Abstract
Expansion microscopy (ExM) is a recently developed technique that enables nanoscale resolution imaging of preserved cells and tissues on conventional diffraction-limited microscopes, via isotropic physical expansion of the specimens before imaging. In ExM, biomolecules and/or fluorescent labels in the specimen are linked to a dense, expandable polymer matrix synthesized evenly throughout the specimen, which undergoes 3-dimensional expansion by ~4.5 fold linearly when immersed in water. Since our first report, versions of ExM optimized for visualization of proteins, RNA, and other biomolecules have emerged. Here, we describe best-practice, step-by-step ExM protocols for performing analysis of proteins (protein retention ExM, or proExM) as well as RNAs (expansion fluorescence in situ hybridization, or ExFISH), using chemicals and hardware found in a typical biology lab. Furthermore, a detailed protocol for handling and mounting expanded samples and for imaging them with confocal and light sheet microscopes is provided.
Keywords: Expansion microscopy, super-resolution microscopy, imaging, tissue clearing, FISH, hydrogel, antibody, immunohistochemistry, immunocytochemistry, fluorescence microscopy, confocal microscopy, light sheet microscopy
INTRODUCTION
In expansion microscopy (ExM), preserved biological samples are embedded within a swellable hydrogel and physically expanded isotropically, leading to optically transparent samples which allow for nanoscale resolution and aberration-free microscopy imaging on conventional diffraction limited microscopes (Chen et al., 2015). Recently, two new ExM versions have been developed, termed protein retention expansion microscopy (proExM) (Tillberg et al., 2016), and expansion fluorescence in situ hybridization (ExFISH) (Chen et al., 2016), where protein (proExM) and RNA (ExFISH) molecules are chemically anchored to the hydrogel and probed upon expansion. This unit describes the most common scenarios for proExM and ExFISH sample preparation, as well as general guidelines for sample handling, and image acquisition.
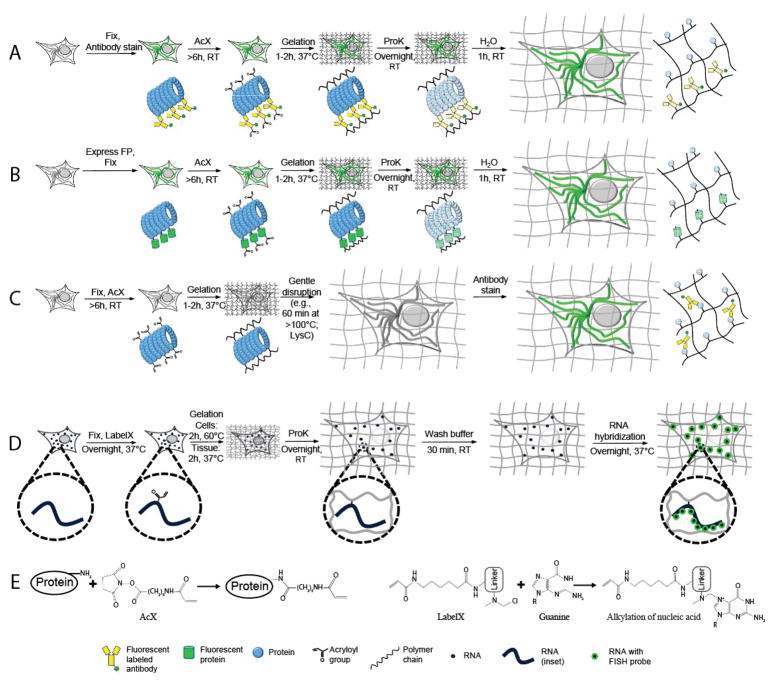
In proExM, proteins are covalently anchored to a hydrogel matrix with a commercially available small molecule (Acryloyl-X SE, or AcX for short) (Tillberg et al., 2016) that binds to primary amine groups on proteins (Fig. 1E, left), and is then incorporated into the hydrogel polymer during the process of polymerization (also known as gelation for a hydrogel) (schematized in Fig. 1A–1C). An enzymatic digestion using proteinase K (ProK) mechanically disrupts the embedded sample, allowing for isotropic expansion in water. One can anchor native proteins to the gel using AcX and apply fluorescent antibodies post-expansion (post-expansion staining, Fig. 1C). This version uses a mechanical homogenization method employing high temperature and detergent, without protease. One can alternatively apply AcX to fixed specimens expressing fluorescent proteins and/or labeled already with fluorescent antibodies, incorporating these directly to the gel (pre-expansion staining, Fig. 1A, or pre-expansion fluorescent protein expression, Fig. 1B). Fluorescent proteins and antibodies are sufficiently resistant to proteolysis that they survive the ProK application. Utilizing a lens with 300 nm diffraction limited resolution, the resulting ~4.5x linear expansion of the embedded samples (~90x volumetric expansion) allows imaging of the sample with ~300 nm / 4.5 ~ 60–70 nm effective resolution.
Figure 1.
Expansion microscopy (ExM) workflows discussed in this unit. (A–C) Protein retention ExM (proExM) workflows. A, Samples are fixed and stained with antibodies using conventional immunostaining protocols, then treated at room temperature (RT) with Acryloyl-X SE (AcX, see panel E, left, for detail), which enables proteins to be anchored to the hydrogel. The samples then undergo gelation, proteinase K (ProK) treatment for mechanical homogenization (also called digestion), and expansion in water. (B) Samples expressing fluorescent proteins are fixed and treated with AcX before going through gelation, mechanical homogenization, and expansion in water. (C) Samples are fixed and treated with AcX before going through gelation, a comparatively gentle mechanical homogenization process (e.g., high temperature denaturing in detergent solution), and expansion, followed by antibody staining. (D) Expansion fluorescence in situ hybridization (ExFISH). Samples are fixed and treated with LabelX (see panel E, right, for detail), which enables RNA to be anchored to the polymer. The samples then go through gelation, mechanical homogenization, and expansion. Finally, FISH probes are hybridized to the anchored RNA. (E) Schematics of AcX binding to a protein (left) and LabelX binding to a guanine base of RNA (right). Modified from (Tillberg et al., 2016) and (Chen et al., 2016).
In ExFISH, RNAs are covalently anchored to the hydrogel matrix with a small molecule, which we call LabelX (which can be easily synthesized by reacting two commercially available molecules), that binds to guanine (in RNA and DNA, Fig. 1E, right) and also to the hydrogel polymer (Chen et al., 2016). The ExFISH pipeline is shown in Fig. 1D: samples are treated with LabelX, the swellable hydrogel polymer formed, ProK applied, and then the specimen is washed with phosphate buffered saline (PBS). Afterwards, RNA-FISH probes are applied to the sample in a hybridization buffer to label RNA molecules of interest. The sample is then expanded and imaged in a low-salt buffer. This nonzero salt concentration, which allows RNA-FISH probes to remain stably bound, results in a ~3x linear expansion (~27x volumetric expansion), or an effective resolution for a 300 nm diffraction limited lens of ~300 / 3 ~ 100 nm.
This unit is structured into the following sections. In the sections BASIC PROEXM PROTOCOL FOR CULTURED CELLS and BASIC PROEXM PROTOCOL FOR INTACT TISSUES, we describe proExM protocols for cells and intact tissues (of <200 μm pre-expansion thickness) that have been immunostained before expansion (pre-expansion staining proExM), or have fluorescent proteins expressed before expansion, as illustrated in Figs. 1A and 1B. These two sections are written independently so that readers can start with either of the two, with minimal reference to the other. The next section PROEXM PROTOCOL FOR THICK INTACT TISSUE SAMPLES (200–500 μm) describes steps that help with processing of thick tissues or tissue slices (e.g., 200–500 μm thick, or potentially thicker), which have been immunostained or had fluorescent proteins expressed before expansion (as illustrated in Figure 1A and 1B). The POST-EXPANSION STAINING section describes the protocol for immunostaining after expansion (post-expansion staining proExM, as illustrated in Figure 1C). We also briefly discuss key considerations when choosing antibodies and dyes for proExM in the section FLUORESCENT PROTEINS, ANTIBODIES AND DYES FOR PROEXM. In the sections ExFISH PROTOCOL FOR CULTURED CELLS and ExFISH PROTOCOL FOR INTACT TISSUES, we describe ExFISH protocols for the named sample types. In the last section IMAGING EXPANDED SAMPLES, we describe different mounting strategies for preparing expanded ExM samples for imaging, on inverted confocal microscopes and the Zeiss Z.1 light sheet microscope. This section on imaging applies to all ExM-processed samples, including products of the proExM and ExFISH protocols here described.
It may be helpful to note that sample preparation and imaging with proExM and ExFISH only require chemicals and microscopes found in a typical biology lab or imaging facility, with the exceptions of anchoring reagents and sodium acrylate, which are available commercially at modest cost. Almost all the protocol steps can be carried out at room temperature on a wetlab bench (unless otherwise noted in the protocol). Basic equipment required includes a benchtop vortex mixer, a shaker, a 37°C incubator (and a 60°C incubator for ExFISH of cultured cells), a desiccator, a 4°C fridge and a −20°C freezer. All the materials and supplies used in this unit are commercially available. Unless otherwise noted, in this unit “water” specifically means “double distilled water.”
BASIC PROEXM PROTOCOL FOR CULTURED CELLS
Materials
For cell culture, fixation and immunostaining:
Cell culture medium suitable for the cell type of choice (e.g., Dulbecco’s Modified Eagle Medium (DMEM) for Human Embryonic Kidney (HEK) cells)
Cell culture wells (e.g., petri dish of sizes and materials suitable for the culture protocol of choice)
Cell culture substrates (e.g., coverslips of sizes, shapes and surface modifications suitable for the culture protocol of choice)
If no preference for the above items: a convenient choice for cell culture, imaging and gelation is a removable-chamber cover glass (e.g., Sigma, catalog no. GBL 112358)
Ethanol
Potassium hydroxide (KOH)
Phosphate buffered saline (PBS)
4% paraformaldehyde (PFA) in PBS or, for stronger fixation, a mixture of 4% PFA and 0.1% glutaraldehyde in PBS
Glycine
Triton X-100
Normal donkey serum
Primary and secondary antibodies for immunostaining
For gelation:
Thin coverslips (e.g., 22 x 22 mm No. 1.5 coverslips)
Thicker coverslips (e.g. 22 x 22 mm No. 2 coverslips)
Microscope slides
Ice bath or cold block at 4°C
Parafilm
Razor blade
Microcentrifuge tubes
Forceps
Paintbrush
6-((Acryloyl)amino)hexanoic acid succinimidyl ester (Acryloyl-X SE or AcX for short)
Dimethylsulfoxide (DMSO, anhydrous, molecular biology grade)
Sodium acrylate
Acrylamide
N,N′-Methylenebisacrylamide
Sodium chloride
10x PBS
Ammonium persulfate (APS)
Tetramethylethylenediamine (TEMED)
For digestion:
Petri dish
Ethylenediaminetetraacetic acid (EDTA)
Triton X-100
Tris (1M), pH 8.0
Proteinase K (ProK)
REAGENTS AND SOLUTIONS
Table 1 Preparation of stock solutions for expansion of cultured cells. Stock solutions are prepared as aqueous solutions unless noted otherwise.
(A) Acryloyl-X SE (AcX)/anhydrous DMSO stock solution. The AcX/anhydrous DMSO stock solution can be aliquotted in 10–20 μL increments and stored at −20°C for at least a month. The aliquots should be stored in a sealed container with drying agents (e.g., Drierite) or in a desiccator to avoid hydration.
| AcX/DMSO stock solution | Stock solution concentration (g/100 mL DMSO) |
|---|---|
| AcX | 1 |
(B) Monomer solution (here referred to as Stock X for short) (total amount 9.4 mL). The Stock X solution can be prepared into aliquots of 1 mL each and stored at −20°C for at least a month. For sodium acrylate, low purity chemical may appear slightly yellow when dissolved in water. In this case, discard the solution and switch to a new batch or a new bottle of sodium acrylate.
| Monomer solution (“Stock X”) | Stock solution concentration (g/100 mL solution) | Amount (mL) | Final concentration (g/100 mL solution) |
|---|---|---|---|
| Sodium acrylate | 38 (33 wt% due to higher density) | 2.25 | 8.6 |
| Acrylamide | 50 | 0.5 | 2.5 |
| N,N′-Methylenebisacrylamide | 2 | 0.75 | 0.15 |
| Sodium chloride | 29.2 (5M) | 4 | 11.7 |
| PBS 10x stock | 10x | 1 | 1x |
| Water | 0.9 | ||
| Total | 9.4 |
(C) Tetramethylethylenediamine (TEMED) and ammonium persulfate (APS) stock solutions. The TEMED and APS stock solutions can be each prepared into aliquots of 1 mL each and stored at −20°C for two weeks or more.
| TEMED and APS stock solutions | Stock solution concentration (g/100 mL solution) |
|---|---|
| TEMED | 10 |
| APS | 10 |
(D) Gelling solution (total amount 200 μl; scale up or down proportionally to the appropriate amount for the sample). The gelling solution should be prepared at 4°C, and used immediately. APS should be mixed into the solution at the end, vortexing right before, to prevent premature polymerization of the gel.
| Gelling solution | Stock solution concentration (g/100 mL solution) | Amount (μL) | Final concentration (g/100 mL solution) |
|---|---|---|---|
| Stock X (Table 1B) | NA | 188 | NA |
| TEMED (Table 1C) | 10 | 4 | 0.2 |
| APS (Table 1C) | 10 | 4 | 0.2 |
| Water | 4 | ||
| Total | 200 |
(E) Digestion buffer (total amount 500 mL; amount can be scaled down if desired). Tris base can be used, instead of Tris (1M) aqueous solution at pH 8. If Tris base is used, the final concentration of Tris needs to be 50 mM, and the pH of the buffer needs to be adjusted to pH 8 (e.g., with hydrochloric acid). The digestion buffer can be stored (minus the Proteinase K) as aliquots at −20°C for a few months. Proteinase K can be stored at −20°C per manufacturer guidelines. Add Proteinase K immediately before the digestion step.
| Digestion buffer | Amount | Final concentration (/100 mL solution) |
|---|---|---|
| Triton-X | 2.50 g | 0.50 g |
| EDTA | 0.146 g | 0.027 g |
| Tris (1M) aqueous solution, pH 8 | 25 mL | 5 mL |
| NaCl | 23.38 g | 4.67 g |
| Water | Add up to a total volume of 500 mL | |
| Proteinase K (800 units/mL) | 1:100 dilution | 800 units (= 8 units/mL) |
| Total | 500 mL |
Cell culture
Cells can be cultured following protocols of your choice. However, if there is no strong preference, we recommended culturing cells on a substrate separable from the culture well (e.g. on coverslips) or on a removable-chamber cover glass (e.g., Sigma, catalog no. GBL 112358) for easy gelation and sample handling in later steps.
The cell culture protocol described below uses adherent cell lines (e.g., HEK or COS7) and has demonstrated consistent results. If removable-chamber cover glasses are used for cell culture, start with the fibronectin modification in step 2.
-
1
12 mm round coverslips are treated with 1M KOH in water for 30 minutes, rinsed with water three times, and stored in 30% ethanol in water.
-
2
The cleaned 12 mm round coverslips are air-dried in a sterile environment (i.e., biosafety cabinet), placed in a 35 mm petri dish, and subsequently treated with 10 mg/mL sterile fibronectin in PBS for 30 minutes at 37°C. The fibronectin solution is removed immediately before cell plating.
-
3
Cells are plated and cultured on the surface of either the 12 mm coverslips or the removable-chamber cover glass to a confluency of 50–80% in a humidified incubator set at 5% CO2 and 37°C.
Cell fixation
Cells can be fixed using protocols of your choice. For instance, 4% paraformaldehyde (PFA) in PBS can be used. For stronger fixation that preserves more ultrastructure, you might try using a mixture of PFA and a small percentage of glutaraldehyde (e.g., 4% PFA + 0.1% glutaraldehyde in PBS), although glutaraldehyde-containing fixatives have been less used in expansion microscopy studies to date, and so you may want to experiment a bit. The 4% PFA in PBS is typically prepared freshly prior to the fixation.
The fixation protocol described below has demonstrated consistent results. Note that all the washing steps should be performed quickly to avoid sample dehydration.
-
4
Exchange the cell culture medium with 4% PFA in PBS for 10 minutes at room temperature (RT; all steps are done at RT unless otherwise indicated). This should be done in a chemical hood, as PFA is considered a potential carcinogen.
-
5
Remove the PFA solution and wash the cells for 5 minutes with 100 mM glycine in PBS to quench the fixation.
-
6
Remove the glycine solution and wash the cells twice for 5 minutes each (abbreviated 2 x 5 minutes) with PBS.
-
7
We recommend moving to the next steps of the protocol immediately.
If necessary, fixed cells can be stored in the dark at 4°C in PBS for up to several weeks before the next steps of the protocol.
Immunostaining/immunohistochemistry (optional)
For immunostaining after fixation, follow protocols of your choice; you can essentially perform conventional histology. The protocol described below has demonstrated consistent results with fixed cells prepared by the protocols described earlier.
-
8
Permeabilize the fixed cells by applying 0.1% Triton X-100 in PBS at RT for 15 minutes. To reduce nonspecific antibody binding, apply 5% normal donkey serum in PBS with 0.1% Triton X-100 (blocking buffer) at RT for 15 minutes. (The serum should be from the animal host of the secondary antibodies, so other types of sera should be used instead of normal donkey serum if the secondary antibodies are from other animals.) The blocking buffer can be stored at 4°C for at least a month.
-
9
Incubate the cells with primary antibodies in the blocking buffer, at the appropriate concentration. The incubation can be done on a shaker at low speed for a period ranging from 1 hour to overnight, at RT or at 4°C, depending on the antibody specifications.
-
10
Remove the antibody solution and wash 4 x 5 minutes with blocking buffer.
-
11
Incubate the cells with secondary antibodies in the blocking buffer on a shaker at low speed for 2–4 hours at RT or overnight at 4°C. If using antibodies conjugated with dyes, see FLUORESCENT PROTEINS, ANTIBODIES AND DYES for a discussion of dye compatibility with pre-expansion staining proExM.
-
12
Remove the antibody solution and wash 4 x 5 minutes with blocking buffer.
-
13
We recommend moving to the next steps of the protocol immediately.
If necessary, the samples can be stored in the dark at 4°C in PBS for up to several weeks before the next steps of the protocol.
Gelation
-
14
Prepare solutions as instructed in Table 1A–C, E.
-
15
Exchange the sample buffer (e.g., the blocking buffer if immunostained following the protocols above) with 0.1 mg/mL Acryloyl-X SE (AcX) in PBS. This AcX solution is prepared by diluting the 10 mg/mL AcX/DMSO stock solution (Table 1A) 1:100 in PBS.
-
16
Leave the cells in AcX solution for 2–3 hours at RT.
Previous protocols suggested >6 hours or overnight, but we routinely find 2–3 hours to be more than enough time with no shaking. -
17
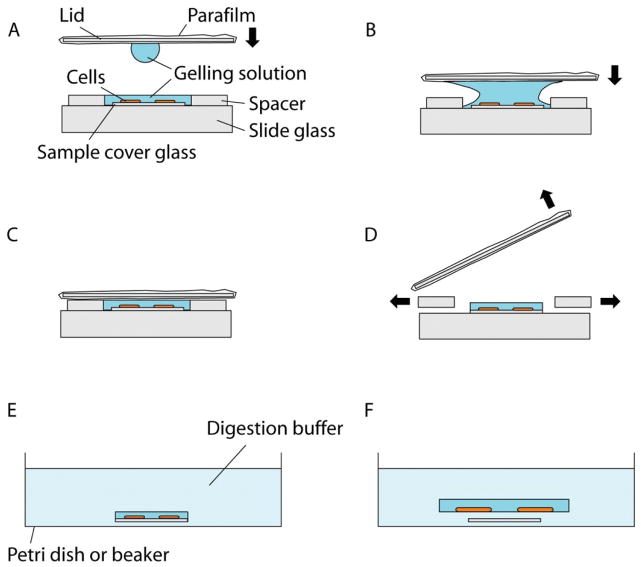
When cells are cultured on a petri dish, the gelation chamber can be meanwhile constructed, by placing two spacers on a slide, separated from each other by a size big enough so that the sample cover glass can fit in between (Figure 2A). Thin and flat objects, such as stacks of coverslips, can serve as the spacers. For example, a stack of two No. 1.5 coverslips has a height of about 0.30–0.33 mm; by changing the number or types of the coverslips, the height of the spacers can be adjusted accordingly to match the cell culture substrate (i.e. the sample cover glass) height. The spacer height should be kept close to the cell culture substrate height to avoid leaving excessive gel on top of the sample, but the total thickness of the gel (i.e., (spacer height) - (cell culture substrate height)) should be at least 0.15 mm for easier gel handling in later steps. A droplet of water (e.g., a few μL) or a very small amount of superglue can be applied to the spacers to adhere them to the slide (and, if multiple spacers, to each other). If a removable-chamber cover glass is used for the cell culture during steps 2–3, the cover glass itself and the gasket can serve as the gelation chamber and the spacer, respectively. In this case, the upper structure of the removable-chamber cover glass can be removed via a removal tool (Sigma, catalog no. GBL 103259). For the gelation chamber lid, a thick and robust coverslip (e.g., a No. 2 coverslip) wrapped in Parafilm can be used, with the Parafilm preventing the gel from sticking to the coverslip when removing the lid in later steps. Make sure the Parafilm surface is flat, clean and free of folds. We recommend finishing preparation of the gelation chamber and the lid before moving to the next step.
-
18
After incubating the cells in AcX solution, wash the cells 2 x 15 minutes with PBS. Begin thawing the components for the gelling solution, the Stock X, APS and TEMED stock solutions (Table 1B and 1C). Keep the solutions chilled in an ice bath or on a cold block (chilled to 4°C).
-
19
Create the gelling solution (Table 1D) now by mixing Stock X, water, TEMED stock solution and APS stock solution in a 47:1:1:1 ratio (gelling solution), in this order (i.e., APS last) in a microcentrifuge tube, and vortex for a few seconds. The amount of gelling solution should be scaled up or down according to the size of the gelation chamber. For instance, 200 μL of gelling solution is enough to fill a gelation chamber with a 22 x 22 x 0.33 mm void. The mixed gelling solution should be kept in an ice bath or on a cold block (chilled to 4°C). Avoid excessive warming of the gelling solution by, for example, holding the lid of the microcentrifuge tube rather the body of the tube. Once the gelling solution is prepared, we recommend completing the next few steps (until placing the gelation chamber in a 37°C incubator in step 24) within 5 minutes to prevent premature gelation.
-
20
Immediately after the previous step, remove the PBS from the cells using a pipette and add a small amount of the mixed gelling solution to the cells on the cell culture substrate. If a removable-chamber cover glass is used, remove the PBS and add the gelling solution to fill in the chamber and continue to step 22.
-
21
Use a pair of forceps to pick up and place the cell culture substrate (i.e., the coverslip upon which cells are cultured) with the cultured cells between the spacers in the gelation chamber. Make sure the cells are facing upward. Slightly press the cell culture substrate down, to hold it in place. If the gelling solution flows away, reapply a small amount of gelling solution to the cells.
-
22
Add a droplet (~20 μL) of gelling solution to one side of the lid, and flip it over so that the droplet hangs from the surface due to surface tension (Figure 2A). Slowly lower the lid so that the droplet will fuse with the gelling solution on the sample coverslip (Figure 2B). Once fused, continue lowering the lid so it rests on the spacers. Make sure no air pockets are introduced inside the solution during this process.
-
23
Using a pipette with a fine tip, keep adding additional gelling solution to both open sides of the chamber until the space between the lid, spacers and the slide is filled with gelling solution (Figure 2C). Capillary forces should cause the gelling solution to evenly fill the void.
-
24
Place the gelation chamber in an incubator at 37°C for 1 hour for polymerization. Be careful not to tilt or shake the chamber during the transfer and the gelation.
Figure 2.
Gelation of cultured cells. (A) Schematic showing the side view of a gelation chamber. The cultured cells are on the top surface of the cell culture substrate. The lid is moved towards the chamber, bearing a droplet of the gelling solution. (B) Schematic showing lid placement. The lid is moved towards the sample so that the droplet merges with the gelling solution in the chamber, which prevents air pocket formation. (C) Schematic showing the gelation chamber ready for polymerization at 37°C. (D) Schematic showing removal of the lid and the spacers. The lid is first pried open and then the gel is trimmed down with a razor blade to remove excess gel. (E) Schematic showing the digestion step. The cell culture substrate with the gel on top is placed in the digestion buffer, which eventually causes the gel to come off the substrate. (F) Schematic showing the gel detached from the substrate and slightly expanded after the digestion step.
Digestion
-
25
Take out the gelation chamber from the incubator.
-
26
Slowly insert a razor blade from the side between the lid and the spacers and pry the lid off. Once the lid is separated from the gel, use the razor blade to remove the spacers (Figure 2D). If a removable-chamber cover glass is used, remove the lid, then use forceps to peel off the black gasket without removing the gel from the cover glass.
-
27
Using a razor blade, trim the gel to a small volume around the sample. This will keep the gel compact and aid in finding cells in later steps. Use the razor blade to remove the cell culture substrate and the gel, together, from the glass slide. If a removable-chamber cover glass is used, trim the gel but do not remove the gel from the cover glass.
You may want to trim the gel to have an asymmetric shape so that its orientation can easily be gauged in later steps simply by examination of the shape (see our video at (proExM for tissues: gelation demonstration)). -
28
Prepare digestion buffer by diluting Proteinase K (ProK) 1:100 in the remaining components in Table 1E (final concentration of ProK, 8 units/mL).
-
29
Since the gel will expand by about 1.5x during digestion, choose a suitably large container (e.g., petri dish) for the digestion. Also, use a separate container for each sample to avoid mix-up. Place the cell culture substrate with the gel in the container and fill it with digestion buffer of volume at least 10-fold excess of that of the gel (Figure 2E). Immerse the gel in digestion buffer for at least 2–3 hours until it naturally peels off from the sample coverslip (Figure 2F). If a removable-chamber cover glass is used, use a diamond scribe to separate each sample and its corresponding piece of the cover glass, by scribing the cover glass and breaking each piece of glass off bearing its piece of gel. Place each piece of cover glass with the gel in a container and immerse in the digestion buffer as described above. A Nunc 4-well rectangular plate (Thermo Fisher, catalog no. 267061), for example, is convenient to contain several pieces of cover glass for digestion. The detached cell substrate can be removed using forceps, to leave only the cell-containing gel in the digestion buffer.
-
30
Leave the gel immersed in the digestion buffer overnight at RT in the dark.
Storage and expansion
-
31
If not done previously, remove the now-detached cell substrate (e.g., coverslip) from the digestion container carefully using forceps without disturbing the sample gel. Remove the digestion buffer and add PBS, and store the gel at 4°C in the dark if storage for later imaging is desired.
Be cautious not to suck up the gel while pipetting. The gel is slightly scattering at this stage, so changing the incident angle of light might help to locate it inside the solution. -
32
If the gel needs to be transferred to another container (e.g., a larger container for trimming, or the sample holder for imaging), you can use a paintbrush of suitable size to pick up and move the gel. To avoid dehydration, add some PBS to the container.
-
33
Before expanding the sample, we recommend trimming the gel to the minimum size necessary. You can use a dissection microscope or a low magnification wide-field microscope to locate the region of interest. To minimize sample movement, most of the surrounding liquid can be temporarily removed during trimming. Under the microscope, use a razor blade to cut and remove gel outside of the region of interest. We advise trimming the gel into an asymmetric shape to easily deduce the orientation. Make sure to trim the gel down to a reasonable size for handling and imaging as the expansion will introduce an additional 2–2.5x (to a total of about 4.5x) linear increase in size. Transfer the gel with the regions of interest to a container (e.g., a petri dish of suitable size for expansion, or the sample holder for imaging) that is large enough to contain the expanded gel.
-
34
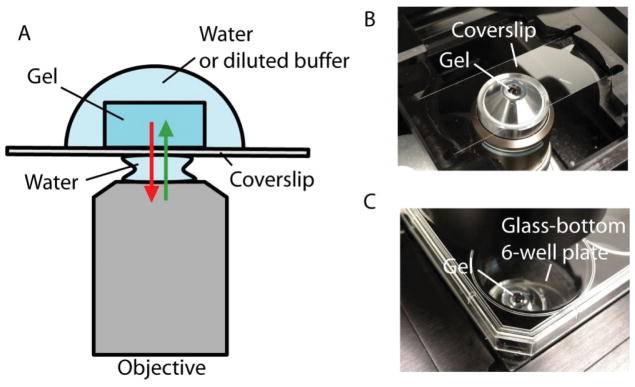
Fill the container and immerse the gel in water and wait for 20 minutes. Exchange the water and wait for another 20 minutes. Redo the exchange one more time (20 minutes in fresh water x 3 in total). The sample should be fully expanded and optically cleared (such as in Figure 3) and ready for imaging (these times are shorter than in previous publications, but reflect numbers that work for routine use in our lab). For long-term imaging, e.g. when imaging a large volume of specimen, sample mounting is necessary after the expansion and before the imaging (see IMAGING EXPANDED SAMPLES). Sample mounting attaches the sample to the sample holder and thus prevents it from drifting, and also can help keep the specimen within the working distance of the objective lens. For a quick check of the expanded sample under the microscope (< 5 minutes), there is typically no need for sample mounting. On an inverted microscope, for example, you can use a pipette to temporarily (<5 minutes) remove the water around the gel to prevent it from sliding away.
-
35
If you need to transfer or flip the expanded gel to a new container, it needs to be handled with care, as it is somewhat delicate in the expanded state. First remove most of the surrounding water and then use a brush to move the gel onto a clean glass coverslip placed in the same container. Next, gently lift the coverslip (with the gel on top) out of the container with forceps. The gel can be transferred by slowly brushing it off the coverslip to the new container. This method can be also used to flip the gel by inverting the coverslip bearing the gel, in case the gel is facing in the wrong direction.
Figure 3.
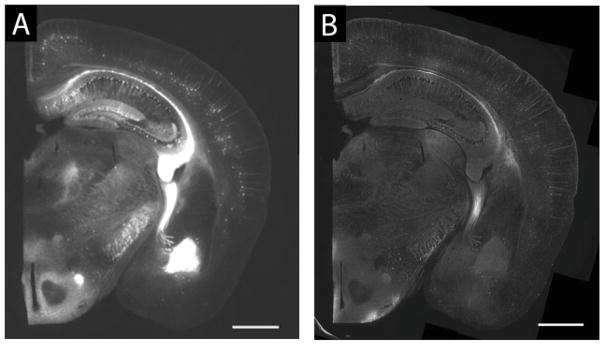
Optical clearing resulting from the expansion process. A 200 μm thick fixed brain hemi-slice is shown left. Post-expansion (right), the gel containing the hemi-slice becomes significantly enlarged and optically clear. Modified from Chen et al., 2015.
BASIC PROEXM PROTOCOL FOR INTACT TISSUES
The basic protocol for intact tissues differs slightly from that for cultured cells. Due to the thickness of the samples, intact tissues require a longer time for AcX diffusion. Furthermore, the gelling solution is supplemented with a polymerization inhibitor to allow for longer diffusion of the gelling solution through the samples before the polymerization begins.
Materials
For tissue fixation and immunostaining:
4% paraformaldehyde (PFA) in PBS or, for stronger fixation, a mixture of 4% PFA and 0.1% glutaraldehyde in PBS
Triton X-100
Normal donkey serum
Primary and secondary antibodies for immunostaining
For cryoslicing (alternative to vibratome slicing):
Sucrose
Glycine
Dry ice
2-methylbutane
Optimal cutting temperature compound (OCT), M-1 or another embedding matrix
For gelation:
Thin coverslips (e.g. 22 x 22 mm No. 1.5 coverslips)
Thicker coverslips (e.g. 22 x 22 mm No. 2 coverslips)
Microscope slides
Ice bath or cold block at 4°C
Parafilm
Razor blade
Microcentrifuge tubes
Forceps
Paintbrush
6-((Acryloyl)amino)hexanoic acid succinimidyl ester (Acryloyl-X SE or AcX for short)
Dimethylsulfoxide (DMSO, molecular biology grade)
Sodium acrylate
Acrylamide
N,N′-Methylenebisacrylamide
Sodium chloride
10x PBS
Ammonium persulfate (APS)
Tetramethylethylenediamine (TEMED)
4-hydroxy-2,2,6,6-tetramethylpiperidin-1-oxyl (4-Hydroxy-TEMPO, or 4HT for short)
For digestion:
Petri dish
Ethylenediaminetetraacetic acid (EDTA)
Triton X-100
Tris (1M), pH 8.0
Proteinase K (ProK)
REAGENTS AND SOLUTIONS
Table 2 Preparation of stock solutions for expansion of intact tissues. Stock solutions are prepared as aqueous solutions unless noted otherwise. The sinking solution is used for cryoslicing on a cryostat. For the AcX/DMSO stock solution, monomer solution (Stock X), TEMED and APS stock solutions, and digestion buffer, refer to Table 1.
(A) Sinking solution (1x PBS + 30% (w/v) sucrose + 100mM glycine) should be stored at 4°C and can be used for at least a month.
| Sinking solution | Amount |
|---|---|
| 10x PBS | 50 mL |
| Sucrose | 150 g |
| Glycine | 3.75 g |
| Water | Add until total volume 500 mL |
(B) 4-Hydroxy-TEMPO (4HT) stock solution. The 4HT stock solution can be prepared in aliquots of 1 mL and stored at −20°C for at least a month.
| 4HT stock solution | Stock solution concentration (g/100 mL solution) |
|---|---|
| 4HT | 0.5 |
(C) Gelling solution for expansion of intact tissues (total amount 200 μl; scale up or down proportionally to the appropriate amount for the sample). The gelling solution should be prepared at 4°C, and used immediately. APS should be mixed into the solution last, before vortexing, to prevent premature polymerization of the gel.
| Gelling solution | Stock solution concentration (g/100 mL solution) | Amount (μL) | Final concentration (g/100 mL solution) |
|---|---|---|---|
| Stock X (Table 1B) | NA | 188 | NA |
| 4HT (Table 2B) | 0.5 | 4 | 0.01 |
| TEMED (Table 1C) | 10 | 4 | 0.2 |
| APS (Table 1C) | 10 | 4 | 0.2 |
| Total | 200 |
Tissue preparation
In this basic proExM tissue protocol, the thickness of the tissue or the tissue slice is assumed to be <200 μm in its thinnest dimension. For tissue samples with thickness beyond 200 μm, see PROEXM PROTOCOL FOR THICK INTACT TISSUE SAMPLES (200–500 μm).
Tissues can be fixed using protocols of your choice. For instance, immersion or perfusion fixation with 4% paraformaldehyde (PFA) in PBS can be used. For stronger fixation that preserves more ultrastructure, you might try a mixture of PFA and a small percentage of glutaraldehyde (e.g., 4% PFA + 0.1% glutaraldehyde in PBS), although glutaraldehyde-containing fixatives have been less used in expansion microscopy protocols to date. The 4% PFA in PBS solution is typically prepared freshly prior to the fixation.
-
1
After fixation, if tissues are thicker than 200 μm, cut them into <200 μm slices using a vibratome. Alternatively, for slicing in a cryostat, the tissue needs first to be cryoprotected by immersing it in PBS containing 30% sucrose and 100 mM glycine (sinking solution, Table 2A) until the tissue sinks to the bottom. The tissue is then taken out and frozen using dry ice and 2-methylbutane. After embedding it in OCT, M-1 or another embedding matrix, the sample is ready to be sliced using a cryotome. After either slicing method, we recommend moving to the next steps of the protocol immediately.
If necessary, fixed tissues or tissue slices can be stored in the dark at 4°C in PBS for up to several weeks before the next steps of the protocol.
Immunostaining/immunohistochemistry (optional)
For immunostaining after fixation, follow protocols of your choice. The protocol described below has demonstrated consistent results with fixed brain tissues prepared by the protocols described earlier.
-
2
Permeabilize the fixed tissue by applying 0.1% Triton X-100 in PBS at RT for 15 minutes, then 5% normal donkey serum in PBS with 0.1% Triton X-100 (blocking buffer) at RT for 6 hours. (The serum should be from the animal host of the secondary antibodies, so other types of sera should be used instead of the normal donkey serum if the secondary antibodies are from other animals.) The blocking buffer can be stored at 4°C for at least a month.
-
3
Incubate the tissues with primary antibodies in the blocking buffer. The incubation can be done on a shaker at low speed overnight at RT or at 4°C.
-
4
Remove the antibody solution and wash 4 x 30 minutes with blocking buffer to remove unbound primary antibodies.
-
5
Incubate the tissues with secondary antibodies in the blocking buffer on a shaker at low speed overnight at RT or at 4°C. If using antibodies conjugated with dyes, see FLUORESCENT PROTEINS, ANTIBODIES AND DYES for a description of dye performance and compatibility with pre-expansion staining proExM.
-
6
Remove the antibody solution and wash 4 x 30 minutes with blocking buffer to remove unbound secondary antibodies.
-
7
We recommend moving to the gelation steps immediately.
The samples can be stored in the dark at 4°C in PBS for up to several weeks before gelation.
Gelation
-
8
Prepare solutions as instructed in Table 1A–C, E, as well as Table 2B.
-
9
Exchange the sample buffer (e.g., the blocking buffer if immunostained following the protocols above) with 0.1 mg/mL Acryloyl-X SE (AcX) in PBS. The AcX solution is prepared by diluting the 10 mg/mL AcX/DMSO stock solution (Table 1A) 1:100 in PBS.
-
10
Leave the tissues in AcX solution for >6 hours (overnight is fine) at RT with no shaking.
-
11
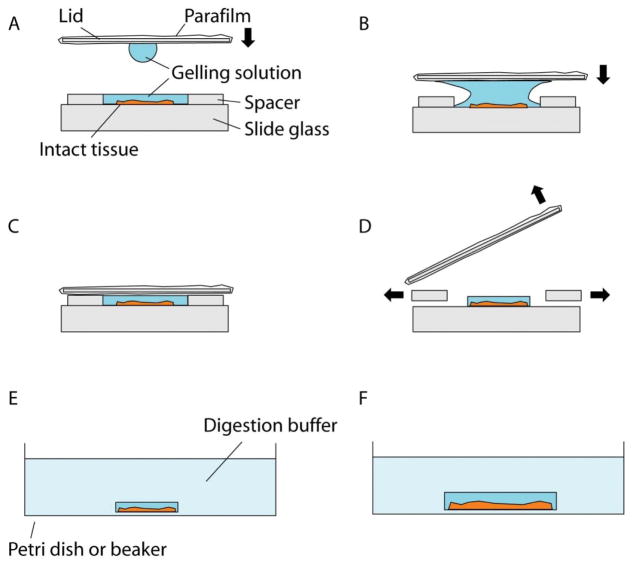
Meanwhile, to construct the gelation chamber, place two spacers on a slide, separated from each other (Figure 4A). Thin and flat objects, such as stacks of coverslips, can serve as the spacers. For example, a stack of two No. 1.5 coverslips has a height of about 0.30–0.33 mm and by changing the number or types of the coverslips, the height of the spaces can be adjusted to accommodate the tissue thickness. The spacer height should be kept close to the tissue thickness to avoid leaving excessive gel on top of the sample, but the total thickness of the gel (i.e., the spacer height) should be at least 0.15 mm for easier gel handling in later steps. A droplet of water (e.g., a few μL) or a very small amount of superglue can be applied to spacers to adhere them to the slide (and, if multiple spacers, to each other). For the gelation chamber lid, a thick and robust coverslip (e.g., a No. 2 coverslip) wrapped in Parafilm can be used, with the Parafilm preventing the gel from sticking to the coverslip when removing the lid in later steps. Make sure the Parafilm surface is flat, clean and free of folds. We recommend finishing preparing the gelation chamber and the lid before moving to the next step.
-
12
After incubating the tissues in AcX, wash the tissues 2 x 15 minutes with PBS. Begin thawing the components for the gelling solution: Stock X, 4HT, APS and TEMED stock solutions (Table 1B, 2B and 1C). Keep the solutions chilled in an ice bath or on a cold block (chilled to 4°C).
-
13
Add Stock X, 4HT, TEMED and APS in a 47:1:1:1 ratio in this order into a microcentrifuge tube (gelling solution), in this order (i.e., APS last) in a microcentrifuge tube, and vortex for a few seconds (Table 2C). The amount of gelling solution should be scaled up or down accordingly to the size of the gelation chamber and the size of the tissue or tissue slice and at least 100-fold excess in volume. For instance, at least 400 μL of gelling solution is used to incubate a 100 μm single coronal slice of mouse brain, enough to fill a gelation chamber with a 22 x 22 x 0.33 mm void, which can fit a full coronal mouse brain slice. To prevent premature polymerization of the gelling solution, APS should be added into the solution last before vortexing. In addition, the mixed gelling solution should be kept in an ice bath or on a cold block (chilled to 4°C). Avoid excessive warming of the gelling solution by, for example, holding the lid of the microcentrifuge tube rather the body of the tube. Finally, we recommend completing the next few steps (until placing the microcentrifuge tube with the gelling solution and the sample at 4°C in step 15) within 5 minutes to prevent premature gelation.
-
14
Immediately after the previous step, use a soft paintbrush to transfer and immerse the tissues in the gelling solution in the microcentrifuge tube.
-
15
Keep the microcentrifuge tube containing the sample and the gelling solution at 4°C in the dark for 30 minutes. After the 30 minutes incubation, we recommend completing the next few steps (until placing the gelation chamber in a 37°C incubator in step 19) within 5 minutes to prevent premature gelation.
-
16
Use the paintbrush to transfer and place the tissue slice between the spacers in the gelation chamber. Make sure the slice is lying flat without wrinkles or distortions. We recommend to first add a droplet of 20 μL of the gelling solution in between the spacers, transfer the tissue slice into the droplet and use the paintbrush to slowly flatten the tissue within the liquid environment, gently pressing it onto the glass surface.
-
17
Add a droplet of 20 μL of the gelling solution to one side of the lid, and flip it over so that the droplet hangs from the surface due to surface tension (Figure 4A). Slowly lower the lid so that the droplet will fuse with the gelling solution on the sample coverslip (Figure 4B). Once fused, continue lowering the lid so it rests on the spacers. Make sure no air pockets are introduced inside the solution during this process.
-
18
Using a pipette and a fine pipette tip, keep adding additional gelling solution to both open sides of the chamber until the space between the lid, spacers and the slide is filled (Figure 4C). Capillary forces will cause the gelling solution to evenly fill the chamber.
-
19
Place the gelation chamber in an incubator at 37°C for 2 hours for polymerization. Be careful not to tilt or shake the chamber during the transfer and the gelation.
Figure 4.
Gelation of intact tissues. (A) Schematic showing the side view of a gelation chamber. The intact tissue is placed in between the spacers, which should be thicker than the tissue. The lid is moved towards the chamber, bearing a droplet of the gelling solution. (B) Schematic showing lid placement. The lid is moved towards the sample so that the droplet merges with the gelling solution in the chamber, which prevents air pocket formation in the gel. (C) Schematic showing the gelation chamber ready for polymerization at 37°C. After the chamber is correctly constructed and filled with gelling solution, polymerization is carried out at 37°C for 2 hours. (D) Schematic showing removal of the lid and the spacers. The lid is first pried open and then the gel is trimmed with a razor blade. (E) Schematic showing the digestion step. The trimmed gel is placed in the digestion buffer. (F) Schematic showing the gel slightly expanded after the digestion step.
Digestion
-
20
Take out the gelation chamber from the incubator.
-
21
Slowly insert a razor blade from the side between the lid and the spacers and pry the lid open. Once the lid is separated from the gel, continue to use the razor blade to remove the spacers in a similar fashion (Figure 4D).
-
22
Using a razor blade, trim off any excessive gel. Trimming the gel will keep it compact and aid in finding the regions of interest in later steps. You may want to trim the gel to have an asymmetric shape so that its orientation can easily be gauged in later steps simply by examination of the shape (see our video at (proExM for tissues: gelation demonstration)).
-
23
Prepare digestion buffer by diluting Proteinase K (ProK) 1:100 with the rest of its components (final concentration of ProK is 8 units/mL) (Table 1E).
We recommend preparing enough digestion buffer to have at least a 10-fold excess in volume compared with that of the gel. -
24
Use a paintbrush to wet the gel and its sides with the digestion buffer. Wait for 10–30 s until the digestion buffer soaks the gel border. This will aid in accessing the gel bottom with the paintbrush, so that the gel can be more easily peeled from the glass substrate.
-
25
Peel off the gel by gently probing the space between the gel and the glass surface with a fine paintbrush. Since the gel will expand to about 1.5x during digestion, choose a suitably large container, such as a petri dish, for the digestion step. For small tissue and tissue slice samples, 1.5 mL microcentrifuge tubes can be used as the container. Use separate containers filled with digestion buffer for each sample. Leave gels immersed in digestion buffer for >8 hours (overnight is fine) at RT in the dark (Figure 4E, F).
Storage and expansion
-
26
Remove the digestion buffer and add PBS, and store the gel at 4°C in the dark if storage for later imaging is desired. Be cautious not to suck up the gel while pipetting.
The embedded tissue is slightly scattering at this stage, so changing the incident angle of light might help to locate it inside the solution. -
27
If the gel needs to be transferred to another container (e.g., a larger container for trimming or the sample holder for imaging), you can use a paintbrush of suitable size to pick up and move the gel. To avoid dehydration, add some PBS to the new container.
-
28
Before expanding the sample, we recommend trimming the gel to the minimum size necessary. You can use a dissection microscope or a low magnification wide-field microscope to locate the region of interest. To minimize sample movement, most of the surrounding liquid can be temporarily removed during trimming. Under the microscope, use a razor blade to cut and remove gel outside of the region of interest. We advise trimming the gel into an asymmetric shape to easily deduce the orientation. Make sure to trim the gel to a reasonable size for handling and imaging as the expansion will introduce an additional 2–2.5x (to a total of about 4.5x) linear increase in size. Transfer the gel with the regions of interest to a container (e.g., a petri dish of suitable size for expansion, or the sample holder for imaging) that is large enough to contain the expanded gel.
-
29
Fill the container and immerse the gel in water and wait for 20 minutes. Exchange the water and wait for another 20 minutes. Redo the exchange one more time (20 minutes in fresh water x 3 in total). The sample should be fully expanded and optically cleared (see Figure 3 for an expanded mouse brain hemi-slice) and ready for imaging (these times are shorter than in previous publications, but reflect numbers that work for routine use in our lab). An example of pre-expansion and post-expansion images of a brain slice containing Brainbow3.0 expressing neurons is shown in Figure 5. For long-term imaging, e.g. when imaging a large volume of specimen, sample mounting is necessary after the expansion and before the imaging (see IMAGING EXPANDED SAMPLES). Sample mounting attaches the sample to the sample holder and thus prevents it from drifting, and also can help keep the specimen within the working distance of the objective lens. For a quick check of the expanded sample under the microscope (< 5 minutes), there is typically no need for sample mounting. On an inverted microscope, for example, you can use a pipette to temporarily (<5 minutes) remove the water around the gel to prevent it from sliding away.
-
30
If you need to transfer or flip the expanded gel to a new container, it needs to be handled with care, as it is somewhat delicate in the expanded state. First remove most of the surrounding water and then use a brush to move the gel onto a clean glass coverslip placed in the same container. Next, gently lift the coverslip (with the gel on top) out of the container with forceps. The gel can be transferred by slowly brushing it off the coverslip to the new container. This method can be also used to flip the gel by inverting the coverslip bearing the gel, in case the gel is facing in the wrong direction.
Figure 5.
Protein retention expansion microscopy (proExM) of virally injected, membrane bound Brainbow3.0 followed by additional antibody staining. A) Maximum intensity projection of a large area of a mouse hippocampus. B) Pre-expansion image of a single optical section of the boxed region in A). C) Post-expansion image of the same optical section as in B). Scale bars: A) 50 μm (physical size post-expansion 198 μm); B) 5 μm; C) 5 μm (19.8 μm). Adapted from Gao et al., 2017 and Tillberg et al., 2016.
PROEXM PROTOCOL FOR THICK INTACT TISSUE SAMPLES (200–500 μm)
For tissues and tissue slices of 200–500 μm thickness, the basic tissue protocol above needs to be modified to allow for deeper penetration of reagents. In addition, the digestion time needs to be prolonged. We have successfully used this protocol to expand samples of up to 500 μm thicknesses; expanding samples thicker than 500 μm may be possible as well.
Materials (in addition to materials used in the BASIC PROEXM PROTOCOL FOR INTACT TISSUES)
2-(N-morpholino)ethanesulfonic acid (MES)
Tissue sample generation is outlined in steps 1–7 in BASIC PROEXM PROTOCOL FOR INTACT TISSUES and can be used for thicker tissue samples.
For the AcX step (step 10 in BASIC PROEXM PROTOCOL FOR INTACT TISSUES), a slightly acidic buffer should be used to suppress the reactivity of the NHS ester of the AcX molecule to allow for deeper penetration into the tissue. We typically use MES buffered saline (MBS) for this slightly acidic buffer, i.e. 100 mM MES + 150 mM NaCl, pH ~6. Dilute the 10 mg/mL AcX/DMSO stock solution 1:100 in the MBS. Leave tissue in the AcX in MBS solution for up to 24 hours at RT, depending on the thickness of the sample. We typically allow for more time for thicker tissues. For instance, we leave a 500 μm brain slice in AcX/MBS solution for 24 hours.
For the gelling solution preparation (step 13 in BASIC PROEXM PROTOCOL FOR INTACT TISSUES), increase the amount of 4HT by 50% (i.e. 47: 1.5 ratio between the Stock X and 4HT) for longer inhibition of polymerization during the incubation. For the incubation (step 15 in BASIC PROEXM PROTOCOL FOR INTACT TISSUES), submerge the sample in the gelling solution for 45–60 minutes (instead of 30 minutes) at 4°C before transferring into the gelation chamber.
For the digestion (step 25 in BASIC PROEXM PROTOCOL FOR INTACT TISSUES), a longer and stronger digestion is necessary. The ProK digestion can be carried out for a longer time and/or at higher temperature (up to 50–60°C), depending on the biological compositions of the tissue and how strongly it is crossed-linked after fixation. For a 250 μm thick mouse brain slice, for example, a 2-day digestion at RT has given adequate digestion of the sample. In this case, the gelled brain slice was digested in digestion buffer (with ProK) at RT for 1 day, then the old buffer was exchanged with fresh digestion buffer (with ProK) and digested for another day.
POST-EXPANSION STAINING PROEXM
Immunostaining can be performed after expansion, as illustrated in Figure 1C. One can perform the mechanical homogenization step using ProK, and then add antibodies afterward. However, only some epitopes, such as YFP, can survive the ProK treatment and can be stained post-expansion; most epitopes do not.
To preserve most epitopes, a milder method, such as protein denaturing by autoclaving in an alkaline detergent-rich buffer (or, alternatively, treatment with a mild protease that cuts at only a small number of sites within proteins, like LysC), can be used for mechanical homogenization (Figure 1C, Supplementary Figure 1 of Tillberg et al., 2016). We describe the protocol for autoclave-mediated disruption below. For examples using various antibodies, see Supplementary Figure 1 of Tillberg et al., 2016.
The following protocol refers to the proExM tissue protocol described earlier, and an example using Thy1-YFP mouse brain slices, and post-expansion anti-GFP staining is included (Figure 6).
Figure 6.
Post-expansion immunostaining. (A) Thy1-YFP-expressing mouse brain hemi-slice before expansion. (B) The brain slice was gelled and mechanically homogenized by autoclaving in an alkaline buffer, followed by anti-GFP primary antibody and Alex Fluor 488 conjugated secondary antibody staining. Scale bar: 1 mm. Adapted from Tillberg et al., 2016.
Infuse a mouse brain slice with AcX and embed in a hydrogel as described in BASIC PROEXM PROTOCOL FOR INTACT TISSUES. After gelation, the sample can be peeled off from the glass substrate and temporarily stored in 1M NaCl solution (e.g., for periods of minutes to hours) before the next step.
Soak the sample in an alkaline buffer (100 mM Tris base, 5% Triton X-100, 1% SDS) for 15 minutes, and then exchange the solution with fresh alkaline buffer of the same composition. Place the sample, immersed in the buffer, in an autoclavable container (e.g., polypropylene bottle), leaving the lid slightly open to allow for pressure relief. Autoclave the samples in the alkaline buffer on liquid sterilization mode, with a temperature of 121°C, for 1 hour. During this and subsequent steps, the sample will expand visibly.
Let the sample cool down to RT and wash 2 x 15 minutes with PBS.
Incubate the sample with primary antibodies in 0.1% Triton X-100 + 5% normal donkey serum in PBS (blocking buffer) on a shaker at low speed overnight at RT or 4°C. (The serum should be from the animal host of the secondary antibodies, so other types of sera should be used instead of the normal donkey serum if the secondary antibodies are from other animals.)
Remove the antibody solution and wash 4 x 30 minutes with blocking buffer.
Incubate the sample with fluorescently labeled secondary antibody of choice in the blocking buffer on a shaker at low speed overnight at RT or 4°C.
Remove the secondary antibody solution and wash 4 x 30 minutes with blocking buffer.
Exchange the buffer with PBS for storage (for up to several weeks), if desired. Expand to the final factor of ~4.5x with 3 x 20 minutes washes in water and then image.
For more detail on how to perform specific steps above, e.g. regarding sample handling, refer to corresponding steps in the section BASIC PROEXM PROTOCOL FOR INTACT TISSUES.
FLUORESCENT PROTEINS, ANTIBODIES AND DYES FOR PROEXM
For the variation of proExM illustrated in Figure 1C (post-expansion staining proExM), any dye conjugated to the secondary antibody should be compatible with the protocol, as the dye is only administered at the end of the process.
For the variations of proExM illustrated in Figure 1A and 1B (pre-expansion staining proExM), most fluorescent proteins are compatible with the basic proExM protocols (see Figure 1b of Tillberg et al., 2016). For example, beta-barrel (e.g., GFP-like) fluorescent proteins are protease resistant and can survive ProK digestion quite well (e.g., >50% fluorescence retention for many fluorescent proteins), but non-beta-barrel fluorescent proteins (e.g., infrared fluorescent proteins based on bacteriophytochromes) are degraded by the ProK step. For antibodies conjugated with fluorescent dyes, most dyes are compatible with the basic proExM protocols (e.g., > 40% fluorescence retention) except for the cyanine-family dyes (Figure 1c of Tillberg et al., 2016). Cyanine-family dyes, such as Cy3, Cy5 and Alexa Fluor 647, are degraded during the polymerization reactions. For far-red dyes, accordingly, we recommend Atto 647N or CF 633.
ExFISH PROTOCOL FOR CULTURED CELLS
ExFISH enables expansion microscopy of RNA by using a small-molecule linker, which we call LabelX, to covalently anchor endogenous RNA molecules within specimens to the swellable gel of ExM. LabelX is analogous to AcX in proExM, but enables RNA molecules to be interrogated after expansion using RNA FISH. For ExFISH-processed cultured cells, single-molecule FISH (smFISH) is used to stain RNA molecules post-expansion.
Materials
For ExM gel preparation:
Sodium acrylate
Acrylamide
N,N′-Methylenebisacrylamide
Ammonium persulfate (APS)
N,N,N′,N′-Tetramethylethylenediamine (TEMED)
4-Hydroxy-TEMPO (4HT)
VA-044
Vacuum desiccator
Tupperware
Nitrogen gas
Nunc 4-well rectangular plates (e.g., Thermo Fisher, catalog no. 267061)
Kapton® Tape or Scotch™ Tape
For hybridization buffer:
Dextran sulfate
20x Saline-sodium citrate (SSC) buffer
Formamide
For fixation and permeabilization:
10x Phosphate buffered saline (PBS)
Tissue-prep buffered 10% formalin
Ethanol
For protease digestion:
Sodium chloride (NaCl)
Ethylenediaminetetraacetic acid (EDTA)
Triton X-100
Tris (1M), pH 8.0
Proteinase K (ProK)
For LabelX preparation:
Label-IT Amine (purchased from Mirus Bio LLC)
Acryloyl-X, SE (AcX)
For LabelX treatment:
3-(N-morpholino)propanesulfonic acid (MOPS)
smFISH Probes:
We primarily use commercially available smFISH probes (eg. Stellaris ® RNA-FISH probes from LGC Biosearch Technologies). Alternatively, it is possible to design custom smFISH probes conjugated to a fluorophore of one’s choice (Raj and Tyagi, 2010). FISH protocols requiring the wash-in of large strands, e.g. RNAscope, are not compatible in our experience, because the DNA strands used for amplification are too large to wash in and out of the gel in a reasonable amount of time.
REAGENTS AND SOLUTIONS
Table 3 Preparation of LabelX and stock solutions for expansion fluorescence in situ hybridization (ExFISH). Stock solutions are prepared as aqueous solutions unless noted otherwise. (A–E) Stock solutions for ExFISH protocol for cultured cells. (A–C and F–J) Stock solutions for ExFISH protocol for tissues. For AcX/DMSO stock solution, gelling solution and digestion buffer, which are needed across multiple protocols, refer to Table 1.
(A) LabelX solution. Resuspend Label-IT amine at 1 mg/mL in the vendor-provided resuspension buffer. Vortex to mix. React the Label-IT/resuspension buffer solution to AcX/DMSO stock solution (Table 1A) at equal mass ratio. For example, add 10 μL of AcX/DMSO stock (at 10 mg/ml) solution to 100 μL of Label-IT solution (both are resuspended in DMSO, so the reaction is carried out in DMSO). React overnight at RT on a benchtop shaker. The AcX is now conjugated to Label-IT, resulting in what we call “LabelX”. Store the LabelX solution at −20°C, in a desiccator, for up to 2 months.
(B) MOPS stock solution. First, prepare the MOPS stock solution (200 mM, 10 times the final concentration) and adjust the pH to 7.7. The MOPS stock solution can be divided into 1mL aliquots and stored at −20°C for up to 12 months. When preparing MOPS buffer, dilute the MOPS stock solution 1:10 in nuclease-free water to bring the final concentration to 20 mM MOPS.
| MOPS stock solution (200 mM) | Stock solution concentration (g/100 mL solution) |
|---|---|
| MOPS | 4.18 |
(C) VA-044 stock solution. The VA-044 stock solution is used to aid gelation. The solution should be prepared 1 mL at a time and used immediately. Keep this stock solution on ice while preparing the gelling solution.
| VA-044 stock solution | Stock solution concentration (g/100 mL solution) |
|---|---|
| VA-044 | 25 g |
(D) Hybridization buffer for single molecule FISH (smFISH). The solution contains 10% (w/v) dextran sulfate, 10% (v/v) formamide, and 2X SSC. First, prepare the hybridization buffer without formamide, which can be stored in 10 ml aliquots at 4°C for up to 2 months.
| Hybridization buffer without formamide | Amount |
|---|---|
| Dextran sulfate | 10 g |
| 20X SSC | 10 mL |
| Nuclease free water | Add until total volume 90 mL |
Add formamide (10% v/v) right before performing FISH.
| Hybridization buffer for smFISH | Amount |
|---|---|
| Formamide | 1 mL |
| Hybridization buffer without formamide | 9 mL |
(E) Wash buffer for smFISH (WA-10). This solution contains 10% (v/v) formamide and 2X SSC, and can be prepared as 10 mL aliquots and stored at RT for up to a week.
| WA-10 | Amount |
|---|---|
| Formamide | 10 mL |
| 20X SSC | 10 mL |
| Nuclease free water | 80 mL |
(F) Hybridization buffer for hybridization chain reaction FISH (HCR-FISH). The solution contains 10% (w/v) dextran sulfate, 20% (v/v) formamide and 2X SSC. First, prepare the hybridization buffer without formamide, which can be stored as 10 ml aliquots at 4°C for up to 2 months.
| Hybridization buffer without formamide | Amount |
|---|---|
| Dextran sulfate | 10 g |
| 20X SSC | 10 mL |
| Nuclease free water | Add until total volume 80 mL |
Add formamide (20% v/v) right before performing HCR-FISH.
| Hybridization buffer for HCR-FISH | Amount |
|---|---|
| Formamide | 2 mL |
| Hybridization buffer without formamide | 8 mL |
(G) Wash buffer for HCR-FISH (WA-20). This solution contains 20% (v/v) formamide and 2X SSC, and can be prepared as 10 mL aliquots and stored at RT for up to a week.
| WA-20 | Amount |
|---|---|
| Formamide | 20 mL |
| 20X SSC | 10 mL |
| Nuclease free water | 70 mL |
(H) 5x SSCT. This solution contains 5x SSC and 0.1% Tween-20, and can be stored at RT for up to a year.
| 5x SSCT | Amount |
|---|---|
| Tween-20 | 0.1 mL |
| 20X SSC | 25 mL |
| Nuclease free water | 75 mL |
(I) 0.05x SSCT. This solution contains 0.05x SSC and 0.1% Tween-20, and can be stored at RT for up to a year.
| 0.05x SSCT | Amount |
|---|---|
| Tween-20 | 0.1 mL |
| 20X SSC | 0.25 mL |
| Nuclease free water | 100 mL |
(J) Amplification buffer. This solution contains 5x SSC, 10% (w/v) dextran sulfate and 0.1% Tween-20, and can be prepared as 1 mL aliquots and stored at 4°C for up to 2 months.
| Amplification buffer | Amount |
|---|---|
| Dextran sulfate | 5 g |
| Tween-20 | 0.05 mL |
| 20X SSC | 12.5 mL |
| Nuclease free water | Fill up to 50 mL |
Cell culture preparation
Cells can be grown as desired to suit experimental needs. However, for ease of gelation and imaging, and handling, we have found 16-well Culturewell removable-chamber cover glasses from Grace Bio Labs (Sigma, catalog no. GBL112358) to be convenient for pre-expansion fluorescence in situ hybridization (ExFISH) imaging, as well as subsequent gelation and digestion. In cases where single molecule FISH (smFISH) is performed before gelation and expansion, we found it convenient to plate cells in 8-well Nunc Lab-Tek chambered cover glasses (Thermo Fisher, catalog no.155411).
Fixation and permeabilization of cultured cells
-
1
Wash cells once with PBS warmed to 37°C to avoid any heat shock.
-
2
Add 10% formalin solution and fix cells for 10 minutes at room temperature (all steps are at RT unless otherwise indicated). Wash with PBS 2 x 2 minutes at RT.
-
3
Replace buffer with 70% ethanol in water for permeabilization. Fixed cells can be stored in 70% ethanol at 4°C for up to 2 weeks. For immediate use, fixed cells should be permeabilized with 70% ethanol for 1 hr at RT.
LabelX treatment of cultured cells
-
4
Rehydrate cells permeabilized with 70% ethanol by washing twice with PBS for 5 minutes per wash at RT. Pre-incubate fixed cultured cells with 20 mM MOPS buffer (Table 3B) for 5 minutes.
-
5
Prepare LabelX (Table 3A) and then dilute in 20 mM MOPS buffer at the desired concentration. We have observed nearly complete RNA retention when using LabelX at a final Label-IT amine concentration of 0.006 mg/mL. For smFISH experiments, we recommend using a final Label-IT amine concentration of 0.006 mg/mL (i.e., a 1:150 dilution from the LabelX stock solution of Table 3A).
-
6
Remove the preincubation MOPS buffer, and add LabelX in MOPS buffer to the fixed cultured cells. For cells grown in the removable-chamber cover glass, use 80 μL of LabelX in MOPS buffer. For cells grown in Nunc Lab-Tek chambered cover glasses, use a volume of 120 μL. Incubate overnight at 37°C. Afterwards, wash twice with PBS at RT for 5 minutes each. At this stage, the cells can be temporarily stored at 4°C for up to a week.
Gelation
The Grace removable-chamber cover glass has an upper structure which can be removed with a removal tool (Sigma, catalog no. GBL 103259). After removal of the chamber, a 1 mm silicone spacer remains with the cover glass and can be used to cast the gel (protocol described below, as well as in BASIC PROEXM PROTOCOL FOR CULTURED CELLS). If cells are grown on a coverslip, then a chamber can be made using coverslip spacers (see BASIC PROEXM PROTOCOL FOR CULTURED CELLS). If cells are grown in Nunc Lab-Tek chambered Cover glasses, we have found that the bottom coverslip along with its 1 mm gasket can be removed from the rest of the plate, and used to cast the gel.
The gelation of fixed cultured cells for ExFISH is different from that of other ExM variants because VA-044, instead of APS, is used as a radical initiator. We found that the use of APS/TEMED yields a dim auto-fluorescent background that is significant in the context of smFISH where the signals themselves are extremely dim. VA-044 is preferable because its use results in minimal auto-fluorescence, which is ideal for smFISH imaging. However, unlike the use of APS with TEMED, VA-044 has a higher decomposition temperature, and initiates radicals at a slower rate. As a result, inhibition due to ambient oxygen can affect gel formation. We have found that degassing followed by perfusion with nitrogen displaces oxygen dissolved in the gelling solution and facilitates gel formation. Therefore, gelation is carried out under a nitrogen atmosphere in a humidified chamber which provides a nitrogen filled environment and minimizes evaporation.
We typically use a Tupperware container, but any container that provides an airtight seal can be used for the humidified chamber (Figure 7). We create two holes in the lid of the Tupperware using a syringe needle. To make the chamber humidified, we add a small amount of water to the bottom of the Tupperware. We position an empty 24-well plastic bottom plate at the very bottom of the Tupperware to provide a raised platform onto which we place the cover glass with the cells and gelling solution; the water level should be much lower than the height of the 24-well plate so that the sample being gelled is kept well above the water.
Figure 7.
Tupperware enclosure setup for nitrogen perfusion and gelling of ExFISH cultured cell samples. (A) Open Tupperware with a plastic platform for supporting the sample and a pool of water at the bottom for humidity. (B) Closed Tupperware with two inlets on the cover sealed with tape. (C) Nitrogen perfusion of sample inside the Tupperware. Tape seals have been removed and a nitrogen line has been inserted through one inlet, with the other remaining open to allow air to escape. (D) Following nitrogen perfusion, the two inlets are sealed with tape, and the Tupperware containing the sample to be gelled is placed in a 60°C incubator.
-
7
For the Grace removable-chamber cover glass: use the removal tool to remove the top wells, leaving behind the black silicone gasket. Leave 40 μL of PBS inside the well to keep the cells hydrated. For the Lab-Tek chambered cover glass, first glue the bottom of the cover glass to a 1 mm thick microscope glass slide for mechanical support using epoxy glue. After the glue has cured, use a razor blade to pry apart the cover glass from the top wells leaving behind the plastic gasket. Leave 100 μL PBS inside the well to keep the cells hydrated.
-
8
Prepare gelling solution by mixing monomer solution (Table 1B) and VA-044 solution (Table 3C) on ice or a cooling block. Dilute the VA-044 to a final concentration of 0.5% (w/v) in monomer solution (i.e., dilute the stock 1:50 in monomer solution). For example, to prepare 1 mL of gelling solution, add 20 μL of VA-044 solution and 40 μL of water to 940 μL of monomer solution. The gelling solution should be prepared at 4°C, and used for the next step immediately.
-
9
In a cold block/rack, or on ice, distribute the gelling solution into 200 μL aliquots in microcentrifuge tubes. Degas for 10 minutes in a vacuum desiccator.
-
10
Remove any remaining PBS from the wells on the cover glasses and add the degassed gelling solution. For the Grace removable-chamber cover glass, add 40 μL of gelling solution. Add 200 μL of gelling solution if using the Lab-Tek chambered cover glass. Place the cover glass in a vacuum desiccator and degas for another 10 minutes. In the meantime, prepare a glass slide covered in Parafilm as the lid (see Fig. 2 and associated text, above). Make sure the Parafilm surface is flat, clean, and free of folds. The Parafilm prevents the gel from sticking to the lid when removing the lid in later steps. Remove the cover glass from the desiccator and immediately place the Parafilm glass slide directly on top of the cover glass. Any excess gelling solution might spill out to the sides. Proceed to the following steps without delay.
-
11
Place the cover glass onto the plastic platform in the humidified chamber. Seal the chamber (Figure 7A, 7B).
-
12
Remove the tape covering the holes. Connect the inlet of the nitrogen line to a syringe tip and insert the tip through one hole (Figure 7C). Slowly, turn on the nitrogen flow regulator to flush nitrogen into the chamber. Increase the flow until you can feel the airflow coming out of the other hole. Flush the chamber with nitrogen for 10 minutes. When done, remove the inlet and immediately seal both holes with tape again (e.g. Kapton® Tape or Scotch™ Tape).
-
13
Place the chamber in a 60°C incubator for 2 hours to initiate the gelation (Figure 7D). Be careful not to tilt or shake the chamber during the transfer and the gelation.
Digestion
-
14
When gelation is finished, take out the humidified chamber/Tupperware from the oven and remove the cover glass. Using a razorblade, gently pry open the Parafilm-covered lid from the top of the wells. For the Grace Biolabs removable-chamber cover glass, gently remove the black silicone gasket by peeling it off using your hands or a pair of forceps. For the Lab-Tek chambered cover glass, do not attempt to remove the plastic gasket. At this point, you will see that gels will have formed in the individual wells on the cover glass.
-
15
For digestion, place the cover glass in a Nunc 4-well rectangular plate. Prepare digestion buffer with Proteinase K (Table 1E). Add 6 mL of digestion buffer into the well containing the cover glass (if using any other container, add digestion buffer of at least 10 times the gel volume to completely cover the cover glass and gels). Make sure the gel is entirely immersed for at least 2–3 hours until it naturally peels off from the sample cover glass (Figure 2F). If a removable-chamber cover glass is used, use a diamond scribe to separate each sample and its corresponding piece of the cover glass, by scribing the cover glass and breaking each piece of glass off bearing its piece of gel. Note that the cell culture substrate is placed so that the gel is on top of the substrate and the cultured cells are at the bottom surface of the gel. Optionally, the detached cell substrate can be carefully removed using forceps, to just leave the cell-containing gel in the digestion buffer.
-
16
Leave the gel immersed in the digestion buffer overnight at RT in the dark.
Storage and expansion
-
17
After digestion, gels can be expanded (temporarily, see below) in a petri dish, or any container large enough to hold the gels. To fully expand, wash gels with excess volume nuclease free water three times with at least an hour per wash. Also see Storage and expansion of proExM samples in BASIC PROEXM PROTOCOL FOR CULTURED CELLS.
smFISH staining of cultured cells after expansion
-
18
Since the gels formed in either of the above chambered cover glasses have a thickness of ~1 mm before expansion, the thickness of the expanded gels needs to be cut down to facilitate efficient diffusion of FISH probes through the gel. To shave down fully expanded gels to a thickness of 1mm, we use 1 mm thick microscope glass slides as spacers as follows. Prepare a large plastic dish to use as a cutting board. We often use the plastic covers of Nunc 4-well rectangular plates, although any similarly sized flat plastic surface will work. Use a small amount of epoxy to glue two 1 mm thick microscope glass slides on their flat slide onto the plastic surface. Position the two glass slides such that they are aligned along their longest dimension with 2.5–3 cm space in between. Allow the epoxy to harden.
-
19
Position a fully expanded gel in between the two glass slides on the plastic cover. The expanded gel should fit in between the two glass slides. If not, trim the gel with a razorblade. Position the gel such that the side of the gel with cultured cells is facing the bottom.
-
20
Place a razor blade across the glass slides so that it bridges the slides. Carefully slide the razor blade along the glass slides and through the gel to shave off everything except the bottom 1 mm of the gel containing the cells.
-
21
Carefully collect the shaved gel at the bottom containing the fixed and expanded cells and move it to PBS. If needed, at this point, shaved gels can be stored in 1x PBS at 4°C for up to a month.
-
22
Proceed to smFISH staining, which can be performed in any container, though we often use glass bottom 24-well plates for the convenience of imaging right after staining.
-
23
Prepare gels by incubating with wash buffer (WA-10) (Table 3E) for 30 minutes at RT.
-
24
Prepare probes by diluting in smFISH hybridization buffer (Table 3D) at the concentration indicated by the vendor for smFISH. We typically carry out the hybridization at a total probe concentration of 100 nM. Vortex to mix.
-
25
Remove the wash buffer from the gels. Add the hybridization buffer with probes onto the gels. Add enough volume to completely cover the gel to be stained (for 24-well plates, we recommend 300 μL). Incubate overnight (or for > 6 hours) at 37°C.
-
26
Wash gels twice with excess volume (e.g. 500 μL for 24-well plates) of WA-10 at 37°C with 30 minutes per wash.
-
27
Wash once with excess volume PBS at 37°C for 30 minutes.
-
28
Imaging can be performed in PBS or any other buffer of choice (expansion factor is determined by salt concentration: using regular PBS results in ~2x expansion; using 0.02x PBS (diluted in water) results in ~3x expansion while still preserving hybridization). An example with HeLa cells is shown in Figure 8. Refer to IMAGING EXPANDED SAMPLES for how to image expanded gels in the most common microscope setups. While the quality of the acquired image will depend on the imaging setup, wide-field imaging is generally recommended over confocal imaging due to the photobleaching of smFISH stains observed during confocal imaging.
Figure 8.
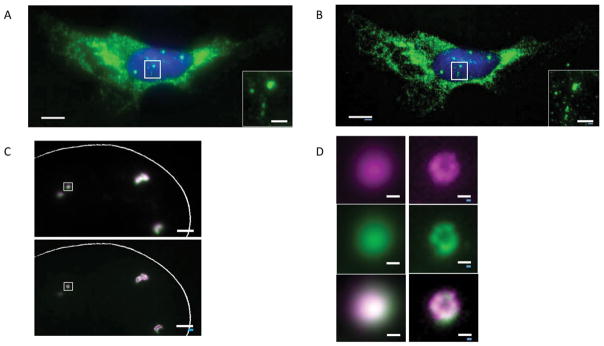
Nanoscale imaging of RNA in cultured cells via expansion fluorescence in situ hybridization (ExFISH). (A) smFISH image of ACTB before expansion of a cultured HeLa cell. Inset shows zoomed in region, highlighting transcription sites in nucleus. (B) As in A, using ExFISH. (C) smFISH image before expansion (top) and using ExFISH (bottom) of NEAT1 lncRNA in the nucleus of a HeLa cell. Magenta and green indicate probe sets binding to different parts of NEAT1. (D) Insets showing a NEAT1 cluster (boxed region of C) with smFISH (left) and ExFISH (right). Scale bars (white, in pre-expansion units; blue scale bars are divided by the expansion factor noted): (A,B) 10 μm (expansion factor, 3.3x), inset 2 μm; (C) 2 μm (3.3x); and (D) 200 nm (3.3x). Adapted from (Chen et al., 2016).
ExFISH PROTOCOL FOR TISSUES
The protocol for performing ExFISH in tissues is different from the cultured cell protocol in a few key aspects. First, the tissue ExFISH protocol requires signal amplification after performing FISH, because conventional smFISH is too dim for confocal imaging and similar 3D imaging techniques needed for tissues. As a result, we use the hybridization chain reaction (HCR) to amplify the signal from FISH probes. Second, the gelling procedure is similar to the proExM protocol for tissues, than to the procedure for cultured cells. Third, higher concentrations of LabelX are used, because better RNA retention yields are observed in tissues at higher concentrations. Finally, after Label-X treatment, it is possible to use AcX to retain fluorescent proteins.
Materials
For fixation and permeabilization:
10x phosphate buffered saline (PBS)
4% paraformaldehyde (PFA) in PBS
Ethanol
For ExM gel or preparation:
Sodium acrylate
Acrylamide
N,N′-Methylenebisacrylamide
Ammonium persulfate (APS)
N,N,N′,N′-Tetramethylethylenediamine (TEMED)
4-Hydroxy-TEMPO (4HT)
Nunc 4-well rectangular plates (e.g., Thermo Fisher, catalog no. 267061)
For hybridization buffer:
Dextran sulfate
20x Saline-sodium citrate (SSC) buffer
Formamide
For protease digestion:
Sodium chloride (NaCl)
Ethylenediaminetetraacetic acid (EDTA)
Triton X-100
Tris (1M), pH 8.0
Proteinase K (ProK)
For hybridization chain reaction (HCR) amplification:
HCR Hairpins (purchased from Molecular Instruments; each hairpin comes at 3μM concentration in a storage buffer)
Amplification buffer (Table 3J)
Tween-20
For LabelX preparation:
Label-IT Amine (purchased from Mirus Bio LLC)
Acryloyl-X, SE (AcX)
For LabelX treatment:
3-(N-morpholino)propanesulfonic acid (MOPS)
HCR-FISH Probe Design:
Probes for HCR-FISH are designed using Stellaris Probe Designer software from LGC Biosearch Technologies. 22bp binding sequences spaced 2bp apart are designed using the software. We often aim for at least 20 sequences targeting each transcript of interest. To design HCR-FISH probes, HCR initiator sequences (Choi et al., 2014) are appended to each binding sequence via a 2-base spacer (AA). The initiator sequences can be appended to either the 5′ end or 3′ end of each binding sequence depending on the orientation of the initiator sequences; I1 initiators are appended to the 5′ end while I2 initiators are appended to the 3′ end. The HCR-FISH probes generated by this procedure are 60bp in final length.
Tissue fixation and slicing
Tissues can be fixed using protocols of your choice. Below is a perfusion fixation and slicing protocol for mouse brain slices that has demonstrated consistent results.
-
1
Transcardially perfuse the mouse brain with ice-cold PBS (5–10 mL) followed by ice-cold 4% paraformaldehyde in PBS (~30 mL).
-
2
Post-fix the brain in 4% paraformaldehyde overnight at 4°C.
-
3
Wash the brain with PBS before cutting slices.
-
4
Cut brain slices on a vibratome in PBS. The thickness of slices should be less than 200 μm to allow for efficient staining with FISH probes after expansion.
-
5
Slices can be stored in PBS if they are used immediately (within 1–2 days), or stored in 70% ethanol for longer times for improved RNA stability at 4°C.
LabelX treatment, gelation, and digestion of tissues and tissue slices
-
6
If tissues or tissue slices were stored in 70% ethanol, rehydrate slices by washing twice with PBS for 15 minutes per wash at room temperature. Otherwise, proceed with the rest of the protocol.
-
7
Pre-incubate tissues or tissue slices with 20 mM MOPS buffer (Table 3B) for 30 minutes.
-
8
Prepare LabelX (Table 3A) by diluting in 20 mM MOPS buffer at a final Label-IT amine concentration of 0.1 mg/mL (i.e. a 1:10 dilution from the LabelX stock solution).
-
9
Remove the pre-incubation MOPS buffer and add to the tissue slices LabelX in MOPS buffer. For convenience, tissues or tissue slices can be processed in 24 well or 48 well plates. (Use enough volume to cover the slices: for example, in a 24 well plate, four 50 μm sections can be incubated together in a 250 μL solution.)
-
10
Incubate tissues or tissue slices with LabelX in MOPS overnight at 37°C. Afterwards, wash twice with PBS for 15 minutes per round.
-
11
(Optional). To retain fluorescent proteins in tissue slices from transgenic specimens, it is possible to do an additional incubation with AcX. Incubate slices with 0.05 mg/ml AcX in 1x PBS at room temperature for at least 6 hours. Wash twice with 1x PBS for 15 minutes per wash.
-
12
Proceed to gelation and digestion as described in the BASIC PROEXM PROTOCOL FOR INTACT TISSUES. Unlike the ExFISH gelation procedure for cultured cells, here gelation with APS/TEMED can be carried out as in the proExM protocol because signal amplification with HCR renders the dim auto-fluorescence that arises from APS/TEMED negligible.
-
13
After digestion, wash gels with PBS twice for 30 minutes each. Gels can be stored in PBS at 4°C after this step for up to two months.
Staining gelled slices with HCR-FISH
-
14
While staining can be performed in any chamber, we often use 24-well plates. Prepare gels by incubating with wash buffer (WA-20) (Table 3G) for 30 minutes at RT.
-
15
Prepare probes by diluting in HCR-FISH hybridization buffer (Table 3F) at the desired concentration (~1 nM per probe). Vortex to mix.
-
16
Remove the wash buffer from the gels. Add the hybridization buffer with probes onto the gels. Add enough volume to completely cover the gel to be stained (for 24-well plates, 300 μl). Incubate overnight at 37°C.
-
17
Wash gels twice with excess volume (e.g., 500 μL for 24-well plates) of WA-20 at 37°C for 30 minutes per wash. For slices thicker than 100 μm before gelation, increase wash times to 45 minutes per wash.
-
18
Wash once with excess volume PBS at 37°C for 2 hours.
-
19
Wash once with excess volume of PBS at RT for 2 hours. For slices thicker than 100 μm before gelation, increase wash time to 4 hours. If needed, after this step, gels stained with FISH probes can be stored overnight in PBS at 4°C before HCR amplification.
HCR amplification (Adapted from (Choi et al., 2014))
-
20
Pre-incubate slices with amplification buffer (Table 3J) for 30 minutes at RT.
-
21
Prepare fluorescently labeled hairpins by snap cooling each hairpin (heat at 95°C for 90 seconds, then cool to room temperature in a drawer for 30 minutes). For each 500 μL of total amplification buffer to be used (in the next step -- we use 150 μL amplification buffer per well of a 48 well plate, or 250 μL per well of a 24 well plate), 10 μL of each hairpin is needed.
-
22
Prepare hairpin solution by adding all snap-cooled hairpins to amplification buffer at RT, using the right volume of amplification buffer to get to the desired final volume. For example, to prepare 500 μL of total amplification buffer with two hairpins, add 10 μL of each hairpin to 480 μL of amplification buffer. Vortex to mix.
-
23
Remove the pre-amplification solution and add the hairpin solution to the gels for 3–6 hours at RT.
-
24
Stop amplification with 4 x 30 minute washes with 5x SSCT (Table 3H).
Expansion and imaging
-
25
At this point the samples can be imaged. To expand the samples, wash 3x 10 minutes with 0.05x SSCT (Table 3I). The expansion factor can be tuned by altering the salt concentration; imaging can also be performed in regular PBS for ~2x expansion. An example mouse brain slice is shown in Figure 9. Refer to IMAGING EXPANDED SAMPLES for how to image expanded gels in the most common microscope setups.
Figure 9.
3D nanoscale imaging of RNA in mouse brain tissue using hybridization chain reaction amplified expansion fluorescence in situ hybridization (HCR-ExFISH). (A) Schematic for HCR-mediated signal amplification. FISH probes bearing HCR initiators are hybridized to a target mRNA, and during amplification, metastable DNA hairpins bearing fluorophores assemble into polymer chains onto the initiators, thereby amplifying the signal downstream of the FISH probe hybridization event. (B) Widefield fluorescence image of Thy1–YFP mouse brain showing HCR-ExFISH of YFP mRNA and Gad1 mRNA (red, YFP protein; cyan, YFP mRNA; magenta, Gad1 mRNA). (C) Confocal image of mouse hippocampal tissue from A, showing single RNA puncta. Inset, one plane of the boxed region. Scale bars (white, in pre-expansion units; blue scale bars are divided by the expansion factor noted): (B) 500 μm (expansion factor 2.9x); (C) 50 μm (2.9x), inset 10 μm. Adapted from (Chen et al., 2016).
IMAGING EXPANDED SAMPLES
Materials
Coverslips (e.g., 24 x 60 mm No. 1.5 coverslips)
Glass-bottom dishes or multi-well plates (e.g., Mattek, catalog no. P35G-0.170-14-C)
Razor blade
Poly-L-lysine
Low melting point agarose
Superglue
Paper wipes (e.g., Kimwipes)
For agarose mounting, 40°C water bath to keep agarose solution liquid until needed
Sample expansion for Imaging
Place the gel-embedded, digested sample in PBS (e.g. step 30 in BASIC PROEXM PROTOCOL FOR CULTURED CELLS, end of step 25 in BASIC PROEXM PROTOCOL FOR INTACT TISSUES, step 17 in ExFISH PROTOCOL FOR CULTURED CELLS, or step 13 in ExFISH PROTOCOL FOR TISSUES) in a petri dish.
Use a dissection scope or a wide-field microscope with a low magnification (e.g., 4x or 10x) objective to find the region of interest. For ExFISH samples, this should be done before staining with FISH probes (i.e. before step 18 in ExFISH PROTOCOL FOR CULTURED CELLS, or before step 14 in ExFISH PROTOCOL FOR TISSUES).
Under the microscope, use a razor blade to remove excess gel (e.g., away from the region of interest). Cultured cell ExFISH samples may also need to be trimmed axially to reduce thickness (see steps 18–21 in ExFISH PROTOCOL FOR CULTURED CELLS).
Fully expand the proExM samples with water. ExFISH samples should be stained with FISH probes and expanded in diluted PBS for cultured cells, or diluted SSC buffer for tissues, following the protocols described earlier (i.e. steps 22–28 in ExFISH PROTOCOL FOR CULTURED CELLS, or steps 14–25 in ExFISH PROTOCOL FOR TISSUES).
Sample mounting
Mounting an expanded ExM sample on a stable surface can help prevent the sample from drifting during imaging, which is important for obtaining high quality images. The best mounting method will depend on the geometries of your microscope and the sample holder (coverslip, Petri dish, or well of a multiwell plate, for example), as well as your imaging requirements such as imaging time and the objective type and magnification.
For a quick inspection (< 5 minutes) of the expanded samples using an inverted or upright microscope with a dry objective, there is typically no need for sample mounting. You can use a pipette to temporarily (< 5 minutes) remove the liquid around the gel to prevent it from floating or sliding away.
For long-term (>5 minutes) imaging or imaging that requires minimal sample drift (e.g., obtaining a z-stack), you might want to mount the sample by physically attaching the expanded samples to the sample holder. In the following sections, we describe three common mounting methods: agarose, poly-L-lysine and superglue. For inverted microscopes, you can use poly-L-lysine to mount the gel to a stable surface. For upright microscopes, you can use agarose, poly-L-lysine, or superglue. For light sheet microscopes, you can use superglue or poly-L-lysine.
The advantage of the poly-L-lysine method is that it provides a transparent interface between the gel and the surface of the sample holder. Therefore, it is suitable for microscope configurations that require imaging through the sample holder, such as an inverted microscope. In comparison, agarose can introduce optical scattering and aberration, and superglue is not transparent after curing. The advantage of the agarose method is that you can retrieve your sample after imaging, as the sample mounting is reversible. The sample can be shrunk (and then separated from the agarose) by placing in high salt buffer after imaging. The advantage of the superglue method is its strong adhesion compared with the other methods. It is suitable for long-term imaging on an upright microscope or a light sheet microscope, even for days.
Sample mounting with poly-L-lysine
Coverslips, glass bottom dishes and glass-bottom multi-well plates can be used as sample holders. Slides or plastic multi-well plates can also be used but the thickness of the sample holder may limit the choice of objectives and/or cause aberrations on inverted microscopes.
Clean the surface of the sample holder by rinsing with water and then ethanol. After rinsing, let the holder dry in air.
Soak the glass surface with 0.1% (w/v) poly-L-lysine aqueous solution for 20 minutes.
Remove the 0.1% poly-L-lysine solution and rinse the coated glass surface with water three times.
Air-dry the glass surface for 1 hour in a clean environment or use dry nitrogen gas to blow-dry the surface.
Use a coverslip and forceps to pick up and transfer the expanded gel to the poly-L-lysine modified glass surface, as follows. First, remove most of the liquid around the gel. Then place a clean coverslip next to the gel and use a paintbrush to slide the gel onto the coverslip. Next, lift the coverslip (with the gel on top) out of its container with a pair of forceps. Before transferring the gel to the poly-L-lysine modified surface, the gel surface that is to be attached needs to be dried for good adhesion. You can use a small piece of a paper wipe to wick away excess liquid. Start absorbing excess liquid from the sides of the gel and then carefully use, for example, the tip of the paper wipe to absorb the liquid in between the bottom of the gel and the coverslip. Use the paintbrush to slide off the gel from the coverslip onto the poly-L-lysine modified surface. Be aware of the orientation of the gel, in the context of your later microscopy work: for example, the side facing toward the sample holder will be more easily reached by the objective (e.g., within the objective working distance) on an inverted setup. In contrast, on an upright microscopy setup, the side facing away from the sample holder will be more accessible to the objective. Therefore, if the region of interest is closer to one particular side of the gel, place that side closer to the objective when mounting the sample. This practice becomes important for expanded samples, as the sample thickness and the gel thickness are scaled up by the expansion factor. This gel transfer should be carried out in a timely manner to avoid dehydration of the gel: we recommend finishing this step within 5 minutes.
After the mounting, immediately add a small amount of water (or diluted PBS for ExFISH samples) to keep the gel hydrated and avoid gel shrinkage.
Sample mounting with agarose
Coverslips, glass bottom dishes and glass-bottom multi-well plates can be used as sample holders. Slides or plastic multi-well plates can also be used but the thickness of the sample holder will limit the choice of objectives and/or cause aberrations on inverted microscopes.
Prepare 0.5% (w/v) low melting agarose solution in the correct imaging medium. For examples, for proExM samples, prepare 0.5% (w/v) agarose in water. For ExFISH samples, prepare 0.5% (w/v) agarose in PBS or other buffer of your choice. Keep the agarose solution warm in a 40°C water bath to avoid premature hardening. If hardened, use a short microwave cycle (10–20 seconds) to melt the agarose solution. Alternatively, heating it to ~60°C will re-melt it as well.
Transfer the expanded gel to the sample holder of choice (e.g., coverslips and glass-bottom dishes). During the transfer, remove and wick away liquid from the expanded gel (see step 5 of Sample mounting with poly-L-lysine, above).
Using a pipette, carefully apply the melted agarose solution on the edges of the gel. If necessary, you can also embed the entire gel in agarose, using the melted agarose solution.
Wait for the agarose to harden at RT or at 4°C and then add a small amount of water (or diluted PBS for ExFISH samples) to prevent dehydration.
After imaging is complete, you can place the mounted sample in high-salt buffer such as PBS. This will shrink the gel but not the agarose. Using paintbrushes or forceps, you can separate the gel from the agarose. The gel can be then stored in PBS for later use.
Sample mounting with superglue
Coverslips, glass bottom dishes and glass-bottom multi-well plates can be used as sample holders. Slides or plastic multi-well plates can also be used. However, since the superglue turns opaque upon curing, this mounting method is not compatible with inverted microscopes which require imaging through the sample holder.
Clean the surface of the sample holder by rinsing with water and then ethanol. After rinsing, let the holder dry in air.
Apply superglue to the surface of the sample holder. Only apply the glue to the area where the gel should be attached. Also make sure excessive superglue is removed by paper wipes, leaving only a thin layer.
Transfer the expanded gel to the sample holder of choice. Before the transfer, remove and wick away liquid from the expanded gel (see step 5 of Sample mounting with poly-L-lysine).
Once the gel is placed on the glue, wait for 20–30 s for the superglue to cure before adding water (or diluted PBS for ExFISH samples) to hydrate the gel. After curing, there should be an opaque interface between the gel and the superglue.
Imaging ExM samples with inverted microscopes (e.g., inverted spinning disk confocal microscopes)
A typical inverted microscope setup is shown in Figure 10. The poly-L-lysine mounting method is recommended for inverted microscopes, since the transparent interface between the gel and the poly-L-lysine modified surface will allow for imaging through the sample holder.
Figure 10.
Imaging expanded ExM samples with inverted microscopes. (A) Schematics showing the configuration of the objective and the expanded sample on an inverted microscope (with a water-immersion objective). The green and red arrows show the excitation and the emission light, respectively. (B) Typical imaging setup on an inverted spinning disk confocal microscope with a coverslip. (C) Typical imaging setup on an inverted spinning disk confocal microscope with a glass-bottom multi-well plate.
For imaging on an inverted microscope, we recommend starting with low magnification (4x or 10x) objectives to find the region of interest first and then switching to high magnification objectives. Air objectives are typically low-magnification objectives (e.g., 4x or 10x) and can be used at the beginning to find the samples and the regions of interest. For high-magnification imaging (>20x), we recommend using water-immersion objectives to avoid aberrations caused by refractive index mismatch between the aqueous mounting medium and the coverslip or oil. Due to the generally short working distances, oil-immersion objectives can be considered for use mainly when the expanded gel is mounted close (within ~10 μm) to the coverslip and when the specimen or object of interest is close to the objective (such as in cultured cells or thin tissues). The working distance of the objective is typically not an issue for thin samples such as cultured cells, if the gel is properly mounted on the sample holder (i.e. the sample side of the gel is mounted facing the objective side using the poly-L-lysine method). For some tissue samples, however, the working distance becomes an issue as it requires imaging deep into the gel (e.g., >300 μm). In this case, long-working distance water-immersion objectives are recommended. As with every microscopy experiment, make sure the correction collar on the objective (if applicable) is adjusted to match the thickness of the coverslips.
Many of the wide-field and confocal microscopes used in biology labs and imaging facilities are inverted microscopes. We found that spinning disk confocal microscopes offer a good combination of resolution and speed and are highly suitable for fast nanoscale imaging of expanded tissues. Laser scanning confocal microscopes can be used most effectively for high signal-to-background imaging of thin and small samples such as cultured cells.
Imaging with upright microscopes
For upright microscopes, all three mounting methods, poly-L-lysine, agarose, and superglue, can be used. If you plan to retrieve your sample after imaging, we recommend the agarose mounting method. In the other two methods, the gel is physically attached to the sample holder, but can potentially be cut free by a razor blade if the sample needs to be retrieved (note: this process can damage the sample, if you are not careful). For long-term imaging (e.g., over a few days), we recommend using the superglue mounting method as it offers the strongest adhesion to the sample holder. The poly-L-lysine method gives a transparent interface between the gel and the sample holder and thus can be imaged on an inverted microscope. For imaging on an upright microscope, we recommend starting with low magnification (4x or 10x) objectives to find the region of interest first and then switching to high magnification objectives, similar to the procedure on an inverted microscope. For high magnification imaging (>20x), we recommend using water-dipping or water-immersion objectives as the gel is immersed in aqueous mounting medium.
Imaging with microscopes with the sample hanging from above (e.g., light sheet microscopes)
Light sheet microscopes are suitable for imaging of expanded tissue samples at high speeds, using long working distance objectives. To accommodate the objectives, some light sheet microscopes hang the samples from above. For example, a typical imaging setup of a light sheet microscope (Zeiss Z.1 light sheet microscope) is shown in Figure 11. The Z.1 light sheet microscope uses a rod to hang samples from above the objectives, perpendicular to the microscope’s light path. The sample is then enclosed in an imaging chamber filed with aqueous mounting medium. For expanded ExM examples, a thin coverslip can be used as the backing to physically support the gel and then the coverslip backing can be attached to the rod through a small 3D-printed adapter, which is available for download at our web resource http://expansionmicroscopy.org. To attach the gel to the coverslip backing, we recommend the superglue mounting method as it most strongly attaches the gel to the coverslip. Poly-L-lysine mounting can be also used for short-term imaging but we have observed detachment of the gel if the mounting is not properly performed (e.g., due to expired poly-L-lysine, fast stage movements, or inadequate water removal from the gel before placement). After mounting the gel to the coverslip backing, the coverslip backing can be attached to the adaptor with superglue. The adaptor is designed to match the dents at the end of the rod so it can be mechanically fixed to the rod.
Figure 11.
Imaging expanded ExM samples with light sheet microscopes. (A) Schematics showing configurations of the objectives and the mounted sample on a commercial light sheet microscope (Zeiss Z.1 light sheet). The green and red arrows show the excitation and emission light, respectively. Note that the gel is facing the objective and is mounted on a coverslip as the backing, which is glued onto a 3D-printed adapter that is connected to the rod-shaped sample holder hanging from the top (the rod is not shown in the schematic). The sample is immersed in water or diluted buffer and the excitation light sheet travels through the sides of the gel perpendicular to the detection objective focal axis. (B) Sample mounting inside the Zeiss Z.1 light sheet microscope, with the imaging chamber removed. Two illumination objectives are on the side to form the excitation light sheets, and a water-immersion objective is located perpendicular to the illumination objectives for the imaging. (C) The same setup as B with the imaging chamber installed. The two side glass windows allow the light sheet to illuminate the sample and the front glass window is used for rough sample alignment using a small camera. Together with the imaging objective at the back, the chamber is sealed and is filled with water (or diluted PBS for ExFISH samples) during the imaging. (D) 3D representation of Zeiss Z.1 light sheet microscopy image of Thy1-YFP (green) brain tissue with ExFISH targeting YFP (red) and Gad1 (blue) mRNAs. (E) Maximum intensity projection (~8 μm in Z) of a subvolume of D demonstrating the capability to resolve single RNA molecules within a cell body after expansion. (F) Volume rendering of a subvolume of D. Scale bars, 10 μm (E, in pre-expansion units) and 20 μm (F, in pre-expansion units). About 3x expansion factor for all samples. Adapted from (Chen et al., 2016). (G) Rendered image of a section of a large scale 3-D multicolor recording of a virally delivered Brainbow3.0 mouse hippocampus section using pre-expansion staining proExM, and imaged on a Zeiss Z.1 light sheet microscope. The expansion factor is ~4.5x and the resulting effective lateral pixel size is 60 nm. Scale bar: 100 μm (in pre-expansion units).
On the Zeiss Z.1 light sheet microscope, we typically use a 1.0 NA, 20x long working distance water immersion objective for high resolution imaging. If the imaging volume is large and requires a long imaging time, we can use lower magnification objectives (5x) to obtain a low-resolution, quick overview of the sample. The Z.1 light sheet microscope is suitable for imaging large, multi-color samples with its two camera set-up, fast image acquisition and capability to capture two color channels simultaneously. In addition, the long working distance of the objective can be used to image large transparent samples. These specifications of the Z.1 light sheet microscope are particularly advantageous for imaging ExM samples.
We first demonstrated these advantages by imaging a ~575 x 575 x 160 μm volume (~6 x 1010 voxels in three colors) of a HCR-ExFISH sample in about ~3 hours with a resolution capable of resolving single RNA puncta within the expanded tissue (Figure 11D–F) (Chen et al., 2016). Even larger samples in the millimeter range have been successfully imaged with this system. One such example is shown in Figure 11G, in which a subregion of a mouse hippocampus infected with a Brainbow3.0 AAV virus (Figure 5) was expanded with a proExM protocol described earlier. The imaging volume was ~1.5 x 0.8 x 0.1 mm in pre-expansion units and the dataset required stitching of 180 individual 3-D image stacks, resulting in a 5 Terabyte, 0.7 Teravoxel dataset with an effective lateral pixel size of about 60 nm.
COMMENTARY
Background Information
We recently discovered that preserved biological samples could be embedded in a polymer hydrogel and physically expanded in an isotropic fashion, thus enabling nanoscale resolution imaging on conventional diffraction limited microscopes, a process we call expansion microscopy (ExM) (Chen et al., 2015). Although preserved biological specimens have been embedded in polymer hydrogels (such as polyacrylamide) since the early 1980s for the purpose of improving microscopy of biological specimens (Hausen and Dreyer, 1981) (Germroth et al., 1995), expansion microscopy utilizes a charged polyelectrolyte gel (i.e., sodium polyacrylate) that is capable of 100x volumetric swelling upon exposure to water (Tanaka et al., 1980), and also explicitly links biomolecules and/or labels to the swellable gel, while decoupling them mechanically from each other, to insure even separation. In addition, the hydrogel is synthesized in a densely cross-linked topology that allows it to expand evenly, preserving the relative organization of the biomolecules/labels.
Our original ExM protocols (Chen et al., 2016; Tillberg et al., 2016; Chen et al., 2015) allowed for an isotropic expansion factor of ~4.5x in each direction (~100x volumetric expansion), enabling a 300 nm diffraction limit objective lens to have an effective resolution of ~300 / 4.5 ~ 60–70 nm. In these forms of ExM, key biomolecules and labels in a cell or tissue are first covalently equipped with anchoring molecules that enable them to be tethered to a polyacrylate network that is synthesized evenly (and densely, with a spacing of perhaps a few nanometers (Cohen et al., 1992)) throughout the specimen. The sample’s mechanical structure is then dissolved and disrupted (e.g., by proteolysis or protein denaturation) followed by uniform expansion by dialysis in water. The expansion results in physical separation of the biomolecules or labels, with their relative positions preserved, so that biomolecules previously spaced within the diffraction limit are moved far enough apart so that they can be resolved with a diffraction-limited microscope. In this unit, we described step-by-step protocols for two versions of ExM, protein retention ExM (proExM) (Tillberg et al., 2016) and expansion fluorescence in situ hybridization (ExFISH) (Chen et al., 2016), in which proteins and RNAs, respectively, are anchored to the hydrogel. One can also combine the two protocols to examine both proteins and RNA in the same sample (e.g., see Fig. 3 of ref. (Chen et al., 2016)). Recently we showed that a sample could be expanded twice, by essentially repeating the ExM procedure, resulting in a ~4.5 x 4.5 ~20x linear expansion (~8000x volumetric expansion), and a corresponding resolution of ~25 nm (Chang et al., 2017). The protocol for this procedure, which we call iterative expansion microscopy (iExM), was not included here since it is evolving, but the current version will continuously be posted at our web resource http://expansionmicroscopy.org. The protocols in this unit are based on the published protocols associated with the aforementioned protocols (and currently posted at http://expansionmicroscopy.org).
Critical Parameters
For the basic proExM protocols (BASIC PROEXM PROTOCOL FOR CULTURED CELLS and BASIC PROEXM PROTOCOL FOR INTACT TISSUES), the timing of each step needs to be followed strictly to prevent premature gelation, which could result in distortion, tearing, or lack of expansion. For example, steps 20–24 in BASIC PROEXM PROTOCOL FOR CULTURED CELLS and steps 14–15 in BASIC PROEXM PROTOCOL FOR INTACT TISSUES should be carried out as quickly as possible, ideally within 5 minutes. Similarly, steps 9–13 in ExFISH PROTOCOL FOR CULTURED CELLS should be carried out with minimal delay. Specific troubleshooting details are listed in the next section.
Troubleshooting
Problems, potential causes and resolutions are listed in Table 4.
Table 4.
Troubleshooting proExM and ExFISH.
| Step | Observed problem | Potential causes & solutions |
|---|---|---|
|
Steps 19–23 in BASIC PROEXM PROTOCOL FOR CULTURED CELLS Steps 13–18 in BASIC PROEXM PROTOCOL FOR INTACT TISSUES Step 3 in PROEXM PROTOCOL FOR THICK INTACT TISSUE SAMPLES Steps 8–12 in ExFISH PROTOCOL FOR CULTURED CELLS |
Sample prematurely forms a gel before the lid can be placed. | The gelation is highly temperature dependent and will initiate prematurely if the mixed gelling solution is warm. Thus, keeping the mixed gelling solution on ice before applying it to the sample, and keeping the handling time (until the lid is placed) less than 5 minutes should usually resolve this problem. If the above does not solve premature gelation, prepare fresh 4-HT solution (Table 2B). Optionally, using 1.5x of the initial 4-HT concentration will help to inhibit the gelation process for a longer time, but the polymerization step might also take longer compared to the time observed with the original 4-HT concentration (refer to step 3 in PROEXM PROTOCOL FOR THICK INTACT TISSUE SAMPLES). |
|
Step 22 in BASIC PROEXM PROTOCOL FOR CULTURED CELLS Step 17 in BASIC PROEXM PROTOCOL FOR INTACT TISSUES Step 10 in ExFISH PROTOCOL FOR CULTURED CELLS |
Air bubble(s) are trapped under the lid. | If the air bubbles are not directly touching the sample, they will not affect the expansion process. If the air bubbles are on top of the sample, or are otherwise touching the sample, they can be gently moved away from the sample by adding more monomer solution from the side (refer to step 23 of BASIC PROEXM PROTOCOL FOR CULTURED CELLS or step 18 of BASIC PROEXM PROTOCOL FOR INTACT TISSUES). To prevent trapping air bubbles, we recommend placing the lid on the sample using the drop method (refer to schematics in Figure 2 and 4). |
|
Step 34–35 in BASIC PROEXM PROTOCOL FOR CULTURED CELLS Step 29–30 in BASIC PROEXM PROTOCOL FOR INTACT TISSUES Step 28 in ExFISH PROTOCOL FOR CULTURED CELLS Step 4 in IMAGING EXPANDED SAMPLES |
Expansion factor is smaller than anticipated | Some small amount of variation is expected from sample to sample. If the expansion is much smaller, first, try more washes with water (ProExM) or diluted PBS (ExFISH) until there is no more change in the gel size (refer to, for example, step 34 in BASIC PROEXM PROTOCOL FOR CULTURED CELLS or step 29 in BASIC PROEXM PROTOCOL FOR INTACT TISSUES). Rinse sample chambers and objectives of the microscope if they have come in contact with imaging medium such as buffers in previous imaging sessions, to remove any possible salt contamination. If the above does not solve the problem, inadequate digestion of the sample might be the cause. Try digesting the sample longer or perform another round of digestion by adding more, or exchanging into new, Proteinase K (ProK) in the digestion buffer (refer to step 4 in PROEXM PROTOCOL FOR THICK INTACT TISSUE SAMPLES). If there is still no improvement in the expansion factor, replace the ProK with a newly purchased batch. |
|
Step 4 in IMAGING EXPANDED SAMPLES Imaging ExM samples with inverted microscopes Imaging with upright microscopes Imaging with microscopes with the sample hanging from above (e.g., light sheet microscopes) |
Cannot locate expanded samples in water (or other diluted buffers used for expansion) | We recommend first illuminating the imaging medium from different angles with a flashlight or a lamp to locate the sample gel. Even in the expanded state, the samples can slightly scatter the incident light. You can also use a fluorescence microscope with a long working distance objective to locate the sample in its respective fluorescence channels (see step 2 of IMAGING EXPANDED SAMPLES). Expanded samples typically have dimmer fluorescence compared to their pre-expansion state due to expansion, and can be located further away from objectives. Therefore, it becomes harder to find the sample with short working distance/high magnification objectives. If possible, start with low magnification objectives (e.g., 4x or 10x objectives) for a larger field of view before switching to objectives of higher magnification (refer to IMAGING EXPANDED SAMPLES). Adjust the field of view and the focus to find the samples, which sometimes requires moving close to the limit of the objective working distance. |
|
Step 4 in IMAGING EXPANDED SAMPLES Imaging ExM samples with inverted microscopes Imaging with upright microscopes Imaging with microscopes with the sample hanging from above (e.g., light sheet microscopes) |
Expanded sample gel breaks during handling | Handling of sample gels becomes increasingly difficult once expanded as the gel becomes more fragile. We recommend using, for example, plastic spatulas and brushes for handling, as described in the sample mounting subsections (Sample mounting with poly-L-lysine, Sample mounting with agarose, Sample mounting with superglue) |
|
Step 4 in IMAGING EXPANDED SAMPLES Imaging ExM samples with inverted microscopes Imaging with upright microscopes Imaging with microscopes with the sample hanging from above (e.g., light sheet microscopes) |
Expanded sample moves too much during imaging | If imaging is performed for a quick inspection or a small image stack (<5 minutes), all surrounding liquid can be removed without significant evaporation and shrinking. In this case, to reduce sample movement, remove as much surrounding liquid as possible. For imaging experiments lasting longer than 5 minutes, we recommend immobilizing the sample in the imaging container to avoid sample drifting during acquisition. The best immobilization method depends on the microscope set-up (refer to Sample Mounting). Use poly-L-lysine treated coverslips or glass-bottom multi-well plates to attach the gel for inverted microscopes (refer to Sample mounting with poly-L-lysine). Use low melting agarose to fix the gel for upright microscopes (refer to Sample mounting with agarose). Use superglue to attach the gel for upright or light sheet microscopes (refer to Sample mounting with superglue). |
|
Step 4 in IMAGING EXPANDED SAMPLES Imaging ExM samples with inverted microscopes Imaging with upright microscopes |
Imaging takes too long | ProExM introduces a linear 4.5x expansion, which translates into a ~90x volumetric increase. Therefore, if volumetric imaging is performed, it requires about 90x the time compared to the pre-expansion sample volume. We therefore recommend using faster imaging methods, such as light sheet, or spinning disc confocal microscopy (such as described in Imaging ExM samples with inverted microscopes and Imaging with microscopes with the sample hanging from above) |
|
Step 4 in IMAGING EXPANDED SAMPLES Imaging ExM samples with inverted microscopes Imaging with upright microscopes Imaging with microscopes with the sample hanging from above (e.g., light sheet microscopes) |
Low or no fluorescence after expansion | Switch to compatible fluorescent dyes for immunostaining and fluorescent proteins that are proExM-optimized (refer to FLUORESCENT PROTEINS, ANTIBODIES AND DYES FOR PROEXM). Furthermore, due the large volumetric expansion, expanded samples are expected to be much dimmer compared to their pre-expansion states. Make sure to use longer exposure time and/or higher laser intensity for imaging. |
| After imaging ExM samples | How to perform image processing and data analyses | Expanded samples usually generate large datasets. There are a few commercial software packages available for large data visualization and image analyses, such as Arivis Vision4D and Bitplane Imaris. Alternatively, the data can be either cropped or divided up and processed more traditionally in smaller data chunks. |
| After imaging ExM samples | Samples lose their fluorescence after few weeks | Fluorescent proteins denature in water over time and other fluorescent probes can degrade or rehybridize in non-buffered environments. Therefore, we do not recommend storing ExM samples in their expanded states. For long-term storage, we recommend storing the samples in a buffered environment, such as PBS. In addition, we keep the samples at 4°C in the dark to prevent bleaching of fluorescence. |
|
Step 4 in IMAGING EXPANDED SAMPLES Imaging ExM samples with inverted microscopes Imaging with upright microscopes Imaging with microscopes with the sample hanging from above (e.g., light sheet microscopes) |
ExFISH: No visible spots after smFISH or FISH-HCR | To avoid RNA degradation, make sure to use nuclease free reagents for the sample and reagent preparation. If working with slices, do not store them in PBS at 4°C for more than 3–4 days - either store them in 70% ethanol at 4°C, or store them frozen with a cryoprotectant. |
|
Step 4 in IMAGING EXPANDED SAMPLES Imaging ExM samples with inverted microscopes Imaging with upright microscopes Imaging with microscopes with the sample hanging from above (e.g., light sheet microscopes) |
ExFISH: smFISH/ FISH-HCR staining too dim after expansion | Increase the probe hybridization time and carry out the hybridization steps for cultured cells and slices for at least overnight. For thick tissue samples (~200 μm), the HCR amplification can be allowed to go overnight. In addition, do not keep gels in low salt buffer for longer than 6 hours, since low salt conditions result in the unbinding of probes which diminish the signal from the staining. After imaging, store smFISH stained gels in 1x PBS at 4°C, and FISH-HCR stained gels in 5x SSCT at 4°C. |
|
Step 4 in IMAGING EXPANDED SAMPLES Imaging ExM samples with inverted microscopes Imaging with upright microscopes Imaging with microscopes with the sample hanging from above (e.g., light sheet microscopes) |
ExFISH: probe wash out is too slow; high background after staining | We recommend to cast a thin gel for ExFISH to reduce background fluorescence and washout times. Trim the fully expanded gel to have a thickness of 1 mm or less (see steps 18–21 in ExFISH PROTOCOL FOR CULTURED CELLS) Alternatively, if working with a thick slice (> 200 μm), cut the slice into thinner sections before starting the ExFISH protocol. |
Understanding Results
ProExM samples from BASIC PROEXM PROTOCOL FOR CULTURED CELLS, BASIC PROEXM PROTOCOL FOR INTACT TISSUES and PROEXM PROTOCOL FOR THICK INTACT TISSUE SAMPLES (200–500 μm) will appear increasingly transparent after mechanical homogenization and expansion. After full expansion to ~4.5x of the initial size, the proExM samples will appear transparent, matching the refractive index of water (Fig. 3). The expanded samples can be fragile to handle and difficult to locate in water (see IMAGING EXPANDED SAMPLES for gel handling and immobilization tips).
Similarly, the digested samples from ExFISH PROTOCOL FOR CULTURED CELLS and ExFISH PROTOCOL FOR INTACT TISSUES will appear transparent. During the RNA-FISH staining steps for both protocols, the gel will shrink and expand because of the different salt contents of the buffers involved. Since the expansion step at the end of the staining protocol is carried out in a low-salt buffer instead of water, the final observed expansion is approximately ~3.3x for both ExFISH protocols.
Fluorescent microscope setups, such as wide-field microscopes, spinning disk and laser scanning confocal microscopes, and light sheet microscopes, can be used to image the expanded proExM and ExFISH samples (see IMAGING EXPANDED SAMPLES, as well as Figures 5, 6, 8, 9, and 11 for representative images of the various protocols described here). Expanded samples typically have lower fluorescence intensities compared with their pre-expansion states due to the physical expansion, and therefore require longer exposure time and/or higher laser intensity (refer to the Troubleshooting section if no or low fluorescence) to acquire images of similar brightness.
Time Considerations
Each procedure’s duration depends on the sample’s parameters, such as thickness and biological composition, as well as the protocol used, but typically falls in the range of one day to a few days, with only a few short periods (1–2 hours each) of manual work required at each defined timepoint in the protocol (the remainder of the time is either allocated for incubation or washing). For example, with BASIC PROEXM PROTOCOL FOR INTACT TISSUES, a tissue slice of 200 μm thickness takes ~2 days to go from the original fixed and sliced tissue to the final mechanically homogenized state embedded in the hydrogel, ready for expansion. With ExFISH PROTOCOL FOR INTACT TISSUES, the protocol similarly takes ~2 days. In both proExM and ExFISH, digested samples can be stored in PBS at 4°C in the dark for weeks before further processing, expansion, and imaging. Staining the gel-embedded ExFISH samples with RNA FISH probes takes one day for cultured cells and two days for tissues, after which the stained samples can be readily expanded and imaged. Fluorescence imaging of the expanded samples can take anywhere from minutes to hours, depending on the imaging volume and image acquisition speed.
SIGNIFICANCE STATEMENT.
Expansion microscopy enables nanoscale imaging on diffraction-limited microscopes via physical expansion of biological specimens. With various expansion microscopy protocols, it is possible to visualize RNA, proteins, and other biomolecules throughout extended 3-dimensional biological specimens with commonly available imaging hardware. In this unit, we explain how to perform expansion microscopy in a variety of contexts of importance to biology.
Acknowledgments
We would like to acknowledge Yosuke Bando and Kazuhiro Hiwada for the image render in Figure 11G, Grace H. Huynh and Linyi Gao for help with sample preparation, Eric Betzig, Jennifer Lippincott-Schwartz and Kathy Schaefer at HHMI Janelia Research Campus for helpful discussions, and Wendy C. Salmon at the W. M. Keck Microscopy Facility at the Whitehead Institute for help with imaging. ESB acknowledges, for funding, the Ludwig Foundation and MIT Aging Brain Initiative, NIH 1R01NS102727, John Doerr, NIH 1R01EB024261, Cancer Research UK Grand Challenge, the HHMI-Simons Faculty Scholars Program, NIH 1R41MH112318, the Open Philanthropy Project, NIH 1R01MH110932, IARPA D16PC00008, U. S. Army Research Laboratory and the U. S. Army Research Office under contract/grant number W911NF1510548, NIH 1RM1HG008525, and NIH Director’s Pioneer Award 1DP1NS087724.
Footnotes
CONFLICTS OF INTEREST
ESB is co-founder of a company seeking to commercialize clinical applications of ExM. ATW, FC, PT, RG, and ESB are inventors on one or more granted or pending patents related to ExM.
References
- Chang J-B, Chen F, Yoon Y-G, Jung EE, Babcock H, Kang JS, Asano S, Suk H-J, Pak N, Tillberg PW, et al. Iterative expansion microscopy. Nature Methods. 2017 doi: 10.1038/nmeth.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Tillberg PW, Boyden ES. Expansion microscopy. Science. 2015;347:543–548. doi: 10.1126/science.1260088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Wassie AT, Cote AJ, Sinha A, Alon S, Asano S, Daugharthy ER, Chang JB, Marblestone A, Church GM, et al. Nanoscale imaging of RNA with expansion microscopy. Nature methods. 2016;13:679–84. doi: 10.1038/nmeth.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HMT, Beck VA, Pierce NA. Next-generation in situ hybridization chain reaction: Higher gain, lower cost, greater durability. ACS Nano. 2014;8:4284–4294. doi: 10.1021/nn405717p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen Y, Ramon O, Kopelman IJ, Mizrahi S. Characterization of inhomogeneous polyacrylamide hydrogels. Journal of Polymer Science Part B: Polymer Physics. 1992;30:1055–1067. [Google Scholar]
- Gao R, Asano SM, Boyden ES. Q&A: Expansion microscopy. BMC Biology. 2017;15:50. doi: 10.1186/s12915-017-0393-3. Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germroth PG, Gourdie RG, Thompson RP. Confocal microscopy of thick sections from acrylamide gel embedded embryos. Microscopy Research and Technique. 1995;30:513–520. doi: 10.1002/jemt.1070300608. Available at: [DOI] [PubMed] [Google Scholar]
- Hausen P, Dreyer C. The use of polyacrylamide as an embedding medium for immunohistochemical studies of embryonic tissues. Stain technology. 1981;56:287–293. doi: 10.3109/10520298109067329. [DOI] [PubMed] [Google Scholar]
- proExM for tissues: gelation demonstration. https://www.youtube.com/watch?v=OksNCAJwxVI.
- Raj A, Tyagi S. Detection of individual endogenous RNA transcripts in situ using multiple singly labeled probes. 2010 doi: 10.1016/S0076-6879(10)72004-8. Available at: [DOI] [PubMed]
- Tanaka T, Fillmore D, Sun ST, Nishio I, Swislow G, Shah A. Phase Transitions in Ionic Gels. Physical Review Letters. 1980;45:1636–1639. [Google Scholar]
- Tillberg PW, Chen F, Piatkevich KD, Zhao Y, Yu C-CJ, English BP, Gao L, Martorell A, Suk H-J, Yoshida F, et al. Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nature biotechnology. 2016:1–9. doi: 10.1038/nbt.3625. [DOI] [PMC free article] [PubMed] [Google Scholar]