Abstract
Spectacular recent progress in structural biology has led to determination of the structures of many integral membrane enzymes that catalyze reactions in which at least one substrate also is membrane bound. A pattern of results seems to be emerging in which the active site chemistry of these enzymes is usually found to be analogous to what is observed for water soluble enzymes catalyzing the same reaction types. However, in light of the chemical, structural, and physical complexity of cellular membranes plus the presence of transmembrane gradients and potentials, it seems likely that these enzymes are subject to membrane-specific regulatory mechanisms that are only now beginning to be uncovered. We review the membrane-specific environmental traits that shape the evolution of membrane-embedded biocatalysts.
Introduction
In the early 1950s Frank Westheimer, William Jencks, and others built on the inspired work of Maud Menten, Leonor Michaelis and other early pioneers of biocatalysis to usher in the classical era of mechanistic enzymology in which the tools of physical organic chemistry were applied to elucidate the chemical and structural basis for enzyme rate acceleration. A high water mark of this era was the mid-1970's introduction of “perfect enzyme theory” by Albery and Knowles, which offered a rigorous conceptual and quantitative reckoning of the energetic hurdles that confront the evolution of enzymes [1,2]. They defined the nature of the energy landscapes in enzyme reaction pathways that, if attained through the process of natural selection, leads to “catalytic perfection”. For a “perfect” enzyme the rate-limiting step of the overall enzyme reaction at physiological substrate concentrations is diffusion of the substrate(s) to the active site. Once this condition is satisfied there is no selective pressure to further evolve the reaction chemistry, substrate affinity, or product release. Understandably, classical enzymology was devoted mainly to the study of water soluble enzymes that catalyze reactions involving soluble substrates. This Opinion is devoted to integral membrane enzymes of biosynthesis, metabolism, and proteolysis that catalyze reactions for which at least one of the substrates is also membrane-associated. The focus is on integral membrane enzymes that function autonomously (rather than as part of multi-protein complexes) and that are not coupled to transport. We also glance at a couple of unusual enzymes where a water soluble catalytic domain is tethered to an integral membrane regulatory domain. We respectfully apologize to our colleagues whose voluminous work on enzymes of the respiratory chain, photosynthesis, and various types of ATPases is not treated herein because of limitations in scope and space.
What unique environmental restraints confront the evolution of membrane enzymes? We offer here a very brief survey, which complements another recent review on membrane biocatalysis. This topic seems timely in light of the remarkable progress in membrane protein structure biology over the past decade, such that there are now more than a mere handful of membrane enzyme structures available (see Stephen White's well-maintained compilation: http://blanco.biomol.uci.edu/mpstruc/). Indeed, the structural biology of membrane enzymes seems, in a great many cases, to have by-passed our knowledge of their mechanisms [3]. In writing this Opinion we acknowledge with admiration that the road from water soluble to integral membrane enzymes is bridged by an impressive body of work on the phospholipases and other soluble enzymes that bind to the membrane surface to execute interfacial catalysis (c.f. [4-9]). An especially important development from these studies is the recognition that enzyme reactions on or in membranes generally conform to the Michaelis-Menten model provided that the concentrations of the enzyme and substrate are treated using membrane mole fraction or related surface concentration units rather than bulk molarity [10].
Membrane enzymes function within a fabulously heterogeneous medium
Integral membrane enzymes can only be removed from the bilayer when it is dissolved. A given biological membrane will contain many dozens, if not hundreds, of chemically distinct lipid molecules [11,12]. This heterogeneity is amplified for the case of eukaryotic membrane enzymes that traffic through more than one organelle, each with a distinct lipid composition. Biological membranes are also asymmetric: the lipid composition on the outer leaflet of a bilayer does not match that on the inner leaflet [11,12]. As is the case for all membrane proteins, membrane enzymes must to some degree be structurally and functionally tolerant of such lipid compositional heterogeneity (see review[13]). At the same time specific protein-lipid interactions may commonly be exploited by evolution as the basis for regulating enzymes. Long-running studies [14-18] of protein-lipid interactions ranging from fleeting solvent-like contacts to stoichiometric complex formation are currently being transformed by mass spectrometry-based approaches for detecting and quantitating specific lipid-protein interactions [19,20].
Lipids and other small molecules in the membrane sometimes play a direct coenzyme role in membrane enzyme chemistry
Membrane-associated small molecules can serve as coenyzmes for membrane enzymes. A key step in chromophore regeneration in the rhodopsin photocycle of vision is the conversion of all-trans 11-retinol back into 11-cis-retinol [21]. Lecithin retinol acyl transferase (LRAT) uses the stored energy of the ester linkage in phosphatidylcholine as the source of the energy that drives this otherwise energetically uphill trans-to-cis double bond isomerization reaction [21,22]. Another example is provided by the lipophilic ubiquinone coenzyme Q, which is used to shuttle electrons across the membrane in various reactions and pathways. These include the reaction in which electrons originating from disulfide bond formation in the periplasm of Gram negative bacteria are transferred from the periplasmic DsbA protein to membrane-embedded DsbB and thence to the freely membrane-diffusible coenzyme Q [23-25]. A similar reaction is catalyzed in the endoplasmic reticulum (ER) membrane by vitamin K epoxide reductase as part of the vitamin K cycle, which is essential for blood coagulation [26-28]. A final example is the exotic isoprenoid lipid dolichol phosphate, with its long polyprenyl tail. This lipid serves as the membrane-anchored covalent scaffold on which oligosaccharides destined for attachment to N-linked glycoproteins are synthesized. The initial reactions to add sugars to the scaffolded glycoside occur on the cytosolic face of the ER membrane. The dolichol/oligosaccharide conjugate is then actively flip-flopped to the luminal face of the membrane, followed by additional reactions to complete biosynthesis of the complex glycoside. This is followed by transferal of the fully elaborated glycoside from the dolichol phosphate head group to the asparagine side chains of nascent N-glycoproteins [29]. The cycle is completed when dolichol phosphate flip-flops back across the membrane for reuse as a scaffold. Undecaprenol phosphate serves an analogous function in related pan-membrane biosynthetic pathways in microbes, such as peptidoglycan biosynthesis [29,30].
Specific lipids are also thought to sometimes play allosteric cofactor roles in regulating membrane enzyme activity (c.f. [31,32]).
Diffusion of membrane enzymes and substrates is quasi-two dimensional, but there are caveats
In an ideal fluid mosaic membrane, proteins bob up and down in the membrane plane and execute rapid axial rotation around their long axes (review in [33]). They typically also undergo lateral 2-D Brownian diffusion in the membrane plane [34,35]. Bulk membrane phases usually approximate the fluid liquid-disordered phase of ideal bilayers, although the effective viscosity of the membrane is higher than in aqueous solution [34,36]. In terms of dictating the rate at which two solutes will bump into each other, the drag of increased viscosity is offset by the reduced dimensionality of the bilayer [34,37,38] such that there is no reason, a priori, to suppose that the rate at which a small molecule substrate in the membrane reaches a membrane enzyme is very different than for a corresponding substrate/enzyme pair in solution.
It is now appreciated that while the fluid mosaic model may apply to a significant fraction of the total area of any real biological membrane, this model is not uniformly applicable across the whole membrane [36,39,40], particularly for the plasma membrane of multicellular organisms. Some enzymes may be associated reversibly or irreversibly with the membrane cytoskeleton and are thereby fixed in the membrane. Even for free molecules, diffusion of both membrane enzymes and substrates may be transiently impeded by barriers or fences in the membrane imposed by the cytoskeleton, connections to the extracellular matrix, tight junctions, large membrane protein complexes, or other fixed molecular assemblies [36,39-41]. Moreover, as noted by Donald Engelman, “membranes are more mosaic than fluid” [42]: as for the cytosol they too represent a protein-crowded milieu. Finally, while the term and actual manifestation of “lipid rafts” in cellular membranes remain controversial [43-46], there seems no doubt that the plasma membranes of many cells contain transient nanodomains composed of certain lipids (particularly cholesterol and sphingolipids and proteins (usually palmitoylated) that do not diffuse or mix freely in the 2-D plane of the membrane. These transient bilayer domains are generally thought to exhibit properties that resemble the liquid-ordered phase, which has been well-characterized in synthetic lipid vesicles [47-49]. If resident in membrane nanodomains, membrane enzymes and substrates are expected to diffuse more slowly than in the bulk membrane phase, exhibit dampened conformational motions, and will interact with a different cohort of lipids and membrane proteins than in the surrounding (fluid phase-like) bulk membrane.
There are reports of various membrane enzymes, including both β-secretase and y-secretase of Alzheimer's disease notoriety, that seem to have an affinity for inclusion in membrane nanodomains [50,51]. In this regard it is notable that the amyloidogenic β-secretase competes for initial cleavage of the full length amyloid precursor protein (APP) with yet another membrane protease, α-secretase, which initiates non-amyloidogenic processing of APP. α-Secretase has been reported to disfavor membrane nanodomains and has been proposed to cleave APP in bulk (disordered) phase membranes [51,52]. The product of the β-secretase reaction, the transmembrane C99 fragment of the APP, was shown to preferentially partition into disordered phase rather than into the liquid-ordered phase of model vesicles [53], but possibly may exhibit more complex preferences in vivo. Combined, these observations suggest that the balance between amyloidogenic and non-amyloidogenic processing of APP may be based, in part, on the differentially-regulated distribution between different membrane domains of full length APP, its C99 fragment, and the three secretases.
Intriguingly, some model membrane studies have also suggested that some membrane proteins appear to preferentially localize to the interface between nanodomains and the surrounding bulk membrane [54,55]. One can speculate that this could be a mechanism sometimes used in nature to generate locally high concentrations of an otherwise surface-dilute enzyme within a membrane.
Membrane enzymes are usually vectorially-oriented
As posited in the original fluid mosaic model, the vast majority of membrane proteins are inserted into the membrane with a near-100% orientational preference [56,57]. The active site of a membrane enzyme with a water soluble substrate will always face either the extracellular/luminal milieu or the cytosol, but almost never both. Lipids and other small molecules often are also preferentially distributed in the inner or outer leaflet, but usually much less exclusively than for membrane protein orientation [11,12]. Depending on the topology of a specific membrane enzyme a good its substrate may have to flip-flop across the bilayer to reach the active site, perhaps as facilitated by a transporter. This scenario appears to apply to microbial diacylglycerol kinase, as will be summarized later in this Opinion. The orientation of a membrane enzyme may also be critical in determining how various transmembrane gradients shape catalysis in cells, as described below.
Membrane enzymes are subject to the influence of transmembrane potentials and gradients
Both the plasma membrane (especially) and the membranes of intracellular organelles delineate transmembrane potentials and gradients, both fixed and variable, that shape the evolution and function of membrane enzymes.
First, most biological membranes serve as boundaries between aqueous phases that have different redox potentials. The cytosol is usually a reducing environment, whereas the lumen of sealed organelles, the periplasm, and the extracellular milieu are usually oxidizing environments. Membrane proteins therefore differ from most soluble proteins in that individual proteins often contain both disulfide bonds and free cysteine residues. Membranes represent an unusually well-suited environment for redox chemistry, in part because membrane enzymes bridge this gradient in redox potential. Membranes are also often subject to gradients in osmolarity and/or pH, gradients that membrane enzymes must, at the very least, adapt to. Finally, some membranes are subject to varying transmembrane electrical potentials that can alter protein structure and function, as is most extensively characterized for voltage-gated ion channels.
The impact of these various transmembrane gradients on membrane enzyme structure, function, and regulation have received scant attention outside of the respiratory chain, active transport, and related bioenergetic systems. In part, this is because it is difficult to establish and manipulate these gradients in the model membrane milieu in which membrane enzymes are often characterized. However, the notion that there may be interesting discoveries waiting to be made in this area is supported by studies of the voltage sensing lipid phosphatase [58]. While the phosphatase domain of this enzyme is water soluble, it is tethered to a transmembrane domain that closely resembles the tetraspan voltage sensor domain present in voltage-gated ion channels [59-61] (Figure 1A). Membrane depolarization activates the phosphatase activity of this enzyme, presumably via electroconformational coupling between the voltage sensor domain and the catalytic domain [60,61]. One wonders how many other enzymes may be regulated by variations in membrane gradients and potentials.
Figure 1.
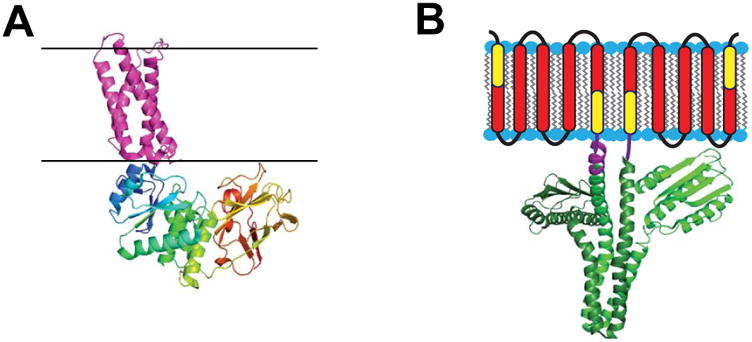
Enzymes for which water soluble catalytic domains are coupled to integral membrane sensor regulatory domains. A) Crystal structure of the voltage-sensor domain of the Ci-VSP phosphatase (4G80) [59] in purple juxtaposed to its cytosolic domain (3AWF) [154] with multicolored chains. Black lines indicate expected membrane boundaries. This figure is adapted from [58]. B) The DesK temperature sensor protein homodimer, with the transmembrane region represented in cartoon form and the cytosolic domain depicted as based on its crystal structure (3GIG) [155], which is that of a non-symmetric dimer. It has been shown that the indicated yellow components of the first and last DesK monomer transmembrane segments can be fused to create a temperature-sensing-functional single pass protein referred to as MS-DesK [78,80]. The linker between the transmembrane and cytosolic domain in each monomer is represented in purple (the crystallographically resolved segment of this linker is longer in the subunit on the left than in the subunit on the right). This figure is adapted from [76].
Membranes have varying lateral pressure, curvature, and thickness
These properties of biological membranes can vary from membrane to membrane or, for curvature and thickness, may vary even within a single membrane. These properties may also be time-dependent and can impact membrane protein structure, folding, and stability [62-68].
It has been shown that the activity of a membrane-associated form of glycerol-3-phosphate dehydrogenase [69] is modulated by membrane lateral pressure. Vmax for this enzyme decreases as the membrane lateral pressure is increased [70]. The activity of leader peptidase has also been shown to vary with changes in membrane lateral pressure [71]. The question of how varying membrane curvature alters integral membrane function has been little-explored. However, given that numerous peripheral membrane enzymes have activities that depend on this membrane trait [72], it will be surprising if many integral enzymes do not as well.
The prokaryotic (integral membrane) diacylglycerol kinase exhibits a catalytic preference for membranes of a particular thickness [73]. Varying membrane thickness appears to regulate the catalytic activity of the microbial DesK histidine kinase, a temperature sensor [74,75] (Figure 1B). DesK has five transmembrane helices, to which is tethered its soluble cytosolic kinase domain. It functions as homodimer. Varying temperature alters the membrane thickness, thereby perturbing the structure of the DesK transmembrane domain in a manner that is transduced to the kinase domain, to regulate its catalytic activity [76,77]. It has been shown that the transmembrane (TM) domain can be whittled down to a single composite TM segment that is able to sense and transduce temperature-dependent changes in membrane thickness to the kinase domain to regulate its catalytic activity [78-80]. Varying membrane thickness is known to shift the distribution of cleavage sites in the amyloid precursor protein by the gamma secretase proteolytic complex, resulting in alteration of the production ratio between the more toxic long forms of the amyloid-beta polypeptide (42-43mers) and the shorter (38-40mer) forms [81-83]. This may be related to the unusual way the substrate for this reaction, the APP C99 domain, adapts topologically to changes in membrane thickness [84]. Major hydrophobic mismatch between the transmembrane domain of a protein and the thickness of the surrounding membrane can be expected to sometimes have an adverse impact on catalytic function (c.f. [85]).
Membrane proteins can distort the membrane
There is a wealth of data that proteins can directly alter membrane curvature and thickness in a regulated manner (c.f. [65,68,86-88]). Among other possible mechanisms, membrane enzymes such as phosphatidylserine decarboxylase likely help to control membrane curvature by altering the ratio between lipids with large head groups such as phosphatidylserine (favoring convex leaflet curvature) and those with smaller head groups, such as phosphatidylethanolamine (c.f. [89]). It has also been proposed that certain membrane enzymes, such as the Gram negative outer membrane phospholipase A, thin the membrane in the vicinity of their transmembrane domains [90], most likely as a mechanism to help promote access of water to active sites in which it participates in the reaction chemistry.
The barriers to water penetration into the interior of the bilayer are anomalously low
Even though water is a highly polar molecule it has a surprisingly high membrane permeability, indicating that the energy barrier for transient excursions deep into the membrane surface is lower than one might guess [11]. This is thought to be because of the very small size of water molecules. Water at or near the membrane surface is thought to have significantly different properties than solvent water [91].
Water-soluble enzymes have active site structures and related dynamic properties that enable them to control water access, excluding from the active site excess water that might otherwise interfere with reaction pathways, while at the same time allowing in water molecules needed to promote substrate binding, reaction chemistry, and product release. It clear that the evolution of membrane enzymes has navigated the same water management issues. Many membrane proteins exhibit bound water molecules in their transmembrane domains [92-94]. Indeed, the introduction of bound water molecules into TM domains appears to be one mechanism used to evolve thermostability in some membrane proteins [95]. At least one membrane enzyme contains a small sealed cavity filled with multiple water molecules that seem to be tapped for use in the reaction cycle (see next section). Some enzymes exhibit aqueous cavities leading to an active site located below the membrane surface (c.f. [96,97]), while still others have aqueous caverns within the transmembrane domain. The Ste24p and ZMPSTE24 CAAX proteases (not to be confused with the architecturally-distinct CAAX protease Rce1) exhibits a barrel-like architecture with transmembrane helices providing the staves [97-101] (Figure 2). Within this barrel is a water-filled cavity large enough for hundreds of water molecules. The active site is located just under an interfacial lid to this barrel, with substrate entry and product exit through fenestrations located near the upper end of the barrel at or just under the water-bilayer interface. The original thinking regarding the substrate specificity of this protease is now in flux [102], but its unusual structure may be related to the facts that (i) it cleaves its substrates near their C-termini but at different numbers of residues from the end from substrate to substrate, (ii) some substrates include attached isoprenoid chains near the cleavage site, and (iii) some substrates are successively cleaved without release of the protein substrate after the first cleavage event, with the two reactions releasing C-terminal peptides of different lengths and different states of posttranslational modification.
Figure 2. The Ste24p CAAX Protease.
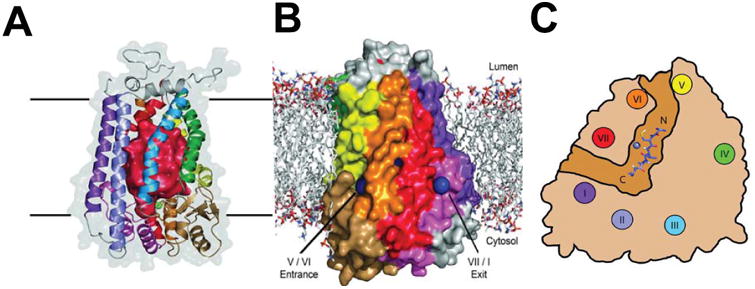
A) Structure of the Ste24p protease shown as a ribbon diagram. The red interior surface illustrates the surfaces of the large water-filled cavity. B) Surface representation of Ste24p in a lipid bilayer). Openings to the substrate binding groove/channel and active site are shown as blue spheres, with the indicated helices corresponding in location and color to those depicted in panel C. C) Cutaway representation of the Ste24p groove/channel viewed looking down on the membrane. The groove contains a 13 residue substrate-derived peptide docked in. These figures are reproduced from Pryor et al. [98] with permission.
We conclude by examining four case studies for membrane enzymes.
Rhomboid protease
Rhomboid is one of several membrane proteases currently under intense study [103,104] and has become a major system for studies of membrane enzyme catalysis, as well as folding and stability [105,106]. Rhomboid proteases are ubiquitous in all domains of life [107,108]. The best characterized forms of rhomboid, such as E. coli GlpG (276 residues) cleave specific single domain membrane protein substrates inside the membrane but near the surface to release the water soluble ectodomain of the substrate. This enzyme usually has 6 transmembrane helices and does not structurally resemble any water soluble protease [109,110], even though its reaction chemistry is similar to that of a classical serine protease [108,111]. Some rhomboids appear to be constitutively active, which implies they are regulated only by substrate availability in the contiguous membrane domain. Rhomboid may thin the adjacent lipid bilayer, facilitating access of water to the active site. Conversely, its activity may in some cases be regulated by hydrophobic mismatch [112-115]. There is also a small cavity in the GlpG rhomboid containing ordered water molecules adjacent to the scissle site. At least one of these ordered waters likely is used during the reaction cycle [116]. Both these internal waters and water contiguous with the aqueous phase are almost certainly subject to controlled access to and exclusion from the active site during the rhomboid reaction.
Recent studies have provided an intriguing body of data supporting a model in which rhomboid appears to have a non-conventional mode of substrate recognition. Potential substrates with a single N-terminal-out topology form a complex with rhomboid, but do not appear to initially engage the active site [117]. If the segment of the bound transmembrane helix located at or near the potential cleavage site transiently unravels (an event that is likely facilitated by water shepherded by rhomboid into the binding site via membrane thinning), the now vulnerable segment is engaged by the actual active site and proteolysis proceeds [117,118]. Transmembrane helices that are completely stable during the lifetime of their complex with rhomboid dissociate from the enzyme unscathed. This “interrogation” of substrate TM helix stability may be the critical determinant of rhomboid substrate specificity. While the lifetime of the interrogation complexes has yet to be measured, the turnover number for cleavage is very slow—an average of 1 cleavage event every 2.5 minutes [119]. Interestingly, good substrates and bad substrates appear to have similar Km, leading one to wonder if non-substrate single span membrane proteins bind to the “interrogation” site with a Kd that equals Km for actual substrates [119]. The substrate Km has been proposed to be orders of magnitude higher than physiological substrate concentrations [119]. Combined, these results suggest that rhomboid binds N-terminal-out single span membrane proteins indiscriminately (with similar Kd values) and only then makes a judgement as to whether or not the bound protein is an actual substrate, a decision based on interrogating helix stability. Rhomboid is therefore unusual as an enzyme in that substrates are distinguished from abundant non-substrates only after complex formation.
It is interesting to note that the seemingly unrelated aspartyl protease gamma-secretase also seems to exhibit an extremely low kcat and substrate Km values that likely are orders of magnitude higher than physiological concentrations [120]. Gamma-secretase functions as a heterotetrameric complex of membrane proteins and has a very broad substrate specificity toward N-terminal-out single span membrane proteins with stubby ectoplasmic domains [121-123]. Like rhomboid, there is evidence that the site of initial engagement between gamma-secretase and its substrates may be adjacent to, but not actually at the active site [124-126]. It also is interesting that addition of hydrophobic small molecules known to modulate the location of the preferred final cleavage site of the APP C99 domain by gamma secretase (“gamma secretase modulators”—GSMs [127,128]) also dramatically expands the substrate specificity of human and Drosophila rhomboids [129]. While the mechanisms underlying these phenomena are not understood, one wonders if some of the unusual mechanistic features of the rhomboids are shared by gamma secretase, despite their vastly different structural properties.
Phosphatidylglycerophosphate phosphatase B (PgpB)
This E. coli form of this enzyme is a member of the type II phosphatidic acid phosphatase (PAP2) family [130,131]. Its common name is a misnomer in that its main function is as a pyrophosphatase that cleaves undecaprenyl pyrophosphate to release the monophosphate form of this carrier lipid to activate its reuse as a membrane-anchored biosynthetic scaffold [132]. PgpB is an integral membrane enzyme with 6 helical transmembrane segments and a V-shaped cleft in its transmembrane domain that leads to an interfacial active site bounded by a small periplasmic domain [133]. What is remarkable is that elements of the PgpB core structure, particularly three transmembrane segments (that include much of the active site), are similar to a trio of helices (and encompassing much of the active site) within the much larger water soluble PAP2 phosphatases, to which PgpB shares weak sequence homology [133] (Figure 3). Nature evidently re-engineered an existing water soluble enzyme into an integral membrane to serve as undecapryl pyrophosphatase rather than evolve a new membrane enzyme for this purpose. This strategy appears to sharply contrast to the approach taken by nature for enzymes such as rhomboid, gamma-secretase, microbial diacylglycerol kinase (below) and the CAAX proteases, which do not appear to have water soluble relatives.
Figure 3. A key structural element of soluble PAP2 phosphatase has been adapted and inserted into the membrane for the PgpB phosphatase.
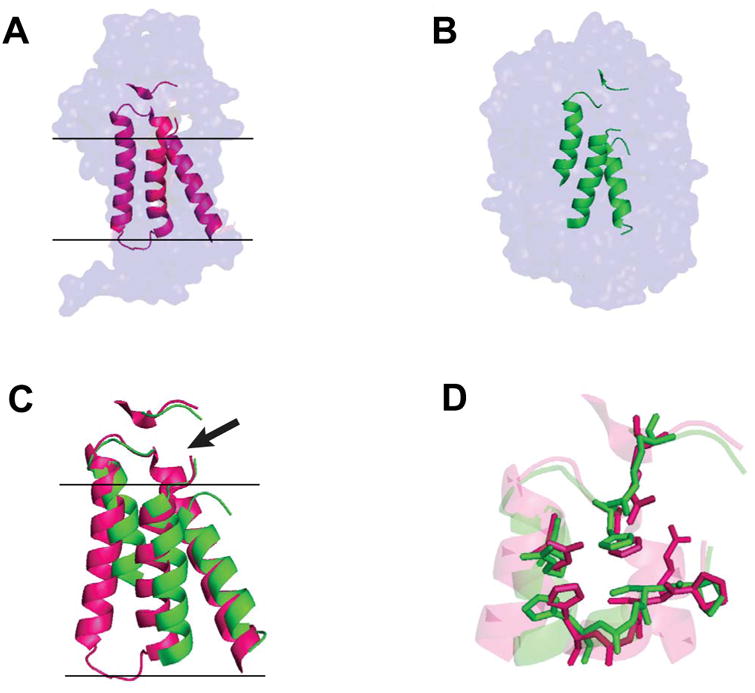
A) Surface representation of the ecPgpB phosphatase crystal structure with key helices highlighted in pink (4PX7) [133]. Black lines indicate estimated membrane boundaries. B) Surface representation of the crystal structure of the water soluble ciCPO PAP2 phosphatase, with key helices highlighted in green. (1VNC) [156]. C) Aligned core helices of membrane-integral ecPgpB (pink) and souble ciCPO (green). Non-core portions of the protein are removed for clarity. The location of the active site is indicated by the arrow. D) Zoom view of the active sites from panel C, with side chains of key catalytic residues now being shown. This figure is adapted from [133].
Monotopic membrane enzymes
Monotopic membrane enzymes have water-exposed catalytic domains that are tightly associated with the membrane via an amphipathic platform that sits deeply in one leaflet of the bilayer. The structures of these platforms vary in details from enzyme to enzyme [134]. These enzymes include prostaglandin H synthase [135,136] (often referred to as cyclooxygenase), fatty acid amide hydrolase [137], and certain cytochrome P450s [138]. The evolutionary strategy underlying the structure-function relationships of these enzymes is to maintain well-evolved water soluble catalytic domains as water-exposed enzymes. However, they have been adapted for catalysis on lipophilic substrate by evolution of a channel leading from the membrane surface up into the enzyme active site located well above the membrane plane in the aqueous phase. These membrane platforms appear not only to anchor the catalytic domain with the correct orientation but in some cases also serves as a septum to seal off water access to the hydrophobic substrate channel leading from the membrane to the active site located well above the membrane surface [134].
Diacylglycerol kinase (DAGK)
Microbial diacylglycerol kinase is the smallest of all kinases—ca. 120 residues—and functions as a homotrimer with three transmembrane domains per subunit and three actives sites—1 at each subunit-subunit interface. It has no homology or structural similarity to the eukaryotic DAGKs, which are water soluble or peripheral membrane enzymes, or to any other kinases [139,140]. Nevertheless its active site chemistry has been shown to be analogous to that observed for water soluble kinases [141]. The convergent evolution of reaction chemistry for water soluble enzymes and for evolutionarily unrelated membrane enzymes seems to be a recurrent theme. The structure of the active form DAGK was determined based on lipidic cubic phase crystallization by the elegant and patient work Li and Caffrey [139,141], superseding the previously published NMR structure of what now is thought to be a heat-inactivated domain-swapped homotrimeric form of the enzyme in DPC micelles [142]. While in some ways DAGK is tolerant of amino acid mutations[143,144], it also has a significant propensity to misfold (c.f. [145] and review in [140]). DAGK has been shown to function after reconstitution into nearly every model membrane system currently available on earth (review in [13,140]). However, it does generally prefer to have at least some lipid present to attain maximal catalytic activity [146-148], with the exception that it is nearly fully active in certain long chain detergents [149]. Activation of DAGK by non-substrate lipid was originally described as reflecting a co-factor role [147,148], but except for its DAG substrate binding site DAGK does not seem to have a discrete high affinity lipid site [139,141].
While the DAGK active site is located at the plasma membrane/cytosol interface, under physiological conditions its substrate, diacylglycerol (DAG), is produced on the other (periplasmic) side of the membrane as a by-product of the membrane-derived oligosaccharide (MDO) cycle of Gram negative bacteria [150] (Figure 4). To reach the active site, DAG must first flip-flop across the membrane to reach the inner leaflet. Because its head group is comprised only of a hydroxyl moiety the spontaneous flip-flop rate of DAG is rather high, roughly 50 per second [151]. DAG flip-flop appears to establish the substrate diffusion rate limit for the DAGK reaction. Based on the physiological concentrations of DAG in E. coli membranes [152] and steady state kinetic analysis of DAGK [153] it has been determined that kcat/Km,DAG DAGK turnover is similar to the DAG flip-flop/diffusion rate to the DAGK active site, indicating that DAGK satisfies the Albery-Knowles definition of being a “perfect” enzyme. However, as such it is still orders of magnitude slower than fully evolved water soluble kinases, which are rate-limited by the much more rapid diffusion of substrates in solution to the active site [153]. It seems that the evolutionary aspirations of DAGK have been limited by the low standard of catalytic perfection imposed by the membrane to its maximum possible reaction rate. As a consequence, except for its sister enzyme, Gram positive undecaprenol kinase, Nature never saw fit to duplicate its gene and then adapt it for paralogous reactions. In a world populated with large and powerful families of kinases, DAGK and undecaprenol kinase remain orphan siblings forever trapped in known metabolic pathways.
Figure 4.
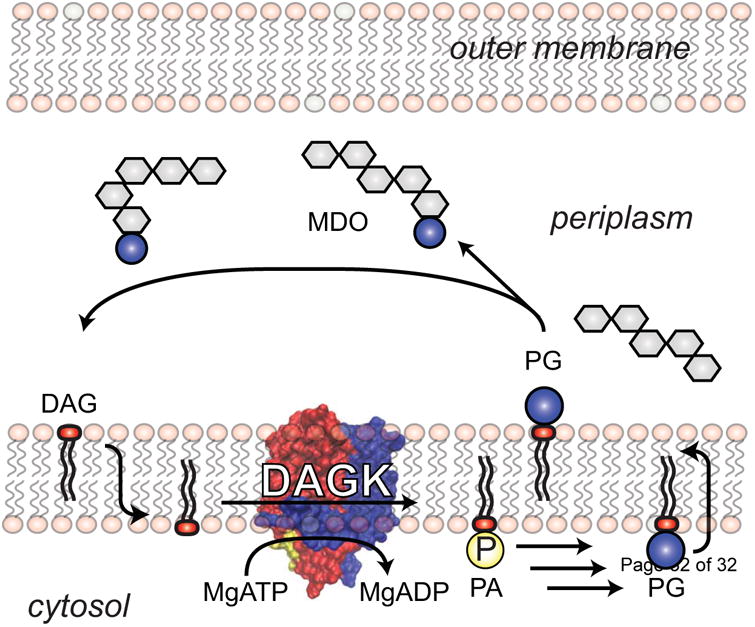
DAGK in the membrane-derived oligosaccharide (MDO) cycle of Gram negative bacteria. Glycerol-1,3-diphosphate is transferred in the periplasm from the head group of phosphatidylglycerol (PG) to decorate the nascent MDOs, with DAG being produced as a byproduct. DAG then flip-flops from the periplasmic to the cytosolic leaflet of the plasma membrane where it is phosphorylated by DAGK to generate phosphatidic acid, which can then be recycled back into PG. The rate of diffusion of DAG to the DAGK active site is thought to be limited by the rate of spontaneous DAG flip-flop to the cytosolic leaflet. DAGK has evolved so as to be able to convert DAG into PA at a rate that roughly matches this lipid translocation event [153]. This figure was adapted from [140].
Conclusions
For many years membrane protein structural biology has been inspired by the notion that the studies of membrane protein structure lag many years behind the study of soluble proteins. But membrane protein structural biology is catching up. We opine that other areas of membrane protein biophysics, which include mechanistic studies of membrane enzymes, may now be critically underdeveloped. While a pattern seems already to have emerged that the active site chemistry of membrane enzymes usually involves variations on themes already established by studies of water soluble enzymes, the frontier most in need of exploration is how membrane enzymes are regulated by varying membrane composition, geometry, biochemical and physical properties, and by transmembrane gradients and potentials. This is especially the case under actual cellular conditions. Given the recent flood of structural information for membrane enzymes and a host of emerging chemical biological and biophysical approaches suitable for examining enzyme function in cells, the time seems ripe for such studies.
Highlights.
Recent progress in the structural biology of integral membrane enzymes has generated a renewal of interest in their mechanisms and modes of regulation.
Membranes contain many distinct traits that shape the evolution of membrane enzymes, in ways that are completely different from water soluble enzymes.
Results so far suggest that membrane enzyme reaction chemistries are largely conventional, but that the membrane-related mechanisms by which these enzymes are regulated are sometimes distinctive and complex.
Our developing understanding of how membrane traits alter membrane enzyme function remains in its infancy.
Acknowledgments
This work was supported by US NIH grants R21 AG026481, R01 GM030910 and R01 AG056147. JMH was support by NIH training grant T32 CA00958229. This Opinion is dedicated to the fond memory of Professor Richard N. Armstrong, who had a long-standing interest in membrane enzymology and was also endearingly opinionated.
Footnotes
Conflict of Interest: The authors declare that they have no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
● of special interest
●● of outstanding interest
- 1.Albery WJ, Knowles JR. Evolution of enzyme function and the development of catalytic efficiency. Biochemistry. 1976;15:5631–5640. doi: 10.1021/bi00670a032. [DOI] [PubMed] [Google Scholar]
- 2.Burbaum JJ, Raines RT, Albery WJ, Knowles JR. Evolutionary optimization of the catalytic effectiveness of an enzyme. Biochemistry. 1989;28:9293–9305. doi: 10.1021/bi00450a009. [DOI] [PubMed] [Google Scholar]
- ●3.Dufrisne MB, Petrou VI, Clarke OB, Mancia F. Structural basis for catalysis at the membrane-water interface. Biochim Biophys Acta. 2017;1862:1368–1385. doi: 10.1016/j.bbalip.2016.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berg OG, Gelb MH, Tsai MD, Jain MK. Interfacial enzymology: the secreted phospholipase A(2)-paradigm. Chem Rev. 2001;101:2613–2654. doi: 10.1021/cr990139w. [DOI] [PubMed] [Google Scholar]
- 5.Cornell RB, Ridgway ND. CTP:phosphocholine cytidylyltransferase: Function, regulation, and structure of an amphitropic enzyme required for membrane biogenesis. Prog Lipid Res. 2015;59:147–171. doi: 10.1016/j.plipres.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Dennis EA, Cao J, Hsu YH, Magrioti V, Kokotos G. Phospholipase A2 enzymes: physical structure, biological function, disease implication, chemical inhibition, and therapeutic intervention. Chem Rev. 2011;111:6130–6185. doi: 10.1021/cr200085w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goni FM, Montes LR, Alonso A. Phospholipases C and sphingomyelinases: Lipids as substrates and modulators of enzyme activity. Prog Lipid Res. 2012;51:238–266. doi: 10.1016/j.plipres.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 8.Hurley JH, Misra S. Signaling and subcellular targeting by membrane-binding domains. Annu Rev Biophys Biomol Struct. 2000;29:49–79. doi: 10.1146/annurev.biophys.29.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roberts MF. Phospholipases: structural and functional motifs for working at an interface. FASEB J. 1996;10:1159–1172. doi: 10.1096/fasebj.10.10.8751718. [DOI] [PubMed] [Google Scholar]
- 10.Carman GM, Deems RA, Dennis EA. Lipid signaling enzymes and surface dilution kinetics. J Biol Chem. 1995;270:18711–18714. doi: 10.1074/jbc.270.32.18711. [DOI] [PubMed] [Google Scholar]
- 11.Gennis RB. Biomembranes : molecular structure and function. Springer-Verlag; New York: 1989. [Google Scholar]
- 12.Luckey M. Membrane structural biology : with biochemical and biophysical foundations. Second. Cambridge University Press; New York: 2014. [Google Scholar]
- 13.Sanders CR, Mittendorf KF. Tolerance to changes in membrane lipid composition as a selected trait of membrane proteins. Biochemistry. 2011;50:7858–7867. doi: 10.1021/bi2011527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee AG. How lipids affect the activities of integral membrane proteins. Biochim Biophys Acta. 2004;1666:62–87. doi: 10.1016/j.bbamem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 15.Lee AG. Biological membranes: the importance of molecular detail. Trends Biochem Sci. 2011;36:493–500. doi: 10.1016/j.tibs.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 16.Marsh D. Protein modulation of lipids, and vice-versa, in membranes. Biochim Biophys Acta. 2008;1778:1545–1575. doi: 10.1016/j.bbamem.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 17.Qin L, Sharpe MA, Garavito RM, Ferguson-Miller S. Conserved lipid-binding sites in membrane proteins: a focus on cytochrome c oxidase. Curr Opin Struct Biol. 2007;17:444–450. doi: 10.1016/j.sbi.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeagle PL. Non-covalent binding of membrane lipids to membrane proteins. Biochim Biophys Acta. 2014;1838:1548–1559. doi: 10.1016/j.bbamem.2013.11.009. [DOI] [PubMed] [Google Scholar]
- ●19.Gupta K, Donlan JAC, Hopper JTS, Uzdavinys P, Landreh M, Struwe WB, Drew D, Baldwin AJ, Stansfeld PJ, Robinson CV. The role of interfacial lipids in stabilizing membrane protein oligomers. Nature. 2017;541:421–424. doi: 10.1038/nature20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laganowsky A, Reading E, Allison TM, Ulmschneider MB, Degiacomi MT, Baldwin AJ, Robinson CV. Membrane proteins bind lipids selectively to modulate their structure and function. Nature. 2014;510:172–175. doi: 10.1038/nature13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sears AE, Palczewski K. Lecithin:Retinol Acyltransferase: A Key Enzyme Involved in the Retinoid (visual) Cycle. Biochemistry. 2016;55:3082–3091. doi: 10.1021/acs.biochem.6b00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rando RR. Membrane phospholipids as an energy source in the operation of the visual cycle. Biochemistry. 1991;30:595–602. doi: 10.1021/bi00217a001. [DOI] [PubMed] [Google Scholar]
- 23.Ito K, Inaba K. The disulfide bond formation (Dsb) system. Curr Opin Struct Biol. 2008;18:450–458. doi: 10.1016/j.sbi.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 24.Zhou Y, Cierpicki T, Jimenez RH, Lukasik SM, Ellena JF, Cafiso DS, Kadokura H, Beckwith J, Bushweller JH. NMR solution structure of the integral membrane enzyme DsbB: functional insights into DsbB-catalyzed disulfide bond formation. Mol Cell. 2008;31:896–908. doi: 10.1016/j.molcel.2008.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sperling LJ, Tang M, Berthold DA, Nesbitt AE, Gennis RB, Rienstra CM. Solid-state NMR study of a 41 kDa membrane protein complex DsbA/DsbB. J Phys Chem B. 2013;117:6052–6060. doi: 10.1021/jp400795d. [DOI] [PubMed] [Google Scholar]
- 26.Li W, Schulman S, Dutton RJ, Boyd D, Beckwith J, Rapoport TA. Structure of a bacterial homologue of vitamin K epoxide reductase. Nature. 2010;463:507–512. doi: 10.1038/nature08720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu S, Cheng W, Fowle Grider R, Shen G, Li W. Structures of an intramembrane vitamin K epoxide reductase homolog reveal control mechanisms for electron transfer. Nat Commun. 2014;5:3110. doi: 10.1038/ncomms4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schulman S, Wang B, Li W, Rapoport TA. Vitamin K epoxide reductase prefers ER membrane-anchored thioredoxin-like redox partners. Proc Natl Acad Sci U S A. 2010;107:15027–15032. doi: 10.1073/pnas.1009972107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hartley MD, Imperiali B. At the membrane frontier: a prospectus on the remarkable evolutionary conservation of polyprenols and polyprenyl-phosphates. Arch Biochem Biophys. 2012;517:83–97. doi: 10.1016/j.abb.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manat G, Roure S, Auger R, Bouhss A, Barreteau H, Mengin-Lecreulx D, Touze T. Deciphering the metabolism of undecaprenyl-phosphate: the bacterial cell-wall unit carrier at the membrane frontier. Microb Drug Resist. 2014;20:199–214. doi: 10.1089/mdr.2014.0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhat BG, Wang P, Coleman RA. Hepatic monoacylglycerol acyltransferase is regulated by sn-1,2- diacylglycerol and by specific lipids in Triton X-100/phospholipid-mixed micelles. J Biol Chem. 1994;269:13172–13178. [PubMed] [Google Scholar]
- 32.Jiang F, Kelly BL, Hagopian K, Greenberg ML. Purification and characterization of phosphatidylglycerolphosphate synthase from Schizosaccharomyces pombe. J Biol Chem. 1998;273:4681–4688. doi: 10.1074/jbc.273.8.4681. [DOI] [PubMed] [Google Scholar]
- 33.Das BB, Park SH, Opella SJ. Membrane protein structure from rotational diffusion. Biochim Biophys Acta. 2015;1848:229–245. doi: 10.1016/j.bbamem.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lenaz G. Lipid fluidity and membrane protein dynamics. Biosci Rep. 1987;7:823–837. doi: 10.1007/BF01119473. [DOI] [PubMed] [Google Scholar]
- 35.Sanderson JM. Resolving the kinetics of lipid, protein and peptide diffusion in membranes. Mol Membr Biol. 2012;29:118–143. doi: 10.3109/09687688.2012.678018. [DOI] [PubMed] [Google Scholar]
- 36.Edidin M. Lipids on the frontier: a century of cell-membrane bilayers. Nat Rev Mol Cell Biol. 2003;4:414–418. doi: 10.1038/nrm1102. [DOI] [PubMed] [Google Scholar]
- 37.Berg OG, von Hippel PH. Diffusion-Controlled Macromolecular Interactions. Annual Review of Biophysics and Biophysical Chemistry. 1985;14:131–160. doi: 10.1146/annurev.bb.14.060185.001023. [DOI] [PubMed] [Google Scholar]
- 38.McCloskey MA, Poo MM. Rates of membrane-associated reactions: reduction of dimensionality revisited. J Cell Biol. 1986;102:88–96. doi: 10.1083/jcb.102.1.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kusumi A, Fujiwara TK, Chadda R, Xie M, Tsunoyama TA, Kalay Z, Kasai RS, Suzuki KG. Dynamic organizing principles of the plasma membrane that regulate signal transduction: commemorating the fortieth anniversary of Singer and Nicolson's fluid-mosaic model. Annu Rev Cell Dev Biol. 2012;28:215–250. doi: 10.1146/annurev-cellbio-100809-151736. [DOI] [PubMed] [Google Scholar]
- 40.Nicolson GL. The Fluid-Mosaic Model of Membrane Structure: still relevant to understanding the structure, function and dynamics of biological membranes after more than 40 years. Biochim Biophys Acta. 2014;1838:1451–1466. doi: 10.1016/j.bbamem.2013.10.019. [DOI] [PubMed] [Google Scholar]
- 41.Ledesma MD, Dotti CG. Peripheral cholesterol, metabolic disorders and Alzheimer's disease. Front Biosci (Elite Ed) 2012;4:181–194. doi: 10.2741/e368. [DOI] [PubMed] [Google Scholar]
- 42.Engelman DM. Membranes are more mosaic than fluid. Nature. 2005;438:578–580. doi: 10.1038/nature04394. [DOI] [PubMed] [Google Scholar]
- 43.Sezgin E, Levental I, Mayor S, Eggeling C. The mystery of membrane organization: composition, regulation and roles of lipid rafts. Nat Rev Mol Cell Biol. 2017;18:361–374. doi: 10.1038/nrm.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bernardino de la Serna J, Schutz GJ, Eggeling C, Cebecauer M. There Is No Simple Model of the Plasma Membrane Organization. Front Cell Dev Biol. 2016;4:106. doi: 10.3389/fcell.2016.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kraft ML. Plasma membrane organization and function: moving past lipid rafts. Mol Biol Cell. 2013;24:2765–2768. doi: 10.1091/mbc.E13-03-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levental I, Veatch S. The Continuing Mystery of Lipid Rafts. J Mol Biol. 2016;428:4749–4764. doi: 10.1016/j.jmb.2016.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Almeida PF. Thermodynamics of lipid interactions in complex bilayers. Biochim Biophys Acta. 2009;1788:72–85. doi: 10.1016/j.bbamem.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 48.Brown DA, London E. Structure and origin of ordered lipid domains in biological membranes. J Membr Biol. 1998;164:103–114. doi: 10.1007/s002329900397. [DOI] [PubMed] [Google Scholar]
- 49.Quinn PJ, Wolf C. The liquid-ordered phase in membranes. Biochim Biophys Acta. 2009;1788:33–46. doi: 10.1016/j.bbamem.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 50.Araki W, Tamaoka A. Amyloid beta-protein and lipid rafts: focused on biogenesis and catabolism. Front Biosci. 2015;20:314–324. doi: 10.2741/4311. [DOI] [PubMed] [Google Scholar]
- 51.Beel AJ, Sakakura M, Barrett PJ, Sanders CR. Direct binding of cholesterol to the amyloid precursor protein: An important interaction in lipid-Alzheimer's disease relationships? Biochim Biophys Acta. 2010;1801:975–982. doi: 10.1016/j.bbalip.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reiss K, Bhakdi S. The plasma membrane: Penultimate regulator of ADAM sheddase function. Biochim Biophys Acta. 2017;1864:2082–2087. doi: 10.1016/j.bbamcr.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 53.Schlebach JP, Barrett PJ, Day CA, Kim JH, Kenworthy AK, Sanders CR. Topologically Diverse Human Membrane Proteins Partition to Liquid-Disordered Domains in Phase-Separated Lipid Vesicles. Biochemistry. 2016;55:985–988. doi: 10.1021/acs.biochem.5b01154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●54.Lorent JH, Levental I. Structural determinants of protein partitioning into ordered membrane domains and lipid rafts. Chem Phys Lipids. 2015;192:23–32. doi: 10.1016/j.chemphyslip.2015.07.022. [DOI] [PubMed] [Google Scholar]
- 55.Yang ST, Kiessling V, Simmons JA, White JM, Tamm LK. HIV gp41-mediated membrane fusion occurs at edges of cholesterol-rich lipid domains. Nat Chem Biol. 2015;11:424–431. doi: 10.1038/nchembio.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bogdanov M, Dowhan W, Vitrac H. Lipids and topological rules governing membrane protein assembly. Biochim Biophys Acta. 2014;1843:1475–1488. doi: 10.1016/j.bbamcr.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee H, Kim H. Membrane topology of transmembrane proteins: determinants and experimental tools. Biochem Biophys Res Commun. 2014;453:268–276. doi: 10.1016/j.bbrc.2014.05.111. [DOI] [PubMed] [Google Scholar]
- 58.Sakata S, Matsuda M, Kawanabe A, Okamura Y. Domain-to-domain coupling in voltage-sensing phosphatase. Biophys Physicobiol. 2017;14:85–97. doi: 10.2142/biophysico.14.0_85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●59.Li Q, Wanderling S, Paduch M, Medovoy D, Singharoy A, McGreevy R, Villalba-Galea CA, Hulse RE, Roux B, Schulten K, et al. Structural mechanism of voltage-dependent gating in an isolated voltage-sensing domain. Nat Struct Mol Biol. 2014;21:244–252. doi: 10.1038/nsmb.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●●60.Murata Y, Iwasaki H, Sasaki M, Inaba K, Okamura Y. Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature. 2005;435:1239–1243. doi: 10.1038/nature03650. [DOI] [PubMed] [Google Scholar]
- 61.Okamura Y, Murata Y, Iwasaki H. Voltage-sensing phosphatase: actions and potentials. J Physiol. 2009;587:513–520. doi: 10.1113/jphysiol.2008.163097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Herrmann M, Danielczak B, Textor M, Klement J, Keller S. Modulating bilayer mechanical properties to promote the coupled folding and insertion of an integral membrane protein. Eur Biophys J. 2015;44:503–512. doi: 10.1007/s00249-015-1032-y. [DOI] [PubMed] [Google Scholar]
- 63.Andersen OS, Koeppe RE., 2nd Bilayer thickness and membrane protein function: an energetic perspective. Annu Rev Biophys Biomol Struct. 2007;36:107–130. doi: 10.1146/annurev.biophys.36.040306.132643. [DOI] [PubMed] [Google Scholar]
- 64.Brown MF. Curvature forces in membrane lipid-protein interactions. Biochemistry. 2012;51:9782–9795. doi: 10.1021/bi301332v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Epand RM. Membrane lipid polymorphism: relationship to bilayer properties and protein function. Methods Mol Biol. 2007;400:15–26. doi: 10.1007/978-1-59745-519-0_2. [DOI] [PubMed] [Google Scholar]
- 66.Jensen MO, Mouritsen OG. Lipids do influence protein function-the hydrophobic matching hypothesis revisited. Biochim Biophys Acta. 2004;1666:205–226. doi: 10.1016/j.bbamem.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 67.Allen SJ, Curran AR, Templer RH, Meijberg W, Booth PJ. Controlling the folding efficiency of an integral membrane protein. J Mol Biol. 2004;342:1293–1304. doi: 10.1016/j.jmb.2004.07.041. [DOI] [PubMed] [Google Scholar]
- 68.McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 69.Yeh JI, Chinte U, Du S. Structure of glycerol-3-phosphate dehydrogenase, an essential monotopic membrane enzyme involved in respiration and metabolism. Proc Natl Acad Sci U S A. 2008;105:3280–3285. doi: 10.1073/pnas.0712331105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Amler E, Jasinska R, Drahota Z, Zborowski J. Membrane lateral pressure as a modulator of glycerol- 3-phosphate dehydrogenase activity. FEBS Lett. 1990;271:165–168. doi: 10.1016/0014-5793(90)80398-3. [DOI] [PubMed] [Google Scholar]
- 71.van den Brink-van der Laan E, Killian JA, de Kruijff B. Nonbilayer lipids affect peripheral and integral membrane proteins via changes in the lateral pressure profile. Biochim Biophys Acta. 2004;1666:275–288. doi: 10.1016/j.bbamem.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 72.Epand RM, D'Souza K, Berno B, Schlame M. Membrane curvature modulation of protein activity determined by NMR. Biochim Biophys Acta. 2015;1848:220–228. doi: 10.1016/j.bbamem.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 73.Pilot JD, East JM, Lee AG. Effects of bilayer thickness on the activity of diacylglycerol kinase of Escherichia coli. Biochemistry. 2001;40:8188–8195. doi: 10.1021/bi0103258. [DOI] [PubMed] [Google Scholar]
- 74.Abriata LA, Albanesi D, Dal Peraro M, de Mendoza D. Signal Sensing and Transduction by Histidine Kinases as Unveiled through Studies on a Temperature Sensor. Acc Chem Res. 2017;50:1359–1366. doi: 10.1021/acs.accounts.6b00593. [DOI] [PubMed] [Google Scholar]
- 75.Saita E, Albanesi D, de Mendoza D. Sensing membrane thickness: Lessons learned from cold stress. Biochim Biophys Acta. 2016;1861:837–846. doi: 10.1016/j.bbalip.2016.01.003. [DOI] [PubMed] [Google Scholar]
- ●76.Inda ME, Vandenbranden M, Fernandez A, de Mendoza D, Ruysschaert JM, Cybulski LE. A lipid-mediated conformational switch modulates the thermosensing activity of DesK. Proc Natl Acad Sci U S A. 2014;111:3579–3584. doi: 10.1073/pnas.1317147111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●77.Saita E, Abriata LA, Tsai YT, Trajtenberg F, Lemmin T, Buschiazzo A, Dal Peraro M, de Mendoza D, Albanesi D. A coiled coil switch mediates cold sensing by the thermosensory protein DesK. Mol Microbiol. 2015;98:258–271. doi: 10.1111/mmi.13118. [DOI] [PubMed] [Google Scholar]
- ●78.Cybulski LE, Ballering J, Moussatova A, Inda ME, Vazquez DB, Wassenaar TA, de Mendoza D, Tieleman DP, Killian JA. Activation of the bacterial thermosensor DesK involves a serine zipper dimerization motif that is modulated by bilayer thickness. Proc Natl Acad Sci U S A. 2015;112:6353–6358. doi: 10.1073/pnas.1422446112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●●79.Inda ME, Oliveira RG, de Mendoza D, Cybulski LE. The Single Transmembrane Segment of Minimal Sensor DesK Senses Temperature via a Membrane-Thickness Caliper. J Bacteriol. 2016;198:2945–2954. doi: 10.1128/JB.00431-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cybulski LE, Martin M, Mansilla MC, Fernandez A, de Mendoza D. Membrane thickness cue for cold sensing in a bacterium. Curr Biol. 2010;20:1539–1544. doi: 10.1016/j.cub.2010.06.074. [DOI] [PubMed] [Google Scholar]
- 81.Holmes O, Paturi S, Ye W, Wolfe MS, Selkoe DJ. Effects of membrane lipids on the activity and processivity of purified gamma-secretase. Biochemistry. 2012;51:3565–3575. doi: 10.1021/bi300303g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lichtenthaler SF, Beher D, Grimm HS, Wang R, Shearman MS, Masters CL, Beyreuther K. The intramembrane cleavage site of the amyloid precursor protein depends on the length of its transmembrane domain. Proc Natl Acad Sci U S A. 2002;99:1365–1370. doi: 10.1073/pnas.032395699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Winkler E, Kamp F, Scheuring J, Ebke A, Fukumori A, Steiner H. Generation of Alzheimer disease-associated amyloid beta42/43 peptide by gamma-secretase can be inhibited directly by modulation of membrane thickness. J Biol Chem. 2012;287:21326–21334. doi: 10.1074/jbc.M112.356659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●84.Song Y, Mittendorf KF, Lu Z, Sanders CR. Impact of bilayer lipid composition on the structure and topology of the transmembrane amyloid precursor C99 protein. J Am Chem Soc. 2014;136:4093–4096. doi: 10.1021/ja4114374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Febo-Ayala W, Morera-Felix SL, Hrycyna CA, Thompson DH. Functional reconstitution of the integral membrane enzyme, isoprenylcysteine carboxyl methyltransferase, in synthetic bolalipid membrane vesicles. Biochemistry. 2006;45:14683–14694. doi: 10.1021/bi061159c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McMahon HT, Boucrot E. Membrane curvature at a glance. J Cell Sci. 2015;128:1065–1070. doi: 10.1242/jcs.114454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Suetsugu S, Kurisu S, Takenawa T. Dynamic shaping of cellular membranes by phospholipids and membrane-deforming proteins. Physiol Rev. 2014;94:1219–1248. doi: 10.1152/physrev.00040.2013. [DOI] [PubMed] [Google Scholar]
- 88.Mitra K, Ubarretxena-Belandia I, Taguchi T, Warren G, Engelman DM. Modulation of the bilayer thickness of exocytic pathway membranes by membrane proteins rather than cholesterol. Proc Natl Acad Sci U S A. 2004;101:4083–4088. doi: 10.1073/pnas.0307332101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chan EY, McQuibban GA. Phosphatidylserine decarboxylase 1 (Psd1) promotes mitochondrial fusion by regulating the biophysical properties of the mitochondrial membrane and alternative topogenesis of mitochondrial genome maintenance protein 1 (Mgm1) J Biol Chem. 2012;287:40131–40139. doi: 10.1074/jbc.M112.399428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wu EL, Fleming PJ, Yeom MS, Widmalm G, Klauda JB, Fleming KG, Im W. E. coli outer membrane and interactions with OmpLA. Biophys J. 2014;106:2493–2502. doi: 10.1016/j.bpj.2014.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Damodaran S. Beyond the hydrophobic effect: Critical function of water at biological phase boundaries--A hypothesis. Adv Colloid Interface Sci. 2015;221:22–33. doi: 10.1016/j.cis.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 92.Miyano M, Ago H, Saino H, Hori T, Ida K. Internally bridging water molecule in transmembrane alpha-helical kink. Curr Opin Struct Biol. 2010;20:456–463. doi: 10.1016/j.sbi.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 93.Orban T, Gupta S, Palczewski K, Chance MR. Visualizing water molecules in transmembrane proteins using radiolytic labeling methods. Biochemistry. 2010;49:827–834. doi: 10.1021/bi901889t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rose A, Theune D, Goede A, Hildebrand PW. MP:PD--a data base of internal packing densities, internal packing defects and internal waters of helical membrane proteins. Nucleic Acids Res. 2014;42:D347–351. doi: 10.1093/nar/gkt1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vaidehi N, Grisshammer R, Tate CG. How Can Mutations Thermostabilize G-Protein-Coupled Receptors? Trends Pharmacol Sci. 2016;37:37–46. doi: 10.1016/j.tips.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●96.Li X, Dang S, Yan C, Gong X, Wang J, Shi Y. Structure of a presenilin family intramembrane aspartate protease. Nature. 2013;493:56–61. doi: 10.1038/nature11801. [DOI] [PubMed] [Google Scholar]
- 97.Manolaridis I, Kulkarni K, Dodd RB, Ogasawara S, Zhang Z, Bineva G, Reilly NO, Hanrahan SJ, Thompson AJ, Cronin N, et al. Mechanism of farnesylated CAAX protein processing by the intramembrane protease Rce1. Nature. 2013;504:301–305. doi: 10.1038/nature12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●●98.Pryor EE, Jr, Horanyi PS, Clark KM, Fedoriw N, Connelly SM, Koszelak-Rosenblum M, Zhu G, Malkowski MG, Wiener MC, Dumont ME. Structure of the integral membrane protein CAAX protease Ste24p. Science. 2013;339:1600–1604. doi: 10.1126/science.1232048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Clark KM, Jenkins JL, Fedoriw N, Dumont ME. Human CaaX protease ZMPSTE24 expressed in yeast: Structure and inhibition by HIV protease inhibitors. Protein Sci. 2017;26:242–257. doi: 10.1002/pro.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Michaelis S, Hrycyna CA. Biochemistry. A protease for the ages. Science. 2013;339:1529–1530. doi: 10.1126/science.1236764. [DOI] [PubMed] [Google Scholar]
- ●●101.Quigley A, Dong YY, Pike AC, Dong L, Shrestha L, Berridge G, Stansfeld PJ, Sansom MS, Edwards AM, Bountra C, et al. The structural basis of ZMPSTE24-dependent laminopathies. Science. 2013;339:1604–1607. doi: 10.1126/science.1231513. [DOI] [PubMed] [Google Scholar]
- ●102.Hildebrandt ER, Arachea BT, Wiener MC, Schmidt WK. Ste24p Mediates Proteolysis of Both Isoprenylated and Non-prenylated Oligopeptides. J Biol Chem. 2016;291:14185–14198. doi: 10.1074/jbc.M116.718197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Langosch D, Scharnagl C, Steiner H, Lemberg MK. Understanding intramembrane proteolysis: from protein dynamics to reaction kinetics. Trends Biochem Sci. 2015;40:318–327. doi: 10.1016/j.tibs.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 104.Sun L, Li X, Shi Y. Structural biology of intramembrane proteases: mechanistic insights from rhomboid and S2P to gamma-secretase. Curr Opin Struct Biol. 2016;37:97–107. doi: 10.1016/j.sbi.2015.12.008. [DOI] [PubMed] [Google Scholar]
- 105.Baker RP, Urban S. Architectural and thermodynamic principles underlying intramembrane protease function. Nat Chem Biol. 2012;8:759–768. doi: 10.1038/nchembio.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Paslawski W, Lillelund OK, Kristensen JV, Schafer NP, Baker RP, Urban S, Otzen DE. Cooperative folding of a polytopic alpha-helical membrane protein involves a compact N-terminal nucleus and nonnative loops. Proc Natl Acad Sci U S A. 2015;112:7978–7983. doi: 10.1073/pnas.1424751112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Freeman M. The rhomboid-like superfamily: molecular mechanisms and biological roles. Annu Rev Cell Dev Biol. 2014;30:235–254. doi: 10.1146/annurev-cellbio-100913-012944. [DOI] [PubMed] [Google Scholar]
- 108.Urban S. A guide to the rhomboid protein superfamily in development and disease. Semin Cell Dev Biol. 2016;60:1–4. doi: 10.1016/j.semcdb.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 109.Brooks CL, Lemieux MJ. Untangling structure-function relationships in the rhomboid family of intramembrane proteases. Biochim Biophys Acta. 2013;1828:2862–2872. doi: 10.1016/j.bbamem.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 110.Wang Y, Zhang Y, Ha Y. Crystal structure of a rhomboid family intramembrane protease. Nature. 2006;444:179–180. doi: 10.1038/nature05255. [DOI] [PubMed] [Google Scholar]
- 111.Vinothkumar KR, Freeman M. Intramembrane proteolysis by rhomboids: catalytic mechanisms and regulatory principles. Curr Opin Struct Biol. 2013;23:851–858. doi: 10.1016/j.sbi.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 112.Reddy T, Rainey JK. Multifaceted substrate capture scheme of a rhomboid protease. J Phys Chem B. 2012;116:8942–8954. doi: 10.1021/jp305077k. [DOI] [PubMed] [Google Scholar]
- 113.Ben-Shem A, Fass D, Bibi E. Structural basis for intramembrane proteolysis by rhomboid serine proteases. Proc Natl Acad Sci U S A. 2007;104:462–466. doi: 10.1073/pnas.0609773104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bondar AN, del Val C, White SH. Rhomboid protease dynamics and lipid interactions. Structure. 2009;17:395–405. doi: 10.1016/j.str.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Foo AC, Harvey BG, Metz JJ, Goto NK. Influence of hydrophobic mismatch on the catalytic activity of Escherichia coli GlpG rhomboid protease. Protein Sci. 2015;24:464–473. doi: 10.1002/pro.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●116.Zhou Y, Moin SM, Urban S, Zhang Y. An internal water-retention site in the rhomboid intramembrane protease GlpG ensures catalytic efficiency. Structure. 2012;20:1255–1263. doi: 10.1016/j.str.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●117.Cho S, Dickey SW, Urban S. Crystal Structures and Inhibition Kinetics Reveal a Two-Stage Catalytic Mechanism with Drug Design Implications for Rhomboid Proteolysis. Mol Cell. 2016;61:329–340. doi: 10.1016/j.molcel.2015.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●●118.Moin SM, Urban S. Membrane immersion allows rhomboid proteases to achieve specificity by reading transmembrane segment dynamics. Elife. 2012;1:e00173. doi: 10.7554/eLife.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●119.Dickey SW, Baker RP, Cho S, Urban S. Proteolysis inside the membrane is a rate-governed reaction not driven by substrate affinity. Cell. 2013;155:1270–1281. doi: 10.1016/j.cell.2013.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●120.Kamp F, Winkler E, Trambauer J, Ebke A, Fluhrer R, Steiner H. Intramembrane proteolysis of beta-amyloid precursor protein by gamma-secretase is an unusually slow process. Biophys J. 2015;108:1229–1237. doi: 10.1016/j.bpj.2014.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Beel AJ, Sanders CR. Substrate specificity of gamma-secretase and other intramembrane proteases. Cell Mol Life Sci. 2008;65:1311–1334. doi: 10.1007/s00018-008-7462-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wolfe MS, Selkoe DJ. gamma-Secretase: a horseshoe structure brings good luck. Cell. 2014;158:247–249. doi: 10.1016/j.cell.2014.06.043. [DOI] [PubMed] [Google Scholar]
- 123.Yang G, Zhou R, Shi Y. Cryo-EM structures of human gamma-secretase. Curr Opin Struct Biol. 2017;46:55–64. doi: 10.1016/j.sbi.2017.05.013. [DOI] [PubMed] [Google Scholar]
- 124.Li S, Zhang W, Han W. Initial Substrate Binding of gamma-Secretase: The Role of Substrate Flexibility. ACS Chem Neurosci. 2017;8:1279–1290. doi: 10.1021/acschemneuro.6b00425. [DOI] [PubMed] [Google Scholar]
- ●125.Li Y, Lu SH, Tsai CJ, Bohm C, Qamar S, Dodd RB, Meadows W, Jeon A, McLeod A, Chen F, et al. Structural interactions between inhibitor and substrate docking sites give insight into mechanisms of human PS1 complexes. Structure. 2014;22:125–135. doi: 10.1016/j.str.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●126.Uemura K, Farner KC, Hashimoto T, Nasser-Ghodsi N, Wolfe MS, Koo EH, Hyman BT, Berezovska O. Substrate docking to gamma-secretase allows access of gamma-secretase modulators to an allosteric site. Nat Commun. 2010;1:130. doi: 10.1038/ncomms1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Crump CJ, Johnson DS, Li YM. Development and mechanism of gamma-secretase modulators for Alzheimer's disease. Biochemistry. 2013;52:3197–3216. doi: 10.1021/bi400377p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Golde TE, Koo EH, Felsenstein KM, Osborne BA, Miele L. gamma-Secretase inhibitors and modulators. Biochim Biophys Acta. 2013;1828:2898–2907. doi: 10.1016/j.bbamem.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●129.Urban S, Moin SM. A subset of membrane-altering agents and gamma-secretase modulators provoke nonsubstrate cleavage by rhomboid proteases. Cell Rep. 2014;8:1241–1247. doi: 10.1016/j.celrep.2014.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sigal YJ, McDermott MI, Morris AJ. Integral membrane lipid phosphatases/phosphotransferases: common structure and diverse functions. Biochem J. 2005;387:281–293. doi: 10.1042/BJ20041771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Stukey J, Carman GM. Identification of a novel phosphatase sequence motif. Protein Sci. 1997;6:469–472. doi: 10.1002/pro.5560060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Touze T, Blanot D, Mengin-Lecreulx D. Substrate specificity and membrane topology of Escherichia coli PgpB, an undecaprenyl pyrophosphate phosphatase. J Biol Chem. 2008;283:16573–16583. doi: 10.1074/jbc.M800394200. [DOI] [PubMed] [Google Scholar]
- ●●133.Fan J, Jiang D, Zhao Y, Liu J, Zhang XC. Crystal structure of lipid phosphatase Escherichia coli phosphatidylglycerophosphate phosphatase B. Proc Natl Acad Sci U S A. 2014;111:7636–7640. doi: 10.1073/pnas.1403097111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bracey MH, Cravatt BF, Stevens RC. Structural commonalities among integral membrane enzymes. FEBS Lett. 2004;567:159–165. doi: 10.1016/j.febslet.2004.04.084. [DOI] [PubMed] [Google Scholar]
- 135.Gupta K, Selinsky BS. Bacterial and algal orthologs of prostaglandin H(2)synthase: novel insights into the evolution of an integral membrane protein. Biochim Biophys Acta. 2015;1848:83–94. doi: 10.1016/j.bbamem.2014.09.011. [DOI] [PubMed] [Google Scholar]
- 136.Smith WL, DeWitt DL, Garavito RM. Cyclooxygenases: structural, cellular, and molecular biology. Annu Rev Biochem. 2000;69:145–182. doi: 10.1146/annurev.biochem.69.1.145. [DOI] [PubMed] [Google Scholar]
- 137.McKinney MK, Cravatt BF. Structure and function of fatty acid amide hydrolase. Annu Rev Biochem. 2005;74:411–432. doi: 10.1146/annurev.biochem.74.082803.133450. [DOI] [PubMed] [Google Scholar]
- 138.Barnaba C, Gentry K, Sumangala N, Ramamoorthy A. The catalytic function of cytochrome P450 is entwined with its membrane-bound nature. F1000Res. 2017;6:662. doi: 10.12688/f1000research.11015.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●139.Li D, Lyons JA, Pye VE, Vogeley L, Aragao D, Kenyon CP, Shah ST, Doherty C, Aherne M, Caffrey M. Crystal structure of the integral membrane diacylglycerol kinase. Nature. 2013;497:521–524. doi: 10.1038/nature12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Van Horn WD, Sanders CR. Prokaryotic diacylglycerol kinase and undecaprenol kinase. Annu Rev Biophys. 2012;41:81–101. doi: 10.1146/annurev-biophys-050511-102330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ●●141.Li D, Stansfeld PJ, Sansom MS, Keogh A, Vogeley L, Howe N, Lyons JA, Aragao D, Fromme P, Fromme R, et al. Ternary structure reveals mechanism of a membrane diacylglycerol kinase. Nat Commun. 2015;6:10140. doi: 10.1038/ncomms10140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Van Horn WD, Kim HJ, Ellis CD, Hadziselimovic A, Sulistijo ES, Karra MD, Tian C, Sonnichsen FD, Sanders CR. Solution nuclear magnetic resonance structure of membrane-integral diacylglycerol kinase. Science. 2009;324:1726–1729. doi: 10.1126/science.1171716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wen J, Chen X, Bowie JU. Exploring the allowed sequence space of a membrane protein. Nat Struct Biol. 1996;3:141–148. doi: 10.1038/nsb0296-141. [DOI] [PubMed] [Google Scholar]
- 144.Zhou Y, Wen J, Bowie JU. A passive transmembrane helix. Nat Struct Biol. 1997;4:986–990. doi: 10.1038/nsb1297-986. [DOI] [PubMed] [Google Scholar]
- 145.Nagy JK, Sanders CR. Destabilizing mutations promote membrane protein misfolding. Biochemistry. 2004;43:19–25. doi: 10.1021/bi035918s. [DOI] [PubMed] [Google Scholar]
- 146.Bohnenberger E, Sandermann H., Jr Lipid dependence of diacylglycerol kinase from Escherichia coli. Eur J Biochem. 1983;132:645–650. doi: 10.1111/j.1432-1033.1983.tb07412.x. [DOI] [PubMed] [Google Scholar]
- 147.Walsh JP, Bell RM. sn-1,2-Diacylglycerol kinase of Escherichia coli. Structural and kinetic analysis of the lipid cofactor dependence. J Biol Chem. 1986;261:15062–15069. [PubMed] [Google Scholar]
- 148.Walsh JP, Bell RM. sn-1,2-Diacylglycerol kinase of Escherichia coli. Mixed micellar analysis of the phospholipid cofactor requirement and divalent cation dependence. J Biol Chem. 1986;261:6239–6247. [PubMed] [Google Scholar]
- 149.Koehler J, Sulistijo ES, Sakakura M, Kim HJ, Ellis CD, Sanders CR. Lysophospholipid micelles sustain the stability and catalytic activity of diacylglycerol kinase in the absence of lipids. Biochemistry. 2010;49:7089–7099. doi: 10.1021/bi100575s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kennedy EP. Sailing to Byzantium. Annu Rev Biochem. 1992;61:1–28. doi: 10.1146/annurev.bi.61.070192.000245. [DOI] [PubMed] [Google Scholar]
- 151.Hamilton JA, Bhamidipati SP, Kodali DR, Small DM. The interfacial conformation and transbilayer movement of diacylglycerols in phospholipid bilayers. J Biol Chem. 1991;266:1177–1186. [PubMed] [Google Scholar]
- 152.Raetz CRH, Newman KF. Diglyceride Kinase Mutants of Escherichia-Coli - Inner Membrane Association of 1,2-Diglyceride and Its Relation to Synthesis of Membrane-Derived Oligosaccharides. Journal of Bacteriology. 1979;137:860–868. doi: 10.1128/jb.137.2.860-868.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Badola P, Sanders CR., 2nd Escherichia coli diacylglycerol kinase is an evolutionarily optimized membrane enzyme and catalyzes direct phosphoryl transfer. J Biol Chem. 1997;272:24176–24182. doi: 10.1074/jbc.272.39.24176. [DOI] [PubMed] [Google Scholar]
- 154.Matsuda M, Takeshita K, Kurokawa T, Sakata S, Suzuki M, Yamashita E, Okamura Y, Nakagawa A. Crystal structure of the cytoplasmic phosphatase and tensin homolog (PTEN)-like region of Ciona intestinalis voltage-sensing phosphatase provides insight into substrate specificity and redox regulation of the phosphoinositide phosphatase activity. J Biol Chem. 2011;286:23368–23377. doi: 10.1074/jbc.M110.214361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Albanesi D, Martin M, Trajtenberg F, Mansilla MC, Haouz A, Alzari PM, de Mendoza D, Buschiazzo A. Structural plasticity and catalysis regulation of a thermosensor histidine kinase. Proc Natl Acad Sci U S A. 2009;106:16185–16190. doi: 10.1073/pnas.0906699106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Messerschmidt A, Wever R. X-ray structure of a vanadium containing enzyme: chloroperoxidase from the fungus Curvularia inaequalis. Proc Natl Acad Sci U S A. 1996;93:392–396. doi: 10.1073/pnas.93.1.392. [DOI] [PMC free article] [PubMed] [Google Scholar]


