Abstract
MicroRNAs (miRNAs) are small non-coding RNAs that regulate multiple physiological and pathological functions through the modulation of gene expression at the post-transcriptional level. Accumulating evidence has established a role for miRNAs in the development and pathogenesis of liver disease. Specifically, a large number of studies have assessed the role of miRNAs in alcoholic liver disease (ALD) and non-alcoholic fatty liver disease (NAFLD), two diseases that share common underlying mechanisms and pathological characteristics. The purpose of the current review is to summarize and update the body of literature investigating the role of miRNAs in liver disease. In addition, the potential use of miRNAs as biomarkers and/or therapeutic targets is discussed. Among all miRNAs analyzed, miR-34a, miR-122 and miR-155 are most involved in the pathogenesis of NAFLD. Of note, these three miRNAs have also been implicated in ALD, reinforcing a common disease mechanism between these two entities and the pleiotropic effects of specific miRNAs. Currently, no single miRNA or panel of miRNAs has been identified for the detection of, or staging of ALD or NAFLD. While promising results have been shown in murine models, no therapeutic based-miRNA agents have been developed for use in humans with liver disease.
Keywords: Alcohol use disorder, Alcoholic liver disease, Non-alcoholic fatty liver disease, Steatosis, Obesity, miRNA, Biomarkers
Core tip: MicroRNAs (miRNAs) are small RNAs that regulate gene expression at a post-transcriptional level. Altered miRNA expression has been found in a variety of liver diseases, including non-alcoholic fatty liver disease and alcoholic liver disease. A group of miRNAs (miR-155, miR-122 and miR-34a) contributes to the pathogenesis of these two diseases and these miRNAs have potential use as biomarkers or therapeutic targets. Several technical limitations and a lack of clinical studies, however, preclude their clinical use.
INTRODUCTION
MicroRNAs (miRNAs), small non-coding RNAs, can modulate gene expression at the post-transcriptional level by targeting messenger RNAs and inhibiting their translation or promoting their degradation[1,2]. Since the discovery of the first miRNA in 1993, lin-4[3], more than 2000 miRNAs have been described in humans and they are believed to regulate up to 60% of protein-coding genes in the human genome[4].
Human miRNAs are involved in virtually all physiological and pathological processes, including cell differentiation and proliferation, signal transduction, inflammation and immune response, metabolism, viral-host interaction, and oncogenesis[1,2]. The expression of a wide variety of miRNAs is potentially regulated by many factors, such as alcohol, but also diet, cigarette smoking and other drugs[5]. Therefore, it is not surprising that miRNAs have been increasingly recognized as key actors in the pathogenesis of a variety of diseases and as potential biomarkers for diagnosis or therapeutic targets[2]. The role of miRNAs in liver inflammation, fibrosis and cirrhosis has been widely described in the last twenty years[6-8]. The current paper reviews the existing literature pertaining to miRNA alteration, function, and the potential clinical application of miRNAs in alcoholic liver disease (ALD) and non-alcoholic fatty liver disease (NAFLD). While ALD and NAFLD differ in some aspects, they also share common features, including underlying mechanisms and clinical and histopathological characteristics[9]. Given the rapid expansion of research in miRNAs in recent years, an updated review on the topic will first be presented, followed by a summary of miRNA alterations that are common to both ALD and NAFLD.
ROLE OF MIRNAS IN ALD
Pathogenic role of miRNAs in ALD
The development of the different forms of ALD (steatosis, alcoholic hepatitis and cirrhosis) requires prolonged and heavy alcohol consumption along with susceptibility to the disease. Pathophysiological mechanisms of ALD are based both on the direct toxic effect of alcohol and also on ethanol-induced alterations in the inflammatory response[10]. A variety of enzymes, such as alcohol dehydrogenase (ADH) and the cytochrome P450 2E1 (CYP2E1), contribute to alcohol metabolism[11], leading to oxygen free radicals, nitric oxide and acetaldehyde, which ultimately can cause cellular damage and liver inflammation[12]. In addition, the toxic effect of acetaldehyde increases intestinal permeability to bacterial lipopolysaccharide (LPS)[13], which binds to toll-like receptors 4 (TLR4) and activates Kupffer and stellate cells through pro-inflammatory cytokines, such as tumour necrosis factor (TNF)-α, production[14]. This inflammatory signal is transmitted via the nuclear factor-κB (NF-κB) pathway, ultimately leading to liver damage[14].
While most immune mechanisms involved in ALD development are related to the TLR4-NF-κB pathway, the activation of TLR4 also triggers the transmission of pro-inflammatory stimuli through other signaling pathways, such us mitogen-activated protein kinases (MAPK) or TIR-domain-containing adapter-inducing interferon-β (TRIF)[14]. miRNAs can regulate this complex interplay between inflammatory signals via the regulation of cytokines and other components of the pathways[15]. Oxidative stress and free oxygen radicals generation involved in ALD development are also regulated by miRNAs through different pathways like Kelch-like ECH-associated protein 1 Kelch-like ECH-associated protein 1 (Keap1) / Nuclear factor-erythroid-2-related factor 2 (Nrf2) pathway[16-20]. In addition to this, miRNAs have also been shown to exert an important modulatory function on macrophage activation and differentiation[21,22]. Moreover, recent studies have shown even broader effects of miRNAs in ALD development, including a role in intercellular communication, in secretion in exosomes[23], in the expression of enzymes directly linked to alcohol metabolism (e.g., regulation of CYP2E1 by miR-214[24]) and in the modulation of pro-inflammatory pathways such as the B-cell translocation gene 2/Yin-yang 1 (BTG2/YY1) signaling pathway by miR-497[25]. Finally, alcohol consumption, with or without concurrent ALD, has also been linked to altered expression of several miRNAs[5,26].
Numerous studies, therefore, have addressed the relationship between ALD development and miRNAs. While animal models have been used in the majority of these studies, there is an increasing number of studies in human cells, tissues and serum, confirming the key role of miRNAs in ALD[27-30]. A summary of all available studies is shown in Table 1. In addition, a summary of the regulatory actions of miRNAs in the inflammatory response according to the different cell types involved, is displayed in Figure 1.
Table 1.
MicroRNA targets involved in alcoholic liver disease pathogenesis
| miRNA | Source of sample | miRNA target |
| let-7[27] | Animal models | Lin28, HMGA2 |
| Human HSCs | ||
| miR-19b[28] | Animal models | TGFβRII, Col1α2, MeCP2 |
| Human HSCs | ||
| miR-21[36,37] | Animal models | FASLG, DR5, Crebl2 |
| miR-26a[35] | Animal models | DUSP4, DUSP5 |
| miR-27a[44,52] | Animal models | Sprouty2, CD206 |
| HMC | ||
| Humans (plasma) | ||
| miR-34a[29,43] | Animal models | SIRT1, CASP2 |
| Human HSCs | ||
| NHH | ||
| HiBECs | ||
| Humans (liver biopsy) | ||
| miR-103 and miR-107[53] | Humans (liver biopsy) | Caveolin-1 |
| miR-122[32,124,125] | Animal models | P4HA1, HO-1, Cyclin G1, Bcl-w, HIF-1α |
| miR-155[38,39,97,126,127] | Animal models | TNFα, SHIP1, SOCS1, IRAKM, C/EBPβ |
| miR-181b-3p[40] | Animal models | Importin α1 |
| miR-182[30] | Animal models | SLC1A1, Cofilin 1, CCL20, CXCL1, IL-8, Cyclin D1, IL-6 |
| Humans (serum samples and liver biopsy) | ||
| miR-199[128] | Animal models | ET-1, ET-BR |
| miR-200a[31] | Animal models | ZEB-2 |
| miR-212[46] | Caco-2 cells | ZO-1 |
| Humans (colon biopsy) | ||
| miR-214[24,34] | Animal models | POR, GSR, CYP2E1 |
| HHCs | ||
| miR-217[41] | Animal models | SIRT-1 |
| miR-223[45] | Animal models | p47phox, IL-6 |
| Humans (serum) | ||
| miR-291b[42] | Animal models | Tollip |
| HPBMs | ||
| miR-378[59] | Animal models | Gli-3 |
| miR-497[25] | Animal models | Btg2, Yy1 |
HSCs: Hepatic stellate cells; HMGA2: High mobility group AT-hook 2; TGFβRII: Transforming growth factor β receptor II; Col1α2: Collagen type I α 2 chain; MeCP2: Methyl-CpG binding protein 2; FASLG: Fas ligand; DR5: Death receptor 5; Crebl2: cAMP responsive element binding protein like 2; DUSP: Dual specificity phosphatase; HMC: Human Monocyte Cells; NHH: Normal Human Hepatocytes; HiBECs: Human intrahepatic Biliary Epithelial Cells; SIRT1: sirtuin 1; CASP2: caspase 2; P4HA1: prolyl 4-hydroxylase subunit α 1; HO-1: heme oxygenase-1; BCL-W: Bcl-2-like protein 2; HIF-1α: Hypoxia inducible factor 1 α; TNFα: Tumor necrosis factor α; SHIP1: Src homology 2 domain-containing inositol phosphatase 1; SOCS1: Suppressor of cytokine signaling 1; IRAKM: Interleukin 1 receptor associated kinase 3; C/EBPβ: CCAAT/enhancer binding protein β; SLC1A1: Solute carrier family 1 member 1; CCL20: C-C motif chemokine ligand 20; CXCL1: C-X-C motif chemokine ligand 1; IL: Interleukin; ET-1: Endothelin-1; ET-BR: Endothelin-B receptor; ZEB-2: Zinc finger E-box binding homeobox 2; ZO-1: Zonula occludens 1; HHCs: Human Hepatoma Cells; POR: Cytochrome P450 oxidoreductase; GSR: Glutathione reductase; CYP2E1: Cytochrome P450 2E1; p47phox: Neutrophil cytosolic factor 1-like; HPBMs: Human Peripheral Blood Monocytes; Tollip: Toll interacting protein; Gli3: GLI Family Zinc Finger 3; Btg2: B-cell translocation gene 2; YY1: Yin yang 1; miRNA: MicroRNA.
Figure 1.
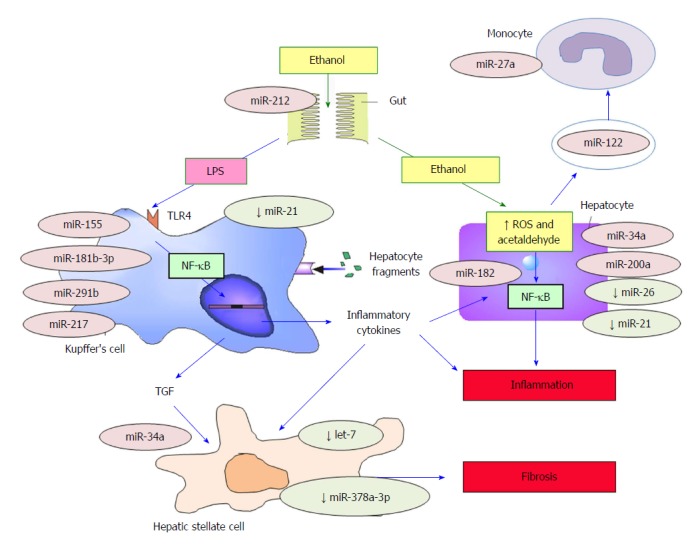
MicroRNAs involved in the pathogenesis of alcoholic liver disease. miRNAs preceded by a ↓ symbol are decreased in ALD or inhibit the development of ALD. The remainder of miRNAs promotes the development of ALD. TLR4: Toll-like receptor 4; TFG: Transforming growth factor; ALD: Alcoholic liver disease; ROS: Reactive oxygen species; NF-κB: Nuclear factor-κB. Figure adapted from Laso et al[10].
Hepatocytes: Some miRNAs (e.g., miR-34a and miR-200a) are responsible for the induction of hepatocytic apoptosis during ALD development[29,31]. In addition, secretion of miRNAs in exosomes (e.g., miR-122) can cause an increase in inflammatory response by targeting monocyte/macrophage cells[32], ultimately leading to hepatocytic injury. MiRNAs action and pleiotropic effects could be different depending on the cell in which they act; thus, miR-122 could have a protective role inside the hepatocyte during alcohol-induced liver damage[33]. Increase in oxidative stress and alterations of enzymatic function in hepatocytes are also regulated by miRNAs[24,34]. Conversely, miRNAs may also have a protective role in ALD. For example miR-26a can increase autophagy[35] and miR-21 can inhibit alcohol-induced apoptosis[36,37].
Kupffer cells (KCs): miR-155, which is increased by chronic alcohol consumption through NF-κB induction, has been shown to be the main regulator of KC activation and function[38]. miR-155 inhibits the expression of multiple TLR4/NF-κB inhibitory regulators such as Src homology 2 domain-containing inositol phosphatase 1 (SHIP1) and Suppressor of cytokine signaling 1 (SOCS1)[38,39] leading to an increase in KC response to LPS and ultimately the development of liver fibrosis[39]. The Keap1/Nrf2 pathway could also be involved in miR-155 role in ALD development and KCs regulation[17]. Other miRNAs, such as miR-181b-3p, are also linked to increased LPS-sensitivity through the TLR4-NF-κB pathway[40]. In addition, miRNAs have been shown to regulate Sirtuin-1-Lipin-1, an inflammatory response mediator, leading to the down-regulation of the NF-κB pathway via de-acetylation. Alcohol consumption increases miR-217 expression, which in turn down-regulates sirtuin-1-Lipin-1[41], consequently leading to more hepatic inflammation[41]. Toll Interacting Protein (Tollip), another down-regulator of the TLR4-NF-κB pathway, is inhibited by miR-291b[42].
Hepatic stellate cells (HSCs): HSCs, responsible for the development of liver fibrosis, are regulated by several miRNAs, including let-7. The downregulation of let-7 by LPS and alcohol use causes an increase in HSCs activation[27]. In addition, chronic alcohol consumption has been linked to an overexpression of miR-34a, which increases the expression of proteins such as transforming growth factor-β1 (TFG-β1), leading to a higher survival of HSCs through apoptosis inhibition[43].
Other cell types: In addition to the cell types described above, other cells involved in ALD development, such as circulating monocytes (by miR-27a[44]), and circulating neutrophils, (by miR-223[45]) are regulated by miRNAs. In addition, miR-212 has been shown to increase permeability to LPS by altering cells of the intestinal mucosa[46].
Due to the role of miRNAs in ALD and the modulatory effects of alcohol consumption on miRNA expression, it is plausible to hypothesize that genetic variations in certain miRNAs may lead to altered miRNA function and an increased risk of liver damage. Consequently, we and others have analyzed the relationship of alcohol-related diseases and polymorphisms within miRNA genes or miRNA targets[47,48]. Interestingly, the miR-146a C>G rs2910164 variant is linked to a susceptibility to alcohol use disorder[47] and the pre-miR-27a A>G rs895819 polymorphism is linked to a higher alcohol intake[49], suggesting a potential relationship between these genetic variants and alcohol-related diseases. The lack of replication studies precludes any conclusions regarding these SNPs, and to date, only rs738409 polymorphism within the PNPLA3 gene is clearly linked to a higher susceptibility to ALD[50].
miRNAs as a target for diagnosis and treatment of ALD
The clinical use of miRNAs as a diagnostic tool or therapeutic agent in ALD has not been well studied[51]. However, over the last years, an increasing number of miRNAs have been proposed as potential biomarkers of ALD. The following is a review of the most promising results.
miR-192 and miR-30a: It has been shown that serum levels of miR-192 and miR-30a are significantly correlated with the diagnosis of alcoholic hepatitis. Therefore, these miRNAs may be useful in the diagnosis, staging, and monitoring of patients with this specific form of ALD[23].
miR-27a: miR-27a has been linked to monocyte differentiation and is increased in extracellular plasmatic vesicles of patients with alcoholic hepatitis, making it a potentially useful diagnostic tool[52].
miR-182: An elevated level of miR-182 has been linked to greater disease severity and liver injury in alcoholic hepatitis. The correlation between miR-182 and disease severity, however, has only been shown in liver biopsies, limiting its application as a diagnostic tool[30].
miR-103 and miR-107: A prior study found that miR-103 and miR-107 were increased in liver from patients with ALD and with NAFLD, but not in healthy livers or in subjects with viral hepatitis[53].
miR-155 and miR-122: Increased blood levels of miR-155[32,54] and miR-122[55] have been found in healthy individuals after binge drinking and in a murine model of liver damage. While these miRNAs could be potential biomarkers of alcohol intake or alcohol liver damage, they are increased in several types of liver disease and therefore are unlikely to be specific to ALD[54].
Therapeutic application of miRNAs in ALD
There are no studies to date supporting a therapeutic role for miRNAs in ALD. Available data, however, suggest a potential role for the inhibition or activation of some miRNAs in the treatment of liver disease. A recent study found that treatment with hyaluronic acid normalized miR-181b-3p and Importin α5 levels in ethanol-fed mice, protecting them from ethanol-induced liver and intestinal damage[40]. In addition, hyaluronic acid normalized the miR-291b/Tollip pathway, leading to a lower sensitization of monocytes/macrophages to ethanol-induced activation via TLR4[42]. While both studies were performed in animal models, taken together they suggest a potential role for hyaluronic acid as a therapeutic regulator of the KC response to ethanol via miRNA modulation.
The role of miR-155 in KC and miR-122 in hepatocytes suggest that these miRNAs may serve as potential targets for treatment of ALD. Miravirsen, an miR-122 inhibitor, has shown promising results in chronic hepatitis C treatment[56,57], suggesting its potential usefulness in ALD. A recent study showed that the restoration of miR-122 in hepatocytes could have a protective role against ALD development[33]. These apparently contradictory results could reflect the ability of miRNAs to develop different actions in different cells and also its relevance in inter-cellular communications[32]. In this sense, the therapeutic action of Miravirsen over viral replication could be explained by the interruption of these communications[57]. In addition, other potential therapeutic miRNAs currently under development for other diseases, such as cardiac fibrosis and remodeling or vascular disease[58], could serve as potential targets for ALD. There is indirect data that inhibition of miR-155, may lead to decreased sensitivity of KC to LPS-mediated activation[39].
In addition to the inhibition of detrimental miRNAs, stimulation of protective miRNAs could also serve as a potential therapeutic target. For example, miR-21, which aids hepatocyte regeneration[36]; miR26a, which protects hepatocytes from fibrosis development[35]; miR-223, which inhibits neutrophil activation and liver infiltration[45]; and miR-378, which exerts a stop-signaling action in HSC[59], are all potential targets for treatment. There are no clinical trials to date involving these miRNAs as therapeutic targets in ALD and further studies will be necessary before clinical application.
ROLE OF MIRNAS IN NAFLD
NAFLD is defined as the accumulation of fat in the liver in the absence of alcohol intake, viral infection or other specific causes of liver disease. NAFLD represents a spectrum of disorders ranging from the simple accumulation of triglycerides in hepatocytes (hepatic steatosis) to steatosis with inflammation [non-alcoholic steatohepatitis (NASH)], fibrosis and cirrhosis[60]. NAFLD and NASH have rapidly become the most common cause of chronic liver disease worldwide in recent decades. The prevalence of these diseases has been estimated between 25% to 45% of the general population[61] with a greater prevalence in patients with obesity, diabetes mellitus or metabolic syndrome, in which case, the prevalence of NAFLD can reach 70% to 90%[62-64]. It is estimated that by 2020 cirrhosis related to NAFLD will be the first indication for liver transplantation[65].
Pathogenic role of miRNAs in NAFLD
The pathogenesis of NAFLD, along with the underlying mechanisms of progression from steatosis to steatohepatitis, has not been fully elucidated. Traditionally, the “two hit” theory[66] has been upheld. The “first hit”, which includes insulin resistance leading to the accumulation of fat in the liver, is followed by a “second hit”, consisting of the interaction of inflammatory cytokines, mitochondrial dysfunction and oxidative stress, leading to hepatocellular injury, inflammation and fibrosis[67]. However, more recently, multiple factors have been implicated in the pathogenesis of NAFLD, such that the “two hit” theory has been replaced by a “multiple-hit” hypothesis[68]. The “multiple-hit” theory includes the involvement of insulin resistance, adipose tissue dysfunction, mitochondrial dysfunction, endoplasmic reticulum stress, dietary factors, fatty acids, iron overload, inflammatory activation, LPS produced by gut microbiota, a chronic inflammatory state, and genetic and epigenetic factors in the pathogenesis and progression of NAFLD[68-70]. Accordingly, the following is a summary of the research implicating several miRNAs in the regulation of key targets in the development of NAFLD[8]. It is of special interest that recent studies have reported differences in miRNA expression between liver samples from patients with NAFLD and controls. Specifically, livers from patients with NAFLD express an upregulation of miR-31, miR-33a, miR-34a, miR-144, miR-146b, miR-150, miR-182, miR-183, miR-200a, miR-224, and miR-301a and a down regulation of miR-17, miR-122, miR-296, miR-373, miR-375 and miR-378c[71-76]. Among these miRNAs, miR-34a, miR-122, and miR-155 have been most often associated with the pathogenesis of NAFLD and as such, the following is a review of these miRNAs in detail. Table 2 displays a list of all miRNAs that have been associated with NAFLD through February 2018.
Table 2.
Summary of microRNAs associated with non-alcoholic fatty liver disease
| miRNA | Source of samples | Change | Main targets |
| miR-9[129] | Human serum; | Upregulated | Onecut2; SIRT1 |
| Human hepatocyte cell line | |||
| miR-10b[130] | Human hepatocyte cell line | Downregulated | PPARα |
| miR-15b[131,132] | Animal models | Upregulated | |
| Human serum | |||
| miR-16[104] | Human serum | Upregulated | |
| miR-17[74] | Human liver | Downregulated | |
| miR-19[84] | Human serum | Upregulated | |
| miR-21[86,87,99,133-136] | Animal models | Upregulated | PPARα; TGF-β |
| Human hepatocyte cell line | PTEN | ||
| Human liver and serum | |||
| miR-21[85,89,137,138] | Animal models | Downregulated | HMGCR; FABP7 |
| Human liver | |||
| Human hepatocyte cell line | |||
| miR-24[139] | Animal models | Upregulated | Insig1; SREBP |
| Human hepatocyte cell line | |||
| miR-26[140] | Animal models | Downregulated | IL-6, IL-7 |
| miR-27a[141] | Animal models | Downregulated | |
| miR-27b[102] | Human serum | Upregulated | |
| miR-29a[142,143] | Animal models | Downregulated | HMGCR; LPL |
| miR-29c[85,89,90] | Animal models | Downregulated | DNMT3A; DNMT3B |
| miR-30b[83] | Human liver | Downregulated | ITGAX; FABP4 |
| Human hepatocyte cell line | |||
| miR-30c[144] | Human serum | Upregulated | |
| miR-31[74,89] | Human liver | Upregulated | |
| Animal models | |||
| miR-33a[73,76] | Human liver | Upregulated | ABCA1; ABCA2 |
| miR-33a[85] | Human liver | Downregulated | |
| MiR-34a[71,81,82,85,87,89,90,92,104,105,145-148] | Animal models | Upregulated | SIRT1; HNF4α; PPARα |
| Human hepatocyte cell line | |||
| Human liver and serum | |||
| miR-99a[149] | Human serum | Downregulated | |
| miR-101[150] | Human hepatocyte cell line | Upregulated | ABCA1 |
| Human monocyte cell line | |||
| miR-103[53,89,151] | Animal models | Upregulated | Cav1 |
| Human liver and serum | |||
| miR-103a[152] | Human liver | Upregulated | |
| Human hepatocyte cell line | |||
| miR-106b[152] | Human liver | Upregulated | |
| miR-107[53,89] | Animal models | Upregulated | Cav1 |
| Human liver | |||
| miR-122[81,84,86,87,101-104,106,153] | Animal models | Upregulated | |
| Human Serum | |||
| miR-122[71,82-85,89,90,99,106,141,154,155] | Animal models | Downregulated | ACC-2; HAMP; FAS; HMGCR; SREBF-1c |
| Human liver | SREPBF-2; HIF-1α; Vimentin; MAP3K3 | ||
| miR-125b[84] | Human serum | Upregulated | |
| miR-125b[156] | Animal models | Downregulated | FAS |
| miR-139-5p[83] | Human liver | Downregulated | TNFα |
| miR-144[76] | Human Liver | Upregulated | ABCA1 |
| miR-144[157] | Animal models | Downregulated | TLR-2 |
| miR-146a[158] | Animal models | Upregulated | |
| Human hepatocyte cell line | |||
| miR-146a[132] | Animal models | Downregulated | Wnt1; Wnt5 |
| Human hepatocyte cell line | |||
| Mir-146b[149,159] | Animal models | Downregulated | IRAK1 |
| Human serum | TRAF6 | ||
| Human hepatocyte cell line | |||
| miR-146b[71,83,158] | Animal models | Upregulated | |
| Human liver | |||
| Human hepatocyte cell line | |||
| miR-149[160] | Animal models | Upregulated | FGF-21 |
| Human hepatocyte cell line | |||
| miR-150[74] | Human liver | Upregulated | |
| miR-152[158] | Animal models | Upregulated | |
| Human hepatocyte cell line | |||
| miR-155[90,97-99,161,162] | Animal models | Upregulated | SOCS1; C/EBP-Β; CES3; PDGF; SMAD3 |
| Human hepatocyte cell line | |||
| miR-155[96] | Animal models | Downregulated | LXRα |
| Human liver and serum | |||
| miR-181a[82] | Animal models | Upregulated | |
| miR-181d[149] | Human serum | Downregulated | |
| miR-182[74] | Human liver | Upregulated | FOXO3 |
| miR-183[74] | Human liver | Upregulated | |
| miR-192[84,90] | Animal models | Downregulated | |
| Human liver | |||
| miR-192-5p[82,84,86,102,106] | Animal models | Upregulated | |
| Human liver and serum | |||
| miR-194[89] | Animal models | Upregulated | |
| miR-197[149] | Human serum | Downregulated | |
| miR-199[163] | Animal models | Upregulated | Cav1; PPARα |
| Human hepatocyte cell line | |||
| Human liver | |||
| miR-200a/b/c[74,82,89,90,141,158,162,164] | Animal models | Upregulated | ZEB1; CDH1; EZH2; IRP1 |
| Human hepatocyte cell line | |||
| miR-203[90,132] | Animal models | Downregulated | |
| miR-212[165] | Animal models | Upregulated | FGF-21 |
| Human hepatocyte cell line | |||
| miR-214[71,166] | Human liver | Upregulated | |
| Animal models | |||
| miR-216[167] | Animal models | Downregulated | |
| miR-219a[74] | Human liver | Downregulated | |
| miR-221[73] | Human liver | Downregulated | |
| miR-221[89,90,99] | Animal models | Upregulated | |
| miR-222[99] | Animal models | Upregulated | |
| miR-223[86,164] | Animals models | Upregulated | IRP1 |
| Human serum | |||
| miR-224[73,74] | Human liver | Upregulated | |
| miR-291b[168] | Animal models | Upregulated | AMPKα1 |
| miR-302a[167] | Animals model | Downregulated | ELOVL6 |
| miR-331[144] | Human serum | Upregulated | |
| miR-335[89] | Animal models | Upregulated | |
| miR-375[84] | Human serum | Upregulated | |
| miR-378i[74] | Human liver | Downregulated | |
| miR-421[169] | Animal models | Upregulated | SIRT-3 |
| miR-422a[83] | Human liver | Downregulated | |
| miR-429[141] | Animal models | Upregulated | |
| miR-451[87] | Human Serum | Upregulated | |
| miR-451[89,141,170] | Animal models | Downregulated | AMPK/AKT |
| Human liver | |||
| miR-467b[171] | Animal models | Downregulated | LPL |
| miR-576[152] | Human liver | Downregulated | RAC1 |
| Human hepatocyte cell line | |||
| miR-590[74] | Human liver | Downregulated | |
| miR-892a[152] | Human liver | Upregulated | |
| Human hepatocyte cell line | |||
| miR-1290[102] | Human serum | Upregulated |
Onecut2: One cut homeobox 2; SIRT: Sirtuin; PPARα: Peroxisome proliferator activated receptor α; TGF-β: Transforming growth factor β; PTEN: Phosphatase and tensin homolog; HMGCR: 3-hydroxy-3-methylglutaryl-CoA reductase; FABP: Fatty acid binding protein; Insig1: Insulin induced gene 1; SREBP: Sterol regulatory element binding protein; IL: Interleukin; LPL: Lipoprotein lipase; DNMT: DNA methyltransferase; ITGAX: Integrin subunit α X; ABCA: ATP binding cassette subfamily A; HNF4α: Hepatocyte nuclear factor 4 α; Cav1: Caveolin 1; ACC-2: Acetyl-CoA carboxylase 2; SREBF: Sterol regulatory element binding transcription factor; HIF-1α: Hypoxia inducible factor 1 α; MAP3K3: Mitogen-activated protein kinase kinase kinase 3; FAS: Fatty acid synthase; TNFα: Tumour necrosis factor α; TLR-2: Toll-like receptor 2; Wnt: Wnt family member; IRAK1: Interleukin 1 receptor associated kinase 1; TRAF6: TNF receptor associated factor 6; FGF-21: Fibroblast growth factor 21; SOCS1: Suppressor of cytokine signaling 1; C/EBPβ: CCAAT/enhancer binding protein β; CES3: Carboxylesterase 3; PDGF: Platelet derived growth factor; SMAD3: SMAD family member 3; LXRα: Liver X receptor; FOXO3: Forkhead box O3; ZEB-1: Zinc finger E-box binding homeobox 1; CDH1: Cadherin 1; EZH2: Enhancer of zeste 2 polycomb repressive complex 2; IRP1: Iron regulatory protein 1; AMPKα1: AMPK: Adenosine monophosphate activated protein kinase α 1; ELOVL6: ELOVL fatty acid elongase 6; AMPK: Adenosine monophosphate activated protein kinase; AKT: AKT serine/threonine kinase 1; RAC1: Ras-related C3 botulinum toxin substrate 1.
miR-122: miR-122 is the most abundant miRNA in the liver and plays a fundamental role in liver physiology[77-79] and lipid metabolism[80]. miR-122 interacts with multiple important lipogenic factors in human NAFLD, such as acetyl coA carboxylase-2 (ACC2) and the sterol regulatory element binding protein (SREBP)[71,81,82]. miR-122 is decreased in liver samples[83-85] but increased in serum[84,86,87] from patients with NAFLD compared to healthy controls. Despite this somewhat paradoxical finding, the association of miR-122 with NAFLD pathogenesis is well established. Inhibition of miR-122 in high-fat fed mice is associated with a significant reduction in hepatic steatosis and plasma cholesterol levels, which was associated with a reduction in hepatic sterol and fatty acid synthesis rates and stimulation of hepatic fatty-acid oxidation mediated by activation of adenosine 5’-monophosphate-activated protein kinase (AMPK)[80]. Moreover, the relationship of miR-122 with the development and progression of hepatic fibrosis has been demonstrated in vitro, through the regulation of HSC proliferation and production of collagen by targeting prolyl 4-hydroxylase subunit α-1 (P4HA1)[88].
miR-34a: miR-34a is overexpressed in both murine models of NAFLD (e.g., mice fed a high-fat diet) and liver and serum from patients with NAFLD[81,87,89,90]. The main target of miR-34a is Sirtuin 1 (SIRT1), which regulates energy homeostasis by activating transcription factors such as peroxisome proliferator activated receptors (PPAR) α and liver X receptor (LXR). In addition, SIRT1 inhibits the co-activator 1α of the PPAR-γ (PGC1-α), the SREBP-1c and the farnesoid X receptor (FXR). SIRT1 is downregulated in the liver of NAFLD patients[91] and the inhibition of miR-34a restores the expression of SIRT1 and PPAR-α, leading to the activation of AMP-activated protein kinase (AMPK) and several target genes of PPAR-α. These findings suggest a fundamental role for miR-34a in the dysregulation of lipid metabolism associated with NAFLD[92].
miR-155: miR-155 is an important regulator of immune cells in both humans and mice and is involved in several inflammatory processes, such as rheumatic diseases[93], lipid metabolism[94] and in ALD (as described above). In patients with NAFLD, miR-155 is dysregulated by adipogenic transcription factors CCAAT/enhancer binding protein (C/EBP)-α, C/EBP-β, PPAR-γ and LXRα[95,96], fibrosis targets platelet derived growth factor (PDGF), Smad3 and C/EBP-β[97], and a tumor suppressor in the liver, SOCS-1[90,98]. However, animal models of NAFLD show contradictory results. For example, miR-155 deficient mice fed a high-fat diet showed a significant increase in hepatic steatosis[98], while miR-155 KO mice fed a methionine-choline-deficient diet showed a decrease in steatosis and expression of genes involved in fatty acid metabolism and fibrosis, with no concomitant liver injury or inflammation[97]. In addition, miR-155 may also be involved in hepatocarcinoma development[99]. These findings suggest that miR-155 may have different roles in fat storage and lipid accumulation in liver diseases and healthy subjects. However, additional research is warranted[97].
miRNAs as biomarkers in the diagnosis of NAFLD
As shown in Table 2, many miRNAs are differentially expressed in patients with NAFLD compared to healthy controls. These miRNAs may serve as potential biomarkers in the diagnosis and staging of NAFLD.
miR-122: Several studies have found that miR-122 is elevated in serum in NAFLD patients[81,86,100-102], even long before an alteration in transaminase levels occurs[103]. The diagnostic potential of miR-122 may extend to an indicator of disease severity and as a predictor of hepatic fibrosis[82,84,87,104].
miR-34a: Similar to miR-122, miR-34a has also been shown to have potential as a biomarker of diagnosis and severity of NAFLD. Several studies have shown that miR-34a is upregulated in the liver and serum of patients with NAFLD[71,81,82,104]. Additionally, elevated serum levels of miR-34a correlate with disease severity from simple steatosis to steatohepatitis, with liver enzyme levels, with fibrosis stage and with inflammation activity[82,104,105].
miR-192: Serum miR-192 levels are positively correlated with the severity of NAFLD-specific liver pathomorphological changes in mice fed a choline and folate deficient diet[82] and miR-192 upregulation in human serum has been demonstrated[82,84,86,102,106]. Interestingly, serum levels of miR-122 and mir-192 have been shown to be strongly correlated[84,86].
Panels: In addition to individual miRNAs, a serum panel comprised of hsa-miR-122-5p, hsa-miR-1290, hsa-miR-27b-3p, and hsa-miR-192-5p has shown high NAFLD diagnostic accuracy, regardless of NAFLD activity score (NAS) status[102]. Another research group found that NAFLD was associated with an miRNA signature based on up-regulation of miR-122, miR-192, miR-19a, miR-19b, miR-125b, and miR-375[84].
It is important to mention that most studies have compared patients with NAFLD to healthy controls or patients with chronic viral hepatitis B[105] or C[104]. However, no comparisons have been performed, to our knowledge, between patients with NAFLD and patients with ALD.
Therapeutic application of miRNAs in NAFLD
As previously mentioned, miRNAs are involved in several stages of NAFLD development (from lipid metabolism or diabetes to liver inflammation), and are therefore potential therapeutic targets[7,107]. The expression of miR-103 and miR-107 is upregulated in obese mice[53,89]. Inactivation of miR-103/107 in murine adipocytes upregulates caveolin-1 (a critical mediator of the insulin receptor) leading to enhanced insulin signaling, decreased adipocyte size and enhanced insulin-stimulated glucose uptake[53,108]. An N-acetylgalactosamine (GalNAc)-conjugated anti-miR-103/107 (RG-125/AZD4076, Regulus Therapeutics) has been developed for the treatment of NAFLD and type 2 diabetes or pre-diabetes[108-110]. Currently, two clinical trials are registered using this drug in patients with NAFLD (ClinicalTrials.gov Identifier: NCT02826525 and NCT02612662), although Regulus has acknowledged that AstraZeneca intends to terminate the clinical development of RG-125/AZD4076[108,111].
miR-122 has also shown promising results as a treatment for NAFLD. There is a high concentration of miR-122 in liver tissue[112] and this miRNA plays an important role in liver development, differentiation, homeostasis and functioning[113]. Over-expression of miR-122 may affect the Ying Yan 1 and Farnesoid X Receptor (YY1-FXR-SHP) regulatory axis leading to a reduction in hepatic triglyceride levels, potentially serving as a target for NAFLD treatment[114]. miR-122 is also an essential host factor for hepatitis C virus (HCV) replication and anti-miR-122 efficiently reduces viral load in chronically infected HCV patients without detectable resistance[108]. The fact that miR-122 has protective effects on NAFLD, while imposing a deleterious impact on HCV infection, emphasizes the importance of cautious targeting of miRNAs therapy since the role of miRNAs can be highly context dependent[115].
circRNA_0046366 antagonizes miR-34a and normalizes PPARα signaling, leading to the amelioration of liver steatosis in a murine model[116]. However, a phase I study on the effects of a miR-34 mimic (MRX34) on primary liver cancer and advanced or metastatic cancer with liver involvement (ClinicalTrials.gov Identifier: NCT01829971) was prematurely terminated due to serious immune-related adverse events[108], highlighting the potential risks of miRNA based-therapies.
CONCLUSION
All except four (miR-199, miR-212, miR-214 and miR-497) of the 21 miRNAs associated with ALD, listed in Table 1, are also related to NAFLD or lipid metabolism (although the four have been associated with other diseases, such as cancer[117]). Conversely, miRNAs that are related to the pathogenesis of NAFLD (miR-122, miR-34a and miR-155) are also clearly linked to ALD. These results reflect the common mechanisms between NAFLD and ALD and also the pleiotropic effects of any particular miRNA.
Due to the lack of specificity of miRNAs, the development of a biomarker or treatment specific to ALD or NAFLD is difficult. It is more feasible that individual miRNAs or a panel of miRNAs would be useful in the staging of liver disease (e.g., distinguishing simple steatosis in ALD or NAFLD from steatohepatitis)[118]. miR-122 is the most promising candidate as a biomarker due to its liver specificity. It is clear however, that miR-122 is also a marker of liver damage regardless of etiology[119]. Technical limitations, such as standardization of techniques and potential costs, add to the difficulties inherent to the development of a validated diagnostic biomarker. Circulating miRNAs are promising as biomarkers due to their stability and potential ability to detect advanced liver disease without a biopsy. However, rigorously validated studied are needed before they can be brought to the clinic[119].
The development of miRNA-targeted interventions for ALD and NAFLD is an intriguing area of research. However, despite the success in animal models and the potential targets described in this review, to the best of our knowledge there are no current clinical trials for miRNA interventions in ALD or NAFLD. The few studies that are being conducted on miRNA treatment in other diseases are phase 1 studies in the field of cancer research (e.g., assessing the activity of miRNA-loaded minicells or TargomiRs in malignant pleural mesothelioma[120]). Theoretical miRNA-based therapies are pharmacologically complex and include miRNA inhibition (e.g., synthetic anti-miRNAs) or miRNA replacement therapy (e.g., lipid vesicles or gold nanoparticles)[121]. One major challenge to the development of miRNA-based therapies is the improvement of drug delivery systems. Due to the biochemical instability of unmodified miRNAs and potential immunogenicity, specific delivery to target organs should be achieved. The high degree of redundancy among miRNAs and the multiple binding sites for any given miRNA must also be taken into account when designing efficacious and safe miRNA-based therapies[122].
To sum up, there is a large body of literature regarding miRNAs in NAFLD and ALD at various stages of the disease. These studies include expression data from microarrays and next generation sequencing from animal models and human studies, and cell-specific data from in situ hybridization and sensor constructs. The role of miRNAs in pathogenesis is well-documented and as such, their potential value as biomarkers or therapeutic targets is warranted. However, most miRNA modifications have a modest phenotypic effect, since miRNAs are unlikely to be the single key factor in chronic and multifactorial diseases such as liver steatosis[123]. Instead, most miRNAs act as fine-tuners in disease pathways and this characteristic, along with their lack of specificity must be considered before use in the clinic. To this end, we must improve our understanding of the interaction of different miRNAs in the development of advanced liver disease.
Footnotes
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Spain
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
Conflict-of-interest statement: No potential conflicts of interest.
Peer-review started: April 14, 2018
First decision: May 21, 2018
Article in press: June 27, 2018
P- Reviewer: Abenavoli L, Gonzalez-Reimers E, Kharbanda KK S- Editor: Wang XJ L- Editor: A E- Editor: Huang Y
Contributor Information
Jorge-Luis Torres, Department of Internal Medicine, University Hospital of Salamanca, Institute of Biomedical Research of Salamanca-IBSAL, Salamanca 37007, Spain; Spanish Working Group on Alcohol and Alcoholism, Spanish Society of Internal Medicine, Madrid 28016, Spain.
Ignacio Novo-Veleiro, Department of Internal Medicine, University Hospital of Santiago de Compostela, A Coruña 15706, Spain; Spanish Working Group on Alcohol and Alcoholism, Spanish Society of Internal Medicine, Madrid 28016, Spain.
Laura Manzanedo, Department of Internal Medicine, University Hospital of Salamanca, Institute of Biomedical Research of Salamanca-IBSAL, Salamanca 37007, Spain.
Lucía Alvela-Suárez, Department of Internal Medicine, HM Rosaleda Hospital, Santiago de Compostela, A Coruña 15701, Spain.
Ronald Macías, Department of Internal Medicine, University Hospital of Salamanca, Institute of Biomedical Research of Salamanca-IBSAL, Salamanca 37007, Spain.
Francisco-Javier Laso, Department of Internal Medicine, University Hospital of Salamanca, Institute of Biomedical Research of Salamanca-IBSAL, Salamanca 37007, Spain; Department of Medicine, Faculty of Medicine, University of Salamanca, Salamanca 37007, Spain; Spanish Working Group on Alcohol and Alcoholism, Spanish Society of Internal Medicine, Madrid 28016, Spain.
Miguel Marcos, Department of Internal Medicine, University Hospital of Salamanca, Institute of Biomedical Research of Salamanca-IBSAL, Salamanca 37007, Spain; Department of Medicine, Faculty of Medicine, University of Salamanca, Salamanca 37007, Spain; Spanish Working Group on Alcohol and Alcoholism, Spanish Society of Internal Medicine, Madrid 28016, Spain. mmarcos@usal.es.
References
- 1.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Hammond SM. An overview of microRNAs. Adv Drug Deliv Rev. 2015;87:3–14. doi: 10.1016/j.addr.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 4.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dolganiuc A, Petrasek J, Kodys K, Catalano D, Mandrekar P, Velayudham A, Szabo G. MicroRNA expression profile in Lieber-DeCarli diet-induced alcoholic and methionine choline deficient diet-induced nonalcoholic steatohepatitis models in mice. Alcohol Clin Exp Res. 2009;33:1704–1710. doi: 10.1111/j.1530-0277.2009.01007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Szabo G, Bala S. MicroRNAs in liver disease. Nat Rev Gastroenterol Hepatol. 2013;10:542–552. doi: 10.1038/nrgastro.2013.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bala S, Marcos M, Szabo G. Emerging role of microRNAs in liver diseases. World J Gastroenterol. 2009;15:5633–5640. doi: 10.3748/wjg.15.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szabo G, Csak T. Role of MicroRNAs in NAFLD/NASH. Dig Dis Sci. 2016;61:1314–1324. doi: 10.1007/s10620-015-4002-4. [DOI] [PubMed] [Google Scholar]
- 9.Sanyal AJ, Mathurin P, Nagy LA. Commonalities and Distinctions Between Alcoholic and Nonalcoholic Fatty Liver Disease. Gastroenterology. 2016;150:1695–1697. doi: 10.1053/j.gastro.2016.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laso FJ, Pastor I, Orfao A. Immune system and alcoholic liver disease. Medic Clin. 2005;125:263–269. doi: 10.1157/13078101. [DOI] [PubMed] [Google Scholar]
- 11.Lieber CS. Hepatic and metabolic effects of ethanol: pathogenesis and prevention. Ann Med. 1994;26:325–330. doi: 10.3109/07853899409148346. [DOI] [PubMed] [Google Scholar]
- 12.Mello T, Ceni E, Surrenti C, Galli A. Alcohol induced hepatic fibrosis: role of acetaldehyde. Mol Aspects Med. 2008;29:17–21. doi: 10.1016/j.mam.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 13.Gao B, Bataller R. Alcoholic liver disease: pathogenesis and new therapeutic targets. Gastroenterology. 2011;141:1572–1585. doi: 10.1053/j.gastro.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mandrekar P, Szabo G. Signalling pathways in alcohol-induced liver inflammation. J Hepatol. 2009;50:1258–1266. doi: 10.1016/j.jhep.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Neill LA, Sheedy FJ, McCoy CE. MicroRNAs: the fine-tuners of Toll-like receptor signalling. Nat Rev Immunol. 2011;11:163–175. doi: 10.1038/nri2957. [DOI] [PubMed] [Google Scholar]
- 16.Boccuto L, Abenavoli L. Genetic and Epigenetic Profile of Patients With Alcoholic Liver Disease. Ann Hepatol. 2017;16:490–500. doi: 10.5604/01.3001.0010.0274. [DOI] [PubMed] [Google Scholar]
- 17.Wan C, Han R, Liu L, Zhang F, Li F, Xiang M, Ding W. Role of miR-155 in fluorooctane sulfonate-induced oxidative hepatic damage via the Nrf2-dependent pathway. Toxicol Appl Pharmacol. 2016;295:85–93. doi: 10.1016/j.taap.2016.01.023. [DOI] [PubMed] [Google Scholar]
- 18.Kurinna S, Werner S. NRF2 and microRNAs: new but awaited relations. Biochem Soc Trans. 2015;43:595–601. doi: 10.1042/BST20140317. [DOI] [PubMed] [Google Scholar]
- 19.Yang JJ, Tao H, Hu W, Liu LP, Shi KH, Deng ZY, Li J. MicroRNA-200a controls Nrf2 activation by target Keap1 in hepatic stellate cell proliferation and fibrosis. Cell Signal. 2014;26:2381–2389. doi: 10.1016/j.cellsig.2014.07.016. [DOI] [PubMed] [Google Scholar]
- 20.Shi L, Wu L, Chen Z, Yang J, Chen X, Yu F, Zheng F, Lin X. MiR-141 Activates Nrf2-Dependent Antioxidant Pathway via Down-Regulating the Expression of Keap1 Conferring the Resistance of Hepatocellular Carcinoma Cells to 5-Fluorouracil. Cell Physiol Biochem. 2015;35:2333–2348. doi: 10.1159/000374036. [DOI] [PubMed] [Google Scholar]
- 21.O’Connell RM, Rao DS, Baltimore D. microRNA regulation of inflammatory responses. Annu Rev Immunol. 2012;30:295–312. doi: 10.1146/annurev-immunol-020711-075013. [DOI] [PubMed] [Google Scholar]
- 22.Wu XQ, Dai Y, Yang Y, Huang C, Meng XM, Wu BM, Li J. Emerging role of microRNAs in regulating macrophage activation and polarization in immune response and inflammation. Immunology. 2016;148:237–248. doi: 10.1111/imm.12608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Momen-Heravi F, Saha B, Kodys K, Catalano D, Satishchandran A, Szabo G. Increased number of circulating exosomes and their microRNA cargos are potential novel biomarkers in alcoholic hepatitis. J Transl Med. 2015;13:261. doi: 10.1186/s12967-015-0623-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Yu D, Tolleson WH, Yu LR, Green B, Zeng L, Chen Y, Chen S, Ren Z, Guo L, et al. A systematic evaluation of microRNAs in regulating human hepatic CYP2E1. Biochem Pharmacol. 2017;138:174–184. doi: 10.1016/j.bcp.2017.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim YD, Hwang SL, Lee EJ, Kim HM, Chung MJ, Elfadl AK, Lee SE, Nedumaran B, Harris RA, Jeong KS. Melatonin ameliorates alcohol-induced bile acid synthesis by enhancing miR-497 expression. J Pineal Res. 2017:62. doi: 10.1111/jpi.12386. [DOI] [PubMed] [Google Scholar]
- 26.Dippold RP, Vadigepalli R, Gonye GE, Patra B, Hoek JB. Chronic ethanol feeding alters miRNA expression dynamics during liver regeneration. Alcohol Clin Exp Res. 2013;37 Suppl 1:E59–E69. doi: 10.1111/j.1530-0277.2012.01852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McDaniel K, Huang L, Sato K, Wu N, Annable T, Zhou T, Ramos-Lorenzo S, Wan Y, Huang Q, Francis H, et al. The let-7/Lin28 axis regulates activation of hepatic stellate cells in alcoholic liver injury. J Biol Chem. 2017;292:11336–11347. doi: 10.1074/jbc.M116.773291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brandon-Warner E, Feilen NA, Culberson CR, Field CO, deLemos AS, Russo MW, Schrum LW. Processing of miR17-92 Cluster in Hepatic Stellate Cells Promotes Hepatic Fibrogenesis During Alcohol-Induced Injury. Alcohol Clin Exp Res. 2016;40:1430–1442. doi: 10.1111/acer.13116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meng F, Glaser SS, Francis H, Yang F, Han Y, Stokes A, Staloch D, McCarra J, Liu J, Venter J, et al. Epigenetic regulation of miR-34a expression in alcoholic liver injury. Am J Pathol. 2012;181:804–817. doi: 10.1016/j.ajpath.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blaya D, Coll M, Rodrigo-Torres D, Vila-Casadesús M, Altamirano J, Llopis M, Graupera I, Perea L, Aguilar-Bravo B, Díaz A, et al. Integrative microRNA profiling in alcoholic hepatitis reveals a role for microRNA-182 in liver injury and inflammation. Gut. 2016;65:1535–1545. doi: 10.1136/gutjnl-2015-311314. [DOI] [PubMed] [Google Scholar]
- 31.Zhao YX, Sun YY, Huang AL, Li XF, Huang C, Ma TT, Li J. MicroRNA-200a induces apoptosis by targeting ZEB2 in alcoholic liver disease. Cell Cycle. 2018;17:250–262. doi: 10.1080/15384101.2017.1417708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Momen-Heravi F, Bala S, Kodys K, Szabo G. Exosomes derived from alcohol-treated hepatocytes horizontally transfer liver specific miRNA-122 and sensitize monocytes to LPS. Sci Rep. 2015;5:9991. doi: 10.1038/srep09991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Satishchandran A, Ambade A, Rao S, Hsueh YC, Iracheta-Vellve A, Tornai D, Lowe P, Gyongyosi B, Li J, Catalano D, et al. MicroRNA 122, Regulated by GRLH2, Protects Livers of Mice and Patients From Ethanol-Induced Liver Disease. Gastroenterology. 2018;154:238–252.e7. doi: 10.1053/j.gastro.2017.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dong X, Liu H, Chen F, Li D, Zhao Y. MiR-214 promotes the alcohol-induced oxidative stress via down-regulation of glutathione reductase and cytochrome P450 oxidoreductase in liver cells. Alcohol Clin Exp Res. 2014;38:68–77. doi: 10.1111/acer.12209. [DOI] [PubMed] [Google Scholar]
- 35.Han W, Fu X, Xie J, Meng Z, Gu Y, Wang X, Li L, Pan H, Huang W. MiR-26a enhances autophagy to protect against ethanol-induced acute liver injury. J Mol Med (Berl) 2015;93:1045–1055. doi: 10.1007/s00109-015-1282-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dippold RP, Vadigepalli R, Gonye GE, Hoek JB. Chronic ethanol feeding enhances miR-21 induction during liver regeneration while inhibiting proliferation in rats. Am J Physiol Gastrointest Liver Physiol. 2012;303:G733–G743. doi: 10.1152/ajpgi.00019.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Francis H, McDaniel K, Han Y, Liu X, Kennedy L, Yang F, McCarra J, Zhou T, Glaser S, Venter J, et al. Regulation of the extrinsic apoptotic pathway by microRNA-21 in alcoholic liver injury. J Biol Chem. 2014;289:27526–27539. doi: 10.1074/jbc.M114.602383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bala S, Marcos M, Kodys K, Csak T, Catalano D, Mandrekar P, Szabo G. Up-regulation of microRNA-155 in macrophages contributes to increased tumor necrosis factor {alpha} (TNF{alpha}) production via increased mRNA half-life in alcoholic liver disease. J Biol Chem. 2011;286:1436–1444. doi: 10.1074/jbc.M110.145870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bala S, Csak T, Kodys K, Catalano D, Ambade A, Furi I, Lowe P, Cho Y, Iracheta-Vellve A, Szabo G. Alcohol-induced miR-155 and HDAC11 inhibit negative regulators of the TLR4 pathway and lead to increased LPS responsiveness of Kupffer cells in alcoholic liver disease. J Leukoc Biol. 2017;102:487–498. doi: 10.1189/jlb.3A0716-310R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saikia P, Bellos D, McMullen MR, Pollard KA, de la Motte C, Nagy LE. MicroRNA 181b-3p and its target importin α5 regulate toll-like receptor 4 signaling in Kupffer cells and liver injury in mice in response to ethanol. Hepatology. 2017;66:602–615. doi: 10.1002/hep.29144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yin H, Liang X, Jogasuria A, Davidson NO, You M. miR-217 regulates ethanol-induced hepatic inflammation by disrupting sirtuin 1-lipin-1 signaling. Am J Pathol. 2015;185:1286–1296. doi: 10.1016/j.ajpath.2015.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saikia P, Roychowdhury S, Bellos D, Pollard KA, McMullen MR, McCullough RL, McCullough AJ, Gholam P, de la Motte C, Nagy LE. Hyaluronic acid 35 normalizes TLR4 signaling in Kupffer cells from ethanol-fed rats via regulation of microRNA291b and its target Tollip. Sci Rep. 2017;7:15671. doi: 10.1038/s41598-017-15760-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wan Y, McDaniel K, Wu N, Ramos-Lorenzo S, Glaser T, Venter J, Francis H, Kennedy L, Sato K, Zhou T, et al. Regulation of Cellular Senescence by miR-34a in Alcoholic Liver Injury. Am J Pathol. 2017;187:2788–2798. doi: 10.1016/j.ajpath.2017.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saha B, Bruneau JC, Kodys K, Szabo G. Alcohol-induced miR-27a regulates differentiation and M2 macrophage polarization of normal human monocytes. J Immunol. 2015;194:3079–3087. doi: 10.4049/jimmunol.1402190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li M, He Y, Zhou Z, Ramirez T, Gao Y, Gao Y, Ross RA, Cao H, Cai Y, Xu M, et al. MicroRNA-223 ameliorates alcoholic liver injury by inhibiting the IL-6-p47 phox -oxidative stress pathway in neutrophils. Gut. 2017;66:705–715. doi: 10.1136/gutjnl-2016-311861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang Y, Banan A, Forsyth CB, Fields JZ, Lau CK, Zhang LJ, Keshavarzian A. Effect of alcohol on miR-212 expression in intestinal epithelial cells and its potential role in alcoholic liver disease. Alcohol Clin Exp Res. 2008;32:355–364. doi: 10.1111/j.1530-0277.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- 47.Novo-Veleiro I, González-Sarmiento R, Cieza-Borrella C, Pastor I, Laso FJ, Marcos M. A genetic variant in the microRNA-146a gene is associated with susceptibility to alcohol use disorders. Eur Psychiatry. 2014;29:288–292. doi: 10.1016/j.eurpsy.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 48.Novo-Veleiro I, Cieza-Borrella C, Pastor I, González-Sarmiento R, Laso FJ, Marcos M. Analysis of the relationship between interleukin polymorphisms within miRNA-binding regions and alcoholic liver disease. Rev Clin Esp. 2018;218:170–176. doi: 10.1016/j.rce.2018.02.005. [DOI] [PubMed] [Google Scholar]
- 49.Barragán R, Coltell O, Asensio EM, Francés F, Sorlí JV, Estruch R, Salas-Huetos A, Ordovas JM, Corella D. MicroRNAs and Drinking: Association between the Pre-miR-27a rs895819 Polymorphism and Alcohol Consumption in a Mediterranean Population. Int J Mol Sci. 2016:1. doi: 10.3390/ijms17081338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chamorro AJ, Torres JL, Mirón-Canelo JA, González-Sarmiento R, Laso FJ, Marcos M. Systematic review with meta-analysis: the I148M variant of patatin-like phospholipase domain-containing 3 gene (PNPLA3) is significantly associated with alcoholic liver cirrhosis. Aliment Pharmacol Ther. 2014;40:571–581. doi: 10.1111/apt.12890. [DOI] [PubMed] [Google Scholar]
- 51.Afonso MB, Rodrigues PM, Simão AL, Castro RE. Circulating microRNAs as Potential Biomarkers in Non-Alcoholic Fatty Liver Disease and Hepatocellular Carcinoma. J Clin Med. 2016:5. doi: 10.3390/jcm5030030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saha B, Momen-Heravi F, Kodys K, Szabo G. MicroRNA Cargo of Extracellular Vesicles from Alcohol-exposed Monocytes Signals Naive Monocytes to Differentiate into M2 Macrophages. J Biol Chem. 2016;291:149–159. doi: 10.1074/jbc.M115.694133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trajkovski M, Hausser J, Soutschek J, Bhat B, Akin A, Zavolan M, Heim MH, Stoffel M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474:649–653. doi: 10.1038/nature10112. [DOI] [PubMed] [Google Scholar]
- 54.Bala S, Petrasek J, Mundkur S, Catalano D, Levin I, Ward J, Alao H, Kodys K, Szabo G. Circulating microRNAs in exosomes indicate hepatocyte injury and inflammation in alcoholic, drug-induced, and inflammatory liver diseases. Hepatology. 2012;56:1946–1957. doi: 10.1002/hep.25873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCrae JC, Sharkey N, Webb DJ, Vliegenthart AD, Dear JW. Ethanol consumption produces a small increase in circulating miR-122 in healthy individuals. Clin Toxicol (Phila) 2016;54:53–55. doi: 10.3109/15563650.2015.1112015. [DOI] [PubMed] [Google Scholar]
- 56.Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 57.Gebert LF, Rebhan MA, Crivelli SE, Denzler R, Stoffel M, Hall J. Miravirsen (SPC3649) can inhibit the biogenesis of miR-122. Nucleic Acids Res. 2014;42:609–621. doi: 10.1093/nar/gkt852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chakraborty C, Sharma AR, Sharma G, Doss CGP, Lee SS. Therapeutic miRNA and siRNA: Moving from Bench to Clinic as Next Generation Medicine. Mol Ther Nucleic Acids. 2017;8:132–143. doi: 10.1016/j.omtn.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hyun J, Wang S, Kim J, Rao KM, Park SY, Chung I, Ha CS, Kim SW, Yun YH, Jung Y. MicroRNA-378 limits activation of hepatic stellate cells and liver fibrosis by suppressing Gli3 expression. Nat Commun. 2016;7:10993. doi: 10.1038/ncomms10993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chalasani N, Younossi Z, Lavine JE, Diehl AM, Brunt EM, Cusi K, Charlton M, Sanyal AJ. The diagnosis and management of non-alcoholic fatty liver disease: practice Guideline by the American Association for the Study of Liver Diseases, American College of Gastroenterology, and the American Gastroenterological Association. Hepatology. 2012;55:2005–2023. doi: 10.1002/hep.25762. [DOI] [PubMed] [Google Scholar]
- 61.Loomba R, Sanyal AJ. The global NAFLD epidemic. Nat Rev Gastroenterol Hepatol. 2013;10:686–690. doi: 10.1038/nrgastro.2013.171. [DOI] [PubMed] [Google Scholar]
- 62.Gholam PM, Kotler DP, Flancbaum LJ. Liver pathology in morbidly obese patients undergoing Roux-en-Y gastric bypass surgery. Obes Surg. 2002;12:49–51. doi: 10.1381/096089202321144577. [DOI] [PubMed] [Google Scholar]
- 63.Praveenraj P, Gomes RM, Kumar S, Karthikeyan P, Shankar A, Parthasarathi R, Senthilnathan P, Rajapandian S, Palanivelu C. Prevalence and Predictors of Non-Alcoholic Fatty Liver Disease in Morbidly Obese South Indian Patients Undergoing Bariatric Surgery. Obes Surg. 2015;25:2078–2087. doi: 10.1007/s11695-015-1655-1. [DOI] [PubMed] [Google Scholar]
- 64.Williamson RM, Price JF, Glancy S, Perry E, Nee LD, Hayes PC, Frier BM, Van Look LA, Johnston GI, Reynolds RM, et al. Prevalence of and risk factors for hepatic steatosis and nonalcoholic Fatty liver disease in people with type 2 diabetes: the Edinburgh Type 2 Diabetes Study. Diabetes Care. 2011;34:1139–1144. doi: 10.2337/dc10-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Charlton MR, Burns JM, Pedersen RA, Watt KD, Heimbach JK, Dierkhising RA. Frequency and outcomes of liver transplantation for nonalcoholic steatohepatitis in the United States. Gastroenterology. 2011;141:1249–1253. doi: 10.1053/j.gastro.2011.06.061. [DOI] [PubMed] [Google Scholar]
- 66.Day CP, James OF. Steatohepatitis: a tale of two “hits”? Gastroenterology. 1998;114:842–845. doi: 10.1016/s0016-5085(98)70599-2. [DOI] [PubMed] [Google Scholar]
- 67.Videla LA, Rodrigo R, Araya J, Poniachik J. Insulin resistance and oxidative stress interdependency in non-alcoholic fatty liver disease. Trends Mol Med. 2006;12:555–558. doi: 10.1016/j.molmed.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 68.Tilg H, Moschen AR. Evolution of inflammation in nonalcoholic fatty liver disease: the multiple parallel hits hypothesis. Hepatology. 2010;52:1836–1846. doi: 10.1002/hep.24001. [DOI] [PubMed] [Google Scholar]
- 69.Buzzetti E, Pinzani M, Tsochatzis EA. The multiple-hit pathogenesis of non-alcoholic fatty liver disease (NAFLD) Metabolism. 2016;65:1038–1048. doi: 10.1016/j.metabol.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 70.Peverill W, Powell LW, Skoien R. Evolving concepts in the pathogenesis of NASH: beyond steatosis and inflammation. Int J Mol Sci. 2014;15:8591–8638. doi: 10.3390/ijms15058591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cheung O, Puri P, Eicken C, Contos MJ, Mirshahi F, Maher JW, Kellum JM, Min H, Luketic VA, Sanyal AJ. Nonalcoholic steatohepatitis is associated with altered hepatic MicroRNA expression. Hepatology. 2008;48:1810–1820. doi: 10.1002/hep.22569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cazanave SC, Mott JL, Elmi NA, Bronk SF, Masuoka HC, Charlton MR, Gores GJ. A role for miR-296 in the regulation of lipoapoptosis by targeting PUMA. J Lipid Res. 2011;52:1517–1525. doi: 10.1194/jlr.M014654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lendvai G, Jármay K, Karácsony G, Halász T, Kovalszky I, Baghy K, Wittmann T, Schaff Z, Kiss A. Elevated miR-33a and miR-224 in steatotic chronic hepatitis C liver biopsies. World J Gastroenterol. 2014;20:15343–15350. doi: 10.3748/wjg.v20.i41.15343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Leti F, Malenica I, Doshi M, Courtright A, Van Keuren-Jensen K, Legendre C, Still CD, Gerhard GS, DiStefano JK. High-throughput sequencing reveals altered expression of hepatic microRNAs in nonalcoholic fatty liver disease-related fibrosis. Transl Res. 2015;166:304–314. doi: 10.1016/j.trsl.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guo Y, Xiong Y, Sheng Q, Zhao S, Wattacheril J, Flynn CR. A micro-RNA expression signature for human NAFLD progression. J Gastroenterol. 2016;51:1022–1030. doi: 10.1007/s00535-016-1178-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vega-Badillo J, Gutiérrez-Vidal R, Hernández-Pérez HA, Villamil-Ramírez H, León-Mimila P, Sánchez-Muñoz F, Morán-Ramos S, Larrieta-Carrasco E, Fernández-Silva I, Méndez-Sánchez N, et al. Hepatic miR-33a/miR-144 and their target gene ABCA1 are associated with steatohepatitis in morbidly obese subjects. Liver Int. 2016;36:1383–1391. doi: 10.1111/liv.13109. [DOI] [PubMed] [Google Scholar]
- 77.Xu H, He JH, Xiao ZD, Zhang QQ, Chen YQ, Zhou H, Qu LH. Liver-enriched transcription factors regulate microRNA-122 that targets CUTL1 during liver development. Hepatology. 2010;52:1431–1442. doi: 10.1002/hep.23818. [DOI] [PubMed] [Google Scholar]
- 78.Laudadio I, Manfroid I, Achouri Y, Schmidt D, Wilson MD, Cordi S, Thorrez L, Knoops L, Jacquemin P, Schuit F, et al. A feedback loop between the liver-enriched transcription factor network and miR-122 controls hepatocyte differentiation. Gastroenterology. 2012;142:119–129. doi: 10.1053/j.gastro.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 79.Deng XG, Qiu RL, Wu YH, Li ZX, Xie P, Zhang J, Zhou JJ, Zeng LX, Tang J, Maharjan A, et al. Overexpression of miR-122 promotes the hepatic differentiation and maturation of mouse ESCs through a miR-122/FoxA1/HNF4a-positive feedback loop. Liver Int. 2014;34:281–295. doi: 10.1111/liv.12239. [DOI] [PubMed] [Google Scholar]
- 80.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 81.Salvoza NC, Klinzing DC, Gopez-Cervantes J, Baclig MO. Association of Circulating Serum miR-34a and miR-122 with Dyslipidemia among Patients with Non-Alcoholic Fatty Liver Disease. PLoS One. 2016;11:e0153497. doi: 10.1371/journal.pone.0153497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tryndyak VP, Latendresse JR, Montgomery B, Ross SA, Beland FA, Rusyn I, Pogribny IP. Plasma microRNAs are sensitive indicators of inter-strain differences in the severity of liver injury induced in mice by a choline- and folate-deficient diet. Toxicol Appl Pharmacol. 2012;262:52–59. doi: 10.1016/j.taap.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Latorre J, Moreno-Navarrete JM, Mercader JM, Sabater M, Rovira Ò, Gironès J, Ricart W, Fernández-Real JM, Ortega FJ. Decreased lipid metabolism but increased FA biosynthesis are coupled with changes in liver microRNAs in obese subjects with NAFLD. Int J Obes (Lond) 2017;41:620–630. doi: 10.1038/ijo.2017.21. [DOI] [PubMed] [Google Scholar]
- 84.Pirola CJ, Fernández Gianotti T, Castaño GO, Mallardi P, San Martino J, Mora Gonzalez Lopez Ledesma M, Flichman D, Mirshahi F, Sanyal AJ, Sookoian S. Circulating microRNA signature in non-alcoholic fatty liver disease: from serum non-coding RNAs to liver histology and disease pathogenesis. Gut. 2015;64:800–812. doi: 10.1136/gutjnl-2014-306996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Braza-Boïls A, Marí-Alexandre J, Molina P, Arnau MA, Barceló-Molina M, Domingo D, Girbes J, Giner J, Martínez-Dolz L, Zorio E. Deregulated hepatic microRNAs underlie the association between non-alcoholic fatty liver disease and coronary artery disease. Liver Int. 2016;36:1221–1229. doi: 10.1111/liv.13097. [DOI] [PubMed] [Google Scholar]
- 86.Becker PP, Rau M, Schmitt J, Malsch C, Hammer C, Bantel H, Müllhaupt B, Geier A. Performance of Serum microRNAs -122, -192 and -21 as Biomarkers in Patients with Non-Alcoholic Steatohepatitis. PLoS One. 2015;10:e0142661. doi: 10.1371/journal.pone.0142661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yamada H, Suzuki K, Ichino N, Ando Y, Sawada A, Osakabe K, Sugimoto K, Ohashi K, Teradaira R, Inoue T, et al. Associations between circulating microRNAs (miR-21, miR-34a, miR-122 and miR-451) and non-alcoholic fatty liver. Clin Chim Acta. 2013;424:99–103. doi: 10.1016/j.cca.2013.05.021. [DOI] [PubMed] [Google Scholar]
- 88.Li J, Ghazwani M, Zhang Y, Lu J, Li J, Fan J, Gandhi CR, Li S. miR-122 regulates collagen production via targeting hepatic stellate cells and suppressing P4HA1 expression. J Hepatol. 2013;58:522–528. doi: 10.1016/j.jhep.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li S, Chen X, Zhang H, Liang X, Xiang Y, Yu C, Zen K, Li Y, Zhang CY. Differential expression of microRNAs in mouse liver under aberrant energy metabolic status. J Lipid Res. 2009;50:1756–1765. doi: 10.1194/jlr.M800509-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pogribny IP, Starlard-Davenport A, Tryndyak VP, Han T, Ross SA, Rusyn I, Beland FA. Difference in expression of hepatic microRNAs miR-29c, miR-34a, miR-155, and miR-200b is associated with strain-specific susceptibility to dietary nonalcoholic steatohepatitis in mice. Lab Invest. 2010;90:1437–1446. doi: 10.1038/labinvest.2010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wu T, Liu YH, Fu YC, Liu XM, Zhou XH. Direct evidence of sirtuin downregulation in the liver of non-alcoholic fatty liver disease patients. Ann Clin Lab Sci. 2014;44:410–418. [PubMed] [Google Scholar]
- 92.Ding J, Li M, Wan X, Jin X, Chen S, Yu C, Li Y. Effect of miR-34a in regulating steatosis by targeting PPARα expression in nonalcoholic fatty liver disease. Sci Rep. 2015;5:13729. doi: 10.1038/srep13729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen JQ, Papp G, Szodoray P, Zeher M. The role of microRNAs in the pathogenesis of autoimmune diseases. Autoimmun Rev. 2016;15:1171–1180. doi: 10.1016/j.autrev.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 94.Chen Y, Siegel F, Kipschull S, Haas B, Fröhlich H, Meister G, Pfeifer A. miR-155 regulates differentiation of brown and beige adipocytes via a bistable circuit. Nat Commun. 2013;4:1769. doi: 10.1038/ncomms2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Virtue A, Johnson C, Lopez-Pastraña J, Shao Y, Fu H, Li X, Li YF, Yin Y, Mai J, Rizzo V, et al. MicroRNA-155 Deficiency Leads to Decreased Atherosclerosis, Increased White Adipose Tissue Obesity, and Non-alcoholic Fatty Liver Disease: A NOVEL MOUSE MODEL OF OBESITY PARADOX. J Biol Chem. 2017;292:1267–1287. doi: 10.1074/jbc.M116.739839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang L, Zhang N, Wang Z, Ai DM, Cao ZY, Pan HP. Decreased MiR-155 Level in the Peripheral Blood of Non-Alcoholic Fatty Liver Disease Patients may Serve as a Biomarker and may Influence LXR Activity. Cell Physiol Biochem. 2016;39:2239–2248. doi: 10.1159/000447917. [DOI] [PubMed] [Google Scholar]
- 97.Csak T, Bala S, Lippai D, Kodys K, Catalano D, Iracheta-Vellve A, Szabo G. MicroRNA-155 Deficiency Attenuates Liver Steatosis and Fibrosis without Reducing Inflammation in a Mouse Model of Steatohepatitis. PLoS One. 2015;10:e0129251. doi: 10.1371/journal.pone.0129251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Miller AM, Gilchrist DS, Nijjar J, Araldi E, Ramirez CM, Lavery CA, Fernández-Hernando C, McInnes IB, Kurowska-Stolarska M. MiR-155 has a protective role in the development of non-alcoholic hepatosteatosis in mice. PLoS One. 2013;8:e72324. doi: 10.1371/journal.pone.0072324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang B, Majumder S, Nuovo G, Kutay H, Volinia S, Patel T, Schmittgen TD, Croce C, Ghoshal K, Jacob ST. Role of microRNA-155 at early stages of hepatocarcinogenesis induced by choline-deficient and amino acid-defined diet in C57BL/6 mice. Hepatology. 2009;50:1152–1161. doi: 10.1002/hep.23100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Akuta N, Kawamura Y, Suzuki F, Saitoh S, Arase Y, Kunimoto H, Sorin Y, Fujiyama S, Sezaki H, Hosaka T, et al. Impact of circulating miR-122 for histological features and hepatocellular carcinoma of nonalcoholic fatty liver disease in Japan. Hepatol Int. 2016;10:647–656. doi: 10.1007/s12072-016-9729-2. [DOI] [PubMed] [Google Scholar]
- 101.Miyaaki H, Ichikawa T, Kamo Y, Taura N, Honda T, Shibata H, Milazzo M, Fornari F, Gramantieri L, Bolondi L, et al. Significance of serum and hepatic microRNA-122 levels in patients with non-alcoholic fatty liver disease. Liver Int. 2014;34:e302–e307. doi: 10.1111/liv.12429. [DOI] [PubMed] [Google Scholar]
- 102.Tan Y, Ge G, Pan T, Wen D, Gan J. A pilot study of serum microRNAs panel as potential biomarkers for diagnosis of nonalcoholic fatty liver disease. PLoS One. 2014;9:e105192. doi: 10.1371/journal.pone.0105192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yamada H, Ohashi K, Suzuki K, Munetsuna E, Ando Y, Yamazaki M, Ishikawa H, Ichino N, Teradaira R, Hashimoto S. Longitudinal study of circulating miR-122 in a rat model of non-alcoholic fatty liver disease. Clin Chim Acta. 2015;446:267–271. doi: 10.1016/j.cca.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 104.Cermelli S, Ruggieri A, Marrero JA, Ioannou GN, Beretta L. Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PLoS One. 2011;6:e23937. doi: 10.1371/journal.pone.0023937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu XL, Pan Q, Zhang RN, Shen F, Yan SY, Sun C, Xu ZJ, Chen YW, Fan JG. Disease-specific miR-34a as diagnostic marker of non-alcoholic steatohepatitis in a Chinese population. World J Gastroenterol. 2016;22:9844–9852. doi: 10.3748/wjg.v22.i44.9844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Povero D, Eguchi A, Li H, Johnson CD, Papouchado BG, Wree A, Messer K, Feldstein AE. Circulating extracellular vesicles with specific proteome and liver microRNAs are potential biomarkers for liver injury in experimental fatty liver disease. PLoS One. 2014;9:e113651. doi: 10.1371/journal.pone.0113651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vienberg S, Geiger J, Madsen S, Dalgaard LT. MicroRNAs in metabolism. Acta Physiol (Oxf) 2017;219:346–361. doi: 10.1111/apha.12681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 109.Regulus Therapeutics I. Regulus announces notice of allowance from US patent office related to microRNA- 103/107 Program in Metabolic Disorders. Press release. 2014
- 110.Regulus Therapeutics I. 2015. RG-125 (AZD4076), a microRNA therapeutic targeting microRNA 103/107 for the treatment of NASH in patients with type 2 diabetes/Pre-Diabetes, selected as clinical candidate by AstraZeneca. Press release. [Google Scholar]
- 111.Astra Zeneca Halts Development of Regulus NASH Candidate. GEN News Highlights, 2017 [Google Scholar]
- 112.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of Tissue-Specific MicroRNAs from Mouse. Current Biology. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 113.Bandiera S, Pfeffer S, Baumert TF, Zeisel MB. miR-122--a key factor and therapeutic target in liver disease. J Hepatol. 2015;62:448–457. doi: 10.1016/j.jhep.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 114.Wu GY, Rui C, Chen JQ, Sho E, Zhan SS, Yuan XW, Ding YT. MicroRNA-122 Inhibits Lipid Droplet Formation and Hepatic Triglyceride Accumulation via Yin Yang 1. Cell Physiol Biochem. 2017;44:1651–1664. doi: 10.1159/000485765. [DOI] [PubMed] [Google Scholar]
- 115.Su Q, Kumar V, Sud N, Mahato RI. MicroRNAs in the pathogenesis and treatment of progressive liver injury in NAFLD and liver fibrosis. Adv Drug Deliv Rev. 2018;129:54–63. doi: 10.1016/j.addr.2018.01.009. [DOI] [PubMed] [Google Scholar]
- 116.Guo XY, Sun F, Chen JN, Wang YQ, Pan Q, Fan JG. circRNA_0046366 inhibits hepatocellular steatosis by normalization of PPAR signaling. World J Gastroenterol. 2018;24:323–337. doi: 10.3748/wjg.v24.i3.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yang G, Xiong G, Cao Z, Zheng S, You L, Zhang T, Zhao Y. miR-497 expression, function and clinical application in cancer. Oncotarget. 2016;7:55900–55911. doi: 10.18632/oncotarget.10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hyun J, Jung Y. MicroRNAs in liver fibrosis: Focusing on the interaction with hedgehog signaling. World J Gastroenterol. 2016;22:6652–6662. doi: 10.3748/wjg.v22.i29.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Loosen SH, Schueller F, Trautwein C, Roy S, Roderburg C. Role of circulating microRNAs in liver diseases. World J Hepatol. 2017;9:586–594. doi: 10.4254/wjh.v9.i12.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.van Zandwijk N, Pavlakis N, Kao SC, Linton A, Boyer MJ, Clarke S, Huynh Y, Chrzanowska A, Fulham MJ, Bailey DL, et al. Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: a first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol. 2017;18:1386–1396. doi: 10.1016/S1470-2045(17)30621-6. [DOI] [PubMed] [Google Scholar]
- 121.Hosseinahli N, Aghapour M, Duijf PHG, Baradaran B. Treating cancer with microRNA replacement therapy: A literature review. J Cell Physiol. 2018;233:5574–5588. doi: 10.1002/jcp.26514. [DOI] [PubMed] [Google Scholar]
- 122.Luck ME, Muljo SA, Collins CB. Prospects for Therapeutic Targeting of MicroRNAs in Human Immunological Diseases. J Immunol. 2015;194:5047–5052. doi: 10.4049/jimmunol.1403146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lai EC. Two decades of miRNA biology: lessons and challenges. RNA. 2015;21:675–677. doi: 10.1261/rna.051193.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Miranda RC, Pietrzykowski AZ, Tang Y, Sathyan P, Mayfield D, Keshavarzian A, Sampson W, Hereld D. MicroRNAs: master regulators of ethanol abuse and toxicity? Alcohol Clin Exp Res. 2010;34:575–587. doi: 10.1111/j.1530-0277.2009.01126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ambade A, Satishchandran A, Szabo G. Alcoholic hepatitis accelerates early hepatobiliary cancer by increasing stemness and miR-122-mediated HIF-1α activation. Scienti Repo. 2016;6:21340. doi: 10.1038/srep21340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bala S, Csak T, Saha B, Zatsiorsky J, Kodys K, Catalano D, Satishchandran A, Szabo G. The pro-inflammatory effects of miR-155 promote liver fibrosis and alcohol-induced steatohepatitis. J Hepatol. 2016;64:1378–1387. doi: 10.1016/j.jhep.2016.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lippai D, Bala S, Catalano D, Kodys K, Szabo G. Micro-RNA-155 deficiency prevents alcohol-induced serum endotoxin increase and small bowel inflammation in mice. Alcohol Clin Exp Res. 2014;38:2217–2224. doi: 10.1111/acer.12483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yeligar S, Tsukamoto H, Kalra VK. Ethanol-induced expression of ET-1 and ET-BR in liver sinusoidal endothelial cells and human endothelial cells involves hypoxia-inducible factor-1alpha and microrNA-199. J Immunol. 2009;183:5232–5243. doi: 10.4049/jimmunol.0901084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ao R, Wang Y, Tong J, Wang BF. Altered microRNA-9 Expression Level is Directly Correlated with Pathogenesis of Nonalcoholic Fatty Liver Disease by Targeting Onecut2 and SIRT1. Med Sci Monit. 2016;22:3804–3819. doi: 10.12659/MSM.897207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zheng L, Lv GC, Sheng J, Yang YD. Effect of miRNA-10b in regulating cellular steatosis level by targeting PPAR-alpha expression, a novel mechanism for the pathogenesis of NAFLD. J Gastroenterol Hepatol. 2010;25:156–163. doi: 10.1111/j.1440-1746.2009.05949.x. [DOI] [PubMed] [Google Scholar]
- 131.Zhang Y, Cheng X, Lu Z, Wang J, Chen H, Fan W, Gao X, Lu D. Upregulation of miR-15b in NAFLD models and in the serum of patients with fatty liver disease. Diabetes Res Clin Pract. 2013;99:327–334. doi: 10.1016/j.diabres.2012.11.025. [DOI] [PubMed] [Google Scholar]
- 132.Du J, Niu X, Wang Y, Kong L, Wang R, Zhang Y, Zhao S, Nan Y. MiR-146a-5p suppresses activation and proliferation of hepatic stellate cells in nonalcoholic fibrosing steatohepatitis through directly targeting Wnt1 and Wnt5a. Sci Rep. 2015;5:16163. doi: 10.1038/srep16163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rodrigues PM, Afonso MB, Simão AL, Carvalho CC, Trindade A, Duarte A, Borralho PM, Machado MV, Cortez-Pinto H, Rodrigues CM, et al. miR-21 ablation and obeticholic acid ameliorate nonalcoholic steatohepatitis in mice. Cell Death Dis. 2017;8:e2748. doi: 10.1038/cddis.2017.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Loyer X, Paradis V, Hénique C, Vion AC, Colnot N, Guerin CL, Devue C, On S, Scetbun J, Romain M, et al. Liver microRNA-21 is overexpressed in non-alcoholic steatohepatitis and contributes to the disease in experimental models by inhibiting PPARα expression. Gut. 2016;65:1882–1894. doi: 10.1136/gutjnl-2014-308883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Dattaroy D, Pourhoseini S, Das S, Alhasson F, Seth RK, Nagarkatti M, Michelotti GA, Diehl AM, Chatterjee S. Micro-RNA 21 inhibition of SMAD7 enhances fibrogenesis via leptin-mediated NADPH oxidase in experimental and human nonalcoholic steatohepatitis. Am J Physiol Gastrointest Liver Physiol. 2015;308:G298–G312. doi: 10.1152/ajpgi.00346.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wu H, Ng R, Chen X, Steer CJ, Song G. MicroRNA-21 is a potential link between non-alcoholic fatty liver disease and hepatocellular carcinoma via modulation of the HBP1-p53-Srebp1c pathway. Gut. 2016;65:1850–1860. doi: 10.1136/gutjnl-2014-308430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sun C, Huang F, Liu X, Xiao X, Yang M, Hu G, Liu H, Liao L. miR-21 regulates triglyceride and cholesterol metabolism in non-alcoholic fatty liver disease by targeting HMGCR. Int J Mol Med. 2015;35:847–853. doi: 10.3892/ijmm.2015.2076. [DOI] [PubMed] [Google Scholar]
- 138.Ahn J, Lee H, Jung CH, Ha T. Lycopene inhibits hepatic steatosis via microRNA-21-induced downregulation of fatty acid-binding protein 7 in mice fed a high-fat diet. Mol Nutr Food Res. 2012;56:1665–1674. doi: 10.1002/mnfr.201200182. [DOI] [PubMed] [Google Scholar]
- 139.Ng R, Wu H, Xiao H, Chen X, Willenbring H, Steer CJ, Song G. Inhibition of microRNA-24 expression in liver prevents hepatic lipid accumulation and hyperlipidemia. Hepatology. 2014;60:554–564. doi: 10.1002/hep.27153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.He Q, Li F, Li J, Li R, Zhan G, Li G, Du W, Tan H. MicroRNA-26a-interleukin (IL)-6-IL-17 axis regulates the development of non-alcoholic fatty liver disease in a murine model. Clin Exp Immunol. 2017;187:174–184. doi: 10.1111/cei.12838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Alisi A, Da Sacco L, Bruscalupi G, Piemonte F, Panera N, De Vito R, Leoni S, Bottazzo GF, Masotti A, Nobili V. Mirnome analysis reveals novel molecular determinants in the pathogenesis of diet-induced nonalcoholic fatty liver disease. Lab Invest. 2011;91:283–293. doi: 10.1038/labinvest.2010.166. [DOI] [PubMed] [Google Scholar]
- 142.Liu MX, Gao M, Li CZ, Yu CZ, Yan H, Peng C, Li Y, Li CG, Ma ZL, Zhao Y, et al. Dicer1/miR-29/HMGCR axis contributes to hepatic free cholesterol accumulation in mouse non-alcoholic steatohepatitis. Acta Pharmacol Sin. 2017;38:660–671. doi: 10.1038/aps.2016.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mattis AN, Song G, Hitchner K, Kim RY, Lee AY, Sharma AD, Malato Y, McManus MT, Esau CC, Koller E, et al. A screen in mice uncovers repression of lipoprotein lipase by microRNA-29a as a mechanism for lipid distribution away from the liver. Hepatology. 2015;61:141–152. doi: 10.1002/hep.27379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zarrinpar A, Gupta S, Maurya MR, Subramaniam S, Loomba R. Serum microRNAs explain discordance of non-alcoholic fatty liver disease in monozygotic and dizygotic twins: a prospective study. Gut. 2016;65:1546–1554. doi: 10.1136/gutjnl-2015-309456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Min HK, Kapoor A, Fuchs M, Mirshahi F, Zhou H, Maher J, Kellum J, Warnick R, Contos MJ, Sanyal AJ. Increased hepatic synthesis and dysregulation of cholesterol metabolism is associated with the severity of nonalcoholic fatty liver disease. Cell Metab. 2012;15:665–674. doi: 10.1016/j.cmet.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Xu Y, Zalzala M, Xu J, Li Y, Yin L, Zhang Y. A metabolic stress-inducible miR-34a-HNF4α pathway regulates lipid and lipoprotein metabolism. Nat Commun. 2015;6:7466. doi: 10.1038/ncomms8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Castro RE, Ferreira DM, Afonso MB, Borralho PM, Machado MV, Cortez-Pinto H, Rodrigues CM. miR-34a/SIRT1/p53 is suppressed by ursodeoxycholic acid in the rat liver and activated by disease severity in human non-alcoholic fatty liver disease. J Hepatol. 2013;58:119–125. doi: 10.1016/j.jhep.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 148.Derdak Z, Villegas KA, Harb R, Wu AM, Sousa A, Wands JR. Inhibition of p53 attenuates steatosis and liver injury in a mouse model of non-alcoholic fatty liver disease. J Hepatol. 2013;58:785–791. doi: 10.1016/j.jhep.2012.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Celikbilek M, Baskol M, Taheri S, Deniz K, Dogan S, Zararsiz G, Gursoy S, Guven K, Ozbakır O, Dundar M, et al. Circulating microRNAs in patients with non-alcoholic fatty liver disease. World J Hepatol. 2014;6:613–620. doi: 10.4254/wjh.v6.i8.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zhang N, Lei J, Lei H, Ruan X, Liu Q, Chen Y, Huang W. MicroRNA-101 overexpression by IL-6 and TNF-α inhibits cholesterol efflux by suppressing ATP-binding cassette transporter A1 expression. Exp Cell Res. 2015;336:33–42. doi: 10.1016/j.yexcr.2015.05.023. [DOI] [PubMed] [Google Scholar]
- 151.Xu Q, Li Y, Shang YF, Wang HL, Yao MX. miRNA-103: molecular link between insulin resistance and nonalcoholic fatty liver disease. World J Gastroenterol. 2015;21:511–516. doi: 10.3748/wjg.v21.i2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Soronen J, Yki-Järvinen H, Zhou Y, Sädevirta S, Sarin AP, Leivonen M, Sevastianova K, Perttilä J, Laurila PP, Sigruener A, et al. Novel hepatic microRNAs upregulated in human nonalcoholic fatty liver disease. Physiol Rep. 2016:4. doi: 10.14814/phy2.12661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Clarke JD, Sharapova T, Lake AD, Blomme E, Maher J, Cherrington NJ. Circulating microRNA 122 in the methionine and choline-deficient mouse model of non-alcoholic steatohepatitis. J Appl Toxicol. 2014;34:726–732. doi: 10.1002/jat.2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Takaki Y, Saito Y, Takasugi A, Toshimitsu K, Yamada S, Muramatsu T, Kimura M, Sugiyama K, Suzuki H, Arai E, et al. Silencing of microRNA-122 is an early event during hepatocarcinogenesis from non-alcoholic steatohepatitis. Cancer Sci. 2014;105:1254–1260. doi: 10.1111/cas.12498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Csak T, Bala S, Lippai D, Satishchandran A, Catalano D, Kodys K, Szabo G. microRNA-122 regulates hypoxia-inducible factor-1 and vimentin in hepatocytes and correlates with fibrosis in diet-induced steatohepatitis. Liver Int. 2015;35:532–541. doi: 10.1111/liv.12633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhang ZC, Liu Y, Xiao LL, Li SF, Jiang JH, Zhao Y, Qian SW, Tang QQ, Li X. Upregulation of miR-125b by estrogen protects against non-alcoholic fatty liver in female mice. J Hepatol. 2015;63:1466–1475. doi: 10.1016/j.jhep.2015.07.037. [DOI] [PubMed] [Google Scholar]
- 157.Li D, Wang X, Lan X, Li Y, Liu L, Yi J, Li J, Sun Q, Wang Y, Li H, et al. Down-regulation of miR-144 elicits proinflammatory cytokine production by targeting toll-like receptor 2 in nonalcoholic steatohepatitis of high-fat-diet-induced metabolic syndrome E3 rats. Mol Cell Endocrinol. 2015;402:1–12. doi: 10.1016/j.mce.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 158.Feng YY, Xu XQ, Ji CB, Shi CM, Guo XR, Fu JF. Aberrant hepatic microRNA expression in nonalcoholic fatty liver disease. Cell Physiol Biochem. 2014;34:1983–1997. doi: 10.1159/000366394. [DOI] [PubMed] [Google Scholar]
- 159.Jiang W, Liu J, Dai Y, Zhou N, Ji C, Li X. MiR-146b attenuates high-fat diet-induced non-alcoholic steatohepatitis in mice. J Gastroenterol Hepatol. 2015;30:933–943. doi: 10.1111/jgh.12878. [DOI] [PubMed] [Google Scholar]
- 160.Xiao J, Lv D, Zhao Y, Chen X, Song M, Liu J, Bei Y, Wang F, Yang W, Yang C. miR-149 controls non-alcoholic fatty liver by targeting FGF-21. J Cell Mol Med. 2016;20:1603–1608. doi: 10.1111/jcmm.12848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lin X, Jia J, Du T, Li W, Wang X, Wei J, Lin X, Zeng H, Yao L, Chen X, et al. Overexpression of miR-155 in the liver of transgenic mice alters the expression profiling of hepatic genes associated with lipid metabolism. PLoS One. 2015;10:e0118417. doi: 10.1371/journal.pone.0118417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Vella S, Gnani D, Crudele A, Ceccarelli S, De Stefanis C, Gaspari S, Nobili V, Locatelli F, Marquez VE, Rota R, et al. EZH2 down-regulation exacerbates lipid accumulation and inflammation in in vitro and in vivo NAFLD. Int J Mol Sci. 2013;14:24154–24168. doi: 10.3390/ijms141224154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Li B, Zhang Z, Zhang H, Quan K, Lu Y, Cai D, Ning G. Aberrant miR199a-5p/caveolin1/PPARα axis in hepatic steatosis. J Mol Endocrinol. 2014;53:393–403. doi: 10.1530/JME-14-0127. [DOI] [PubMed] [Google Scholar]
- 164.Shpyleva S, Pogribna M, Cozart C, Bryant MS, Muskhelishvili L, Tryndyak VP, Ross SA, Beland FA, Pogribny IP. Interstrain differences in the progression of nonalcoholic steatohepatitis to fibrosis in mice are associated with altered hepatic iron metabolism. J Nutr Biochem. 2014;25:1235–1242. doi: 10.1016/j.jnutbio.2014.06.012. [DOI] [PubMed] [Google Scholar]
- 165.Xiao J, Bei Y, Liu J, Dimitrova-Shumkovska J, Kuang D, Zhou Q, Li J, Yang Y, Xiang Y, Wang F, et al. miR-212 downregulation contributes to the protective effect of exercise against non-alcoholic fatty liver via targeting FGF-21. J Cell Mol Med. 2016;20:204–216. doi: 10.1111/jcmm.12733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Jin X, Chen YP, Kong M, Zheng L, Yang YD, Li YM. Transition from hepatic steatosis to steatohepatitis: unique microRNA patterns and potential downstream functions and pathways. J Gastroenterol Hepatol. 2012;27:331–340. doi: 10.1111/j.1440-1746.2011.06864.x. [DOI] [PubMed] [Google Scholar]
- 167.Hoekstra M, van der Sluis RJ, Kuiper J, Van Berkel TJ. Nonalcoholic fatty liver disease is associated with an altered hepatocyte microRNA profile in LDL receptor knockout mice. J Nutr Biochem. 2012;23:622–628. doi: 10.1016/j.jnutbio.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 168.Meng X, Guo J, Fang W, Dou L, Li M, Huang X, Zhou S, Man Y, Tang W, Yu L, et al. Liver MicroRNA-291b-3p Promotes Hepatic Lipogenesis through Negative Regulation of Adenosine 5’-Monophosphate (AMP)-activated Protein Kinase α1. J Biol Chem. 2016;291:10625–10634. doi: 10.1074/jbc.M116.713768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Cheng Y, Mai J, Hou T, Ping J. MicroRNA-421 induces hepatic mitochondrial dysfunction in non-alcoholic fatty liver disease mice by inhibiting sirtuin 3. Biochem Biophys Res Commun. 2016;474:57–63. doi: 10.1016/j.bbrc.2016.04.065. [DOI] [PubMed] [Google Scholar]
- 170.Hur W, Lee JH, Kim SW, Kim JH, Bae SH, Kim M, Hwang D, Kim YS, Park T, Um SJ, et al. Downregulation of microRNA-451 in non-alcoholic steatohepatitis inhibits fatty acid-induced proinflammatory cytokine production through the AMPK/AKT pathway. Int J Biochem Cell Biol. 2015;64:265–276. doi: 10.1016/j.biocel.2015.04.016. [DOI] [PubMed] [Google Scholar]
- 171.Ahn J, Lee H, Chung CH, Ha T. High fat diet induced downregulation of microRNA-467b increased lipoprotein lipase in hepatic steatosis. Biochem Biophys Res Commun. 2011;414:664–669. doi: 10.1016/j.bbrc.2011.09.120. [DOI] [PubMed] [Google Scholar]


