Abstract
Periodontitis (PD) is a widespread chronic inflammatory disease in the human population. Porphyromonas gingivalis is associated with PD and can citrullinate host proteins via P. gingivalis peptidyl arginine deiminase (PPAD). Here, we hypothesized that infection of human dental follicle stem cells (hDFSCs) with P. gingivalis and subsequent interaction with neutrophils will alter the neutrophil phenotype. To test this hypothesis, we established and analyzed a triple-culture system of neutrophils and hDFSCs primed with P. gingivalis. Mitogen-activated pathway blocking reagents were applied to gain insight into stem cell signaling after infection. Naïve hDFSCs do not influence the neutrophil phenotype. However, infection of hDFSCs with P. gingivalis prolongs the survival of neutrophils and increases their migration. These phenotypic changes depend on direct cellular contacts and PPAD expression by P. gingivalis. Active JNK and ERK pathways in primed hDFSCs are essential for the phenotypic changes in neutrophils. Collectively, our results confirm that P. gingivalis modifies hDFSCs, thereby causing an immune imbalance.
Keywords: Periodontitis, Bacterial infection, Stem cells, Neutrophils, Viability, Immune modulation, Porphyromonas gingivalis, Peptidyl arginine deiminase, Citrullination
Introduction
Periodontitis (PD) is a prevalent chronic inflammatory condition. Apart from individual consequences for quality of life, affected patients and their treatment create a significant burden for healthcare systems. PD-induced inflammation is sustained by anaerobic bacterial pathogens, e.g., Porphyromonas gingivalis, Aggregatibacter actinomycetemcomitans, and Tannerella forsythia [1, 2]. These species colonize the gingival sulcus and proliferate in the subgingival plaque [3]. Chronic inflammatory host responses in the presence of these bacteria subsequently lead to progressive loss of alveolar bone. If left untreated, this condition can culminate in the loss of teeth [4]. Local and systemic treatment of bacterial infection and inflammation in patients with PD reduces systemic inflammatory markers [5].
The composition of bacterial species is altered in PD, and the Gram-negative bacterium P. gingivalis is especially established in the oral cavity. Dental stem cells are present in the soft oral tissue [6]. The ability of P. gingivalis to adhere to and internalize into host cells, e.g., epithelial cells and stem cells, has been thoroughly described [7, 8]. During adherence and internalization, P. gingivalis modifies host cells via Toll-like receptors or miRNAs [9, 10]. However, the relevance of a peptidyl arginine deiminase (PAD) expressed by P. gingivalis, P. gingivalis peptidyl arginine deiminase (PPAD), as a virulence factor is currently unclear.
Thus far, P. gingivalis is unique due to the presence of an active prokaryotic PPAD; the substrates for its enzymatic activity are bacterial and host proteins [11, 12]. Citrullination is a physiological process in humans and is performed by 5 members of the PAD family. The combination of human and bacterial PAD activity increases the risk for high titers of antibodies against citrullinated peptide antigens (ACPAs). In vivo mouse infection models revealed that infection with the PPAD-deficient strain was associated with significantly attenuated periodontal inflammation and a reduced level of ACPAs in the serum as markers of experimental arthritis [13]. However, the exact molecular mechanisms behind these observations remain unclear.
PPAD is located on the bacterial surface and is anchored into the outer membrane. As a suspected virulence factor, PPAD could be important in interactions with eukaryotic cells, e.g., neutrophils, macrophages, and epithelial cells [14]. Marchant et al. demonstrated that monocytes and macrophages exposed to vital P. gingivalis showed an increase in citrulline in the supernatant after 2 and 24 h [15]. This effect was not based on the influence of PPAD on endogenous PAD expression in infected cells. The exact molecular and cellular mechanisms of PPAD activity in human cells in the oral cavity and subsequent effects on immune cells remain unknown.
P. gingivalis interacts with various types of cells, including stem cells in the oral cavity, which can migrate to the site of infection during tissue inflammation to modulate immune responses [16]. Raffaghello et al. showed that the presence of human bone marrow stem cells (hBMSCs) reduces the apoptosis of neutrophils [17]. Lipopolysaccharide (LPS) stimulation of hBMSCs results in the secretion of various cytokines, chemokines, and growth factors leading to neutrophil recruitment [18]. In an anaerobic co-culture system, we demonstrated that compared with gingival epithelial cells, human dental follicle stem cells (hDFSCs) survive anaerobic conditions and are less affected by infection with oral pathogenic anaerobic microorganisms. The interaction of host cells with periodontal pathogenic bacteria in our previous studies did not result in massive proinflammatory cytokine responses, suggesting an immunomodulatory function of the stem cells [7, 19]. Moreover, reduced neutrophil activity is observed in vitro after infection of dental stem cells with Prevotella intermedia or T. forsythia [8].
Apparently, the presence of vital oral bacteria influences the activity of stem cells and immune cells, although the mechanisms of this action are not fully understood [20]. This lack of information leads to the question of how hDFSCs are affected by PPAD and whether this interaction has an impact on neutrophils.
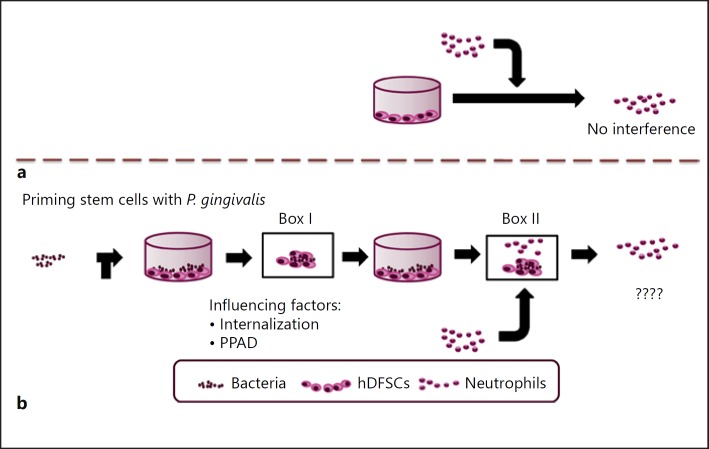
In the periodontal pocket of PD patients, vital bacteria are in close contact with stem cells and immune cells. However, how these interactions work on the cellular and molecular level is currently unclear. There are no convincing data demonstrating how these 3 interacting partners influence each other. As shown in Figure 1, which summarizes our study design and experimental setup, we know that neutrophils are unaffected by stem cells alone (Fig. 1a). Thus, we asked whether an interaction between bacteria and stem cells (Fig. 1b, Box I) modifies the interaction of neutrophils and stem cells primed by bacteria (Fig. 1b, Box II). Various microbial pathobionts present in the oral cavity were analyzed concerning their effects on neutrophils. Thus, the main focus of this study was to elucidate how P. gingivalis influences host stem cells via PPAD expression on the cellular level (Fig. 1b, Box I) and which effects on neutrophils, if any, are detectable (Fig. 1b, Box II).
Fig. 1.
Outline of the study design and setup for investigating the interaction of P. gingivalis with hDFSCs and the resulting effects on neutrophils in a newly established triple-culture system. a Direct interaction of neutrophils and stem cells. b Direct interaction of living P. gingivalis and hDFSCs (Box I) is required for downstream effects on neutrophils (Box II). The question is, which factors in Box I are essential to modify the interaction in Box II?
Materials and Methods
Cells and Culture Conditions
hDFSCs were isolated as described by Haddouti et al. [21]. Wisdom teeth from young volunteers were extracted by the Department of Oral and Maxillofacial Plastic Surgery, University of Rostock. After extraction, dental follicles were washed immediately several times with cold phosphate-buffered saline (PBS) solution containing an increasing concentration of 2–10% penicillin/streptomycin (GIBCO, Carlsbad, CA, USA). The dental follicles were dissected and digested in serum-free Dulbecco's Modified Eagle Medium Nutrient Mixture F-12 (DMEM F-12; Invitrogen, Karlsruhe, Germany) supplemented with 1 mg/mL of Dispase II (Sigma-Aldrich Chemie GmbH, Steinheim, Germany) for 2 h at 37°C. The enzyme mixture was removed, and the tissue was cultured in DMEM F-12 medium containing 10% fetal calf serum (FCS; PAA Laboratories, Cölbe, Germany) at 37°C, 5% CO2.
Comparatively, hBMSCs, isolated as described by Gaebel et al. [22], were provided by the Department of Cardiac Surgery of the University of Rostock. Additionally, the gingival epithelial cell line Ca9–22 was provided by the Japanese Collection of Research Bioresources Cell Bank, Osaka, Japan.
Neutrophils were isolated from healthy young volunteer donors utilizing Erythrocyte Lysis Buffer (EL Buffer; Qiagen, Venlo, The Netherlands) and density gradient centrifugation with Ficoll-Paque PLUS (GE Healthcare Life Sciences, Little Chalfont, UK). The isolation was performed at room temperature. Whole blood (9 mL) was collected from the donors into heparin-coated vacutainers (S-Monovette, Sarstedt, Germany). Venous blood was mixed with EL buffer at a ratio of 1: 5 and incubated for 15 min. Lysed erythrocytes were removed, and cells were washed with fresh EL buffer and PBS. The pellet was resuspended in PBS, layered over 5 mL of Ficoll and centrifuged for 40 min at 400 g. Ficoll-Paque PLUS reagent was removed, and the neutrophils were washed twice with PBS. The cell viability and cell yield were evaluated by Trypan blue staining. The isolation was analyzed for the purity of neutrophils using anti-CD15. Samples were centrifuged (10 min, 300 g), and the pellet was resuspended in 40 µL of neutrophil fluorescence-activated cell sorting (FACS) buffer (1 × PBS supplemented with 0.5% bovine serum albumin and 0.1% NaN3) containing 5 µL of CD15 antibody (ImmunoTools, Friesoythe, Germany). Excess staining was removed, and the samples were resuspended in FACS buffer and analyzed with a flow cytometer, BD Accuri C6 (BD Biosciences, Heidelberg, Germany). The experiments were performed in DMEM (Invitrogen, Karlsruhe, Germany) or Roswell Memorial Park Institute (RPMI) 1640 (Invitrogen, Karlsruhe, Germany) with 10% FCS.
Neutrophils were routinely cultured in RPMI 1640. Nevertheless, a previously established co-culture system demonstrated the suitability of DMEM for stem cell bacteria cultivation [7, 8, 19]. In our experiments, the results of RPMI 1640 incubation of neutrophils were comparable to those of DMEM incubation (data not shown).
Ethics Statement
The experimental protocol and further experiments concerning the isolation of hDFSCs were reviewed and approved by the Ethics Committee of the University of Rostock (https://ethik.med.uni-rostock.de/) using reference number A 2011 91. The isolation of hBMSCs is listed with reference number A 2011 119. The experimental protocols and further use of neutrophils were also approved by the Ethics Committee of the University of Rostock (No. A 2013 0127, No. A 2014 0131). All donors gave their written approval for use of their donated materials in the research performed in this study.
Cultivation of Bacteria
The microorganisms A. actinomycetemcomitans DSMZ 11123, P. gingivalis W83, P. intermedia ATCC 25611, and T. forsythia ATCC 43037 were cultivated in peptone-yeast-glucose broth (PYG) medium at 37°C in an anaerobic atmosphere (10% CO2, 10% H2, 80% N2). PYG medium was supplemented with 5 µg/mL hemin and 1% vitamin K. P. gingivalis W83, Δppad and Δ::ppad were kindly provided by Wegner et al. [12].
Cellular Viability Assay
Cellular counts and the viability of hDFSCs and neutrophils were determined by Trypan blue dye enumeration. Cells were seeded into 24-well plates and exposed to various conditions. After the desired incubation time, hDFSCs were trypsinized and suspended in DMEM with 10% heat-inactivated FCS. hDFSCs and neutrophils were stained with Trypan blue, and viable cells were counted using a Neubauer counting chamber and an inverted light microscope. Additionally, neutrophil apoptosis was measured after Annexin V and 7-amino-actinomycin D (7-AAD) staining by flow cytometry analysis. The viability of neutrophils was analyzed after isolation and the indicated time points of incubation. Neutrophil samples were centrifuged (10 min, 300 g, room temperature) and suspended in 100 µL of 1 × Binding Buffer (eBioscience, Frankfurt, Germany) and 5 µL of Annexin V. Samples were incubated for 15 min in the dark at room temperature. The staining of the samples was removed by centrifugation, and the pellet was resuspended in 1 × PBS supplemented with 0.5% bovine serum albumin and 0.1% NaN3. 7-AAD staining solution was added immediately before measurement.
Co-Culture of Cells and Neutrophils
After isolation, 3 × 105 neutrophils per mL were seeded in 1 mL of DMEM, DMEM/hDFSCs, DMEM/hBMSCs, or DMEM/Ca9–22. Co-cultivation of neutrophils with cells was performed at a ratio of 1: 10. The incubation was performed in an aerobic (5% CO2) or anaerobic atmosphere (10% CO2, 10% H2, and 80% N2) for 24 h. The cell count was analyzed via Trypan blue staining, and the viability of neutrophils was tested via Annexin V/7-AAD staining combined with flow cytometry analysis.
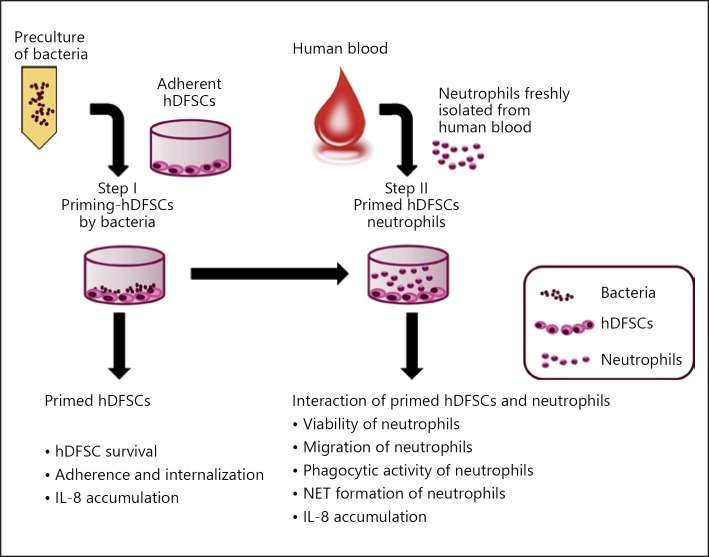
Triple Culture of Oral Microorganism, hDFSCs, and Neutrophils
The concept of triple culture is shown in Figure 2. hDFSCs were preincubated with oral bacteria under anaerobic conditions at a multiplicity of infection equal to 100 (MOI = 100) in DMEM with or without 10% FCS (Step I). For control purposes, cells and oral bacteria were separately incubated in DMEM, respectively. Subsequently, freshly isolated neutrophils were incubated in conditioned media (Step II). Neutrophils were added at a ratio of 1: 10 to hDFSCs.
Fig. 2.
General scheme of the triple-cultivation system setup and the measured parameters at different steps. The triple culture consists of 2 steps of interaction. Step I: hDFSC interaction with bacteria. In a second step, primed hDFSCs were allowed to interact with neutrophils in direct and indirect (2-chamber system) contact experiments (Step II). Unprimed hDFSCs, bacteria, fresh medium, or conditioned DMEM media from the initial co-culture (Step I), which was carried over in the second step, were used as controls. Stem cells were preincubated with oral bacteria under anaerobic conditions at MOI = 100 (Step I). For the direct contact experiments, freshly isolated neutrophils were incubated with hDFSCs at a ratio of 1: 10 under anaerobic conditions (Step II). At each experimental level, several phenotypes were analyzed as denoted in the figure.
Viability of Neutrophils in Triple Culture
To measure the influence of hDFSCs on neutrophil survival, we preincubated hDFSCs with oral bacteria for 24 h under anaerobic conditions (MOI = 100) in DMEM supplemented with 10% FCS. Cells and oral bacteria alone incubated in DMEM over 24 h were used as internal controls. Next, supernatants were sterile filtered by Sterifix 0.2 µm and utilized as conditioned medium. Prior to the co-culture of hDFSCs with neutrophils, the cells were treated with antibiotics for 4 h. After these preparatory steps, freshly isolated neutrophils (I) were incubated in the conditioned media (II) in the presence of untreated and primed stem cells, and (III) in co-culture with primed stem cells using fresh medium. The addition of neutrophils was performed at a ratio of 1: 10 to hDFSCs.
Application of Inhibitors in the Triple-Culture System
After the attachment of hDFSCs in culture plates, inhibitors of the mitogen-activated protein kinase (MAPK) pathway, i.e., polymyxin B (50 µg/mL), SB203580 (10 µM), U0126 (25 µM), SP600125 (25 µM), and Pepinh-MYD (10 µM), were added to the supernatant. After 24 h, a second dose of inhibitor was added 60 min before the infection of hDFSCs. Next, cells were washed, the medium was exchanged, and the cells were primed as described above. Since dimethyl sulfoxide (DMSO) was used as solvent for the inhibitors, control experiments used DMSO only.
Measurement of PPAD Activity
The enzymatic deamination of the substrate N-α-benzoyl-L-arginine ethyl ester (BAEE) was analyzed as described by Takahara et al. [23]. Cells were washed and resuspended in 0.2 M Tris-HCl (pH 8.0), 1 mM EDTA, 1 µM flavin mononucleotide, and 10 mM cysteine. A 10-µL aliquot of BAEE (30 mM) was added to 50-µL samples and incubated at 37 °C for 30 min before the addition of 200 µL of detection reagent. The detection reagent consisted of part A (0.5% diacetyl monoxime and 0.01% thiosemicarbazide) and 2 parts of B (FeCl3 0.25 mg/mL in 24.5% sulfuric acid and 17% phosphoric acid). The reaction plate was heated at 99°C for 25 min, cooled, and optically measured at 490 nm using a spectrophotometer. The standard curve was generated using L-citrulline (0–1,400 µM) to quantify citrulline in all the experimental samples.
Adherence and Internalization
The adherence and internalization assay experiments were performed with various P. gingivalis strains under anaerobic conditions as previously described [7]. hDFSCs were co-cultured with the strains for 2 h. Adherent bacteria were quantified on agar plates after treatment of hDFSCs in deionized water. For internalization, cells were washed twice with PBS after 2 h of infection and treated with antibiotics (penicillin 100 U/mL and streptomycin 0.1 mg/mL) to kill extracellular bacteria. After another 2 h, the cells were treated as described for adherence.
To analyze the involvement of hDFSC cytoskeletal proteins in the observed effects, we performed inhibition experiments using the pharmacological inhibitor latrunculin B. The inhibitor was solved in DMSO. hDFSCs were preincubated with the inhibitor 60 min before infection. Pure DMSO was used in negative control experiments in parallel setups. Subsequently, the cells were washed with PBS, the bacteria were added in fresh medium, and adherence and internalization assays were performed as described above.
Interleukin Concentration in the Supernatant
hDFSCs were co-cultivated with the P. gingivalis strains in DMEM with 10% FCS, and the Δ::ppad strain was further supplemented with 2.5 µg PPAD in a parallel experiment. Unprimed cells with or without PPAD supplementation served as the control. After 24 h, neutrophils were applied to the co-culture. Supernatants were collected after 24 and 48 h and stored at −20°C for analysis of the IL-8 content via an OptEIA Human IL-8 ELISA Kit (BD, Franklin Lakes, NJ, USA). Performance and calibration were accomplished according to the manufacturer's instructions.
Transwell Migration Assay
The Transwell migration assay was modified from Smith et al. [24]. Briefly, Transwell Permeable Support plates (3-µm polyester membrane; Corning) were coated with 2.5 µg/mL of fibrinogen for 1 h, washed twice with sterile PBS, and dried overnight. hDFSCs and P. gingivalis strains were co-cultured (MOI = 100) for 24 h in anaerobic conditions in DMEM. Unstimulated hDFSCs were used as controls. The sterile-filtered bacterial supernatants were placed into the lower wells, and a suspension of 106 freshly isolated neutrophils per milliliter was added into the inserts. After incubation (2 h, aerobic, 37°C), neutrophil migration into the lower well was counted using a Neubauer counting chamber.
Quantitative Phagocytosis
The quantitative phagocytosis assay was adapted from Laheij et al. [25]. Overnight cultures of various P. gingivalis strains were adjusted to 107 microorganisms in DMEM. This suspension was added to adherent hDFSCs (MOI = 100). Freshly isolated neutrophils were added and incubated for 2 h. Viable counts of inoculated bacteria without neutrophils (control) and in the supernatant (extracellular bacteria) of the co-culture setup were determined.
NET Quantification
Freshly isolated neutrophils were incubated anaerobically for 24 h and adjusted to 106 cells per milliliter. The neutrophils were stimulated with 107 microorganisms per milliliter and incubated for 165 min at 37°C in a black 96-well microplate. Neutrophils stimulated with 20 mU/mL glucose oxidase served as a positive control. The quantification of extracellular DNA was performed with Sytox Green (Life Technologies, Carlsbad, CA, USA). The samples were stained with 30 µL of 50 µM Sytox Green and incubated for 15 min at 37°C. The fluorescence was quantified at 485/520 nm in a SpectraMax plate reader (Molecular Devices, Sunnyvale, CA, USA).
Data Analysis
Values are presented as the median, and biological replicates, equal to the number of shown data points, are displayed. The p values were determined by the Mann-Whitney U test or by analysis of variance (ANOVA) for more than 2 groups. Statistical analysis was performed using GraphPad 6.
Results
Stem Cells Do Not Induce Neutrophil Apoptosis
Prior to applying the triple-system setup (bacteria-stem cells-neutrophils) to investigate cellular interactions, numerous control experiments were performed to establish the basic system parameters. We first asked whether stem cells and anaerobic conditions alone interfere with the viability of neutrophils in the co-culture system. hDFSCs and hBMSCs were used as representatives of undifferentiated cells. The gingival epithelial cell line Ca9–22 was used as a differentiated control. Freshly isolated neutrophils were co-cultivated in DMEM for 24 h alone, together with hDFSCs, and with hBMSCs or epithelial cells under aerobic and anaerobic conditions. Signs of apoptosis or necrosis were assessed to document neutrophil viability using flow cytometry. Cells with no indication for either or both conditions were considered viable. The co-cultivation was performed at a ratio of 1: 10 neutrophils to adherent cells.
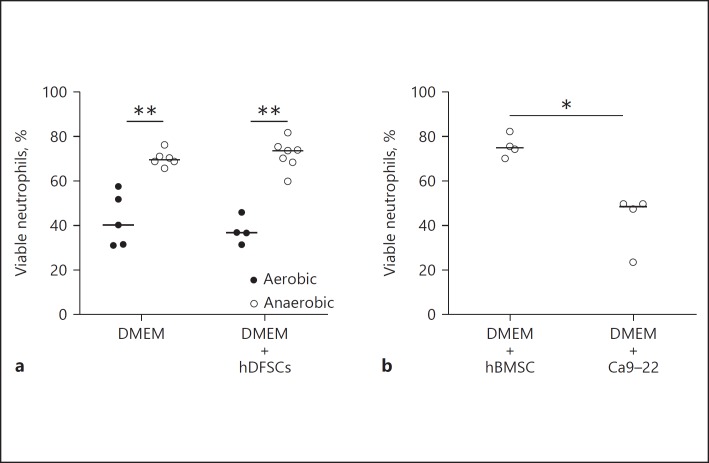
As shown in Figure 3a, 40% of neutrophils survived for 24 h when cultivated under aerobic conditions in the absence of another cell type. Anaerobic incubation in the same setup significantly increased neutrophil viability, reaching 70% viable neutrophils after 24 h. hDFSC-neutrophil co-culture (Fig. 3a) and hBMSC-neutrophil co-culture (Fig. 3b) under anaerobic conditions had no effect on neutrophil viability. By contrast, Ca9–22-neutrophil co-culture (Fig. 3b) significantly attenuated neutrophil viability.
Fig. 3.
Viability of neutrophils under aerobic and anaerobic conditions and co-cultivation with stem cells and epithelial cells. Neutrophils were cultivated for 24 h in DMEM under aerobic or anaerobic conditions. The co-cultivation with cells was performed at a ratio of 1: 10 PMNs. Neutrophil apoptosis was analyzed via Annexin V/7-AAD staining and flow cytometry analysis. The percentage of cells without signs of apoptosis was plotted as the percentage in relation to the total cell numbers. Comparison between aerobic and anaerobic conditions in the presence or absence of hDFSCs (a) and co-culture of neutrophils with hBMSCs and Ca9–22 in anaerobic conditions (b) are demonstrated. The results are displayed as the median; * p < 0.05, ** p < 0.01 were considered significant (Mann-Whitney U test), n ≥ 4 (each data point represents 1 independent biological replicate).
In summary, previous results [7, 8, 19] showed that oxygen limitation does not affect stem cells. The data presented here demonstrate that anaerobiosis had beneficial effects for neutrophils. Our results demonstrate that the setup is suitable for interaction studies with obligate anaerobic oral pathogenic bacteria. Furthermore, naïve stem cells do not affect neutrophil viability under anaerobic conditions. This result was a prerequisite for all follow-up experiments investigating the influence of stem cells on neutrophils after the initial infection of stem cells with bacteria.
P. gingivalis Infection of hDFSCs Increased Neutrophil Viability
Next, we tested whether bacterial infection of stem cells prior to neutrophil contact, as it occurs during the course of periodontal pocket inflammation, affects the behavior of neutrophils in co-culture. To address this question experimentally, we precultured stem cells with oral pathogenic microorganisms (allowing potential adherence and internalization, as well as the influence of secreted bacterial factors). Subsequently, primed stem cells were further co-cultured in conditioned medium with neutrophils to analyze their influence on neutrophil viability. Due to the complexity of the experiments and possible influencing factors, several additional controls were performed. These controls included: (1) co-culture of fresh neutrophils with non-primed hDFSCs (see online suppl. Fig. S1A, for all online suppl. material, see www.karger.com/doi/10.1159/00489020), (2) co-culture of fresh neutrophils with primed cells but in fresh DMEM medium instead of conditioned medium (see online suppl. Fig. S1B), and finally, (3) co-culture of fresh neutrophils in the supernatants of bacteria cultured in DMEM only (see online suppl. Fig. S1A). The results from the control experiment (2) with fresh medium were similar to those with conditioned medium and are shown in the supplementary material (see online suppl. Fig. S1). Control experiments (1) and (3) were used as the basis for data evaluation and statistical calculations of the data points shown in Figure 4.
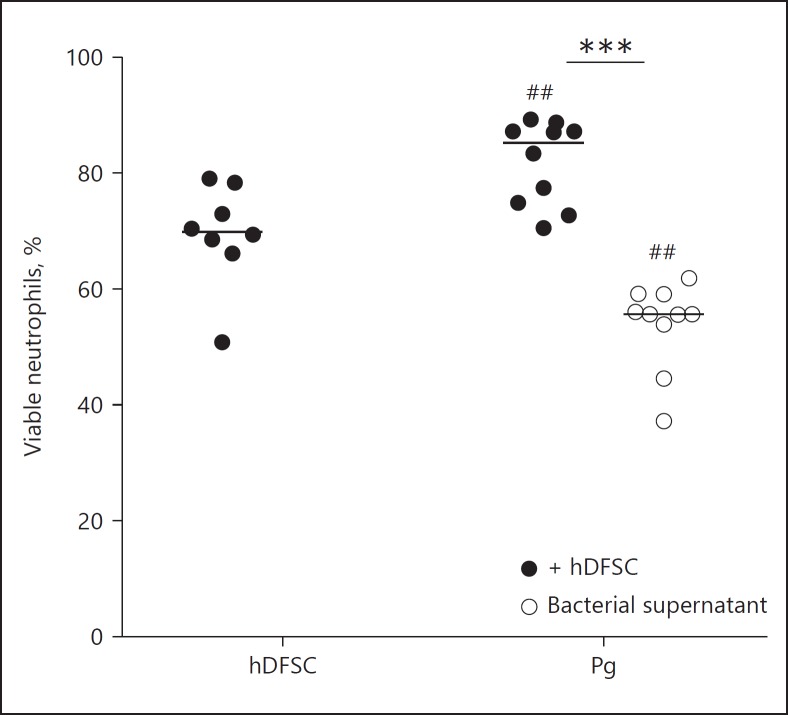
Fig. 4.
hDFSCs primed by P. gingivalis prolong the viability of neutrophils. Analysis of neutrophil viability after co-culture with primed stem cells or in bacterial DMEM supernatant. hDFSCs were co-cultivated with P. gingivalis W83 (Pg) over 24 h under anaerobic conditions, and unprimed hDFSCs and bacteria only were used as a control. After sterile filtration of supernatants and antibiotic treatment, neutrophils were co-cultivated with primed hDFSCs for another 24 h in anaerobic conditions. The apoptosis of neutrophils was analyzed via Annexin V/7-AAD staining and flow cytometry analysis. The percentage of viable cells without signs of apoptosis or necrosis is plotted in the diagram. The results are displayed as the median; *** p < 0.001 was considered significant (ANOVA). Significance of unprimed stem cell control to the co-culture condition: ## p < 0.01, n ≥ 4 in all experiments (each data point represents 1 independent biological replicate).
The results in Figure 4 show that the incubation of neutrophils with hDFSCs (previously primed by P. gingivalis) resulted in similar significantly higher percentages of viable neutrophils (80%). Remarkably, the bacterial supernatant of P. gingivalis without stem cells drastically decreased neutrophil viability (50%; Fig. 4). Apparently, the interaction between P. gingivalis and hDFSCs is required for prolonged neutrophil viability.
Furthermore, compared with the viability of neutrophils co-cultured with unprimed hDFSCs (70%; see online suppl. Fig. S1), the viability of neutrophils co-cultured with the bacterial culture supernatant of A. actinomycetemcomitans was significantly attenuated (55%). Of note, hDFSCs that could preinteract with A. actinomycetemcomitans caused a further reduction in neutrophil viability (33%) in hDFSC-neutrophil co-cultures. Surprisingly, compared with incubation of neutrophils in the hDFSC control, incubation of neutrophils in sterile-filtered DMEM supernatants of T. forsythia or P. intermedia cultures, as well as neutrophil co-culture with hDFSC primed with T. forsythia or P. intermedia, resulted in similar positive effects on neutrophil viability, with significantly increasing numbers of viable neutrophils (85%). Apparently, compared with bacterial supernatants alone, primed hDFSCs do not have further influence on the latter 2 species (see online suppl. Fig. S1).
Together, these experiments revealed that hDFSCs primed with P. gingivalis exerted positive effects on neutrophil viability. The direct interaction of stem cells and P. gingivalis was essential for this positive effect, and similar effects could not be reached by secreted factors from stem cells (conditioned DMEM medium) or bacteria (bacterial DMEM supernatant). hDFSCs primed with A. actinomycetemcomitans decreased neutrophil viability. Moreover, DMEM-conditioned medium from A. actinomycetemcomitans and P. gingivalis significantly decreased neutrophil viability when stem cells were not involved. In summary, bacterially primed hDFSCs can differentially interact with neutrophils, depending on the oral pathogenic bacterial species.
P. gingivalis PAD Supports Neutrophil Viability
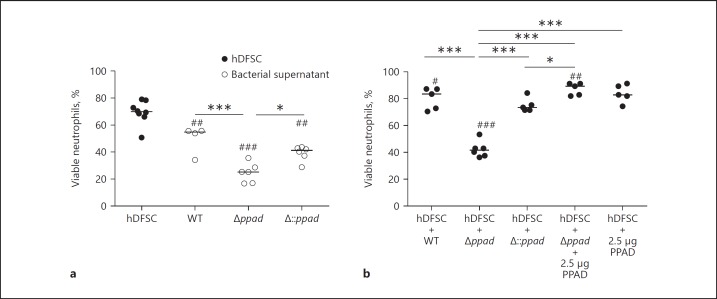
P. gingivalis is a periodontal pathogen and has unique PPAD activity. Thus, we next investigated whether PPAD expression plays a direct role in hDFSC stimulation and ultimately leads to the protective effects on neutrophil survival described in the previous section. To approach this question, we used the P. gingivalis W83 (wild-type; WT) strain, the deletion mutant strain P. gingivalis W83 Δppad (Δppad), and the complemented strain P. gingivalis W83 Δ::ppad (Δ::ppad) in the triple-culture system. Again, several control experiments were required. To this end, neutrophils were cultured with untreated hDFSCs in conditioned medium of all 3 strains. The results are shown in Figure 5a (represented by open circles). Conditioned medium from the P. gingivalis Δ::ppad strain decreased neutrophil viability to 40% and showed comparable results to those of WT P. gingivalis (compare with Fig. 4). In Figure 5a, the deletion of PPAD and the incubation of neutrophils in the mutant supernatant also resulted in significant reductions in neutrophil viability (Δppad 25%). Of note, a significant PPAD-dependent effect was observed in the comparison of the conditioned medium from all 3 P. gingivalis strains (Fig. 4, 5a).
Fig. 5.
P. gingivalis PAD significantly influences the viability of neutrophils. a Control experiments show the viability of neutrophils cultivated with hDFSCs or sterile-filtered P. gingivalis DMEM supernatant. b Viability of neutrophils cultivated with P. gingivalis-primed stem cells. hDFSCs were incubated with P. gingivalis WT, P. gingivalis Δppad, or P. gingivalis Δ::ppad as described in Figure 2 (Step I). Infection experiments of hDFSCs with Δppad strain were supplemented with recombinant PPAD (2.5 µg). The conditioned medium was obtained from hDFSCs primed with respective P. gingivalis strains and sterile filtered. The incubation of neutrophils with untreated hDFSCs as well as the sterile-filtered DMEM supernatant of the various P. gingivalis strains served as controls. The controls were equally incubated over 24 h under anaerobic conditions. The results are displayed as the median; * p < 0.05, *** p < 0.001 were considered significant (ANOVA), #p < 0.05, ## p < 0.01, ### p < 0.001 and both variables refer to the significance of primed stem cells to the respective bacterial supernatant, n ≥ 4 (each data point represents 1 independent biological replicate).
Next, we analyzed the influence of PPAD expression during the presence and interaction of P. gingivalis strains with hDFSCs and elucidated their effects on neutrophil viability. The results of these experiments are presented in Figure 5b (represented by filled circles). Infection of hDFSCs with the WT strain resulted in 80% viable neutrophils after 24 h of anaerobic incubation (as illustrated in Fig. 4). By contrast, the PPAD deletion mutant strain Δppad caused a highly significant and almost 50% reduction in neutrophil viability (down to 40% viable neutrophils). The WT phenotype was restored by genetic complementation. Infection of hDFSCs with the Δ::ppad strain prior to neutrophil contact significantly increased neutrophil viability almost back to 80% (Fig. 5b). This result suggested that PPAD is an important stimulus in the P. gingivalis-hDFSC interaction. This finding led us to ask whether the WT phenotype could be restored by adding recombinant purified PPAD enzyme instead of using a fully genetically complemented strain. To address this question, we supplemented hDFSCs primed with Δppad with recombinant PPAD (2.5 µg). This supplementation led to a full recovery of the WT phenotype (85% viable neutrophils, Fig. 5b). A PPAD concentration-dependent effect was tested beforehand (see online suppl. Fig. S2), and 2.5 µg was sufficient. Recombinant PPAD enzyme in the absence of P. gingivalis did not have any effect on the hDFSC-neutrophil axis (Fig. 5b).
Collectively, our experiments have shown that the viability of neutrophils was significantly higher in co-culture with primed hDFSCs than that in co-culture with bacterial P. gingivalis supernatants. Again, this finding confirms the results shown in Figure 4. More importantly, the presence of PPAD during hDFSC infection with P. gingivalis (Fig. 2, Step I) positively affected neutrophil viability (Fig. 2, Step II), and the lack of PPAD expression in the mutants and its associated phenotypic effect was fully restored by the presence of recombinant PPAD in the initial P. gingivalis-hDFSC interaction.
PPAD Supports Neutrophil Migration
Compared with the presence of the PPAD deletion mutant strain, the presence of PPAD during stem cell infection led to increased viability of neutrophils in primed culture supernatants. This result prompted us to question whether other conceivable neutrophil phenotypes (Fig. 2, Step II) could be influenced by P. gingivalis hDFSC infection (Fig. 2, Step I). Consequently, we next analyzed the differences in neutrophil phagocytic clearance, neutrophil extracellular trap (NET) formation, and chemokine secretion and migration. The results are collectively displayed in Figure 6.
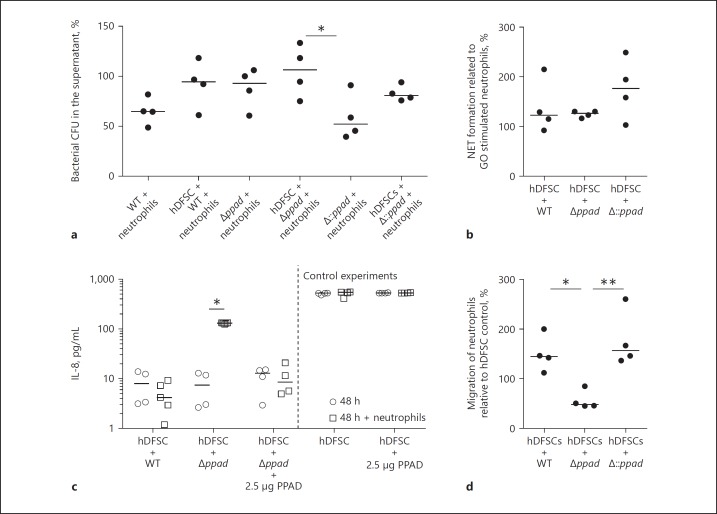
Fig. 6.
Interaction between P. gingivalis, hDFSCs and neutrophils in the triple culture. a Phagocytic activity of neutrophils. Analysis of the ability of neutrophils to phagocytose P. gingivalis strains in the presence of primed hDFSCs. The hDFSCs were incubated with WT, Δppad, or Δ::ppad for 24 h under anaerobic conditions. The bacterial control representing 100% is the amount of bacteria without neutrophils. b Analysis of the NET formation of neutrophils after 24 h of incubation with primed stem cells. Subsequently, the supernatants were sterile filtered, and freshly isolated neutrophils were incubated for 24 h under anaerobic conditions. Neutrophils incubated with untreated hDFSCs and activated with glucose oxidase for 2 h served as a control. Neutrophils activated by P. gingivalis strains were incubated for 2 h, and the formation of NETs was determined. The results were correlated with the glucose oxidase stimulation, which was defined as 100%. c The IL-8 concentration was measured in the culture supernatant. hDFSCs were primed with P. gingivalis WT, Δppad, or Δ::ppad under anaerobic conditions (open circles, compare with Fig. 2, Step I). After 24 h, neutrophils were co-cultured in sterile-filtered supernatants for another 24 h (open squares, compare with Figure 2, Step II). As a control, the IL-8 levels were quantified via ELISA after 48 h from the supernatant of the primed cells without neutrophils (open circles). Unprimed stem cells served as controls. d Analysis of neutrophil migration in a Transwell system. To this end, hDFSCs were primed with WT, Δppad or Δ::ppad. The supernatant was sterile filtered into the wells, and a suspension of freshly isolated neutrophils was added into the insert. The number of neutrophils in the wells was counted and compared with the medium control, which was defined as 100%. The results are displayed as the median; * p < 0.05, ** p < 0.01 were considered significant (ANOVA), n ≥ 4 (each data point represents 1 independent biological replicate).
Phagocytic clearance of bacteria from the supernatant (Fig. 6a) and NET formation of neutrophils (Fig. 6b) could be affected by hDFSC infection. As displayed in the figures, both neutrophil phenotypic activities were unchanged and not different between the strains regardless of the presence of hDFSCs and PPAD (Fig. 6a, b).
Compared with the unprimed control, infection of hDFSCs with the 3 P. gingivalis strains resulted in surprisingly low IL-8 accumulation after 24 and 48 h postinfection (Fig. 6c). IL-8 concentrations after 24 h were comparable to those after 48 h (data not shown). This result supported the notion of an anti-inflammatory environment created during infection. Of note, infection of hDFSCs with the P. gingivalis PAD deletion mutant provoked significantly increased IL-8 concentrations in the presence of neutrophils. Controls of primed hDFSCs anaerobically cultivated for 48 h without neutrophils did not show any increase compared with those cultivated for 24 h (Fig. 6c). Therefore, increases in IL-8 levels were solely triggered by neutrophil activity.
The migration of neutrophils to the site of infection is another important immunological feature during their lifespan. Thus, we next tested the consequences of P. gingivalis-hDFSC interaction (Fig. 2, Step I) on the hDFSC-neutrophil axis (Fig. 2, Step II) by studying neutrophil migration activity in our experimental triple-culture system (Fig. 6d). Neutrophil Transwell experiments revealed significant migration of neutrophils toward the supernatants of unprimed hDFSCs (value set to 100%). Compared with these controls, infection of hDFSCs with the P. gingivalis WT strain led to increased neutrophil migratory activity. Compared with the medium control, infection of hDFSCs with the P. gingivalis deletion strain (Δppad) resulted in significantly reduced neutrophil migratory activity (down to 60%). The P. gingivalis WT as well as the P. gingivalis complemented strain Δ::ppad induced the same migratory levels in the neutrophils.
Collectively, these data suggest that PPAD not only supports the viability of neutrophils, but also leads to the increased migration activity of neutrophils.
Internalization of P. gingivalis Is Necessary to Prime hDFSCs
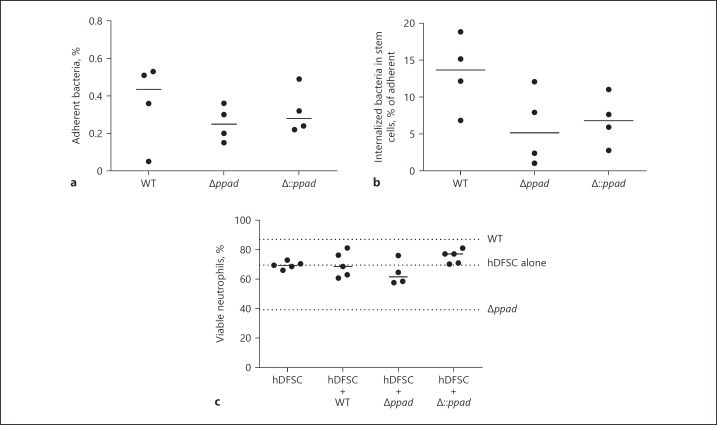
The next series of experiments explored the molecular mechanisms that explain the phenotypes observed. First, we determined whether P. gingivalis adherence to and internalization into hDFSCs is a prerequisite for the effects on neutrophils. As shown in the control experiments depicted in Figure 7a, b, no significant differences in adherence to and internalization into hDFSCs by the P. gingivalis WT strain, deletion mutant strain Δppad and complementation strain Δ::ppad were observed. Thus, attenuated host cell interaction behavior does not explain hDFSC effects on neutrophils. This result further indicated that PPAD is not a bacterial virulence factor involved in host stem cell adherence and internalization.
Fig. 7.
Internalization of P. gingivalis into hDFSCs is necessary for stem cell priming and altered neutrophil viability. hDFSCs were primed with P. gingivalis WT, Δppad, or Δ::ppad under anaerobic conditions for 2 h. Adherent (a) and internalized (b) bacteria were quantified. Adherent bacteria were related to the reference bacteria present in the inoculum, and internalized bacteria were related to the adherent bacteria. c The viability of neutrophils after inhibition of bacterial internalization into hDFSCs. hDFSCs were incubated with latrunculin B 60 min prior to hDFSC priming with P. gingivalis W83 WT strain, deletion mutant strain Δppad, or control strain Δ::ppad. Prior to infection, the inhibitor was completely removed and the hDFSCs were washed. The analysis of bacteria-hDFSC interaction was performed over 24 h in anaerobic conditions. Subsequently, the bacteria were removed from the culture system, and experiments were performed as described for the triple-culture system. Finally, the viability of neutrophils was analyzed, and the dotted lines show a comparable median to that depicted in Figure 5 without inhibitor. The results are displayed as the median; n ≥ 4 (each data point represents 1 independent biological replicate).
We next tested whether the blocking of adherence to and internalization into hDFSCs by P. gingivalis contributes to the downstream phenotypic effects on neutrophils. For these experiments, hDFSCs were preincubated with latrunculin B (10 µM) for 60 min to block cellular actin polymerization, thus preventing bacterial internalization into host cells. Internalization was completely abolished (data not shown). Subsequently, we determined neutrophil viability in the supernatants after blocking the internalization of the P. gingivalis PAD deletion strain into hDFSCs, which apparently eliminated the reduction in neutrophil viability (Fig. 7c). Together, these results suggest that bacterial internalization is a critical molecular mechanism in this triple-culture neutrophil phenotype setup.
JNK and ERK Pathways Are Exploited by P. gingivalis and Involved in Prolonged Neutrophil Viability
The previous experiments established that PPAD and direct interaction of P. gingivalis with hDFSCs (Fig. 2, Step I) are necessary to prime hDFSCs for the induction of prolonged neutrophil viability (Fig. 2, Step II). However, the following major question remains: what occurs on the molecular level during the bacteria-hDFSC interaction that leads to the observed downstream effects? To further address this question, we explored which pathways in hDFSCs are activated. In rheumatoid arthritis, MAPKs are activated in patient tissues, which led us to hypothesize that MAPKs could also be involved in the cellular regulatory processes observed in our study [26].
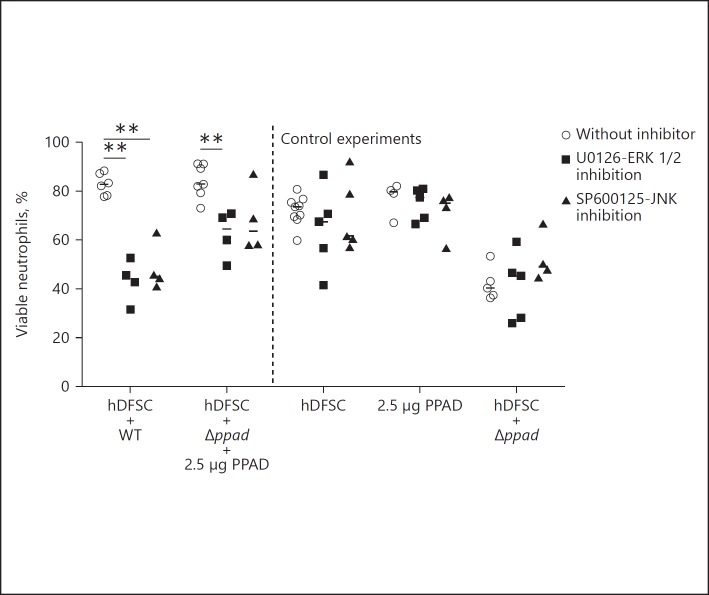
Therefore, MAPKs JNK (c-Jun N-terminal kinase) and ERK (extracellular signal-regulated kinase) were pharmacologically inhibited to reveal affected pathways involved in the mediation of neutrophil viability. Inhibitors were applied 1 h before infection of the stem cells with P. gingivalis strains. Subsequently, inhibitors were completely removed from the system, and the triple-culture system was used as described in Figure 2. Compared with the corresponding untreated control, the inhibition of JNK or ERK exclusively reduced the viability of neutrophils (Fig. 8), whereas p38 inhibition had no effect (data not shown). The effects of P. gingivalis WT and Δppad supplemented with recombinant enzyme were altered by blocking these pathways. Collectively, this result suggests that the effects of P. gingivalis-primed hDFSCs on neutrophils somehow involve ERK and JNK pathway activation in stem cells.
Fig. 8.
MAP kinases JNK and ERK are involved in prolonged neutrophil viability. The viability of neutrophils after pathway inhibition of PPAD in hDFSCs. The hDFSCs were incubated with the inhibitor of ERK (U0126) and JNK (SP600125) before hDFSCs were primed with the P. gingivalis WT strain and deletion mutant strain Δppad with the addition of recombinant PPAD. Prior to infection, the inhibitor was completely removed and hDFSCs were washed. The interaction of bacteria and hDFSCs was performed over 24 h in anaerobic conditions. Subsequently, bacteria were removed from the culture system and freshly isolated neutrophils were incubated for another 24 h under anaerobic conditions. Finally, the apoptosis of neutrophils was analyzed via flow cytometric analysis, and the percentage of viable cells was plotted in the diagram. Data points labeled “without inhibitor” refer to DMSO solvent control experiments. The results are displayed as the median; ** p < 0.01 was considered significant (ANOVA), n ≥ 4 (each data point represents 1 independent biological replicate).
Discussion
PD is a chronic inflammatory disease involving neutrophil activity. PD could be affected by PAD citrullination activity expressed by a periodontal pathogen such as P. gingivalis (PPAD).
In particular, our understanding of the basic mechanisms leading to the chronicity of the inflammatory response in periodontal pockets is limited. Therefore, this study analyzed the effects of an oral bacterial infection of dental stem cells on neutrophils. To accomplish this goal, we established a triple-culture system (Fig. 1; online suppl. Fig. S3). The influence of PPAD on hDFSCs (online suppl. Fig. S3, Box I), and the downstream effect on neutrophils (online suppl. Fig. S3, Box II) was studied. The focus of this analysis was the direct interaction of hDFSCs with P. gingivalis and the respective PPAD mutants. The effects were monitored on a second level; namely, neutrophil viability was used as a surrogate marker for sustained inflammation. Direct interaction of stem cells and P. gingivalis exclusively results in activation of a MAPK pathway, which is essential for the effects on neutrophils.
PPAD is required for the prolonged viability of neutrophils. In detail, the activity of bacterial PPAD and direct P. gingivalis/stem cell interaction influences neutrophils via the exploitation of the ERK and JNK signaling pathways in P. gingivalis-primed stem cells. This interaction leads to the prolonged viability of neutrophils as an important indicator of chronic inflammation and influences neutrophil migration.
Before the influence of PPAD could be investigated, the co-culture system had to be evaluated. Co-cultivation was performed in anaerobic conditions because P. gingivalis remains viable, and hypoxia itself is apparently a signal prolonging the viability of neutrophils, whereas stem cells are largely unaffected by these conditions [7, 27]. Stem cells do not notably influence neutrophils in anaerobic conditions. Raffaghello et al. demonstrated the prolonged viability of neutrophils after co-culture with hBMSCs over 40 h [17]. Furthermore, Cianci et al. determined that human periodontal ligament stem cells, hPDLSCs, inhibit the apoptosis of neutrophils [28]. In our experimental setup, compared with incubation in medium, incubation in the presence of hDFSCs or hBMSCs did not prolong the viability of neutrophils. Furthermore, the immune modulatory functions of hBMSCs and hDFSCs on immune cells were previously shown [29, 30]. For example, interleukin-6 (IL-6) influenced the viability rate of neutrophils and inhibited apoptosis [31]. hBMSCs continuously expressed IL-6, and hDFSCs secreted IL-6 after mechanical stress [32]. Furthermore, anti-inflammatory IL-10 was secreted by hBMSCs. The secretion of IL-10 in our test system under the chosen conditions was previously described by Kriebel et al. [7]. Additionally, IL-10 can influence apoptosis of neutrophils [33] but was secreted in low concentrations in this anaerobic system [7]. A wide range of other factors can influence the viability of neutrophils. However, in the co-cultivation system presented in our study, previously published results could not be verified and supported since neutrophils remain unaffected by stem cell co-cultivation. Nevertheless, neutrophils show a higher viability rate after incubation with undifferentiated cells than after incubation with gingival epithelial cells.
Our study demonstrated that a direct interaction between bacteria and hDFSCs affected neutrophil apoptosis. Infection of stem cells with various vital bacterial species caused changes in the viability rates of neutrophils in the triple-culture system. Stimulation of hBMSCs or hDFSCs with LPS activated stem cells and resulted in the recruitment of neutrophils [34]. Neutrophils showed a prolonged lifespan and increased expression of inflammatory chemokines [18]. We demonstrated that primed hDFSCs modify the viability of neutrophils. Of note, hDFSC infection with A. actinomy cetemcomitans, P. intermedia, and T. forsythia did not result in a uniform phenotype. Interaction between hDFSCs and A. actinomycetemcomitans reduced the number of viable neutrophils, a phenotype that could depend on the secretion of a leukotoxin. Leukotoxins activate neutrophils and lead to the release of proteolytic enzymes [35]. The infection of hDFSCs with P. intermedia and T. forsythia had no influence on neutrophil apoptosis. Thus, infection of hDFSCs can influence the viability of neutrophils. However, the results are bacterial species specific.
P. gingivalis-hDFSC interaction prolonged the viability of neutrophils in the triple-culture system. The ability of metabolically active P. gingivalis to protect epithelial cells and not induce apoptosis in neutrophils was previously shown [36]. The aforementioned study revealed that stem cell survival was less influenced by P. gingivalis. In our current study, P. gingivalis manipulated hDFSCs (online suppl. Fig. S3, Box I). These manipulated stem cells modified the viability of neutrophils without the direct interaction of neutrophils and P. gingivalis (online suppl. Fig. S3, Box II). In direct interaction assays, LPS and lipid A of P. gingivalis can delay apoptosis of neutrophils in direct co-culture [37]. In addition, P. gingivalis LPS can promote neutrophil migration [38]. P. gingivalis prolongs the viability of neutrophils via Toll-like receptor 3 activation in direct contact [39]. Furthermore, a whole range of hDFSC-secreted factors, e.g., IL-1β, IL-6, and IL-8, can influence the viability of neutrophils [40]. However, the modification of neutrophils via IL secretion is unlikely. P. gingivalis is capable of degrading cytokines via protease activity postsecretion, which is supported by the results presented in Figure 6a [41].
In the current study, we demonstrated that the presence of PPAD during the time of hDFSC infection (online suppl. Fig. S3, Box I) played a crucial role in prolonging the viability of neutrophils. Among bacteria, P. gingivalis is thus far unique in having demonstrated functional and enzymatic PAD activity [11]. The cultivation of hDFSCs (online suppl. Fig. S3, Box I) with the deletion mutant Δppad increased the percentage of apoptotic neutrophils, whereas the WT strain, control strain, and the addition of recombinant PPAD (online suppl. Fig. S3, Box I) prolonged the viability of neutrophils. Importantly, the combination of a direct interaction of P. gingivalis with hDFSCs and the presence of PPAD is crucial to prime hDFSCs. Human cells encode and express autologous PAD enzymes, and in particular, PAD4 is involved in the NET formation of neutrophils [42]. This study analyzed and showed that bacterial PPAD has no influence on the NET formation or phagocytosis capacity of neutrophils via hDFSCs. This result leads to the question of how the presence of PPAD (online suppl. Fig. S3, Box I) influenced neutrophils in this culture system. Wegner et al. [12] described PPAD-dependent human fibrinogen citrullination in vitro. Marchant et al. [15] showed that PPAD expression does not influence the expression of the host PAD but may influence the viability of neutrophils. Stem cells are apparently modified during the initial bacterial infection, and these modifications were responsible for the effects on neutrophils (online suppl. Fig. S3, Box II). To further address this question, we studied several signal transduction pathways in primed stem cells.
MAPKs are involved in various cellular response processes, e.g., apoptosis, cell growth, or innate immunity. The MAPKs p38, JNK, and ERK are extensively described in the literature [43]. The stimulation of hBMSCs with LPS resulted in increased activation of NFκB and ERK but not JNK [44]. p38 MAPK signaling is involved in cytokine production and can influence periodontal disease progression [45]. Our study showed direct interaction of hDFSCs and P. gingivalis that relied on intact JNK and ERK pathways in hDFSCs. This finding suggested that the signaling events via these pathways could explain the phenotype seen in the stem cell-neutrophil interaction (online suppl. Fig. S3, Box II). Blocking these pathways with an ERK or JNK inhibitor resulted in the apoptosis of co-cultured neutrophils. These results indicate that a cellular response prolonged the viability of neutrophils. The PPAD deletion mutant strain is attenuated in exploiting the JNK and ERK pathways in hDFSCs.
In conclusion, expression of PPAD by P. gingivalis activates the JNK/ERK pathways in primed stem cells, resulting in the prolonged viability of neutrophils. Apparently, these pathways are not activated if the P. gingivalis PPAD deletion strain is used. Overall, the interaction of P. gingivalis and hDFSCs was intensively studied (online suppl. Fig. S3, Box I), but the mechanism behind the downstream prolonged viability effect of neutrophils was not completely elucidated in this preliminary study.
During inflammation, the balance between the production, recruitment, and death of neutrophils is important for a consequent immune response. Our study shows that P. gingivalis can create an imbalance and prolong the viability of neutrophils indirectly via stem cell infection. The sustained presence of immune cells could extend inflammation and enable P. gingivalis to colonize deeper in the periodontal pocket. However, vital immune cells increase the clearance of bacteria. The function of PPAD is a double-edged sword for P. gingivalis.
The role of hDFSCs in this interaction remains unclear because their presence can increase the survival rates of neutrophils. Our results collectively suggest that PPAD is a virulence factor of P. gingivalis that sustains inflammation in periodontal pockets.
Disclosure Statement
The authors have no competing financial interests and declare no conflicts of interest.
Supplementary Material
Supplementary data
Acknowledgments
We would like to thank Patrick J. Venables from the Kennedy Institute of Rheumatology Division, Imperial College London, for providing the P. gingivalis strains. C.H. kindly acknowledges funding by the University of Rostock Landesgraduiertenförderung of Mecklenburg Vorpommern, Germany scholarship program. J.P. acknowledges support by grant DE 023207 from US NIH/NIDCR. Purchase of the BD Accuri C6 flow cytometer (BD Biosciences) was kindly supported by funds provided by the European Union Program for Regional Development (EFRE, UHROM 25) and Rostock University Medical Center.
References
- 1.Michalowicz BS, Ronderos M, Camara-Silva R, Contreras A, Slots J. Human herpesviruses and Porphyromonas gingivalis are associated with juvenile periodontitis. J Periodontol. 2000;71:981–988. doi: 10.1902/jop.2000.71.6.981. [DOI] [PubMed] [Google Scholar]
- 2.Paster BJ, Olsen I, Aas JA, Dewhirst FE. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontology 2000. 2006;42:80–87. doi: 10.1111/j.1600-0757.2006.00174.x. [DOI] [PubMed] [Google Scholar]
- 3.Darveau RP, Tanner A, Page RC. The microbial challenge in periodontitis. Periodontology 2000. 1997;14:12–32. doi: 10.1111/j.1600-0757.1997.tb00190.x. [DOI] [PubMed] [Google Scholar]
- 4.Nanci A, Bosshardt DD. Structure of periodontal tissues in health and disease. Periodontology 2000. 2006;40:11–28. doi: 10.1111/j.1600-0757.2005.00141.x. [DOI] [PubMed] [Google Scholar]
- 5.D'Aiuto F, Parkar M, Andreou G, Suvan J, Brett PM, Ready D, Tonetti MS. Periodontitis and systemic inflammation: control of the local infection is associated with a reduction in serum inflammatory markers. J Dent Res. 2004;83:156–160. doi: 10.1177/154405910408300214. [DOI] [PubMed] [Google Scholar]
- 6.Seo B-M, Miura M, Gronthos S, Bartold PM, Batouli S, Brahim J, Young M, Robey PG, Wang CY, Shi S. Investigation of multipotent postnatal stem cells from human periodontal ligament. Lancet. 2004;364:149–155. doi: 10.1016/S0140-6736(04)16627-0. [DOI] [PubMed] [Google Scholar]
- 7.Kriebel K, Biedermann A, Kreikemeyer B, Lang H. Anaerobic co-culture of mesenchymal stem cells and anaerobic pathogens-a new in vitro model system. PLoS One. 2013;8:e78226. doi: 10.1371/journal.pone.0078226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hieke C, Kriebel K, Engelmann R, Müller-Hilke BLH, Kreikemeyer B. Human dental stem cells suppress PMN activity after infection with the periodontopathogens Prevotella intermedia and Tannerella forsythia. Sci Rep. 2016;6:39096. doi: 10.1038/srep39096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sugawara Y, Uehara A, Fujimoto Y, Kusumoto S, Fukase K, Shibata K, Sugawara S, Sasano T, Takada H. Toll-like receptors, NOD1, and NOD2 in oral epithelial cells. J Dent Res. 2006;85:524–529. doi: 10.1177/154405910608500609. [DOI] [PubMed] [Google Scholar]
- 10.Benakanakere MR, Li Q, Eskan MA, Singh AV, Zhao J, Galicia JC, Stathopoulou P, Knudsen TB, Kinane DF. Modulation of TLR2 protein expression by miR-105 in human oral keratinocytes. J Biol Chem. 2009;284:23107–23115. doi: 10.1074/jbc.M109.013862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McGraw WT, Potempa J, Farley D, Travis J. Purification, characterization, and sequence analysis of a potential virulence factor from Porphyromonas gingivalis, peptidylarginine deiminase. Infect Immun. 1999;67:3248–3256. doi: 10.1128/iai.67.7.3248-3256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wegner N, Wait R, Sroka A, Eick S, Nguyen K-A, Lundberg K, Kinloch A, Culshaw S, Potempa J, Venables PJ. Peptidylarginine deiminase from Porphyromonas gingivalis citrullinates human fibrinogen and α-enolase: implications for autoimmunity in rheumatoid arthritis. Arthritis Rheum. 2010;62:2662–2672. doi: 10.1002/art.27552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gully N, Bright R, Marino V, Marchant C, Cantley M, Haynes D, Butler C, Dashper S, Reynolds E, Bartold M. Porphyromonas gingivalis peptidylarginine deiminase, a key contributor in the pathogenesis of experimental periodontal disease and experimental arthritis. PLoS One. 2014;9:e100838. doi: 10.1371/journal.pone.0100838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quirke A-M, Lugli EB, Wegner N, Hamilton BC, Charles P, Chowdhury M, Ytterberg AJ, Zubarev RA, Potempa J, Culshaw S, et al. Heightened immune response to autocitrullinated Porphyromonas gingivalis peptidylarginine deiminase: a potential mechanism for breaching immunologic tolerance in rheumatoid arthritis. Ann Rheum Dis. 2014;73:263–269. doi: 10.1136/annrheumdis-2012-202726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marchant C, Smith MD, Proudman S, Haynes DR, Bartold PM. Effect of Porphyromonas gingivalis on citrullination of proteins by macrophages in vitro. J Periodontol. 2013;84:1272–1280. doi: 10.1902/jop.2012.120103. [DOI] [PubMed] [Google Scholar]
- 16.Shi Y, Su J, Roberts AI, Shou P, Rabson AB, Ren G. How mesenchymal stem cells interact with tissue immune responses. Trends Immunol. 2012;33:136–143. doi: 10.1016/j.it.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raffaghello L, Bianchi G, Bertolotto M, Montecucco F, Busca A, Dallegri F, Ottonello L, Pistoia V. Human mesenchymal stem cells inhibit neutrophil apoptosis: a model for neutrophil preservation in the bone marrow niche. Stem Cells. 2008;26:151–162. doi: 10.1634/stemcells.2007-0416. [DOI] [PubMed] [Google Scholar]
- 18.Brandau S, Jakob M, Hemeda H, Bruderek K, Janeschik S, Bootz F, Lang S. Tissue-resident mesenchymal stem cells attract peripheral blood neutrophils and enhance their inflammatory activity in response to microbial challenge. J Leukoc Biol. 2010;88:1005–1015. doi: 10.1189/jlb.0410207. [DOI] [PubMed] [Google Scholar]
- 19.Biedermann A, Kriebel K, Kreikemeyer B, Lang H. Interactions of anaerobic bacteria with dental stem cells: an in vitro study. PLoS One. 2014;9:e110616. doi: 10.1371/journal.pone.0110616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kriebel K, Hieke C, Müller-Hilke B, Nakata M, Kreikemeyer B. Oral biofilms from symbiotic to pathogenic interactions and associated disease - connection of periodontitis and rheumatoid arthritis by peptidylarginine deiminase. Front Microbiol. 2018;9:53. doi: 10.3389/fmicb.2018.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haddouti E-M, Skroch M, Zippel N, Müller C, Birova B, Pansky A, Kleinfeld C, Winter M, Tobiasch E. Human dental follicle precursor cells of wisdom teeth: isolation and differentiation towards osteoblasts for implants with and without scaffolds. Materialwiss Werkstofftech. 2009;40:732–737. [Google Scholar]
- 22.Gaebel R, Ma N, Liu J, Guan J, Koch L, Klopsch C, Gruene M, Toelk A, Wang W, Mark P, et al. Patterning human stem cells and endothelial cells with laser printing for cardiac regeneration. Biomaterials. 2011;32:9218–9230. doi: 10.1016/j.biomaterials.2011.08.071. [DOI] [PubMed] [Google Scholar]
- 23.Takahra H, Okamoto H, Sugawara K. Affinity chromatography of peptidylarginine deiminase from rabbit skeletal muscle on a column of soybean trypsin inhibitor (Kunitz)-Sepharose. J Biochem. 1986;99:1417–1424. doi: 10.1093/oxfordjournals.jbchem.a135611. [DOI] [PubMed] [Google Scholar]
- 24.Smith WB, Clark-Lewis I, Vadas MA. Interleukin-8 induces neutrophil transendothelial migration. Immunology. 1991;72:65. [PMC free article] [PubMed] [Google Scholar]
- 25.Laheij AM, Soet JJ de, Veerman ECI, Bolscher JGM, van Loveren C. The influence of oral bacteria on epithelial cell migration in vitro. Mediators Inflamm. 2013;2013:154532. doi: 10.1155/2013/154532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sundarrajan M, Boyle DL, Chabaud-Riou M, Hammaker D, Firestein GS. Expression of the MAPK kinases MKK-4 and MKK-7 in rheumatoid arthritis and their role as key regulators of JNK. Arthritis Rheum. 2003;48:2450–2460. doi: 10.1002/art.11228. [DOI] [PubMed] [Google Scholar]
- 27.Hannah S, Mecklenburgh K, Rahman I, Bellingan GJ, Greening A, Haslett C, Chilvers ER. Hypoxia prolongs neutrophil survival in vitro. FEBS Lett. 1995;372:233–237. doi: 10.1016/0014-5793(95)00986-j. [DOI] [PubMed] [Google Scholar]
- 28.Cianci E, Recchiuti A, Trubiani O, Diomede F, Marchisio M, Miscia S, Colas RA, Dalli J, Serhan CN, Romano M. Human periodontal stem cells release specialized proresolving mediators and carry immunomodulatory and prohealing properties regulated by lipoxins. Stem Cells Transl Med. 2016;5:20–32. doi: 10.5966/sctm.2015-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Q, Shi S, Liu Y, Uyanne J, Shi Y, Shi S, Le AD. Mesenchymal stem cells derived from human gingiva are capable of immunomodulatory functions and ameliorate inflammation-related tissue destruction in experimental colitis. J Immunol. 2009;183:7787–7798. doi: 10.4049/jimmunol.0902318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wada N, Gronthos S, Bartold PM. Immunomodulatory effects of stem cells. Periodontology 2000. 2013;63:198–216. doi: 10.1111/prd.12024. [DOI] [PubMed] [Google Scholar]
- 31.Ottonello L, Frumento G, Arduino N, Bertolotto M, Dapino P, Mancini M, Dallegri F. Differential regulation of spontaneous and immune complex-induced neutrophil apoptosis by proinflammatory cytokines: role of oxidants, Bax and caspase-3. J Leukoc Biol. 2002;72:125–132. [PubMed] [Google Scholar]
- 32.Kim DH, Yoo KH, Choi KS, Choi J, Choi S-Y, Yang S-E, Yang Y-S, Im HJ, Kim KH, Jung HL, et al. Gene expression profile of cytokine and growth factor during differentiation of bone marrow-derived mesenchymal stem cell. Cytokine. 2005;31:119–126. doi: 10.1016/j.cyto.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 33.Keel M, Ungethüm U, Steckholzer U, Niederer E, Hartung T, Trentz O, Ertel W. Interleukin-10 counterregulates proinflammatory cytokine-induced inhibition of neutrophil apoptosis during severe sepsis. Blood. 1997;90:3356–3363. [PubMed] [Google Scholar]
- 34.Chatzivasileiou K, Lux CA, Steinhoff G, Lang H. Dental follicle progenitor cells responses to Porphyromonas gingivalis LPS. J Cell Mol Med. 2013;17:766–773. doi: 10.1111/jcmm.12058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johansson A, Kalfas S. Virulence mechanisms of leukotoxin from Aggregatibacter actinomycetemcomitans. In: Virdi M, Oral Health Care - Prosthodontics, Periodontology, Biology, Research and Systemic Conditions, editor. Rijeka: InTech; 2012. [Google Scholar]
- 36.Nakhjiri SF, Park Y, Yilmaz O, Chung WO, Watanabe K, El-Sabaeny A, Park K, Lamont RJ. Inhibition of epithelial cell apoptosis by Porphyromonas gingivalis. FEMS Microbiol Lett. 2001;200:145–149. doi: 10.1111/j.1574-6968.2001.tb10706.x. [DOI] [PubMed] [Google Scholar]
- 37.Johansson A. Aggregatibacter actinomycetemcomitans leukotoxin: a powerful tool with capacity to cause imbalance in the host inflammatory response. Toxins. 2011;3:242–259. doi: 10.3390/toxins3030242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olsen I, Hajishengallis G. Major neutrophil functions subverted by Porphyromonas gingivalis. J Oral Microbiol. 2016;8:30936. doi: 10.3402/jom.v8.30936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cassatella MA, Mosna F, Micheletti A, Lisi V, Tamassia N, Cont C, Calzetti F, Pelletier M, Pizzolo G, Krampera M. Toll-like receptor-3-activated human mesenchymal stromal cells significantly prolong the survival and function of neutrophils. Stem Cells. 2011;29:1001–1011. doi: 10.1002/stem.651. [DOI] [PubMed] [Google Scholar]
- 40.Akgul C, Moulding DA, Edwards SW. Molecular control of neutrophil apoptosis. FEBS Lett. 2001;487:318–322. doi: 10.1016/s0014-5793(00)02324-3. [DOI] [PubMed] [Google Scholar]
- 41.Scheres N, Laine ML, Vries TJ de, Everts V, van Winkelhoff AJ. Gingival and periodontal ligament fibroblasts differ in their inflammatory response to viable Porphyromonas gingivalis. J Periodont Res. 2010;45:262–270. doi: 10.1111/j.1600-0765.2009.01229.x. [DOI] [PubMed] [Google Scholar]
- 42.Hensen SMM, Pruijn GJM. Methods for the detection of peptidylarginine deiminase (PAD) activity and protein citrullination. Mol Cell Proteomics. 2014;13:388–396. doi: 10.1074/mcp.R113.033746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson GL, Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science. 2002;298:1911–1912. doi: 10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- 44.Crisostomo PR, Wang Y, Markel TA, Wang M, Lahm T, Meldrum DR. Human mesenchymal stem cells stimulated by TNF-α, LPS, or hypoxia produce growth factors by an NFκB-but not JNK-dependent mechanism. Am J Physiol Cell Physiol. 2008;294:C675–C682. doi: 10.1152/ajpcell.00437.2007. [DOI] [PubMed] [Google Scholar]
- 45.Li Q, Valerio MS, Kirkwood KL. MAPK usage in periodontal disease progression. J Signal Transduct. 2012;2012:308943. doi: 10.1155/2012/308943. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data