Abstract
Purpose
Expression of RNA-binding motif protein 3 (RBM3) is induced by hypoxia and hypothermia. Recently, high expression of RBM3 was reported to be associated with a good prognosis in colon cancer, prostate cancer, ovarian cancer, and malignant melanoma. Studies on RBM3 in invasive breast carcinoma (IBC), however, are limited.
Methods
RBM3 expression was examined using a tissue microarray from 361 patients with IBC. Immunohistochemistry was performed for estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (HER2), and Ki-67 to compare the expression of these markers. For scoring of RBM3 expression, NF (nuclear staining fraction)×NI (nuclear staining intensity) was used. The RBM3 expression score was considered indicative of either low (≤4) or high (>4) expression. Western blot analysis was performed on breast cancer cell lines to evaluate RBM3 expression.
Results
Of the total 361 samples, 240 (66.5%) exhibited high RBM3 expression. High RBM3 expression was significantly associated with positivity for ER (p<0.001), PR (p<0.001), T stage (p<0.001), histologic grade (p<0.001), and % Ki-67 staining (p=0.004). Multivariate analysis revealed that high RBM3 expression was closely associated with prolonged disease-free survival (DFS) (p<0.001) and overall survival (OS) (p<0.001). Western blot analysis revealed reduced RBM3 expression in HCC1954 (HER2-enriched) and BT-20 (basal-like) cells with an aggressive phenotype.
Conclusion
High nuclear RBM3 expression is strongly associated with a prolonged DFS and OS. Furthermore, RBM3 expression is closely associated with good prognostic markers such as ER and PR in IBC. High nuclear RBM3 expression is, therefore, a critical biomarker of favorable clinical outcomes in IBC.
Keywords: Breast neoplasms, Immunohistochemistry, Prognosis, RNA-binding proteins
INTRODUCTION
Invasive breast carcinoma (IBC) is the most common malignancy and the second leading cause of cancer death in women [1]. In Korea, IBC is the second most common malignancy and the fifth most common cause of cancer death in women [2]. Due to significant improvements in the diagnosis and treatment of IBC, the survival rate of the malignancy has increased over the past 30 years. IBC is subdivided into four molecular/intrinsic subtypes: luminal A, luminal B, human epidermal growth factor receptor 2 (HER2)-enriched type, and basal-like subtype, according to the gene expression pattern [3,4]. The basal-like subtype overlaps with “triple-negative” subtypes and yields high-grade tumors with a heavy infiltration of lymphocytes [5,6].
Two mammalian cold-shock proteins have been reported to date: cold-inducible RNA binding protein and RNA-binding motif protein 3 (RBM3) [7,8]. RNA-binding proteins with RNA-binding motifs (RBMs) have been identified through specific subgroups containing single-stranded RNA binding proteins (SBPs). These SBPs are involved in posttranslational regulation of RNA and gene transcription via alterations of microRNA complexes to stimulate protein synthesis, proliferation, and anti-apoptosis [9]. The RBM3 gene, one of three X chromosome-associated RBM genes (RBMX, RBM3, and RBM10), encodes a 157-amino acid protein of 17 kDa [7]. Initially, the RBM3 gene was discovered in a cDNA library from human fetal brain tissues, and it is located in sub-band Xp11.23 [7]. Martínez-Arribas et al. [10] reported that RBM proteins regulate apoptosis and thus, there is a positive correlation between the RBMX, RBM3, and RBM10-encoding genes and the Bax gene in breast cancers. At first, RBM3 expression was found to be upregulated in various cancer tissues in the Human Protein Atlas (HPA); therefore, it was considered to have oncogenic potential [11]. However, high RBM3 expression has recently been associated with a good prognosis in several malignancies, including breast [12], ovary [13], prostate [14], urinary bladder [15], and colon [16] cancer. Nevertheless, the role of RBM3 in malignancies has not yet been clearly identified. In addition, the prognostic implications of tumor-specific RBM3 expression in IBC have not been investigated in a large cohort study. In the present study, the prognostic consequences of RBM3 expression in IBC were investigated by immunohistochemistry (IHC) from the tissue microarray (TMA) of 361 patients, as well as by western blot analysis of human breast cancer cell lines.
METHODS
Patients
This study included 361 cases of primary IBC in patients that had undergone surgical resection in Keimyung University Dongsan Medical Center between 2003 and 2007. Cases with distant metastasis at the time of diagnosis as well patients who received neoadjuvant chemotherapy were excluded. All tissues were fixed in 10% buffered formalin and embedded in paraffin. Patient and tumor characteristics, including patient age at the time of initial diagnosis, tumor size, lymph node status, histologic grade, and follow-up data were obtained from pathology reports and patients' medical records. Tumor stage was determined based on the 7th American Joint Committee on Cancer criteria [17]. Overall survival (OS) was defined as the time interval between the date of the cancer diagnosis and the date of death from any cause. Disease-free survival (DFS) was defined as the number of months from surgical resection to the development of documented relapse, including locoregional recurrence or distant metastasis. The requirement for informed consent from the patients was waived by the ethics committee; this study was approved by the Institutional Review Board of Dongsan Medical Center (DSMC No. 2012-16).
Construction of tissue microarrays
Before TMA construction, all cases were histopathologically reviewed on hematoxylin and eosin-stained slides by a specialized breast pathologist. After reviewing the slides, the pathologist selected a representative block for each case with which to construct the TMA. A pair of tissue cores 2 mm in diameter were retrieved from each tumor block and transferred to the recipient block using a Quick-Ray Manual Tissue Microarrayer (Unitma, Seoul, Korea) and Quick-Ray recipient blocks with 2-mm cores (Unitma). All TMAs were provided by the Department of Pathology, Dongsan Medical Center.
Immunohistochemical assessment and scoring
IHC was performed using the automated Benchmark platform (Ventana Medical Systems, Tucson, USA) according to the manufacturer's recommendations. Sections 4 µm in width were immunostained for RBM3, estrogen receptor (ER), progesterone receptor (PR), HER2, and Ki-67 using an UltraView™ Universal DAB detection kit (Ventana Medical Systems). Antigen retrieval using cell conditioning solution (CC1; Ventana Medical Systems) was performed for all cases using the Benchmark platform, and slides were counterstained with hematoxylin. The antibodies and staining conditions used in this study are detailed in Table 1. ER and PR staining were scored according to the American Society of Clinical Oncology (ASCO) and the College of American Pathologists (CAP) guidelines. The guidelines recommend classifying all cases with at least 1% positive cells as receptor positive [18]. HER2 staining was also scored according to the ASCO and the CAP guidelines [19], which include four scores from 0 to 3: no or incomplete staining, faint/barely perceptible membrane staining in ≤10% of invasive tumor cells (score 0); incomplete, faint/barely perceptible membrane staining in >10% of invasive tumor cells (score 1); incomplete and/or weak to moderate circumferential membrane staining in >10% of invasive tumor cells or complete, intense, circumferential membrane staining in ≤10% of invasive tumor cells (score 2); and complete, intense, circumferential membrane staining in >10% of invasive tumor cells (score 3). A score of 3 for HER2 is regarded as positive. For the equivocal (score 2) cases of HER2, HER2 status was determined using silver in situ hybridization. Quantification of Ki-67 was performed in 1,000 tumor cells, and Ki-67 positivity was defined as ≥14%. RBM3 protein was found to be expressed primarily in the nuclei of tumor cells. The nuclear staining fraction (NF) was assigned a score of 0 (0%–1%), 1 (2%–25%), 2 (26%–50%), 3 (51%–75%), or 4 (>75%), and nuclear staining intensity (NI) was noted as 0 (negative), 1 (weak), 2 (msoderate), and 3 (strong). Subsequently, a combined nuclear score (NS) was calculated by multiplying NF and NI (range of 0 to 12). For statistical analyses, the cutoff values for RBM3 expression were chosen on the basis of heterogeneity using the log-rank test for OS. The optimal cutoff value was determined as low (NS ≤4) or high (NS >4) RBM3 expression. The distribution of the intrinsic subtypes of IBC according to IHC was divided into five subtypes: luminal A (ER+ and/or PR+, HER2−, Ki-67 <14%); luminal B, HER2(−) (ER+ and/or PR+, HER2−, Ki-67≥14%); luminal B, HER2(+) (ER+ and/or PR+, HER2+); HER2-enriched (ER−, PR−, HER2+); and triple-negative (ER−, PR−, HER2−).
Table 1. Antibodies and staining conditions used for this study.
| Antibody | Company | Clone | Dilution | Antigen retrieval* | Incubation |
|---|---|---|---|---|---|
| RBM3 | Sigma-Aldrich (St. Louis, USA) | Polyclonal | 1:100 | Standard | 37℃, 32 min |
| ER | Ventana Medical Systems (Tucson, USA) | SP1 | Predilution | Standard | 37℃, 16 min |
| PR | Ventana Medical Systems | 1E2 | Predilution | Standard | 37℃, 16 min |
| HER2 | Ventana Medical Systems | 4B5 | Predilution | Mild | 37℃, 16 min |
| Ki-67 | Ventana Medical Systems | 30-9 | Predilution | Standard | 37℃, 16 min |
RBM3=RNA-binding motif protein 3; ER=estrogen receptor; PR=progesterone receptor; HER2=human epidermal growth factor receptor 2.
*Standard: 100℃, 60 minutes; Mild: 100℃, 30 minutes.
Cell culture
Human breast cancer cell lines MCF-7, T47D, HCC1954, BT-20, and ZR-75-1 were obtained from the Korean Cell Line Bank (Seoul, Korea). All cell lines were maintained in RPMI-1640 medium (WelGENE, Daegu, Korea) and were supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, USA) and 1% penicillin/streptomycin (Sigma-Aldrich, St. Louis, USA). Cells were grown as monolayers under standard conditions (37℃ in a humidified atmosphere containing 5% CO2). The cells were cultured in BD Falcon™ (BD Biosciences, Bedford, USA) 250-mL 75-cm2 cell culture flasks for gene expression analysis.
Western blot analysis
Cells were washed twice with cold phosphate-buffered saline (pH 7.0) and scraped in radioimmunoprecipitation assay lysis buffer (150 mM NaCl, 1% Nonidet P40, 0.5% deoxycholic acid, 0.1% sodium dodecyl sulfate [SDS], and 50 mM Tris pH 8.0) supplemented with 40.0 µg/mL leupeptin, 1 µM pepstatin, and 0.1 mM phenylmethylsulfonyl-fluoride. Cell lysates were incubated on ice for 30 minutes before being subjected to centrifugation for 15 minutes at 15,000×g at 4℃. Proteins in the supernatant were then extracted and quantified using the BAP assay (Pierce, Rockford, USA). Subsequently, samples containing 40 µg of protein were loaded onto 10% SDS-polyacrylamide gel electrophoresis gels with 4× loading dye (Tris-HCl pH 7.4, 1% SDS, glycerol, dithiothreitol, and bromophenol blue), electrophoresed, and transferred onto nitrocellulose membrane (Bio-Rad, Richmond, USA). After being blocked with 5% nonfat milk in Tris-buffered saline (25 mM Tris base and 150 mM NaCl) for 2 hours at room temperature, the membranes were incubated with primary antibodies overnight at 4℃. Next, the membranes were incubated with horseradish peroxidase-conjugated secondary antibody (SC-2005, 1:5,000; Santa Cruz, Dallas, USA) for 2 hours at room temperature. The primary antibodies used included rabbit polyclonal anti-RBM3 antibody (HPA003624, 1:1,000; Sigma-Aldrich) and anti-actin antibody (MA5-11866, 1:5,000; Thermo Fisher Scientific, Rockford, USA).
Statistical analyses
Statistical analyses were performed using SPSS version 20.0 for Windows (IBM Corp., Armonk, USA). The significance of correlations between RBM3 expression and clinicopathological characteristics was evaluated using the chi-square test. Kaplan-Meier analysis with a log-rank test were used to estimate the effects of RBM3 expression on patient OS and DFS. Variables found to be significant in univariate and multivariate analyses were assessed using a Cox regression proportional hazards model. Adjusted hazard ratios (HRs) and associated 95% confidence intervals (CIs) were estimated for each variable. All statistical tests were two-sided, and a p-value of <0.05 was considered statistically significant.
RESULTS
Patient characteristics, histological features, and intrinsic subtypes
Patients ranged in age from 24 to 84 years (median, 51 years). Of the 361 cases, 311 (86.1%) were invasive ductal carcinoma, not otherwise specified; 15 (4.2%) were invasive lobular carcinoma; and 35 (9.7%) were a variety of other histological types (6 mucinous, 6 micropapillary, 4 papillary, 4 metaplastic, 4 medullary, 2 tubular, 1 cribriform, and 8 mixed carcinoma). Of the 361 patients, 164 (45.4%) received radiotherapy, and 250 (69.3%) received hormone therapy. A total of 272 (75.1%) patients received adjuvant chemotherapy. The median follow-up period was 113.2 months (range, 2.3–181.6 months). The frequencies of the intrinsic subtypes were as follows: luminal A (120, 33.2%); HER2(−) luminal B (113, 31.3%); HER2(+) luminal B (45, 12.5%); HER2-enriched (28, 7.8%); and triple-negative (55, 15.2%).
High nuclear RBM3 expression is associated with favorable clinicopathological characteristics
To evaluate the potential association between nuclear RBM3 expression and clinicopathological characteristics, the immunohistochemical results of the TMAs were assessed. Immunohistochemical findings of low and high RBM3 expression are shown in Figures 1 and 2, respectively. Typical clinicopathological parameters and immunohistochemical staining of low and high RBM3 expression are shown in Table 2. Among the 361 IBC samples, high nuclear RBM3 protein expression (combined NS >4) was detected in 240 samples (66.5%), and low RBM3 protein expression (combined NS ≤4) was observed in 121 samples (33.5%) (Table 2). As summarized in Table 2, there was no significant correlation between high expression of RBM3 protein and patient age. High RBM3 protein expression, however, was markedly associated with all other clinicopathological parameters except adjuvant radiotherapy, including T stage (p<0.001), N stage (p=0.004), histologic grade (p<0.001), expression of ER and PR (p<0.001), HER2 status (p=0.028), Ki-67 % staining (p=0.004), and adjuvant chemotherapy (p<0.001). However, no difference was observed in the distribution of adjuvant radiotherapy between the low and high RBM3 protein expression groups. Regarding the intrinsic subtypes [20], high RBM3 nuclear expression was more frequently shown in luminal A and luminal B subtypes than in HER2 and triple-negative subtypes (p<0.001).
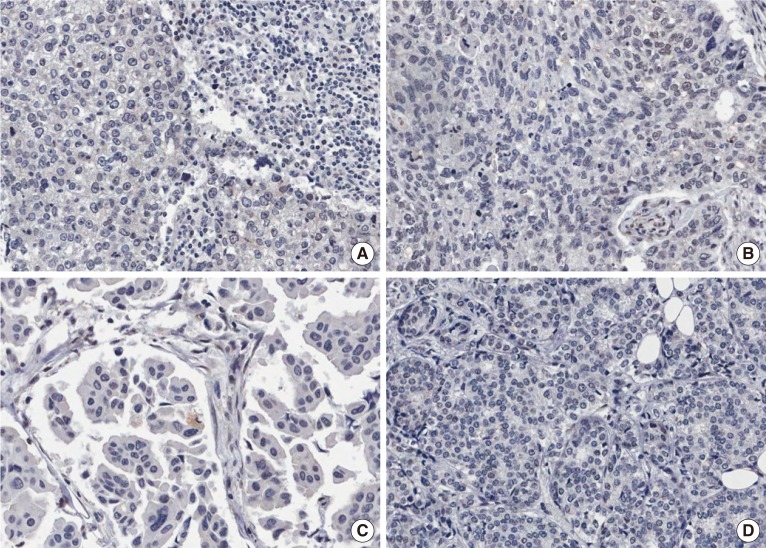
Figure 1. Immunohistochemical findings of low RNA-binding motif protein 3 (RBM3) nuclear expression (RBM3≤4). (A, B) No expression of RBM3 in poorly differentiated invasive ductal carcinomas. (C) Weakly expressed RBM3 in invasive micropapillary carcinoma. (D) No expression of RBM3 in moderately differentiated invasive ductal carcinoma (immunohistochemistry for RBM3, ×200).
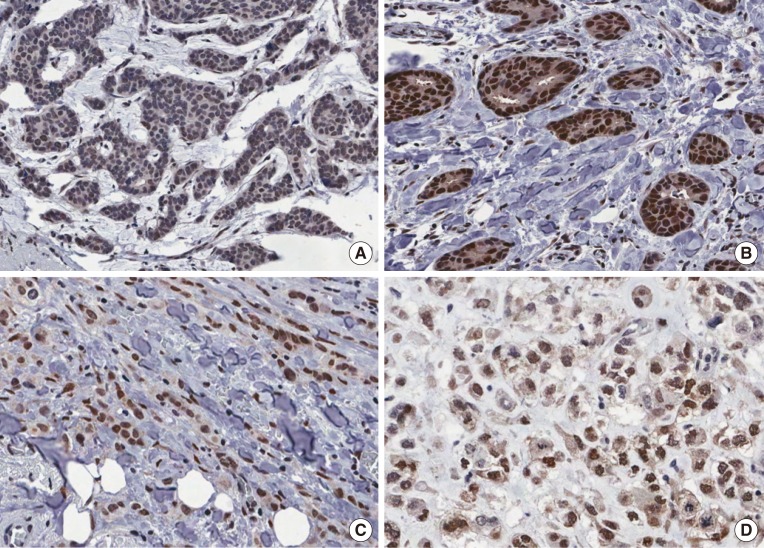
Figure 2. Immunohistochemical findings of high RNA-binding motif protein 3 (RBM3) nuclear expression (RBM3>4). (A) Moderately expressed RBM3 in mucinous carcinoma. (B) Strongly expressed RBM3 in well differentiated invasive ductal carcinoma. (C) Strongly expressed RBM3 in classic invasive lobular carcinoma. (D) Moderately expressed RBM3 in metaplastic carcinoma (immunohistochemistry for RBM3, ×200).
Table 2. Correlation of RBM3 expression with clinicopathologic characteristics of invasive breast carcinomas.
| Characteristic | RBM3 expression | p-value | |
|---|---|---|---|
| Low expression No. (%) | High expression No. (%) | ||
| Age (yr)* | 49 (24–84) | 50 (26–80) | 0.723 |
| T stage | < 0.001 | ||
| T1 | 28 (7.8) | 115 (31.9) | |
| T2 | 81 (22.4) | 113 (31.3) | |
| T3 | 10 (2.8) | 11 (3.0) | |
| T4 | 2 (0.6) | 1 (0.3) | |
| N stage | 0.004 | ||
| N0 | 50 (13.9) | 131 (36.3) | |
| N1 | 33 (9.1) | 58 (16.1) | |
| N2 | 18 (5.0) | 33 (9.1) | |
| N3 | 20 (5.5) | 18 (5.0) | |
| Histologic grade | < 0.001 | ||
| 1 | 5 (1.4) | 30 (8.3) | |
| 2 | 24 (6.6) | 80 (22.2) | |
| 3 | 92 (25.5) | 130 (36.0) | |
| Estrogen receptor | < 0.001 | ||
| Positive | 52 (14.4) | 194 (53.7) | |
| Negative | 69 (19.1) | 46 (12.7) | |
| Progesterone receptor | < 0.001 | ||
| Positive | 64 (17.7) | 196 (54.3) | |
| Negative | 57 (15.8) | 44 (12.2) | |
| Hormone receptor | < 0.001 | ||
| Positive | 70 (19.4) | 207 (57.3) | |
| Negative | 51 (14.1) | 33 (9.1) | |
| HER2 status | 0.028 | ||
| Positive | 32 (8.9) | 40 (11.1) | |
| Negative | 89 (24.7) | 200 (55.4) | |
| Ki-67 (%) | 0.004 | ||
| < 14 | 36 (10.0) | 109 (30.2) | |
| ≥ 14 | 85 (23.5) | 131 (36.3) | |
| Intrinsic subtype | < 0.001 | ||
| Luminal A | 22 (6.1) | 98 (27.1) | |
| Luminal B, HER2(–) | 29 (8.0) | 84 (23.3) | |
| Luminal B, HER2(+) | 19 (5.3) | 26 (7.2) | |
| HER2-enriched | 15 (4.2) | 13 (3.6) | |
| Triple-negative | 36 (10.0) | 19 (5.3) | |
| Adjuvant chemotherapy | 0.001 | ||
| Yes | 103 (28.5) | 169 (46.9) | |
| No | 18 (5.0) | 71 (19.7) | |
| Adjuvant radiotherapy | 0.600 | ||
| Yes | 52 (14.4) | 112 (31.0) | |
| No | 69 (19.1) | 128 (35.5) | |
RBM3=RNA-binding motif protein 3; HER2=human epidermal growth factor receptor 2.
*Median (range).
Reduced expression of RBM3 protein in phenotypically aggressive human breast cancer cell lines
Western blot analysis using the anti-RBM3 antibody yielded a single band at 17 kDa in several breast cancer cell lines: MCF-7 and T47D (luminal A subtypes), ZR-75-1 (luminal B subtype), BT-20 (triple-negative subtype), and HCC1954 (HER2-enriched type) (Figure 3). The differential expression of RBM3 between different cell types was evident, with the highest levels being observed in MCF-7. In contrast, HCC1954 (HER2-enriched) and BT-20 (triple-negative) exhibited markedly lower RBM3 expression.
Figure 3. Differences of RNA-binding motif protein 3 (RBM3) protein expression in various human breast cancer cell lines by western blot analysis. RBM3 protein is markedly increased in MCF-7 and T47D (luminal A subtype) and ZR-75-1 (luminal B subtype). On the other hand, RBM3 protein is significantly reduced in HCC1954 (HER2-enriched type) and BT-20 (basal-like subtype).
High nuclear RBM3 expression is an independent predictor of patient survival
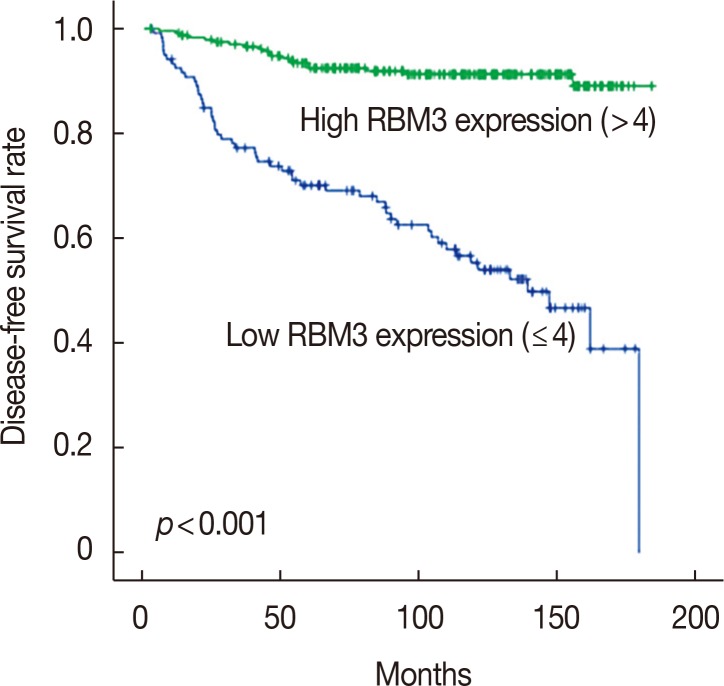
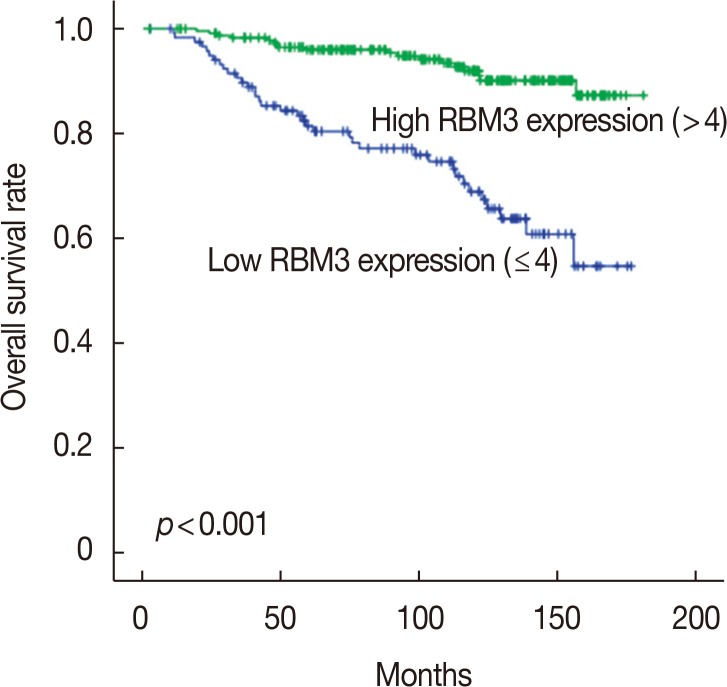
To investigate the prognostic significance of high RBM3 protein expression levels in IBC, OS, and DFS were analyzed using the Kaplan-Meier analysis with a log-rank test. The Kaplan-Meier analysis demonstrated that high nuclear RBM3 expression was significantly associated with prolonged DFS (Figure 4) and OS (Figure 5) compared with low nuclear RBM3 expression. The expression of RBM3 protein may, therefore, affect the prognosis of IBC patients, and high RBM3 expression levels may serve as an independent indicator for improved patient survival. This was further demonstrated when the clinicopathological characteristics and RBM3 expression levels were assessed by Cox univariate and multivariate regression models in this study (Table 3). Univariate analysis revealed that improved OS and DFS in IBC were significantly related to high RBM3 expression correlated with T stage, N stage, histologic grade, ER and PR expression, and % of Ki-67 staining. T stage (p=0.025 only for DFS), N stage (p<0.001 for DFS and OS), and Ki-67 (p=0.005 only for OS) were significant independent prognostic factors for poor DFS and OS in IBC patients (Table 3). Multivariate analysis indicated that high expression of RBM3 protein was a significant independent factor for good DFS (HR, 0.199; 95% CI, 0.144–0.346; p<0.001) and OS (HR, 0.245; 95% CI, 0.133–0.451; p<0.001) in IBC patients (Table 3).
Figure 4. The comparison of disease-free survival between patients with high RNA-binding motif protein 3 (RBM3) expression (RBM3>4) and low RBM3 expression (RBM3≤4) in invasive ductal carcinoma.
Figure 5. The comparison of overall survival between patients with high RNA-binding motif protein 3 (RBM3) expression (RBM3>4) and low RBM3 expression (RBM3≤4) in invasive ductal carcinoma.
Table 3. Multivariate analysis of the DFS and OS in invasive breast carcinomas.
| Characteristic | DFS | p-value | OS | p-value | ||
|---|---|---|---|---|---|---|
| HR | 95% CI | HR | 95% CI | |||
| ER (positive vs. negative) | 0.876 | 0.453–1.694 | 0.694 | 2.022 | 0.918–4.455 | 0.081 |
| PR (positive vs. negative) | 1.031 | 0.561–1.892 | 0.922 | 0.540 | 0.257–1.134 | 0.103 |
| Ki-67 (< 14% vs. ≥ 14%) | 1.354 | 0.787–2.328 | 0.274 | 2.689 | 1.338–5.403 | 0.005 |
| Histologic grade (grade 1 vs. grade 2, 3) | 1.354 | 0.795–2.307 | 0.264 | 1.328 | 0.727–2.425 | 0.356 |
| T stage ( ≤ 5 cm vs. > 5 cm) | 1.606 | 1.061–2.431 | 0.025 | 1.349 | 0.830–2.195 | 0.227 |
| N stage (N0 vs. N1–3) | 1.732 | 1.406–2.134 | < 0.001 | 1.845 | 1.441–2.362 | < 0.001 |
| RBM3 (> 4 vs. ≤ 4) | 0.199 | 0.114–0.346 | < 0.001 | 0.245 | 0.133–0.451 | < 0.001 |
DFS=disease-free survival; OS=overall survival; HR=hazard ratio; CI=confidence interval; ER=estrogen receptor; PR=progesterone receptor; RBM3=RNA-binding motif protein 3.
DISCUSSION
In the present study, the prognostic value of RBM3 expression was demonstrated using TMAs of human IBC and human breast cancer cell lines assessed by IHC and western blot analysis. RBM3 protein levels were dichotomized into low and high nuclear expression. High RBM3 expression was associated with improved DFS and OS compared to low RBM3 expression. Consequently, high RBM3 expression was found to act as an independent prognostic indicator according to univariate and multivariate analyses. Additionally, tumors with high nuclear RBM3 expression were strongly associated with smaller tumor size, decreased N stage, ER and PR positivity, low Ki-67 proliferation index, and adjuvant chemotherapy. Based on our aforementioned data, high RBM3 expression can be considered a significantly favorable prognostic biomarker of IBC with prolonged survival.
In addition, RBM3 expression was markedly reduced in phenotypically aggressive breast cancer cell lines, such as HCC1954 (HER2-enriched) and BT-20 (triple-negative) cells, according to the western blot analysis. In contrast, increased levels of nuclear RBM3 expression were noted in lower grade breast cancer cell lines, specifically in luminal A (MCF-7 and T47D) and luminal B subtypes (ZR-75-1). These findings, which suggest that high nuclear RBM3 expression in IBC is distinctly associated with less aggressive phenotypes and a meaningfully improved patient survival, agree with other studies on RBM3 and favorable clinical outcomes.
SBPs with RBMs are involved in many aspects of RNA metabolism as well as in the regulation of gene transcription [9,21]. The RBM3 gene directly regulates microRNAs posttranscriptionally via Dicer activity [22]. RBM3 protein is a well-known cold-shock protein, which is activated by mild cold stress (32℃) [8]; however, the role of RBM3 has not been fully elucidated yet. An analysis of RBM3 protein and mRNA expression confirmed that RBM3 is rarely expressed in normal tissues; moreover, RBM3 protein was expressed at higher levels in various tumor tissues than in normal tissues [12,16,23,24]. However, a number of studies have found that upregulated RBM3 expression is closely associated with an improved OS and DFS in some tumors, including breast cancer [12], ovarian cancer [25], colon cancer [16], prostate cancer [23], and bladder tumors [15], possibly due to the link between RBM3 expression and DNA repair [25]. The results of these previous studies are similar to those of our study.
We observed that nuclear RBM3 expression was significantly higher in luminal A and luminal B subtypes than in the HER2-enriched and triple-negative subtypes. To our best knowledge, this study is the first to report on the expression of RBM3 according the intrinsic breast cancer subtypes.
In vitro models have shown that RBM3 plays a role in cancer progression-associated processes such as apoptosis [9,26] and tumor proliferation [27]. Furthermore, under moderate hypoxia conditions, tumor angiogenesis was accompanied by increased levels of RBM3 similar to the increase in vascular endothelial growth factor under moderate hypoxia conditions [27]. Wellmann et al. [27] explained the role of RBM3 as an on/off switch under resource-poor conditions such as hypoxia, hypothermia, or serum starvation.
The HPA program (http://www.proteinatlas.org/), a biomarker discovery strategy using antibody-based proteomics, initially reported high nuclear RBM3 expression in breast cancer to be associated with favorable clinicopathologic parameters in two different breast cancer cohorts [12]. In addition, Ehlén et al. [13] showed that silencing of RBM3 in ovarian cancer cell lines resulted in reduced cisplatin (platinum-based chemotherapy) sensitivity. In malignant melanoma, low RBM3 protein expression reportedly correlated with tumor progression and metastasis [28]. Gene set enrichment analysis (GSEA) was previously carried out on 267 epithelial ovarian carcinoma samples to assess whether each case tended to be associated with an up- or a downregulation of RBM3; RBM3 was demonstrated to be involved in the maintenance of DNA integrity, including DNA-dependent replication, chromatin remodeling, and damage checkpoints [25]. The findings reported for this GSEA indicate that RBM3 overexpression can attenuate a number of DNA damage checkpoint kinases, such as checkpoint kinase 1 (CHK1) and checkpoint kinase 2 (CHK2), as well as minichromosome maintenance protein 3 to promote tumorigenesis [25]. RBM8A and RBM8B, members of the extended RBM family, interact with OVCA1, a candidate tumor suppressor and exert protective functions in carcinogenesis similar to RBM3 [27]. Contrary to these studies, including the current study, it was reported that high expression levels of RBM8A predict poor patient prognosis and promote tumor cell invasion in hepatocellular carcinoma [29]. Additionally, Sureban et al. [30] described RBM3 as a protooncogene that prevents mitotic catastrophe in colorectal carcinomas. High RBM3 expression was shown in one study to be an independent poor prognostic marker linked to erythroblast transformation specific-related gene (ERG) positivity and PTEN deletions in prostate cancer [23]. This contradicts the data from Jonsson et al. [14] which showed that high nuclear RBM3 expression is closely related to favorable clinical outcomes. The discrepancies in the reported roles of RBM warrant further investigation with variable steps to fully elucidate the precise function of RBM3 as a biomarker in various types of cancers.
Some limitations should be considered in our study. First, this was a retrospective study carried out at a single institution; therefore, selection bias may have been present. Patients who underwent various adjuvant chemo- and radiotherapies may have shown favorable prognoses unrelated to RBM3 expression. Second, the use of TMA may have under- or overestimated RBM3 protein expression due to the tumoral heterogeneity of breast cancer. Third, our study included limited breast cancer cell lines. Therefore, we plan to include more breast cancer cell lines according to molecular subtypes in future studies. Despite these limitations, the strengths of our study were the large number of included breast cancer cases and the follow-up duration (up to 14 years), which was the longest among studies of RBM3 expression.
In conclusion, we assessed the clinicopathological parameters related to and the prognostic impact of RBM3 expression using IHC in IBC. The findings reported here show that high nuclear RBM3 expression in IBC is strongly associated with a significantly prolonged DFS and OS. Furthermore, high nuclear expression of RBM3 may act as an independent predictor and a potential biomarker of favorable clinical outcomes in IBC.
Footnotes
This work was supported by the research promoting grant from the Keimyung University Dongsan Medical Center in 2013 and also supported by Korea Breast Cancer Foundation in 2013.
CONFLICT OF INTEREST: The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Oh CM, Won YJ, Jung KW, Kong HJ, Cho H, Lee JK, et al. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2013. Cancer Res Treat. 2016;48:436–450. doi: 10.4143/crt.2016.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prat A, Perou CM. Deconstructing the molecular portraits of breast cancer. Mol Oncol. 2011;5:5–23. doi: 10.1016/j.molonc.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eroles P, Bosch A, Pérez-Fidalgo JA, Lluch A. Molecular biology in breast cancer: intrinsic subtypes and signaling pathways. Cancer Treat Rev. 2012;38:698–707. doi: 10.1016/j.ctrv.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 5.Diaz LK, Cryns VL, Symmans WF, Sneige N. Triple negative breast carcinoma and the basal phenotype: from expression profiling to clinical practice. Adv Anat Pathol. 2007;14:419–430. doi: 10.1097/PAP.0b013e3181594733. [DOI] [PubMed] [Google Scholar]
- 6.Rakha EA, Ellis IO. Triple-negative/basal-like breast cancer: review. Pathology. 2009;41:40–47. doi: 10.1080/00313020802563510. [DOI] [PubMed] [Google Scholar]
- 7.Derry JM, Kerns JA, Francke U. RBM3, a novel human gene in Xp11.23 with a putative RNA-binding domain. Hum Mol Genet. 1995;4:2307–2311. doi: 10.1093/hmg/4.12.2307. [DOI] [PubMed] [Google Scholar]
- 8.Danno S, Nishiyama H, Higashitsuji H, Yokoi H, Xue JH, Itoh K, et al. Increased transcript level of RBM3, a member of the glycine-rich RNA-binding protein family, in human cells in response to cold stress. Biochem Biophys Res Commun. 1997;236:804–807. doi: 10.1006/bbrc.1997.7059. [DOI] [PubMed] [Google Scholar]
- 9.Sutherland LC, Rintala-Maki ND, White RD, Morin CD. RNA binding motif (RBM) proteins: a novel family of apoptosis modulators? J Cell Biochem. 2005;94:5–24. doi: 10.1002/jcb.20204. [DOI] [PubMed] [Google Scholar]
- 10.Martínez-Arribas F, Agudo D, Pollán M, Gómez-Esquer F, Díaz-Gil G, Lucas R, et al. Positive correlation between the expression of X-chromosome RBM genes (RBMX, RBM3, RBM10) and the proapoptotic Bax gene in human breast cancer. J Cell Biochem. 2006;97:1275–1282. doi: 10.1002/jcb.20725. [DOI] [PubMed] [Google Scholar]
- 11.Uhlén M, Björling E, Agaton C, Szigyarto CA, Amini B, Andersen E, et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics. 2005;4:1920–1932. doi: 10.1074/mcp.M500279-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Jögi A, Brennan DJ, Rydén L, Magnusson K, Fernö M, Stål O, et al. Nuclear expression of the RNA-binding protein RBM3 is associated with an improved clinical outcome in breast cancer. Mod Pathol. 2009;22:1564–1574. doi: 10.1038/modpathol.2009.124. [DOI] [PubMed] [Google Scholar]
- 13.Ehlén A, Brennan DJ, Nodin B, O’Connor DP, Eberhard J, Alvarado-Kristensson M, et al. Expression of the RNA-binding protein RBM3 is associated with a favourable prognosis and cisplatin sensitivity in epithelial ovarian cancer. J Transl Med. 2010;8:78. doi: 10.1186/1479-5876-8-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jonsson L, Gaber A, Ulmert D, Uhlén M, Bjartell A, Jirström K. High RBM3 expression in prostate cancer independently predicts a reduced risk of biochemical recurrence and disease progression. Diagn Pathol. 2011;6:91. doi: 10.1186/1746-1596-6-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Florianova L, Xu B, Traboulsi S, Elmansi H, Tanguay S, Aprikian A, et al. Evaluation of RNA-binding motif protein 3 expression in urothelial carcinoma of the bladder: an immunohistochemical study. World J Surg Oncol. 2015;13:317. doi: 10.1186/s12957-015-0730-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jang HH, Lee HN, Kim SY, Hong S, Lee WS. Expression of RNA-binding motif protein 3 (RBM3) and cold-inducible RNA-binding protein (CIRP) is associated with improved clinical outcome in patients with colon cancer. Anticancer Res. 2017;37:1779–1785. doi: 10.21873/anticanres.11511. [DOI] [PubMed] [Google Scholar]
- 17.Edge SB American Joint Committee on Cancer. AJCC cancer staging handbook: from the AJCC cancer staging manual. 7th ed. New York: Springer; 2010. [DOI] [PubMed] [Google Scholar]
- 18.Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer (unabridged version) Arch Pathol Lab Med. 2010;134:e48–e72. doi: 10.5858/134.7.e48. [DOI] [PubMed] [Google Scholar]
- 19.Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013;31:3997–4013. doi: 10.1200/JCO.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 20.Goldhirsch A, Wood WC, Coates AS, Gelber RD, Thürlimann B, Senn HJ, et al. Strategies for subtypes: dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol. 2011;22:1736–1747. doi: 10.1093/annonc/mdr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 22.Pilotte J, Dupont-Versteegden EE, Vanderklish PW. Widespread regulation of miRNA biogenesis at the Dicer step by the cold-inducible RNA-binding protein, RBM3. PLoS One. 2011;6:e28446. doi: 10.1371/journal.pone.0028446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grupp K, Wilking J, Prien K, Hube-Magg C, Sirma H, Simon R, et al. High RNA-binding motif protein 3 expression is an independent prognostic marker in operated prostate cancer and tightly linked to ERG activation and PTEN deletions. Eur J Cancer. 2014;50:852–861. doi: 10.1016/j.ejca.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 24.Boman K, Segersten U, Ahlgren G, Eberhard J, Uhlén M, Jirström K, et al. Decreased expression of RNA-binding motif protein 3 correlates with tumour progression and poor prognosis in urothelial bladder cancer. BMC Urol. 2013;13:17. doi: 10.1186/1471-2490-13-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ehlén Å, Nodin B, Rexhepaj E, Brändstedt J, Uhlén M, Alvarado-Kristensson M, et al. RBM3-regulated genes promote DNA integrity and affect clinical outcome in epithelial ovarian cancer. Transl Oncol. 2011;4:212–221. doi: 10.1593/tlo.11106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Al-Astal HI, Massad M, AlMatar M, Ekal H. Cellular functions of RNA-binding motif protein 3 (RBM3): clues in hypothermia, cancer biology and apoptosis. Protein Pept Lett. 2016;23:828–835. doi: 10.2174/0929866523666160628090340. [DOI] [PubMed] [Google Scholar]
- 27.Wellmann S, Truss M, Bruder E, Tornillo L, Zelmer A, Seeger K, et al. The RNA-binding protein RBM3 is required for cell proliferation and protects against serum deprivation-induced cell death. Pediatr Res. 2010;67:35–41. doi: 10.1203/PDR.0b013e3181c13326. [DOI] [PubMed] [Google Scholar]
- 28.Jonsson L, Bergman J, Nodin B, Manjer J, Pontén F, Uhlén M, et al. Low RBM3 protein expression correlates with tumour progression and poor prognosis in malignant melanoma: an analysis of 215 cases from the Malmö Diet and Cancer Study. J Transl Med. 2011;9:114. doi: 10.1186/1479-5876-9-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wellmann S, Bührer C, Moderegger E, Zelmer A, Kirschner R, Koehne P, et al. Oxygen-regulated expression of the RNA-binding proteins RBM3 and CIRP by a HIF-1-independent mechanism. J Cell Sci. 2004;117(Pt 9):1785–1794. doi: 10.1242/jcs.01026. [DOI] [PubMed] [Google Scholar]
- 30.Sureban SM, Ramalingam S, Natarajan G, May R, Subramaniam D, Bishnupuri KS, et al. Translation regulatory factor RBM3 is a proto-oncogene that prevents mitotic catastrophe. Oncogene. 2008;27:4544–4556. doi: 10.1038/onc.2008.97. [DOI] [PMC free article] [PubMed] [Google Scholar]