The capsid-binding small molecule PF74 inhibits HIV-1 infection at early stages and perturbs uncoating. However, the mechanism by which PF74 alters capsid stability and reduces viral infection is presently unknown. We recently introduced time-lapse atomic force microscopy to study the morphology and physical properties of HIV-1 cores during the course of reverse transcription. Here, we apply this AFM methodology to show that PF74 prevented the complete disassembly of HIV-1 cores normally observed during 24 h of reverse transcription. Specifically, cores with PF74 only partially disassembled: the main body of the capsid remained intact and stiff, but a cap-like structure dissociated from the narrow end of the core HIV-1. Our result provides direct evidence that PF74 directly stabilizes the HIV-1 capsid lattice.
KEYWORDS: HIV-1, PF74, atomic force microscopy, capsid, reverse transcription, uncoating
ABSTRACT
The RNA genome of human immunodeficiency virus type 1 (HIV-1) is enclosed in a cone-shaped capsid shell that disassembles following cell entry via a process known as uncoating. During HIV-1 infection, the capsid is important for reverse transcription and entry of the virus into the target cell nucleus. The small molecule PF74 inhibits HIV-1 infection at early stages by binding to the capsid and perturbing uncoating. However, the mechanism by which PF74 alters capsid stability and reduces viral infection is presently unknown. Here, we show, using atomic force microscopy (AFM), that binding of PF74 to recombinant capsid-like assemblies and to HIV-1 isolated cores stabilizes the capsid in a concentration-dependent manner. At a PF74 concentration of 10 μM, the mechanical stability of the core is increased to a level similar to that of the intrinsically hyperstable capsid mutant E45A. PF74 also prevented the complete disassembly of HIV-1 cores normally observed during 24 h of reverse transcription. Specifically, cores treated with PF74 only partially disassembled: the main body of the capsid remained intact and stiff, and a cap-like structure dissociated from the narrow end of the core. Moreover, the internal coiled structure that was observed to form during reverse transcription in vitro persisted throughout the duration of the measurement (∼24 h). Our results provide direct evidence that PF74 directly stabilizes the HIV-1 capsid lattice, thereby permitting reverse transcription while interfering with a late step in uncoating.
IMPORTANCE The capsid-binding small molecule PF74 inhibits HIV-1 infection at early stages and perturbs uncoating. However, the mechanism by which PF74 alters capsid stability and reduces viral infection is presently unknown. We recently introduced time-lapse atomic force microscopy to study the morphology and physical properties of HIV-1 cores during the course of reverse transcription. Here, we apply this AFM methodology to show that PF74 prevented the complete disassembly of HIV-1 cores normally observed during 24 h of reverse transcription. Specifically, cores with PF74 only partially disassembled: the main body of the capsid remained intact and stiff, but a cap-like structure dissociated from the narrow end of the core HIV-1. Our result provides direct evidence that PF74 directly stabilizes the HIV-1 capsid lattice.
INTRODUCTION
The human immunodeficiency virus type I (HIV-1) genome is encapsulated in a viral core with a capsid shell. The shell consists of capsid (CA) protein monomers organized in a hexameric lattice that contains 12 pentamers located at the ends, where the curvature of the surface is greatest (1–3). Following fusion between the viral envelope and the cellular membrane, the core enters the cell and undergoes controlled disassembly in the cytoplasm in a process commonly termed uncoating (4, 5). Several studies have shown that uncoating is correlated with the stability of the capsid (3, 6–9), which in turn affects reverse transcription (10–15) and viral infectivity (15–17). Moreover, there is an optimal capsid stability required for proper capsid disassembly (18) and successful infection (10). All of the above make the HIV-1 capsid an attractive target for antiviral drugs. One of these drugs is PF-3450074 (PF74), which was shown to dramatically decrease HIV-1 infectivity (19). PF74 binds directly to the core's capsid shell and preferentially interacts with the hexamer rather than the pentamer form of CA (20–23). The binding of PF74 impacts both intra- and inter-CA–hexamer interactions to alter the stability of the capsid (21).
According to several studies, PF74 is proposed to attenuate HIV-1 infection via a concentration-dependent bimodal mechanism (20, 22, 24, 25). At concentrations of 2 μM and lower, PF74 directly competes with binding of cleavage and polyadenylation specific factor 6 (CPSF6) and nucleoporin 153 (NUP153) (5, 22, 24, 25). Both factors were shown to play a role in HIV-1 infection (5, 26–29). At higher concentrations (∼10 μM), PF74 accelerates uncoating and blocks reverse transcription as well as virus assembly (19–23, 25). The above effects of PF74 at higher concentrations were initially proposed to be the outcome of capsid destabilization (23). However, a more recent study showed that in infected cells, high concentrations of PF74 do not alter the integrity of the capsid during infection (30). Thus, the mechanism underlying the interplay between PF74 binding, capsid stability, and HIV-1 infection is currently unclear.
The study of HIV-1 cores using commonly available imaging techniques is challenging owing to the relatively small size of the HIV-1 core (∼80 nm) (31). Atomic force microscopy (AFM) provides a unique means to analyze isolated viral cores under physiological conditions. In a previous study, we applied AFM to characterize the physical properties of assembled capsids, both wild-type (WT) and hyperstable CA mutants, in vitro (32). More recently, using time-lapse AFM, we analyzed the morphology and physical properties of isolated cores during the course of reverse transcription (18). We found that during reverse transcription the pressure inside the capsid increases until the internal stress exceeds the strength of the capsid structure, resulting in capsid rupture near the narrow end of the conical structure.
In the present study, we applied similar AFM technologies to characterize the effect of PF74 on the stiffness of in vitro-assembled capsid and HIV-1 isolated cores. In addition, using time-lapse AFM, we analyzed the morphology and physical properties of isolated HIV-1 cores treated with PF74 during the course of reverse transcription in vitro. We find that binding of PF74 to capsids assembled from CA proteins in vitro and to HIV-1 isolated cores increases the stability of the capsid in a concentration-dependent manner. At a PF74 concentration of 10 μM, the mechanical stability of the core was elevated to a level similar to that of the hyperstable capsid mutant E45A. However, during reverse transcription, the core underwent only partial disassembly: specifically, the main body of the capsid remained intact and stiff, and a cap-like structure dissociated from the narrow end of the core. Moreover, the internal coiled structure that forms during reverse transcription persisted throughout the duration of the measurement (∼24 h) instead of disappearing. Our results provide direct evidence that PF74 can directly stabilize the HIV-1 capsid lattice, resulting in perturbation of uncoating steps that follow reverse transcription.
RESULTS
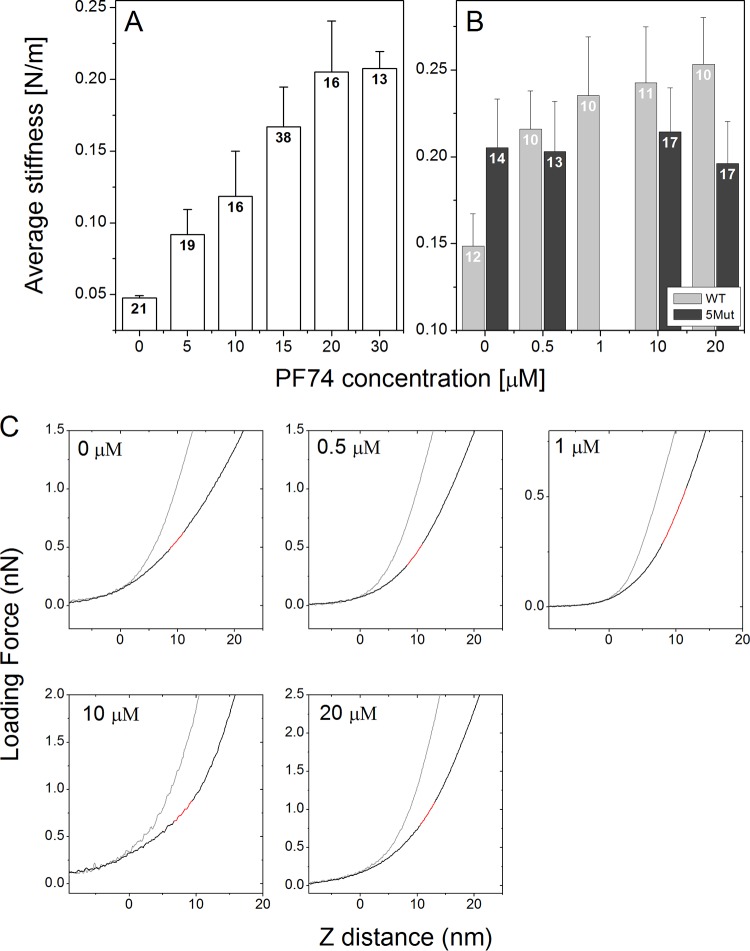
To study the effect of PF74 binding on the stability of capsid-like CA assemblies and native HIV-1 cores, we quantified their point stiffness at several PF74 concentrations, using AFM that was operated in the nano-indentation mode. The measured point stiffness values of recombinant CA protein assemblies and of isolated cores are summarized in Fig. 1A and B, respectively. Uncertainty values represent the standard error of the mean. All measurements were carried out in buffer: capsid assembly buffer (CAB) for capsid assemblies and 3-(N-morpholino)propanesulfonic acid (MOPS) for HIV-1 cores.
FIG 1.
PF74 binding increases the stiffness of recombinant CA assemblies (A) and isolated WT and 5Mut HIV-1 cores (B). Each value was calculated as the average of ∼100 or ∼400 force-distance curves obtained from individual capsids or cores, respectively. The average stiffness values derived from all of the measurements were plotted. Error bars represent the standard errors of the means. The number in each bar indicates the number of capsid particles analyzed. (C) Representative averaged force-distance curves for isolated cores at five different PF74 concentrations are shown (black lines). The cantilever deflection curves (gray line) were obtained by acquiring force-distance curves of the glass substrate. All stiffness analyses were calculated at a maximal loading force that corresponds to ∼4-nm indentation depths. Stiffness values were calculated by fitting a linear function to the force curve region bounded by 3- and 4-nm indentation depths. The corresponding line fit is plotted in red.
CA assemblies without PF74 treatment had the lowest averaged stiffness values of 0.048 ± 0.002 N/m (n = 21). This value is similar to our previously reported stiffness of CA assemblies (32). As the PF74 concentration increased, the stiffness values of the capsids gradually rose (the increase in stiffness was confirmed to be statistically significant using a Mann-Whitney U test for a P value of <0.05), until they reached a maximal value of 0.208 ± 0.012 N/m (n = 13) at a PF74 concentration of 20 μM (Fig. 1A). At this concentration, the capsid stiffness values plateaued such that capsid stiffness remained unchanged at higher PF74 levels (changes in stiffness were not statistically significant according to a Mann-Whitney U test for a P value of <0.05). In agreement with our previously reported findings (18, 32), isolated cores were stiffer than CA assemblies, with an averaged stiffness value of 0.148 ± 0.002 N/m (n = 12) (Fig. 1B).
Similarly to its effects on CA assemblies, the binding of PF74 to isolated HIV-1 cores increased their stiffness in a concentration-dependent manner. However, the stability of cores reached a similar maximal averaged stiffness value of 0.235 ± 0.034 N/m (n = 10) at a PF74 concentration of 1 μM (confirmed to be statistically significant using a Mann-Whitney U test for P value of <0.05) and remained unchanged at higher PF74 concentrations (Fig. 1B). To confirm that the observed increased stability is the consequence of PF74 binding to the capsid shell, we measured cores isolated from the PF74-resistant mutant HIV-1 virus 5Mut (33) (Fig. 1B). 5Mut particles contain a stable core (23) which exhibited an averaged stiffness value of 0.205 ± 0.028 N/m (n = 14). Importantly, however, increasing the concentration of PF74 had no statistically significant effect on the averaged stiffness values of 5Mut cores according to a Mann-Whitney U test for a P value of <0.05, thus demonstrating that their stability is independent of PF74.
To study the effect of PF74 on morphological and mechanical changes in the HIV-1 core occurring during reverse transcription, we prepared samples for AFM analysis using a previously described method (18). All measurements were carried out in MOPS buffer containing 10 μM PF74. Initially, we analyzed the average stiffness of cores during the time course of reverse transcription (Fig. 2), where the average stiffness value prior to reverse transcription (0.261 ± 0.038 N/m; n = 11) was positioned at time zero. We then initiated reverse transcription by adding reverse transcription buffer (to achieve a final total concentration of 100 μM deoxyribonucleoside triphosphates [dNTPs] and 1 mM MgCl2) (34) to 10 μM PF74 and monitored core stiffness values as a function of time. Measurements were performed in real time without stopping the reaction. At each time point, stiffness values of 3 to 5 cores were measured, and average core stiffness was calculated. Overall, the reverse transcription experiment was repeated at least three times. The average core stiffness increased to a maximum of 0.371 ± 0.069 N/m (n = 8) at 5 h, which was almost 1.5-fold higher than the initial stiffness. The maximum stiffness value was reached earlier than that for cores without PF74 (5 h compared to 7 h) (18). Of the total of eight cores examined at this time point, three failed to exhibit a significant increase in their measured stiffness values. Subsequently, core stiffness dropped back to its initial value (0.249 ± 0.026 N/m; n = 10) and remained unchanged over the duration of the reaction, which was terminated after 24 h.
FIG 2.
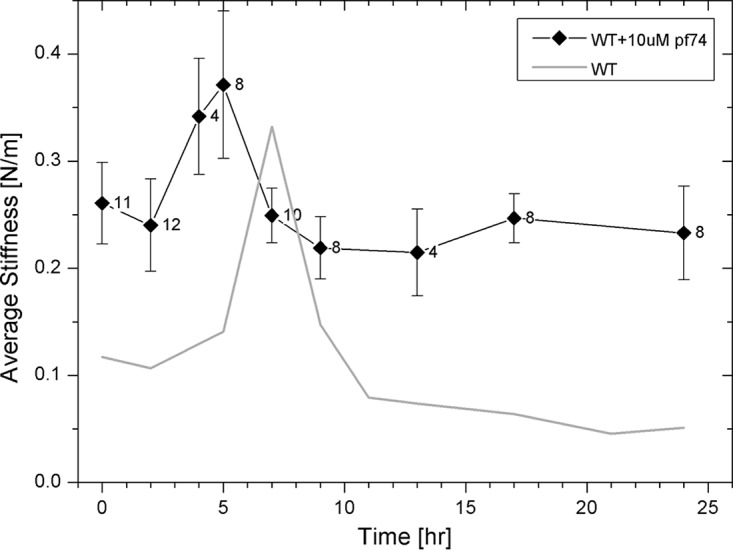
Measured stiffness values of HIV-1 cores treated with 10 μM PF74 as a function of the progress of reverse transcription. Cores were adhered to HMDS-coated glass slides and kept in MOPS buffer. The reaction was initiated by the addition of dNTPs and MgCl2 to the cores, and their stiffness values were measured using AFM. The initial average stiffness values of cores prior to reverse transcription are represented as time zero. At each time point, stiffness of 3 to 5 cores was measured without stopping the reaction, and average core stiffness was calculated. The reverse transcription experiment was repeated at least three times. WT data (taken from reference 18) is presented as a gray line for comparison. The error bars represent the standard errors of the means, and the number of cores analyzed is indicated beside each data point.
In parallel with the mechanical analysis, we characterized the morphology of cores incubated with 10 μM PF74 during the course of reverse transcription by AFM operated in the quantitative imaging (QI) mode. Prior to reverse transcription, cores had a well-defined conical structure. Two representative cores (out of a total of 18 cores that were imaged) are shown in Fig. 3A and B. After 9 h, a relatively small opening in the capsid appeared (Fig. 3C). As the reaction proceeded, the size of the opening grew (Fig. 3D and E). Similarly to our previous findings (18), the opening was localized exclusively at or near the narrow end of the core (in all of the 32 cores visualized during reverse transcription). In contrast to HIV-1 cores, which underwent complete disassembly after 24 h of reverse transcription (18), about half of the PF74-reinforced cores (8 of the 17 cores imaged) remained intact. The other 9 cores underwent partial disassembly, with the main body of the capsid remaining intact and with dissociation of a cap-like structure from the narrow end of the core (Fig. 3F). The reduction in the length of the cores at 24 h into reverse transcription (average length of 160 ± 6 nm compared with 195 ± 6 nm prior to reverse transcription) provides further support to their observed partial disassembly. It should be noted that the length of the core appears to be larger than expected due to convolution between the AFM tip and the core.
FIG 3.
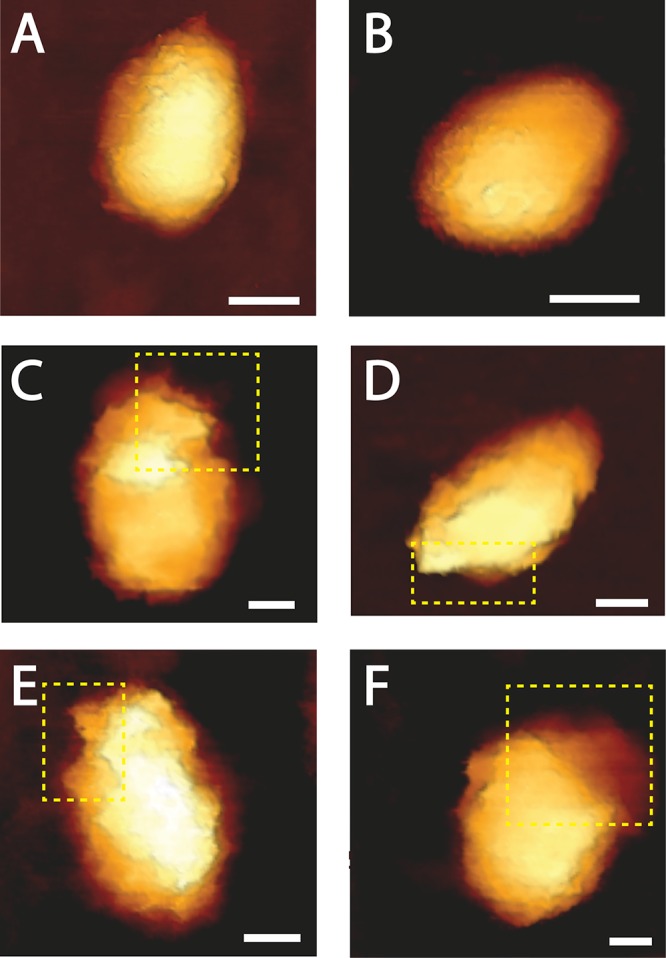
Topographic AFM images of the morphological changes that HIV-1 cores treated with 10 μM PF74 undergo during reverse transcription. Cores were adhered to HMDS-coated glass slides and kept in MOPS buffer. Transcription was initiated by adding dNTPs and MgCl2 to the cores. Images were acquired using the QI mode. (A and B) Typical cone-shaped cores observed prior to reverse transcription (out of a total of 18 cores that were imaged). (C to F) Deformed and opened cores visualized after 9 to 24 h of reverse transcription. For clarity, openings in the cores are shown within a dashed yellow rectangle. Scale bar, 50 nm. A total of 49 cores were visualized during reverse transcription.
Finally, we acquired high-lateral-resolution mechanical maps of the core surface (QI mode; JPK Instruments) (Fig. 4). In contrast to the more typical AFM topographic images, which provide a height distribution map of the image, mechanical maps provide a qualitative view of the lateral distribution of core stiffness. In these images, a darker color represents a higher stiffness value. Prior to reverse transcription, the core surfaces appeared to be mechanically homogenous without any defined pattern in the surface distribution of stiffness measurements (Fig. 4A). A similar uniform stiffness distribution was observed on the surface of cores following 2 h of reverse transcription (Fig. 4B). Such a smooth pattern was observed on all cores that were imaged at these two time points (n = 21 cores). Remarkably, 5 h into reverse transcription, a clear pattern appeared beneath the capsid (Fig. 4C). As we proposed elsewhere (18), this pattern may represent the existence of a stiff coiled filament located in the inner periphery underneath the core surface. Comparable striations in the capsid's mechanical maps were observed in 6 of 13 analyzed cores. We previously reported that during reverse transcription in the absence of PF74, the pattern begins to disappear (18). Strikingly, in cores treated with PF74, the pattern persisted (in 15 of 26 analyzed cores) during the course of the experiment (Fig. 4D to F). At the end of the experiment (24 h of reverse transcription), a spiral pattern in the core surface was observed in 11 of 17 analyzed cores. We conclude that during reverse transcription in the presence of PF74, the coiled structure that is formed persists during the reaction course, and the capsid remains mostly intact.
FIG 4.
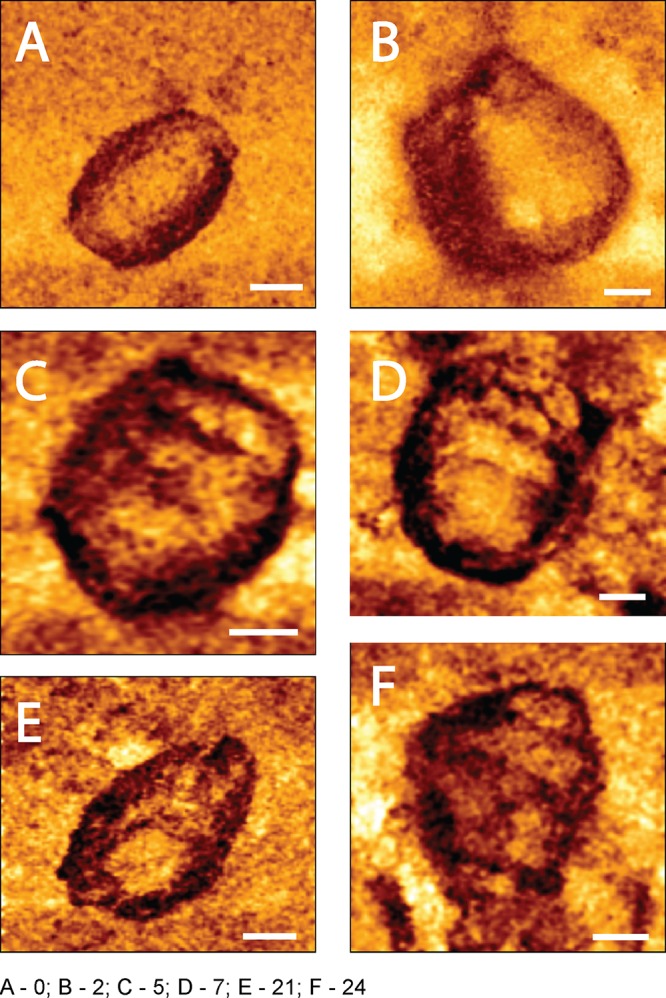
High-resolution spatial mechanical mapping of an HIV-1 core surface treated with 10 μM PF74 during reverse transcription. Cores were adhered to HMDS-coated glass slides and kept in MOPS buffer. Reactions were initiated by adding dNTPs and MgCl2 to the cores. Images were acquired using the QI mode. (A) The mechanical map of a core before the beginning of reverse transcription. (B to F) Mechanical maps of cores after 2, 5, 7, 21, and 24 h of reverse transcription, respectively. Scale bar, 50 nm.
DISCUSSION
In this study, we examined the effect of the CA-targeting inhibitor PF74 on the mechanical stability of HIV-1 capsid and consequently on capsid disassembly. Previous studies reported conflicting effects of PF74. Addition of PF74 to HIV-1 cores reduced their stability in vitro, resulting in accelerated capsid disassembly (19, 20, 23). However, Hulme et al. observed no effect of PF74 on the integrity of the capsid in infected cells (30). Conversely, PF74 was shown to increase the stability of HIV-1 CA assemblies in vitro (20, 24).
Here, we employed AFM to evaluate the effect of PF74 binding on the mechanical stability of in vitro CA assemblies and isolated HIV-1 cores by measuring their stiffness. We observed that binding of PF74 to capsids assembled from CA protein in vitro and to purified HIV-1 cores increased the stiffness of the capsid in a concentration-dependent manner. As we previously showed (32), the stiffness value of the in vitro-assembled capsids is significantly lower than that of the isolated viral core. Similarly, the maximal averaged stiffness, observed upon PF74 binding, of in vitro-assembled capsids was slightly lower than that of isolated cores. Interestingly, a 50% increase in the core stiffness is obtained at a PF74 concentration (∼0.4 μM; interpolated from data shown in Fig. 1B) that is similar to the antiviral 50% inhibitory concentration (IC50) for the inhibitor (23). The quantitative concordance between the effects of PF74 on capsid stiffness and infectivity supports the suggestion that stiffening the capsid inhibits viral infectivity. When PF74 was added to cores isolated from the 5Mut CA mutant, which is resistant to PF74 (33), the averaged stiffness remained roughly unchanged. However, the 5Mut cores exhibited elevated stiffness in the absence of the inhibitor, suggesting that stiffness is only one factor affecting HIV-1 capsid function. We conclude that the observed reinforcement in WT capsids is the result of PF74 binding to the capsid shell.
Recently, we monitored the morphological and mechanical changes in isolated HIV-1 cores during the course of reverse transcription (18). We found that the core stiffness increased during reverse transcription, reaching a maximum after 7 h, followed by complete disassembly of the capsid. Interestingly, the rupture of the capsid occurred exclusively near the narrow end of the cone. Further, we showed that in the hyperstable CA mutant, E45A, the peak stiffness appeared sooner, after 4 h of transcription, but did not result in capsid disassembly. We proposed that the kinetic difference is related to the observation that E45A cores appear to be more permeable to small molecules than WT cores (33), which enables enhanced dNTP influx, potentially resulting in accelerated reverse transcription. In the current study, we observed that at a PF74 concentration of 10 μM, the stiffness of the core rose to a level similar to that of the E45A mutant. We observed that the pressure inside the capsid with PF74 increased during reverse transcription, with a peak appearing earlier (after 5 h of transcription) than with the WT. In parallel to the stiffness elevation, high-resolution stiffness distribution maps of cores reveal the formation of a stiff coiled filamentous structure beneath the capsid surface. However, in contrast to results with WT cores, when PF74 is added, the coiled structure remains detectable during the entire length of the measurement (24 h). This observation suggests that the transcription complex remained associated with the hexamer lattice. Recent data suggest that nucleotides are transported into the core through electrostatic channels that were found in CA hexamers (35). These channels were shown to have an open and a closed state. Binding of PF74 to CA hexamers may stabilize the open conformation, which increases the permeability of the capsid shell to dNTP molecules, resulting in more rapid reverse transcription.
The stiffness profile of WT cores treated with PF74 during reverse transcription is similar to that of the E45A mutant observed in the absence of the inhibitor. The cores' initial, maximal, and final stiffness values were nearly identical. It is therefore surprising that an analysis of the core morphology during reverse transcription reveals striking differences between WT cores and E45A cores treated with PF74. In contrast to E45A cores with PF74, which remained fully intact during the entire length of the measurement (24 h), WT cores treated with PF74 undergo partial disassembly. We find that after 24 h of reverse transcription, the main body of the capsid remains fully intact and stiff (with a value similar to the initial stiffness value). However, at this time point, a cap-like structure appears to dissociate from the narrow end of the core. In a previous study, we observed that WT cores broke open in the proximity of the core's narrow end (18), where the local density of CA pentamers is highest (1–3, 36). A structural study conducted using high-resolution electron microscopy showed that CA hexamers and pentamers expose different regions of the CA protein surface, resulting in different interactions with cellular factors or capsid-targeting inhibitors such as PF74 (36). Indeed, PF74 was found to bind CA hexamers with higher affinity than unassembled CA (20–23). Based on the above, we propose that PF74 binding increases the stability of CA hexamers and of the intersubunit contacts within hexamers. As a result, the overall stiffness of the capsid shell is increased, but near the narrow end of the core, local weak regions exist. We propose that the increased stiffness, induced by reverse transcription, imposes mechanical stress on the capsid lattice. The failure of the core structure is expected to begin at the least stable region of the capsid (the narrow end). However, the main body of the capsid, which is primarily composed of CA hexamers, is mechanically reinforced, which prevents further disassembly of the core. This consequent impairment of uncoating enables PF74 to reduce HIV-1 infectivity. PF74's antiviral activity is also modulated by capsid-binding host factors, including cyclophilin A and NUP153. Thus, PF74's antiviral activity profile depends on the interplay of its effects on multiple HIV-1 capsid functions.
MATERIALS AND METHODS
Capsid assembly.
Lyophilized purified recombinant WT HIV-1 capsid proteins were resuspended in storage buffer (20 mM Tris-HCl at pH 8.0, 40 mM NaCl, 60 mM β-mercaptoethanol) to a final concentration of 160 μM. Resuspended CA proteins were self-assembled using a previously described method (3). Briefly, CA suspension was placed in dialysis minicups (Slyde-A-Lyzer Mini; Thermo Scientific) and dialyzed overnight against CAB (100 mM Tris-HCl at pH 8.0, 200 mM NaCl) at 4°C. CA assemblies were then characterized by AFM.
Isolation of HIV-1 cores.
HIV-1 pseudovirions were used for isolation of HIV-1 cores. Pseudovirus particles were produced by a previously described protocol (18). Briefly, approximately 106 human embryonic kidney (HEK) 293T cells were transfected with 2.5 μg of ΔEnv IN− HIV-1 plasmid (DHIV3-GFP-D116G) (37) or the PF74-resistant proviral clone R9ΔE-5Mut using 10 μg of polyethylenimine (PEI) (branched, molecular weight [MW] of ∼25,000; Sigma-Aldrich). The Env-defective, PF74-resistant close was constructed by transferring an SalI-BamHI fragment from R9ΔE (38) into the corresponding region of R9.5Mut (23). After 20 h, the medium was replaced with fresh medium (Dulbecco's modified Eagle medium supplemented with 10% heat-inactivated bovine serum, 1% penicillin-streptomycin, and 1% glutamine), and the cells were incubated at 37°C in 5% CO2. After 6 h, the supernatant was harvested, centrifuged at 1,000 rpm for 10 min, and filtered through a 0.45-μm-pore-size filter. The virus-containing supernatant was concentrated by ultracentrifugation in an SW-28 rotor (25,000 rpm, 2 h, 4°C) using OptiPrep density gradient medium (Sigma-Aldrich). Part of the supernatant containing pelleted viruses was collected, mixed with 10 ml of TNE buffer (50 mM Tris-HCl, 100 mM NaCl, 0.1 mM EDTA [pH 7.4]), and added to the 100,000-molecular-weight-cutoff [MWCO] Vivaspin 20 centrifugal concentrators (Sartorius AG, Germany). The mixture was centrifuged twice at 2,500 × g for 25 to 30 min at 4°C until the supernatant level in the concentrators reached 200 to 300 μl.
From concentrated virus-containing supernatant, viral cores were isolated using a previously described protocol (31), with modifications. Briefly, ∼40 μl of purified HIV-1 pseudovirus particles was mixed with an equal amount of 1% Triton-X diluted in 100 mM 3-(N-morpholino)propanesulfonic acid (MOPS) buffer (pH 7.0) and incubated for 2 min at 4°C. The mixture was centrifuged at 13,800 × g for 8 min at 4°C. After the supernatant was removed, the pellet was washed twice by the addition of ∼80 μl of MOPS buffer and centrifugation at 13,800 × g for 8 min at 4°C. The pellet was resuspended in 10 μl of MOPS buffer.
AFM measurements and analysis.
AFM measurements and analysis were performed as previously described (18). Briefly, 10 μl of assembled CA in CAB buffer or of isolated HIV-1 cores resuspended in MOPS buffer solution was incubated for 30 min at room temperature on hexamethyldisilazane (HMDS)-coated microscope glass slides. Measurements were carried out in CAB buffer for capsid assemblies or in MOPS buffer for cores, without sample fixation. Every experiment was repeated at least three times, each time with independently purified HIV-1 cores. PF-3450074 (PF74) (19) was synthesized by the Vanderbilt Institute of Chemical Biology Synthesis Core, Vanderbilt University, Nashville, TN. Measurements were carried out with a JPK Nanowizard Ultra-Speed atomic force microscope (JPK Instruments, Berlin, Germany) mounted on an inverted optical microscope (Axio Observer; Carl Zeiss, Heidelberg, Germany). Silicon nitride probes (mean cantilever spring constant, kcant, of 0.12 N/m [DNP, Bruker]) were used. Height topographic and mechanical map images were acquired in quantitative imaging (QI) mode at a rate of 0.5 lines/s and a loading force of 300 pN.
Capsid or core stiffness was obtained by the nanoindentation method, as previously described (18, 39–41). Briefly, the stiffness value of each capsid was determined by acquiring ∼100 force-distance (F-D) curves. To determine the stiffness value of cores, 20 F-D curves were acquired at a rate of 20 Hz at each of 24 different points on the core surface. To confirm that the capsid or the core remained stable during the entire indentation experiment, we analyzed each experiment by plotting the individual measured point stiffness as a histogram (Fig. 5A) and as a function of the measurement count (Fig. 5B). Samples whose point stiffness values decreased consistently during experimentation were discarded since they underwent irreversible deformation.
FIG 5.
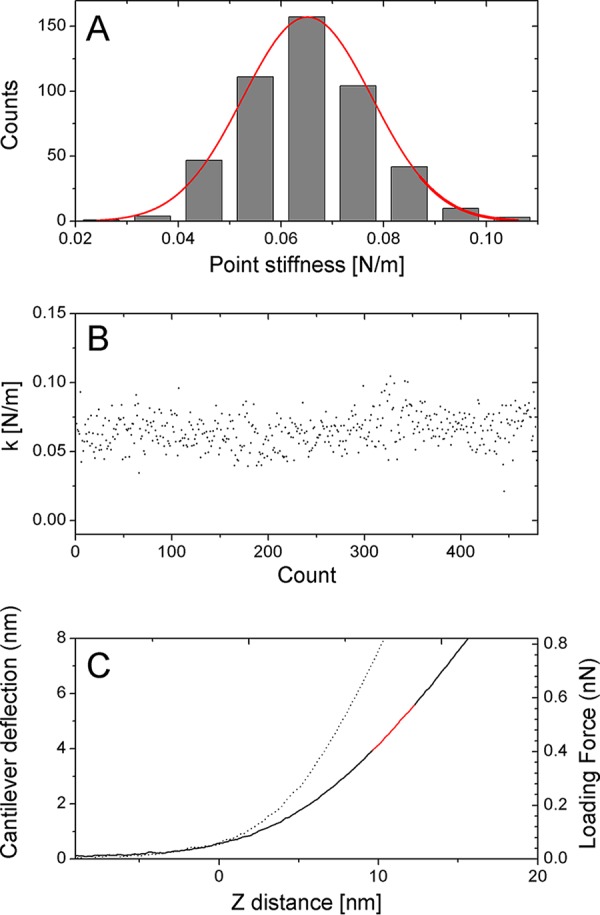
Measuring the point stiffness of the capsid or core by indentation-type experiment. (A) Histogram and Gaussian-fitted curves of the individual measured point stiffness values derived from the consecutive force-distance curves of a single core attached to a microscope's glass slide. (B) The individual measured point stiffness values obtained for the experiment shown in panel A against the experiment number (count) during a single experiment. (C) Typical force-distance curves of cantilever deflection (dotted line) and isolated core attached to an HMDS-pretreated glass slide (solid line). The corresponding line fit used to calculate sample stiffness is plotted in red. Such plots together with an analysis of the narrow distribution of the individual measured spring constants demonstrate that the core did not undergo a significant irreversible deformation during the indentation measurements.
The maximum indentation of the sample was 4 nm, which corresponds to a maximum loading force of 0.2 to 1.5 nN. Prior to analysis, each curve was shifted to set the deflection in the noncontact section to zero. The set of force-distance curves was then averaged (Fig. 5C). From the slope of the averaged F-D curve, measured stiffness was derived mathematically. The stiffness of the capsid was computed using Hooke's law on the assumption that the experimental system may be modeled as two springs (the capsid and the cantilever) arranged in series. The spring constant of the cantilever was determined during experiment by measuring thermal fluctuation (42). To reduce the error in the calculated point stiffness, we chose cantilevers such that the measured point stiffness was <70% of the cantilever spring constant. Data analysis was carried out using MATLAB software (The Math Works, Natick, MA).
ACKNOWLEDGMENTS
S.R. was supported by a fellowship from the Kreitman School of Advanced Studies (Ben-Gurion University of the Negev). This work was supported in part by NIH grant R01 AI114339 (C.A.) and by the Israel Science Foundation (grant 234/17).
REFERENCES
- 1.Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. 1999. Assembly and analysis of conical models for the HIV-1 core. Science 283:80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- 2.Sundquist WI, Krausslich HG. 2012. HIV-1 assembly, budding, and maturation. Cold Spring Harb Perspect Med 2:a006924. doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao GP, Perilla JR, Yufenyuy EL, Meng X, Chen B, Ning JY, Ahn J, Gronenborn AM, Schulten K, Aiken C, Zhang PJ. 2013. Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature 497:643–646. doi: 10.1038/nature12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lelek M, Di Nunzio F, Henriques R, Charneau P, Arhel N, Zimmer C. 2012. Superresolution imaging of HIV in infected cells with FlAsH-PALM. Proc Natl Acad Sci U S A 109:8564–8569. doi: 10.1073/pnas.1013267109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matreyek KA, Yucel SS, Li X, Engelman A. 2013. Nucleoporin NUP153 phenylalanine-glycine motifs engage a common binding pocket within the HIV-1 capsid protein to mediate lentiviral infectivity. PLoS Pathog 9:e1003693. doi: 10.1371/journal.ppat.1003693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ambrose Z, Aiken C. 2014. HIV-1 uncoating: connection to nuclear entry and regulation by host proteins. Virology 454:371–379. doi: 10.1016/j.virol.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hulme AE, Perez O, Hope TJ. 2011. Complementary assays reveal a relationship between HIV-1 uncoating and reverse transcription. Proc Natl Acad Sci U S A 108:9975–9980. doi: 10.1073/pnas.1014522108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, Kar AK, Sodroski J. 2009. Target cell type-dependent modulation of human immunodeficiency virus type 1 capsid disassembly by cyclophilin A. J Virol 83:10951–10962. doi: 10.1128/JVI.00682-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shah VB, Shi J, Hout DR, Oztop I, Krishnan L, Ahn J, Shotwell MS, Engelman A, Aiken C. 2013. The host proteins transportin SR2/TNPO3 and cyclophilin A exert opposing effects on HIV-1 uncoating. J Virol 87:422–432. doi: 10.1128/JVI.07177-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forshey BM, von Schwedler U, Sundquist WI, Aiken C. 2002. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J Virol 76:5667–5677. doi: 10.1128/JVI.76.11.5667-5677.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. 2004. The cytoplasmic body component TRIM5 alpha restricts HIV-1 infection in Old World monkeys. Nature 427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 12.Tang SX, Murakami T, Agresta BE, Campbell S, Freed EO, Levin JG. 2001. Human immunodeficiency virus type 1 N-terminal capsid mutants that exhibit aberrant core morphology and are blocked in initiation of reverse transcription in infected cells. J Virol 75:9357–9366. doi: 10.1128/JVI.75.19.9357-9366.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warrilow D, Stenzel D, Harrich D. 2007. Isolated HIV-1 core is active for reverse transcription. Retrovirology 4:77. doi: 10.1186/1742-4690-4-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warrilow D, Warren K, Harrich D. 2010. Strand transfer and elongation of HIV-1 reverse transcription is facilitated by cell factors in vitro. PLoS One 5:e13229. doi: 10.1371/journal.pone.0013229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu XL, Anderson JL, Campbell EM, Joseph AM, Hope TJ. 2006. Proteasome inhibitors uncouple rhesus TRIM5 alpha restriction of HIV-1 reverse transcription and infection. Proc Natl Acad Sci U S A 103:7465–7470. doi: 10.1073/pnas.0510483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ambrose Z, Lee K, Ndjomou J, Xu HZ, Oztop I, Matous J, Takemura T, Unutmaz D, Engelman A, Hughes SH, KewalRamani VN. 2012. Human immunodeficiency virus type 1 capsid mutation N74D alters cyclophilin A dependence and impairs macrophage infection. J Virol 86:4708–4714. doi: 10.1128/JVI.05887-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamashita M, Perez O, Hope TJ, Emerman M. 2007. Evidence for direct involvement of the capsid protein in HIV infection of nondividing cells. PLoS Pathog 3:1502–1510. doi: 10.1371/journal.ppat.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rankovic S, Varadarajan J, Ramalho R, Aiken C, Rousso I. 2017. Reverse transcription mechanically initiates HIV-1 capsid disassembly. J Virol 91:e00289-. doi: 10.1128/JVI.00289-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blair WS, Pickford C, Irving SL, Brown DG, Anderson M, Bazin R, Cao JA, Ciaramella G, Isaacson J, Jackson L, Hunt R, Kjerrstrom A, Nieman JA, Patick AK, Perros M, Scott AD, Whitby K, Wu H, Butler SL. 2010. HIV capsid is a tractable target for small molecule therapeutic intervention. PLoS Pathog 6:e1001220. doi: 10.1371/journal.ppat.1001220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhattacharya A, Alam SL, Fricke T, Zadrozny K, Sedzicki J, Taylor AB, Demeler B, Pornillos O, Ganser-Pornillos BK, Diaz-Griffero F, Ivanov DN, Yeager M. 2014. Structural basis of HIV-1 capsid recognition by PF74 and CPSF6. Proc Natl Acad Sci U S A 111:18625–18630. doi: 10.1073/pnas.1419945112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gres AT, Kirby KA, KewalRamani VN, Tanner JJ, Pornillos O, Sarafianos SG. 2015. X-ray crystal structures of native HIV-1 capsid protein reveal conformational variability. Science 349:99–103. doi: 10.1126/science.aaa5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price AJ, Jacques DA, McEwan WA, Fletcher AJ, Essig S, Chin JW, Halambage UD, Aiken C, James LC. 2014. Host cofactors and pharmacologic ligands share an essential interface in HIV-1 capsid that is lost upon disassembly. PLoS Pathog 10:e1004459. doi: 10.1371/journal.ppat.1004459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi J, Zhou J, Shah VB, Aiken C, Whitby K. 2011. Small-molecule inhibition of human immunodeficiency virus type 1 infection by virus capsid destabilization. J Virol 85:542–549. doi: 10.1128/JVI.01406-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fricke T, Buffone C, Opp S, Valle-Casuso J, Diaz-Griffero F. 2014. BI-2 destabilizes HIV-1 cores during infection and prevents binding of CPSF6 to the HIV-1 capsid. Retrovirology 11:120. doi: 10.1186/s12977-014-0120-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng K, Muranyi W, Glass B, Laketa V, Yant SR, Tsai L, Cihlar T, Muller B, Krausslich HG. 2014. Quantitative microscopy of functional HIV post-entry complexes reveals association of replication with the viral capsid. Elife 3:e04114. doi: 10.7554/eLife.04114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. 2008. Identification of host proteins required for HIV infection through a functional genomic screen. Science 319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 27.Konig R, Zhou YY, Elleder D, Diamond TL, Bonamy GMC, Irelan JT, Chiang CY, Tu BP, De Jesus PD, Lilley CE, Seidel S, Opaluch AM, Caldwell JS, Weitzman MD, Kuhen KL, Bandyopadhyay S, Ideker T, Orth AP, Miraglia LJ, Bushman FD, Young JA, Chanda SK. 2008. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell 135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee K, Ambrose Z, Martin TD, Oztop I, Mulky A, Julias JG, Vandegraaff N, Baumann JG, Wang R, Yuen W, Takemura T, Shelton K, Taniuchi I, Li Y, Sodroski J, Littman DR, Coffin JM, Hughes SH, Unutmaz D, Engelman A, KewalRamani VN. 2010. Flexible use of nuclear import pathways by HIV-1. Cell Host Microbe 7:221–233. doi: 10.1016/j.chom.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matreyek KA, Engelman A. 2011. The requirement for nucleoporin NUP153 during human immunodeficiency virus type 1 infection is determined by the viral capsid. J Virol 85:7818–7827. doi: 10.1128/JVI.00325-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hulme AE, Kelley Z, Foley D, Hope TJ. 2015. Complementary assays reveal a low level of CA associated with viral complexes in the nuclei of HIV-1-infected cells. J Virol 89:5350–5361. doi: 10.1128/JVI.00476-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Welker R, Hohenberg H, Tessmer U, Huckhagel C, Krausslich HG. 2000. Biochemical and structural analysis of isolated mature cores of human immunodeficiency virus type 1. J Virol 74:1168–1177. doi: 10.1128/JVI.74.3.1168-1177.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramalho R, Rankovic S, Zhou J, Aiken C, Rousso I. 2016. Analysis of the mechanical properties of wild type and hyperstable mutants of the HIV-1 capsid. Retrovirology 13:17. doi: 10.1186/s12977-016-0250-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu HZ, Franks T, Gibson G, Huber K, Rahm N, De Castillia CS, Luban J, Aiken C, Watkins S, Sluis-Cremer N, Ambrose Z. 2013. Evidence for biphasic uncoating during HIV-1 infection from a novel imaging assay. Retrovirology 10:70. doi: 10.1186/1742-4690-10-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang H, Dornadula G, Pomerantz RJ. 1996. Endogenous reverse transcription of human immunodeficiency virus type 1 in physiological microenvironments: an important stage for viral infection of nondividing cells. J Virol 70:2809–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jacques DA, McEwan WA, Hilditch L, Price AJ, Towers GJ, James LC. 2016. HIV-1 uses dynamic capsid pores to import nucleotides and fuel encapsidated DNA synthesis. Nature 536:349–353. doi: 10.1038/nature19098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mattei S, Glass B, Hagen WJH, Krausslich HG, Briggs JAG. 2016. The structure and flexibility of conical HIV-1 capsids determined within intact virions. Science 354:1434–1437. doi: 10.1126/science.aah4972. [DOI] [PubMed] [Google Scholar]
- 37.DeHart JL, Andersen JL, Zimmerman ES, Ardon O, An DS, Blackett J, Kim B, Planelles V. 2005. The ataxia telangiectasia-mutated and Rad3-related protein is dispensable for retroviral integration. J Virol 79:1389–1396. doi: 10.1128/JVI.79.3.1389-1396.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou J, Aiken C. 2001. Nef enhances human immunodeficiency virus type 1 infectivity resulting from intervirion fusion: Evidence supporting a role for Nef at the virion envelope. J Virol 75:5851–5859. doi: 10.1128/JVI.75.13.5851-5859.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kol N, Gladnikoff M, Barlam D, Shneck RZ, Rein A, Rousso I. 2006. Mechanical properties of murine leukemia virus particles: effect of maturation. Biophys J 91:767–774. doi: 10.1529/biophysj.105.079657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kol N, Shi Y, Tsvitov M, Barlam D, Shneck RZ, Kay MS, Rousso I. 2007. A stiffness switch in human immunodeficiency virus. Biophys J 92:1777–1783. doi: 10.1529/biophysj.106.093914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pang HB, Hevroni L, Kol N, Eckert DM, Tsvitov M, Kay MS, Rousso I. 2013. Virion stiffness regulates immature HIV-1 entry. Retrovirology 10:4. doi: 10.1186/1742-4690-10-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hutter JL, Bechhoefer J. 1993. Calibration of atomic-force microscope tips. Rev Sci Instrum 64:1868–1873. doi: 10.1063/1.1143970. [DOI] [Google Scholar]