one of the major targets of oxidation 31. Interestingly, two kinetically distinguishable modes of killing by H2O2 seem to exist as shown with the model organism Escherichia coli 32,33. Cells treated with H2O2 concentrations lower than 3 mM were more susceptible compared to H2O2 concentrations between 5 to 20 mM. Above 20 mM H2O2, the survival rate is inversely proportional to the H2O2 concentration, as would be expected. The dual range of H2O2 susceptibility is attributed to separate killing mechanisms. Lower concentrations of H2O2 will mostly damage DNA due to Fenton reaction hydroxyl radicals inducing a lethal mutation rate 27,34. The ability of Fe to bind to DNA is a key feature of this mechanism 35. The second mode of killing at higher H2O2 concentrations seems to be the result of damage to other target(s) 33,34, including proteins and lipids. However, the biologically relevant H2O2 concentration bacteria normally encounter is usually in the nM to lower mM range 36–39. Hydrogen peroxide concentrations up to 7 mM have been reported for streptococci 37, although those measurements were mostly performed in batch cultures that may not be reflective of the normal H2O2 production capacity of streptococcal biofilms. Regardless, it seems unlikely that E. coli would encounter >20 mM H2O2 during growth in bacterial communities. A similar dual susceptibility to H2O2 has also been reported for Streptococcus thermophilus 40, which is intriguing since both organisms utilize distinct mechanisms to detoxify H2O2. E. coli directly degrades H2O2 via the catalase enzyme 41, whereas streptococci typically lack catalase and rely on other mechanisms 42,43. It is worth noting that S. mutans is equally susceptible to killing when treated with 1 to 5 mM H2O2 44. Thus, the dual susceptibility to H2O2 killing might not be generalizable for all species and/or growth conditions.
H2O2 mediated damage can also occur to enzymes with iron-sulfur clusters abolishing enzyme activity, as well as enzymes that require a single Fe2+ ion as cofactor for enzymatic activity 31. Other potential targets are lipids in the cell membrane of bacterial cells. However, this has not been studied with oral biofilm bacteria and the lack of the main target for lipid peroxidation, polyunsaturated fatty acid chains in most bacterial membranes, makes it unlikely that this mode of killing action occurs 27,38.
Sources of microbial hydrogen peroxide production in the oral biofilm
In the oral biofilm, naturally produced H2O2 crosses the bacterial membrane of producer cells and is released into the environment where it can influence neighboring cells and ultimately oral biofilm ecology. Our group and others have demonstrated that abundant H2O2 production in the pioneer colonizer S. sanguinis is critically dependent upon the activity of the pyruvate oxidase, SpxB. SpxB knock-out mutants do not produce sufficient extracellular H2O2 to inhibit competing H2O2-susceptible species like Streptococcus mutans 45. SpxB-dependent H2O2 production has also been shown experimentally for S. gordonii 45 and Streptococcus parasanguinis 46 as well as Streptococcus infantis, Streptococcus oralis, Streptococcus cristatus and Streptococcus mitis as shown on Prussian Blue H2O2 indicator plates 47,48 (Fig. 1, unpublished).
Fig. 1:
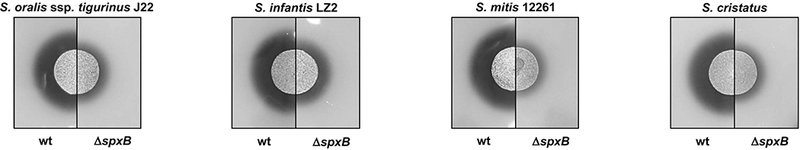
Comparison of H2O2 production by S. oralis ssp. tigurinus J22, S. infantis LZ2, S. mitis 12261, and S. cristatus wild type strains and their respective isogenic spxB mutants. H2O2 production from all strains was assessed on H2O2 indicator plates after overnight incubation at 37ºC aerobically. H2O2 production leads to the precipitation of Prussian blue during aerobic growth, which forms a blue halo around the producer 47,48.
i). Pyruvate oxidase, SpxB
The initial characterization of the pyruvate oxidase from S. sanguinis was reported by Jan Carlsson and colleagues. Permeabilized cells exhibited pyruvate oxidase activity dependent upon thiamine pyrophosphate (TPP), flavin adenine dinucleotide (FAD), Mg2+, and orthophosphate 49. Catalytic activity was also significantly increased in cells grown aerobically 49, while the reaction itself is oxygen-dependent 50. SpxB catalyzes the oxidative decarboxylation of pyruvate to the high energy metabolite acetyl-phosphate in addition to CO2, H2O2, and ATP. Thus, the reaction provides an energetic growth advantage for the producer 15. Our group confirmed the presence of SpxB under anaerobic conditions, suggesting that the enzyme is present in cells even when H2O2 production is not detectable 51. Consistent with the results from Carlsson, we observed S. sanguinis spxB expression to increase about 25-fold in aerobically grown cultures with the highest spxB expression occurring in late log phase 51. Even so, S. sanguinis still produces small amounts of SpxB under anaerobic conditions when pyruvate is primarily metabolized via the pyruvate formate lyase 49. The reason for this is not entirely clear, but it could potentially provide a growth advantage once oxygen becomes available. Alternatively, SpxB might have additional moonlighting functions that have yet to be discovered.
The biochemical mechanism of SpxB catalysis has not been elucidated in oral streptococci, but this has been investigated using the Lactobacillus plantarum pyruvate oxidase (LpPox) ortholog. Several characteristics between LpPox and S. sanguinis SpxB are conserved and they share 49% identity (Table 1) 52. Molecular modeling using the automated protein structure homology-modelling server SWISS-MODEL (https://swissmodel.expasy.org) 53 predicted a high conservation of the S. sanguinis SpxB protein structure compared to the known structures of the pyruvate oxidases from L. plantarum 54 and Aerococcus viridans 55 (Fig. 2).
Table 1:
Comparison of SpxB among different species
| Species | Strain | number of amino acids | % identity |
% positives |
|---|---|---|---|---|
| Streptococcus sanguinis | SK36 | 591 | 100 | 100 |
| Streptococcus gordonii | CH1 | 591 | 98 | 99 |
| Streptococcus mitis | B6 | 591 | 98 | 98 |
| Streptococcus peroris | ATCC 700780 | 591 | 98 | 98 |
| Streptococcus infantis | ATCC 700779 | 591 | 98 | 98 |
| Streptococcus oralis | Uo5 | 591 | 98 | 98 |
| Streptococcus pseudopneumoniae | IS7493 | 591 | 98 | 98 |
| Streptococcus pneumoniae | R6 | 591 | 98 | 98 |
| Streptococcus parasanguinis | CC87K | 591 | 98 | 98 |
| Streptococcus vestibularis | ATCC 49124 | 591 | 98 | 99 |
| Streptococcus salivarius | HSISS4 | 591 | 97 | 98 |
| Streptococcus cristatus | AS1.3089 | 591 | 98 | 99 |
| Streptococcus anginosus | C1051 | 591 | 98 | 99 |
| Streptococcus australis | ATCC 700641 | 591 | 96 | 98 |
| Streptococcus himalayensis | HTS2 | 593 | 89 | 94 |
| Streptococcus acidominimus | NCTC11291 | 593 | 85 | 92 |
| 583 (pseudogene)* | 70 | 83 | ||
| Aerococcus viridans | CCUG4311 | 592 | 69 | 82 |
| Lactobacillus plantarum | ZJ316 | 603 (PoxB) ** | 49 | 66 |
% identity: identity on the amino acid level to the query sequence of spxB from S. sanguinis.
% positives: amino acids with a conservative exchange were physiochemical properties are preserved when compared to the query sequence of spxB from S. sanguinis.
The second pyruvate oxidase gene in S. acidominimus possesses an internal stop codon.
L. plantarum encodes several pyruvate oxidases, shown here is the relevant pyruvate oxidase used for molecular modeling.
Fig. 2:

Protein structure homology modeling of SpxB from S. sanguinis using the automated protein structure homology-modelling server SWISS-MODEL. The reference proteins used were from Aerococcus viridans and Lactobacillus plantarum. The quaternary structure quality estimate (QSQE) based upon the modeling with Aerococcus viridans SpxB is 0.78. This protein has 69% sequence identity to the S. sanguinis SpxB. The QSQE score ranges from 0 to 1 with higher numbers indicating higher reliability.
From the LpPox crystal structure, several aspects of the catalytic process have been revealed, including the roles of the enzyme-bound co-factors TPP and FAD 56,57. The reaction occurs in several steps beginning with a nucleophilic attack on the bound pyruvate molecule by TPP leading to acetyl-TPP and FADH2. In a subsequent oxygen-dependent mechanism, FADH2 is oxidized to FAD generating H2O2, the acetyl-TPP reacts with inorganic phosphate to create acetyl-phosphate, and TPP is regenerated 57,58 (Fig. 3). A similar catalytic mechanism would be expected for S. sanguinis SpxB. However, LpPox can substitute any of the divalent metal ions Mg2+, Mn2+, and Ca2+ to fix the di-phosphate moiety of TPP, while the S. sanguinis SpxB seems to favor Mg2+ 52.
Fig. 3:

Generalized overview of pyruvate oxidase mechanism, Fenton reaction, and DNA damage. The pyruvate oxidase catalyzes the oxidative decarboxylation of pyruvate and requires the cofactors TPP and FAD. During the multistep reaction, two reducing equivalents are transferred from TPP to FAD, yielding 2-acetyl-thiamine pyrophosphate and FADH2. FADH2 is re-oxidized by O2 which generates H2O2 and FAD. Ac-TPP is then cleaved phosphorolytically to yield acetyl phosphate and TPP 146. Fe2+ is able to non-specifically bind to DNA and react with the generated H2O2 in the Fenton reaction to cause DNA damage in the form of DNA strand breakage (as shown here) or base alterations in the DNA.
i.i). Distribution and chromosomal location of spxB in oral streptococci
SpxB-dependent H2O2 generation is conserved among most of the oral streptococci. Using the amino acid sequence of SpxB from S. sanguinis SK36 as a reference, we performed a BlastP search of the entire Human Oral Microbiome Database (http://www.homd.org/) 59. A surprisingly high conservation of the amino acid composition (≥95 identity/≥98% similarity) and protein length (591 amino acids) was observed among oral streptococci (Table 1).
Among streptococci, spxB is typically expressed as a single gene controlled by its own promoter (Fig. 4). In several species, the immediate upstream open reading frame encodes a P-type ATPase copper transporter. Copper is a trace metal that can participate in the Fenton reaction and is also able to disrupt the iron sulfur clusters of various proteins, several of which participate in a variety of cellular processes influenced by H2O2. Accordingly, exposure of Streptococcus pyogenes to H2O2 leads to the induction of genes with predicted roles in iron sulfur cluster assembly 60. Therefore, a coordinated regulation of copper homeostasis and hydrogen peroxide production could be one mechanism employed as a strategy to mitigate the potential toxicity triggered by iron-sulfur cluster protein oxidation.
Fig. 4:
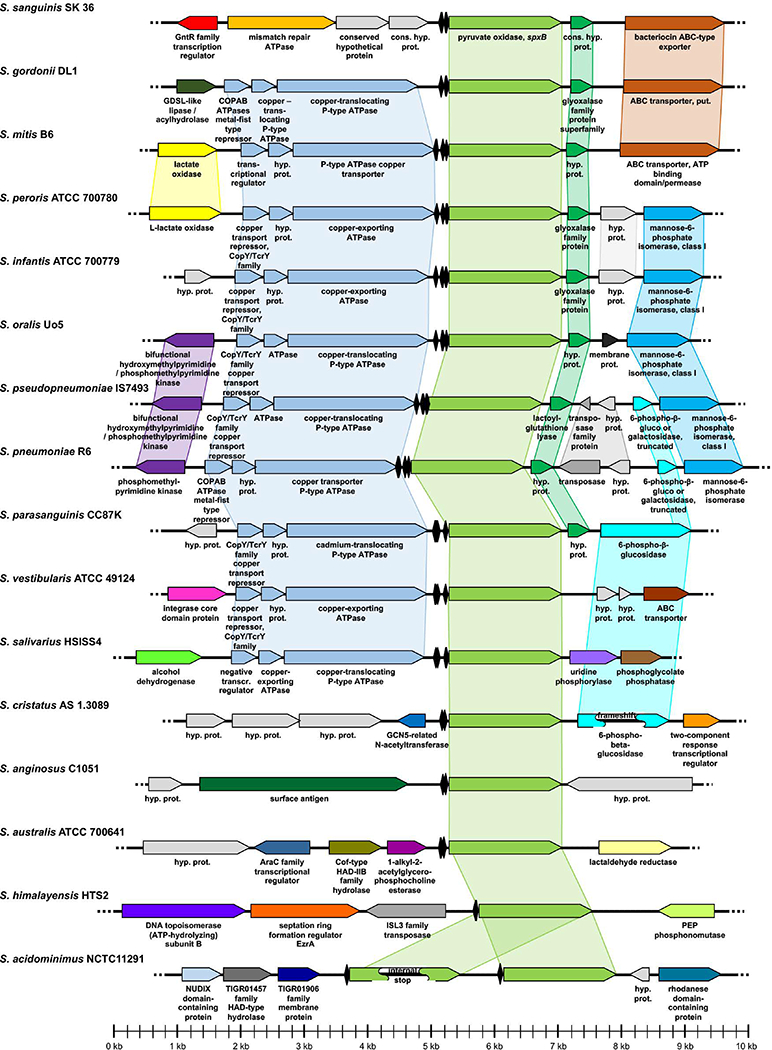
Chromosomal context of spxB in multiple oral streptococci. The sequences of the given species and strains as well as the displayed gene names and functions were obtained from NCBI. Species were sorted with respect to their analogy in gene assortment. Related or functionally connected genes are shown in matching colors. In case the gene function was not already given in the NCBI database (“conserved hypothetical protein” or “hypothetical protein”), related or functionally connected genes were identified by BLAST sequence comparison. Figures were manually generated using Microsoft Powerpoint. Catabolite responsive elements (cre boxes) are indicated (♦) and were predicted as previously described 121. The respective cre boxes of S. sanguinis and S. gordonii were experimental validated 123.
i.ii). Evidence of in vivo expression
In an effort to demonstrate the in vivo relevance of pyruvate oxidase, spxB gene expression was quantified in clinical plaque samples from different individuals 61. Universal spxB primers were developed for streptococci and could readily amplify spxB from isolates identified as H2O2 producers on Prussian Blue agar plates 47,48. Conversely, isolates exhibiting no H2O2 production also failed to yield spxB PCR amplicons 61. The expression characteristics of spxB was also determined in these clinical plaque samples. While spxB transcript levels were quite variable between the 9 subjects, the day to day variability was quite low and seemed relatively constant over time for one selected subject 61. The ability to detect spxB expression in oral plaque samples points to its active role in oral streptococcal physiology. Moreover, spxB is expected to play a particularly important role in early biofilm development, since SpxB-positive streptococci comprise significant fractions of newly formed oral biofilms 62 and these biofilms also contain significantly higher oxygen concentrations compared to older, mature biofilms 63.
ii). Other sources of streptococcal hydrogen peroxide production
Another commonly encoded H2O2-producing enzyme of oral streptococci is the FMN (Flavin mononucleotide)-dependent lactate oxidase (LctO; sometimes referred to as Lox) 64,15. The metabolic function of LctO is the aerobic oxidation of L-lactate to generate pyruvate. Since pyruvate is also the substrate for SpxB, both LctO and SpxB are linked in the central metabolism of oral streptococci during aerobic growth. The catalytic mechanisms of SpxB and LctO are different, since no TPP is involved in LctO oxidation of L-lactate. However, for both enzymes the cofactor FMN is initially reduced to FMNH2 and then subsequently oxidized to regenerate FMN and produce H2O2 65.
The LctO-dependent inhibitory ability of Streptococcus oligofermentans toward S. mutans has been studied in detail 66–68. (Note: Streptococcus oligofermentans has been recently re-classified as Streptococcus cristatus 69, but for the reminder of the review we adhere to its old designation). Overall, the H2O2-dependent inhibition of S. mutans by S. oligofermentans seems to be a coordinated event between SpxB and LctO, with SpxB producing the majority of H2O2 during early and logarithmic growth and LctO dominating during stationary phase. This might be partially due to the fact that the lactate concentration is initially limited, thus precluding LctO from producing much H2O2. However, lactate will accumulate during growth and can later be used by LctO to produce pyruvate and H2O2. Interestingly, inactivation of SpxB in S. oligofermentans has a negative effect on the activity of LctO, further suggesting these two enzymes are coordinately controlled. Furthermore, in the context of a multispecies biofilm, LctO activity would presumably play an important ecological role in the presence of copious lactate producers like S. mutans. It has been suggested that this lactate-dependent oxidase activity ensures successful competition with S. mutans and may partially explain the high prevalence of S. oligofermentans in certain caries-free individuals 68,70. Other than S. oligofermentans, the role of LctO has only been studied in S. pyogenes 71 and S. pneumoniae 72. Therefore, it remains to be determined whether other oral streptococci can exploit LctO activity to inhibit cariogenic species.
In addition to SpxB and LctO, S. oligofermentans may utilize another oxidase to produce competitive, S. mutans-inhibiting quantities of H2O2 73–75. This enzyme was initially classified as an L-amino acid oxidase (LAAO) because of its ability to use peptone and amino acids such as L-aspartic acid, L-tryptophan, L-lysine, L-isoleucine, L-arginine, L-asparagine and L-glutamine to produce significant amounts of H2O2. Tong and colleagues suggested LAAO activity could be important under specific growth conditions 74. For example, when saliva is used as sole nutrient source, polypeptides might be catabolized into free amino acids that could serve as LAAO substrates to yield α-keto acids, ammonia, and H2O2. However, the activity of the enzyme was later revised and aminoacetone was reported to be the preferred substrate for purified LAAO 75. This was independently confirmed and the structure of the enzyme solved. Thus, the enzyme was renamed to aminoacetone oxidase or AAO. Its activity previously measured using amino acids substrates is therefore likely the result of either promiscuous catalysis on non-preferred substrates or an artifact of the in vitro reaction conditions 73. Nonetheless, AAO seems to be a unique enzyme in oral streptococci and there is strong evidence to suggest that S. oligofermentans acquired this gene via horizontal gene transfer 76. Although LctO-mediated inhibition of S. mutans is ten-fold stronger as compared to AAO 74, it might still provide a selective advantage for S. oligofermentans under certain ecological conditions. Unlike SpxB and LctO, AAO also has the added advantage of producing ammonia as a byproduct 73, which, like the arginine deiminases of other commensal oral streptococci, might similarly provide protection against plaque acidification 77.
Other fundamental redox reactions in the cell can also yield modest amounts of H2O2. For example, the regeneration of NAD+ via NADH oxidase can yield H2O2 78. In general, NADH oxidases reduce molecular oxygen to either H2O2 (via Nox-1) or H2O (via Nox-2) using NADH as a substrate 78. Among streptococci, differences exist on the distribution of the nox-1/2 genes. S. mutans encodes nox-1 and nox-2 79, while S. sanguinis only encodes nox-2 78. The H2O2 production capacity of nox-1 is apparently limited, since S. mutans does not produce inhibiting amounts of H2O2 45. In a recent study, Ge et al. demonstrated that a nox-2 knock-out mutant in S. sanguinis loses its ability to compete with S. mutans due to a decrease in extracellular H2O2 production 78. Interestingly, this effect on H2O2 production was suggested to be indirect since the intracellular concentration of H2O2 was actually slightly increased and no deleterious impacts were observed upon SpxB activity. The authors also reported that the nox-2 deletion affects membrane fluidity, which has led to the speculation that the observed lack of S. mutans inhibition might result from a decrease in H2O2 diffusion across the membrane 78.
Self-compatibility and the response to hydrogen peroxide stress
When comparing the aerobic growth of H2O2-producing streptococci in the presence and absence of externally added catalase, it is evident that H2O2 production yields self-toxicity 80. Typically, streptococci do not encode catalase, suggesting that the benefit from excreting relatively large amounts of H2O2 outweighs the potential negative effects its production might pose on the producer itself. The absence of catalase or other commonly employed H2O2-detoxifying enzymes in oral streptococci also suggests that other important, perhaps uncharacterized mechanisms are involved in protection from H2O2 toxicity. Certainly, it is also possible that in the oral cavity H2O2 toxicity might not be as pronounced as under laboratory conditions. Saliva has an inherent capacity to deplete H2O2 via the salivary lactoperoxidase system, which catalyzes the oxidation of thiocyanate by H2O2 to produce the antimicrobial compound hypothiocyanite 81–83. It is currently unclear to what extent, if any, that lactoperoxidase might influence the local H2O2 concentrations in oral biofilms. Furthermore, saliva flow aides in the dilution of H2O2 84. However, saliva flow can be limited, for example, in biofilms or interproximal spaces. The actual concentration of H2O2 in saliva or at the biofilm-tooth interface has not been determined in vivo. The detection is hindered by several problems, including the presence of the salivary lactoperoxidase system which can eliminate H2O2 before its reliable detection. The lactoperoxidase substrates (thiocyanate) and products (hypothiocyanite) can be measured in saliva however, and this allows salivary H2O2 concentrations to be extrapolated to approximately 10 µM 85. We currently lack the sensor technologies required to directly measure H2O2 at the biofilm interface of patients, but one would expect this value to be significantly higher than the 10 µM value estimated for saliva. Recent real-time measurements of the H2O2 concentration 100 μm above an in vitro polymicrobial biofilm found up to 1.4 mM of H2O2 can be generated 86. This suggests that during early biofilm development, in which there is an abundance of SpxB positive streptococci, considerable amounts of H2O2 can be produced that presumably requires efficient cellular defense mechanisms to avoid H2O2-dependent toxicity.
A key bacterial mechanism to prevent H2O2 toxicity is through the avoidance of the Fenton reaction. We have previously demonstrated in S. sanguinis and S. gordonii that mutants lacking Dps, a ferritin-like protein involved in iron sequestration from DNA, are about 104-fold more susceptible to exogenous H2O2 compared to their respective wild types 43. A similar hypersensitivity was also reported for S. mutans as well as other streptococci, suggesting that Dps is a broadly conserved and critical protein for the defense against H2O2 87,88. In S. mutans, superoxide dismutase (Sod) mutants exhibit a similar hypersensitivity as Dps mutants 89, while the same Sod mutations in S. sanguinis and S. gordonii only result in about two to three-fold increases in H2O2 sensitivity 43. TrxB mutants of S. sanguinis and S. gordonii also exhibit dramatically different H2O2 sensitivity phenotypes. TrxB is a thioredoxin reductase that catalyzes the reduction of thioredoxin and plays important role in the formation of disulfide bonds in oxidized proteins. An S. sanguinis TrxB mutant is severely impaired in its H2O2 resistance, whereas the S. gordonii TrxB mutant exhibits no obvious survival defects after H2O2 challenge 43.
Studies of the S. pneumoniae pyruvate oxidase SpxB demonstrated an unexpected direct role for SpxB in the protection against H2O2 toxicity. S. pneumoniae SpxB deletion mutants are 102 to 103-fold more susceptible to exogenously added H2O2 90. The source of this change in H2O2 susceptibility was determined to be dependent upon post translational modifications of SpxB itself, in which sulfenylation of SpxB occurs during endogenous H2O2 production as part of a reactive oxygen species adaptation mechanism 91. Although not entirely understood, the sulfenylation of SpxB is postulated to serve as an “H2O2 sink” that scavenges intracellular H2O2 91. Thus, S. pneumoniae has apparently evolved an intriguing mechanism to use SpxB to mitigate the oxidative stress that is created through its own catalytic activity. However, this mechanism does not appear to be a conserved characteristic of SpxB orthologs, as spxB mutations do not impact H2O2 sensitivity for either S. sanguinis or S. gordonii 43. Based upon our current knowledge of oxidative stress tolerance, it would appear that the basic machinery employed by oral streptococci are part of a common toolbox. Yet, the observed differences among streptococci for proteins like SpxB, Sod, and TrxB suggest that there are significant aspects of H2O2 resistance that have yet to be discovered in these species. This is particularly evident from a recent publication illustrating a wide variety of oxidative stress tolerance phenotypes among different strains of S. mutans that all exhibit similar expression patterns for known components of the ROS response machinery 92. For further information about streptococcal ROS response mechanisms, see 93.
Hydrogen peroxide production and biofilm ecology
Our view of oral disease development has evolved in the last two decades due to advances in our understanding of the ecology within oral biofilms 94. It is now widely accepted that caries and periodontal disease are byproducts of oral dysbiosis 95. However, due to a historical focus upon pathogenic bacteria, a major aspect of oral health has been relatively understudied: the protective abilities of the commensal flora. Currently, there is a paucity of studies revealing the biological mechanisms that support symbiosis, a research area that we refer to as molecular commensalism 20. A better understanding of such mechanisms could provide valuable new strategies to prevent dysbiotic diseases. In fact, there is already evidence that such an approach is feasible. By promoting the growth of alkali-producing oral streptococci via arginine supplementation, it is possible to achieve a net increase in dental plaque pH and lower caries scores 77,96. H2O2 production is another aspect that could be exploited, as H2O2 is specifically associated with the commensal flora. Cariogenic species like S. mutans and most periodontopathogens are all exquisitely sensitive to H2O2 toxicity 15,97.
i). H2O2 and interspecies competition in the oral biofilm
To better understand how H2O2 production is influenced by the immediate biofilm environment, scanning electrochemical microscopy (SECM) has been used to monitor H2O2 production in real-time at the surface of an S. gordonii-S. mutans dual species biofilm 98. In combination with a pH probe, Joshi and colleagues identified the Achilles heel of S. gordonii H2O2 production. In an in vitro dual species biofilm assay grown in artificial saliva, S. gordonii is able to produce a considerable amount of H2O2 at a neutral pH. In fact, S. gordonii can slow the process of biofilm acidification by increasing spxB expression and H2O2 production once the environmental pH starts to fall due to sugar fermentation. This adjustment in H2O2 production provides a temporary check on S. mutans growth. However, once the environmental pH falls below the physiological buffering capacity of saliva, both spxB expression and H2O2 production diminish, which allows S. mutans to eventually dominate 98. This clearly shows that a prolonged reduction of the salivary pH not only promotes the growth of aciduric species, but also impairs the ability of some commensal species to effectively antagonize H2O2-sensitive pathobionts like S. mutans 98.
The effect of the environmental pH on the H2O2 production capability of oral streptococci seems to be a key issue in their competitive abilities. Contrary to the observation with S. gordonii, S. sanguinis exhibited no change in the production of H2O2 when growing at pH6, demonstrating that the response to a lower pH is not a universal decrease in H2O2 production 99. Although the observed phenotypic response showed consistency among different strains, the authors would like to emphasize that the ability of streptococci to adjust to environmental perturbations needs to be assessed on the species and sometimes strain level and should not be generalized 92,99. Interestingly, the observed reduction of spxB expression and H2O2 production in S. gordonii leads to a redirection of the metabolic flux at the pyruvate node 99. Since less pyruvate is metabolized through SpxB catalytic activity, an increase in lactic acid production was observed, which is also reflected in an increased expression of the lactate dehydrogenase gene ldh. The increase in lactic acid production itself is an adaptation mechanism improving the survival of S. gordonii at lower pH 99. S. sanguinis in general produces less H2O2 than S. gordonii and is more susceptible towards lower pH. Furthermore, the reduced H2O2 production at lower pH observed in S. gordonii still exceeds S. sanguinis production ability 99. Perhaps, the production of H2O2 serves a slightly different ecological purpose since S. sanguinis is one of the most-abundant early colonizers 20 and biofilm mediated protection mechanisms may protect the S. sanguinis population against low pH 100. At the same time, higher abundance means less energy needs to be channeled towards H2O2 production in general, while S. gordonii as a species with lower abundance needs to be more aggressive to ensure competitiveness.
The impact of environmental pH upon competitive H2O2 production has also been examined in other species 99,101,102. S. oligofermentans, a potent H2O2 producer, is able to inhibit the growth of S. mutans at pH6 in deferred antagonism assays, which is quite surprising, since S. oligofermentans growth at pH6 is compromised to a much greater degree compared to S. mutans. Even with its impaired growth at pH6, S. oligofermentans still produces up to 1.5-fold more H2O2 compared to pH7 101. At a pH of 5.5, S. oligofermentans growth defects are too severe to remain competitive with S. mutans, despite its aggressive H2O2 production. The inhibitory spectrum of commensal oral streptococcal H2O2-production was also confirmed for periodontopathogens like Aggregatibacter actinomycetemcomitans, Prevotella intermedia, and Porphyromonas gingivalis 97,103, suggesting that H2O2 production might similarly influence the composition of subgingival biofilms.
ii). Tipping the balance – influencing the production of H2O2
A key outcome for studies of molecular commensalism would be to develop new approaches to sustain a symbiotic biofilm community, even under conditions that otherwise would lead to dysbiosis. Conceivably, this could be achieved either by increasing the number of health-associated bacteria, such as SpxB-encoding streptococci, and/or by increasing the H2O2 output of producer species. Clinically, it has been shown that S. sanguinis isolates from caries-free individuals have increased H2O2 production relative to high caries individuals, which further supports the health-protective role of H2O2 104. In addition, our previous studies of an S. gordonii CcpA mutant have demonstrated the conceptual utility of exogenously manipulating H2O2 production to achieve a desired ecological outcome. CcpA is a transcriptional regulator of spxB and its deletion increases spxB expression and H2O2 production 105. Consequently, CcpA mutants also exhibit an enhanced ability to antagonize the growth of S. mutans. Similarly, in vitro studies of a 3-species S. gordonii, S. mutans, and A. naeslundii biofilm model have demonstrated that L-arginine supplementation can prevent S. mutans dominance and favor S. gordonii growth even in the presence of sucrose 106, a carbohydrate that strongly promotes the dysbiotic overgrowth of S. mutans 107. In this system, L-arginine was able to increase the pH of the biofilm, as expected, but it had the added benefit of increasing the production of H2O2 by S. gordonii as well. Conversely, in a 14-species biofilm model containing both H2O2-producing streptococci and periodontopathogens, the addition of clinically relevant concentrations of the H2O2-inactivating enzymes myeloperoxidase, lactoperoxidase, and erythrocyte catalase strongly favored the growth of the periodontopathogens at the expense of the commensals 108. Overall, there is ample clinical and experimental evidence establishing a clear inverse correlation between the abundance of H2O2 produced in an oral biofilm community and the prevalence of pathobionts.
When comparing the oral biofilms of different people, it is evident that considerable variability exists not only at the species level, but even among different strains of the same species 109. This has important ecological implications for one’s individual likelihood of developing oral dysbiosis. For example, differences in the H2O2 susceptibility of an individual’s pathobionts can influence how much H2O2 production is required to sustain symbiosis. This is evident from a recent study of S. mutans clinical isolates, which demonstrated how variability among the oxidative stress tolerance phenotypes of these strains has a dramatic influence upon their competitiveness in a multispecies biofilm setting 92. S. mutans clinical isolates were mixed with S. oralis and A. naeslundii and grown as biofilms for up to 91 hours. One particular S. mutans strain (Smu81) that was isolated from subgingival plaque 110 exhibited poor competitive abilities after 67 and 91 hours of incubation in the presence of sucrose, while all other S. mutans strains eventually became the dominant biofilm species under the same growth conditions 92. In a deferred antagonism assay, S. oralis was able to severely inhibit the growth of Smu81, but this effect was completely abolished in the presence of catalase 92. It is still a mystery why different strains of S. mutans can exhibit such a wide range of oxidative stress tolerance.
Similarly, clinical isolates of H2O2-producing streptococci also exhibit a range of H2O2-production phenotypes. Thus far, we have identified three distinct groups of H2O2-producers: group I – high-level H2O2 production, carbon catabolite repression (CCR) insensitive; group II – low-level H2O2 production, CCR insensitive; and group III – CCR-sensitive H2O2 production (Fig. 5). Importantly, like S. mutans oxidative stress tolerance, the H2O2-production phenotypes of these isolates were largely determined at the strain level 121. Further details regarding the role of CCR-dependent regulation of H2O2-production will be described later.
Fig. 5:
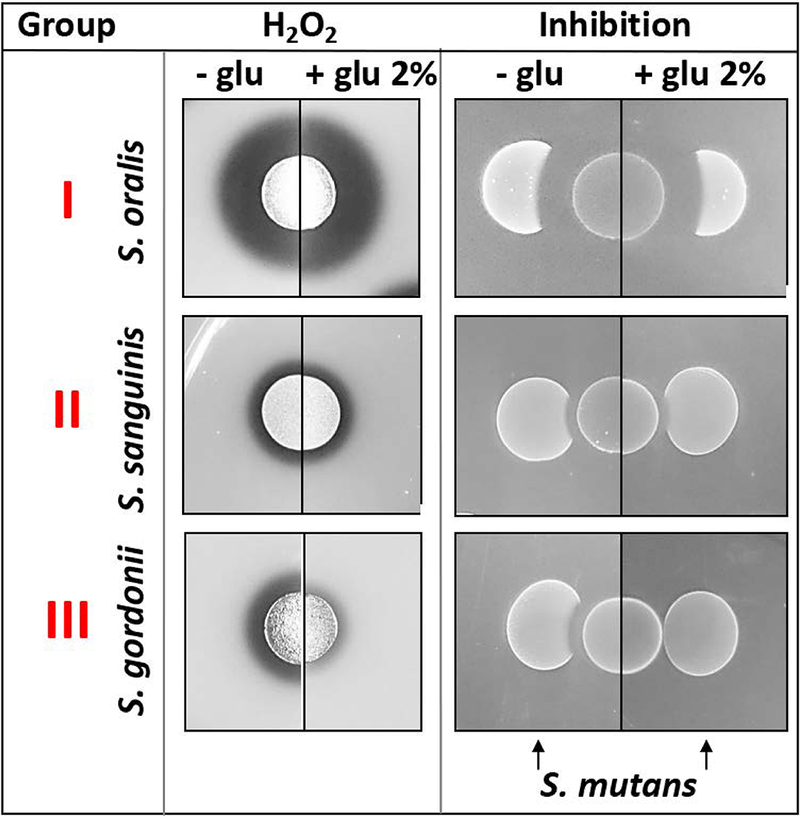
Representative species for the three identified H2O2-production groups and their ability to inhibit S. mutans ± glucose.
As mentioned previously, environmental pH is another factor that can have a profound impact upon H2O2 production. In a recent study by Huang and colleagues, alkali-generating arginolytic species were isolated from subjects with and without caries 111. Overall, 6 different Streptococcus species were detected with S. sanguinis being predominant (67.9%) followed by S. gordonii (8.9%), S. intermedius (8.9%), Streptococcus cristatus (8.9%), S. australis (3.6%), and S. parasanguinis (1.8%) 111. Each of these species also encodes spxB (see Table 1) and further analysis determined H2O2 production to be their principal mechanism of S. mutans antagonism 112. While H2O2 production is negatively impacted by acidic growth conditions for some streptococci 99, the arginine deiminase system is highly acid tolerant 113,114 and could serve to bolster SpxB activity to prolong competitive H2O2 production.
iii). Beyond competition – H2O2 signaling, biofilm formation, and community development
Another emerging aspect of H2O2 production is its influence on bacterial synergistic interactions, potentially functioning as a signal molecule. The role of streptococcal H2O2 in interspecies interactions with A. actinomycetemcomitans has been the subject of several excellent publications 115–117. Initial investigations with S. gordonii and A. actinomycetemcomitans revealed that the latter is able to utilize the lactic acid produced by S. gordonii as a carbon source, thus providing metabolic complementation and potentially promoting virulence 116. The dilemma for A. actinomycetemcomitans is its susceptibility to the H2O2 produced by S. gordonii. To minimize its H2O2 exposure, A. actinomycetemcomitans increases the production of its catalase gene katA as well as a novel gene dspB 115. DspB is an extracellular enzyme that promotes the dispersal of A. actinomycetemcomitans biofilms by hydrolyzing the major component of its extracellular matrix, poly-N-acetylglucosamine. This allows A. actinomycetemcomitans to keep a “safe” distance from S. gordonii in mixed species murine abscesses. Normally, A. actinomycetemcomitans and S. gordonii both form small aggregates of about 250–1000 cells in experimental abscesses, while those aggregates increased about two-fold in an A. actinomycetemcomitans DspB mutant 115. Moreover, the DspB mutant aggregates were either contiguous to or within 4 µm of those of S. gordonii, while the wild type maintained a distance between 4 and 13 µm from the S. gordonii aggregates. Presumably, this distance provides the optimal balance between the benefits provided by carbon source acquisition and the limitations of its catalase activity 115. In a separate mixed species study of A. actinomycetemcomitans and S. parasanguinis, it was demonstrated that a close proximity between both species can be maintained by triggering a decrease in spxB expression 118. In fact, dual species biofilms exhibited a significant increase in bio-volume and biofilm thickness as compared to mono-species biofilms, while both species remained intimately associated. Interestingly, A. actinomycetemcomitans catalase did not play an important role in this assay, presumably because the down regulation of spxB limited H2O2 production to a level at which katA expression is unnecessary.
A similar phenomenon has been reported for mixed species biofilms of S. gordonii and A. naeslundii 119. Like A. actinomycetemcomitans, A. naeslundii is very susceptible to inhibition by H2O2 and encodes catalase. Interestingly, in co-cultures with A. naeslundii, S. gordonii exhibits reduced oxidative damage to its surface proteins, which is likely a benefit provided in trans from A. naeslundii catalase 119. Thus, certain combinations of species might prove beneficial to H2O2 producers by minimizing the self-toxicity associated with their H2O2 production. Taken together, multiple lines of evidence support the role of H2O2 in shaping the ecology within the polymicrobial dental biofilm through both pathobiont inhibition and interspecies interactions. Further knowledge of the biogeographical landscape of the numerous polymicrobial physical 12 and chemical interactions 120 will be required to fully understand the dynamics of these interactions and how they may influence the balance between symbiosis and dysbiosis.
Genetic regulation of streptococcal spxB expression
The expression of spxB is under the control of several transcriptional regulators that are conserved in oral streptococci and respond to either metabolic or environmental stimuli. Some species- and strain-specific variations exist in their mode of control and it is currently unknown how these different control elements are coordinated in a hierarchy of transcriptional regulation. In addition, a specific regulatory mechanism has yet to be established for the decreased spxB expression observed during the switch from aerobic to anaerobic growth conditions. Likewise, it is currently unclear whether the previously described pH dependence of H2O2 production is the result of transcriptional regulation, post-transcriptional regulation, or an indirect consequence of a metabolic adjustment.
i). CcpA
Oral streptococci have access to and metabolize a large variety of carbohydrates. Glucose is the most important and preferred carbon source, but its concentration in the oral cavity fluctuates as a consequence of the host diet. In the presence of glucose, most streptococci will suppress the expression of genes involved in the utilization of other carbon sources, a process referred to as carbon catabolite repression (CCR) 121. We previously identified distinct spxB transcriptional responses to glucose from S. sanguinis and S. gordonii. S. gordonii follows the classic carbon catabolite repression response and decreases spxB expression in the presence of glucose, while spxB expression remains unchanged in S. sanguinis, although its expression is consistently lower than in S. gordonii overall 105,122,123. Given this difference in regulation, it was surprising to find a consensus sequence for the catabolite control protein A (CcpA) (cre) in both species. CcpA is the main global regulator for carbohydrate metabolism in Gram-positives 121. Interestingly, S. pneumoniae spxB expression is also controlled by CcpA, but is not affected by glucose similar to S. sanguinis 124. The regulatory role of CcpA has been confirmed by mutagenesis, and as expected, spxB expression is increased in the CcpA mutants of both S. sanguinis and S. gordonii, suggesting that S. sanguinis CcpA-dependent control of spxB expression is glucose independent 123. To further explore the regulatory role of CcpA in both species, promoter binding studies were performed and confirmed to depend upon the two cre sites for both species 123. Despite this, it is still not clear why the two species differ in their CCR response for spxB, but we have some evidence indicating that a regulatory mechanism upstream of CcpA is most likely responsible. Furthermore, we observed similar carbohydrate-dependent and carbohydrate-independent spxB control mechanisms from a variety of clinical isolates of different oral streptococci, which indicates that the distinctions between S. gordonii and S. sanguinis are commonly found in other species as well 123.The ecological implications of this differential regulation of spxB are still unclear and this is a subject of active investigation.
ii). SpxR
One of the first spxB transcriptional regulators identified in S. pneumoniae is SpxR. Mutational studies confirmed the role of SpxR as a transcription activator of spxB gene expression. SpxR mutants exhibit decreased H2O2 production and are severely impaired for virulence in a murine infection model 124. The regulatory role of SpxR was also demonstrated in S. sanguinis 125. The domain architecture of SpxR contains a helix-turn-helix motif for DNA binding as well as CBS and HotDog domains. CBS and HotDog domains seem to be ancient and ubiquitous motifs with potential sensory functions monitoring the cellular energy status by binding to AMP, ATP, or S-adenosyl methionine 126. Thus, it has been suggested that SpxR connects spxB expression to the energy and metabolic status of the cell 124,125, but this prediction has yet to be experimentally confirmed and the signal sensed by SpxR remains unknown.
iii). SpxA1 and SpxA2
SpxA1 and SpxA2 are both transcriptional regulators implicated in the control of spxB expression. Both regulators appear conserved among streptococci and fulfill several important functions in oxidative stress tolerance as well as biofilm and competence development. A detailed analysis in S. sanguinis demonstrated that a deletion of spxA1 decreased the production of H2O2 threefold along with a similar reduction in spxB expression. In the spxA2 deletion background, H2O2 production was not found to be significantly affected 127, but this result has been recently challenged 128. Interestingly, SpxA1/2 exhibit significant homology, including a Cys-X-X-Cys motif that has previously been shown to sense the intracellular redox state 129. Further studies are required to better understand the mechanistic role of both transcriptional regulators for spxB regulation.
iv). Two-Component Systems (TCS) VicRK and SptRS
Environmental information is commonly transmitted via TCS signaling in bacteria 130,131. Two S. sanguinis TCS have been previously demonstrated to influence spxB gene expression 132,133. The initial discovery that the TCS VicRK positively regulates spxB expression as well as biofilm formation and the release of extracellular DNA demonstrates that H2O2 production is involved in important cellular developmental processes 133. This is consistent with our previous studies of the connection between spxB expression, H2O2 production, and the induction of extracellular DNA release 134. More recently, the S. sanguinis SptSR TCS has also been demonstrated to control spxB expression 132. In contrast to the VicRK TCS, a deletion of either sptS or sptR increases the production of eDNA and H2O2. While VicR was shown to bind the spxB promoter 133, this was not possible for SptR, although a putative consensus binding site was identified in the spxB promoter 132. Therefore, it is currently unknown whether the regulatory effect of the SptRS TCS is direct or indirect. The latter is a distinct possibility, since the sptR and sptS mutations trigger significant transcriptional changes in the regulatory genes vicR and spxR 132.
Streptococcal H2O2 production at extra-oral locations
The reservoir of oral streptococci is not confined to the oral cavity, as S. dentisani, S. tigurinus, S. oralis, S. oligofermentans, S. mitis, S. infantis, and S. gordonii have all been isolated from a various areas of nasopharynx from healthy adults 135. In addition, it has been suggested that streptococci are generally protective for paranasal sinuses to certain diseases such as chronic rhinosinusitis 136. Although there is currently no experimental evidence of H2O2 production as a key facet of this protection, the availability of oxygen in the nasopharynx would certainly allow for its production.
Oral streptococci are also commonly associated with the lung microbiome, and their presence in patients with either chronic obstructive pulmonary disease (COPD) or cystic fibrosis (CF) has been shown 137,138. For COPD patients, the streptococcal community in the lungs was shown to originate primarily from the oral cavity through aspiration based upon comparisons of the overall similarity of the bronchial, peripheral lung, nasal, and oral microbiomes 138. This suggests that aspiration and colonization by oral streptococci is likely a common occurrence. A recent clinical study found that a pediatric control cohort had a higher percentage of Streptococcus in their broncho-alveolar lavage fluid compared to an age-matched cohort of pediatric CF patients, while the study failed to detect any Streptococcus in adult CF patients 137. Over time, the non-traditional CF taxa like the streptococci seem to be replaced by the traditional CF taxa including Pseudomonas, Staphylococcus, and Stenotrophomonas 137. This observation may indicate that the non-traditional taxa including Streptococcus can colonize better and initially provide some level of protection. It has also been hypothesized that oral streptococci may play an important role in increasing the diversity of the cystic fibrosis microbiome, thus promoting patient stability 139. Mechanistically, this is consistent with the findings by Scoffield and Wu, which demonstrated that oral streptococci successfully interfere with Pseudomonas aeruginosa through H2O2 production 46. Surprisingly, multiple streptococci including S. parasanguinis, S. sanguinis and S. gordonii all failed to inhibit the growth of P. aeruginosa in antagonism assays when using regular growth medium, but exhibited inhibition when nitrite was supplemented. This effect was also SpxB-dependent. In a Drosophila melanogaster infection model, the authors demonstrated that H2O2-producing oral streptococci are similarly protective against P. aeruginosa in the presence of nitrite 46. Nitrite concentrations seem to be elevated in the sputum of CF patients 140 and can be converted to peroxynitrite in the presence of H2O2. It has been suggested that peroxynitrite is potentially internalized through a permease to exert its antimicrobial effect upon P. aeruginosa 46. It is also worth noting that multiple CF studies have detected oral streptococci from the Streptococcus anginosus group (formerly Milleri group) as being strongly associated with CF pathogenesis, namely S. intermedius and S. constellatus 141,142. Coincidently, after searching the annotated genomes available in the Human Oral microbiome Database (HOMD), both S. intermedius and S. constellatus were not found to encode spxB and are thus not expected to be significant H2O2 producers.
In conclusion, oral streptococci are present in multiple sites of the human host. Most likely, their dissemination originates from the oral cavity. In future studies, it would be interesting to determine whether commensal oral streptococci exert similar health protective benefits outside of the oral cavity via their H2O2 production.
Outlook
Hydrogen peroxide production plays an important role in the metabolism of oral streptococci and effects the ecology of dental biofilms. Experimental evidence over the last decade supports a critical role for H2O2 in maintaining oral symbiotic homeostasis. With our deeper understanding of the ecological aspects of oral diseases, it has been suggested that future treatments are likely to remodel dysbiotic communities back to a state of symbiosis with the host 19,20,143. This concept is a promising alternative to conventional therapies, but its translation is only likely to occur with a more thorough understanding of molecular commensalism 20. The manipulation of H2O2 production among the commensal flora is certainly one conceivable strategy to prevent and/or reverse dysbiosis.
Furthermore, a recent systematic review about the diagnostic accuracy of current Caries Risk Assessment (CRA) methods concluded “the analyzed methods may lead to patients with increased risk not being identified” 144. Clinical CRA guidelines approved by the American Dental Association fail to consider the polymicrobial nature of caries and lack any molecular markers sampled directly at the tooth surface where caries occurs 145. The identification of such microbial biomarkers to monitor changes at the tooth surface before the clinical manifestation of caries is detectable would greatly improve early interventions. Currently, the caries disease process cannot be accurately determined in the absence of observable demineralization. Therefore, predictive preclinical biomarkers are clearly desirable and much needed. H2O2 production might prove to be a useful biomarker of the underlying ecology within the oral cavity.
Acknowledgements
This project was supported by NIH grants DE018893, DE022083, and DE023850 to JM and an NIH grant DE021726 to JK.
Footnotes
CONFLICTS OF INTEREST
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- 1.Lloyd-Price J, Abu-Ali G, Huttenhower C. The healthy human microbiome. Genome Med. 2016;8(1):51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marsh PD, Devine DA. How is the development of dental biofilms influenced by the host? J Clin Periodontol. 2011;38 Suppl 11:28–35. [DOI] [PubMed] [Google Scholar]
- 3.Jakubovics NS. Talk of the town: interspecies communication in oral biofilms. Mol Oral Microbiol. 2010;25(1):4–14. [DOI] [PubMed] [Google Scholar]
- 4.Lang ML, Zhu L, Kreth J. Keeping the bad bacteria in check: interactions of the host immune system with oral cavity biofilms. Endodontic Topics. 2012;22(1):17–32. [Google Scholar]
- 5.Chiu L, Bazin T, Truchetet ME, Schaeverbeke T, Delhaes L, Pradeu T. Protective Microbiota: From Localized to Long-Reaching Co-Immunity. Front Immunol. 2017;8:1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zaura E, ten Cate JM. Towards understanding oral health. Caries Res. 2015;49 Suppl 1:55–61. [DOI] [PubMed] [Google Scholar]
- 7.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43(11):5721–5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simon-Soro A, Belda-Ferre P, Cabrera-Rubio R, Alcaraz LD, Mira A. A tissue-dependent hypothesis of dental caries. Caries Res. 2013;47(6):591–600. [DOI] [PubMed] [Google Scholar]
- 9.Sanz M, Beighton D, Curtis MA, et al. Role of microbial biofilms in the maintenance of oral health and in the development of dental caries and periodontal diseases. Consensus report of group 1 of the Joint EFP/ORCA workshop on the boundaries between caries and periodontal disease. J Clin Periodontol. 2017;44 Suppl 18:S5–S11. [DOI] [PubMed] [Google Scholar]
- 10.Valm AM, Mark Welch JL, Rieken CW, et al. Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proc Natl Acad Sci U S A. 2011;108(10):4152–4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zijnge V, van Leeuwen MB, Degener JE, et al. Oral biofilm architecture on natural teeth. PLoS One. 2010;5(2):e9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mark Welch JL, Rossetti BJ, Rieken CW, Dewhirst FE, Borisy GG. Biogeography of a human oral microbiome at the micron scale. Proc Natl Acad Sci U S A. 2016;113(6):E791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diaz PI, Hoare A, Hong BY. Subgingival Microbiome Shifts and Community Dynamics in Periodontal Diseases. J Calif Dent Assoc. 2016;44(7):421–435. [PubMed] [Google Scholar]
- 14.Wake N, Asahi Y, Noiri Y, et al. Temporal dynamics of bacterial microbiota in the human oral cavity determined using an in situ model of dental biofilms. NPJ Biofilms Microbiomes. 2016;2:16018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu L, Kreth J. The role of hydrogen peroxide in environmental adaptation of oral microbial communities. Oxidative medicine and cellular longevity. 2012;2012:717843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hannig M, Joiner A. The structure, function and properties of the acquired pellicle. Monogr Oral Sci. 2006;19:29–64. [DOI] [PubMed] [Google Scholar]
- 17.Wright CJ, Burns LH, Jack AA, et al. Microbial interactions in building of communities. Mol Oral Microbiol. 2013;28(2):83–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kreth J, Merritt J, Qi F. Bacterial and host interactions of oral streptococci. DNA Cell Biol. 2009;28(8):397–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoare A, Marsh PD, Diaz PI. Ecological Therapeutic Opportunities for Oral Diseases. Microbiol Spectr. 2017;5(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kreth J, Giacaman RA, Raghavan R, Merritt J. The road less traveled - defining molecular commensalism with Streptococcus sanguinis. Mol Oral Microbiol. 2017;32(3):181–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wood PM. The potential diagram for oxygen at pH 7. Biochem J. 1988;253(1):287–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahaseth T, Kuzminov A. Potentiation of hydrogen peroxide toxicity: From catalase inhibition to stable DNA-iron complexes. Mutat Res. 2017;773:274–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pryor WA. Oxy-radicals and related species: their formation, lifetimes, and reactions. Annu Rev Physiol. 1986;48:657–667. [DOI] [PubMed] [Google Scholar]
- 24.Goldstein S, Meyerstein D, Czapski G. The Fenton reagents. Free Radic Biol Med. 1993;15(4):435–445. [DOI] [PubMed] [Google Scholar]
- 25.Halliwell B, Gutteridge JMC. Free radicals in biology and medicine. 5th Edition ed: Oxford University Press; 2015. [Google Scholar]
- 26.Winterbourn CC. Toxicity of iron and hydrogen peroxide: the Fenton reaction. Toxicol Lett. 1995;82–83:969–974. [DOI] [PubMed] [Google Scholar]
- 27.Imlay JA. Pathways of oxidative damage. Annu Rev Microbiol. 2003;57:395–418. [DOI] [PubMed] [Google Scholar]
- 28.Mello Filho AC, Hoffmann ME, Meneghini R. Cell killing and DNA damage by hydrogen peroxide are mediated by intracellular iron. Biochem J. 1984;218(1):273–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iqbal M, Sharma SD, Mizote A, Fujisawa M, Okada S. Differential role of hydrogen peroxide and organic hydroperoxides in augmenting ferric nitrilotriacetate (Fe-NTA)-mediated DNA damage: implications for carcinogenesis. Teratog Carcinog Mutagen. 2003;Suppl 1:13–21. [DOI] [PubMed] [Google Scholar]
- 30.Storz G, Imlay JA. Oxidative stress. Curr Opin Microbiol. 1999;2(2):188–194. [DOI] [PubMed] [Google Scholar]
- 31.Imlay JA. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat Rev Microbiol. 2013;11(7):443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brandi G, Sestili P, Pedrini MA, Salvaggio L, Cattabeni F, Cantoni O. The effect of temperature or anoxia on Escherichia coli killing induced by hydrogen peroxide. Mutat Res. 1987;190(4):237–240. [DOI] [PubMed] [Google Scholar]
- 33.Imlay JA, Linn S. Bimodal pattern of killing of DNA-repair-defective or anoxically grown Escherichia coli by hydrogen peroxide. J Bacteriol. 1986;166(2):519–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Linley E, Denyer SP, McDonnell G, Simons C, Maillard JY. Use of hydrogen peroxide as a biocide: new consideration of its mechanisms of biocidal action. J Antimicrob Chemother. 2012;67(7):1589–1596. [DOI] [PubMed] [Google Scholar]
- 35.Rai P, Cole TD, Wemmer DE, Linn S. Localization of Fe(2+) at an RTGR sequence within a DNA duplex explains preferential cleavage by Fe(2+) and H2O2. J Mol Biol. 2001;312(5):1089–1101. [DOI] [PubMed] [Google Scholar]
- 36.Gonzalez-Flecha B, Demple B. Homeostatic regulation of intracellular hydrogen peroxide concentration in aerobically growing Escherichia coli. J Bacteriol. 1997;179(2):382–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barnard JP, Stinson MW. The alpha-hemolysin of Streptococcus gordonii is hydrogen peroxide. Infect Immun. 1996;64(9):3853–3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pesakhov S, Benisty R, Sikron N, et al. Effect of hydrogen peroxide production and the Fenton reaction on membrane composition of Streptococcus pneumoniae. Biochim Biophys Acta. 2007;1768(3):590–597. [DOI] [PubMed] [Google Scholar]
- 39.Seaver LC, Imlay JA. Hydrogen peroxide fluxes and compartmentalization inside growing Escherichia coli. J Bacteriol. 2001;183(24):7182–7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thibessard A, Fernandez A, Gintz B, Leblond-Bourget N, Decaris B. Hydrogen peroxide effects on Streptococcus thermophilus CNRZ368 cell viability. Res Microbiol. 2001;152(6):593–596. [DOI] [PubMed] [Google Scholar]
- 41.Clark JB. Catalase activity in Escherichia coli. J Bacteriol. 1952;64(4):527–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henningham A, Dohrmann S, Nizet V, Cole JN. Mechanisms of group A Streptococcus resistance to reactive oxygen species. FEMS Microbiol Rev. 2015;39(4):488–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu Y, Itzek A, Kreth J. Comparison of genes required for H2O2 resistance in Streptococcus gordonii and Streptococcus sanguinis. Microbiology. 2014;160(Pt 12): 2627–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomas EL, Milligan TW, Joyner RE, Jefferson MM. Antibacterial activity of hydrogen peroxide and the lactoperoxidase-hydrogen peroxide-thiocyanate system against oral streptococci. Infect Immun. 1994;62(2):529–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kreth J, Merritt J, Shi W, Qi F. Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. J Bacteriol. 2005;187(21):7193–7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scoffield JA, Wu H. Oral streptococci and nitrite-mediated interference of Pseudomonas aeruginosa. Infect Immun. 2015;83(1):101–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saito M, Seki M, Iida K, Nakayama H, Yoshida S. A novel agar medium to detect hydrogen peroxide-producing bacteria based on the prussian blue-forming reaction. Microbiol Immunol. 2007;51(9):889–892. [DOI] [PubMed] [Google Scholar]
- 48.Sumioka R, Nakata M, Okahashi N, et al. Streptococcus sanguinis induces neutrophil cell death by production of hydrogen peroxide. PLoS One. 2017;12(2):e0172223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carlsson J, Edlund MB. Pyruvate oxidase in Streptococcus sanguis under various growth conditions. Oral Microbiol Immunol. 1987;2(1):10–14. [DOI] [PubMed] [Google Scholar]
- 50.Carlsson J, Kujala U. Pyruvate oxidase activity dependent on thiamine pyrophosphate, flavin adenine dinucleotide and orthophosphate in Streptococcus sanguis. FEMS Microbiology Letters. 1984;25(1):53–56. [Google Scholar]
- 51.Zheng LY, Itzek A, Chen ZY, Kreth J. Oxygen dependent pyruvate oxidase expression and production in Streptococcus sanguinis. Int J Oral Sci. 2011;3(2):82–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carlsson J, Edlund MB, Lundmark SK. Characteristics of a hydrogen peroxide-forming pyruvate oxidase from Streptococcus sanguis. Oral Microbiol Immunol. 1987;2(1):15–20. [DOI] [PubMed] [Google Scholar]
- 53.Biasini M, Bienert S, Waterhouse A, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42(Web Server issue):W252–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muller YA, Schumacher G, Rudolph R, Schulz GE. The refined structures of a stabilized mutant and of wild-type pyruvate oxidase from Lactobacillus plantarum. J Mol Biol. 1994;237(3):315–335. [DOI] [PubMed] [Google Scholar]
- 55.Juan EC, Hoque MM, Hossain MT, et al. The structures of pyruvate oxidase from Aerococcus viridans with cofactors and with a reaction intermediate reveal the flexibility of the active-site tunnel for catalysis. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007;63(Pt 11): 900–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meyer D, Neumann P, Koers E, et al. Unexpected tautomeric equilibria of the carbanion-enamine intermediate in pyruvate oxidase highlight unrecognized chemical versatility of thiamin. Proc Natl Acad Sci U S A. 2012;109(27):10867–10872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wille G, Meyer D, Steinmetz A, Hinze E, Golbik R, Tittmann K. The catalytic cycle of a thiamin diphosphate enzyme examined by cryocrystallography. Nat Chem Biol. 2006;2(6):324–328. [DOI] [PubMed] [Google Scholar]
- 58.Tittmann K, Golbik R, Ghisla S, Hubner G. Mechanism of elementary catalytic steps of pyruvate oxidase from Lactobacillus plantarum. Biochemistry. 2000;39(35):10747–10754. [DOI] [PubMed] [Google Scholar]
- 59.Chen T, Yu WH, Izard J, Baranova OV, Lakshmanan A, Dewhirst FE. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford). 2010;2010:baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grifantini R, Toukoki C, Colaprico A, Gryllos I. Peroxide stimulon and role of PerR in group A Streptococcus. J Bacteriol. 2011;193(23):6539–6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu L, Xu Y, Ferretti JJ, Kreth J. Probing oral microbial functionality--expression of spxB in plaque samples. PLoS One. 2014;9(1):e86685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Belstrom D, Constancias F, Liu Y, et al. Metagenomic and metatranscriptomic analysis of saliva reveals disease-associated microbiota in patients with periodontitis and dental caries. NPJ Biofilms Microbiomes. 2017;3:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stewart PS, Franklin MJ. Physiological heterogeneity in biofilms. Nat Rev Microbiol. 2008;6(3):199–210. [DOI] [PubMed] [Google Scholar]
- 64.Lockridge O, Massey V, Sullivan PA. Mechanism of action of the flavoenzyme lactate oxidase. J Biol Chem. 1972;247(24):8097–8106. [PubMed] [Google Scholar]
- 65.Maeda-Yorita K, Aki K, Sagai H, Misaki H, Massey V. L-lactate oxidase and L-lactate monooxygenase: mechanistic variations on a common structural theme. Biochimie. 1995;77(7–8):631–642. [DOI] [PubMed] [Google Scholar]
- 66.Bao X, de Soet JJ, Tong H, et al. Streptococcus oligofermentans Inhibits Streptococcus mutans in Biofilms at Both Neutral pH and Cariogenic Conditions. PLoS One. 2015;10(6):e0130962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu L, Tong H, Dong X. Function of the pyruvate oxidase-lactate oxidase cascade in interspecies competition between Streptococcus oligofermentans and Streptococcus mutans. Appl Environ Microbiol. 2012;78(7):2120–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tong H, Chen W, Merritt J, Qi F, Shi W, Dong X. Streptococcus oligofermentans inhibits Streptococcus mutans through conversion of lactic acid into inhibitory H2O2: a possible counteroffensive strategy for interspecies competition. Mol Microbiol. 2007;63(3):872–880. [DOI] [PubMed] [Google Scholar]
- 69.Jensen A, Scholz CF, Kilian M. Re-evaluation of the taxonomy of the Mitis group of the genus Streptococcus based on whole genome phylogenetic analyses, and proposed reclassification of Streptococcus dentisani as Streptococcus oralis subsp. dentisani comb. nov., Streptococcus tigurinus as Streptococcus oralis subsp. tigurinus comb. nov., and Streptococcus oligofermentans as a later synonym of Streptococcus cristatus. Int J Syst Evol Microbiol. 2016;66(11):4803–4820. [DOI] [PubMed] [Google Scholar]
- 70.Tong H, Gao X, Dong X. Streptococcus oligofermentans sp. nov., a novel oral isolate from caries-free humans. Int J Syst Evol Microbiol. 2003;53(Pt 4): 1101–1104. [DOI] [PubMed] [Google Scholar]
- 71.Seki M, Iida K, Saito M, Nakayama H, Yoshida S. Hydrogen peroxide production in Streptococcus pyogenes: involvement of lactate oxidase and coupling with aerobic utilization of lactate. J Bacteriol. 2004;186(7):2046–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Taniai H, Iida K, Seki M, et al. Concerted action of lactate oxidase and pyruvate oxidase in aerobic growth of Streptococcus pneumoniae: role of lactate as an energy source. J Bacteriol. 2008;190(10):3572–3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Molla G, Nardini M, Motta P, D’Arrigo P, Panzeri W, Pollegioni L. Aminoacetone oxidase from Streptococcus oligofermentans belongs to a new three-domain family of bacterial flavoproteins. Biochem J. 2014;464(3):387–399. [DOI] [PubMed] [Google Scholar]
- 74.Tong H, Chen W, Shi W, Qi F, Dong X. SO-LAAO, a novel L-amino acid oxidase that enables Streptococcus oligofermentans to outcompete Streptococcus mutans by generating H2O2 from peptone. J Bacteriol. 2008;190(13):4716–4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhou P, Liu L, Tong H, Dong X. Role of operon aaoSo-mutT in antioxidant defense in Streptococcus oligofermentans. PLoS One. 2012;7(5):e38133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Boggs JM, South AH, Hughes AL. Phylogenetic analysis supports horizontal gene transfer of L-amino acid oxidase gene in Streptococcus oligofermentans. Infect Genet Evol. 2012;12(5):1005–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nascimento MM, Browngardt C, Xiaohui X, Klepac-Ceraj V, Paster BJ, Burne RA. The effect of arginine on oral biofilm communities. Mol Oral Microbiol. 2014;29(1):45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ge X, Yu Y, Zhang M, et al. Involvement of NADH Oxidase in Competition and Endocarditis Virulence in Streptococcus sanguinis. Infect Immun. 2016;84(5):1470–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Higuchi M, Yamamoto Y, Poole LB, et al. Functions of two types of NADH oxidases in energy metabolism and oxidative stress of Streptococcus mutans. J Bacteriol. 1999;181(19):5940–5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gualdi L, Hayre JK, Gerlini A, et al. Regulation of neuraminidase expression in Streptococcus pneumoniae. BMC Microbiol. 2012;12:200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ashby MT. Inorganic chemistry of defensive peroxidases in the human oral cavity. J Dent Res. 2008;87(10):900–914. [DOI] [PubMed] [Google Scholar]
- 82.Ashby MT, Kreth J, Soundarajan M, Sivuilu LS. Influence of a model human defensive peroxidase system on oral streptococcal antagonism. Microbiology. 2009;155(Pt 11):3691–3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Welk A, Rudolph P, Kreth J, Schwahn C, Kramer A, Below H. Microbicidal efficacy of thiocyanate hydrogen peroxide after adding lactoperoxidase under saliva loading in the quantitative suspension test. Arch Oral Biol. 2011;56(12):1576–1582. [DOI] [PubMed] [Google Scholar]
- 84.Britse A, Lagerlof F. The diluting effect of saliva on the sucrose concentration in different parts of the human mouth after a mouth-rinse with sucrose. Arch Oral Biol. 1987;32(10):755–756. [DOI] [PubMed] [Google Scholar]
- 85.Pruitt KM, Tenovuo J, Mansson-Rahemtulla B, Harrington P, Baldone DC. Is thiocyanate peroxidation at equilibrium in vivo? Biochim Biophys Acta. 1986;870(3):385–391. [DOI] [PubMed] [Google Scholar]
- 86.Liu X, Ramsey MM, Chen X, Koley D, Whiteley M, Bard AJ. Real-time mapping of a hydrogen peroxide concentration profile across a polymicrobial bacterial biofilm using scanning electrochemical microscopy. Proc Natl Acad Sci U S A. 2011;108(7):2668–2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tsou CC, Chiang-Ni C, Lin YS, et al. An iron-binding protein, Dpr, decreases hydrogen peroxide stress and protects Streptococcus pyogenes against multiple stresses. Infect Immun. 2008;76(9):4038–4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yamamoto Y, Higuchi M, Poole LB, Kamio Y. Role of the dpr product in oxygen tolerance in Streptococcus mutans. J Bacteriol. 2000;182(13):3740–3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fujishima K, Kawada-Matsuo M, Oogai Y, Tokuda M, Torii M, Komatsuzawa H. dpr and sod in Streptococcus mutans are involved in coexistence with S. sanguinis, and PerR is associated with resistance to H2O2. Appl Environ Microbiol. 2013;79(5):1436–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pericone CD, Park S, Imlay JA, Weiser JN. Factors contributing to hydrogen peroxide resistance in Streptococcus pneumoniae include pyruvate oxidase (SpxB) and avoidance of the toxic effects of the fenton reaction. J Bacteriol. 2003;185(23):6815–6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lisher JP, Tsui HT, Ramos-Montanez S, et al. Biological and Chemical Adaptation to Endogenous Hydrogen Peroxide Production in Streptococcus pneumoniae D39. mSphere. 2017;2(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liu Y, Palmer SR, Chang H, Combs AN, Burne RA, Koo H. Differential oxidative stress tolerance of Streptococcus mutans isolates affects competition in an ecological mixed-species biofilm model. Environ Microbiol Rep. 2018;10(1):12–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yesilkaya H, Andisi VF, Andrew PW, Bijlsma JJ. Streptococcus pneumoniae and reactive oxygen species: an unusual approach to living with radicals. Trends Microbiol. 2013;21(4):187–195. [DOI] [PubMed] [Google Scholar]
- 94.Hajishengallis G, Lamont RJ. Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol. 2012;27(6):409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Marsh PD. In Sickness and in Health-What Does the Oral Microbiome Mean to Us? An Ecological Perspective. Adv Dent Res. 2018;29(1):60–65. [DOI] [PubMed] [Google Scholar]
- 96.Zheng X, He J, Wang L, et al. Ecological Effect of Arginine on Oral Microbiota. Sci Rep. 2017;7(1):7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Herrero ER, Slomka V, Bernaerts K, et al. Antimicrobial effects of commensal oral species are regulated by environmental factors. J Dent. 2016;47:23–33. [DOI] [PubMed] [Google Scholar]
- 98.Joshi VS, Sheet PS, Cullin N, Kreth J, Koley D. Real-Time Metabolic Interactions between Two Bacterial Species Using a Carbon-Based pH Microsensor as a Scanning Electrochemical Microscopy Probe. Anal Chem. 2017;89(20):11044–11052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cheng X, Redanz S, Cullin N, et al. Plasticity of the Pyruvate Node Modulates Hydrogen Peroxide Production and Acid Tolerance in Multiple Oral Streptococci. Appl Environ Microbiol. 2018;84(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fux CA, Costerton JW, Stewart PS, Stoodley P. Survival strategies of infectious biofilms. Trends Microbiol. 2005;13(1):34–40. [DOI] [PubMed] [Google Scholar]
- 101.Liu Y, Chu L, Wu F, et al. Influence of pH on inhibition of Streptococcus mutans by Streptococcus oligofermentans. Eur J Oral Sci. 2014;122(1):57–61. [DOI] [PubMed] [Google Scholar]
- 102.Bao X, Yang J, de Soet JJ, et al. Factors Influencing the Competition between Streptococcus oligofermentans and Streptococcus mutans in Dual-Species Biofilms. Caries Res. 2017;51(5):507–514. [DOI] [PubMed] [Google Scholar]
- 103.Kreth J, Zhang Y, Herzberg MC. Streptococcal antagonism in oral biofilms: Streptococcus sanguinis and Streptococcus gordonii interference with Streptococcus mutans. J Bacteriol. 2008;190(13):4632–4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Giacaman RA, Torres S, Gomez Y, Munoz-Sandoval C, Kreth J. Correlation of Streptococcus mutans and Streptococcus sanguinis colonization and ex vivo hydrogen peroxide production in carious lesion-free and high caries adults. Arch Oral Biol. 2015;60(1):154–159. [DOI] [PubMed] [Google Scholar]
- 105.Zheng L, Chen Z, Itzek A, Ashby M, Kreth J. Catabolite control protein A controls hydrogen peroxide production and cell death in Streptococcus sanguinis. J Bacteriol. 2011;193(2):516–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.He J, Hwang G, Liu Y, et al. l-Arginine Modifies the Exopolysaccharide Matrix and Thwarts Streptococcus mutans Outgrowth within Mixed-Species Oral Biofilms. J Bacteriol. 2016;198(19):2651–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kreth J, Zhu L, Merritt J, Shi W, Qi F. Role of sucrose in the fitness of Streptococcus mutans. Oral Microbiol Immunol. 2008;23(3):213–219. [DOI] [PubMed] [Google Scholar]
- 108.Herrero ER, Boon N, Bernaerts K, et al. Clinical concentrations of peroxidases cause dysbiosis in in vitro oral biofilm s. J Periodontal Res. 2018. [DOI] [PubMed] [Google Scholar]
- 109.Sato Y, Yamagishi J, Yamashita R, et al. Inter-Individual Differences in the Oral Bacteriome Are Greater than Intra-Day Fluctuations in Individuals. PLoS One. 2015;10(6):e0131607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Palmer SR, Miller JH, Abranches J, et al. Phenotypic heterogeneity of genomically-diverse isolates of Streptococcus mutans. PLoS One. 2013;8(4):e61358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Huang X, Schulte RM, Burne RA, Nascimento MM. Characterization of the arginolytic microflora provides insights into pH homeostasis in human oral biofilms. Caries Res. 2015;49(2):165–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Huang X, Browngardt CM, Jiang M, Ahn SJ, Burne RA, Nascimento MM. Diversity in Antagonistic Interactions between Commensal Oral Streptococci and Streptococcus mutans. Caries Res. 2018;52(1–2):88–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Curran TM, Lieou J, Marquis RE. Arginine deiminase system and acid adaptation of oral streptococci. Appl Environ Microbiol. 1995;61(12):4494–4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Marquis RE, Bender GR, Murray DR, Wong A. Arginine deiminase system and bacterial adaptation to acid environments. Appl Environ Microbiol. 1987;53(1):198–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Stacy A, Everett J, Jorth P, Trivedi U, Rumbaugh KP, Whiteley M. Bacterial fight-and-flight responses enhance virulence in a polymicrobial infection. Proc Natl Acad Sci U S A. 2014;111(21):7819–7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ramsey MM, Rumbaugh KP, Whiteley M. Metabolite cross-feeding enhances virulence in a model polymicrobial infection. PLoS pathogens. 2011;7(3):e1002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ramsey MM, Whiteley M. Polymicrobial interactions stimulate resistance to host innate immunity through metabolite perception. Proc Natl Acad Sci U S A. 2009;106(5):1578–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Duan D, Scoffield JA, Zhou X, Wu H. Fine-tuned production of hydrogen peroxide promotes biofilm formation of Streptococcus parasanguinis by a pathogenic cohabitant Aggregatibacter actinomycetemcomitans. Environ Microbiol. 2016;18(11):4023–4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jakubovics NS, Gill SR, Vickerman MM, Kolenbrander PE. Role of hydrogen peroxide in competition and cooperation between Streptococcus gordonii and Actinomyces naeslundii. FEMS microbiology ecology. 2008;66(3):637–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Edlund A, Garg N, Mohimani H, et al. Metabolic Fingerprints from the Human Oral Microbiome Reveal a Vast Knowledge Gap of Secreted Small Peptidic Molecules. mSystems. 2017;2(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Deutscher J The mechanisms of carbon catabolite repression in bacteria. Curr Opin Microbiol. 2008;11(2):87–93. [DOI] [PubMed] [Google Scholar]
- 122.Zheng L, Itzek A, Chen Z, Kreth J. Environmental influences on competitive hydrogen peroxide production in Streptococcus gordonii. Appl Environ Microbiol. 2011;77(13):4318–4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Redanz S, Masilamani R, Cullin N, Giacaman RA, Merritt J, Kreth J. Distinct Regulatory Role of Carbon Catabolite Protein A (CcpA) in Oral Streptococcal spxB Expression. J Bacteriol. 2018;200(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ramos-Montanez S, Tsui HC, Wayne KJ, et al. Polymorphism and regulation of the spxB (pyruvate oxidase) virulence factor gene by a CBS-HotDog domain protein (SpxR) in serotype 2 Streptococcus pneumoniae. Mol Microbiol. 2008;67(4):729–746. [DOI] [PubMed] [Google Scholar]
- 125.Chen L, Ge X, Dou Y, Wang X, Patel JR, Xu P. Identification of hydrogen peroxide production-related genes in Streptococcus sanguinis and their functional relationship with pyruvate oxidase. Microbiology. 2011;157(Pt 1): 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dillon SC, Bateman A. The Hotdog fold: wrapping up a superfamily of thioesterases and dehydratases. BMC Bioinformatics. 2004;5:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chen L, Ge X, Wang X, Patel JR, Xu P. SpxA1 involved in hydrogen peroxide production, stress tolerance and endocarditis virulence in Streptococcus sanguinis. PLoS One. 2012;7(6):e40034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Li T, Xu M, Zheng L. Is SpxA2 involved in hydrogen peroxide production and competence development in Streptococcus sanguinis? J Med Microbiol. 2017;66(7):981–989. [DOI] [PubMed] [Google Scholar]
- 129.Nakano S, Erwin KN, Ralle M, Zuber P. Redox-sensitive transcriptional control by a thiol/disulphide switch in the global regulator, Spx. Mol Microbiol. 2005;55(2):498–510. [DOI] [PubMed] [Google Scholar]
- 130.Mattos-Graner RO, Duncan MJ. Two-component signal transduction systems in oral bacteria. J Oral Microbiol. 2017;9(1):1400858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. [DOI] [PubMed] [Google Scholar]
- 132.Camargo TM, Stipp RN, Alves LA, Harth-Chu EN, Hofling JF, Mattos-Graner RO. Novel Two-Component System of Streptococcus sanguinis Affecting Functions Associated with Viability in Saliva and Biofilm Formation. Infect Immun. 2018;86(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Moraes JJ, Stipp RN, Harth-Chu EN, Camargo TM, Hofling JF, Mattos-Graner RO. Two-component system VicRK regulates functions associated with establishment of Streptococcus sanguinis in biofilms. Infect Immun. 2014;82(12):4941–4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Itzek A, Zheng L, Chen Z, Merritt J, Kreth J. Hydrogen peroxide-dependent DNA release and transfer of antibiotic resistance genes in Streptococcus gordonii. J Bacteriol. 2011;193(24):6912–6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.De Boeck I, Wittouck S, Wuyts S, et al. Comparing the Healthy Nose and Nasopharynx Microbiota Reveals Continuity As Well As Niche-Specificity. Front Microbiol. 2017;8:2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wilson MT, Hamilos DL. The nasal and sinus microbiome in health and disease. Curr Allergy Asthma Rep. 2014;14(12):485. [DOI] [PubMed] [Google Scholar]
- 137.Zemanick ET, Wagner BD, Robertson CE, et al. Airway microbiota across age and disease spectrum in cystic fibrosis. Eur Respir J. 2017;50(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Pragman AA, Lyu T, Baller JA, et al. The lung tissue microbiota of mild and moderate chronic obstructive pulmonary disease. Microbiome. 2018;6(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Filkins LM, Hampton TH, Gifford AH, et al. Prevalence of streptococci and increased polymicrobial diversity associated with cystic fibrosis patient stability. J Bacteriol. 2012;194(17):4709–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Linnane SJ, Keatings VM, Costello CM, et al. Total sputum nitrate plus nitrite is raised during acute pulmonary infection in cystic fibrosis. Am J Respir Crit Care Med. 1998;158(1):207–212. [DOI] [PubMed] [Google Scholar]
- 141.Sibley CD, Parkins MD, Rabin HR, Duan K, Norgaard JC, Surette MG. A polymicrobial perspective of pulmonary infections exposes an enigmatic pathogen in cystic fibrosis patients. Proc Natl Acad Sci U S A. 2008;105(39):15070–15075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Whelan FJ, Heirali AA, Rossi L, Rabin HR, Parkins MD, Surette MG. Longitudinal sampling of the lung microbiota in individuals with cystic fibrosis. PLoS One. 2017;12(3):e0172811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Nascimento MM. Oral microbiota transplant: a potential new therapy for oral diseases. J Calif Dent Assoc. 2017;45(10):565–568. [PMC free article] [PubMed] [Google Scholar]
- 144.Tellez M, Gomez J, Pretty I, Ellwood R, Ismail AI. Evidence on existing caries risk assessment systems: are they predictive of future caries? Community Dent Oral Epidemiol. 2013;41(1):67–78. [DOI] [PubMed] [Google Scholar]
- 145.American Dental Association. 2017; https://www.ada.org/en/member-center/oral-health-topics/caries-risk-assessment-and-management.
- 146.Tittmann K, Wille G, Golbik R, Weidner A, Ghisla S, Hubner G. Radical phosphate transfer mechanism for the thiamin diphosphate- and FAD-dependent pyruvate oxidase from Lactobacillus plantarum. Kinetic coupling of intercofactor electron transfer with phosphate transfer to acetyl-thiamin diphosphate via a transient FAD semiquinone/hydroxyethyl-ThDP radical pair. Biochemistry. 2005;44(40):13291–13303. [DOI] [PubMed] [Google Scholar]


