Abstract
The OpenWorm Project is an international open-source collaboration to create a multi-scale model of the organism Caenorhabditis elegans. At each scale, including subcellular, cellular, network and behaviour, this project employs one or more computational models that aim to recapitulate the corresponding biological system at that scale. This requires that the simulated behaviour of each model be compared with experimental data both as the model is continuously refined and as new experimental data become available. Here we report the use of SciUnit, a software framework for model validation, to attempt to achieve these goals. During project development, each model is continuously subjected to data-driven ‘unit tests’ that quantitatively summarize model-data agreement, identifying modelling progress and highlighting particular aspects of each model that fail to adequately reproduce known features of the biological organism and its components. This workflow is publicly visible via both GitHub and a web application and accepts community contributions to ensure that modelling goals are transparent and well-informed.
This article is part of a discussion meeting issue ‘Connectome to behaviour: modelling C. elegans at cellular resolution’.
Keywords: modelling, informatics, unit-testing, Python
1. Introduction
The OpenWorm Project (http://openworm.org) is an initiative to develop a multi-scale model of the roundworm Caenorhabditis elegans with contributions from more than 80 individuals working in over a dozen countries.
The computer code defining the model is spread across sub-projects that each defines specific aspects of the organism, such as cellular ion channels or motor behaviours. Each of these spatial and conceptual scales is typically modelled independently, with multi-scale integration being a long-term goal of the project. These scales vary in spatial extent (nanometre to millimetre) and in level of abstraction (tracking of ion concentrations versus body wall dynamics). Each sub-project is organized as a public repository on GitHub, a web-based software development platform that serves as the de facto location for collaborative, open-source software projects, including those defining biological models. Each sub-project also has distinct success criteria, which all have in common a goal of reproducing empirical stylized facts associated with a biological subsystem using biologically inspired dynamical models.
(a). Unit tests
In practice, formalizing these success criteria requires producing a checklist of experimental observations and then assessing how well a single model can reproduce them when simulated with the appropriate parameters and stimulus conditions. Here we implicitly assume that the ability to reproduce experimental results is an indicator of model fitness, and that models which reproduce more such results are, ceteris paribus, superior to those that reproduce fewer. This is analogous to the ubiquitous practice of unit-testing in software development, where the progress and status of a piece of software is determined by how many unit tests are passed. These unit tests are mini-programs that report whether the larger piece of software successfully implements a particular operation or functionality [1].
For example, a program aiming to list prime numbers between user defined values a and b, and to throw an appropriate exception when a or b has an unexpected value, might be assessed using a suite of unit tests. Some of these tests would check the response of the software to different possible values of entries for variables a and b against a known list of true prime number values. Other tests might check for edge cases such as negative values of a, values where a > b, or non-numeric values for a and b, and verify that the program returns the correct list of primes (or none at all). When the program passes all of these tests (i.e. when each test confirms that program output is as expected), the program is considered to be valid and to provide a satisfactory solution for the task.
Unit-testing is widely considered to be essential for the development of any program of sufficient complexity. One strong argument in its favour is that formalizing and automating success criteria makes it easy to identify when any change to the program or its external dependencies causes one of those criteria to no longer be met—known as a ‘regression’ in the program. This enables developers to repair the offending sections of the program to make it meet the success criteria once again.
(b). SciUnit
The analogy to scientific modelling is clear and compelling: a successful scientific model should pass unit tests which encode specific empirical outcomes that investigators believe adequately characterize the experimental system. A successful model for classical mechanics, for example, should pass unit tests corresponding to the measured trajectories of fired projectiles, collisions of billiard balls and rotations of spinning tops. While the number of potential, reasonable unit tests might be vast, it may be sufficient to deploy a few suites of tests that together cover a broad number of decisive experimental results and that collectively summarize the state of empirical knowledge in the field.
SciUnit is a Python framework which realizes this analogy and enables the rapid construction and deployment of data-driven unit tests for scientific models [2]. In SciUnit, each type of scientific model is a class (in the programming language sense), where an instance of the class represents a particular realization of the same kind of model but with specific parameters. Sustaining the classical mechanics analogy, Newton's Laws could be encoded as a model class, and instances of that class might represent different values of parameters such the gravitational constant. Each type of test is also a class corresponding to a kind of empirical observation, and an instance of the class represents a particular value for that observation. In classical mechanics, one test class might generically encode the path of a projectile, and specific instances would encode specific projectile attributes and path observations. When a test instance judges a model instance, it determines whether the output of the model under those parameters adequately matches the observation. This judgement is represented as a test score, which can be qualitative (e.g. pass/fail) or quantitative (e.g. a Z-score of 1.26). The ensemble of test scores across all the tests in a suite summarizes the validity of one particular parameterization of a scientific model.
Importantly, SciUnit is designed so that testing does not require specific knowledge about how a particular model implements the generation of predictions (i.e. model output). In other words, tests do not need to be written to match the specific implementations of models. Instead, each test simply enumerates which capabilities (i.e. methods with specific signatures) a model is required to have, and then the model must implement those capabilities in whatever way it can. For example, a projectile path test might require models to implement a function that takes an initial velocity vector as input and returns a time series of projectile locations as output. How the model does so, internally, is irrelevant to the test. More details on this model/test interface and how it is realized are given elsewhere [2,3].
2. Material and methods
We describe partial testing of three OpenWorm sub-projects: ChannelWorm for cellular ion channels (http://github.com/openworm/ChannelWorm), c302 for neural circuits (http://github.com/openworm/c302), and Sibernetic for worm body motion (http://github.com/openworm/sibernetic). Testing involves installation of part or all of the corresponding sub-project and its dependencies, and is described in the documentation in the ‘test repository’ (http://github.com/openworm/tests).
(a). Tools
Python 3.6 is used for execution of all code except where described below. Simulations are controlled directly from tests, and use a variety of simulator backends (depending on the sub-project), including jNeuroML (https://github.com/NeuroML/jNeuroML) and NEURON [4] for ion channel and neuron simulations, and Sibernetic [5], for calculating the dynamics of the muscle fibres and the elastic body and interactions with the surrounding fluid. In order to test the robustness of the SciUnit framework, we deliberately re-run model tests where the model, observations and simulation protocols are held constant, but the simulator backend varies. Under these conditions, for example simulating with NEURON versus jNeuroML, test results do not depend on the choice of simulator.
SciUnit is a discipline-independent framework; however, an extensible library for a particular scientific discipline is needed to provide an interface to the simulators, data repositories and analysis tools specific to that discipline. NeuronUnit is one such SciUnit-driven library, developed for the investigation of ion channel, neuron and neural circuit models [6]. NeuronUnit was used to organize models and tests for ChannelWorm and c302. Through its interactions with the tools above, NeuronUnit generates simulator code for these models from their original simulator-independent representations in NeuroML [7,8], an extensible markup language for simulator-independent descriptions of neuroscience models. While SciUnit is agnostic to model representation, and NeuronUnit does not impose any hard model description requirements, several convenience functions are available in NeuronUnit for models described using NeuroML.
For worm movement models, Sibernetic was used to produce whole-worm movement trajectories due to muscle contractions driven by activity modelled using c302. In other words, the motor output was controlled by a neural circuit model acting on a musculoskeletal model, representing a multi-scale simulation. Output trajectories were encoded using Worm Commons Object Notation (WCON) files (https://github.com/openworm/tracker-commons), and movement features were extracted from these files using the Open Worm Analysis Toolbox (http://github.com/openworm/open-worm-analysis-toolbox). Corresponding features were obtained for experimental data from the Open Worm Movement Database (http://movement.openworm.org), which contains over 4 TB of C. elegans behavioural data, including videos, extracted movement trajectories and corresponding movement features.
(b). Reproducibility
Software described here is available in the test repository on GitHub at http://github.com/openworm/tests (OpenWorm specific tests and infrastructure) and at the SciDash website (http://www.scidash.org). Jupyter notebooks [9] (http://jupyter.org) reproducing all figures shown here are available in the test repository. Installation and execution of all code is continuously tested on Travis-CI (https://travis-ci.org), which verifies that all components of the test repository (and their dependencies) are running correctly. We also provide a Docker image in the test repository, which implements all tests in all sub-projects without the need for local installation of SciUnit, NEURON and the NeuroML tool chain. Analogous to an image on a virtual machine, a Docker image (http://www.docker.com) is a compartmentalized operating system which is frozen in a pre-configured state. Docker removes a large technical burden from the user by shifting package management complexity from users to developers [10]. Consequently, the Docker image is guaranteed to run and reproduce all results on any machine with no additional configuration.
3. Results
SciUnit is currently applied to three OpenWorm sub-projects: (i) ChannelWorm, for ion channel models in excitable cells; (ii) c302, for models of neurons, myocytes and their connectivity; and (iii) Sibernetic, for simulating the locomotor behaviour of the intact organism. Here we describe the infrastructure common to these projects and then describe tests and test performance in each project individually. It is important to note that this manuscript is not a demonstration of commendable performance on these tests, nor of exhaustive testing of each project, but rather of the existence and integration of this test infrastructure into the project. We expect that new tests will be added, and performance on all tests will improve as each model continues to develop, now that quantitative project goals, represented by test scores, are being made concrete.
(a). Common infrastructure
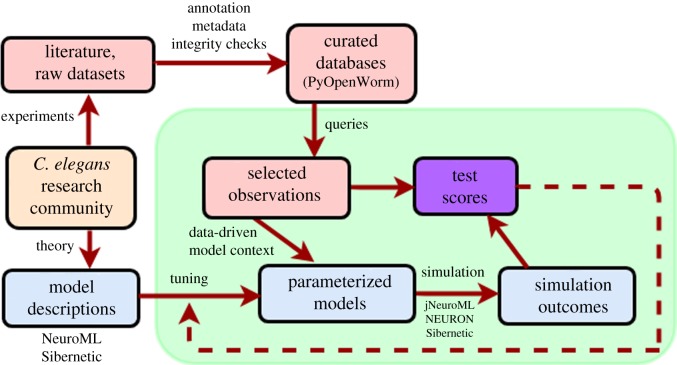
The testing workflow is schematized in figure 1. The research community (much of which is actively involved in the OpenWorm project) produces both models and datasets. A great deal of standardization, annotation and integrity checking is performed by OpenWorm contributors via both manual curation and automated tools, yielding curated databases of empirical observations (from experiments on the biological worm) and structured, machine-readable models at several scales. Summaries of observations are extracted from these databases, representing stylized facts about experiments. Models are simulated to produce corresponding predictions, and these predictions are compared with observations to yield test scores. Models may then be tuned based on the adequacy of these scores in order to produce better-performing models.
Figure 1.
Testing overview. The C. elegans research community, including members of OpenWorm, produce models using standards such as NeuroML and optionally tune their parameters. The community also produces abundant experimental data, which OpenWorm has taken the lead in annotating and curating into easily accessible databases. The testing procedure (green box) invokes SciUnit and related packages to query these databases for observations that parameterize tests, to simulate these models and judge their output according to its correspondence with these empirical observations, and to generate test scores summarizing this correspondence.
(i). Testing
All common infrastructure for OpenWorm model validation (at any scale) is handled in the test repository (see 2). Installation of this repository's package makes testing and examining test scores for specific OpenWorm sub-projects possible. This common infrastructure includes SciUnit, NeuronUnit, dozens of others software tools and several other OpenWorm sub-projects. All project tests can be executed and visualized either by installing the testing package and running the main script or, to guarantee platform independence and avoid configuration hassles, by launching the provided Docker container. Subsets of projects can also be tested with the appropriate configuration options. Testing is realized using three layers as shown in figure 2.
Figure 2.
Layers of testing for the OpenWorm Project. In the first, outer-most layer, the Python unittest module is invoked from the command line or a continuous integration script, where it can run elements in the second, middle layer. This layer consists of simple blocks of code that can test models, or any project components more generally, but also can run Jupyter notebooks that contain the configuration and execution of most of the SciUnit tests. The notebooks can be explored locally or interactively or simply viewed remotely on GitHub. SciUnit test execution represents the third, inner-most layer and relies on code in the sciunit package or a variety of SciUnit-based packages such as neuronunit. Test metadata and test results, including scores, optionally can be uploaded to http://dash.scidash.org to add to a public record of the history of model performance on the tests.
(1) The outer layer invokes Python's unittest module from the console to run all tests on demand or whenever changes to the underlying models are made.
(2) The middle layer consists of either simple code blocks (less common) or graphical Jupyter notebooks (more common). Jupyter has emerged as the premier approach to producing reproducible, interactive workflows, and supports embedded graphical elements such as figures, tables and HTML [9]. Through the core Jupyter technology of notebooks that can be edited, executed and viewed in the browser, interested parties can examine the test workflow and any artefacts (e.g. membrane potential traces) it produces without diving deep into the code. Jupyter notebooks can also be executed, with possible exploratory modifications in the browser, independently from the outer layer. These notebooks can provide graphical insights into the causes of successful or failed tests, as well as a reproduction workflow for a single test or test suite that can be shared independently of the outer layer. This middle layer involves querying databases for experimental observations, loading or downloading model files and importing libraries needed for test execution and visualization, such as SciUnit and NeuronUnit. Any of these tasks can be performed using simple code blocks, but is enhanced when using Jupyter due to its support for inline HTML and javascript outputs that can produce, for example, sortable tables of test scores. A grand summary of test results is displayed at the end, and an output file is generated containing these results as well as metadata about the current software environment. This information can also be optionally uploaded back to the test repository or to a test tracking dashboard (3aii, see below).
(3) The inner layer is the execution of the SciUnit test. All such tests use SciUnit, and, depending on the sub-project being tested, they may also use NeuronUnit, MuscleUnit, MovementUnit, or some other auxiliary SciUnit-based helper library. From the perspective of the middle layer, they are all invoked with a simple method invocation on a test or test suite like suite.judge(models), which subjects a list of models to a suite of tests. Most of the runtime complexity lies in this step, as it may require that large simulations be executed. A subset of the tools used here provide features to accelerate these simulations, such as parallelization in NeuronUnit or GPU-support in Sibernetic.
(ii). SciDash test dashboard
The SciUnit developers provide a web-based test dashboard (http://dash.scidash.org) to which SciUnit test scores can be uploaded and then searched, filtered, sorted, and visualized. By filtering for OpenWorm related tests, this dashboard can be used as a master project progress tracker, allowing the community to evaluate progress towards a model of C. elegans that is concordant with experimental data (figure 3). In some cases, satisfactory agreement between model output and experimental data may be measured using least square error comparisons of waveforms. In other cases, such as tests based on higher-order features like interspike interval histograms, the mean and standard deviation of interspike intervals are compared across model and experiment. Selection of appropriate tests is an important step in the validation process.
Figure 3.
The SciDash web dashboard for test scores. Each time a test is run, the results are optionally programatically uploaded to http://dash.scidash.org, where they can be viewed. In this example, the scores shown represent Z-Scores, the normalized deviation between the model output and the experimental data distribution. Test scores are colour coded from green (good) to red (bad), to visualize comparison of models on a given test (but not necessarily across tests). Additional metadata (not shown) is also viewable by clicking on links associated with a score's entry. This dashboard can be filtered, sorted and searched to allow anyone to check the progress of the various models, or to compare different versions of the model on their test performance.
(b). Ion channels
Several ion channel models have been developed that aim to mimic channels expressed in C. elegans neurons. These channel models are based on known physics of ion channels, with voltage-gated conductances modulated by the states of multiple gating state variables. Channel dynamics can be probed in a number of ways, including measuring kinetics in response to step changes in trans-membrane voltage or measuring steady-state currents at multiple voltages (IV curves). Experimental data for the corresponding biological channels can be found in a variety of journal articles, and many such data are curated in a database associated with the ChannelWorm repository. This database admits easy programmatic access to its contents via a Python API. Consequently, ion channel tests can be constructed by programatically extracting channel data corresponding to a given experiment from this database, and then using the data to parameterize the test. Each test can subject a model for the corresponding ion channel to the same experiments, extract model output (the prediction) and compare it to the experimental observation drawn from the database. An example of an IV Curve test for the EGL-19 ion channel is shown in figure 4.
Figure 4.
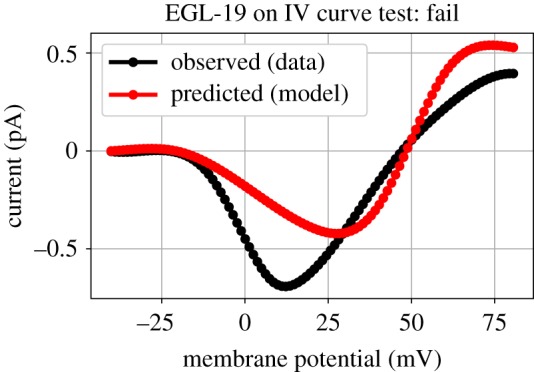
Data-driven validation testing of a C. elegans ion channel model: ChannelWorm. The black and red current-voltage relationships (IV curves) were produced by the real and simulated worm EGL-19 ion channels, respectively. The model is judged to fail this test due to the deviation between the two. This output is also displayed in the corresponding Jupyter notebook for this test.
While many tests could be made, here we show the results for one test based on IV curve measurements for the biological channel reported in [11]. With the corresponding model simulated using the same parameters as in the experiment, the agreement between model and data IV curves can be assessed. In this test, goodness-of-fit is assessed using the sum-squared-error between the curves, with lower numbers representing better fit. Two values are reported: first, the raw goodness-of-fit; and second, a pass/fail Boolean score (the default SciUnit score type) indicating whether this value is above a goodness-of-fit threshold. While this threshold is arbitrary, for practical purposes a particular goodness-of-fit may be considered ‘good enough’, and so model variants which all have values on the ‘passing’ side of the threshold might be considered equally good.
This example represents the performance of one parameterization of the EGL-19 channel model; alternatively, one may wish to retain exact goodness-of-fit information as part of an explicit model optimization routine. Optimization of the model parameters using this test (i.e. changing parameters until the model and data IV curves were very similar) could produce a version of the EGL-19 model that more closely reproduces experimental data collected from the biological channel. NeuronUnit supports this kind of optimization [12]; however, it may be better to develop many more tests (based on other measurements from the EGL-19 ion channel) first, and then execute a multi-objective optimization of this test ensemble. NeuronUnit also supports this kind of multi-objective optimization [13], yielding a manifold of parameter sets, but this kind of optimization has not yet been performed for the ChannelWorm project. At press time, the project has designated ChannelWorm to be the OpenWorm ‘Project of the Month’ for August 2018, where an effort to perform these optimizations and update ten such ion channel models is planned.
(c). Neurons and neural circuits
c302 is a sub-project of OpenWorm, so named because C. elegans has 302 neurons, that aims to model these neurons and their synaptic connectivity. Here we show how one model in this sub-project is tested. This model contains one modulating or command motor neuron (AVAL) and a synaptically connected myocyte (body wall muscle cell, MDR01). The model is simulated in a fashion similar to the ion channel model in the previous section, although the repertoire of simulation parameters is far greater. Several groups of simulation parameters (each group based on specific experimental conditions or assumptions about the system, and representing one point in ‘parameter space’) are available for use. Here we show several tests applied to the model under one of these simulation parameter groups (figure 5).
Figure 5.
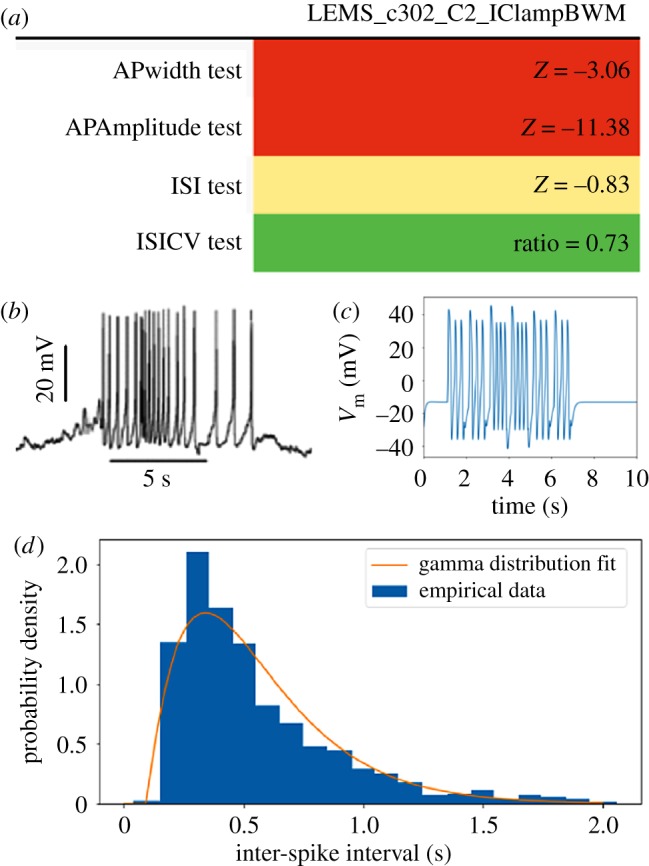
Data-driven validation testing of a C. elegans muscle cell and the motor neuron that drives it: c302. (a) Results on a suite of four tests, two that compare the distributions of inter-spike-intervals (ISIs) and two that compare the shapes of the action potentials. This exact output is also displayed in the corresponding Jupyter notebook for this test suite. (b) A segment of the experimental membrane potential traces. (c) A segment of the simulated model membrane potential trace. (d) Normalized ISI histogram observed in the experiment and a fit to a theoretical gamma distribution. The test computes and uses parameters from such a fit to assess agreement to a similarly fit model ISI histogram.
In contrast with the ion channel model in the previous section where only one test was made, here we apply a suite of several tests reflecting a variety of measurements of the experimental system. Each of these measurements is associated with the membrane potential dynamics of the myocyte under the selected conditions. Many of these reflect spiking behaviour, including firing patterns and action potential waveforms. Corresponding tests of the motor neuron are also possible, but since that neuron is non-spiking, these would need to reflect sub-threshold dynamics. The tests are parameterized using electrophysiological data from body-wall muscle cells reported by Liu et al. [14].
In figure 5, we show the performance of the model on this test suite. In contrast with the previous section, here scores are reported as Z-Scores (another built-in SciUnit score type). These Z-Scores can be interpreted as a measurement of model output (e.g. mean action potential width) normalized to the mean and standard deviation of the corresponding experimental measurement. This allows the results of tests corresponding to heterogeneous measurement types with a variety of units to be compared on a single principle: how well do they conform to the empirical data distribution?
These Z-Scores show that the inter-spike-interval (ISI) distributions in the model are within the corresponding empirical data distribution, but the shapes of the action potentials are not. This shortcoming is probably due to the use of a minimal complement of ion channels in the model. Improvements to the ion channel models and an emphasis on including channels that can produce the rich dynamics observed during the action potential may lead to improved action potential shape in future versions of the model.
(d). Locomotor behaviour
Sibernetic is a sub-project of OpenWorm that aims to model the movement of C. elegans via detailed fluid mechanics simulations. Among its outputs are time series corresponding to every position on the external surface of the worm's body. These can be reduced to the two-dimensional coordinates (as viewed dorsally) of dozens or hundreds of body segments, summarizing moment-to-moment changes in body position and orientation over time. When coupled with simulations from c302 (used to determine the activity of the muscles that drive the body), a multi-scale model can be realized that could in principle produce realistic worm-driven motor activity.
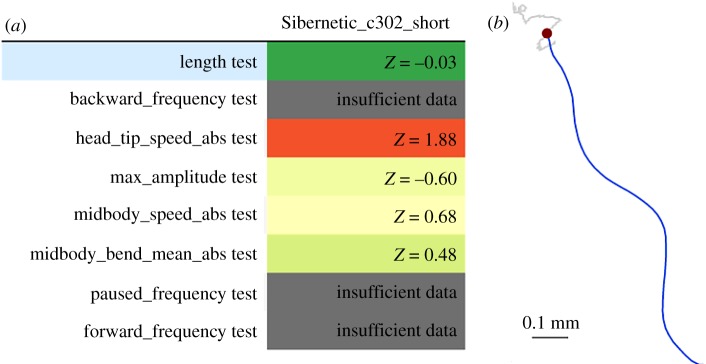
In order to determine if this simulated motor activity corresponds to that observed in the real organism, many thousands of video recordings of swimming C. elegans have been curated into a large database (greater than 4 TB) available at http://movement.openworm.org, and corresponding time series of body segment positions have been extracted. Sibernetic model validity is assessed by comparing features of those time series to corresponding features in simulation output. The OpenWorm movement validation team identified ten ‘core” features that provide especially important information about real or simulated worm posture and movement. We extracted eight of these features from a representative Sibernetic model simulation and compared them to distributions of the same features extracted from the video recordings and curated in the database (figure 6).
Figure 6.
Data-driven validation testing of a C. elegans motor behaviour model: Sibernetic. (a) Scores for each of eight tests representing core movement and posture features are shown. In some cases, test results are unavailable because the corresponding features cannot be computed for simulation output in the current state of the model. In all other cases, scores indicate that model output is within two standard deviations of the experimental data distribution. (b) Graphical depiction of the contents of a WCON file constructed from segmentation of a video of C. elegans locomotor behaviour. One frame is shown, with the body orientation in blue, the head location in red and head locations from previous frames shown in grey.
For some of these features, functions for feature extraction on the model do not yet exist, and so there is insufficient data to make a comparison with experimental data. However, other model output features were not substantially different from those computed based on experiments on real worms.
4. Discussion
Here we demonstrate the beginnings of data-driven validation testing at three model scales for a large, collaborative effort to model C. elegans. The results of such tests inform current modelling efforts by pointing developers to model deficiencies, as judged by discordance with observations from laboratory experiments on C. elegans. While agreement with empirical studies is an end goal of The OpenWorm Project, this report is not meant to demonstrate that this goal has been achieved, or that it is close at hand. Rather it is meant to report that the infrastructure for assessing model validity has been developed, that these assessments are on-going, and that model refinement towards a realistic simulation of C. elegans now has a clear task list: achieving passing scores on several suites of SciUnit tests. Future model development will proceed accordingly.
(a). OpenWorm as an experiment in test-driven model development
As far as we are aware, OpenWorm is the largest (and perhaps only) open source, community-driven, biological modelling project that has begin to utilize such a test-driven infrastructure. The completely open nature of the project facilitates such test integration, for at least four reasons. First, the OpenWorm model development cycle is one of continuous public delivery, rather than infrequent public versions released after internal, private development. This means that regressions—changes to models that might lead to decreased model fitness—are likely to be both common and publicly visible. Identifying these regressions by means of continuous validation tests can help direct modelling effort towards repairing them. Second, OpenWorm consists of several largely separate modelling projects produced by independent teams which do not have the same level of design integration as they might in, say, a single research lab. When these separate models are linked (e.g. when neural circuit models drive behaviour models), validation tests provide the only measure of successful integration. Third, all modelling tools and validation datasets are based on open source components, so anyone from inside or outside the project can add and execute validation tests without the need to access proprietary code. Fourth, OpenWorm does not have any public relations requirements that would discourage the public advertisement of test failures during development. Other large projects that currently receive substantial funding (and media scrutiny) might be reluctant to be so transparent with provisional test failures, even though they are a natural part of any modelling project.
The decision to model the nervous system of C. elegans, as opposed to that of a more complex organism (e.g. a mammal), may also have resulted in synergies with test-driven development. Caenorhabditis elegans offers experimental tractability, a superior understanding (relative to other organisms) of its wiring diagram, and the ability to link stereotyped anatomy and physiology to a set of simple behaviours. These features should lead to both multi-scale models with relatively few free parameters and rich, decisive, reproducible experimental datasets that can—through validation testing—adjudicate between competing realizations of such models.
(b). Model failures
Poor performance on SciUnit tests can result from misparameterized models, from models that lack important components of the corresponding experimental system, from incomplete metadata leading to context-less simulations, or from summary statistics that fail to adequately capture the essence of the observation. Several of these factors are likely in play in some of the model failures shown here. For example, the EGL-19 ion channel model is probably misparameterized, whereas the c302 myocyte model lacks ‘realistic looking’ action potentials due to insufficient model complexity; in particular, it was not built from a complement of validated ion channel models.
(c). Perils of test-driven development
Are there any drawbacks to using test scores to drive development of scientific models? There is a risk in overemphasizing small test score improvements at the expense of conceptual simplicity. This is one reason why we have avoided blindly optimizing models in advance, instead letting domain experts work on models first, and only then assessing (and hopefully improving) their performance on a suite of validation tests. Other features like computational complexity, cyclomatic complexity of code, memory consumption, etc. could also be incorporated into the objective function to privilege simplicity against over-fitting. None of these have currently been implemented. Another concern is that models might be engineered to pass tests without concern for the realism of the underlying components. This can be avoided by ensuring that models are based upon known physical laws—so-called ab initio modelling—and by ultimately building models hierarchically from validated lower-level components, as discussed in the next section.
(d). Hierarchical model validation
In a multi-scale model, high-level outputs such as behaviour may be under the control of lower-level model components such as neural circuits. One limitation of testing these high-level outputs is that, when they do not match corresponding experimental observations, it is difficult to know for certain which system in the multi-scale hierarchy has been modelled incorrectly, or whether the coupling between models is implemented incorrectly (see below). Although all successfully tested C. elegans locomotor model features were within two standard deviations of the corresponding experimental data distribution, it is not clear how to change the model components to further improve the correspondence. Should the next step be to model the neural activity more accurately, or should the hydrodynamics simulation be further improved? We believe that improving test scores for Sibernetic worm movement behaviour output will require the former rather than the latter, in part because the physiological data to constrain the neural activity is incomplete, whereas the physics to implement the hydrodynamics simulation is well-understood.
In any case, if the multi-scale model is constructed hierarchically, such that the higher-level model components depend directly on the output of the lower-level components (i.e. ion channel activity determines neuron activity and neuron activity determines motor output), then optimizing the lower-level components will help reduce test uncertainty in higher-level components. In other words, ion channel models which pass all tests can be used to construct neuron models which have a chance to pass all tests; similarly, neural circuit models which pass all tests can be used to drive motor behaviour models which have a chance to pass all tests.
Coupling scales or domains (e.g. inserting validated ion channels into neurons, or using neural activity to drive muscle contraction) is itself also modelling; however, this is not yet tested in the same way as the models being coupled. Instead, decisions about coupling implementation have been based either on qualitative knowledge about C. elegans anatomy and physiology, such as which channels have been identified in which cells. Technically, we could construct SciUnit tests that validate these coupling implementations. For example, we could construct a test to see if direct stimulation of a motor neuron produced the expected pattern of body wall movement. This would require a corresponding biological dataset to parameterize the test, for example, from an experiment in which that neuron is under optogenetic control.
5. Conclusion
Inverting the usual workflow of computational modelling—using test scores to steer model development rather than simply assessing the entire model post hoc—constitutes a paradigm shift. Continuous, automated validation has the potential to accelerate the successful development of large, collaborative, multi-scale models of biological systems. The Royal Society's motto Nullius in verba – ‘take nobody's word for it’ can be realized only when the validity of each model component can be validated transparently against publicly available experimental data. Applying SciUnit to the OpenWorm Project allows model developers to assess their progress and identify key areas where models can be improved. As SciUnit testing of the project expands, it will become possible to assess the epistemic state of each subproject—and the project as a whole—at a glance, regardless of its complexity.
Acknowledgements
The authors wish to thank The Royal Society for supporting the presentation of these results, The Open Worm Project members for creating models, curating the data and writing analysis code upon which this work was built, and MetaCell, LLC for technical support on the SciDash website.
Data accessibility
Instructions for reproducing the content described here are available at http://github.com/openworm/tests as described in §2b.
Author's contributions
All authors conceived of and designed this study. R.C.G. and R.J.J. wrote the software. R.C.G. drafted the manuscript. All authors read, edited and approved the manuscript. S.M.C. presented an initial version of this work at The Royal Society meeting Connectome to behaviour: modelling C. elegans at cellular resolution.
Competing interests
The authors declare that they have no competing interests.
Funding
This research was funded in part by R01MH106674 from NIMH of the National Institutes of Health and R01EB021711 from NIBIB of the National Institutes of Health.
References
- 1.Beck K. 2003. Test-driven development: by example. Boston, MA: Addison-Wesley Professional. [Google Scholar]
- 2.Omar C, Aldrich J, Gerkin RC. 2014. Collaborative Infrastructure for Test-driven Scientific Model Validation. In Companion Proc. of the 36th International Conf. on Software Engineering, ICSE Companion 2014, pp. 524–527, New York, NY, USA: ACM; http://doi.acm.org/10.1145/2591062.2591129. [Google Scholar]
- 3.Sarma GP, Jacobs TW, Watts MD, Ghayoomie SV, Larson SD, Gerkin RC. 2016. Unit testing, model validation, and biological simulation. F1000Research 5, 1946 ( 10.12688/f1000research.9315.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hines ML, Carnevale NT. 1997. The NEURON simulation environment. Neural Comput. 9, 1179–1209. [DOI] [PubMed] [Google Scholar]
- 5.Yu Palyanov A, Khayrulin SS. 2015. Sibernetic: a software complex based on the PCI SPH algorithm aimed at simulation problems in biomechanics. Russ. J. Genet Appl. Res. 5, 635–641. ( 10.1134/S2079059715060052) [DOI] [Google Scholar]
- 6.Gerkin RC, Omar C. 2013. NeuroUnit: validation tests for neuroscience models. Front. Neuroinf. 7 ( 10.3389/conf.fninf.2013.09.00013) [DOI] [Google Scholar]
- 7.Gleeson P. et al. 2010. NeuroML: a language for describing data driven models of neurons and networks with a high degree of biological detail. PLoS Comput. Biol. 6, e1000815 ( 10.1371/journal.pcbi.1000815) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gleeson P, Crook S, Silver A, Cannon R. 2011. Development of NeuroML version 2.0: greater extensibility, support for abstract neuronal models and interaction with Systems Biology languages. BMC Neurosci. 12, P29 ( 10.1186/1471-2202-12-S1-P290) [DOI] [Google Scholar]
- 9.Kluyver T. et al. 2016. Jupyter Notebooks - a publishing format for reproducible computational workflows. In Positioning and Power in Academic Publishing: Players, Agents and Agendas (eds Loizides F, Schmidt B), pp. 87–90. IOS Press. [Google Scholar]
- 10.Merkel D. 2014. Docker: lightweight Linux containers for consistent development and deployment. Linux J. 2014, 2. [Google Scholar]
- 11.Jospin M, Jacquemond V, Mariol MC, Segalat L, Allard B. 2002. The L-type voltage-dependent Ca2+ channel EGL-19 controls body wall muscle function in Caenorhabditis elegans. J. Cell Biol. 159, 337-–348. ( 10.1083/jcb.200203055) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jarvis RJ, Crook SM, Gerkin RC. 2017. Optimization of Reduced Models against Diverse Experimental Neuron Physiology Datasets with NeuronUnit, CRCNS PI Meeting, Brown University, 2017. [Google Scholar]
- 13.Jarvis RJ, Crook SM, Gerkin RC. 2017. Parallel Model Optimization against Experimental Neuron Physiology Data with DEAP and NeuronUnit. [Google Scholar]
- 14.Liu P, Ge Q, Chen B, Salkoff L, Kotlikoff MI, Wang ZW. 2011. Genetic dissection of ion currents underlying all-or-none action potentials in C. elegans body-wall muscle cells. J. Physiol. 589, 101–117. ( 10.1113/jphysiol.2010.200683) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Instructions for reproducing the content described here are available at http://github.com/openworm/tests as described in §2b.