Although E3 region-encoded proteins are involved in the modulation of leukocyte functions (N. Arnberg, Proc Natl Acad Sci U S A 110:19976–19977, 2013) and inducing a lytic infection of lymphocytes (V. K. Murali, D. A. Ornelles, L. R. Gooding, H. T. Wilms, W. Huang, A. E. Tollefson, W. S. Wold, and C. Garnett-Benson, J Virol 88:903–912, 2014), none of the E3 proteins appear to be a component of virion capsid or required for replication of adenovirus. Here, we demonstrate that the 13.7K protein encoded by the E3 region of porcine adenovirus type 3 is a component of progeny virion capsids and appears to be essential for maintaining the integrity of virion capsid and production of infectious progeny virions. To our knowledge, this is the first report to suggest that an adenovirus E3-encoded protein is an essential structural protein.
KEYWORDS: 13.7K, E3, essential protein, PAdV-3, porcine adenovirus, structural protein
ABSTRACT
The adenovirus E3 region encodes proteins that are not essential for viral replication in vitro. The porcine adenovirus type 3 (PAdV-3) E3 region encodes three proteins, including 13.7K. Here, we report that 13.7K is expressed as an early protein, which localizes to the nucleus of infected cells. The 13.7K protein is a structural protein, as it is incorporated in CsCl-purified virions. The 13.7K protein appears to be essential for PAdV-3 replication, as mutant PAV13.73A expressing a mutated 13.7K could be isolated only in VIDO AS2 cells expressing the 13.7K protein. Analysis of PAV13.73A suggested that even in the presence of reduced levels of some late viral proteins, there appeared to be no effect on virus assembly and production of mature virions. Further analysis of CsCl-purified PAV13.73A by transmission electron microscopy revealed the presence of disrupted/broken capsids, suggesting that inactivation of 13.7K protein expression may produce fragile capsids. Our results suggest that the PAdV-3 E3 region-encoded 13.7K protein is a capsid protein, which appears to be essential for the formation of stable capsids and production of infectious progeny virions.
IMPORTANCE Although E3 region-encoded proteins are involved in the modulation of leukocyte functions (N. Arnberg, Proc Natl Acad Sci U S A 110:19976–19977, 2013) and inducing a lytic infection of lymphocytes (V. K. Murali, D. A. Ornelles, L. R. Gooding, H. T. Wilms, W. Huang, A. E. Tollefson, W. S. Wold, and C. Garnett-Benson, J Virol 88:903–912, 2014), none of the E3 proteins appear to be a component of virion capsid or required for replication of adenovirus. Here, we demonstrate that the 13.7K protein encoded by the E3 region of porcine adenovirus type 3 is a component of progeny virion capsids and appears to be essential for maintaining the integrity of virion capsid and production of infectious progeny virions. To our knowledge, this is the first report to suggest that an adenovirus E3-encoded protein is an essential structural protein.
INTRODUCTION
Porcine adenovirus (PAdV) is a member of the genus Mastadenovirus within the family Adenoviridae. It was first isolated from a rectal swab of a piglet with diarrhea (1) and subsequently has been isolated from healthy pig feces, piglet kidney cell cultures, and encephalitic piglet brains (2, 3). PAdV type 3 (PAdV-3), one of the five known serotypes, is characterized by low virulence and the ability to grow to high titers in cell culture (4, 5); therefore, it has been chosen as a candidate vector for vaccine design (6, 7). Molecular characterization has identified its nucleotide sequence and transcriptional map (8). The overall genome organization of PAdV-3 exhibits generally a colinear gene arrangement similar to that of human adenoviruses (HAdVs), with minor differences (8, 9). In contrast, significant differences were reported in proteins encoded by the PAdV-3 E1, E3, and E4 regions and proteins encoded by the E1, E2, E3, and E4 regions of different members of the Mastadenovirus genus (10–12).
The E3 regions of different animal adenoviruses are variable in size and range from 1 kb to 5.6 kb (8, 12–16). The adenovirus E3 region is complex and produces several overlapping mRNAs, encoding different proteins (17). Although the E3 transcription unit is nonessential for replication of adenovirus (18) and can be used for insertion of transgenes (6, 7, 17, 18), the efficient expression of some transgenes requires the presence of some E3 genes (18).
The E3 region of PAdV-3 is 1,179 bp (8, 12) and produces three overlapping open reading frames (ORFs) (13.7K, 23K, and 13.1K). Analysis of the amino acid sequence of PAdV-3 E3 13.7K does not show any significant amino acid identity with proteins encoded by the E3 region of nonporcine adenoviruses (data not shown). Since the adenovirus E3 transcription unit is nonessential for viral replication (17), we hypothesized that deletion of the E3 region may not affect PAdV-3 replication. Although we successfully isolated a mutant PAdV-3 containing deletions of 23K and 13.1K ORFs (546 bp) in the E3 region (7), surprisingly, repeated attempts to isolate PAdV-3 containing a complete deletion of 13.7K, 23K, and 13.1K ORFs proved futile.
The E3-encoded 13.7K is 117 amino acids long and shows over 95% amino acid identity to homologs encoded by E3 regions of PAdV-1 and PAdV-2 (19), suggesting that 13.7K may have an important role in virus life cycle. To the best of our knowledge, currently, no data are available on the role of the 13.7K protein encoded by the E3 region of PAdV-3 in the replication process of the PAdV-3.
In this report, we demonstrate that 13.7K is expressed early in infection and localizes to the nucleus of the infected cell. Moreover, 13.7K appears to be incorporated in purified virion capsids and plays an essential role in stabilizing the capsid integrity and production of infectious progeny PAdV-3 virions.
RESULTS
Sequence analysis of PAdV-3 13.7K.
Amino acid sequence analysis of 13.7K (19) demonstrated homology to E3 proteins encoded by other members of Mastadenovirus, including ORF2 of PAdV-5 and the 12.5-kDa protein of human adenoviruses (20). The 13.7K protein and its homologs are predicted to be nonmembrane proteins, which do not contain a signal sequence, a transmembrane domain(s), or a potential nuclear localization signal. Although 13.7K contains an N-linked glycosylation motif (105NRS107), it may not be utilized due to the absence of a signal sequence. Moreover, compared to other proteins, 13.7K protein is rich in arginine residues. The arginine-rich viral proteins have been reported to be involved in different aspects of viral replication (21). Interestingly, 13.7K and its homologs are rich in cysteine residues. Cysteine-rich proteins have been speculated to be involved in the regulation of virus gene expression (22).
Expression of 13.7K protein.
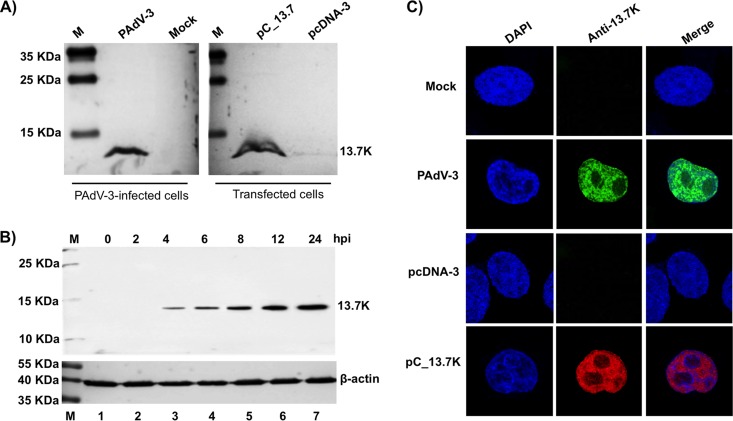
To characterize the 13.7K protein, the protein-specific antiserum raised in rabbits using the glutathione S-transferase (GST)-13.7K fusion protein was analyzed using Western blotting. As seen in Fig. 1A, anti-13.7K serum (this study) detected a protein of 13.7 kDa in PAdV-3-infected cells (Fig. 1A, lane 2) or in plasmid pC-13.7-transfected cells (lane 4). No such protein could be detected in mock-infected (lane 1) or plasmid pCDNA3 DNA-transfected cells (lane 3). Moreover, 13.7K could be detected as early as 2 to 4 h after PAdV-3 infection (Fig. 1B, lanes 3 to 7). Anti-β-actin was used as a loading control.
FIG 1.
Expression of 13.7K. (A, B) Western blots. Proteins from the lysates of mock-infected cells (lanes 1), PAdV-3-infected cells (panel A, lane 2; panel B, lanes 2 to 7), plasmid pcDNA-3 DNA-transfected cells (panel A, lane 3), or plasmid pC_13.7 DNA-transfected VIDO R1 cells (6) (panel A, lane 4) were separated by 12% SDS-PAGE, transferred to nitrocellulose, and analyzed by Western blotting using anti-13.7K serum. Expression of β-actin using an anti-β-actin MAb was used as a loading control. The positions of molecular weight markers in kilodaltons are shown on the left. The molecular weights (in kilodaltons) of observed proteins are shown on the right. (C) Subcellular localization of PAdV-3 13.7K. Monolayers of VIDO R1 (6) cells mock infected (top row), infected with PAdV-3 (second row), transfected with plasmid pcDNA-3 DNA (third row), or transfected with plasmid pC_13.7K DNA (bottom row) were analyzed by indirect immunofluorescence using anti 13.7K serum and Alexa Fluor 488-conjugated (top and second rows) or TRITC-conjugated (third and bottom rows) goat anti-rabbit serum. The cells were mounted in a Vectashield mounting medium with DAPI (Vector Laboratories) to stain nuclei and monitored by confocal microscope.
Subcellular localization of PAdV-3 13.7K.
To determine the subcellular localization of 13.7K in infected cells, VIDO R1 cells (6), either mock infected or infected with PAdV-3, were analyzed by indirect immunofluorescence using anti-13.7K serum as a primary antibody and Alexa Fluor 488-conjugated goat anti-rabbit antibody (for infected cells) or tetramethyl rhodamine (TRITC)-conjugated goat anti-rabbit antibody (for transfected cells) as a secondary antibody. Our results (Fig. 1C) revealed that 13.7K protein appears to be localized predominantly in the nucleus of the PAdV-3-infected (Fig. 1C, second row) or plasmid pC-13.7 DNA-transfected (Fig. 1C, bottom row) cells. No such protein could be detected in mock-infected (Fig. 1C, top row) or plasmid pCDNA3 DNA-transfected (Fig. 1C, third row) cells.
Detection of 13.7K in purified empty/mature PAdV-3 capsids.
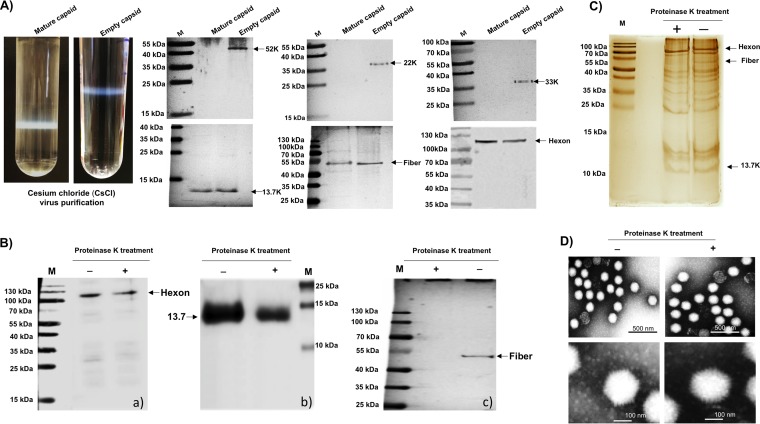
To determine whether 13.7K is a part of the virions, empty and mature capsids were purified from PAdV-3-infected cells using CsCl density gradient ultracentrifugation. Two distinct bands representing empty and mature capsids were collected separately and subjected to the second round of CsCl density gradient purification (Fig. 2A). The incorporation of 13.7K was detected by Western blotting using anti-13.7K serum. As seen in Fig. 2A, nonstructural proteins 52K, 22K, and 33K could be detected in purified empty capsids but not in mature virions. However, like hexon and fiber, 13.7K protein could be detected in both purified empty and mature capsids of PAdV-3.
FIG 2.
Analysis of CsCl-purified PAdV-3 virions. (A) CsCl density gradient purification of PAdV-3. The upper band contains empty capsids (EC), and the lower band contains mature capsid (MC) (upper panels). Two to five micrograms of PAdV-3 empty capsids or mature capsids was used with anti-13.7K, anti-52K (23), antifiber (23), antihexon (23) anti-22K (23), or anti-33K (23) serum. (B) Proteinase K treatment. Proteins from purified PAdV-3 untreated (lane −) or treated (lane +) with 20 μg of proteinase K were separated by 10% to 12% SDS-PAGE, transferred to nitrocellulose, and probed with antihexon serum (a), anti-13.7K serum (b), and antifiber serum (c). The molecular weight markers (M) in kilodaltons are shown. (C) Silver staining. Proteins from purified PAdV-3 untreated (lane −) or treated (lane +) with 20 μg of proteinase K were separated by 12% SDS-PAGE, and the gel was stained with silver staining. The positions of the molecular weights (M) in kilodaltons are shown on the left. (D) Transmission electron microscopy. Purified PAdV-3 virions untreated (lane −) or treated (lane +) with 20 μg proteinase K were negatively stained with 2% aqueous phosphotungstic acid and analyzed by transmission electron microscopy.
Detection of 13.7K protein in proteinase K-treated PAdV-3.
In order to determine whether 13.7K is loosely associated or actually incorporated in CsCl-purified empty or mature PAdV-3 capsids, the purified PAdV-3 virions were treated with proteinase K and purified by CsCl density gradient purification. Initially, proteinase K-treated PAdV-3 virions were analyzed by Western blotting using protein-specific antibodies. As seen in Fig. 2B, anti-hexon (23) serum detected a 103-kDa protein in both proteinase K-treated (lane “+”) and untreated (lane “−”) PAdV-3 (Fig. 2Ba). Similarly, anti-13.7K serum detected a 13.7-kDa protein in both proteinase-treated (+) and untreated (−) purified PAdV-3 (Fig. 2Bb). In contrast, anti-fiber serum (23) detected a 50-kDa protein in untreated (lane −) but not in proteinase-treated (lane +) PAdV-3 (Fig. 2Bc).
Next, the purity of proteinase K-treated purified virions was determined using silver staining of SDS-PAGE-separated viral proteins. The same amounts of proteinase K-treated and untreated purified PAdV-3 virions were separated on 12% SDS-PAGE and subjected to silver staining (24–28). As expected, several viral proteins bands, including 13.7K, hexon, and fiber proteins (Fig. 2C) could be resolved in untreated virions (lane −), However, similar results were observed in proteinase K-treated virions (lane +), except that fiber protein was absent (Fig. 2C).
To determine if proteinase K treatment affected the capsid stability, transmission electron microscopy (TEM) analysis was carried out. The proteinase K-treated and untreated purified PAdV-3 virions were analyzed by TEM. As seen in Fig. 2D, intact virions with icosahedral shape could be visualized in both proteinase K-treated (+) and untreated (−) purified PAdV-3.
To confirm the incorporation of 13.7K in mature PAdV-3 virions, the proteinase K-treated or untreated purified PAdV-3 virions were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). The entire sample was subjected to sequential one-dimensional reversed-phase chromatography coupled online to MS/MS analysis (one-dimensional [1D]-nanospray-LC-MS/MS). The proteomic results were statistically analyzed with Scaffold Q+S software package (Proteome Software, Inc., Portland, OR, USA). The LC-MS/MS analysis of untreated purified PAdV-3 virions revealed the presence of a unique PAdV-3-encoded 13.7K protein and 11 virus-encoded proteins (Table 1), which have been detected in other adenoviruses (26, 28). Similar results were obtained using proteinase K-treated purified PAdV-3, except peptides representing PAdV-3 fiber protein could be not detected (Table 2).
TABLE 1.
Proteins identified in CsCl-purified PAdV-3 (without proteinase K treatment) by LC-MS/MS
| Protein name | No. of identified peptides | Sequence coverage (%) | Mascot score |
|---|---|---|---|
| p13.7 | 44 | 33 | 659 |
| Fiber | 18 | 27 | 893 |
| Hexon | 289 | 69 | 1,012 |
| pVIII | 165 | 74 | 977 |
| pV | 18 | 23 | 614 |
| pX | 12 | 21 | 690 |
| pVI | 119 | 67 | 1,032 |
| pIIIa | 12 | 21 | 690 |
| pIII (penton) | 11 | 17 | 543 |
| pIX | 8 | 9 | 443 |
| DBP | 15 | 18 | 752 |
TABLE 2.
Proteins identified in protein K-treated purified PAdV-3 by LC-MS/MS after proteinase K treatment
| Protein name | No. of identified peptides | Sequence coverage (%) | Mascot score |
|---|---|---|---|
| p13.7 | 12 | 9 | 83 |
| Fiber | 0 | ||
| Hexon | 196 | 61 | 975 |
| pVIII | 123 | 67 | 840 |
| pV | 17 | 21 | 553 |
| pX | 2 | 4 | 74 |
| pVI | 75 | 54 | 866 |
| pIIIa | 4 | 3 | 59 |
| pIII (penton) | 9 | 15 | 445 |
| pIX | 5 | 6 | 96 |
| DBP | 4 | 7 | 123 |
Isolation of mutant PAV13.73A using VIDO R1 cells.
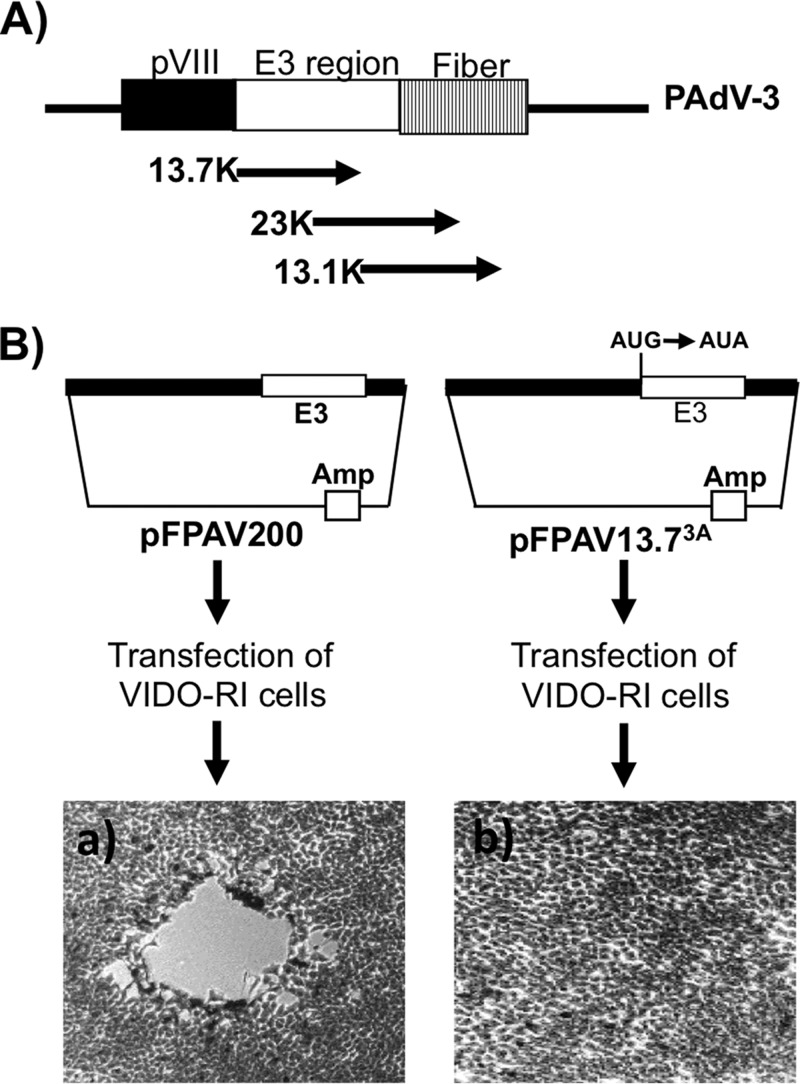
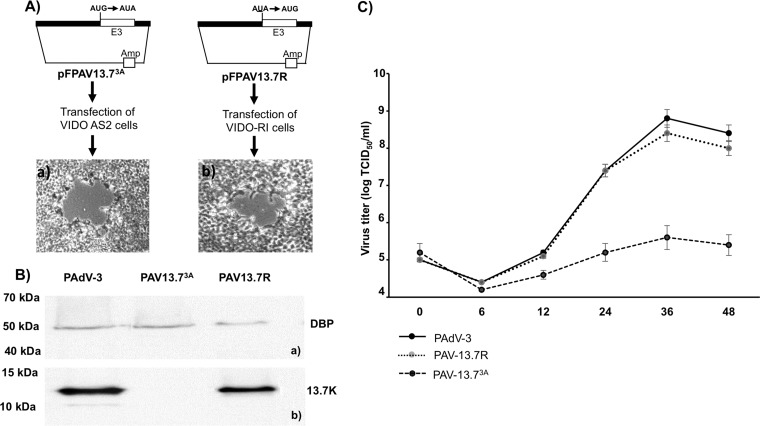
The genomic organization of the E3 region encoding three overlapping ORFs (13.7K, 23K, and 13.1K) located between pVIII and fiber ORFs is shown in the Fig. 3A. In order to determine whether the 13.7K gene is essential for PAdV-3 replication in VIDO R1 cells (not expressing 13.7K), we constructed a plasmid, pFPAV13.73A, containing the PAdV-3 genome with a point mutation (in bold and underlined) in the start codon AUG (AUG to AUA) of 13.7K ORF. The change resulted in the abrogation of 13.7K expression without affecting the transcription of the overlapping gene (23K). VIDO R1 cells (6) transfected with PacI-digested individual plasmid pFPAV200 DNA (containing the full-length PAdV-3 genome) (7) or pFPAV13.73A DNA (containing the full-length PAdV-3 genome with a point mutation in start codon of 13.7K AUG to AUA) were observed for the appearance of cytopathic effects (CPEs). After 7 days posttransfection, the CPEs were clearly visible in the cells transfected with pFPAV200 (Fig. 3B, left). However, no cytopathic effects could be observed in cells transfected with plasmid pFPAV13.73A DNA even after 15 days posttransfection. (Fig. 3B, right).
FIG 3.
Isolation of PAV13.73A in VIDO R1 cells (6). (A) Schematic illustration of the genomic organization of pVIII, early (E) region 3, and fiber in the PAdV-3 genome (8, 12). The E3 region encoding three overlapping proteins (13.7K, 23K, and 13.1K) is shown. (B) Schematic representation of plasmids pFPAV200 (7) and pFPAV13.73A. PAdV-3 genomic DNA (black filled box). E3 region (unfilled box). Plasmid DNA is represented by a thin line). Amp, ampicillin resistance gene. Mutation of 13.7K start codon AUG to AUA in plasmid pFPAV13.73A DNA is depicted. Micrographs represent cells showing cytopathic effect (a) and no cytopathic effect (b).
Generation of VIDO AS2 cells stably expressing 13.7K protein.
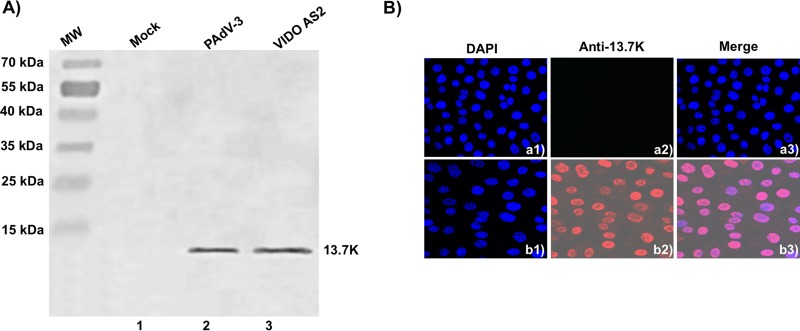
To develop a stable cell line expressing the PAdV-3 13.7K protein, VIDO R1 cells (6) were transduced with lentivirus expressing 13.7K as described previously (29). After incubation of transduced cells in medium containing hygromycin for 5 to 7 days, selected hygromycin-resistant clones were expanded and analyzed for the expression of 13.7K protein by Western blotting and immunofluorescence (IF) assay using the anti-13.7K-specific serum. As seen in Fig. 4A, anti-13.7K serum detected a protein of 13.7 kDa in the lysates of PAdV-3-infected cells (lane 2) or hygromycin-resistant clone VIDO AS2 (lane 3) expressing 13.7K. No such protein could be detected in mock-infected cells (lane 1). Furthermore, the expression of 13.7K was also evaluated by IF assay using anti-13.7K serum and Alexa Fluor 488-conjugated goat anti-rabbit antibody as a primary and a secondary antibody, respectively. As seen in Fig. 4B, more than 95% of the cells showed fluorescence in VIDO AS2 cells (Fig. 4Bb1 to b3). No such fluorescence could be observed in VIDO R1 cells (Fig. 4Ba1 to a3).
FIG 4.
Expression of 13.7K in VIDO AS2 cell line. (A) Western blot. Proteins from the lysates (10 μl) of VIDO R1 cells (6) mock infected (lane 1) or infected with PAdV-3 at an MOI of 2 (lane 2) and of VIDO AS2 cells (lane 3) were analyzed by Western blotting using anti-13.7K serum. (B) Indirect immunofluorescence. Monolayers of VIDO-R1 cells (a1 to a3) or VIDO AS2 cells (expressing protein 13.7K) (b1 to b3) were fixed with 3.7% paraformaldehyde and visualized by indirect immunostaining using anti-13.7K serum followed by TRITC-conjugated goat anti-rabbit IgG using a confocal microscope. The nuclei were stained by DAPI.
Isolation of mutant PAV13.73A using VIDO AS2 cells.
To isolate mutant PAdV-3 containing a defect in 13.7K expression, VIDO AS2 cells (expressing 13.7K) were transfected with PacI-digested plasmid pFPAV13.73A DNA and observed daily for development of cytopathic effects. After 10 days posttransfection, the cells showing cytopathic effects (Fig. 5Aa) were collected, freeze-thawed, and analyzed.
FIG 5.
Isolation of PAV13.73A in VIDO R1 cells. (A) Schematic representation of plasmid pFPAV13.73A and plasmid pFPAV13.7R DNAs as described for Fig. 3B. (B) Proteins from the lysates of cells infected with indicated viruses were analyzed by Western blotting using anti-DBP serum (24) as described for Fig. 1A and B. (C) Virus growth. Confluent monolayers of VIDO R1 were infected with PAdV-3 (wild-type), PAV13.73A (13.7K point mutation), or PAV13.7R (PAV13.73A revertant). At indicated times postinfection, the cell pellets were collected, freeze-thawed, and virus titrated on VIDO R1 (6) cells using TCID50 assay (6, 7). Values represent averages from two independent experiments, each with triplicate samples, and error bars represent standard deviations.
To confirm that PAV3 13.7K is responsible for the replication defect of PAV13.73A, VIDO R1 cells (6), not expressing 13.7K, were transfected with PacI-digested plasmid pFPAV13.7R (revertant) DNA and observed for the development of cytopathic effects. The cells showing cytopathic effect (Fig. 5Ab) at 7 days posttransfection were collected, freeze-thawed, and analyzed.
The identity of the recombinant virus, designated PAV13.73A (pFPV13.73A), and that of revertant PAV13.7R (pFPAV13.7R) were confirmed by Western blotting using anti-13.7K serum. As seen in Fig. 5B, anti-DNA binding protein (anti-DBP) serum (30) detected a 50-kDa protein in VIDO R1 (6) cells infected with PAdV-3, PAV13.73A, or PAV13.7R (Fig. 5Ba). Similarly, anti-13.7K serum detected 13.7K protein in cells infected with PAdV-3 or PAV13.7R (Fig. 5Bb). However, no such protein could be detected by anti-13.7K serum in cells infected with PAV13.73A (Fig. 5Bb).
Growth of PAV13.73A in VIDO R1 cells.
Since adenovirus E3-encoded proteins are not essential for virus replication, we determined whether PAV-13.73A can produce infectious virus in 13.7K-negative cells. VIDO R1 (6) cells were infected individually with PAV13.73A, PAV13.7R, or PAdV-3 at a multiplicity of infection (MOI) of 1. At different times (0, 6, 12, 24, 36, and 48 h) postinfection, the infected cells were collected, freeze-thawed, and used for titration by 50% tissue culture infectious dose (TCID50) assay in VIDO R1 cells. As seen in Fig. 5C, PAdV-3 and PAV13.7R (PAV.13.73A revertant) grew to a titer of 108.5 TCID50/ml at 36 h postinfection. However, PAV13.73A showed a negligible increase in the titer.
Protein expression in PAV13.73A-infected cells.
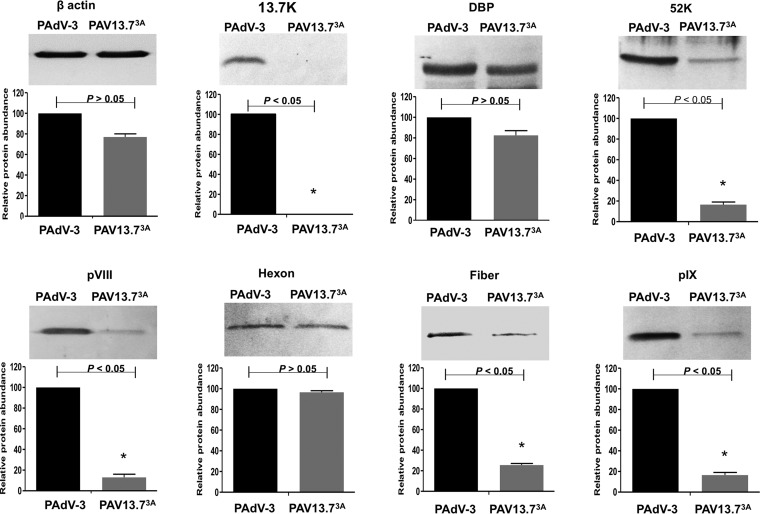
In order to assess whether restricting the expression of 13.7K protein alters the expression of other PAdV-3 proteins in infected cells, VIDO R1 (6) cells were infected with PAdV-3 or PAV13.73A at an MOI of 1. At 24 h postinfection, the cells were harvested, lysed, and subjected to Western blot analysis using protein-specific antisera. As expected, anti-13.7K serum detected a 13.7K protein in the cells infected with PAdV-3 but not in the cells infected with PAV13.73A (Fig. 6). Compared to PAdV-3, there appears to be a modest reduction in the level of expression of DBP and hexon. However, compared to PAdV-3, a significant reduction in the levels of expression of intermediate (pIX) and late viral (52K, pVIII, fiber) proteins could be observed in PAdV-13.73A-infected VIDO R1 cells (Fig. 6).
FIG 6.
Analysis of viral protein expression in virus-infected cells. Proteins from the lysates of PAdV-3- or PAV13.73A-infected cells were analyzed by Western blotting using anti-pIX (23), anti-DBP (30), anti-13.7K (this study), anti-52K (23), antihexon (23), anti-pVIII (50), and anti-fiber (22) antibodies followed by alkaline phosphatase-conjugated secondary antibodies. The β-actin was detected using anti-β-actin MAb followed by alkaline phosphatase-conjugated secondary antibodies. The BCIP/NBT solution was used as a substrate to visualize proteins. The results were quantified using ImageJ software (http://rsb.info.nih.gov/ij/). Values represent the means from two independent experiments, and error bars indicate SD. Significant differences (P < 0.05) are indicated with an asterisk (*).
Virus DNA replication.
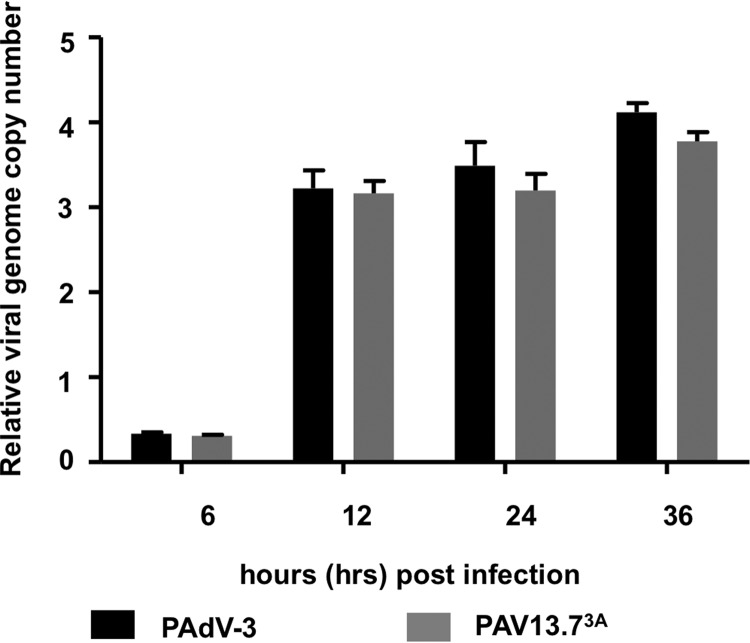
To determine if restricting 13.7K protein expression alters DNA replication, we analyzed DNA replication at different time points postinfection (6, 12, 24, and 36 h) by quantitative real-time PCR. As seen in Fig. 7, there was no significant difference in the viral genomic DNA replication of PAdV-3 or PAV13.73A in infected cells.
FIG 7.
Analysis of mutant PAV13.73A. Analysis of genome replication in virus-infected cells. VIDO R1 cells were infected with PAdV-3 or PAV13.73A at an MOI of 2 and harvested at indicated times postinfection. The cell pellets were collected, and genomic DNA was extracted. The viral genome copy number was determined by quantitative PCR using primers (Table 3) as described previously (46).
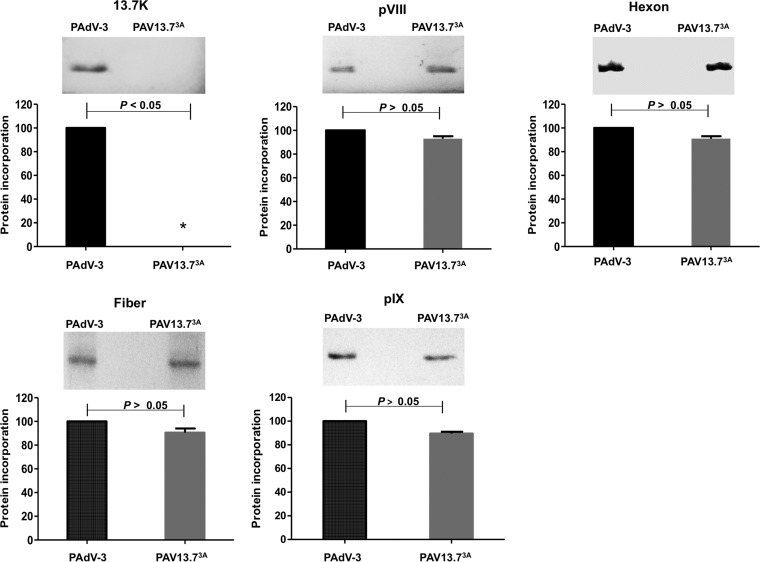
Protein incorporation in PAV13.73A virions.
To determine the incorporation of viral proteins in progeny virions, CsCl-purified virions were separated by 10 to 12% SDS-PAGE, transferred to nitrocellulose, and analyzed by Western blotting using protein-specific sera. As seen in Fig. 8, compared to PAdV-3, no significant difference could be observed in the incorporation of the viral proteins (pVIII, hexon, fiber, and pIX) in purified PAV13.73A virions. However, as expected, a protein of 13.7K could be detected in purified PAdV-3 virions but not in purified PAV13.73A virions.
FIG 8.
Analysis of viral protein incorporation in purified virions. Proteins from CsCl-purified PAdV-3 or PAV13.73A were separated on 10 to 12% SDS-PAGE, transferred to nitrocellulose, and probed by Western blotting using anti-13.7K (this study), anti-pVIII (50), anti-hexon (23), anti-fiber (23), and anti-pIX (23) sera. All data were analyzed using GraphPad Prism, version 6 (GraphPad Software, Inc., La Jolla, CA, USA). The values represent averages from three independent experiments, and errors bars represent standard deviations.
TEM of PAV13.73A-infected cells.
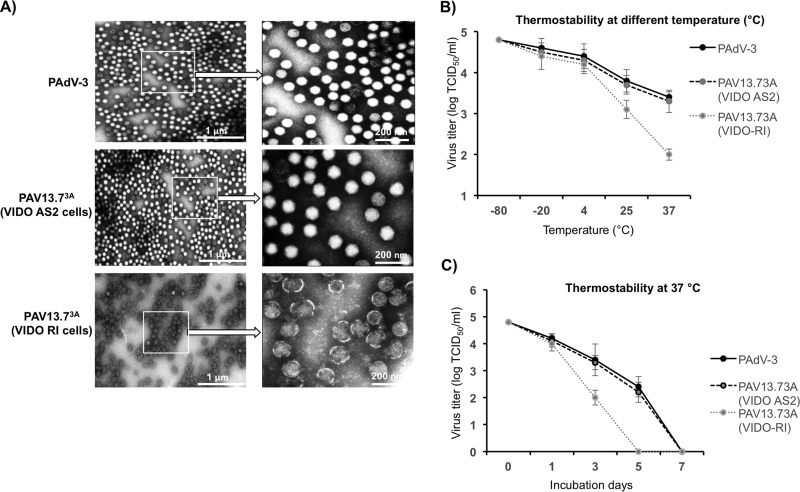
To investigate if restricting the expression of 13.7K protein affects the structure of the virions, PAdV-3 and PAV13.73A individually propagated in VIDO R1 (6) cells or PAV13.73A propagated in VIDO AS2 cells (VIDO R1 cells expressing 13.7K protein; this study) were purified by CsCl density gradient. The purified virus particles were observed by negative-staining electron microscopy. As seen in Fig. 9A, most of the virions of PAdV-3 or PAV13.73A cultivated in VIDO AS2 cells were observed to have intact capsids with a typical icosahedral shape. In contrast, PAV13.73A virions cultivated in VIDO R1 (6) cells depicted a more circular shape with disrupted capsids.
FIG 9.
Purified virus analysis and interaction of protein 13.7K with other PADV-3 protein(s). (A) Electron microscopic analysis. Purified PAdV-3 and PAV13.73A grown in VIDO AS2 cells and PAV13.73A grown in VIDO R-1 cells (6) are shown as indicated at a magnification of ×30,000. The arrows indicate enlargements of selected boxed regions of each virus (magnification, ×1,000,000). (B, C) Thermostability assay. Purified virions (105 TCID50) of PAdV-3 and PAV13.73A grown in either VIDO R1 or VIDO AS2 (expressing protein 13.7K) cells were incubated at different temperatures for 3 days, and the residual viral infectivity was determined by TCID50 (B). Purified virions (105 TCID50) of PAdV-3 and PAV13.73A grown in either VIDO R1 or VIDO AS2 cells were incubated at 37°C for the indicated time points (0, 1, 3, or 7 days postinfection), and the residual viral infectivity was determined by TCID50 (C).
Thermostability of PAV13.73A.
The thermostability of purified virions was determined as described earlier (30). Initially, the infectivity of 105 purified viral particles was determined by TCID50 assay after incubating the individual viruses at different temperatures for 3 days in phosphate-buffered saline (PBS) containing 10% glycerol. As seen in Fig. 9B, the thermostability or dynamics of viral inactivation of PAV13.73A grown in VIDO R1 cells (6) appears to be different from that of PAdV-3 or PAV13.73A grown in VIDO AS2 cells. Next, the infectivity of 105 purified virus particles was determined by TCID50 assay after incubating the individual viruses at 37°C for different numbers of days in PBS containing 10% glycerol. As seen in Fig. 9C, compared to PAdV-3 or PAV13.73A (grown in VIDO AS2 cells), there was rapid loss of infectivity of PAV13.73A grown in VIDO R1 cells (6).
DISCUSSION
Despite divergence in the size and number of genes carried by the adenovirus E3 region (10, 15–18), a positional homolog of the E3 region appears to be conserved in members of Mastadenovirus (10–16), suggesting an important role for E3 in the virus life cycle (17, 31–33). The proteins encoded by the E3 region not only help in immune evasion by adenovirus (32), including modulation of leukocyte functions (31), but are also involved in inducing lytic infection of lymphocytes (33) and helping in the stable expression of toxic transgene products in adenovirus vectors (18). In spite of the preservation of the E3 region and its important role in the adenovirus life cycle, none of the proteins encoded by adenovirus E3 regions appear to be a component of the progeny virion or required for adenovirus replication (18). In the present study, we demonstrate that 13.7K protein encoded by the PAdV-3 E3 region is incorporated in the mature and immature virions and appears to be essential for the replication of PAdV-3 and stability of virion capsid. To our knowledge, this is the first report to suggest that adenovirus E3-encoded 13.7K protein is a structural protein required for efficient viral replication.
The PAdV-3 E3 region 13.7K gene, encoding a protein of 117 amino acids, is expressed as a 13.7-kDa protein early in infection, which translocates to the nucleus of PAdV-3-infected cells. To date, reports suggest that proteins encoded by the adenovirus E3 region are not incorporated in the viral capsid (26, 28). Our observations demonstrate that 13.7K protein is a component of mature PAdV-3 virions. First, Western blot analysis of CsCl-purified empty and mature capsids detected a protein of 13.7K using anti-13K serum. Second, Western blot analysis of proteinase K-treated CsCl gradient-purified mature PAdV-3 virions also detected a protein of 13.7K using anti-13.7K serum. Finally, proteomic analysis of protein K-treated CsCl-purified mature PAdV-3 but not PAV13.7K3A virions using liquid chromatography-mass spectrometry detected a protein of 13.7K.
Earlier, a mutant PAdV-3 containing a deletion of E3 ORFs including 13.7K could not be isolated (7). Similarly, a mutant PAdV-5 containing a deletion of 12.5K, a positional homolog of 13.7K, could not be isolated (20). These results suggested that 13.7K may be essential for the replication of PAdV-3. The isolation of a viable mutant PAV13.7K3A (not expressing 13.7K) using the VIDO AS2 cell line (providing 13.7K protein in trans) confirms that 13.7K is essential for efficient replication of PAdV-3. However, the HAdV type 5 E3-encoded 12.5K protein showing limited homology (40%) to PAdV-3 13.7K does not appear to be essential for viral replication (18). Interestingly, homologs of PAdV-3 ORF2 in PAdV-1 and PAdV-2 show amino acid identities of 97.4% and 94%, respectively, with PAdV-3 ORF2 (19), suggesting that PAdVs may have a unique feature of encoding an E3 protein, which appears to be essential for virus replication.
While deletion of some adenovirus proteins does not affect capsid formation and genome packaging (37), it may affect virus entry in naive cells (39). In contrast, deletion of some adenovirus proteins may lead to capsid formation but may affect genome packaging (34–38). Inactivation of 13.7K expression did not affect (i) the virus infectivity and DNA replication of PAV13.73A (grown in VIDO AS2 cells) or (ii) early gene expression in PAV13.73A-infected cells. However, in spite of a reduction in the expression of some late genes, capsid formation and virion assembly do not appear to be severely affected, as there appears to be no difference in the incorporation of proteins in mutant PAV13.7K3A virus capsids. Moreover, the purification of both empty and mature capsids by CsCl gradient analysis of PAV13.73A-infected VIDO R1 (6) cells suggests that neither capsid formation nor genome packaging is mainly affected by inactivation of 13.7K expression.
The interaction of different structural proteins in adenovirus capsid is critical for the stability of mature virions (40–42). Earlier reports analyzing the atomic models of adenovirus have predicted the association of different structural proteins, which may be required for proper assembly of adenovirus and formation of stable virions (42–45). Further analysis of mutant adenoviruses suggested that deletion/mutation of adenovirus proteins, including IVa2 (34), VIII (46), and pV (40, 47), leads to the production of disrupted/aberrant capsids. Our results demonstrate that inactivation of E3 13.7K protein expression results in the production of disfigured capsids, suggesting that 13.7K appears to be essential for maintaining the stability of PAdV-3 virions.
Adenovirus cement proteins (pIX, IIIa, VI, V) are arranged in two layers, viz., an outer layer (pIX, IIIa) and an inner layer (VI, VIII, V), which are involved mainly in stabilizing the hexon capsid (42–44). Protein pIX stabilizes the adenovirus virion by cementing the group of nine hexons (42–44). Protein IIIa appears to cement the penton base with peripentonal hexons. Protein IV along with other proteins (V and VIII) stabilizes adjacent peripentonal hexons (42–44). It is possible that inactivation of E3 13.7K protein expression disrupts the stability of the hexon shell, leading to production of fragile capsids, which may be broken due to harsh treatment during CsCl/TEM procedures. Alternatively, it is possible that production of virions with fragile capsids could be due to the inefficient packaging of virion DNA (48). Several lines of evidence suggest that the capsid instability could be due to the loss of interaction of 13.7K with other adenovirus cement proteins. First, coimmunoprecipitation experiments suggested that 13.7K interacts with IIIa (23). Second, biomolecular fluorescence complementation (BiFC) assays detected interactions of 13.7K with pIX, IIIa, and pIV (A. Said and S. K. Tikoo, unpublished data). Finally, analysis of PAV13.7K3A-infected cells produced virion banding at a CsCl gradient consistent with the DNA packaging and formation of mature virions.
The E3 region of subgroup C HAdVs encodes a membrane protein named adenovirus death protein (ADP), which is modified by addition of oligosaccharides and palmitic acid (49). ADP, predominantly a late protein, promotes lysis of infected cells (49). Although homologs of ADP have been identified in other HAdV serotypes, none of them appear to be involved in lysis of infected cells (49).
Mutant PAV13.73A produces progeny virions without producing cytopathic effects. It is possible that 13.7K may be involved in facilitating the lysis of infected cells. However, the absence of amino acid homology, signal/transmembrane regions, and potential protein modifications (absence of multiple bands in Western blots) suggests that 13.7K may not perform function(s) similar to those of ADP. Alternatively, it is possible that the absence of viral plaque formation is due to the presence of a low number of progeny virus. Further experiments are required to test these speculations.
In summary, we have demonstrated that PAdV-3 13.7K protein (i) is incorporated into purified PAdV-3 virions, (ii) is essential for virus replication in vitro, and (iii) may not be required for virus assembly but appears to be required for maintaining the capsid integrity.
MATERIALS AND METHODS
Viruses and cells.
VIDO R1 (6), VIDO AS2 (VIDO R1 expressing PAdV-3 E3 13.7K; this study), and 293T (ATCC CRL-3216) cells were propagated at 37°C and 5% CO2 in minimal essential medium (MEM; Sigma), supplemented with 5% fetal bovine serum (FBS; Life Sciences), 0.1 mM nonessential amino acids (NEAA; Life Sciences), and 50 μg/ml gentamicin (HyClone Laboratories, Inc.). Wild-type and mutant PAdV-3 viruses were propagated as described earlier (6, 7, 9).
Antibodies.
Production of PAdV-3 anti-DBP serum has been described previously (30). The production and characterization of PAdV-3 anti-hexon serum and PAdV-3 anti-fiber serum will be described elsewhere. Briefly, anti-pIX, anti-52K, anti-hexon, anti-fiber, and anti-VIII sera recognize proteins of 22 kDa, 40 kDa, 103 kDa, 50 kDa (23), and 25/12/8 kDa (50), respectively, in PAdV-3-infected cells. The anti-β-actin mouse monoclonal antibody (MAb; Sigma-Aldrich), Alexa Fluor 488 goat anti-rabbit goat, TRITC-conjugated anti-rabbit and anti-mouse IgG alkaline phosphatase antibodies were purchased from Jackson ImmunoResearch.
Antiserum against PAdV3 13.7K was produced using GST-13.7K fusion protein. A 250-bp fragment encoding amino acids 5 to 117 of PAdV-3 E3 13.7K was amplified by PCR using primers (5′-CGGGATCCCCCAGGGTCAGCA-3′ and 5′-GCGTCGACTCAGCGGTCTCC-3′) and plasmid pFPAV200 (7) as the DNA template. The PCR fragment was digested with BamHI-SalI and ligated to BamHI-SalI-digested pGEX5X3, creating plasmid pGST-13.7. The GST alone or GST-13.7K fusion protein was induced and purified as described previously (30). Rabbit procedures were approved by the University Committee of Animal Care and Supply (protocol no. 19990212) at the University of Saskatchewan, Saskatoon, Canada, according to guidelines set by the Canadian Council of Animal Care. Rabbits were injected with purified fusion protein subcutaneously in Freund's complete adjuvant followed by three immunizations 3 weeks apart in Freund's incomplete adjuvant. One week after the last immunization, sera were collected and tested by Western blotting.
Construction of recombinant plasmids.
The construction of plasmid pFPAV200 (7), plasmid pMCS1 (51), and plasmid pTRIP-hygro (29) has been described earlier. Plasmids pMD2 and pSPAX2 were gifts from R. Brownlie (VIDO, University of Saskatchewan, Saskatoon, SK, Canada). The following plasmids were constructed using standard procedures (52). The identity of the plasmids was confirmed by restriction enzyme analysis and DNA sequencing.
(i) Plasmid pC-13.7.
A 354-bp DNA fragment containing PAdV-3 13.7K gene was amplified by PCR using plasmid pFPAV200 (7) as the DNA substrate and primers P1 and P2 (Table 3). The PCR products were digested with NotI and XhoI and ligated to Not1-Xho1-digested plasmid pcDNA3 (Invitrogen), creating plasmid pC-13.7K.
TABLE 3.
Oligonucleotides used in plasmid constructions
| Primer | Sequence (5′–3′)a |
|---|---|
| P1 | TTAGCGGCCGCATGACTGACGGTCCCCAG |
| P2 | ATTCTCGAGCTACGAGCGTTGTACAG |
| P3 | TTATCTAGAGCCGCCACCATGACTGACGGTCCCCAGGG |
| P4 | TTACTCGAGCTACGAGCGGTTGTACAGCTCA |
| P5 | TTAGCATGCAATTAGCCCGGGGGCGGAGCCCTC |
| P6 | GGCCCTGCAGGAGCCTGCTACAGAGTCCG |
| P7 | GGCCCTGCAGGCGTCAGCCCCCTACACCTC |
| P8 | GGCTACGTATTGCCCTACGGTTGGCTC |
| P9 | GTTCAATTGCAATTGCATGACATCAAAAAG |
| P10 | TGACCCTGGGGACCGTCAGTTATAGCCTGCTACAGAGTCCGTT |
| P11 | AACGGACTCTGTAGCAGGCTATAACTGACGGTCCCCAGGGTCA |
| P12 | TGACCCTGGGGACCGTCAGTTATGGCCTGCTACAGAGTCCGTT |
| P13 | AACGGACTCTGTAGCAGGCTGTAACTGACGGTCCCCAGGGTCA |
| β-Actin FW | TGTCATGGACTCTGGGGATG |
| β-Actin RV | GGGCAGCTCGTAGCTCTTCT |
| Hexon FW | GGCGACGGAGACCTACTTTA |
| Hexon RV | TGAGTGTCCTCCTTGTCCAC |
Sequences in italics indicate additional bases that are not present in the target sequences; restriction enzyme nucleotide sequences are in bold letters. Underlined sequences indicate the start codon ATG (methionine) of 13.7K changed to ATA (isoleucine) and in PAV13.7R, ATA (isoleucine) changed to ATG (methionine).
(ii) Plasmid pTrip-13.7-hygromycin.
A 354-bp DNA fragment containing PAdV-3 13.7K gene was amplified by PCR using plasmid pFPAV200 (7) DNA as the template and primers P3 and P4 (Table 3) and ligated to NotI- and XhoI-digested plasmid pTrip-hygro (containing a hygromycin resistance marker), creating plasmid pTrip-CMV-13.7-hygromycin.
(iii) Plasmid pMCS-d13.7.
A 2,653-bp MfeI-SnaBI DNA fragment of plasmid pFPAV200 was isolated and ligated to a 2.1-kb MfeI-SnaBI fragment of plasmid pMCS1 (51), creating plasmid pMCS-13.7. Next, a 351-bp SphI-SbFI fragment was amplified using plasmid pMCS-13.7 DNA as a template and primers P5 and P6 (Table 3). Similarly, a 609-bp SbFI-SnaBI fragment was amplified by PCR using plasmid pMCS-13.7 DNA as a template and primers P7 and P8 (Table 3). In the third PCR, these two PCR fragments were annealed and used as a DNA template to amplify the 960-bp SphI-SnaBI DNA fragment using primers P7 and P8 (Table 3). Finally, the 960-bp SphI-SnaBI DNA was ligated to SphI-SnaBI-digested plasmid pMCS-13.7 DNA, creating plasmid pMCS-d13.7.
(iv) Plasmid pFPAVd13.7-Kan.
A 1.6-kb SbfI fragment (containing a kanamycin resistance gene) of plasmid pUC4K was isolated and ligated to SbfI-digested plasmid pMCS.d13.7 DNA, creating plasmid pMCS-d13.7-Kan. The recombinant plasmid pFPAVd13.7-Kan was generated by homologous recombination in Escherichia coli BJ5183 between plasmid pFPAV200 DNA and a 3,899-bp MfeI-SnaBI DNA fragment of plasmid pMCS-d13.7-Kan.
(v) Plasmid pFPAV13.73A.
A 1,713-bp fragment was amplified by PCR using plasmid pMCS-13.7 DNA as a template and primers P9 and P10 (Table 3). Similarly, a 983-bp fragment was amplified by PCR using plasmid pMCS-13.7 DNA as a template and primers P8 and P11 (Table 3). In a third PCR, two PCR fragments were annealed and external primers P8 and P9 (Table 3) were used for a PCR across to give a 2,696-bp MfeI-SnaBI fragment (containing a point mutation in start codon AUG of 13.7K, AUG to AUA). This PCR product was digested with MfeI-SnaBI and ligated to MfeI-SnaBI-digested plasmid pMCS1 DNA (51), creating plasmid pMCS-13.73A. The recombinant plasmid pFPAV13.73A was generated by homologous recombination in Escherichia coli BJ5183 between the SbF-I-digested plasmid pFPAVd13.7-KAN DNA and a 2,696-bp MfeI-SnaBI fragment (containing a point mutation in start codon AUG of 13.7K, AUG to AUA) of plasmid pMCS-13.73A.
(vi) Plasmid pFPAV13.7R.
A 1,713-bp fragment was amplified by PCR using plasmid pMCS-13.73A DNA as a template and primers P9 to P12 (Table 3). Similarly, a 983-bp fragment was amplified using plasmid pMCS-13.73A DNA as a template and by PCR using primers P8 and P13 (Table 3). In a third PCR, two PCR fragments were annealed and external primers P8 and P9 (Table 3) were used to amplify the 2,696-bp fragment (containing a change of AUA to AUG). This PCR product was digested with MfeI-SnaBI and ligated to MfeI-SnaBI-digested plasmid pMCS1 (51) DNA, creating plasmid pMCS-13.7R. The recombinant plasmid pFPAV13.7R was generated by homologous recombination in Escherichia coli BJ5183 between the SbF-I-digested plasmid pFPAVd13.7-Kan DNA and a 2,696-bp MfeI-SnaBI fragment of plasmid pMCS-13.7R.
Immunoblotting, silver staining, and indirect immunofluorescence (IF) analysis.
Expression of proteins was detected by Western blotting as described earlier (53). Briefly, the cells seeded in 6-well plates were infected with virus (wild-type or mutant PAdV-3) at an MOI of 1 or transfected with individual plasmids' DNA (2 to 3 μg per 106 cells). At indicated times postinfection or posttransfection, the lysates of the cells were analyzed by Western blotting using protein-specific primary antibodies and alkaline phosphatase conjugated as a secondary antibody. The BCIP/NBT solution (nitroblue tetrazolium with 5-bromo-4-chloro-3-indolyl phosphate; Sigma) was used as a substrate to visualize target protein bands.
For IF analysis, the cells grown in two-well chamber slides were infected with wild-type PAdV-3 or transfected with plasmid DNA (2 to 3 μg per 106 cells). After 24 h of infection or 36 h of transfection, the cells were processed as described earlier (40) using anti-13.7K serum as a primary antibody and Alexa Fluor 488- or TRITC-conjugated goat anti-rabbit antibody as a secondary antibody. Finally, cells were mounted by mounting buffer (Vector Laboratories) containing DAPI (4′,6-diamidino-2-phenylindole) and inspected under a confocal microscope (Leica SP5, Zeiss Two-Photon).
For silver staining analysis, purified PAdV-3 virions untreated or treated with proteinase K were processed and stained as described earlier (24–27).
Construction of VIDO AS2 cells (VIDO R1 cell line expressing PAdV-3 13.7K).
A lentivirus system was used to isolate VIDO R1 cells (6) expressing 13.7K as described earlier (29, 40). Briefly, 293T cells were cotransfected with DNAs from plasmids pTrip-CMV-13.7-hygromycin, pMD2.G (expressing vesicular stomatitis virus G protein), and pSPAX2 (expressing human immunodeficiency virus Gag/Pol protein) using Lipofectamine 2000 (Life Technologies). After 48 h posttransfection (h p.t.), the supernatant containing the lentivirus was harvested and used for lentivirus transduction of VIDO R1 (6) cells. After 24 h postransduction, medium was replaced by fresh medium containing 500 μg/ml hygromycin for selection. The hygromycin-resistant cell clones were selected, propagated in hygromycin-containing medium, and analyzed for expression of PAdV-3 13.7K using Western blot assay.
Isolation of mutant PAdV-3.
VIDO R1 (6) cells or VIDO AS2 cells (this report) in 6-well plates were transfected with PacI-digested plasmid DNA using Lipofectamine 2000 (Life Technologies). After 5 to 10 days posttransfection, the cells showing cytopathic effects were collected and freeze-thawed three times. The recombinant virus, named PAV13.73A, was propagated in VIDO AS2 cells (6, 7) and purified using CsCl density gradient centrifugation.
For isolation of revertant PAV13.7R, VIDO R1 (6) cells in 6-well plates were transfected with plasmid pFPAV13.7R DNA (2 to 3 μg per 106 cells). After 3 to 6 days posttransfection, the cells showing cytopathic effects were collected, and freeze-thawed three times. The revertant PAdV-3 virus, named PAV13.7R, was propagated in VIDO R1 (6) cells and purified using cesium chloride (CsCl) density gradient centrifugation.
Proteinase K treatment.
The purified PAdV-3 virions were treated with proteinase K exactly as described earlier (26). Briefly, the CsCl gradient-purified PAdV-3 virions were incubated in 1 ml of MNT buffer (20 μg proteinase K [AmbionTM; Thermo Fisher Scientific], 10 mM NaCl, 20 mM Tris-HCl [pH 7.4], and 30 mM morpholineethanesulfonic acid) for 1 h at room temperature (RT). Treated virions were purified by CsCl gradient density purification.
Growth of recombinant viruses.
The virus titers were determined as described earlier (6, 7). In brief, the VIDO R1 or VIDO AS2 cells were cultured in 96-well plates at 37°C and 5% CO2. Tenfold dilutions of PAdV-3, mutant PAV13.73A, and revertant PAV13.7R were prepared in MEM supplemented with 2% FBS. The cultured cells infected with the indicated viruses were incubated at 37°C and observed daily for cytopathic effects (CPEs). The titer of the virus was determined using the 50% tissue infectious dose (TCID50) assay. For virus growth kinetics, VIDO R1 cells were infected with either PAdV-3, PAV13.73A, or PAV-13.7R separately at an MOI of 1. The infected cells were harvested at different times (0, 6, 12, 24, 36, and 48 h) postinfection and analyzed using the TCID50 assay.
Virus DNA replication.
VIDO R1 cells (6) infected with PAdV-3 or PAV13.73A were harvested at indicated times postinfection. After centrifugation, the cell pellets were collected, genomic DNA was extracted, and the viral genome copy number was determined by quantitative real-time PCR as described previously (46). Specifically, two sets of oligonucleotide primers (β-actin FW and β-actin RV; Hexon FW and Hexon RV), listed in Table 3, were used in quantitative PCR. Viral genomic replication was calculated as the value of viral genome copy number divided by actin copy number. The experiment was performed independently three times, and each sample was tested in triplicate.
Transmission electron microscopy.
TEM analysis was carried out using CsCl gradient-purified PAdV-3 either treated or untreated with proteinase K as described before (26). For negative-staining preparation, the purified viruses were adsorbed for 1 min to a Formvar film on carbon-coated 300-mesh grids (Ted Pella, Inc.). After being washed three times with distal water, the grids were stained with 0.5% phosphotungstic acid (J. B. Em Services Inc., Dorval, QC, Canada) for 1 min. Finally, the grids with negatively stained virus particles were photographed at 80 kV using a Philips CM10 TEM at the EM facility of the veterinary school, University of Saskatchewan, Canada.
For TEM analysis, wild-type or mutant PAdV-3-infected cells were collected after 30 h postinfection, fixed in 2.5% glutaraldehyde, and postfixed with 1% OsO4 in 0.1 M PBS. The cells were rapidly dehydrated with a graded ethanol series and propylene oxide, followed by embedding of the specimens in a mixture of propylene oxide and Epon-812 (45345-250ML-F; Sigma) for 24 h. Ultrathin sections were obtained and stained with a Reichert Ultracut ultramicrotome and counterstained with alkaline lead citrate. Finally, the stained sections were imaged using a Philips CM10 TEM.
Liquid chromatography-tandem mass spectrometry analysis.
The LC-MS/MS analysis was performed at the University of Victoria Genome British Columbia Proteomics Centre, using CsCl gradient-purified PAdV-3. Briefly, a 100-μg sample of each protein was solubilized in 25 mM ammonium bicarbonate prior to reduction with 15 μl of 200 mM dithiothreitol (DTT) and incubated 45 min at 37°C. Alkylated samples were quenched and digested with 30 μl of 200 mM DTT and 10 μg of trypsin (Promega) at 37°C for 16 h. Prior to LC-MS/MS analysis, samples were desalted using an Oasis HLB 1cc cartridge (10 mg) and then rehydrated with 2% acetonitrile, 98% water, and 0.1% formic acid. Online reverse-phase chromatography using a Thermo Scientific EASY-nLC II system with a reversed-phase Magic C18AQ precolumn (Michrom BioResources Inc., Auburn, CA) was used to separate the peptide mixture. The coupled online chromatography system with an LTQ OrbitrapVelos mass spectrometer and equipped with a NanosprayFlex source (Thermo Fisher Scientific) was used to analyze the resulted peptides.
Raw files were created by Fusion Control software (Thermo Scientific) and analyzed with the Proteome Discoverer software suite (Thermo Scientific). Parameters for the spectrum selection to generate peak lists of the higher-energy collisional dissociation (HCD) spectra were as follows: activation type, HCD; s/n cutoff, 1.5; total intensity threshold, 0; minimum peak count, 1; precursor mass, 350 to 5,000 Da). The peak lists were submitted to an in-house Mascot 2.4.1 server against database Uniprot-Swissprot 20161219 (592,628 sequences; 221,445,742 residues). Statistical analyses of the Proteome Discover result files were performed with the Scaffold Q+S software package (Proteome Software, Inc., Portland, OR). The identified protein sequences with a mascot score greater than 30 were considered significant. Moreover, identified proteins were accepted when the protein probability was greater than 99.0% and contained at least 2 identified proteins or the peptide probability was greater than 95.0% peptides (26, 28). The manual systemic evaluation was used to identify PAdV-3 peptides.
Thermostability of PAdV-3.
The thermostability of purified virions was determined as described previously (40, 46).
Statistical analysis.
All data were analyzed using GraphPad Prism, version 6 (GraphPad Software, Inc., La Jolla, CA, USA). Statistical differences among the groups were calculated using an unpaired t test. P values are indicated in the figures.
ACKNOWLEDGMENTS
We are thankful to other members of the Tikoo laboratory for helpful suggestions and to Caron Pyne and members of the VIDO Animal Care Unit for their help in generating the 13.7K-specific sera.
This work was supported by a grant from the Natural Sciences and Engineering Research Council of Canada to S.K.T.
Footnotes
This is contribution 837 from VIDO-InterVac.
REFERENCES
- 1.Haig DA, Clarke MC, Pereira MS. 1964. Isolation of an adenovirus from a pig. J Compar Pathol 74:81–84. doi: 10.1016/S0368-1742(64)80009-6. [DOI] [PubMed] [Google Scholar]
- 2.Kadoi K, Inoue Y, Ikeda T, Kamata H, Yukawa M, Iwabuchi M, Inaba Y. 1995. Isolation of porcine adenovirus as a candidate of 5th serotype. J Basic Microbiol 35:195–204. doi: 10.1002/jobm.3620350314. [DOI] [PubMed] [Google Scholar]
- 3.Kadoi K, Iwabuchi M, Satoh T, Katase T, Kawaji T, Morichi T. 1997. Adenovirus isolation from spleen lymphocytes of apparently healthy pigs. New Microbiol 20:215–220. [PubMed] [Google Scholar]
- 4.Hirahara T, Yasuhara H, Matsui O, Yamanaka M, Tanaka M, Fukuyama S, Izumida A, Yoshiki K, Kodama K, Nakai M, et al. 1990. Isolation of porcine adenovirus from the respiratory tract of pigs in Japan. Nihon juigaku zasshi. Jpn J Vet Sci 52:407–409. doi: 10.1292/jvms1939.52.407. [DOI] [PubMed] [Google Scholar]
- 5.Derbyshire JB, Clarke MC, Collins AP. 1975. Serological and pathogenicity studies with some unclassified porcine adenoviruses. J Compar Pathol 85:437–443. doi: 10.1016/0021-9975(75)90031-6. [DOI] [PubMed] [Google Scholar]
- 6.Reddy PS, Idamakanti N, Babiuk LA, Mehtali M, Tikoo SK. 1999. Porcine adenovirus-3 as a helper dependent expression vector. J Gen Virol 80:2909–2916. doi: 10.1099/0022-1317-80-11-2909. [DOI] [PubMed] [Google Scholar]
- 7.Reddy PS, Idamakanti N, Hyun BH, Tikoo SK, Babiuk LA. 1999. Development of porcine adenovirus-3 as an expression vector. J Gen Virol 80(Part 3):563–570. doi: 10.1099/0022-1317-80-3-563. [DOI] [PubMed] [Google Scholar]
- 8.Reddy PS, Idamakanti N, Song JY, Lee JB, Hyun BH, Park JH, Cha SH, Bae YT, Tikoo SK, Babiuk LA. 1998. Nucleotide sequence and transcription map of porcine adenovirus type 3. Virology 251:414–426. doi: 10.1006/viro.1998.9418. [DOI] [PubMed] [Google Scholar]
- 9.Reddy PS, Tuboly T, Dennis JR, Derbyshire JB, Nagy E. 1995. Comparison of the inverted terminal repetition sequences from five porcine adenovirus serotypes. Virology 212:237–239. doi: 10.1006/viro.1995.1475. [DOI] [PubMed] [Google Scholar]
- 10.Davison AJ, Benko M, Harrach B. 2003. Genetic content and evolution of adenoviruses. J Gen Virol 84:2895–2908. doi: 10.1099/vir.0.19497-0. [DOI] [PubMed] [Google Scholar]
- 11.Kovacs ER, Benko M. 2009. Confirmation of a novel siadenovirus species detected in raptors: partial sequence and phylogenetic analysis. Virus Res 140:64–70. doi: 10.1016/j.virusres.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 12.Reddy PS, Nagy E, Derbyshire JB. 1995. Sequence analysis of putative pVIII, E3 and fibre regions of porcine adenovirus type 3. Virus Res 36:97–106. doi: 10.1016/0168-1702(94)00105-L. [DOI] [PubMed] [Google Scholar]
- 13.Kleiboeker SB. 1994. Sequence analysis of putative E3, pVIII, and fiber genomic regions of a porcine adenovirus. Virus Res 31:17–25. doi: 10.1016/0168-1702(94)90067-1. [DOI] [PubMed] [Google Scholar]
- 14.Linne T. 1992. Differences in the E3 regions of the canine adenovirus type 1 and type 2. Virus Res 23:119–133. doi: 10.1016/0168-1702(92)90072-H. [DOI] [PubMed] [Google Scholar]
- 15.Mittal SK, Prevec L, Babiuk LA, Graham FL. 1993. Sequence analysis of bovine adenovirus type 3 early region 3 and fibre protein genes. J Gen Virol 74(Part 12):2825. doi: 10.1099/0022-1317-74-12-2825. [DOI] [PubMed] [Google Scholar]
- 16.Tan B, Yang XL, Ge XY, Peng C, Zhang YZ, Zhang LB, Shi ZL. 2016. Novel bat adenoviruses with an extremely large E3 gene. J Gen Virol 97:1625–1635. doi: 10.1099/jgv.0.000470. [DOI] [PubMed] [Google Scholar]
- 17.Wold WS, Tollefson AE, Hermiston TW. 1995. E3 transcription unit of adenovirus. Curr Top Microbiol Immunol 199(Part 1):237–274. [DOI] [PubMed] [Google Scholar]
- 18.Haut LH, Gill AL, Kurupati RK, Bian A, Li Y, Giles-Davis W, Xiang Z, Zhou XY, Ertl HC. 2016. A partial E3 deletion in replication-defective adenoviral vectors allows for stable expression of potentially toxic transgene products. Hum Gene Ther Methods 27:187–196. doi: 10.1089/hgtb.2016.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reddy PS, Idamakanti N, Derbyshire JB, Nagy E. 1996. Porcine adenovirus types 1,2 and 3 have short and simple early E3 regions. Virus Res 43:99–109. doi: 10.1016/0168-1702(96)01336-6. [DOI] [PubMed] [Google Scholar]
- 20.Tuboly T, Nagy E. 2000. Sequence analysis and deletion of porcine adenovirus serotype 5 E3 region. Virus Res 68:109–117. doi: 10.1016/S0168-1702(00)00157-X. [DOI] [PubMed] [Google Scholar]
- 21.Chandana T, Venkatesh YP. 2016. Occurrence, functions and biological significance of arginine rich proteins. Curr Protein Pept Sci 17:507–516. doi: 10.2174/1389203717666151201192348. [DOI] [PubMed] [Google Scholar]
- 22.Koonin KV, Boyko VP, Dolja VV. 1991. Small cysteine rich proteins of different RNA viruses are related to different families of nucleic acid binding proteins. Virology 181:395–398. doi: 10.1016/0042-6822(91)90512-A. [DOI] [PubMed] [Google Scholar]
- 23.Kessel JAV. 2006. Molecular and functional characterization of the IIIA protein of porcine adenovirus type 3. MSc thesis. University of Saskatchewan, Saskatoon, SK, Canada: https://ecommons.usask.ca/handle/10388/etd-04252006-105057. [Google Scholar]
- 24.Kramer T, Greco TM, Enquist LW, Cristea IM. 2011. Proteomic characterization of pseudorabies virus extracellular virions. J Virol 85:6427–6441. doi: 10.1128/JVI.02253-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lete C, Palmeira L, Leroy B, Mast J, Machiels B, Wattiez R, Vanderplasschen A, Gillet L. 2012. Proteomic characterization of bovine herpesvirus 4 extracellular virions. J Virol 86:11567–11580. doi: 10.1128/JVI.00456-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar P, van den Hurk J, Ayalew LE, Gaba A, Tikoo SK. 2015. Proteomic analysis of purified turkey adenovirus 3 virions. Vet Res 46:79. doi: 10.1186/s13567-015-0214-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morrissey JH. 1981. Silver stain for proteins in polyacrylamide gels: a modified procedure with enhanced uniform sensitivity. Anal Biochem 117:307–310. doi: 10.1016/0003-2697(81)90783-1. [DOI] [PubMed] [Google Scholar]
- 28.Chelius D, Huhmer AF, Shieh CH, Lehmberg E, Traina JA, Slattery TK, Pungor E Jr. 2002. Analysis of the adenovirus type 5 proteome by liquid chromatography and tandem mass spectrometry methods. J Proteome Res 1:501–513. doi: 10.1021/pr025528c. [DOI] [PubMed] [Google Scholar]
- 29.Du E, Tikoo SK. 2010. Efficient replication and generation of recombinant bovine adenovirus-3 in non-bovine cotton rat lung cells expressing I-Sce_I endonuclease. J Gen Med 12:840–847. doi: 10.1002/jgm.1505. [DOI] [PubMed] [Google Scholar]
- 30.Zhou Y, Pyne C, Tikoo SK. 2001. Characterization of DNA binding protein of porcine adenovirus type 3. Intervirology 44:350–354. doi: 10.1159/000050070. [DOI] [PubMed] [Google Scholar]
- 31.Windheim M, Southcombe JH, Kramer E, Chaplin L, Urlaub D, Falk CS, Claus M, Mihm J, Braithwaite M, Dennehy K, Renz H, Sester M, Watzl C, Burgert HG. 2013. A unique secreted adenovirus E3 protein binds to the leukocyte common antigen CD45 and modulates leukocyte functions. Proc Natl Acad Sci U S A 110:E4884–E4993. doi: 10.1073/pnas.1312420110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Windheim M, Hilgendorf A, Burgert HG. 2004. Immune evasion by adenovirus E3 proteins: exploitation of intracellular trafficking pathways. Curr Top Microbiol Immunol 273:29–85. [DOI] [PubMed] [Google Scholar]
- 33.Murali VK, Ornelles DA, Gooding LR, Wilms HT, Huang W, Tollefson AE, Wold WS, Garnett-Benson C. 2014. Adenovirus death protein (ADP) is required for lytic infection of human lymphocytes. J Virol 88:903–912. doi: 10.1128/JVI.01675-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ostapchuk P, Almond M, Hearing P. 2011. Characterization of empty adenovirus particles assembled in the absence of a functional adenovirus IVa2 protein. J Virol 85:5524–5531. doi: 10.1128/JVI.02538-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu K, Orozco D, Hearing P. 2012. The adenovirus L4-22K protein is multifunctional and is an integral component of crucial aspects of infection. J Virol 86:10474–10483. doi: 10.1128/JVI.01463-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gustin KE, Imperiale MJ. 1998. Encapsidation of viral DNA requires the adenovirus L1 52/55-kilodalton protein. J Virol 72:7860–7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahi YS, Vemula SV, Hassan AO, Costakes G, Stauffacher C, Mittal SK. 2015. Adenoviral L4 33K forms ring-like oligomers and stimulates ATPase activity of IVa2: implications in viral genome packaging. Front Microbiol 6:318. doi: 10.3389/fmicb.2015.00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu K, Guimet D, Hearing P. 2013. The adenovirus L4-33K protein regulates both late gene expression patterns and viral DNA packaging. J Virol 87:6739–6747. doi: 10.1128/JVI.00652-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ostapchuk P, Suomalainen M, Zheng Y, Boucke K, Greber UF, Hearing P. 2018. The adenovirus major core protein VII is dispensable for virion assembly but is essential for lytic infection. PLoS Pathog 13:e1006455. doi: 10.1371/journal.ppat.1006455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao X, Tikoo SK. 2016. Deletion of pV affects integrity of capsid causing defect in the infectivity of bovine adenovirus-3. J Gen Virol 97:2657–2667. doi: 10.1099/jgv.0.000570. [DOI] [PubMed] [Google Scholar]
- 41.Perez-Vargas J, Vaughan RC, Houser C, Hastie KM, Kao CC, Nemerow GR. 2014. Isolation and characterization of the DNA and protein binding activities of adenovirus core protein V. J Virol 88:9287–9296. doi: 10.1128/JVI.00935-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reddy VS, Nemerow GR. 2014. Structures and organization of adenovirus cement proteins provide insights into the role of capsid maturation in virus entry and infection. Proc Natl Acad Sci U S A 111:11715–11720. doi: 10.1073/pnas.1408462111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dai X, Wu L, Sun R, Zhou ZH. 2017. Atomic structures of minor proteins VI and VII in the human adenovirus. J Virol 91:e00850-. doi: 10.1128/JVI.00850-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu H, Jin L, Koh SB, Atanasov I, Schein S, Wu L, Zhou ZH. 2010. Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science 329:1038–1043. doi: 10.1126/science.1187433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reddy VS, Natchiar SK, Stewart PL, Nemerow GR. 2010. Crystal structure of human adenovirus at 3.5 A resolution. Science 329:1071–1075. doi: 10.1126/science.1187292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gaba A, Ayalew L, Makadiya N, Tikoo S. 2017. Proteolytic cleavage of bovine adenovirus 3-encoded pVIII. J Virol 91:e00211-. doi: 10.1128/JVI.00211-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ugai H, Borovjagin AV, Le LP, Wang M, Curiel DT. 2007. Thermostability defect caused by deletion of the core protein V gene in human adenovirus type 5 is rescued by thermos stable mutations in the core protein X precursor. J Mol Biol 366:1142–1160. doi: 10.1016/j.jmb.2006.11.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saha B, Wong CM, Parks RJ. 2014. The adenovirus genome contributes to the structural stability of the virion. Viruses 6:3563–3583. doi: 10.3390/v6093563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lichtenstein DL, Toth K, Doronin K, Tollefson AE, Wold WSM. 2004. Functions and mechanisms of action of the adenovirus E3 proteins. Int Rev Immunol 23:111. doi: 10.1080/08830180490265556. [DOI] [PubMed] [Google Scholar]
- 50.Singh M, Shmulevitz M, Tikoo SK. 2005. A newly identified interaction between IVa2 and pVIII proteins during porcine adenovirus type 3 infection. Virology 336:60–69. doi: 10.1016/j.virol.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 51.Thanbichler M, Iniesta AA, Shapiro L. 2007. A comprehensive set of plasmids for vanillate- and xylose-inducible gene expression in Caulobacter crescentus. Nucleic Acids Res 35(20):e137. doi: 10.1093/nar/gkm818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 53.Said A, Damiani A, Osterrieder N. 2014. Ubiquitination and degradation of the ORF34 gene product of equine herpesvirus type 1 (EHV-1) at late times of infection. Virology 460-461:11–22. doi: 10.1016/j.virol.2014.05.009. [DOI] [PubMed] [Google Scholar]