Middle East respiratory syndrome coronavirus (MERS-CoV) causes respiratory failure with a high case fatality rate in patients, yet effective antivirals and vaccines are currently not available. Stress granule (SG) formation is one of the cellular stress responses to virus infection and is generally triggered as a result of stress-induced translation arrest. SGs can be beneficial or detrimental for virus replication, and the biological role of SGs in CoV infection is unclear. The present study showed that the MERS-CoV 4a accessory protein, which was reported to block SG formation in cells in which it was expressed, inhibited SG formation in infected cells. Our data suggest that 4a-mediated inhibition of SG formation facilitates the translation of viral mRNAs, resulting in efficient virus replication. To our knowledge, this report is the first to show the biological significance of SG in CoV replication and provides insight into the interplay between MERS-CoV and antiviral stress responses.
KEYWORDS: MERS coronavirus, accessory protein, stress granules
ABSTRACT
Stress granule (SG) formation is generally triggered as a result of stress-induced translation arrest. The impact of SG formation on virus replication varies among different viruses, and the significance of SGs in coronavirus (CoV) replication is largely unknown. The present study examined the biological role of SGs in Middle East respiratory syndrome (MERS)-CoV replication. The MERS-CoV 4a accessory protein is known to inhibit SG formation in cells in which it was expressed by binding to double-stranded RNAs and inhibiting protein kinase R (PKR)-mediated phosphorylation of the α subunit of eukaryotic initiation factor 2 (eIF2α). Replication of MERS-CoV lacking the genes for 4a and 4b (MERS-CoV-Δp4), but not MERS-CoV, induced SG accumulation in MERS-CoV-susceptible HeLa/CD26 cells, while replication of both viruses failed to induce SGs in Vero cells, demonstrating cell type-specific differences in MERS-CoV-Δp4-induced SG formation. MERS-CoV-Δp4 replicated less efficiently than MERS-CoV in HeLa/CD26 cells, and inhibition of SG formation by small interfering RNA-mediated depletion of the SG components promoted MERS-CoV-Δp4 replication, demonstrating that SG formation was detrimental for MERS-CoV replication. Inefficient MERS-CoV-Δp4 replication was not due to either the induction of type I and type III interferons or the accumulation of viral mRNAs in the SGs. Rather, it was due to the inefficient translation of viral proteins, which was caused by high levels of PKR-mediated eIF2α phosphorylation and likely by the confinement of various factors that are required for translation in the SGs. Finally, we established that deletion of the 4a gene alone was sufficient for inducing SGs in infected cells. Our study revealed that 4a-mediated inhibition of SG formation facilitates viral translation, leading to efficient MERS-CoV replication.
IMPORTANCE Middle East respiratory syndrome coronavirus (MERS-CoV) causes respiratory failure with a high case fatality rate in patients, yet effective antivirals and vaccines are currently not available. Stress granule (SG) formation is one of the cellular stress responses to virus infection and is generally triggered as a result of stress-induced translation arrest. SGs can be beneficial or detrimental for virus replication, and the biological role of SGs in CoV infection is unclear. The present study showed that the MERS-CoV 4a accessory protein, which was reported to block SG formation in cells in which it was expressed, inhibited SG formation in infected cells. Our data suggest that 4a-mediated inhibition of SG formation facilitates the translation of viral mRNAs, resulting in efficient virus replication. To our knowledge, this report is the first to show the biological significance of SG in CoV replication and provides insight into the interplay between MERS-CoV and antiviral stress responses.
INTRODUCTION
Middle East respiratory syndrome (MERS), which was first reported in 2012, is a zoonotic disease caused by MERS coronavirus (MERS-CoV) (1). It has been suspected that the virus is of bat origin, while dromedaries serve as a reservoir and transmit the virus to humans primarily in Middle Eastern countries (2–5). MERS patients suffer from fever, cough, and pneumonia, which can lead to respiratory failure (1, 6). The reported case fatality rate is approximately 36% (http://www.who.int/emergencies/mers-cov/en/). Hence, MERS-CoV represents a serious public health threat.
CoVs are enveloped viruses carrying a single-stranded positive-sense RNA genome of ∼30 kb in length (7–9) and are classified into four genera, Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus. Replication of MERS-CoV, a Betacoronavirus, starts with binding of the virus to a specific viral receptor, CD26 (also known as dipeptidyl peptidase 4) (10). After the fusion of virus and host cell membranes, the incoming genomic RNA undergoes translation of two large polyproteins from open reading frame (ORF) 1a and from ORFs 1a and 1b, both of which are located in gene 1 (Fig. 1A). The polyproteins are processed into 16 mature nonstructural proteins (nsp1 to nsp16), most of which are essential for the synthesis of viral RNAs (11–13), including genome-length mRNA 1 and subgenomic mRNAs, mRNAs 2 to 8. Viral mRNAs have a common 3′ end, constituting a 3′-coterminal nested set structure, and the 5′ ends of all viral mRNAs carry a common leader sequence (14–17). Subgenomic mRNAs encode viral structural proteins and accessory proteins, including 3, 4a, 4b, and 5, but the latter are not essential for virus replication, yet they do affect viral pathogenicity (18–22).
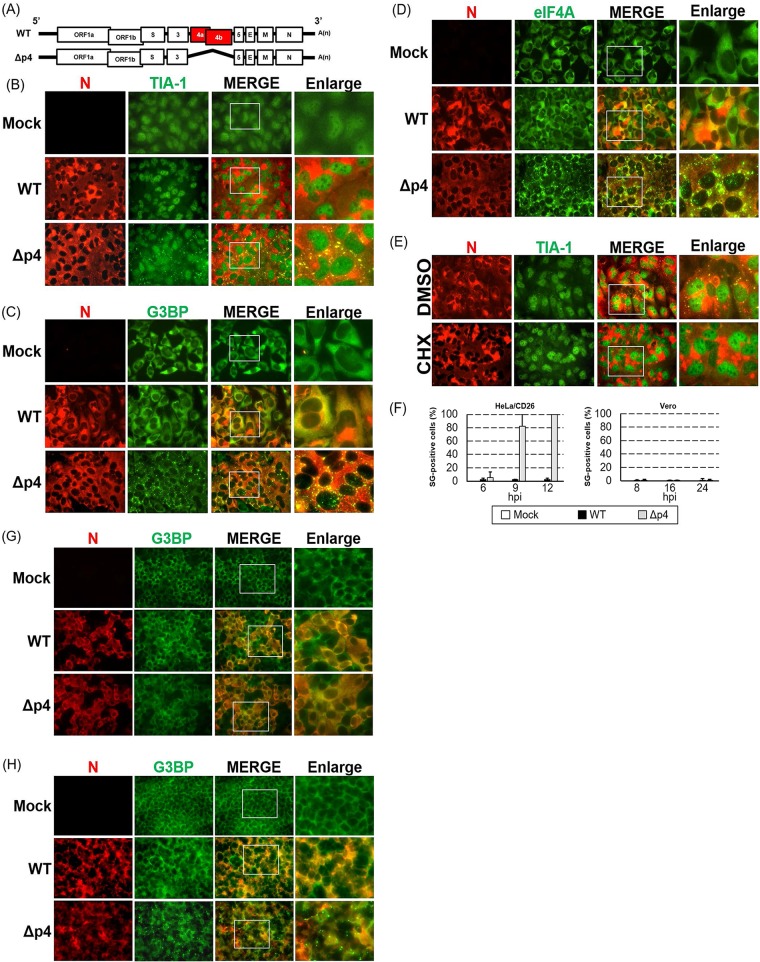
FIG 1.
Induction of SGs in MERS-CoV-Δp4-infected cells. (A) Schematic diagrams of genomes of MERS-CoV-WT (WT) and MERS-CoV-Δp4 (Δp4). The 5′ and 3′ untranslated regions (bars) and viral open reading frames (boxes) are not drawn according to their lengths. (B to D) HeLa/CD26 cells were infected with MERS-CoV-WT or MERS-CoV-Δp4 at an MOI of 3. At 9 h p.i., the infected cells were fixed with 4% formaldehyde and stained for TIA-1 (B), G3BP (C), or eIF4A (D), shown in green, together with the MERS-CoV N protein, shown in red. Right panels show enlarged images of the regions shown in white boxes in the merged image panels. (E) HeLa/CD26 cells were infected with MERS-CoV-Δp4 at an MOI of 3. At 8 h p.i., the infected cells were treated with 100 μg/ml of CHX or DMSO for 1 h. After fixing, the cells were stained for TIA-1 (green) and N protein (red). (F) HeLa/CD26 cells (left) or Vero cells (right) were infected with MERS-CoV-WT or MERS-CoV-Δp4 at an MOI of 3. At the indicated times p.i., the cells were fixed and stained for TIA-1 and MERS-CoV N protein. Among the N protein-positive cells, those carrying at least a single TIA-1-postive granule were counted as SG positive, while SG-negative calls lacked any TIA-1-postive granules. The percentage of SG-positive cells was calculated by counting the number of SG-positive cells out of 25 to 34 N protein-positive cells per field. A total of 20 fields were counted for each sample. Each bar represents the mean (±standard deviation). hpi, hours postinfection. (G, H) Vero cells (G) or 293/CD26 cells (H) were mock infected (Mock) or infected with MERS-CoV-WT or MERS-CoV-Δp4 at an MOI of 3. At 24 h p.i., the infected cells were fixed with 4% formaldehyde and were stained for G3BP (green), together with MERS-CoV N protein (red).
Virus replication, which represents a stress to the cells, activates several signaling pathways, including those triggered by activated protein kinase R (PKR). Upon binding viral double-stranded RNAs (dsRNAs), PKR undergoes an autophosphorylation reaction that activates the kinase. Activated PKR phosphorylates the α subunit of eukaryotic initiation factor 2 (eIF2α) (23–25), which prevents the recycling of ternary complex tRNA[stack][low]i[high]Met[/stack]-GTP-eIF2 and inhibits 43S translation complex formation, leading to the inhibition of translation (23, 24, 26, 27). Hence, PKR activation inhibits viral gene expression, contributing to host cell survival from infection.
The translation inhibition by eIF2α phosphorylation also leads to polysome disassembly and the subsequent accumulation of the mRNAs associated with stalled ribosome complexes in cytoplasmic structures called stress granules (SGs) (27, 28). SGs act as dynamic microdomains; once translation activities are restored, SGs are disassembled and mRNAs that have been stored in the SGs can rapidly resume translation (27, 29, 30). SGs contain mRNAs bound to translation factors, such as eIF4A and eIF3, and the 40S ribosomal subunit, plus many additional proteins affecting mRNA functions. SG assembly is driven by aggregation-prone cellular RNA-binding proteins, such as T cell internal antigen 1 (TIA-1) and Ras-GTPase-activating SH3 domain binding protein 1 (G3BP1) (27). Recent studies have shown the localization of RIG-I-like receptors (RLRs) and PKR in SGs during viral infection (31–33). It has been proposed that SGs exert specific antiviral effects (34) by providing a critical platform for interactions between antiviral proteins and non-self RNA ligands (31, 35, 36).
The impact of SG formation on virus replication varies among different viruses (37, 38). Some viruses accomplish efficient replication by inhibiting SG formation via various mechanisms. For instance, influenza A virus blocks SG formation by using the NS1 protein, which sequesters dsRNAs from PKR (39). Alphaherpesviruses block SG formation by impairing the activation of eIF2α through the virion host shutoff protein, Us11, ICP34.5, and glycoprotein B (40–43). The vaccinia virus E3L protein suppresses SG formation by binding to dsRNAs and preventing PKR activation (44). Finally, picornaviruses disassemble SG by cleaving G3BP1 via the activity of the viral 3C proteinase (45). Some viruses induce or modulate SG formation for their replication. For example, Newcastle disease virus triggers the stable formation of SGs, which benefit viral protein translation and virus replication by arresting cellular mRNAs (46). Vesicular stomatitis virus induces SG-like structures, which contain viral RNAs and viral proteins necessary for RNA synthesis, suggesting that SG-like structures are important for virus replication (47, 48). Hepatitis C virus induces the assembly and disassembly of SGs in an eIF2α-dependent manner, and some components of SGs play a pivotal role in several steps of the virus life cycle (49, 50).
Several past studies reported either SG formation or the absence of it in the context of CoV infection. Transmissible gastroenteritis coronavirus (TGEV), an alphacoronavirus, induces the aggregation of granules containing viral mRNAs associated with the polypyrimidine tract-binding protein and SG markers TIA-1 and TIAR late in infection (51). Mouse hepatitis virus (MHV), a betacoronavirus, induces SGs when the phosphorylation of eIF2α and host translational shutoff occur (52). Rabouw et al. reported that expression of the MERS-CoV 4a protein, an accessory protein, impedes PKR phosphorylation and SG formation, whereas MERS-CoV as well as MERS-CoV lacking both ORFs 4a and 4b (Fig. 1A) did not induce SGs in infected cells (53). The absence of SG formation in the mutant MERS-CoV-infected cells led the authors to speculate that MERS-CoV encodes at least one other stress response antagonist with a mode of action that differs from that of 4a (53). Currently, the biological significance of SG formation or the inhibition of it during CoV replication is unclear.
Rabouw et al. tested SG formation in MERS-CoV-infected Vero cells, but they used HeLa cells to study 4a protein-induced SG formation (53). As PKR expression levels are low in Vero cells (54), the extent of the PKR-induced eIF2α phosphorylation caused by the MERS-CoV mutant in Vero cells might have been too low for induction of SG formation. If this is the case, the MERS-CoV mutant would induce SGs in other cells, where PKR expression levels are high enough for inducing eIF2α phosphorylation and the extent of PKR expression is sufficient for SG formation. In the present study, we explored this possibility and found that the replication of a MERS-CoV mutant lacking ORFs 4a and 4b (MERS-CoV-Δp4) but not that of wild-type (WT) MERS-CoV (MERS-CoV-WT) induced SG formation in HeLa cell-derived susceptible cells. Our study further demonstrates that inhibition of SG formation facilitates the translation of viral proteins, leading to efficient virus replication, and that depletion of MERS-CoV 4a protein alone is sufficient for inducing SG formation in infected cells. To our knowledge, this is the first study revealing the biological role of SGs in CoV replication and identifying a CoV protein that suppresses SG accumulation in infected cells.
RESULTS
MERS-CoV-Δp4 infection induces SGs in HeLa/CD26 cells.
To determine whether the replication of MERS-CoV-Δp4 (Fig. 1A) induces SG formation, we established a HeLa cell line stably expressing the MERS-CoV receptor human CD26 (HeLa/CD26 cells). HeLa cells express significantly higher levels of PKR than Vero cells (54). We inoculated MERS-CoV-WT or MERS-CoV-Δp4, both of which were rescued by using a reverse genetics system (55), into HeLa/CD26 cells at a multiplicity of infection (MOI) of 3; our MERS-CoV-Δp4 and the MERS-CoV mutant described by Rabouw et al. (53) have the same deletion in gene 4. The cells were subjected to immunofluorescence analysis using antibodies specific for SG marker TIA-1, G3BP, or eIF4A, together with the MERS-CoV N protein. Granules containing these SG markers accumulated in MERS-CoV-Δp4-infected cells but not in MERS-CoV-WT-infected cells (Fig. 1B to D).
SGs are dynamic structures and disperse without a source of new translation initiation complexes (27–30). Cycloheximide (CHX) stalls translation, leading to dismantling of SGs (28); hence, if these granules carrying the SG markers in MERS-CoV-Δp4-infected cells are indeed SGs, CHX treatment would disperse them. CHX treatment, but not dimethyl sulfoxide (DMSO) treatment, caused dispersion of TIA-1-positive granules in MERS-CoV-Δp4-infected cells, establishing that MERS-CoV-Δp4, but not MERS-CoV-WT, induced SGs in infected HeLa/CD26 cells (Fig. 1E).
We next determined the kinetics of accumulation of SG-positive cells by counting cells positive for both TIA-1 and N proteins (SG-positive cells) and those positive for only N protein (SG-negative cells) at different times postinfection (p.i.); cells showing at least one SG were considered to be SG positive. Approximately 5% of MERS-CoV-Δp4-infected cells were SG positive at 6 h p.i., and the number of SG-positive cells increased as infection progressed, with ∼80% and ∼100% of cells being SG positive at 9 h p.i. and at 12 h p.i., respectively (Fig. 1F, left). In contrast, SG-positive cells represented a very minor population in MERS-CoV-WT-infected cells throughout the infection.
We also tested MERS-CoV-Δp4-induced SG formation in other cells. Replication of MERS-CoV-WT and MERS-CoV-Δp4 did not induce SGs in Vero cells (Fig. 1F, right, and G), confirming the data shown in a previous report (53). In contrast, replication of MERS-CoV-Δp4, but not MERS-CoV-WT, in 293 cells stably expressing CD26 (293/CD26 cells) (56) induced SG accumulation (Fig. 1H), demonstrating that MERS-CoV-Δp4-induced SG formation was not limited to HeLa/CD26 cells. These data show that MERS-CoV-Δp4-induced SG formation is cell type dependent.
Growth kinetics of MERS-CoV-WT and MERS-CoV-Δp4 in Vero and HeLa/CD26 cells.
To glean whether SG formation affects virus replication, we next examined the growth kinetics of MERS-CoV-WT and MERS-CoV-Δp4 in HeLa/CD26 and Vero cells. In HeLa/CD26 cells, the titers of MERS-CoV-WT were significantly higher than those of MERS-CoV-Δp4 at 18 and 24 h p.i. at an MOI of 0.01 and were also higher at 12, 18, and 24 h p.i. at an MOI of 3 (Fig. 2A). In Vero cells, both viruses replicated similarly at an MOI of 0.01, while the titers of MERS-CoV-WT were higher than those of MERS-CoV-Δp4 at 24, 36, and 48 h p.i. at an MOI of 3 (Fig. 2B). As SG formation did not occur in MERS-CoV-Δp4-infected Vero cells, the differences in virus titers between the two viruses in Vero cells were not due to SG formation. At a high MOI, the difference in the maximum virus titers between the two viruses in Vero cells (∼3.3 times at 48 h p.i.) was less prominent than that in HeLa/CD26 cells (∼12 times at 18 h p.i.). Considering the fact that MERS-CoV-Δp4 replication induced SGs in HeLa/CD26 cells but not in Vero cells, these results suggest that the SG formation negatively affects virus replication.
FIG 2.
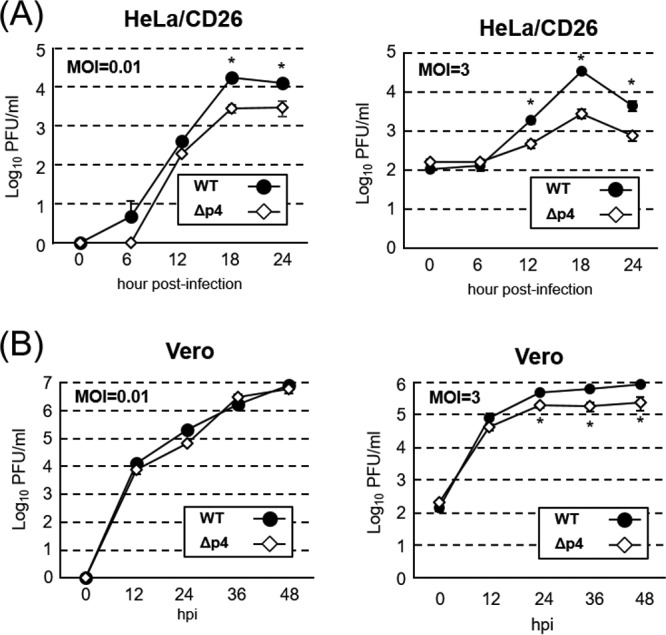
Growth kinetics of MERS-CoV-WT and MERS-CoV-Δp4 in HeLa/CD26 and Vero cells. HeLa/CD26 cells (A) or Vero cells (B) were infected with MERS-CoV-WT (WT) or MERS-CoV-Δp4 (Δp4) at an MOI of 0.01 (left) or 3 (right). Culture supernatants were collected at the indicated times p.i., and virus titers were determined by plaque assay. Each dot represents the mean virus titer (±standard deviation) for three wells. Asterisks represent statistically significant differences in virus titers (P < 0.05).
Phosphorylation status of PKR and eIF2α and translation activities in infected cells.
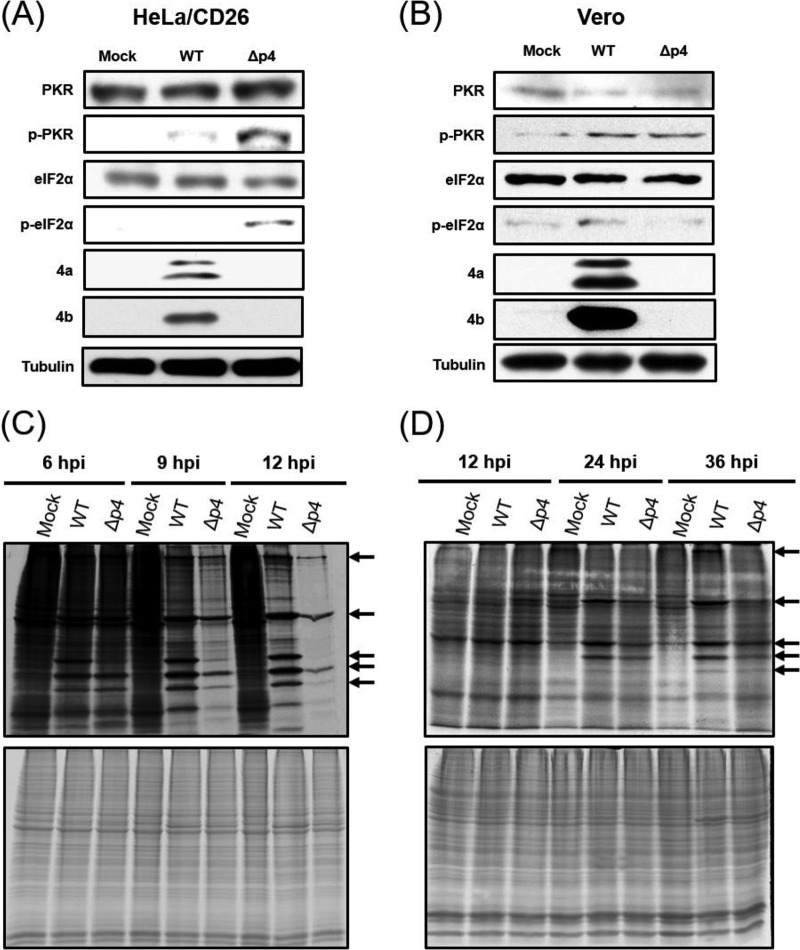
The MERS-CoV 4a protein inhibits PKR phosphorylation by binding to dsRNAs and sequestering dsRNAs from PKR (53), yet the effects of 4a on PKR activation and eIF2α phosphorylation in infected cells are unknown. We found that the phosphorylation levels of PKR and eIF2α were clearly higher in HeLa/CD26 cells infected with MERS-CoV-Δp4 than in those infected with MERS-CoV-WT (Fig. 3A). In contrast, both viruses induced low levels of PKR phosphorylation and eIF2α phosphorylation in Vero cells (Fig. 3B). As expected, the 4a and 4b proteins accumulated in MERS-CoV-WT-infected cells but not in MERS-CoV-Δp4-infected cells (Fig. 3A and B). The appearance of two 4a protein bands suggests that the 4a accessory protein undergoes modification, the nature of which is unknown, in infected cells.
FIG 3.
Phosphorylation statuses of PKR and eIF2α and efficiencies of host and viral protein synthesis in infected cells. HeLa/CD26 cells or Vero cells were either mock infected (Mock) or infected with MERS-CoV-WT (WT) or MERS-CoV-Δp4 (Δp4) at an MOI of 3. (A and B) Whole-cell lysates were prepared at 9 h p.i. for HeLa/CD26 cells (A) and 24 h p.i. for Vero cells (B) and subjected to Western blot analysis to detect PKR, phosphorylated PKR (p-PKR), eIF2α, phosphorylated eIF2α (p-eIF2α), the MERS-CoV 4a protein, the MERS-CoV 4b protein, and tubulin. (C and D) HeLa/CD26 cells (C) or Vero cells (D) were radiolabeled for 1 h with 100 μCi of Tran35S-label, and cell lysates were prepared at the indicated times p.i. Cell lysates were subjected to SDS-PAGE analysis, followed by autoradiography (top) and colloid Coomassie brilliant blue staining (bottom). Arrows, virus-specific proteins.
We next investigated the extent of host and viral protein synthesis by pulse radiolabeling of the cells with [35S]methionine-cysteine. In HeLa/CD26 cells, both viruses clearly induced translation suppression after 9 h p.i., with stronger inhibition in MERS-CoV-Δp4-infected cells than in MERS-CoV-WT-infected cells (Fig. 3C). Also, the synthesis of virus-specific proteins was lower in MERS-CoV-Δp4-infected cells than in MERS-CoV-WT-infected cells after 9 h p.i. Thus, there was an inverse correlation between the extent of phosphorylation of PKR and eIF2α and translation activities in infected HeLa/CD26 cells. In Vero cells, the synthesis of virus-specific proteins was notable after 24 h p.i., and the levels of host protein synthesis were similar among mock-infected cells, MERS-CoV-WT-infected cells, and MERS-CoV-Δp4-infected cells (Fig. 3D). These data imply that low levels of eIF2α phosphorylation did not inhibit host and viral protein synthesis in infected Vero cells.
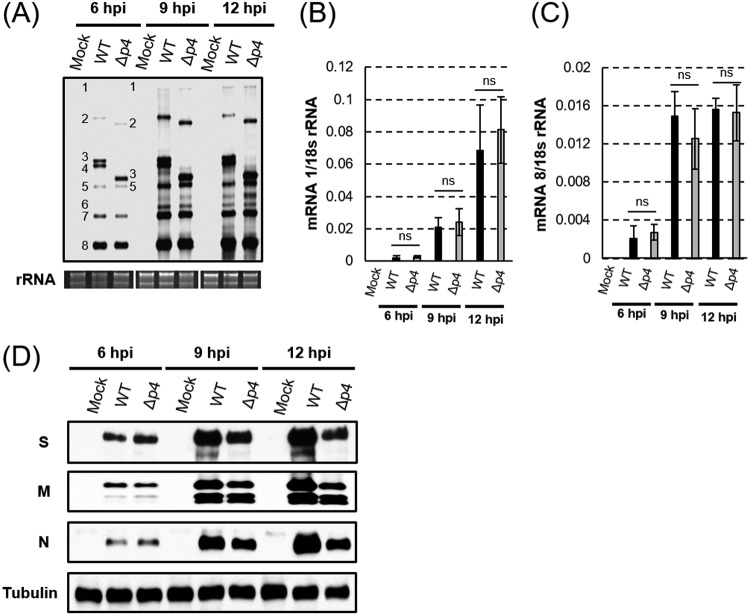
To further establish that MERS-CoV-Δp4 inhibited efficient viral protein synthesis in HeLa/CD26 cells, we examined the abundance of viral mRNAs and proteins in infected HeLa/CD26 cells. Northern blot analysis showed similar levels of viral mRNA accumulation between MERS-CoV-WT- and MERS-CoV-Δp4-infected cells at different times p.i. (Fig. 4A). Due to the deletion of ORFs 4a and 4b, mRNAs 1 to 3 of MERS-CoV-Δp4 migrated faster than those of MERS-CoV-WT in the gel. Quantitative reverse transcription-PCR (qRT-PCR) showed that mRNA 1 and mRNA 8 encoding N protein accumulated to similar levels in MERS-CoV-WT- and MERS-CoV-Δp4-infected cells (Fig. 4B and C). These studies established that the depletion of ORFs 4a and 4b had little impact on the accumulation of viral mRNAs.
FIG 4.
Accumulation of viral mRNAs and proteins in infected HeLa/CD26 cells. HeLa/CD26 cells were mock infected (Mock) or infected with MERS-CoV-WT (WT) or MERS-CoV-Δp4 (Δp4) at an MOI of 3. At the indicated times p.i., total intracellular RNAs and proteins were prepared. (A) Northern blot analysis of viral mRNAs using a riboprobe that binds to all viral mRNAs. The numbers 1 to 8 represent viral mRNA species. The 28S and 18S rRNAs were detected by ethidium bromide staining (rRNA). (B and C) The amounts of mRNA 1 (B) and subgenomic mRNA 8 (C) were quantified by qRT-PCR. The expression levels of mRNAs were normalized to the levels of 18S rRNA. Each bar represents the mean (±standard deviation) for three independent samples. ns, not significant (P > 0.05). (D) Western blot analysis of intracellular accumulation of the MERS-CoV S, M, and N proteins and tubulin.
Western blot analysis showed that the viral structural proteins S, M, and N accumulated similarly in both viruses at 6 h p.i., while at 9 h p.i., when PKR-mediated eIF2α phosphorylation had occurred (Fig. 3A), the viral structural proteins accumulated to higher levels in MERS-CoV-WT-infected cells than in MERS-CoV-Δp4-infected cells (Fig. 4D). Similar results were noted at 12 h p.i. (Fig. 4D). These data establish that the translation of viral proteins was indeed inefficient in MERS-CoV-Δp4-infected cells.
We interpreted these data as follows: (i) MERS-CoV-Δp4 replication in HeLa/CD26 cells activated PKR, which induced eIF2α phosphorylation and strong translational suppression, leading to SG formation; (ii) in MERS-CoV-WT-infected HeLa/CD26 cells, 4a-mediated inhibition of PKR activation likely prevented efficient eIF2α phosphorylation, allowing viral and host translation and preventing SG formation; (iii) the PKR antagonistic function of 4a failed to show significant biological effects in Vero cells, probably due to the low expression levels of PKR; and (iv) due to the low levels of PKR expression, the PKR-mediated eIF2α phosphorylation was inefficient in infected Vero cells, allowing unimpeded translation of viral and host proteins and inhibiting SG formation.
Innate antiviral responses in MERS-CoV-Δp4-infected HeLa/CD26 cells.
Our data support a notion that inhibition of viral translation, which was induced by phosphorylated eIF2α, caused inefficient MERS-CoV-Δp4 replication in HeLa/CD26 cells. In addition, we suspected that SG formation itself also contributed to the inefficient MERS-CoV-Δp4 replication by one of the following mechanisms: (i) SG formation induces activation of innate immune responses, leading to suppression of viral replication; (ii) viral mRNAs are stored in the SGs and not available for translation, leading to inefficient viral translation; and (iii) SGs store various factors required for translation, e.g., translation factors and the 40S ribosomal subunit, and restrict the availability of these factors for translation, leading to inefficient viral translation. The following experiments tested these possibilities.
SGs provide a platform for the interaction of RIG-I-like receptors and viral mRNAs, leading to interferon (IFN) production (31, 35, 36). Additionally, antiviral proteins, e.g., PKR, 2′-5′-oligoadenylate synthetase (OAS), and RNase L, are recruited to SGs and exert antiviral functions (31, 32). We explored whether SG formation could trigger innate immune responses by examining the expression levels of the beta interferon (IFN-β), IFN-λ1, OAS, and interferon-stimulated gene 56 (ISG56) mRNAs in MERS-CoV-WT- and MERS-CoV-Δp4-infected HeLa/CD26 cells (Fig. 5). Sendai virus (SeV), a positive control, induced efficient expression of these mRNAs, while mock infection did not (Fig. 5). MERS-CoV-WT or MERS-CoV-Δp4 did not induce efficient expression of these mRNAs at any time p.i. Also, there were no significant differences in the expression levels of these mRNAs between MERS-CoV-WT- and MERS-CoV-Δp4-infected cells. These data suggest that the SGs do not play significant roles in innate immune gene expression in MERS-CoV-Δp4-infected HeLa/CD26 cells.
FIG 5.
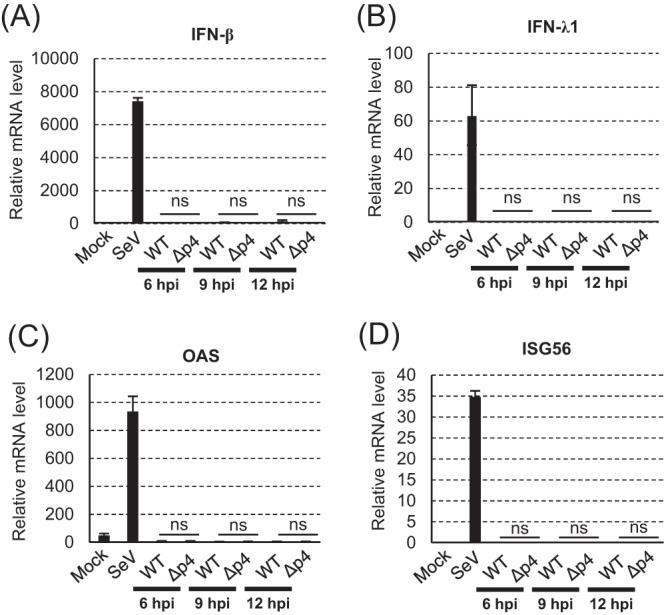
Expression of host mRNAs involved in innate immune responses in infected HeLa/CD26 cells. HeLa/CD26 cells were mock infected (Mock) or infected with MERS-CoV-WT (WT) or MERS-CoV-Δp4 (Δp4) at an MOI of 3. SeV was used as a positive control. Total intracellular RNAs were extracted at the indicated times p.i., and the amounts of endogenous IFN-β (A), IFN-λ1 (B), OAS (C), and ISG56 (D) mRNAs were determined by qRT-PCR analysis. The expression levels of the genes were normalized to the expression levels of 18S rRNA. Each bar represents the mean (±standard deviation) for three independent samples. ns, not significant (P > 0.05).
MERS-CoV mRNAs are not confined in the SGs.
The absence of innate immune gene expression in MERS-CoV-Δp4-infected HeLa/CD26 cells may be due to the possibility that the viral mRNAs are not confined in the SGs, thus escaping recognition by RIG-I-like receptors in the SGs. Alternatively, viral mRNAs might have been efficiently stored in the SGs and contributed to inefficient viral translation, yet they were unable to trigger innate immune gene expression.
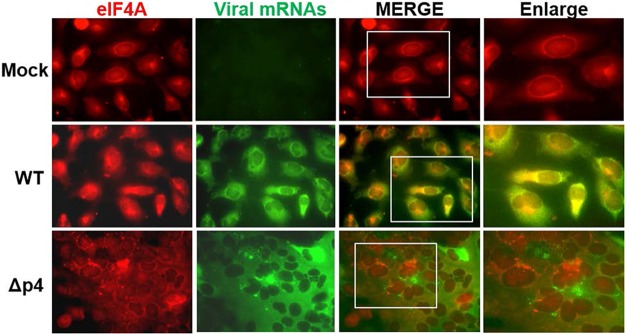
To determine whether viral mRNAs are trapped in the SGs, we visualized viral mRNAs by using fluorescent in situ hybridization (FISH) analysis with a probe binding to all viral mRNAs. SGs were visualized by immunofluorescence assay staining of an SG marker, eIF4A, in mock-infected HeLa/CD26 cells, MERS-CoV-WT-infected cells, and MERS-CoV-Δp4-infected cells. eIF4A showed a diffuse distribution in mock-infected cells and MERS-CoV-WT-infected cells, demonstrating the absence of SG formation. Although eIF4A accumulated within SGs in MERS-CoV-Δp4-infected cells (Fig. 6), viral mRNAs were diffusely distributed in the cytoplasm in infected cells (Fig. 6). The absence of clear colocalization of viral mRNAs and eIF4A demonstrated that most of the viral mRNAs were not confined in the SGs.
FIG 6.
Subcellular localization of viral mRNAs and an SG marker, eIF4A, in MERS-CoV-Δp4-infected cells. HeLa/CD26 cells were mock infected (top) or infected with MERS-CoV-WT (middle) or MERS-CoV-Δp4 (bottom) at an MOI of 3. At 9 h p.i., viral mRNAs were detected by riboprobe binding to all viral mRNAs (green), and SGs were detected by anti-eIF4A antibody (red). Samples were subjected to fluorescence microscopic examination.
SG formation interferes with MERS-CoV replication.
We next explored a possibility that various factors involved in translation are accumulated in the SGs, causing a reduction of their abundance in the cytoplasm and leading to the inefficient translation of viral proteins. If this is the case, inhibition of SG accumulation would lead to the promotion of viral gene expression, as those factors involved in translation are not confined in the SGs and are available for translation. We tested this possibility by examining the replication and gene expression of MERS-CoV-Δp4 in HeLa/CD26 cells that were depleted of TIA-1 or both G3BP1 and G3BP2 (G3BP1/2) by small interfering RNA (siRNA) treatment. The siRNAs targeting TIA-1 and those targeting G3BP1/2 efficiently reduced the levels of TIA-1 and G3BP1/2, respectively (Fig. 7A). Depletion of TIA-1 or G3BP1/2 did not prevent SG formation in MERS-CoV-Δp4-infected cells, whereas it reduced the average numbers of SGs (Fig. 7A, top). MERS-CoV-WT replicated less efficiently in cells depleted of TIA-1 or those depleted of G3BP1/2 than in control siRNA-treated cells (Fig. 7B, top), implying that TIA-1 and G3BP1/2 facilitated MERS-CoV replication. In contrast, MERS-CoV-Δp4 replicated more efficiently in TIA-1-depleted cells than in control siRNA-treated cells (Fig. 7B, bottom left). Likewise, the replication of MERS-CoV-Δp4 was also better in G3BP1/2-depleted cells than in control siRNA-treated cells (Fig. 7B, bottom right). These data show that the reduction in the number of SGs facilitated MERS-CoV-Δp4 replication and also suggest that inhibition of SG formation has effects on promoting virus replication more pronounced than the positive effects of TIA-1 or G3BP1/2 for virus replication.
FIG 7.
SG formation interferes with efficient MERS-CoV replication. HeLa/CD26 cells were transfected with control siRNA (siCont) or siRNA targeting TIA-1 (siTIA-1) or G3BP1/2 (siG3BP1/2). (A) At 24 h after siRNA transfection, cells were infected with MERS-CoV-Δp4 at an MOI of 3. At 9 h p.i., the cells were fixed and stained for G3BP or TIA-1, together with N protein. The numbers of SGs in each N protein-positive cell were counted, and the average numbers of SGs per cell were calculated. Each bar represents the mean (±standard deviation) for 20 cells infected with MERS-CoV-Δp4. Whole-cell lysates were prepared at 24 h posttransfection and subjected to Western blot analysis to detect TIA-1, G3BP1, G3BP2, or tubulin. (B) At 24 h after siRNA transfection, cells were infected with MERS-CoV-WT or MERS-CoV-Δp4 at an MOI of 3. The titers of the released viruses at the indicated times p.i. were determined by plaque assay. Filled symbols represent virus titers in control siRNA-transfected cells, while empty symbols represent virus titers in TIA-1-specific siRNA-transfected cells or G3BP1/2-specific siRNA-transfected cells. Each box represents the mean (±standard deviation) for three wells. Asterisks represent statistically significant differences (P < 0.05). (C and D) At 24 h after siRNA transfection, cells were mock infected (Mock) or infected with MERS-CoV-WT (WT) or MERS-CoV-Δp4 (Δp4) at an MOI of 3. These cells were radiolabeled for 1 h with 100 μCi of Tran35S-label, and cell lysates were prepared at the indicated times p.i. Cell lysates were subjected to SDS-PAGE analysis, followed by autoradiography (left) and colloid Coomassie brilliant blue staining (right). (E and F) At 24 h after siRNA transfection, cells were infected with MERS-CoV-Δp4 at an MOI of 3, and total cell lysates and RNAs were prepared at 9 h p.i. Western blot analysis of viral S protein, M protein, N protein, and tubulin (E). The amounts of mRNA 1 and subgenomic mRNA 8 were quantified by qRT-PCR (F). Expression levels of mRNAs were normalized to the expression levels of 18S rRNA. Each bar represents the mean (±standard deviation) for three independent samples. ns, not significant (P > 0.05). (G and H) At 24 h siRNA after transfection, cells were mock infected or infected with MERS-CoV-WT or MERS-CoV-Δp4 at an MOI of 3. Cell lysates were prepared at 9 h p.i. and subjected to Western blot analysis to detect eIF2α and phosphorylated eIF2α (p-eIF2α).
The effects of inefficient SG accumulation on translation activities were examined next. Control siRNA-treated cells and TIA-1-depleted cells were first mock infected or infected with MERS-CoV-Δp4 or MERS-CoV-WT. Translational activities were then examined by metabolic radiolabeling. In control siRNA-treated cells, MERS-CoV-WT and MERS-CoV-Δp4 induced translational inhibition, with the latter virus showing stronger inhibition at 9 and 12 h p.i. (Fig. 7C). In TIA-1-depleted cells, the translation-inhibitory effects caused by MERS-CoV-Δp4 were less prominent at 9 and 12 h p.i. (Fig. 7C). We performed similar experiments by using cells depleted of G3BP1/2 and found that depletion of G3BP1/2 induced less pronounced translation-inhibitory effects at 9 and 12 h p.i. in MERS-CoV-Δp4-infected cells (Fig. 7D). Western blot analysis showed that the accumulation of structural proteins of MERS-CoV-Δp4 was stronger in cells depleted of TIA-1 or G3BP1/2 than in control siRNA-treated cells (Fig. 7E), whereas depletion of TIA-1 or G3BP1/2 did not affect the accumulation of viral mRNAs, mRNA 1, and mRNA 8 (Fig. 7F). These data demonstrate that inhibition of SG accumulation promotes the translation of viral and host proteins.
Because SG formation occurs downstream of eIF2α phosphorylation, we anticipated that inhibition of SG formation by depletion of TIA-1 or G3BP1/2 would not affect the status of eIF2α phosphorylation in infected cells. Consistent with this anticipation, depletion of TIA-1 or G3BP1/2 did not alter eIF2α phosphorylation levels in MERS-CoV-Δp4-infected HeLa/CD26 cells or MERS-CoV-WT-infected HeLa/CD26 cells (Fig. 7G and H). These data reveal that the increased viral gene expression of MERS-CoV-Δp4 in cells depleted of TIA-1 or G3BP1/2 is not due to changes in eIF2α phosphorylation.
Taken together, these data are consistent with a notion that SG formation prevents translation by sequestering various factors that are involved in translation to SGs and preventing them from engaging in translation, leading to inefficient virus replication.
Deletion of the 4a gene alone is sufficient for suppression of SG formation in infected cells.
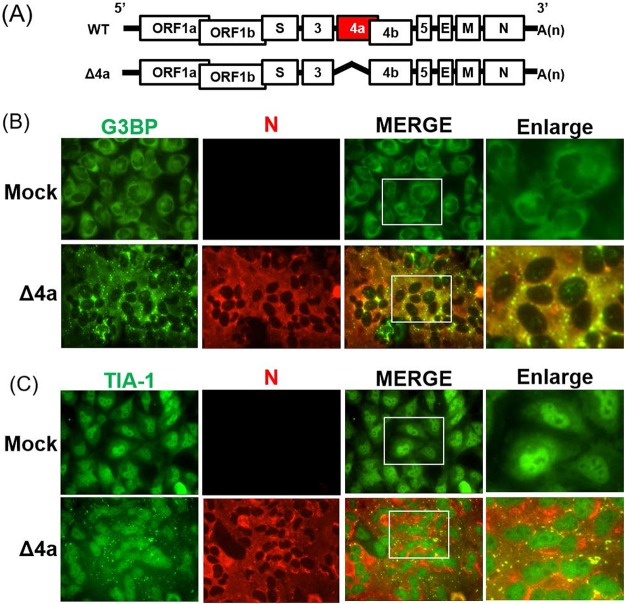
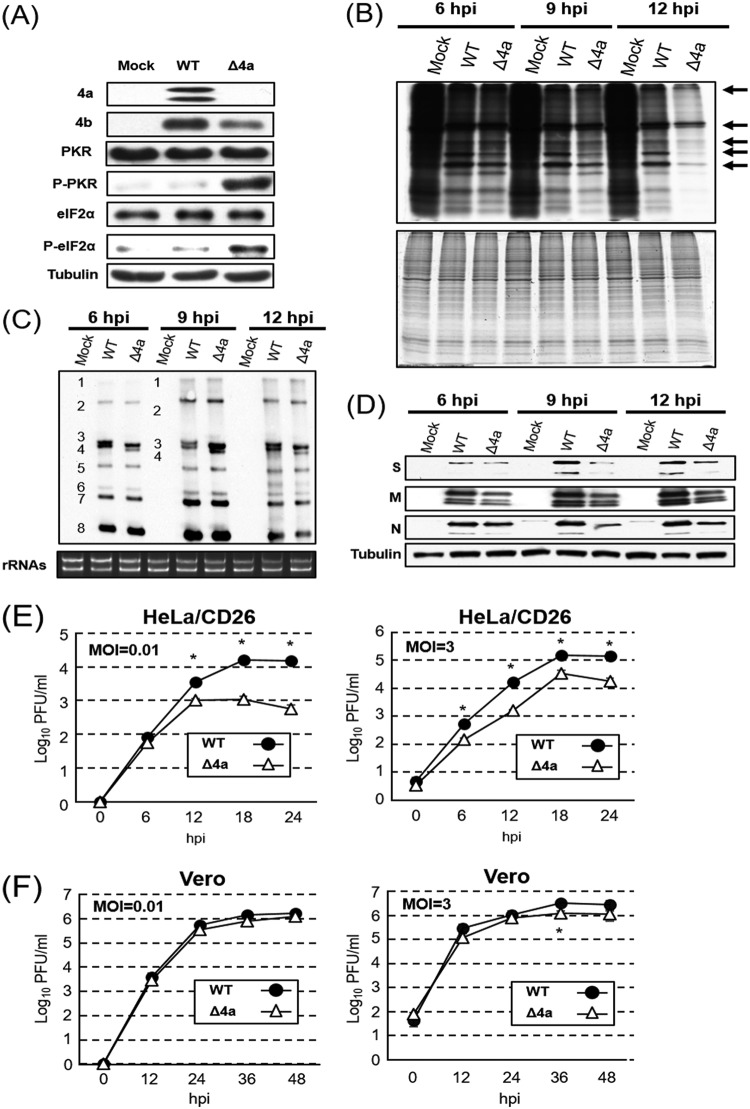
To establish that the 4a accessory protein inhibits SG formation in MERS-CoV infection, we examined SG formation in HeLa/CD26 cells that were infected with MERS-CoV lacking ORF 4a (MERS-CoV-Δ4a) (Fig. 8A). Immunofluorescence analysis using antibodies specific for SG marker TIA-1 or G3BP, together with the MERS-CoV N protein, showed the accumulation of SGs in MERS-CoV-Δ4a-infected cells (Fig. 8B and C). There were no notable differences in the quantity of SGs per cell or the number of SG-positive cells between MERS-CoV-Δp4-infected and MERS-CoV-Δ4a-infected cells. As expected, accumulation of the 4b protein, but not that of the 4a protein, occurred in MERS-CoV-Δ4a-infected cells (Fig. 9A), demonstrating that the 4a accessory protein alone was sufficient for inhibiting SG formation in infected cells. The efficient phosphorylation of PKR and eIF2α occurred in MERS-CoV-Δ4a-infected cells (Fig. 9A), establishing that the 4a protein alone was sufficient for inhibition of PKR-mediated eIF2α phosphorylation in infected cells. Pulse radiolabeling experiments showed less efficient translational activities in MERS-CoV-Δ4a-infected cells than in MERS-CoV-WT-infected cells after 9 h p.i. (Fig. 9B), suggesting that a combined effect of SG formation and efficient eIF2α phosphorylation causes strong translational suppression in MERS-CoV-Δ4a-infected cells. There were no substantial differences in the accumulation levels of viral mRNAs between MERS-CoV-Δ4a-infected cells and MERS-CoV-WT-infected cells, except that the former had slightly lower levels of mRNA 4 than the latter (Fig. 9C). The accumulation of viral structural proteins was higher in MERS-CoV-WT-infected cells than in MERS-CoV-Δ4a-infected cells (Fig. 9D), demonstrating that the 4a protein inhibited viral translation without affecting viral RNA accumulation. We suspect that inefficient translational activity and lower levels of mRNA 4 accumulation in MERS-CoV-Δ4a-infected cells contributed to the lower level of 4b protein accumulation in MERS-CoV-Δ4a-infected cells than in MERS-CoV-WT-infected cells (Fig. 9A). MERS-CoV-Δ4a replicated less efficiently than MERS-CoV-WT in HeLa/CD26 cells, regardless of the MOI (Fig. 9E). In contrast, both viruses replicated to comparable levels with similar kinetics in Vero cells at both high and low MOIs, except that the titers of MERS-CoV-Δ4a were statistically significantly lower than those of MERS-CoV-WT at 36 h p.i. at an MOI of 3 (Fig. 9F). Taken together, these data support the notion that, due to the absence of the 4a protein, MERS-CoV-Δ4a induced efficient PKR-mediated eIF2α phosphorylation and SG formation, both of which caused inhibition of viral and host translation, resulting in inefficient virus replication in HeLa/CD26 cells. Overall, MERS-CoV-Δ4a and MERS-CoV-Δp4 showed similar virological properties and induced analogous changes in translational activities and phosphorylation statuses of PKR and eIF2α, strongly suggesting that the observed biological differences between MERS-CoV-Δp4 and MERS-CoV-WT in infected HeLa/CD26 cells are primarily due to the absence of the 4a protein in MERS-CoV-Δp4-infected cells.
FIG 8.
MERS-CoV 4a accessory protein alone is sufficient for inhibiting SG formation in infected cells. (A) Schematic diagrams of the genomes of MERS-CoV-WT (WT) and MERS-CoV-Δ4a (Δ4a). Boxes represent open reading frames derived from MERS-CoV-WT. The 5′ and 3′ untranslated regions (bars) and viral open reading frames (boxes) are not drawn according to their lengths. (B and C) HeLa/CD26 cells were infected with MERS-CoV-WT or MERS-CoV-Δ4a at an MOI of 3. At 9 h p.i., the infected cells were fixed and stained for TIA-1 (C) or G3BP (B) (green), together with MERS-CoV N protein (red).
FIG 9.
Characterization of MERS-CoV-Δ4a replication in HeLa/CD26 cells. HeLa/CD26 cells were either mock infected (Mock) or infected with MERS-CoV-WT (WT) or MERS-CoV-Δ4a (Δ4a) at an MOI of 3. (A) Whole-cell lysates were prepared at 9 h p.i. and subjected to Western blot analysis to detect PKR, phosphorylated PKR (p-PKR), eIF2α, phosphorylated eIF2α (p-eIF2α), the MERS-CoV 4a protein, the MERS-CoV 4b protein, and tubulin. (B) HeLa/CD26 cells were radiolabeled for 1 h with 100 μCi of Tran35S-label, and cell lysates were prepared at the indicated times p.i. Cell lysates were subjected to SDS-PAGE analysis, followed by autoradiography (top) and colloid Coomassie brilliant blue staining (bottom). Arrows depict virus-specific proteins. (C) Northern blot analysis of viral mRNAs using a riboprobe that binds to all viral mRNAs. The numbers 1 to 8 represent viral mRNA species. The 28S and 18S rRNAs were detected by ethidium bromide staining (rRNA). (D) Western blot analysis of intracellular accumulation of the MERS-CoV S, M, and N proteins and tubulin. (E and F) HeLa/CD26 (E) or Vero cells (F) were infected with MERS-CoV-WT or MERS-CoV-Δ4a at an MOI of 0.01 (left) or 3 (right). Culture supernatants were collected at the indicated times p.i., and virus titers were determined by plaque assay. Each dot represents the mean virus titer (±standard deviation) for three wells. Asterisks represent statically significant differences in virus titers (P < 0.05).
DISCUSSION
SG formation or inhibition has been reported for various viruses, including CoVs (37, 38). TGEV and MHV induce SG or SG-like granules (51, 52), and MERS-CoV did not induce SGs in Vero cells (53), yet the biological roles of SGs or the absence of SGs in CoV replication are unknown. The present study demonstrated that replication of MERS-CoV-Δp4 and MERS-CoV-Δ4a, but not MERS-CoV-WT, induced SG formation in HeLa/CD26 cells. MERS-CoV-Δp4 and MERS-CoV-Δ4a replicated less efficiently than MERS-CoV-WT (Fig. 2 and 9), and inhibition of SG formation promoted MERS-CoV-Δp4 replication in this cell line (Fig. 7), demonstrating that SG formation is detrimental for MERS-CoV replication. The less efficient replication of MERS-CoV-Δp4 was at least partly due to the attenuation of viral protein synthesis, which was not caused by the induction of type I and type III IFNs or by the sequestration of viral mRNAs to the SGs. Rather, our data suggest that high levels of PKR-mediated eIF2α phosphorylation and the sequestration of host factors that are required for translation to the SGs prevent viral translation in MERS-CoV-Δp4 replication. To our knowledge, this is the first report uncovering the biological role of SGs in CoV infection and identifying a CoV protein that suppresses SG accumulation in infected cells.
We observed cell type-dependent SG formation in MERS-CoV-Δp4 infection; SG formation occurred in HeLa/CD26 cells and 293/CD26 cells but not in Vero cells (Fig. 1). Our data were consistent with those from a past study reporting the absence of SG formation in Vero cells infected with MERS-CoV and its mutant lacking the 4a and 4b ORFs (53). Because the translational suppression induced by phosphorylated eIF2α triggers SG formation (27, 28, 30), efficient eIF2α phosphorylation-mediated translational suppression, which occurred in MERS-CoV-Δp4-infected HeLa/CD26 cells but not in MERS-CoV-Δp4-infected Vero cells, most likely contributed to SG formation in HeLa/CD26 cells. PKR expression is low in Vero cells (54), and the levels of phosphorylated PKR were similar between MERS-CoV-Δp4-infected Vero cells and MERS-CoV-WT-infected Vero cells (Fig. 3B). In contrast, MERS-CoV-Δp4 replication, but not MERS-CoV-WT replication, induced efficient PKR phosphorylation in HeLa/CD26 cells (Fig. 3A). MERS-CoV-Δ4a replication also induced efficient PKR phosphorylation in HeLa/CD26 cells (Fig. 9A). These data suggest that the low-level expression of PKR in Vero cells masks the effects of the 4a protein for inhibiting eIF2α-mediated SG formation in MERS-CoV-Δp4 replication. Likewise, replication of a herpes simplex virus 1 mutant activates PKR and induces eIF2α phosphorylation in HeLa cells but not in Vero cells (57). In addition to the presence of a genetic defect for IFN production (58, 59), low expression levels of PKR, which were insufficient for PKR-induced eIF2α phosphorylation and SG formation in MERS-CoV-Δp4 replication and MERS-CoV-Δ4a replication, might have contributed to the efficient replication of various viruses in Vero cells.
The translation of viral proteins was less efficient in MERS-CoV-Δp4-infected HeLa/CD26 cells than in MERS-CoV-WT-infected HeLa/CD26 cells (Fig. 4D). Likewise, inefficient viral protein synthesis occurred in MERS-CoV-Δ4a replication (Fig. 9B). In contrast, the amounts of viral mRNAs were similar in these cells (Fig. 4B and C and 9C). As the nsp3 to nsp16 and N proteins (13, 60, 61) drive viral RNA synthesis, it is somewhat puzzling that the differences in the translational efficiency of viral proteins did not result in differences in the accumulation of the viral mRNAs. We noted that these three viruses showed similar levels of translational activities (Fig. 3C and 9B) and N protein accumulation (Fig. 4D and 9D) at 6 h p.i., implying that they accumulated similar levels of nsp3 to nsp16 and N proteins early in infection. If this is the case and if the nsp3 to nsp16 and N proteins, which are synthesized early in infection, primarily determine viral mRNA synthesis efficiency, it would result in similar levels of viral mRNA accumulation in cells infected with these three viruses.
The absence of SG formation in MERS-CoV-Δp4-infected Vero cells led Rabouw et al. to suspect that MERS-CoV encodes at least one other stress response antagonist with a mode of action that differs from that of 4a (53). MERS-CoV-Δp4 replication in HeLa/CD26 cells induced SGs in ∼100% of infected cells by 12 h p.i., while SGs were detected in very small numbers of MERS-CoV-WT-infected HeLa/CD26 cells (Fig. 1F). Furthermore, MERS-CoV-Δ4a induced SGs in HeLa/CD26 cells (Fig. 8B and C). These data suggest that the 4a accessory protein is the stress response antagonist that inhibits the formation of SGs and that MERS-CoV does not encode another protein(s) that efficiently suppresses SG formation. It should be also noted that the number of SG-positive cells increased according to the progression of MERS-CoV-Δp4 replication (Fig. 1F). The steady SG accumulation during MERS-CoV-Δp4 replication differs from the transient induction and disruption of SGs seen during the replication of some viruses (49, 62). These data imply that MERS-CoV does not encode a protein that disrupts preexisting SGs.
Several past studies reported that SGs serve as a platform for the sensing of non-self RNA by RLRs and that SG formation is important for the innate immune response during viral infection (31, 35, 36), whereas MERS-CoV-Δp4 infection did not induce the accumulation of IFN-β, IFN-λ1, OAS, and ISG56 mRNAs in HeLa/CD26 cells (Fig. 5). Likewise, Rabouw et al. reported the absence of IFN-β production in MERS-CoV-Δp4-infected Huh-7 cells (53). Several possible reasons for the lack of accumulation of those mRNAs involved in the innate immune responses in MERS-CoV-Δp4 infection are conceivable. One is that the poor accumulation of viral mRNAs in the SGs (Fig. 6) prevents the recognition of viral mRNAs by RLRs in the SGs. If this is true, there is a possibility that MERS-CoV alters the cellular environment to actively prevent the accumulation of viral mRNAs in SGs. Another possibility is that the SGs cannot serve as a platform for sensing of viral mRNAs due to the low concentration of RLRs in the SGs. Alternatively, SGs might serve as a platform for the sensing of viral mRNAs by RLRs, yet another viral protein(s) prevents the accumulation of mRNAs involved in the innate immune response by inhibiting the signaling pathways that induce them, suppressing their transcription/processing or promoting their degradation.
Depletion of TIA-1 or G3BP1/2 reduced the number of SGs and promoted viral and host translation and MERS-CoV-Δp4 replication, without affecting the levels of eIF2α phosphorylation (Fig. 7G and H). These data strongly suggest that SG formation itself interferes with translation. As viral mRNAs are not confined in the SGs (Fig. 6), we suspect that the confinement of various factors that are required for translation, e.g., translation factors and 40S ribosomes, in the SGs interferes with efficient translation in MERS-CoV-Δp4-infected HeLa/CD26 cells. Although the depletion of TIA-1 or G3BP1/2 promoted the replication of MERS-CoV-Δp4, it inhibited the replication of MERS-CoV-WT (Fig. 7). These data suggest that the inhibition of SG formation by depleting TIA-1 or G3BP1/2 is more pronounced than the effect of TIA-1 and G3BP1/2 for promoting virus replication. Some viruses, e.g., flaviviruses and alphaviruses, inhibit SG accumulation by using certain SG components for viral RNA synthesis (63–65). Accordingly, MERS-CoV may also prevent SG formation to exploit TIA-1 and G3BP1/2 for virus replication. As the levels of accumulation of viral mRNAs were similar between MERS-CoV-WT-infected HeLa/CD26 cells and MERS-CoV-Δp4-infected HeLa/CD26 cells (Fig. 4A to C), TIA-1 and G3BP1/2 may promote virus replication at the post-viral transcription step.
MERS-CoV accessory proteins encoded from ORF 3 to ORF 5 have major implications for viral replication and pathogenesis in the mouse model (18), while the role of the 4a accessory protein in viral pathogenicity is unclear. Unlike Vero cells, many cells in animal hosts likely express PKR, the levels of which are sufficient for eIF2α phosphorylation and inhibition of viral translation; otherwise, cells expressing low levels of PKR would be highly susceptible to virus infection, and this would be detrimental for host survival against virus infection. Accordingly, we speculate that MERS-CoV-Δp4 and MERS-CoV-Δ4a induce SGs, which suppress viral translation, in most of the susceptible cells/organs in infected animals. Testing of the pathogenesis of MERS-CoV-Δp4 and MERS-CoV-Δ4a in a mouse model would provide a clue as to the role of SGs in MERS-CoV pathogenesis.
MATERIALS AND METHODS
Cells.
Vero cells were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum. HeLa/CD26 cells and 293/CD26 cells were generated by transfecting pCAGGS-CD26-blasticidinR, which expresses the blasticidin resistance gene and human CD26, into HeLa cells and 293 cells (ATCC), respectively, and incubating the transfected cells in selection medium containing blasticidin (12 μg/ml) for 2 weeks. The stable expression of human CD26 in 293/CD26 and HeLa/CD26 cells was confirmed by Western blotting with anti-human DPP4 antibody (R&D Systems).
Viruses.
MERS-CoV-WT, MERS-CoV-Δp4 lacking ORFs 4a and 4b, and MERS-CoV-Δ4a lacking ORF 4a were rescued by using a reverse genetics system of MERS-CoV (55). Briefly, a recombinant PCR procedure was used to delete both ORFs 4a and 4b or only ORF 4a in fragment F of the MERS-CoV reverse genetics system (55). After assembly of the MERS-CoV full-length cDNA by ligating fragments A through F, we synthesized the full-length RNA transcripts and rescued the recombinant viruses according to established protocols, as described previously (55). MERS-CoV-Δp4 and MERS-CoV-Δ4a had a deletion from nucleotides (nt) 25852 to 26833 and from nt 25852 to 26181, respectively, in MERS-CoV genomic RNA (GenBank accession number JX869059.2). We passaged the rescued viruses once in Vero cells, confirmed the presence of the expected mutation in MERS-CoV-Δp4 and MERS-CoV-Δ4a and the absence of the mutation at the corresponding region in MERS-CoV-WT, and used the viruses for the experiments. All experiments with infectious MERS-CoV were performed in an approved biosafety level 3 laboratory at The University of Texas Medical Branch at Galveston. Sendai virus (SeV; Cantell strain), obtained from Charles River Laboratory (Wilmington, MA), was used to infect cells at 400 hemagglutination units/ml.
Immunofluorescence staining and CHX treatment.
Cells were fixed overnight with 4% paraformaldehyde and permeabilized for 15 min with 0.1% Triton X-100 in phosphate-buffered saline (PBS). After blocking with 1% bovine serum albumin in PBS for 1 h, the cells were incubated with primary antibodies (anti-MERS-CoV N protein antibody and goat anti-TIA-1 antibody [Santa Cruz], mouse anti-G3BP antibody [BD Biosciences], or mouse anti-eIF4AI/II antibody [Santa Cruz]) overnight at 4°C, washed, and incubated for 1 h at room temperature with Alexa Fluor-conjugated secondary antibodies (Thermo Fisher Scientific). Anti-MERS-CoV N protein antibody was generated by immunizing rabbits with a synthetic peptide (NDITNTNLSRGRGRNPKPR). Fluorescence was visualized by imaging using a Zeiss Axioplan 2 microscope. For CHX treatment, HeLa/CD26 cells were infected with virus at an MOI of 3. At 8 h p.i., medium was replaced with growth medium containing 100 μg/ml CHX (Sigma) dissolved in DMSO. An equal volume of DMSO was added to the control medium. At 9 h p.i., cells were fixed and processed for immunofluorescence staining for anti-MERS-CoV N protein antibody and TIA-1 antibody, as described above.
Fluorescent in situ hybridization.
MERS-CoV mRNAs were detected by using a digoxigenin (DIG)-labeled riboprobe binding to nt 29084 to 29608 of the MERS-CoV genomic RNA. The probe was denatured at 100°C for 10 min, diluted 1:1 in hybridization buffer (2% SSC [0.15 M NaCl plus 0.015 M sodium citrate], 10% dextran sulfate), and hybridized to the cells at 45°C for 18 h. After two washes using wash buffer I (50% formamide, 0.1% sodium dodecyl sulfate [SDS], 0.1% SSC) and one subsequent wash with wash buffer II (50% formamide, 0.2% SSC), cells were incubated with anti-digoxigenin-fluorescein (Sigma-Aldrich) and anti-eIF4AI/II antibodies overnight at 4°C. Then, the cells were washed in wash buffer III (8% formamide, 2% SSC) and incubated for 1 h at room temperature with Alexa Fluor-conjugated secondary antibodies (Thermo Fisher Scientific). Images were captured by a Zeiss Axiophot 2 fluorescence microscope and processed with ImageJ software (http://rsbweb.nih.gov/ij/).
Virus growth in Vero and HeLa/CD26 cells.
Monolayer cultures of Vero cells or HeLa/CD26 cells were infected with MERS-CoV-WT, MERS-CoV-Δp4, or MERS-CoV-Δ4a at an MOI of 0.01 or 3 for 1 h at 37°C. After virus adsorption, the cells were washed twice with PBS and incubated with the appropriate medium. Viruses in the culture supernatants were harvested at different times p.i. The infectious virus titers were determined by plaque assay by using Vero cells and are expressed in PFU per milliliter.
Western blot analysis.
Confluent cells were infected with MERS-CoV-WT, MERS-CoV-Δp4, or MERS-CoV-Δ4a at an MOI of 3. At different times p.i., the cells were lysed in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. Anti-PKR antibody (Abcam), anti-phospho-PKR antibody (Abcam), anti-eIF2α antibody (Cell Signaling Technology), anti-phosphorylated eIF2α antibody (Cell Signaling Technology), anti-α-tubulin antibody (Calbiochem), anti-MERS-CoV N protein antibody, or antibodies for MERS-CoV proteins (anti-MERS-CoV S protein antibody, anti-MERS-CoV M protein antibody, anti-MERS-CoV 4a protein antibody, and anti-MERS-CoV 4b protein antibody, which were generated by immunizing rabbits with the synthetic peptides DDRTEVPQLVNANQYSPCVSIVC, CDYDRLPNEVTVAK, QRIAWLLHKDGGIPD, and RKARKRSHSPTKKLRYVKRR, respectively) were used as primary antibodies. The peptide antibodies were produced by ProSci Inc. The secondary antibodies consisted of goat anti-mouse immunoglobulin G-horseradish peroxidase and goat anti-rabbit immunoglobulin G-horseradish peroxidase (Santa Cruz).
Metabolic radiolabeling of intracellular proteins in virus-infected cells.
Vero or HeLa/CD26 cells were mock infected or infected with MERS-CoV-WT, MERS-CoV-Δp4, or MERS-CoV-Δ4a at an MOI of 3. Cells were radiolabeled with 100 μCi of the Tran35S-label labeling reagent/ml (PerkinElmer) for 1 h at different times p.i. The cell extracts were prepared by lysing the cells in SDS-PAGE sample buffer. Cell lysates were subjected to SDS-PAGE analysis, followed by autoradiography and colloid Coomassie brilliant blue staining.
Total RNA extraction and qRT-PCR.
Total cellular RNAs were extracted from cells by using the TRIzol LS reagent (Invitrogen) and a Direct-zol RNA MiniPrep kit (Zymo Research) following the instructions in the manufacturers' product manuals. cDNAs were synthesized using SuperScript III reverse transcriptase (Invitrogen) and random primers (Invitrogen). To specifically detect MERS-CoV mRNAs, a MERS-CoV gene-specific primer (5′-TTTTTTTTCTAATCAGTGTTAACATCAATCATTGG-3′) was used for cDNA synthesis. qRT-PCR was performed using a Bio-Rad CFX96 real-time PCR apparatus and SYBR green master mix (Bio-Rad). PCR conditions were as follows: preincubation at 95°C for 30 s and amplification with 40 cycles of 95°C for 15 s and 60°C for 20 s. The purity of the amplified PCR products was confirmed by the dissociation melting curves obtained after each reaction. The primers used for human IFN-β mRNA were 5′-AAGGCCAAGGAGTACAGTC-3′ (forward) and 5′-ATCTTCAGTTTVGGAGGTAA-3′ (reverse), the primers for IFN-λ1 mRNA were 5′-CGCCTTGGAAGAGTCACTCA-3′ (forward) and 5′-GAAGCCTCAGGTCCCAATTC-3′ (reverse), the primers for OAS mRNA were 5′-GCCCTGGGTCAGTTGACTGG-3′ (forward) and 5′-TGAAGCAGGTGGAGAACTCGC-3′ (reverse), the primers for ISG56 mRNA were 5′-CAGCAACCATGAGTACAAAT-3′ (forward) and 5′-AAGTGACATCTCAATTGCTC-3′ (reverse), the primers for 18S rRNA were 5′-CCGGTACAGTGAAACTGCGAATG-3′ (forward) and 5′-GTTATCCAAGTAGGAGAGGAGCGAG-3′ (reverse), the primers for MERS-CoV mRNA 1 were 5′-AATACACGGTTTCGTCCGGTG-3′ (forward) and 5′-ACCACAGAGTGGCACAGTTAG-3′ (reverse), and the primers for MERS-CoV mRNA 8 were 5′-CTCGTTCTCTTGCAGAACTTTG-3′ (forward) and 5′-TGCCCAGGTGGAAAGGT-3′ (reverse). The relative expression level of each gene mRNA was normalized to 18S rRNA expression levels. All of the assays were performed in triplicate, and the results are expressed as the means ± standard deviations.
Northern blot analysis.
Total cellular RNAs were extracted from mock-infected cells or infected cells as described above. A DIG-labeled riboprobe, corresponding to nt 29084 to 29608 of the MERS-CoV genomic RNA, was used to detect MERS-CoV mRNAs as described previously (56).
siRNA treatment.
HeLa/CD26 cells were transfected with the indicated siRNAs using RNAiMAX according to the manufacturer's protocol (Invitrogen). Nontargeting siRNA (Dharmacon) and siRNA targeting G3BP1 (Thermo Fisher Scientific), G3BP2 (Thermo Fisher Scientific), or TIA-1 (Dharmacon) were used. At 24 h posttransfection, cells were either mock infected or infected with MERS-CoV-WT or MERS-CoV-Δp4 at an MOI of 3.
Statistical analysis.
A two-tailed Student's t test was conducted to determine statistical significance. Statistical significance was defined as a P value of <0.05.
ACKNOWLEDGMENTS
We thank Ralph Baric for kindly providing the MERS-CoV reverse genetics system.
This study was supported by Public Health Service grants AI99107 and AI114657 from the National Institutes of Health and a grant from the Institute for Human Infections and Immunity at The University of Texas Medical Branch. K. Nakagawa was supported by the James W. McLaughlin fellowship fund.
REFERENCES
- 1.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. 2012. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 2.Alagaili AN, Briese T, Mishra N, Kapoor V, Sameroff SC, Burbelo PD, de Wit E, Munster VJ, Hensley LE, Zalmout IS, Kapoor A, Epstein JH, Karesh WB, Daszak P, Mohammed OB, Lipkin WI. 2014. Middle East respiratory syndrome coronavirus infection in dromedary camels in Saudi Arabia. mBio 5:e00884-. doi: 10.1128/mBio.00884-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wernery U, Lau SK, Woo PC. 2017. Middle East respiratory syndrome (MERS) coronavirus and dromedaries. Vet J 220:75–79. doi: 10.1016/j.tvjl.2016.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chu DK, Poon LL, Gomaa MM, Shehata MM, Perera RA, Abu Zeid D, El Rifay AS, Siu LY, Guan Y, Webby RJ, Ali MA, Peiris M, Kayali G. 2014. MERS coronaviruses in dromedary camels, Egypt. Emerg Infect Dis 20:1049–1053. doi: 10.3201/eid2006.140299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drosten C, Kellam P, Memish ZA. 2014. Evidence for camel-to-human transmission of MERS coronavirus. N Engl J Med 371:1359–1360. doi: 10.1056/NEJMc1409847. [DOI] [PubMed] [Google Scholar]
- 6.Drosten C, Gunther S, Preiser W, van der Werf S, Brodt HR, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier RA, Berger A, Burguiere AM, Cinatl J, Eickmann M, Escriou N, Grywna K, Kramme S, Manuguerra JC, Muller S, Rickerts V, Sturmer M, Vieth S, Klenk HD, Osterhaus AD, Schmitz H, Doerr HW. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 7.Lee HJ, Shieh CK, Gorbalenya AE, Koonin EV, La Monica N, Tuler J, Bagdzhadzhyan A, Lai MM. 1991. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology 180:567–582. doi: 10.1016/0042-6822(91)90071-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lomniczi B. 1977. Biological properties of avian coronavirus RNA. J Gen Virol 36:531–533. doi: 10.1099/0022-1317-36-3-531. [DOI] [PubMed] [Google Scholar]
- 9.Lomniczi B, Kennedy I. 1977. Genome of infectious bronchitis virus. J Virol 24:99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raj VS, Mou H, Smits SL, Dekkers DH, Muller MA, Dijkman R, Muth D, Demmers JA, Zaki A, Fouchier RA, Thiel V, Drosten C, Rottier PJ, Osterhaus AD, Bosch BJ, Haagmans BL. 2013. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hurst-Hess KR, Kuo L, Masters PS. 2015. Dissection of amino-terminal functional domains of murine coronavirus nonstructural protein 3. J Virol 89:6033–6047. doi: 10.1128/JVI.00197-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graham RL, Sims AC, Brockway SM, Baric RS, Denison MR. 2005. The nsp2 replicase proteins of murine hepatitis virus and severe acute respiratory syndrome coronavirus are dispensable for viral replication. J Virol 79:13399–13411. doi: 10.1128/JVI.79.21.13399-13411.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neuman BW, Chamberlain P, Bowden F, Joseph J. 2014. Atlas of coronavirus replicase structure. Virus Res 194:49–66. doi: 10.1016/j.virusres.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai MM, Baric RS, Brayton PR, Stohlman SA. 1984. Characterization of leader RNA sequences on the virion and mRNAs of mouse hepatitis virus, a cytoplasmic RNA virus. Proc Natl Acad Sci U S A 81:3626–3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai MM, Patton CD, Baric RS, Stohlman SA. 1983. Presence of leader sequences in the mRNA of mouse hepatitis virus. J Virol 46:1027–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leibowitz JL, Wilhelmsen KC, Bond CW. 1981. The virus-specific intracellular RNA species of two murine coronaviruses: MHV-a59 and MHV-JHM. Virology 114:39–51. doi: 10.1016/0042-6822(81)90250-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spaan WJ, Rottier PJ, Horzinek MC, van der Zeijst BA. 1982. Sequence relationships between the genome and the intracellular RNA species 1, 3, 6, and 7 of mouse hepatitis virus strain A59. J Virol 42:432–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Menachery VD, Mitchell HD, Cockrell AS, Gralinski LE, Yount BL Jr, Graham RL, McAnarney ET, Douglas MG, Scobey T, Beall A, Dinnon K III, Kocher JF, Hale AE, Stratton KG, Waters KM, Baric RS. 2017. MERS-CoV accessory ORFs play key role for infection and pathogenesis. mBio 8:e00665-. doi: 10.1128/mBio.00665-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thornbrough JM, Jha BK, Yount B, Goldstein SA, Li Y, Elliott R, Sims AC, Baric RS, Silverman RH, Weiss SR. 2016. Middle East respiratory syndrome coronavirus NS4b protein inhibits host RNase L activation. mBio 7:e00258-. doi: 10.1128/mBio.00258-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matthews KL, Coleman CM, van der Meer Y, Snijder EJ, Frieman MB. 2014. The ORF4b-encoded accessory proteins of Middle East respiratory syndrome coronavirus and two related bat coronaviruses localize to the nucleus and inhibit innate immune signalling. J Gen Virol 95:874–882. doi: 10.1099/vir.0.062059-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niemeyer D, Zillinger T, Muth D, Zielecki F, Horvath G, Suliman T, Barchet W, Weber F, Drosten C, Muller MA. 2013. Middle East respiratory syndrome coronavirus accessory protein 4a is a type I interferon antagonist. J Virol 87:12489–12495. doi: 10.1128/JVI.01845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Canton J, Fehr AR, Fernandez-Delgado R, Gutierrez-Alvarez FJ, Sanchez-Aparicio MT, Garcia-Sastre A, Perlman S, Enjuanes L, Sola I. 2018. MERS-CoV 4b protein interferes with the NF-kappaB-dependent innate immune response during infection. PLoS Pathog 14:e1006838. doi: 10.1371/journal.ppat.1006838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holcik M, Sonenberg N. 2005. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol 6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 24.Garcia MA, Meurs EF, Esteban M. 2007. The dsRNA protein kinase PKR: virus and cell control. Biochimie 89:799–811. doi: 10.1016/j.biochi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 25.Farrell PJ, Balkow K, Hunt T, Jackson RJ, Trachsel H. 1977. Phosphorylation of initiation factor eIF-2 and the control of reticulocyte protein synthesis. Cell 11:187–200. doi: 10.1016/0092-8674(77)90330-0. [DOI] [PubMed] [Google Scholar]
- 26.Wek RC, Jiang HY, Anthony TG. 2006. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans 34:7–11. doi: 10.1042/BST0340007. [DOI] [PubMed] [Google Scholar]
- 27.Anderson P, Kedersha N. 2008. Stress granules: the Tao of RNA triage. Trends Biochem Sci 33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 28.Anderson P, Kedersha N. 2009. Stress granules. Curr Biol 19:R397–R398. doi: 10.1016/j.cub.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 29.Kedersha N, Anderson P. 2009. Regulation of translation by stress granules and processing bodies. Prog Mol Biol Transl Sci 90:155–185. doi: 10.1016/S1877-1173(09)90004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kedersha NL, Gupta M, Li W, Miller I, Anderson P. 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol 147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Onomoto K, Jogi M, Yoo JS, Narita R, Morimoto S, Takemura A, Sambhara S, Kawaguchi A, Osari S, Nagata K, Matsumiya T, Namiki H, Yoneyama M, Fujita T. 2012. Critical role of an antiviral stress granule containing RIG-I and PKR in viral detection and innate immunity. PLoS One 7:e43031. doi: 10.1371/journal.pone.0043031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reineke LC, Lloyd RE. 2015. The stress granule protein G3BP1 recruits protein kinase R to promote multiple innate immune antiviral responses. J Virol 89:2575–2589. doi: 10.1128/JVI.02791-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langereis MA, Feng Q, van Kuppeveld FJ. 2013. MDA5 localizes to stress granules, but this localization is not required for the induction of type I interferon. J Virol 87:6314–6325. doi: 10.1128/JVI.03213-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Onomoto K, Yoneyama M, Fung G, Kato H, Fujita T. 2014. Antiviral innate immunity and stress granule responses. Trends Immunol 35:420–428. doi: 10.1016/j.it.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okonski KM, Samuel CE. 2013. Stress granule formation induced by measles virus is protein kinase PKR dependent and impaired by RNA adenosine deaminase ADAR1. J Virol 87:756–766. doi: 10.1128/JVI.02270-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ng CS, Jogi M, Yoo JS, Onomoto K, Koike S, Iwasaki T, Yoneyama M, Kato H, Fujita T. 2013. Encephalomyocarditis virus disrupts stress granules, the critical platform for triggering antiviral innate immune responses. J Virol 87:9511–9522. doi: 10.1128/JVI.03248-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White JP, Lloyd RE. 2012. Regulation of stress granules in virus systems. Trends Microbiol 20:175–183. doi: 10.1016/j.tim.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poblete-Duran N, Prades-Perez Y, Vera-Otarola J, Soto-Rifo R, Valiente-Echeverria F. 2016. Who regulates whom? An overview of RNA granules and viral infections. Viruses 8:E180. doi: 10.3390/v8070180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khaperskyy DA, Hatchette TF, McCormick C. 2012. Influenza A virus inhibits cytoplasmic stress granule formation. FASEB J 26:1629–1639. doi: 10.1096/fj.11-196915. [DOI] [PubMed] [Google Scholar]
- 40.Sciortino MT, Parisi T, Siracusano G, Mastino A, Taddeo B, Roizman B. 2013. The virion host shutoff RNase plays a key role in blocking the activation of protein kinase R in cells infected with herpes simplex virus 1. J Virol 87:3271–3276. doi: 10.1128/JVI.03049-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cassady KA, Gross M. 2002. The herpes simplex virus type 1 U(S)11 protein interacts with protein kinase R in infected cells and requires a 30-amino-acid sequence adjacent to a kinase substrate domain. J Virol 76:2029–2035. doi: 10.1128/jvi.76.5.2029-2035.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He B, Gross M, Roizman B. 1997. The gamma(1)34.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1alpha to dephosphorylate the alpha subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc Natl Acad Sci U S A 94:843–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mulvey M, Arias C, Mohr I. 2007. Maintenance of endoplasmic reticulum (ER) homeostasis in herpes simplex virus type 1-infected cells through the association of a viral glycoprotein with PERK, a cellular ER stress sensor. J Virol 81:3377–3390. doi: 10.1128/JVI.02191-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simpson-Holley M, Kedersha N, Dower K, Rubins KH, Anderson P, Hensley LE, Connor JH. 2011. Formation of antiviral cytoplasmic granules during orthopoxvirus infection. J Virol 85:1581–1593. doi: 10.1128/JVI.02247-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.White JP, Cardenas AM, Marissen WE, Lloyd RE. 2007. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe 2:295–305. doi: 10.1016/j.chom.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 46.Sun Y, Dong L, Yu S, Wang X, Zheng H, Zhang P, Meng C, Zhan Y, Tan L, Song C, Qiu X, Wang G, Liao Y, Ding C. 2017. Newcastle disease virus induces stable formation of bona fide stress granules to facilitate viral replication through manipulating host protein translation. FASEB J 31:1337–1353. doi: 10.1096/fj.201600980R. [DOI] [PubMed] [Google Scholar]
- 47.Heinrich BS, Cureton DK, Rahmeh AA, Whelan SP. 2010. Protein expression redirects vesicular stomatitis virus RNA synthesis to cytoplasmic inclusions. PLoS Pathog 6:e1000958. doi: 10.1371/journal.ppat.1000958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dinh PX, Beura LK, Das PB, Panda D, Das A, Pattnaik AK. 2013. Induction of stress granule-like structures in vesicular stomatitis virus-infected cells. J Virol 87:372–383. doi: 10.1128/JVI.02305-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garaigorta U, Heim MH, Boyd B, Wieland S, Chisari FV. 2012. Hepatitis C virus (HCV) induces formation of stress granules whose proteins regulate HCV RNA replication and virus assembly and egress. J Virol 86:11043–11056. doi: 10.1128/JVI.07101-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ruggieri A, Dazert E, Metz P, Hofmann S, Bergeest JP, Mazur J, Bankhead P, Hiet MS, Kallis S, Alvisi G, Samuel CE, Lohmann V, Kaderali L, Rohr K, Frese M, Stoecklin G, Bartenschlager R. 2012. Dynamic oscillation of translation and stress granule formation mark the cellular response to virus infection. Cell Host Microbe 12:71–85. doi: 10.1016/j.chom.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sola I, Galan C, Mateos-Gomez PA, Palacio L, Zuniga S, Cruz JL, Almazan F, Enjuanes L. 2011. The polypyrimidine tract-binding protein affects coronavirus RNA accumulation levels and relocalizes viral RNAs to novel cytoplasmic domains different from replication-transcription sites. J Virol 85:5136–5149. doi: 10.1128/JVI.00195-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raaben M, Groot Koerkamp MJ, Rottier PJ, de Haan CA. 2007. Mouse hepatitis coronavirus replication induces host translational shutoff and mRNA decay, with concomitant formation of stress granules and processing bodies. Cell Microbiol 9:2218–2229. doi: 10.1111/j.1462-5822.2007.00951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rabouw HH, Langereis MA, Knaap RC, Dalebout TJ, Canton J, Sola I, Enjuanes L, Bredenbeek PJ, Kikkert M, de Groot RJ, van Kuppeveld FJ. 2016. Middle East respiratory coronavirus accessory protein 4a inhibits PKR-mediated antiviral stress responses. PLoS Pathog 12:e1005982. doi: 10.1371/journal.ppat.1005982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park SH, Choi J, Kang JI, Choi SY, Hwang SB, Kim JP, Ahn BY. 2006. Attenuated expression of interferon-induced protein kinase PKR in a simian cell devoid of type I interferons. Mol Cells 21:21–28. [PubMed] [Google Scholar]
- 55.Scobey T, Yount BL, Sims AC, Donaldson EF, Agnihothram SS, Menachery VD, Graham RL, Swanstrom J, Bove PF, Kim JD, Grego S, Randell SH, Baric RS. 2013. Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc Natl Acad Sci U S A 110:16157–16162. doi: 10.1073/pnas.1311542110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lokugamage KG, Narayanan K, Nakagawa K, Terasaki K, Ramirez SI, Tseng CT, Makino S. 2015. Middle East respiratory syndrome coronavirus nsp1 inhibits host gene expression by selectively targeting mRNAs transcribed in the nucleus while sparing mRNAs of cytoplasmic origin. J Virol 89:10970–10981. doi: 10.1128/JVI.01352-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dauber B, Poon D, Dos Santos T, Duguay BA, Mehta N, Saffran HA, Smiley JR. 2016. The herpes simplex virus virion host shutoff protein enhances translation of viral true late mRNAs independently of suppressing protein kinase R and stress granule formation. J Virol 90:6049–6057. doi: 10.1128/JVI.03180-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Emeny JM, Morgan MJ. 1979. Regulation of the interferon system: evidence that Vero cells have a genetic defect in interferon production. J Gen Virol 43:247–252. doi: 10.1099/0022-1317-43-1-247. [DOI] [PubMed] [Google Scholar]
- 59.Wathelet MG, Berr PM, Huez GA. 1992. Regulation of gene expression by cytokines and virus in human cells lacking the type-I interferon locus. Eur J Biochem 206:901–910. doi: 10.1111/j.1432-1033.1992.tb16999.x. [DOI] [PubMed] [Google Scholar]
- 60.Baric RS, Nelson GW, Fleming JO, Deans RJ, Keck JG, Casteel N, Stohlman SA. 1988. Interactions between coronavirus nucleocapsid protein and viral RNAs: implications for viral transcription. J Virol 62:4280–4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Compton SR, Rogers DB, Holmes KV, Fertsch D, Remenick J, McGowan JJ. 1987. In vitro replication of mouse hepatitis virus strain A59. J Virol 61:1814–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Panas MD, Varjak M, Lulla A, Eng KE, Merits A, Karlsson Hedestam GB, McInerney GM. 2012. Sequestration of G3BP coupled with efficient translation inhibits stress granules in Semliki Forest virus infection. Mol Biol Cell 23:4701–4712. doi: 10.1091/mbc.e12-08-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Emara MM, Liu H, Davis WG, Brinton MA. 2008. Mutation of mapped TIA-1/TIAR binding sites in the 3′ terminal stem-loop of West Nile virus minus-strand RNA in an infectious clone negatively affects genomic RNA amplification. J Virol 82:10657–10670. doi: 10.1128/JVI.00991-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li W, Li Y, Kedersha N, Anderson P, Emara M, Swiderek KM, Moreno GT, Brinton MA. 2002. Cell proteins TIA-1 and TIAR interact with the 3′ stem-loop of the West Nile virus complementary minus-strand RNA and facilitate virus replication. J Virol 76:11989–12000. doi: 10.1128/JVI.76.23.11989-12000.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Scholte FE, Tas A, Albulescu IC, Zusinaite E, Merits A, Snijder EJ, van Hemert MJ. 2015. Stress granule components G3BP1 and G3BP2 play a proviral role early in Chikungunya virus replication. J Virol 89:4457–4469. doi: 10.1128/JVI.03612-14. [DOI] [PMC free article] [PubMed] [Google Scholar]