In order to make protein, mRNA transcribed from DNA in the nucleus must enter the cytoplasm. Nuclear export of mRNA requires correct processing of mRNAs by enzymes that function in splicing and nuclear export. During the Epstein-Barr virus (EBV) lytic cycle, nuclear export of cellular mRNAs is blocked, yet export of viral mRNAs is facilitated. Here we report the dispersal and dramatic reorganization of cellular (SC35, SON, SRp20, Y14, ALY, and NXF1) and viral (BMLF1 and BGLF5) proteins that play key roles in pre-mRNA processing and export of mRNA. These virus-induced nuclear changes culminate in formation of VINORCs, novel nodular structures composed of viral and cellular RNA splicing and export factors. VINORCs localize to the periphery of viral replication compartments, where viral mRNAs reside. These EBV-induced changes in nuclear organization may contribute to blockade of nuclear export of host mRNA, while enabling selective processing and export of viral mRNA.
KEYWORDS: BMLF1, Epstein-Barr virus, NXF1, nuclear organization, RNA export, RNA processing, SC35, SON, SRp20, viral host shutoff
ABSTRACT
Profound alterations in host cell nuclear architecture accompany the lytic phase of Epstein-Barr virus (EBV) infection. Viral replication compartments assemble, host chromatin marginalizes to the nuclear periphery, cytoplasmic poly(A)-binding protein translocates to the nucleus, and polyadenylated mRNAs are sequestered within the nucleus. Virus-induced changes to nuclear architecture that contribute to viral host shutoff (VHS) must accommodate selective processing and export of viral mRNAs. Here we describe additional previously unrecognized nuclear alterations during EBV lytic infection in which viral and cellular factors that function in pre-mRNA processing and mRNA export are redistributed. Early during lytic infection, before formation of viral replication compartments, two cellular pre-mRNA splicing factors, SC35 and SON, were dispersed from interchromatin granule clusters, and three mRNA export factors, Y14, ALY, and NXF1, were depleted from the nucleus. During late lytic infection, virus-induced nodular structures (VINORCs) formed at the periphery of viral replication compartments. VINORCs were composed of viral (BMLF1 and BGLF5) and cellular (SC35, SON, SRp20, and NXF1) proteins that mediate pre-mRNA processing and mRNA export. BHLF1 long noncoding RNA was invariably found in VINORCs. VINORCs did not contain other nodular nuclear cellular proteins (PML or coilin), nor did they contain viral proteins (BRLF1 or BMRF1) found exclusively within replication compartments. VINORCs are novel EBV-induced nuclear structures. We propose that EBV-induced dispersal and depletion of pre-mRNA processing and mRNA export factors during early lytic infection contribute to VHS; subsequent relocalization of these pre-mRNA processing and mRNA export proteins to VINORCs and viral replication compartments facilitates selective processing and export of viral mRNAs.
IMPORTANCE In order to make protein, mRNA transcribed from DNA in the nucleus must enter the cytoplasm. Nuclear export of mRNA requires correct processing of mRNAs by enzymes that function in splicing and nuclear export. During the Epstein-Barr virus (EBV) lytic cycle, nuclear export of cellular mRNAs is blocked, yet export of viral mRNAs is facilitated. Here we report the dispersal and dramatic reorganization of cellular (SC35, SON, SRp20, Y14, ALY, and NXF1) and viral (BMLF1 and BGLF5) proteins that play key roles in pre-mRNA processing and export of mRNA. These virus-induced nuclear changes culminate in formation of VINORCs, novel nodular structures composed of viral and cellular RNA splicing and export factors. VINORCs localize to the periphery of viral replication compartments, where viral mRNAs reside. These EBV-induced changes in nuclear organization may contribute to blockade of nuclear export of host mRNA, while enabling selective processing and export of viral mRNA.
INTRODUCTION
The eukaryotic nucleus is highly organized into subnuclear domains and nonmembranous structures that fulfill specialized roles in gene expression. During the course of herpesvirus infection, the host intranuclear environment is extensively modified and adapted toward a milieu conducive to efficient survival and replication of the virus. Thus, investigation of virally induced alterations to nuclear organization, specifically alterations in the composition, localization, and architecture of nuclear substructures, is essential to understanding mechanisms of viral gene expression and replication.
Initial studies of herpesvirus-induced alterations to nuclear architecture focused on formation and structure of viral replication compartments (1–5). These globular structures form during the onset of viral DNA synthesis within the lytic programs of all three herpesvirus subclasses. Coincident with the maturation of viral replication compartments, host chromatin is marginalized to the nuclear periphery (6–9). Viral replication compartments, sites of viral DNA synthesis and viral gene transcription, are enriched in proteins needed for lytic viral DNA replication, newly synthesized viral DNA, and viral early and late mRNAs (5, 10).
A number of changes in nuclear organization and architecture, in addition to the formation of viral replication compartments, during herpesviral infection have been described. PML bodies, which serve an antiviral function, are disrupted during lytic replication of herpes simplex virus 1 (HSV-1), cytomegalovirus (CMV), and Epstein-Barr virus (EBV) (11–14). During lytic replication of HSV-1, the nuclear lamina is partially disassembled to allow nuclear egress of viral capsids (15–17). As part of a viral host shutoff (VHS) mechanism, cytoplasmic poly(A)-binding protein (PABPC) is translocated to the nucleus by ICP27 and UL47 during lytic HSV-1 infection (18, 19), by SOX during lytic KSHV infection (20), and by BGLF5, a viral nuclease, and ZEBRA, a viral transcription factor and replication protein, during lytic EBV infection (21). Translocated PABPC is diffusely distributed throughout the nucleus yet is excluded from viral replication compartments. In EBV infection, ZEBRA controls the intranuclear distribution of translocated PABPC (21). Nuclear translocation of PABPC during herpesvirus-mediated host shutoff is accompanied by a block to nuclear export of poly(A) mRNAs (20).
Nuclear export of mRNA is a highly regulated process requiring successful cotranscriptional processing (capping, splicing, cleavage, and polyadenylation) of pre-mRNA and incorporation of mRNA into export-competent messenger ribonucleoproteins (mRNPs) containing export adapters and export receptors (22–24). RNA-binding export adapter proteins (i.e., Y14, Magoh, ALY-REF, SRp20, and 9G8) monitor correctly processed pre-mRNAs (25–28) and recruit export receptor proteins (NXF1:NXT1 and CRM1). Nuclear export receptor proteins recruited to mRNPs bind the nuclear pore complex (NPC) and enable transit of mature mRNPs through the NPC (29, 30).
Interchromatin granule clusters (IGCs), one of the most conspicuous and widely studied nuclear subcompartments, serve as dynamic repositories of small nuclear ribonucleoproteins (snRNPs), SR protein splicing factors, spliceosome subunits, RNA polymerase II (Pol II) large subunit, and SR protein kinases and phosphatases; these essential splicing and transcription factors are supplied by IGCs to sites of transcription. IGCs are located near sites of highly active transcription; they respond morphologically to perturbations in transcription and influence the nucleocytoplasmic export of mRNAs (31–33). Despite their dynamic nature and important roles in processing and export of cellular mRNAs, alterations in the architecture of IGCs during the course of EBV lytic infection have not been described.
Like cellular mRNA, EBV mRNAs are transcribed by RNA Pol II, capped, spliced, cleaved, and polyadenylated. Yet unlike cellular mRNAs, which are sequestered in the nucleus as result of a VHS-induced block to mRNA export, EBV mRNAs are efficiently exported to the cytoplasm. EBV BMLF1 protein (EB2; Mta; SM) regulates the export of most early and late viral mRNAs from the nucleus (34–36). Previous studies have characterized the cellular interacting partners of BMLF1 and the mechanisms by which BMLF1 regulates nuclear export of mRNA. BMLF1 binds RNA via its arginine-rich region (37), shuttles between the nucleus and cytoplasm (38), and interacts with RNA nuclear export factors ALY and NXF1 (37, 39, 40). BMLF1 also regulates the abundance of specific mRNAs (41), stimulates translation of target mRNAs (42), and interacts with cellular splicing factor SRp20 to modulate splice site selection (43).
In this study, we investigated progressive changes to the nuclear architecture of viral and cellular factors with roles in pre-mRNA processing and nuclear export of mRNA during successive stages of lytic EBV infection. We report the following: during the early stage of EBV lytic infection, two pre-mRNA splicing factors, SC35 and SON, were dispersed from IGCs, and three key mRNA export factors, Y14, ALY, and NXF1, were depleted from the nucleus. During late lytic infection, virally induced nodular structures selectively composed of viral (BMLF1 and BGLF5) and cellular (SC35, SON, SRp20, and NXF1) proteins that mediate pre-mRNA processing and mRNA export were assembled at the periphery of viral replication compartments. These nodules, termed VINORCs, are novel EBV-induced nuclear structures. A viral long noncoding RNA, BHLF1, was also a component of VINORCs. We hypothesize that EBV-induced dispersal and depletion of pre-mRNA processing and mRNA export factors early during lytic infection contribute to a VHS-associated block in nuclear export of cellular mRNA. Recruitment of these pre-mRNA processing and mRNA export proteins to VINORCs and viral replication compartments during late lytic infection may facilitate selective processing and export of viral mRNAs.
RESULTS
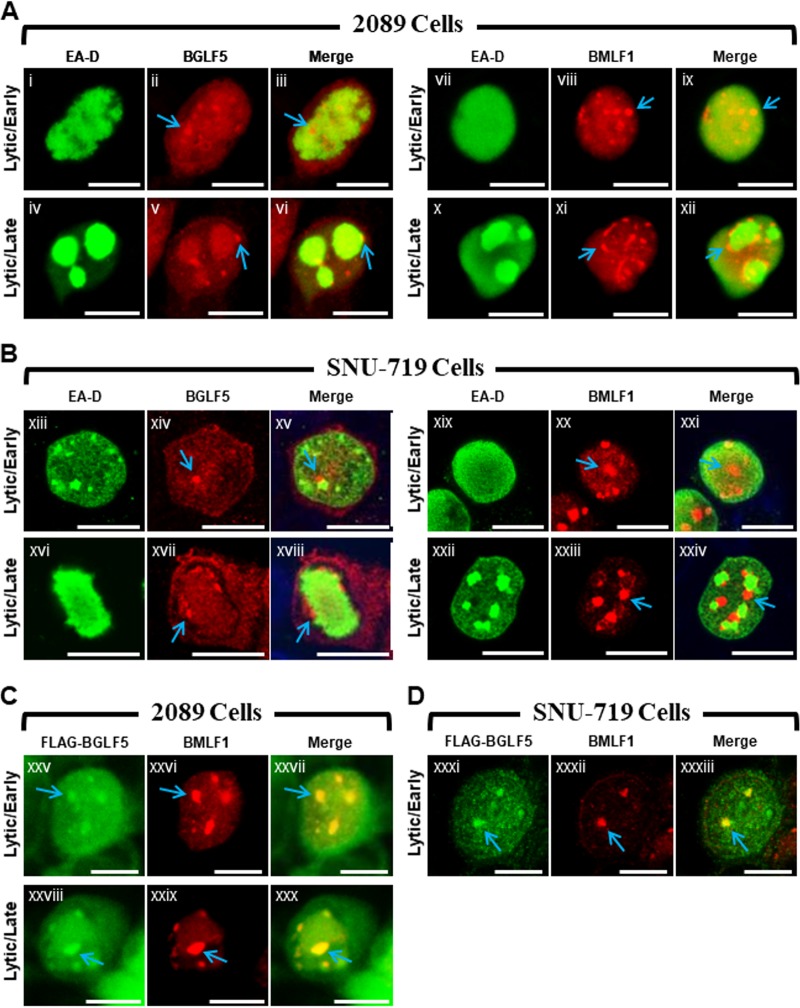
BGLF5 and BMLF1 colocalize within nodules located at the periphery of viral replication compartments (Fig. 1).
FIG 1.
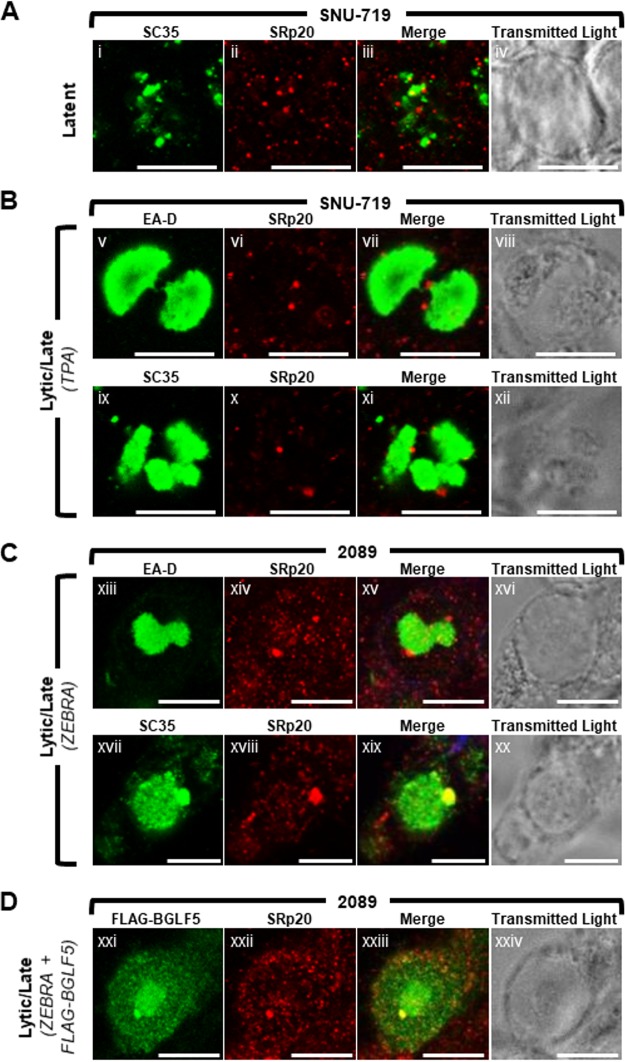
EBV BGLF5 and BMLF1 proteins colocalize within nodular structures in the nucleus. 2089 cells (A and C) and EBV-infected gastric carcinoma cells (SNU-719) (B and D) were induced into the lytic phase by transfection with ZEBRA (A and B) or cotransfection with ZEBRA and FLAG-BGLF5 (C and D). Cells were fixed and stained with antibodies specific for EA-D (A and B), BGLF5 (A and B), BMLF1 (A to D), and FLAG (C and D) as indicated and fluorophore-conjugated secondary antibodies. Each row of three adjacent panels depicts the same field of view. Blue arrows indicate nodular structures containing BGLF5 or BMLF1. The reference bar in each image represents 10 μm.
We previously reported that two EBV lytic proteins, BMLF1, a protein involved in processing and export of viral mRNAs, and BGLF5, a nuclease that functions in DNA replication and viral host shutoff, were similarly localized in lytically induced 293HEK epithelial cells containing wild-type EBV bacmids (2089 cells) (21). Both proteins localized to discrete nodular intranuclear structures; in 2089 cells containing globular viral replication compartments, foci of BMLF1 and BGLF5 localized to the external surface of viral replication compartments. Despite their similar localization patterns, we had not provided evidence that nodules of BMLF1 were distinct from or coincided with nodules of BGLF5. Nor had we demonstrated the nodular localization pattern of BMLF1 or BGLF5 in more physiologically relevant cell lines infected with EBV, rather than with bacmid DNA.
The distribution patterns of BMLF1 and BGLF5 were compared in 2089 cells and in SNU-719 cells, a gastric carcinoma cell line derived from a human tumor biopsy specimen naturally infected with EBV (Fig. 1) (44). The localizations of BMLF1 and BGLF5 were examined 45 h after lytic induction in cells lacking viral replication compartments, indicative of the EBV early lytic stage, and in cells containing globular viral replication compartments, characteristic of the EBV late lytic stage. In 2089 cells in the early lytic stage, nodules of BMLF1 and BGLF5 were present in cells lacking viral replication compartments, marked by the diffuse distribution of EBV polymerase processivity factor EA-D (Fig. 1Ai to Aiii and Avii to Aix), and in cells in the late lytic stage containing viral replication compartments in which the EA-D protein was globular (Fig. 1Aiv to Avi and Ax to Axii). In 2089 cells containing viral replication compartments, nodules of BGLF5 (Fig. 1Av and Avi, blue arrows) and nodules of BMLF1 (Fig. 1Axi and Axii, blue arrows) were concentrated at the periphery of viral replication compartments.
During the EBV lytic cycle in nuclei of SNU-719 cells, BGLF5 (Fig. 1Bxiv, Bxv, Bxvii, and Bxviii, blue arrows) and BMLF1 (Fig. 1Bxx, Bxxi, Bxxiii, and Bxxiv, blue arrows) were concentrated in nodular structures that were similar in number, size, shape, and distribution to nodules in lytic 2089 cells. As seen in 2089 cells, nodules of BGLF5 (Fig. 1Bxvi to Bxviii) and BMLF1 (Fig. 1Bxxii to Bxxiv) localized to the periphery of viral replication compartments in SNU-719 cells.
To determine whether the nodules containing BMLF1 also contained BGLF5, 2089 cells (Fig. 1C) and SNU-719 cells (Fig. 1D) were cotransfected with ZEBRA to induce the lytic cycle and FLAG-BGLF5. In both cell types costained for BMLF1 using rabbit polyclonal antisera, and for FLAG-BGLF5 with anti-FLAG antibody, nodules containing BMLF1 precisely colocalized with nodules containing FLAG-BGLF5. We conclude that both proteins were present within the same nodular structures.
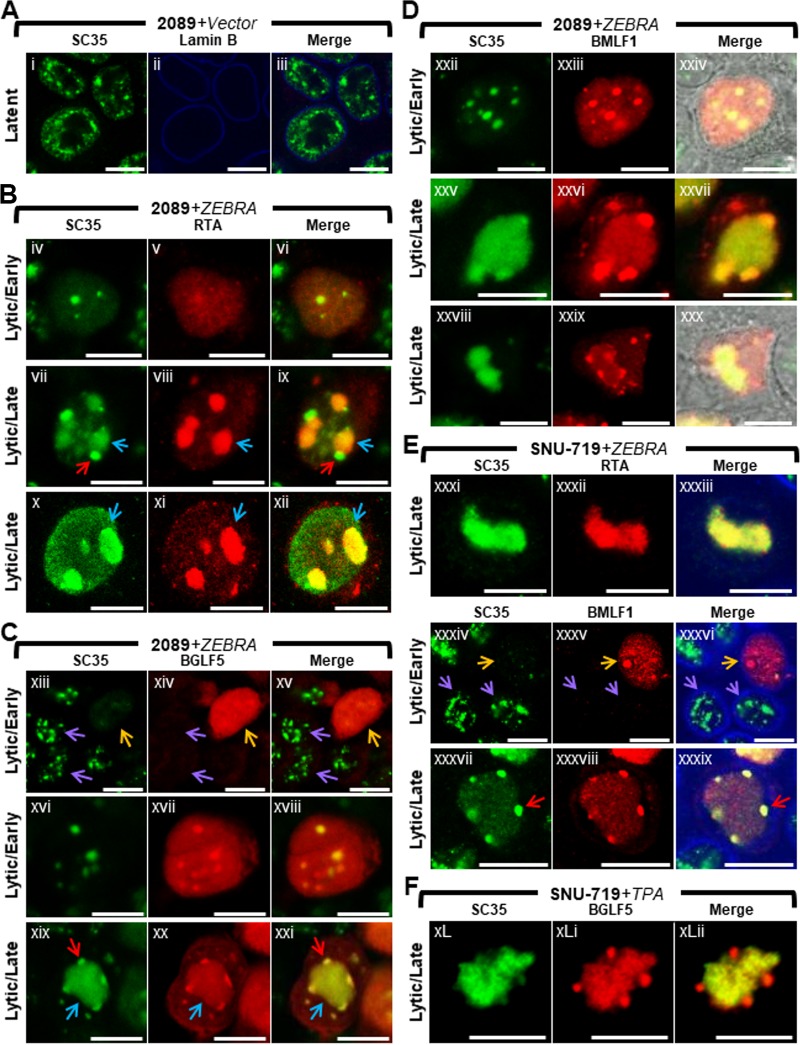
Nuclear reorganization during lytic EBV infection is accompanied by dispersal of SC35 from IGCs, colocalization of SC35 with BMLF1 and BGLF5 at virally induced nodules on replication compartments (VINORCs), and redistribution of SC35 to viral replication compartments (Fig. 2).
FIG 2.
Nuclear reorganization during lytic EBV infection is accompanied by dispersal of SC35 from IGCs, colocalization of SC35 with BMLF1 and BGLF5 at virus-induced nodules on replication compartments (VINORCs) and redistribution of SC35 to viral replication compartments. 2089 cells (A to D) and SNU-719 cells (E and F) were transfected with control vector (A) or induced into the lytic phase by transfection with ZEBRA (B to E). SNU-719 cells were also induced into the lytic phase by treatment with TPA (F). Cells were fixed and stained with antibodies specific for SC35 (A to F), RTA (B and E), BGLF5 (C and F), BMLF1 (D and E), and lamin B (A and E) as indicated and fluorophore-conjugated secondary antibodies. Transmitted light images are shown in panels Dxxiv and Dxxx. Blue arrows (B and C) indicate viral replication compartments, red arrows (B, C, and E) indicate VINORCs containing SC35, orange arrows (C and E) indicate cells undergoing lytic EBV infection, and purple arrows indicate cells not expressing BGLF1 (C) or BMLF1 (E). Each row of three adjacent panels depicts the same field of view. The reference bar in each image represents 10 μm.
EBV BMLF1 regulates alternative splicing and export of viral early and late mRNAs from viral replication compartments to the cytoplasm during lytic induction (38, 45, 46). The recruitment of BMLF1 to discrete VINORCs prompted us to examine alterations in distribution of cellular pre-mRNA splicing factors during lytic induction of EBV. The SR protein SC35 (serine/arginine-rich splicing factor 2 [SRSF2]) (47) is an essential constitutive splicing factor during the early steps of spliceosome assembly (48–50); SC35 also functions as a regulator of 5′ splice site selection during alternative splicing (51, 52) and as a transcription elongation factor (53). SC35 localizes to, and is a well-characterized marker for, interchromatin granule clusters (IGCs). IGCs are dynamic reservoirs and repositories of cellular pre-mRNA splicing factors and transcription factors; often located in close proximity to sites of active transcription, IGCs provide splicing and transcription factors to nearby sites of transcription (54–57).
We examined EBV-induced changes in IGCs by observing changes in localization of SC35 in lytically induced 2089 cells and SNU-719 cells (Fig. 2). During latent EBV infection (Fig. 2A), SC35 was present exclusively within the nucleus, where it was distributed in a coarse granular pattern interspersed with numerous (10 to >30) nondiscrete foci and fibrils of heterogeneous shape that ranged in size from 0.2 μm to 1 μm in diameter. This distribution resembled previously published images of SC35 within IGCs (47). Using EBV replication and transcription activator (RTA) as an indicator of lytic induction and as a marker for viral replication compartments, we observed changes in localization of SC35. Forty-five hours after lytic induction of EBV in 2089 cells (Fig. 2B, C, and D) and SNU-719 cells (Fig. 2E and F), the localization of SC35 was markedly altered from its typical distribution pattern seen in latency. 2089 and SNU-719 cells showed the same altered distribution patterns of SC35 during lytic induction, whether the inducing stimulus was transfection of ZEBRA or treatment of SNU719 cells with tetradecanoyl phorbol acetate (TPA).
In cells lacking viral replication compartments where diffuse RTA was indicative of an early lytic stage, SC35 was dispersed from IGCs and distributed in an evenly diffuse pattern that was faintly visible throughout the nucleus. In some cells containing diffuse RTA, the finely diffuse pattern of SC35 also showed 1 to 8 discrete, round, intranuclear nodules of SC35, measuring 0.2 μm to 2 μm in diameter (Fig. 2Biv to Bvi). In a majority of cells containing viral replication compartments, SC35 colocalized with RTA in a globular mass encompassing the viral replication compartment (Fig. 2Bvii to Bix, blue arrows) and was also present as discrete nodules on the surface of viral replication compartments (Fig. 2Bvii and Bix, red arrows). These nodules contained SC35 but did not contain RTA. In a significant proportion of cells containing viral replication compartments, SC35 localized only within viral replication compartments as a globular, speckled mass that colocalized with RTA (Fig. 2Bx to Bxii and Exxxi to Exxxiii).
Costaining of SC35 with BGLF5 or BMLF1 in lytically induced cells lacking viral replication compartments showed dispersal of SC35 from IGCs. SC35 was present in a faintly visible diffuse distribution that either lacked nodules of SC35 (Fig. 2Cxiii to Cxv, orange arrows, and Exxxiv to Exxxvi, orange arrows) or contained 1 to 8 discrete round nodules of SC35 that colocalized with nodules containing BGLF5 (Fig. 2Cxvi to Cxviii) and BMLF1 (Fig. 2Dxxii to Dxxiv). Interestingly, nodules containing SC35 invariably contained BGLF5 and/or BMLF1; however, nodules containing BGLF5 (Fig. 2Cxiii to Cxv) and BMLF1 (Fig. 2Exxxiv to Exxxvi) could be seen in nuclei lacking nodules of SC35. This observation suggests that the two viral proteins are recruited to nodular structures independently of recruitment of SC35.
In cells in which SC35 was recruited both to viral replication compartments and to VINORCs, SC35 colocalized with BGLF5 (Fig. 2Cxix to Cxxi) and BMLF1 (Fig. 2Dxxv to Dxxvii and Exxxvii to Exxxix) within VINORCs. In cells where SC35 was restricted to globular viral replication compartments, VINORCs containing BMLF1 (Fig. 2Dxxviii to Dxxx) or BGLF5 (Fig. 2FxL to FxLii) did not costain with SC35. In such cells, SC35 costained with RTA within viral replication compartments (Fig. 2Bx to Bxii).
In summary, in cells undergoing lytic induction of EBV, SC35 assumed four distinct patterns of redistribution. In the absence of viral replication compartments, SC35 was (i) diffuse despite the presence of discrete nodules containing BMLF1 and BGLF5 or (ii) diffuse and enriched in several round foci that colocalized with nodules of BMLF1 and BGLF5. (iii) In nuclei containing viral replication compartments SC35 localized both to VINORCs containing BMLF1 and BGLF5 and to viral replication compartments. (iv) SC35 localized only to viral replication compartments and was not present in VINORCs.
The cellular protein SON is dispersed from IGCs and relocalized to nodules at the periphery of viral replication compartments during lytic EBV replication.
SON plays a structural role within IGCs by maintaining localization of pre-mRNA splicing factors (58). To learn whether the redistribution of SC35 to VINORCs and viral replication compartments is limited to SC35, or whether lytic induction of EBV alters other components of the IGC, we studied changes in localization of SON during lytic EBV replication. EBV-induced changes in SON localization were assessed in 2089 cells (Fig. 3A and B, 4A and B, and 5B), SNU-719 cells (Fig. 3C and 4C), and the Burkitt lymphoma cell line HH514-16 (Fig. 3D and 4D). The distribution of SON in these 3 cell types was compared to those of EA-D (Fig. 3) and SC35 (Fig. 4).
FIG 3.
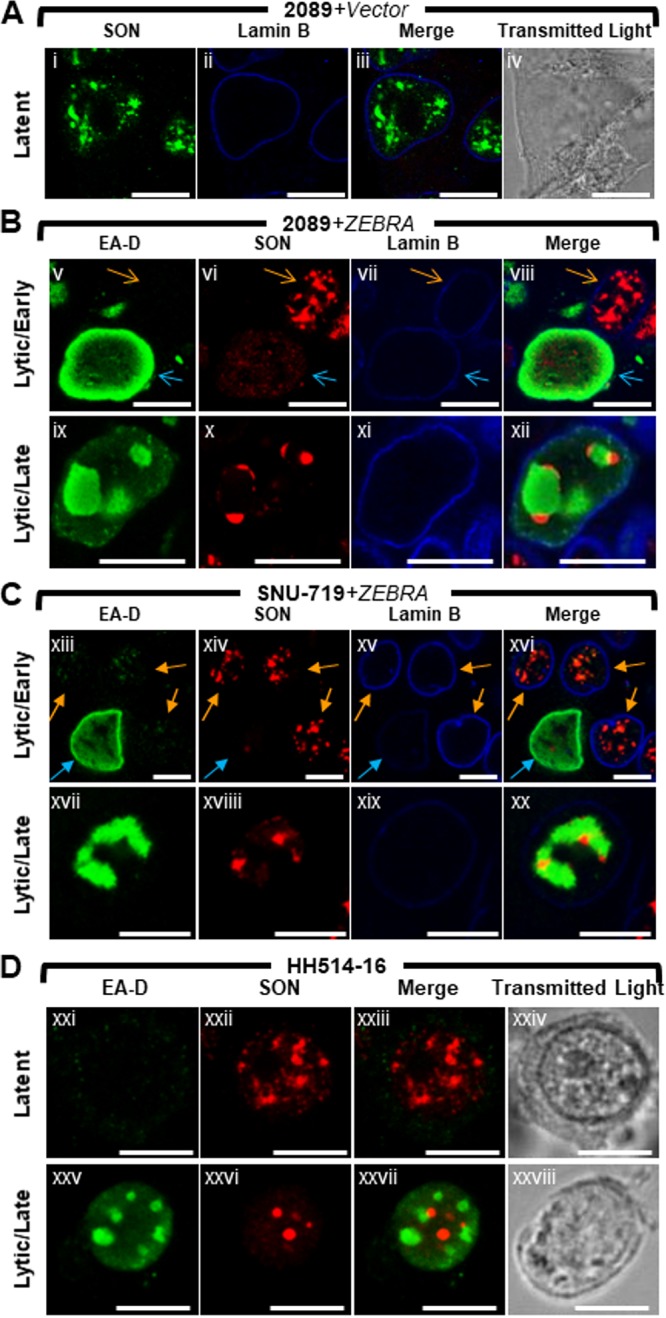
The cellular splicing factor SON is redistributed to nodular structures at the periphery of viral replication compartments during lytic EBV replication. 2089 cells (A and B) and SNU-719 cells (C) were transfected with empty vector (A) or induced into the lytic phase by transfection with ZEBRA (B and C). Burkitt lymphoma cells (HH514-16) were left untreated (Dxxi to Dxxiv) or induced into the lytic phase by treatment with sodium butyrate (Dxxv to Dxxviii). Cells were fixed and stained with antibodies specific for SON (A to D), EA-D (B to D), and lamin B (A to C) as indicated and fluorophore-conjugated secondary antibodies. Transmitted light images are shown in panels Aiv, Dxxiv, and Dxxviii. Blue arrows (B and C) indicate cells that expressed EA-D; orange arrows (B and C) indicate cells that did not express EA-D. Each row depicts the same field of view. The reference bar in each image represents 10 μm.
FIG 4.
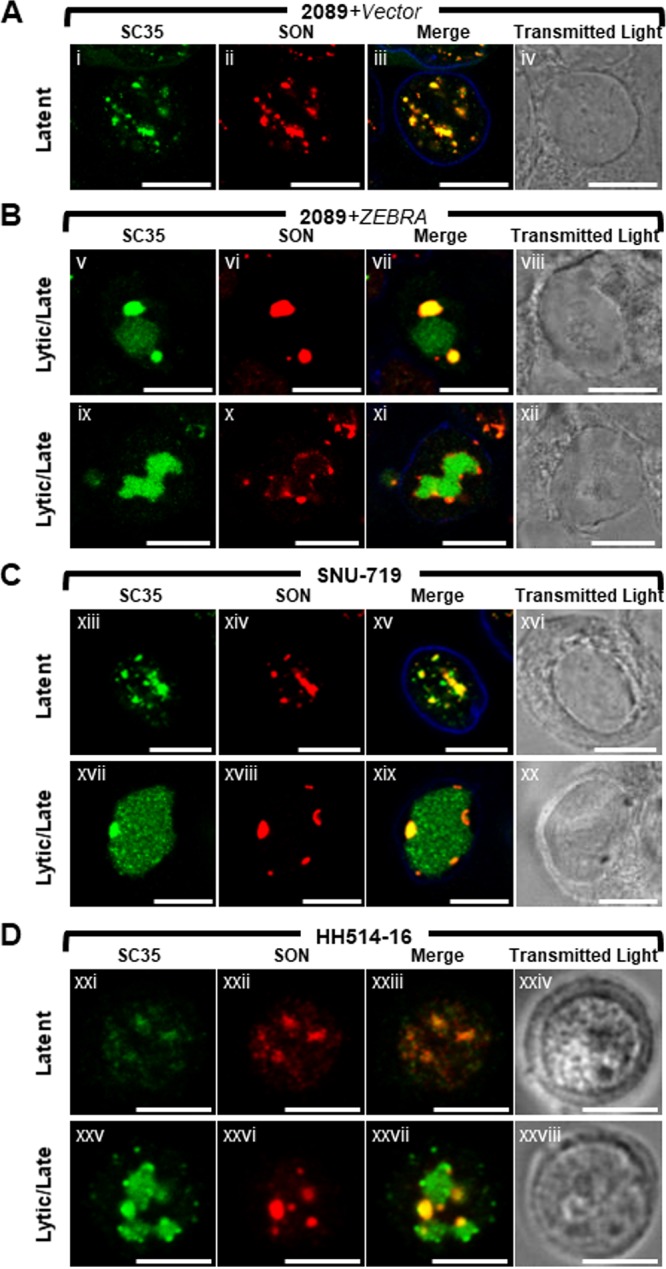
SON colocalizes with SC35 in VINORCs. SC35, but not SON, is recruited to viral replication compartments. 2089 cells (A and B) and SNU-719 cells (C) were transfected with empty vector (A) or ZEBRA (Bv to Bxii and Cxvii to Cxx) or left untreated (Cxiii to Cxvi). HH514-16 cells were left untreated (Dxxi to Dxxiv) or induced into the lytic phase of EBV infection by treatment with sodium butyrate (Dxxv to Dxxviii). Cells were fixed and stained with primary antibodies specific for SC35 (A to D), SON (A to D), and lamin B (A to C) as indicated, followed by fluorophore-conjugated secondary antibodies. Transmitted light images are also shown. Each row depicts the same field of view. The reference bar in each image represents 10 μm.
FIG 5.
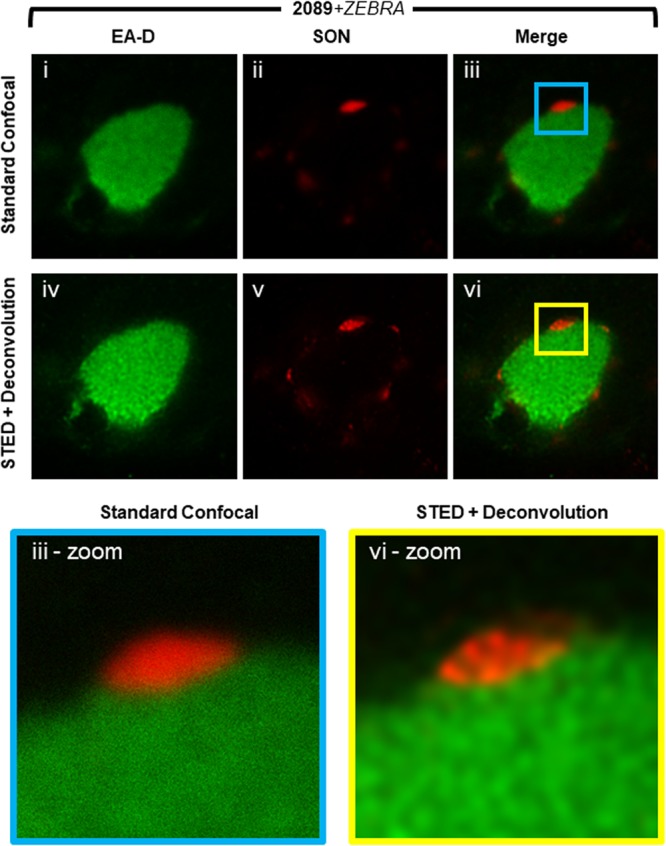
Super-resolution microscopy of SON in VINORCs. 2089 cells transfected with ZEBRA were fixed and stained with primary antibodies specific for EA-D and SON followed by fluorophore-conjugated secondary antibodies. A viral replication compartment and adjacent VINORCs were imaged by standard confocal microscopy (i to iii) and STED microscopy (iv to vi). STED images were deconvoluted using Huygens deconvolution software. Enlarged images of a representative VINORC by standard confocal (iii-zoom) and STED (vi-zoom) microscopy are shown.
In cells latently infected with EBV (Fig. 3A and Dxxi to Dxxiv), SON was present exclusively within the nucleus, where it was distributed as numerous (10 to >30) nondiscrete foci and fibrils of heterogeneous shape that ranged in size from 0.2 μm to 2 μm. Costaining for SC35 and SON showed precise colocalization of these two proteins in latently infected cells (Fig. 4Ai to Aiv, Cxiii to Cxvi, and 4Dxxi to Dxxiv). This colocalization is consistent with SC35 and SON both being integral components of IGCs (47, 58).
Lytic induction of EBV caused redistributions of SON that were similar in all 3 cell types. In nuclei where EA-D was diffuse and viral replication compartments were not present, SON was completely dispersed from IGCs (Fig. 3Bv to Bviii, blue arrows, and 3Cxiii to Cxvi, blue arrows) and, like SC35, was distributed diffusely at a low level throughout the nucleus; this diffuse distribution of SON was sometimes accompanied by 1 to 8 discrete intranuclear nodules, roughly spherical and 0.2 μm to 2 μm in diameter. In cells containing viral replication compartments (Fig. 3Bix to Bxii, Cxvii to Cxx, and 3Dxxv to Dxxvii), SON localized strongly to VINORCs at the periphery of viral replication compartments, but unlike SC35, SON was not recruited to the interior of viral replication compartments.
To confirm differences between SC35 and SON with respect to localization in VINORCs and viral replication compartments, cells were costained for the two proteins and their distributions within single cells were compared (Fig. 4). During EBV latency in 2089 cells, SNU-719 cells, and HH514-16 cells, SC35 and SON precisely colocalized within IGCs (Fig. 4Ai to Aiv, 4Cxiii to Cxvi, and Dxxi to Dxxiv). During lytic induction of all 3 cell types, SC35 was present either in both VINORCs and viral replication compartments (Fig. 4Bv and Bvii, Cxvii and Cxix, and Dxxv and Dxxvii) or exclusively in viral replication compartments (Fig. 4Bix and Bxi). However, within cells containing SC35 in viral replication compartments, SON was present exclusively in VINORCs and was not recruited to the interior of viral replication compartments (Fig. 4Bvi, Bvii, Bx, and Bxi, 4Cxviii and Cxix, and Dxxvi and Dxxvii).
Super-resolution imaging of SON during lytic EBV replication.
With conventional confocal microscopy, SON appeared to be distributed evenly and diffusely throughout VINORCs, with no discernible pattern of organization within VINORCs (Fig. 5i to iii and iii, outlined area [iii-zoom]). However, the limit of resolution attainable by conventional light microscopy is diffraction limited to approximately 200 nm, which is insufficient to resolve the ultrastructure of the interior of VINORCs. To observe the ultrastructure of VINORCs, we used stimulated emission depletion (STED) super-resolution microscopy combined with deconvolution by Huygens software. In contrast to the even distribution of SON in VINORCs seen with conventional confocal microscopy, SON in VINORCs visualized by STED and deconvolution was present in clusters of multiple subunits each measuring 40 nm to 80 nm in diameter (Fig. 5iv to vi and vi-zoom). Using STED-enhanced resolution and a higher (100×) objective lens, a relatively thin layer of SON was also often seen encompassing the outer periphery of viral replication compartments.
Frequency of distinct distribution patterns of SC35 during latency and lytic stages of EBV infection (Fig. 6 and 7).
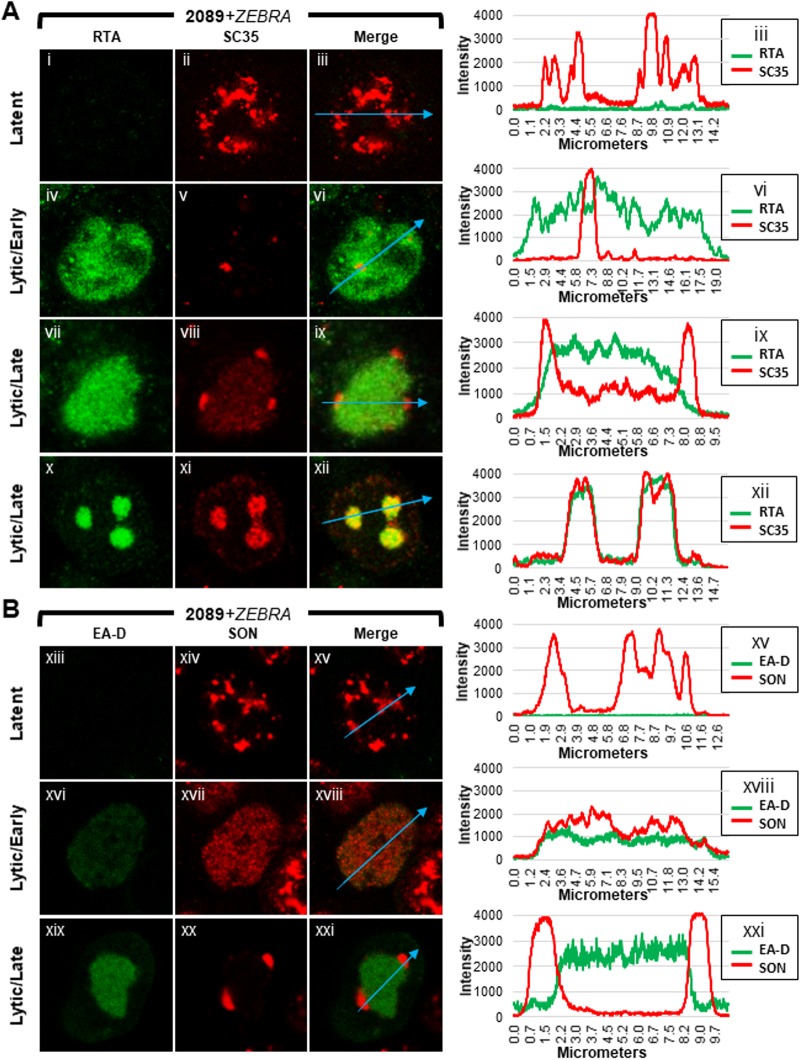
FIG 6.
Distributions of components of VINORCs and of viral replication compartments during EBV latency and lytic replication. 2089 cells transfected with ZEBRA were fixed and stained with primary antibodies specific for RTA and SC35 (A) or EA-D and SON (B) as indicated, followed by fluorophore-conjugated secondary antibodies. Each row depicts the same field of view. Fluorescence intensities of each protein pair were measured along intranuclear cross sections indicated by blue arrows in merged panels. Fluorescence intensity values are plotted on accompanying graphs.
FIG 7.
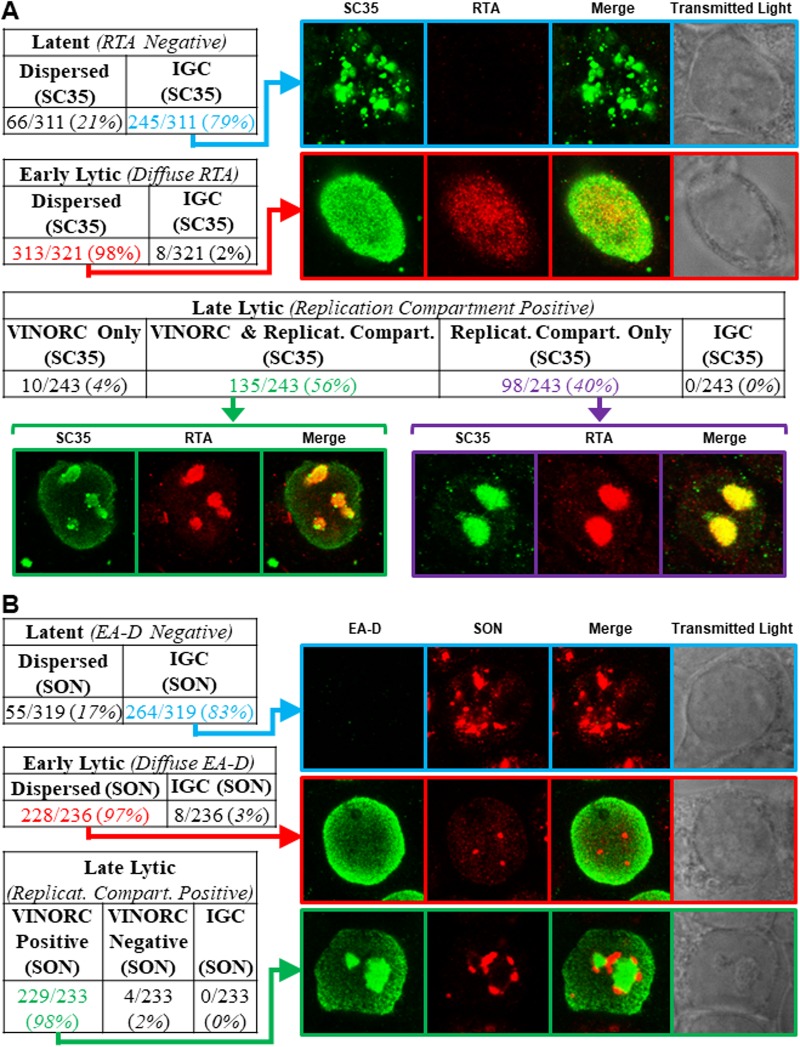
Frequency of different distributions of SC35 and SON during three phases of the EBV life cycle. 2089 cells transfected with ZEBRA were fixed 45 h posttransfection and then stained with primary antibodies specific for RTA and SC35 (A) or EA-D and SON (B) followed by fluorophore-conjugated secondary antibodies. Each row depicts the same field of view. Cells were scored for different distributions of RTA (A) or EA-D (B) characteristic of latent, early lytic, or late lytic stages. Cells in each of these viral stages were scored for distributions of SC35 and SON in interchromatin granule clusters (IGCs), through the nucleus, in VINORCs, and in viral replication compartments.
Distribution patterns of SC35 were analyzed by measuring fluorescence intensities of each protein along cross sections of nuclei during latency, early, and late stages of lytic induction; measurements made using ImageJ software were portrayed graphically (Fig. 6). Frequencies of each distribution pattern of SC35 and SON were scored for each viral stage within a population of 2089 cells at 45 h following transfection with ZEBRA (Fig. 7).
Plots of measurements of SC35 along cross sections of latently infected cells (Fig. 6Ai to Aiii) showed multiple peaks and troughs of heterogeneous heights, depths, and widths distributed unevenly across the nucleus (Fig. 6Aiii, blue arrow, and corresponding graph). These peaks correspond to the IGCs. Cell counts showed SC35 was distributed within IGCs in the majority (79%) of latently infected cells (Fig. 7A, highlighted in blue); a minority of cells (21%) showed SC35 dispersed diffusely through the nucleus.
Measurements along cross sections of early lytic stage cells marked by diffuse RTA (Fig. 6Aiv to Avi) showed SC35 distributed in a uniform, low-level pattern across the nucleus (Fig. 6Avi, blue arrow, and corresponding graph); measurements along cross sections of early lytic cells that intersected a nodule of SC35 appeared graphically as a spike. In contrast to latently infected cells, in which SC35 was predominantly found in IGCs, in cells at the early stage of lytic infection SC35 was dispersed throughout the nucleus in nearly all cells (98%) (Fig. 7A, highlighted in red).
When SC35 was present in VINORCs and in viral replication compartments (Fig. 6Avii to Aix), ImageJ measurements showed spikes of SC35 corresponding to VINORCs at the periphery of viral replication compartments and an elevated level of SC35 uniformly distributed within viral replication compartments (Fig. 6Aix, blue arrow and corresponding graph). In cells where SC35 was present only within viral replication compartments (Fig. 6Ax to Axii), SC35 precisely colocalized with RTA (Fig. 6Axii, blue arrow and corresponding graph). SC35 within viral replication compartments often appeared speckled. During the late stage of lytic infection, 56% of cells showed SC35 localized to both VINORCs and viral replication compartments (Fig. 7A, highlighted in green), 40% of cells showed SC35 localizing only to viral replication compartments (Fig. 7A, highlighted in purple), a small minority of cells (4%) showed SC35 localized to VINORCs but not to replication compartments, and no cells showed SC35 distributed within IGCs.
Frequency of distinct distribution patterns of SON during latency and lytic stages of EBV infection.
Measurements of SON taken along cross sections of latently infected cells showed peaks and troughs of heterogeneous heights, depths, and widths distributed unevenly across the nucleus (Fig. 6Bxiii to Bxv, blue arrow, and corresponding graph); this pattern, indicative of IGCs, was similar to that in graphs of SC35 in latently infected nuclei. Cell counts showed that 83% of latently infected cells contained SON localized to IGCs (Fig. 7B, highlighted in blue); 17% showed SON dispersed diffusely through the nucleus.
During the early stage of lytic infection, measurements of SON along nuclear cross sections (Fig. 6Bxvi to Bxviii) showed SON distributed in a uniform, low-level pattern across the nucleus (Fig. 6Bxviii, blue arrow, and corresponding graph); cross sections of early lytic cells occasionally intersected a discrete focus of SON that appeared graphically as a spike (data not shown). Counts of cells undergoing early lytic infection showed that SON was dispersed from IGCs and distributed diffusely throughout the nucleus in nearly all (97%) cells (Fig. 7B, highlighted in red).
In cells undergoing late lytic infection (Fig. 6Bxix to Bxxi), plotted measurements of SON taken along nuclear cross sections showed spikes corresponding to VINORCs at the periphery of viral replication compartments (Fig. 6Bxxi, blue arrow; and corresponding graph). Quantitation of SON across viral replication compartments marked by EA-D showed no enrichment of SON within viral replication compartments (Fig. 6Bxxi graph).
These studies indicate similar distribution patterns of SC35 and SON during latency and early EBV lytic cycle. In the late lytic cycle, SC35, but not SON, was enriched in viral replication compartments. SON was invariably present in VINORCs; SC35 was variably present in VINORCs. Nearly all (98%) of cells containing viral replication compartments also possessed VINORCs marked by SON (Fig. 7B, highlighted in green).
Redistribution of SON and SC35 occurs during the early stage of lytic infection.
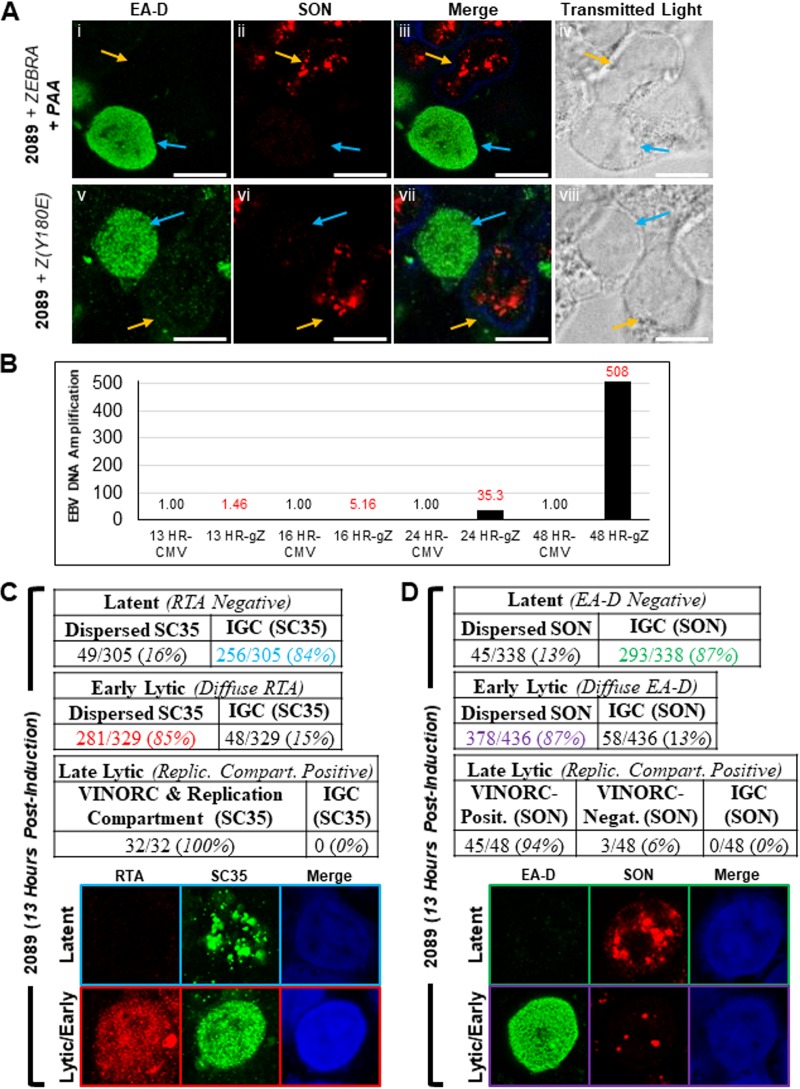
In lytically induced cells lacking viral replication compartments, SC35 and SON were dispersed from IGCs and were diffusely distributed throughout the nucleus (Fig. 2Biv to Bvi, 3Bv to Bviii, and Cxiii to xiv). These observations indicate that redistribution of both proteins occurs during an early stage of lytic induction, prior to the formation of viral replication compartments. We verified this hypothesis using three different approaches: (i) lytically induced cells were arrested at the prereplication stage by treatment with phosphonoacetic acid (PAA), an inhibitor of viral DNA replication (Fig. 8Ai to 9iv); (ii) lytic cycle was inducted by a ZEBRA missense point mutant, Z(Y180E), that is defective in driving progression of the EBV lytic cycle beyond the early stage (59) (Fig. 8Av to Aviii); and (iii) changes in distribution of SC35 and SON were observed at an early time point prior to lytic viral DNA replication (Fig. 8B to D).
FIG 8.
Dispersal of SC35 and SON occurs during the early lytic stage of EBV infection. (A) 2089 cells were induced into the EBV lytic phase by transfection with wild-type ZEBRA and treated with the viral DNA replication inhibitor PAA (Ai to Aiv) or transfected with a replication-defective point mutant of ZEBRA, Z(Y180E) (Av to Aviii). Cells were fixed and stained with antibodies specific for EA-D and SON and fluorophore-conjugated secondary antibodies. Transmitted light images are shown as indicated. Each row depicts the same field of view. The reference bar in each image represents 10 μm. (B) Time course of DNA amplification. 2089 cells were transfected with plasmids expressing ZEBRA (gZ) or the CMV immediate early promoter (CMV). Samples harvested at the indicated times were assessed for EBV content by quantitative PCR. (C and D) Cells harvested 13 h after transfection with ZEBRA were analyzed for frequency of distribution pattern of SC35 (C) or SON (D). Cells during the early lytic phase were distinguished from those in latency by the presence of absence of a diffuse pattern of expression by RTA (C) or EA-D (D).
FIG 9.
The cellular splicing factor and mRNA export factor SRp20 is recruited to VINORCs during lytic EBV replication. SNU-719 cells were left untreated (A) or induced into the lytic phase of EBV infection by treatment with TPA (B). 2089 cells were induced into the lytic phase by transfection with ZEBRA (C) or cotransfection with ZEBRA and FLAG-BGLF5 (D). Cells were fixed and stained with primary antibodies specific for SRp20 (A to D; red), SC35 (A to C; green), EA-D (B and C; green), and FLAG (D; green) as indicated and fluorophore-conjugated secondary antibodies. Transmitted light images are shown as indicated. Each row depicts the same field of view. The reference bar in each image represents 10 μm.
Treatment with PAA arrested lytically induced 2089 cells at the early lytic stage, as indicated by a diffuse distribution of EA-D and absence of viral replication compartments in most cells. PAA-treated cells expressing EA-D invariably showed dispersal of SON from IGCs (Fig. 8Ai to Aiv, blue arrows). In contrast, in surrounding latently infected cells (Fig. 8Ai to Aiv, orange arrows), SON was present in IGCs. Use of ZEBRA mutant Z(Y180E) excludes possible nonspecific effects of PAA on SON localization. The majority of 2089 cells transfected with Z(Y180E) were positive for diffuse EA-D expression but lacked viral replication compartments. In these cells, SON was dispersed from IGCs and, similar to the case with PAA-treated cells, was distributed diffusely throughout the nucleus (Fig. 8Av to Aviii, blue arrows). Latent cells negative for expression of EA-D showed SON localized to IGCs (Fig. 8Av to Aviii, orange arrows).
Using quantitative PCR analysis with primers specific for OriLyt, 13 h following transfection of 2089 cells with ZEBRA was determined to be a time point of the early lytic stage prior to significant lytic viral DNA replication (Fig. 8B). At 13 h after transfection with ZEBRA, 91% (329 of 361) of RTA-positive cells showed a diffuse distribution of RTA, and 9% (32 of 361) showed RTA in viral replication compartments. Of cells containing diffuse RTA, 85% (281 of 329) showed dispersal of SC35 from IGCs (Fig. 8C, highlighted in red). In contrast, 84% (256 of 305) of latent cells contained SC35 distributed within IGCs (Fig. 8C, highlighted in blue). At 13 h after transfection with ZEBRA, 90% (436 of 484) of cells showed a diffuse distribution of EA-D, and 10% (48 of 484) showed EA-D in viral replication compartments. Of those cells containing diffuse EA-D, 87% (328 of 436) showed dispersal of SON from IGCs (Fig. 8D, highlighted in purple), whereas 87% (293 of 338) of cells in latency showed SON distributed within IGCs. The results of these experiments, using the three methods described above indicate that dispersal of SC35 and SON from IGCs occurs during the EBV early lytic stage, prior to formation of replication compartments or formation of VINORCs.
The cellular splicing factor and mRNA export factor SRp20 localizes to VINORCs during lytic EBV replication.
IGCs, found in close proximity to sites of active transcription (31–33, 60–62), are believed to function as repositories and reservoirs of transcription factors and splicing factors (63–66). The recruitment of BMLF1 and IGC components SC35 and SON to VINORCs, as well as the localization of VINORCs at the periphery of viral replication compartments, suggested a possible role for VINORCs in facilitating viral mRNA processing and/or nuclear export. To investigate this hypothesis, we assessed whether SRp20 (serine/arginine-rich splicing factor 3 [SRSF3]) was recruited to VINORCs. SRp20 is a cellular pre-mRNA splicing factor (67–71) and mRNA export factor (27, 28). Like SC35, SRp20 belongs to the SR family of splicing factors; however, unlike SC35 and SON, SRp20 often does not colocalize with IGCs. Assaying for recruitment of SRp20 to VINORCs is particularly relevant to VINORC function, as interaction of SRp20 with BMLF1 mediates the ability of BMLF1 to direct alternative splicing (43).
During latency (Fig. 9A), SRp20 was distributed throughout the nucleus in a finely speckled pattern; SRp20 and SC35 did not colocalize well. In lytically induced SNU-719 cells (Fig. 9B) and 2089 cells (Fig. 9C) containing viral replication compartments, SRp20 localized to nodules at the surface of viral replication compartments. Costaining of SRp20 with VINORC components SC35 (Fig. 9Cxvii to Cxix) and FLAG-BGLF5 (Fig. 9Dxxi to Dxxiv) showed colocalization within VINORCs; this result indicates recruitment of SRp20 to VINORCs. However, unlike SC35, SRp20 was not enriched within the interior of viral replication compartments (Fig. 9Bv to Bviii, Bix to Bxii, Cxiii to Cxvi, and Cxvii to Cxix). In cells with SC35 localized exclusively to the interior of viral replication compartments, SRp20 was recruited to VINORCs that lacked SC35 (Fig. 9Bix to Bxii). In summary, during lytic replication of EBV, SRp20 was recruited to VINORCs and colocalized with BGLF5, an invariant marker of VINORCs.
EBV long noncoding RNA BHLF1 is a component of VINORCs.
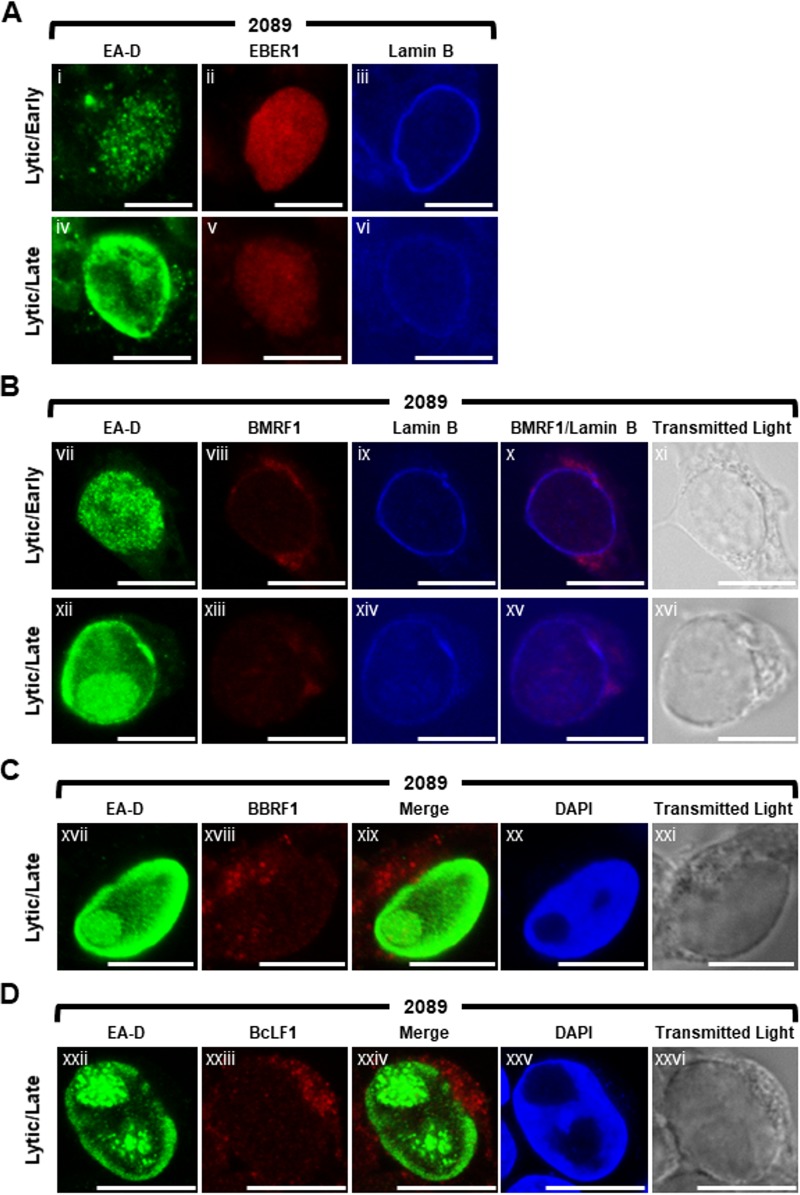
Since all VINORC components identified thus far were RNA-binding proteins, we hypothesized that VINORCs might also contain an RNA component. To identify possible viral RNA components of VINORCs, using fluorescence in situ hybridization (FISH) we examined the localization of four distinct types of EB viral RNAs: (i) a short noncoding RNA, EBER-1, 167 nucleotides in length, that is present during the latent (72) and lytic phases; (ii) an early lytic mRNA, BMRF1; (iii) two late lytic mRNAs, BBRF1 and BcLF1; and (iv) a lytic long noncoding RNA (lncRNA), BHLF1, 1,980 nucleotides in length. The appearance of EBER-1 and the three EBV mRNAs was distinctly different from that of the BHLF1 lncRNA.
In both latent (data not shown) and lytically induced 2089 cells probed for EBER-1 RNA and EA-D protein, EBER-1 was diffusely distributed within the nucleus and lacked any discrete foci (Fig. 10A). In lytically induced 2089 cells probed for BMRF1 mRNA and EA-D protein, BMRF1 mRNA localized primarily to the cytoplasm (Fig. 10B). During early lytic infection, a low level of BMRF1 mRNA was visible diffusely throughout lytically induced nuclei; however, intranuclear foci of BMRF1 mRNA were not seen. During late lytic stage, a slight enrichment of BMRF1 mRNA was seen colocalized with viral replication compartments, and faintly stained foci of BMRF1 were observed at the periphery of viral replication compartments (Fig. 10Bxii to Bxvi). BBRF1 mRNA, which does not require BMLF1 for nuclear export (35), was concentrated in subterritories of the cytoplasm as distinct foci. A low level of BBRF1 mRNA was visible diffusely throughout nucleus and in foci within viral replication compartments marked by EA-D and the absence of 4′,6-diamidino-2-phenylindole (DAPI) staining (Fig. 10C). Foci of BBRF1 mRNA that could correspond to VINORCs were not seen at the periphery of viral replication compartments. BcLF1 mRNA, which is dependent on BMLF1 for cytoplasmic accumulation (35), was enriched in the cytoplasm as discrete foci and visible at a low level diffusely throughout the nucleus. However, intranuclear foci of BcLF1 mRNA were not seen (Fig. 10D).
FIG 10.
Distribution patterns of four EBV RNAs during lytic cycle of EBV infection: EBER-1 (a short noncoding RNA), BMRF1 (an early lytic mRNA), and BBRF1 and BcLF1 (two late lytic mRNAs). 2089 cells were induced into the lytic phase of EBV infection by transfection with ZEBRA. EBER-1 RNA (A), BMRF1 mRNA (B), BBRF1 mRNA (C), and BcLF1 mRNA (D) were detected by RNA FISH using antisense strand oligonucleotide probes. Cells were costained for viral proteins by indirect immunofluorescence using primary antibodies specific for EA-D and lamin B (A and B) and fluorophore-conjugated secondary antibodies. Cells were stained with DAPI (C and D) as indicated. Transmitted light images (B to D) are shown as indicated. Each row depicts the same field of view. The reference bar in each image represents 10 μm.
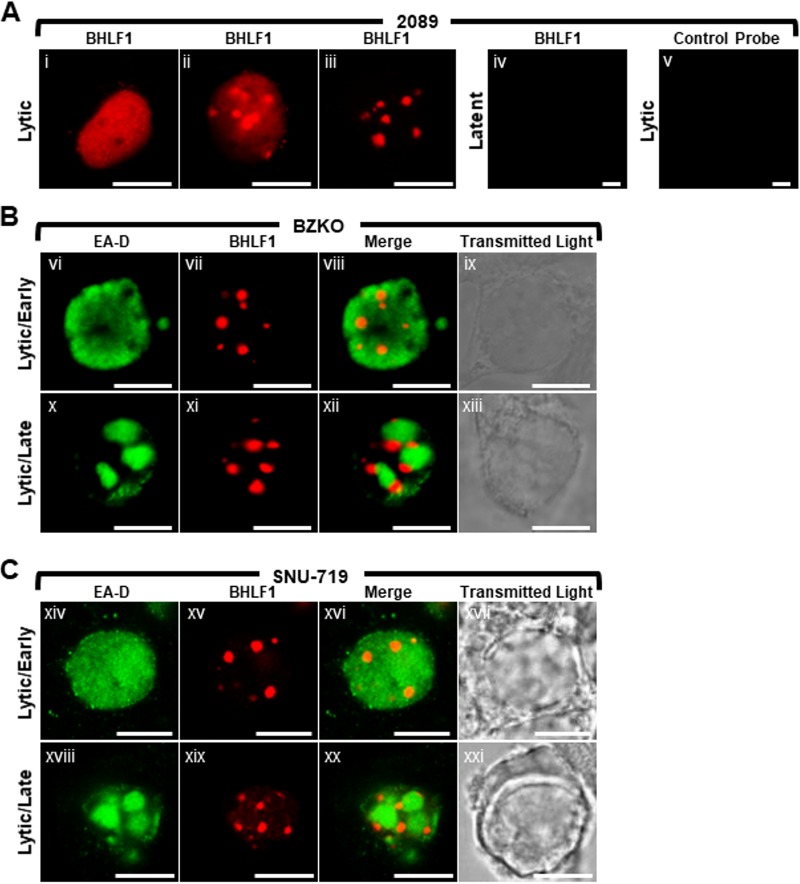
2089 cells (Fig. 11Ai to Aiii and Av), BZKO cells (Fig. 11B), and SNU-719 cells (Fig. 11C) were induced into the lytic phase by transfection with ZEBRA and probed for BHLF1 lncRNA by RNA FISH using antisense oligonucleotide probes. BHLF1 lncRNA was not detected in latent cells (Fig. 11Aiv); no nonspecific signal was seen in lytic cells probed with BHLF1 sense strand oligonucleotide control probes (Fig. 11Av). In lytically induced 2089, BZKO, and SNU-719 cells, BHLF1 lncRNA was distributed in the nucleus in 3 different patterns: diffusely (Fig. 11Ai), diffusely and enriched in 1 to 10 discrete foci 0.2 μm to 2 μm in diameter (Fig. 11Aii), or present exclusively within 3 to 10 discrete foci 0.2 μm to 2 μm in diameter (Fig. 11Aiii, Bvii and Bxi, and Cxv and Cxix). In cells expressing diffuse EA-D but lacking viral replication compartments, characteristic of the early lytic phase, BHLF1 lncRNA was found in all 3 distributions (Fig. 11Bvi to Bix and Cxiv to Cxvii). However, in cells containing viral replication compartments (Fig. 11Bx to Bxiii and Bxviii to Bxxi), BHLF1 lncRNA was localized to nodules situated at the surface of viral replication compartments; BHLF1 lncRNA was not found within viral replication compartments.
FIG 11.
EBV long noncoding RNA BHLF1 forms nodules at the periphery of viral replication compartments. 2089 (Ai to Aiii and Av), BZKO (B), and SNU-719 (C) cells were induced into the lytic phase of EBV infection by transfection with ZEBRA or not transfected (Aiv). BHLF1 lncRNA (Ai to Aiv, B, and C) was detected by RNA FISH using antisense strand oligonucleotide probes. As a control for specificity of BHLF1 probes, lytically induced cells were probed with complementary sense strand oligonucleotides probes of BHLF1 (Av). Cells were costained for EA-D by indirect immunofluorescence. Transmitted light images are shown as indicated. Each row depicts the same field of view. The reference bar in each image represents 10 μm.
To test whether nodules of BHLF1 lncRNA colocalized with known protein components of VINORCs, lytically induced BZKO (Fig. 12A), 2089 (Fig. 12B), and SNU-719 (Fig. 12C) cells were probed for BHLF1 lncRNA by FISH and costained for BMLF1 (Fig. 12A), SON (Fig. 12B and C), and SC35 (Fig. 12B and C). In all 3 cell types, nodules of BHLF1 lncRNA invariably and precisely colocalized with three VINORC components, BMLF1, SON, and SC35. These results indicate that BHLF1 lncRNA is a viral RNA component of VINORCs.
FIG 12.
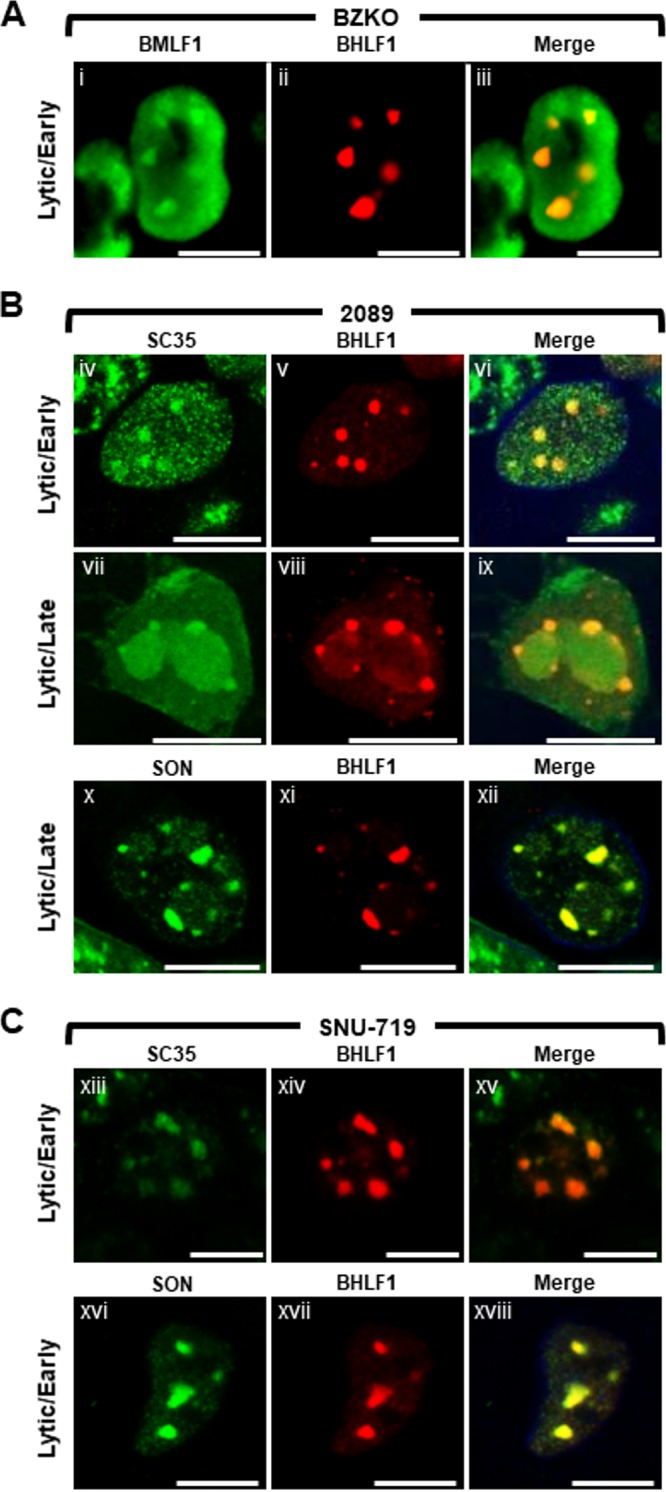
BHLF1 colocalizes with other components of VINORCs. BZKO (A), 2089 (B), and SNU-719 (C) cells were induced into the lytic phase of EBV infection by transfection with ZEBRA. BHLF1 lncRNA was detected by RNA FISH (A to C; red) and costained by indirect immunofluorescence for BMLF1 (A; green), SC35 (B and C; green), and SON (B and C; green) proteins, as indicated. Each row depicts the same field of view. The reference bar in each image represents 10 μm.
Three-dimensional appearance of VINORCs.
Two-dimensional images are inadequate for addressing certain questions regarding the morphology and distribution of VINORCs at the periphery of viral replication compartments. Are VINORCs spherical, ovoid, or cylindrical? Where are VINORCs located in the nucleus with respect to the apical-basal (Z) axis? What are the relative positions of VINORCs and viral replication compartments with respect to the Z axis? To better visualize the distribution of VINORCs within a cell nucleus undergoing lytic EBV replication, a 3-dimensional Z-stack reconstruction of a series of X-Y planar images was generated by confocal microscopy (Fig. 13). VINORCs and viral replication compartments in lytically induced BZKO cells were marked by BHLF1 lncRNA and EA-D, respectively. VINORCs, globular and ovoid in shape, were positioned on the exterior surface of viral replication compartments. Unlike PML bodies, which are adjacent to but physically separate from viral replication compartments, no intervening space was visible between VINORCs and viral replication compartments at a resolution limit of 200 nm to 300 nm. Overlap between the green fluorescence of EA-D and red fluorescence of BHLF1 lncRNA produced a sliver of yellow fluorescence at the interface of VINORCs and viral replication compartments; this overlap suggests synaptic contact between the two distinct structures. Most VINORCs were situated at the basal side of viral replication compartments (90° and 270° [Fig. 13]).
FIG 13.
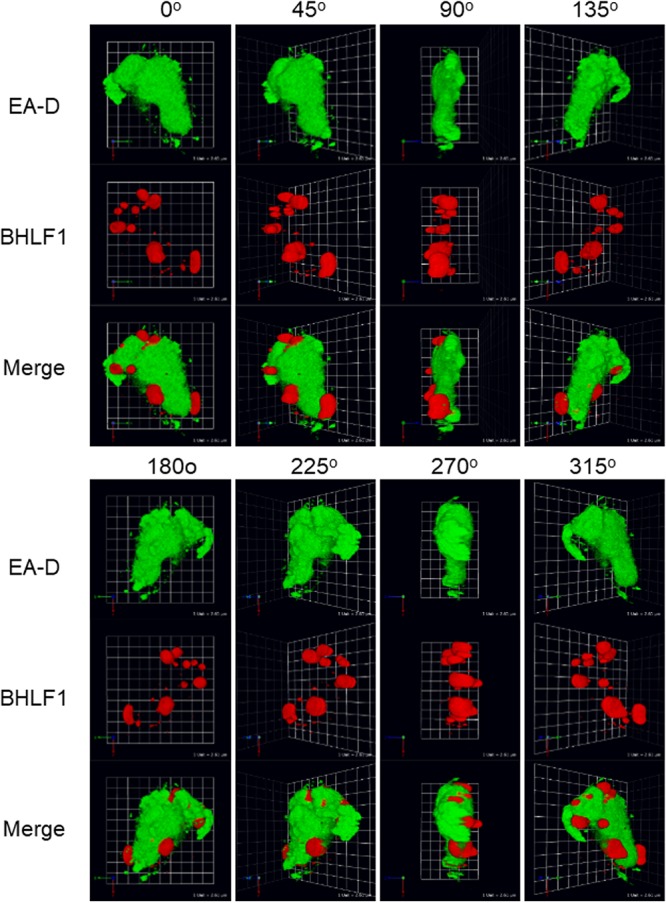
Z-stack reconstruction of VINORCs containing BHLF1 at the periphery of viral replication compartments. BZKO cells were induced into the lytic phase of EBV infection by transfection with ZEBRA. BHLF1 lncRNA and EA-D protein were detected by simultaneous RNA FISH and indirect immunofluorescence analysis. A 3-dimensional representation was reconstructed from Z-stack micrographs obtained by confocal microscopy. Images were taken of the Z-stack reconstruction rotated around the y axis at 45° intervals.
Neither coilin nor PML is redistributed or recruited to VINORCs during lytic EBV induction.
VINORC components, including BMLF1, BGLF5, SC35, and SON, each possess RNA processing or mRNA export activity, whereas viral and cellular proteins that are not recruited to VINORCs, such as EA-D, RTA, ZEBRA, and lamin B, lack known RNA processing or export activity. To further investigate the specificity of recruitment to VINORCs, two cellular nuclear proteins with no known RNA processing or export activity, namely, coilin, a major component of Cajal bodies, and PML, were observed during lytic EBV replication.
In latently infected 2089 cells (Fig. 14Ai, Aii, Bix, and Bx) and SNU-719 cells (Fig. 14Aiii, Aiv, Bxi, and Bxii), coilin localized to 1 to 10 discrete punctate intranuclear foci, ranging from 0.2 μm to 0.5 μm in diameter. Costaining showed that during latency coilin did not colocalize with SON in IGCs (Fig. 14Bix and Bxi). In cells undergoing lytic induction, there was no change in the number, size, shape, or appearance of coilin foci (Fig. 14Av to Aviii and Bxiii to Bxvi) compared to latently infected cells. Costaining with RTA showed that coilin foci, unlike VINORCs, did not localize to the periphery of viral replication compartments (Fig. 14Av and Avii). Costaining of coilin and SON showed that during lytic EBV induction, coilin did not colocalize with nodules of SON (Fig. 14Bxiii to Bxvi).
FIG 14.
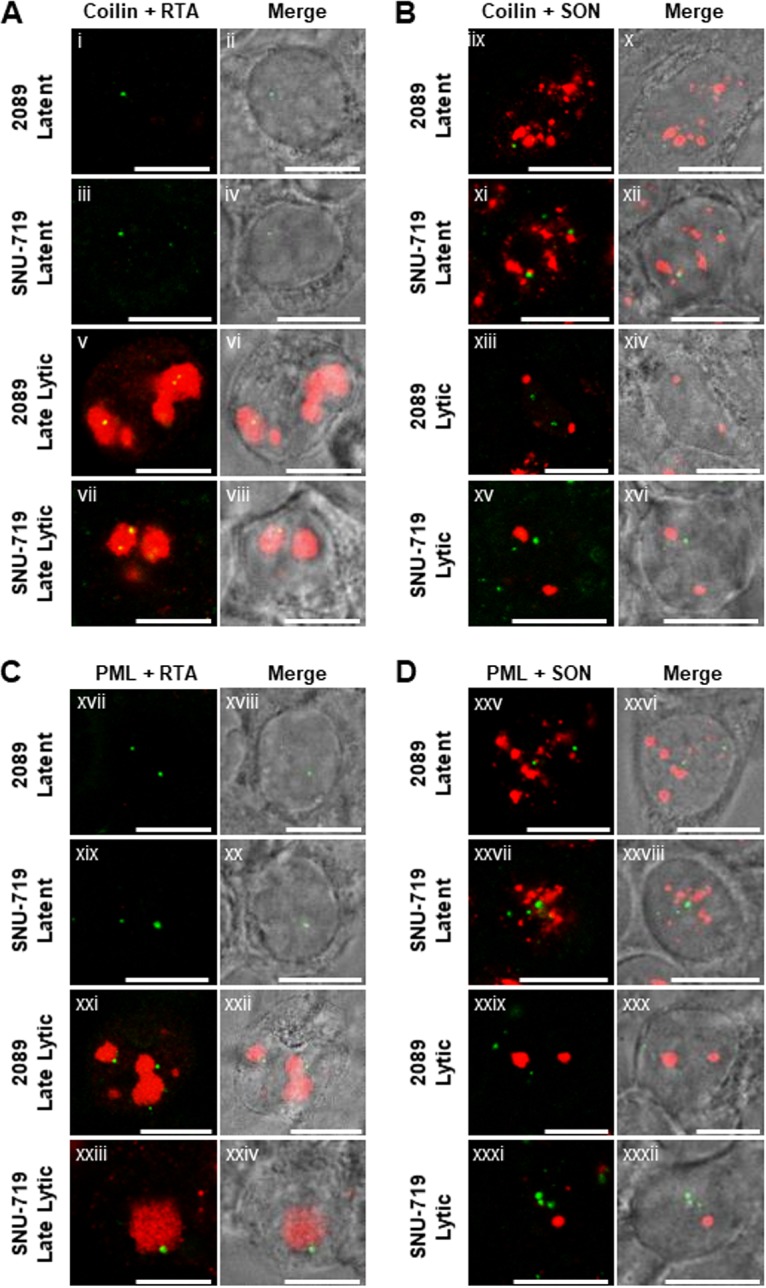
Coilin and PML protein are not recruited to VINORCs. 2089 cells or SNU-719 cells were left untreated (Ai to Aiv, Bix to Bxii, Cxvii to Cxx, and Dxxv to Dxxviii) or induced into the lytic phase of EBV infection by transfection with ZEBRA (Av to Aviii, Bxiii to Bxvi, Cxxi to Cxxiv, and Dxxix to Dxxxii). Cells were fixed then stained with primary antibodies specific for coilin (A and B; green), PML (C and D; green), RTA (A and C; red) and SON (B and D; red), and fluorophore-conjugated secondary antibodies. Transmitted light images are shown as indicated. Each row of two adjacent images depicts the same field of view. The reference bar in each image represents 10 μm.
PML, a component of PML nuclear bodies, has been implicated in a wide range of cellular functions, including p53-mediated apoptosis, cellular senescence, antiviral mechanisms, and DNA damage responses (73–78). In latently infected 2089 cells (Fig. 14Cxvii, Cxviii, Dxxv, and Dxxvi) and SNU-719 cells (Fig. 14Cxix, Cxx, Dxxvii, and Dxxviii), PML localized to 1 to 10 discrete punctate foci per nucleus, ranging 0.2 to 1 μm in diameter. Costaining with antibody to SON showed that during latency PML did not colocalize with SON in IGCs (Fig. 14Dxxv and Dxxvii). In cells undergoing lytic induction, there was no change in the number, size, shape, or appearance of PML foci (Fig. 14Cxxi to Cxxiv and Dxxix to Dxxxii) compared to latently infected cells. Costaining with RTA showed that PML foci localized near the periphery of viral replication compartments (Fig. 14Cxxi and Cxxiii). However, costaining with SON showed that during lytic EBV induction, foci of PML did not colocalize with nodules of SON (Fig. 14Dxxix and Dxxxi). In summary, neither coilin nor PML was recruited to VINORCs or viral replication compartments.
Cellular mRNA export adapters Y14 and ALY are depleted from the nucleus and recruited to VINORCs.
SC35 and SON are dispersed from IGCs during early lytic EBV infection; during late stages of lytic infection, these proteins are recruited to VINORCs and viral replication compartments. Dispersal of SC35 and SON from IGCs could be an important event in VHS. Since EBV replication compartments are enriched in viral mRNA and coincide with subnuclear regions devoid of translocated PABPC, recruitment of viral (BMLF1) and cellular (SC35, SON, and SRp20) proteins that mediate pre-mRNA splicing and mRNA nuclear export strongly suggests that VINORCs and viral replication compartments are involved in selective nuclear export of viral mRNA. To further investigate this hypothesis, we examined alterations in the intranuclear distribution of key cellular regulators of mRNA export.
Nuclear export of mRNA requires cotranscriptional processing (capping, splicing, and 3′-end formation) of pre-mRNA and recruitment of export adapters, such as Y14 and ALY, to RNA to form large messenger ribonucleoprotein (mRNP) complexes. Export adapters regulate nuclear export of mRNA by ensuring correct processing of mRNA prior to export and by mediating recruitment of nuclear export receptors (NXF1:NXT and CRM1) that dock the mature mRNP with the nuclear pore complex (NPC) and facilitate transit through the NPC to the cytoplasm (26, 79–83). We examined EBV-induced changes in nuclear organization of Y14 and ALY, particularly with respect to VINORCs and viral replication compartments, during progression of the lytic cycle.
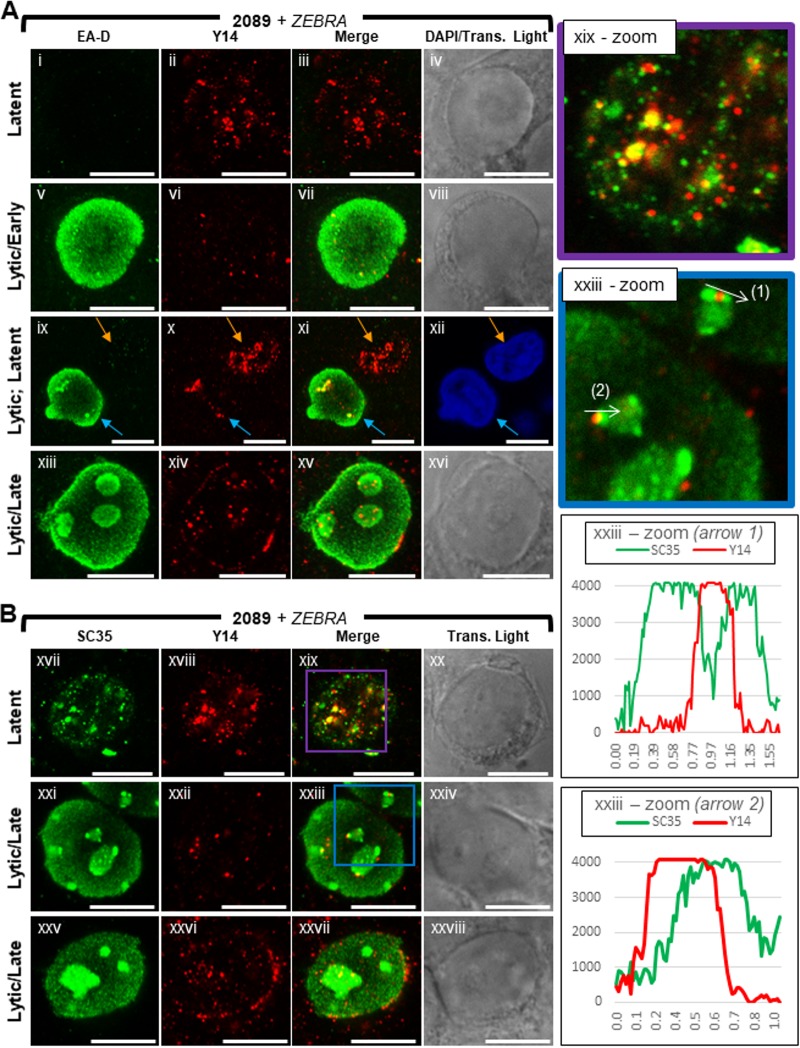
Y14, a core component of the exon junction complex, binds mRNA at a late stage of splicing and remains bound to mRNA until translation in the cytoplasm (84, 85). In 2089 cells latently infected with EBV (Fig. 15Ai to Aiv, Bxvii to Bxx, and Bxix-zoom), Y14 was present primarily in the nucleus, where it was distributed throughout the nuclear volume as numerous (>30) discrete round foci of various size <0.4 μm in diameter; few foci of Y14 were also present in the cytoplasm. Costaining of Y14 with SC35 showed a general enrichment of Y14 at IGCs marked by SC35. Most foci of Y14 either colocalized with SC35, partially overlapped with foci of SC35, or were immediately adjacent to foci of SC35. Enrichment of Y14 at IGCs marked by SC35 supports previously published analyses of Y14 localization (86).
FIG 15.
Y14 is depleted from the nucleus and recruited to viral replication compartments and the nuclear periphery during lytic EBV replication. 2089 cells were induced into the lytic phase of EBV infection by transfection with ZEBRA. Cells were fixed and stained with antibodies specific for EA-D (A), SC35 (B), and Y14 (A and B) and fluorophore-conjugated secondary antibodies. Each row of adjacent images depicts the same field of view. The reference bar in each image represents 10 μm. Enlarged images of latent stage (xix-zoom; purple outline) and late lytic stage (xxiii-zoom; blue outline) cells stained for SC35 and Y14 are shown. Fluorescence intensities of SC35 and Y14 were measured along VINORCs, indicated by white arrows (xxxiii-zoom, arrows 1 and 2). Fluorescence intensity values are plotted on the accompanying graphs.
Lytic induction of EBV caused a comprehensive loss of Y14 foci from nuclei. In most early lytic cells marked by diffuse EA-D (Fig. 15Av to Aviii), few (<10) foci of Y14 remained in the nucleus. Even though the abundance of Y14 was decreased, the size and round morphology of individual Y14 foci seen during latency were unchanged following lytic induction. In late lytic cells (Fig. 15Axiii to Axvi and Bxxi to Bxxviii), foci of Y14 were consistently enriched within viral replication compartments and at segments of the nuclear membrane.
Depletion of Y14 foci persisted into the late lytic stage. Despite recruitment of Y14 to viral replication compartments and the nuclear membrane, the subnuclear region surrounding viral replication compartments was depleted of Y14 foci. Nonetheless, some foci of Y14 were present at the periphery of viral replication compartments. Costaining of Y14 with SC35 in cells at the late lytic stage showed recruitment of Y14 to VINORCs; however, foci of Y14 foci did not precisely colocalize with VINORCs marked by SC35. Y14 foci often partially overlapped or were immediately adjacent to VINORCs marked by SC35 (Fig. 15Bxxi to Bxxiv and Bxxiii-zoom, arrows 1 and 2 and graphs).
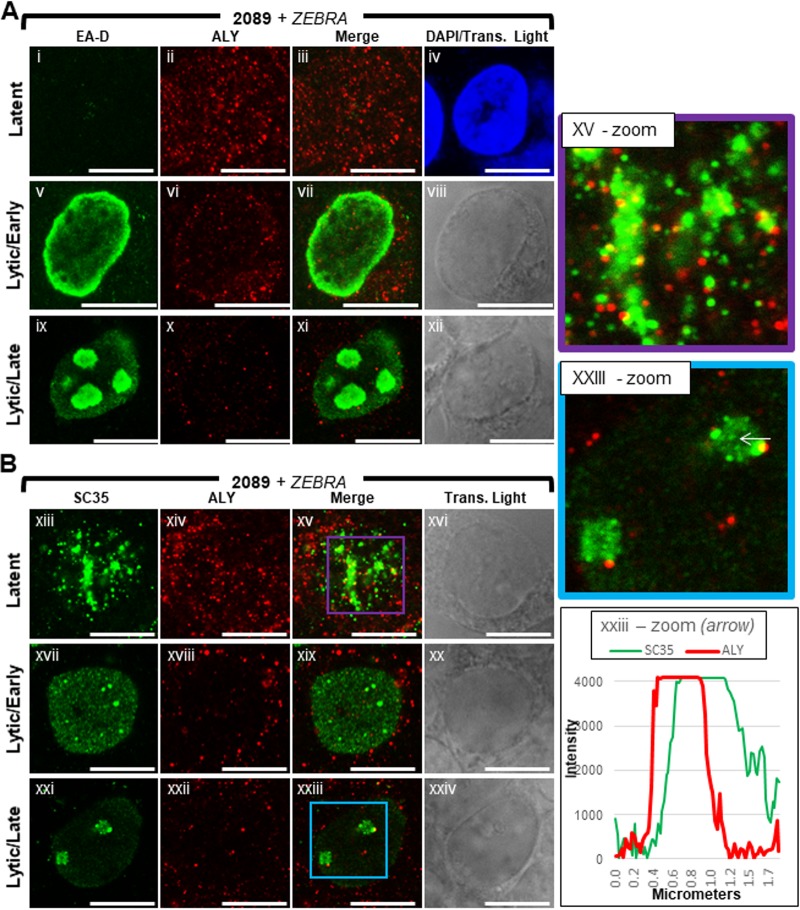
ALY, a well-characterized export adapter, is a core component of TREX, the key regulator of NXF1-mediated nuclear export of mRNA (26). ALY, as part of TREX, recruits the primary mRNA export receptor heterodimer, NXF1:NXT, to mRNPs (81). Binding of TREX to the 5′ end of mRNA is mediated by the cap binding complex and splicing machinery, thereby linking processing of mRNA to nuclear export (25, 87, 88).
In latently infected 2089 cells (Fig. 16Ai to Aiv, Bxiii to xvi, and Bxv-zoom), ALY was present throughout the nucleus and cytoplasm. Unlike for Y14, which was rarely seen in the cytoplasm, the abundance of ALY was often elevated in the cytoplasm. ALY was distributed throughout the cell as numerous (>100) discrete foci of various sizes <0.3 μm in diameter. Similarly to Y14, lytic induction of EBV resulted in a dramatic loss of ALY foci throughout the nucleus; the abundance of ALY foci was also reduced in the cytoplasm. The size and shape of individual ALY foci seen during latency appeared unchanged during lytic induction. In both early lytic and late lytic cells (Fig. 16Av to Axii and Bxvii to Bxxiv), few (<12) foci of ALY were present in each nucleus. Unlike for Y14, enrichment of ALY within viral replication compartments or at the nuclear membrane was not seen during lytic replication. Costaining of ALY with SC35 often showed recruitment of foci of ALY to VINORCs; however, foci of ALY did not precisely colocalize with VINORCs marked by SC35. ALY foci often partially overlapped or were immediately adjacent to VINORCs marked by SC35 (Fig. 16Bxxiii-zoom, arrow, graph).
FIG 16.
ALY is depleted from the nucleus during lytic EBV replication. 2089 cells were induced into the lytic phase of EBV infection by transfection with ZEBRA. Cells were fixed and stained with antibodies specific for EA-D (A), SC35 (B), and ALY (A and B) and fluorophore-conjugated secondary antibodies. Each row of adjacent images depicts the same field of view. The reference bar in each image represents 10 μm. Enlarged images of latent stage (xv-zoom; purple outline) and late lytic stage (xxiii-zoom; blue outline) cells stained for SC35 and ALY are shown. Fluorescence intensities of SC35 and ALY were measured along a VINORC, indicated by a white arrow (xxxiii-zoom). Fluorescence intensity values are plotted on the accompanying graph.
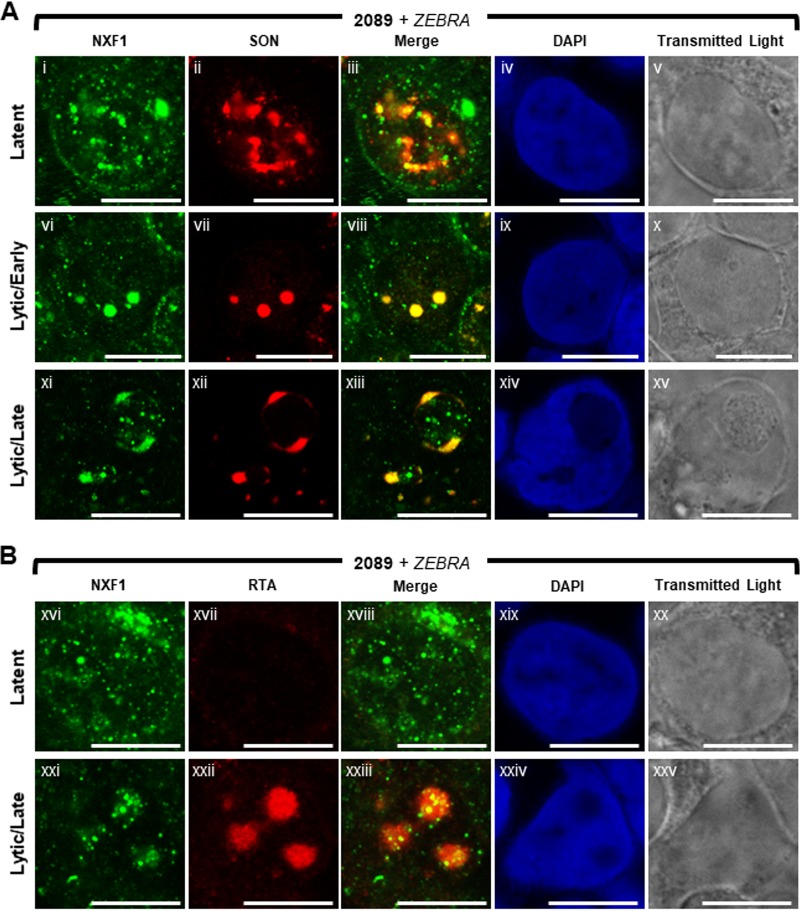
The major mRNA export receptor, NXF1, is recruited to VINORCs and viral replication compartments.
NXF1 is the principal export receptor of mRNA from mammalian cell nuclei (79, 82, 83). Export adapters, particularly ALY and SR proteins, such as SRp20, recruit NXF1 to mRNPs (28, 81, 83). Association of mature mRNP with NXF1 enables docking with NPC basket nucleoporins (i.e., TPR and Nup153) and subsequent transit through the NPC (82, 89). In latently infected 2089 cells (Fig. 17Ai to Av and Bxvi to Bxx), NXF1 was present throughout the nucleus and cytoplasm. NXF1 was distributed as numerous (>50) discrete small foci, < 0.2 μm to 1 μm in diameter, and as fewer (1 to 5) larger aggregations, 1 μM to 2 μm in width. Costaining of NXF1 with SON (Fig. 17Ai to Av) showed that although foci of NXF1 did not colocalize with SON in IGCs, NXF1 foci were enriched at IGCs. Enrichment of NXF1 in the vicinity of IGCs supports previously published analyses of NXF1 localization (86).
FIG 17.
NXF1 is redistributed and recruited to VINORCs and viral replication compartments during lytic EBV replication. 2089 cells were induced into the lytic phase of EBV infection by transfection with ZEBRA. Cells were fixed and stained with antibodies specific for SON (A), RTA (B), and NXF1 (A and B), fluorophore-conjugated secondary antibodies, and DAPI. Each row of adjacent images depicts the same field of view. The reference bar in each image represents 10 μm.
Lytic induction of EBV caused a decline in the abundance of foci of NXF1 (Fig. 17Avi to Ax, Axi to Axv, and Bxxi to Bxxv). During early lytic stage, costaining of NXF1 and SON (Fig. 17Avi to Ax) showed that nodules of NXF1 precisely colocalized with nodules of SON. Most strikingly, during late lytic infection, NXF1 was strongly recruited to VINORCs and precisely colocalized with SON in VINORCs (Fig. 17Axi to Axv). Within VINORCs, NXF1 was distributed as a globular mass and as small foci. During the late lytic stage, foci of NXF1 were also recruited to the interior of viral replication compartments marked by RTA (Fig. 17Bxxi to Bxxv). Viral replication compartments characterized by an absence of DAPI staining were enriched in foci of NXF1 (Fig. 17Axi to Axv and Bxxi to Bxxv); DAPI-positive intranuclear regions outside viral replication compartments were depleted of NXF1 foci. In summary, NXF1 is a component of VINORCs and is also recruited to the interior of viral replication compartments; concurrently, NXF1 is depleted from intranuclear regions surrounding VINORCs and viral replication compartments.
DISCUSSION
The host cell nucleus is profoundly reorganized and restructured to facilitate essential processes in the lytic phase of the viral life cycle of herpesviruses. These processes include viral DNA synthesis, virus-induced host shutoff (VHS), nuclear egress of viral capsids, and evasion of host antiviral responses. In this report we describe additional previously unrecognized changes in nuclear architecture in which viral and cellular factors involved in mRNA splicing and nuclear export are redistributed during EBV lytic replication. These changes, including the formation of VINORCs, are likely to contribute additional mechanisms responsible for VHS and selective processing and export of herpesviral mRNAs.
Novel observations.
We describe several novel observations about changes in nuclear architecture during progression of EBV lytic infection. During the early lytic stage, (i) nodular structures containing EBV proteins, BMLF1 and BGLF5, and EBV lncRNA, BHLF1, were formed; (ii) cellular splicing factors, SC35 and SON, were dispersed from IGCs; and (iii) cellular mRNA export factors, Y14, ALY, and NXF1, were depleted from the nucleus. During the late lytic stage following assembly of viral replication compartments, (iv) viral (BMLF1 and BGLF5) and cellular (SC35, SON, SRp20, Y14, ALY, and NXF1) proteins that mediate pre-mRNA processing and mRNA export and an EBV lncRNA (BHLF1) were recruited to nodular structures (VINORCs) situated at the periphery of viral replication compartments; cellular (PML and coilin) and viral (RTA and EA-D) nuclear proteins with no known direct role in RNA processing or nuclear export were not recruited to VINORCs. (v) Some viral (BMLF1 and BGLF5) and cellular (SC35, Y14, and NXF1) proteins were recruited to the interior of viral replication compartments. (vi) Nuclear regions outside VINORCs and viral replication compartments remained depleted of SC35, SON, Y14, ALY, and NXF1.
Dispersal of SC35 and SON during the early lytic stage and recruitment to VINORCs during the late lytic stage occurred in nearly every cell undergoing lytic EBV induction (Fig. 7). Although previously unrecognized, both processes represent invariable, fundamental changes to host cell nuclear architecture during lytic EBV replication. VINORCs are novel EBV-induced nuclear structures. VINORCs were consistently observed during lytic replication of EBV in all cell types analyzed: epithelial cell lines containing EBV-bacmids (2089, BZKO), an EBV-infected gastric carcinoma cell line (SNU-719), an EBV-infected Burkitt lymphoma cell line (HH514-16), and an EBV-infected marmoset B-lymphoblastoid cell line (B95-8 [data not shown]). Selective recruitment to VINORCs of viral and cellular proteins that mediate pre-mRNA processing and mRNA export, as well as the location of VINORCs at the surface of viral replication compartments containing EBV mRNA, strongly suggest that VINORCs participate in the processing and nuclear export of viral mRNAs in the context of VHS.
Comparison and contrast of mechanisms of vhs and viral mRNA export by different herpesviruses.
Global degradation of host mRNA, nuclear translocation of PABPC, and a block to nuclear export of host mRNA are events that are common to the host shutoff programs of EBV, Kaposi's sarcoma-associated herpesvirus (KSHV), and HSV-1 (20, 21, 90–92). Host shutoff during lytic infection of HSV-1 is mediated by two viral proteins: vhs protein, a riboendonuclease belonging to the FEN-1 family of nucleases (93), and ICP27, a multifunctional immediate early protein that is also the principal regulator of nuclear export of HSV-1 mRNA (94, 95). Host shutoff during KSHV infection is activated by a single protein, SOX, a conserved viral alkaline endonuclease (96). Host shutoff during EBV infection is mediated by two viral proteins: BGLF5, a homolog of SOX, and ZEBRA, an immediate early protein that also functions as a transcription factor and viral replication protein (21, 91). Two unrelated proteins, the vhs protein and BGLF5/SOX, stimulate turnover of host mRNA via activation of the cellular endonuclease Xrn1 (97).
As the principal virally encoded regulator of nuclear export of viral mRNA, EBV BMLF1 is the functional homolog of HSV-1 ICP27 (98). Both proteins bind RNA, shuttle between the nucleus and the cytoplasm, interact with SR proteins, and regulate splicing of specific mRNAs (38, 45, 99–101). BMLF1 and ICP27 differ in localization and in their roles in redistribution of SC35. BMLF1 is an invariable component of VINORCs, yet ICP27 does not localize to nodules of SC35 during lytic HSV-1 infection.
During lytic HSV-1 infection, ICP27 contributes to host shutoff by inhibiting pre-mRNA splicing and reducing levels of cytoplasmic cellular spliced mRNAs (102). ICP27 mediates inhibition of splicing through interactions with SR proteins and by modulating phosphorylation of SR proteins via interaction with SR protein kinase 1 (SRPK1) (103). ICP27 reorganizes SC35 and snRNPs within IGCs into discrete, nodular structures outside viral replication compartments (104, 105). ICP27-mediated inhibition of splicing and reorganization of cellular splicing factors during HSV-1 infection concurrently fulfill dual functions, namely, inhibition of nuclear export of host mRNAs that are predominantly spliced and selective nuclear export of HSV-1 mRNAs that are mainly intronless (101, 106).
Nuclear reorganization in EBV- and HSV-1-infected cells.
In a manner similar to ICP27-mediated inhibitory effects on splicing, EBV-induced changes may contribute to host shutoff and selective nuclear export of EBV mRNAs. During early lytic EBV infection, SC35 and SON were dispersed from IGCs. In approximately 55% of cells undergoing early lytic infection, this dispersed distribution was accompanied by discrete nodules containing SC35, SON, BMLF1, BGLF5, and BHLF1; these EBV-induced nodules resembled ICP27-induced nodules of SC35 seen during lytic HSV-1 infection. However, in approximately 45% of cells undergoing early EBV lytic infection, SC35 and SON were completely dispersed and no nodules were present. At the subsequent late lytic stage of EBV infection, nodules containing SC35 and SON, as well as SRp20, BMLF1, BGLF5, BHLF1, and NXF1, were reconstituted as VINORCs at the periphery of viral replication compartments. In contrast, complete dispersal of SC35 from IGCs has not been reported during lytic HSV-1 infection. ICP27-induced nodules of SC35 may result from condensation of SC35 within pre-existing IGCs rather than dispersal of SC35 from IGCs and reconstitution as VINORCs as observed in EBV infection.
The most obvious differences between HSV-1 and EBV are the lack of enrichment of SC35 in the interior of HSV-1 replication compartments and the strong recruitment of SC35 to the interior of EBV replication compartments (96% of late lytic EBV-infected cells [Fig. 7]). Efficient recruitment of SC35 into EBV replication compartments is a unique distribution not shared by other VINORC components, including the related SR protein splicing factor SRp20. Unlike other SR proteins, SC35 promotes transcriptional elongation by Pol II (53). Recruitment of SC35 to EBV replication compartments may therefore indicate an additional transcriptional regulatory role for SC35 within viral replication compartments. By dispersal from IGCs followed by recruitment to VINORCs and replication compartments, SC35 may selectively promote viral gene expression through both splicing and transcriptional elongation.
Recruitment of NXF1 to cellular mRNPs requires that particular combinations of export adapters associate with mRNA (83). During lytic induction of EBV, nuclear export adapters ALY and Y14 and the major export receptor, NXF1, are cleared from intranuclear regions outside viral replication compartments. NXF1 is strongly recruited to VINORCS, and foci of ALY and Y14 often overlap VINORCs. NXF1 and Y14 are recruited to the interior of EBV-1 replication compartments, whereas ALY is not. In contrast, ALY is strongly recruited to the interior of HSV-1 replication compartments by ICP27, whereas NXF1 is neither recruited to HSV-1 replication compartments nor depleted from nuclear regions outside viral replication compartments.
Assembly of VINORCs.
SON and the EBV BHLF1 lncRNA may play important roles in mediating assembly of EBV-induced nodules and VINORCs. SON maintains proper organization of pre-mRNA processing factors in IGCs (58). Analogous to its structural role in IGCs, SON may play a role in assembly and structural maintenance of VINORCs. A long stretch of repetitive amino acids and an RS domain mediating protein-protein interactions with other SR splicing factors may facilitate this role. In VINORCs SON may function to recruit and organize cellular and viral proteins involved in viral mRNA processing and export.
The GC-rich EBV lncRNA BHLF1 may also assume a structural role in VINORCs. A series of 12 125-bp tandem repeat sequences within BHLF1 suggests that the lncRNA may play structural and scaffolding roles in recruitment of RNA-binding proteins to VINORCs. A stable population of polyadenylated cellular noncoding RNAs was identified within IGCs by electron microscopy (107). The nuclear cellular lncRNA MALAT1 localizes to IGCs, interacts with SR proteins, and controls the cellular levels of hyperphosphorylated active forms of SR proteins (108). MALAT1 affects the distribution of a subset of splicing factors within IGCs and regulates alternative splicing (108). lncRNAs, both viral and cellular, may function as structural platforms for the recruitment of particular RNA-binding splicing factors and SR protein kinases. SON and BHLF1 may have redundant structural functions in VINORC assembly; conversely, SON and BHLF1 may both be required to maintain VINORC structure: SON and BHLF1 may each recruit different splicing factors to VINORCs.
Selectivity of recruitment to VINORCs.
Certain viral and cellular proteins are recruited to VINORCs, whereas others are not (Table 1). Selective recruitment of proteins of known function to VINORCs provides clues to possible functional roles for VINORCs during lytic infection. All eight proteins recruited to VINORCs are known to bind RNA. Seven proteins (BMLF1, SC35, SON, SRp20, Y14, ALY, and NXF1) have well-characterized roles in pre-mRNA splicing and mRNA nuclear export. The fifth protein, BGLF5, is a nuclease that induces cleavage of cellular mRNAs during VHS (91, 97, 109, 110). Recruitment of BMLF1 to VINORCs is significant; BMLF1 is a nucleocytoplasmic shuttling protein (38, 98) that plays well-characterized and essential roles in nuclear export of EBV mRNAs (34, 37, 38, 40, 46, 98) and regulates alternative splicing (39, 43, 46, 111). Recruitment of SRp20 to VINORCs, where it colocalizes with BMLF1, is also significant, since regulation of alternative splicing by BMLF1 requires direct interaction with SRp20 (43). Recruitment of NXF1, the principal cellular nuclear export receptor of mRNA, to VINORCs is strong evidence of a role for VINORCs in nuclear export of mRNA.
TABLE 1.
Summary of the selective recruitment of viral and cellular factors to VINORCs and viral replication compartments during lytic EBV replicationa
| Protein/RNA name | Gene name | Type of factor | Subnuclear recruitment |
Function(s) | |
|---|---|---|---|---|---|
| Nodule/VINORC | Replication compartment | ||||
| BMLF1 | BMLF1 | Viral | + | + | Splicing factor; RNA export factor |
| BGLF5 | BGLF5 | Viral | + | + | Nuclease; host shutoff |
| BHLF1 | BHLF1 | Viral | + | − | Long noncoding RNA |
| SC35 | SRSF2 | Cellular | +/− | + | Splicing factor; transcription elongation |
| SON | SON | Cellular | + | − | Splicing factor |
| SRp20 | SRSF3 | Cellular | + | − | Splicing factor; RNA export factor |
| Y14 | Y14 | Cellular | + | + | RNA processing; RNA export |
| ALY | ALY | Cellular | + | − | RNA processing; RNA export |
| NXF1 | NXF1 | Cellular | + | + | RNA export |
| ZEBRA | BZLF1 | Viral | − | + | Transcription factor; viral DNA replication |
| RTA | BRLF1 | Viral | − | + | Transcription factor; viral DNA replication |
| EA-D | BMLF1 | Viral | − | + | Viral DNA replication |
| EBER-1 | Viral | − | − | Noncoding RNA | |
| Coilin | COIL | Cellular | − | − | Cajal body structure |
| PML | PML | Cellular | − | − | Stress response; DNA damage response; apoptosis |
| Lamin B | LMNB1/2 | Cellular | − | − | Nucleus structure |
Nuclear viral and cellular proteins and viral nontranslated RNAs are classified according to their recruitment to VINORCs and viral replication compartments during lytic EBV infection. The known functions of each factor is indicated.
Other viral and cellular proteins that were excluded from VINORCs, namely, ZEBRA (viral transcription factor and replication protein), RTA (viral transcription factor and replication protein), EA-D (polymerase processivity factor), PML (regulator of transcription), coilin (Cajal body structural protein), and lamin B (nucleus structural protein), have no known roles in pre-mRNA splicing or mRNA nuclear export. Since these proteins are not recruited to VINORCs, recruitment of certain nuclear proteins to VINORCs is specific. Two other discrete subnuclear nodular structures, PML bodies and Cajal bodies, that have no direct functions in pre-mRNA splicing or nucleocytoplasmic mRNA export were distinct from VINORCs. The selective incorporation of RNA splicing and nuclear export factors into VINORCs, the exclusion of other nuclear proteins and nuclear RNAs from VINORCs with functions other than RNA splicing and nuclear export, and the location of VINORCs at the periphery of viral replication compartments containing viral mRNAs strongly suggest a possible role for VINORCs in the splicing and nuclear export of viral mRNAs.
IGC reorganization, redistribution of mRNA export factors, and VINORC assembly during EBV lytic induction may all contribute to host shutoff.
IGCs serve as dynamic repositories and recycling centers that supply essential pre-mRNA splicing factors, such as SC35 and snRNPs, and transcription factors, such as RNA Pol II large subunit, to sites of active transcription (66). SON is required to maintain the structural integrity of IGCs; SC35 is essential for constitutive splicing of pre-mRNAs. The dispersal of SC35 and SON during the early lytic stage of EBV infection may thus impair splicing and nuclear export of host pre-mRNA on a global level. EBV mRNAs are mostly intronless and thus may be resistant to the reduced supply of splicing factors caused by dispersal of SC35 and SON from IGCs.
The mRNA export adapters ALY and Y14 and the major mRNA export receptor, NXF1, are essential for nuclear export of bulk cellular mRNA. Intranuclear depletion of these factors may significantly contribute to VHS; recruitment of NXF1 to VINORCs and viral replication compartments may enable selective nuclear export of EBV mRNAs. Dispersal of IGCs and depletion of mRNA export factors may be two additional mechanisms that act in concert with translocation of PABPC and degradation of host mRNA by Xrn1 to effect host shutoff.
Following dispersal of SC35 and SON during the early viral lytic stage, both proteins, as well as SRp20, a cellular splicing factor and NXF1-binding export adapter (27, 28), BMLF1, the principal EBV mRNA export factor, and NXF1 itself, were recruited to VINORCs at the periphery of EBV replication compartments. EBV mRNAs localize to viral replication compartments (112) that reside within subnuclear regions devoid of VHS-induced translocated PABPC; viral mRNAs present in these nuclear subregions would not be susceptible to nuclear retention mediated by binding of translocated PABPC. VINORCs situated at the periphery of viral replication compartments may supply viral and cellular splicing factors and mRNA export factors specifically to sites of active viral transcription in a manner similar to the function of IGCs. Alternatively, VINORCs may be specialized structures that function as channels or hubs of viral mRNA processing and nuclear export. VINORCs and viral replication compartments may function as privileged nuclear subdomains that allow selective pre-mRNA processing and nuclear export of EBV mRNAs in the context of VHS-mediated block to expression of host mRNAs.
MATERIALS AND METHODS
Cell lines.
293HEK is a human embryonic kidney cell line immortalized by the early region of adenovirus (113). 2089 is a 293HEK cell line stably transfected with a bacmid containing the B95-8 EBV genome and a hygromycin B resistance gene (114). BZKO is a 293HEK cell line containing an EBV bacmid in which the BZLF1 gene has been inactivated by insertion of a kanamycin resistance cassette (115). SNU-719 is a gastric carcinoma cell line infected with EBV (44). HH514-16 is a subclone of the P3J-HR1K Burkitt lymphoma cell line (116). 293HEK cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS). 2089 and BZKO cells were maintained in DMEM supplemented with 10% FBS and 100 μg/ml hygromycin B. SNU-719 and HH514-16 cells were maintained in RPMI medium supplemented with 10% FBS. All cell cultures contained 50 units/ml penicillin-streptomycin and 1 μg/ml amphotericin B.
Expression vectors.
Control empty vector, pHD1013, containing the CMV immediate early promoter, and expression vectors containing wild-type BZLF1 or mutant Z(Y180E) BZLF1 genes in pHD1013 have been described previously (59, 117, 118). pFLAG-BGLF5 was constructed as follows: the full-length BGLF5 gene was amplified by PCR using pEGFP-CPO-IP-EBV-DNase plasmid (kindly provided by Su-Fang Lin) as a template and then ligated into pCMV-FLAG vector cleaved by HindIII.
Antibodies.
In immunofluorescence experiments, RTA was detected using rabbit polyclonal antisera raised to a purified 320-amino-acid N-terminal fragment of RTA that was generated using the pET expression system (Novagen) (119). EA-D was detected using the mouse monoclonal antibody R3.1 (120). BGLF5 was detected using rabbit polyclonal antisera raised against nearly full-length (amino acids 2 to 469) BGLF5 protein expressed in Escherichia coli using pET22 vector containing the corresponding encoding gene sequences. BMLF1 was detected using rabbit polyclonal antisera raised against full-length BMLF1 protein expressed in E. coli using pET22 vector containing the BMLF1 gene. SC35, PML, β-actin, FLAG tag, and coilin were detected using commercially available mouse monoclonal antibodies (ab11826 [Abcam], sc-966 [Santa Cruz], A5316 [Sigma], F1804 [Sigma], and ab87913 [Abcam]). SON, SRp20, Y14, ALY, and CBP20 were detected using commercially available rabbit polyclonal antibodies (ab121759 [Abcam], LS-C352832 [LifeSpan BioSciences], ab229573[Abcam], ab202894 [Abcam], and ab224569 [Abcam]). Lamin B was detected using a goat polyclonal antibody purchased from Santa Cruz Biotechnology (sc-6216). Secondary antibodies used in immunofluorescence experiments were purchased from Jackson ImmunoResearch Labs: Alexa Fluor 488-donkey anti-mouse (number 715-545-150), Cy3-donkey anti-rabbit (number 711-165-152), and Alexa Fluor 647-donkey anti-goat (number 805-605-180).
Indirect immunofluorescence.
Cells grown on glass coverslips were transfected with plasmid DNA using Lipofectamine 2000 reagent (Invitrogen) according to manufacturer's recommended protocol. Forty-five hours after transfection, a time previously determined to be adequate for detection of lytic viral DNA replication, cells were fixed in chilled methanol for 30 min at −20°C, washed with phosphate-buffered saline (PBS), and incubated in blocking solution (10% human serum in PBS) for 1 h at room temperature. Cells were stained with primary antibody diluted in blocking solution for 1 h at room temperature in humidified chambers. Cells were washed with PBS and then incubated with secondary antibody in blocking solution for 1 h at room temperature. Cells were washed with PBS, briefly rinsed in distilled H2O, and then mounted on glass slides using Vectashield mounting medium (Vector Laboratories). Digital images of fluorescence and transmitted light were acquired on a Zeiss LSM510 confocal laser scanning microscope using a 63× objective lens. Quantitative measurements of fluorescence intensities of indirect immunofluorescently labeled SC35, SON, RTA, and EA-D were made using ImageJ software. A 3-dimensional representation of nodules of BHLF1 lncRNA and a viral replication compartment marked by EA-D within a lytically induced BZKO nucleus was reconstructed from a Z-stack of digital micrographs using Volocity 3-D imaging software (version 6.3; PerkinElmer).
Super-resolution microscopy.
Cells grown on coverslips were transfected, fixed, stained, and mounted on coverslips as described above. Super-resolution images were obtained by stimulated emission depletion (STED) confocal microscopy using a Leica SP8 gated STED 3× microscope equipped with Leica LAS-X software. STED images were acquired via a 100× objective lens and enhanced by Huygens STED deconvolution software.
DNA replication assay.
A total of 2 × 106 cells were resuspended in 400 μl lysis buffer containing 50 mM Tris-HCl (pH 8.1), 1% SDS, and 10 mM EDTA. Cells were lysed by sonication. Cell lysate was centrifuged. Lysate was diluted in 16.7 mM Tris-HCl (pH 8.1), 0.01% SDS, 1.1% Triton X-100, 167 mM NaCl, and 1.2 mM EDTA.
RNA FISH analysis.
Cells grown on glass coverslips were fixed in a solution of 3.7% paraformaldehyde in PBS, washed several times in PBS, and then permeabilized overnight in 70% ethanol at 4°C. Cells were washed twice with SSCF wash buffer (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 30% formamide), prehybridized for 1 h at 37°C in hybridization solution, and then hybridized overnight at 37°C with biotinylated oligonucleotide probes diluted in hybridization solution. All probes used in RNA FISH analyses were DNA oligonucleotides purchased from IDT. Probe sequences are listed in Table S1 in the supplemental material. Oligonucleotide probes were biotinylated using a commercially available oligonucleotide 3′-end labeling kit (Roche; number 03353575910) and biotin-16-ddUTP (Roche; number 11427598910). Cells were washed twice in SSCF buffer at 37°C, followed by 2 more washes in 2× SSC. Cells were incubated in blocking solution (5% bovine serum albumin [BSA] in PBS) for 1 h, followed by a 1-h incubation with Cy3-streptavidin (Jackson ImmunoResearch). When proteins were also analyzed by immunofluorescence, cells were incubated for 1 h with primary antibody and 1 h with fluorophore-conjugated secondary antibody following the blocking step. Cells were washed several times with PBS, briefly rinsed in distilled H2O, and then mounted on glass slides using Vectashield mounting medium (Vector Laboratories). A Zeiss LSM510 confocal laser scanning microscope was used to obtain digital images of fluorescence and transmitted light.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by NIH grants CA16038, CA12055, and S10 OD020142.
We thank Joan Steitz, Tenaya Vallery, Karla Neugebauer, Ayman El-Guindy, and Danielle Lyons for helpful discussions. We thank Henri-Jacques Delecluse for generously providing 2089 and BZKO cells. We thank Jae Gahb Park and the Korean Cell Line Bank for generously providing SNU-719 cells. We are grateful to Susan Prisley and Karen Lavery for help in preparation of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01254-18.
REFERENCES
- 1.Quinlan MP, Chen LB, Knipe DM. 1984. The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell 36:857–868. doi: 10.1016/0092-8674(84)90035-7. [DOI] [PubMed] [Google Scholar]
- 2.Knipe DM, Senechek D, Rice SA, Smith JL. 1987. Stages in the nuclear association of the herpes simplex virus transcriptional activator protein ICP4. J Virol 61:276–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takagi S, Takada K, Sairenji T. 1991. Formation of intranuclear replication compartments of Epstein-Barr virus with redistribution of BZLF1 and BMRF1 gene products. Virology 185:309–315. doi: 10.1016/0042-6822(91)90778-A. [DOI] [PubMed] [Google Scholar]
- 4.Bush M, Yager DR, Gao M, Weisshart K, Marcy AI, Coen DM, Knipe DM. 1991. Correct intranuclear localization of herpes simplex virus DNA polymerase requires the viral ICP8 DNA-binding protein. J Virol 65:1082–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Bruyn Kops A, Uprichard SL, Chen M, Knipe DM. 1998. Comparison of the intranuclear distributions of herpes simplex virus proteins involved in various viral functions. Virology 252:162–178. doi: 10.1006/viro.1998.9450. [DOI] [PubMed] [Google Scholar]
- 6.Myllys M, Ruokolainen V, Aho V, Smith EA, Hakanen S, Peri P, Salvetti A, Timonen J, Hukkanen V, Larabell CA, Vihinen-Ranta M. 2016. Herpes simplex virus 1 induces egress channels through marginalized host chromatin. Sci Rep 6:28844. doi: 10.1038/srep28844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monier K, Armas JC, Etteldorf S, Ghazal P, Sullivan KF. 2000. Annexation of the interchromosomal space during viral infection. Nat Cell Biol 2:661–665. doi: 10.1038/35023615. [DOI] [PubMed] [Google Scholar]
- 8.Simpson-Holley M, Baines J, Roller R, Knipe DM. 2004. Herpes simplex virus 1 U(L)31 and U(L)34 gene products promote the late maturation of viral replication compartments to the nuclear periphery. J Virol 78:5591–5600. doi: 10.1128/JVI.78.11.5591-5600.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin TE, Barghusen SC, Leser GP, Spear PG. 1987. Redistribution of nuclear ribonucleoprotein antigens during herpes simplex virus infection. J Cell Biol 105:2069–2082. doi: 10.1083/jcb.105.5.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daikoku T, Kudoh A, Fujita M, Sugaya Y, Isomura H, Shirata N, Tsurumi T. 2005. Architecture of replication compartments formed during Epstein-Barr virus lytic replication. J Virol 79:3409–3418. doi: 10.1128/JVI.79.6.3409-3418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Everett RD, Maul GG. 1994. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J 13:5062–5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahn JH, Hayward GS. 1997. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J Virol 71:4599–4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Müller S, Dejean A. 1999. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J Virol 73:5137–5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adamson AL, Kenney S. 2001. Epstein-Barr virus immediate-early protein BZLF1 is SUMO-1 modified and disrupts promyelocytic leukemia bodies. J Virol 75:2388–2399. doi: 10.1128/JVI.75.5.2388-2399.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scott ES, O'Hare P. 2001. Fate of the inner nuclear membrane protein lamin B receptor and nuclear lamins in herpes simplex virus type 1 infection. J Virol 75:8818–8830. doi: 10.1128/JVI.75.18.8818-8830.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reynolds AE, Liang L, Baines JD. 2004. Conformational changes in the nuclear lamina induced by herpes simplex virus type 1 require genes U(L)31 and U(L)34. J Virol 78:5564–5575. doi: 10.1128/JVI.78.11.5564-5575.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park R, Baines JD. 2006. Herpes simplex virus type 1 infection induces activation and recruitment of protein kinase C to the nuclear membrane and increased phosphorylation of lamin B. J Virol 80:494–504. doi: 10.1128/JVI.80.1.494-504.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dobrikova E, Shveygert M, Walters R, Gromeier M. 2010. Herpes simplex virus proteins ICP27 and UL47 associate with polyadenylate-binding protein and control its subcellular distribution. J Virol 84:270–279. doi: 10.1128/JVI.01740-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salaun C, MacDonald AI, Larralde O, Howard L, Lochtie K, Burgess HM, Brook M, Malik P, Gray NK, Graham SV. 2010. Poly(A)-binding protein 1 partially relocalizes to the nucleus during herpes simplex virus type 1 infection in an ICP27-independent manner and does not inhibit virus replication. J Virol 84:8539–8548. doi: 10.1128/JVI.00668-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee YJ, Glaunsinger BA. 2009. Aberrant herpesvirus-induced polyadenylation correlates with cellular messenger RNA destruction. PLoS Biol 7:e1000107. doi: 10.1371/journal.pbio.1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park R, El-Guindy A, Heston L, Lin SF, Yu KP, Nagy M, Borah S, Delecluse HJ, Steitz J, Miller G. 2014. Nuclear translocation and regulation of intranuclear distribution of cytoplasmic poly(A)-binding protein are distinct processes mediated by two Epstein Barr virus proteins. PLoS One 9:e92593. doi: 10.1371/journal.pone.0092593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lei EP, Krebber H, Silver PA. 2001. Messenger RNAs are recruited for nuclear export during transcription. Genes Dev 15:1771–1782. doi: 10.1101/gad.892401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Köhler A, Hurt E. 2007. Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol 8:761–773. doi: 10.1038/nrm2255. [DOI] [PubMed] [Google Scholar]
- 24.Elbarbary RA, Maquat LE. 2016. Coupling pre-mRNA splicing and 3′ end formation to mRNA export: alternative ways to punch the nuclear export clock. Genes Dev 30:487–488. doi: 10.1101/gad.278937.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou Z, Luo MJ, Straesser K, Katahira J, Hurt E, Reed R. 2000. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature 407:401–405. doi: 10.1038/35030160. [DOI] [PubMed] [Google Scholar]
- 26.Stutz F, Bachi A, Doerks T, Braun IC, Seraphin B, Wilm M, Bork P, Izaurralde E. 2000. REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA 6:638–650. doi: 10.1017/S1355838200000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang Y, Steitz JA. 2001. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol Cell 7:899–905. doi: 10.1016/S1097-2765(01)00233-7. [DOI] [PubMed] [Google Scholar]
- 28.Huang Y, Gattoni R, Stevenin J, Steitz JA. 2003. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol Cell 11:837–843. doi: 10.1016/S1097-2765(03)00089-3. [DOI] [PubMed] [Google Scholar]
- 29.Izaurralde E. 2002. A novel family of nuclear transport receptors mediates the export of messenger RNA to the cytoplasm. Eur J Cell Biol 81:577–584. doi: 10.1078/0171-9335-00273. [DOI] [PubMed] [Google Scholar]
- 30.Lei EP, Silver PA. 2002. Protein and RNA export from the nucleus. Dev Cell 2:261–272. doi: 10.1016/S1534-5807(02)00134-X. [DOI] [PubMed] [Google Scholar]
- 31.Huang S, Spector DL. 1991. Nascent pre-mRNA transcripts are associated with nuclear regions enriched in splicing factors. Genes Dev 5:2288–2302. doi: 10.1101/gad.5.12a.2288. [DOI] [PubMed] [Google Scholar]
- 32.Brown JM, Green J, das Neves RP, Wallace HA, Smith AJ, Hughes J, Gray N, Taylor S, Wood WG, Higgs DR, Iborra FJ, Buckle VJ. 2008. Association between active genes occurs at nuclear speckles and is modulated by chromatin environment. J Cell Biol 182:1083–1097. doi: 10.1083/jcb.200803174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moen PT Jr, Johnson CV, Byron M, Shopland LS, de la Serna IL, Imbalzano AN, Lawrence JB. 2004. Repositioning of muscle-specific genes relative to the periphery of SC-35 domains during skeletal myogenesis. Mol Biol Cell 15:197–206. doi: 10.1091/mbc.e03-06-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gruffat H, Batisse J, Pich D, Neuhierl B, Manet E, Hammerschmidt W, Sergeant A. 2002. Epstein-Barr virus mRNA export factor EB2 is essential for production of infectious virus. J Virol 76:9635–9644. doi: 10.1128/JVI.76.19.9635-9644.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Batisse J, Manet E, Middeldorp J, Sergeant A, Gruffat H. 2005. Epstein-Barr virus mRNA export factor EB2 is essential for intranuclear capsid assembly and production of gp350. J Virol 79:14102–14111. doi: 10.1128/JVI.79.22.14102-14111.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han Z, Marendy E, Wang YD, Yuan J, Sample JT, Swaminathan S. 2007. Multiple roles of Epstein-Barr virus SM protein in lytic replication. J Virol 81:4058–4069. doi: 10.1128/JVI.02665-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hiriart E, Bardouillet L, Manet E, Gruffat H, Penin F, Montserret R, Farjot G, Sergeant A. 2003. A region of the Epstein-Barr virus (EBV) mRNA export factor EB2 containing an arginine-rich motif mediates direct binding to RNA. J Biol Chem 278:37790–37798. doi: 10.1074/jbc.M305925200. [DOI] [PubMed] [Google Scholar]
- 38.Farjot G, Buisson M, Duc Dodon M, Gazzolo L, Sergeant A, Mikaelian I. 2000. Epstein-Barr virus EB2 protein exports unspliced RNA via a Crm-1-independent pathway. J Virol 74:6068–6076. doi: 10.1128/JVI.74.13.6068-6076.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hiriart E, Gruffat H, Buisson M, Mikaelian I, Keppler S, Meresse P, Mercher T, Bernard OA, Sergeant A, Manet E. 2005. Interaction of the Epstein-Barr virus mRNA export factor EB2 with human Spen proteins SHARP, OTT1, and a novel member of the family, OTT3, links Spen proteins with splicing regulation and mRNA export. J Biol Chem 280:36935–36945. doi: 10.1074/jbc.M501725200. [DOI] [PubMed] [Google Scholar]
- 40.Juillard F, Hiriart E, Sergeant N, Vingtdeux-Didier V, Drobecq H, Sergeant A, Manet E, Gruffat H. 2009. Epstein-Barr virus protein EB2 contains an N-terminal transferable nuclear export signal that promotes nucleocytoplasmic export by directly binding TAP/NXF1. J Virol 83:12759–12768. doi: 10.1128/JVI.01276-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Juillard F, Bazot Q, Mure F, Tafforeau L, Macri C, Rabourdin-Combe C, Lotteau V, Manet E, Gruffat H. 2012. Epstein-Barr virus protein EB2 stimulates cytoplasmic mRNA accumulation by counteracting the deleterious effects of SRp20 on viral mRNAs. Nucleic Acids Res 40:6834–6849. doi: 10.1093/nar/gks319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ricci EP, Mure F, Gruffat H, Decimo D, Medina-Palazon C, Ohlmann T, Manet E. 2009. Translation of intronless RNAs is strongly stimulated by the Epstein-Barr virus mRNA export factor EB2. Nucleic Acids Res 37:4932–4943. doi: 10.1093/nar/gkp497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verma D, Bais S, Gaillard M, Swaminathan S. 2010. Epstein-Barr virus SM protein utilizes cellular splicing factor SRp20 to mediate alternative splicing. J Virol 84:11781–11789. doi: 10.1128/JVI.01359-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oh ST, Seo JS, Moon UY, Kang KH, Shin DJ, Yoon SK, Kim WH, Park JG, Lee SK. 2004. A naturally derived gastric cancer cell line shows latency I Epstein-Barr virus infection closely resembling EBV-associated gastric cancer. Virology 320:330–336. doi: 10.1016/j.virol.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 45.Boyle SM, Ruvolo V, Gupta AK, Swaminathan S. 1999. Association with the cellular export receptor CRM 1 mediates function and intracellular localization of Epstein-Barr virus SM protein, a regulator of gene expression. J Virol 73:6872–6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buisson M, Morand P, Peoc'h M, Bouchard O, Bourgeat MJ, Seigneurin JM. 1999. Development of an Epstein-Barr virus type 2 (EBV-2)-associated hepatic B cell non-Hodgkin's lymphoma in an HIV-1-infected patient following a change in the EBV dominant type. Leukemia 13:298–301. [DOI] [PubMed] [Google Scholar]
- 47.Wansink DG, Schul W, van der Kraan I, van Steensel B, van Driel R, de Jong L. 1993. Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J Cell Biol 122:283–293. doi: 10.1083/jcb.122.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu JY, Maniatis T. 1993. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell 75:1061–1070. doi: 10.1016/0092-8674(93)90316-I. [DOI] [PubMed] [Google Scholar]
- 49.Kohtz JD, Jamison SF, Will CL, Zuo P, Luhrmann R, Garcia-Blanco MA, Manley JL. 1994. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature 368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- 50.Krainer AR, Conway GC, Kozak D. 1990. Purification and characterization of pre-mRNA splicing factor SF2 from HeLa cells. Genes Dev 4:1158–1171. doi: 10.1101/gad.4.7.1158. [DOI] [PubMed] [Google Scholar]
- 51.Zahler AM, Roth MB. 1995. Distinct functions of SR proteins in recruitment of U1 small nuclear ribonucleoprotein to alternative 5′ splice sites. Proc Natl Acad Sci U S A 92:2642–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Long JC, Caceres JF. 2009. The SR protein family of splicing factors: master regulators of gene expression. Biochem J 417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 53.Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD. 2008. The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol 15:819–826. doi: 10.1038/nsmb.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lerner EA, Lerner MR, Janeway CA Jr, Steitz JA. 1981. Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc Natl Acad Sci U S A 78:2737–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Perraud M, Gioud M, Monier JC. 1979. Intranuclear structures of monkey kidney cells recognised by immunofluorescence and immuno-electron microscopy using anti-ribonucleoprotein antibodies (author's transl). Ann Immunol (Paris) 130C:635–647. (In French.) [PubMed] [Google Scholar]
- 56.Thiry M. 1995. The interchromatin granules. Histol Histopathol 10:1035–1045. [PubMed] [Google Scholar]
- 57.Misteli T, Caceres JF, Spector DL. 1997. The dynamics of a pre-mRNA splicing factor in living cells. Nature 387:523–527. doi: 10.1038/387523a0. [DOI] [PubMed] [Google Scholar]
- 58.Sharma A, Takata H, Shibahara K, Bubulya A, Bubulya PA. 2010. Son is essential for nuclear speckle organization and cell cycle progression. Mol Biol Cell 21:650–663. doi: 10.1091/mbc.e09-02-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heston L, El-Guindy A, Countryman J, Dela Cruz C, Delecluse HJ, Miller G. 2006. Amino acids in the basic domain of Epstein-Barr virus ZEBRA protein play distinct roles in DNA binding, activation of early lytic gene expression, and promotion of viral DNA replication. J Virol 80:9115–9133. doi: 10.1128/JVI.00909-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith KP, Moen PT, Wydner KL, Coleman JR, Lawrence JB. 1999. Processing of endogenous pre-mRNAs in association with SC-35 domains is gene specific. J Cell Biol 144:617–629. doi: 10.1083/jcb.144.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xing Y, Johnson CV, Dobner PR, Lawrence JB. 1993. Higher level organization of individual gene transcription and RNA splicing. Science 259:1326–1330. doi: 10.1126/science.8446901. [DOI] [PubMed] [Google Scholar]
- 62.Xing Y, Johnson CV, Moen PT Jr, McNeil JA, Lawrence J. 1995. Nonrandom gene organization: structural arrangements of specific pre-mRNA transcription and splicing with SC-35 domains. J Cell Biol 131:1635–1647. doi: 10.1083/jcb.131.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jiménez-García LF, Spector DL. 1993. In vivo evidence that transcription and splicing are coordinated by a recruiting mechanism. Cell 73:47–59. doi: 10.1016/0092-8674(93)90159-N. [DOI] [PubMed] [Google Scholar]
- 64.Cmarko D, Verschure PJ, Martin TE, Dahmus ME, Krause S, Fu XD, van Driel R, Fakan S. 1999. Ultrastructural analysis of transcription and splicing in the cell nucleus after bromo-UTP microinjection. Mol Biol Cell 10:211–223. doi: 10.1091/mbc.10.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zeng C, Kim E, Warren SL, Berget SM. 1997. Dynamic relocation of transcription and splicing factors dependent upon transcriptional activity. EMBO J 16:1401–1412. doi: 10.1093/emboj/16.6.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spector DL, Lamond AI. 2011. Nuclear speckles. Cold Spring Harb Perspect Biol 3:a000646. doi: 10.1101/cshperspect.a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jumaa H, Guenet JL, Nielsen PJ. 1997. Regulated expression and RNA processing of transcripts from the Srp20 splicing factor gene during the cell cycle. Mol Cell Biol 17:3116–3124. doi: 10.1128/MCB.17.6.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jumaa H, Nielsen PJ. 1997. The splicing factor SRp20 modifies splicing of its own mRNA and ASF/SF2 antagonizes this regulation. EMBO J 16:5077–5085. doi: 10.1093/emboj/16.16.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Galiana-Arnoux D, Lejeune F, Gesnel MC, Stevenin J, Breathnach R, Del Gatto-Konczak F. 2003. The CD44 alternative v9 exon contains a splicing enhancer responsive to the SR proteins 9G8, ASF/SF2, and SRp20. J Biol Chem 278:32943–32953. doi: 10.1074/jbc.M301090200. [DOI] [PubMed] [Google Scholar]
- 70.de la Mata M, Kornblihtt AR. 2006. RNA polymerase II C-terminal domain mediates regulation of alternative splicing by SRp20. Nat Struct Mol Biol 13:973–980. doi: 10.1038/nsmb1155. [DOI] [PubMed] [Google Scholar]
- 71.Wong J, Garner B, Halliday GM, Kwok JB. 2012. Srp20 regulates TrkB pre-mRNA splicing to generate TrkB-Shc transcripts with implications for Alzheimer's disease. J Neurochem 123:159–171. doi: 10.1111/j.1471-4159.2012.07873.x. [DOI] [PubMed] [Google Scholar]
- 72.Howe JG, Steitz JA. 1986. Localization of Epstein-Barr virus-encoded small RNAs by in situ hybridization. Proc Natl Acad Sci U S A 83:9006–9010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Terris B, Baldin V, Dubois S, Degott C, Flejou JF, Henin D, Dejean A. 1995. PML nuclear bodies are general targets for inflammation and cell proliferation. Cancer Res 55:1590–1597. [PubMed] [Google Scholar]
- 74.Everett RD, Chelbi-Alix MK. 2007. PML and PML nuclear bodies: implications in antiviral defence. Biochimie 89:819–830. doi: 10.1016/j.biochi.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 75.Everett RD, Parada C, Gripon P, Sirma H, Orr A. 2008. Replication of ICP0-null mutant herpes simplex virus type 1 is restricted by both PML and Sp100. J Virol 82:2661–2672. doi: 10.1128/JVI.02308-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jiang WQ, Ringertz N. 1997. Altered distribution of the promyelocytic leukemia-associated protein is associated with cellular senescence. Cell Growth Differ 8:513–522. [PubMed] [Google Scholar]
- 77.Pearson M, Carbone R, Sebastiani C, Cioce M, Fagioli M, Saito S, Higashimoto Y, Appella E, Minucci S, Pandolfi PP, Pelicci PG. 2000. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature 406:207–210. doi: 10.1038/35018127. [DOI] [PubMed] [Google Scholar]
- 78.de Stanchina E, Querido E, Narita M, Davuluri RV, Pandolfi PP, Ferbeyre G, Lowe SW. 2004. PML is a direct p53 target that modulates p53 effector functions. Mol Cell 13:523–535. doi: 10.1016/S1097-2765(04)00062-0. [DOI] [PubMed] [Google Scholar]
- 79.Kang Y, Cullen BR. 1999. The human Tap protein is a nuclear mRNA export factor that contains novel RNA-binding and nucleocytoplasmic transport sequences. Genes Dev 13:1126–1139. doi: 10.1101/gad.13.9.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Neville M, Rosbash M. 1999. The NES-Crm1p export pathway is not a major mRNA export route in Saccharomyces cerevisiae. EMBO J 18:3746–3756. doi: 10.1093/emboj/18.13.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Viphakone N, Hautbergue GM, Walsh M, Chang CT, Holland A, Folco EG, Reed R, Wilson SA. 2012. TREX exposes the RNA-binding domain of Nxf1 to enable mRNA export. Nat Commun 3:1006. doi: 10.1038/ncomms2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Braun IC, Herold A, Rode M, Izaurralde E. 2002. Nuclear export of mRNA by TAP/NXF1 requires two nucleoporin-binding sites but not p15. Mol Cell Biol 22:5405–5418. doi: 10.1128/MCB.22.15.5405-5418.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Delaleau M, Borden KL. 2015. Multiple export mechanisms for mRNAs. Cells 4:452–473. doi: 10.3390/cells4030452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kataoka N, Dreyfuss G. 2004. A simple whole cell lysate system for in vitro splicing reveals a stepwise assembly of the exon-exon junction complex. J Biol Chem 279:7009–7013. doi: 10.1074/jbc.M307692200. [DOI] [PubMed] [Google Scholar]
- 85.Dostie J, Dreyfuss G. 2002. Translation is required to remove Y14 from mRNAs in the cytoplasm. Curr Biol 12:1060–1067. doi: 10.1016/S0960-9822(02)00902-8. [DOI] [PubMed] [Google Scholar]
- 86.Schmidt U, Richter K, Berger AB, Lichter P. 2006. In vivo BiFC analysis of Y14 and NXF1 mRNA export complexes: preferential localization within and around SC35 domains. J Cell Biol 172:373–381. doi: 10.1083/jcb.200503061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tutucci E, Stutz F. 2011. Keeping mRNPs in check during assembly and nuclear export. Nat Rev Mol Cell Biol 12:377–384. doi: 10.1038/nrm3119. [DOI] [PubMed] [Google Scholar]
- 88.Katahira J. 2012. mRNA export and the TREX complex. Biochim Biophys Acta 1819:507–513. doi: 10.1016/j.bbagrm.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 89.Bachi A, Braun IC, Rodrigues JP, Pante N, Ribbeck K, von Kobbe C, Kutay U, Wilm M, Gorlich D, Carmo-Fonseca M, Izaurralde E. 2000. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA 6:136–158. doi: 10.1017/S1355838200991994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Glaunsinger B, Ganem D. 2004. Lytic KSHV infection inhibits host gene expression by accelerating global mRNA turnover. Mol Cell 13:713–723. doi: 10.1016/S1097-2765(04)00091-7. [DOI] [PubMed] [Google Scholar]
- 91.Rowe M, Glaunsinger B, van Leeuwen D, Zuo J, Sweetman D, Ganem D, Middeldorp J, Wiertz EJ, Ressing ME. 2007. Host shutoff during productive Epstein-Barr virus infection is mediated by BGLF5 and may contribute to immune evasion. Proc Natl Acad Sci U S A 104:3366–3371. doi: 10.1073/pnas.0611128104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kumar GR, Glaunsinger BA. 2010. Nuclear import of cytoplasmic poly(A) binding protein restricts gene expression via hyperadenylation and nuclear retention of mRNA. Mol Cell Biol 30:4996–5008. doi: 10.1128/MCB.00600-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kwong AD, Frenkel N. 1989. The herpes simplex virus virion host shutoff function. J Virol 63:4834–4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hardwicke MA, Sandri-Goldin RM. 1994. The herpes simplex virus regulatory protein ICP27 contributes to the decrease in cellular mRNA levels during infection. J Virol 68:4797–4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sandri-Goldin RM. 1998. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev 12:868–879. doi: 10.1101/gad.12.6.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Glaunsinger B, Chavez L, Ganem D. 2005. The exonuclease and host shutoff functions of the SOX protein of Kaposi's sarcoma-associated herpesvirus are genetically separable. J Virol 79:7396–7401. doi: 10.1128/JVI.79.12.7396-7401.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gaglia MM, Covarrubias S, Wong W, Glaunsinger BA. 2012. A common strategy for host RNA degradation by divergent viruses. J Virol 86:9527–9530. doi: 10.1128/JVI.01230-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Semmes OJ, Chen L, Sarisky RT, Gao Z, Zhong L, Hayward SD. 1998. Mta has properties of an RNA export protein and increases cytoplasmic accumulation of Epstein-Barr virus replication gene mRNA. J Virol 72:9526–9534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Phelan A, Clements JB. 1997. Herpes simplex virus type 1 immediate early protein IE63 shuttles between nuclear compartments and the cytoplasm. J Gen Virol 78(Part 12):3327–3331. [DOI] [PubMed] [Google Scholar]
- 100.Mears WE, Rice SA. 1998. The herpes simplex virus immediate-early protein ICP27 shuttles between nucleus and cytoplasm. Virology 242:128–137. doi: 10.1006/viro.1997.9006. [DOI] [PubMed] [Google Scholar]
- 101.Soliman TM, Sandri-Goldin RM, Silverstein SJ. 1997. Shuttling of the herpes simplex virus type 1 regulatory protein ICP27 between the nucleus and cytoplasm mediates the expression of late proteins. J Virol 71:9188–9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hardy WR, Sandri-Goldin RM. 1994. Herpes simplex virus inhibits host cell splicing, and regulatory protein ICP27 is required for this effect. J Virol 68:7790–7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sciabica KS, Dai QJ, Sandri-Goldin RM. 2003. ICP27 interacts with SRPK1 to mediate HSV splicing inhibition by altering SR protein phosphorylation. EMBO J 22:1608–1619. doi: 10.1093/emboj/cdg166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sandri-Goldin RM, Hibbard MK, Hardwicke MA. 1995. The C-terminal repressor region of herpes simplex virus type 1 ICP27 is required for the redistribution of small nuclear ribonucleoprotein particles and splicing factor SC35; however, these alterations are not sufficient to inhibit host cell splicing. J Virol 69:6063–6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Phelan A, Carmo-Fonseca M, McLaughlan J, Lamond AI, Clements JB. 1993. A herpes simplex virus type 1 immediate-early gene product, IE63, regulates small nuclear ribonucleoprotein distribution. Proc Natl Acad Sci U S A 90:9056–9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sandri-Goldin RM. 2008. The many roles of the regulatory protein ICP27 during herpes simplex virus infection. Front Biosci 13:5241–5256. doi: 10.2741/3078. [DOI] [PubMed] [Google Scholar]
- 107.Huang S, Deerinck TJ, Ellisman MH, Spector DL. 1994. In vivo analysis of the stability and transport of nuclear poly(A)+ RNA. J Cell Biol 126:877–899. doi: 10.1083/jcb.126.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, Blencowe BJ, Prasanth SG, Prasanth KV. 2010. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zuo J, Thomas W, van Leeuwen D, Middeldorp JM, Wiertz EJ, Ressing ME, Rowe M. 2008. The DNase of gammaherpesviruses impairs recognition by virus-specific CD8+ T cells through an additional host shutoff function. J Virol 82:2385–2393. doi: 10.1128/JVI.01946-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Horst D, Burmeister WP, Boer IG, van Leeuwen D, Buisson M, Gorbalenya AE, Wiertz EJ, Ressing ME. 2012. The “bridge” in the Epstein-Barr virus alkaline exonuclease protein BGLF5 contributes to shutoff activity during productive infection. J Virol 86:9175–9187. doi: 10.1128/JVI.00309-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Verma D, Swaminathan S. 2008. Epstein-Barr virus SM protein functions as an alternative splicing factor. J Virol 82:7180–7188. doi: 10.1128/JVI.00344-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sugimoto A, Sato Y, Kanda T, Murata T, Narita Y, Kawashima D, Kimura H, Tsurumi T. 2013. Different distributions of Epstein-Barr virus early and late gene transcripts within viral replication compartments. J Virol 87:6693–6699. doi: 10.1128/JVI.00219-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Graham FL, van der Eb AJ, Heijneker HL. 1974. Size and location of the transforming region in human adenovirus type 5 DNA. Nature 251:687–691. doi: 10.1038/251687a0. [DOI] [PubMed] [Google Scholar]
- 114.Delecluse HJ, Hilsendegen T, Pich D, Zeidler R, Hammerschmidt W. 1998. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc Natl Acad Sci U S A 95:8245–8250. doi: 10.1073/pnas.95.14.8245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Feederle R, Kost M, Baumann M, Janz A, Drouet E, Hammerschmidt W, Delecluse HJ. 2000. The Epstein-Barr virus lytic program is controlled by the co-operative functions of two transactivators. EMBO J 19:3080–3089. doi: 10.1093/emboj/19.12.3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Heston L, Rabson M, Brown N, Miller G. 1982. New Epstein-Barr virus variants from cellular subclones of P3J-HR-1 Burkitt lymphoma. Nature 295:160–163. doi: 10.1038/295160a0. [DOI] [PubMed] [Google Scholar]
- 117.Davis MG, Huang ES. 1988. Transfer and expression of plasmids containing human cytomegalovirus immediate-early gene 1 promoter-enhancer sequences in eukaryotic and prokaryotic cells. Biotechnol Appl Biochem 10:6–12. [PubMed] [Google Scholar]
- 118.Francis AL, Gradoville L, Miller G. 1997. Alteration of a single serine in the basic domain of the Epstein-Barr virus ZEBRA protein separates its functions of transcriptional activation and disruption of latency. J Virol 71:3054–3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ragoczy T, Heston L, Miller G. 1998. The Epstein-Barr virus Rta protein activates lytic cycle genes and can disrupt latency in B lymphocytes. J Virol 72:7978–7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pearson GR, Vroman B, Chase B, Sculley T, Hummel M, Kieff E. 1983. Identification of polypeptide components of the Epstein-Barr virus early antigen complex with monoclonal antibodies. J Virol 47:193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.