Abstract
Brucella ovis is a non-zoonotic Brucella species lacking specific vaccine. It presents a narrow host range, a unique biology relative to other Brucella species, and important distinct surface properties. To increase our knowledge on its peculiar surface and virulence features, and seeking to develop a specific vaccine, multiple mutants for nine relevant cell-envelope-related genes were investigated. Mutants lacking Omp10 plus Omp19 could not be obtained, suggesting that at least one of these lipoproteins is required for viability. A similar result was obtained for the double deletion of omp31 and omp25 that encode two major surface proteins. Conversely, the absence of major Omp25c (proved essential for internalization in HeLa cells) together with Omp25 or Omp31 was tolerated by the bacterium. Although showing important in vitro and in vivo defects, the Δomp10Δomp31Δomp25c mutant was obtained, demonstrating that B. ovis PA survives to the simultaneous absence of Omp10 and four out seven proteins of the Omp25/Omp31 family (i.e., Omp31, Omp25c, Omp25b, and Omp31b, the two latter naturally absent in B. ovis). Three multiple mutants were selected for a detailed analysis of virulence in the mouse model. The Δomp31Δcgs and Δomp10Δomp31Δomp25c mutants were highly attenuated when inoculated at 106 colony forming units/mouse but they established a persistent infection when the infection dose was increased 100-fold. The Δomp10ΔugpBΔomp31 mutant showed a similar behavior until week 3 post-infection but was then totally cleared from spleen. Accordingly, it was retained as vaccine candidate for mice protection assays. When compared to classical B. melitensis Rev1 heterologous vaccine, the triple mutant induced limited splenomegaly, a significantly higher antibody response against whole B. ovis PA cells, an equivalent memory cellular response and, according to spleen colonization measurements, better protection against a challenge with virulent B. ovis PA. Therefore, it would be a good candidate to be evaluated in the natural host as a specific vaccine against B. ovis that would avoid the drawbacks of B. melitensis Rev1. In addition, the lack in this attenuated strain of Omp31, recognized as a highly immunogenic protein during B. ovis infection, would favor the differentiation between infected and vaccinated animals using Omp31 as diagnostic target.
Keywords: Brucella ovis, outer membrane, virulence, recombinant vaccine, Omp31, Omp25/Omp31 family, cyclic glucans, lipoprotein
Introduction
Brucella ovis is a Gram-negative bacterial species belonging to the genus Brucella. It is a non-zoonotic species mainly provoking epididymitis and other genital lesions in rams, although it has also been associated with increased perinatal mortality in lambs and placentitis, abortions, and infertility in ewes (OIE, 2017b). It causes significant economic losses worldwide and lacks commercially available specific vaccine. B. ovis lipopolysaccharide (LPS) is devoid of O-polysaccharide (O-PS) chains and is defined as rough (R) LPS (R-LPS). B. ovis and B. canis are the sole species of the Brucella genus constituted exclusively by R strains that are virulent for their natural hosts. This characteristic differentiates them from smooth (S) brucellae that require O-PS for full virulence (e.g., B. melitensis, B. abortus, or B. suis). B. melitensis Rev1, currently used for vaccination against ovine and caprine brucellosis caused by B. melitensis, is considered the best available vaccine against B. ovis (OIE, 2017a). However, this vaccine is banned in countries or regions where infection by B. melitensis is eradicated because, among other drawbacks, it induces antibodies that interfere with the serological diagnosis of infections caused by S brucellae. Therefore, the development of a specific vaccine for the prophylaxis of B. ovis infection is a matter of interest. Considering that the best available vaccines against brucellosis caused by S Brucella strains are homologous S attenuated strains (Nicoletti, 2010), the search for a B. ovis attenuated vaccine strain seems an interesting approach. The first step to achieve this goal is the identification of virulence factors that can be removed from B. ovis, to minimize its deleterious effects on the host, but without compromising its immunogenicity.
Comparatively to its S counterparts, little is known about the virulence of B. ovis, although a species-specific ABC transporter (Silva et al., 2011, 2013) and some classical virulence factors described in S species have been identified as necessary for its virulence (Martín-Martín et al., 2012; Sá et al., 2012; Macedo et al., 2015). O-PS chains mask other outer membrane (OM) components in S strains (Cloeckaert et al., 2002), hindering their interaction with host cells, antibodies, and other elements of the immune system. According to the surface exposure of OM molecules other than LPS in R B. ovis, a more important role in the interaction with host cells and virulence than in S strains would be expected. However, several OM proteins (OMPs) and OM-related genes necessary for full virulence in S strains seem not required in B. ovis experimental infection models (Martín-Martín et al., 2012; Sidhu-Muñoz et al., 2016). This observation reveals differences among the brucellae regarding the role of the OM molecules in host–pathogen interactions, differences that might be associated with their heterogeneity regarding OM-related properties (Martín-Martín et al., 2011; Vizcaíno and Cloeckaert, 2012), host-preference, and pathogenicity. Although the Brucella species share a high level of DNA homology, an increased number of pseudogenes and insertion sequences has been detected in B. ovis, when compared to zoonotic S Brucella (Tsolis et al., 2009). This feature led to hypothesize that its narrow host-range and tissue tropism (almost exclusively restricted to ovine male genital tract) is in part consequence of genome degradation (Tsolis et al., 2009). However, despite this genome degradation, that among others affects O-PS biosynthetic genes and several OMPs (Tsolis et al., 2009), B. ovis causes a chronic infection in its natural host and in laboratory animals (Caro-Hernández et al., 2007; Silva et al., 2011; OIE, 2017b), which would also support a specific pattern of interaction between the host and the bacterial OM.
With the aim of increasing our knowledge about the contribution of cell envelope components to OM-related properties and virulence of B. ovis and as a tool to develop a specific live attenuated vaccine, in this work we have constructed and characterized a panel of B. ovis multiple mutants in genes related to the cell envelope that either code for major OMPs or either are individually required in S Brucella strains, but not in B. ovis, for virulence (Caro-Hernández et al., 2007; Martín-Martín et al., 2012; Sidhu-Muñoz et al., 2016). Genes targeted in multiple mutations were: (i) omp31, omp25, and omp25c that code for major OMPs in B. ovis (Cloeckaert et al., 2002; Martín-Martín et al., 2009); (ii) omp10 and omp19 that encode two minor OM lipoproteins (Tibor et al., 1999) required in B. abortus 544 for full virulence (Tibor et al., 2002); (iii) bepC that encodes a TolC-homolog protein necessary in B. suis 1330 for full virulence (Posadas et al., 2007); (iv) bacA that encodes an integral inner membrane protein involved in lipid A acylation, cell-envelope properties, and virulence in B. abortus 2308 (LeVier et al., 2000; Roop et al., 2002; Ferguson et al., 2004; Parent et al., 2007); and (v) ugpB that encodes SP41, a surface protein involved in invasion of B. suis 1330 to HeLa cells (Castañeda-Roldán et al., 2006). Beside these genes that are not individually required for virulence in B. ovis PA, multiple mutations also included cgs, involved in the synthesis of periplasmic cyclic β-1,2-glucans (CβGs) (Iñón de Iannino et al., 1998; Haag et al., 2010) and necessary for full virulence in B. abortus 2308 (Briones et al., 2001). The Δcgs mutant of B. ovis PA was also highly attenuated when it was intraperitoneally inoculated at a dose of 106 colony forming units (CFU)/mouse (Martín-Martín et al., 2012), but when the dose usually employed for protection experiments (108 CFU/mouse) (Sancho et al., 2014; Soler-Lloréns et al., 2014; Silva et al., 2015b) was used, the bacterial counts in spleen increased to levels that were close to those of the parental strain (unpublished results). Several combinations of deleted genes were tempted in the panel of multiple mutants, although, keeping in mind the development of an attenuated vaccine, the inclusion of Δcgs and Δomp31 mutations was prioritized for two reasons: (i) to study how the addition of new mutations could increase the attenuation of the Δcgs single mutant to appropriate levels for an attenuated vaccine, and (ii) Omp31 is an abundant OMP in the OM of B. ovis PA (Martín-Martín et al., 2009), it is the most immunogenic OMP in the course of B. ovis infection (Kittelberger et al., 1998) and its interest for a serological diagnosis favoring the differentiation between infected and vaccinated animals (DIVA diagnosis) has also been evidenced (Vizcaíno et al., 2001b).
Materials and Methods
Bacterial Strains, Culture Conditions, and Plasmids
Brucella ovis PA and B. melitensis Rev1 were obtained from BCCN (Brucella Culture Collection Nouzilly, maintained at the Institut National de la Recherche Agronomique, Nouzilly, France), and the other B. ovis strains are listed in Table 1. Recombinant plasmids used for mutagenesis of cgs (BOV_RS00 535), bacA (BOV_RS01960), omp10 (BOV_RS10700), omp19 (BOV_RS09115), bepC (BOV_RS04655), and ugpB (BOV_RS13470), and the corresponding single mutants derived from parental B. ovis PA have previously been described (Martín-Martín et al., 2012; Sidhu-Muñoz et al., 2016). B. ovis Δomp31 (BOV_RS12205) and Δomp25 (BOV_RS3460) single nonpolar mutants (Table 1) were obtained from parental B. ovis PA as described below. Multiple mutants (Table 1) were constructed from initial single mutants where one or two of the mentioned genes were additionally deleted. The Δomp31-k and Δomp25c-k mutant strains (omp31 or omp25c replaced by a kanamycin resistance cassette) and the same mutants complemented with the corresponding omp31 or omp25c (BOV_RS00575) wild type genes (Δomp31-k com and Δomp25c-k com strains) were previously obtained (Caro-Hernández et al., 2007).
Table 1.
Most relevant bacterial strains used in this work, growth characteristics, and preliminary evaluation of virulence.
| Log CFU/spleen at week (W) p.i.c |
||||||
|---|---|---|---|---|---|---|
| Deleted gene/s and strain | Log CFU/ml | (dose 106 CFU) |
(dose 108 CFU) |
|||
| Brucella ovis strainsa | abbreviation in the text | OD600 = 0.2b | W3 | W7 | W3 | W7 |
| B. ovis PA (BCCN 76-250) | Parental strain, PA | 9.09 0.04 | 6.90 | 5.85 | 7.62 | 6.31 |
| Single mutants | ||||||
| B. ovis-pPS31OVL02M | Δomp31 | 8.80 0.10* | 5.44 | 5.79 | – | – |
| B. ovis-pNV25OVL02M | Δomp25 | 9.08 0.07 | 6.87 | 4.92 | – | – |
| B. ovis-PNV25cA | Δomp25c-k | 8.93 0.03* | 7.20 | 5.61 | – | – |
| B. ovis-pNVcgs03M | Δcgs | 8.88 0.08* | 0.45 | 0.53 | 6.20 | 5.53 |
| B. ovis-pNVbacA03M | ΔbacA | 9.12 0.07 | 6.48 | 5.64 | – | – |
| B. ovis-pNV1002M | Δomp10 | 8.84 0.04* | 6.91 | 5.79 | – | – |
| B. ovis-pNV1902M | Δomp19 | 8.92 0.05* | 6.51 | 5.90 | – | – |
| B. ovis-pNVBepC02M | ΔbepC | 9.15 0.08 | 6.17 | 5.28 | – | – |
| B. ovis-pNVSP4102M | ΔugpB | 9.10 0.03 | 7.72 | 6.30 | – | – |
| Double mutants | ||||||
| B. ovis Δomp31-pNV1902M | Δomp31Δomp19 | 8.78 0.04* | 5.25 | 5.92 | – | – |
| B. ovis Δomp31-pNVBepC02M | Δomp31ΔbepC | 8.80 0.04* | 6.78 | 5.75 | – | – |
| B. ovis Δomp31-pNVSP4102M | Δomp31ΔugpB | 8.82 0.06* | 5.90 | 6.32 | – | – |
| B. ovis Δomp31-pNVcgs03M | Δomp31Δcgs | 8.49 0.09* | – | – | 5.16 | 4.95 |
| B. ovis Δomp25-pNVcgs03M | Δomp25Δcgs | 8.56 0.06* | – | – | 5.52 | 5.15 |
| B. ovis Δomp25-pNV25cOVL02M | Δomp25Δomp25c | 8.82 0.06* | 6.91 | 6.32 | – | – |
| B. ovis Δcgs-pNVSP4102M | ΔcgsΔugpB | 8.89 0.04* | – | – | 5.24 | 4.80 |
| B. ovis Δcgs-pNVBepC02M | ΔcgsΔbepC | 8.87 0.07* | – | – | 5.45 | 4.63 |
| B. ovis Δomp10-pPS31OVL02M | Δomp10Δomp31 | 8.89 0.07* | 6.72 | 6.66 | – | – |
| B. ovis Δomp10-pNVSP4102M | Δomp10ΔugpB | 8.91 0.06* | 7.29 | 5.66 | – | – |
| B. ovis Δomp10-pNVcgs03M | Δomp10Δcgs | 8.82 0.09* | – | – | 5.21 | 5.39 |
| B. ovis Δomp19-pNVcgs03M | Δomp19Δcgs | 8.79 0.03* | – | – | 6.07 | 5.99 |
| B. ovis Δomp19-pNVSP4102M | Δomp19ΔugpB | 8.88 0.03* | 6.57 | 5.36 | – | – |
| B. ovis ΔbepC-pNVSP4102M | ΔbepCΔugpB | 9.10 0.05 | 7.37 | 5.63 | – | – |
| B. ovis ΔbacA-pPS31OVL02M | ΔbacAΔomp31 | 8.81 0.02* | 6.05 | 5.36 | – | – |
| Triple mutants | ||||||
| B. ovis Δomp10ΔugpB-pPS31OVL02M | Δomp10ΔugpBΔomp31 | 8.76 0.18* | 0.47 | 0.57 | 5.53 | 0.52 |
| B. ovis Δomp10Δomp31-pNV25cOVL02M | Δomp10Δomp31Δomp25c | 8.49 0.04* | 2.57 | 3.26 | 5.30 | 5.70 |
| B. ovis Δomp31Δomp10-pNV25cOVL02M | Δomp31Δomp10Δomp25c | 8.39 0.03* | 2.42 | 2.03 | – | – |
| B. ovis Δomp31Δcgs-pNV1002M | Δomp31ΔcgsΔomp10 | 8.61 0.07* | – | – | 5.53 | 5.73 |
| B. ovis Δomp31Δcgs-pNV1902M | Δomp31ΔcgsΔomp19 | 8.71 0.05* | – | – | 5.07 | 4.61 |
| B. ovis Δomp31ΔbepC-pNVSP4102M | Δomp31ΔbepCΔugpB | 8.86 0.12* | 7.65 | 6.00 | – | – |
| Other previous mutants used as controls | ||||||
| B. ovis PNV31A | Δomp31-k | 8.85 0.03* | – | – | – | – |
| B. ovis PNV31A-com | Δomp31-k com | 8.89 0.03* | – | – | – | – |
| B. ovis PNV25c-com | Δomp25c-k com | 8.99 0.06 | – | – | – | – |
aLog CFU/spleen 3 and 7 weeks after intraperitoneal infection with 1 × 106 or 1 × 108 CFU/mouse. One mouse was used per time point in this preliminary assay of virulence. –, not determined. bThe B. ovis mutants derive from B. ovis PA, which was obtained from BCCN (Brucella Culture Collection Nouzilly, Institut National de la Recherche Agronomique, Nouzilly, France). B. ovis Δcgs, ΔbacA, Δomp10, Δomp19, ΔbepC, and ΔugpB were previously described (Martín-Martín et al., 2012; Sidhu-Muñoz et al., 2016). B. ovis PA Δomp25c-k and Δomp31-k were also previously obtained replacing omp25c and omp31, respectively, by a kanamycin resistance cassette; they were complemented with wild-type omp25c and omp31, respectively, to give B. ovis Δomp25c-k com and B. ovis Δomp31-k com (Caro-Hernández et al., 2007). The other B. ovis mutants were obtained in this work. cThe asterisk (∗) indicates statistically significant differences (P < 0.05), compared to the parental strain. Log CFU/ml of bacterial suspensions adjusted to an OD600 value of 0.2. The results are expressed as the mean ± SD of three assays.
Brucella ovis strains and the B. melitensis Rev1 attenuated vaccine were cultured in tryptic soy agar or tryptic soy broth (Pronadisa-Laboratorios Conda, Torrejón de Ardoz, Spain) both supplemented with 0.3% yeast extract (Pronadisa-Laboratorios Conda, Torrejón de Ardoz, Spain) and 5% horse serum (Gibco-Life Technologies, Grand Island, NY, United States) (TSA-YE-HS and TSB-YE-HS, respectively). Incubations were performed at 37°C in a 5% CO2 atmosphere and, in the case of TSB-YE-HS liquid medium, under agitation at 120 rpm. When required, streptomycin (Strep; 50 μg/ml) (Sigma-Aldrich, St. Louis, MO, United States) was added to the culture medium of B. melitensis Rev1 (Strep-resistant strain). Similarly, when necessary for the selection of the recombinant B. ovis strains, kanamycin (50 μg/ml) or 5% sucrose (Sigma-Aldrich, St. Louis, MO, United States) was added. Assays with the B. ovis strains, including Δomp31-k, Δomp25c-k, Δomp31-k com, and Δomp25c-k com strains, were performed in the absence of antibiotics.
Plasmid pGEM-T Easy (Promega, Madison, WI, United States) was used to clone PCR-amplified fragments and pCVD-KanD (Martín-Martín et al., 2012) was the suicide plasmid employed to insert the mutant genes into parental B. ovis PA. They were propagated in Escherichia coli JM109 or CC118 (λpir), respectively, that were incubated at 37°C in Luria Bertani medium supplemented, when required, with ampicillin or kanamycin (50 μg/ml). Their derived recombinant plasmids constructed during this work are mentioned below.
DNA Primers and Mutagenesis
DNA primers (IDT, Leuven, Belgium) used for the construction and verification of the B. ovis Δomp31, Δomp25, and Δomp25c single and multiple nonpolar mutants are listed in Table 2. The additional primers used to check the multiple mutants were previously described (Martín-Martín et al., 2012; Sidhu-Muñoz et al., 2016).
Table 2.
Primers used in this work for the construction and verification of omp25, omp25c, and omp31 single and multiple mutantsa.
| Primer name | Nucleotide sequence 5′–3′b | Target gene or plasmidc |
|---|---|---|
| Construction of B. ovis PA mutants | ||
| 25MUTZ-F | CGACCTTATCCTCCTGAA | omp25 |
| 25OVL-R | GACGATTACGAGAGACTT | omp25 |
| 25OVL-F | AAGTCTCTCGTAATCGTCAAGCTGGACACGCAGGAT | omp25 |
| 25MUTZ-R | TTTGCGACGTTTTGCTGG | omp25 |
| 25cdMUT-F | TGCGTGGTTCAGATTTCG | omp25c |
| 25cOVL-R | AGCCTTGAGCTTCATGAT | omp25c |
| 25cOVL-F | ATCATGAAGCTCAAGGCTGCTTACAAGTTCTGATAG | omp25c |
| 25cMUT-R | AGCCGTAACCAACCTGAC | omp25c |
| 31MUT-F | AGAATAAAACACATGCCC | omp31 |
| 31OVL-R | GATGGACGCCAAAATTAC | omp31 |
| 31OVL-F | GTAATTTTGGCGTCCATCGTCGGTCTGAACTACAAG | omp31 |
| 31MUT-R | GCTGAATGCGGAGATGGT | omp31 |
| Additional primers for the verification of recombinant plasmids | ||
| and mutants | ||
| Universal-F | GTTTTCCCAGTCACGAC | pGEM-T Easy |
| Universal-R | CAGGAAACAGCTATGAC | pGEM-T Easy |
| 25-Sec | GGACCGCGCAAAACGTAATT | omp25 |
| 25-MAT | GCCGACGCCATCCAGGAA | omp25 |
| 25c-MAT | GCTGACGCCGTCATTGAA | omp25c |
| 31-MAT | GCCGACGTGGTTGTTTCT | omp31 |
aPrimers for the verification of proper deletion of other genes have been previously described (Martín-Martín et al., 2012; Sidhu-Muñoz et al., 2016). bUnderlined sequences in 25OVL-F, 25cOVL-F, and 31OVL-F correspond to regions overlapping with 25OVL-R, 25cOVL-R, and 31OVL-R, respectively. cTarget gene is the B. ovis gene to be deleted or PCR-amplified for the verification of mutant strains. Primers Universal-F and Universal-R target pGEM-T Easy and its derived recombinant plasmids at both sides of the cloned insert and were used for sequencing of the DNA insert. The remaining primers target the B. ovis genome and were designed according to the published genome sequence of B. ovis 63/290 (ATCC 25840) (accession numbers NC_009505 and NC_009504 for chromosome I and II, respectively).
For the construction of the recombinant plasmids used in the mutagenesis process, inactivation of omp31, omp25, and omp25c was performed by in-frame deletion with overlapping PCR (Martín-Martín et al., 2012). Briefly, the 5′- and 3′-ends of each target gene, together with about 300–700 pb upstream or downstream, respectively, were separately amplified by PCR with two pairs of primers (31MUT-F + 31OVL-R and 31OVL-F + 31MUT R for omp31, 25MUTZ-F + 25OVL-R and 25OVL-F + 25MUTZ-R for omp25, 25cdMUT-F + 25cOVL-R and 25cOVL-F + 25cMUT-R) (Table 2). The two amplified fragments were fused, through the overlapping sequences located in the internal primers (primers OVL-F and OVL-R) (Table 2), in a PCR reaction with the two external primers of each fragment (31MUT-F + 31MUT-R for omp31, 25MUTZ-F + 25MUTZ-R for omp25, and 25cdMUT-F + 25cMUT-R for omp25c). The resulting mutant genes were cloned in pGEM-T Easy, verified by DNA sequencing, and subsequently cloned in pCVD-KanD to give pPS31OVL02, pNV25OVL02, and pNV25cOVL02, respectively. Plasmids were introduced in B. ovis PA by electroporation and the selection of bacteria with the corresponding plasmid integrated in the chromosome (intermediate strains), through a single homologous recombination event, was performed on TSA-YE-HS plates with kanamycin. The selected intermediate strains were plated onto TSA-YE-HS supplemented with 5% sucrose to give either the desired mutant strain (wild-type gene replaced by the inactivated gene) or a strain reverting to the parental genotype (Rv). They were differentiated by PCR amplification with the two primers external to each side of the deleted gene (amplified fragment with higher size in Rv strains than in mutant strains) and a second PCR with an external primer and a primer annealing inside the deleted fragment (primers 31-MAT, 25-MAT, and 25c-MAT) (Table 2). The latter PCR reaction produces no amplification in mutant strains.
For multiple gene mutagenesis, single mutants listed in Table 1 were subjected to a second mutagenesis round with a recombinant plasmid containing another inactivated gene. A third round of mutagenesis was conducted on some selected double mutants to inactivate a third gene. The selection of mutant strains was performed with specific PCRs targeting each inactivated locus.
Growth Pattern, Autoagglutination, and Susceptibility Assays
Growth characteristics of the mutant strains in TSA-YE-HS plates and TSB-YE-HS liquid medium were compared to those of parental B. ovis PA. Numbers of CFU/ml corresponding to bacterial suspensions in PBS with optical density scores at 600 nm (OD600) of 0.2 were determined for each mutant by triplicate plating on TSA-YE-HS of the properly diluted suspensions. The initial suspensions were prepared from bacteria cultured in TSA-YE-HS plates for 44 h. Colony size was photographed 5 days after plating and colonies enumerated 8 days after plating. Growth curves were established for triplicate bacterial suspensions in TSB-YE-HS medium (30 ml) with initial OD600 readings of 0.05 that were incubated at 37°C under agitation (120 rpm) and a 5% CO2 atmosphere. OD600 scores were measured through a 120-h period, and CFU/ml numbers were evaluated at the beginning of the experiment (t0), and after 24, 52, and 77 h of incubation (t24, t52, and t77, respectively), by plating the properly diluted cultures on TSA-YE-HS.
The autoagglutination assay was performed as described previously (Caro-Hernández et al., 2007; Martín-Martín et al., 2012) by measuring the evolution, over 48 h of static incubation, of the OD600 values of bacterial suspensions with initial readings of 0.8 (100% OD600) in TSB-YE-HS. Susceptibility to 1 mg/ml of polymyxin B (Sigma-Aldrich, St. Louis, MO, United States) and 0.1 mg/ml of sodium deoxycholate (Sigma-Aldrich, St. Louis, MO, United States) in PBS was expressed as the percentage of survival after 80 min of exposure. It was determined by comparison of the numbers of CFU in untreated (incubation in PBS, 100% survival) and treated bacterial suspensions (Caro-Hernández et al., 2007; Martín-Martín et al., 2011). The results were expressed as means ± standard deviation (SD) of three assays.
Mapping of Cell Envelope Antigens
Reactivity of B. ovis mutants with MAbs specific for cell-envelope antigens (Table 3) was measured by indirect enzyme-linked immunosorbent assay (iELISA) as previously described (Soler-Lloréns et al., 2014). MAbs specific for Brucella peptidoglycan (PG), R-LPS, S-LPS, Omp2b, Omp10, Omp16, Omp19, Omp25, Omp31, or periplasmic BP26 were used (Cloeckaert et al., 1990, 1991, 1996a,b; Vizcaíno et al., 2001b; Seco-Mediavilla et al., 2003). Briefly, 96-well plates were coated overnight at room temperature with sonicated (mild homogenization for 10 s at 40% intensity in a Sonic Dismembrator model 120, Thermo Fisher Scientific, Waltham, Ma, United States) bacterial suspensions in PBS (OD600 = 1), which were prepared from cultures in TSA-YE-HS plates. MAbs (hybridoma supernatant) diluted 1/2 and a goat anti-mouse IgG-horseradish peroxidase conjugate (Bio-Rad, Hercules, CA, United States) diluted 1:9000 were used as primary and secondary antibodies, respectively. Antigen–antibody binding was revealed by incubation for 20 min with TMB as substrate for peroxidase and subsequent addition of a 1 M HCl-based stop solution (Interchim, Montluçon, France). The results were expressed as means ± SD of the values recorded at 450 nm (OD450) in a Labsystems Multiskan Ascent microplate reader (Thermo Fisher Scientific) for three repeats by MAb and strain.
Table 3.
Main characteristics of the monoclonal antibodies used in this work.
| Monoclonal antibodya | Specificity | Abbreviation |
|---|---|---|
| A68/15B06/C08 | Omp2b | C08 |
| A68/07G11/C10 | Omp10 | C10 |
| A76/08C03/G03 | Omp16 | G03 |
| A76/18B02/D06 | Omp19 | D06 |
| A59/10F09/G10 | Omp31 | G10 |
| A59/05F01/C09 | Omp25 | C09 |
| A18/13D02/F05 | Omp25 | F05 |
| A76/08H09/A02 | Omp25 | A02 |
| V78/09B12/B02 | BP26 | B02 |
| V78/02E08/F03 | BP26 | F03 |
| V78/04D01/A10 | BP26 | A10 |
| V78B/04G07/H05 | BP26 | H05 |
| A76/03D06/A09 | PG | A09 |
| A76/12G12/F12 | S-LPS | F12 |
| A68/03F03/D05 | R-LPS | D05 |
aThe MAbs were obtained and characterized previously (Cloeckaert et al., 1990, 1991, 1992, 1996a,b; Zygmunt et al., 1994; Seco-Mediavilla et al., 2003).
Additionally, sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) and immunoblot were also performed and carried out as previously described (Vizcaíno et al., 2001b; Martín-Martín et al., 2009). Briefly, bacterial suspensions concentrated to OD600 values of 20 were prepared in H2O with the different B. ovis strains. They were submitted to SDS–PAGE on a Protean II xi cell (Bio-Rad, Hercules, CA, United States) and either stained with Coomassie blue (Bio-Rad, Hercules, CA, United States) or transferred to a nitrocellulose membrane in a semidry electroblotter (Amersham, GE Healthcare, Little Chalfont, United Kingdom). Prestained protein marker VI (Applichem-Panreac, Barcelona, Spain) was used as protein standard. Nitrocellulose strips were saturated with skim milk and then incubated with sera obtained, as described before (Martín-Martín et al., 2009), by immunization of rabbits with Omp31b, Omp25c, Omp25d, and Omp22 purified recombinant proteins. Binding of the secondary antibody – a goat anti-rabbit IgG-peroxidase conjugate (Sigma-Aldrich, St. Louis, MO, United States) – was detected with a 4-chloro-1-naphthol substrate solution.
Infection Assays on Murine Macrophage and HeLa Cells
Infection assays of murine macrophage-like J774.A1 cells (DSMZ ACC170) and epithelial HeLa (ATCC CCL-2TM) cells were performed as described previously (Sidhu-Muñoz et al., 2016). Briefly, 2 × 104 J774.A1 macrophages or 1.5 × 104 HeLa cells/well were seeded on 96-well sterile microplates and incubated for 24 h at 37°C under a 5% CO2 atmosphere. After incubation for 2 h with the B. ovis strains (4 × 106 CFU/well for macrophages or 8 × 106 CFU/well for HeLa cells) and killing of extracellular bacteria by incubation with gentamycin for 1 h, intracellular bacteria were enumerated in three wells per strain after lysis of the phagocytes with H2O [t0 post-infection (p.i.)] (Sidhu-Muñoz et al., 2016). The remaining wells were maintained in the presence of gentamycin and intracellular bacteria were similarly determined at 20 (t20) and 44 h (t44) p.i. The results were expressed as means ± SD of the log CFU/well at each selected p.i. time point (t0, t20, and t44) and are representative of at least two experiments.
Mice and Ethics Statement
Female 6-week-old BALB/c mice (Charles River Laboratories, Chatillon-sur-Chalaronne, France), received 1 week previously, were used. They were randomly distributed into experimental groups and kept with water and food ad libitum in the animal experimentation facilities of the University of Salamanca (registration number PAE SA-001) or the Unidad de Producción y Sanidad Animal, Instituto Agroalimentario de Aragón-IA2 (CITA-Universidad de Zaragoza) (registration number ES502970012005).
Procedures with mice were designed according to Spanish and European legislation regarding the use of animals in research (RD 53/2013 and directive 2010/63/UE). Microbiological practices and animal experimentation were approved by the Biosecurity and Bioethics Committees of the University of Salamanca, and authorized by the competent authority of Junta de Castilla y León, Spain. The Animal Welfare Committee of the CITA (Spain) also reviewed and approved the protocols.
Virulence Assays and Antibody Response in Mice
Preliminary evaluation of virulence in mice was performed by intraperitoneal inoculation of 106 or 108 CFU/mouse depending on the previous information about each single mutant (Martín-Martín et al., 2012; Sidhu-Muñoz et al., 2016). One mouse was used per strain, dose and time of analysis and splenic colonization was evaluated 3 or 7 weeks (W3 and W7) p.i. as described previously (Sancho et al., 2014). These time points in parental B. ovis PA correspond to the acute and chronic phase of infection, respectively (Caro-Hernández et al., 2007; Martín-Martín et al., 2012; Sidhu-Muñoz et al., 2016).
Spleen colonization of the selected mutants – according to the results of the preliminary analysis – was evaluated at W3 and W7 p.i. in mice inoculated intraperitoneally with 106 CFU, and at W1, W3, W5, W7, and W11 p.i. in mice inoculated intraperitoneally with 108 CFU. Five mice per strain and time point were used. The results were expressed as means ± SD (n = 5) of the log of CFU/spleen for each strain and time point. The identity of the recovered colonies was checked by PCR. Antibodies specific for B. ovis PA were determined by iELISA (Sancho et al., 2014) in sera obtained from submandibular blood from the same mice. Briefly, a suspension in PBS of heat-inactivated B. ovis PA whole cells (OD600 = 1) was used as the coating antigen of 96-well plates and a goat antimouse IgG-peroxidase conjugate (Sigma–Aldrich, St. Louis, MO, United States) was used as the secondary antibody. OD405 readings were recorded on a Multiskan Go Microplate Reader (Thermo Fisher Scientific) after 30 min incubation at room temperature with the substrate solution constituted by 1 mM 2,2′-azino-di-(3-3-ethylbenzothiazoline-sulfonic acid) (ABTS; Sigma-Aldrich, St. Louis, MO, United States) and 2 mM H2O2 (Sigma-Aldrich, St. Louis, MO, United States) in 0.1 M citrate, pH 4.2. Antibody titers in serum were defined as the inverse of the highest serum dilution scoring an OD405 value twice as high as that obtained with the blank (mean OD405 of six wells in which serum was replaced by dilution buffer). The results were represented as means ± SD of the log of the titers obtained with five mice analyzed individually.
Vaccine Efficacy and Immune Response of B. ovis Δomp10ΔugpBΔomp31
BALB/c mice were inoculated intraperitoneally with PBS, 105 CFU of the classical vaccine B. melitensis Rev1, or 108 CFU of the attenuated mutant B. ovis Δomp10ΔugpBΔomp31. Seven weeks later, corresponding with the time point where the B. ovis vaccine was cleared from spleen, five mice per group were either challenged with 2 × 105 CFU of B. ovis PA, or processed for the evaluation of the antibody and cellular immune responses specific for B. ovis PA. Determination of virulent B. ovis PA in spleen was evaluated 3 weeks after experimental challenge (Sancho et al., 2014). The CFU number of virulent B. ovis PA in mice vaccinated with B. melitensis Rev 1 was obtained by subtracting the values obtained in TSA-YE-HS-Strep medium from those obtained in the same medium without antibiotic. Results were expressed as means ± SD (n = 5) of the log CFU/spleen of B. ovis PA for each vaccination group.
IgG titers in serum were analyzed, as described above, in five mice per group 7 weeks after vaccination by using heat-inactivated B. ovis PA whole cells as the coating antigen in a iELISA test. Additionally, IgG isotypes were determined under the same conditions but using goat anti-mouse IgG1-, IgG2a-, or IgG2b-peroxidase conjugates (Santa Cruz Biotechnology, Dallas, TX, United States). The same mice were processed as previously described (Sancho et al., 2014) to evaluate the cytokine response of splenocytes to a second stimulus with B. ovis PA. Briefly, spleen cells from immunized mice were cultured in 24-well sterile plates and stimulated by exposure to heat-inactivated (1 h at 65°C) B. ovis PA whole cells (107 CFU/well), 10 μg/ml of the mitogen concanavalin A (Sigma-Aldrich, St. Louis, MO, United States) as a positive control of cell proliferation, or culture medium as a negative control. After 72 h of incubation, the culture supernatants were harvested to evaluate the levels of interferon-γ (IFN-γ), tumor necrosis factor-α (TNF-α), IL-10, and IL-12(p40). They were determined by sandwich ELISA with OptEIATM Mouse Sets specific to each cytokine, as instructed by the manufacturer (BD Biosciences, San Diego, CA, United States). Two wells were used for each experimental condition and mouse. The results for each vaccination group were expressed as means ± SD of the cytokine amount (ng/well) in the supernatants of splenocytes obtained from five individual mice. The results obtained with the positive and negative controls (concanavalin A and RPMI as stimulating agents, respectively) were as expected and are not shown.
Statistical Analysis
Statistical comparisons between means were performed using analysis of variance. The significance of the differences (P < 0.05) between the experimental groups was determined with the post hoc Fisher’s protected least significant differences (PLSD) test. To simplify the figures and tables, no ranking of P-values has been established and all significant differences are marked as P < 0.05.
Results
Genotypic Characterization of the Mutants
The B. ovis mutants (Table 1) were genotypically characterized as described in the section “Materials and Methods.” Two PCR reactions were settled for each inactivated gene, one reaction with external primers to each side of the deleted locus (amplified fragment of lower size in mutant strains than in parental strain), and a second reaction with an external primer to the deleted loci and a primer annealing inside the deleted region (no amplification in mutant strains). All single and multiple mutants gave the expected results for each inactivated gene (data not shown).
Although not all combinations of single genes shown in Table 1 were tempted in multiple mutants, two combinations were specially assayed without success: a double mutant in lipoproteins Omp10 and Omp19 and a double mutant in major Omp25 and Omp31 proteins. Despite multiple attempts, the intermediate strains for these double mutants always reverted to the single mutant initial genotype. Similarly, two independent omp25c intermediate strains always reverted to the parental genotype and, consequently, the non-polar Δomp25c single mutant was not obtained. However, the non-polar deletion of omp25c, with plasmid pNV25cOVL02, was successful in the Δomp25Δomp25c double mutant and in the Δomp10Δomp31Δomp25c and Δomp31Δomp10Δomp25c triple mutants. The Δomp25c-k mutant, where omp25c was replaced by a kanamycin resistance cassette (Caro-Hernández et al., 2007), was used in this work for comparisons with the non-polar Δomp25c double and triple mutants.
Growth Characteristics of the Mutants
The numbers of CFU in bacterial suspensions with OD600 values of 0.2 were determined for each mutant in TSA-YE-HS plates. Bacterial counts obtained for Δomp25, ΔbepC, ΔugpB, and ΔbacA single mutants and for ΔbepCΔugpB double mutant did not significantly differ from those observed with the parental strain. On the contrary, Δomp31, Δomp25c-k, Δomp10, Δomp19, Δcgs, and the remaining multiple mutants showed, in different degrees, lower CFU values than B. ovis PA (Table 1). The Δomp31Δcgs double mutant and its derived triple mutants, the Δomp25Δcgs mutant and the Δomp10Δomp31Δomp25c and Δomp31Δomp10Δomp25c triple mutants (bearing the same three genes inactivated but in a different order) showed the lowest CFU/ml values in TSA-YE-HS plates (Table 1). The numbers of CFU/ml obtained for each mutant were used for the calculation of the bacterial doses in further experiments.
In addition, the size of the colonies in TSA-YE-HS was monitored over time and recorded 5 days after inoculation (see some representative results in Figure 1A). Colony size of the Δomp25, ΔbacA, ΔbepC, ΔugpB, Δomp25c-k, ΔbepCΔugpB, and Δomp25Δomp25c mutants was undistinguishable from that of the parental strain (see B. ovis PA and the Δomp25Δomp25c mutant in Figure 1A). Growth of the Δomp10 and Δomp19 mutants was slightly delayed when compared to the parental strain but the initial differences in colony size were not apparent after 5 days of incubation (data not shown). Colonies of the Δomp31 and Δcgs single mutants appeared hardly visible 72 h after inoculation, 24 h later than those of the B. ovis PA parental strain, and were smaller than those of the parental strain after 5 days (see Δomp31 in Figure 1A). A similar or higher delay, depending on the strain, was detected with colonies of their multiple mutants. Despite the triple gene deletion, the Δomp10ΔugpBΔomp31 mutant showed a colony growth pattern undistinguishable from that of the single Δomp31 mutant (Figure 1A). The most important growth defects were observed with the Δomp31Δcgs mutant and its derived triple mutants (with the additional deletion of omp10 or omp19), whose colonies started to be detected 96 h after inoculation and showed the smallest size after 5 days of incubation (Figure 1A). Colonies of the Δomp10Δomp31Δomp25c and Δomp31Δomp10Δomp25c triple mutants were also detected after 96 h of incubation but, at day 5 post-inoculation, they were bigger (see colonies of the Δomp10Δomp31Δomp25c mutant that are representative for both mutants) than those of B. ovis Δomp31Δcgs (Figure 1A).
FIGURE 1.
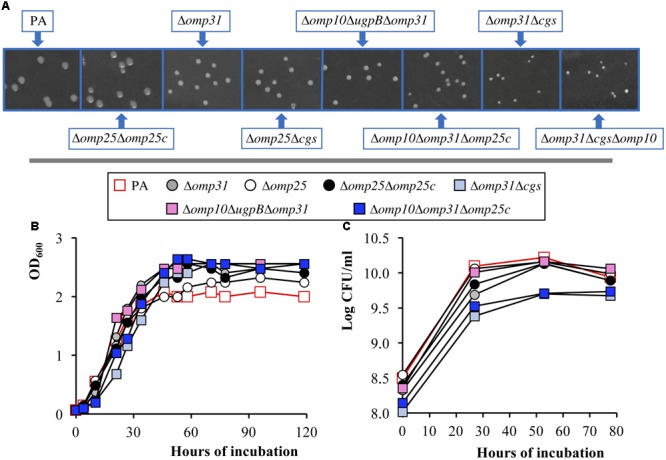
Growth pattern of B. ovis PA and selected mutants in TSA-YE-HS (A) and TSB-YE-HS (B,C). Colony size in TSA-YE-HS (A) was photographed 5 days after inoculation. Plates were taken in the same picture and a section of each strain was extracted to compose the final panel. Growth in TSB-YE-HS liquid medium was determined by the evolution with time of the OD600 values (B) and of the bacterial CFU/ml numbers (C). The SD of three assays, which was always lower than 5% of the mean, is not shown. To simplify the figure, relevant statistically significant differences (P < 0.05) are mentioned in the text.
The Δomp31Δcgs, Δomp10Δomp31Δomp25c, and Δomp10ΔugpBΔomp31 mutants (selected according to the results obtained in the evaluation of virulence described below), the Δomp25Δomp25c mutant (selected because it lacks two major OMPs in B. ovis PA) and their corresponding single mutants were also analyzed regarding growth in TSB-YE-HS liquid medium (Figures 1B,C). Cultures were adjusted to OD600 values of 0.05 and incubated at 37°C under agitation (120 rpm) and a 5% CO2 atmosphere. As expected, according to the correlation OD600-CFU/ml in solid medium previously determined, values of CFU/ml at t0 for mutants with inactivated omp31 or cgs were lower than those of B. ovis PA (P < 0.05), the Δomp31Δcgs and Δomp10Δomp31Δomp25c mutants presenting the lowest values (P < 0.05) (Figure 1C). However, the CFU/ml values of the Δomp31 and Δomp10ΔugpBΔomp31 mutants at the beginning of the stationary phase (approximately t52) were similar to those of B. ovis PA (P > 0.05), which correlates with the higher OD600 scores observed with these mutants by this time (P < 0.05). Although the Δomp31Δcgs and Δomp10Δomp31Δomp25c mutants, those showing the most important growth defects in solid medium (Figure 1A), also had higher OD600 values at this moment (P < 0.05), their CFU/ml counts never reached the maximum level of B. ovis PA (detected at t52) (Figures 1B,C). The Δcgs, Δomp25, Δomp25c-k, and Δomp25Δomp25c mutants had slightly higher OD600 scores in the stationary phase, when compared to the parental strain, but not important differences in the CFU/ml values (Figures 1B,C and data not shown). The Δomp10 and ΔugpB mutants behaved similarly to the parental strain (data not shown).
Virulence in the Mouse Model
For a preliminary assay of virulence, aiming to select the most relevant mutants, mice were inoculated with 106 CFU of the parental strain or its derived mutants and bacteria were enumerated in spleen at W3 and W7 p.i. The multiple cgs mutants were excluded from this first analysis, since the single Δcgs mutant showed poor spleen colonization in a previous study (Martín-Martín et al., 2012). Only those mutants that, when compared to the parental strain, showed bacterial counts lower than 1.5 logarithmic units were considered as probably attenuated and they were later inoculated in a dose of 108 CFU/mouse. Uniquely the Δcgs, Δomp10ΔugpBΔomp31, Δomp10Δomp31Δomp25c, and Δomp31Δomp10Δomp25c mutants met this requirement and, together with the Δcgs multiple mutants, were evaluated with the increased dose (except B. ovis Δomp31Δomp10Δomp25c that bears the same mutations as the Δomp10Δomp31Δomp25c but in a different order). All these mutants were present in spleen at W3 p.i. but the CFU were between 1 and 2.5 log units lower than those obtained with the parental strain (Table 1). All cgs mutants and B. ovis Δomp10Δomp31Δomp25c had similar splenic counts at W7 p.i., which were not drastically different from those observed at W3 p.i., while the Δomp10ΔugpBΔomp31 mutant was not detected in spleen at W7 p.i. (Table 1).
According to these results, the Δomp31Δcgs, Δomp10 ΔugpBΔomp31, and Δomp10Δomp31Δomp25c mutants were retained for a detailed evaluation of virulence and the selection of vaccine candidates. First, to statistically verify the previous results, the spleen colonization was evaluated in five mice per group at W3 and W7 after intraperitoneal infection with 106 CFU (Figure 2A). The spleen weight (Figure 2B) and the levels in serum of IgG reacting against B. ovis PA whole cells (Figure 2C) were also determined. As expected, spleen infection of B. ovis PA was noteworthy at both sampling points – with mean values ranging between log 6 and 7 of CFU/spleen – while the three mutants presented a poor colonization under these conditions (Figure 2A). Spleen infection of the mutants was undetectable in several mice per group, although two out five mice inoculated with the Δomp10Δomp31Δomp25c mutant gave values of about log 5 CFU/spleen at W7 p.i. (Figure 2A). Spleen weight and IgG titers in serum were also significantly higher (P < 0.05) in mice inoculated with the parental strain (Figures 2B,C).
FIGURE 2.
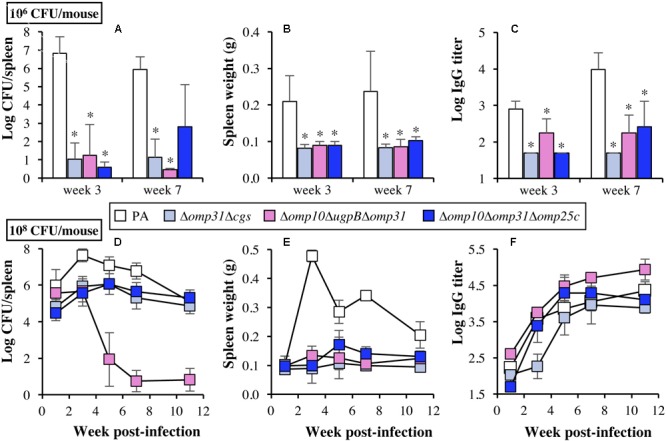
Bacterial spleen colonization (A,D), spleen weight (B,E), and IgG response in serum against B. ovis PA whole cells (C,F) in BALB/c mice. Mice (n = 5 per group and time point) were inoculated intraperitoneally with 106 (upper panels) or 108 (lower panels) CFU/mouse of B. ovis PA or Δomp31Δcgs, Δomp10ΔugpBΔomp31, and Δomp10Δomp31Δomp25c mutants. Statistically significant differences in upper panels, compared to parental B. ovis PA, are marked with an asterisk. To simplify the figure, relevant statistically significant differences in lower panels (P < 0.05) are mentioned in the text. Detection limit for antibody titers is log 1.7. As a reference for panels B and E, spleen weight of mice inoculated with PBS (n = 5) for the protection experiment described in Figure 8 was 0.096 ± 0.005 (W7 post-inoculation).
The same parameters described above were evaluated at several time points over a 11-week period in mice inoculated with 108 CFU of each strain (Figures 2D–F), the dose usually employed for evaluation of attenuated strains as vaccines (Sancho et al., 2014; Soler-Lloréns et al., 2014; Silva et al., 2015b). The parental strain showed high levels of spleen infection through the entire experiment with mean log CFU/spleen values of 5.99 at W1 p.i. and a peak at W3 p.i. of 7.64. B. ovis PA counts decreased thereafter but were still high at the end of the experiment (mean log CFU/spleen values of 5.01 at W11) (Figure 2D). The Δomp31Δcgs and Δomp10Δomp31Δomp25c mutants displayed lower levels of splenic infection at W1, with mean CFU/spleen values 1–1.5 log lower than those of the parental strain (P < 0.05). However, although their maximum CFU/spleen scores (about log 6 at W3–W5 p.i.) did never reach those of B. ovis PA (log 7.64 at W3 p.i.), they persisted in spleen with similar values to those of the parental strain at W11 p.i. (P > 0.05) (Figure 2D). The spleen colonization of the Δomp10ΔugpBΔomp31 mutant did not show statistically significant differences with the parental strain at W1 p.i., but the bacterial counts did not increase at W3 p.i. and decreased thereafter until the detection limit of infection at W7 p.i. (P < 0.05) (Figure 2D).
At W1 p.i., the spleen weight in mice infected with 108 CFU of the parental strain (0.102 ± 0.030 g) was analogous to that currently obtained with mice inoculated with PBS and that was also observed in the PBS group used to determine the production of cytokines by splenocytes in the experiment described below (0.096 ± 0.005 g) (P > 0.05). However, a prominent splenomegaly was detected at W3 p.i. (0.478 ± 0.020 g/spleen) which, although decreased after W3 p.i., persisted until the end of the experiment (0.205 ± 0.046 g/spleen) (P < 0.05) (Figure 2E). On the contrary, the B. ovis mutants did never reach mean values of 0.200 g/spleen (Figure 2E). At W1 p.i., all groups of mice had low levels of IgG reactive with whole cells of B. ovis PA. An important increase of the antibody response was detected at W3 p.i. in all groups (P < 0.05), except in mice vaccinated with the Δomp31Δcgs mutant (P > 0.05). At W5 p.i. and thereafter, mice inoculated with the parental strain or the Δomp31Δcgs and Δomp10Δomp31Δomp25c mutants displayed IgG titers of about log 4, while mice inoculated with B. ovis Δomp10ΔugpBΔomp31 gave titers in the order of 0.5 logarithmic units higher (P < 0.05) than those detected in the B. ovis PA group (Figure 2F).
Autoagglutination and Susceptibility Assays
In addition to the three selected mutants, other relevant mutants were included in the remaining studies by their interest to increase the knowledge about the B. ovis cell envelope. Several tests related to the OM properties of Brucella spp. (i.e., autoagglutination and resistance to polymyxin B and sodium deoxycholate) (Martín-Martín et al., 2011; Vizcaíno and Cloeckaert, 2012) were performed. In the autoagglutination assay, most of the single mutants behaved as the B. ovis PA parental strain (P > 0.05), remaining in suspension until the end of the experiment (Figure 3 and data not shown). The exceptions were B. ovis Δomp31 – with %OD600 values of about 30 and 10% after 24 and 48 h of static incubation, respectively – and the Δcgs and Δomp25c-k mutants, whose %OD600 values gradually decreased until 70% at the end of the experiment (P < 0.05 when compared to the parental strain) (Figure 3). The omp31 multiple mutants behaved similarly to the single mutant, although the Δomp10Δomp31Δomp25c and Δomp31Δomp10Δomp25c mutants agglutinated more quickly (they were almost completely settled after 10 h) (P < 0.05) (Figure 3). B. ovis Δomp25Δcgs behaved like the single Δcgs mutant while B. ovis Δomp25Δomp25c did not show differences with the parental strain (P > 0.05) (Figure 3).
FIGURE 3.
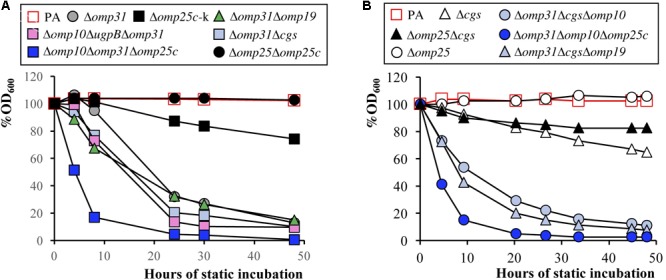
Autoagglutination assay with bacterial suspensions of initial OD600 readings of 0.8 (100% OD600). The percentage of the OD600 values was determined over 48 h of static incubation. Panels A and B represent results of two independent experiments. The SD of three assays, which was always lower than 5% of the mean, is not shown. To simplify the figure, relevant statistically significant differences (P < 0.05) are mentioned in the text.
Brucella ovis ΔbacA and the B. ovis ΔbacAΔomp31 double mutant were more resistant than B. ovis PA to polymyxin B exposure for 80 min (Table 4). On the contrary, B. ovis Δomp25Δomp25c, and the Δomp31 single and multiple mutants – except the previously mentioned ΔbacAΔomp31 mutant – showed higher susceptibility to polymyxin B than the parental strain (Table 4). The lowest survival percentages were obtained with the Δomp10Δomp31Δomp25c and Δomp31Δomp10Δomp25c mutants (less than 10% survival). Only the Δomp31Δcgs double mutant and its derived Δomp31ΔcgsΔomp10 and Δomp31ΔcgsΔomp19 triple mutants were more susceptible to sodium deoxycholate than the parental strain (about 30% survival versus 86% survival, respectively) (Table 4).
Table 4.
Susceptibility of B. ovis PA and selected mutants to polymyxin B and Na deoxycholate.
| % Survival after exposure toa: |
||
|---|---|---|
| Brucella ovis strainsb | Polymyxin B (1 mg/ml) | Na deoxycholate (0.1 mg/ml) |
| B. ovis PA | 69.03 6.12 | 86.25 5.74 |
| Single mutants | ||
| Δomp31 | 27.47 3.04* | 83.74 3.09 |
| Δomp25 | 65.84 7.17 | 92.94 4.06 |
| Δomp25c-k | 65.99 4.72 | 95.89 7.15 |
| Δcgs | 77.96 11.80 | 79.82 9.07 |
| ΔbacA | 84.10 5.49* | 87.38 8.28 |
| Δomp10 | 62.04 3.46 | 91.20 4.22 |
| Δomp19 | 67.27 7.05 | 83.19 7.75 |
| ΔugpB | 77.59 11.67 | 90.66 9.42 |
| Double mutants | ||
| Δomp31Δomp19 | 25.68 2.94* | 90.49 7.79 |
| Δomp31ΔugpB | 20.66 0.57* | 92.65 3.64 |
| Δomp31Δcgs | 31.20 14.30* | 32.14 1.27* |
| Δomp25Δcgs | 61.71 7.30 | 92.35 5.18 |
| Δomp25Δomp25c | 45.23 5.10* | 91.36 9.59 |
| Δomp10Δomp31 | 33.31 4.58* | 84.66 6.36 |
| Δomp10ΔugpB | 64.22 5.36 | 77.93 9.26 |
| ΔbacAΔomp31 | 88.06 18.61* | 86.90 5.51 |
| Triple mutants | ||
| Δomp10ΔugpBΔomp31 | 30.50 9.94* | 88.52 6.22 |
| Δomp10Δomp31Δomp25c | 1.65 0.36* | 76.74 9.37 |
| Δomp31Δomp10Δomp25c | 7.19 4.38* | 80.40 7.49 |
| Δomp31ΔcgsΔomp10 | 37.84 9.43* | 33.63 1.78* |
| Δomp31ΔcgsΔomp19 | 33.30 3.68* | 34.39 2.94* |
aResults are expressed as the mean ± SD of three assays. Statistically significant differences (P < 0.05), compared to the parental strain, are marked with an asterisk. bSee legend to Figure 1 for strain characteristics.
Mapping of Cell Envelope Antigens
The reactivity of the B. ovis mutants with MAbs raised against Brucella S-LPS, R-LPS, PG, major and minor OMPs, and periplasmic BP26 was analyzed by iELISA (Figure 4 and data not shown). As expected, the MAb-specific against S-LPS did not react with any mutant or with the parental strain (data not shown) and omp31, omp25, omp10, or omp19 single and multiple mutants did not react with the respective MAbs (see Figure 4 for representative results). No relevant differences were observed between the parental strain and the mutants regarding reactivity with MAbs specific against Omp31 (except with mutants lacking omp31), Omp10 (except with mutants lacking omp10), Omp16, Omp2b, PG, and R-LPS. On the contrary, all omp31 mutants showed a stronger reaction with MAb C09 (specific against Omp25) and omp19 mutants showed a stronger reaction with MAb B02, specific against BP26 (Figure 4, upper panels and data not shown). Accordingly, several omp31 mutants were tested in iELISA with three MAbs specific against Omp25, while omp19 mutants were tested with four anti-BP26 MAbs. All the Δomp31 and Δomp19 mutants showed a stronger reactivity with the anti-Omp25 and anti-BP26 MAbs, respectively (Figures 4G,H), which confirmed the results obtained in the previous analysis.
FIGURE 4.
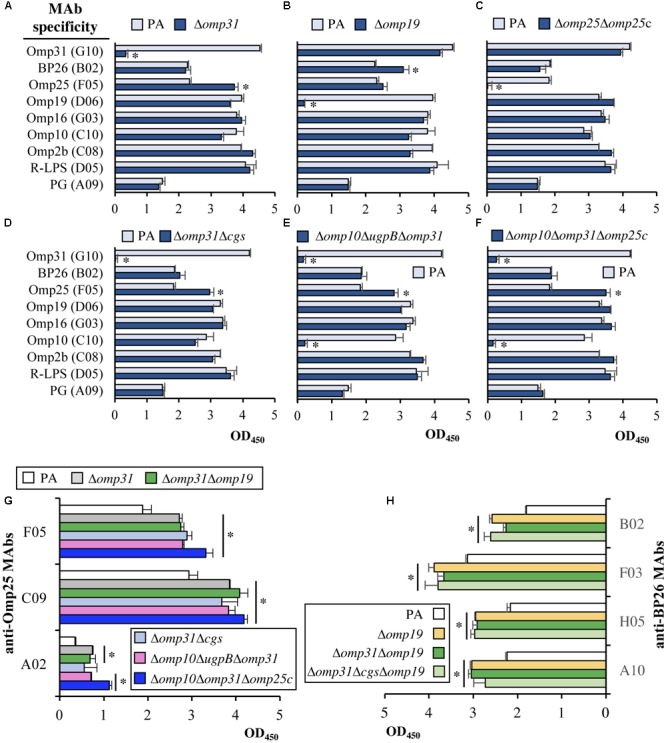
Reactivity by iELISA of MAbs against cell envelope antigens with B. ovis PA and selected mutants. The reactivity of MAbs specific against PG, R-LPS, the BP26 periplasmic protein, the Omp2b, Omp31, and Omp25 major OMPS, and the Omp10, Omp16, and Omp19 OM lipoproteins (A–F) was tested with most of the mutant strains obtained in this work. Representative results are shown in panels A–F and others are commented in the text. Differences between the mutant strains and the parental strain were considered relevant for further analysis when, in addition to being statistically significant (P < 0.05), the mean values differed more than 25% (marked with an asterisk). Reactivity of Δomp31 single and multiple mutants with several anti-Omp25 MAbs (G), and of Δomp19 mutants with several anti-BP26 MAbs (H), is also shown. Statistically significant differences between the mutant strains and the parental strain (P < 0.05) in panels G and H, are marked with an asterisk.
The protein profile of the most relevant strains was evaluated by SDS–PAGE followed by Coomassie blue straining (Figure 5A). Proteins were also transferred to nitrocellulose to assess the reactivity with sera raised against the proteins of the Omp25/Omp31 family. A serum against Omp25c was used to detect both Omp25c and Omp25 since these proteins display cross-reacting epitopes (Martín-Martín et al., 2009), while the detection of Omp31 was performed with a serum raised against Omp31b (an OMP absent in B. ovis) that strongly cross-react with Omp31 (Martín-Martín et al., 2009). Omp25d and Omp22 were detected by reactivity with their respective anti-sera (Martín-Martín et al., 2009).
FIGURE 5.
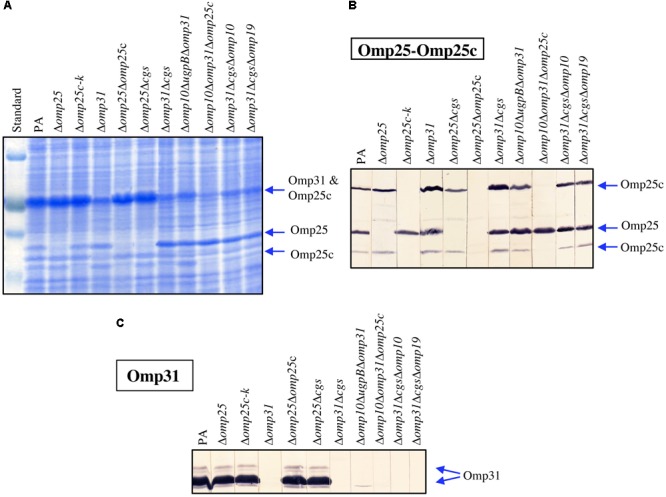
Profiles of B. ovis PA and selected mutants in SDS–PAGE (A) and immunoblot with sera of rabbits immunized with purified recombinant Omp25c (B) or Omp31b (C). The prestained protein marker VI (Applichem-Panreac) is shown in (A) and positions of known OMP bands are marked with arrows (A–C).
A multiple band pattern, dependent on the electrophoretic conditions, is frequently observed in SDS–PAGE for Omp25, Omp25c, and Omp31 (Cloeckaert et al., 1990; Martín-Martín et al., 2009). Two protein bands corresponding to Omp25c were detected by immunoblot in parental B. ovis PA and were absent in all omp25c mutants (Figure 5B). Additionally, the intensity of the upper band in omp31 mutants was higher than that observed with the parental strain (Figure 5B). Absence of the lower band of Omp25c in the Δomp25c mutants was also evident in the SDS–PAGE gel (Figure 5A, lanes Δomp25c-k, Δomp25Domp25c, and Δomp10Δomp31Δomp25c) while the absence of the upper band was only apparent in the Δomp10Δomp31Δomp25c triple mutant (Figure 5A). This result can be explained by the fact that this mutant also lacks Omp31, which is a major OMP that exhibits an electrophoretic mobility similar to that of Omp25c (Figure 5A) and, consequently, masks the absence of the upper Omp25c band in the other Δomp25c mutants.
Omp25 was detected between the two Omp25c bands except in the omp25 mutants. Absence of Omp25 in these mutants was evidenced in both the SDS–PAGE gel (Figure 5A) and the immunostained nitrocellulose (Figure 5B) (see Δomp25, Δomp25Δcgs, and Δomp25Δomp25c lanes in comparison with PA lanes). With both techniques, the Omp25 band of the Δomp31 mutants was more intense than in B. ovis PA. Omp31 was detected in immunoblot and with the same intensity in all the strains, except those lacking the encoding gene (Figure 5C). Absence of Omp31 in these latter mutants was also evident in the SDS–PAGE gel (Figure 5A). The Omp22 band was revealed in B. ovis PA and the analyzed mutants, while Omp25d – that has only been observed in a complemented mutant overexpressing the protein (Martín-Martín et al., 2009) – was not detected (data not shown).
Cellular Models of Infection
The intracellular behavior of B. ovis PA and the most relevant mutants was evaluated in J774.A1 murine macrophages and HeLa cells. No significant differences between strains (P > 0.05) were observed regarding internalization in J774.A1 macrophages (Figure 6, upper panels). However, although the intracellular bacterial numbers decreased at t20 with all the strains, the reduction was more pronounced with strains Δomp25Δomp25c (Figure 6A), Δomp25Δcgs, Δomp31Δcgs, and its derived triple mutants (Figure 6C), and with B. ovis Δomp10ΔugpBΔomp31 and Δomp10Δomp31Δomp25c (Figure 6B) (P < 0.05). After this moment, all strains were able to replicate, reaching intracellular CFU numbers at t44 about 1 log unit higher than those detected at t20 for each strain (P < 0.05) (Figure 6, upper panels).
FIGURE 6.
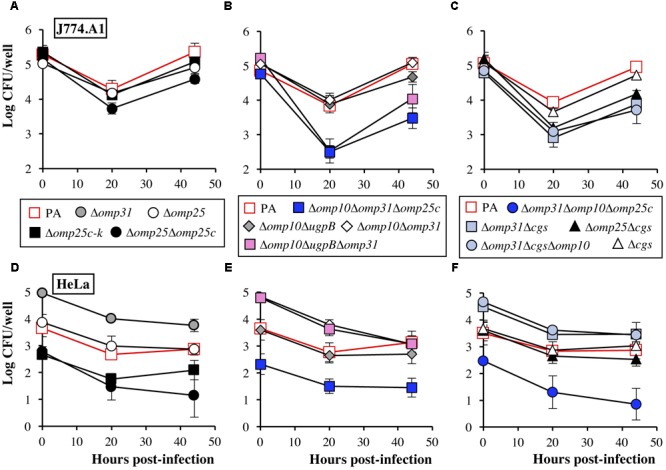
Internalization and intracellular behavior of B. ovis PA and selected mutants in J774.A1 murine macrophages (A–C) and HeLa cells (D–F). The results are expressed as the means ± SD (n = 3) of the log CFU/well at each time point. To simplify the figure, relevant statistically significant differences (P < 0.05) are mentioned in the text.
In HeLa cells, the Δomp25c mutants (Δomp25c-k, Δomp25Δomp25c, Δomp10Δomp31Δomp25c, and Δomp31 Δomp10Δomp25c) showed a deficient internalization, with intracellular CFU at t0 in the order of 1–1.5 log units lower than those determined for B. ovis PA (P < 0.05). On the contrary, all mutants bearing the omp31 deletion, except those with the Δomp25c genotype (B. ovis Δomp10Δomp31Δomp25c and Δomp31Δomp10Δomp25c), showed an increased internalization in HeLa cells, with CFU at t0 about 1 log higher than those of the parental strain (P < 0.05) (Figure 6, lower panels). All strains suffered a reduction (P < 0.05) of intracellular CFU a t20 while at t44 the bacterial numbers were, in general, similar to those obtained at t20 (Figure 6, lower panels).
Searching for an explanation to the internalization differences observed in HeLa cells, the Δomp25c-k mutant complemented with the wild-type gene and the Δomp31-k mutant together with its complemented strain (Caro-Hernández et al., 2007) were also analyzed regarding internalization in HeLa cells (CFU determined at t0) and their behavior in immunoblot with sera reacting with Omp31 and Omp25c (Figure 7). Complementation of the Δomp25c-k mutant restored its ability to enter HeLa cells like the parental strain (Figure 7A) and the production of Omp25c (Figures 6B,C). The complemented Δomp31-k mutant recovered the ability to produce Omp31, but its level was lower than that of the parental strain (Figures 7B,D). This fact was concomitant with a higher intensity of Omp25c bands, when compared to B. ovis PA that, however, did not reach the intensity observed with the Δomp31 and Δomp31-k mutants (Figure 7C). Internalization of the complemented Δomp31-k mutant in HeLa cells was similar to that of the Δomp31 mutant and significantly higher than that observed with the parental strain (Figure 7A).
FIGURE 7.
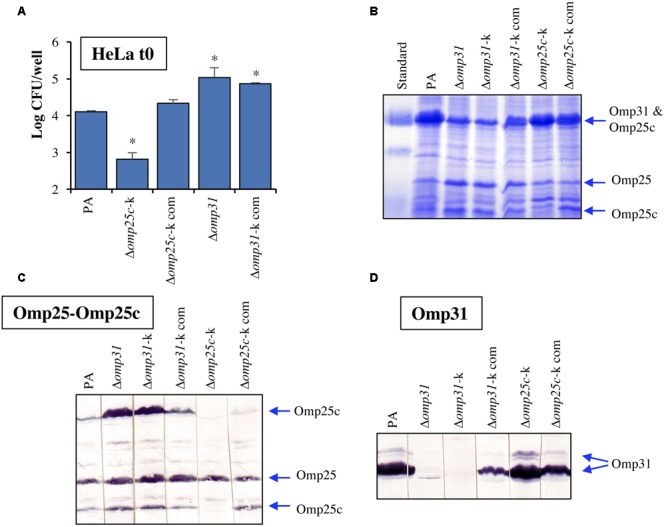
(A) Internalization in HeLa cells of parental B. ovis PA, the Δomp25c and Δomp31 mutants and mutant strains complemented with wild-type omp25c or omp31. SDS–PAGE profiles (B) and immunoblot detection of Omp25, Omp25c (C), and Omp31 (D) in the same strains. Strains Δomp31-k and Δomp25c-k were previously obtained by replacement of omp31 or omp25c by a kanamycin-resistance cassette (Caro-Hernández et al., 2007). They were complemented with the corresponding wild-type genes to give strains identified as Δomp31-k com and Δomp25c-k com, respectively (Caro-Hernández et al., 2007). The prestained protein marker VI (Applichem-Panreac) is shown in (B) and positions of known OMP bands are marked with arrows. Statistically significant differences when compared to the parental strain are marked with an asterisk.
Efficacy as Vaccine of B. ovis Δomp10ΔugpBΔomp31
The usefulness as vaccine of B. ovis Δomp10ΔugpBΔomp31 against B. ovis PA was evaluated in mice and compared to that of the B. melitensis Rev1 heterologous vaccine. Spleen colonization of the challenge strain was evaluated 3 weeks after infection (Figure 8F) and the humoral (Figure 8E) and cellular (Figures 8A–D) immune response was evaluated at the time point selected for the challenge with the virulent strain (W7 post-vaccination).
FIGURE 8.
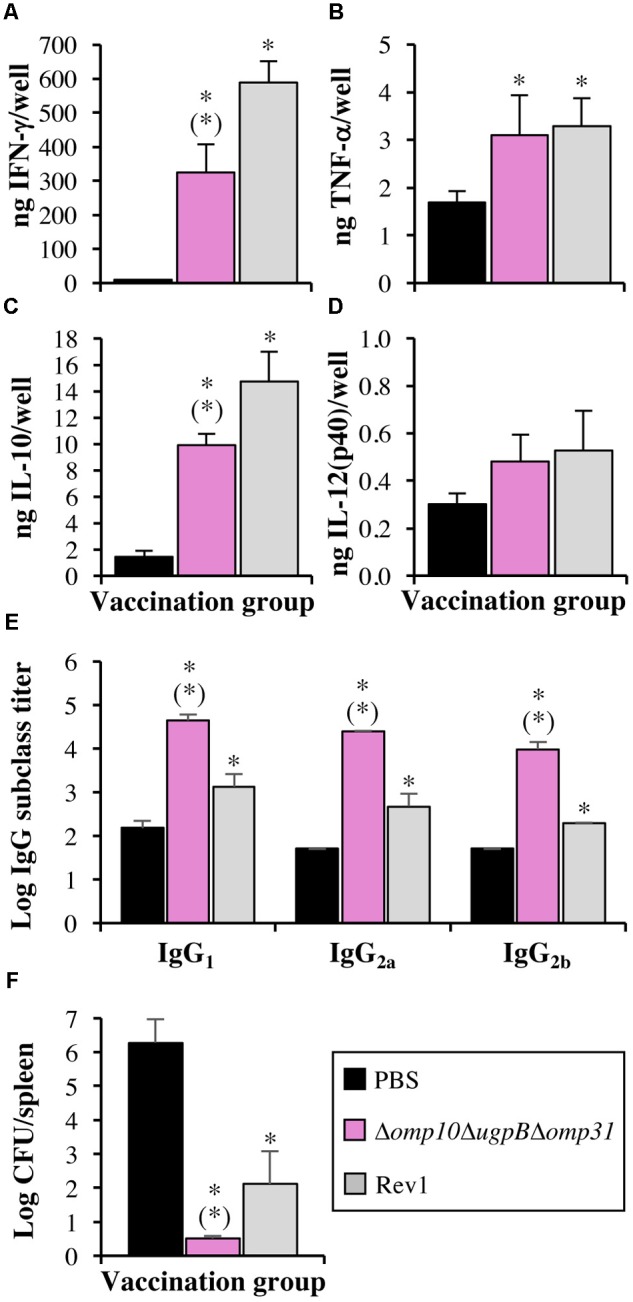
Cytokine secretion by stimulated splenocytes (A–D), levels in serum of IgG subclasses reactive with B. ovis PA whole cells (E), and protective efficacy against B. ovis PA infection (F) in mice vaccinated with B. ovis Δomp10ΔugpBΔomp31 or B. melitensis Rev1 and in mice inoculated with PBS. Splenocytes (A–D) and sera (E) were obtained 7 weeks after inoculation (five mice per group). At that time, five mice per group were inoculated with virulent B. ovis PA and bacterial colonization of spleen was determined 3 weeks later to evaluate the protective efficacy of the vaccines (F). Splenocytes were stimulated with heat-killed B. ovis PA whole cells for 72 h (A–D) before the evaluation of secreted cytokines. Statistically significant differences, compared to mice inoculated with PBS, are marked with an asterisk. Statistically significant differences of the Δomp10ΔugpBΔomp31 group compared to the B. melitensis Rev1 group are marked with an asterisk in brackets.
Weight of spleens obtained at W7 post-vaccination from mice inoculated with the Δomp10ΔugpBΔomp31 mutant (0.104 ± 0.012 g) did not show statistically significant differences with that of mice inoculated with PBS (0.096 ± 0.005 g), while splenomegaly (P < 0.05) was detected in mice vaccinated with B. melitensis Rev1 (0.136 ± 0.011 g) (data not shown). Additionally, B. melitensis Rev1 was detected in spleen (log CFU/spleen values of 3.66 ± 0.07), while the Δomp10ΔugpBΔomp31 mutant was cleared by this time (data not shown).
Splenocytes were stimulated with B. ovis PA whole cells to evaluate, by specific ELISA tests, the production of cytokines. Splenocytes of mice inoculated with PBS secreted limited amounts of IFN-γ, while about 30 and 60 times higher levels (P < 0.05) were detected in the groups vaccinated with the Δomp10ΔugpBΔomp31 mutant and B. melitensis Rev1, respectively (Figure 8A). In a lesser extent, splenocytes from vaccinated mice also produced more IL-10 (P < 0.05) than those obtained from mice of the PBS group (mean values of 14.7, 9.9, and 1.4 ng/well for the B. melitensis Rev1, B. ovis Δomp10ΔugpBΔomp31, and PBS groups, respectively) (Figure 8C). Regarding the production of TNF-α, splenocytes from mice vaccinated with the two Brucella attenuated strains secreted about double amounts (mean values of 3 ng/well) than those obtained from the unvaccinated control group (P < 0.05) (Figure 8B). No statistically significant differences were observed between groups concerning the production of IL-12(p40) (Figure 8D).
At the time of challenge, mice inoculated with PBS 7 weeks earlier showed low titers (close to the detection limit of log 1.7) of serum antibodies of the IgG1, IgG2a, and IgG2b subclasses able to react with B. ovis PA whole cells in iELISA (Figure 8E). On the contrary, mice vaccinated with B. ovis Δomp10ΔugpBΔomp31 had titers of the three IgG subclasses (ranging from log 3.99 ± 0.16 for IgG2b to log 4.65 ± 0.13 for IgG1) that were about 1.5 log units higher (P < 0.05) than those detected in mice vaccinated with the B. melitensis Rev1 heterologous vaccine. Within each vaccination group, no relevant differences were observed between the three IgG subclasses (Figure 8E).
The homologous B. ovis Δomp10ΔugpBΔomp31 vaccine was not detected in spleen 3 weeks after the experimental challenge while it prevented spleen colonization of virulent B. ovis PA (CFU/spleen counts were under the detection limit) (Figure 8F). When compared to control mice inoculated with PBS, vaccination with heterologous B. melitensis Rev1 also drastically reduced the spleen colonization of B. ovis PA (Figure 8F), but both the vaccine strain and the challenge strain were detected in some mice.
Discussion
With the aim to increase our knowledge about the OM and virulence of B. ovis and to develop a specific homologous attenuated vaccine, multiple deletion mutants targeting nine genes related to the OM were obtained and characterized. Two of these genes encode Omp10 and Omp19, two lipoproteins that are conserved in Brucella (Cloeckaert et al., 1990; Tibor et al., 1994, 1996) and have orthologs in other phylogenetically related bacteria able to establish interactions with eukaryotic cells either as symbionts or as pathogens (e.g., Ochrobactrum, Bartonella, Rhizobium, Ensifer, Sinorhizobium) (Cloeckaert et al., 1999; Barquero-Calvo et al., 2009). This observation suggests their relevant role for the bacterium and, in fact, both lipoproteins have been linked to the virulence of B. abortus 544 (Tibor et al., 2002). Surprisingly, the individual absence of Omp10 and Omp19 in the OM of B. ovis PA did not affect its virulence or modified notably its OM-related properties (Sidhu-Muñoz et al., 2016). On the contrary, the multiple assays we have performed to achieve the simultaneous deletion of omp10 and omp19 in B. ovis PA were unsuccessful. Since OM lipoproteins are thought to anchor the OM through the lipid moiety and contribute to the stability of the cell envelope (Goolab et al., 2015), Omp10 and Omp19 could have an interchangeable role necessary to maintain the OM integrity. Nevertheless, the amount of Omp10 in B. ovis Δomp19 and the amount of Omp19 in the Δomp10 mutant does not seem to be increased (Figure 4B and data not shown), although the absence of one of these lipoproteins, in the presence of the other, could also be compensated by an increase in other cell envelope proteins. In this respect, a higher reactivity with MAbs against BP26 suggestive of increased levels of this periplasmic protein was detected in the mutants bearing the omp19 deletion (Figure 4H). Contrasting with the impossibility to obtain the double Δomp10Δomp19 mutant, the simultaneous absence of major Omp31 together with Omp10 or Omp19 lipoprotein was well tolerated in B. ovis PA, since the respective double mutants do not have prominent defects in virulence (Table 1) and the other phenotypic changes are associated to the deletion of omp31 (Table 4, Figures 3, 5, 6, and data not shown). Similarly, deletion of ugpB on the Δomp10 or Δomp19 genetic background did not affect the virulence (Table 1), and the alterations detected after deletion of cgs on the same mutants resembled those detected with the B. ovis Δcgs single mutant (Table 1).
Other target surface antigens were Omp31, Omp25, and Omp25c that are abundant OMPs in B. ovis (Cloeckaert et al., 2002; Martín-Martín et al., 2009; Vizcaíno and Cloeckaert, 2012) and the main members of the Omp25/Omp31 family that is constituted by seven homologous OMPs (Salhi et al., 2003; Vizcaíno et al., 2004; Vizcaíno and Cloeckaert, 2012). Orthologs of this family of OMPs have been found in Bartonella quintana (Minnick et al., 2003) and other Rhizobiales (e.g., Rhizobium leguminosarum, Agrobacterium tumefaciens, or O. anthropi), which suggests that the redundancy of these OMPs provides an advantage that could be related with the compensatory effect between these proteins previously reported (Salhi et al., 2003; Caro-Hernández et al., 2007; Martín-Martín et al., 2009; Vizcaíno and Cloeckaert, 2012). This compensatory effect is in accordance with the apparent increase of Omp25 and Omp25c that was observed with all omp31 mutants, except those also lacking omp25c where Omp25c was not detected (Figures 4, 5). On the contrary, omp25 and omp25c mutants and even the double Δomp25Δomp25c mutant did not show higher levels of Omp31 (Figures 4, 5). In addition, this latter double mutant did not suffer important defects regarding in vitro growth (Figure 1) or virulence in mice (Table 1). These observations could be indicative of a more relevant role for Omp31, whose absence in the OM would require a compensation with another paralog that would not be necessary when Omp25 or Omp25c are missing.
However, considering that our attempts to obtain a B. ovis PA double mutant defective in Omp31 and Omp25 were fruitless, it seems probable that the presence in the bacterial surface of one of these two major OMPs is a requirement at least for in vitro survival. This assertion is in contradiction with the availability of Δomp25 mutants in B. abortus (Edmonds et al., 2002; Manterola et al., 2007), a Brucella species naturally lacking Omp31 due to a 25-kb deletion involving its encoding gene (Vizcaíno et al., 2001a). Nevertheless, unlike B. ovis where omp31b is a pseudogene (Vizcaíno et al., 2004), B. abortus strains synthetize Omp31b (Martín-Martín et al., 2009), an OMP sharing 67% of amino acid identity with Omp31 (Salhi et al., 2003) and that could compensate for the absence of Omp31 in this species. Research aiming to construct the Δomp25Δomp31b double mutant in the B. abortus genetic background would help to a better understanding of the relationships between the members of the Omp25/Omp31 family.
According to the availability of the Δomp10Δomp31Δomp25c and Δomp31Δomp10Δomp25c mutants (with the same deleted genes in a different order), B. ovis PA survives to the simultaneous absence of Omp10 lipoprotein and major Omp31 and Omp25c, together with the natural lack of Omp31b and Omp25b (Martín-Martín et al., 2009). However, both mutants suffered important phenotypic changes, such as strong in vitro growth defects, quick autoagglutination, remarkable susceptibility to polymyxin B, increased killing by murine macrophages in the first 20 h of infection, defective internalization in HeLa cells, and limited spleen colonization in mice inoculated with 106 CFU. Nevertheless, not only both strains replicate in murine macrophages after 20 h of infection (Figure 6B and data not shown) but at least B. ovis Δomp10Δomp31Δomp25c can establish a chronic infection in mice, with important levels of spleen colonization, when inoculated at 108 CFU/mouse (Figure 2D).
Another finding regarding the Omp25/Omp31 family, and that was further confirmed by confocal microscopy (data not shown), is that all omp25c mutants are impaired for internalization in HeLa cells (Figures 6D–F), while all strains bearing the Δomp31 mutation (except the triple mutants also bearing the omp25c deletion) internalize more efficiently than the parental strain (Figures 6D–F and data not shown). Since immunoblot assays showed that Δomp31 mutants had higher levels of Omp25c, we postulated that Omp25c is involved in the internalization of B. ovis PA in HeLa cells. This hypothesis is supported by the immunoblots and HeLa cells assays performed with omp31 and omp25c mutant and complemented strains (Figure 7). Whether this characteristic is specific for HeLa cells, for human cells or for non-professional phagocytes independently of the animal species remains to be studied.
As expected (Martín-Martín et al., 2012) the Δcgs mutant did not suffer drastic changes in the in vitro characteristics, but was the only single mutant that was attenuated in the murine model when it was inoculated at 106 CFU/mouse (Table 1). However, a 100-fold increase of the inoculation dose led to high splenic infection levels at both the acute and chronic phase of infection (Table 1) and to a kinetics of spleen colonization resembling that observed in mice experimentally infected with 106 CFU of the parental strain (data not shown; see the equivalent spleen colonization profile of the Δomp31Δcgs mutant in Figure 2). According to the similarities between the parental strain and the Δcgs mutant regarding their behavior in the cellular models of professional and non-professional phagocytes used in this work (Figures 6C,F) the attenuation of the Δcgs mutant of B. ovis PA does not seem to be due to killing inside phagocytic cells. However, since we have used individual cell lines, a defective interaction with one or more types of professional and/or non-professional phagocytes of BALB/c mice cannot be discarded. In fact, Δcgs mutants of B. abortus 2308 and S19 are defective in both HeLa cells (Briones et al., 2001; Arellano-Reynoso et al., 2005; Roset et al., 2006, 2014) and peritoneal macrophages of C57BL/6 mice (Arellano-Reynoso et al., 2005) but, on the contrary, they behave as the parental strains in bone marrow cells also obtained from C57BL/6 mice (Salcedo et al., 2008; Roset et al., 2014). Another possible explanation for the attenuation of B. ovis Δcgs could be related to the association that has been established between CβGs and a dual pro- and anti-inflammatory response that transiently recruits neutrophils and suggests a controlled local inflammatory response (Degos et al., 2015). This controlled inflammatory response might be important for the establishment of the Brucella infection, since survival and replication inside phagocytes is a characteristic trait of Brucella that provides the bacterium with a safe environment. A strong inflammatory process would trigger a detrimental immune response against the pathogen but, on the contrary, a diminished inflammatory response in the absence of CβGs would reduce the presence of suitable target cells for Brucella replication and, therefore, could influence the outcome of infection.
The Δomp31Δcgs double mutant, defective in a major OMP and in periplasmic CβGs, maintained or exacerbated the most severe phenotype of the corresponding single mutants. Thus, the susceptibility to DOC (Table 4) and in vitro growth defects were more prominent in the Δomp31Δcgs mutant than in the single mutants, which would be in accordance with cell envelope modifications compromising the permeability of the bacterial cell to nutrients and toxic compounds. Strikingly, despite its important in vitro growth impairment and its increased killing during the first stage of infection in murine macrophages (Figure 6C), the Δomp31Δcgs mutant behaved similarly to the Δcgs mutant in the mouse model, except for a lower spleen colonization at W3 p.i. (Table 1, Figure 2, and data not shown). Additionally, a third deletion of omp10 or omp19 did not produce apparent differences with respect to the Δomp31Δcgs mutant in any of the characteristics evaluated and attenuation of the Δcgs mutant was not drastically intensified by the deletion of omp10, omp19, bepC, ugpB, or omp25 (Table 1).
Similarly to B. ovis Δomp31Δcgs, the Δomp10Δomp31 Δomp25c mutant suffered a higher decrease of the intracellular CFU during the first 20 h of macrophage infection but both mutants multiplied properly after that moment (Figures 6B,C). This ability to replicate intracellularly might explain why, despite their impairment to infect mice at doses of 106 CFU/mouse, they persisted at least until week 11 p.i. – with splenic counts equivalent to those of the parental strain at that moment – when 108 CFU were inoculated (Figure 2). Increased killing inside professional phagocytes at the onset of infection could have dramatic effects on the outcome of infection when the lower dose of infection is used. However, higher doses of infection would allow some bacteria to escape to the bactericidal mechanisms of phagocytes and reach the replicative niche to establish a persistent infection thereafter. At least in part, the higher reduction of intracellular CFU at t20 could be related to a defective growth inside the phagocytes – mimicking that observed in vitro for both mutants (Figure 1) – and that might be due to alterations in OM permeability to nutrients and/or toxic compounds. On the other side, both mutants showed an opposed internalization pattern in HeLa non-professional phagocytes (Figures 6E,F) that does not correlate with their similar attenuation in mice. Since, in addition, other single and multiple mutants with analogous profiles in HeLa cells were not attenuated in mice (e.g., B. ovis Δomp10Δomp31 and Δomp25Δomp25c) (Table 1 and Figure 6), an altered behavior in HeLa cells of a B. ovis mutant does not imply attenuation in the mouse model.
The Δomp10ΔugpBΔomp31 mutant behaved in macrophages similarly to the Δomp31Δcgs and Δomp10Δomp31Δomp25c mutants (Figure 6B). However, although during the first 3 W p.i. the three mutants showed a similar pattern of spleen colonization in mice (Figure 2), the Δomp10ΔugpBΔomp31 mutant was progressively cleared from spleen thereafter (Figure 2D). Therefore, additional defects, preventing the establishment of a chronic infection, must be present in the Δomp10ΔugpBΔomp31 mutant. It must be noted that after W5 p.i., concomitantly with the splenic dampening of CFU, the antibody response induced by B. ovis PA Δomp10ΔugpBΔomp31 was higher than that observed in mice infected with the parental strain (Figure 2F) (P < 0.05), despite the higher spleen colonization of the latter (Figure 2A). This increased humoral immunity might contribute to the clearance of the mutant but, at the same time, be a relevant characteristic for an attenuated vaccine. Clearing of the mutant after W3 p.i. could also be related with defects impairing its colonization and/or survival inside reservoir cells involved in sustaining the chronic phase of infection. In the case of B. abortus, alternatively activated macrophages – more abundant during the chronic phase of infection – have been described as preferential target cells for survival and replication in mice (Xavier et al., 2013). This preference has been related with an increase in glucose availability in these cells (Xavier et al., 2013), which adds new evidence about the relevant role of metabolism in the acute phase of infection and/or in sustaining persistent Brucella infection (Hong et al., 2000; Ronneau et al., 2016; Barbier et al., 2018). Since transport through membranes of metabolic substrates, metals, and other compounds is a key mechanism for bacterial homeostasis, the changes in the bacterial surface suffered by the Δomp10ΔugpBΔomp31 mutant might lead to an altered transport of essential molecules that could impair survival in the reservoir cells. However, these changes would not have dramatic effects under in vitro growth conditions, at least in the culture medium used in this work (Figure 1).
According to its attenuation profile, the Δomp10ΔugpB Δomp31 mutant was considered the best candidate to evaluate its vaccine properties against in the mouse model. Three weeks after the challenge, when B. ovis PA reaches its peak of infection in unvaccinated mice (Figure 2), neither the virulent strain nor the vaccine strain were detected in spleen, while both strains were detected – although in low levels – in some mice vaccinated with B. melitensis Rev1 (Figure 8F). The protective activity of B. ovis Δomp10ΔugpBΔomp31 was accompanied by a good antibody response against B. ovis PA whole cells that was significantly higher than that obtained with B. melitensis Rev1 by the time of challenge (Figure 8F) and even higher (P < 0.05) than that observed in mice infected with the parental strain (Figure 2F). The development of specific antibodies is critical for the protective activity against B. melitensis of both killed and live strains (Vitry et al., 2014) and was described as even more relevant than cell-mediated immunity in protecting against B. ovis infection (Jiménez de Bagüés et al., 1994). Accordingly, the strong humoral immune response elicited by B. ovis Δomp10ΔugpBΔomp31 could account for its remarkable protective activity against B. ovis. The similar levels of specific IgG1, IgG2a, and IgG2b suggest a mixed Th1/Th2 response, also observed with other B. ovis PA attenuated mutants (Sancho et al., 2014), that is considered as an advantage for attenuated live vaccines in controlling both early and late events in Brucella infections (Vitry et al., 2014). B. ovis Δomp10ΔugpBΔomp31 also induced a memory immune response evidenced by the profile of cytokine secretion obtained with splenocytes of vaccinated mice after re-stimulation with B. ovis PA whole cells (Figure 8). The cytokine response, at least regarding the parameters evaluated in this work, resembled that obtained with the classical vaccine B. melitensis Rev1, the best available vaccine against B. ovis with proved protective activity in the natural host (OIE, 2017b). Both vaccination groups showed remarkable levels of IFN-g, key cytokine for the control of Brucella infections (Zhan and Cheers, 1993; Murphy et al., 2001; Figure 8). Important secretion of IL-10, described as an anti-inflammatory cytokine that also suppresses phagolysosome fusion in macrophages contributing to Brucella survival in the host (Corsetti et al., 2013; Hop et al., 2018), was also detected in both vaccination groups (Figure 8). However, since both vaccines provided good protection against infection, this increase in IL-10 levels could reflect a strong activation of the immune response after a second exposure, that would neutralize the pathogen but that could also have detrimental effects for the host and would need to be controlled.
Although mutants should be evaluated simultaneously to establish accurate comparisons, and other experimental conditions in the murine model would provide a more accurate information (e.g., different challenge doses, interval vaccination-challenge), the level of protection conferred by the Δomp10ΔugpBΔomp31 mutant against B. ovis infection seems better than that previously described for other B. ovis attenuated vaccines inoculated by the i.p. route (Sancho et al., 2014; Soler-Lloréns et al., 2014; Silva et al., 2015b). Additionally, a ΔabcAB mutant of B. ovis, which is defective in a species-specific ABC transporter (Silva et al., 2011) and that even encapsulated did not provide good protection level in mice (Silva et al., 2015b), protected against B. ovis in rams (Silva et al., 2015a). Accordingly, the results obtained in this work encourage the evaluation of B. ovis Δomp10ΔugpBΔomp31 as vaccine in the natural host. Moreover, the usefulness of B. ovis Δomp31Δcgs and Δomp10Δomp31Δomp25c in rams should not be discarded, since the mouse model – although considered a good approach to evaluate the virulence of Brucella strains – has limitations (Kahl-McDonagh and Ficht, 2006; Grilló et al., 2012) and some virulent mutants in the mouse model were found attenuated in the natural host (Bellaire et al., 2003). Additionally, considering that they are avirulent in mice at low doses of infection, that they do not reach the maximum levels of infection at higher infection doses (Figure 2) and that they barely induce inflammation in spleen (Figure 2), the Δomp31Δcgs and Δomp10Δomp31Δomp25c mutants might be unable to produce epididymitis in rams. In addition to the protective properties, the three attenuated mutants would provide diagnostic advantages. They would not interfere with the serological diagnosis of infections caused by smooth Brucella and, since they are defective in major Omp31 that has been proposed as an interesting diagnostic antigen for B. ovis infection (Vizcaíno et al., 2001b), they would favor the differentiation between vaccinated and infected animals by using Omp31 as diagnostic antigen.
In addition to the development of a vaccine candidate against B. ovis, the results obtained in this work provide new information about the relationships among cell-envelope molecules of B. ovis and the peculiar characteristics of its OM. This knowledge could help to explain why B. ovis, despite its reported genome degradation (Tsolis et al., 2009), establishes persistent infections in its natural host whereas rough mutants derived from S Brucella do not or why its pathogenicity differs from that of B. melitensis in the same preferred host.
Author Contributions
NV, RS-M, and PS conceived the study and wrote the paper. RS-M, PS, NV, MM, CT, AC, and MZ performed the experimental work. All authors participated in the presentation and discussion of the results and in the revision of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the staff of the animal experimentation and DNA sequencing facilities of the University of Salamanca for their helpful collaboration.
Footnotes
Funding. Financial support to RS-M (FPI Grant BES-2012-057056) and this work was provided by the Ministerio de Economía y Competitividad (MINECO) of Spain (Grants AGL2008-04514-C03-03, AGL2011-30453-C04-02, AGL2014-58795-C4-4-R – cofinanced with FEDER funds – and AGL2014-58795-C4-3-R). Funding was also provided by Aragon Government (Grupo de investigación en desarrollo A13-17D).
References
- Arellano-Reynoso B., Lapaque N., Salcedo S., Briones G., Ciocchini A. E., Ugalde R., et al. (2005). Cyclic ß-1,2-glucan is a brucella virulence factor required for intracellular survival. Nat. Immunol. 6 618–625. 10.1038/ni1202 [DOI] [PubMed] [Google Scholar]
- Barbier T., Zúñiga-Ripa A., Moussa S., Plovier H., Sternon J. F., Lázaro-Antón L., et al. (2018). Brucella central carbon metabolism: an update. Crit. Rev. Microbiol. 44 182–211. 10.1080/1040841X.2017.1332002 [DOI] [PubMed] [Google Scholar]
- Barquero-Calvo E., Conde-Álvarez R., Chacón-Díaz C., Quesada-Lobo L., Martirosyan A., Guzmán-Verri C., et al. (2009). The differential interaction of Brucella and Ochrobactrum with innate immunity reveals traits related to the evolution of stealthy pathogens. PLoS One 4:e5893. 10.1371/journal.pone.0005893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellaire B. H., Elzer P. H., Hagius S., Walker J., Baldwin C. L., Roop R. M., II (2003). Genetic organization and iron-responsive regulation of the Brucella abortus 2,3-dihydroxybenzoic acid biosynthesis operon, a cluster of genes required for wild-type virulence in pregnant cattle. Infect. Immun. 71 1794–1803. 10.1128/IAI.71.4.1794-1803.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briones G., Iñón de Iannino N., Roset M., Vigliocco A., Paulo P. S., Ugalde R. A. (2001). Brucella abortus cyclic ß-1,2-glucan mutants have reduced virulence in mice and are defective in intracellular replication in HeLa cells. Infect. Immun. 69 4528–4535. 10.1128/IAI.69.7.4528-4535.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caro-Hernández P., Fernández-Lago L., de Miguel M. J., Martín-Martín A. I., Cloeckaert A., Grilló M. J., et al. (2007). Role of the Omp25/Omp31 family in outer membrane properties and virulence of Brucella ovis. Infect. Immun. 75 4050–4061. 10.1128/IAI.00486-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castañeda-Roldán E. I., Ouahrani-Bettache S., Saldaña Z., Avelino F., Rendón M. A., Dornand J., et al. (2006). Characterization of SP41, a surface protein of Brucella associated with adherence and invasion of host epithelial cells. Cell. Microbiol. 8 1877–1887. 10.1111/j.1462-5822.2006.00754.x [DOI] [PubMed] [Google Scholar]
- Cloeckaert A., Debbarh H. S., Zygmunt M. S., Dubray G. (1996a). Production and characterisation of monoclonal antibodies to Brucella melitensis cytosoluble proteins that are able to differentiate antibody responses of infected sheep from rev. 1 vaccinated sheep. J. Med. Microbiol. 45 206–213. [DOI] [PubMed] [Google Scholar]
- Cloeckaert A., de Wergifosse P., Dubray G., Limet J. N. (1990). Identification of seven surface-exposed Brucella outer membrane proteins by use of monoclonal antibodies: immunogold labeling for electron microscopy and enzyme-linked immunosorbent assay. Infect. Immun. 58 3980–3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloeckaert A., Jacques I., Bosseray N., Limet J. N., Bowden R., Dubray G., et al. (1991). Protection conferred on mice by monoclonal antibodies directed against outer-membrane-protein antigens of Brucella. J. Med. Microbiol. 34 175–180. 10.1099/00222615-34-3-175 [DOI] [PubMed] [Google Scholar]
- Cloeckaert A., Tibor A., Zygmunt M. S. (1999). Brucella outer membrane lipoproteins share antigenic determinants with bacteria of the family Rhizobiaceae. Clin. Diagn. Lab. Immunol. 6 627–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloeckaert A., Verger J. M., Grayon M., Zygmunt M. S., Grépinet O. (1996b). Nucleotide sequence and expression of the gene encoding the major 25-kilodalton outer membrane protein of Brucella ovis: evidence for antigenic shift, compared with other Brucella species, due to a deletion in the gene. Infect. Immun. 64 2047–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloeckaert A., Vizcaíno N., Paquet J. Y., Bowden R. A., Elzer P. H. (2002). Major outer membrane proteins of Brucella spp.: past, present and future. Vet. Microbiol. 90 229–247. 10.1016/S0378-1135(02)00211-0 [DOI] [PubMed] [Google Scholar]
- Cloeckaert A., Zygmunt M. S., de Wergifosse P., Dubray G., Limet J. N. (1992). Demonstration of peptidoglycan-associated Brucella outer-membrane proteins by use of monoclonal antibodies. J. Gen. Microbiol. 138 1543–1550. 10.1099/00221287-138-7-1543 [DOI] [PubMed] [Google Scholar]
- Corsetti P. P., de Almeida L. A., Carvalho N. B., Azevedo V., Silva T. M., Teixeira H. C., et al. (2013). Lack of endogenous IL-10 enhances production of proinflammatory cytokines and leads to Brucella abortus clearance in mice. PLoS One 8:e74729. 10.1371/journal.pone.0074729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degos C., Gagnaire A., Banchereau R., Moriyón I., Gorvel J. P. (2015). Brucella CβG induces a dual pro- and anti-inflammatory response leading to a transient neutrophil recruitment. Virulence 6 19–28. 10.4161/21505594.2014.979692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmonds M. D., Cloeckaert A., Elzer P. H. (2002). Brucella species lacking the major outer membrane protein Omp25 are attenuated in mice and protect against Brucella melitensis and Brucella ovis. Vet. Microbiol. 88 205–221. 10.1016/S0378-1135(02)00110-4 [DOI] [PubMed] [Google Scholar]
- Ferguson G. P., Datta A., Baumgartner J., Roop R. M., II, Carlson R. W., Walker G. C. (2004). Similarity to peroxisomal-membrane protein family reveals that Sinorhizobium and Brucella BacA affect lipid-A fatty acids. Proc. Natl. Acad. Sci. U.S.A. 101 5012–5017. 10.1073/pnas.0307137101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goolab S., Roth R. L., van Heerden H., Crampton M. C. (2015). Analyzing the molecular mechanism of lipoprotein localization in Brucella. Front. Microbiol. 6:1189. 10.3389/fmicb.2015.01189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grilló M. J., Blasco J. M., Gorvel J. P., Moriyón I., Moreno E. (2012). What have we learned from brucellosis in the mouse model? Vet. Res. 43:29. 10.1186/1297-9716-43-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag A. F., Myka K. K., Arnold M. F., Caro-Hernández P., Ferguson G. P. (2010). Importance of lipopolysaccharide and cyclic ß-1,2-glucans in Brucella-mammalian infections. Int. J. Microbiol. 2010:124509. 10.1155/2010/124509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong P. C., Tsolis R. M., Ficht T. A. (2000). Identification of genes required for chronic persistence of Brucella abortus in mice. Infect. Immun. 68 4102–4107. 10.1128/IAI.68.7.4102-4107.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hop H. T., Reyes A. W. B., Huy T. X. N., Arayan L. T., Min W., Lee H. J., et al. (2018). Interleukin 10 suppresses lysosome-mediated killing of Brucella abortus in cultured macrophages. J. Biol. Chem. 293 3134–3144. 10.1074/jbc.M117.805556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iñón de Iannino N., Briones G., Tolmasky M., Ugalde R. A. (1998). Molecular cloning and characterization of cgs, the Brucella abortus cyclic ß(1-2) glucan synthetase gene: genetic complementation of Rhizobium meliloti ndvB and Agrobacterium tumefaciens chvB mutants. J. Bacteriol. 180 4392–4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez de Bagüés M. P., Elzer P. H., Blasco J. M., Marín C. M., Gamazo C., Winter A. J. (1994). Protective immunity to Brucella ovis in BALB/c mice following recovery from primary infection or immunization with subcellular vaccines. Infect. Immun. 62 632–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahl-McDonagh M. M., Ficht T. A. (2006). Evaluation of protection afforded by Brucella abortus and Brucella melitensis unmarked deletion mutants exhibiting different rates of clearance in BALB/c mice. Infect. Immun. 74 4048–4057. 10.1128/IAI.01787-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittelberger R., Diack D. S., Vizcaíno N., Zygmunt M. S., Cloeckaert A. (1998). Characterization of an immuno-dominant antigen in Brucella ovis and evaluation of its use in an enzyme-linked immunosorbent assay. Vet. Microbiol. 59 213–227. 10.1016/S0378-1135(97)00196-X [DOI] [PubMed] [Google Scholar]
- LeVier K., Phillips R. W., Grippe V. K., Roop R. M., II, Walker G. C. (2000). Similar requirements of a plant symbiont and a mammalian pathogen for prolonged intracellular survival. Science 287 2492–2493. 10.1126/science.287.5462.2492 [DOI] [PubMed] [Google Scholar]
- Macedo A. A., Silva A. P., Mol J. P., Costa L. F., Garcia L. N., Araújo M. S., et al. (2015). The abcEDCBA-encoded ABC transporter and the virB operon-encoded type IV secretion system of Brucella ovis are critical for intracellular trafficking and survival in ovine monocyte-derived macrophages. PLoS One 10:e0138131. 10.1371/journal.pone.0138131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manterola L., Guzmán-Verri C., Chaves-Olarte E., Barquero-Calvo E., de Miguel M. J., Moriyón I., et al. (2007). BvrR/BvrS-controlled outer membrane proteins Omp3a and Omp3b are not essential for Brucella abortus virulence. Infect. Immun. 75 4867–4874. 10.1128/IAI.00439-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Martín A. I., Caro-Hernández P., Sancho P., Tejedor C., Cloeckaert A., Fernández-Lago L., et al. (2009). Analysis of the occurrence and distribution of the Omp25/Omp31 family of surface proteins in the six classical Brucella species. Vet. Microbiol. 137 74–82. 10.1016/j.vetmic.2008.12.003 [DOI] [PubMed] [Google Scholar]
- Martín-Martín A. I., Sancho P., de Miguel M. J., Fernández-Lago L., Vizcaíno N. (2012). Quorum-sensing and BvrR/BvrS regulation, the type IV secretion system, cyclic glucans, and BacA in the virulence of Brucella ovis: similarities to and differences from smooth brucellae. Infect. Immun. 80 1783–1793. 10.1128/IAI.06257-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Martín A. I., Sancho P., Tejedor C., Fernández-Lago L., Vizcaíno N. (2011). Differences in the outer membrane-related properties of the six classical Brucella species. Vet. J. 189 103–105. 10.1016/j.tvjl.2010.05.021 [DOI] [PubMed] [Google Scholar]
- Minnick M. F., Sappington K. N., Smitherman L. S., Andersson S. G., Karlberg O., Carroll J. A. (2003). Five-member gene family of Bartonella quintana. Infect. Immun. 71 814–821. 10.1128/IAI.71.2.814-821.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E. A., Sathiyaseelan J., Parent M. A., Zou B., Baldwin C. L. (2001). Interferon-gamma is crucial for surviving a Brucella abortus infection in both resistant C57BL/6 and susceptible BALB/c mice. Immunology 103 511–518. 10.1046/j.1365-2567.2001.01258.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoletti P. (2010). Brucellosis: past, present and future. Prilozi 31 21–32. [PubMed] [Google Scholar]
- OIE (2017a). “Chapter 2.1.4. Brucellosis (Brucella abortus, B. melitensis and B. suis) (infection with B. abortus, B. melitensis and B. suis),” in Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. Paris: OIE; Available at: http://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/2.01.04_BRUCELLOSIS.pdf [Google Scholar]
- OIE (2017b). “Chapter 2.7.8. Ovine epididymitis (Brucella ovis),” in Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. Paris: OIE; Available at: http://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/2.07.08_OVINE_EPID.pdf [Google Scholar]
- Parent M. A., Goenka R., Murphy E., Levier K., Carreiro N., Golding B., et al. (2007). Brucella abortus bacA mutant induces greater pro-inflammatory cytokines than the wild-type parent strain. Microbes Infect. 9 55–62. 10.1016/j.micinf.2006.10.008 [DOI] [PubMed] [Google Scholar]
- Posadas D. M., Martín F. A., Sabio y García J. V., Spera J. M., Delpino M. V., Baldi P., et al. (2007). The TolC homologue of Brucella suis is involved in resistance to antimicrobial compounds and virulence. Infect. Immun. 75 379–389. 10.1128/IAI.01349-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronneau S., Moussa S., Barbier T., Conde-Álvarez R., Zúñiga-Ripa A., Moriyón I., et al. (2016). Brucella, nitrogen and virulence. Crit. Rev. Microbiol. 42 507–525. 10.3109/1040841X.2014.962480 [DOI] [PubMed] [Google Scholar]
- Roop R. M., II, Robertson G. T., Ferguson G. P., Milford L. E., Winkler M. E., Walker G. C. (2002). Seeking a niche: putative contributions of the hfq and bacA gene products to the successful adaptation of the brucellae to their intracellular home. Vet. Microbiol. 90 349–363. 10.1016/S0378-1135(02)00220-1 [DOI] [PubMed] [Google Scholar]
- Roset M. S., Ciocchini A. E., Ugalde R. A., Iñón de Iannino N. (2006). The Brucella abortus cyclic ß-1,2-glucan virulence factor is substituted with O-ester-linked succinyl residues. J. Bacteriol. 188 5003–5013. 10.1128/JB.00086-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roset M. S., Ibañez A. E., de Souza Filho J. A., Spera J. M., Minatel L., Oliveira S. C., et al. (2014). Brucella cyclic β-1,2-glucan plays a critical role in the induction of splenomegaly in mice. PLoS One 9:e101279. 10.1371/journal.pone.0101279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sá J. C., Silva T. M., Costa E. A., Silva A. P., Tsolis R. M., Paixão T. A., et al. (2012). The virB-encoded type IV secretion system is critical for establishment of infection and persistence of Brucella ovis infection in mice. Vet. Microbiol. 159 130–140. 10.1016/j.vetmic.2012.03.029 [DOI] [PubMed] [Google Scholar]
- Salcedo S. P., Marchesini M. I., Lelouard H., Fugier E., Jolly G., Balor S., et al. (2008). Brucella control of dendritic cell maturation is dependent on the TIR-containing protein Btp1. PLoS Pathog. 4:e21. 10.1371/journal.ppat.0040021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salhi I., Boigegrain R. A., Machold J., Weise C., Cloeckaert A., Rouot B. (2003). Characterization of new members of the group 3 outer membrane protein family of Brucella spp. Infect. Immun. 71 4326–4332. 10.1128/IAI.71.8.4326-4332.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho P., Tejedor C., Sidhu-Muñoz R. S., Fernández-Lago L., Vizcaíno N. (2014). Evaluation in mice of Brucella ovis attenuated mutants for use as live vaccines against B. ovis infection. Vet. Res. 45:61. 10.1186/1297-9716-45-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seco-Mediavilla P., Verger J. M., Grayon M., Cloeckaert A., Marín C. M., Zygmunt M. S., et al. (2003). Epitope mapping of the Brucella melitensis BP26 immunogenic protein: usefulness for diagnosis of sheep brucellosis. Clin. Diagn. Lab. Immunol. 10 647–651. 10.1128/CDLI.10.4.647-651.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu-Muñoz R. S., Sancho P., Vizcaíno N. (2016). Brucella ovis PA mutants for outer membrane proteins Omp10, Omp19, SP41, and BepC are not altered in their virulence and outer membrane properties. Vet. Microbiol. 186 59–66. 10.1016/j.vetmic.2016.02.010 [DOI] [PubMed] [Google Scholar]
- Silva A. P., Macêdo A. A., Costa L. F., Rocha C. E., Garcia L. N., Farias J. R., et al. (2015a). Encapsulated Brucella ovis lacking a putative ATP-binding cassette transporter (ΔabcBA) protects against wild type Brucella ovis in rams. PLoS One 10:e0136865. 10.1371/journal.pone.0136865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva A. P., Macêdo A. A., Costa L. F., Turchetti A. P., Bull V., Pessoa M. S., et al. (2013). Brucella ovis lacking a species-specific putative ATP-binding cassette transporter is attenuated but immunogenic in rams. Vet. Microbiol. 167 546–553. 10.1016/j.vetmic.2013.09.003 [DOI] [PubMed] [Google Scholar]
- Silva A. P., Macêdo A. A., Silva T. M., Ximenes L. C., Brandão H. M., Paixão T. A., et al. (2015b). Protection provided by an encapsulated live attenuated ΔabcBA strain of Brucella ovis against experimental challenge in a murine model. Clin. Vaccine Immunol. 22 789–797. 10.1128/CVI.00191-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva T. M., Paixão T. A., Costa E. A., Xavier M. N., Sá J. C., Moustacas V. S., et al. (2011). Putative ATP-binding cassette transporter is essential for Brucella ovis pathogenesis in mice. Infect. Immun. 79 1706–1717. 10.1128/IAI.01109-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler-Lloréns P., Gil-Ramírez Y., Zabalza-Baranguá A., Iriarte M., Conde-Álvarez R., Zúñiga-Ripa A., et al. (2014). Mutants in the lipopolysaccharide of Brucella ovis are attenuated and protect against B. ovis infection in mice. Vet. Res. 45:72. 10.1186/PREACCEPT-4884962711194249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibor A., Decelle B., Letesson J. J. (1999). Outer membrane proteins Omp10, Omp16, and Omp19 of Brucella spp. are lipoproteins. Infect. Immun. 67 4960–4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibor A., Saman E., de Wergifosse P., Cloeckaert A., Limet J. N., Letesson J. J. (1996). Molecular characterization, occurrence, and immunogenicity in infected sheep and cattle of two minor outer membrane proteins of Brucella abortus. Infect. Immun. 64 100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibor A., Wansard V., Bielartz V., Delrue R. M., Danese I., Michel P., et al. (2002). Effect of omp10 or omp19 deletion on Brucella abortus outer membrane properties and virulence in mice. Infect. Immun. 70 5540–5546. 10.1128/IAI.70.10.5540-5546.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibor A., Weynants V., Denoel P., Lichtfouse B., De Bolle X., Saman E., et al. (1994). Molecular cloning, nucleotide sequence, and occurrence of a 16.5-kilodalton outer membrane protein of Brucella abortus with similarity to PAL lipoproteins. Infect. Immun. 62 3633–3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsolis R. M., Seshadri R., Santos R. L., Sangari F. J., García Lobo J. M., de Jong M. F., et al. (2009). Genome degradation in Brucella ovis corresponds with narrowing of its host range and tissue tropism. PLoS One 4:e5519. 10.1371/journal.pone.0005519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitry M. A., Hanot Mambres D., De Trez C., Akira S., Ryffel B., Letesson J. J., et al. (2014). Humoral immunity and CD4 + Th1 cells are both necessary for a fully protective immune response upon secondary infection with Brucella melitensis. J. Immunol. 192 3740–3752. 10.4049/jimmunol.1302561 [DOI] [PubMed] [Google Scholar]
- Vizcaíno N., Caro-Hernández P., Cloeckaert A., Fernández-Lago L. (2004). DNA polymorphism in the omp25/omp31 family of Brucella spp.: identification of a 1.7-kb inversion in Brucella cetaceae and of a 15.1-kb genomic island, absent from Brucella ovis, related to the synthesis of smooth lipopolysaccharide. Microbes Infect. 6 821–834. 10.1016/j.micinf.2004.04.009 [DOI] [PubMed] [Google Scholar]
- Vizcaíno N., Cloeckaert A. (2012). “Biology and genetics of the Brucella outer membrane,” in Brucella Molecular Microbiology and Genomics eds López-Goñi I., O’Callaghan D. (Norfolk: Caister Academic Press; ) 133–161. [Google Scholar]
- Vizcaíno N., Cloeckaert A., Zygmunt M. S., Fernández-Lago L. (2001a). Characterization of a Brucella species 25-kilobase DNA fragment deleted from Brucella abortus reveals a large gene cluster related to the synthesis of a polysaccharide. Infect. Immun. 69 6738–6748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno N., Kittelberger R., Cloeckaert A., Marín C. M., Fernández-Lago L. (2001b). Minor nucleotide substitutions in the omp31 gene of Brucella ovis result in antigenic differences in the major outer membrane protein that it encodes compared to those of the other Brucella species. Infect. Immun. 69 7020–7028. 10.1128/IAI.69.11.7020-7028.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier M. N., Winter M. G., Spees A. M., den Hartigh A. B., Nguyen K., Roux C. M., et al. (2013). PPARγ-mediated increase in glucose availability sustains chronic Brucella abortus infection in alternatively activated macrophages. Cell Host Microbe 14 159–170. 10.1016/j.chom.2013.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan Y., Cheers C. (1993). Endogenous gamma interferon mediates resistance to Brucella abortus infection. Infect. Immun. 61 4899–4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zygmunt M. S., Cloeckaert A., Dubray G. (1994). Brucella melitensis cell envelope protein and lipopolysaccharide epitopes involved in humoral immune responses of naturally and experimentally infected sheep. J. Clin. Microbiol. 32 2514–2522. [DOI] [PMC free article] [PubMed] [Google Scholar]


