Herpes simplex virus (HSV) and other alphaherpesviruses, such as varicella-zoster virus, depend upon the capacity to navigate in neuronal axons. To do this, virus particles tether onto dyneins and kinesins that motor along microtubules from axon tips to neuronal cell bodies (retrograde) or from cell bodies to axon tips (anterograde). Following reactivation from latency, alphaherpesviruses absolutely depend upon anterograde transport of virus particles in axons in order to reinfect peripheral tissues and spread to other hosts. Which of the many axonal kinesins transport HSV in axons is not clear. We characterized fluorescent cellular cargo molecules and kinesins to provide evidence that HSV enveloped particles are ferried by kinesin-1 proteins KIF5A, -5B, and -5C and their light chains, KLC1 and KLC2, in axons. Moreover, we obtained evidence that kinesin-1 proteins are functionally important in anterograde transport of HSV virions by silencing these proteins.
KEYWORDS: neurons, kinesins, anterograde, KIF5, KIF1, axons, silencing, miRNAs, baculoviruses
ABSTRACT
Herpes simplex virus (HSV) and other alphaherpesviruses must spread from sites of viral latency in sensory ganglia to peripheral tissues, where the viruses can replicate to higher titers before spreading to other hosts. These viruses move in neuronal axons from ganglia to the periphery propelled by kinesin motors moving along microtubules. Two forms of HSV particles undergo this anterograde transport in axons: (i) unenveloped capsids that become enveloped after reaching axon tips and (ii) enveloped virions that are transported within membrane vesicles in axons. Fundamental to understanding this axonal transport is the question of which of many different axonal kinesins convey HSV particles. Knowing which kinesins promote axonal transport would provide clues to the identity of HSV proteins that tether onto kinesins. Prominent among axonal kinesins are the kinesin-1 (KIF5A, -5B, and -5C) and kinesin-3 (e.g., KIF1A and -1B) families. We characterized fluorescent forms of cellular cargo molecules to determine if enveloped HSV particles were present in the vesicles containing these cargos. Kinesin-1 cargo proteins were present in vesicles containing HSV particles, but not kinesin-3 cargos. Fluorescent kinesin-1 protein KIF5C extensively colocalized with HSV particles, while fluorescent kinesin-1 KIF1A did not. Silencing of kinesin-1 proteins KIF5A, -5B, and -5C or light chains KLC1 and KLC2 inhibited the majority of HSV anterograde transport, while silencing of KIF1A had little effect on HSV transport in axons. We concluded that kinesin-1 proteins are important in the anterograde transport of the majority of HSV enveloped virions in neuronal axons and kinesin-3 proteins are less important.
IMPORTANCE Herpes simplex virus (HSV) and other alphaherpesviruses, such as varicella-zoster virus, depend upon the capacity to navigate in neuronal axons. To do this, virus particles tether onto dyneins and kinesins that motor along microtubules from axon tips to neuronal cell bodies (retrograde) or from cell bodies to axon tips (anterograde). Following reactivation from latency, alphaherpesviruses absolutely depend upon anterograde transport of virus particles in axons in order to reinfect peripheral tissues and spread to other hosts. Which of the many axonal kinesins transport HSV in axons is not clear. We characterized fluorescent cellular cargo molecules and kinesins to provide evidence that HSV enveloped particles are ferried by kinesin-1 proteins KIF5A, -5B, and -5C and their light chains, KLC1 and KLC2, in axons. Moreover, we obtained evidence that kinesin-1 proteins are functionally important in anterograde transport of HSV virions by silencing these proteins.
INTRODUCTION
Herpes simplex viruses (HSVs) and other alphaherpesviruses, such as varicella-zoster virus, infect mucosal epithelium or skin and spread into neuronal axons. They travel in the retrograde direction in axons to neuron cell bodies that are in ganglia, where lifelong latency is established. Alphaherpesviruses periodically reactivate from latency and are transported in the anterograde direction in neuronal axons to mucosal surfaces or skin, where they replicate and produce ulcers in the epithelium. Anterograde transport involves transport of virus particles by kinesin motors that move virus particles by fast axonal transport on microtubules (reviewed in reference 1). Anterograde transport is an indispensable process for alphaherpesviruses, permitting periodic egress from neurons, spread within epithelial tissues, and production of larger quantities of infectious virus, promoting dissemination to other hosts.
There are over 45 different mammalian kinesin proteins arranged into 14 different families (2–6). The founding members of the kinesin superfamily are the kinesin-1 proteins, which consist of 3 heavy chains—KIF5A, -5B, and -5C—and 2 light chains—KLC1 and KLC2. KIF5A and -5C tend to be neuron specific, while KIF5B (classic kinesin) is expressed in most cells. Another important kinesin family in neuronal axons consists of the kinesin-3 proteins, including KIF1A and -1B, KIF13A and -13B, and KIF16A and -16B. However, several other kinesins, including KIF2, KIF4, KIF17, and KIF21, are also important in axonal transport (6–9). Anterograde transport of HSV requires kinesins, but it is currently not clear which kinesins promote this essential process. Specifically, there has been no functional evidence that any kinesin protein is important for HSV anterograde transport. For the related pig alphaherpesvirus pseudorabies virus (PRV) there was a report that a PRV membrane protein, US9, binds kinesin-3 KIF1A, a fluorescent protein colocalized with enveloped PRV particles moving anterograde in neuronal axons (10). Significantly, a dominant negative KIF1A reduced PRV axonal transport by approximately 75% in this study, supporting the conclusion that kinesin-3 proteins are important for PRV anterograde transport. There was a report that HSV US9 derived from bacteria can bind kinesin-1 proteins also produced in bacteria in in vitro pulldown experiments (11). However, with these studies of HSV anterograde transport, there were no experiments performed with neurons, no characterization of the binding to other kinesin family members, and no evidence that kinesin-1 motors were functionally important.
The identity of the kinesins that transport HSV in axons has important implications for several reasons. First, this is a central part of the biology of this virus and targeting the transport of reactivating HSV from ganglia to mucosa might have important therapeutic implications. Second, knowing which kinesins transport HSV in axons advances efforts to identify and characterize the viral proteins that participate in this transport. While it has been suggested that PRV and HSV US9 tether virus particles onto kinesins, recent studies by us and others have brought these conclusions into question. Loss of both HSV US9 and another membrane protein, gE/gI, leads to highly diminished axonal transport: only a very few virus particles enter axons, about 5% of the amount observed with wild-type (WT) HSV (12). Importantly, the few capsids that do enter axons with gE− US9− double mutants are transported with normal velocities and without delays (12). Instead of defects in transport within axons, as one might expect if US9 or gE/gI tethers virus particles onto kinesins during transport, gE− US9− mutants exhibited major defects in the assembly of enveloped particles in the cytoplasm and sorting of particles into axons (13). We concluded that HSV gE/gI and US9 are required in the cytoplasm and not after virus particles enter axons. There is also good evidence that PRV US9 is important for PRV anterograde transport (10, 14–16). However, again a PRV US9-null mutant was transported with normal kinetics in axons (17). Therefore, we believe that there are viral proteins other than US9 and gE/gI that function to tether HSV and PRV particles onto kinesins during transport in axons. Identifying these viral proteins will benefit from first knowing which cellular kinesins transport HSV in axons.
We elected to characterize a panel of fluorescent forms of cellular cargo molecules that are transported in neuronal axons as part of vesicles that might also transport HSV virions. Transport of certain of these cargo molecules into axons revealed discrete puncta that also contained fluorescent HSV enveloped particles. There was colocalization of several kinesin-1 cargo molecules with HSV particles but no colocalization between HSV and several kinesin-3 cargos. HSV particles also colocalized with fluorescent kinesin-1 molecules (KIF5C) and not with kinesin-3 proteins (KIF1A). Silencing of kinesin-1 light chains KLC1 and KLC2 and KIF5A and -5C reduced HSV axonal transport, but this was not the case when kinesin-3 KIF1A was silenced. We concluded that HSV is dependent upon kinesin-1 proteins for anterograde transport of virus particles in neuronal axons.
RESULTS
HSV particles in axons are transported in vesicles that contain kinesin-1 cargo molecules.
One of the most powerful methods for determining which of the many kinesins are active in some specific form of axonal transport is to determine which cargo molecules are present. Identifying these cargos frequently produces solid support for the role of specific kinesins or kinesin families in axonal transport. For example, certain synaptic vesicle proteins are ferried specifically by either kinesin-1 or kinesin-3 motors (3, 6, 8, 18). We previously showed that HSV enveloped particles are the major form of axonal transport of virus particles in mouse CAD neurons (13). These enveloped virions are apparently present within membrane vesicles, vesicles that are similar to those that ferry synaptic proteins or plasma membrane proteins (so-called axonal cargo molecules) toward axon tips. In contrast, in superior cervical (SCG) primary rat neurons and human SK-N-SH neurons, we observed a mixture of unenveloped capsids and enveloped virions in axons (12, 13, 19). The studies described here entirely focused on transport of enveloped virions in CAD neurons because enveloped virions present in membrane vesicles can be readily characterized for the presence of cellular cargos. Differentiation of CAD cells over 7 to 10 days produces relatively long axons, frequently of 40 to 80 μm, and HSV anterograde transport in these axons is robust, involving speeds of 0.5 to 2 μm/s, similar to the kinetics of transport we observed in rat embryonic SCG neurons (12, 13, 19).
To express fluorescent cargo molecules in CAD neurons, we used recombinant baculoviruses that express the vesicular stomatitis virus G protein (VSV-G), which mediates efficient entry into the neurons. These baculoviruses have major advantages for transducing neurons, which are difficult to transfect. Previously, we used nonreplicating adenovirus (Ad) and lentivirus vectors to deliver foreign genes into neurons but found negative effects of Ad vectors on HSV gene expression in neurons and that HSV extinguished lentivirus gene expression. Baculoviruses do not replicate in CAD or rat SCG neurons and do not harm the neurons appreciably; i.e., neurons remained bound to substrate for long periods after baculovirus infection and continued to transport fluorescent cargo molecules for many days after baculovirus infection. Using these baculoviruses, we could transduce as many as 85% of CAD neurons to produce intense fluorescence in cell bodies and observed discrete fluorescent puncta representing axonal transport of vesicles within axons. However, in most of the studies we used lesser quantities of baculoviruses in order to transduce 33 to 50% of the CAD neurons with strong fluorescence in cell bodies in order to reduce chances of overexpression of cargos. The intensities of some of the fluorescent puncta were similar to those we previously observed with single fluorescent capsids in neuronal axons (12, 13, 19; also see below), though other cargo puncta were substantially dimmer. It is also important to note that we produced a significant list of baculoviruses that expressed cargo molecules, and many were not amenable to imaging (Table 1). While about half of the baculovirus-expressed fluorescent cargo molecules produced discrete puncta, in some cases these puncta were very concentrated and difficult to compare to virus particles. Other cargos produced diffuse fluorescence in axons or puncta that were not discrete (well separated), or the cargo molecules did not efficiently enter axons. For example, vesicle-associated membrane protein 1 (VAMP1) fused to blue fluorescent protein (BFP) remained in the cytoplasm and did not enter axons efficiently, and the nuclear distribution protein nudE-like 1 protein (NDEL1-BFP) produced diffuse fluorescence and no puncta in axons. These defects may reflect fusion of the cargo molecules onto green fluorescent protein (GFP), red fluorescent protein (RFP), or BFP. Note that we could adjust the quantities of these fluorescent cargos in neurons by using different doses of baculoviruses or delivering the baculoviruses for various times (12 to 36 h) before HSV infection. We did not observe any gross defects in the transport of effects of HSV capsids in axons following baculovirus infection, compared with neurons not infected by baculoviruses (data not shown), and cargo molecules were obviously transported well into axons (see below).
TABLE 1.
List of cargo molecules expressed as fluorescent proteins using baculoviruses
| Cargoa | Fluorescent tag | Expressionb | Transport into axonsc | Diffuse or punctate localization in axonsd |
|---|---|---|---|---|
| APP-BFP | BFP | Yes | Yes | Punctate |
| BDNF-BFP | BFP | Yes | Yes | Punctate |
| SNAP25-GFP | GFP | Yes | Yes | Punctate |
| VAMP1-BFP | BFP | Yes | No | |
| LRP8-BFP | BFP | Yes | Yes | Diffuse |
| NDEL1-BFP | BFP | Yes | Diffuse | |
| PRP-BFP | BFP | Yes | Yes | Diffuse |
| GAP43-GFP | GFP | Yes | Yes | Punctate |
| TGOLN-2-GFP | GFP | Yes | Yes | Punctate |
| LAMP2-RFP | RFP | No | NAe | NA |
| TfnR-GFP | GFP | Yes | No | NA |
| CgA-RFP | RFP | Yes | Yes | Punctate |
| SYP-BFP | BFP | Yes | Yes | Punctate |
| NTRK-GFP | GFP | No | ||
| Rab5a-GFP | GFP | Yes | Yes | Punctate |
| Rab7a-GFP | GFP | Yes | Yes | Punctate |
| Calreticulin-RFP | RFP | Yes | No | NA |
APP, amyloid precursor protein; BDNF, brain-derived neurotrophic factor; SNAP25, synaptosome-associated protein (25 kDa); VAMP1, vesicle-associated membrane protein (synaptobrevin 1); LRP8, low-density lipoprotein receptor-related protein 8; NDEL1, nuclear distribution protein nudE-like; PRP, prion protein; GAP-43, growth-associated protein; TGOLN-2, trans-Golgi network protein 2; LAMP2, lysosome-associated membrane glycoprotein 2; TfnR, transferrin receptor; CgA, chromagranin A; SYP, synaptophysin; NTRK, neurotrophic tyrosine kinase receptor type 1; Rab5a, Ras-related protein Rab-5a; Rab7a, Ras-related protein Rab-7a.
Expression of the fluorescent protein was assessed by infecting CAD neurons with baculoviruses and performing immunofluorescence studies.
Transport into axons was determined by imaging CAD neuron axons following infection with baculoviruses.
CAD neurons expressing these fluorescent proteins were imaged to determine whether there was punctate or diffuse fluorescence in axons.
NA, not applicable.
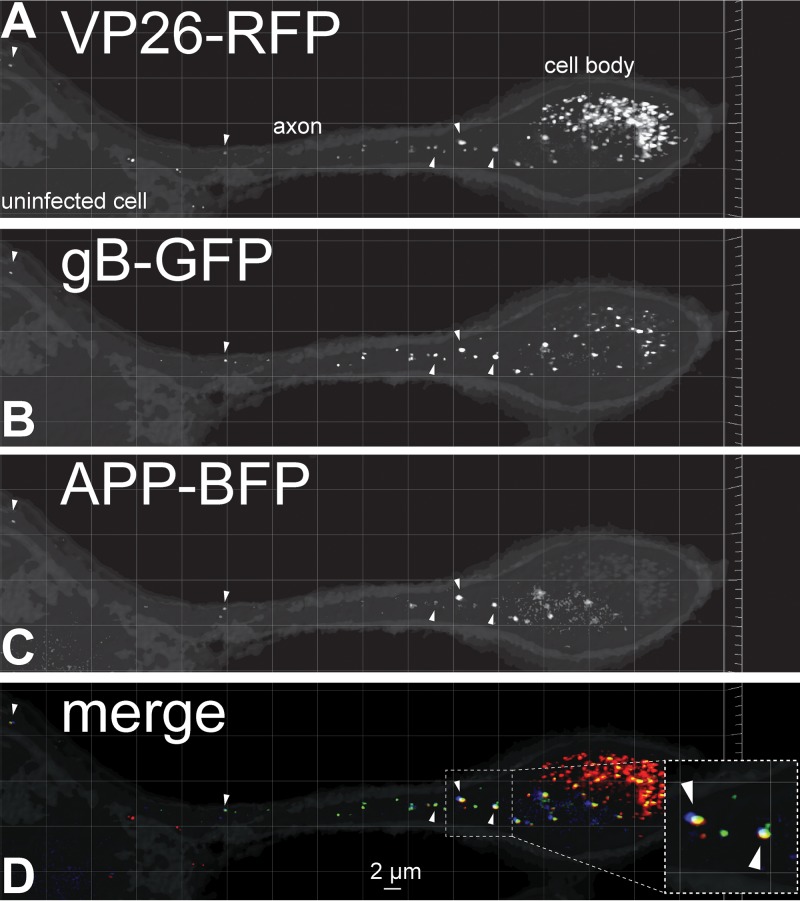
A baculovirus expressing the amyloid precursor protein (APP) fused to BFP was constructed and used to transduce CAD neurons for 22 h producing discrete puncta in axons (Fig. 1). The neurons were subsequently infected with an HSV recombinant virus, GS2843, that expresses gB (a component of the virion envelope) fused to GFP and VP26 (small capsid protein) fused to monomeric RFP (mRFP) (20). The image shown in Fig. 1 depicts a neuron cell body on the right (with intense capsid fluorescence) and an axon extending to the left that travels over top of another neuron that lacks appreciable HSV proteins. APP puncta were observed in membrane vesicles within the HSV-infected neuronal axon that also contained HSV enveloped virions exhibiting both gB-GFP and VP26-RFP. We counted HSV gB and VP26 puncta and APP puncta by constructing three-dimensional (3D) images composed of multiple 0.2-μm-thick fluorescent image stacks encompassing the entire axon depth. These 3D images were then rotated in order to eliminate out-of-plane false positives and separate overlapping signals from those that were adjacent or close by. These analyses indicated that 69% of capsid+ vesicles contained detectable APP and 41% of vesicles contained HSV capsids showing APP fluorescence (Table 2). APP is a transmembrane protein with a cytoplasmic domain which is a well-defined cargo molecule ferried by kinesin-1 proteins, including KIF5C (21). Thus, these studies supported the hypothesis that HSV capsids can be transported in vesicles that contain other cargo molecules ferried by kinesin-1 motors.
FIG 1.
Fluorescent imaging of capsids, gB, and amyloid precursor protein (APP) in CAD neurons. Differentiated CAD neurons growing on poly-lysine–laminin-coated glass coverslips were infected for 22 h with a baculovirus expressing APP-BFP, a cargo transported by kinesin-1 proteins. The neurons were subsequently infected with HSV recombinant GS2843, which expresses gB-GFP and VP26-RFP (small capsid protein), for 18 h before being fixed with 4% PFA and imaged. The right side of the image shows a neuron with a cell body (labeled), which was filled with capsids and extended an axon toward the left over top of an uninfected neuron with a cell body that showed no capsids in the nucleus (uninfected cell). Arrowheads in panels A and B indicate enveloped virions decorated with VP26-RFP (capsids in panel A) and gB-GFP (glycoproteins and virion envelope in panel B) in the infected CAD neuronal axon. Panel C shows APP-BFP puncta in the same axon, with Arrowheads pointing to puncta that also showed VP26-RFP and gB-GFP fluorescence. Most APP+ vesicles contained HSV enveloped particles as shown in panel D, which represents the merging of GFP, RFP, and BFP signals. The inset in panel D is a magnified image showing colocalization of APP-BFP with VP26-RFP and gB-GFP signals indicated with arrows.
TABLE 2.
HSV enveloped virions and capsids transported in vesicles containing cargo molecules and kinesin motors
| Molecule or motor | Kinesin | No. of axons counted | No. of: |
% of: |
|||
|---|---|---|---|---|---|---|---|
| Capsid+ vesiclesa | Cargo+ vesiclesb | Cargo+ vesicles with capsidc | Cargo+ vesicles with capsidd | Capsid+ vesicles with cargoe | |||
| Cargo | |||||||
| APP | Kinesin-1 | 6 | 108 | 64 | 44 | 68.8 | 40.7 |
| GAP43 | Kinesin-1 | 4 | 114 | 68 | 42 | 61.8 | 36.8 |
| CgA | Kinesin-3 | 7 | 130 | 69 | 0 | 0.0 | 0.0 |
| Rab5a | Kinesin-3 | 4 | 36 | 40 | 0 | 0.0 | 0.0 |
| Rab7a | Kinesin-3 | 5 | 67 | 58 | 0 | 0.0 | 0.0 |
| Kinesin | No. of axons counted | Capsid+ vesiclesa | Kinesin+ vesiclesb | Kinesin+ vesicles with capsidc | % kinesin+ with capsidd | % capsid+ vesicles with kinesine | |
| KIF5C tail | Kinesin-1 | 6 | 131 | 103 | 73 | 70.9 | 55.7 |
| KIF1A | Kinesin-3 | 5 | 126 | 66 | 3 | 4.5 | 2.4 |
| Cargo + kinesin | No. of axons counted | Cargo+ vesiclesa | Kinesin+ vesiclesb | Cargo+ vesicles with kinesin | % kinesin+ with cargof | % cargo+ vesicles with kinesing | |
| CgA + KIF1A | 4 | 106 | 98 | 67 | 68.4 | 63.2 | |
| APP + KIF5C tail | 4 | 54 | 51 | 39 | 76.5 | 72.2 | |
Total number of vesicles in axons that contained capsids.
Total number of vesicles in axons that contained cargo molecules or kinesins.
Vesicles containing cargo (cargo+) or kinesins (kinesin+) that also contained capsids.
Percentage of vesicles containing cargo or kinesins that also contained capsids.
Percentage of vesicles containing capsids that also contained cargo or kinesins.
Percentage of vesicles containing kinesins that also contained cargos.
Percentage of vesicles containing cargo that also contained kinesins.
We note that in every experiment involving fluorescent cargo molecules (described above and below), the cargo+ membrane vesicles were less numerous (frequently 50%) than vesicles containing HSV particles (Table 1). The fewer puncta containing detectable cargo molecules might be explained by our observations that there were highly variable rates of incorporation of these fluorescent cargos into transport vesicles; i.e., there were large differences in the fluorescence intensity of most cargo+ vesicles. These observations strongly suggested that we could not detect a substantial fraction of vesicles containing fluorescent cargos because the numbers of fluorescent molecules were below the levels of detection. In contrast, most vesicles containing capsids showed more uniform, stronger fluorescence. It appears that the assembly of numerous fluorescent VP26 copies into the capsid structure produced more uniform fluorescent signals in more vesicles than with fluorescent cargos. Based on these considerations, we believe that it makes more sense to emphasize the numbers of cargo+ vesicles that contain capsids rather than capsid+ vesicles that contain fluorescent cargos. Another way of putting this is as follows. One might detect 100 fluorescent capsids and 50 fluorescent cargos that are, in every case, colocalized with a capsid. Thus, 100% of the cargos are found in a vesicle with a capsid, but only 50% of the capsids would be in vesicles with detectable cargo. To us, the more relevant representation is the 100% colocalization. In this study, we observed APP+ vesicles that were less numerous or limiting, so the important question is as follows: what is the fraction of APP+ vesicles that contain a capsid? However, Table 2 shows the data presented in both ways.
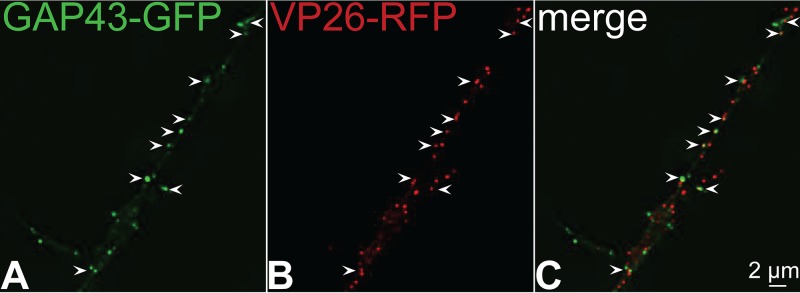
A second kinesin-1 cargo, growth-associated protein 43 (GAP43), which is a peripheral membrane protein bound to membranes via palmitate, was also characterized (22, 23). GAP43 was fused to GFP and expressed using a baculovirus and CAD neurons infected with a different HSV recombinant, GS2822, which expresses VP26-RFP but not fluorescent gB. There was also extensive colocalization of GAP43 with HSV capsids (Fig. 2). In images such as Fig. 2, it is necessary to carefully compare the image of GFP fluorescence representing GAP43 alone with other panels showing VP26-RFP alone because the intensities of fluorescence of GAP43 varied substantially among different transport vesicles. Vesicles containing a capsid or capsids were less variable in this respect. Thus, it can be difficult to discern whether an HSV capsid is present in a vesicle that also contains GAP43 and vice versa unless the GFP and RFP channels are evaluated separately as well as simultaneously (merge channel). As with the data in Fig. 1, we constructed 3D images in order to quantify whether HSV capsids and fluorescent GAP43 were present in the same membrane vesicles. This analysis showed that 62% of GAP43+ vesicles contained an HSV capsid and 37% of capsid+ vesicles contained GAP43 (Table 2). A third kinesin-1 cargo, synaptosome-associated protein 25 (SNAP-25), also colocalized extensively with HSV capsids (data not shown). These results supported the hypothesis that kinesin-1 proteins KIF-5A, -5B, and -5C participate in anterograde transport of HSV particles.
FIG 2.
Imaging of fluorescent growth-associated protein 43 (GAP43) and capsids in CAD neuron axons. CAD neurons were infected for 22 h with a baculovirus expressing GAP43-GFP, a cargo molecule transported by kinesin-1 family members, and then infected with GS2822, which expresses VP26-RFP (small capsid protein fused to RFP), for 18 h. The neurons were fixed with 4% PFA and imaged. Arrows indicate sites of colocalization of VP26-RFP containing capsids with GAP43-GFP puncta in the axon.
Cargos and Rab proteins transported by kinesin-3 motors do not colocalize with HSV particles in neuronal axons.
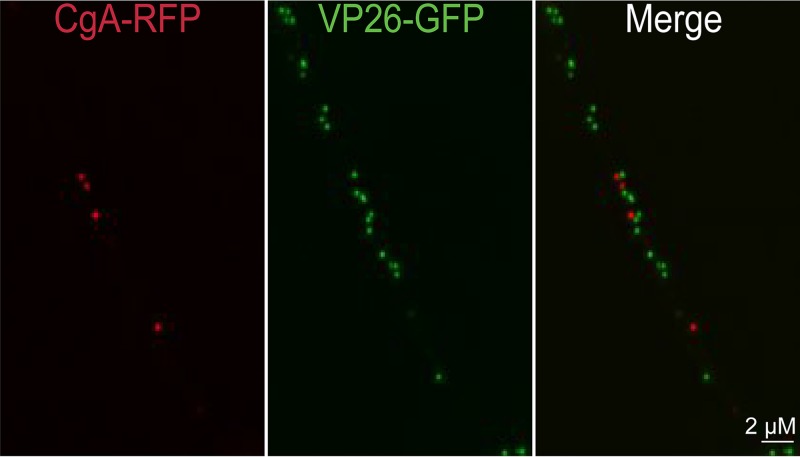
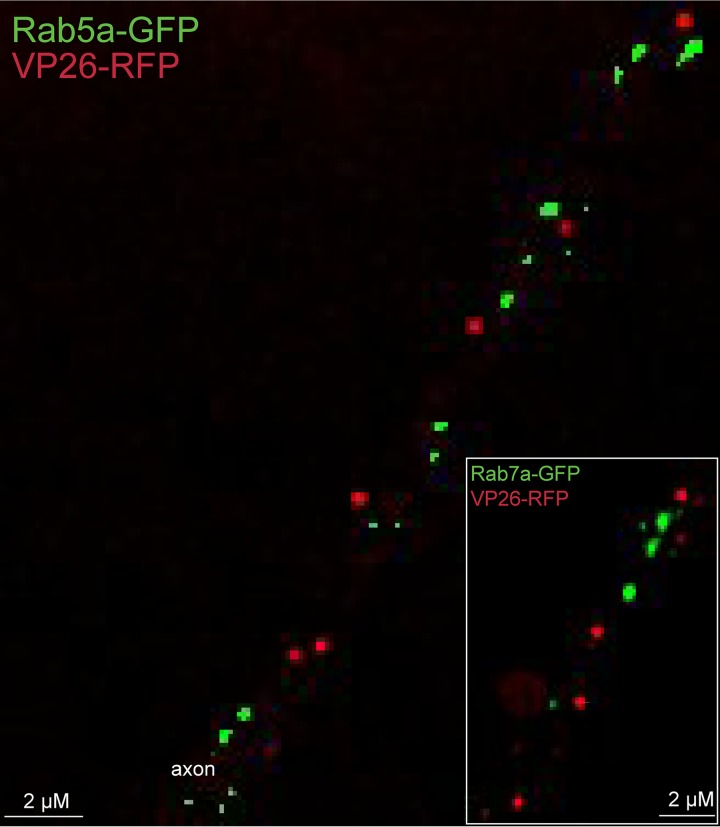
In the introduction, we described the compelling evidence that PRV is transported anterograde in the form of enveloped virions by kinesin-3 KIF1A (10). Given this, it was important to characterize whether HSV particles could be found in vesicles containing kinesin-3 cargo molecules. Chromagranin A (CgA) is ferried by kinesin-3 proteins, including KIF1A and KIF1B (24). There were fewer CgA puncta in CAD neuron axons (compared with APP and GAP43), but there was no colocalization of CgA puncta and HSV capsids in axons (Fig. 3). We counted 69 CgA+ puncta (note that it was necessary to count more neurons to attain this number of CgA puncta) and found that there were no cases of HSV capsids present in CgA+ vesicles and no cases of capsid+ puncta that contained CgA (Table 2). Similarly, synaptophysin (SYP) is transported by kinesin-3 proteins, including KIF1A (25), and no SYP+ vesicles contained HSV capsids, although the numbers of SYP+ capsids were low and are not included in Table 2. To further analyze whether kinesin-3 proteins are involved in HSV anterograde transport, we expressed two Rab small GTPases, Ras-related protein Rab-5a (Rab5a) and Ras-related protein Rab-7a (Rab7a). Rab5a is preferentially found in early endosomes but efficiently transported into axons and to axon tips by KIF13A and KIF13B, kinesin-3 family members (3, 18, 26–29). Rab7a is preferentially found in late endosomes that are transported into axons by KIF1A and -1B (29). Rab5a-GFP+ or Rab7a-GFP+ vesicles did not contain any HSV capsids (Fig. 4; Table 2). These results supported the hypothesis that HSV particles are not transported by kinesin-3 motors.
FIG 3.
Imaging of fluorescent chromagranin A (CgA) and capsids in axons. CAD neurons were infected for 20 h with a baculovirus expressing CgA-RFP, a cargo transported by kinesin-3 family members, and then infected with F-VP26/GFP, which expresses VP26-GFP (small capsid protein fused to GFP), for 18 h. The neurons were fixed with 4% PFA and imaged. None of the capsid+ puncta contained detectable CgA-RFP fluorescence and vice versa.
FIG 4.
Imaging of fluorescent Rab5a-GFP and Rab7a-GFP with HSV capsids in axons. CAD neurons were infected for 22 h with baculoviruses expressing Rab5a-GFP or Rab7a-GFP (inset), which are both endosomal markers and cargo molecules that are transported by kinesin-3 family members. Neurons were subsequently infected with GS2822, which expresses VP26-RFP, for 18 h and then fixed with 4% PFA. There was little or no colocalization of Rab5a-GFP or Rab7a-GFP with VP26-RFP containing capsids within axons.
HSV capsids colocalize with kinesin-1 motors but not kinesin-3 motors.
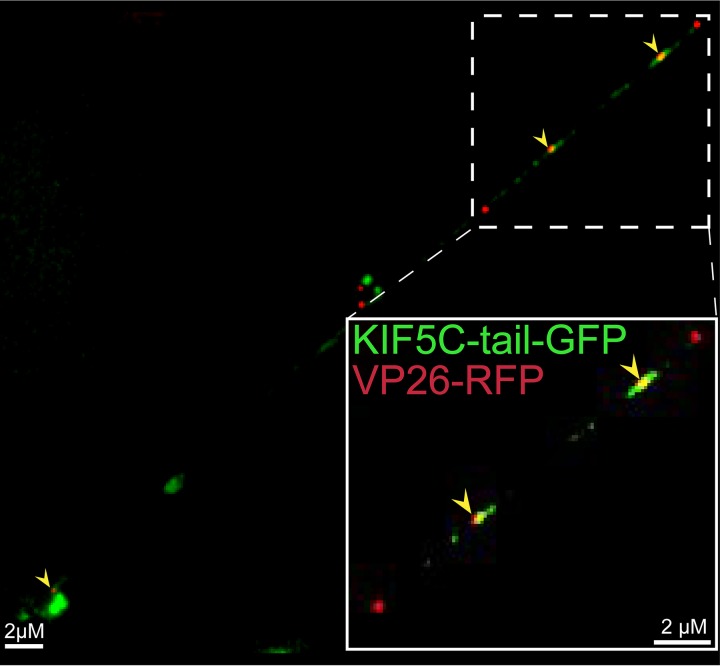
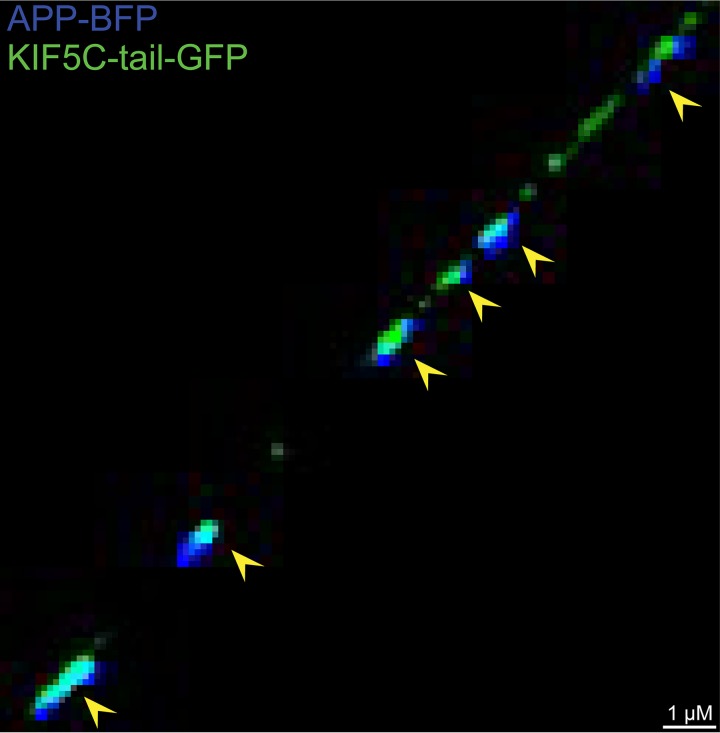
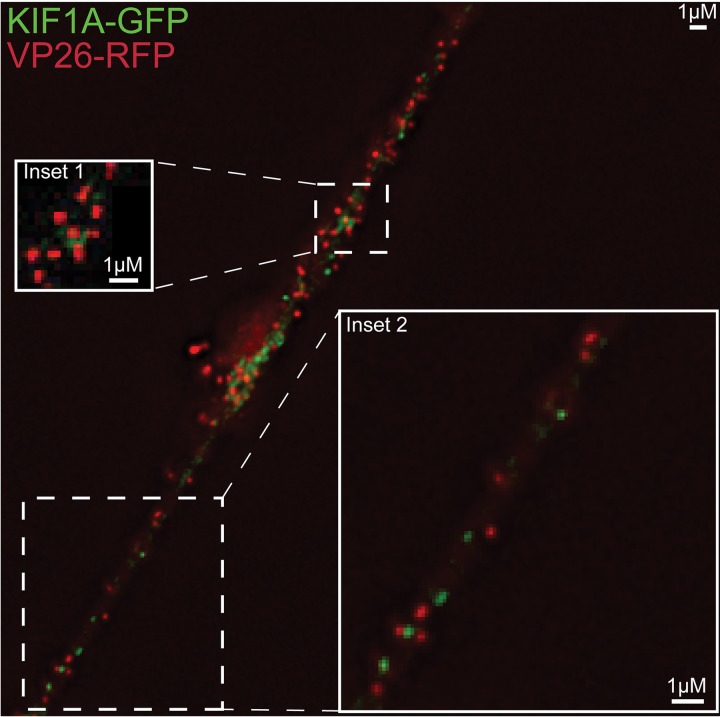
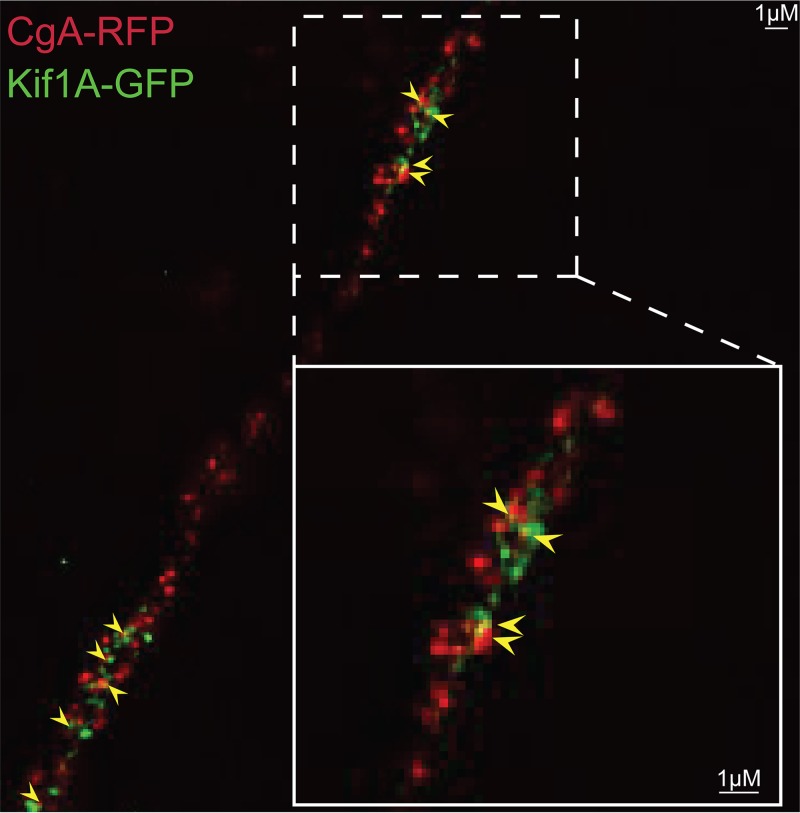
To extend these results, we constructed fluorescent forms of kinesin-1 and kinesin-3 proteins. Kinesin-1 protein KIF5C is principally expressed in neurons, unlike classic kinesin KIF5B, which is found in most cells. We expressed a truncated form of KIF5C with the motor domain replaced with GFP, leaving behind the “tail” domain that binds adaptors and cargo molecules (30). Kinesin tail domains do not bind to microtubules but are transported by other, full-length kinesins (containing motor domains) that are bound to the same membrane vesicles with cargo (29). The KIF5C tail-GFP protein was expressed in CAD neurons using a recombinant baculovirus, and KIF5C tail-GFP produced discrete puncta that frequently overlapped with HSV VP26-RFP capsids (Fig. 5). Approximately 71% of KIF5C+ vesicles contained an HSV capsid (Table 2). The KIF5C tail-GFP protein also colocalized extensively with APP-BFP (Fig. 6), so 77% of the KIF5C tail+ vesicles contained APP (Table 2). In other experiments, full-length KIF5C-GFP also extensively colocalized with HSV capsids, although full-length KIF5C-GFP produced lower levels of fluorescence than KIF5C tail-GFP (data not shown). A full-length version of KIF1A, a kinesin-3 protein, was fused to GFP, and vesicles containing this fusion infrequently contained HSV capsids (Fig. 7). The image in Fig. 7 again shows regions of relatively dense HSV capsids, e.g., in the region of inset 1, but 3D reconstruction of the capsids rarely showed that KIF1A colocalized with these capsids. Figure 7, inset 2, shows less dense capsids and clear separation from KIF1A puncta. Counts showed that <5% of KIF1A+ vesicles contained HSV particles (Table 2). The KIF1A-GFP+ vesicles were frequently (68%) positive for CgA (Fig. 8; Table 2). We hypothesized from the data involving fluorescent cargos and kinesins that HSV enveloped particles are transported in axons by kinesin-1 proteins and not by kinesin-3 proteins.
FIG 5.
Imaging of fluorescent kinesin KIF5C and HSV capsids in axons. CAD neurons were infected for 22 h with a baculovirus expressing kinesin KIF5C tail-GFP, a kinesin heavy-chain 1 (KIF5C) protein in which the motor domain was replaced by GFP sequences. Subsequently, the neurons were infected for an additional 18 h with GS2822, which expresses VP26-RFP, and then fixed with 4% PFA. Arrows indicate colocalization of KIF5C tail-GFP and VP26-RFP containing capsids within an axon. The kinesin-1 KIF5C tail extensively colocalized with HSV particles.
FIG 6.
Imaging of APP and KIF5C tail in axons. CAD neurons were infected with two baculoviruses, one expressing APP-BFP and a second expressing KIF5C tail-GFP, for 22 h and then fixed with 4% PFA and imaged. Arrows indicate colocalization of APP-BFP and KIF5C tail-GFP within an axon. The fluorescent kinesin-1 cargo APP was extensively colocalized with fluorescent kinesin-1 KIF5C tail.
FIG 7.
Imaging of fluorescent KIF1A and HSV capsids in axons. CAD neurons were infected for 22 h with a baculovirus expressing KIF1A-GFP (a kinesin-3 family member) and then infected with GS2822, expressing VP26-RFP, for 18 h. The neurons were fixed with 4% PFA and then imaged. Inset 1 shows a more congested region of the axon that has many capsids and fewer KIF1A puncta, and inset 2 shows a less congested region of the axon. There was little colocalization of KIF1A-GFP with VP26+ HSV capsids.
FIG 8.
Fluorescent imaging of CgA and KIF1A in CAD neurons. CAD neurons were infected with two different baculoviruses: one expressing CgA-RFP, a cargo transported by kinesin-3 family members, and one expressing KIF1A, a kinesin-3 family member, for 20 h. The neurons were then fixed with 4% PFA. Arrows indicate colocalization of CgA-RFP and KIF1A-GFP in the axon. The kinesin-3 cargo CgA-RFP extensively colocalized with the kinesin-3 KIF1A-GFP.
HSV anterograde transport following the silencing of kinesins.
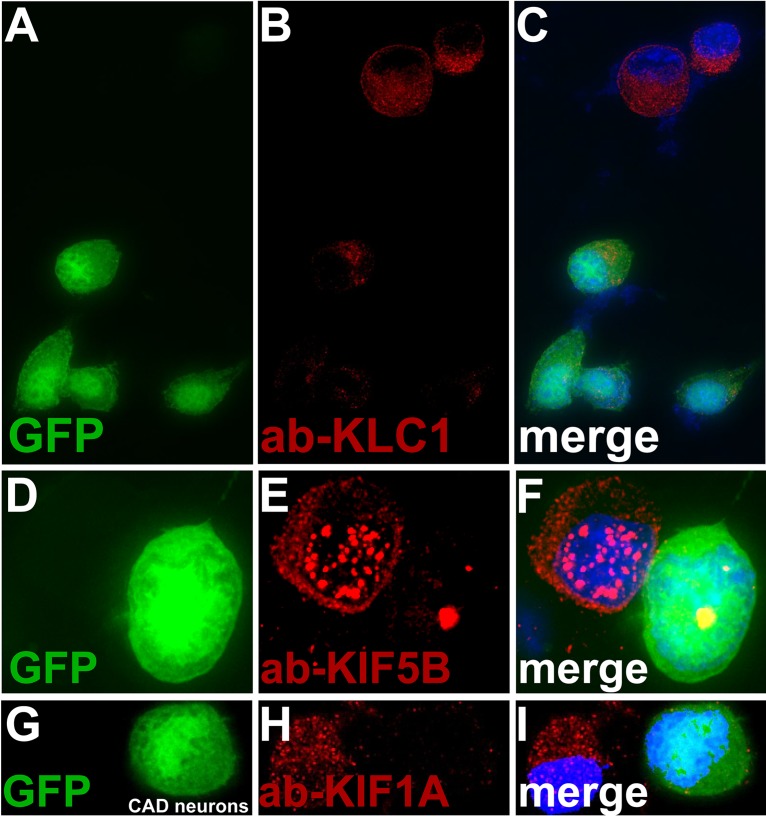
To determine whether kinesin-1 proteins are necessary or important for HSV anterograde transport, we constructed baculoviruses expressing microRNAs (miRNAs) that reduced the expression of various kinesins. Specifically, there were four baculoviruses that expressed miRNAs targeting (i) KIF5A and -5C, kinesin-1 heavy-chain proteins that are primarily neuron specific, (ii) KIF5A, -5B (classic kinesin), and -5C, i.e., all 3 kinesin-1 heavy-chain proteins, (iii) KLC1 and KLC2, both kinesin-1 light chains, and (iv) kinesin-3 protein KIF1A. Each of these baculoviruses expressed two different miRNAs that targeted one of the kinesin proteins; e.g., a total of 6 miRNAs were expressed in the baculovirus targeting KIF5A, -5B, and -5C. Every baculovirus also expressed GFP, so we could focus on axons of neurons that moderately to strongly expressed miRNAs. There was evidence for efficient silencing by the baculoviruses expressing these miRNAs in neurons. Specifically, we observed little or no KLC1 stained using an antibody in neurons infected with the baculovirus expressing KLC1 and KLC2 miRNAs (Fig. 9A to C). Similarly, the baculovirus expressing miRNAs targeting KIF5A, -5B, and -5C reduced expression of KIF5B stained with an antibody (Fig. 9D to F). The baculovirus expressing miRNAs targeting KIF1A also dramatically reduced KIF1A expression (Fig. 9G to I). In other studies, Western blots also showed reductions in expression of KLC2, KIF5A, KIF5C, and KIF1, although these reductions were not as profound as those involving immunofluorescence because the blot experiments did not specifically focus on GFP+ neurons. CAD neurons transduced with a baculovirus expressing GFP but no miRNA displayed HSV capsids that efficiently entered axons (Fig. 10A). Note that in our images we show just the RFP-labeled capsids, although these neurons were selected for strong GFP expression. Neurons infected with a baculovirus expressing the KIF5A- and -5C-specific miRNAs showed fewer capsids in axons (Fig. 10B). We counted 4 to 8 neurons for each of the sets of miRNAs in order to quantify 200 to 400 capsids in cell bodies and axons. Silencing with KIF5A and -5C produced 67% inhibition of the numbers of capsids in axons compared with neurons expressing no miRNA (Table 3). Expression of KIF5A-, 5B- and 5C-specific miRNAs reduced HSV capsids in axons further (Fig. 10C), and counts revealed 84% inhibition compared with the control (Table 3). Expression of kinesin light chain KLC1- and KLC2-specific miRNAs also reduced HSV capsids in axons (Fig. 10D), in this case to 87% of the control (Table 3). In contrast, expression of KIF1A-specific miRNAs in neurons produced only a small reduction in capsids in axons (Fig. 10E), 6.3% of the control (Table 3). We concluded that reducing kinesin-1 heavy chains KIF5A, -5B, and -5C and kinesin-1 light chains had a substantial negative effect on anterograde transport of HSV particles into axons, while silencing of kinesin-3 KIF1A had much less effect.
FIG 9.
Fluorescent imaging of kinesins present in axons following expression of miRNAs targeting kinesins. CAD neurons were infected with baculoviruses expressing miRNAs targeting kinesin-1 light chains KLC1 and KLC2, as well as GFP (A to C), miRNAs targeting kinesin-1 heavy chains KIF5A, -5B, and -5C and GFP (D to F), and miRNAs targeting kinesin-1 KIF1A and GFP (G to I). After 72 h, the neurons were fixed with 4% PFA, permeabilized, and immunostained with antibodies specific for KLC1 (A to C), KIF5B (D to F), or KIF1A (G to I) and also with 300 nM DAPI. CAD neurons with strong GFP expression exhibited little KLC1 (red) (A to C), little KIF5B (D to F), and little KIF1A (G to I).
FIG 10.
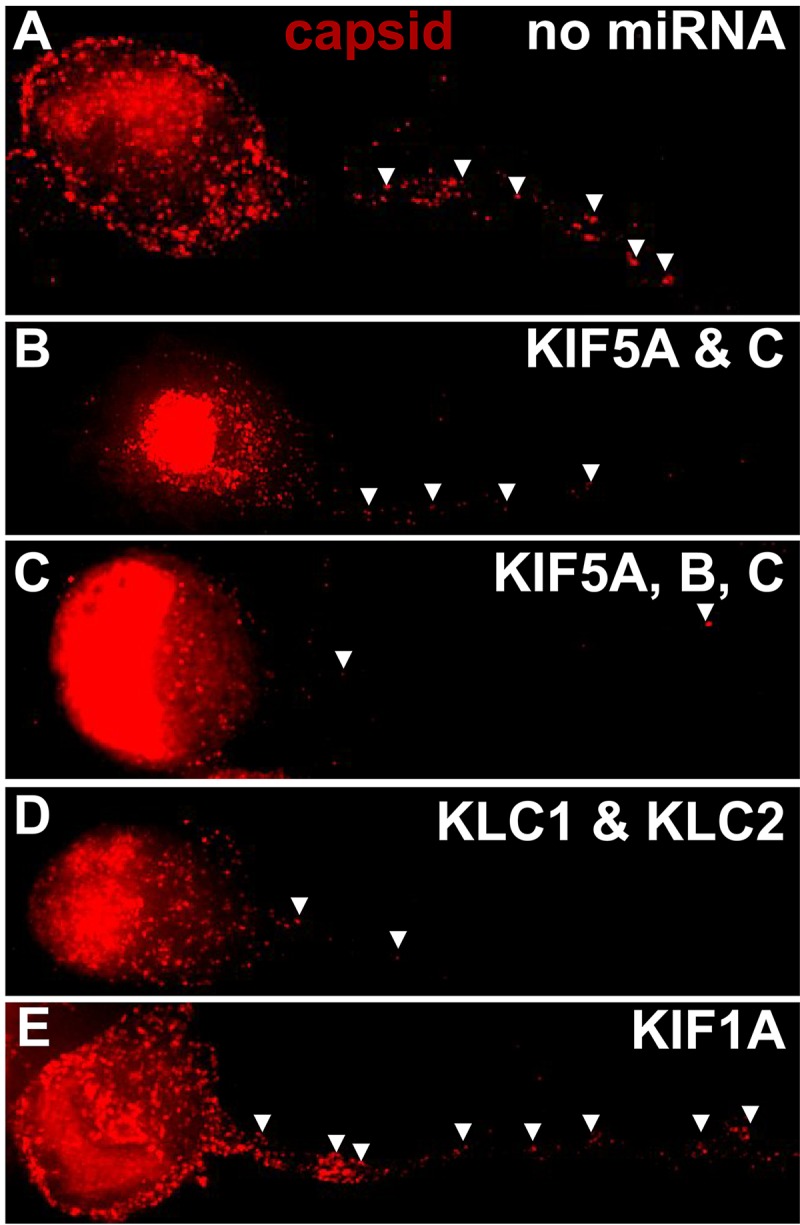
Fluorescent imaging of HSV capsids in neurons expressing kinesin-specific miRNAs. CAD neurons were infected with baculoviruses expressing GFP and also no miRNA (A) and miRNAs targeting KIF5A and -5C (B), KIF5A, -5B, and -5C (C), KLC1 and KLC2 (D), or KIF1A (E) for 54 h. Subsequently, the neurons were infected for 21 h with HSV GS2822, which expresses VP26-RFP, and then fixed with 4% PFA. CAD neurons expressing strong GFP signal were selected for imaging of RFP-labeled capsids in axons, which are indicated by arrowheads. (A) CAD neurons that were transduced with a baculovirus expressing GFP but no miRNA (control) displayed numerous HSV capsids in axons. (B) Neurons expressing KIF5A- and -5C-specific miRNAs showed fewer capsids in axons than the control. (C) Neurons expressing KIF5A-, -5B-, and -5C-specific miRNAs showed further reduced HSV capsids in axons. (D) Neurons expressing KLC1- and KLC2-specific miRNAs also showed reduced HSV capsids in axons. (E) In contrast, neurons expressing KIF1A-specific miRNAs showed only a small reduction in capsid numbers in axons.
TABLE 3.
Inhibition of HSV axonal transport by silencing of kinesin proteins
| miRNA(s) | No. of: |
% inhibition | ||
|---|---|---|---|---|
| Cytoplasmic capsids | Axonal capsids | Axonal capsids/100 cytoplasmic capsids | ||
| No miRNA | 180 | 105 | 58.3 | 0.0 |
| KIF5A and KIF5C | 369 | 71 | 19.2 | 67.0 |
| KIF5A, -5B, and -5C | 206 | 19 | 9.2 | 84.2 |
| KLC1 and KLC2 | 201 | 15 | 7.5 | 87.2 |
| KIF1A | 313 | 171 | 54.6 | 6.3 |
DISCUSSION
The HSV proteins that promote anterograde transport in neuronal axons are, as of yet, not all well characterized. There is good evidence that HSV and PRV mutants lacking gE/gI and US9 have defects in this transport. However, as described above, there is now good evidence that these proteins do not function in axons but instead act in the cytoplasm of neuron cell bodies to promote entry of virus particles into axons (12, 13). It is difficult to understand how gE/gI and US9 could be necessary for tethering onto kinesins within axons when these mutants behave this way. Of course, gE/gI and US9 may influence the initial interactions between HSV and kinesins in the cytoplasm before transport into axons. However, we have hypothesized that HSV proteins, other than gE/gI and US9, function to tether vesicles containing enveloped virions onto kinesin motors in axons. Efforts to identify these viral proteins that interact with kinesins would be markedly advanced by the identification of the specific kinesins that transport virus particles in neuronal axons.
We began to characterize the kinesins that transport HSV in axons by using CAD neurons that primarily transport enveloped virions inside membrane vesicles in axons. Fluorescent forms of cellular cargo molecules were expressed in these neurons to determine which kinesins were involved. This is a classic method for characterizing kinesin motors. In CAD neurons the membrane vesicles that contained HSV virions (13) displayed fluorescent APP. A total of 69% of APP+ vesicles contained fluorescent capsids, while 41% of the virions were present in vesicles that contained APP. APP is a cargo ferried primarily by kinesin-1 proteins, KIF5A, -5B, and -5C (21). As noted above, there were fewer vesicles with detectable APP (∼60%) than HSV capsids. The relatively lower numbers of vesicles containing detectable fluorescent cargos may relate to the presence of native (nonfluorescent) molecules in axons that reduced fluorescent signals below detectable levels in some vesicles. The levels of fluorescence in different vesicles varied substantially, so it is likely that there were axonal vesicles with quantities of fluorescent cargos below our level of detection. HSV capsids were more uniformly fluorescent, likely because these structures are composed of numerous copies of VP26-RFP. Results similar to those for APP were obtained with fluorescent GAP43: over half the GAP43+ vesicles contained capsids, and many of the capsids were found in vesicles that contained detectable GAP43. Like APP, GAP43 is ferried by kinesin-1 proteins in axons, including KIF5A, -5B, and -5C (22, 23). In contrast, HSV capsids were not found in any vesicles containing CgA, and no CgA+ vesicles contained HSV capsids. CgA is a well-characterized cargo transported by kinesin-3 proteins, including KIF1A (24). Two proteins, Rab5a and Rab7a, which are transported in axons by kinesin-3 motors (3, 29), were also not colocalized with HSV capsids and vice versa. Rab5a is also transported with vesicles containing SYP, which is a well-characterized kinesin-3 cargo molecule (31), and we found that SYP+ vesicles did not contain HSV capsids (data not shown). Together these results provided evidence that kinesin-1 motors transport HSV enveloped particles in CAD neuronal axons and kinesin-3 proteins do not.
The second approach that we used was to express fluorescent kinesin molecules. A KIF5C tail extensively colocalized with HSV capsids, 73% of KIF5C tail+ vesicles contained capsids, and 71% of capsid+ vesicles contained KIF5C tail. We noted that the levels of fluorescent KIF5C tail in puncta within axons was much less variable than fluorescent cargos. This may explain the higher frequencies of finding KIF5C tail in vesicles containing HSV capsids than with APP. Full-length KIF5C behaved very similarly to KIF5C tail. Together, these studies of both fluorescent cargos and kinesins suggest that the majority of HSV anterograde transport involves kinesin-1 proteins. In contrast, KIF1A was rarely detected with vesicles containing HSV capsids: 4.5% of KIF1A+ vesicles contained capsids, and 2.4% of vesicles containing capsids also displayed detectable KIF1A. It is possible that a small fraction of HSV particles are ferried by KIF1A, but the cargo experiments showed that no CgA, Rab5a, or Rab7a colocalized with HSV particles. The large fraction of vesicles that have associated kinesin-1 motors and kinesin-1 cargo that also have capsids or enveloped virions with them supports the conclusion that kinesin-1 proteins play a major role in HSV anterograde transport in axons. In contrast, kinesin-3 cargos and KIF1A seldom colocalized with HSV. These approaches suffer from their indirect nature, which assumes that each vesicle that contains a given cargo or kinesin family member does not also have other cargos or kinesin family members present. There may be several kinesin families all bound to the same vesicle and several cargos present. However, the silencing data discussed below shows that kinesin-1 proteins are required for the vast majority of HSV anterograde transport.
There have been other reports linking kinesin-1 motors to the anterograde transport of HSV particles. HSV was injected into squid neurons in studies showing that virus particles colocalized with APP and kinesin-1 (32). Observations that HSV US9 binds kinesin-1 proteins were coupled with the suggestion that kinesin-1 proteins are involved in anterograde transport. We consider the evidence that US9 binds kinesin-1 in neurons weak, as it is entirely based on in vitro pulldown experiments, with no studies showing these interactions in neurons and no other kinesins as negative controls. However, even if US9 binds kinesins, two points argue against this interaction having a meaningful role in HSV anterograde transport. First, we demonstrated that an HSV gE− US9− double mutant has defects in virus assembly in the cytoplasm but the few virus particles that enter axons move with normal kinetics and do not exhibit delays in movement (12, 13). These observations and similar results with a PRV US9− mutant (17) argued that HSV and PRV US9 are not required for axonal transport. Second, the group that proposed that US9 binds kinesin-1 has reported that only unenveloped capsids are transported in axons of their neurons (33–35). Thus, it was difficult for us to understand how a membrane protein such as US9, which would not be present with unenveloped capsids, could tether onto kinesin motors to promote anterograde transport. There was also a report showing immunoelectron microscopy (immune-EM) images suggesting that virus particles in neurons were associated with Rab3a, SNAP25 (a kinesin-1 cargo), GAP43, and kinesin-1 (36). These EM images showed that the GAP43, Rab3a, SNAP25, and kinesin-1 antibodies decorated enveloped virions that were present in varicosities or growth cones. The antibodies used to detect cargo molecules preferentially or entirely bound to virus particles themselves, including extracellular virions, rather than to the contents of the vesicles that surrounded virions. Again, it was difficult to understand how these observations on membrane-bound cargos can account for transport of unenveloped capsids in this system.
We silenced kinesin-1 proteins to determine the functional importance of these kinesins in HSV anterograde transport. A baculovirus expressing miRNAs that reduced expression of KIF5A and -5C, kinesins that are largely neuron specific, reduced capsids in axons by 67%. Silencing of all of the kinesin-1 heavy chains—KIF5A, -5B (classic kinesin), and -5C—reduced HSV transport by 84%. Kinesin-1 heavy chains can transport some cargos without light chains, but other cargos require the presence of kinesin-1 light chains KLC1 and KLC2 (7, 23, 37). Consistent with an important role for KLC1 and/or KLC2, silencing of these proteins reduced HSV capsids in axons by 87%. Silencing of KIF1A, which functions to transport PRV particles in axons, had little effect: transport was reduced by 6%. Together these data provided strong support for the conclusion that kinesin-1 heavy and light chains are important for the majority of the anterograde transport of HSV enveloped virions in axons. Kinesin-3 motor KIF1A apparently plays little or no role in this transport. However, our data do not eliminate the possibility that other kinesins also contribute, likely in a less substantial way, to HSV anterograde transport.
MATERIALS AND METHODS
Viruses.
HSV-1 strain F recombinant GS2843 (wild type [WT]), which expresses VP26-mRFP and gB-GFP, and recombinant GS2822, which expresses VP26-mRFP (20), were kind gifts from Greg Smith (Northwestern University Medical School). The HSV-1 strain F recombinant that expresses VP26-GFP was described previously (38). These viruses were propagated and titers were determined on Vero cells.
Neuronal cell cultures.
CAD cells are a derivative of a mouse catecholaminergic central nervous system cell line and were a kind gift from Greg Smith at Northwestern University Medical School, Chicago, IL (39). They were maintained in Dulbecco modified Eagle medium–F-12 (DMEM–F-12) containing 8% fetal bovine serum (FBS) and passaged by gentle dissociation with sodium citrate buffer (134 mM KCl, 15 mM sodium citrate [pH 7.3 to 7.4]). Differentiation of CAD cells into neurons was achieved by plating cells on poly-d-lysine (30 μg/ml)- and laminin (2 μg/ml)-coated glass coverslips in differentiation medium (DMEM–F-12 containing 0.5% FBS, 10 μM 3-isobutyl-1-methylxanthine [IBMX], 150 μM dibutyryl-cAMP [dbcAMP], and 1 ng/ml nerve growth factor [NGF; 2.5S; Invitrogen]). After 2 days the IBMX and dbcAMP were removed. The cells were differentiated for 7 to 10 days before being infected with recombinant baculoviruses and HSV.
Baculoviruses expressing fluorescent forms of cellular cargo and kinesin proteins.
Human cDNAs encoding the entire cargo proteins GAP43, SNAP25, and APP all fused at the N terminus of GFP, mRFP, or BFP were all obtained from Origene. A cDNA encoding CgA fused to the N terminus of mRFP was a gift from L. Taupenot, NIH. The entire rat KIF1A cDNA was fused onto the GFP with KIF1A at the N terminus of the fusion protein and GFP C terminus; this construct was a kind gift from Gary Banker (Oregon Health and Sciences University [OHSU]). The KIF5C tail-GFP recombinant protein consisted of a rat KIF5C cDNA with the N-terminal motor domain replaced by GFP before the coil-coiled domain (at amino acid [aa] 379) via insertion into pEGFP-C2. These fusion proteins were subsequently subcloned into the BacMam pCMV-DEST vector (using the ViraPower BacMam expression system; Thermo Fisher), and these genes were coupled to cytomegalovirus (CMV) promoter, the woodchuck hepatitis virus posttranscriptional regulatory element (WPRE), and simian virus 40 (SV40) poly(A) sequence. This vector also contains a gene encoding the vesicular stomatitis virus G protein (VSV-G), which allows efficient entry into mammalian cells. BacMam pCMV-Dest vectors containing cargo or kinesin fusion protein sequences were then transformed into MaxEfficiency DH10Bac cells (Thermo Fisher) for transposition into a bacmid that contains the baculovirus genome. After selection, recombinant bacmid DNA was purified and introduced into Sf9 insect cells (obtained from Eric Gouaux, OHSU) by transfection with Cellfectin (Thermo Fisher). Transfected insect cells produced recombinant infectious baculoviruses capable of expressing fluorescent kinesins or cargo molecules. Baculoviruses were propagated and amplified using Sf9 suspension cultures (106 cells/ml) in Sf-900 III SFM medium (100 U/ml penicillin-streptomycin [HyClone]) with incubation at 130 rpm and 27°C for 3 days. Baculoviruses were concentrated by ultracentrifugation of 40 ml supernatant for 1 h at 96,000 × g and 4°C. Pellets were resuspended in 0.4 ml DMEM. Titers for concentrated baculovirus preparations were determined by infecting differentiated CAD cells for 16 to 24 h and determining the quantities of the virus preparations (dilutions of virus stocks) required to produce GFP, RFP, or BFP expression in 33 to 50% of the CAD cells.
Baculoviruses expressing miRNAs to silence kinesins.
miRNA RNA interference (RNAi) sequences targeting the open reading frames of mouse KLC1, KLC2, KIF5A, KIF5B, KIF5C, and KIF1A were generated using the BLOCK-iT RNAi Designer (Thermo Fisher). Primer pairs used to silence these cellular proteins were ordered from IDT technologies. Following annealing, these pre-miRNA sequences were ligated into the pcDNA 6.2-GW/EmGFP-miR vector (Block-it miRNA kit; Thermo Fisher). Plasmid DNA was obtained from bacteria containing these miRNA constructs, and these DNAs were transfected into CAD cells in order to verify whether individual miRNAs reduced specific kinesins by using immunoblotting and/or immunofluorescence to assess kinesins in neuronal axons. Those miRNAs that demonstrated silencing were then “chained together” to produce cocistronic expression of multiple miRNAs following the manufacturer's protocol. In all cases at least two miRNAs were used to target each kinesin, and where three kinesins (KIF5A, -5B, and -5C) were targeted in one baculovirus, a total of 6 miRNAs were chained together. In brief, this consisted of using BamHI and XhoI to excise an individual miRNA coding DNA and BglII and XhoI to excise a second miRNA, followed by ligation into a pcDNA 6.2-GW/EmGFP-miR vector. For example, this involved chaining together 6 miRNAs in order to produce miRNAs targeting all of KIF5A, -5B, and -5C in one vector. These miRNA pcDNA 6.2-GW/EmGFP-miR constructs were then introduced into the BacMam pCMV-Dest vector using the Invitrogen Gateway technology BP/LR reaction (Thermo Fisher). In brief, Gateway BP Clonase II enzyme mix was used to transfer EmGFP-miR sequences into the pDON222 vector, and then these sequences were transferred into the BacMam pCMV-Dest vector to produce baculoviruses expressing miRNAs. The BacMam pCMV-Dest vectors were transferred into MaxEfficiency DH10Bac bacterial cells and baculovirus DNA was produced and used to transfect Sf9 cells as described in the previous section. Baculoviruses derived from these Sf9 cells were expanded and concentrated as described above. These baculoviruses expressed both miRNAs and also GFP so that the virus titers could be determined by infecting differentiated CAD neurons, followed by determination of the quantities of virus required to produce GFP expression in 30 to 100% of the CAD cells. Normally in the experiments shown a dilution of each of these miRNA-expressing baculoviruses was used so that ∼33 to 50% of CAD neurons exhibited relatively intense GFP expression.
Fluorescence imaging of CAD neurons and cargo or kinesins.
Differentiated CAD neurons were infected with recombinant baculovirus containing fluorescent cargo or kinesin proteins for 22 h and then subsequently infected with HSV for 18 h. In other cases, CAD neurons were coinfected with two recombinant baculoviruses—one expressing a fluorescent cargo molecule and another expressing fluorescent kinesin molecule—for 22 h, and no HSV infection was initiated. Following these infections, neurons were washed with phosphate-buffered saline (PBS; pH 7.4) and then fixed with PBS containing 4% paraformaldehyde (PFA) for 20 min at 20°C. For some samples, 300 nM 4′,6-diamidino-2-phenylindole (DAPI) was added for 5 min to stain nuclei. Fluorescence microscopy was performed in the Oregon Health and Sciences University's Advanced Light Microscopy Core using a Deltavision CoreDV high-resolution wide-field deconvolution system (Applied Precision). This system includes an Olympus IX71 inverted microscope with a proprietary XYZ stage, a solid-state module for fluorescence, and a Nikon Coolsnap ES2 HQ camera. The system was built on an Olympus IX71 inverted microscope and features a light-emitting diode (LED)-transmitted light source for differential interference contrast and a 7-color solid-state illumination unit for exciting fluorophores across the visible spectrum. Polychroic beam splitters, optimized for imaging with blue-green-red-far red fluorophore combinations in fixed cells or cyan fluorescent protein (CFP)-yellow fluorescent protein (YFP) and GFP-RFP combinations of expressed proteins in living cells, were combined with emission switching through fast filter wheels. The motorized stage was controlled by XYZ nanomotors for accurate z-stack and point-visiting functions. Images were acquired as z-stacks in a 1,024-by-1,024 format with a 60× (numerical aperture, 1.42) Plan Apo N objective in three fluorescence channels: 435, 549, and 649 nm. The images were deconvolved with the appropriate optical transfer function (OTF) using an iterative algorithm of 10 iterations. After deconvolution, images were processed with FIJI (ImageJ) software. For visualization of puncta representing fluorescent capsids, viral glycoproteins, cargo molecules, and kinesins, at least 4 images (10,560 μm2) per slide were captured with a minimum of 5 0.2-μm z-sections per axon. These images were analyzed using a minimum intensity threshold chosen based on matched uninfected control cells without VP26 or cargo/kinesin fluorescence. Puncta in axons representing capsids, cargo molecules, or kinesins were manually counted from images representing 4 to 10 different axons.
Immunofluorescence imaging of kinesins following miRNA silencing.
To assess silencing-induced reductions in kinesins by using immunofluorescence, CAD neurons were infected with recombinant baculovirus expressing miRNAs for 72 h and then the cells were permeabilized using 0.5% sodium deoxycholate for 5 min and fixed with 4% PFA. The neurons were incubated for 20 min in 5% normal goat serum and for 1 h in 0.3 mg/ml sheep anti-mouse IgG to reduce nonspecific IgG binding; then the cells were incubated with primary kinesin-specific antibodies in PBS containing 0.1% Tween 20 and 5% normal goat serum for 1 h. The cells were washed in PBS containing 0.1% Tween 20 and incubated with secondary fluorescent antibodies in PBS containing 0.1% Tween 20 for 1 h. For some samples, 300 nM DAPI was added for 5 min to stain nuclei. Silencing of kinesins was characterized in neurons expressing baculovirus-expressed GFP.
Imaging and miRNA capsid quantification.
To quantify the capsids in axons following in miRNA silencing, CAD neurons were infected with baculovirus expressing GFP and miRNAs for 54 h, and then the neurons were subsequently infected with wild-type HSV GS2822 expressing VP26-RFP. Twenty-one hours later, neurons were fixed with 4% PFA, z-stacks of 0.2-μm z-sections of axons from neurons that expressed GFP were imaged, and capsids (VP26-mRFP) were counted. Nuclear capsids were excluded from the cytoplasmic count using DAPI staining to define the nucleus.
ACKNOWLEDGMENTS
We thank Gary Banker (OHSU) for a great deal of helpful advice, many reagents, and kinesin constructs. We are grateful to Aurelie Snyder for excellent support for deconvolution experiments in the Advanced Light Microscopy Core at the Jungers Center, OHSU.
These studies were supported by National Institutes of Health grant R01 EY018755 (to D.C.J.).
REFERENCES
- 1.Smith G. 2012. Herpesvirus transport to the nervous system and back again. Annu Rev Microbiol 66:153–176. doi: 10.1146/annurev-micro-092611-150051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldstein AY, Wang X, Schwarz TL. 2008. Axonal transport and the delivery of pre-synaptic components. Curr Opin Neurobiol 18:495–503. doi: 10.1016/j.conb.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hirokawa N, Noda Y, Tanaka Y, Niwa S. 2009. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol 10:682–696. doi: 10.1038/nrm2774. [DOI] [PubMed] [Google Scholar]
- 4.Verhey KJ, Kaul N, Soppina V. 2011. Kinesin assembly and movement in cells. Annu Rev Biophys 40:267–288. doi: 10.1146/annurev-biophys-042910-155310. [DOI] [PubMed] [Google Scholar]
- 5.Hirokawa N, Tanaka Y. 2015. Kinesin superfamily proteins (KIFs): various functions and their relevance for important phenomena in life and diseases. Exp Cell Res 334:16–25. doi: 10.1016/j.yexcr.2015.02.016. [DOI] [PubMed] [Google Scholar]
- 6.Klinman E, Holzbaur EL. 2016. Comparative analysis of axonal transport markers in primary mammalian neurons. Methods Cell Biol 131:409–424. doi: 10.1016/bs.mcb.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 7.Hirokawa N, Nitta R, Okada Y. 2009. The mechanisms of kinesin motor motility: lessons from the monomeric motor KIF1A. Nat Rev Mol Cell Biol 10:877–884. doi: 10.1038/nrm2807. [DOI] [PubMed] [Google Scholar]
- 8.Muresan V. 2000. One axon, many kinesins: what's the logic? J Neurocytol 29:799–818. doi: 10.1023/A:1010943424272. [DOI] [PubMed] [Google Scholar]
- 9.Yang R, Bentley M, Huang CF, Banker G. 2016. Analyzing kinesin motor domain translocation in cultured hippocampal neurons. Methods Cell Biol 131:217–232. doi: 10.1016/bs.mcb.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kramer T, Greco TM, Taylor MP, Ambrosini AE, Cristea IM, Enquist LW. 2012. Kinesin-3 mediates axonal sorting and directional transport of alphaherpesvirus particles in neurons. Cell Host Microbe 12:806–814. doi: 10.1016/j.chom.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diefenbach RJ, Davis A, Miranda-Saksena M, Fernandez MA, Kelly BJ, Jones CA, LaVail JH, Xue J, Lai J, Cunningham AL. 2016. The basic domain of herpes simplex virus 1 pUS9 recruits kinesin-1 to facilitate egress from neurons. J Virol 90:2102–2111. doi: 10.1128/JVI.03041-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howard PW, Howard TL, Johnson DC. 2013. Herpes simplex virus membrane proteins gE/gI and US9 act cooperatively to promote transport of capsids and glycoproteins from neuron cell bodies into initial axon segments. J Virol 87:403–414. doi: 10.1128/JVI.02465-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DuRaine G, Wisner TW, Howard P, Williams M, Johnson DC. 2017. Herpes simplex virus gE/gI and US9 promote both envelopment and sorting of virus particles in the cytoplasm of neurons, two processes that precede anterograde transport in axons. J Virol 91:e00050-. doi: 10.1128/JVI.00050-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brideau AD, Eldridge MG, Enquist LW. 2000. Directional transneuronal infection by pseudorabies virus is dependent on an acidic internalization motif in the Us9 cytoplasmic tail. J Virol 74:4549–4561. doi: 10.1128/JVI.74.10.4549-4561.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lyman MG, Feierbach B, Curanovic D, Bisher M, Enquist LW. 2007. PRV Us9 directs axonal sorting of viral capsids. J Virol 81:11363–11371. doi: 10.1128/JVI.01281-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomishima MJ, Enquist LW. 2001. A conserved alpha-herpesvirus protein necessary for axonal localization of viral membrane proteins. J Cell Biol 154:741–752. doi: 10.1083/jcb.200011146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daniel GR, Sollars PJ, Pickard GE, Smith GA. 2015. Pseudorabies virus fast axonal transport occurs by a pUS9-independent mechanism. J Virol 89:8088–8091. doi: 10.1128/JVI.00771-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirokawa N, Takemura R. 2005. Molecular motors and mechanisms of directional transport in neurons. Nat Rev Neurosci 6:201–214. doi: 10.1038/nrn1624. [DOI] [PubMed] [Google Scholar]
- 19.Wisner TW, Sugimoto K, Howard PW, Kawaguchi Y, Johnson DC. 2011. Anterograde transport of herpes simplex virus capsids in neurons by both separate and married mechanisms. J Virol 85:5919–5928. doi: 10.1128/JVI.00116-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Antinone SE, Smith GA. 2010. Retrograde axon transport of herpes simplex virus and pseudorabies virus: a live-cell comparative analysis. J Virol 84:1504–1512. doi: 10.1128/JVI.02029-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szodorai A, Kuan YH, Hunzelmann S, Engel U, Sakane A, Sasaki T, Takai Y, Kirsch J, Muller U, Beyreuther K, Brady S, Morfini G, Kins S. 2009. APP anterograde transport requires Rab3A GTPase activity for assembly of the transport vesicle. J Neurosci 29:14534–14544. doi: 10.1523/JNEUROSCI.1546-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferreira A, Niclas J, Vale RD, Banker G, Kosik KS. 1992. Suppression of kinesin expression in cultured hippocampal neurons using antisense oligonucleotides. J Cell Biol 117:595–606. doi: 10.1083/jcb.117.3.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamal A, Stokin GB, Yang Z, Xia CH, Goldstein LS. 2000. Axonal transport of amyloid precursor protein is mediated by direct binding to the kinesin light chain subunit of kinesin-I. Neuron 28:449–459. doi: 10.1016/S0896-6273(00)00124-0. [DOI] [PubMed] [Google Scholar]
- 24.Lo KY, Kuzmin A, Unger SM, Petersen JD, Silverman MA. 2011. KIF1A is the primary anterograde motor protein required for the axonal transport of dense-core vesicles in cultured hippocampal neurons. Neurosci Lett 491:168–173. doi: 10.1016/j.neulet.2011.01.018. [DOI] [PubMed] [Google Scholar]
- 25.Okada Y, Yamazaki H, Sekine-Aizawa Y, Hirokawa N. 1995. The neuron-specific kinesin superfamily protein KIF1A is a unique monomeric motor for anterograde axonal transport of synaptic vesicle precursors. Cell 81:769–780. doi: 10.1016/0092-8674(95)90538-3. [DOI] [PubMed] [Google Scholar]
- 26.Huckaba TM, Gennerich A, Wilhelm JE, Chishti AH, Vale RD. 2011. Kinesin-73 is a processive motor that localizes to Rab5-containing organelles. J Biol Chem 286:7457–7467. doi: 10.1074/jbc.M110.167023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoepfner S, Severin F, Cabezas A, Habermann B, Runge A, Gillooly D, Stenmark H, Zerial M. 2005. Modulation of receptor recycling and degradation by the endosomal kinesin KIF16B. Cell 121:437–450. doi: 10.1016/j.cell.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 28.Seidel C, Moreno-Velásquez SD, Riquelme M, Fischer R. 2013. Neurospora crassa NKIN2, a kinesin-3 motor, transports early endosomes and is required for polarized growth. Eukaryot Cell 12:1020–1032. doi: 10.1128/EC.00081-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bentley M, Decker H, Luisi J, Banker G. 2015. A novel assay reveals preferential binding between Rabs, kinesins, and specific endosomal subpopulations. J Cell Biol 208:273–281. doi: 10.1083/jcb.201408056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bentley M, Banker G. 2015. A novel assay to identify the trafficking proteins that bind to specific vesicle populations. Curr Protoc Cell Biol 69:13.8.1–13.8.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Hoop MJ, Huber LA, Stenmark H, Williamson E, Zerial M, Parton RG, Dotti CG. 1994. The involvement of the small GTP-binding protein Rab5a in neuronal endocytosis. Neuron 13:11–22. doi: 10.1016/0896-6273(94)90456-1. [DOI] [PubMed] [Google Scholar]
- 32.Satpute-Krishnan P, DeGiorgis JA, Bearer EL. 2003. Fast anterograde transport of herpes simplex virus: role for the amyloid precursor protein of Alzheimer's disease. Aging Cell 2:305–318. doi: 10.1046/j.1474-9728.2003.00069.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Penfold ME, Armati P, Cunningham AL. 1994. Axonal transport of herpes simplex virions to epidermal cells: evidence for a specialized mode of virus transport and assembly. Proc Natl Acad Sci U S A 91:6529–6533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saksena MM, Wakisaka H, Tijono B, Boadle RA, Rixon F, Takahashi H, Cunningham AL. 2006. Herpes simplex virus type 1 accumulation, envelopment, and exit in growth cones and varicosities in mid-distal regions of axons. J Virol 80:3592–3606. doi: 10.1128/JVI.80.7.3592-3606.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cunningham A, Miranda-Saksena M, Diefenbach R, Johnson D. 2013. Letter in response to: Making the case: married versus separate models of alphaherpes virus anterograde transport in axons. Rev Med Virol 23:414–418. doi: 10.1002/rmv.1760. [DOI] [PubMed] [Google Scholar]
- 36.Miranda-Saksena M, Boadle RA, Aggarwal A, Tijono B, Rixon FJ, Diefenbach RJ, Cunningham AL. 2009. Herpes simplex virus utilizes the large secretory vesicle pathway for anterograde transport of tegument and envelope proteins and for viral exocytosis from growth cones of human fetal axons. J Virol 83:3187–3199. doi: 10.1128/JVI.01579-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rahman A, Kamal A, Roberts EA, Goldstein LS. 1999. Defective kinesin heavy chain behavior in mouse kinesin light chain mutants. J Cell Biol 146:1277–1288. doi: 10.1083/jcb.146.6.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Snyder A, Wisner TW, Johnson DC. 2006. Herpes simplex virus capsids are transported in neuronal axons without an envelope containing the viral glycoproteins. J Virol 80:11165–11177. doi: 10.1128/JVI.01107-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Antinone SE, Zaichick SV, Smith GA. 2010. Resolving the assembly state of herpes simplex virus during axon transport by live-cell imaging. J Virol 84:13019–13030. doi: 10.1128/JVI.01296-10. [DOI] [PMC free article] [PubMed] [Google Scholar]