Abstract
The successful implementation of synthetic biology for chemicals biosynthesis relies on the availability of large libraries of well-characterized enzymatic building blocks. Here we present a scalable pipeline that applies the methodology of synthetic biology itself to bootstrap the creation of such a library. By designing and building a cytochrome P450 enzyme collection and testing it in a custom-made untargeted GC/MS-metabolomics-based approach, we were able to rapidly create and characterize a comprehensive enzyme library for the controlled oxyfunctionalisation of terpene scaffolds with a wide range of activities and selectivities towards several monoterpenes. This novel resource can now be used to access the extensive chemical diversity of terpenoids by pathway engineering and the assembly of biocatalytic cascades to subsequently produce libraries of oxygenated terpenoids and their derivatives for diverse applications, including drug discovery.
Introduction
Biosynthetic expansion of the chemical diversity of terpenes is a major challenge for biocatalysis and synthetic biology programmes in order to access new chemical space. Establishing a biocatalyst toolkit that performs site-selective oxyfunctionalisation of monoterpene hydrocarbons is required to make added-value products or to produce intermediates for subsequent tailoring by other enzymes. Oxyfunctionalisation of terpene scaffolds based on cytochrome P450 (CYP) dependent catalysis has been proposed as a route for expansion of chemical diversity, enabling the biosynthesis of high value-added end products1,2. CYPs offer several advantages as biocatalysts. They can, for example, oxidise inert C-H bonds and they have a broad substrate scope, regio- and stereo-selectivities3,4. Some CYPs were reported to be capable of oxygenating a few monoterpene scaffolds2,5,6. This foundational information was the starting point to develop a comprehensive CYP toolbox for the enzymatic tailoring of monoterpenes. Our CYP toolkit was carefully designed to overcome limiting factors (low protein solubility, cofactor incorporation, co-expression of redox partners) typically associated with CYPs7.
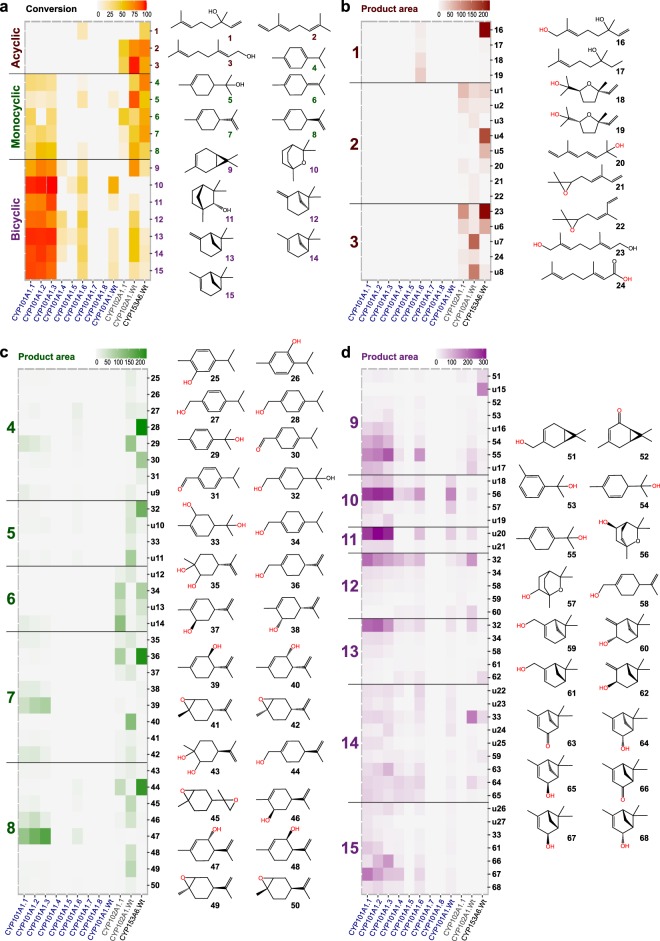
Reactivities and selectivities for each member in our CYP toolbox were assayed across 15 different substrates representative of acyclic ((+/−)-linalool, (E/Z)-β-ocimene, geraniol; 1–3), monocyclic (γ-terpinene, α-terpienol, terpinolene, (R)- and (S)-limonene; 4–8) and bicyclic ((+)-carene, 1,8-cineole, (+)-fenchol, (−)- and (+)-β-pinene, (−)- and (+)-α-pinene; 9–15) monoterpene scaffolds (Fig. 1). All enzymes were tested using a cell-free extract in the presence of high substrate concentrations and a glucose dehydrogenase NADH/NADPH recycling system to ensure multiple turnover conditions (Methods Section). The majority of CYPs selected for our toolbox are reported to have differential degrees of promiscuity and relaxed substrate specificity, which prevents anticipating product formation upon oxyfunctionalisation. Thus, a non-directed GC/MS-based metabolomics approach was used to investigate the chemical nature and regioselectivity of the CYP oxyfunctionalisation reactions (Methods Section). Our non-directed GC/MS screening detected a wide variety of oxyfunctionalised products. (Supplementary Tables S1–3 from which up to 70% could be conveniently identified (Supplementary Tables S4–5).
Figure 1.
Untargeted GC/MS screening of the CYPs library. (a) Conversion of monoterpene substrates by the CYP library. Acyclic (brown), monocyclic (green) and bicyclic monoterpenes (lilac). (b–d) Product landscapes from biotransformation of acyclic, monocyclic and bicyclic substrates, respectively. Product conversions shown are calculated as described in the methods section. Structures of identified products are shown. Mass spectra of unidentified products are described in the online Supplementary Table S16.
The activity of the CYP toolbox is summarized in Fig. 1. Conversion of the 15 different monoterpene substrates (1–15) studied is displayed in Fig. 1a and Supplementary Fig. S1–15. Figure 1b–d summarises the product landscape upon oxyfunctionalisation, including identified (16–68) and non-identified products (u1–u27). The entire list of product names can be found in the online Supplementary Tables S1–15. Our toolbox showed a remarkable range of activities against different monoterpene scaffolds with the best toolbox catalysts showing a wide range of product profiles and differential substrate selectivity: CYP101A1.1-3 (4, 6–8 and 9–15), CYP102A1.Wt (2–9 and 12–14) and CYP153A6.Wt (1–8). This emphasises the capacity of these enzymes to form a comprehensive toolbox to widen monoterpenoid chemical diversity through oxyfunctionalisation. The substrate selectivity of the toolbox members can be rationalised by structural correlations between monoterpene scaffolds and the natural substrates of these CYPs: D-camphor (CYP101A1.Wt), fatty acids (CYP102A1.Wt) and n-alkanes (CYP153A6.Wt).
Initial studies focused on assessing the functional expression and activity of several bacterial CYPs, including: Pseudomonas putida P450cam (CYP101A1), Pseudomonas sp. P450terp (CYP108), Bacillus megaterium P450BM3 (CYP102A1), Bacillus licheniformis CYP102A7 and Mycobacterium HXN-1500. CYP153A6. CYP101A1 and CYP108 systems comprise the cytochrome P450 enzyme and two redox partners (redoxin and redoxin reductase). To investigate cofactor incorporation these components were expressed separately and purified. CYP108 was eliminated at this stage, as the redoxin 2S-2Fe cofactor was not incorporated. CYP101A1.Wt was reconstituted in vitro from purified proteins and was active with D-camphor as substrate. Functional expression of the CYP101A1 system in E. coli was achieved by constructing a polycistronic vector based on the JBEI-6411 backbone2,8 and previous studies9,10. Reconstituted CYP102A1.Wt and CYP102A7.Wt were active using substrate (S)-limonene 8 but CYP102A7 was subsequently excluded because of poor substrate conversion and functional stability11.
The CYP toolbox was diversified by addition of active site variants of CYP102A1.Wt and CYP101A1.Wt (see Table 1). The CYP102A1.1 triple variant (A264V/A328V/L437F) was selected from a small library reported previously6 as it can convert (R)-limonene (7) to (R)-perillyl alcohol 36. CYP101A1 variants were designed based on structural information and previous engineering studies5. These variants combine different mutations (see Table 1) at four active site residues (Tyr96, Phe87, Leu244 and Val247). The Y96F mutation was included in all variants to better accommodate non-polar monoterpenes in the active site. Residues close to the heme cofactor (Leu244 and Val247), or on top of the binding pocket (Phe87), were altered with the aim of modifying reaction regioselectivity and substrate binding. Eight CYP101A1 variants were constructed. Among those incorporating the L244A mutation (CYP101A.4–8) only CYP101A1.6 showed enhanced conversion of monoterpenes compared to CYP101A1.Wt. The CYP101A1.1-3 variants showed high monoterpene conversion. The above variants showed small product regioselectivity differences, revealed by the ratio of (−)-cis-carveol (46) vs (−)-trans-isopiperitenol (47) or of (+)-trans-verbenol (67) vs (+)-myrtenol (61) after conversion of (S)-limonene (8) and (+)-α-pinene (15), respectively. Therefore, the diversity of oxyfunctionalised products achieved by the CYP toolbox relies mainly on the differential regioselectivity between CYP classes rather than through protein engineering within a specific class.
Table 1.
CYP450 samples and variants employed in this study.
| CYP450 family | Mutation | Name |
|---|---|---|
| CYP101A1 | — | CYP101A1.Wt |
| Y96F | CYP101A1.1 | |
| Y96F, V247L | CYP101A1.2 | |
| F87W, Y96F, V247L | CYP101A1.3 | |
| Y96F, L244A | CYP101A1.4 | |
| F87W, Y96F, L244A | CYP101A1.5 | |
| Y96F, L244A, V247A | CYP101A1.6 | |
| Y96F, L244A, V247F | CYP101A1.7 | |
| Y96F, L244A, V247L | CYP101A1.8 | |
| CYP102A1 | — | CYP102A1.Wt |
| A264V, A328V, L437F | CYP102A1.1 | |
| CYP153A6 | — | CYP153A6Wt |
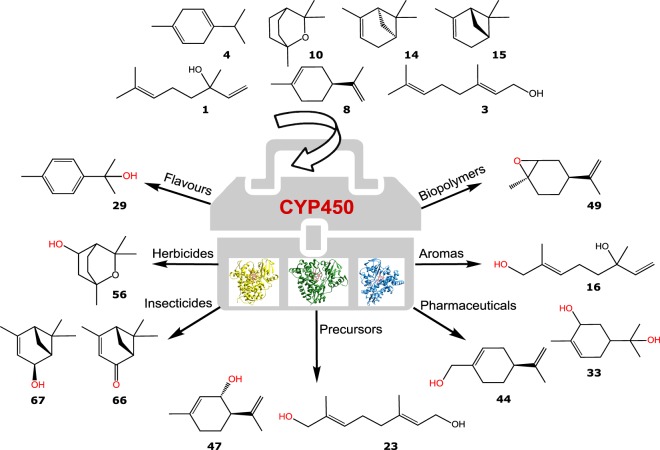
The observed products from monoterpene oxyfunctionalisation catalysed by the CYP toolbox include a variety of known pharmaceuticals, precursors, herbicides, flavours and aromas (Fig. 2). For example, we observed the production of sobrerol 33, a known clinical expectorant and potential anti-cancer drug12. This product was obtained by biotransformations of CYP101A1 variants and CYP102A1.Wt with two structurally different scaffolds, namely (−)-α-pinene 14 and α-terpineol 5, with the combination of CYP102A1.Wt and 14 being the best sobrerol 33 producing system. The reactions are thought to proceed via a conventional CYP-like monooxygenation mechanism, however the reaction with (−)-α-pinene likely includes the formation of an epoxide intermediate, followed by non-enzymatic water-mediated decomposition (Fig. 3)13.
Figure 2.
Key products formed by the CYP toolbox and their applications. Chemical structures from substrate scaffolds and products are shown. Miniatures of crystal structures from CYP101A1.Wt (blue) CYP102A1.Wt (yellow) and CYP153A7.Wt (similar sequence to CYP153A6.Wt, green) are shown (from PDB entries: 3wrj, 1bvy and 3rwl, respectively).
Figure 3.
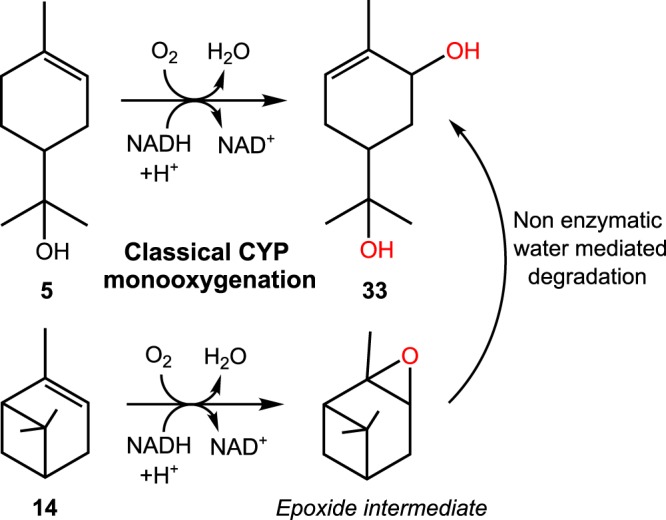
Putative routes of the CYP102A1.Wt-mediated conversion of α-terpineol 5 and (−)-α-pinene 14 to sobrerol 33. The route from 5 to 33 is a classical CYP-like monooxygenation mechanism. The two step process from 14 includes a non-enzymatic water-mediated decomposition stage.
Other pharmaceutical products obtained were (4R)- and (4S)-perillyl alcohol (36 and 44), which have known analgesic properties and a potential role in preventing Alzheimer’s disease14,15. The relaxed substrate enantioselectivity of CYP153A6.Wt was observed by the generation of the different enantiomeric products (36 and 44) during biotransformations with (R)- and (S)-limonene (7 and 8), respectively. The production in vivo of (4R)-perillyl alcohol 44 by metabolically engineering E. coli has been demonstrated previously via incorporation of a heterologous mevalonate pathway coupled with the M. spicata limonene synthase and CYP153A62,8.
Biotransformations of CYP101A1.3 with 14 generated relatively small amounts of (−)-verbenone 63 and (−)-trans-verbenol 64. These cytotoxic compounds are known to decrease the viability of either colon tumour cells or of normal cells, respectively, suggesting a potential role of 63 as a colon cancer drug16. Interestingly, reactions of CYP101A1.1 with the related isomer 15 generated high yields of (+)-verbenone 66 and (+)-trans-verbenol 67, in line with previous studies5,17. However, it is not known whether 66 has a similar anti-tumour effect as its isomer 63. These results are in line with studies using whole cells of Picea abies, where constitutive CYP enzyme(s) transformed racemic pinene into cis/trans- verbenol enantiomers (64/65, 67/68), with further transformations to 63 and 6618. The latter compound is a well-known pheromone used to protect pine trees from attack by the mountain pine beetle19.
Biotransformations of CYP102A1.Wt with (S)-8 generated trans-limonene-1,2-epoxide 49, which is useful commercially as a monomer for copolymerisation with CO2 to form biodegradable polycarbonate20. In addition, both cis-carveol 46 and cis-isopiperitenol 48 were found in relatively small amounts. This is analogous to previous studies of CYP102A1.Wt with (R)-7, which showed a mixture of (4R)-limonene-1,2-epoxide, (4R)-limonene-8,9-epoxide and carveol6,21. Studies have shown that some terpene-derived polymers based on 7 and 8 have shown excellent properties for optoelectronic applications22. Therefore this CYP toolbox approach could provide valuable routes to existing and de novo biodegradable polymers.
The oxidation of (S)-8 by CYP101A1.3 generated (−)-trans-isopiperitenol 47, as reported before17, an intermediate in the Mentha piperita (peppermint) pathway to menthol isomers. Recent synthetic biology approaches towards the in vitro biosynthesis of (−)-menthol by recombinant E. coli extracts have been successful starting from pathway intermediates 47 and (R)-pulegone23,24. Therefore, the inclusion of a CYP generating 47 from (S)-8 into existing recombinant pathways is a potentially important step forward towards complete microbial in vivo production of (−)-menthol from simple carbon sources25. 8-hydroxygeraniol 23 was also generated through the action of CYP153A6.Wt on geraniol 3, in line with the terminal oxidation of aliphatic hydrocarbons observed previously26,27. This compound is part of the indole alkaloids loganin and secologanin biosynthesis routes in Catharanthus roseus28. These alkaloids are important due to their known antibacterial and anti-imflammatory properties29. Differently, 2,3- and 6,7-epoxy-geraniols were found after conversion of geraniol 3 by other CYP102A1 variants studied before30.
A variety of other monoterpenoid derivatives generated from the CYP toolbox are known to have diverse potential commercial applications. For example, the biotransformation of (+/−)-linalool 1 by CYP153A6.Wt generated 8-hydroxylinalool 16, a known precursor of valuable fragrance additives such as lilac alcohols29. This activity was reported before for CYP111 as the first committed step for linalool 1 utilisation as sole carbon source in Pseudomonas incognita31. The oxidation of monoterpenoids γ-terpinene 4 generated p-cymen-8-ol 29, a flavouring agent which contributes to the off-flavors in lemon juice32. Other products have known phytotoxic properties, such as 3-exo-hydroxy-1,8-cineole 56, generated from the action of enzymes CYP101A1.1-3 on 1,8-cineole 10. The hydroxylation of 1,8-cineole 10 by CYPs introduces several chiral centres which makes difficult the identification of reaction products33–35.
Our CYP toolbox provides an important repository of biocatalytic parts for synthetic biology, metabolic engineering and biocatalysis projects where the expansion of chemical diversity of monoterpene hydrocarbon scaffolds is required. Several studies have reported the evaluation of different CYP libraries for oxyfunctionalisation of monoterpene substrates such as (R)-limonene (7) or (+)-α-pinene (15) using traditional targeted GC5 or GC/MS analysis6 developed on a case-by-case basis. However, such tailored methods do not allow one to conduct a comprehensive and systematic study of different CYPs with diverse substrates thus limiting the access to a broad CYP catalysts landscape. Here we present an advanced approach for broad profiling of enzyme activities36 and enzyme function discovery37, combining the power of biocatalysis and non-directed metabolomics. Metabolomics or the systematic investigation of the comprehensive profile of all metabolites in a biological substrate is particularly well suited for impartially monitoring metabolic transformations, defined as consumption of substrates and generation of products. Non-directed metabolomics is of particular interest when products derived from such biotransformations cannot be anticipated, as in the case of the CYP library studied here. However, non-directed GC-MS metabolomics experiments produce large volumes of spectral data requiring essential data processing steps to uncover biological findings enclosed in such raw data. In this regard, an automated open-data and open-source GG-MS data analysis workflow has been implemented for rapid unbiased comparison of chromatograms. This consists of five automated sequential steps: (i) peak detection at the total ion count level for each chromatogram; (ii) matching and aligning homologous peaks across chromatograms; (iii) peak integration and normalization to corresponding internal standard; (iv) assessment of both substrate conversion and product landscape formation; (v) extraction of consensus spectra and computation of retention indices for each peak; and finally (vi) compound identification which requires manual curation and intensive interpretation of the observed mass spectra. In conclusion we report, for the first time, a comprehensively characterised CYP toolkit for metabolic pathway engineering and cascade biocatalysis to produce value added terpenoid products and terpene-based small molecule libraries for diverse applications.
Methods
Cell cultures, chemicals and commercial enzymes
Culture media for E. coli was obtained from Formedium. Cells were routinely grown in Lysogenic Broth (LB) or on LB agar plates including antibiotics, which were supplemented as appropriate (ampicillin, 100 μg·mL−1; kanamycin, 50 μg·mL−1). Cloning and plasmid propagation was performed using E. coli DH5α (New England Biolabs, NEB). The following chemicals and enzymes were purchased from Sigma: linalool, geraniol, (E/Z)-β-ocimene, (R)- and (S)-limonene, γ-terpinene, terpinolene, α-terpineol, (+)- and (−)-α-pinene, (+)- and (−)-β-pinene, (+)-fenchol, sec-butyl-benzene, ethyl-acetate, and glucose dehydrogenase (Pseudomonas sp.). (+)-car-3-ene and 1,8-cineole were purchased from TCI Chemicals. Information about the authentic standards used can be found in Supplementary Table S17.
General molecular cloning
Cytochrome CYP153A6 and its redox partners encoded on the plasmid pJBEI-6411 (Addgene plasmid #47050) were a gift from Prof. Taek Soon Lee2. CYP450cam (CYP101A1, variant C334A, referred to as Wt in this manuscript) and its redox partners PdR and PdX (variant C73S/C85S) were synthesised, codon optimised for E. coli and cloned into pETM11 or pET21b expression vectors (GeneArt, Life Technologies). Self-sufficient CYP450BM3 (CYP102A1) encoded in pET15b was kindly donated by Prof. Andrew Munro (The University of Manchester). A plasmid carrying CYP101A1.Wt together with its redox partners, PdR and PdX, was constructed and organized in an operon under the control of a single PBAD promoter. Each gene was amplified via PCR from the corresponding expression vectors, while pJBEI-6411 was linearized by inverse PCR. The assembly of three genes/parts into one operon was performed using overlap-extension PCR (OE-PCR)38. Finally, pJBEI-6411 backbone and the fused OE-PCR products were assembled into one vector by In-Fusion® cloning (Takara). The same procedure was employed to assemble CYP102A1.Wt into the pJBEI-6411 backbone. CYP101A1 and CYP102A1 variants were prepared using QuikChange® site-directed mutagenesis (Stratagene). All constructs were confirmed by sequencing. All primers, PCR details, variants details, and pJBEI-6411-CYP101A1 map can be found in online Supplementary Methods.
Protein expression and characterization
Single colonies were used to inoculate 50 mL of LB medium supplemented with the corresponding antibiotic. Those overnight cultures were used as starting material to inoculate large volume media (500 mL). All CYPs were grown on TB media supplemented with 15 μM FeCl3 at 37 °C until the optical density at 600 nm (OD600) reached 0.8–1 at which point 500 μM δ-aminolevulinic and 50 mM arabinose were added to support protein expression. After induction, cells were grown for 20–24 hours at 25 °C (CYP101A1 and CYP102A1) or 20 °C (CYP153A6) and collected by centrifugation at 4 °C. Cell pellets were resuspended (200 mg·mL−1) in 50 mM potassium phosphate buffer pH 7, 200 mM NaCl and 10% glycerol. Cells were lysed using a constant cell disruptor system (Constant Systems Ltd, UK). Protease inhibitor tablets (Sigma) were added to the cell lysates to avoid enzymatic degradation of over-expressed proteins. Cell lysates were immediately used for biotransformation or kept frozen at −80 °C. Heme iron thiolate coordination was established by formation of the Fe(II)CO complex at 450 nm after dithionite-reduction and bubbling with carbon monoxide gas. CYPs concentrations were calculated using the extinction coefficient ε450 = 96,000, as described previously39.
Enzymatic biotransformations
Reactions were performed in 4 mL screw PTFE liner cap vials. The following 1 mL biotransformation mixtures (contained in 50 mM potassium phosphate buffer pH 7, 200 mM NaCl and 10% glycerol) were used: cell lysate (1.5 μM P450 final concentration), 0.2 mM NADH/NADPH, 5 mM monoterpene substrate, 30 mM glucose and 10 U·mL−1 glucose dehydrogenase. Enzymatic biotransformations were left shaking overnight at 25 °C.
Oxyfunctionalisation activity-based metabolomic profiling
In order to detect and quantify unexpected products formed upon oxyfunctionalisation, an untargeted GC/MS metabolomics analysis was applied for each one of the individual fifteen substrates. Thus, for each substrate, we created an experimental batch containing vials from triplicate biotransformations of each CYP in our toolkit together with their respective blank (lysate free) and control (CYPs free) reactions. Following biotransformations, vials were cooled for 15 min at 4 °C and subsequently mixed with ethyl acetate (1:1) (containing 0.01% sec-butyl benzene used as internal standard for GC/MS analysis). Then, they were vigorously vortexed (1 min), centrifuged (14,000 rpm, 5 min, 4 °C) and dried over anhydrous MgSO4. 1 μL of the resulting mixture was automatically injected into a GC/MS system (Agilent Technologies 7890B GC and 5977 A MSD) equipped with DB-WAX column (30 m × 0.32 mm i.d., 0.25 μM film; Agilent Technologies). For each experimental batch an external standard containing C8-C20 alkanes was injected to further determine retention indexes (RI). GC/MS conditions were as follows: the injector temperature was set at 240 °C with a split ratio of 20:1 and the helium carrier flow-rate was kept constant at 2 mL·min−1; the oven temperature was 50 °C (1 min held), increased to 68 °C at 5 °C·min−1 rate (2 min held), and increased to 240 °C at 25 °C·min−1 (2 min held); the ion source temperature was set to 230 °C and it was operated in the electron impact ionization mode (70 eV); mass spectra were recorded at 6.6 scans·s−1 at mass scanning range from m/z 50–250 after a solvent delay of 2.20 min. For each batch an external standard containing C8-C20 alkanes was injected to further determine Kovats RI. Raw GC/MS files were converted to open-data format mzXML40 using Proteowizard41 and further data analysis was conducted using open-source R packages42,43. Peak peaking was performed at the total ion count (TIC) level in each chromatogram. Subsequently, homologous peaks were matched and aligned across samples within the same batch. Peaks were integrated and normalized to their corresponding internal standard peak area. To guarantee biological consistency, only those product peaks appearing in all sample triplicates and which median area was higher than the corresponding area in negative controls or blank reactions, were further considered. Product identification was conducted by comparison to reference spectra on the MS NIST library (level 2 annotation44) and/or using authentic standards (level 1 annotation44) as shown in online Supplementary Table S18. Substrate conversion (%) was calculated as per equation (1), where Sx and Sc represent the substrate peak area from each biotransformation and from the negative control, respectively.
| 1 |
Electronic supplementary material
Acknowledgements
This work was funded by the UK Biotechnology and Biological Sciences Research Council (BBSRC; awards BB/M000354/1 & BB/M017702/1) and the Engineering and Physical Sciences Research Council (EPSRC; award EP/J020192/1). This is a contribution from the BBSRC/EPSRC Manchester Synthetic Biology Research Centre (SYNBIOCHEM).
Author Contributions
A.H.-O. performed the protein expression and characterisation experiments, enzymatic biotransformations, GC/MS experiments, MS data interpretation and wrote the main paper and the Supplementary Information.; M.V. designed the GC/MS screening strategy, performed the numerical data analysis and supported the MS data interpretation; Z.Z. designed and performed the molecular cloning experiments; E.T. commented on the manuscript and supervised the work; N.S.S. designed the project and supervised the work. All authors edited the manuscript, discussed the results and commented on the manuscript at all stages.
Data Availability
All data generated or analysed during this study are included in this published article and its Supplementary Information.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-32816-1.
References
- 1.Chang MC, Eachus RA, Trieu W, Ro DK, Keasling JD. Engineering Escherichia coli for production of functionalized terpenoids using plant P450s. Nat Chem Biol. 2007;3:274–7. doi: 10.1038/nchembio875. [DOI] [PubMed] [Google Scholar]
- 2.Alonso-Gutierrez J, et al. Metabolic engineering of Escherichia coli for limonene and perillyl alcohol production. Metab Eng. 2013;19:33–41. doi: 10.1016/j.ymben.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 3.Yeston J. Guiding an enzyme all around a ring. Science. 2018;359:44–44. [Google Scholar]
- 4.Gilbert MM, et al. Synthesis of Diverse 11- and 12-Membered Macrolactones from a Common Linear Substrate Using a Single Biocatalyst. ACS Cent Sci. 2017;3:1304–1310. doi: 10.1021/acscentsci.7b00450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell SG, et al. Molecular recognition in (+)-alpha-pinene oxidation by cytochrome P450cam. J Am Chem Soc. 2003;125:705–14. doi: 10.1021/ja028460a. [DOI] [PubMed] [Google Scholar]
- 6.Seifert A, Antonovici M, Hauer B, Pleiss J. An efficient route to selective bio-oxidation catalysts: an iterative approach comprising modeling, diversification, and screening, based on CYP102A1. Chembiochem. 2011;12:1346–51. doi: 10.1002/cbic.201100067. [DOI] [PubMed] [Google Scholar]
- 7.Bernhardt R, Urlacher VB. Cytochromes P450 as promising catalysts for biotechnological application: chances and limitations. Appl Microbiol Biotechnol. 2014;98:6185–203. doi: 10.1007/s00253-014-5767-7. [DOI] [PubMed] [Google Scholar]
- 8.van Beilen JB, et al. Biocatalytic production of perillyl alcohol from limonene by using a novel Mycobacterium sp. cytochrome P450 alkane hydroxylase expressed in Pseudomonas putida. Appl Environ Microbiol. 2005;71:1737–44. doi: 10.1128/AEM.71.4.1737-1744.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bell SG, Harford-Cross CF, Wong LL. Engineering the CYP101 system for in vivo oxidation of unnatural substrates. Protein Eng. 2001;14:797–802. doi: 10.1093/protein/14.10.797. [DOI] [PubMed] [Google Scholar]
- 10.Kim D, Ortiz de Montellano PR. Tricistronic overexpression of cytochrome P450cam, putidaredoxin, and putidaredoxin reductase provides a useful cell-based catalytic system. Biotechnol Lett. 2009;31:1427–31. doi: 10.1007/s10529-009-0016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dietrich M, et al. Cloning, expression and characterisation of CYP102A7, a self-sufficient P450 monooxygenase from Bacillus licheniformis. Appl Microbiol Biotechnol. 2008;79:931–40. doi: 10.1007/s00253-008-1500-8. [DOI] [PubMed] [Google Scholar]
- 12.Lee Tae Kyoung, Roh Hyun-Soo, Yu Jae Sik, Baek Jiwon, Lee Seul, Ra Moonjin, Kim Sun Young, Baek Kwan-Hyuck, Kim Ki Hyun. Pinecone ofPinus koraiensisInducing Apoptosis in Human Lung Cancer Cells by Activating Caspase-3 and its Chemical Constituents. Chemistry & Biodiversity. 2017;14(4):e1600412. doi: 10.1002/cbdv.201600412. [DOI] [PubMed] [Google Scholar]
- 13.Vespermann KA, et al. Biotransformation of alpha- and beta-pinene into flavor compounds. Appl Microbiol Biotechnol. 2017;101:1805–1817. doi: 10.1007/s00253-016-8066-7. [DOI] [PubMed] [Google Scholar]
- 14.Tomaz-Morais JF, et al. Orofacial antinociceptive activity of (S)-(−)-perillyl alcohol in mice: a randomized, controlled and triple-blind study. Int J Oral Maxillofac Surg. 2017;46:662–667. doi: 10.1016/j.ijom.2017.01.024. [DOI] [PubMed] [Google Scholar]
- 15.Zafeer, M. F. et al. Perillyl alcohol alleviates amyloid-beta peptides-induced mitochondrial dysfunction and cytotoxicity in SH-SY5Y cells. Int J Biol Macromol (2017). [DOI] [PubMed]
- 16.Paduch R, et al. Biological activity of terpene compounds produced by biotechnological methods. Pharm Biol. 2016;54:1096–107. doi: 10.3109/13880209.2015.1103753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bell, S. G., Sowden, R. J. & Wong, L. -L. Engineering the haem monooxygenase cytochrome P450cam for monoterpene oxidation. Chemical Communications, 635–636 (2001).
- 18.Vanek T, Halik J, Vankova R, Valterova I. Formation of trans-verbenol and verbenone from alpha-pinene catalysed by immobilised Picea abies cells. Biosci Biotechnol Biochem. 2005;69:321–5. doi: 10.1271/bbb.69.321. [DOI] [PubMed] [Google Scholar]
- 19.Gillette NE, et al. Efficacy of verbenone and green leaf volatiles for protecting whitebark and limber pines from attack by mountain pine beetle (Coleoptera: Curculionidae: Scolytinae) Environ Entomol. 2014;43:1019–26. doi: 10.1603/EN12330. [DOI] [PubMed] [Google Scholar]
- 20.Byrne CM, Allen SD, Lobkovsky EB, Coates GW. Alternating copolymerization of limonene oxide and carbon dioxide. J Am Chem Soc. 2004;126:11404–5. doi: 10.1021/ja0472580. [DOI] [PubMed] [Google Scholar]
- 21.Seifert A, et al. Rational design of a minimal and highly enriched CYP102A1 mutant library with improved regio-, stereo- and chemoselectivity. Chembiochem. 2009;10:853–61. doi: 10.1002/cbic.200800799. [DOI] [PubMed] [Google Scholar]
- 22.Winnacker M, Rieger B. Recent progress in sustainable polymers obtained from cyclic terpenes: synthesis, properties, and application potential. ChemSusChem. 2015;8:2455–71. doi: 10.1002/cssc.201500421. [DOI] [PubMed] [Google Scholar]
- 23.Toogood HS, et al. Enzymatic menthol production: one-pot approach using engineered Escherichia coli. ACS Synth Biol. 2015;4:1112–23. doi: 10.1021/acssynbio.5b00092. [DOI] [PubMed] [Google Scholar]
- 24.Currin A, et al. Engineering the “missing link” in biosynthetic (−)-menthol production: bacterial isopulegone isomerase. ACS Catal. 2018;8:2012–2020. doi: 10.1021/acscatal.7b04115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leferink NGH, et al. A ‘plug and play’ platform for the production of diverse monoterpene hydrocarbon scaffolds in Escherichia coli. ChemistrySelect. 2016;1:1893–1896. doi: 10.1002/slct.201600563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Funhoff EG, Bauer U, Garcia-Rubio I, Witholt B, van Beilen JB. CYP153A6, a soluble P450 oxygenase catalyzing terminal-alkane hydroxylation. J Bacteriol. 2006;188:5220–7. doi: 10.1128/JB.00286-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Beilen JB, et al. Cytochrome P450 alkane hydroxylases of the CYP153 family are common in alkane-degrading eubacteria lacking integral membrane alkane hydroxylases. Appl Environ Microbiol. 2006;72:59–65. doi: 10.1128/AEM.72.1.59-65.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krithika R, et al. Characterization of 10-hydroxygeraniol dehydrogenase from Catharanthus roseus reveals cascaded enzymatic activity in iridoid biosynthesis. Sci Rep. 2015;5:8258. doi: 10.1038/srep08258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ilc T, Parage C, Boachon B, Navrot N, Werck-Reichhart D. Monoterpenol oxidative metabolism: role in plant adaptation and potential applications. Front Plant Sci. 2016;7:509. doi: 10.3389/fpls.2016.00509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watanabe Y, et al. Oxidation of acyclic monoterpenes by P450 BM-3 monooxygenase: influence of the substrate E/Z-isomerism on enzyme chemo-and regioselectivity. Tetrahedron. 2007;63:9413–9422. doi: 10.1016/j.tet.2007.06.104. [DOI] [Google Scholar]
- 31.Ropp JD, Gunsalus IC, Sligar SG. Cloning and expression of a member of a new cytochrome P-450 family: cytochrome P-450lin (CYP111) from Pseudomonas incognita. J Bacteriol. 1993;175:6028–37. doi: 10.1128/jb.175.18.6028-6037.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peacock VE, Kuneman DW. Inhibition of the formation of. alpha.-p-dimethylstyrene and p-cymen-8-ol in a carbonated citral-containing beverage system. Journal of Agricultural and Food Chemistry. 1985;33:330–335. doi: 10.1021/jf00063a003. [DOI] [Google Scholar]
- 33.Unterweger B, et al. CYP101J2, CYP101J3, and CYP101J4, 1,8-Cineole-Hydroxylating Cytochrome P450 Monooxygenases from Sphingobium yanoikuyae Strain B2. Appl Environ Microbiol. 2016;82:6507–6517. doi: 10.1128/AEM.02067-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hawkes DB, Adams GW, Burlingame AL, Ortiz de Montellano PR, De Voss JJ. Cytochrome P450(cin) (CYP176A), isolation, expression, and characterization. J Biol Chem. 2002;277:27725–32. doi: 10.1074/jbc.M203382200. [DOI] [PubMed] [Google Scholar]
- 35.Stok JE, et al. In vivo and in vitro hydroxylation of cineole and camphor by cytochromes P450CYP101A1, CYP101B1 and N242A CYP176A1. Journal of Molecular Catalysis B: Enzymatic. 2016;128:52–64. doi: 10.1016/j.molcatb.2016.03.004. [DOI] [Google Scholar]
- 36.Hunerdosse D, Nomura DK. Activity-based proteomic and metabolomic approaches for understanding metabolism. Curr Opin Biotechnol. 2014;28:116–26. doi: 10.1016/j.copbio.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prosser GA, Larrouy-Maumus G, de Carvalho LP. Metabolomic strategies for the identification of new enzyme functions and metabolic pathways. EMBO Rep. 2014;15:657–69. doi: 10.15252/embr.201338283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zebec Z, Zink IA, Kerou M, Schleper C. Efficient CRISPR-mediated post-transcriptional gene silencing in a hyperthermophilic archaeon using multiplexed crRNA expression. G3 (Bethesda) 2016;6:3161–3168. doi: 10.1534/g3.116.032482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Omura T, Sato R. The carbon monoxide-binding pigment of liver microsomes. II. solubilization, purification, and properties. J Biol Chem. 1964;239:2379–85. [PubMed] [Google Scholar]
- 40.Pedrioli PG, et al. A common open representation of mass spectrometry data and its application to proteomics research. Nat Biotechnol. 2004;22:1459–66. doi: 10.1038/nbt1031. [DOI] [PubMed] [Google Scholar]
- 41.Kessner D, Chambers M, Burke R, Agus D, Mallick P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 2008;24:2534–6. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chambers MC, et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotechnol. 2012;30:918–20. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ottensmann M, Stoffel MA, Nichols HJ, Hoffman JI. GCalignR: An R package for aligning gas-chromatography data for ecological and evolutionary studies. PLoS One. 2018;13:e0198311. doi: 10.1371/journal.pone.0198311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sumner LW, et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI) Metabolomics. 2007;3:211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article and its Supplementary Information.