Even though the loss of DNA, caused by heat treatment, destabilizes the T7 phage, its capsid is remarkably able to withstand high temperatures with a more or less intact global topographical structure. Thus, partial denaturation within the global structural constraints of the viral capsid may have a stabilizing effect. Understanding the structural design of viruses may help in constructing artificial nanocapsules for the packaging and delivery of materials under harsh environmental conditions.
KEYWORDS: atomic force microscopy, nanoindentation, thermally induced structural change, CD spectroscopy
ABSTRACT
Viruses are nanoscale infectious agents which may be inactivated by heat treatment. The global molecular mechanisms of virus inactivation and the thermally induced structural changes in viruses are not fully understood. In this study, we measured the heat-induced changes in the properties of T7 bacteriophage particles exposed to a two-stage (65°C and 80°C) thermal effect, by using atomic force microscopy (AFM)-based nanomechanical and topographical measurements. We found that exposure to 65°C led to the release of genomic DNA and to the loss of the capsid tail; hence, the T7 particles became destabilized. Further heating to 80°C surprisingly led to an increase in mechanical stability, due likely to partial denaturation of the capsomeric proteins kept within the global capsid arrangement.
IMPORTANCE Even though the loss of DNA, caused by heat treatment, destabilizes the T7 phage, its capsid is remarkably able to withstand high temperatures with a more or less intact global topographical structure. Thus, partial denaturation within the global structural constraints of the viral capsid may have a stabilizing effect. Understanding the structural design of viruses may help in constructing artificial nanocapsules for the packaging and delivery of materials under harsh environmental conditions.
INTRODUCTION
Viruses are remarkable nanoscale machineries that harbor a piece of genetic material within a proteinaceous capsule. As obligatory parasites, they are capable of efficiently fooling the host organism into manufacturing the viral structural elements, which spontaneously reproduce the virus particle by self-assembly. Because of their biological, medical, and even economic importance, the properties of viruses have been investigated by a wide array of experimental approaches. It has long been known that most viruses can be thermally inactivated (1–4). It is hypothesized that thermal virus inactivation is caused by the release of the genetic material or some kind of capsid disruption (5–8), but little is known about the exact nature of the thermally driven structural transitions within the viruses. Differential scanning calorimetry and cryo-electron microscopy experiments have revealed a reversible structural transition at 53°C limited to the hexamers of the HK97 bacteriophage (9). Heating the HK97 phage further results in the release of genomic DNA by not precisely known mechanisms, and heating even further to 80°C results in an irreversible transition of thermal melting (10). In the case of bacteriophage λ, heat-induced transitions at 68°C and 87°C have been assigned to the escape of DNA and irreversible melting, respectively (11). Simultaneous observations of capsid- and DNA-related events, however, have not so far been possible at the level of the individual virus particles.
Previously, a distinct thermal melting of the bacteriophage T7 has been documented (12–15). T7 is a nonenveloped, short-tailed icosahedral Escherichia coli phage that contains a 40-kbp genomic DNA (16). Thermal melting, measured by following the optical density at 260 nm (OD260) as a function of temperature, involves two major transitions related to DNA. The first transition occurs between 50 and 60°C, and it is thought to correspond to the release of DNA from the capsid. This transition is accompanied by a marked loss of infectivity (17). A second transition is detected in the sample at temperatures above 80°C, and it is related to DNA denaturation. Temperature-dependent circular dichroism (CD) spectroscopic measurements indicated that both of these transitions are likely to be accompanied by structural changes in the capsid proteins as well (12). Although high-resolution structural information on the protein capsid of T7 is available (18, 19), the details of the thermally induced transitions within the protein components of the capsid remain unclear.
In recent years, atomic force microscopy (AFM)-based nanomechanical experiments emerged as a sensitive tool to explore the properties of viruses (20–29). It has been shown that nanomechanical parameters, such as stiffness and capsid breaking force, may reveal molecular mechanisms underlying capsid maturation and the packaging, storage, and release of genetic material.
In this study, we employed AFM to explore the nanomechanical and topographical changes in T7 bacteriophages exposed to two-stage thermal treatment (65°C and 80°C). We show that distinct changes in the nanomechanical properties of T7 occur upon heat treatment. Topographical analysis revealed the structural alterations that underlie the nanomechanical changes: 65°C treatment leads to the release of genomic DNA and the loss of the tail complex, and further heating to 80°C leads, on one hand, to the appearance of large globular particles that likely correspond to disassembled capsids and, on the other hand, to a partial structural stabilization of the remaining capsids, due most likely to rearrangements via partial denaturation of the capsomeric gp10A proteins.
RESULTS AND DISCUSSION
Nanomechanics of heat-treated T7 phages.
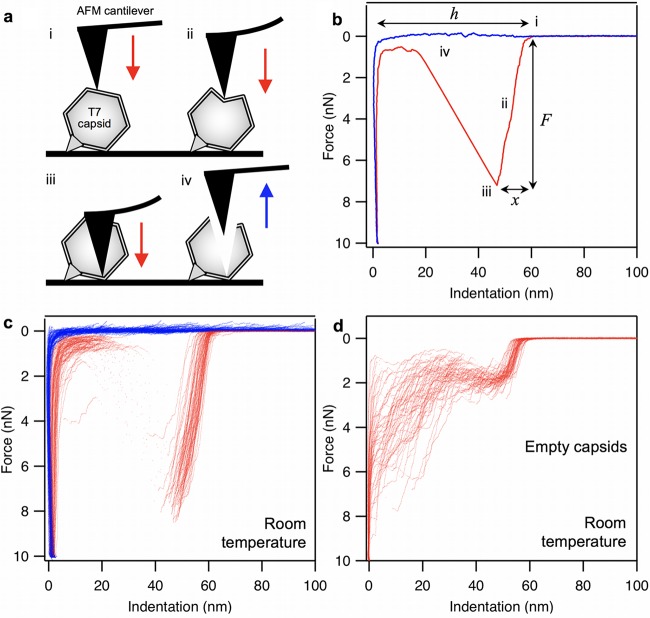
In the present work, individual, surface-adsorbed T7 phage particles exposed to different temperatures (room temperature [RT], 65°C, and 80°C) were manipulated with AFM to reveal their nanomechanical properties and the thermally induced changes in these properties. Figure 1 shows our results obtained on phages at RT. After landing the AFM tip on the capsid surface, force increased linearly to about 8 nN, where a sharp transition marked by a sudden drop of force and corresponding to capsid breakage occurred. Upon pressing the AFM tip further, force fluctuated below 2 nN; then it began to rise sharply upon approaching the substrate surface. The retraction force trace was essentially featureless; therefore, a large force hysteresis was present, indicating that the mechanical manipulation resulted in an irreversible conformational change (breakage) of the capsid. Although for the majority of the capsids similar force traces were recorded (Fig. 1c), in a fraction of them we obtained traces with a significantly different, but reproducible, appearance (Fig. 1d). In these traces the initial linear regime ended at about 2 nN (we refer to these as putative empty capsids; see below).
FIG 1.
Nanomechanics of T7 phages. (a) Schematics of mechanical manipulation. The tip of the AFM cantilever is first brought into contact with the T7 phage surface (i), which is then pressed (ii) with a preadjusted velocity to 10 nN maximal force, during which the capsid eventually ruptures (iii). Finally, the cantilever is lifted (iv). AFM cantilever and T7 phage are not to scale. (b) Representative force versus indentation curve obtained at room temperature. Data collected during the indentation half-cycle are displayed in red, whereas those during retraction are in blue. Notable stages of the nanomechanics experiments are shown with Roman numerals (i to iv). Variables extracted from the data (breaking force F, maximal indentation distance x, and capsid height h) are shown with italic letters. Capsid stiffness (k) is obtained by fitting a line in the initial linear regime of the indentation data (ii). (c) Data set containing 80 similar, overlaid force versus indentation curves collected in independent experiments on different phage particles at room temperature (29). Red and blue traces are indentation and retraction half cycles, respectively. (d) Data set containing 55 similar, overlaid force versus indentation curves (indentation half-cycle only), collected at room temperature in independent experiments, which are similar to each other but are distinctively different from the data set in panel c (putatively called empty-capsid curves).
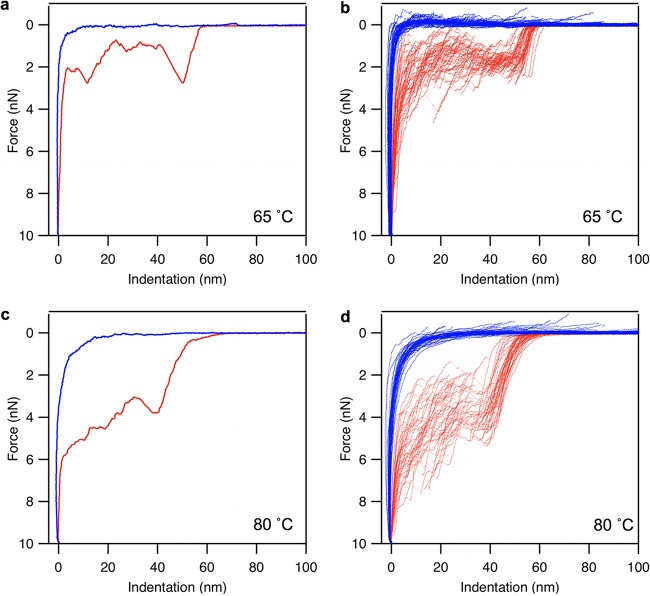
In T7 capsids treated at 65°C (Fig. 2a and b), the force traces were similar to those seen in Fig. 1d: capsid breakage occurred at about 2 nN, and then force fluctuated around 2 nN before increasing abruptly upon approaching the substrate surface. In T7 capsids heated further to 80°C (Fig. 2c and d), the overall appearance of the force traces was similar to that seen for the 65°C samples, but capsid breakage and the following force fluctuation occurred at greater force levels.
FIG 2.
Nanomechanics data of heat-treated T7 phages. Red and blue indicate indentation and retraction half-cycles, respectively. (a) Representative force versus indentation curve measured on a T7 phage particle that has been exposed to a temperature of 65°C for 15 min. (b) Data set containing 45 similar, overlaid force versus indentation curves collected in independent experiments on different phage particles heat-treated at 65°C. (c) Representative force versus indentation curve measured on a T7 phage particle that has been exposed to a temperature of 80°C for 15 min. (d) Data set containing 41 similar, overlaid force versus indentation curves collected in independent experiments on different phage particles heat treated at 80°C.
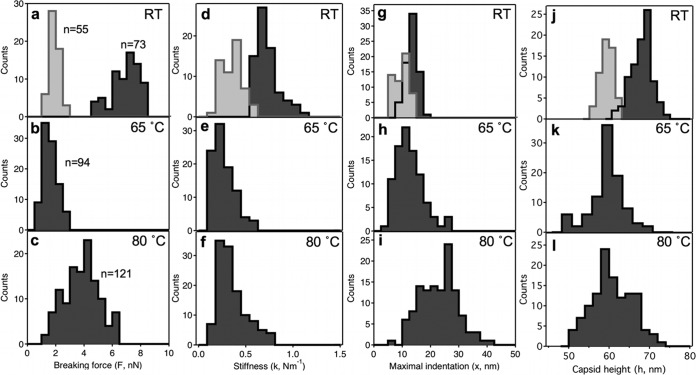
Figure 3 displays the distribution of the parameters extracted from the force traces (see also Table 1). The breaking force values in the RT samples (Fig. 3a) partition into two modes, according to the distinct types of force curves (Fig. 1c and d). The low-force mode of the RT sample aligns well with the histogram peak of the 65°C data (Fig. 3b). Considering that T7 phages heated to temperatures above 60°C are thought to lose their DNA (12), we tentatively conclude that the low-force peak in the RT data may correspond to empty capsids. The presence of empty capsids in the RT sample indicates that the spontaneous or artificially induced DNA ejection of T7 phages is not negligible (30). The breaking force is severely reduced in 65°C-treated capsids (from 6.90 nN to 1.61 nN [Table 1]). Since, according to AFM imaging results (see Fig. 5), the 65°C treatment indeed resulted in the release of DNA from the capsids, our findings indicate that the presence of packaged DNA within the phage contributes to its mechanical stability. Quite interestingly, the breaking force was increased in the 80°C-treated T7 phages relative to the ones treated at 65°C (Fig. 3c). Conceivably, structural rearrangements occurred in the capsomeric proteins between 65 and 80°C, which resulted in a stabilization of their interactions, hence to an increased mechanical stability of the phage particle.
FIG 3.
Distribution of variables obtained from nanomechanics data. Breaking force (a, b, and c), stiffness (d, e, and f), maximal indentation distance (g, h, and i) and capsid height (j, k, and l) histograms for T7 phage particles at room temperature (RT) and ones treated at 65°C and 80°C are shown. Light gray bars correspond to data obtained on empty capsids at room temperature. The numbers (n) refer to the number of force curves analyzed to obtain the nanomechanical parameters.
TABLE 1.
Nanomechanical and topographical parameters of T7 bacteriophage capsids and globular particles
| Parameter | Value at temp (mean ± SD) |
|||
|---|---|---|---|---|
| RT (DNA filled) | RT (empty) | 65°C | 80°C | |
| Nanomechanics | ||||
| Breaking force (F, nN) | 6.90 ± 0.97 | 1.90 ± 0.34 | 1.61 ± 0.52 | 3.83 ± 1.20 |
| Capsid stiffness (k, nm−1) | 0.73 ± 0.12 | 0.36 ± 0.10 | 0.27 ± 0.11 | 0.35 ± 0.15 |
| Maximal indentation (x, nm) | 13.24 ± 2.13 | 9.51 ± 2.41 | 12.56 ± 4.70 | 22.97 ± 6.61 |
| Capsid height (h, nm) | 60.4 ± 1.71 | 59.93 ± 1.56 | 59.41 ± 4.00 | 60.71 ± 4.87 |
| Capsid topography | ||||
| Ratio of tailed capsids | 0.83 ± 0.1 | NAa | 0.19 ± 0.12 | 0.05 ± 0.06 |
| Peak capsid height (nm) | 61.8 ± 3.6 | NA | 58.2 ± 1.7 | 63.3 ± 5.5 |
| Capsomer diameter (nm) | 11.0 ± 0.8 | NA | 9.8 ± 1.0 | 13.9 ± 2.2 |
| Globular particle parameters | ||||
| Particle height (nm) | 7.6 ± 3.2 | NA | 6.9 ± 2.5 | 10.0 ± 5.4 |
NA, not available.
FIG 5.
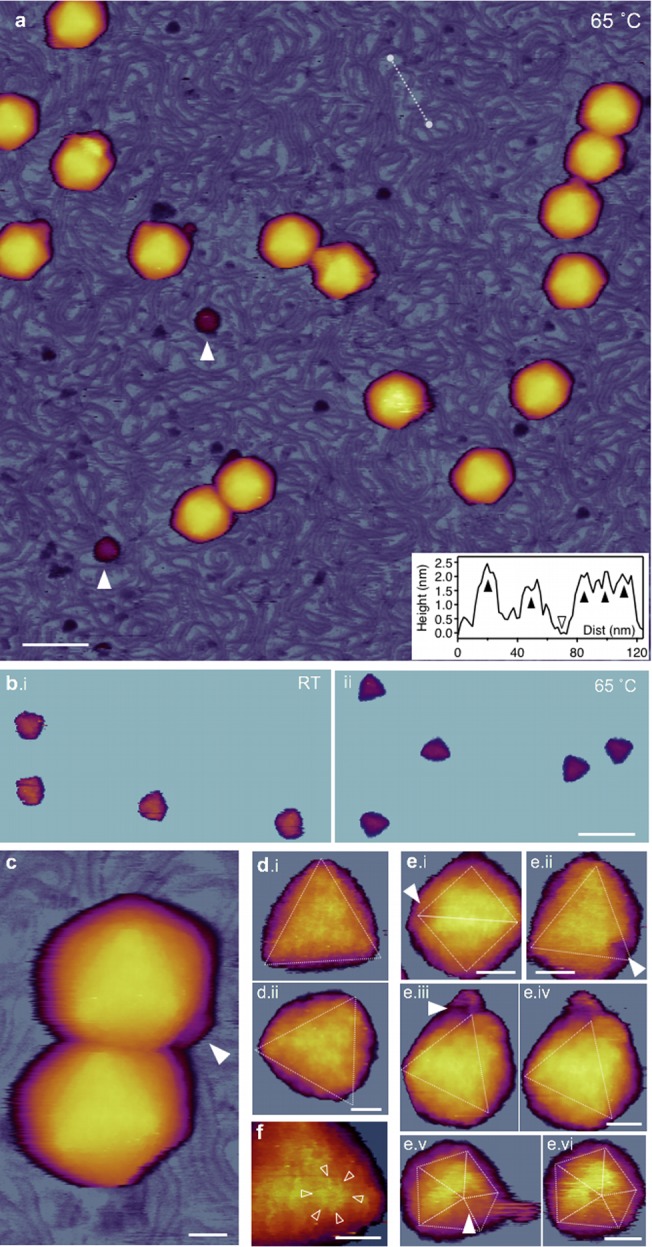
AFM of T7 phages treated at 65°C. (a) Overview of a 1-μm by 1-μm sample area. White arrowheads point at large (>10 nm) globular particles. Scale bar, 100 nm. (Inset) Topographical height map along an arbitrarily chosen line in the background (white dashed line). Black arrowheads point at DNA cross sections, whereas the empty arrowhead at the substrate (mica) surface. (b) Comparison of icosahedral facets of room temperature (i) and 65-degree (ii) capsids. AFM images were contrast enhanced with identical color-scale offset (48 nm) and range (20 nm). (c) AFM image of two T7 particles. The white arrowhead points at the short, stubby tail complex visible on one of the particles, whereas there is no visible tail on the other one. Scale bar, 20 nm. (d) High-resolution AFM images of 65°C-treated T7 phage particles with resolvable capsomeres on their surfaces. Views are along the 3-fold symmetry axes. Because of contrast enhancement, only the top facets are visible and the rest of the capsid is hidden. Scale bar, 10 nm. (e) T7 particles with resolvable DNA exit holes (white arrowheads). The exit hole appears as a gap in the location of a missing pentagonal capsomere at one of the icosahedron vertices. Images viewed along the 2-fold (i), 3-fold (ii, iii, and iv), and 5-fold (v and vi) symmetry axes are shown. Images iii and v are reconstructed from the rightward fast AFM scanlines, whereas images iv and vi are from leftward (reverse) scanlines from the same sample area. Scale bars, 20 nm. (f) Magnified view of a cogwheel-shaped hexagonal capsomere. Arrowheads point at the spokes of the cogwheel. Scale bar, 10 nm.
Stiffness was largest in the intact T7 phage, and the reduced stiffness values were similar in the RT empty capsids and the heat-treated ones (Fig. 3d to f). Thus, the presence of packaged DNA contributes to the stiffness of the T7 phage.
The maximal indentation values progressively increased as a result of heat treatment (Fig. 3g to i), which is a combined effect of the underlying changes in breaking forces and stiffness. Thus, even though the stiffness of 80°C-treated T7 capsids is reduced, because of the increased breaking forces, they withstand greater indentations prior to breakage.
The mean capsid height became slightly reduced upon 65°C treatment (Fig. 3j and k). Notably, the mean capsid height of the empty-capsid RT phages was essentially identical to that of the 65°C-treated ones, indicating that the presence of packaged DNA within the phage increases its diameter (by about 10 nm). Conceivably, the DNA pressure inside the phage causes the expansion of the icosahedral phage structure. We note that the 10-nm difference in mean capsid height between the RT and 65°C-treated capsids is only partly due to the DNA pressure; since capsid height was obtained from mechanical measurements with a pyramidal AFM tip, the presence of upward-oriented phage tails likely shifted the average height to greater values in the RT samples. The capsid height was slightly increased in the 80°C-treated sample relative to 65°C, which, as judged from the histogram shape (Fig. 3l), is probably due to the emergence of a subpopulation of capsids with a larger diameter.
AFM structure of heat-treated T7 phages.
To reveal the structural detail and mechanisms behind the heat-induced nanomechanical changes in T7 capsids, we carried out high-resolution AFM measurements on phage particles exposed to 65°C and 80°C (Fig. 4a). In an overview AFM image of a typical RT sample (Fig. 4b), the characteristic T7 phage particles could be visualized against a nearly featureless substrate background. Occasionally, a DNA molecule released from the capsid upon mechanical perturbation could be observed. The mechanically induced DNA ejection is characterized by the sudden, within-one-scanline appearance of the DNA chain (30). We note that there were a few globular particles in the background, which may correspond to the core T7 phage proteins that become ejected simultaneously with DNA (30). Importantly, the conical tail complex could be observed on most of the phage particles. Depending on the surface binding of the phages, the tail complex was oriented in different directions (Fig. 4c). In high-resolution AFM images, even the size, the cogwheel shape, and the central pore of the capsomeres could be resolved (Fig. 4d and e).
FIG 4.
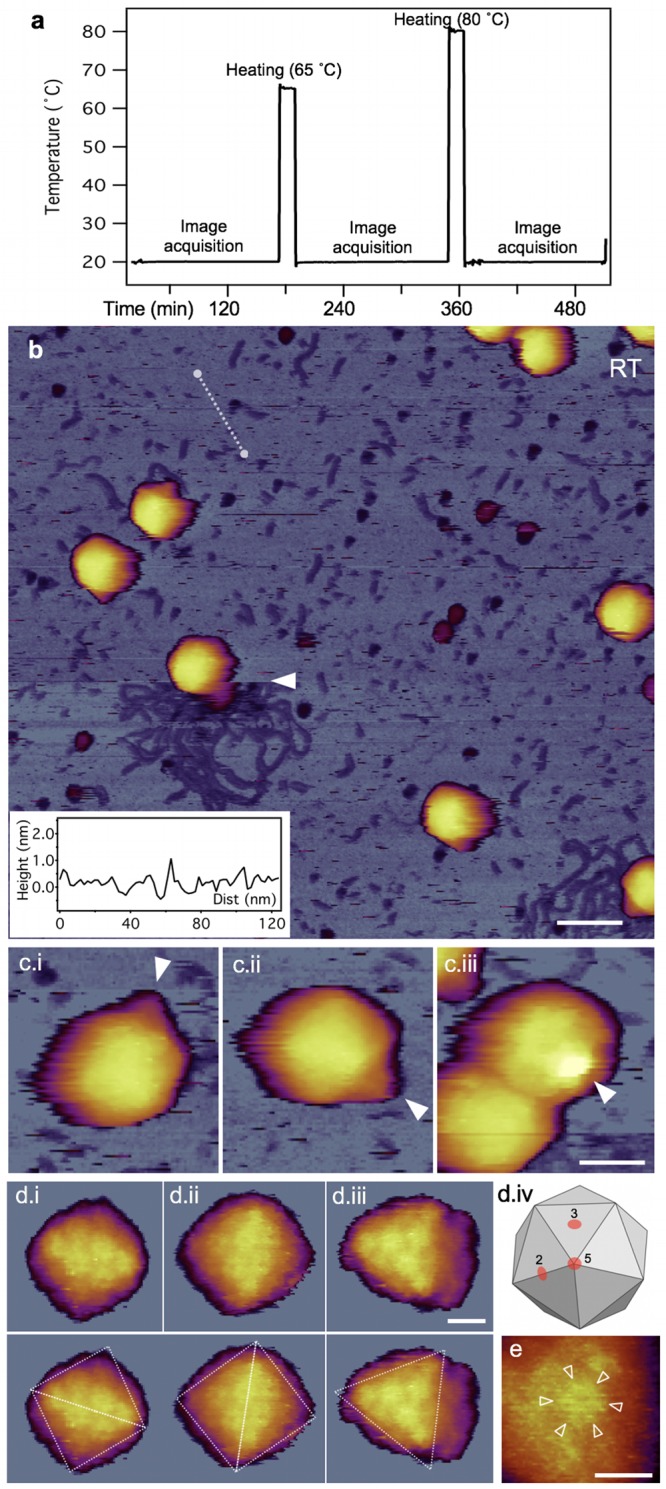
Temperature-dependent AFM measurements on T7 phage particles. (a) Thermal treatment protocol. Shown is sample temperature versus time trace recorded in a typical experiment. The same sample was exposed to consecutive heating (for 15 min), cooling (to 20°C), and image acquisition (at 20°C) cycles. (b) Overview of a 1-μm by 1-μm sample area at room temperature (20°C). Slow AFM raster scan direction is from top to bottom of the image. The white arrowhead points at the nearly instantaneous event of mechanically induced DNA ejection. Scale bar, 100 nm. (Inset) Topographical height map along an arbitrarily chosen line in the background (white dashed line). (c) AFM images of T7 phage particles displaying their conical tail in different orientations. White arrowheads point at the tail apices. Scale bar, 30 nm. (d) High-resolution AFM images of the T7 phage surfaces with resolvable capsomeres. Views along the 2-fold (I and ii) and 3-fold (iii) symmetry axes, which are explained in image iv, are shown. Scale bar, 10 nm. In the bottom row dashed guiding lines are superimposed on the respective images to indicate the symmetries. (e) Magnified view of a cogwheel-shaped hexagonal capsomere. Arrowheads point at the spokes of the cogwheel. Scale bar, 10 nm.
Upon 65°C treatment, the topography of the background, and to some extent the capsids, became different (Fig. 5). The most striking feature is that the substrate became covered with a meshwork of DNA chains. A height profile of a section of the background (Fig. 5a, inset) shows that the cross-sectional height of the individual strands is about 2 nm, which demonstrates that they are indeed DNA. Thus, the 65°C treatment, as suggested earlier (12), indeed resulted in the release of DNA from the T7 capsids. The second notable feature in the AFM images is that in most of the capsids the conical tail complex is not visible. Even if a tail can be seen, its structure is usually stubby, quite different from a cone (Fig. 5c). Thus, DNA has been released from the phage particles, because of a separation of the tail complex from the capsid. Because the gp8 protein plays an important role in the connecting the tail complex to the capsid, we hypothesize that it might be a thermally sensitive component of T7. As a result, large (>10 nm) globular particles can be identified in the background, which may correspond to the remnants of the broken-off tail complexes. Although DNA release and the loss of tail were clearly observed in the samples treated at 65°C, we do not exclude the possibility that these structural transitions may begin to occur at lower temperatures already (31, 32). We note that we were unable to detect the presence of L-shaped tail fibers on the substrate surface. Possibly, the poly-l-lysine-coated surface and the large amount of DNA precluded the binding of the tail fibers in proper orientation. The third striking feature is that the capsid surface became more faceted, and the icosahedron edges and faces emerged more distinctively (Fig. 5b). Such a faceted appearance can be well explained by the shrinkage of the capsids upon DNA release (Fig. 3j and k). In high-resolution AFM images, the cogwheel shape of the individual capsomeres could be well identified (Fig. 5d and f). In a few capsids, we noticed gaps in the position of the pentameres, which are most likely the exit holes through which DNA escaped (Fig. 5e).
In T7 samples exposed to 80°C, the background was also densely populated with DNA strands (Fig. 6a). A notable feature is the large number of globular particles scattered in the background. Even large aggregates of the particles could be observed (Fig. 6b). Considering that the size of the aggregates far exceeds that of the tail complex, we hypothesize that the aggregates, hence their component globular particles, originate from the capsid wall. In high-resolution AFM images (Fig. 6c and d), the capsomeres appeared swollen, and they displayed a less distinct cogwheel structure according to visual inspection. Altogether, the major transitions of T7 upon heating to 65°C are the release of DNA rearrangements in the capsid. The transitions upon further heating to 80°C entail structural changes within the capsid wall.
FIG 6.
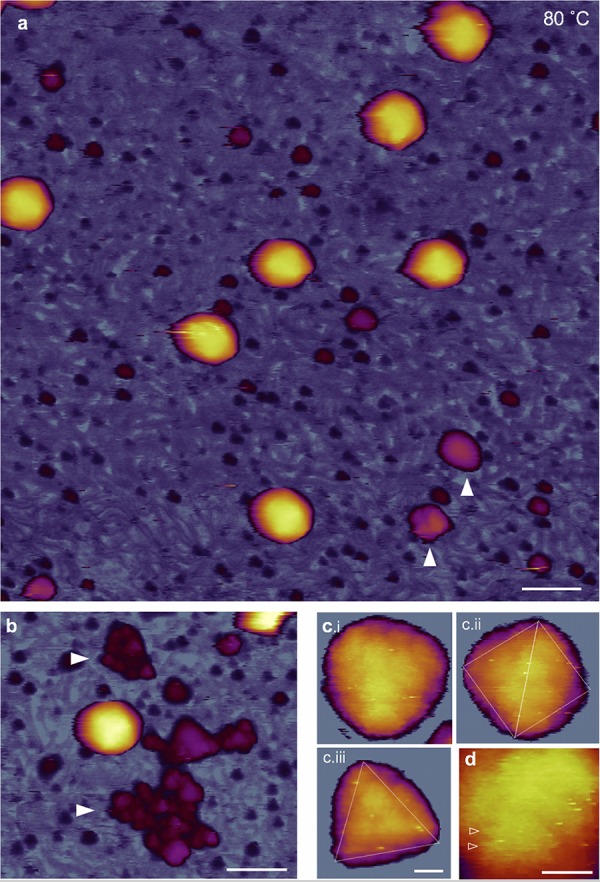
AFM of T7 phages treated at 80°C. (a) Overview of a 1-μm by 1-μm sample area. White arrowheads point at large (>10 nm) globular particles. Scale bar, 100 nm. (b) AFM image showing large aggregates of globular particles (white arrowheads) (c) High-resolution AFM images of 80-degree-treated T7 phage particles with resolvable capsomeres on their surface. Views along the 3-fold (i and iii) and 2-fold (ii) symmetry axes are shown. Scale bar, 10 nm. (d) Magnified view of the capsomeric structure. Arrowheads point at putative spokes of the originally cogwheel-shaped capsomere. Note that the central pore cannot be resolved, most likely due to the swelling of the protein matrix. Scale bar, 10 nm.
Analysis of topographical data.
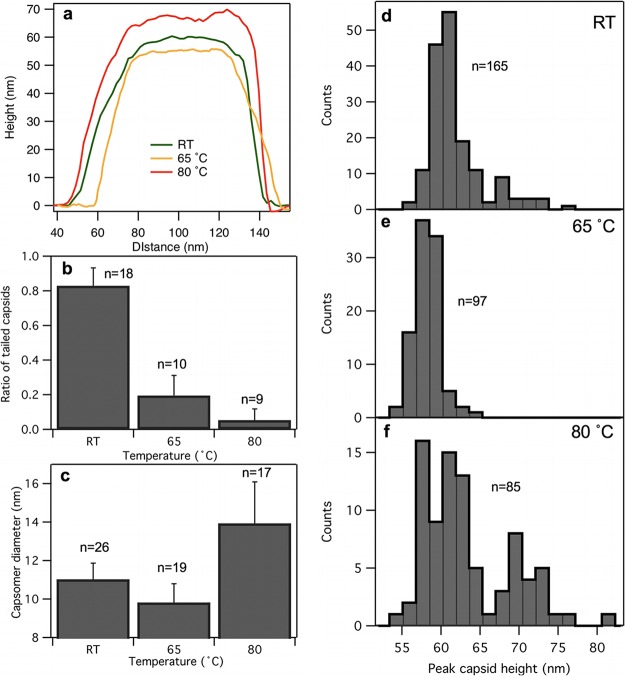
Representative height profiles across individual capsids (Fig. 7a) demonstrate the heat-induced topographical changes in T7. Upon 65°C treatment, the capsid slightly shrunk and its faces became flattened and its edges more distinct. The 80°C-treated capsid whose height map is shown here became swollen and its surface rugged. The ratio of capsids with visible tail complexes was progressively reduced with heat treatment (Fig. 7b). The capsomere diameter was considerably increased after 80°C treatment (Fig. 7c). We hypothesize that thermally induced conformational changes, most likely partial denaturation, occurred in the gp10A capsomeric proteins, which resulted in an increase of their apparent volume. It might well be possible that the partial denaturation exposed the hydrophobic core of the capsomere proteins, and a hydrophobic interaction occurred in between neighboring capsomeres. Such an interaction may explain the increase in breaking force between 65°C and 80°C observed in the nanomechanical experiments (Fig. 3b and c). The peak capsid height decreased slightly upon 65°C treatment (Fig. 7d and e), but the 80°C treatment resulted in the emergence of a subpopulation with larger height values (Fig. 7f). We hypothesize that this subpopulation corresponds to capsids with swollen wall structure. The peak capsid height analysis more or less reflects the tendencies observed in the nanomechanical experiments. However, because AFM images allow us to select the tallest topographical point on the capsids, the peak height analysis is more sensitive to local variations, which are hidden or averaged out in the nanomechanics experiment.
FIG 7.
Analysis of capsid topography data. (a) Topographical height map along the cross section of either a capsid at room temperature (green trace) or ones treated at 65°C (orange) or 80°C (red). (b) Ratio of capsids with tails as a function of temperature. The numbers above the bars represent the number of fields analyzed for every T7 particle. Error bars represent SDs. (c) Capsomer diameter as a function of temperature. The numbers above the bars represent the numbers of capsomers measured. Error bars represent SDs. (d, e, and f) Histograms of peak capsid heights for room temperature-, 65°C-, and 80°C-treated T7 phages, respectively. Peak height refers to the tallest topographical point in the capsid image. The numbers refer to the numbers of T7 phage particles analyzed.
The number of globular particles progressively increased in the samples upon heat treatment (Fig. 8a). The height of the major population of the particles is centered around 6 nm regardless of heat treatment (Fig. 8b to d). In the 80°C-treated samples, particle populations with much larger heights emerged (Fig. 8c). While the ∼6-nm particles may correspond to the ejected core proteins, the large globular particles are most likely capsomeric proteins and their aggregates, which appear due to the complete disassembly of some of the capsids.
FIG 8.
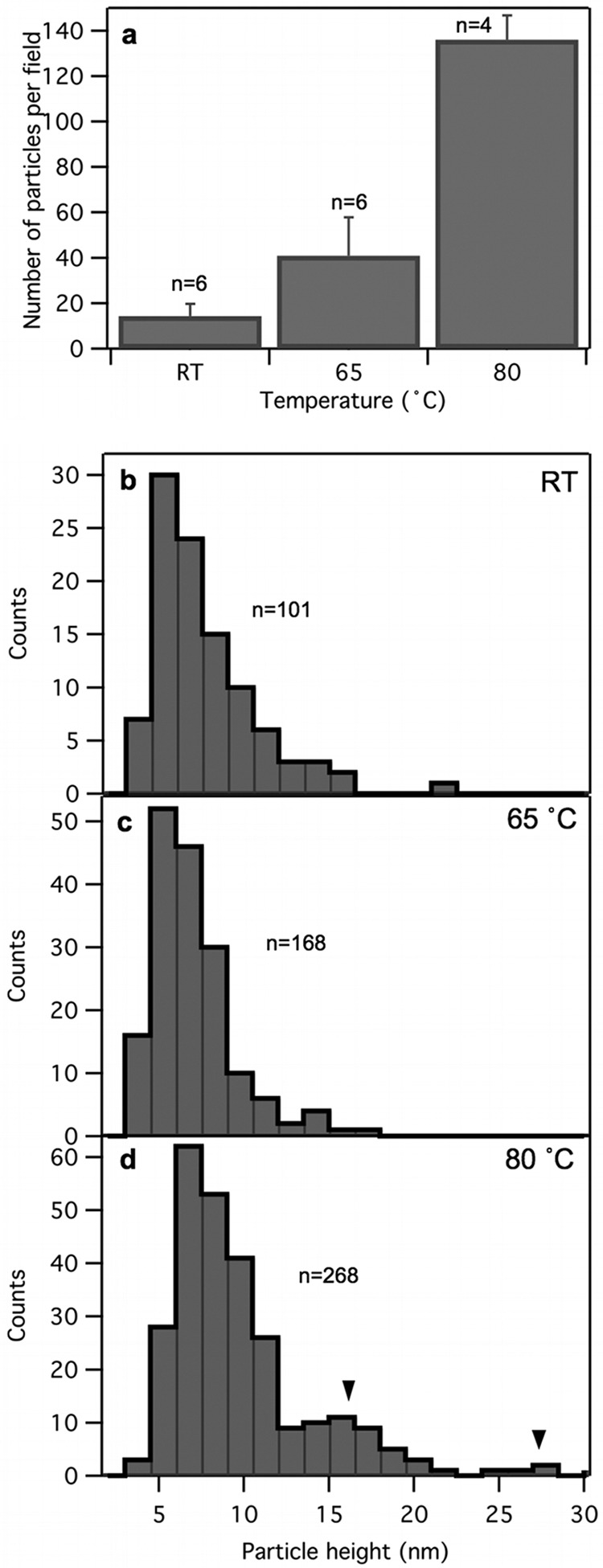
Analysis of topographical data of globular particles. (a) Number of globular particles per field as a function of temperature. The numbers above the bars represent the numbers of fields analyzed for every particle. Error bars represent SDs. (b, c, and d) Histograms of globular particle height for samples at room temperature and ones treated at 65°C and 80°C, respectively. Black arrowheads point at populations of large globular particles. The numbers refer to the numbers of globular particles analyzed.
CD spectroscopic analysis.
To investigate the structural changes in heat-treated T7 further, CD spectroscopic measurements were performed at room temperature (25°C) and progressively increased temperatures (61°C and 83°C) (Fig. 9). The CD spectra, recorded at 25, 61, and 83°C, display differences in the short-wavelength regime (at 225 nm), indicating that capsid protein-associated structural transitions take place not only between 25 and 61°C but also between 61 and 83°C. The latter appears to be part of a broad temperature-dependent transition that begins slightly below 80°C and peaks at 92°C and corresponds most likely to the loss of protein α-helix content in a denaturing transition (12). In the AFM experiments, due to the brief (15 min) exposure to high temperature (80°C) followed by cooling to room temperature, only partial denaturation may have taken place.
FIG 9.
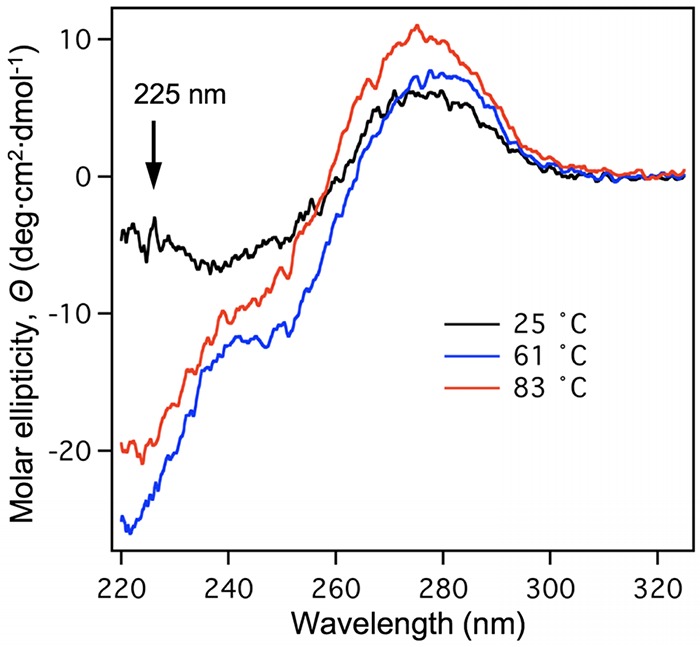
CD spectroscopy of heat-treated T7. CD spectra of T7 samples recorded at room temperature (25°C, black line), 61°C (blue line), and 83°C (red line) are shown. The arrow marks the wavelength of 225 nm, at which the protein-dependent changes were monitored.
Model of thermally induced structural changes in T7.
We propose the following phenomenological model to explain our observations (Fig. 10). At room temperature (Fig. 10a), T7 displays a characteristic icosahedral structure with a distinctive conical tail complex. The icosahedron is slightly swollen due to the DNA pressure inside the capsid. The DNA-filled T7 phages have a high stiffness and withstand (instantaneous) forces up to about 8 nN prior to breakage. Upon heating to 65°C (Fig. 10b), the genomic DNA is ejected from the capsid. The release of DNA is likely caused by the conical tail complex breaking off the capsid, rather than by activating the natural DNA ejection machinery. It is currently unclear whether the entire genome of T7 exits the capsid or a portion remains inside. It is hypothesized that during its natural DNA ejection, only part of the T7 genome is driven out of the capsid due to the DNA pressure. The remaining DNA is thought to be pulled into the E. coli by an active process (33–35). However, because the thermally induced changes involve the loss of the entire tail complex, there might be enough room for the nearly complete release of the T7 genome. Regardless of how much DNA exits during this process, the resulting drop in DNA pressure is accompanied by a shrinkage and a more faceted appearance of the capsid. Upon further heating to 80°C (Fig. 10c) the capsids do not disappear, but they are still present with a maintained global structure. A partial denaturation likely takes place in the gp10A proteins that form the capsomeres and hence the capsid wall. The partial protein denaturation within the global confinement of the capsid architecture and the resulting exposure of hydrophobic protein regions result in capsomere swelling and a new set of intercapsomeric interactions, most probably via hydrophobic protein regions. Facilitated folding and misfolding following repetitive partial or complete denaturation have been observed in proteins confined either in a chaperonin system (36) or in a force field (37). Notably, capturing a protein in the misfolded state results in a considerable conformational expansion (38). We envision that similar processes may occur in the gp10A proteins heated to 80°C and then cooled to room temperature. In the end, the capsid wall becomes thicker, and the entire capsid surface becomes rugged. It is quite conceivable that the hydrophobic stabilization is the result not of the heating per se but of the relaxation from the thermal exposure. That is, capsids that did not completely fall apart during the 80°C treatment may relax into a stabilized structure upon cooling back to room temperature. By adjusting the time of exposure to the high temperature, the capsid stabilization may conceivably be tuned.
FIG 10.
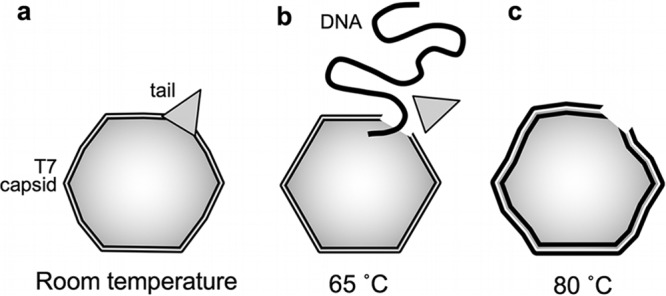
Schematic model of thermally induced changes in the T7 bacteriophage. At room temperature (a) the capsid is slightly swollen because of the DNA pressure inside. The bulging of the capsid wall as shown in the scheme is not to scale. Upon heating to and incubation at a temperature of 65°C (b), the tail complex is broken off, resulting in the release of the genomic DNA. The capsid becomes more faceted due to the relaxation of the capsid pressure. Finally, at 80°C (c), the capsid becomes swollen and its surface irregular, and the capsids may become fragmented into large globular particles (this step is not shown).
Conclusions.
We have directly shown that exposing T7 to a thermal treatment at 65°C caused the release of its genomic DNA due to the tail complex breaking off the capsid. The loss of DNA and/or thermally driven changes in capsomeric protein structure result in a reduced capsid stiffness and breaking force. Further heating to 80°C leads to rearrangements within the capsid wall, caused most likely by partial denaturation of the component gp10A proteins. Even though the capsids are destabilized, they are remarkably able to still withstand high temperatures with a more or less intact global topographical structure. Thus, partial denaturation within the global structural constraints of the viral capsid may have a stabilizing effect. Understanding the structural design of viruses may help in constructing artificial nanocapsules for the packaging and delivery of materials under harsh environmental conditions. By tuning capsid stability, these nanocapsules may, in principle, be tailored for specific applications.
MATERIALS AND METHODS
T7 preparation.
T7 (ATCC 11303-B7) was grown in Escherichia coli (ATCC 11303) host cells and purified according established methods (39). Briefly, the phage suspension was concentrated on a CsCl gradient and dialyzed against buffer (20 mM Tris-HCl, 50 mM NaCl [pH 7.4]) (14). T7 bacteriophage concentration was determined from optical density by using an extinction coefficient (ε260) of 7.3 × 103 (mol nucleotide bases/liter/cm). The dialyzed T7 samples were kept at 4°C for a few months without significant loss of activity. Prior to further use the T7 samples were diluted with PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4 [pH 7.4]).
Atomic force microscopy and nanomanipulation.
T7 samples properly diluted in PBS were applied to freshly cleaved mica functionalized with glutaraldehyde (29, 40). The dilution was adjusted so that an approximate surface density of 10 phage particles per μm2 was achieved. Freshly cleaved mica was first incubated with poly-l-lysine (0.01% aqueous solution) for 20 min at room temperature, then rinsed extensively with Milli-Q water, and dried with a stream of high-purity N2 gas. Subsequently, the surface was incubated with 10% aqueous glutaraldehyde for 30 min at room temperature, then rinsed extensively with Milli-Q water, and dried with a stream of high-purity N2 gas. Finally, a sample of T7 phage was loaded onto the substrate surface and incubated for 40 min on ice. Unbound viruses were removed by gentle washing with PBS. Non-contact mode AFM images were acquired with an Asylum Research Cypher instrument (Asylum Research, Santa Barbara, CA) by using silicon-nitride cantilevers (Olympus BL-AC40TS-C2 or Nanoworld PNP-TR). Images (512 by 512 pixels) were collected at a typical scanning frequency of 0.3 to 1.5 Hz and with a mean indentation force of about 30 pN. All of the images presented in this work were collected on nonfixed samples under aqueous buffer conditions. For temperature-dependent measurements we used the cooler/heater stage of the AFM instrument. Temperature was kept constant with a precision of 0.1°C. Evaporation of water was prevented by the sealed container housing the AFM scanner. For nanomechanical measurements the surface-bound viruses were manipulated by first pressing the cantilever (Nanoworld PNP-TR, lever 1) tip against the apex of the virus and then pulling the cantilever away with a constant, preadjusted rate (29). Typical cantilever movement rate was 1 μm/s except where noted otherwise. Stiffness was determined for each cantilever by using the thermal method (41).
Image processing and data analysis.
AFM images and force spectra were analyzed using algorithms built in the Cypher controller software (Asylum Research, Santa Barbara, CA). Indentation distance (z) was calculated from cantilever displacement (s), force (F), and cantilever stiffness (k) as
| (1) |
AFM images were corrected for flatness of field (within a few angstroms), and their color contrast was adjusted in order to better communicate the relevant features. No additional image processing was carried out.
CD spectroscopy.
Circular dichroism measurements were carried out on a Jasco J-810 dichrograph in a 1-cm quartz cell. The solvent reference spectra were automatically subtracted from the CD spectra of the samples. The solvent was a buffer containing 20 mM Tris and 50 mM NaCl (pH 7.4). Temperature was controlled with a PFD-425S-type Peltier heating system. For temperature-controlled measurements, the sample cell was equilibrated for 3 min at the target temperature prior to data acquisition. CD band intensities were expressed in molar ellipticity, ϴ (deg · cm2/dmol). Spectra were smoothed by the Savitzky-Golay algorithm.
Statistics.
The numbers of nanomechanical curves, images, and particles (T7 or globular) analyzed are shown in the relevant figures. The results shown were collected in 15 independent nanomechanics and 9 independent AFM imaging experiments. The CD spectra are the averages from three scans. CD data were analyzed by using Microcal Origin software (OriginLab, Northampton, MA). Statistical analyses and graph plotting were carried out by using either the KaleidaGraph (v.4.5.1; Synergy Software, Reading, PA) or IgorPro (v. 6.34A; Wavementrics, Lake Oswego, OR) program.
ACKNOWLEDGMENTS
This work was supported by grants from the Hungarian National Research, Development and Innovation Office (K109480, K124966, VKSZ_14-1-2015-0052, and NVKP-16-1-2016-0017 [National Heart Program]). The research leading to these results has received funding from the European Union's Seventh Framework Program (FP7/2007-2013) under grant agreement no. HEALTH-F2-2011-278850 (INMiND).
Z.V. performed research, analyzed data, and wrote the paper; G.C. performed research, contributed analytic tools, and wrote the paper; L.H. analyzed data and wrote the paper; and M.K. designed research, performed research, analyzed data, and wrote the paper.
REFERENCES
- 1.Foster RAC, Johnson FH, Miller VK. 1949. The influence of hydrostatic pressure and urethane on the thermal inactivation of bacteriophage. J Gen Physiol 33:1–16. doi: 10.1085/jgp.33.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pollard EC, Solosko W. 1971. The thermal inactivation of T4 and lambda bacteriophage. Biophys J 11:66–74. doi: 10.1016/S0006-3495(71)86195-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamagishi H, Ozeki H. 1972. Comparative study of thermal inactivation of phage phi 80 and lambda. Virology 48:316–322. doi: 10.1016/0042-6822(72)90042-6. [DOI] [PubMed] [Google Scholar]
- 4.Wetzel R, Perry LJ, Baase WA, Becktel WJ. 1988. Disulfide bonds and thermal stability in T4 lysozyme. Proc Natl Acad Sci U S A 85:401–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brié A, Bertrand I, Meo M, Boudaud N, Gantzer C. 2016. The effect of heat on the physicochemical properties of bacteriophage MS2. Food Environ Virol 8:251–261. doi: 10.1007/s12560-016-9248-2. [DOI] [PubMed] [Google Scholar]
- 6.Wigginton KR, Kohn T. 2012. Virus disinfection mechanisms: the role of virus composition, structure, and function. Curr Opin Virol 2:84–89. doi: 10.1016/j.coviro.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pfaender S, Brinkmann J, Todt D, Riebesehl N, Steinmann J, Steinmann J, Pietschmann T, Steinmann E. 2015. Mechanisms of methods for hepatitis C virus inactivation. Appl Environ Microbiol 81:1616–1621. doi: 10.1128/AEM.03580-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wigginton KR, Pecson BM, Sigstam T, Bosshard F, Kohn T. 2012. Virus inactivation mechanisms: impact of disinfectants on virus function and structural integrity. Environ Sci Technol 46:12069–12078. doi: 10.1021/es3029473. [DOI] [PubMed] [Google Scholar]
- 9.Conway JF, Cheng N, Ross PD, Hendrix RW, Duda RL, Steven AC. 2007. A thermally induced phase transition in a viral capsid transforms the hexamers, leaving the pentamers unchanged. J Struct Biol 158:224–232. doi: 10.1016/j.jsb.2006.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duda RL, Ross PD, Cheng N, Firek BA, Hendrix RW, Conway JF, Steven AC. 2009. Structure and energetics of encapsidated DNA in bacteriophage HK97 studied by scanning calorimetry and cryo-electron microscopy. J Mol Biol 391:471–483. doi: 10.1016/j.jmb.2009.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qiu X. 2012. Heat induced capsid disassembly and DNA release of bacteriophage λ. PLoS One 7:e39793. doi: 10.1371/journal.pone.0039793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Csík G, Egyeki M, Herényi L, Majer Z, Tóth K. 2009. Role of structure-proteins in the porphyrin-DNA interaction. J Photochem Photobiol B 96:207–215. doi: 10.1016/j.jphotobiol.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 13.Tóth K, Rontó G. 1987. Salt effects on bacteriophage T7-I. Physiol Chem Phys Med NMR 19:59–66. [PubMed] [Google Scholar]
- 14.Tóth K, Csik G, Rontó GY. 1987. Salt effects on the bacteriophage T7-II structure and activity changes. Physiol Chem Phys Med NMR 19:67–74. [PubMed] [Google Scholar]
- 15.Zupán K, Herényi L, Tóth K, Majer Z, Csík G. 2004. Binding of cationic porphyrin to isolated and encapsidated viral DNA analyzed by comprehensive spectroscopic methods. Biochemistry 43:9151–9159. doi: 10.1021/bi0495477. [DOI] [PubMed] [Google Scholar]
- 16.Cerritelli ME, Cheng N, Rosenberg AH, McPherson CE, Booy FP, Steven AC. 1997. Encapsidated conformation of bacteriophage T7 DNA. Cell 91:271–280. doi: 10.1016/S0092-8674(00)80409-2. [DOI] [PubMed] [Google Scholar]
- 17.Fekete A, Rontó G, Feigin LA, Tikhonychev VV, Módos K. 1982. Temperature dependent structural changes of intraphage T7 DNA. Biophys Struct Mech 9:1–9. doi: 10.1007/BF00536011. [DOI] [PubMed] [Google Scholar]
- 18.Agirrezabala X, Velázquez-Muriel JA, Gómez-Puertas P, Scheres SHW, Carazo JM, Carrascosa JL. 2007. Quasi-atomic model of bacteriophage T7 procapsid shell: insights into the structure and evolution of a basic fold. Structure 15:461–472. doi: 10.1016/j.str.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 19.Guo F, Liu Z, Fang PA, Zhang Q, Wright ET, Wu W, Zhang C, Vago F, Ren Y, Jakana J, Chiu W, Serwer P, Jiang W. 2014. Capsid expansion mechanism of bacteriophage T7 revealed by multistate atomic models derived from cryo-EM reconstructions. Proc Natl Acad Sci U S A 111:E4606–E4614. doi: 10.1073/pnas.1407020111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michel JP, Ivanovska IL, Gibbons MM, Klug WS, Knobler CM, Wuite GJL, Schmidt CF. 2006. Nanoindentation studies of full and empty viral capsids and the effects of capsid protein mutations on elasticity and strength. Proc Natl Acad Sci U S A 103:6184–6189. doi: 10.1073/pnas.0601744103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kol N, Gladnikoff M, Barlam D, Shneck RZ, Rein A, Rousso I. 2006. Mechanical properties of murine leukemia virus particles: effect of maturation. Biophys J 91:767–774. doi: 10.1529/biophysj.105.079657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ivanovska IL, Miranda R, Carrascosa JL, Wuite GJL, Schmidt CF. 2011. Discrete fracture patterns of virus shells reveal mechanical building blocks. Proc Natl Acad Sci U S A 108:12611–12616. doi: 10.1073/pnas.1105586108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurland NE, Drira Z, Yadavalli VK. 2012. Measurement of nanomechanical properties of biomolecules using atomic force microscopy. Micron 43:116–128. doi: 10.1016/j.micron.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 24.Castellanos M, Pérez R, Carrasco C, Hernando-Perez M, Gómez-Herrero J, de Pablo PJ, Mateu MG. 2012. Mechanical elasticity as a physical signature of conformational dynamics in a virus particle. Proc Natl Acad Sci U S A 109:12028–12033. doi: 10.1073/pnas.1207437109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mateu MG. 2012. Mechanical properties of viruses analyzed by atomic force microscopy: a virological perspective. Virus Res 168:1–22. doi: 10.1016/j.virusres.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 26.Cieplak M, Robbins MO. 2013. Nanoindentation of 35 virus capsids in a molecular model: relating mechanical properties to structure. PLoS One 8:e63640. doi: 10.1371/journal.pone.0063640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hernando-Pérez M, Lambert S, Nakatani-Webster E, Catalano CE, de Pablo PJ. 2014. Cementing proteins provide extra mechanical stabilization to viral cages. Nat Commun 5:4520. doi: 10.1038/ncomms5520. [DOI] [PubMed] [Google Scholar]
- 28.Ramalho R, Rankovic S, Zhou J, Aiken C, Rousso I. 2016. Analysis of the mechanical properties of wild type and hyperstable mutants of the HIV-1 capsid. Retrovirology 13:17. doi: 10.1186/s12977-016-0250-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vörös Z, Csík G, Herényi L, Kellermayer MSZ. 2017. Stepwise reversible nanomechanical buckling in a viral capsid. Nanoscale 9:1136–1143. doi: 10.1039/C6NR06598H. [DOI] [PubMed] [Google Scholar]
- 30.Kellermayer MSZ, Vörös Z, Csík G, Herényi L. 2018. Forced phage uncorking: viral DNA ejection triggered by a mechanically sensitive switch. Nanoscale 10:1898–1904. doi: 10.1039/C7NR05897G. [DOI] [PubMed] [Google Scholar]
- 31.Serwer P. 1976. Internal proteins of bacteriophage T7. J Mol Biol 107:271–291. doi: 10.1016/S0022-2836(76)80005-8. [DOI] [PubMed] [Google Scholar]
- 32.Steven AC, Trus BL, Maizel JV, Unser M, Parry DA, Wall JS, Hainfeld JF, Studier FW. 1988. Molecular substructure of a viral receptor-recognition protein. The gp17 tail-fiber of bacteriophage T7. J Mol Biol 200:351–365. [DOI] [PubMed] [Google Scholar]
- 33.Molineux IJ. 2001. No syringes please, ejection of phage T7 DNA from the virion is enzyme driven. Mol Microbiol 40:1–8. doi: 10.1046/j.1365-2958.2001.02357.x. [DOI] [PubMed] [Google Scholar]
- 34.Molineux IJ, Panja D. 2013. Popping the cork: mechanisms of phage genome ejection. Nat Rev Microbiol 11:194–204. doi: 10.1038/nrmicro2988. [DOI] [PubMed] [Google Scholar]
- 35.Molineux IJ. 2006. Fifty-three years since Hershey and Chase; much ado about pressure but which pressure is it? Virology 344:221–229. doi: 10.1016/j.virol.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 36.Lin Z, Puchalla J, Shoup D, Rye HS. 2013. Repetitive protein unfolding by the trans ring of the GroEL-GroES chaperonin complex stimulates folding. J Biol Chem 288:30944–30955. doi: 10.1074/jbc.M113.480178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oberhauser AF, Marszalek PE, Carrion-Vazquez M, Fernandez JM. 1999. Single protein misfolding events captured by atomic force microscopy. Nat Struct Biol 6:1025–1028. [DOI] [PubMed] [Google Scholar]
- 38.Hammarstrom P, Persson M, Carlsson U. 2001. Protein compactness measured by fluorescence resonance energy transfer. Human carbonic anhydrase ii is considerably expanded by the interaction of GroEL. J Biol Chem 276:21765–21775. [DOI] [PubMed] [Google Scholar]
- 39.Strauss JH, Sinsheimer RL. 1963. Purification and properties of bacteriophage MS2 and of its ribonucleic acid. J Mol Biol 7:43–54. [DOI] [PubMed] [Google Scholar]
- 40.Wang H, Bash R, Yodh JG, Hager GL, Lohr D, Lindsay SM. 2002. Glutaraldehyde modified mica: a new surface for atomic force microscopy of chromatin. Biophys J 83:3619–3625. doi: 10.1016/S0006-3495(02)75362-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matei GA, Thoreson EJ, Pratt JR, Newell DB, Burnham NA. 2006. Precision and accuracy of thermal calibration of atomic force microscopy cantilevers. Rev Sci Instrum 77:083703. doi: 10.1063/1.2336115. [DOI] [Google Scholar]