Avian and human influenza A viruses (IAVs) preferentially bind α2,3- and α2,6-linked sialic acids (SIAs), respectively. A functional balance between the hemagglutinin (HA) attachment and neuraminidase (NA) proteins is thought to be important for host tropism. What this balance entails at the molecular level is, however, not well understood. We now show that N1 proteins of both avian and human viruses prefer cleaving avian- over human-type receptors although human viruses were relatively better in cleavage of the human-type receptors. In addition, we show that substitutions at different positions in the second SIA-binding site found in NA proteins of human IAVs have a profound effect on binding and cleavage of multivalent, but not monovalent, receptors and affect virus replication. Our results indicate that the HA-NA balance can be tuned via modification of substrate binding via this site and suggest an important role of the second SIA-binding site in host tropism.
KEYWORDS: second SIA-binding site, influenza A virus, neuraminidase, sialic acid
ABSTRACT
The influenza A virus (IAV) neuraminidase (NA) protein plays an essential role in the release of virus particles from cells and decoy receptors. The NA enzymatic activity presumably needs to match the activity of the IAV hemagglutinin (HA) attachment protein and the host sialic acid (SIA) receptor repertoire. We analyzed the enzymatic activities of N1 NA proteins derived from avian (H5N1) and human (H1N1) IAVs and analyzed the role of the second SIA-binding site, located adjacent to the conserved catalytic site, therein. SIA contact residues in the second SIA-binding site of NA are highly conserved in avian, but not human, IAVs. All N1 proteins preferred cleaving α2,3- over α2,6-linked SIAs even when their corresponding HA proteins displayed a strict preference for α2,6-linked SIAs, indicating that the specificity of the NA protein does not need to fully match that of the corresponding HA protein. NA activity was affected by substitutions in the second SIA-binding site that are observed in avian and human IAVs, at least when multivalent rather than monovalent substrates were used. These mutations included both SIA contact residues and residues that do not directly interact with SIA in all three loops of the second SIA-binding site. Substrate binding via the second SIA-binding site enhanced the catalytic activity of N1. Mutation of the second SIA-binding site was also shown to affect virus replication in vitro. Our results indicate an important role for the N1 second SIA-binding site in binding to and cleavage of multivalent substrates.
IMPORTANCE Avian and human influenza A viruses (IAVs) preferentially bind α2,3- and α2,6-linked sialic acids (SIAs), respectively. A functional balance between the hemagglutinin (HA) attachment and neuraminidase (NA) proteins is thought to be important for host tropism. What this balance entails at the molecular level is, however, not well understood. We now show that N1 proteins of both avian and human viruses prefer cleaving avian- over human-type receptors although human viruses were relatively better in cleavage of the human-type receptors. In addition, we show that substitutions at different positions in the second SIA-binding site found in NA proteins of human IAVs have a profound effect on binding and cleavage of multivalent, but not monovalent, receptors and affect virus replication. Our results indicate that the HA-NA balance can be tuned via modification of substrate binding via this site and suggest an important role of the second SIA-binding site in host tropism.
INTRODUCTION
Influenza A virus (IAV) particles contain a hemagglutinin (HA) protein that binds sialic acid (SIA)-containing receptors and a neuraminidase (NA) protein that cleaves SIA from sialosides. The NA protein is essential for release of virus particles from infected cells and decoy receptors (e.g., in mucus) and for preventing virion aggregation. The NA protein is also an important target of the immune response and antiviral drugs (1–3). The IAV NA protein appears, however, understudied compared to the HA protein.
The IAV NA protein is a tetrameric, type II transmembrane protein, of which nine subtypes are known (N1 to N9). Its globular head domain is linked to the transmembrane domain via a thin stalk. Tetramerization of NA is required for enzymatic activity (4). The head domain contains the active site, which is composed of several highly conserved catalytic residues that contact SIA and structural residues that keep the catalytic residues in place (5, 6). In addition, the NA proteins of some IAVs have been shown to bind sialosides via a 2nd SIA-binding site that consists of three loops and is located next to the catalytic site (5, 7, 8). Binding of substrates via the 2nd SIA-binding site enhances the catalytic activity of NA against these substrates (9, 10) although this has so far been studied only for N2 and N9 proteins. In general, most residues that contact SIA in the 2nd SIA-binding site are highly conserved in avian IAVs but much less so in human IAVs (9, 11).
A functional balance between the IAV HA and NA proteins is thought to be important for efficient replication and transmission (12–14) although it is not clear what this functional balance entails at the molecular level. The optimal balance between the HA and NA proteins is probably adapted to the receptor repertoire of a specific host (13) and needs tuning when IAVs encounter the SIA repertoire of a new host or when IAVs acquire altered HA receptor-binding properties. The HA proteins of avian and human viruses generally prefer binding to α2,3- and α2,6-linked sialosides, respectively. The few studies that addressed the substrate specificity of NA proteins of human and avian viruses generally indicate preferred cleavage by all NAs of α2,3-linked SIAs (15–17). In these studies, however, monovalent soluble synthetic substrates or resialylated red blood cells were used, which are not representative of the multivalent substrates found in mucus or on host cells.
The enzymatic activity of the NA protein may be modified via mutation of the catalytic site residues although these residues are generally highly conserved (5, 6). In addition, residues have been identified that affect the enzymatic activity of NA via long-range interactions (17, 18). The overall NA enzymatic activity can also be adapted via modification of NA expression levels or NA folding and oligomerization (17). Alternatively, the NA enzymatic activity may be decreased by shortening the stalk domain, thereby reducing the substrate accessibility of the NA proteins in the context of virus particles (reviewed in reference 19). More recently, it was shown for N2 and N9 that the enzymatic activity for multivalent substrates may also be adapted by manipulating the binding of these substrates to NA via the 2nd SIA-binding site (9, 10, 20). N2 proteins of human pandemic viruses were shown to display decreased SIA binding and cleavage due to mutation of an SIA contact residue in the 370 loop of the 2nd SIA-binding site (9). For the N9 protein, which preferentially binds α2,3-linked sialosides, it was shown that substitution of a residue lacking SIA contact (here, a noncontact residue) in the 400 loop could also affect binding and cleavage of substrates (10). To what extent other substitutions in the 2nd SIA-binding site and the 2nd SIA-binding site in other NA subtypes affect/contribute to binding and cleavage of multivalent substrates is not known.
N1 proteins have also been shown to bind SIA, presumably via their 2nd SIA-binding site, using saturation transfer difference nuclear magnetic resonance (STD-NMR) (21). N1 proteins of two human viruses displayed much lower affinity than N1 of an avian virus. The functional consequences of these differences in SIA binding are, however, not known. Furthermore, it is not known which substitutions in the 2nd SIA-binding site are responsible for the observed differences. In the present study, we made a comparative analysis of the enzymatic activity of NA proteins derived from highly pathogenic avian IAV H5N1, a new pandemic human IAV H1N1 (H1N1pdm09; also referred to as CA/09 here), and a laboratory-adapted human IAV H1N1 (WSN) by using a recombinant soluble expression approach in combination with different mono- and multivalent substrates. The avian and human N1 proteins were shown to differ in their activities and specificities although all NA proteins preferred cleavage of α2,3-linked SIAs when the glycoprotein fetuin was used as the substrate. The N1 proteins derived from human viruses were, however, relatively better in cleaving α2,6-linked sialosides. Mutations were identified in the 2nd SIA-binding site, involving both SIA contact and noncontact residues in all three loops, that affected activity and specificity of the N1 proteins. By using recombinant viruses, the 2nd SIA-binding site in N1 was also shown to affect virus replication in vitro. Our results indicate that the enzymatic activity and/or specificity of IAV N1 proteins may be modified by mutation of residues at different positions in the 2nd SIA-binding site and that binding of substrates via the 2nd SIA-binding site plays an important role in N1 activity and virus replication.
RESULTS
Previously, we expressed recombinant soluble tetrameric versions of the N1 proteins derived from different highly pathogenic H5N1 viruses (A/duck/Hunan/795/2002, A/Vietnam/1194/04, A/turkey/Turkey/1/2005, and A/Hubei/1/2010, referred to as HN, VN, TK and HB viruses, respectively) and analyzed their enzymatic activities using the monovalent substrate MUNANA [2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid] (22). In the current study, we analyzed the enzymatic activity of these proteins by enzyme-linked lectin assays (ELLAs) (10) using the glycoproteins fetuin and transferrin. Multivalent sialylated glycoproteins better mimic the substrates that NA proteins encounter in vivo than the monovalent MUNANA. Furthermore, the contribution of SIA binding via the 2nd SIA-binding site to NA catalytic activity cannot be observed with MUNANA but only with multivalent substrates (9, 10). Fetuin contains mono-, bi-, and triantennary glycans with α2,3- and α2,6-linked SIAs in a 2:1 ratio (23), while transferrin contains two biantennary N-linked glycan chains with only α2,6-linked SIAs (24, 25). Cleavage of SIAs from fetuin and transferrin by serially diluted NA proteins was quantified by analyzing the binding of lectins with different binding specificities. Erythrina crista-galli lectin (ECA) specifically recognizes glycans containing terminal Galβ1-4GlcNAc, which generally correspond to desialylated N-linked sugars (26), while peanut agglutinin (PNA) binds to terminal Galβ1-3GalNAc corresponding with desialylated O-linked glycans (27). Maackia amunrensis lectin 1 (MALI) and Sambucus nigra lectin (SNA) specifically bind α2,3- and α2,6-linked SIAs, respectively (28, 29).
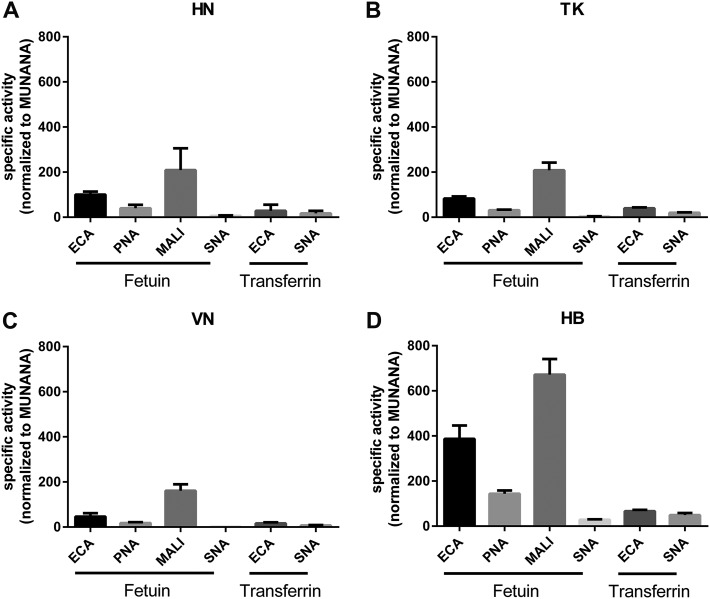
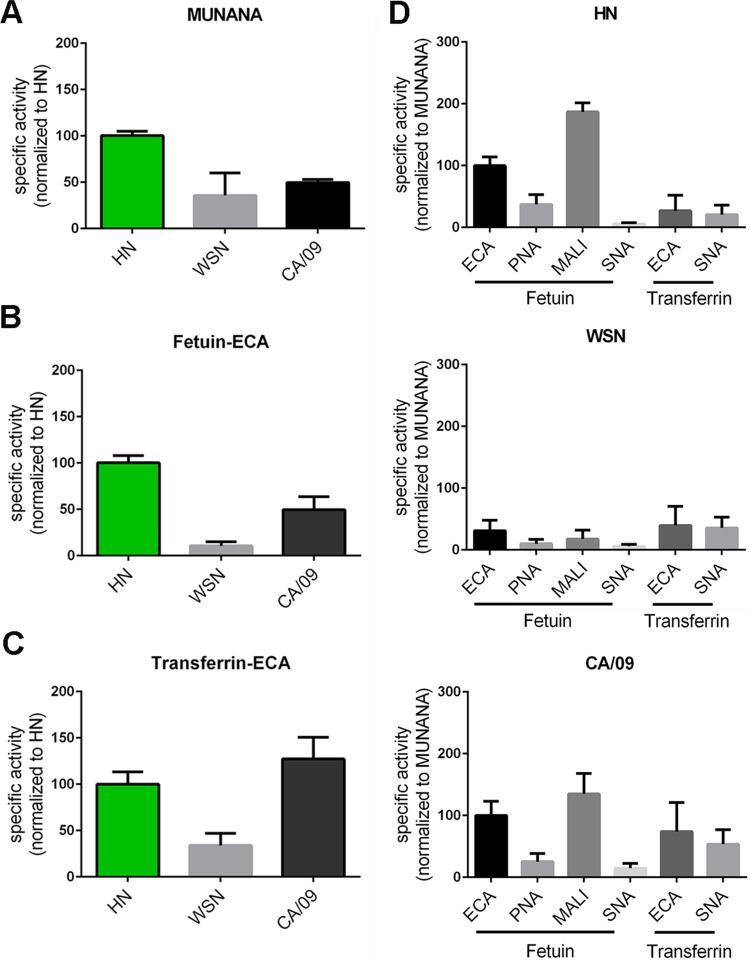
Analysis of the relative specific activities of the different proteins when the MUNANA substrate was used indicated that the HN, TK, and HB proteins cleave this substrate 2- to 3-fold more efficiently than the VN protein (Fig. 1A), in agreement with previous results (22). A remarkably different result was obtained when the relative specific activity was determined with fetuin using ECA staining (Fig. 1B). While the specific activities of the HN, VN, and TK proteins reflected the specific activities as determined with MUNANA, the specific activity of the HB protein was much higher with this substrate. This increase was smaller with transferrin, which contains only α2,6-linked sialosides (Fig. 1C), while similarly increased specific activities for the HB protein were obtained when other lectins in combination with fetuin were used (Fig. 2). Graphing the specific activities of each NA protein determined for each glycoprotein-lectin combination normalized to its MUNANA specific activity showed that all proteins preferred cleaving α2,3-linked (determined with fetuin-MALI) over α2,6-linked (determined with fetuin-SNA) SIAs (Fig. 2). In agreement with this finding, the specific activities were higher when determined with fetuin-ECA than with transferrin-ECA. The HB protein displayed the largest preference for the cleavage of α2,3- over α2,6-linked SIAs, in agreement with the much higher increase in the specific activity of this protein when fetuin-ECA was used than when transferrin-ECA was used (Fig. 1B and C and 2).
FIG 1.
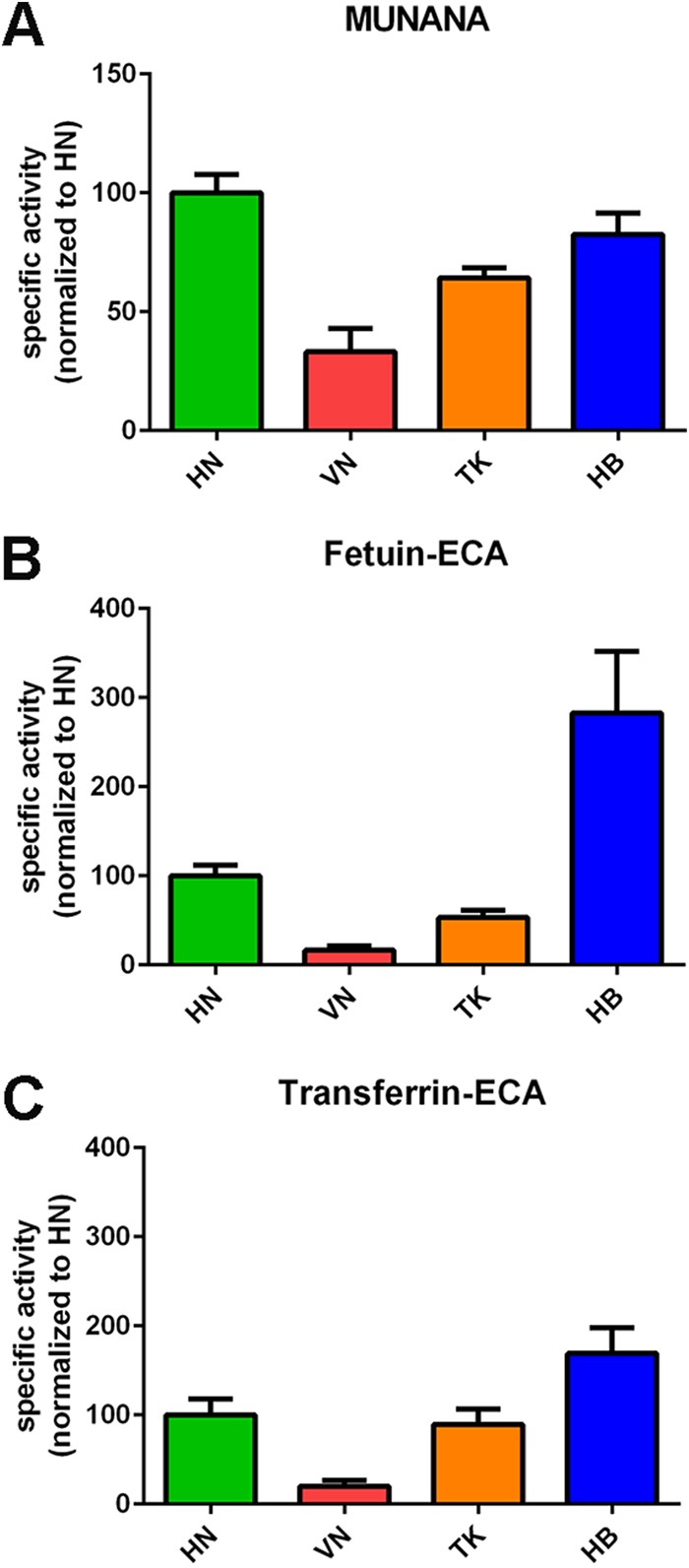
Specific activity of H5N1 NA proteins using monovalent and multivalent substrates. (A) Specific activities of indicated H5N1 NA proteins using the substrate MUNANA are graphed normalized to the specific activity of NA HN. (B and C) Specific activities of the indicated NA proteins were determined by ELLA using different glycoprotein-lectin combinations, as indicated, and graphed normalized to the activity of NA HN. Means of three independent experiments performed in triplicate are shown. Standard deviations are indicated.
FIG 2.
Specificity of H5N1 NA proteins. The specific activities of the indicated H5N1 NA proteins were determined by ELLA using different glycoprotein-lectin combinations and graphed normalized to the specific activity determined with MUNANA. Means of three independent experiments performed in triplicate are shown. Standard deviations are indicated.
Increased cleavage of sialosides present on fetuin, but not of MUNANA, by HB is indicative of increased binding of this NA protein to fetuin via its 2nd SIA-binding site. Sequence analysis of the H5N1 NA proteins showed that the HB protein differs from the others by the mutation N369H in the 370 loop of the 2nd SIA-binding site (Table 1 and Fig. 3). Although this substitution does not involve an SIA contact residue according to available crystal structures (7, 8) (Fig. 4A and B), we analyzed whether the observed increased cleavage of fetuin by the HB protein could be attributed to this mutation. To this end, we introduced the N369H mutation in the HN protein. As expected, this mutation did not appreciably affect the ability of the resulting protein (HN-N369H) to cleave MUNANA or transferrin (Fig. 5A and C). In contrast, this mutation resulted in a higher specific activity, which was similar to that of the HB protein, when fetuin was used as the substrate (Fig. 5B).
TABLE 1.
Sequence alignment of 2nd SIA-binding site of N1 proteins used in this study
| N1 protein | 2nd SIA-binding site sequencea |
||
|---|---|---|---|
| 370 loop | 400 loop | 430 loop | |
| Avian IAV | |||
| HN/TK/VN | KSTNSRSG | AITDWS | PKE |
| HB | KSTHSRSG | AITDWS | PKE |
| Human IAV | |||
| WSN | KSDSSRHG | AMTDRS | PEE |
| CA/09 | KSISSRNG | GINEWS | PKE |
| HN mutants | |||
| N369H | KSTHSRSG | AITDWS | PKE |
| TN-IS | KSISSRSG | AITDWS | PKE |
| TN-DS | KSDSSRSG | AITDWS | PKE |
| S372H | KSTNSRHG | AITDWS | PKE |
| S372N | KSTNSRNG | AITDWS | PKE |
| I400 M | KSTNSRSG | AMTDWS | PKE |
| W403R | KSTNSRSG | AITDRS | PKE |
| HN K432E | KSTNSRSG | AITDWS | PEE |
| Other | KSTNSRSG | AITDWS | PKE |
Residues corresponding to the SIA contact residues in the N9 protein (Fig. 4) (S367, S370, S372, I400, W403, and K432) are underlined. Residues that differ between the avian HN and HB, WSN, and CA/09 proteins in the 2nd SIA-binding site are in boldface. Substitutions in the HN mutants are also in boldface.
FIG 3.
Alignment of N1 proteins. Alignment of several N1 proteins analyzed in this study is shown. The start of the N1 protein ectodomain expressed as a recombinant soluble fusion protein is indicated. Catalytic and framework residues in the active site are shown in red and yellow, respectively (5, 6). SIA contact residues in the 2nd SIA-binding site, based on the N9 crystal structure (7), are shown in light blue. Residues introduced in HN N1 are shown in purple. N2 numbering is indicated.
FIG 4.
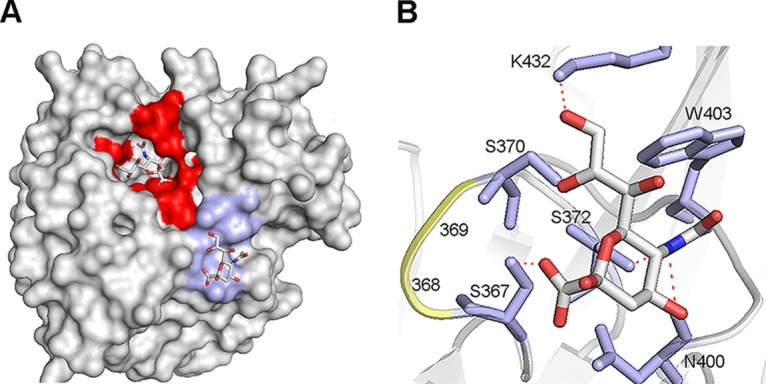
Structure of the N9 2nd SIA-binding site. (A and B) Crystal structure of N9 from A/tern/Australia/G70C/75 in complex with SIA (N-acetylneuraminic acid) (PDB 1MWE) (7). (A) Surface representation. The NA active site and the 2nd SIA-binding site (SIA contact residues) are shown in red and light blue, respectively. The SIA moieties in these sites are shown as sticks (oxygen in red, nitrogen in blue, and carbon in gray). (B) Structure of the 2nd SIA-binding site. SIA is shown as sticks (oxygen in red, nitrogen in blue, and carbon in gray). Residues in the 2nd SIA-binding site that directly contact SIA (S367, S370, S372, N400, W403, and K432) are shown in stick representation (light blue), amino acids at positions 368 and 369 that differ between HN and WSN and CA/09 are shown in cartoon representation (yellow). Hydrogen bonds between SIA and residues in the 2nd SIA-binding site are shown as dashed red lines. Figures were made using PyMOL.
FIG 5.
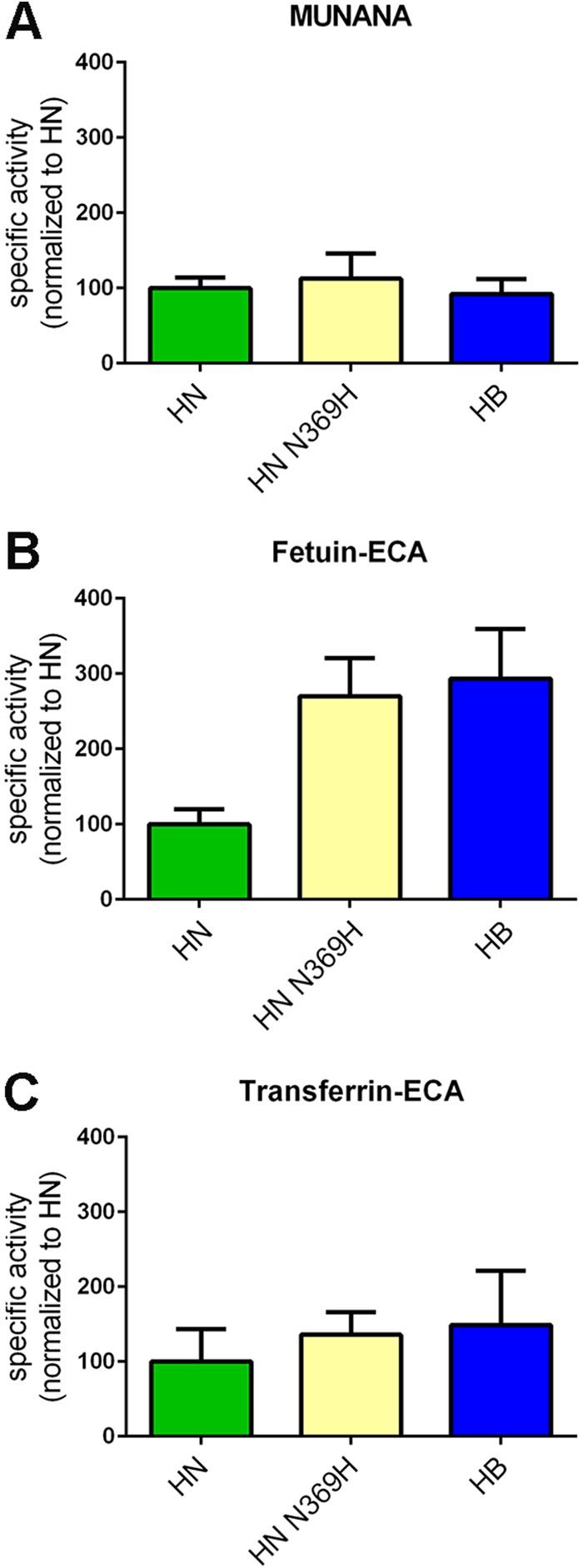
Substitution N369H affects H5N1 NA enzymatic activity. Specific activities of the indicated (mutant) H5N1 NA proteins determined by MUNANA assay (A) or by ELLA using fetuin (B) or transferrin (C) in combination with lectin ECA were graphed normalized to the activity of N1 HN. Means of two independent experiments performed in triplicate are shown. Standard deviations are indicated.
These results may be explained by the mutant HN and the HB proteins displaying increased binding to fetuin via their 2nd SIA-binding sites. We were not able, however, to demonstrate binding of the N1 proteins to fetuin in a solid-phase binding assay or to chicken or human erythrocytes in a hemagglutination assay when the N1 proteins were complexed using antibodies as similarly described previously (10). The N1 proteins were also negative in the hemagglutination assays when the antibody-NA complexes were linked to lumazine synthase nanoparticles (Fig. 6A). In this assay, which is more sensitive, the N9 protein of A/Anhui01/2013 (H7N9) was shown to be hemagglutination positive although it was previously found to be binding negative (10). As an alternative assay, we expressed full-length, transmembrane domain-containing HB and HN NA proteins and generated membrane vesicles (30). Previously, we showed that these full-length NA proteins display similar expression levels and have the same specific activities for MUNANA (22). The membrane vesicles were analyzed in a biolayer interferometry binding experiment using similar amounts of NA protein based on MUNANA activity (22) (Fig. 6B). Streptavidin sensors were coated with biotinylated synthetic glycans containing either α2,3- or α2,6-linked sialoglycans (3′-SLNLNLN or 6′-SLNLNLN, respectively). In the absence of the NA inhibitor oseltamivir carboxylate (OsC), no binding was observed. In the presence of OsC, which occupies the NA active site, HB-containing but not HN-containing vesicles bound to sensors coated with 3′-SLNLNLN but not with 6′-SLNLNLN. Also, the HN-N369H protein-containing vesicles displayed binding specifically to 3′-SLNLNLN, thereby confirming the importance of residue 369 for binding via the 2nd SIA-binding site.
FIG 6.
Substrate binding of NA proteins via the 2nd SIA-binding site. (A). HN and HB NA proteins and N9 proteins with different binding properties (10) were complexed to lumazine synthase nanoparticles displaying domain B of protein A using anti-Strep-tag monoclonal antibodies. Limiting dilutions of these complexes were incubated with red blood cells in the presence of OsC, and hemagglutination titers were determined. Standard deviations are indicated. (B and C) Membrane vesicles containing full-length HN, HN N369H, or HB protein were analyzed for their ability to bind 3′-SLNLNLN or 6′-SLNLNLN in the presence of OsC using biolayer interferometry. Representative experiments are shown.
SIA contact residues in the 2nd SIA-binding site are highly conserved in N2 proteins of avian IAVs but much less so in human IAVs (9, 11). Also for N1, the conservation of the 2nd SIA-binding site appears lost in human IAVs (9, 11, 14) although a detailed analysis of the conservation of the 2nd SIA-binding site of avian and human viruses has not been performed. Therefore, we studied the conservation of the three loops of the 2nd SIA-binding site of N1 by generating sequence logos based on multiple sequence alignments using N1 sequences of avian and human viruses available via the Influenza Research Database (https://www.fludb.org/). While the SIA contact residues are highly conserved in the N1 protein of avian viruses, it is clear that this conservation is lost for both the seasonal and the H1N1pdm09 viruses (Fig. 7). Both for the seasonal and the H1N1pdm09 viruses, the contact residue at position 372 is essentially invariably mutated. Interestingly, the residue at position 369 that is mutated in the HB protein compared to sequence of the HN protein is much less conserved than all other residues in the loops that constitute the 2nd SIA-binding site in N1 of avian viruses.
FIG 7.
Sequence logos of the 2nd SIA-binding site of avian and human N1 proteins. Sequence logos were generated for the three loops that constitute the 2nd SIA-binding site using the WebLogo website (http://weblogo.berkeley.edu/) (40) using all sequences available of avian N1 viruses, human N1 viruses prior to 2009 (seasonal human N1), and human H1N1pdm09 viruses from 2009 to 2014 (pandemic human N1) via the Influenza Research Database (https://www.fludb.org/). Asterisks indicate SIA contact residues according to the N9 crystal structure (7).
As a next step, we compared the enzymatic activity and specificity of the HN N1 protein with the those of the N1 proteins of the H1N1pdm09 virus (referred to as CA/09) and the human (laboratory-adapted) H1N1 WSN virus (referred to as WSN) (Fig. 8A). The CA/09 and the WSN proteins displayed 2- to 3-fold lower specific activity than that of the HN protein when MUNANA was used. A similar (CA/09) or even larger (WSN) decrease in specific activity was observed with fetuin-ECA (Fig. 8B). The specific activities of CA/09 and WSN NA proteins were relatively higher with transferrin as the substrate (Fig. 8C), with the CA/09 protein even displaying specific activity similar to that of the HN protein. When the different specific activities are graphed relative to the MUNANA specific activity (Fig. 8D), it is clear that particularly the WSN protein displayed a decreased ability to cleave α2,3-linked SIAs (determined with fetuin-MALI). The CA/09 protein was better in cleaving α2,6-linked SIAs than the HN protein, which was most apparent when transferrin was used as the substrate.
FIG 8.
Enzymatic activity of HN, WSN, and CA/09 NA proteins using monovalent and multivalent substrates. Specific activities of indicated N1 proteins determined by MUNANA assay (A) or by ELLA using fetuin (B) and transferrin (C) in combination with lectin ECA were graphed normalized to the activity of N1 HN. (D) Substrate specificities of the indicated N1 proteins were determined by ELLA using different glycoprotein-lectin combinations and graphed normalized to the specific activity determined with MUNANA. Means of 2 to 3 independent experiments performed in triplicate are shown for the MUNANA and ELLA assays. Standard deviations are indicated.
Subsequently, we analyzed to what extent the differences in the enzymatic activities between the different NA proteins could be attributed to changes in the 2nd SIA-binding site. In the 370 loop, HN NA differs from WSN NA at positions 368/369 (TN versus DS), while again other residues (IS) are found in the CA/09 protein at these positions (Table 1 and Fig. 3). In addition, S372, which unlike residues at positions 368/369 is a SIA contact residue (Fig. 4), is replaced with H in the WSN and with N in CA/09 protein. In the 400 and 430 loops, positions that are SIA contact residues in N9 (Fig. 2) are substituted. In the 400 loop, residues I400 and W403 of HN are replaced in WSN NA with M and R, respectively. In the 430 loop, the K432E substitution is observed in WSN NA.
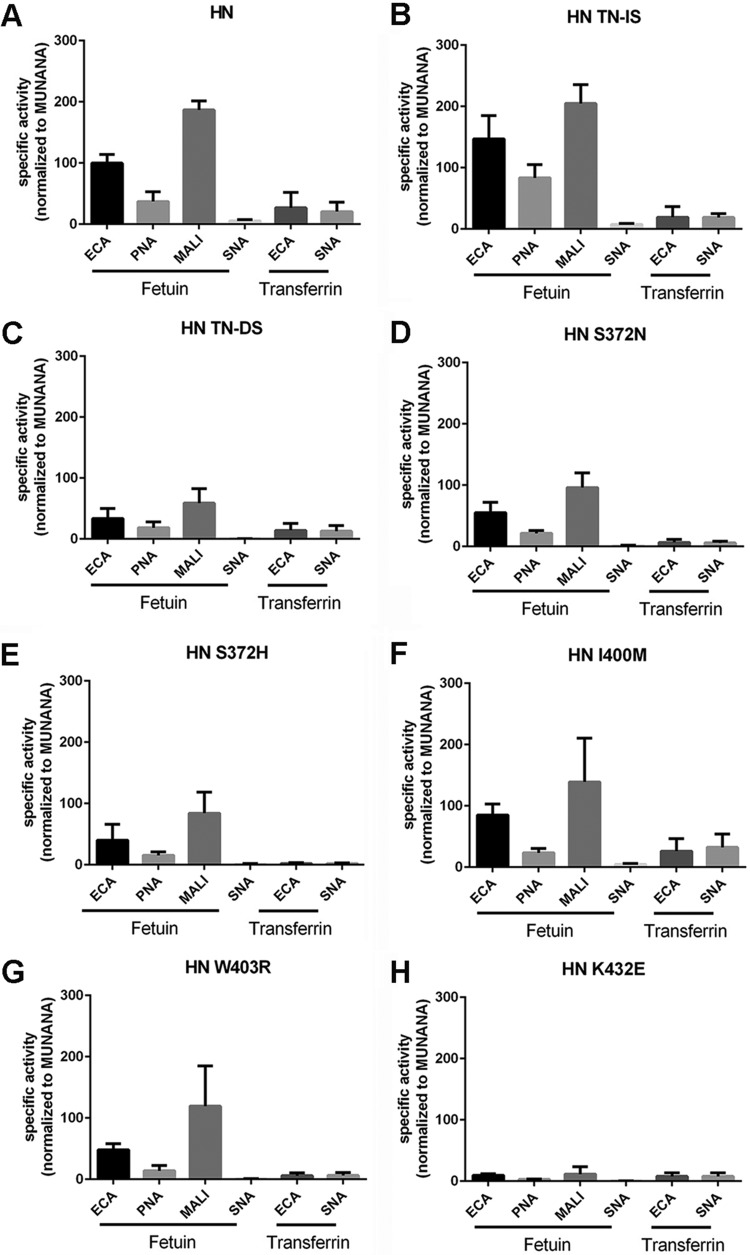
All of the substitutions mentioned above were introduced in the background of the HN protein, and the enzymatic activities of the resulting proteins were investigated using MUNANA, fetuin, and transferrin as substrates. Mutation of the TN residues at position 368 and 369 in the 370 loop to IS or DS had only a limited effect on the cleavage of MUNANA (Fig. 9A). While the TN-to-IS substitution had a small positive effect on the cleavage of both fetuin and transferrin, the TN-to-DS substitution had a large negative effect on the cleavage of sialosides linked to fetuin. This negative effect was smaller when transferrin was used (Fig. 9B and C). As a result, the HN TN-DS protein was relatively better in cleaving α2,6-linked SIAs linked to transferrin (transferrin-ECA/SNA) than α2,3-linked SIAs linked to fetuin (fetuin-ECA/MALI) compared to the wild-type HN and the HN TN-IS proteins (Fig. 10A to C). These results indicate that, similar to the N369H mutation, substitutions of residues in the 370 loop of the 2nd SIA-binding site not directly contacting SIA can affect cleavage of glycoprotein-linked sialosides. Most likely, this is not explained by these mutations affecting cleavage per se (as determined with MUNANA) but, rather, by their effect on the interaction of the NA protein with multivalent substrates via the 2nd SIA-binding site. Mutation of the SIA contact residue at position 372 into N or H (found in the CA/09 or WSN N1 protein, respectively) had no significant negative effects on the cleavage of MUNANA (Fig. 9A), while cleavage of both glycoproteins was consistently negatively affected (Fig. 9B and C), with the S372H mutation, found in WSN NA, having the largest negative effect.
FIG 9.
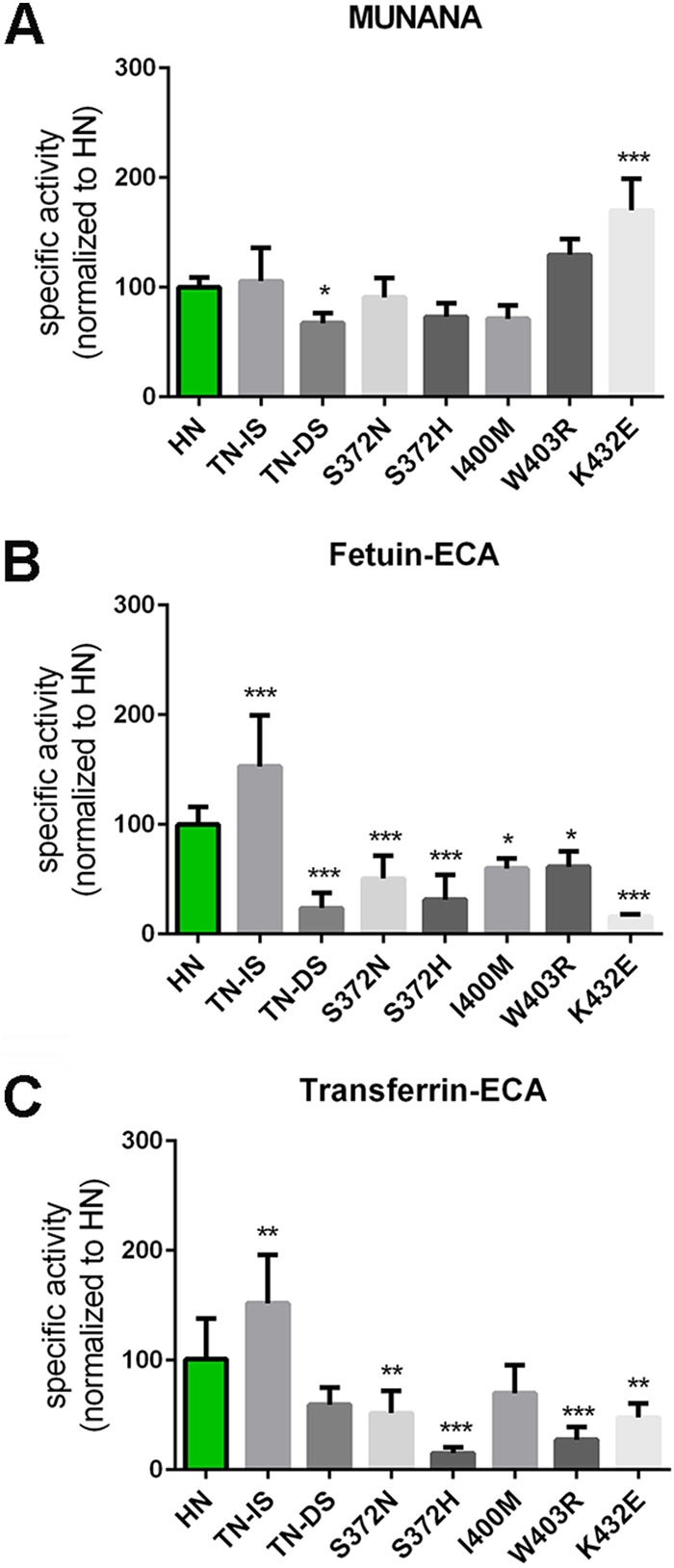
Effect of substitutions in the 2nd SIA-binding site on NA enzymatic activity. Specific activities of wild-type and mutant HN determined by MUNANA assay (A) or by ELLA using fetuin (B) and transferrin (C) in combination with lectin ECA were graphed normalized to that of wild-type HN. Means of 2 to 3 independent experiments performed in triplicate are shown. Standard deviations are indicated. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
FIG 10.
Substrate specificity of mutant HN NA proteins. Substrate specificities of the indicated mutant HN proteins (same as those shown in Fig. 5) were determined by ELLA using different glycoprotein-lectin combinations and graphed normalized to the specific activity determined with MUNANA. Means of 2 to 3 independent experiments performed in triplicate are shown. Standard deviations are indicated.
Substitution of residues in the 400 loop either had a small negative effect on cleavage of all substrates including MUNANA (I400M) or specifically had a negative effect on the cleavage of sialosides linked to fetuin or transferrin (W403R) (Fig. 9A and B). While an N at position 400 was also shown to be an SIA contact residue in the N9 protein (Fig. 4), this interaction may already be affected by the presence of an I in the HN protein, thereby explaining why the I400M mutation did not result in a specific negative effect on the cleavage of glycoprotein-linked glycans. The negative effect of the substitution at position 403 is in agreement with the W being an SIA contact residue (Fig. 4). Interestingly, the negative effect of this substitution on the cleavage of transferrin-linked sialosides appeared larger than the effect on cleavage of fetuin-linked sialosides (Fig. 9 and 10), suggesting that N1 proteins may also bind to some extent to α2,6-linked SIAs and that mutations in the 2nd SIA-binding site may differentially affect binding to α2,3- or α2,6-linked SIAs. The largest negative effect on the cleavage of fetuin-linked sialosides was observed when the SIA contact residue in the 430 loop (i.e., K432) was replaced by E, as observed in WSN NA (Fig. 9B). This substitution, which even resulted in increased cleavage of MUNANA (Fig. 9A), also decreased cleavage of transferrin-linked glycans (Fig. 9C) but to a smaller extent than the cleavage of fetuin-linked sialosides. In agreement with this observation, the HN K432E NA protein displayed a much less pronounced preference for the cleavage of fetuin- over transferrin-linked sialosides (Fig. 10H).
The CA/09 and the WSN proteins displayed much lower expression levels than the N1 proteins of the H5N1 viruses. For unknown reasons, these expression levels were reduced even more by the introduction of reciprocal mutations in the 2nd SIA-binding site of these proteins, which precluded further analysis in the solid-phase cleavage assays. To determine the specificity of the observed effects of the substitutions in or close to the 2nd SIA-binding site, several other mutations found in WSN NA compared to the sequence of the HN protein at positions that are also substituted in the CA/09 protein (Fig. 3) were introduced in the HN protein, and the specific activities of the resulting proteins were determined using MUNANA as well as fetuin and transferrin. With the exception of the H155Y and the S434D mutations, these single and double substitutions (V99I, TS186/187MG, M257I, A284T, Y347N, and S389R; N2 numbering) did not appreciably affect the specific activities of the mutant HN proteins regardless of the substrates used (Fig. 11). The H155Y substitution, located next to a framework residue (Fig. 3), resulted in reduced specific activity for all three substrates (Fig. 11A to C). The S434D mutation, which is located two residues downstream of the K432 residue (Fig. 3), reduced cleavage of sialosides linked to fetuin and transferrin (Fig. 11B and C), while the specific activity against MUNANA was not significantly affected (Fig. 11A). The S434D mutation may very well affect the conformation of the 430 loop and therefore substrate binding and cleavage.
FIG 11.
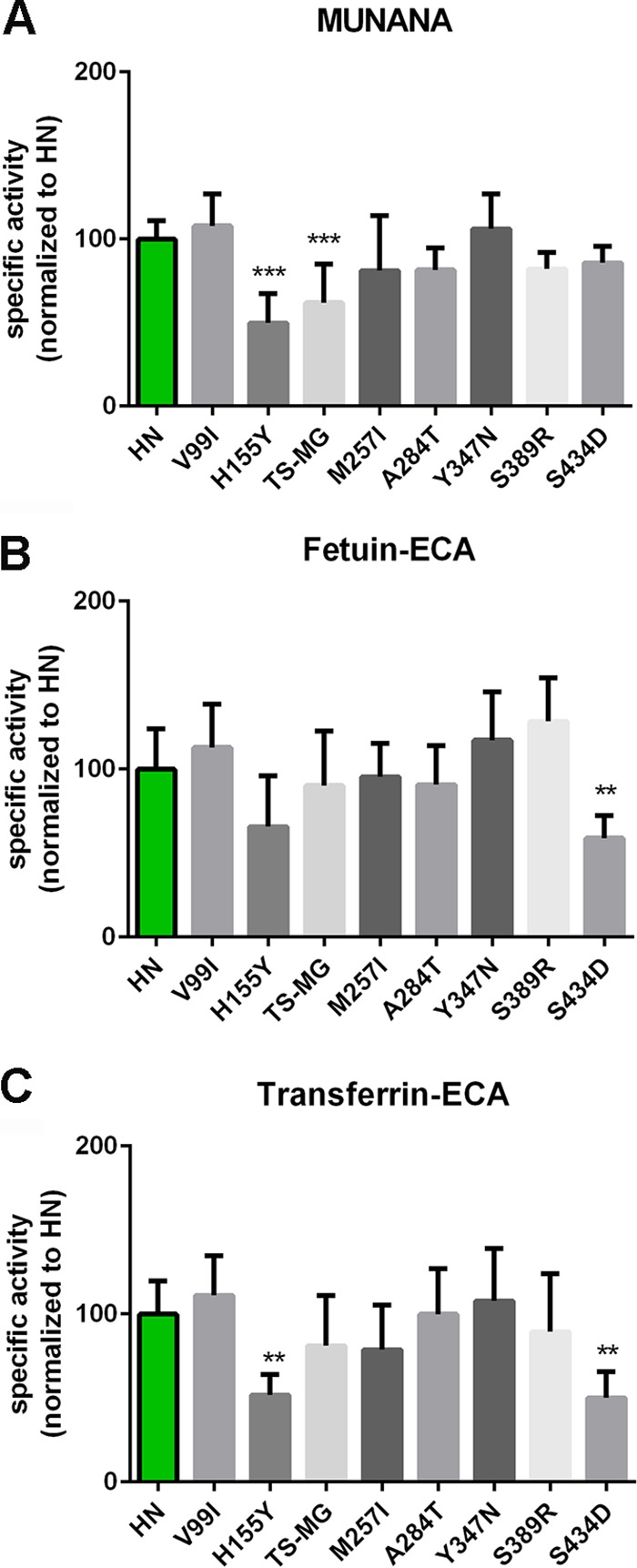
Effect of substitutions outside the 2nd SIA-binding site on NA enzymatic activity. Specific activities of wild-type and mutant HN determined by MUNANA assay (A) or by ELLA using fetuin (B) or transferrin (C) in combination with lectin ECA were graphed normalized to the activity of wild-type HN. Means of at least two independent experiments performed in triplicate are shown. Standard deviations are indicated. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
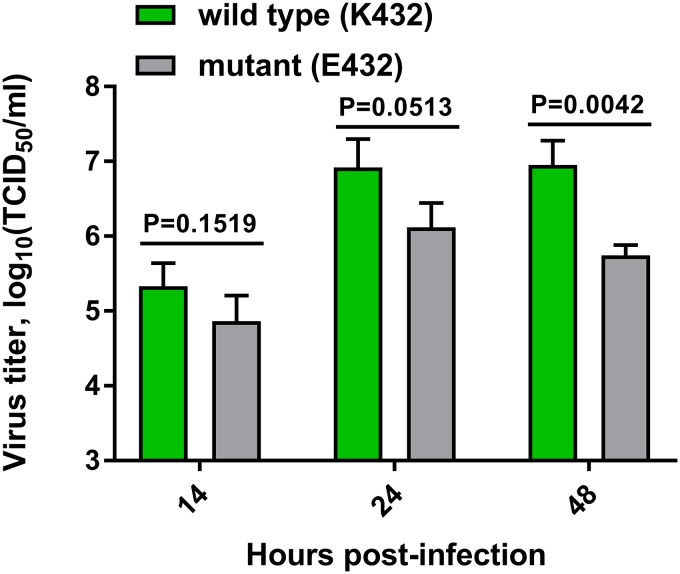
In order to analyze the importance of the 2nd SIA-binding site for replication in vitro, recombinant H5N1 viruses were generated in the background of PR8 virus with and without a functional 2nd SIA-binding site. These viruses carried the HN HA and NA genes. Plasmids used to generate viruses with the wild-type and mutant N1 genes contained the AAA (K432) and GAG (E432) codons, respectively. The K432E substitution had a large negative effect on the cleavage of fetuin- and transferrin-linked sialosides but not on MUNANA (Fig. 9). The wild-type H5N1 virus was readily generated, and its identity was confirmed by sequence analysis of the HA and NA genes. For the mutant H5N1 virus, however, already after one passage a large proportion contained the AAG codon encoding K432, as determined by sequence analysis of cloned PCR products. While the rapid reversion of the H5N1 mutant virus precluded detailed analysis of the in vitro growth kinetics, it indicates that the presence of a functional 2nd SIA-binding site in N1 is beneficial for in vitro growth. We also generated recombinant PR8 viruses carrying the wild type (K432) and mutant (E432) HN NA genes combined with the PR8 HA gene. The identity of these viruses was confirmed by sequence analysis. The virus carrying the wild-type HN protein grew to significantly higher titers than the one with the mutant HN protein (Fig. 12), indicating that when combined with PR8 HA, the 2nd SIA-binding site in N1 also affects growth in vitro.
FIG 12.
Replication of viruses with and without a 2nd SIA-binding site. MDCK-II cells were infected a multiplicity of infection of 0.001 with PR8 (7+1) viruses carrying wild-type (K432) or mutant (E432) HN NA proteins. Virus in the supernatant at the indicated time points postinfection was titrated. Means of three independent experiments are graphed. Standard deviations are indicated. Significant differences were analyzed using a Student t test, and the P values are shown. TCID50, 50% tissue culture infective dose.
DISCUSSION
In general, avian and human IAVs, including the H5N1 and H1N1 viruses analyzed in this study (31–33), prefer binding to α2,3- and α2,6-linked sialosides, respectively. Nevertheless, all corresponding NA proteins preferentially cleaved α2,3-linked SIAs when fetuin, containing both types of sialosides, was used as the substrate. The human H1N1 viruses were relatively better, however, in cleaving α2,6-linked SIAs than the avian H5N1 viruses, which was also apparent when transferrin, which contains only α2,6-linked SIAs, was used. Assuming that IAVs evolve an optimal HA-NA balance for their replication, these results indicate that such a balance does not require the NA proteins to fully mimic the specificity of the corresponding HA protein. Our results furthermore indicate that the enzymatic activity of IAV N1 is considerably affected by the 2nd SIA-binding site. Most substitutions in this site had only a very limited effect on NA activity per se, as measured with the monovalent MUNANA substrate. However, the effects were much larger when the N1 proteins were analyzed with solid-phase cleavage assays using multivalent glycoproteins, which better mimic the substrates found in mucus or on the cell surface. Our results indicate that binding of N1 to glycoprotein substrates is an important determinant of NA enzymatic activity, and as this feature is strictly conserved in avian, but not human, viruses and was shown to affect virus replication in vitro, the 2nd SIA-binding site is likely to play an important role in host tropism.
Different mutations in the 2nd SIA-binding site were shown to have different effects on NA cleavage activity and specificity. While some mutations particularly affected cleavage of fetuin-linked glycans (N369H and TN368/369DS in the 370 loop and K432E in the 430 loop), other mutations appeared to similarly affect cleavage of fetuin- and transferrin-linked sialosides (S372N/H and S434D) or even to have a larger effect on the cleavage of sialosides linked to transferrin (W403R). Furthermore, NA activity could be modified not only by mutation of SIA contact residues in the 370 (9; this study), 400 (9; this study), and 430 (this study) loops but also by substitution of noncontact residues in the 370 loop (this study), the 400 loop (10), and the 430 loop (this study). Surprisingly, substitution of noncontact residues in the 370 loop (position 368/369) resulted in even larger effects than mutation of a contact residue (position 372) in this loop. Possibly, these noncontact residues interact with the penultimate galactose in the glycan chain. The most dramatic effects, however, were observed after substitution of the K432 residue in the 430 loop. Strikingly, while the residue at position 432 appears to be very important for binding and enzymatic activity of N1, for other NA subtypes this residue is less conserved than the other SIA contact residues in the 2nd SIA-binding site of the NA proteins of avian IAVs. This finding may be related to the apparent lack of hydrogen bond interaction between the residue at position 432 and SIA in N2, N5, and N6 (8). Collectively, these results demonstrate the large arsenal available to IAVs to modulate the activity of their NA proteins via the 2nd SIA-binding site, which thus far has been an underappreciated feature.
Binding via the 2nd SIA-binding site enhances the catalytic activity of N1. The higher specific activity of the HB protein for fetuin than that of the HN protein can be attributed to a single mutation in the 2nd SIA-binding site (N369H), which specifically enhances binding to α2,3-linked sialosides (Fig. 6). In agreement with this observation, this mutation has only a marginal effect on the cleavage of sialosides linked to transferrin, which contains only α2,6-linked SIAs (Fig. 5). Other mutations in the 2nd SIA-binding site, however, were shown to significantly affect cleavage of sialosides linked to transferrin (e.g., S372H or W403R) (Fig. 8), suggesting that the HN N1 protein is also able to bind to some extent to α2,6-linked sialosides. The low receptor-binding avidity of the N1 proteins via their 2nd SIA-binding site, however, precluded detailed analysis of the N1 receptor-binding preference. Previously, we showed that N9 proteins display appreciable binding only to α2,3-linked sialosides via their 2nd SIA-binding site using glycan array analysis (10). In contrast, an N2 protein with a functional 2nd SIA-binding site was shown to hemadsorb erythrocytes containing either type of sialoside (9). To determine whether the receptor specificity of the 2nd SIA-binding site of different NA subtypes differs, a head-to-head comparison and more sensitive binding assays are needed. It is clear, however, that NA proteins of different subtypes may differ remarkably in their receptor binding avidities, as demonstrated by the differences observed in the hemagglutination assay for the N9 and N1 proteins (Fig. 6B). Whether N1 proteins generally display a lower avidity than N9 proteins remains to be determined.
Both the H1N1pdm09 and the WSN viruses contain several mutations in the 2nd SIA-binding site that negatively affect cleavage of specifically multivalent substrates. Substitutions in the 2nd SIA-binding site, which are observed in all pandemic human viruses (9, 11), may be selected as they tune the balance between the (novel) receptor repertoire of humans, the (altered) receptor-binding properties of HA, and the enzymatic activity of NA. Alternatively, these mutations may be selected as a result of immune pressure on the surface-exposed 2nd SIA-binding site. The selective advantage of being able to bind to avian-type receptors via the 2nd SIA-binding site may be lost for human viruses carrying HA proteins that bind only to human-type receptors. The high conservation of the SIA contact residues in the 2nd SIA-binding site of NA proteins of avian viruses suggests an important role for this binding site for virus replication and/or for transmission of viruses carrying HA proteins preferring binding to avian-type receptors. Indeed, recombinant H5N1 virus carrying a mutated 2nd SIA-binding site, as a result of a K432E substitution, rapidly reverted to a functional 2nd SIA-binding site through back mutation of the residue at position 432. The rapid reversion of this virus precluded detailed growth analysis, but also in combination with PR8 HA, which displays dual specificity for both human and avian receptors (34, 35), mutation of the 2nd SIA-binding site significantly affected in vitro growth. The importance of the 2nd SIA-binding site for replication and transmission in vivo remains to be established. Mutation of the 2nd SIA-binding site in an N2 protein, which reduced NA hemadsorption activity, negatively affected replication of a recombinant virus in chicken embryo fibroblasts but, strikingly, not in ducks compared to the level of the control virus (11). Of note, in these recombinant viruses, the NA proteins were combined with noncognate HA proteins. Clearly, more research on this topic is needed to irrefutably demonstrate the importance of the 2nd SIA-binding site for virus replication and transmission in vivo.
MATERIALS AND METHODS
Protein expression and purification.
Expression plasmids encoding the NA ectodomain (head plus stalk domain) of A/duck/Hunan/795/2002 (H5N1) (GenBank accession no. BAM85820.1; referred to as N1 HN), A/Vietnam/1194/04 (H5N1) (GenBank accession no. AAT73327; referred to as N1 VN), A/turkey/Turkey/1/2005 (H5N1) (GenBank accession no. ABQ58915.1; referred to as N1 TK), A/Hubei/1/2010 (H5N1) (GenBank accession no. AEO89183.1; referred to as N1 HB), A/Anhui/1/2013 (H7N9) (Global Initiative on Sharing All Influenza Data [GISAID] isolate EPI439507; referred to as N9 Anhui), and of A/Anas crecca/Spain/1460/2008 (H7N9) (GenBank accession no. HQ244409.1; referred to as N9 Spain) fused to a Staphylothermus marinus tetrabrachion tetramerization domain and a double Strep-tag have been described previously (10, 22). Similar expression plasmids were constructed for the N1 proteins of A/WSN/1933 (H1N1) (GenBank accession no. AAA91328.1; referred to as N1 WSN) and A/California/04/2009 (H1N1) (GenBank accession no. ACP41107.1; referred to as N1 CA/09). Mutations of interest were introduced into the corresponding NA genes by using a Q5 site-directed mutagenesis kit (New England BioLabs) and confirmed by sequencing. NA proteins were expressed by transfection of HEK293T cells (ATCC) with NA gene-containing plasmids, and the proteins were purified from the cell culture supernatants as similarly described previously (36). Quantification of the purified proteins was performed by comparative Coomassie gel staining using standard bovine serum albumin (BSA) samples (Sigma-Aldrich) with known concentrations as a reference.
Enzyme activity assays.
The activity of N1 proteins toward the synthetic monovalent substrate MUNANA (Sigma-Aldrich) was determined by using a fluorometric assay as similarly described previously (22). The activities of the NA proteins toward multivalent glycoprotein substrates fetuin and transferrin were analyzed by ELLA as similarly described previously (10). While NA activity results in increased binding of the ECA and PNA lectins, which bind to desialylated glycans, decreased binding of MAL I and SNA is observed after NA cleavage as these two lectins bind to sialylated glycans. Lectin binding and MUNANA cleavage were measured after incubation of the substrates with limiting dilutions of NA. The amount of NA protein corresponding to half-maximum lectin binding or MUNANA cleavage was determined from the resulting curves by fitting the data by nonlinear regression analysis using Prism, version 6.05, software (GraphPad) as described previously (10, 22). The reciprocal of this amount is a measure for the relative specific activity (activity per amount of protein).
Hemagglutination assay.
The SIA-binding ability of NA proteins was analyzed using hemagglutination assays. Purified NA protein (4 μg) was precomplexed with anti-Strep-tag mouse antibody (2 μg) (IBA) and with lumazine synthase nanoparticles genetically fused to domain B of protein A (0.25 μg) (37) on ice for half an hour prior to incubation of limiting dilutions of these complexes with 0.5% human erythrocytes for 2 h at 4°C in the presence of 5 μM zanamivir (GlaxoSmithKline). The hemagglutination assays were performed twice in duplicate. The mean values of these experiments are graphed.
Biolayer interferometry.
HEK293T cells were transfected with expression plasmids encoding full-length N1 HN and HB proteins (22) as described above. At 72 h posttransfection, cells were vesiculated as similarly described previously (30), and vesicle preparations were purified using Capto Core 700 beads (GE Healthcare Life Sciences) according to the manufacturer's instructions. NA activity in the purified vesicle preparations was determined using the MUNANA assay described above. Similar amounts of NA activity, and thus NA protein (22), were applied in the Biolayer interferometry assays using an Octet RED384 instrument (Fortebio) with streptavidin biosensors loaded to saturation with biotinylated 3′-SLNLNLN or 6′-SLNLNLN. Association of the NA-containing vesicles was analyzed for 45 min in the absence or presence of 10 μM OsC (Roche).
Recombinant viruses.
Recombinant IAV containing the HN NA gene [from A/duck/Hunan/795/2002 (H5N1)] combined with either its cognate HA gene (GenBank accession number CY028963; 6+2 virus) or with the HA gene of strain A/Puerto Rico/8/34 H1N1 (PR8; 7+1 virus) in the genetic background of PR8 were generated by means of reverse genetics, using PR8 plasmids provided by E. Hoffmann and R. G. Webster (St. Jude Children's Research Hospital, Memphis) as described previously (38, 39). Ready-to-use plasmids consisting of synthetic H5 or N1 genes in vector pHW2000 were obtained from GenScript. The H5 and N1 genes were flanked by the 3′ and 5′ untranslated regions of the corresponding segments from PR8. The AAA codon encoding K432 in wild-type HN N1 was replaced by the GAG codon encoding E432 in the mutant HN N1. The nucleotide sequence encoding the wild-type multibasic amino acid sequence was modified to encode a low-pathogenic cleavage site, as described previously (39). Viruses were grown in MDCK-II cells.
Statistical analysis.
The mean values of at least two experiments performed in duplicate/triplicate with independently generated protein preparations are graphed. All statistical analyses were performed by one-way analysis of variance (ANOVA) using Tukey's multiple-comparison test (Graph Pad Prism, version 6.05). The levels of significance were determined as P values of <0.05, <0.01, and <0.001, as indicated in the figure legends.
ACKNOWLEDGMENTS
W.D. and M.D. were supported by grants from the Chinese Scholarship Council. C.A.M.D.H. was supported by the Dutch Ministry of Economic Affairs, Agriculture, and Innovation, within the Castellum Project Zoonotic Avian Influenza. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We thank Roche for kindly providing OsC and Olav de Leeuw for technical assistance.
REFERENCES
- 1.Yen HL, Herlocher LM, Hoffmann E, Matrosovich MN, Monto AS, Webster RG, Govorkova EA. 2005. Neuraminidase inhibitor-resistant influenza viruses may differ substantially in fitness and transmissibility. Antimicrob Agents Chemother 49:4075–4084. doi: 10.1128/AAC.49.10.4075-4084.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wohlbold TJ, Nachbagauer R, Xu H, Tan GS, Hirsh A, Brokstad KA, Cox RJ, Palese P, Krammer F. 2015. Vaccination with adjuvanted recombinant neuraminidase induces broad heterologous, but not heterosubtypic, cross-protection against influenza virus infection in mice. mBio 6:e02556-14. doi: 10.1128/mBio.02556-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang L, Fantoni G, Couzens L, Gao J, Plant E, Ye Z, Eichelberger MC, Wan H. 2016. Comparative efficacy of monoclonal antibodies that bind to different epitopes of the 2009 pandemic H1N1 influenza virus neuraminidase. J Virol 90:117–128. doi: 10.1128/JVI.01756-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saito T, Taylor G, Webster RG. 1995. Steps in maturation of influenza A virus neuraminidase. J Virol 69:5011–5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Air GM. 2012. Influenza neuraminidase. Influenza Other Respir Viruses 6:245–256. doi: 10.1111/j.1750-2659.2011.00304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burmeister WP, Ruigrok RW, Cusack S. 1992. The 2.2 Å resolution crystal structure of influenza B neuraminidase and its complex with sialic acid. EMBO J 11:49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Varghese JN, Colman PM, van Donkelaar A, Blick TJ, Sahasrabudhe A, McKimm-Breschkin JL. 1997. Structural evidence for a second sialic acid binding site in avian influenza virus neuraminidases. Proc Natl Acad Sci U S A 94:11808–11812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun X, Li Q, Wu Y, Wang M, Liu Y, Qi J, Vavricka CJ, Gao GF. 2014. Structure of influenza virus N7: the last piece of the neuraminidase “jigsaw” puzzle. J Virol 88:9197–9207. doi: 10.1128/JVI.00805-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uhlendorff J, Matrosovich T, Klenk H, Matrosovich M. 2009. Functional significance of the hemadsorption activity of influenza virus neuraminidase and its alteration in pandemic viruses. Arch Virol 154:945–957. doi: 10.1007/s00705-009-0393-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dai M, McBride R, Dortmans JC, Peng W, Bakkers MJ, de Groot RJ, van Kuppeveld FJ, Paulson JC, de Vries E, de Haan CA. 2017. Mutation of the second sialic acid-binding site resulting in reduced neuraminidase activity preceded emergence of H7N9 influenza A virus. J Virol 91:e00049-17. doi: 10.1128/JVI.00049-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobasa D, Rodgers ME, Wells K, Kawaoka Y. 1997. Neuraminidase hemadsorption activity, conserved in avian influenza A viruses, does not influence viral replication in ducks. J Virol 71:6706–6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yen HL, Liang CH, Wu CY, Forrest HL, Ferguson A, Choy KT, Jones J, Wong DD, Cheung PP, Hsu CH, Li OT, Yuen KM, Chan RW, Poon LL, Chan MC, Nicholls JM, Krauss S, Wong CH, Guan Y, Webster RG, Webby RJ, Peiris M. 2011. Hemagglutinin-neuraminidase balance confers respiratory-droplet transmissibility of the pandemic H1N1 influenza virus in ferrets. Proc Natl Acad Sci U S A 108:14264–14269. doi: 10.1073/pnas.1111000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu R, Zhu X, McBride R, Nycholat CM, Yu W, Paulson JC, Wilson IA. 2012. Functional balance of the hemagglutinin and neuraminidase activities accompanies the emergence of the 2009 H1N1 influenza pandemic. J Virol 86:9221–9232. doi: 10.1128/JVI.00697-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gambaryan A, Matrosovich M. 2015. What adaptive changes in hemagglutinin and neuraminidase are necessary for emergence of pandemic influenza virus from its avian precursor? Biochemistry (Mosc) 80:872–880. doi: 10.1134/S000629791507007X. [DOI] [PubMed] [Google Scholar]
- 15.Franca de Barros J Jr, Sales Alviano D, da Silva MH, Dutra Wigg M, Sales Alviano C, Schauer R, dos Santos Silva Couceiro JN. 2003. Characterization of sialidase from an influenza A (H3N2) virus strain: kinetic parameters and substrate specificity. Intervirology 46:199–206. doi: 10.1159/000072428. [DOI] [PubMed] [Google Scholar]
- 16.Baum LG, Paulson JC. 1991. The N2 neuraminidase of human influenza virus has acquired a substrate specificity complementary to the hemagglutinin receptor specificity. Virology 180:10–15. doi: 10.1016/0042-6822(91)90003-T. [DOI] [PubMed] [Google Scholar]
- 17.Gerlach T, Kühling L, Uhlendorff J, Laukemper V, Matrosovich T, Czudai-Matwich V, Schwalm F, Klenk HD, Matrosovich M. 2012. Characterization of the neuraminidase of the H1N1/09 pandemic influenza virus. Vaccine 30:7348–7352. doi: 10.1016/j.vaccine.2012.09.078. [DOI] [PubMed] [Google Scholar]
- 18.Kobasa D, Kodihalli S, Luo M, Castrucci MR, Donatelli I, Suzuki Y, Suzuki T, Kawaoka Y. 1999. Amino acid residues contributing to the substrate specificity of the influenza A virus neuraminidase. J Virol 73:6743–6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wagner R, Matrosovich M, Klenk H. 2002. Functional balance between haemagglutinin and neuraminidase in influenza virus infections. Rev Med Virol 12:159–166. doi: 10.1002/rmv.352. [DOI] [PubMed] [Google Scholar]
- 20.Benton DJ, Wharton SA, Martin SR, McCauley JW. 2017. Role of neuraminidase in influenza A(H7N9) virus receptor binding. J Virol 91:e02293-16. doi: 10.1128/JVI.02293-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lai JC, Garcia J, Dyason JC, Böhm R, Madge PD, Rose FJ, Nicholls JM, Peiris J, Haselhorst T, Von Itzstein M. 2012. A secondary sialic acid binding site on influenza virus neuraminidase: fact or fiction? Angew Chemie Int Ed Engl 51:2221–2224. doi: 10.1002/anie.201108245. [DOI] [PubMed] [Google Scholar]
- 22.Dai M, Guo H, Dortmans J, Dekkers J, Nordholm J, Daniels R, van Kuppeveld FJ, de Vries E, de Haan CA. 2016. Identification of residues that affect oligomerization and/or enzymatic activity of influenza virus H5N1 neuraminidase proteins. J Virol 90:9457–9470. doi: 10.1128/JVI.01346-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baenziger JU, Fiete D. 1979. Structure of the complex oligosaccharides of fetuin. J Biol Chem 254:789–795. [PubMed] [Google Scholar]
- 24.Spik G, Bayard B, Fournet B, Strecker G, Bouquelet S, Montreuil J. 1975. Studies on glycoconjugates. LXIV. Complete structure of two carbohydrate units of human serotransferrin. Studies on glycoconjugates XIV. FEBS Lett 50:296–299. [DOI] [PubMed] [Google Scholar]
- 25.von Bonsdorff L, Tölö H, Lindeberg E, Nyman T, Harju A, Parkkinen J. 2001. Development of a pharmaceutical apotransferrin product for iron binding therapy. Biologicals 29:27–37. doi: 10.1006/biol.2001.0273. [DOI] [PubMed] [Google Scholar]
- 26.Wu AM, Wu JH, Tsai M, Yang Z, Sharon N, Herp A. 2007. Differential affinities of Erythrina cristagalli lectin (ECL) toward monosaccharides and polyvalent mammalian structural units. Glycoconj J 24:591–604. doi: 10.1007/s10719-007-9063-y. [DOI] [PubMed] [Google Scholar]
- 27.Sharma V, Srinivas VR, Adhikari P, Vijayan M, Surolia A. 1998. Molecular basis of recognition by Gal/GalNAc specific legume lectins: influence of Glu 129 on the specificity of peanut agglutinin (PNA) towards C2-substituents of galactose. Glycobiology 8:1007–1012. doi: 10.1093/glycob/8.10.1007. [DOI] [PubMed] [Google Scholar]
- 28.Geisler C, Jarvis DL. 2011. Effective glycoanalysis with Maackia amurensis lectins requires a clear understanding of their binding specificities. Glycobiology 21:988–993. doi: 10.1093/glycob/cwr080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shibuya N, Goldstein IJ, Broekaert WF, Nsimba-Lubaki M, Peeters B, Peumans WJ. 1987. The elderberry (Sambucus nigra L.) bark lectin recognizes the Neu5Ac(α2-6)Gal/GalNAc sequence. J Biol Chem 262:1596–1601. [PubMed] [Google Scholar]
- 30.Del Piccolo N, Placone J, He L, Agudelo SC, Hristova K. 2012. Production of plasma membrane vesicles with chloride salts and their utility as a cell membrane mimetic for biophysical characterization of membrane protein interactions. Anal Chem 84:8650–8655. doi: 10.1021/ac301776j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Vries RP, de Vries E, Moore KS, Rigter A, Rottier PJ, de Haan CA. 2011. Only two residues are responsible for the dramatic difference in receptor binding between swine and new pandemic H1 hemagglutinin. J. Biol Chem 286:5868–5875. doi: 10.1074/jbc.M110.193557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wickramasinghe IN, de Vries RP, Grone A, de Haan CA, Verheije MH. 2011. Binding of avian coronavirus spike proteins to host factors reflects virus tropism and pathogenicity. J Virol 85:8903–8912. doi: 10.1128/JVI.05112-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leung HS, Li OT, Chan RW, Chan MC, Nicholls JM, Poon LL. 2012. Entry of influenza A Virus with a α2,6-linked sialic acid binding preference requires host fibronectin. J Virol 86:10704–10713. doi: 10.1128/JVI.01166-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gamblin SJ, Haire LF, Russell RJ, Stevens DJ, Xiao B, Ha Y, Vasisht N, Steinhauer DA, Daniels RS, Elliot A, Wiley DC, Skehel JJ. 2004. The structure and receptor binding properties of the 1918 influenza hemagglutinin. Science 303:1838–1842. doi: 10.1126/science.1093155. [DOI] [PubMed] [Google Scholar]
- 35.Rogers GN, D'Souza BL. 1989. Receptor binding properties of human and animal H1 influenza virus isolates. Virology 173:317–322. doi: 10.1016/0042-6822(89)90249-3. [DOI] [PubMed] [Google Scholar]
- 36.Bosch BJ, Bodewes R, de Vries RP, Kreijtz JH, Bartelink W, van Amerongen G, Rimmelzwaan GF, de Haan CA, Osterhaus AD, Rottier PJ. 2010. Recombinant soluble, multimeric HA and NA exhibit distinctive types of protection against pandemic swine-origin 2009 A(H1N1) influenza virus infection in ferrets. J Virol 84:10366–10374. doi: 10.1128/JVI.01035-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li W, Hulswit RJG, Widjaja I, Raj VS, McBride R, Peng W, Widagdo W, Tortorici MA, van Dieren B, Lang Y, van Lent JWM, Paulson JC, de Haan CAM, de Groot RJ, van Kuppeveld FJM, Haagmans BL, Bosch BJ. 2017. Identification of sialic acid-binding function for the Middle East respiratory syndrome coronavirus spike glycoprotein. Proc Natl Acad Sci U S A 114:E8508–E8517. doi: 10.1073/pnas.1712592114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoffmann E, Krauss S, Perez D, Webby R, Webster RG. 2002. Eight-plasmid system for rapid generation of influenza virus vaccines. Vaccine 20:3165–3170. doi: 10.1016/S0264-410X(02)00268-2. [DOI] [PubMed] [Google Scholar]
- 39.Peeters B, Reemers S, Dortmans J, de Vries E, de Jong M, van de Zande S, Rottier PJM, de Haan CAM. 2017. Genetic versus antigenic differences among highly pathogenic H5N1 avian influenza A viruses: consequences for vaccine strain selection. Virology 503:83–93. doi: 10.1016/j.virol.2017.01.012. [DOI] [PubMed] [Google Scholar]
- 40.Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]