Abstract
Oxygen limitation is regarded as a useful strategy to improve enzyme production by mycelial fungus like Aspergillus niger. However, the intracellular metabolic response of A. niger to oxygen limitation is still obscure. To address this, the metabolism of A. niger was studied using multi-omics integrated analysis based on the latest GEMs (genome-scale metabolic model), including metabolomics, fluxomics and transcriptomics. Upon sharp reduction of the oxygen supply, A. niger metabolism shifted to higher redox level status, as well as lower energy supply, down-regulation of genes for fatty acid synthesis and a rapid decrease of the specific growth rate. The gene expression of the glyoxylate bypass was activated, which was consistent with flux analysis using the A. niger GEMs iHL1210. The increasing flux of the glyoxylate bypass was assumed to reduce the NADH formation from TCA cycle and benefit maintenance of the cellular redox balance under hypoxic conditions. In addition, the relative fluxes of the EMP pathway were increased, which possibly relieved the energy demand for cell metabolism. The above multi-omics integrative analysis provided new insights on metabolic regulatory mechanisms of A. niger associated with enzyme production under oxygen-limited condition, which will benefit systematic design and optimization of the A. niger microbial cell factory.
Introduction
With a GRAS (generally regarded as safe) status, Aspergillus niger is widely applied in the biosynthesis of organic acids and enzymes1. A. niger has an excellent ability of protein expression and secretion, such as for the industrial production of glucoamylase2. In 2007, the genome sequence and annotation information of A. niger was published3, which became an important basis for systems biology studies of A. niger. Genome annotation of A. niger showed its huge potential as an efficient cell factory3 for the production of different enzymes and secondary metabolites. A genome-scale metabolic model (GEMs) was also reconstructed based on the genome annotation of A. niger4. Recently, the GEMs of A. niger was further updated in our lab5. Centering on enzyme and protein production, separate studies on A. niger metabolomics, transcriptomics and proteomics have been conducted6.
In the industrial enzyme production by A. niger, the poor solubility of oxygen limits mass transfer in bioreactors. This is further aggravated by the complex mycelial morphology, resulting in oxygen limitation for the cell metabolism. As the growth of A. niger is strictly aerobic7, a limited oxygen supply has a strong impact on the fermentation process. For citric acid production, the limited oxygen supply leads to a significant increase in productivity8. Although the specific production rate of glucoamylase was decreased due to the oxygen limitation, its yield per unit of substrate was increased2. The similar results could be found in exogenous protein production by Pichia pastoris9 and other microorganisms. It has been found that large amounts of organic acids (like oxalic acid and citric acid) and polyols (like mannitol and erythritol) were secreted by A. niger under oxygen limited conditions10,11, indicating a high intracellular redox level. These microbial physiology phenomena indicate that the metabolic balance between cell growth and product synthesis is sensitive to oxygen limitation in A. niger. However, details of the global metabolic changes and their interpretation in terms of regulatory mechanisms has only been subject of little systematic research.
Omics studies play an increasingly important role in investigation of the cell metabolic response and regulation mechanisms, and there are a few studies using these approaches to study how cells adapt to oxygen limited conditions. Via transcriptome analysis of Trichoderma reesei, it was found that the expression of genes from metabolic pathways related to the energy consumption were significantly down-regulated in response to the limited oxygen supply12. Using proteomics analysis, it was shown that the expression of 117 proteins in A. fumigatus, involved in the PP pathway, TCA pathway and EMP pathway, was up-regulated to adapt to the hypoxic environment13. With the aid of transcriptome analysis, Choi et al.14 found that the genes involved in the sterol regulatory synthesis pathways were activated under oxygen limited conditions, which facilitated the synthesis of sterols and maintained the cell mycelial growth capabilities.
Compared with single omics analysis, a multi-omics integrative analysis could help to reveal interactions among different metabolic regulation levels. Based on the evidence from transcriptome analysis and molecular experiments, Kroll et al.15 found that the electron transport chain plays an important role in sensing the extracellular oxygen concentration and transmitting the hypoxia signal to the mitochondria. The metabolic characteristics of glucoamylase production by integration of 13C metabolic flux analysis and metabolomics16 was recently carried out in our lab. The results showed that the intracellular metabolic fluxes were redistributed to respond the enzyme synthesis and redox balance. Baumann et al.17 studied the metabolic mechanism of Pichia pastoris under oxygen limited conditions with integrative analysis of metabolomics, transcriptomics and proteomics, and found that flux changes in the PP, TCA and EMP pathways were mainly regulated at a transcriptional level.
To better understand the mechanisms supporting a high yield of glucoamylase production and global metabolic regulation under oxygen limitation, the multi-omics integrative analysis based on GEMs is employed, which provides holistic views for the rational optimization of industrial bioprocess and strain performance.
Materials and Methods
Strains and cultivations
The glucoamylase high-producing strain Aspergillus niger DS03043, donated by DSM (Netherlands) was used in all the cultivations in this work. To obtain spores, Petri dishes containing PDA (Potato Dextrose Agar) medium were incubated with spores from a frozen stock (stored in 50% glycerin at −80 °C). During seed culture, 500 mL shake flasks with baffles were inoculated with 107 spores per 100 mL broth. A 5 L fermentor with an electronic balance was used for the fed-batch cultivations with the agitation rate at 375 rpm and the aeration at 1 vvm. During the cultivation, the overpressure was maintained at 0.05 MPa and the temperature was at 34 °C. The broth pH was maintained at 4.5 by addition of NH3 solution (5% w/w). The working volume for the 5 L fermenter during batch cultivation was 3 L. When the glucose concentration reduced to 5 g/L (after about 36 h of the fermentation) during the batch cultivation, the feed was started and the glucose concentration was kept at around 5 g/L by adjusting the feed rate. Concentrations of oxygen and carbon dioxide in the exhaust gas were determined by process mass spectrometers (MAX300-LG, Extrel) during the fermentation and the dissolved oxygen concentration in the broth was determined with a low-drift polarographic electrode (Mettler Toledo).
The medium for the seed and fed-batch fermentations can be found in the literature reported by Lu18.
Quantification of biomass and enzyme activity
10 mL fermentation broth was filtered by filter paper, pre-weighed and pre-dried to a constant weight (at 80 °C for 24 h). Biomass was rinsed three times with deionized water and dried at 80 °C for 24 h. Dried biomass was re-weighed immediately. Enzyme activity of all samples was determined by a standard procedure18.
Quantification of extracellular sugar and organic acids
Residual sugar from the fermentation broth was determined by a glucose analyser (Shandong Academy of Sciences, China). Extracellular organic acids (acetic acid, citric acid, oxalic acid, malic acid, fumaric acid, pyruvic acid and succinic acid) were determined by high performance liquid chromatography (HPLC). The HPLC was equipped with an ion exclusion column and an absorption detector spectrophotometer. 10 mM H2SO4 was used to wash the ion exclusion chromatography column with the flow rate of 0.5 mL/min at 50 °C and the wavelength of the spectrophotometer was set at 210 nm.
Sampling and quantitative analysis of intracellular metabolites
The protocol for quantitative analysis of intracellular metabolites was modified based on Douma et al.19. Using fast sampling equipment, 1–2 ml broth was pumped from the 5 L fermenter into a 10 ml precooled quenching solution (40% v/v methanol solution at −27.6 °C) at 18 h, 24 h, 36 h, 48 h, 60 h, 72 h and 96 h, respectively. The tubes were weighed before and after the sampling procedure to estimate the exact amount of broth. Then, extracellular metabolites were removed by vacuum filtration and filter cake was washed by 120 ml precooled quenching solution. Isotope dilution mass spectrometry (IDMS)20 was used in this work for the quantification of metabolite concentrations. Washed filter cake, as well as 13C internal standard solution was added to 25 ml pre-warmed 75% (v/v) ethanol solution and the extraction continued for 3 minutes at 95 °C. The metabolites concentration was determined with UPLC-MS/MS (Thermo Fisher Scientific Corporation) and GC-MS.
As for metabolomics data, the principal component analysis (PCA) and partial least square discriminant analysis (PLS-DA) were conducted based on the R programming language. If the variable importance of the projection (VIP) score of one metabolite is above 1, it means that the pool size of this metabolite changes significantly in different fermentation phases. The Heatmap analysis of the metabolomics data from different fermentation phases was conducted using the superheat package of the R language (https://cran.r-project.org/web/packages/superheat/).
Transcriptome analysis
According to the online DO profile, the sampling time for RNA-seq analysis was set at 18 h, 24 h, 42 h and 66 h, which corresponding to the logarithmic phase and early, middle, late phases of oxygen limitation, respectively. After sampling, the broth was immediately frozen in liquid nitrogen and stored at −80 °C. These frozen samples were sent to Sangon Biotech for RNA extraction and RNA samples were sent to the Beijing Genomics Institute (BGI) for sequencing. Qualified RNA samples of each sampling time were ensured with at least 2 replicate samples for sequencing. Gene expression data of different phases was clustered by the Mfuzz package based on R language21. The DAVID database was used for GO enrichment analysis of the interested gene sets22. The Piano package based on the R language was used for KEGG pathway and GO function enrichment analysis of differentially expressed genes23. For gene set analysis, a mapping, established between genes of A. niger and the KEGG pathway and the GO function, as well as gene expression data of different phases, were used as inputs of the Piano package for statistical analysis. For more details, please refer to the literature23.
Flux simulation using GEMs
Flux balance analysis (FBA) based on constraints is widely exploited in the fields of genome-scale metabolic network reconstruction and cellular phenotypic prediction24. FBA is used for prediction and analysis of intracellular fluxes with the optimization of an objective function under constraints. Constraints25 could be an intracellular metabolite balance (Equation 2), reaction reversibility, a maximum enzyme reaction rate and an exchange reaction rate (Equation 3). An objective function is for example maximization of cell growth or optimization of cell energy utilization26 (Equation 1). The rate of each reaction in the model has a limit. For reversible reactions, the upper and lower bound is set to 1000 and −1000 mmol/gBiomass.h, respectively. For irreversible reactions, the lower bound is set to zero. The COBRA toolbox and the Gurobi 5 linear optimization algorithm were used for FBA analysis in this study.
| 1 |
| 2 |
| 3 |
where S is a m × n sparse matrix, in which m refers to the number of metabolites and n refers to the number of reactions. v represents the rate vector of all reactions. lb and ub defines the lower and upper bound of each reaction, respectively. In equation 1, CT refers to the coefficient of each metabolite in the objective function.
Intracellular metabolic fluxes were predicted by the parsimonious FBA (pFBA)27. The model used in this study, A. niger GEMs iHL1210 5 that was updated recently by our laboratory, contains 1727 mass and proton balanced reactions and 1210 ORFs (see Supplementary File 3). Maximization of cell growth was set as the objective function and the measured values of qS, qby-product, qP, qO2 and mATP as constraints during simulation for the fed-batch fermentations (see Supplementary File 3). The prediction performance using iHL1210 was assessed by comparing the predicted values of μ and qCO2 with the measured values.
Results
In order to investigate the global regulating mechanisms of cell metabolism under oxygen-limited condition, the integrative analysis of physiological phenotypic data, metabolomics, transcriptomics and fluxomics was adopted in this work (Fig. 1).
Figure 1.
Framework of multi-omics integration analysis used in this work.
Quantitative analysis of physiological parameters
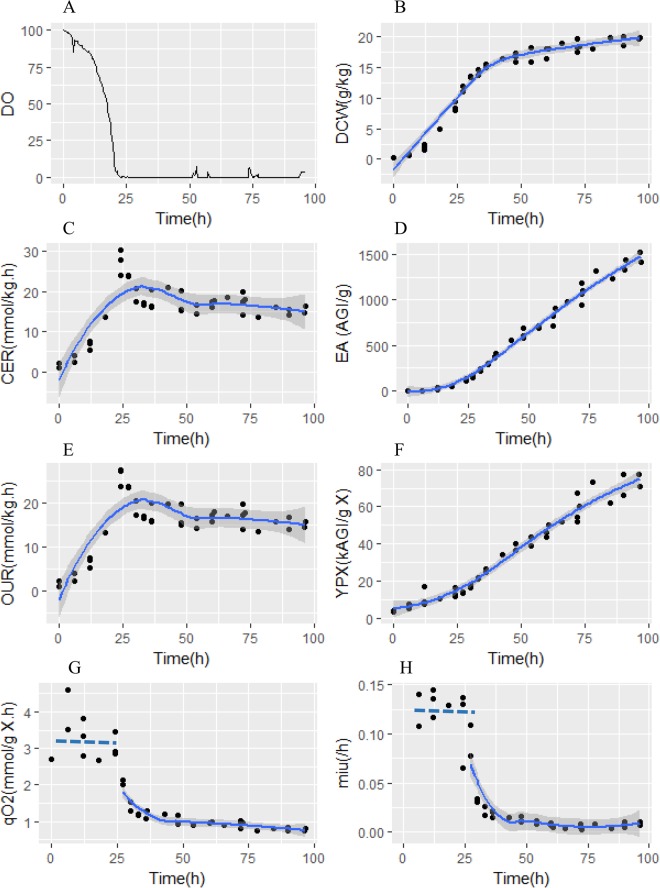
Mimicking the industrial production using A. niger, fed-batch cultivations applying oxygen limited strategy were conducted in this work. The fed-batch fermentation process could be initially divided into 2 main phases, i.e. aerobic phase (0–20 h) and oxygen limited phase (20–72 h) according to profiles of the dissolved oxygen concentration (DO) (Fig. 2A). The changes in the profiles of oxygen uptake rate (OUR), carbon dioxide emission rate (CER) and dry cell weight (DCW) were determined by oxygen supply (OTR). When the oxygen supply was limited, the specific growth rate (μ) quickly decreased (Fig. 2H). Meanwhile, the OUR and CER decreased sharply to a stable level (Fig. 2C and E). By-products analysis showed that organic acids and polyols were slightly excreted by cell (see Supplementary File 1, Fig. S1). The detailed calculation showed that the total carbon ratio of these by-products (Yby-prodcuts/s) is about 5%, thus it can be concluded that most of carbon source fluxed into the biomass, CO2 and product. During the oxygen limited phase, in contrast to the increased yield of glucoamylase (Fig. 2F), μ was decreased continuously and lower than 0.02 h−1 (Fig. 2H) at the end of fermentation.
Figure 2.
Profiles of DO (A), dry cell weight (DCW) (B), CO2 production rate (CER) (C), glucoamylase enzyme activity (D), O2 uptake rate (OUR) (E), yield of glucoamylase per gram biomass (YPX) (F), specific oxygen uptake rate (qO2) (G) and specific growth rate (µ) (H) for A. niger DS03043 with high glucoamylase production during fed-batch cultivations.
Profiling of key metabolites in core carbon metabolism
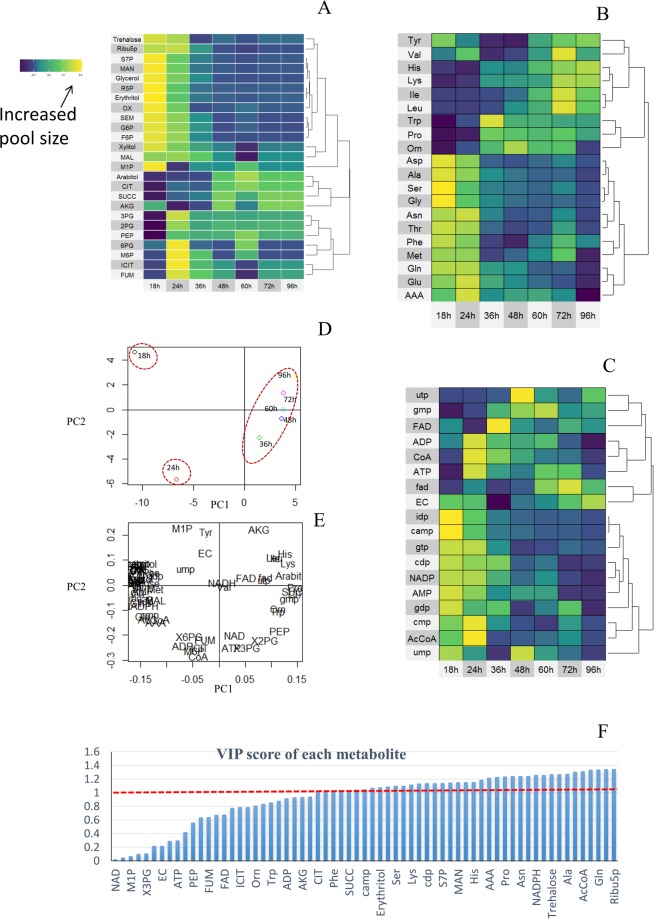
In the first time, the pool sizes of 65 intracellular metabolites (amino acids, organic acids, sugar phosphates, nucleotides and coenzymes) from different fermentation phases were determined by LC-MS/MS or GC-MS. As shown in Fig. 3A–C, the pool sizes of most intracellular metabolites decreased sharply when A. niger entered into the oxygen limited phase. However, it was also found that some amino acids (like Tyr and Val) and organic acids (like SUCC and CIT) still accumulated over time (Fig. 4). The accumulation of organic acids was consistent with the high intracellular redox level under limited O2 supply10,28.
Figure 3.
Data analysis of metabolomics at different fermentation phases (18 h, 24 h, 36 h, 48 h, 60 h, 72 h and 96 h. Heatmap of organic acid and sugar phosphates (A), amino acids (B), nucleotide and coenzymes (C), scores plot for samples in PCA analysis (D), loadings plot for metabolites in PCA analysis (E) VIP score of 69 intracellular metabolites calculated using PLS model (F). The detailed VIP scores for each metabolites can be found in Supplementary File 2.
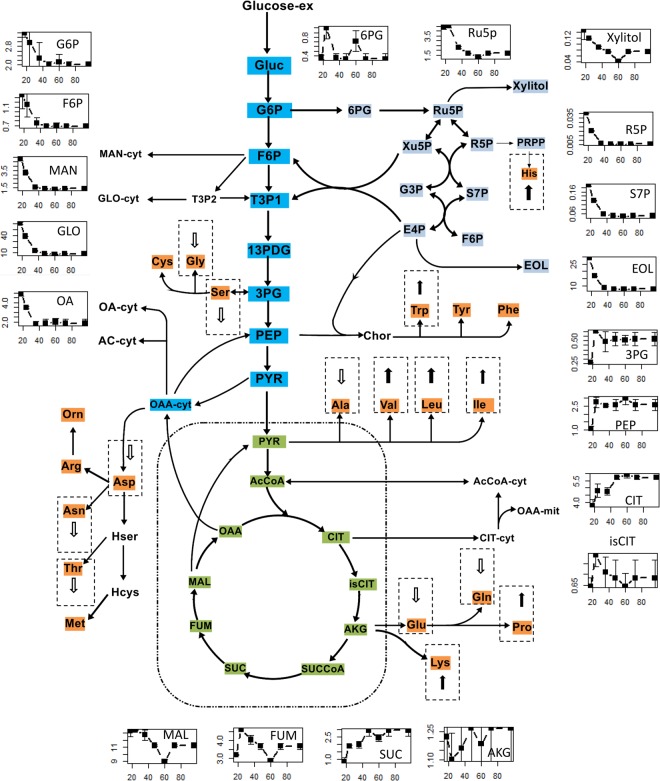
Figure 4.
Schematic representation of the changes in the pool sizes of organic acids, sugar phosphates and amino acids during different fermentation phases onto the core carbon metabolism network. All metabolite pool sizes were determined in at least triplicate measurements. The fermentation time (h) is on the x-axis and the metabolites concentration (μmol/gDCW) is on the y-axis. The arrows in the small gridlines represent the increase or decrease for the pool sizes of intracellular amino acids during the oxygen limited phase compared to that in oxygen excess phase. The detailed profiles of intracellular amino acids pool sizes could be found in Supplementary File 1, Fig. S2.
The changes in intracellular amino acid pool sizes during the oxygen limited phase exhibited two different tendencies (Figs 3B and 4). The pool sizes of Ala, Gly, Asp, Glu and Ser decreased sharply when the cells entered into the oxygen limited phase, while the pool sizes of Val, Leu, Ile and His increased significantly (see Supplementary File 1, Fig. S2), consistent with the extracellular accumulation of these amino acids (see Supplementary File 1, Fig. S3).
Furthermore, the principal component analysis (Fig. 3E) showed that all the samples can be categorized into three groups, which is difficult observed from physiological profiles (Fig. 2). On the other hand, the relation between changes in pool sizes of metabolites and qO2 was studied by partial least squares (PLS) analysis. The VIP of 45 metabolites was above 1 (Fig. 3F), indicating that the changes of most intracellular metabolites concentration were sensitive to the external environment perturbations.
Systematic analysis of gene and typical transcription factors (TFs) expression pattern related to external environmental changes
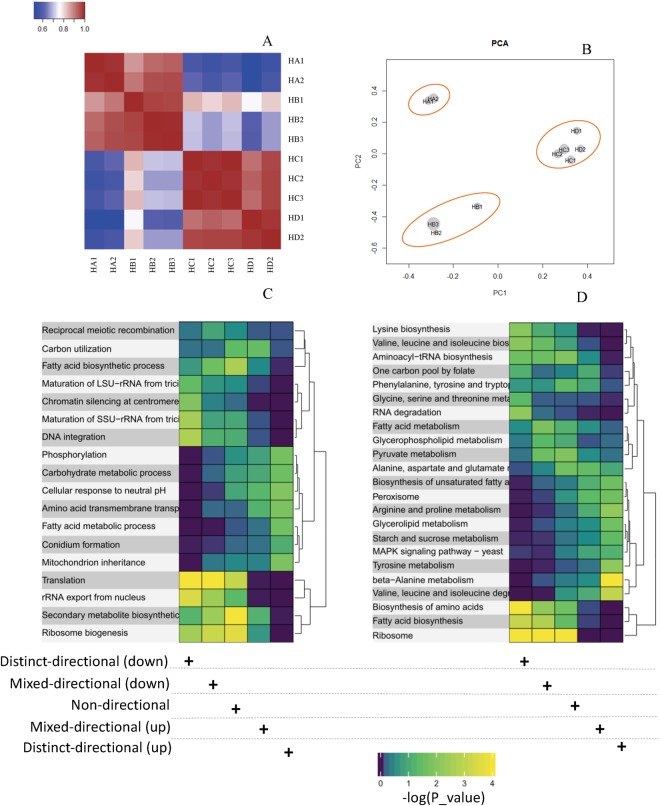
The expression data of 10,445 genes from different fermentation phases (16 h, 24 h, 42 h and 66 h) was determined using RNA-seq. According to PCA analysis based on FPKM values of genes (Fig. 5B), it is shown that the replicate samples from the same time point could be clustered together. All the samples can be clustered into three groups, consistent with that using metabolomics analysis. To obtain the main metabolic characteristics of A. niger under oxygen limited environment, the gene expression pattern analysis along the fed-batch process was firstly carried out, followed by the gene set analysis of differentially expressed genes in two distinct fermentation phases (oxygen sufficient phase 16 h and oxygen limitation phase 42 h).
Figure 5.
Gene set analysis of differentially expressed genes in the aerobic (16 h) and oxygen limited phases (42 h). Correlation coefficients of samples (A), scores plot for samples in PCA analysis using gene expression values (B), Gene set analysis based on GO function annotation (C) and KEGG pathway enrichment analysis (D). The non-directional class disregards the direction of change. The distinct-directional class takes direction of change into account. The mixed-directional class considers the up-regulated subset and the down-regulated subset of a gene set separately. Each subset is scored according to the proportion of significant genes.
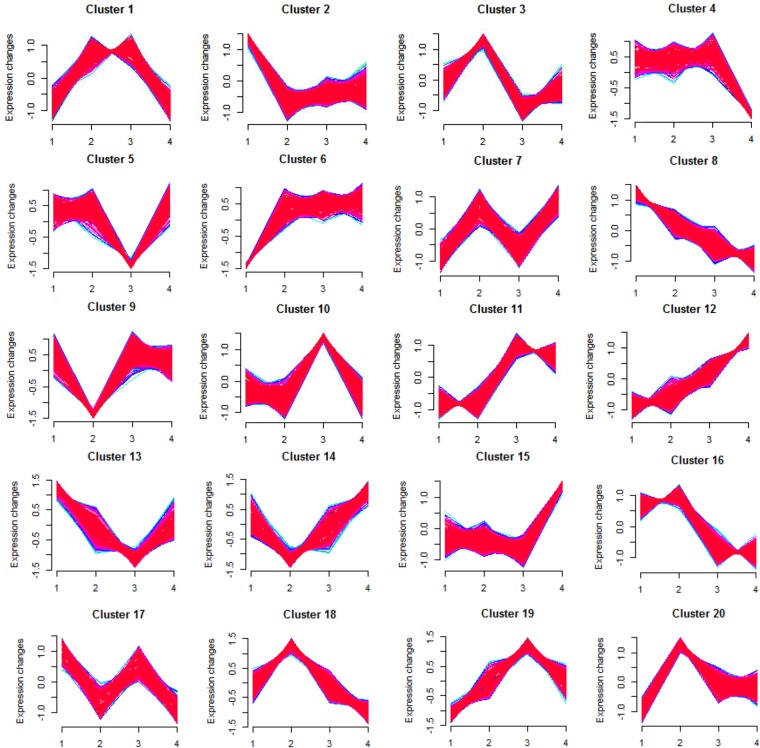
With the aid of the Mfuzz package21 based on the R language, those genes, with little expression (even no expression) at least two or more time points during the fermentation, were firstly removed and finally 6,662 genes were screened from the total 10,445 genes. Then the cluster analysis of gene expression profiles was conducted for the remaining 6,662 genes. The result showed that the expression pattern of all 6,662 genes could be divided into 20 clusters (Fig. 6). Among the 20 clusters, there exist several clusters in which the expression of genes was consistent to the changes of qO2 (Fig. 2G). For example, the expression of 408 genes in cluster 8 decreased with the decrease of qO2, while in cluster 12, the expression of 373 genes increased with the decrease of qO2. In addition, in cluster 2, the expression of 592 genes decreased sharply from the aerobic phase (16 h) to the transition phase (24 h), and remained stable in the mid and later oxygen limitation phases (42 h and 66 h). In contrast, 201 genes in cluster 6 showed an opposite tendency.
Figure 6.
Clusters of expressed genes during different fermentation phases. The number of 1, 2, 3 and 4 in each small graph represents 16 h, 24 h, 42 h and 66 h respectively.
DAVID, an online gene function annotation tool22, was exploited to carry out gene function annotation and enrichment analysis of the genes from the up-regulation group (cluster 12 and cluster 6) (Table 1) and the down-regulation group (cluster 8 and cluster 2) (Table 2). There were about 423 transcription factors (TFs) in A. niger genome according to the Aspergillus Genome Database29. In the above cluster analysis, the expression of TFs in different clusters was extracted and studied. Cluster 12, where the gene expression was continuously up-regulated, contained 10 TFs, among which flbA (An02g03160, related to morphological development) and riaA (An16g05550, related to NADPH oxidation regulation) were included. Another TF, brlA (An01g10540), related to the formation of spores in cluster 2, was down-regulated under oxygen limited condition. The expression of 9 TFs in cluster 8 was significantly down-regulated. Among them, An12g00130, which is thought to play a role in regulating the mitochondrial respiratory chain complex IV biogenesis. Under oxygen limited conditions, the cell could decrease the biosynthesis of complex IV in the electron transport chain by down-regulating the expression of An12g00130.
Table 1.
GO function enrichment analysis of genes from the up-regulation group.
| Category | GO Number | Term | P-Value |
|---|---|---|---|
| cluster 12 | |||
| GOTERM_BP_FAT | GO:0006913 | nucleocytoplasmic transport | 0.001 |
| GOTERM_BP_FAT | GO:0051169 | nuclear transport | 0.001 |
| GOTERM_CC_FAT | GO:0005643 | nuclear pore | 0.002 |
| GOTERM_CC_FAT | GO:0046930 | pore complex | 0.002 |
| GOTERM_CC_FAT | GO:0005635 | nuclear envelope | 0.002 |
| GOTERM_BP_FAT | GO:0046907 | intracellular transport | 0.024 |
| GOTERM_CC_FAT | GO:0031967 | organelle envelope | 0.029 |
| GOTERM_CC_FAT | GO:0012505 | endomembrane system | 0.030 |
| GOTERM_CC_FAT | GO:0031975 | envelope | 0.032 |
| GOTERM_BP_FAT | GO:0051170 | nuclear import | 0.034 |
| GOTERM_BP_FAT | GO:0034504 | protein localization in nucleus | 0.034 |
| GOTERM_BP_FAT | GO:0006606 | protein import into nucleus | 0.034 |
| GOTERM_CC_FAT | GO:0016021 | integral to membrane | 0.035 |
| GOTERM_CC_FAT | GO:0031224 | intrinsic to membrane | 0.040 |
| GOTERM_MF_FAT | GO:0004857 | enzyme inhibitor activity | 0.050 |
| INTERPRO | Hypoxia induced protein conserved region | 0.057 | |
| GOTERM_BP_FAT | GO:0017038 | protein import | 0.076 |
| SP_PIR_KEYWORDS | tricarboxylic acid cycle | 0.077 | |
| GOTERM_BP_FAT | GO:0006979 | response to oxidative stress | 0.084 |
| GOTERM_MF_FAT | GO:0004022 | alcohol dehydrogenase (NAD) activity | 0.088 |
| cluster 6 | |||
| INTERPRO | Acyl-CoA oxidase/dehydrogenase, type1/2, C-terminal | 0.012 | |
| INTERPRO | Acyl-CoA oxidase/dehydrogenase, central region | 0.014 | |
| GOTERM_MF_FAT | GO:0003995 | acyl-CoA dehydrogenase activity | 0.015 |
| GOTERM_MF_FAT | GO:0009055 | electron carrier activity | 0.034 |
| GOTERM_BP_FAT | GO:0043087 | regulation of GTPase activity | 0.043 |
| GOTERM_BP_FAT | GO:0051336 | regulation of hydrolase activity | 0.043 |
| GOTERM_MF_FAT | GO:0008336 | gamma-butyrobetaine dioxygenase activity | 0.048 |
| GOTERM_BP_FAT | GO:0006091 | generation of precursor metabolites and energy | 0.077 |
| GOTERM_MF_FAT | GO:0030695 | small GTPase regulator activity | 0.095 |
Table 2.
GO function enrichment analysis of genes from the down-regulation group.
| Category | GO Number | Term | P-Value |
|---|---|---|---|
| cluster 8 | |||
| GOTERM_BP_FAT | GO:0006259 | DNA metabolic process | 0.009 |
| GOTERM_BP_FAT | GO:0033554 | cellular response to stress | 0.011 |
| GOTERM_BP_FAT | GO:0006260 | DNA replication | 0.013 |
| GOTERM_MF_FAT | GO:0000036 | acyl carrier activity | 0.014 |
| GOTERM_MF_FAT | GO:0016597 | amino acid binding | 0.014 |
| GOTERM_MF_FAT | GO:0043176 | amine binding | 0.014 |
| GOTERM_BP_FAT | GO:0006281 | DNA repair | 0.022 |
| GOTERM_BP_FAT | GO:0006974 | response to DNA damage stimulus | 0.023 |
| GOTERM_MF_FAT | GO:0004386 | helicase activity | 0.028 |
| GOTERM_MF_FAT | GO:0031177 | phosphopantetheine binding | 0.035 |
| GOTERM_MF_FAT | GO:0031406 | carboxylic acid binding | 0.049 |
| GOTERM_MF_FAT | GO:0048037 | cofactor binding | 0.050 |
| GOTERM_BP_FAT | GO:0019748 | secondary metabolic process | 0.067 |
| GOTERM_BP_FAT | GO:0051276 | chromosome organization | 0.089 |
| GOTERM_MF_FAT | GO:0016879 | ligase activity, forming carbon-nitrogen bonds | 0.096 |
| GOTERM_MF_FAT | GO:0004842 | ubiquitin-protein ligase activity | 0.097 |
| cluster 2 | |||
| GOTERM_BP_FAT | GO:0030163 | protein catabolic process | 0.047 |
| GOTERM_CC_FAT | GO:0044430 | cytoskeletal part | 0.049 |
| GOTERM_MF_FAT | GO:0008171 | O-methyltransferase activity | 0.055 |
| GOTERM_BP_FAT | GO:0009057 | macromolecule catabolic process | 0.064 |
| GOTERM_CC_FAT | GO:0015630 | microtubule cytoskeleton | 0.067 |
| GOTERM_MF_FAT | GO:0000166 | nucleotide binding | 0.085 |
| GOTERM_CC_FAT | GO:0005856 | Cytoskeleton | 0.096 |
| GOTERM_MF_FAT | GO:0004386 | helicase activity | 0.097 |
| GOTERM_MF_FAT | GO:0016405 | CoA-ligase activity | 0.098 |
The gene set analysis of differentially expressed genes at 42 h and 16 h was further performed by the Piano package23 based on the R language (Fig. 5C,D). The results showed that in order to adapt to the limited oxygen supply, the biosynthesis of fatty acids and secondary metabolites, ribosome biogenesis and translation were significantly down-regulated (P_value < 0.05), while the fatty acid catabolism was up-regulated, which can further be validated by changes in tendencies of genes expression from the related synthesis pathway (see Supplementary File 1, Fig. S6).
Flux simulation based on newly updated GEMs
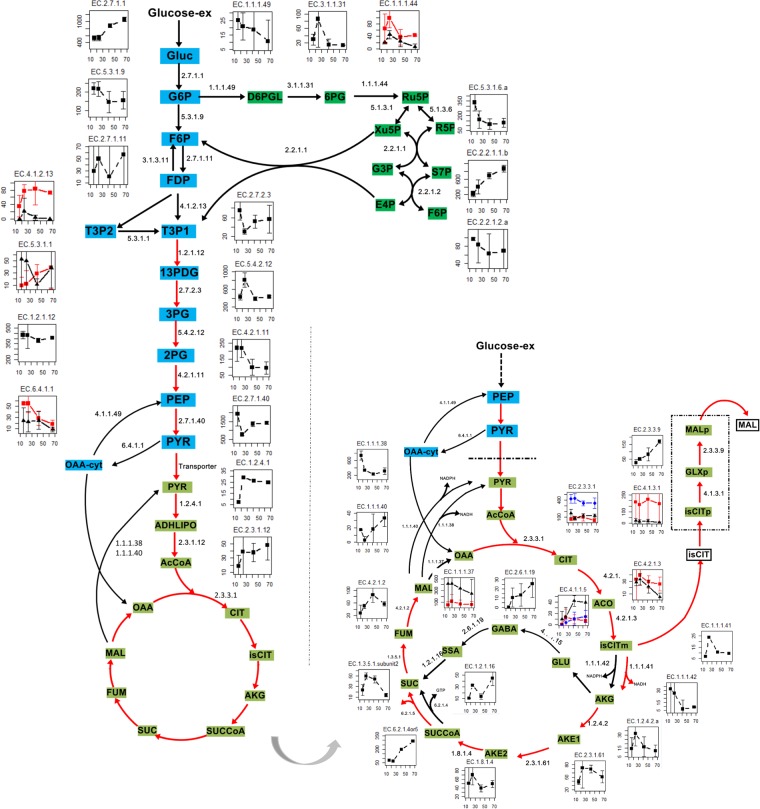
To further investigate how the cell adapted to the external environmental changes, the flux distribution predicted by pFBA was exploited. The predicted μ and qCO2 are consistent with the measured values (see Supplementary File 3), indicating the good performances of iHL1210. The flux simulation using iHL12105 showed that the relative fluxes through the EMP pathway increased when the cells entered into the oxygen limited phase (Fig. 7, Supplementary File 3). Consistent with the increased relative flux through EMP pathway, the expression of the corresponding genes was maintained at a stable level, such as fructose-bisphosphate aldolase (EC 4.1.2.13) and pyruvate dehydrogenase (EC 1.2.4.1, EC 2.3.1.12). In addition, the pool sizes of some amino acids from the aromatic and pyruvate families increased accordingly, which might provide precursors for enzyme production.
Figure 7.
Trends of genes expression and relative flux of the EMP and PP and TCA pathways under different fermentation phases. The red arrows represent the reactions with an obvious increase in the relative flux predicted by iHL1210 in the oxygen limited phase. The fermentation time (h) is on the x-axis and the gene expression value (FPKM) is on the y-axis. The black and red lines in each small graph represent independent isogenies encoding the specific enzymes. The detailed flux distribution by pFBA at different time points could be found in Supplementary File 3.
Flux simulation of GEMs iHL1210 showed that relative fluxes of the PP pathway decreased slowly along the fermentation (see Supplementary File 3), consistent with the reduction tendency in the gene expression values for glucose-6-phosphate dehydrogenase (EC 1.1.1.49). However, it was notable that gene expression of transketolase (EC 2.2.1.1) in the PP pathway was significantly up-regulated.
Consistent with higher pool sizes of organic acids, the flux simulation with iHL1210 showed that the relative flux through the TCA cycle was increased upon oxygen limitation, which was related to the lower demand for the anabolic metabolism from the cell growth. To maintain the redox balance, the cell metabolic activities were regulated in transcriptional level. Firstly, the expression of genes encoding the key enzymes of the TCA cycle, including citrate synthase (EC 2.3.3.1) and aconitate hydratase (EC 4.2.1.3), were down-regulated. Meanwhile, the expression of genes (EC 2.3.3.9 and EC 2.6.1.19) from the glyoxylate and GABA cycles were significantly up-regulated (Fig. 7). Consistent with this observation, the flux simulation indicated that the relative flux through the glyoxylate cycle increased (see Supplementary File 3).
Discussion
Cell metabolism based on gene expression pattern analysis under oxygen limited conditions
According to the cluster analysis in the Results section, there exist two obvious gene expression patterns, namely up-regulation (cluster 12 and cluster 6) and down-regulation (cluster 8 and cluster 2). Under oxygen limited conditions, the energy supply could become a main bottleneck for complex cell metabolic functions including membrane synthesis. The enrichment analysis of genes in cluster 6 revealed that the energy production was up-regulated during the oxygen limited phase, helping to relieve the shortage in energy supply. Genes in cluster 8 were enriched in the biological processes closely related to DNA unwinding, replication and transcription, indicating that oxygen limitation could weaken the transcription and translation to reduce energy demand. Parts of the genes in cluster 2 were also enriched in DNA unwinding, indicating that the DNA replication was obviously weakened, which was highly consistent with the decreased specific growth rate during the oxygen limited phase (Fig. 2H). Meanwhile, the synthesis of macromolecules slowed down and the expression of genes encoding degradation pathways of macromolecules (like proteins) was up-regulated accordingly (cluster 2) to strengthen the turnover of intracellular metabolites. According to the genome annotation29, transcription factor flbA in cluster 12 is the regulator of the G-protein signaling protein. It has been reported that the deletion of this gene displayed the phenotype of long thin mycelium without arthrospore, and promoted protein secretion30. In this study, the expression of flbA increased continuously, which might thicken the cell wall and hinder the protein secretion. Sterol regulatory element-binding proteins (SREBPs) are important TFs for filamentous fungi to adapt to an anaerobic environment as the decrease of the intracellular sterol concentration in anaerobic environment will activate the expression of SREBPs31. In A. niger, the corresponding genes are srbA (An03g05170) and srbB (An14g02540), respectively32. In this work, the expression of the former was down-regulated first and then was up-regulated (in cluster 14) while the expression of the latter was continuously elevated (in cluster 12), indicating that SREBPs are conserved for fungi to adapt to the oxygen limited environment.
Potential advantages of the oxygen limited strategy used for enzyme production by A. niger
Similar to P. pastoris17, an appropriate oxygen limitation favors glucoamylase production by A. niger. However, the detailed mechanisms behind this phenomenon are not clear. Generally, there is an inverse correlation between protein production and cell growth33 as the protein synthesis usually needs the competitive precursors for the cell growth. During the oxygen limited phase, μ was lower than 0.02 h−1 (Fig. 2H). It could be speculated that the low specific growth rate can be a possible reason of high yield of glucoamylase due to the fact that once the growth was limited, more NADPH, NADH and precursors could be fluxed into synthesis of glucoamylase. Especially, the increased relative flux in the EMP pathway, along with the accumulation of amino acids from the pyruvate and aromatic families, is beneficial for protein synthesis. Furthermore, the transcription factors relating to sterol synthesis, one important composition of the cell membrane, was activated during the oxygen limitation, which possibly helped to maintain the integrity of the cell membrane14, favoring an efficient secretion of glucoamylase.
It has been reported that the exogenous addition of the limited amino acids could effectively alleviate the shortage in supply of amino acids, energy and reducing power34. In this work, the addition of Ala, Gly, Asp, Glu and Ser could obviously promote the enzyme production (see Supplementary File 1, Fig. S4). Coincidentally, these amino acids have a decreased tendency in intracellular pool sizes along with the fermentation. In combination with the fact that the four amino acids are the main compositions of glucoamylase (Table 3), it can be inferred that they might be the limiting precursors for the synthesis of the target protein, which provides new clues for metabolic engineering to promote enzyme production efficiency using molecular biology. It should be also noted that some amino acids, like His, Lys, Val, Ile, etc., were accumulated within the cell. For the accumulation of aromatic amino acids and part of the pyruvate family, a driver could be the increased flux through the EMP pathway. Moreover, the pool sizes of amino acids were closely related to the changes in the expression levels of genes from the corresponding pathways. Taking Ala and Gly as an example, the decrease in pool sizes of these two amino acids was accompanied with the observation of a prominent decrease in gene expression levels of the Ala and Gly synthesis pathways (see Supplementary File 1, Fig. S5), initially indicating that the pool sizes of amino acids were rigidly controlled. In terms of energy requirements, Lys, Met, Ile, Trp and His can be regarded as expensive amino acids35. It is reported that under environmental stress conditions the cell could secure some of the expensive amino acids while at the same time decreasing the pool sizes of the cheap amino acids36. As a result, when the environment changes again to more favorable conditions, the cell could realize fast growth by mobilizing the energy expensive amino acids. The precise mechanisms for the amino acids accumulation under oxygen limited conditions still need further experimental validation.
Table 3.
Amino acid composition of glucoamylase.
| animo acid | content | amino acid | content |
|---|---|---|---|
| Ala | 10.17% | Met | 0.47% |
| Cys | 1.56% | Asn | 3.91% |
| Asp | 6.89% | Pro | 3.44% |
| Glu | 3.91% | Gln | 2.66% |
| Phe | 3.44% | Arg | 3.13% |
| Gly | 7.20% | Ser | 13.77% |
| His | 0.63% | Thr | 11.58% |
| Ile | 3.76% | Val | 6.57% |
| Lys | 2.03% | Trp | 3.13% |
| Leu | 7.51% | Tyr | 4.23% |
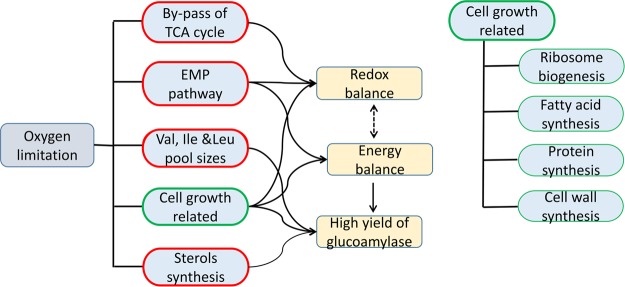
The possible mechanisms for high yield of glucoamylase during the oxygen limited phase are summarized in Fig. 8.
Figure 8.
Possible coordinated regulation mechanisms for maintenance of the redox (energy) balance, as well as for the high enzyme production by A. niger upon oxygen limitation. The contents in red and green frames represent the up- and down-regulated metabolic activities, respectively.
How the cell maintains redox and energy balance under hypoxic conditions?
Under hypoxic conditions, the ways for different species to maintain intracellular energy and redox balance may be different. Unlike fungal strains, S. cerevisiae and mammalian cells could regenerate NADH via the formation of ethanol and lactic acid to maintain the intracellular energy and redox balance. With 13C labeling experiments, the P/O ratio of S. cerevisiae was still found high under hypoxic conditions37, which indicates that the oxidative phosphorylation remained efficient. A high formation rate of ATP helps to promote the oxidative phosphorylation under hypoxic conditions38. Also, the oxygen limitation conditions could reduce leakage of protons and uncoupled respiration38. In this work, the marginal secretion of polyols and reduced organic acids was not enough to sustain the regeneration of NAD+. Therefore, it could be speculated that, similar to A. fumigatus13, oxidative phosphorylation is the main metabolic pathway for maintaining the intracellular balance of energy and redox. The transcriptomics and flux simulation indicated that the flux through the glyoxylate cycle was increased, reducing NADH formation from the TCA cycle, which helps to maintain the redox balance. In addition, as reported in the literature, during oxygen limited conditions, the strain could maintain the redox balance by the reduction of nitrate, as well as the secretion of branched amino acids39. As ammonium sulfate was used as nitrogen source in the work, the reduction of nitrate could be excluded. As for the secretion of branched amino acids, like Val, Ile, Leu, etc., they are slightly accumulated within the cell and the extracellular secretion was marginal, although still insufficient to maintain the intracellular redox balance. Therefore, we concluded that mainly the enforcements of glyoxylate cycle (the GABA shunt is also possible) and oxidative phosphorylation help to maintain the intracellular redox balance.
On the other hand, the oxygen limitation could lead to the shortage in supply of ATP and NADPH. To maintain the cell normal metabolic activities, the cell can reduce anabolism and strengthen the catabolism, which was validated by a decrease in the gene expression levels of the fatty acid synthesis pathway, as well as the increase in gene expression levels of the fatty acid catabolism pathway. The enforcement of the EMP pathway could increase the formation of ATP, alleviating the energy demand. As for NADPH supply, there are mainly three sources according to transcriptomics data: the PP pathway, ICIT + NADP = > AKG + NADPH and MAL + NADP = > PYR + NADPH. The gene expression levels of the former two sources were decreased while they were increased in the third source. With 13C labeling flux analysis40, it was found that in the A. niger high-producing strain, the flux through MAL + NADP = > PYR + NADPH was higher than that in the wild type strain. So it could be concluded that under oxygen limited conditions, MAL + NADP = > PYR + NADPH might be a potential gene target for metabolic engineering to provide more NADPH supply. The supposed metabolic regulation mechanisms for the cell to maintain the intracellular redox and energy balance can be found in Fig. 8.
Conclusion
The multi-omics integrative analysis provides us new insights on the mechanisms of A. niger metabolic regulation under fed-batch process conditions for enzyme production. To maintain the intracellular redox and energy balance under hypoxic condition, the cell metabolism was regulated at different aspects. The pool sizes of most intermediate metabolites from the upper EMP and PP pathways decreased along the fermentation. Meanwhile the gene expression was reduced for the fatty acid and ribosome synthesis pathways accordingly to weaken the cell anabolic metabolism. On the contrary, the EMP pathway and glyoxylate pathway were activated, which can be validated by the association analysis of transcriptomics and fluxomics. The possible reasons for a high yield of glucoamylase during the oxygen limited phase can be summarized as follows. Firstly, the increased relative flux through the EMP pathway could provide more precursors for enzyme synthesis. Secondly, the down regulations in fatty acid and ribosome biogenesis could also channel more precursors towards glucoamylase synthesis. Thirdly, the up-regulation in gene expression for sterols synthesis might favor the enzyme secretion. The multi-omics integrative analysis illustrated, in a systematic view, the potential of an oxygen limited strategy used in the industrial fed-batch fermentation. Furthermore, a rational optimization of the A. niger metabolic network in terms of precursors and NADPH supply, as well as re-balancing the NADH and sterol biosynthesis may further help A. niger adapt to hypoxic conditions, as well as improvement of the enzyme productivity.
Electronic supplementary material
Acknowledgements
This work was financially supported by Royal DSM (Delft, the Netherlands) and partially supported by National Basic Research Program (973 Program 2013CB733600), NWO-MoST Joint Program (2013DFG32630), Open Funding Project of the State Key Laboratory of Bioreactor Engineering.
Author Contributions
H.L. and W.C. carried out the experiments. X.L. and Y.S. analyzed the data. J.C. and L.O. participated in the design and coordination of the study. M.H., J.X., Y.Z., S.Z. and N.H. helped to draft the manuscript. All authors read and approved the final manuscript.
Data Availability
All data generated or analysed during this study are included in this article (and its Supplementary Information Files).
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Hongzhong Lu and Weiqiang Cao contributed equally.
Contributor Information
Liming Ouyang, Email: ouyanglm@ecust.edu.cn.
Ju Chu, Email: juchu@ecust.edu.cn.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-32341-1.
References
- 1.Knuf C, Nielsen J. Aspergilli: Systems biology and industrial applications. Biotechnology Journal. 2012;7:1147–1155. doi: 10.1002/biot.201200169. [DOI] [PubMed] [Google Scholar]
- 2.Pedersen L, Hansen K, Nielsen J, Lantz AE, Thykaer J. Industrial glucoamylase fed-batch benefits from oxygen limitation and high osmolarity. Biotechnol. Bioeng. 2011;109:116–124. doi: 10.1002/bit.23287. [DOI] [PubMed] [Google Scholar]
- 3.Stam H, et al. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat. Biotechnol. 2007;25:221–231. doi: 10.1038/nbt1282. [DOI] [PubMed] [Google Scholar]
- 4.Andersen, M. R., Nielsen, M. L. & Nielsen, J. Metabolic model integration of the bibliome, genome, metabolome and reactome of Aspergillus niger. Mol. Syst. Biol. 4, http://www.nature.com/msb/journal/v4/n1/suppinfo/msb200812_S1.html (2008). [DOI] [PMC free article] [PubMed]
- 5.Lu, H. et al. Comprehensive reconstruction and in silico analysis of Aspergillus niger genome‐scale metabolic network model that accounts for 1210 ORFs. Biotechnol. Bioeng (2016). [DOI] [PubMed]
- 6.Meyer, V., Fiedler, M., Nitsche, B. & King, R. Ch. 297, 1–42 (Springer Berlin Heidelberg, 2015).
- 7.Hall LA, Denning DW. Oxygen requirements of Aspergillus species. J. Med. Microbiol. 1994;41:311–315. doi: 10.1099/00222615-41-5-311. [DOI] [PubMed] [Google Scholar]
- 8.Show PL, et al. Overview of citric acid production from Aspergillus niger. Frontiers in Life Science. 2015;8:271–283. doi: 10.1080/21553769.2015.1033653. [DOI] [Google Scholar]
- 9.Baumann KMM, Dragosits M, Cos O, Ferrer P, Mattanovich D. Hypoxic fed-batch cultivation of Pichia pastoris increases specific and volumetric productivity of recombinant proteins. Biotechnol. Bioeng. 2008;100:177–183. doi: 10.1002/bit.21763. [DOI] [PubMed] [Google Scholar]
- 10.Diano A, Peeters J, Dynesen J, Nielsen J. Physiology of Aspergillus niger in oxygen-limited continuous cultures: Influence of aeration, carbon source concentration and dilution rate. Biotechnol. Bioeng. 2009;103:956–965. doi: 10.1002/bit.22329. [DOI] [PubMed] [Google Scholar]
- 11.Diano A, Bekker-Jensen S, Dynesen J, Nielsen J. Polyol synthesis in Aspergillus niger: Influence of oxygen availability, carbon and nitrogen sources on the metabolism. Biotechnol. Bioeng. 2006;94:899–908. doi: 10.1002/bit.20915. [DOI] [PubMed] [Google Scholar]
- 12.Bonaccorsi ED, et al. Transcriptional response of the obligatory aerobe Trichoderma reesei to hypoxia and transient anoxia: Implications for energy production and survival in the absence of oxygen. Biochemistry. 2006;45:3912–3924. doi: 10.1021/bi052045o. [DOI] [PubMed] [Google Scholar]
- 13.Vödisch M, et al. Analysis of the Aspergillus fumigatus proteome reveals metabolic changes and the activation of the pseurotin A biosynthesis gene cluster in response to hypoxia. Journal of proteome research. 2011;10:2508–2524. doi: 10.1021/pr1012812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi J, et al. Genome-wide analysis of hypoxia-responsive genes in the rice blast fungus, Magnaporthe oryzae. PloS one. 2015;10:1–19. doi: 10.1371/journal.pone.0134939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kroll K, et al. Identification of hypoxia-inducible target genes of Aspergillus fumigatus by transcriptome analysis reveals cellular respiration as an important contributor to hypoxic survival. Eukaryot. Cell. 2014;13:1241–1253. doi: 10.1128/EC.00084-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu H, et al. Integrated isotope-assisted metabolomics and 13C metabolic flux analysis reveals metabolic flux redistribution for high glucoamylase production by Aspergillus niger. Microbial Cell Factories. 2015;14:1–14. doi: 10.1186/s12934-015-0329-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baumann K, et al. A multi-level study of recombinant Pichia pastoris in different oxygen conditions. BMC systems biology. 2010;4:1. doi: 10.1186/1752-0509-4-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu H, et al. Dependence of fungal characteristics on seed morphology and shear stress in bioreactors. Bioprocess Biosyst Eng. 2015;38:917–928. doi: 10.1007/s00449-014-1337-8. [DOI] [PubMed] [Google Scholar]
- 19.Douma RD, et al. Intracellular Metabolite Determination in the Presence of Extracellular Abundance: Application to the Penicillin Biosynthesis Pathway in Penicillium chrysogenum. Biotechnol. Bioeng. 2010;107:105–115. doi: 10.1002/bit.22786. [DOI] [PubMed] [Google Scholar]
- 20.Wu L, et al. Quantitative analysis of the microbial metabolome by isotope dilution mass spectrometry using uniformly C-13-labeled cell extracts as internal standards. Anal. Biochem. 2005;336:164–171. doi: 10.1016/j.ab.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 21.Kumar L, Futschik ME. Mfuzz: a software package for soft clustering of microarray data. Bioinformation. 2007;2:5. doi: 10.6026/97320630002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dennis G, Jr, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome biology. 2003;4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 23.Väremo L, Nielsen J, Nookaew I. Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Res. 2013;41:4378–4391. doi: 10.1093/nar/gkt111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orth JD, Thiele I, Palsson BO. What is flux balance analysis? Nat. Biotechnol. 2010;28:245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bordbar A, Monk JM, King ZA, Palsson BO. Constraint-based models predict metabolic and associated cellular functions. Nature Reviews Genetics. 2014;15:107–120. doi: 10.1038/nrg3643. [DOI] [PubMed] [Google Scholar]
- 26.Schuetz, R., Kuepfer, L. & Sauer, U. Systematic evaluation of objective functions for predicting intracellular fluxes in Escherichia coli. Mol. Syst. Biol. 3, http://www.nature.com/msb/journal/v3/n1/suppinfo/msb4100162_S1.html (2007). [DOI] [PMC free article] [PubMed]
- 27.Schellenberger J, et al. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolboxv2.0. Nat. Protocols. 2011;6:1290–1307. doi: 10.1038/nprot.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meijer S, Panagiotou G, Olsson L, Nielsen J. Physiological characterization of xylose metabolism in Aspergillus niger under oxygen-limited conditions. Biotechnol. Bioeng. 2007;98:462–475. doi: 10.1002/bit.21397. [DOI] [PubMed] [Google Scholar]
- 29.Arnaud MB, et al. The Aspergillus Genome Database (AspGD): recent developments in comprehensive multispecies curation, comparative genomics and community resources. Nucleic Acids Res. 2012;40:D653–659. doi: 10.1093/nar/gkr875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krijgsheld P, et al. Deletion of flbA Results in Increased Secretome Complexity and Reduced Secretion Heterogeneity in Colonies of Aspergillus niger. Journal of Proteome Research. 2013;12:1808–1819. doi: 10.1021/pr301154w. [DOI] [PubMed] [Google Scholar]
- 31.Butler G. Hypoxia and gene expression in eukaryotic microbes. Annu. Rev. Microbiol. 2013;67:291–312. doi: 10.1146/annurev-micro-092412-155658. [DOI] [PubMed] [Google Scholar]
- 32.Sui, Y.-f. et al. Global transcriptional response of Aspergillus niger in the process of glucoamylase fermentation. Bioresources and Bioprocessing4, 10.1186/s40643-017-0160-x (2017).
- 33.Melzer G, et al. Metabolic flux analysis using stoichiometric models for Aspergillus niger: Comparison under glucoamylase-producing and non-producing conditions. J. Biotechnol. 2007;132:405–417. doi: 10.1016/j.jbiotec.2007.08.034. [DOI] [PubMed] [Google Scholar]
- 34.Korneli C, Bolten CJ, Godard T, Franco-Lara E, Wittmann C. Debottlenecking recombinant protein production in Bacillus megaterium under large-scale conditions—targeted precursor feeding designed from metabolomics. Biotechnol. Bioeng. 2012;109:1538–1550. doi: 10.1002/bit.24434. [DOI] [PubMed] [Google Scholar]
- 35.Barton MD, Delneri D, Oliver SG, Rattray M, Bergman CM. Evolutionary Systems Biology of Amino Acid Biosynthetic Cost in Yeast. PLOS ONE. 2010;5:e11935. doi: 10.1371/journal.pone.0011935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Link H, Fuhrer T, Gerosa L, Zamboni N, Sauer U. Real-time metabolome profiling of the metabolic switch between starvation and growth. Nat. Methods. 2015;12:1091–1097. doi: 10.1038/nmeth.3584. [DOI] [PubMed] [Google Scholar]
- 37.Jouhten P, et al. Oxygen dependence of metabolic fluxes and energy generation of Saccharomyces cerevisiae CEN. PK113-1A. BMC systems biology. 2008;2:1–19. doi: 10.1186/1752-0509-2-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gnaiger E, Méndez G, Hand SC. High phosphorylation efficiency and depression of uncoupled respiration in mitochondria under hypoxia. Proceedings of the National Academy of Sciences. 2000;97:11080–11085. doi: 10.1073/pnas.97.20.11080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shimizu M, Fujii T, Masuo S, Takaya N. Mechanism of de novo branched-chain amino acid synthesis as an alternative electron sink in hypoxic Aspergillus nidulans cells. Appl. Environ. Microbiol. 2010;76:1507–1515. doi: 10.1128/AEM.02135-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Driouch H, Melzer G, Wittmann C. Integration of in vivo and in silico metabolic fluxes for improvement of recombinant protein production. Metab. Eng. 2012;14:47–58. doi: 10.1016/j.ymben.2011.11.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this article (and its Supplementary Information Files).