During HIV-1 infection, the virus must travel across the cytoplasm to enter the nucleus. The host cell motor protein complex dynein has been implicated in HIV-1 intracellular transport. We show that expression of the dynein heavy chain, components of the dynein-associated dynactin complex, and the dynein adaptor BICD2 in target cells are important for HIV-1 infection and nuclear entry. BICD2 interacts with the HIV-1 capsid in vitro, suggesting that it functions as a capsid-specific adaptor for HIV-1 intracellular transport. Our work identifies specific host proteins involved in microtubule-dependent HIV-1 intracellular transport and highlights the BICD2-capsid interaction as a potential target for antiviral therapy.
KEYWORDS: BICD2, dynactin, dynein, HIV-1, transport
ABSTRACT
Human immunodeficiency virus type 1 (HIV-1) infection depends on efficient intracytoplasmic transport of the incoming viral core to the target cell nucleus. Evidence suggests that this movement is facilitated by the microtubule motor dynein, a large multiprotein complex that interacts with dynactin and cargo-specific adaptor proteins for retrograde movement via microtubules. Dynein adaptor proteins are necessary for activating dynein movement and for linking specific cargoes to dynein. We hypothesized that HIV-1 engages the dynein motor complex via an adaptor for intracellular transport. Here, we show that small interfering RNA depletion of the dynein heavy chain, components of the dynactin complex, and the dynein adaptor BICD2 reduced cell permissiveness to HIV-1 infection. Cell depletion of dynein heavy chain and BICD2 resulted in impaired HIV-1 DNA accumulation in the nucleus and decreased retrograde movement of the virus. Biochemical studies revealed that dynein components and BICD2 associate with capsid-like assemblies of the HIV-1 CA protein in cell extracts and that purified recombinant BICD2 binds to CA assemblies in vitro. Association of dynein with CA assemblies was reduced upon immunodepletion of BICD2 from cell extracts. We conclude that BICD2 is a capsid-associated dynein adaptor utilized by HIV-1 for transport to the nucleus.
IMPORTANCE During HIV-1 infection, the virus must travel across the cytoplasm to enter the nucleus. The host cell motor protein complex dynein has been implicated in HIV-1 intracellular transport. We show that expression of the dynein heavy chain, components of the dynein-associated dynactin complex, and the dynein adaptor BICD2 in target cells are important for HIV-1 infection and nuclear entry. BICD2 interacts with the HIV-1 capsid in vitro, suggesting that it functions as a capsid-specific adaptor for HIV-1 intracellular transport. Our work identifies specific host proteins involved in microtubule-dependent HIV-1 intracellular transport and highlights the BICD2-capsid interaction as a potential target for antiviral therapy.
INTRODUCTION
For human immunodeficiency virus type 1 (HIV-1) to successfully infect cells, it must undergo a series of early events, including cellular fusion, reverse transcription, uncoating, nuclear entry, and integration. Fusion occurs at or near the plasma membrane; therefore, HIV-1 must traverse the cytoplasm from the cell periphery to reach the nucleus. The process by which the virus is transported to the nucleus has not been thoroughly investigated. Genome-wide small interfering RNA (siRNA) screens have revealed microtubule-associated proteins as necessary for HIV-1 infection (1–4). Recent studies suggest that the cellular transport of HIV-1 is dependent on the microtubule network and on the molecular motor proteins dynein and kinesin (5–9).
Dynein is a microtubule motor complex that utilizes ATP to walk along microtubules and move cargo toward the negative end of microtubules, toward the cell nucleus. Dynein functions in vesicle and endosomal transport and helps position organelles and the mitotic spindle during cell division (reviewed in references 10, 11, 12, and 13). The dynein complex consists of two copies of the heavy chain (DYNC1H1 cytoplasmic and DYNC2H1 flagella), which contains the ATPase activity required for movement; two intermediate chains (DYNC1I1 and DYNC1I2) and two light-intermediate chains (DYNC1LI1 and DYNC1LI2) that stabilize the structure of the complex. The complex also contains multiple copies of various light chains (DYNLT1, DYNLT3, DYNLRB1, DYNLRB2, and DYNLL1/2) that link dynein to other proteins and to cargo. Previous studies have indicated that dynein light chain 2 in yeast may be involved in HIV-1 integrase transport (14) and that DYNLL1 may interact with HIV-1 capsid and affect HIV-1 reverse transcription (RT) (15).
Dynein interacts with dynactin, another multisubunit adaptor complex that links dynein to cargo and acts as a regulator for movement. Engagement of dynactin increases dynein's processivity while walking along microtubules (16–21). The dynactin complex has at least five components, including (i) an Arp1 backbone (ACTR1A) (22, 23), (ii) a p150 side-arm (DCTN1) that aids in microtubule processivity (16, 18), (iii) a shoulder complex (DCTN2/DCTN3) that attaches the side arm to the backbone and binds additional interacting proteins (24–26), (iv) a pointed end complex (DCTN4, DCTN5, and DCTN6) that binds the nuclear envelope prior to mitosis (27), and (v) a barbed end complex (CAPZA and CAPZB) that aids in structure stabilization (22). The function of each of these components depends on interactions with the other components. In contrast to dynein, dynactin proteins do not function redundantly. Together, the five components interact to form the dynactin complex. Although there is compelling evidence that HIV-1 infection depends on dynein-dependent transport, the role of dynactin and of dynein adaptors in infection is not well understood.
Owing to the variety of protein cargoes that require cellular motors for movement, the cell faces the problem of cargo specificity. To solve this problem, dynein interacts with numerous cargo-specific adaptor proteins. Adaptor proteins actively link dynein to dynactin (28–30), resulting in dynein activation and processive walking along microtubules. Some dynein adaptors also act in specific cellular functions through their interactions with dynein, playing roles in interphase transport and organelle and microtubule positioning before and during mitosis (31–37). The number of known dynein adaptor proteins has expanded in recent years, with new adaptors being discovered and new functions being assigned for previously known adaptors (reviewed in reference 38). Some of the best-characterized adaptors are LIS1, NUDE, and NUDEL, which form complexes between themselves and dynein to function in nuclear and spindle positioning, and in organelle and mRNA transport. The adaptors ROD-ZW10-Zwilch and Spindly also interact with dynein to aid in docking dynein and other dynein adaptors to the kinetochores. Bicaudal D has been shown to be important for organelle and mRNA transport. There are two homologues of Bicaudal D: BICD1 and BICD2. Although both serve similar functions in the cell, BICD2 is more abundant. In mammalian tissue culture cells, BICD2 has a role in dynein-mediated transport from the endoplasmic reticulum to the Golgi compartment and within the Golgi compartment via its transport RAB6-positive endosomes. Prior to mitotic entry, BICD2 interacts with RANBP2 at the nuclear pores and recruits dynein and dynactin to ensure proper positioning of the nucleus (reviewed in reference 38). Recently, an independent study reported that BICD2 facilitates HIV-1 intracellular transport and promotes infection (39).
Because HIV-1 infection depends on efficient traversal of the cytoplasm to the nucleus, we hypothesized that the virus exploits a specific adaptor protein and the dynactin complex for dynein-dependent transport. We utilized an siRNA-based screen to identify dynein, dynactin, and known and potential adaptors that facilitate HIV-1 infection. We show that HIV-1 transport and infection are facilitated by the dynactin backbone and shoulder complex (ACTR1A and DCTN2/DCTN3) and the dynein adaptor BICD2. Furthermore, we show that the HIV-1 capsid can bind to the dynein complex and to BICD2 in vitro and that BICD2 acts to facilitate binding between the capsid and dynein. Our results indicate that HIV-1 utilizes a dynein-dynactin-BICD2 complex for infection and suggest that BICD2 functions as a capsid-specific dynein adaptor protein.
RESULTS
Dynein heavy chain and dynactin component depletion inhibits HIV-1 infection.
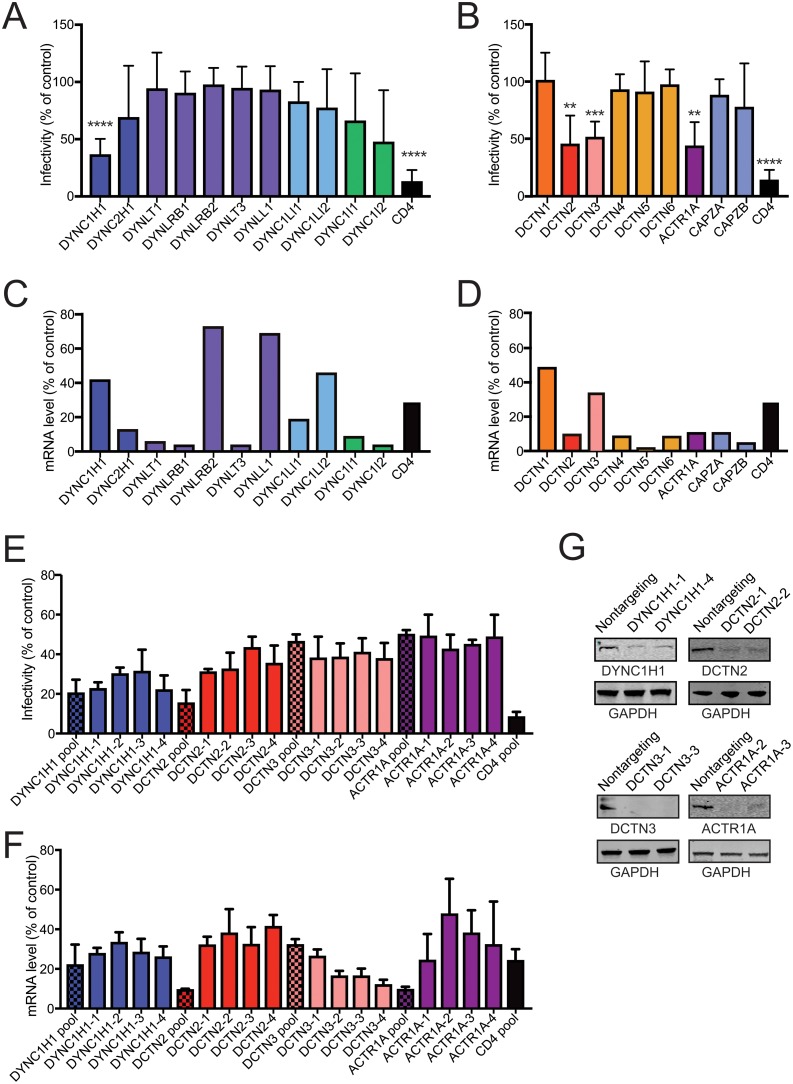
Previous studies have indicated that dynein plays a role in HIV-1 infection and intracellular transit (5, 7). However, a systematic analysis of the components of dynein and the associated dynactin complex required for HIV-1 infection has not yet been reported. To determine the contribution of dynein and dynactin to HIV-1 infection, we analyzed the effects of depleting components of the dynein and dynactin complex on cell permissiveness to HIV-1 infection. TZM-bl cells were transfected with pooled siRNAs specific to individual genes of the dynein or dynactin complex, or a nontargeting siRNA control, then inoculated with the GFP-encoding HIV-1 reporter virus NL43-GFP (Fig. 1A and B), corresponding to the full-length NL4-3 virus encoding GFP in place of Nef. An siRNA targeting of the HIV-1 cell receptor CD4 was used as a positive control for reduction in HIV-1 infection. Effects on expression of the targeted mRNAs were analyzed by quantitative RT-PCR (Fig. 1C and D). The dynein complex is composed of two heavy chains (cytoplasmic DYNC1H1 or ciliary DYNC2H1), two intermediate chains (DYNC1I1 and DYNC1I2), two light intermediate chains (DYNC1LI1 and DYNC1LI2), and multiple sets of light chains (DYNLT1, DYNRB1, DYNRB2, DYNLT3, and DYNLL1) (Fig. 1A). The various chains in dynein and dynactin can exhibit functional redundancy. We observed that depletion of the dynein heavy chain significantly reduced the extent of HIV-1 infection, consistent with previous reports (5, 7). In contrast, we observed no significant effect of depleting other dynein components on HIV-1 infection, despite efficient knockdown of most of these components as assessed by mRNA quantification (Fig. 1A and C). As expected, depletion of cellular CD4 markedly reduced cell permissiveness to HIV-1.
FIG 1.
Depletion of DYNC1H1 and some dynactin components inhibits HIV-1 infection. (A to D) TZM-bl cells were pretreated with indicated pooled siRNAs and then inoculated with GFP-expressing HIV-1 (A and B) or harvested for knockdown efficiency by qPCR analysis (C and D). Infection was assessed by flow cytometry for GFP expression. The values shown represent the extent of infection relative to nontargeting siRNA treatment (A and B). Infection results are the means of three independent determinations. Error bars represent standard deviations. Statistical significance was calculated by a Student t test for each siRNA treatment compared to nontargeting control siRNA treatment. (**, P < 0.01; ***, P < 0.001; ****, P < 0.0001). (C and D) mRNA analyses are from a single experiment. (E) TZM-bl cells were pretreated with indicated pooled siRNA, or individual siRNAs from the pool for 72 h, and then analyzed as described previously. All samples had equal significance values (****, P < 0.0001). (F) mRNA analysis represents measurements from three independent experiments, normalized to the control. (G) Immunoblot analysis of the effects of two individual siRNAs for four targets on the corresponding target protein levels.
The dynactin complex associates with dynein, which increases the processivity of dynein movement along microtubules (17). Dynactin can also act as an adaptor for some cargoes. The dynactin complex is composed of the p150 sidearm (DCTN1), dynamitin/shoulder (DCTN2 and DCTN3), pointed-end complex (DCTN4, DCTN5, and DCTN6), Arp1 rod (ACTR1A), and the barbed end complex (CAPZA and CAPZB) (Fig. 1B). Depletion of DCTN2, DCTN3, and ACTR1A resulted in decreased HIV-1 infection, consistent with a role for the dynactin complex in HIV-1 infection. To validate these results, we tested the effects of individual siRNAs from the four siRNA pools in Fig. 1A and B that showed a significant effect on HIV-1 infection (Fig. 1E). Each single siRNA also significantly reduced the extent of infection of TZM-bl cells (P < 0.0001 for all samples), thus validating the results of the pooled siRNAs. Depletion of the corresponding mRNAs was confirmed by RT-PCR (Fig. 1F). The observation that each of the individual siRNAs had a similar effect as the pooled siRNAs argues against the possibility of off-target effects being responsible for the observed outcome. In addition to RT-PCR analysis, protein depletion was confirmed by immunoblot analysis of extracts of cells transfected with each of two individual siRNAs for four targets for which antibodies were available (Fig. 1G). Collectively, the results shown in Fig. 1 indicate that HIV-1 infection is dependent on the dynein heavy chain and on components of the dynactin complex.
Analysis of the effects of depletion of dynein adaptors on HIV-1 infection.
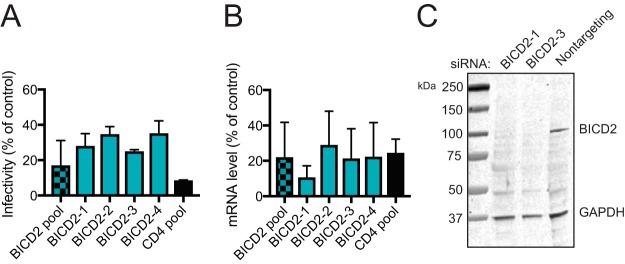
Dynein adaptors are host proteins that associate with dynein to provide cargo specificity and to activate dynein movement along microtubules. We hypothesized that HIV-1 exploits one or more dynein adaptors for infection and transport to the nucleus. To identify dynein adaptors utilized during HIV-1 infection, we screened a panel of host proteins that represented possible candidates for HIV dynein adaptors. These included known and putative dynein adaptors, including proteins with both dynein and HIV-1 interactions, and other host proteins that had previously been shown to be important for HIV-1 transport (see Table S4 in the supplemental material). TZM-bl cells were treated with pooled siRNAs for 48 h and then challenged with HIV-1. Targets that had the most pronounced and reproducible effect on HIV-1 infection at 48 h of siRNA treatment were analyzed under 72 h of siRNA treatment (see Table S4 in the supplemental material). Bicaudal D homolog 2 (BICD2) depletion consistently reduced HIV-1 infection (Fig. 2A), similar to a recent finding in BICD2 knockout cells (39). To confirm that BICD2 depletion reduces cell permissiveness to HIV-1 infection, we repeated the experiments using individual siRNA duplexes present in the siRNA pool (Fig. 2A). We found that the individual siRNAs also reduced HIV-1 infection. The knockdown efficiency of the individual siRNAs was analyzed by RT-PCR (Fig. 2B). To confirm that the siRNAs reduced BICD2 protein levels, we analyzed extracts of cells transfected with either of two of the individual siRNAs by immunoblotting (Fig. 2C). We observed a marked decrease in BICD2 protein levels in the cell extracts transfected with either of the two BICD2 siRNAs.
FIG 2.
Analysis of the effects of depletion of dynein adaptors on HIV-1 infection. (A) TZM-bl cells were pretreated with BICD2 pooled siRNA or individual siRNAs and then inoculated with GFP-expressing HIV-1 (A) or harvested for knockdown efficiency by qPCR analysis (B). After infection with an HIV-1 reporter virus, the cells were fixed and analyzed by fluorescence-activated cell sorting for GFP expression, and the infectivity relative to nontargeting siRNA treatment was determined (A). The results shown are mean values from three independent determinations. Error bars represent standard deviations. Statistical significance was calculated was performed as in the legend to Fig. 1. All samples had equal significance values (****, P < 0.0001). (C) Two of the single siRNAs were selected for BICD2 protein knockdown analysis by immunoblotting.
Depletion of components of the dynein-dynactin-BICD2 complex results in impaired HIV-1 nuclear import.
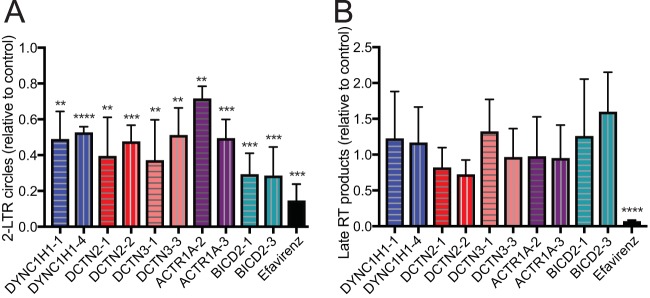
After determining that the dynein-dynactin-BICD2 complex is important for HIV-1 infection, we sought to determine which stage of HIV-1 infection the complex affects. Before infection occurs, HIV-1 must enter at the cell membrane and traverse the cytoplasm to enter the nucleus for integration. During this period, the virus undergoes reverse transcription and partial uncoating (40). We hypothesized that if the dynein-dynactin-BICD2 complex is important for transport of the virus to the nucleus, depletion of the components would result in reduced accumulation of HIV-1 DNA in the nucleus. After entry of HIV-1 into the nucleus, a fraction of the viral DNA is not integrated but instead forms 2-LTR circles within the nucleus, rendering 2-LTR circles a convenient PCR-based readout of nuclear entry. To determine whether the dynein-dynactin-BICD2 complex is important for HIV-1 intracellular transit, we tested the effects of host proteins on the levels of accumulated 2-LTR circles. Two individual siRNAs were chosen for each gene of interest. TZM-bl cells were treated with siRNAs and subsequently challenged with GFP-expressing HIV-1 pseudotyped with vesicular stomatitis virus G protein (VSV-G). 2-LTR circles were quantified, and the reduction relative to nontargeting control was determined. Efavirenz treatment, which blocks reverse transcription and therefore downstream 2-LTR production, was used as a positive control. Depletion of the dynein heavy chain, dynactin components, and BICD2 each led to a reduction in HIV-1 2-LTR circles (Fig. 3A), suggesting that HIV-1 does not efficiently reach the nucleus when components of the complex are depleted. Prior to nuclear entry, HIV-1 undergoes reverse transcription in the cytoplasm. Because formation of 2-LTR circles depends on viral DNA synthesis, a reduction in reverse transcription would be expected to result in a decreased level of 2-LTR circles. To test whether depletion of the dynein-dynactin-BICD2 complex affects HIV-1 reverse transcription, we assayed late reverse transcription products by quantitative PCR using virus-specific primers (Fig. 3B). Efavirenz treatment was used as a control to confirm that the assay signals resulted from bona fide reverse transcription versus contaminating plasmid DNA. We observed no significant effect of siRNA treatment on the production of late RT products relative to the nontargeting siRNA control. Our results are concordant with one, and differ with another, previous report of dynein heavy chain function in HIV-1 infection (41, 42); they are also concordant with a recent study of the dependence of HIV-1 infection on BICD2 expression (39). Our data suggest that the reduction in HIV-1 infection by depletion of the dynein-dynactin-BICD2 complex results from intracellular transport and/or impaired nuclear entry.
FIG 3.
Analysis of HIV-1 DNA synthesis and nuclear entry in cells depleted for components of the dynein-dynactin-BICD2 complex. TZM-bl cells were pretreated with individual siRNA duplexes for 72 h and then inoculated with GFP-expressing HIV-1 pseudotyped with VSV-G. Cells were harvested for quantitative analysis of 2-LTR circles (A) and for analysis of second strand transfer reverse transcripts (B). The effects relative to nontargeting siRNA treatment are shown. The results shown are mean values from three independent determinations. Error bars represent standard deviations. Statistical significance was calculated by an unpaired parametric t test for each siRNA treatment versus nontargeting control siRNA treatment (**, P < 0.01; ***, P < 0.001; ****, P < 0.0001). No significant effects of the siRNAs on late reverse transcripts were observed.
The dynein-dynactin-BICD2 complex facilitates MLV and SIV infection.
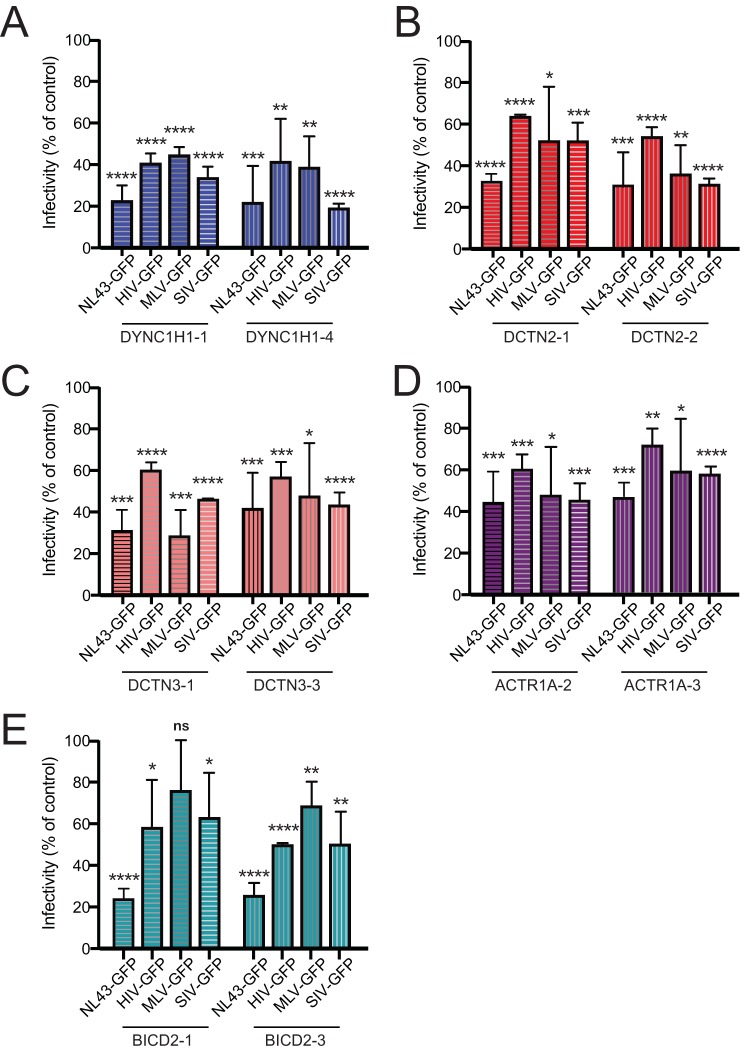
Previous studies of murine leukemia virus (MLV) infection have suggested that the dynein heavy chain, the dynein intermediate chain, the dynein light chain DYNLRB2, and the dynactin component DCTN2/p50/dynamitin are involved in MLV infection (43, 44). Although we did not observe an effect of depletion of either dynein intermediate chain (DYNC1I1, DYNC1I2), or DYNLRB2 on HIV-1 infection, we did observe that the dynein heavy chain (DYNC1H1), and dynactin DCTN2 promote infection (Fig. 1; Table S4 in the supplemental material). To determine whether the dependence of infection on these proteins extends to other retroviruses, we tested the effect of depletion of DYNC1H1, DCTN2, DCTN3, ACTR1A, and BICD2 (Fig. 4, panels A to E, respectively) on cell permissiveness to MLV and simian immunodeficiency virus (SIV) infection. To ensure that the viruses use a common mechanism for cell entry, we also used green fluorescent protein (GFP) reporter particles that were pseudotyped with VSV-G. Interestingly, pseudotyping HIV-1 led to a decrease in the effects on infection compared to HIV-1 particles bearing native Env for all dynein-dynactin-BICD2 targets. The observed effect of VSV-G pseudotyping is consistent with another study of the effects of BICD2 depletion on HIV-1 infection (39). As with HIV-1, both MLV and SIV infection were reduced when DYNC1H1 (Fig. 4A) and DCTN2 (Fig. 4B) were depleted, consistent with a previous study of MLV (44). Similar observations were obtained for the other dynactin components DCTN3 (Fig. 4C) and ACTR1A (Fig. 4D), which were not previously reported to play a role in MLV or SIV infection. We also observed that BICD2 depletion also reduced MLV and SIV infection, though the effect on MLV infection appeared to be less pronounced (Fig. 4E). Together, our results suggest that the dynein-dynactin-BICD2 complex is exploited by both MLV and SIV for infection.
FIG 4.
Effects of depletion of components of the dynein-dynactin-BICD2 complex on MLV and SIV infection. (A to E) TZM-bl cells were pretreated with individual siRNA duplexes for 72 h and inoculated with GFP-expressing HIV-1 (NL43-GFP), as well as VSV-G-pseudotyped HIV-1 (HIV-GFP), murine leukemia virus (MLV-GFP), and simian immunodeficiency virus (SIV-GFP). Inoculated cells were fixed and analyzed by fluorescence-activated cell sorting for GFP expression, and the extent of infection relative to nontargeting siRNA treatment was determined. Shown are mean values and standard deviations from three independent experiments. The statistical significance was calculated by an unpaired parametric t test for each siRNA treatment compared to nontargeting control siRNA treatment (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
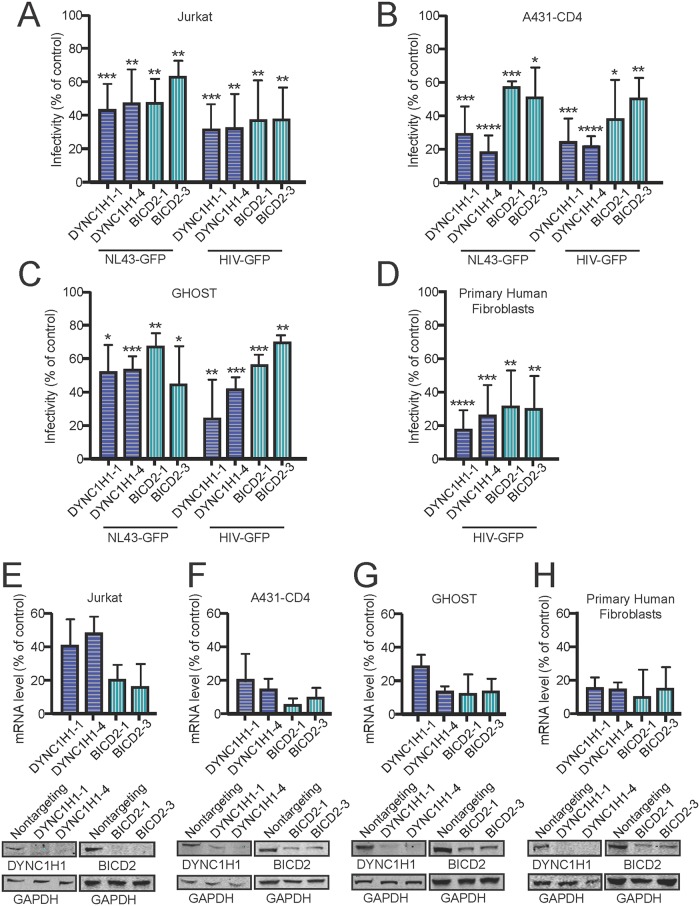
DYNC1H1 and BICD2 facilitate HIV-1 infection in multiple cell lines.
To determine whether the dependence of HIV-1 infection on DYNC1H1 and BICD2 extends to other cell types, we tested the effect of depletion of DYNC1H1 and BICD2 on cell permissiveness to infection in Jurkat, A431-CD4, GHOST, and primary human fibroblast cultures. Cells were transfected with two individual siRNAs for DYNC1H1 and BICD2 for 72 h before challenge with HIV-1. For the infection assays, we used GFP reporter particles that had either native HIV-1 envelope (NL43-GFP) or particles that were pseudotyped with VSV-G (HIV-GFP). DYNC1H1 and BICD2 depletion reduced HIV-1 infection in all of the cells (Fig. 5A to D). Interestingly, pseudotyping HIV-1 did not lead to a decrease in the effects on infection compared to native HIV-1 Env as was previously seen in TZM-bl cells. To confirm that DYNC1H1 and BICD2 were depleted in the siRNA-treated cells, mRNA levels were quantified by RT-PCR, and protein levels were assessed by immunoblotting (Fig. 5E to H). Together, our results suggest that dynein and BICD2 facilitate HIV-1 for infection in a range of cell types, including the Jurkat T cell line.
FIG 5.
DYNC1H1 and BICD2 facilitate HIV-1 infection in various human cell lines. Cells were pretreated with individual siRNA duplexes and either inoculated with GFP-expressing HIV-1 (NL43-GFP) or VSV-G pseudotyped HIV-1 (HIV-GFP) for infection analysis (A to D) or harvested for knockdown efficiency analysis by qPCR and immunoblotting (E to H). Inoculated cells were fixed and analyzed by fluorescence-activated cell sorting for GFP expression, and the extent of infection relative to nontargeting siRNA treatment was determined. Shown are mean values and standard deviations from three independent experiments. Statistical significance was evaluated by an unpaired parametric t test for each siRNA treatment compared to nontargeting control siRNA treatment (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
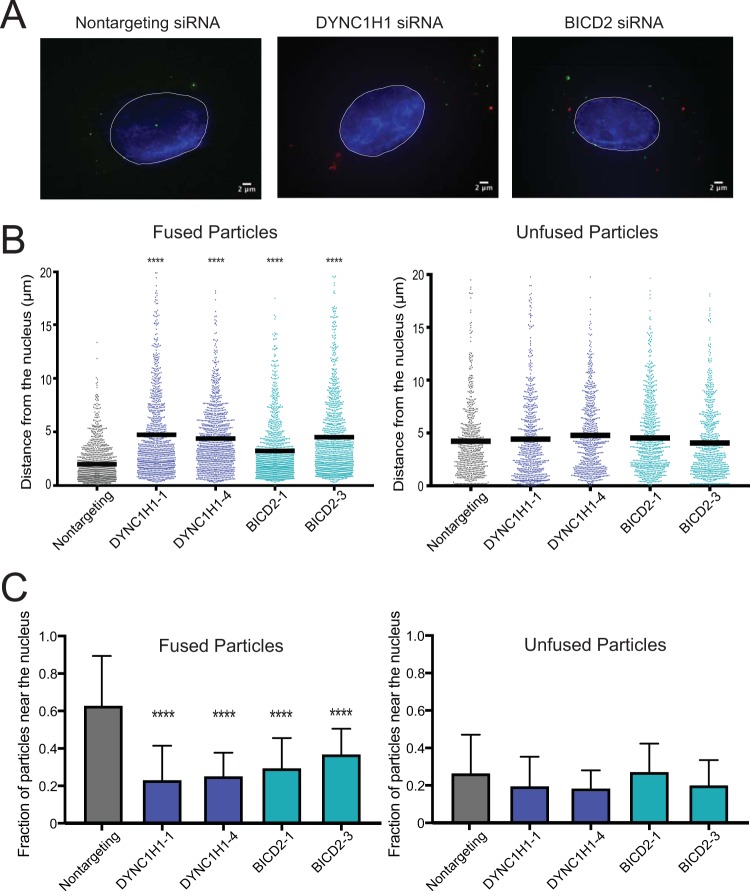
BICD2 promotes HIV-1 retrograde trafficking to the nucleus.
By definition, a dynein adaptor engages both cargo and dynein and promotes intracellular transport. To determine whether BICD2 promotes HIV-1 trafficking to the nucleus, we employed fluorescence deconvolution microscopy to analyze the intracellular location of HIV-1 cores in the target cell cytoplasm 1 h postinfection in BICD2-depleted cells and in nontargeting siRNA-treated cells (Fig. 6). HIV-1 particles, pseudotyped with VSV-G, were generated containing two labels: the core-associated marker GFP-Vpr and the membrane label S15-mCherry (45). Upon fusion of the viral and cell membranes, the membrane-bound S15-mCherry separates from the viral core, thus permitting identification of fused particles via loss of the associated mCherry fluorescence. The dynein heavy chain, DYNC1H1, was employed as a positive control, as previous studies have shown that depletion of this dynein component results in accumulation of HIV-1 particles at the cell periphery (5, 7). Consistent with those reports, we observed a significant reduction in the average HIV-1 particle distance traveled toward the nucleus from the cell periphery in cells depleted of DYNC1H1 (Fig. 6A and B, left). BICD2 depletion also significantly increased the average HIV-1 distance from the nucleus (Fig. 6A and B, left), suggesting that BICD2 facilitates HIV-1 movement to the nucleus. We also analyzed the fraction of HIV-1 particles that reached the nucleus within 1 h in siRNA DYNC1H1- and BICD2-treated cells (Fig. 6C). Consistent with a previous study (5), the fraction of fused HIV-1 particles that reached the nucleus in DYNC1H1-depleted cells was significantly lower than that in to control siRNA-transfected cells. Similarly, fewer particles reached the nucleus in BICD2-depleted versus control cells (Fig. 6C, left), consistent with a role of BICD2 in intracellular HIV-1 trafficking to the nucleus. For both analyses, movement of the unfused particles (Fig. 6B and C, right panels) was unaffected by DYNC1H1 or BICD2 depletion compared to nontargeting control, indicating that the role of dynein and BICD2 in transport is specific to fused HIV-1 particles.
FIG 6.
BICD2 promotes HIV-1 movement to the nucleus. (A to C) TZM-bl cells were pretreated with siRNAs and inoculated with VSV-G-pseudotyped HIV-1 particles labeled with GFP-Vpr and S15-mCherry, which permits detection of particles that have fused with the cell. (A) Representative images of cells. (B) Quantitative analysis of HIV-1 particle distance from the nucleus. Each dot represents the measurement of an individual HIV-1 particle. The results shown are from two independent experiments, each with >20 cells analyzed and >400 particles analyzed per condition. Fused particle analysis (left) and unfused particle analysis (right) results are both shown. A one-way ANOVA was performed for statistical analysis, followed by a Dunnett's multiple-comparison test, and all treatment groups in the fused particle analysis exhibited P values of <0.0001. No treatment groups in the unfused particle analysis exhibited a significant difference compared to nontargeting control. (C) The average and standard deviation of the fraction of HIV-1 particles within 2 μm of the nucleus for each cell were analyzed. Analysis results for fused particles (left) and unfused particles (right) are shown. Significance was determined by one-way ANOVA, as in panel B.
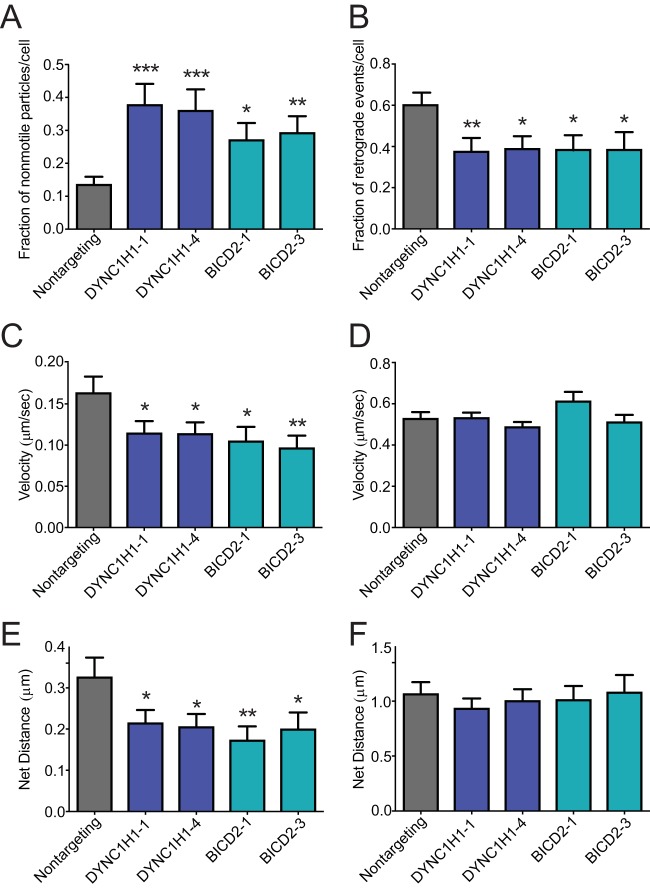
To test for a role of DYNC1H1 and BICD2 in the bidirectional motions of HIV-1 particles, we employed continuous imaging of live cells. Virions were dually labeled with GFP-Vpr and mCherry-S15 to distinguish fused from unfused particles (45). Immediately before movies were captured, the S15-mCherry signal was imaged. In subsequent analysis, particles displaying mCherry signal were excluded from analysis. In addition, particles entering the field during the imaging were excluded since their fusion status was unknown. Infected cells were imaged at a rate of 2 frames per second over a 2.5-min period (see Movies S1 to S3 in the supplemental material). The velocity, net distance traveled, and directionality of individual virus particles were analyzed. In cells depleted of DYNC1H1 or BICD2, we observed a high fraction (30 to 40%) of nonmotile or slow-moving particles (<4 μm total distance over the 2.5-min imaging period), relative to control nontargeting siRNA-treated cells (∼13%) (Fig. 7A). Within the population of moving particles, we analyzed the net directionality of the particles. Particles moving toward the nucleus were classified as retrograde, and particles moving away from the nucleus were classified as anterograde. Depletion of DYNC1H1 or BICD2 resulted in a significantly lower fraction of HIV-1 retrograde particles versus control siRNA treatment (Fig. 7B). These findings suggest that dynein and BICD2 are utilized to activate HIV-1 particle movement in the retrograde direction, consistent with BICD2's function as a dynein adaptor.
FIG 7.
BICD2 promotes retrograde intracellular trafficking of HIV-1. (A and B) TZM-bl cells were pretreated with indicated single siRNAs and subsequently inoculated with pseudotyped HIV-1 labeled with GFP-Vpr and S15-mCherry. (A) Fraction of nonmotile particles per each cell. (B) Fraction of retrograde events per each cell for the motile population of particles. (C) Velocity of particle movement in the entire particle population. (D) Velocity of particle movement in the population of motile particles. (E) Net distance traveled in the entire particle population. (F) Net distance traveled in the population of motile particles. Results shown are from two independent experiments, each with >20 cells analyzed and >400 particles analyzed per condition. Shown are the mean values and standard deviations of the particle events/cell for the cell population. Statistical significance was calculated by an unpaired parametric t test for each siRNA treatment relative to nontargeting control siRNA treatment (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
Quantification of HIV-1 intracellular velocity also revealed a reduction in DYNC1H1- or BICD2-depleted cells relative to control cells (Fig. 7C), in agreement with a recent report (39). However, when nonmotile particles (described in Fig. 7A) were excluded from the analysis, thus quantifying only moving particles, we observed that the effect of DYNC1H1 or BICD2 depletion on velocity was no longer significant (Fig. 7D). Comparing the net distance of HIV-1 particle movement, we also observed a reduction in cells depleted of DYNC1H1 or BICD2 (Fig. 7E). As with the velocity measurements, this difference was not detected when the nonmotile population of HIV-1 particles was excluded from the analysis (Fig. 7F). Taken together, these results indicate that DYNC1H1 and BICD2 promote HIV-1 particle movement in the retrograde direction, likely by promoting the initiation of transport.
The HIV-1 capsid binds the dynein-dynactin complex.
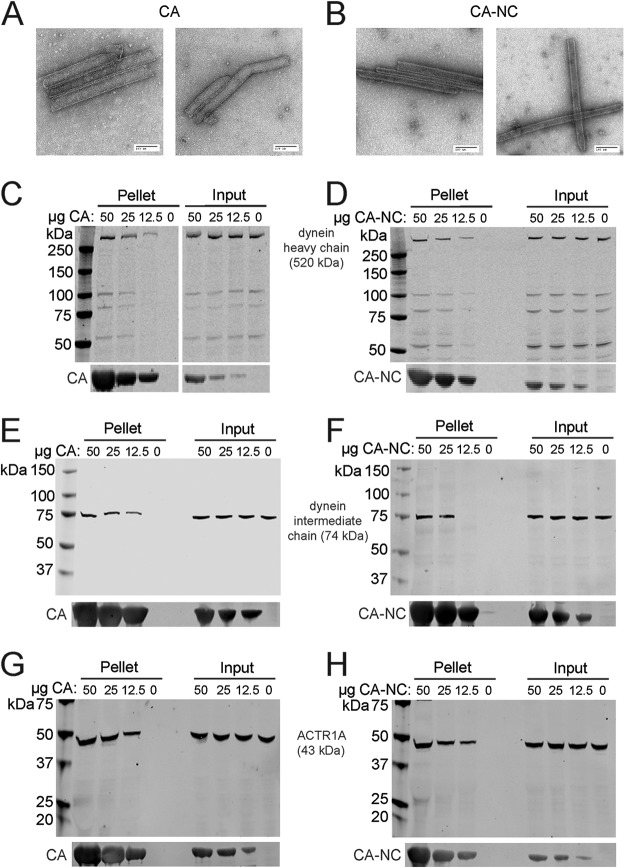
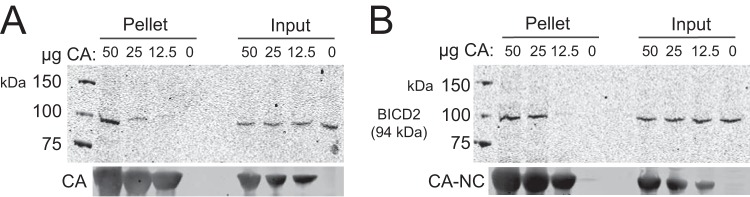
In order for HIV-1 to be transported to the nucleus by dynein, the virus should associate with the components of the dynein complex. The HIV-1 capsid encapsulates the internal core components, making it the most easily accessible component of HIV-1 for an interaction with dynein. The kinesin-1 adaptor FEZ1 also interacts with the HIV-1 capsid, suggesting that the capsid is important for molecular motor engagement (7). Therefore, we hypothesized that dynein interacts with the HIV-1 capsid to facilitate viral intracellular transport. To test this, we assayed association of dynein components in cell extracts with assembled CA tubes in vitro. The CA protein assembles into a hexameric lattice structure that can be stabilized by engineered disulfide cross-links at amino acid positions 14 and 45 in the CA protein or by linking CA to NC. These assemblies structurally resemble the native capsid lattice and are often used as surrogates in capsid-binding assays. We confirmed the tubular morphology of the assemblies by negative-stain electron microscopy (Fig. 8A, CA; Fig. 8B, CA-NC). Assembled tubes can be readily pelleted by low-speed centrifugation, thus facilitating the detection of proteins that associate with the HIV-1 capsid. To study capsid binding to dynein and dynactin components, we incubated 293T cell extracts with various quantities of CA assemblies, pelleted the complexes, and analyzed the associated proteins by immunoblotting (Fig. 8). Dynein heavy chain (Fig. 8C and D), dynein intermediate chain (Fig. 8E and F), and dynactin component ACTR1A (Fig. 8G and H) were all enriched in pellets from reaction mixtures containing CA tubes, suggesting that these dynein and dynactin components can associate with assembled HIV-1 capsids in vitro.
FIG 8.
Association of dynein and dynactin with capsid-like HIV-1 assemblies in vitro. (A and B) Recombinant CA and CA-NC proteins were assembled into tubes and examined by negative-stain electron microscopy to confirm tubular assembly. CA tubes were incubated with 293T cell extracts. The reactions were then centrifuged, the pellets were washed twice, and the pelleted proteins were analyzed by SDS-PAGE and immunoblotting. Blots were probed with antibodies to dynein heavy chain (C, CA; D, CA-NC), dynein intermediate chain (E, CA; F, CA-NC), and dynactin component ACTR1A (G, CA; H, CA-NC) or CA (C to H). Input samples corresponding to 10% of each reaction were removed prior to pelleting. Representative immunoblots from three independent experiments are shown.
BICD2 binds the HIV-1 capsid.
Our results indicated that the HIV-1 capsid can associate with the dynein complex in vitro. We hypothesized that the adaptor protein BICD2 binds to the HIV-1 capsid and functions as a capsid-specific dynein adaptor. To test this, CA (Fig. 9A) and CA-NC (Fig. 9B) tubes were incubated with cell extracts and pelleted, and proteins in the pelleted complexes were separated by SDS-PAGE and analyzed by immunoblotting with antibody to BICD2. We observed that cellular BICD2 associates with both CA and CA-NC tubes in vitro (Fig. 9, panels A and B, respectively).
FIG 9.
BICD2 binds to the HIV-1 capsid. (A) Capsid-like protein assemblies were titrated and incubated with 293T cell extracts. The reactions were then centrifuged, and the pelleted material was washed once and subjected to immunoblot analysis. Endogenous cellular BICD2 was probed for interaction with CA (A, CA tubes; B, CA-NC tubes).
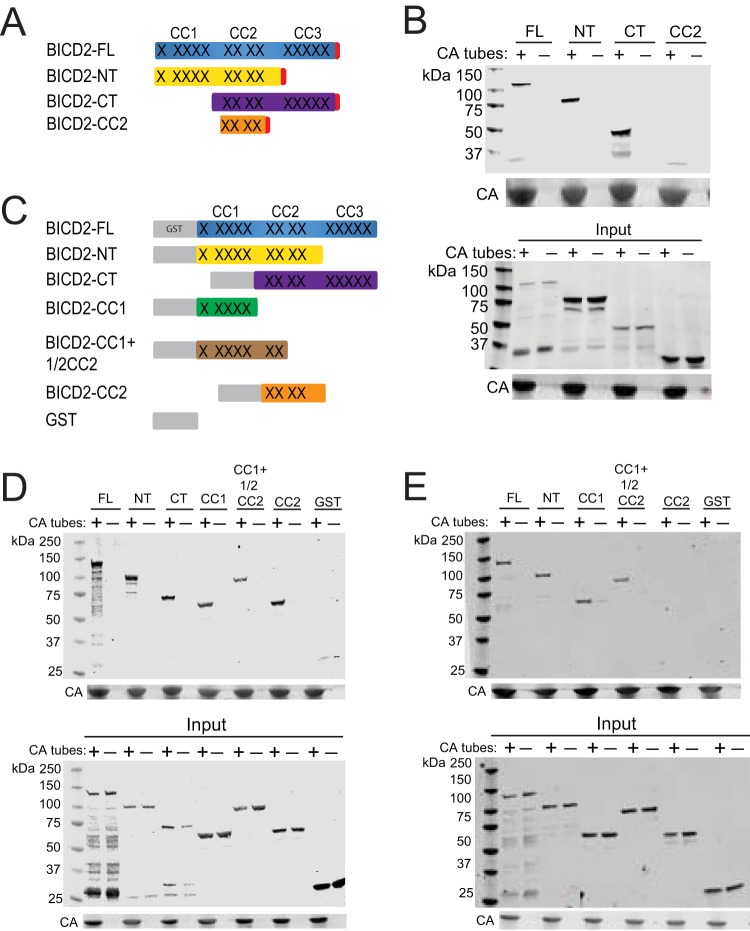
We next sought to identify the capsid-binding domain in BICD2. BICD2 contains two major domains: an N-terminal (NT) domain that links dynein to dynactin to activate processive movement, and a C-terminal (CT) domain that normally binds to cargo (Fig. 10A) (31, 46, 47). There are three regions in BICD2 containing coiled coils: CC1 on the N-terminal end, a C-terminal region (CC3), and a central CC2 region that overlaps the NT and CT domains. To localize the region of BICD2 to which the HIV-1 capsid binds, we ectopically expressed full-length BICD2 and portions corresponding to the individual NT and CT domains and a region encoding only CC2 as N-terminal HA-tagged proteins in 293T cells. Cell extracts were prepared and utilized in CA tube-binding assays (Fig. 10B). Surprisingly, we observed that NT and CT domain fragments, as well as the full-length BICD2 protein, cosedimented with assembled CA, suggesting that both BICD2 domains can associate with the viral capsid. However, we observed less association of the protein containing only CC2 relative to the NT and CT proteins, consistent with recently published results (39). These results suggest that BICD2 may contain two capsid-binding sites. The C-terminal domain represents the canonical binding site for other cargo (46), but a binding site in the N-terminal domain for cargo seemed unexpected to us.
FIG 10.
Analysis of BICD2 regions contributing to capsid binding. (A) Schematic representation of the HA-tagged BICD2 constructs used in panel B. Coiled-coil regions of the protein are represented by X's. (B) BICD2-HA proteins were expressed in 293T cells, and cell extracts were prepared and incubated with assembled CA tubes. Pelleted material was analyzed by immunoblotting. An antibody against HA was used to detect the proteins. (C) Schematic of GST-BICD2 fusion proteins used to study BICD2-capsid interactions. (D and E) Purified recombinant GST-BICD2 fusion proteins and GST were incubated with CA tubes, and the pelleted complexes were analyzed for association of the recombinant proteins by immunoblotting with antibody to GST. Binding reactions were performed with 150 mM NaCl (D) or 750 mM NaCl (E). Input samples correspond to 10% of each reaction removed prior to pelleting. Representative immunoblots from three independent experiments are shown.
The capsid-binding assays described above were performed with cell extracts; thus, it was possible that one or both BICD2 domains had bound the CA tubes indirectly, e.g., via association with dynactin. To assess whether both domains can directly bind to CA tubes, we expressed and purified the recombinant N-terminal GST-BICD2 fusion proteins from Escherichia coli and tested them for capsid binding in vitro. To further define the binding domain in the NT, we engineered an additional construct encoding only CC1, and an extended CC1 protein containing approximately one half of CC2 (Fig. 10C). We also made a GST-CC2 protein (Fig. 10C).
Binding assays with the purified recombinant proteins revealed that at physiological salt concentrations (150 mM NaCl), all of the GST-BICD2 fusion proteins copelleted with CA tubes, but the GST control protein did not (Fig. 10D). The observed association of the GST-CC2 with CA tubes was surprising, as we had not seen binding with the CC2 in HA-lysates (Fig. 10B). However, the GST-CC2 construct extended beyond the portion of the HA-CC2 (336-595 versus 340-539, respectively). To examine the relative stability of the capsid associations of the recombinant proteins, we assayed binding of several of the GST-BICD2 fusions under elevated salt conditions (750 mM NaCl). Under these conditions, the capsid association of the GST-BICD2 fusions was retained, except that of the GST-CC2 protein (Fig. 10E). Collectively, these results indicate that BICD2 can bind the HIV-1 capsid directly via both its N-terminal and C-terminal domains, likely utilizing the CC1 and CC3 coiled-coil regions.
BICD2 promotes the association of dynein with capsid-like HIV-1 assemblies in vitro.
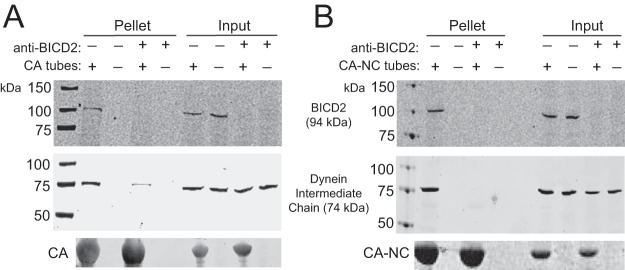
As a dynein adaptor, BICD2 may link dynein to the viral capsid. To test this hypothesis, we analyzed the effects of BICD2 depletion on the binding of dynein present in cell extracts to capsid-like assemblies in vitro. We immunodepleted BICD2 from 293T cell extracts and incubated the extracts with CA (Fig. 11A) or CA-NC (Fig. 11B) tubes. The tubes were then pelleted, and proteins in the pelleted complexes were separated by SDS-PAGE and analyzed by immunoblotting. We observed reduced binding of the dynein intermediate chain (middle) in BICD2-depleted extracts to CA and CA-NC tubes, suggesting that BICD2 links the HIV-1 capsid to dynein. Analysis of the input samples demonstrated that BICD2 was effectively immunodepleted from the extracts, and that levels of dynein intermediate chain were unaltered by BICD2 depletion.
FIG 11.
BICD2 facilitates association of dynein with capsid-like protein assemblies in vitro. (A and B) 293T lysates were immunodepleted for BICD2 before incubation with CA tubes (A, CA; B, CA-NC). The reactions were then centrifuged, the pellets were washed once, and the proteins were separated by SDS-PAGE and analyzed by immunoblotting. Blots were probed with antibodies to BICD2 to test for depletion (top), dynein intermediate chain to test for binding (middle), and CA (bottom). Input samples correspond to 10% of each reaction removed prior to pelleting. Representative immunoblots from three independent experiments are shown.
DISCUSSION
In this study, we performed a systematic analysis of the components of the dynein and dynactin complexes and of known dynein adaptors for their effects on HIV-1 infection. Consistent with earlier reports (5, 7), we observed that depletion of DYNC1H1 reduced cell permissiveness to HIV-1 infection. In addition to dynein heavy chain, depletion of the dynactin components DCTN2, DCTN3, and ACTR1A decreased cell permissiveness to HIV-1 infection, indicating that the dynactin complex also facilitates HIV-1 infection. Finally, we observed that BICD2 depletion reduced cell permissiveness to infection. Our results suggest that a dynein-dynactin-BICD2 complex promotes HIV-1 infection (Fig. 1 and 2).
Analysis of the stage at which infection is inhibited and virus imaging experiments indicate that the dynein-dynactin-BICD2 complex promotes the intracellular transport of HIV-1 to the nuclear envelope. Depletion of DYNC1H1, dynactin components, and BICD2 each resulted in a decrease in the levels of nuclear HIV-1 DNA (2-LTR circles) without a reduction in viral DNA levels, suggesting that the virus is impaired at or prior to nuclear entry (Fig. 3). While an effect of dynein depletion on reverse transcription was previously observed in one study (42), another study found the opposite (41). Our observation that dynein depletion reduced nuclear HIV-1 DNA without altering reverse transcription suggested that dynein accelerates HIV-1 transport to the nucleus or nuclear entry itself. Consistent with our findings, previous studies reported that depletion of DYNC1H1 by antibody or siRNA results in a build-up of HIV-1 particles at the cell periphery (5, 7).
We performed two types of cell imaging analysis, the results of which indicate that the dynein-dynactin-BICD2 complex promotes intracytoplasmic movement to the nucleus (Fig. 6 and 7). First, using fixed cell imaging, we showed that HIV-1 particles exhibit a greater average distance from the nuclear envelope in cells depleted of DYNC1H1 or BICD2. Second, our live imaging studies indicated that of the fraction of virus particles that exhibit movement within the cell was reduced in the depleted cells, and that particles that were moving exhibited reduced retrograde motion. Combined with the viral DNA analysis, these observations indicate that the failure of the virus to enter the nucleus can be mainly attributed to a trafficking defect versus a specific impairment in nuclear import. Nonetheless, we have not excluded a direct effect of the dynein complex on HIV-1 nuclear entry.
We observed that, relative to nonpseudotyped HIV-1, infection by HIV-1 particles pseudotyped by VSV-G was less affected by depletion of DYNC1H1, dynactin components, and BICD2 in TZM-bl cells (Fig. 4). Similar effects of pseudotyping on BICD2 dependence of HIV-1 were also observed in a recent study (39). Cell entry by HIV-1 infection normally occurs via direct fusion at the plasma membrane; in contrast, HIV-1 (VSV-G) enters cells by endocytosis and requires the low pH of the endosome for fusion. We speculate that this alters the location at which the dynein machinery engages the virus, thereby reducing the requirement for the dynein-dynactin-BICD2 complex. Interestingly, however, this difference in the dependence of HIV-1 infection on dynein and BICD2 between pseudotyped and nonpseudotyped viruses was not observed in other cell lines tested (Fig. 5), suggesting that this effect may be specific to TZM-bl cells.
In addition to HIV-1, infection by VSV-G-pseudotyped SIV and MLV was dependent on dynein, dynactin, and BICD2 (Fig. 4), indicating that a common mechanism of dynein transport can be utilized by a variety of retroviruses. A previous report showed that infection by ecotropic but not amphotropic MLV depends on the adaptor NudEL (44), further suggesting that dynein adaptor utilization depends on the cell receptor and/or entry pathway utilized by the virus. The apparent broad dependence of diverse retroviruses on BICD2 suggests the possibility that convergent virus evolution adopted a common solution for engagement of the intracellular transport machinery. It will be interesting to determine whether nonprimate lentiviruses and avian retroviruses also utilize BICD2 for infection.
Our results indicate that BICD2 interacts directly with the viral capsid. We observed an association of native cellular BICD2 with CA assemblies in vitro, and recombinant purified BICD2 also bound to the assemblies. Surprisingly, we observed that both the N-terminal and C-terminal regions of BICD2 interacted with HIV-1 capsid assemblies (Fig. 10). The C-terminal region of BICD2 normally is responsible for cargo binding, while the N-terminal region engages dynein-dynactin. These two regions can interact with one another for autoinhibition, and previous models posit that BICD2 must sequentially interact with cargo after engaging the dynein-dynactin complex, thus ensuring efficient coupling (34). The presence of capsid-binding sites at both the C terminus and the N terminus of BICD2 suggests that the capsid may bind in a canonical manner to the BICD2 C-terminal site but that it also can bind in a noncanonical way to the BICD2 N terminus. Since the N-terminal site would not normally be available for binding until cargo is bound, it is possible that the HIV-1 capsid hijacks an open complex of BICD2 bound to a cellular cargo via its C-terminal domain.
Dynein adaptors link specific cargo to the dynein-dynactin complex, resulting in dynein activation and efficient movement along microtubules. Our data indicate that the adaptor protein BICD2 is important for HIV-1 infection. HIV-1 capsid-like assemblies bound to dynein chains, the dynactin component ACTR1A, and BICD2 in cell extracts (Fig. 8 and 9) and to recombinant BICD2 in vitro (Fig. 9). These observations suggested that BICD2 links the viral capsid to the dynein complex. In support of the adaptor model for BICD2 function in HIV-1 infection, we also observed that immunodepletion of BICD2 in lysates reduced the binding of dynein intermediate chain to HIV-1 capsid-like assemblies (Fig. 11). BICD2 interacts directly with DCTN2/dynamitin (31, 34), and a current model proposes that dynactin links BICD2 to dynein and activates movement, thereby promoting cargo recruitment and transport. It will be of interest to characterize the interactions of additional dynein and dynactin components with HIV-1 capsid assemblies and to determine whether the associations depend on BICD2.
MATERIALS AND METHODS
Plasmids.
HIV-GFP is an Env-defective HIV-1 proviral construct encoding GFP in place of nef (48). The plasmid NL43-GFP was constructed by replacing the SalI-BamHI region of HIV-GFP with that from pNL4-3, restoring the Env open reading frame. The VSV-G expression plasmid pHCMV-G was provided by J. Burns (University of California, San Diego) (49). The plasmids GFP-Vpr and S15-mCherry were obtained from Thomas Hope (45). The plasmid psPAX2 was obtained through the NIH AIDS Reagent Program from Didier Trono. The plasmid pVPack-GP was obtained from Agilent Technologies. The SIV plasmid SIV 239IG was provided by P. Bieniasz (50). The MLV transfer vector construct pBABE-EGFP was constructed by inserting a BamHI-EcoRI fragment from pHR′-CMV-GFP (51) into pBABEpuro (52).
A human BICD2 cDNA (Dharmacon) was cloned into EGFP-C1 (Clontech) via Gibson assembly at sites EcoRI and KpnI to make EGFP-C1-BICD2. This plasmid was a gift from Ryoma Ohi. For construction of CMX-HA, a hemagglutinin (HA) epitope tag sequence was inserted into the CMX-PL1 plasmid at the KpnI and NheI sites. BICD2, BICD2-NT, BICD2-CT, and BICD2-CC2 sequences, encoding amino acids 1 to 824, 1 to 580, 485 to 824, and 340 to 539, respectively, were PCR amplified from EGFP-C1-BICD2 and inserted into the CMX-HA plasmid at the EcoRI and KpnI sites in frame with the hemagglutinin epitope tag sequence. GST-BICD2 fusion proteins were constructed as follows: GST-BICD2 encoding full-length BICD2 (amino acids 1 to 824 [1–824]); GST-NT (1–580); GST-CT (485–824); GST-CC1 (1–271); GST-CC1.5 (1–468); and GST-CC2 (336–595). GST-BICD2 fusion proteins were PCR amplified from BICD2-CMX-PL1 and inserted into pGEX-6P-3 at the BamHI and EcoRI sites. All primer sequences used for cloning are listed in Table S1 in the supplemental material.
The recombinant CA protein 14C/45C was made by introduction of cysteine mutations into pET21a(+) HIV-1 CA at positions 14 and 45 (53). The HIV-1 CA-NC expression plasmid WISP-98-68 (54) was kindly provided by Wes Sundquist.
Cells and viruses.
TZM-bl cells, Jurkat cells, and GHOST CXCR4+ CCR5+ cells were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH, from John C Kappes, Xiaoyun Wu, and Tranzyme, Inc. (55–59); Arthur Weiss (60); and Vineet N. KewalRamani and Dan R. Littman, respectively (61). A431-CD4 cells were generated from A431 cells (ATCC) by transduction with pBABE-CD4 and selection in puromycin. Primary human fibroblasts were purchased from Advanced BioMatrix, Inc. 293T cells were purchased from the ATCC. 293T cells, A431-CD4 cells, GHOST CXCR4+ CCR5+ cells, primary human fibroblasts, and TZM-bl cells were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 IU/ml penicillin, and 100 μg/ml streptomycin. Jurkat cells were cultured in Roswell Park Memorial Institute (RPMI) medium supplemented with 10% FBS, 100 IU/ml penicillin, and 100 μg/ml streptomycin. Viruses for fixed cell imaging studies were produced by cotransfection of HIV-GFP (8 μg) with pHCMV-G (2 μg), GFP-Vpr (2 μg), and S15-mCherry (1 μg). Viruses for use in live cell imaging studies were produced by cotransfection of psPax2 with pHCMV-G, GFP-Vpr, and S15-mCherry. HIV-1 stocks were clarified through 0.45-μm-pore size filters, assayed for p24 antigen using an enzyme-linked immunosorbent assay, and stored at −80°C until use. Murine leukemia virus (MLV-GFP) was produced by cotransfection of pVPack-GP (Agilent Technologies) with pBABE-EGFP and pHCMV-G. SIV-GFP was produced by cotransfection with SIV 239IG and pHCMV-G.
Recombinant proteins.
GST-BICD2 fusion proteins, recombinant HIV-1 CA protein encoding substitutions of Cys for positions 14 and 45 (14C/45C), and recombinant HIV-1 CA-NC were expressed in E. coli using induction by using 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside; Gold Biotechnology). Cells were lysed by sonication, using a Sonic Dismembrator ultrasonic processor (Fisher Scientific), and lysates cleared by centrifugation for 1 h at 15,000 rpm at 4°C. GST-BICD2 constructs were purified by batch method on glutathione agarose beads as recommended by the manufacturer (Pierce). CA and CA-NC constructs were purified by anion-exchange chromatography as previously described (53, 54).
siRNA transfection and knockdown efficiency screening.
All siRNAs were purchased from Dharmacon (ON-TARGETplus siRNAs). Pools of four individual siRNA duplexes (SMARTpool) were used for initial screening of targets, and specific targets were verified using the four individual siRNA duplexes from the pool. The siRNA sequences are listed in Table S2 in the supplemental material. TZM-bl cells were transfected with 40 nM siRNA and DharmaFECT 1 transfection reagent and washed after 16 h. Cells were cultured for either 48 h (only in initial screening) or 72 h before virus inoculation or cell harvesting for knockdown efficiency analysis. To quantify mRNA levels, cDNA was synthesized (Applied Biosystems), and SYBR green qPCR was performed for each gene of interest, standardized against nontargeting siRNA control, with GAPDH (glyceraldehyde-3-phosphate dehydrogenase) levels as an internal control. The primers used for mRNA quantification analysis are listed in Table S3 in the supplemental material. For protein expression analysis, cytoplasmic extracts were prepared by lysing cells in NP-40 buffer (150 mM NaCl, 50 mM Tris-HCl [pH 8.0], 1% NP-40) containing protease inhibitor cocktail (Roche) for 30 min on ice. Samples were centrifuged at 19,000 × g for 5 min at 4°C to remove nuclei, and proteins in the supernatants were separated by SDS-PAGE on 4 to 20% gradient gels (GenScript) and then analyzed by immunoblotting.
Infection assays.
Cells were seeded in a 96-well plate at 10,000 cells per well. After 1 day of culture, cells were inoculated with GFP-encoding virus (native Env HIV-1, 200 ng/well; VSV-G-pseudotyped viruses 0.5 ng/well plus 8 μg/ml Polybrene) and cultured for 48 h. No Polybrene treatment was utilized with native enveloped HIV-1 in order to ensure that the route of viral entry was not affected. Cells were detached with trypsin-EDTA (100 μl) and fixed with an equal volume of 4% paraformaldehyde. The extent of infection was assayed by flow cytometry for GFP expression using an Accuri C6 flow cytometer. At least 7,000 cells were analyzed for each sample.
PCR quantitation of nuclear entry and reverse transcription in target cells.
TZM-bl cells were transfected with siRNA for 72 h and then inoculated with 40 ng of DNase I-treated HIV-1 pseudotyped with VSV-G. Cells were harvested 8 h after infection for second strand-transfer analysis or 24 h after infection for 2-LTR circle analysis. For harvesting, cells were washed with 1× PBS and then treated with qPCR lysis buffer (50 mM KCl, 1.5 mM MgCl2, 1.5 mM Tris-HCl [pH 8.0], 0.45% NP-40, and 0.45% Tween 20) containing proteinase K (1 mg/ml) for 1 h at 57°C. Samples were then incubated at 95°C for 15 min to inactivate proteinase K and stored at −80°C. HIV-1 DNA in the samples was quantified by real-time PCR with SYBR green detection on a Stratagene MX3000p instrument. GAPDH DNA levels quantified by real-time PCR were used to normalize the total DNA in sample values. To control for possible contaminating plasmid DNA in the virus inocula, the reverse transcriptase inhibitor efavirenz (1 μM) was added to some cultures at the time of infection.
Trafficking analysis, fixed-cell imaging, and live-cell imaging.
TZM-bl cells were transfected for with 40 nM siRNA and cultured for 72 h prior to virus inoculation. One day before infection, cells were detached with trypsin, reseeded onto coverslips (fixed cell) or MatTek dishes (live cell), and allowed to adhere overnight. For fixed cell analysis, the cells were spinoculated with HIV-GFP(VSV-G) dually labeled with GFP-Vpr and S15-mCherry at 800 × g for 30 min at 15°C to allow virus adhesion but not entry. After spinoculation, media on the cells was replaced, and the cultures were incubated for an additional 1 h at 37°C. Cells were washed and fixed with methanol at −20°C for 10 min and stained with 500 nM DAPI (4′,6′-diamidino-2-phenylindole) to visualize nuclei. Stained cells were mounted in Gelvatol (13.3 mg/ml polyvinyl alcohol, 0.67 M Tris-HCl [pH 8.6], 0.33 mg/ml glycerol, 2.5% 1,4-diazabicyclo[2.2.2]octane [DABCO]). Images were acquired with a DeltaVision Elite image restoration system (GE Healthcare) equipped with a 60× 1.4 numerical aperture lens (Olympus) and a Cool SnapHQ2 charge-coupled device camera (Photometrics). Z-sections spaced 200 nm apart were acquired and deconvolved with SoftWorx (GE Healthcare). Maximum intensity z-projections were generated with Fiji (62), and the distance of particles from the nucleus was measured. Particles exhibiting detectable mCherry fluorescence were omitted from analysis.
Two independent experiments were performed, each with >20 cells/condition analyzed, and >400 particles were analyzed per condition. For live cell analysis, cells were spinoculated with psPax2 VSVG dually labeled with GFP-Vpr and S15-mCherry for 30 min and then cultured at 37°C for 20 min before imaging. Live cell imaging was performed in the presence of CO2 at 37°C in FluoroBrite DMEM (Thermo Fisher) supplemented with 10% FBS and 7 mM HEPES-KOH (pH 7.7), using the DeltaVision Elite system. Images were captured every 0.5 s for a period of 2.5 min. Images to test for mCherry particle signal were taken immediately before the 2.5-min time frame. Particles displaying mCherry signal were considered to be unfused and were excluded from track analysis. In addition, any particles that were not present at the start of imaging were excluded because their fusion status could not be determined. Images were taken of various individual cells over the course of 1 h. Tracking was analyzed by TrackMate (63) in Fiji, and tracks were classified as retrograde or anterograde trajectories. The distances of these trajectories and the number of trajectories were used for analysis. Two independent experiments were performed, each with >20 cells/condition analyzed, and >300 tracks were analyzed for each condition. We observed efficient virion labeling efficiency, with 95 to 99% of particles containing both labels and an average of 98%. At the 1-h time point, a fusion rate (the loss of S15-mCherry signal) of 44 to 68% was observed, with no significant fusion differences occurring between the nontargeting and knockdown conditions.
In vitro binding assay.
To prepare protein extracts, 293T cells were detached with trypsin-EDTA, resuspended in complete medium, washed in PBS, resuspended in 5× volume hypotonic buffer (10 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 10 mM NaCl), Dounce homogenized with 25 strokes with the tight pestle, and precleared by centrifugation for 15 min at 4°C in a microcentrifuge (18,800 × g). Extracts were stored at −80°C until further use. HIV-1 CA (14C/45C) protein was assembled under high-salt conditions (1 M NaCl, 50 mM Tris-HCl [pH 8]), as previously described (64). HIV-1 CA-NC was assembled by dilution to 0.3 mM at 37°C for 1 h (500 mM NaCl, 50 mM Tris-HCl [pH 8], 60 μM TG50 (purchased from IDT). Cross-linked recombinant CA assemblies were incubated with cell extracts or purified GST proteins in CA tube-binding buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 5 mM MgCl2, 0.5 mM EDTA) or CA-NC tube-binding buffer (50 mM Tris [pH 8.0], 500 mM NaCl, 5 mM MgCl2, 0.5 mM EDTA) for 1 h at room temperature. The higher salt concentration was required to prevent CA-NC tube disassembly. Reactions were then centrifuged for 5 min at 10,000 × g, and the pellets were rinsed once with binding buffer to remove unbound proteins. Pelleted proteins were separated by SDS-PAGE and analyzed by immunoblotting.
Immunodepletion of BICD2 from cell extracts.
Protein A-agarose (Santa Cruz Biotechnology, 40 μl packed beads) was washed twice in 3× volume binding buffer (10 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 10 mM NaCl) and centrifuged for 5 min at 10,000 × g. Beads were resuspended in a 1× volume of binding buffer and bound to BICD2 antibody (15 μg) for 1 h rotating at 4°C or unbound beads as a negative control. Beads were then washed two more times in binding buffer to remove unbound antibody. Freshly prepared 293T cell extract (2.5 mg total protein) was then incubated with the BICD2 antibody-bound beads or beads alone for mock depletion overnight at 4°C with rotation. Beads were pelleted for 5 min at 10,000 × g, and the supernatant was recovered for use in binding reactions. The BICD2-depleted extracts were analyzed for BICD2 and dynein by immunoblotting.
Visualization of CA tube assembly by electron microscopy.
Cross-linked recombinant CA assemblies were visualized by negative-stain electron microscopy to check for tube assembly. Samples were stained with 0.7% uranyl formate and visualized on a FEI Morgagni electron microscope at 100 kV at a magnification of ×22000. Images were recorded on an ATM 1000×1000 charge-coupled device camera.
Antibodies and immunoblotting.
To the binding assay pellets, an equal volume of 2× Laemmli buffer was added, followed by heating at 100°C for 5 min. Samples were loaded onto a 4 to 20% polyacrylamide gel (GenScript) for SDS-PAGE. After separation, the proteins were transferred to nitrocellulose membranes (Perkin-Elmer) and probed with the following antibodies: BICD2 (Abcam, 117818; 5 μg/ml), DYNC1H1 (Sigma, D1667; 1:1,000 dilution), DIC (Abcam, 23905; 1 μg/ml), ACTR1A (Abcam, 227425; 1:2,000 dilution), HA (Sigma 3F10; 200 ng/ml), GST (Abcam, 3416; 1:5,000 dilution), and CA (183-H12-5C, NIH AIDS Research and Reference Reagent Program; 1:5,000 dilution). For protein expression analysis, the antibodies described above were used, as well as dynamitin/DCTN2 (Abcam, 133492; 1:2,000 dilution), DCTN3 (Abcam, 124674; 1:2,000 dilution), and GAPDH (Thermo Fisher, A21994; 5 μg/ml). Antibody complexes were detected with the appropriate IR dye-conjugated secondary antibodies (LI-COR Biosciences) and detected by scanning with a LI-COR Odyssey imaging system.
Statistical analysis.
Statistical significance was assessed by Student t test compared individual targets to the nontargeting siRNA control. The results are represented as mean values and standard deviations. A one-way analysis of variance (ANOVA) was performed for statistical analysis of the imaging results, followed by a Dunnett's multiple-comparison test. Statistical analysis was performed using GraphPad Prism software.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ryoma Ohi and Stephen Norris for the EGFP-C1-BICD2 plasmid and for instrument use and training for the microscopy work. We thank Elad Binshtein and the Vanderbilt University Center for Structural Biology for electron microscopy analysis.
This research was supported by NIH R01 AI076121 (C.A.) and NIH F31 AI129747-01A1 (S.K.C.).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00358-18.
REFERENCES
- 1.Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. 2008. Identification of host proteins required for HIV infection through a functional genomic screen. Science 319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 2.Konig R, Zhou Y, Elleder D, Diamond TL, Bonamy GM, Irelan JT, Chiang CY, Tu BP, De Jesus PD, Lilley CE, Seidel S, Opaluch AM, Caldwell JS, Weitzman MD, Kuhen KL, Bandyopadhyay S, Ideker T, Orth AP, Miraglia LJ, Bushman FD, Young JA, Chanda SK. 2008. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell 135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou H, Xu M, Huang Q, Gates AT, Zhang XD, Castle JC, Stec E, Ferrer M, Strulovici B, Hazuda DJ, Espeseth AS. 2008. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe 4:495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Fernandez J, Portilho DM, Danckaert A, Munier S, Becker A, Roux P, Zambo A, Shorte S, Jacob Y, Vidalain PO, Charneau P, Clavel F, Arhel NJ. 2015. Microtubule-associated proteins 1 (MAP1) promote human immunodeficiency virus type I (HIV-1) intracytoplasmic routing to the nucleus. J Biol Chem 290:4631–4646. doi: 10.1074/jbc.M114.613133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McDonald D, Vodicka MA, Lucero G, Svitkina TM, Borisy GG, Emerman M, Hope TJ. 2002. Visualization of the intracellular behavior of HIV in living cells. J Cell Biol 159:441–452. doi: 10.1083/jcb.200203150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sabo Y, Walsh D, Barry DS, Tinaztepe S, de Los Santos K, Goff SP, Gundersen GG, Naghavi MH. 2013. HIV-1 induces the formation of stable microtubules to enhance early infection. Cell Host Microbe 14:535–546. doi: 10.1016/j.chom.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malikov V, da Silva ES, Jovasevic V, Bennett G, de Souza Aranha Vieira DA, Schulte B, Diaz-Griffero F, Walsh D, Naghavi MH. 2015. HIV-1 capsids bind and exploit the kinesin-1 adaptor FEZ1 for inward movement to the nucleus. Nat Commun 6:6660. doi: 10.1038/ncomms7660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dharan A, Talley S, Tripathi A, Mamede JI, Majetschak M, Hope TJ, Campbell EM. 2016. KIF5B and Nup358 cooperatively mediate the nuclear import of HIV-1 during infection. PLoS Pathog 12:e1005700. doi: 10.1371/journal.ppat.1005700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaudin R, de Alencar BC, Arhel N, Benaroch P. 2013. HIV trafficking in host cells: motors wanted! Trends Cell Biol 23:652–662. doi: 10.1016/j.tcb.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 10.Hook P, Vallee RB. 2006. The dynein family at a glance. J Cell Sci 119:4369–4371. doi: 10.1242/jcs.03176. [DOI] [PubMed] [Google Scholar]
- 11.Kato Y, Miyakawa T, Tanokura M. 2017. Overview of the mechanism of cytoskeletal motors based on structure. Biophys Rev 10:571–581. doi: 10.1007/s12551-017-0368-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu JJ. 2017. Regulation of dynein-dynactin-driven vesicular transport. Traffic 18:336–347. doi: 10.1111/tra.12475. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt H, Carter AP. 2016. Review: Structure and mechanism of the dynein motor ATPase. Biopolymers 105:557–567. doi: 10.1002/bip.22856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desfarges S, Salin B, Calmels C, Andreola ML, Parissi V, Fournier M. 2009. HIV-1 integrase trafficking in S. cerevisiae: a useful model to dissect the microtubule network involvement of viral protein nuclear import. Yeast 26:39–54. doi: 10.1002/yea.1651. [DOI] [PubMed] [Google Scholar]
- 15.Jayappa KD, Ao Z, Wang X, Mouland AJ, Shekhar S, Yang X, Yao X. 2015. Human immunodeficiency virus type 1 employs the cellular dynein light chain 1 protein for reverse transcription through interaction with its integrase protein. J Virol 89:3497–3511. doi: 10.1128/JVI.03347-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ayloo S, Lazarus JE, Dodda A, Tokito M, Ostap EM, Holzbaur EL. 2014. Dynactin functions as both a dynamic tether and brake during dynein-driven motility. Nat Commun 5:4807. doi: 10.1038/ncomms5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.King SJ, Schroer TA. 2000. Dynactin increases the processivity of the cytoplasmic dynein motor. Nat Cell Biol 2:20–24. doi: 10.1038/71338. [DOI] [PubMed] [Google Scholar]
- 18.Culver-Hanlon TL, Lex SA, Stephens AD, Quintyne NJ, King SJ. 2006. A microtubule-binding domain in dynactin increases dynein processivity by skating along microtubules. Nat Cell Biol 8:264–270. doi: 10.1038/ncb1370. [DOI] [PubMed] [Google Scholar]
- 19.Ross JL, Wallace K, Shuman H, Goldman YE, Holzbaur EL. 2006. Processive bidirectional motion of dynein-dynactin complexes in vitro. Nat Cell Biol 8:562–570. doi: 10.1038/ncb1421. [DOI] [PubMed] [Google Scholar]
- 20.Tripathy SK, Weil SJ, Chen C, Anand P, Vallee RB, Gross SP. 2014. Autoregulatory mechanism for dynactin control of processive and diffusive dynein transport. Nat Cell Biol 16:1192–1201. doi: 10.1038/ncb3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kobayashi T, Miyashita T, Murayama T, Toyoshima YY. 2017. Dynactin has two antagonistic regulatory domains and exerts opposing effects on dynein motility. PLoS One 12:e0183672. doi: 10.1371/journal.pone.0183672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Urnavicius L, Zhang K, Diamant AG, Motz C, Schlager MA, Yu M, Patel NA, Robinson CV, Carter AP. 2015. The structure of the dynactin complex and its interaction with dynein. Science 347:1441–1446. doi: 10.1126/science.aaa4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheong FK, Feng L, Sarkeshik A, Yates JR III, Schroer TA. 2014. Dynactin integrity depends upon direct binding of dynamitin to Arp1. Mol Biol Cell 25:2171–2180. doi: 10.1091/mbc.e14-03-0842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Melkonian KA, Maier KC, Godfrey JE, Rodgers M, Schroer TA. 2007. Mechanism of dynamitin-mediated disruption of dynactin. J Biol Chem 282:19355–19364. doi: 10.1074/jbc.M700003200. [DOI] [PubMed] [Google Scholar]
- 25.Goodson HV, Skube SB, Stalder R, Valetti C, Kreis TE, Morrison EE, Schroer TA. 2003. CLIP-170 interacts with dynactin complex and the APC-binding protein EB1 by different mechanisms. Cell Motil Cytoskeleton 55:156–173. doi: 10.1002/cm.10114. [DOI] [PubMed] [Google Scholar]
- 26.Jacquot G, Maidou-Peindara P, Benichou S. 2010. Molecular and functional basis for the scaffolding role of the p50/dynamitin subunit of the microtubule-associated dynactin complex. J Biol Chem 285:23019–23031. doi: 10.1074/jbc.M110.100602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yeh TY, Quintyne NJ, Scipioni BR, Eckley DM, Schroer TA. 2012. Dynactin's pointed-end complex is a cargo-targeting module. Mol Biol Cell 23:3827–3837. doi: 10.1091/mbc.e12-07-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schroeder CM, Vale RD. 2016. Assembly and activation of dynein-dynactin by the cargo adaptor protein Hook3. J Cell Biol 214:309–318. doi: 10.1083/jcb.201604002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKenney RJ, Huynh W, Tanenbaum ME, Bhabha G, Vale RD. 2014. Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science 345:337–341. doi: 10.1126/science.1254198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schlager MA, Hoang HT, Urnavicius L, Bullock SL, Carter AP. 2014. In vitro reconstitution of a highly processive recombinant human dynein complex. EMBO J 33:1855–1868. doi: 10.15252/embj.201488792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Splinter D, Razafsky DS, Schlager MA, Serra-Marques A, Grigoriev I, Demmers J, Keijzer N, Jiang K, Poser I, Hyman AA, Hoogenraad CC, King SJ, Akhmanova A. 2012. BICD2, dynactin, and LIS1 cooperate in regulating dynein recruitment to cellular structures. Mol Biol Cell 23:4226–4241. doi: 10.1091/mbc.e12-03-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamada M, Toba S, Yoshida Y, Haratani K, Mori D, Yano Y, Mimori-Kiyosue Y, Nakamura T, Itoh K, Fushiki S, Setou M, Wynshaw-Boris A, Torisawa T, Toyoshima YY, Hirotsune S. 2008. LIS1 and NDEL1 coordinate the plus-end-directed transport of cytoplasmic dynein. EMBO J 27:2471–2483. doi: 10.1038/emboj.2008.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lam C, Vergnolle MA, Thorpe L, Woodman PG, Allan VJ. 2010. Functional interplay between LIS1, NDE1 and NDEL1 in dynein-dependent organelle positioning. J Cell Sci 123:202–212. doi: 10.1242/jcs.059337. [DOI] [PubMed] [Google Scholar]
- 34.Hoogenraad CC, Akhmanova A, Howell SA, Dortland BR, De Zeeuw CI, Willemsen R, Visser P, Grosveld F, Galjart N. 2001. Mammalian Golgi-associated Bicaudal-D2 functions in the dynein-dynactin pathway by interacting with these complexes. EMBO J 20:4041–4054. doi: 10.1093/emboj/20.15.4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gassmann R, Essex A, Hu JS, Maddox PS, Motegi F, Sugimoto A, O'Rourke SM, Bowerman B, McLeod I, Yates JR III, Oegema K, Cheeseman IM, Desai A. 2008. A new mechanism controlling kinetochore-microtubule interactions revealed by comparison of two dynein-targeting components: SPDL-1 and the Rod/Zwilch/Zw10 complex. Genes Dev 22:2385–2399. doi: 10.1101/gad.1687508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams BC, Li Z, Liu S, Williams EV, Leung G, Yen TJ, Goldberg ML. 2003. Zwilch, a new component of the ZW10/ROD complex required for kinetochore functions. Mol Biol Cell 14:1379–1391. doi: 10.1091/mbc.e02-09-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J, Qiu R, Arst HN Jr, Penalva MA, Xiang X. 2014. HookA is a novel dynein-early endosome linker critical for cargo movement in vivo. J Cell Biol 204:1009–1026. doi: 10.1083/jcb.201308009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kardon JR, Vale RD. 2009. Regulators of the cytoplasmic dynein motor. Nat Rev Mol Cell Biol 10:854–865. doi: 10.1038/nrm2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dharan A, Opp S, Abdel-Rahim O, Keceli SK, Imam S, Diaz-Griffero F, Campbell EM. 2017. Bicaudal D2 facilitates the cytoplasmic trafficking and nuclear import of HIV-1 genomes during infection. Proc Natl Acad Sci U S A 114:E10707–E10716. doi: 10.1073/pnas.1712033114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Campbell EM, Hope TJ. 2015. HIV-1 capsid: the multifaceted key player in HIV-1 infection. Nat Rev Microbiol 13:471–483. doi: 10.1038/nrmicro3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pawlica P, Berthoux L. 2014. Cytoplasmic dynein promotes HIV-1 uncoating. Viruses 6:4195–4211. doi: 10.3390/v6114195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lukic Z, Dharan A, Fricke T, Diaz-Griffero F, Campbell EM. 2014. HIV-1 uncoating is facilitated by dynein and kinesin 1. J Virol 88:13613–13625. doi: 10.1128/JVI.02219-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Opazo T, Garces A, Tapia D, Barraza F, Bravo A, Schwenke T, Cancino J, Arriagada G. 2017. Functional evidence of the involvement of the dynein light chain DYNLRB2 in murine leukemia virus infection. J Virol 91:e00129-. doi: 10.1128/JVI.00129-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valle-Tenney R, Opazo T, Cancino J, Goff SP, Arriagada G. 2016. Dynein regulators are important for ecotropic murine leukemia virus infection. J Virol 90:6896–6905. doi: 10.1128/JVI.00863-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Campbell EM, Perez O, Melar M, Hope TJ. 2007. Labeling HIV-1 virions with two fluorescent proteins allows identification of virions that have productively entered the target cell. Virology 360:286–293. doi: 10.1016/j.virol.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Y, Salter HK, Holding AN, Johnson CM, Stephens E, Lukavsky PJ, Walshaw J, Bullock SL. 2013. Bicaudal-D uses a parallel, homodimeric coiled coil with heterotypic registry to coordinate recruitment of cargos to dynein. Genes Dev 27:1233–1246. doi: 10.1101/gad.212381.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoogenraad CC, Wulf P, Schiefermeier N, Stepanova T, Galjart N, Small JV, Grosveld F, de Zeeuw CI, Akhmanova A. 2003. Bicaudal D induces selective dynein-mediated microtubule minus end-directed transport. EMBO J 22:6004–6015. doi: 10.1093/emboj/cdg592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He J, Chen Y, Farzan M, Choe H, Ohagen A, Gartner S, Busciglio J, Yang X, Hofmann W, Newman W, Mackay CR, Sodroski J, Gabuzda D. 1997. CCR3 and CCR5 are coreceptors for HIV-1 infection of microglia. Nature 385:645–649. doi: 10.1038/385645a0. [DOI] [PubMed] [Google Scholar]
- 49.Yee JK, Friedmann T, Burns JC. 1994. Generation of high-titer pseudotyped retroviral vectors with very broad host range. Methods Cell Biol 43(Part A):99–112. doi: 10.1016/S0091-679X(08)60600-7. [DOI] [PubMed] [Google Scholar]
- 50.Zhang YJ, Hatziioannou T, Zang T, Braaten D, Luban J, Goff SP, Bieniasz PD. 2002. Envelope-dependent, cyclophilin-independent effects of glycosaminoglycans on human immunodeficiency virus type 1 attachment and infection. J Virol 76:6332–6343. doi: 10.1128/JVI.76.12.6332-6343.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miyoshi H, Takahashi M, Gage FH, Verma IM. 1997. Stable and efficient gene transfer into the retina using an HIV-based lentiviral vector. Proc Natl Acad Sci U S A 94:10319–10323. doi: 10.1073/pnas.94.19.10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morgenstern JP, Land H. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res 18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Du S, Betts L, Yang R, Shi H, Concel J, Ahn J, Aiken C, Zhang P, Yeh JI. 2011. Structure of the HIV-1 full-length capsid protein in a conformationally trapped unassembled state induced by small-molecule binding. J Mol Biol 406:371–386. doi: 10.1016/j.jmb.2010.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lemke CT, Titolo S, von Schwedler U, Goudreau N, Mercier JF, Wardrop E, Faucher AM, Coulombe R, Banik SS, Fader L, Gagnon A, Kawai SH, Rancourt J, Tremblay M, Yoakim C, Simoneau B, Archambault J, Sundquist WI, Mason SW. 2012. Distinct effects of two HIV-1 capsid assembly inhibitor families that bind the same site within the N-terminal domain of the viral CA protein. J Virol 86:6643–6655. doi: 10.1128/JVI.00493-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, Kappes JC. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother 46:1896–1905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Platt EJ, Bilska M, Kozak SL, Kabat D, Montefiori DC. 2009. Evidence that ecotropic murine leukemia virus contamination in TZM-bl cells does not affect the outcome of neutralizing antibody assays with human immunodeficiency virus type 1. J Virol 83:8289–8292. doi: 10.1128/JVI.00709-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takeuchi Y, McClure MO, Pizzato M. 2008. Identification of gammaretroviruses constitutively released from cell lines used for human immunodeficiency virus research. J Virol 82:12585–12588. doi: 10.1128/JVI.01726-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Derdeyn CA, Decker JM, Sfakianos JN, Wu X, O'Brien WA, Ratner L, Kappes JC, Shaw GM, Hunter E. 2000. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J Virol 74:8358–8367. doi: 10.1128/JVI.74.18.8358-8367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol 72:2855–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weiss A, Wiskocil RL, Stobo JD. 1984. The role of T3 surface molecules in the activation of human T cells: a two-stimulus requirement for IL 2 production reflects events occurring at a pre-translational level. J Immunol 133:123–128. [PubMed] [Google Scholar]
- 61.Morner A, Bjorndal A, Albert J, Kewalramani VN, Littman DR, Inoue R, Thorstensson R, Fenyo EM, Bjorling E. 1999. Primary human immunodeficiency virus type 2 (HIV-2) isolates, like HIV-1 isolates, frequently use CCR5 but show promiscuity in coreceptor usage. J Virol 73:2343–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tinevez JY, Perry N, Schindelin J, Hoopes GM, Reynolds GD, Laplantine E, Bednarek SY, Shorte SL, Eliceiri KW. 2017. TrackMate: an open and extensible platform for single-particle tracking. Methods 115:80–90. doi: 10.1016/j.ymeth.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 64.Burse M, Shi J, Aiken C. 2017. Cyclophilin A potentiates TRIM5alpha inhibition of HIV-1 nuclear import without promoting TRIM5alpha binding to the viral capsid. PLoS One 12:e0182298. doi: 10.1371/journal.pone.0182298. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.