HCV is a major public health problem worldwide. The virion harbors two envelope proteins, E1 and E2, which are involved at different steps of the viral life cycle. Whereas E2 has been extensively characterized, the function of E1 remains poorly defined. We characterized here the function of the putative fusion peptide and the region containing α helices of the E1 ectodomain, which had been previously suggested to be important for virus entry. We could confirm the importance of these regions for the virus infectivity. Interestingly, we found several residues modulating the virus's dependence on several HCV receptors, thus highlighting the role of E1 in the interaction of the virus with cellular receptors. Whereas mutations in the putative fusion peptide affected HCV infectivity and morphogenesis, several mutations in the α2-helix region led to a loss of infectivity with no effect on assembly, indicating a role of this region in virus entry.
KEYWORDS: hepatitis C virus, glycoprotein, envelope proteins, viral entry, viral assembly
ABSTRACT
In the hepatitis C virus (HCV) envelope glycoproteins E1 and E2, which form a heterodimer, E2 is the receptor binding protein and the major target of neutralizing antibodies, whereas the function of E1 remains less characterized. To investigate E1 functions, we generated a series of mutants in the conserved residues of the C-terminal region of the E1 ectodomain in the context of an infectious clone. We focused our analyses on two regions of interest. The first region is located in the middle of the E1 glycoprotein (between amino acid [aa] 270 and aa 291), which contains a conserved hydrophobic sequence and was proposed to constitute a putative fusion peptide. The second series of mutants was generated in the region from aa 314 to aa 342 (the aa314-342 region), which has been shown to contain two α helices (α2 and α3) by nuclear magnetic resonance studies. Of the 22 generated mutants, 20 were either attenuated or noninfectious. Several mutations modulated the virus's dependence on claudin-1 and the scavenger receptor BI coreceptors for entry. Most of the mutations in the putative fusion peptide region affected virus assembly. Conversely, mutations in the α-helix aa 315 to 324 (315-324) residues M318, W320, D321, and M322 resulted in a complete loss of infectivity without any impact on E1E2 folding and on viral assembly. Further characterization of the W320A mutant in the HCVpp model indicated that the loss of infectivity was due to a defect in viral entry. Together, these results support a role for E1 in modulating HCV interaction with its coreceptors and in HCV assembly. They also highlight the involvement of α-helix 315-324 in a late step of HCV entry.
IMPORTANCE HCV is a major public health problem worldwide. The virion harbors two envelope proteins, E1 and E2, which are involved at different steps of the viral life cycle. Whereas E2 has been extensively characterized, the function of E1 remains poorly defined. We characterized here the function of the putative fusion peptide and the region containing α helices of the E1 ectodomain, which had been previously suggested to be important for virus entry. We could confirm the importance of these regions for the virus infectivity. Interestingly, we found several residues modulating the virus's dependence on several HCV receptors, thus highlighting the role of E1 in the interaction of the virus with cellular receptors. Whereas mutations in the putative fusion peptide affected HCV infectivity and morphogenesis, several mutations in the α2-helix region led to a loss of infectivity with no effect on assembly, indicating a role of this region in virus entry.
INTRODUCTION
With 70 million people infected worldwide, hepatitis C virus infection is a major health problem (1). With a high propensity for establishing chronic infections, HCV is considered the major cause of chronic hepatitis, fibrosis, cirrhosis, and hepatocellular carcinoma. The development of direct-acting antivirals against HCV has been a milestone in the treatment of hepatitis C. This treatment shows high efficacy against all HCV genotypes, reaching high HCV clearance rates. However, the high cost of these antiviral therapies precludes their accessibility to the large majority of HCV-infected patients (2). In this context, the development of a preventive HCV vaccine would constitute the most cost-effective means to limit HCV spread. Therefore, a thorough understanding of the contribution of HCV glycoproteins E1 and E2 to viral entry and assembly is still required for the development of therapeutic and preventive vaccines.
HCV is an enveloped virus with a positive-strand RNA genome that belongs to the Hepacivirus genus of the Flaviviridae family (3). Its genome encodes a single polyprotein that is processed by cellular and viral proteases into 10 polypeptides, which include seven nonstructural proteins (p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) and three structural proteins (the core protein and the two envelope glycoproteins E1 and E2) that are the components of the viral particle (4). Thus, the viral particle is composed of a nucleocapsid constituted of the genomic RNA and the core protein, which is surrounded by a lipid membrane in which the heterodimer formed by the two envelope glycoproteins E1 and E2 is anchored. E1 and E2 glycoproteins are type I transmembrane proteins with well-conserved C-terminal transmembrane domains and highly glycosylated N-terminal ectodomains (5). As components of the viral particle, E1 and E2 are involved in virion morphogenesis and constitute the major viral determinant of HCV entry (6, 7). HCV entry into hepatocytes is a complex process that involves several cell surface molecules. Among them, the contributions to HCV entry of the scavenger receptor BI (SR-BI), the tetraspanin CD81, and the tight-junction proteins claudin-1 (CLDN1) and occludin (OCLN) have been the most characterized (8).
For a long time, E2 was the most-studied HCV envelope protein. Indeed, E2 is the major target of neutralizing antibodies, it mediates the interaction between the virus and SR-BI and CD81 receptors, and it was postulated to be the fusion protein of the virus (9). However, the structure of E2 does not harbor the features of a fusion protein, which led to the hypothesis that the fusion step would rather rely on E1 (10, 11). Recently, the N-terminal part of E1 (residues 192 to 270) was crystallized. The characterization of the role of the conserved residues of this region during the HCV life cycle showed that it is important for the virus infectivity and E1E2 heterodimer formation (12). Moreover, this region was shown to contain residues that mediate the dependence of the virus on the claudin-1 receptor for entry. In addition, a cross talk between HCV glycoprotein E1 and the viral genomic RNA was identified (12).
To further characterize the contribution of E1 to the different steps of the HCV life cycle, we generated a new series of mutants in the conserved residues of the C-terminal region of the E1 ectodomain. We focused our analysis on two regions of interest. The first region is located in the middle of the polypeptide (putative fusion peptide [PFP], located between amino acid [aa] 270 and aa 291), which contains conserved hydrophobic sequences that could potentially act as a fusion peptide (13, 14). The second series of mutants was generated in the aa314-342 region, which has been shown to contain two α helices (α2 and α3) by nuclear magnetic resonance (NMR) and X-ray crystallography studies (15, 16). We thus took advantage of these data to further investigate the functional role of the C-terminal part of the E1 ectodomain by alanine replacement of residues in the context of an infectious clone. Of the 22 generated mutants, only 2 exhibited a wild-type (wt) phenotype, while 9 were no longer infectious. Several mutations modulated the dependence of the virus on claudin-1 and/or SR-BI receptors for entry. Importantly, mutations in the α2 residues M318, W320, D321, and M322 resulted in a complete loss of infectivity without any impact on E1E2 folding or on viral assembly. Further characterization of the W320A mutant in the context of the HCV pseudoparticle (HCVpp) system indicated that the loss of infectivity is due to a defect in viral entry.
RESULTS
Amino acid conservation in the second half of the E1 ectodomain and mutated residues.
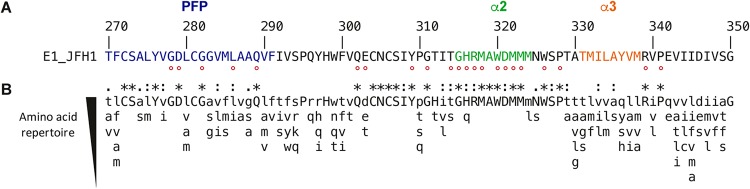
Amino acid conservation in E1 region from aa 270 to 350 among HCV genotypes is represented in Fig. 1. This region contains the putative fusion peptide aa270-291 (PFP) (13) and two α helices, α2 (aa315-324) and α3 (aa331-338), as revealed by NMR and X-ray crystallography studies performed on the aa314-342 and aa314-324 E1 peptides (15). The less-variable residues of this region among HCV genotypes were individually replaced by alanine in the context of the JFH1 infectious clone. This led to the generation of 22 mutants. Among them, 5 belong to the PFP region, whereas 8 are located in the α2-helix region. Unexpectedly, the residues of the α3 helix were more variable, which suggested a less important role for this region in the HCV life cycle. Mutations were introduced in a modified version of the plasmid encoding the full-length JFH1 genome in which the N-terminal E1 sequence has been modified to reconstitute the A4 epitope, which is present in E1 of genotype 1a (17), and therefore allows for the identification of this modified E1 of genotype 2a for which there is no antibody readily available. It is worth noting that introduction of the A4 epitope does not affect HCV infectivity and thus does not interfere with the characterization of the phenotypes of E1 mutants.
FIG 1.
E1 C-terminal region sequence analyses. (A) The E1 aa270-350 sequence from the HCV JFH1 strain (AB047639; genotype 2a) is indicated with respect to the polyprotein numbering. Amino acids mutated in this study are indicated by a red dot. (B) Amino acid repertoires of the C-terminal region of E1. The amino acid (aa) repertoire was deduced from the ClustalW multiple alignment of the 28 representative E1 sequences from confirmed genotypes and subtypes in the European HCV database (https://euhcvdb.ibcp.fr/euHCVdb/jsp/nomen_tab1.jsp). Amino acids observed at a given position in fewer than two distinct sequences were not included. Amino acids observed at a given position in more than 25 distinct sequences are shown in capital letters. The degree of amino acid conservation at each position can be inferred from the extent of variability (with the observed amino acids listed in decreasing order of frequency from top to bottom), together with the similarity index according to ClustalW convention (asterisk [*], invariant; colon [:], highly similar; dot [.], similar).
Effect of E1 mutations on HCV replication.
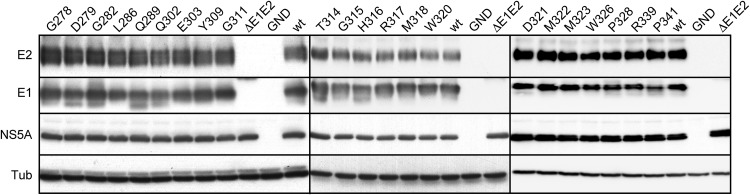
In a first step, we assessed the ability of the produced mutants to replicate. For this purpose, the expression of several HCV proteins (E1, E2, and NS5A) was examined at 48 h postelectroporation of Huh-7 hepatoma cells with wt and mutant HCV RNA. For all mutants, similar levels of protein expression could be observed; hence, any effect of the mutations on viral replication could be excluded (Fig. 2). We included in our analysis the GND nonreplicative HCV mutant and the ΔE1E2 assembly-deficient mutant that carries an in-frame deletion in E1E2 coding region. Interestingly, D279 and Q289 mutations in the PFP and Q302 in the region between PFP and α2 led to the detection of an additional band of lower molecular weight, which likely corresponds to an alternative glycoform of E1 as previously observed when E1 is expressed as a recombinant protein (18).
FIG 2.
Effect of E1 mutations on the expression of viral proteins. Viral RNA transcribed from JFH1-derived mutants was electroporated into Huh-7 cells that were lysed 48 h later. Viral proteins were separated by SDS-PAGE and revealed by Western blotting with MAbs A4 (anti-E1), 3/11 (anti-E2), and anti-NS5A, as well as anti-beta-tubulin antibody, to verify loading of equal amounts of cell lysates. The protein detected by MAb A4 in cells expressing ΔE1E2 mutant corresponds to a fusion protein between the N terminus of E1 and the C terminus of E2.
Effect of E1 mutations on HCV infectivity.
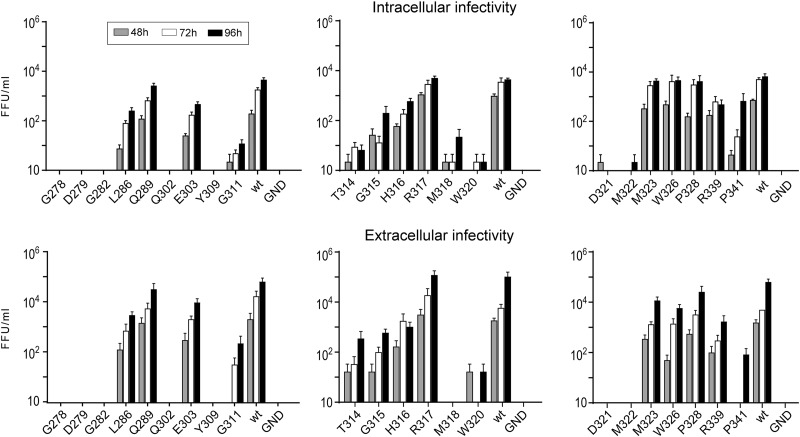
Since the introduced mutations did not affect viral replication, we assessed their impact on the production of infectious virus. To do so, we determined the intracellular and extracellular infectivity after electroporation of Huh-7 cells with viral RNAs. We observed different phenotypes of virus infectivity: (i) complete loss of infectivity for mutants G278A, D279A, G282A, Q302A, Y309A, M318A, W320A, D321A, and M322A; (ii) severe attenuation of infectivity for mutants G311A, T314A, G315A, H316A, R339A, and P341A; (iii) slight attenuation of infectivity for mutants L286A, E303A, M323A, W326A, and P328A; and (iv) no effect on infectivity for mutants Q289A and R317A.
In most cases, intra- and extracellular infectivity profiles were similar, suggesting that the mutations did not affect infectious virus release. However, the P341A mutant showed a 3-log decrease in its extracellular infectivity level compared to wild-type infectivity, while its intracellular infectivity was reduced by only 1 log at 96 h postelectroporation. This result is in favor of an effect of this mutation on the secretion of infectious virus.
These initial data show that most mutations in the putative fusion peptide and α2-helix regions result in a loss of infectivity or a severe attenuation, suggesting that these regions are important for the HCV life cycle (Fig. 3).
FIG 3.
Effect of mutations on extracellular and intracellular infectivities. Viral RNA transcribed from JFH1-derived mutants was electroporated into Huh-7 cells. The infectivities of the supernatants and intracellular viruses were determined at 48, 72, and 96 h postelectroporation by titration. Error bars indicate standard errors of the means from at least three independent experiments. Values were compared to the wild-type virus. Differences were considered statistically significant for the extracellular infectivity of mutants G278A, D279A, G282A, L286A, Q302A, E303A Y309A, G311A, T314A, G315A, H316A, M318A, W320A, D321A, M322A, W326A, R339A, and P341A (P < 0.05) and for the intracellular infectivity of mutants G278A, D279A, G282A, L286A, Q302A, E303A Y309A, G311A, T314A, G315A, H316A, M318A, W320A, D321A, M322A, R339A, and P341A (P < 0.05) at 96 h postinfection.
Effect of E1 mutations on virion release.
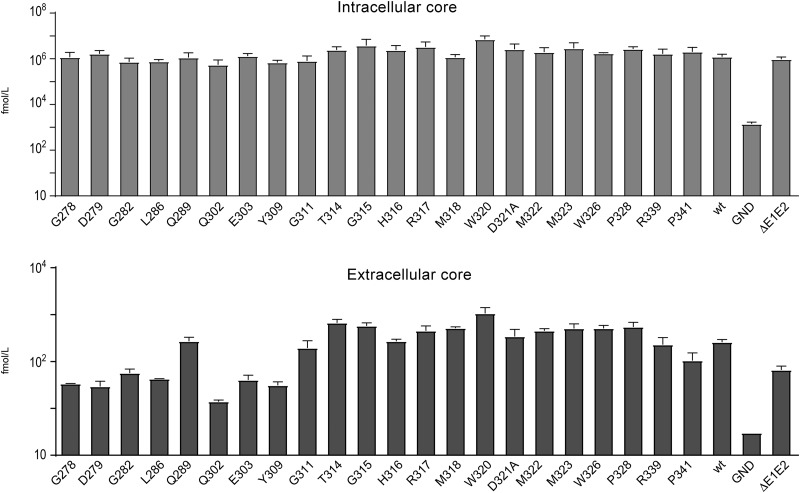
To determine whether the mutations affected the release of viral particles, the intra- and extracellular levels of HCV core protein at 48 h postelectroporation were quantified. For all mutants, the level of intracellular core protein was similar to the wild-type virus, confirming the absence of effect of E1 mutations on viral replication. In contrast, the levels of extracellular core proteins were reduced for most mutants in the potential fusion peptide region that presented impaired infectivity, as well as for the severely attenuated P341A mutant, indicating a defect in the secretion or assembly of viral particles (Fig. 4). Interestingly, most of the mutations in or close to the E1 α2 helix (G311A, T314A, G315A, H316A, M318A, W320A, D321A, and M322A) affecting virus infectivity had no impact on the secretion of core protein, suggesting that these mutations led to the release of noninfectious viral particles.
FIG 4.
Effects of E1 mutations on HCV core protein secretion. Huh-7 cells were electroporated with wild-type or mutant viral RNAs. The levels of core protein in supernatants and cell lysates were determined at 48 h postelectroporation. Error bars indicate standard error of the means from at least three independent experiments. Values of core protein were compared to the wild-type value. Differences were considered statistically significant for extracellular mutants G278A, D279A, G282A, L286A, Q302A, E303A Y309A, and P341A (P < 0.05).
Effect of E1 mutations on HCV glycoprotein folding and E1E2 heterodimerization.
Since E1 and E2 cooperate for their respective folding, we analyzed the effect of the mutations on the formation of E1E2 heterodimers (19). For this purpose, we performed pulldown assays using the CD81 large extracellular loop (CD81-LEL), which recognizes correctly folded E2 (Fig. 5A). E2 protein from all mutants could be precipitated by CD81-LEL, indicating that the E1 mutations had no effect on E2-folding. In the PFP segment and the downstream E1 region (aa274-309), nearly all mutations impacting infectivity affected the coprecipitation of E1. Thus, for the attenuated mutants (L286A and E303A), a lower signal on the E1 Western blot was observed after CD81-LEL pulldown. For the noninfectious mutants (G278A, D279A, Q302A, and Y309A), the E1 protein was not detectable. The impairment in E1 coprecipitation indicates that these mutations affect the interaction between E1 and E2, at least in the context of properly folded E2. These results suggest that the potential fusion peptide and the downstream E1 region (aa274-309) are involved in E1E2 interaction. Thus, the loss of infectivity of the mutants in this region might be due to the associated alteration in protein folding.
FIG 5.
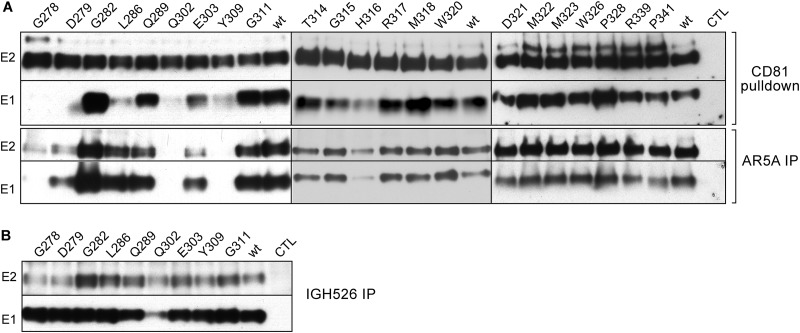
Effect of E1 mutations on E1E2 conformation. (A, upper panel) Interaction of HCV glycoproteins and CD81 (HCV entry factor). E1 and E2 from cell lysates were analyzed by GST pulldown at 48 h postelectroporation using a CD81-LEL-GST fusion protein. Pulled-down E1 and E2 were separated by SDS-PAGE and revealed by Western blotting with MAbs anti-E1 (A4) and anti-E2 (3/11). (A, lower panel, and B) Recognition of HCV E1 and E2 glycoproteins by conformation-sensitive anti-E1E2 MAb AR5A and anti-E1 MAb IGH526, as indicated. At 48 h postelectroporation, E1 and E2 proteins from cell lysates were analyzed by immunoprecipitation with MAbs AR5A and IGH526. Immunoprecipitated proteins were revealed by Western blotting using MAbs A4 and 3/11.
The G282A mutation in the PFP and the majority of the mutations in α2 and the downstream region (aa325-341) of E1 had no impact on the formation of the E1E2 heterodimer since E1 could be efficiently precipitated with E2 in CD81-LEL pulldown for all mutants, except for one (H316A) in this region. Thus, mutations G311A, T314A, G315A, M318A, W320A, D321A, M322A, M323A, W326A, P328A, R339A, and P341A, which led to the production of attenuated or noninfectious virus, had no impact on E1E2 interaction. These mutants do not present conformational or heterodimerization defects that could explain their loss of infectivity.
To further characterize the effect of E1 mutations on the folding of the E1E2 heterodimer, we performed immunoprecipitation experiments with conformation-sensitive antibodies. In a first step, we used the human monoclonal antibody (MAb) AR5A, which recognizes an epitope shared by E1 and E2 (20) (Fig. 5A). Data obtained in this assay correlated with the results of the CD81 pulldown assay. Indeed, for the noninfectious or attenuated mutants in the potential fusion peptide and downstream region (G278A, D279A, Q302A, E303A, and Y309A), E1E2 glycoproteins were either weakly or not recognized by MAb AR5A. These findings confirm that these mutations affected E1E2 conformation and that the loss of infectivity or attenuation was due to an alteration in protein folding. Unexpectedly, E1 and E2 were well recognized by the AR5A MAb for the mutant L286A, for which CD81 pulldown assay showed a weak signal for E1 coprecipitation, suggesting an effect on the interaction between E1 and E2. This might be due to a partial alteration of the affinity between E1 and E2, which is in agreement with its attenuated infectivity.
In the case of the G282A mutant in the PFP region and most of the mutants in the α2 and downstream region (G311A, T314A, G315A, M318A, W320A, D321A, M322A, M323A, W326A, P328A, R339A, and P341A), which are either attenuated or noninfectious, E1 and E2 were recognized by AR5A MAb, excluding any effect on the heterodimerization or folding for these mutations. The absence of effects on E1E2 heterodimerization and folding is in agreement with the unimpaired virus secretion observed for the mutants in the α2 helix and downstream region. Altogether, these results suggest that these mutations lead to the production of noninfectious viral particles.
For the mutants in the PFP region, E1 folding was further characterized by immunoprecipitation with the E1-specific antibody IGH-526 (Fig. 5B), which recognizes a discontinuous epitope that includes a linear region spanning residues 313 to 327 (16). Due to the overlap between α2-helix residues and the IGH-526 epitope, we could not use this antibody to characterize E1 folding for α2-helix mutants. For all tested mutants except for the mutant Q302A, the glycoprotein E1 was recognized by the MAb IGH-526, indicating that these mutations have no drastic effect on the conformation of the E1 glycoprotein. E2 coprecipitated with E1 for most mutants. However, the signal for E2 was lower for G278A, D279A, Q302A, and Y309A mutants compared to the wild type, which is in agreement with the defect in the heterodimerization of the envelope proteins observed in the CD81 pulldown assay and AR5A immunoprecipitation. In addition, this test further confirms a remaining interaction between E1 and E2 of the mutant L286A, which might be responsible for the attenuated infection observed for this mutant.
Whereas the defects in infectivity of most PFP and aa292-309 downstream region mutants can be attributed to impairment in virus assembly and envelope protein folding, the loss of infectivity of most mutants in the α2 helix and downstream region remains unexplained.
Effect of E1 mutations on HCV neutralization and inhibition by CD81.
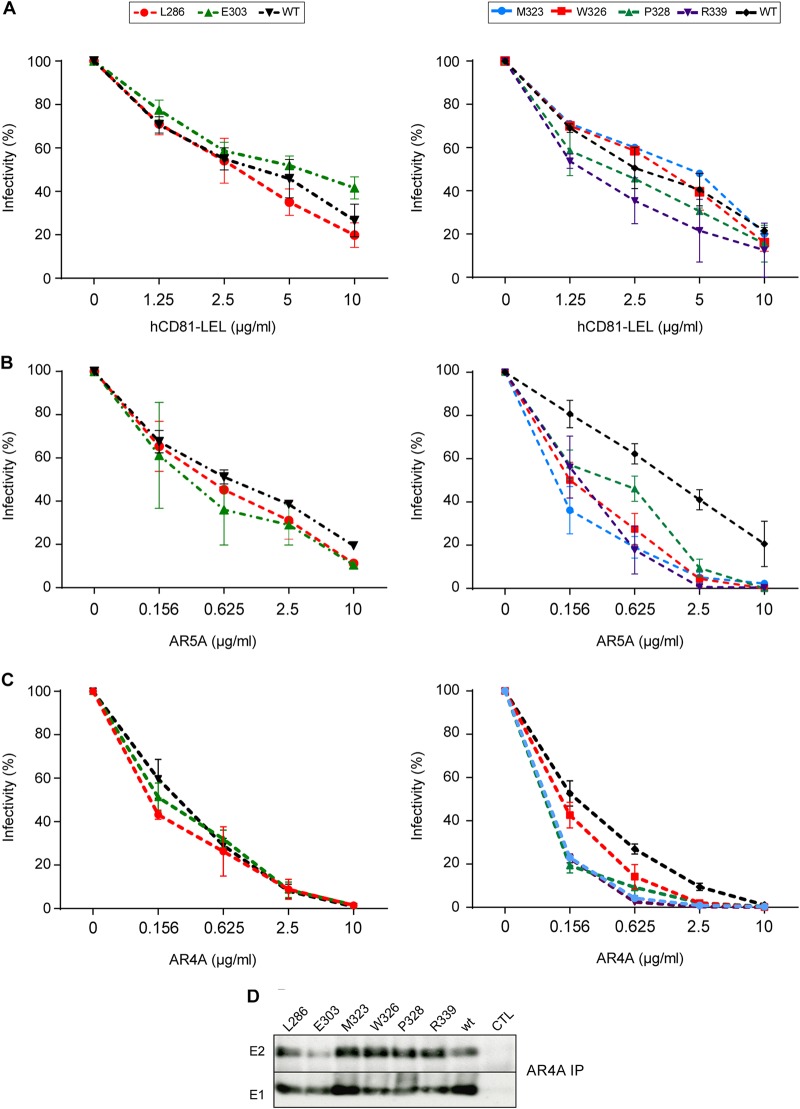
During their incorporation at the surface of viral particles, envelope glycoproteins undergo structural changes (19, 21). However, due to the low particle production yield of the HCV cell culture system, biochemical analyses of particle-associated envelope proteins are difficult to implement. Alternatively, the effect of the mutations on the folding of virus-associated envelope proteins can be determined by the analysis of the sensitivity of the virus to neutralization with the help of conformational neutralizing antibodies or CD81-LEL. However, this approach is only possible for the characterization of attenuated viruses. Thus, neutralization assays were performed with mutants showing a decrease in infectivity of ≤1 log10 (L286A, E303A, M323A, W326A, P328A, and R339A) (Fig. 6).
FIG 6.
Effect of E1 mutations on E1E2 interaction with HCV neutralizing antibodies and CD81. CD81 inhibition assays (A) and AR5A (B) and AR4A (C) neutralization experiments were carried out by incubating E1 mutants or wild-type virus with increasing concentrations of human CD81-LEL, MAb AR5A, or MAb AR4A at 37°C for 2 h. The mixture was then added to naive Huh-7 cells that were plated 1 day before. At 72 h postinfection, infectivity was determined by immunofluorescence. The values are the combined data from three independent experiments. The error bars represent standard errors of the means. Results were compared to those of the wild type and a P value of <0.05 was obtained for mutants M323A, W326A, P328A, and R339A in the AR5A and AR4A neutralization experiments. (D) Recognition of HCV E1 and E2 glycoproteins by conformation-sensitive anti-E1E2 MAb AR4A. At 48 h postelectroporation, E1 and E2 proteins from cell lysates were analyzed by immunoprecipitation with MAb AR4A. Immunoprecipitated proteins were revealed by Western blotting with MAbs A4 and 3/11.
The L286A and E303A mutants did not show any difference in sensitivity to inhibition by CD81-LEL and AR5A. This result contrasts with the effect of the L286A and E303A mutations on the heterodimer formation observed in biochemical interactions assays. Thus, although these mutations affect intracellular envelope protein heterodimerization, they have no major impact on E1E2 folding at the surfaces of the viral particles.
Conversely, α2-region mutations that had no impact on intracellular E1E2 recognition by AR5A or CD81-LEL led to an increase in virus sensitivity to inhibition by AR5A. Thus, the M323A, W326A, P328A, and R339A mutations likely induce a conformational change of virion-associated E1E2, leading to a better access of the AR5A epitope.
To further confirm the results obtained with AR5A, we used AR4A MAb, which recognizes a discontinuous epitope on E1 and E2, in neutralization and immunoprecipitation assays (20). As shown on Fig. 6D, AR4A MAb could precipitate E1 and E2 from M323A, W326A, P328A, and R339A mutants with the same efficiency as wt E1E2. As found in AR5A-mediated neutralization, AR4A inhibited the infectivity of M323A, P328A, W326A, and R339A mutants with a higher efficiency (Fig. 6C). This result further supports a specific impact of these mutations on the virus-associated E1E2 conformation.
On another hand, the recognition of E1E2 of E303A mutant by AR4A in immunoprecipitation assays was slightly affected, while L286A mutation had no impact on E1E2 recognition (Fig. 6D). This confirms the effect of the E303A mutation on the conformation of intracellular E1E2 glycoproteins, as shown by the results obtained in AR5A immunoprecipitation. As observed in AR5A neutralization experiments, E303A and L286A do not significantly affect the neutralization efficiency of AR4A (Fig. 6C). These findings support a specific impact of E303A mutation on intracellular forms of E1E2 glycoproteins.
Effect of E1 mutations on the recognition of HCV receptors.
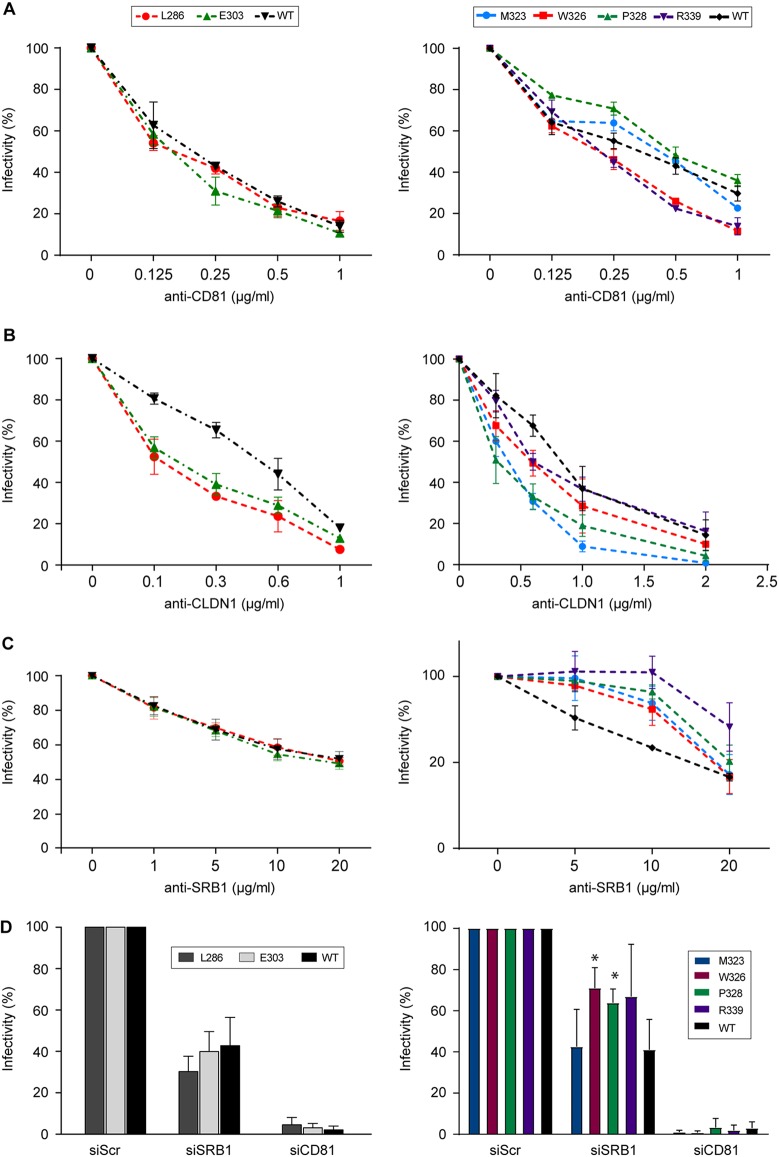
We further characterized the phenotypes of attenuated mutants by analyzing their dependence on the main known HCV receptors. For this, we determined their sensitivity to inhibition by anti-receptor MAbs, previously reported to affect HCV entry (Fig. 7).
FIG 7.
Effect of E1 mutations on the recognition of HCV receptors. Huh-7 cells were preincubated at 37°C for 2 h with increasing concentrations of antibodies targeting HCV receptors: anti-CD81 MAb JS81 (A), anti-CLDN1 MAb OM8A9-A3 (B), and anti-SR-BI MAb Cla-I (C). E1 mutants or wild-type virus were then inoculated onto the cells. At 72 h postinfection, the residual infectivity was determined by immunofluorescence. The values are the combined data from three independent experiments. The error bars represent standard errors of the means. Results were compared to those of the wild type. A P value of <0.05 was determined for mutants L286A, E303A, M323A, and P328A in the presence of anti-CLDN1 MAbs and for mutants M323A, W326A, P328A, and R339A in the presence of anti-SR-BI MAbs. (D) SRB1 or CD81 expression was downregulated by siRNA targeting SRB1 or CD81 mRNA. Infectivity is expressed as the percentage of infection performed in the presence of the control siRNA. Mean values and standard deviations from three independent experiments are shown. The unpaired t test was used to compare the infectivities of the wild-type and mutant viruses. Differences were considered statistically significant if the P value was <0.05.
No significant difference in sensitivity to inhibition by the anti-CD81 antibody was observed for the mutants. The absence of effect of the mutations on the dependence on CD81 for entry was confirmed by silencing the expression of CD81 with small interfering RNA (siRNA).
Interestingly, mutants in the α2 helix or downstream region (M323A, W326A, P328A, and R339A) were less sensitive to inhibition by the SR-BI-specific antibody, whereas the sensitivity to the SR-BI antibody was not affected for L286A and E303A mutants. Moreover, for the W326A and P328A mutants, infection was significantly less inhibited by the downregulation of SR-BI expression with siRNA. This suggests that residues M323, W326, P328, and R339 modulate HCV dependence on the SR-BI receptor.
Similar phenotypes were obtained regarding CLDN1 dependence for mutants in the PFP and α2-helix regions. Indeed, L286A (PFP), E303A (PFP downstream region), M323A (α2 helix), and P328A (α2-helix downstream region) were more sensitive to inhibition by the anti-CLDN1 MAb than the wild type, suggesting that they are more dependent on CLDN1 for entry. However, the W326A and R339A mutants showed the same sensitivity as the wild-type virus to anti-CLDN1 inhibition, suggesting that only specific residues in the α2 helix and downstream region modulate the dependence of the virus on CLDN1.
Due to the absence of an anti-OCLN MAb capable of neutralizing HCV infection, the dependence on the OCLN receptor was tested using a knockout cell line (OKH4) (22). As found for the wild-type virus, all of the mutants failed to infect the cells, indicating that the mutations have no effect on the dependence on the OCLN receptor.
Characterization of SR-BI independent mutants.
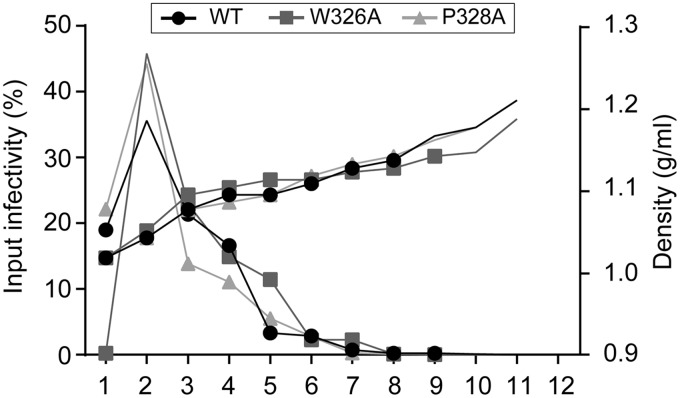
HCV associates with lipoproteins to form lipo-viro particles (23). Moreover, HCV-associated lipoproteins modulate HCV infectivity and play a role in virus interaction with SR-BI (24, 25, 26). Accordingly, E2 mutations that modulate HCV dependence on SR-BI have been associated with a shift in virion density (27, 28, 29). In this context, we sought to determine whether E1 mutations that led to a decrease in SR-BI dependence were also associated with a change in viral particle density. For this, we analyzed the density of infectious viral particles obtained for the W326A and P328A mutants. After ultracentrifugation, the distribution of infectious particles in density gradients was determined by quantification of infectivity in the different fractions. As shown in Fig. 8, no difference was observed between the distribution of infectious wild-type virus and mutant W326A and P328A viruses, which were mainly concentrated in the 1.05 density fraction. Thus, W326A and P328A mutations that affect SR-BI dependence of the virus do not appear to affect virus association with lipoproteins.
FIG 8.
Density gradient analyses of SRBI independent mutants. Concentrated supernatants of cells electroporated with HCV RNA were separated by sedimentation through a 10 to 50% iodixanol gradient. Fractions were collected from the top and analyzed for their infectivity by titration and for their density.
Characterization of noninfectious, assembly-competent E1 mutants.
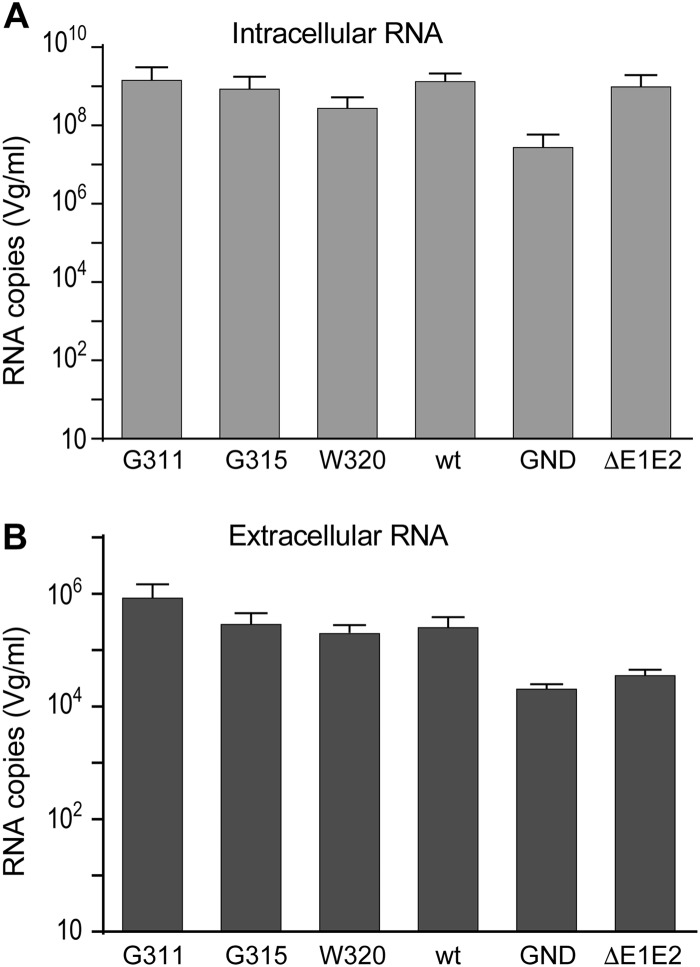
In this study, we identified several mutants that either lost their infectivity (M318A, W320A, D321A, and M322A) or were severely attenuated (G311A, T314A, and G315A) but showed a level of core release similar to the wild-type virus. Thus, these mutations did not affect viral assembly and led to the secretion of noninfectious particles. Moreover, they had no impact on E1 folding or on E1E2 heterodimerization. Since specific mutations in E1 can induce the release of viral particles devoid of genomic RNA (12), we sought to determine whether the impaired infectivity of these mutants was due to similar defects. We chose to quantify the RNA content of the particles released for the two severely attenuated mutants, G315A in the α2 helix and G311A in the upstream region, and the noninfectious mutant W320A in the α2 helix. After electroporation of viral RNA in Huh-7 cells, viral particles released in the supernatant were precipitated with polyethylene glycol, which allowed the removal of the free RNA present in the medium after electroporation. Following this step, viral RNA was extracted and quantified by quantitative reverse transcription-PCR. In parallel, intracellular viral RNA content was determined. As expected, lower levels of extracellular viral RNA were obtained for the nonreplicative GND mutant and the assembly-deficient ΔE1E2 mutant. On the opposite, viral RNA of G311A, G315A, and W320A mutants accumulated at similar levels as the wild type, both intra- and extracellularly (Fig. 9). These findings indicate that the loss of infectivity of these mutants was not due to a defect in RNA uptake in the viral particle.
FIG 9.
Effect of E1 mutations on viral RNA incorporation. Huh-7 cells were electroporated with mutants and wt RNA. (A) At 48 h postelectroporation, intracellular viral RNA was extracted and quantified by quantitative RT-PCR. In parallel, the viral particles were precipitated from the supernatant with polyethylene glycol and concentrated by ultracentrifugation. (B) Extracellular viral RNA contained in the concentrated virus was extracted and quantified by quantitative RT-PCR.
During morphogenesis, HCV glycoprotein E1 assembles to form noncovalent trimers which is essential for infectivity (21). Therefore, we determined the effect of the G311A, G315A, and W320A mutations on the capacity of E1 to form trimers in infected cells. Thus, after electroporation of viral RNA into Huh7 cells, envelope proteins from the lysate were concentrated by pulldown with Galanthus nivalis lectin and analyzed by Western blotting without thermal denaturation as previously described (21). Similar amounts of E1 trimers could be observed for the G311A, G315A, and W320A mutants than for the wt virus, indicating that the mutations do not alter E1 trimerization (data not shown).
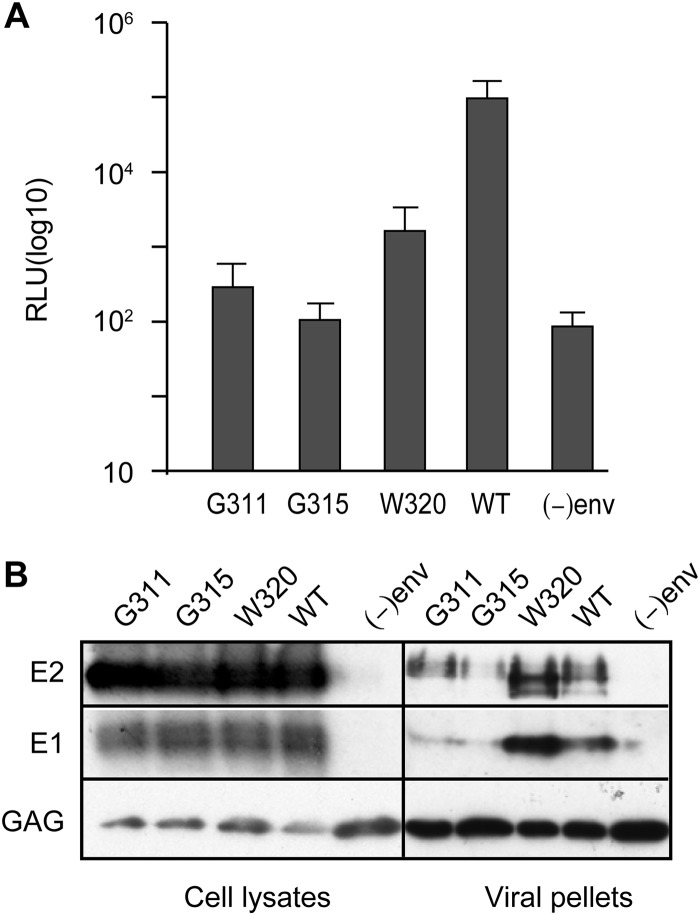
Since they did not affect envelope protein folding, core secretion, or RNA encapsidation, the G311A, G315A, and W320A mutations lead to the release of noninfectious viral particles that may be deficient in viral entry. To confirm this hypothesis, we produced retroviral particles pseudotyped with HCV envelope proteins bearing the corresponding mutations in E1. As observed in Fig. 10, infectivity of HCVpp carrying E1 mutations was reduced by 2 log compared to HCVpp wild-type infectivity. This result could be due either to a defect in particle production or to the production of noninfectious particles. We thus assessed the efficiency of the incorporation of envelope proteins in HCVpp particles. To do so, extracellular particles were concentrated on a sucrose cushion by ultracentrifugation and analyzed by Western blotting. Although similar levels of retroviral capsid proteins were observed for all viruses, smaller amounts of E1 and E2 were detected for the G311A and G315A mutants, indicating that these mutations affect E1 and E2 incorporation in HCVpp viral particles. In contrast, the W320A mutation was associated with an increased incorporation of E1 and E2 in the particles, as shown by the detection of larger amounts of E1 and E2 in the concentrated particles samples. Thus, the loss of infectivity of G311A and G315A might be due to an impaired incorporation of the envelope proteins in the viral particles, while the absence of infectivity of the W320A mutant indicates an effect of this mutation on virus entry.
FIG 10.
Effect of E1 mutations on HCVpp infectivity. Infectivity of HCVpp harboring E1 with G311A, G315A, or W320A mutation. (A) HCVpp infectivity was determined by measuring the activity of the luciferase reporter gene in infected Huh7 cells. Pseudotyped particles produced in the absence of envelope proteins were used as negative controls. The results are reported as means ± the standard deviations (error bars) of three independent experiments. A P value of <0.05 was obtained for mutants G311A, G315A, and W320A. (B) Effect of E1 mutations on the incorporation of envelope proteins in HCVpp particles. Cells producing HCVpp were lysed and analyzed by Western blotting. HCVpps contained in the supernatants of transfected 293T cells were concentrated on a 20% sucrose cushion by ultracentrifugation and analyzed by Western blotting. E1, E2, and capsid were detected using MAbs A6, 3/11, and CRL1912, respectively.
DISCUSSION
Although the precise role of E1 in the HCV life cycle has been poorly characterized until recently, the structure of E2 suggests that E1 might play a key role in the fusion process (10, 11). Moreover, the recent characterization of the role of conserved residues in the N-terminal part of the protein highlighted a role for E1 in the virus interplay with CLDN-1 and in the incorporation of viral RNA into the nucleocapsid (12). To further characterize the role played by the E1 protein during the HCV life cycle, we investigated the functional role of the C-terminal region of the ectodomain, which contains the putative fusion peptide and two α helices. A summary of the mutant phenotypes is presented in Table 1. Briefly, our results show that several residues in the PFP and the downstream region play a role in E1E2 heterodimerization, as well as in the assembly and release of infectious viral particles. Several mutations in the PFP and the α2-helix region decreased the sensitivity of the virus to neutralization by CLDN1 or SR-BI-specific antibodies, indicating a functional role for E1 in HCV-CLDN1 and HCV-SR-BI interaction. Furthermore, most mutations in the α2 region affected virus infectivity without any effect on virion assembly, suggesting that they led to the secretion of noninfectious or attenuated viruses, likely defective in virus entry.
TABLE 1.
Summary of the phenotypes of E1 mutants
| Wild type or mutant | E1 region | Infectivitya | Core secretionb | E1E2 heterodimerization and foldingc |
Infection inhibition assaysd |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CD81 PD |
IP AR5A | IP AR4A | hCD81 LEL | Anti-E1E2 |
Anti-CD81 | Anti-CLDN1 | Anti-SRBI | ||||||
| E1 | E2 | AR5A | AR4A | ||||||||||
| Wild type | +++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | |
| G278A | FP | – | – | – | ++ | – | NDe | ND | ND | ND | ND | ND | ND |
| D279A | FP | – | – | – | ++ | + | ND | ND | ND | ND | ND | ND | ND |
| G282A | FP | – | – | ++ | ++ | ++ | ND | ND | ND | ND | ND | ND | ND |
| L286A | FP | ++ | – | + | ++ | ++ | ++ | ++ | ++ | ++ | ++ | +++ | ++ |
| Q289A | FP | +++ | ++ | ++ | ++ | ++ | ND | ND | ND | ND | ND | ND | ND |
| Q302A | – | – | – | ++ | – | ND | ND | ND | ND | ND | ND | ND | |
| E303A | ++ | – | + | ++ | + | + | ++ | ++ | ++ | ++ | +++ | ++ | |
| Y309A | – | – | – | ++ | – | ND | ND | ND | ND | ND | ND | ND | |
| G311A | + | ++ | ++ | ++ | ++ | ND | ND | ND | ND | ND | ND | ND | |
| T314A | + | ++ | ++ | ++ | ++ | ND | ND | ND | ND | ND | ND | ND | |
| G315A | α2 | + | ++ | ++ | ++ | ++ | ND | ND | ND | ND | ND | ND | ND |
| H316A | α2 | + | ++ | + | ++ | + | ND | ND | ND | ND | ND | ND | ND |
| R317A | α2 | +++ | ++ | ++ | ++ | ++ | ND | ND | ND | ND | ND | ND | ND |
| M318A | α2 | – | ++ | ++ | ++ | ++ | ND | ND | ND | ND | ND | ND | ND |
| W320A | α2 | – | ++ | ++ | ++ | ++ | ND | ND | ND | ND | ND | ND | ND |
| D321A | α2 | – | ++ | ++ | ++ | ++ | ND | ND | ND | ND | ND | ND | ND |
| M322A | α2 | – | ++ | ++ | ++ | ++ | ND | ND | ND | ND | ND | ND | ND |
| M323A | α2 | ++ | ++ | ++ | ++ | ++ | ++ | ++ | +++ | +++ | ++ | +++ | – |
| W326A | ++ | ++ | ++ | ++ | ++ | ++ | ++ | +++ | +++ | ++ | ++ | – | |
| P328A | +++ | ++ | ++ | ++ | ++ | ++ | ++ | +++ | +++ | ++ | +++ | – | |
| R339A | ++ | ++ | ++ | ++ | ++ | ++ | + | +++ | +++ | ++ | ++ | – | |
| P341A | – | + | ++ | ++ | ++ | ND | ND | ND | ND | ND | ND | ND | |
The infectivity of HCVcc harboring the different E1E2 glycoproteins in the supernatant of electroporated Huh-7 cells was quantified 96 h postelectroporation (Fig. 3). +++, infectious titers higher than 104 FFU/ml; ++, infectious titers higher than 103 FFU/ml; +, infectious titers higher than 102 FFU/ml; −, titers between 0 and 50 FFU/ml.
Secretion of core Ag in the supernatant quantified at 48 h postelectroporation of Huh-7 cells (Fig. 4). ++, concentration greater than or equal to the wild type; +, concentration reduced by <1 log.; −, concentration reduced by ≥1 log.
The recognition of E1E2 proteins by AR4A and AR5A conformational antibodies and their interaction with hCD81 LEL were determined by precipitation experiments (Fig. 5). ++, similar amount of E1E2 precipitated to that of the wild type; +, lower amount of E1E2 precipitated; −, no E1E2 precipitated.
The sensitivity of the mutants to inhibition of infectivity by different antibodies or the hCD81 LEL was assessed (Fig. 6 and 7). ++, wild-type sensitivity to neutralization; +++, higher sensitivity to inhibition than the wild type; +, lower sensitivity to inhibition than the wild type.
ND, not determined.
Most mutations in the PFP region and the downstream region (G278A, D279A, L286A, Q302A, E303A, and Y309A) affect the interaction between E1 and E2. These results confirm the involvement of E1 region from aa 290 to 306 in E1E2 interaction, as observed in the computational prediction of the E1E2 heterodimer structure proposed by Freedman at al. (30). Thus, although interactions between E1 and E2 transmembrane domains are essential for E1E2 heterodimerization, several residues in their ectodomains also contribute to the interaction. Indeed, N-terminal residues 201 to 206 are essential for the structure of the AR5A and AR4A antibody epitope, which spans the E1 and E2 proteins (20). Recently, Gopal et al. generated a library of alanine scanning mutants covering the full E1E2 sequence of the genotype 1a H77 strain (31). Using a high-throughput flow cytometry-based assay, these researchers probed the mutants library for binding to a collection of antibodies. In agreement with our data, this approach revealed a decrease in the recognition of mutants G278A, D279A, Q302A, and D303A by AR5A, whereas mutants G282A, L286A, Q289A, and G311A were unaffected. Moreover, the characterization of the functionality of chimeric heterodimers derived from different genotypes allowed the identification of residues at position 308, 330, and 345 as being involved in the functional interaction between E1 and E2 (32). In addition, we previously identified three residues (W239, I262, and D263) located in the β-sheet structure of the N-terminal part of E1 that are involved in E1E2 interaction (12). Among the mutants in the PFP region that presented a deficiency in infectivity, only G282A was not affected in its capacity to interact with E2. However, this mutation led to an impairment in virion assembly that might explain the loss of infectivity of this mutant. It is worth noting that the functionality of this residue was characterized in the HCVpp system of genotypes 2a and 1a (33, 34), and in a cell culture model allowing the transcomplementation of E1 in the HCV genome lacking the E1 coding sequence. In these contexts, the mutation led to a severe decrease in infectivity. Furthermore, in the E1 transcomplementation HCV system, lower levels of extracellular viral RNA were obtained for this mutant, which supports an effect of the mutation on virus assembly.
Several attenuated mutants in the PFP (L286A), in the downstream region (E303A), and in the α2 region (M323A and P328A) exhibited an increased sensitivity to neutralization of infectivity by CLDN1-specific antibodies, suggesting that these mutants are more dependent on CLDN1 for cellular entry. This result is in line with the potential involvement of E1 in HCV-CLDN1 interplay, as previously observed (12, 35). However, the E1 mutations reported until now (T213A, I262A, and H316N) had an opposite effect on CLDN1 dependence, since they led to a decreased sensitivity of the virus to the inhibitory effect of anti-CLDN1 antibodies while increasing its dependence on CLDN6 for entry. Thus, it seems that different regions of E1 have opposite effects on the requirement of CLDN1 for HCV entry. Moreover, these results strengthen the hypothesis of the involvement of E1 in HCV particle interaction with the CLDN1 coreceptor. Whereas some E1 mutations have been shown to affect the binding of HCVpp to CLDN1-expressing cells, no direct interaction between E1 and CLDN1 has been reported until now. Alternatively, E1 could modulate the affinity of E2 for CLDN1 as was reported for the E2-CD81 interaction (12, 19).
Several mutations in the α2 region (M323A, W326A, and P328A) affect the dependence of HCV on SR-BI for entry. Indeed, these mutants were found to be partially resistant to inhibition of the infection by an SR-BI-specific antibody or by siRNA treatment. Several mutations in E2 (G451R, V514A, and the murine CD81-adapted HCV mutant Jc1/mCD81), as well as the deletion of HVR1 have been previously shown to decrease the dependence of the virus on SR-BI (7, 27, 28). At the same time, these variants exhibited an increased affinity for CD81. In contrast, the M323A, W326A, and P328A E1 mutants had no significant effect on the dependence of the virus on CD81, but M323A and P328A presented an increased dependence on CLDN1. Therefore, it seems that the alteration of the interaction between the envelope proteins and a receptor frequently impacts their affinity for other entry receptors. These findings are in agreement with the finely regulated process of HCV entry that sequentially involves SR-BI, CD81, CLDN1, and OCLN (reviewed in reference 32). SR-BI plays a role at several steps of the entry process (27, 36). It is notably thought to contribute to virus attachment through interaction with virus-associated lipoproteins. Its lipid transfer activity has been also shown to be important for productive viral entry. In agreement with the functional interaction of SR-BI with HCV-associated lipoproteins, several E2 mutations that reduce SR-BI dependence were found to be associated with an increase in viral particle density (27). This density shift is supposed to reflect a change in the lipid content of HCV particles and has been suggested to explain the lower dependence of this mutant on SR-BI for entry. Interestingly, cysteine mutations in E1 (C207A and C272A) have also been shown to affect the density of infectious viral particles, suggesting that the E1E2 heterodimer influences the interplay between HCV and lipoproteins. In line with this result, E1 and E2 have been shown to interact with ApoE, whose expression is essential to the production of infectious viral particles (37–39). In the case of the W326A and P328A mutants, no change in the density of the particle could be observed, which suggests that other parameters might influence the interaction of the virus with SR-BI. Alternatively, E1 mutations might modulate the interaction of E2 with SR-BI, as previously reported for its interaction with CD81 (19).
Most mutations in the α2-helix region of E1 led to a severe decrease or a loss of infectivity without affecting E1E2 heterodimerization nor the viral particle assembly. It is worth noting that the absence of effect of most mutations in α2-helix region on E1E2 conformation had been previously observed in the characterization of the binding of E1E2-specific antibodies to an alanine scanning E1E2 mutants library (31). Further characterization of the attenuated mutants (M323A, W326A, and P328A) revealed a change in their dependence on CLDN1 and SR-BI for entry. These results suggest that the E1 α2-helix region is important for the interplay of HCV with SR-BI and CLDN1 receptors during the entry process. For the severely attenuated mutants G311A and G315A, we observed a deficiency in the incorporation of the envelope proteins in the HCVpp model that could explain the effect of these mutations on infectivity. Due to the difficulties of producing noninfectious cell culture-derived HCV (HCVcc) particles in great amounts, we could not determine the level of E1E2 incorporation in HCVcc particles for these mutants. In contrast, the characterization of the noninfectious W320A mutant did not reveal any deficiency that could explain its loss of infectivity. Indeed, this mutation had no effect on E1E2 heterodimerization, particle assembly, E1 trimerization, or viral RNA encapsidation. These results and the absence of infectivity observed for this mutant in the HCVpp system support an effect of the mutation on the virus's entry step. Unfortunately, our attempts to further determine the step of entry that was impaired were unsuccessful due to the low production of particles. Finally, our data would be in agreement with the direct or indirect involvement of the α2 region in the fusion step of the entry, as suggested by the membranotropic properties of this region (15).
Interestingly, the functional characterization of the IGH526 epitope (16) revealed that, similarly to what we found in HCVcc, the G315A, M318A, D321A, and M322A mutations dramatically affected the infectivity of genotype 1a HCVpp. However, introduction of W320A mutation in HCVpp of genotype 1a only resulted in a 30% decrease of infectivity. The more pronounced decrease of infectivity observed for this mutant in HCVcc and HCVpp 2a could be due to the difference of sequences between genotypes 1a and 2a envelope proteins that may modulate the functional impact of the mutations. The potential involvement of α2-helix region in fusion would suggest that IGH526 inhibits HCV infection by targeting the fusion step as it has been reported for some HIV and influenza viruses neutralizing antibodies (16, 40).
On the other hand, our data revealed that the PFP region is important for E1 and E2 interaction, as well as for virus assembly, which does not exclude an involvement of this region in the fusion step between the viral and cell membrane. Indeed, while mutations in the fusion peptide of the viral fusion protein lead to a loss of activity and a consequent loss of virus entry, they can also affect additional steps of the viral life cycle. For instance, mutations in the fusion peptide of the Semliki Forest virus have been shown to affect the interaction between the two envelope proteins, as well as the assembly of the virus (41, 42). In addition, mutation of one glycine residue in the fusion peptide of the influenza virus can result in a variety of phenotypes, depending on the nature of the substituted residue (43). However, the fact that mutations in the PFP region affect different steps of the HCV life cycle complicates the characterization of the role of this region in the entry and fusion steps. Thus, in addition to the potential involvement of PFP in fusion suggested in previous studies (13, 14), our results suggest that the E1 α2 helix also participates in this entry step. The contribution of several E1 regions to fusion is in line with the fact that fusion is a complex process that involves several membranotopic segments of the envelope proteins. While the fusion peptide is responsible for the first step of fusion, additional envelope proteins segments have been shown to be involved at later stages of fusion, such as pore formation or enlargement (44–46). As found for other enveloped viruses, the resolution of the E1 crystal structure would provide crucial elements to definitively identify the fusion peptide of HCV.
To conclude, our results exemplify the important roles played by E1 at different stages of the HCV life cycle, including involvement in E1E2 assembly, in virus morphogenesis, in the interplay with HCV receptors, and potentially in the fusion step.
MATERIALS AND METHODS
Cell culture.
Huh-7 human hepatoma cells (47) were grown in Dulbecco modified essential medium (Thermo Fisher), supplemented with GlutaMAX, 10% fetal calf serum, and nonessential amino acids.
Antibodies.
Anti-HCV MAbs A4 (anti-E1) (48) and 3/11 (anti-E2; kindly provided by J. A. McKeating, University of Birmingham, Birmingham, United Kingdom) (49) were generated in vitro by using a MiniPerm apparatus (Heraeus) as recommended by the manufacturer. Anti-E1E2 MAbs AR4A and AR5A (20) and anti-E1 MAb IGH526 (16) were kindly provided by M. Law (Scripps Research Institute, La Jolla, CA). The anti-NS5A MAb 9E10 (50) was a gift from C. M. Rice (Rockefeller University, New York, NY) and a polyclonal antibody raised against NS5A (51) was kindly provided by M. Harris (University of Leeds, Leeds, United Kingdom). The MAb A6 (anti-E1) has been previously described (52). Anti-CLDN1 MAb OM8A9-A3 has been described before (53). Anti-CD81 MAb JS81 (BD Pharmingen), anti-SR-BI MAb Cla-I (BD Biosciences), and anti-tubulin (Sigma) are commercially available, as well as the secondary antibodies used for immunofluorescence, which were purchased from Jackson ImmunoResearch. Anti-capsid of murine leukemia virus (ATCC CRL1912) was produced in vitro by using a MiniPerm apparatus (Heraeus) as recommended by the manufacturer.
Mutagenesis and virus production.
The virus used in the present study is a modified version of the JFH1 isolate (genotype 2a; GenBank accession number AB237837) (54), kindly provided by T. Wakita (National Institute of Infectious Diseases, Tokyo, Japan). It was engineered to reconstitute the A4 epitope in E1 (pJFH1-CS-A4) (17) and titer-enhancing mutations (55). Mutants were generated by site-directed mutagenesis and the selected conserved residues were replaced by alanine. Viral RNAs were produced by in vitro transcription as previously reported (56). Viruses were produced by electroporation of viral RNA into Huh7 cells as previously described (12). The controls used in this study are the GND mutant, a nonreplicative control of the HCV genome containing a GND mutation in the NS5B active site (54) and the ΔE1E2 mutant, an assembly-deficient control, which contains an in-frame deletion introduced into the E1E2 regions (54).
Infectivity assays.
Intra- and extracellular infectivities were determined as described previously (56). In brief, viral RNAs were electroporated into Huh-7 cells. Supernatants containing extracellular virus were collected at different time points (48, 72, and 96 h) after electroporation, and cell debris was eliminated by centrifugation for 5 min at 10,000 × g. To obtain intracellular viral particles, infected cells were washed with phosphate-buffered saline (PBS) and harvested after trypsinization, which was then followed by four freeze-thaw cycles. Cell lysates were then clarified by centrifugation at 10,000 × g for 7 min. The clarified supernatants containing extracellular virus or intracellular virus were used for infection of naive Huh-7 cells. Infected cells were later fixed with ice-cold methanol (100%) and immunostained with A4 anti-E1 antibody.
Immunofluorescence.
Immunofluorescence analyses were performed as previously described (57). Briefly, after fixation of Huh-7 infected cells with cold methanol (100%) for 10 min, cells were washed twice with PBS and incubated in 10% goat serum for 10 min. The primary anti-E1 antibody A4 was diluted in 10% goat serum, and the coverslips were incubated with the antibody for 25 min at room temperature. The cells were then washed three times with PBS. The secondary Cy3-conjugated antibody diluted in goat serum (1/500) was incubated with the cells for 20 min. The cells were washed again with PBS. Nuclei were stained with DAPI (4′,6′-diamidino-2-phenylindole). Images were processed by using ImageJ software.
Equilibrium density gradient analysis.
Equilibrium density gradient analyses were performed as described previously (19) after polyethylene glycol (PEG) precipitation of viral preparations (58). Briefly, supernatants were collected at 48 h after electroporation of Huh7 cells. Approximately 80 ml of viral supernatant were precipitated using PEG 6000 at a final concentration of 8%. The mixture was incubated overnight at 4°C and centrifuged for 25 min at 8,000 rpm (Beckman JLA-10.5 rotor), and the pellet was resuspended in 1 ml of sterile PBS. The concentrated viral solution was then loaded onto a 10 to 50% continuous iodixanol gradient. The gradients were spun for 16 h at 36,000 rpm in a SW41 rotor (Beckman). Fractions (1 ml) were collected from the top of each tube and analyzed for their infectivity and density.
HCV core protein quantification.
HCV core protein was quantified by a fully automated chemiluminescent microparticle immunoassay according to the manufacturer's instructions (Architect HCVAg; Abbott, Germany) (59).
Western blotting.
Western blotting experiments were performed as previously described (12). Cells were lysed in PBS lysis buffer (1% Triton X-100 and protease inhibitor cocktail [Roche]). Cell lysates were then precleared by centrifugation at 14,000 × g for 10 min at 4°C. Protein samples were heated for 7 min at 70°C in Laemmli sample buffer, followed by separation by SDS-PAGE. The proteins were then transferred onto nitrocellulose membranes (Hybond-ECL; Amersham) and detected with specific primary antibodies, which was followed by incubation with the corresponding peroxidase-conjugated anti-rat (Jackson), anti-sheep (Amersham), or anti-human or anti-mouse (Dako) antibodies. Detection of proteins was done by using enhanced chemiluminescence (Amersham) as recommended by the manufacturer.
CD81 interaction and immunoprecipitation assays.
CD81 pulldown and immunoprecipitation experiments were performed as previously described (7). Cells were lysed in lysis buffer (1% Triton X-100 and protease inhibitor cocktail [Roche] in PBS). Cell lysates were then cleared by centrifugation at 14,000 × g for 15 min at 4°C. For CD81 pulldown, glutathione-Sepharose beads (glutathione-Sepharose 4B; Amersham Bioscience) were washed twice with cold PBS to remove the storage buffer. For each cell lysate sample, 50 μl of glutathione beads was incubated with 10 μg of human CD81 (hCD81) large extracellular loop (LEL) glutathione S-transferase (GST) recombinant protein in 1 ml of cold PBS containing 1% Triton X-100 for 2 h at 4°C. After incubation, glutathione-Sepharose beads were washed with cold PBS. Cell lysates containing E1E2 proteins were then incubated with the glutathione beads-CD81-LEL complex overnight at 4°C. The next day, the beads were washed five times with cold PBS–1% Triton X-100 and then finally resuspended in 30 μl of Laemmli buffer and heated at 70°C for 10 min. Samples were loaded onto 10% SDS-PAGE gels, and HCV envelope glycoproteins were revealed by Western blotting. For immunoprecipitation assays, 70 μl of protein A-agarose beads were incubated with 10 μg of rabbit anti-human IgG (Dako) in 1 ml of cold PBS–1% Triton X-100 for 2 h at 4°C. Meanwhile, 100 μl of cell lysates were incubated with 2 μg of MAb AR5A (anti-E1E2) or MAb IGH526 (anti-E1) in 400 μl of cold PBS–1% Triton X-100 for 2 h at 4°C. After incubation, the agarose beads were washed twice with cold PBS–1% Triton X-100 and added to cell lysates. The mixture was incubated for 90 min at 4°C, which was followed by washing the beads five times with cold PBS–1% Triton X-100. Finally, the beads were resuspended in 30 μl of Laemmli buffer. The presence of HCV envelope glycoproteins was then detected by Western blotting.
Entry inhibition assays and neutralization assays.
Viruses or cells were preincubated with human CD81-LEL, MAb AR5A, or anti-receptor antibody for 2 h at 37°C. The viruses were then inoculated onto Huh-7 cells. At 6 h postinfection, the inoculum was removed, and the cells were further incubated for 72 h with complete medium. The cells were then processed for immunofluorescence to measure residual infectivity.
HCVpp assay.
HCVpp were produced as described previously (6). Briefly, 293T cells seeded for 1 day in 6-well plates were cotransfected with 300 ng/well of a pcDNA plasmid expressing HCV envelope glycoproteins (JFH1), 300 ng of a murine leukemia virus Gag-Pol expression packaging vector, and 400 ng of a firefly luciferase reporter transfer vector. Plasmid containing no envelope protein sequence was used as a negative control. After transfection, the cells were incubated for 48 h at 37°C. Supernatants containing the pseudoparticles were then harvested and filtered through 0.45-μm-pore-size membranes to be used as HCVpps in infection assays or pelleted by ultracentrifugation through a 20% sucrose cushion at 27,000 rpm (Beckman, type SW41 rotor) for 4 h at 4°C and analyzed by Western blotting. To minimize artifacts that might be caused by differences in the quality of preparations, each experiment was performed using concurrently produced pseudoparticles (60). Infectivity of HCVpp on target Huh-7 cells was assessed after 72 h by using a firefly luciferase reporter gene activity kit (Promega), as recommended by the manufacturer. The results are presented as means ± the standard deviations of results of three independent experiments.
Graphs and statistics.
Prism software (version 5.0c; GraphPad, Inc., La Jolla, CA) was used for creating graphs and to determine statistical significance of differences between data sets using a Mann-Whitney test.
ACKNOWLEDGMENTS
We thank F. L. Cosset, M. Harris, M. Law, J. McKeating, C. Rice, and T. Wakita for providing essential reagents. We also thank Sophana Ung for help in preparing the figures. The immunofluorescence analyses were performed with the help of the imaging core facility of the BioImaging Center Lille Nord-de-France.
This study was supported by the French National Agency for Research on AIDS and Viral Hepatitis (ANRS) and the ANR through the ERA-NET Infect-ERA program (ANR-13-IFEC-0002-01). J.G.H. was successively supported by a fellowship from the Lebanese Development Association and from the ANRS. R.I.M. was supported by a fellowship from the Ministry of Higher Education of Egypt.
R.I.M., J.G.H., L.L., X.H., T.F.B., G.D., P.M., J.D., and M.L. conceived and designed the experiments. R.I.M., J.G.H., L.L., X.H., V.D., A.A.M., and M.L. performed the experiments. A.A.M., T.F.B., and P.M. provided reagents. R.I.M., J.G.H., X.H., J.D., and M.L. analyzed the data. R.I.M., J.G.H., J.D., and M.L. wrote the paper.
REFERENCES
- 1.Messina JP, Humphreys I, Flaxman A, Brown A, Cooke GS, Pybus OG, Barnes E. 2015. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology 61:77–87. doi: 10.1002/hep.27259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taherkhani R, Farshadpour F. 2017. Global elimination of hepatitis C virus infection: progresses and the remaining challenges. World J Hepatol 9:1239–1252. doi: 10.4254/wjh.v9.i33.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simmonds P. 2013. The origin of hepatitis C virus, p 1–15. In Bartenschlager R. (ed), Hepatitis C virus: from molecular virology to antiviral therapy. Springer, Berlin, Germany. [Google Scholar]
- 4.Moradpour D, Penin F. 2013. Hepatitis C virus proteins: from structure to function, p 113–142. In Bartenschlager R. (ed), Hepatitis C virus: from molecular virology to antiviral therapy. Springer, Berlin, Germany. [Google Scholar]
- 5.Cocquerel L, Wychowski C, Minner F, Penin F, Dubuisson J. 2000. Charged residues in the transmembrane domains of hepatitis C virus glycoproteins play a major role in the processing, subcellular localization, and assembly of these envelope proteins. J Virol 74:3623–3633. doi: 10.1128/JVI.74.8.3623-3633.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartosch B, Dubuisson J, Cosset F-L. 2003. Infectious hepatitis C virus pseudo-particles containing functional E1–E2 envelope protein complexes. J Exp Med 197:633–642. doi: 10.1084/jem.20021756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lavie M, Sarrazin S, Montserret R, Descamps V, Baumert TF, Duverlie G, Séron K, Penin F, Dubuisson J. 2014. Identification of conserved residues in hepatitis C virus envelope glycoprotein E2 that modulate virus dependence on CD81 and SRB1 entry factors. J Virol 88:10584–10597. doi: 10.1128/JVI.01402-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Douam F, Lavillette D, Cosset F-L. 2015. The mechanism of HCV entry into host cells, p 63–107. In Klasse PJ. (ed), Progress in molecular biology and translational science. Academic Press, Inc, New York, NY. [DOI] [PubMed] [Google Scholar]
- 9.Lavie M, Penin F, Dubuisson J. 2015. HCV envelope glycoproteins in evirion assembly and entry. Future Virol 10:297–312. doi: 10.2217/fvl.14.114. [DOI] [Google Scholar]
- 10.Kong L, Giang E, Nieusma T, Kadam RU, Cogburn KE, Hua Y, Dai X, Stanfield RL, Burton DR, Ward AB, Wilson IA, Law M. 2013. Hepatitis C virus E2 envelope glycoprotein core structure. Science 342:1090–1094. doi: 10.1126/science.1243876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khan AG, Whidby J, Miller MT, Scarborough H, Zatorski AV, Cygan A, Price AA, Yost SA, Bohannon CD, Jacob J, Grakoui A, Marcotrigiano J. 2014. Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2. Nature 509:381–384. doi: 10.1038/nature13117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haddad JG, Rouillé Y, Hanoulle X, Descamps V, Hamze M, Dabboussi F, Baumert TF, Duverlie G, Lavie M, Dubuisson J. 2017. Identification of novel functions for hepatitis C virus envelope glycoprotein E1 in virus entry and assembly. J Virol 91:e00048-. doi: 10.1128/JVI.00048-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drummer HE, Boo I, Poumbourios P. 2007. Mutagenesis of a conserved fusion peptide-like motif and membrane-proximal heptad-repeat region of hepatitis C virus glycoprotein E1. J Gen Virol 88:1144–1148. doi: 10.1099/vir.0.82567-0. [DOI] [PubMed] [Google Scholar]
- 14.Garry RF, Dash S. 2003. Proteomics computational analyses suggest that hepatitis C virus E1 and pestivirus E2 envelope glycoproteins are truncated class II fusion proteins. Virology 307:255–265. doi: 10.1016/S0042-6822(02)00065-X. [DOI] [PubMed] [Google Scholar]
- 15.Spadaccini R, D'Errico G, D'Alessio V, Notomista E, Bianchi A, Merola M, Picone D. 2010. Structural characterization of the transmembrane proximal region of the hepatitis C virus E1 glycoprotein. Biochim Biophys Acta Biomembr 1798:344–353. doi: 10.1016/j.bbamem.2009.10.018. [DOI] [PubMed] [Google Scholar]
- 16.Kong L, Kadam RU, Giang E, Ruwona TB, Nieusma T, Culhane JC, Stanfield RL, Dawson PE, Wilson IA, Law M. 2015. Structure of hepatitis C virus envelope glycoprotein E1 antigenic site 314-324 in complex with antibody IGH526. J Mol Biol 427:2617–2628. doi: 10.1016/j.jmb.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goueslain L, Alsaleh K, Horellou P, Roingeard P, Descamps V, Duverlie G, Ciczora Y, Wychowski C, Dubuisson J, Rouillé Y. 2010. Identification of GBF1 as a cellular factor required for hepatitis C virus RNA replication. J Virol 84:773–787. doi: 10.1128/JVI.01190-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duvet S, Op De Beeck A, Cocquerel L, Wychowski C, Cacan R, Dubuisson J. 2002. Glycosylation of the hepatitis C virus envelope protein E1 occurs posttranslationally in a mannosylphosphoryldolichol-deficient CHO mutant cell line. Glycobiology 12:95–101. doi: 10.1093/glycob/12.2.95. [DOI] [PubMed] [Google Scholar]
- 19.Wahid A, Helle F, Descamps V, Duverlie G, Penin F, Dubuisson J. 2013. Disulfide bonds in hepatitis C virus glycoprotein E1 control the assembly and entry functions of E2 glycoprotein. J Virol 87:1605–1617. doi: 10.1128/JVI.02659-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giang E, Dorner M, Prentoe JC, Dreux M, Evans MJ, Bukh J, Rice CM, Ploss A, Burton DR, Law M. 2012. Human broadly neutralizing antibodies to the envelope glycoprotein complex of hepatitis C virus. Proc Natl Acad Sci U S A 109:6205–6210. doi: 10.1073/pnas.1114927109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falson P, Bartosch B, Alsaleh K, Tews BA, Loquet A, Ciczora Y, Riva L, Montigny C, Montpellier C, Duverlie G, Pécheur E-I, le Maire M, Cosset F-L, Dubuisson J, Penin F. 2015. Hepatitis C virus envelope glycoprotein E1 forms trimers at the surface of the virion. J Virol 89:10333–10346. doi: 10.1128/JVI.00991-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shirasago Y, Shimizu Y, Tanida I, Suzuki T, Suzuki R, Sugiyama K, Wakita T, Hanada K, Yagi K, Kondoh M, Fukasawa M. 2016. Occludin-knockout human hepatic Huh7.5.1-8-derived cells are completely resistant to hepatitis C virus infection. Biol Pharm Bull 39:839–848. doi: 10.1248/bpb.b15-01023. [DOI] [PubMed] [Google Scholar]
- 23.Bartenschlager R, Penin F, Lohmann V, André P. 2011. Assembly of infectious hepatitis C virus particles. Trends Microbiol 19:95–103. doi: 10.1016/j.tim.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 24.Andréo U, Maillard P, Kalinina O, Walic M, Meurs E, Martinot M, Marcellin P, Budkowska A. 2007. Lipoprotein lipase mediates hepatitis C virus (HCV) cell entry and inhibits HCV infection. Cell Microbiol 9:2445–2456. doi: 10.1111/j.1462-5822.2007.00972.x. [DOI] [PubMed] [Google Scholar]
- 25.Chang K-S, Jiang J, Cai Z, Luo G. 2007. Human apolipoprotein E is required for infectivity and production of hepatitis C virus in cell culture. J Virol 81:13783–13793. doi: 10.1128/JVI.01091-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maillard P, Huby T, Andréo U, Moreau M, Chapman J, Budkowska A. 2006. The interaction of natural hepatitis C virus with human scavenger receptor SR-BI/Cla1 is mediated by ApoB-containing lipoproteins. FASEB J 20:735–737. doi: 10.1096/fj.05-4728fje. [DOI] [PubMed] [Google Scholar]
- 27.Grove J, Nielsen S, Zhong J, Bassendine MF, Drummer HE, Balfe P, McKeating JA. 2008. Identification of a residue in hepatitis C virus E2 glycoprotein that determines scavenger receptor BI and CD81 receptor dependency and sensitivity to neutralizing antibodies. J Virol 82:12020–12029. doi: 10.1128/JVI.01569-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bankwitz D, Steinmann E, Bitzegeio J, Ciesek S, Friesland M, Herrmann E, Zeisel MB, Baumert TF, Keck Z, Foung SKH, Pécheur E-I, Pietschmann T. 2010. Hepatitis C virus hypervariable region 1 modulates receptor interactions, conceals the CD81 binding site, and protects conserved neutralizing epitopes. J Virol 84:5751–5763. doi: 10.1128/JVI.02200-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prentoe J, Jensen TB, Meuleman P, Serre SBN, Scheel TKH, Leroux-Roels G, Gottwein JM, Bukh J. 2011. Hypervariable region 1 differentially impacts viability of hepatitis C virus strains of genotypes 1 to 6 and impairs virus neutralization. J Virol 85:2224–2234. doi: 10.1128/JVI.01594-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freedman H, Logan MR, Hockman D, Koehler Leman J, Law JLM, Houghton M. 2017. Computational prediction of the heterodimeric and higher-order structure of gpE1/gpE2 envelope glycoproteins encoded by hepatitis C virus. J Virol 91:e02309-. doi: 10.1128/JVI.02309-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gopal R, Jackson K, Tzarum N, Kong L, Ettenger A, Guest J, Pfaff JM, Barnes T, Honda A, Giang E, Davidson E, Wilson IA, Doranz BJ, Law M. 2017. Probing the antigenicity of hepatitis C virus envelope glycoprotein complex by high-throughput mutagenesis. PLoS Pathog 13:e1006735. doi: 10.1371/journal.ppat.1006735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Douam F, Dao Thi VL, Maurin G, Fresquet J, Mompelat D, Zeisel MB, Baumert TF, Cosset F-L, Lavillette D. 2014. Critical interaction between E1 and E2 glycoproteins determines binding and fusion properties of hepatitis C virus during cell entry. Hepatology 59:776–788. doi: 10.1002/hep.26733. [DOI] [PubMed] [Google Scholar]
- 33.Lavillette D, Pécheur E-I, Donot P, Fresquet J, Molle J, Corbau R, Dreux M, Penin F, Cosset F-L. 2007. Characterization of fusion determinants points to the involvement of three discrete regions of both E1 and E2 glycoproteins in the membrane fusion process of hepatitis C virus. J Virol 81:8752–8765. doi: 10.1128/JVI.02642-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tong Y, Chi X, Yang W, Zhong J. 2017. Functional analysis of hepatitis C virus (HCV) envelope protein E1 using a trans-complementation system reveals a dual role of a putative fusion peptide of E1 in both HCV entry and morphogenesis. J Virol 91:e02468-16. doi: 10.1128/JVI.02468-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hopcraft SE, Evans MJ. 2015. Selection of a hepatitis C virus with altered entry factor requirements reveals a genetic interaction between the E1 glycoprotein and claudins. Hepatology 62:1059–1069. doi: 10.1002/hep.27815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dao Thi VL, Granier C, Zeisel MB, Guérin M, Mancip J, Granio O, Penin F, Lavillette D, Bartenschlager R, Baumert TF, Cosset F-L, Dreux M. 2012. Characterization of hepatitis C virus particle subpopulations reveals multiple usage of the scavenger receptor BI for entry steps. J Biol Chem 287:31242–31257. doi: 10.1074/jbc.M112.365924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boyer A, Dumans A, Beaumont E, Etienne L, Roingeard P, Meunier J-C. 2014. The association of hepatitis C virus glycoproteins with apolipoproteins E and B early in assembly is conserved in lipoviral particles. J Biol Chem 289:18904–18913. doi: 10.1074/jbc.M113.538256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee J-Y, Acosta EG, Stoeck IK, Long G, Hiet M-S, Mueller B, Fackler OT, Kallis S, Bartenschlager R. 2014. Apolipoprotein E Likely contributes to a maturation step of infectious hepatitis C virus particles and interacts with viral envelope glycoproteins. J Virol 88:12422–12437. doi: 10.1128/JVI.01660-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mazumdar B, Banerjee A, Meyer K, Ray R. 2011. Hepatitis C virus E1 envelope glycoprotein interacts with apolipoproteins in facilitating entry into hepatocytes. Hepatology 54:1149–1156. doi: 10.1002/hep.24523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hashem AM, Van Domselaar G, Li C, Wang J, She Y-M, Cyr TD, Sui J, He R, Marasco WA, Li X. 2010. Universal antibodies against the highly conserved influenza fusion peptide cross-neutralize several subtypes of influenza A virus. Biochem Biophys Res Commun 403:247–251. doi: 10.1016/j.bbrc.2010.11.030. [DOI] [PubMed] [Google Scholar]
- 41.Duffus WA, Levy-Mintz P, Klimjack MR, Kielian M. 1995. Mutations in the putative fusion peptide of Semliki Forest virus affect spike protein oligomerization and virus assembly. J Virol 69:2471–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gibbons DL, Vaney M-C, Roussel A, Vigouroux A, Reilly B, Lepault J, Kielian M, Rey FA. 2004. Conformational change and protein–protein interactions of the fusion protein of Semliki Forest virus. Nature 427:320. doi: 10.1038/nature02239. [DOI] [PubMed] [Google Scholar]
- 43.Qiao H, Armstrong RT, Melikyan GB, Cohen FS, White JM. 1999. A specific point mutant at position 1 of the influenza hemagglutinin fusion peptide displays a hemifusion phenotype. Mol Biol Cell 10:2759–2769. doi: 10.1091/mbc.10.8.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Epand RM. 2003. Fusion peptides and the mechanism of viral fusion. Biochim Biophys Acta 1614:116–121. [DOI] [PubMed] [Google Scholar]
- 45.Kielian M. 2014. Mechanisms of virus membrane fusion proteins. Annu Rev Virol 1:171–189. doi: 10.1146/annurev-virology-031413-085521. [DOI] [PubMed] [Google Scholar]
- 46.Kielian M, Rey FA. 2006. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat Rev Microbiol 4:67. doi: 10.1038/nrmicro1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakabayashi H, Taketa K, Miyano K, Yamane T, Sato J. 1982. Growth of human hepatoma cell lines with differentiated functions in chemically defined medium. Cancer Res 42:3858. [PubMed] [Google Scholar]
- 48.Dubuisson J, Hsu HH, Cheung RC, Greenberg HB, Russell DG, Rice CM. 1994. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J Virol 68:6147–6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Flint M, Maidens C, Loomis-Price LD, Shotton C, Dubuisson J, Monk P, Higginbottom A, Levy S, McKeating JA. 1999. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J Virol 73:6235–6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lindenbach BD, Evans MJ, Syder AJ, Wölk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 51.Macdonald A, Crowder K, Street A, McCormick C, Saksela K, Harris M. 2003. The hepatitis C virus nonstructural NS5A protein inhibits activating protein-1 function by perturbing Ras-ERK pathway signaling. J Biol Chem 278:17775–17784. doi: 10.1074/jbc.M210900200. [DOI] [PubMed] [Google Scholar]
- 52.Mesalam AA, Desombere I, Farhoudi A, Van Houtte F, Verhoye L, Ball J, Dubuisson J, Foung SKH, Patel AH, Persson MAA, Leroux-Roels G, Meuleman P. 2018. Development and characterization of a human monoclonal antibody targeting the N-terminal region of hepatitis C virus envelope glycoprotein E1. Virology 514:30–41. doi: 10.1016/j.virol.2017.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fofana I, Krieger SE, Grunert F, Glauben S, Xiao F, Fafi-Kremer S, Soulier E, Royer C, Thumann C, Mee CJ, McKeating JA, Dragic T, Pessaux P, Stoll-Keller F, Schuster C, Thompson J, Baumert TF. 2010. Monoclonal anti-claudin 1 antibodies prevent hepatitis C virus infection of primary human hepatocytes. Gastroenterology 139:953–964. doi: 10.1053/j.gastro.2010.05.073. [DOI] [PubMed] [Google Scholar]
- 54.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Kräusslich H-G, Mizokami M, Bartenschlager R, Liang TJ. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med 11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Delgrange D, Pillez A, Castelain S, Cocquerel L, Rouillé Y, Dubuisson J, Wakita T, Duverlie G, Wychowski C. 2007. Robust production of infectious viral particles in Huh-7 cells by introducing mutations in hepatitis C virus structural proteins. J Gen Virol 88:2495–2503. doi: 10.1099/vir.0.82872-0. [DOI] [PubMed] [Google Scholar]
- 56.Rouillé Y, Helle F, Delgrange D, Roingeard P, Voisset C, Blanchard E, Belouzard S, McKeating J, Patel AH, Maertens G, Wakita T, Wychowski C, Dubuisson J. 2006. Subcellular localization of hepatitis C virus structural proteins in a cell culture system that efficiently replicates the virus. J Virol 80:2832–2841. doi: 10.1128/JVI.80.6.2832-2841.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alsaleh K, Delavalle P-Y, Pillez A, Duverlie G, Descamps V, Rouillé Y, Dubuisson J, Wychowski C. 2010. Identification of basic amino acids at the N-terminal end of the core protein that are crucial for hepatitis C virus infectivity. J Virol 84:12515–12528. doi: 10.1128/JVI.01393-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Calland N, Albecka A, Belouzard S, Wychowski C, Duverlie G, Descamps V, Hober D, Dubuisson J, Rouillé Y, Séron K. 2012. (−)-Epigallocatechin-3-gallate is a new inhibitor of hepatitis C virus entry. Hepatology 55:720–729. doi: 10.1002/hep.24803. [DOI] [PubMed] [Google Scholar]
- 59.Mederacke I, Wedemeyer H, Ciesek S, Steinmann E, Raupach R, Wursthorn K, Manns MP, Tillmann HL. 2009. Performance and clinical utility of a novel fully automated quantitative HCV-core antigen assay. J Clin Virol 46:210–215. doi: 10.1016/j.jcv.2009.08.014. [DOI] [PubMed] [Google Scholar]
- 60.Lavillette D, Tarr AW, Voisset C, Donot P, Bartosch B, Bain C, Patel AH, Dubuisson J, Ball JK, Cosset F-L. 2005. Characterization of host-range and cell entry properties of the major genotypes and subtypes of hepatitis C virus. Hepatology 41:265–274. doi: 10.1002/hep.20542. [DOI] [PubMed] [Google Scholar]