Abstract
Here we show that laboratory of genetics and physiology 2 (LGP2) virus sensor protein regulates gene expression network of endogenous genes mediated by TAR-RNA binding protein (TRBP)-bound microRNAs (miRNAs). TRBP is an enhancer of RNA silencing, and functions to recruit precursor-miRNAs (pre-miRNAs) to Dicer that processes pre-miRNA into mature miRNA. Viral infection activates the antiviral innate immune response in mammalian cells. Retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), including RIG-I, melanoma-differentiation-associated gene 5 (MDA5), and LGP2, function as cytoplasmic virus sensor proteins during viral infection. RIG-I and MDA5 can distinguish between different types of RNA viruses to produce antiviral cytokines, including type I interferon. However, the role of LGP2 is controversial. We found that LGP2 bound to the double-stranded RNA binding sites of TRBP, resulting in inhibition of pre-miRNA binding and recruitment by TRBP. Furthermore, although it is unclear whether TRBP binds to specific pre-miRNA, we found that TRBP bound to particular pre-miRNAs with common structural characteristics. Thus, LGP2 represses specific miRNA activities by interacting with TRBP, resulting in selective regulation of target genes. Our findings show that a novel function of LGP2 is to modulate RNA silencing, indicating the crosstalk between RNA silencing and RLR signaling in mammalian cells.
INTRODUCTION
RNA silencing is a conserved mechanism from plants to mammals, and regulates gene expression through post-transcriptional regulation and/or translational repression directed by microRNAs (miRNAs), a class of approximately 21–22-nucleotide (nt)-long non-coding RNAs. The human genome encodes ∼2000 miRNAs that target multiple genes to regulate endogenous gene expression (1). Primary miRNAs (pri-miRNAs) are transcribed from the genome and processed into precursor-miRNAs (pre-miRNAs) by Drosha, a dsRNA cleaving enzyme found in the nucleus. From the nucleus, pre-miRNAs are exported to the cytoplasm by exportin-5 (2,3) and further processed by Dicer, another dsRNA cleaving enzyme, to yield dsRNA with 2-nt 3′-overhangs (4). miRNA duplexes are unwound into single-stranded miRNAs on Argonaute (AGO), which is found in the RNA-induced silencing complex (RISC). Once unwound, the single-stranded miRNAs, which are the guide strands, function to repress target RNAs with sequence complementarity (4–6). The trans-activation-responsive region (TAR)-RNA binding protein (TRBP) functions to recruit pre-miRNA into Dicer, which processes the pre-miRNA (7,8). TRBP was originally identified as a protein that interacted with human immunodeficiency virus (HIV)-1 TAR RNA (9) and was later demonstrated to regulate IFN-induced protein kinase R (PKR), a kinase involved in the cellular response to viral infection (10,11).
During RNA virus infection, viral dsRNAs are recognized by host pattern recognition receptors (PRRs) in mammalian cells (12,13). One of the Toll-like receptor family proteins (TLRs), TLR3 (14), and three retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), RIG-I (also known as DDX58), melanoma-differentiation-associated gene 5 (MDA5 or IFIH1), and laboratory of genetics and physiology 2 (LGP2 or DHX58) (15,16), serve as RNA virus receptors in mammalian cells. RIG-I is activated by 5′-triphosphate- or 5′-diphosphate-containing RNA and small RNA duplexes (17–21), and MDA5 is activated by long RNA duplexes (22). Both RIG-I and MDA5 have caspase recruitment domains (CARDs), which are essential for signal transfer to downstream molecules (12,13). Although LGP2 is an RLR, it does not have a CARD, rendering it incapable of transferring signals downstream (12,13). Thus, its function has remained unclear. Both TLR3 and RLRs are able to induce the transcription of antiviral cytokines, including type I interferons (IFNs), in response to viral dsRNAs, although their respective signal transduction pathways differ. TLR3 is localized in the endosomal membrane and activates TANK-binding kinase 1 (TBK1) and inducible IκB kinase (IKKi or IKKϵ) through Toll-interleukin (IL)-1-resistance (TIR) domain-containing adaptor-inducing IFN-β (TRIF) (23). RLRs are cytoplasmic DExD/H box-containing RNA helicases, which recognize extrinsic viral ribonucleic acids (12,15) and induce type I IFNs through IFN-β-promoter stimulator 1 (IPS-1; or MAVS, VISA or Cardif) on the mitochondrial outer membrane (24–26). The secreted IFNs transmit secondary signals via type I IFN receptors, and activate the expression of hundreds of IFN-stimulated genes (ISGs), including TLR3 and RLRs, establishing a potent antiviral state.
Both RNA silencing and RLR signaling are triggered by dsRNA in the cytoplasm. However, the relationship between these two pathways is not well understood in mammalian cells (27–30). In this study, we show for the first time that LGP2, an RLR with previously unknown function, regulates miRNA-mediated gene expression of endogenous genes by interacting with the RNA silencing enhancer, TRBP. Our results revealed a primary function of LGP2, a cytoplasmic viral sensor protein. Furthermore, we showed that crosstalk between RNA silencing and RLR signaling occurs in mammalian cells.
MATERIALS AND METHODS
Cell culture
Human HeLa WT, LGP2−/− or TRBP−/− cells were cultured in Dulbecco's Modified Eagle's Medium (Wako) containing 10% fetal bovine serum (Gibco) at 37°C with 5% CO2. Flp-In 293 TRBP cells were cultured in Dulbecco's Modified Eagle's Medium (Wako) containing 10% Tet system approved FBS (Clontech) and 10 ng/ml hygromycin at 37°C with 5% CO2.
Plasmid construction
The expression plasmids of C-terminal Myc-tagged TRBP (TRBP-Myc), dsRBDmt1, dsRBDmt2, and dsRBDmt1+2 were constructed as described previously (31). Plasmids expressing various parts of TRBP (L1+D1+L2, L2+ D2+L3, D1, D2) were generated by amplification of each fragment by PCR with primers containing the restriction enzyme sites of HindIII or NotI using the KOD-Plus-Mutagenesis Kit (TOYOBO). The amplified fragments were digested with HindIII and NotI and cloned into the pcDNA3.1 vector (Invitrogen) digested with the same enzymes. The expression plasmid of N-terminal FLAG-tagged TRBP (FLAG-TRBP) was amplified from TRBP-Myc expression plasmid by PCR with primers containing the restriction enzyme sites of NheI or HindIII. The amplified fragment was digested with NheI and HindIII and cloned into pcDNA5 vector (Invitrogen) digested with the same enzymes. The expression plasmids of pre-miRNA like RNA targeting firefly luciferase, pre-miLuc (pSilencer-FL774) was constructed as described previously (32). Plasmids encoding the LGP2 or deletion mutants (WT, ΔCTD, CTD) were kindly provided from Dr T. Fujita (33) and plasmids encoding miRNA inhibitor (Tough Decoy, TuD) was kindly provided form Dr H. Iba and Dr T. Haraguchi (34). The primer sequences are shown in Supplementary Table S1.
Immunoprecipitation
A HeLa cell suspension (1.2 × 106 cells/dish) was plated into a 6-cm dish prior to transfection 1 day before transfection. Cells were transfected with each combination of the plasmids. Cells were washed with PBS and lysed in cold lysis buffer (10 mM Hepes-NaOH [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 140 mM NaCl, 1 mM EDTA, 1 mM Na3VO4, 10 mM NaF, 0.5% NP-40 and complete protease inhibitor) 24 h following transfection. RNase V1 (0.3 U/ml; Ambion) was added to the lysis buffer to remove dsRNAs. For immunoprecipitation, the cell lysates were mixed with 30 μl of Protein G Sepharose 4B (Sigma) and rotated at 4°C for 2 h with 2.5 μg of mouse anti-FLAG antibody (Sigma), anti-Myc (Calbiochem), or 2.5 μg of mouse IgG (Santa Cruz Biotechnology) as a negative control. The cell lysates were then added to antibody-bound protein G Sepharose and rotated at 4°C for 2 h. The beads were washed twice with wash buffer containing 300 mM NaCl and once with lysis buffer. To elute the bound proteins, 2 × SDS-PAGE sample buffer (30 μl) was added and the beads were boiled for 5 min.
Western blot
The samples were separated by SDS-PAGE and transferred to a polyvinylidene fluoride membrane using the Trans-Blot Turbo Transfer System (Bio-Rad). The membrane was blocked for 1 h in Tris-buffered saline-Triton X-100 or Tween 20 (TBS-T; 20 mM Tris–HCl [pH 7.5], 150 mM NaCl, 0.2% Triton X-100 or 0.1% Tween) supplemented with 5% Difco skim milk (Becton, Dickinson and Company), and incubated with specific antibodies in Can Get Signal immunoreaction enhancer solution (TOYOBO) at 4°C overnight. Anti-TRBP (AbFrontier), anti-FLAG or anti-Myc (Cell Signaling), and anti-α-tubulin antibodies (ICN/CAPPEL Biomedicals) were used. Anti-human RIG-I, MDA5, LGP2 and Dicer antibodies were generated by immunizing rabbits with a synthetic peptide (4,35). The membranes were washed three times with TBS-T and reacted with HRP-linked anti-rabbit or anti-mouse antibody (GE Healthcare) at room temperature for 1 h. After being washed three times with TBS-T, the membrane was incubated with ECL Prime Western Blotting Detection Reagent (GE Healthcare) and visualized with ImageQuant LAS4000 mini (GE Healthcare).
Northern blot
A HeLa cell suspension (1.2 × 106 cells/dish) was plated into a 6-cm dish with or without human type I IFN (IFN-α1; Cell Signaling). RNA extraction was performed by ISOGEN (NIPPON GENE). Ten micrograms of total RNA are loaded on 10% denatured polyacrylamide gel containing 8 M Urea and transferred to Hybond-N+ membrane (Amersham) for 25 V, 45 min. The membrane was hybridized with 32P-labeled DNA probe overnight following crosslink and pre-hybridization treatment. The membranes were washed three times and visualized with Typhoon FLA 9500 (GE Healthcare). The probe sequences are shown in Supplementary Table S2.
RNA silencing activity assay for firefly luciferase gene
RNA silencing activity was measured with a luciferase reporter assay. A HeLa cell suspension (1.0 × 105 cells/ml) was inoculated in 24-well plates with or without IFN 1 day before transfection. Cells were transfected with 0.5 μg of pGL3-Control vector (Promega) encoding the firefly luciferase gene, 0.1 μg of pRL-SV40 vector (Promega) encoding the Renilla luciferase gene, 5 ng of pSilencer-3.1-H1-puro vector encoding pre-miLuc against the firefly luciferase with Lipofectamine 2000 reagent (Invitrogen). The transfected cells were lysed with 1 × passive lysis buffer (Promega) 24 h following transfection. Luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega), and the firefly luciferase activity normalized to Renilla luciferase (firefly luciferase activity/Renilla luciferase activity) was determined.
Quantitative RT-PCR
Total RNA was extracted by RNeasy mini kit (QIAGEN) and treated with DNase I. A half μg of the total RNA was used for cDNA synthesis by Transcriptor High Fidelity cDNA kit (Roche). Quantitative RT-PCR was performed by KAPA SYBR Fast qPCR Master Mix ABI Prism (Kapa Biosystems) by StepOnePlus Real-Time PCR system (Applied Biosystems). The primer sequences are shown in Supplementary Table S3.
RT-PCR
Total RNA was extracted by FastGene RNA Premium kit (Nippon Genetics). A half μg of the total RNA was used for cDNA synthesis of mature miRNA with miRNA-specific RT-primer by Transcriptor High Fidelity cDNA kit. RT-PCR was performed by AmpliTaq Gold PCR Master Mix 2x (Applied Biosystems). The primer sequences are shown in Supplementary Table S3.
Generation of LGP2, and TRBP knockout cell lines by CRISPR genome editing
Single guide RNAs (sgRNA) for cleavage and non-homologous end joining were designed for dsRBD1 in exon 2 of TRBP and CARD in exon 1 of LGP2 by CRISPRdirect (36). Exon2 of TRBP is a common region in the transcription variants 1 and 2 of TRBP. The oligonucleotide sequences were digested with BbsI and cloned into the pSUPER-CRISPR guide RNA vector (37). The sequences are shown in Supplementary Table S4. A HeLa cell suspension (6.0 × 105 cells/ml) was inoculated in a 6-well plate 1 day prior to transfection. Cells were transfected with the pSUPER-CRISPR guide RNA vector (2 μg) and plasmid encoding the Cas9 protein (2 μg) (Addgene #41815). Puromycin selection was performed 30 h following transfection and limiting dilution was performed twice for the isolation of knockout cells. Sequence analysis of genomic DNA and western blot analysis were performed to confirm knocked out of the targeted gene.
Generation of doxycycline-inducible FLAG-tagged TRBP expression stable cell line
Flp-In 293 Host Cell Line was purchased from Invitrogen. The pcDNA5/FRT expression plasmid containing N-terminal FLAG-tagged TRBP was co-transfected with pOG44 which express Flp recombinase into the Flp-In 293 Host Cell Line. Cell selection was performed with 100 μg/ml hygromycin 48 hours after the transfection. The expression of FLAG-TRBP protein was confirmed by western blot with anti-FLAG antibody.
RNA sequencing analysis
Flp-In 293 TRBP expression cell line suspension was plated into a 9-cm dish with 1 μg/ml doxycycline and immunoprecipitated by anti-FLAG antibody. Extracted RNAs were measured by Qubit2.0 Fluorometer (Life Technologies) and the qualities were confirmed by Bioanalyzer (Agilent). Ribosomal RNA was removed by Ribo-Zero rRNA Removal kit (AR BROWN). RNA-seq was carried out using cDNA libraries prepared from small RNA fraction (15–100 nt) by illumina Truseq small RNA kit and sequenced 36 nt by HiSeq2500 in single-end mode.
All of the 36 nt reads were mapped to the RefSeq sequences (GRCh38) using Bowtie 1.1.2 allowing for 2 nt mismatches and annotated according to the miRNA database (miRBase release 21). After counting raw reads using HTseq (38), reads corresponding to pre-miRNAs were normalized by RPM (Reads per million mapped reads). Enrichment was calculated by the following formula: (IPRPM + 1)/(inputRPM + 1). Base-pairing probability of each pre-miRNA was calculated by CentroidFold (39) using pre-miRNA sequences. In miRBase, pre-miRNAs are registered with additional sequences at the 5′- or 3′-end on actual pre-miRNA sequence. Then pre-miRNA sequences were prepared by cleaving off the additional sequence at the 5′- or 3′-end, based on registered mature miRNA sequence. The pre-miRNAs whose sequence reads were zero in input RNAs or immunoprecipitated RNAs were excluded for the calculation of sequence read enrichment by immunoprecipitation.
Microarray analysis
The qualities of total RNA extracted from HeLa WT or LGP2−/−cells were confirmed by Bioanalyzer (Agilent Technologies), and cDNA and Cy-3 labeled RNA were synthesized by Quick Amp Labeling kit for One color (Agilent Technologies). Cy-3 labeled RNAs were fragmented by Gene Expression Hybridization kit (Agilent Technologies) and hybridized to SurePrint G3 Human GE microarray version 3 (Agilent Technologies) at 65°C for 17 h. After being washed, microarray slide was scanned by DNA Microarray Scanner (Agilent Technologies) and quantified signals by Feature Extraction 10.5.1.1 (Agilent Technologies). Data analysis was performed on probes passed filters.
RESULTS
LGP2 interacts with TRBP, an RNA silencing enhancer
The core factor of RNA silencing (Dicer) and the RLR cytoplasmic viral sensor proteins (LGP2, RIG-I and MDA5) are members of the ATPase/helicase family proteins (Figure 1A). The Protein-BLAST Expect (E)-value is a parameter that describes the number of hits that can be expected by chance when searching a database. E-values showed that the amino acid sequences of ATPase/helicase domains are very similar; in particular, those of Dicer and LGP2 showed strong similarity (Figure 1A). Dicer interacts with the RNA silencing enhancer, TRBP, via its ATPase/helicase domain to receive pre-miRNA to process into mature miRNA (40). Such notable similarity between Dicer and LGP2 led us to examine whether LGP2 could interact with TRBP.
Figure 1.
Laboratory of genetics and physiology 2 (LGP2) interacts with the RNA silencing enhancer, trans-activation-responsive region-RNA binding protein (TRBP). (A) The domain architecture of TRBP, Dicer, LGP2, retinoic acid-inducible gene I (RIG-I), and melanoma-differentiation-associated gene 5 (MDA5) proteins. E-values were calculated by Protein-BLAST compared to the ATPase/helicase domain of Dicer as a basis. (B) Immunoprecipitation of LGP2 with TRBP. Plasmids encoding FLAG-TRBP or FLAG-EGFP were transfected into interferon (IFN)-non-treated or IFN-treated HeLa cells and immunoprecipitation was performed with anti-FLAG antibody. Endogenous LGP2, RIG-I, MDA5 and Dicer were detected with specific antibodies. (C) Indirect interaction between LGP2 and Dicer detected by immunoprecipitation. Empty vector, or plasmids encoding FLAG-LGP2 were transfected into HeLa wild-type (WT) or TRBP−/−cells and immunoprecipitation was performed with anti-FLAG antibody. Endogenous TRBP, Dicer and actin were detected with specific antibodies.
We examined the interaction between TRBP and LGP2 by immunoprecipitation analysis using HeLa cells in the presence or absence of IFN (Figure 1B), which is known to increase the protein levels of RIG-I, MDA5, and LGP2 (Supplementary Figure S1). The protein levels of RLRs were certainly increased, while little or no changes were observed in the protein levels of AGO2, Dicer, TRBP, and tubulin (control) (Supplementary Figure S1). FLAG-tagged expression plasmids encoding TRBP or EGFP (negative control) were transfected into cells with or without IFN treatment, and immunoprecipitation was performed with anti-FLAG antibody. Dicer is known to interact with TRBP (40), and it was clearly immunoprecipitated with TRBP regardless of whether cells were treated with IFN (Figure 1B). Endogenous LGP2 protein was specifically co-immunoprecipitated with TRBP with or without IFN treatment, although the amount of immunoprecipitated LGP2 protein was obviously upregulated by IFN treatment (Figure 1B). However, RIG-I and MDA5 were not co-immunoprecipitated. Conversely, immunoprecipitation of FLAG-LGP2 revealed that LGP2 interacted with both TRBP and Dicer (Figure 1C). To evaluate if the LGP2-Dicer interaction occurs through direct or indirect interaction with TRBP, we performed an immunoprecipitation assay using TRBP knockout (TRBP−/−) cells that were generated using the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) genome engineering method (Supplementary Figure S2) (41). The results showed that TRBP was indispensable for LGP2-Dicer interaction (Figure 1C), indicating that the LGP2-Dicer interaction is indirect and occurs via TRBP.
LGP2 interacts with TRBP via its ATPase/helicase domain
To determine the region of LGP2 that interacts with TRBP, plasmids encoding FLAG-tagged full length wild-type LGP2 (WT LGP2), its helicase region (ΔC-terminal domain (CTD)) or its CTD region (Figure 2A) were transfected into HeLa cells expressing Myc-TRBP, and immunoprecipitation was performed with anti-Myc antibody (Figure 2B). Although the ΔCTD region, as well as WT LGP2, were unambiguously immunoprecipitated with WT TRBP (TRBP-WT), the CTD region was not, indicating that LGP2 interacts with TRBP via its ATPase/helicase domain (Figure 2B).
Figure 2.
LGP2 interacts with TRBP via its ATPase/helicase domain. (A) Domain architecture of LGP2-WT protein and the generated mutant constructs. (B) Immunoprecipitation of LGP2-WT protein and its mutants with the TRBP-WT protein. The anti-Myc antibody was used for immunoprecipitation of TRBP, and LGP2 was detected with the anti-FLAG antibody.
LGP2 interacts with TRBP via its dsRNA binding sites
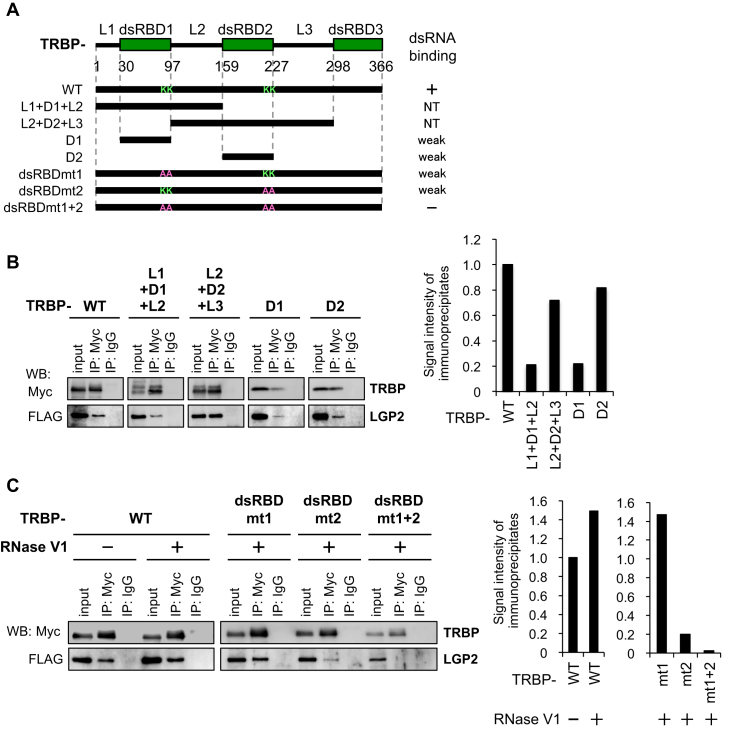
TRBP consists of three dsRNA-binding domains (dsRBDs) (Figure 3A) (7,8,31,42). Among them, the N-terminal domains dsRBD1 and dsRBD2 bind dsRNA or small interfering RNA (siRNA) (31,43), and the C-terminal domain dsRBD3 is necessary for interaction with Dicer (40). To identify the region of TRBP necessary for interaction with LGP2, we constructed several Myc-tagged plasmids expressing different TRBP domain(s): linker region 1 (L1)+dsRBD1 (D1)+L2, L2+D2+L3, D1 or D2 (Figure 3A). Each plasmid was transfected into HeLa cells expressing FLAG-LGP2 and immunoprecipitated with anti-Myc antibody. A significant amount of LGP2 was immunoprecipitated with TRBP-WT and L2+D2+L3, but only a small amount of LGP2 was detected with L1+D1+L2 (Figure 3B). Furthermore, a greater amount of LGP2 was immunoprecipitated with D2 than D1, indicating that dsRBD2 and dsRBD1 function as major and minor sites of interaction with LGP2, respectively.
Figure 3.
LGP2 interacts with the dsRNA-binding site of TRBP. (A) Domain architecture of TRBP-WT and its mutant proteins. The lysines (K) at positions 80 and 81 in dsRBD1 were substituted with alanines (A) in the TRBP-dsRBDmt1 and TRBP-dsRBDmt1+2 mutants, and the lysines at positions 210 and 211 in dsRBD2 were substituted with alanines in the TRBP-dsRBDmt2 and TRBP-dsRBDmt1+2 mutants. The previously reported dsRNA binding affinities are summarized on the right. (B) Immunoprecipitation of LGP2 with TRBP-WT protein and its mutants. The anti-Myc antibody was used for immunoprecipitation of TRBP, and LGP2 was detected with anti-FLAG antibody. The bar graph shows the quantified signal intensities of the immunoprecipitates. (C) Immunoprecipitation of LGP2 with TRBP-WT protein and its mutants. RNase V1 was added to the immunoprecipitation buffer to remove dsRNA. The bar graph shows the quantified signal intensities of the immunoprecipitates.
We previously reported that the active sites of TRBP that are required for siRNA binding contain two sets of two lysine residues (KK) located in dsRBD1 and dsRBD2 (31). A double KK TRBP mutant, in which both dsRBD1 and dsRBD2 were mutated (dsRBDmt1+2), could not bind siRNA, but each of the single KK mutants (dsRBDmt1 or dsRBDmt2) showed weak binding, and dsRBD2 had higher affinity to siRNA than dsRBD1 (31). Immunoprecipitation assays using the KK mutants in the presence or absence of RNase V1, a non-sequence specific dsRNA nuclease, were performed to examine if dsRNA is necessary for the interaction between TRBP and LGP2 (Figure 3C). TRBP-WT was evidently immunoprecipitated with LGP2 in the presence and absence of RNase V1 treatment (Figure 3C, left panel), but the amount of LGP2 immunoprecipitated with TRBP was increased by RNase V1 treatment, indicating that LGP2-TRBP interaction is disrupted by dsRNA (Figure 3C, left panel). Furthermore, little or no interaction between LGP2 and dsRBDmt1+2 was observed, while dsRBDmt1 interacted with LGP2 strongly and dsRBDmt2 interacted weakly (Figure 3C, right panel). Thus, the lysine residues in dsRBD2 and dsRBD1 function as the major and minor sites for LGP2 binding, respectively. Furthermore, the extent of their contributions to LGP2 binding is consistent with their affinities to siRNA, suggesting that LGP2 binds to TRBP to replace dsRNA bound on the same active site.
LGP2 represses pre-miRNA recruitment by TRBP to inhibit miRNA maturation
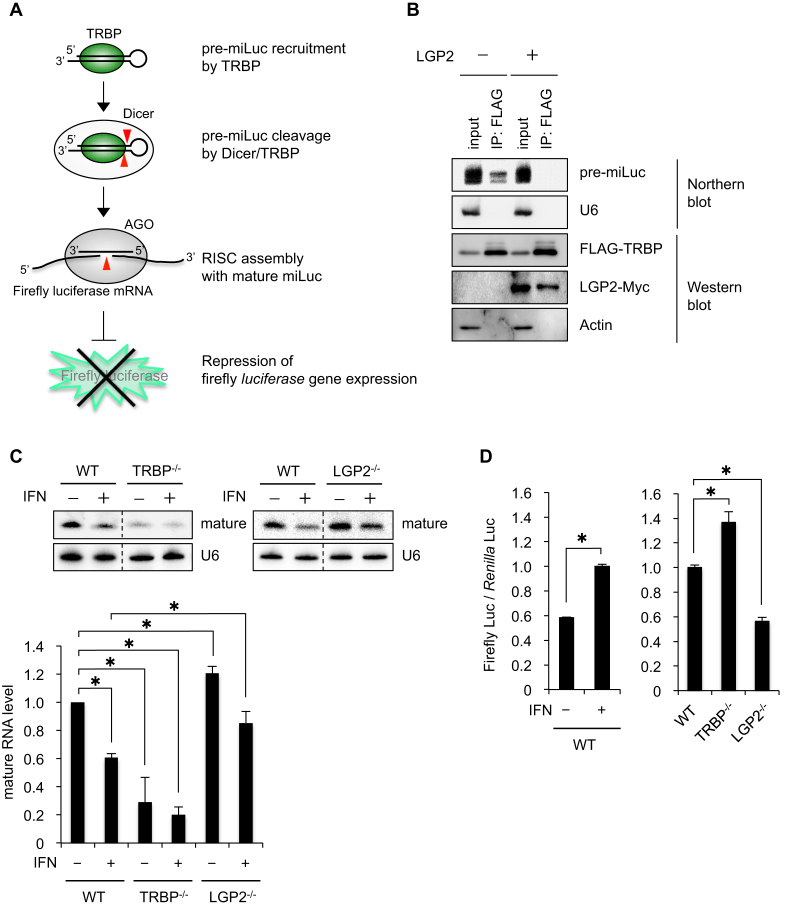
TRBP binds to pre-miRNA to recruit it into Dicer, to promote miRNA maturation (Figure 4A) (7,8). To determine if the interaction between TRBP and LGP2 inhibits pre-miRNA binding, the amount of pre-miRNA bound to TRBP was determined with or without overexpression of LGP2 (Figure 4B). Plasmids encoding stem-loop-structured RNA resembling pre-miRNA targeting firefly luciferase (pre-miLuc) and FLAG-TRBP were co-transfected into HeLa cells. FLAG-TRBP was then immunoprecipitated with anti-FLAG antibody. Pre-miLuc RNA that immunoprecipitated with TRBP was clearly detected in the absence of LGP2. However, little or no pre-miLuc RNA was observed when LGP2 was overexpressed, suggesting that pre-miRNA binding to TRBP is efficiently disturbed by LGP2 expression through competition for the dsRNA-binding sites of TRBP.
Figure 4.
LGP2 represses pre-miRNA recruitment by TRBP to inhibit microRNA (miRNA) maturation. (A) Schematic diagram of the pre-miRNA-mediated RNA silencing pathway. TRBP recruits pre-miRNA into Dicer to promote miRNA maturation. The mature miRNA is loaded on Argonaute (AGO) in the RNA-induced silencing complex (RISC), and represses target RNA with sequence complementarity. (B) Immunoprecipitation of TRBP and Northern blot of pre-miRNA targeting firefly luciferase (pre-miLuc). Plasmids encoding FLAG-TRBP, pre-miLuc, and LGP2-Myc were co-transfected to HeLa cells and immunoprecipitation was performed with anti-FLAG-antibody. Total RNA purified from each immunoprecipitate was subjected to Northern blot analysis to detect pre-miLuc. U6 was used as a control. (C) Northern blots of mature miLuc in WT, TRBP−/−, or LGP2−/− cells with or without IFN. Plasmids encoding pre-miLuc were transfected into WT, TRBP−/− or LGP2−/− cells in the absence and presence of IFN. The bar graph shows the quantified signal intensities of mature miLuc relative to the levels in HeLa-WT cells without IFN (mature RNA level of 1). The experiments were performed in triplicate and P-values were determined by Student's t-test (*P < 0.05). (D) RNA silencing activity assay using dual luciferase reporter assay in WT, TRBP−/− or LGP2−/− cells. Plasmids encoding pre-miLuc, firefly luciferase, and Renilla luciferase were co-transfected into WT, TRBP−/− or LGP2−/− cells, and relative luciferase activity was measured. The experiments were performed in triplicate and P-values were determined by Student's t-test (*P < 0.05).
To further examine whether miRNA maturation is inhibited when pre-miRNA recruitment by TRBP is disturbed by LGP2, northern blot analysis was performed using TRBP or LGP2 knockout (TRBP−/− or LGP2−/−) cells, which were generated with the CRISPR/Cas system (Figure 4C, Supplementary Figures S2 and S3). Plasmids encoding pre-miLuc were transfected into WT, TRBP−/− or LGP2−/− cells and Northern blotting was performed to detect mature miLuc RNA. The amount of mature miLuc RNA was greatly decreased in TRBP−/- cells, and IFN treatment also decreased the amount of mature miLuc RNA in WT cells, consistent with previous reports (7,8,44). In contrast, the amount of mature miLuc RNA was increased in LGP2−/− cells compared with WT cells, both in the presence and absence of IFN. These results suggest that the repression of miRNA maturation is mediated by LGP2, and essentially depends on its expression level. That is, LGP2 is upregulated by IFN treatment, leading to suppression of miLuc maturation by inhibiting TRBP function.
Next, the RNA silencing activities of mature miLuc, which is processed from pre-miLuc, were examined by luciferase reporter assay (Figure 4D). Plasmids encoding the firefly luciferase gene and Renilla luciferase gene were co-transfected into WT, TRBP−/− or LGP2−/− cells expressing pre-miLuc expression plasmids and the firefly luciferase activity normalized to Renilla luciferase activity (firefly luciferase activity/Renilla luciferase activity) was determined. IFN treatment in WT and TRBP−/− cells repressed RNA silencing, consistent with a decrease in the level of mature miLuc (Figure 4C). In addition, the RNA silencing activity in LGP2−/− cells was enhanced, consistent with an increase in the level of mature miLuc (Figure 4C). These results suggest that LGP2 disturbs pre-miRNA recruitment by TRBP, resulting in the repression of miRNA maturation followed by a reduction in RNA silencing activity.
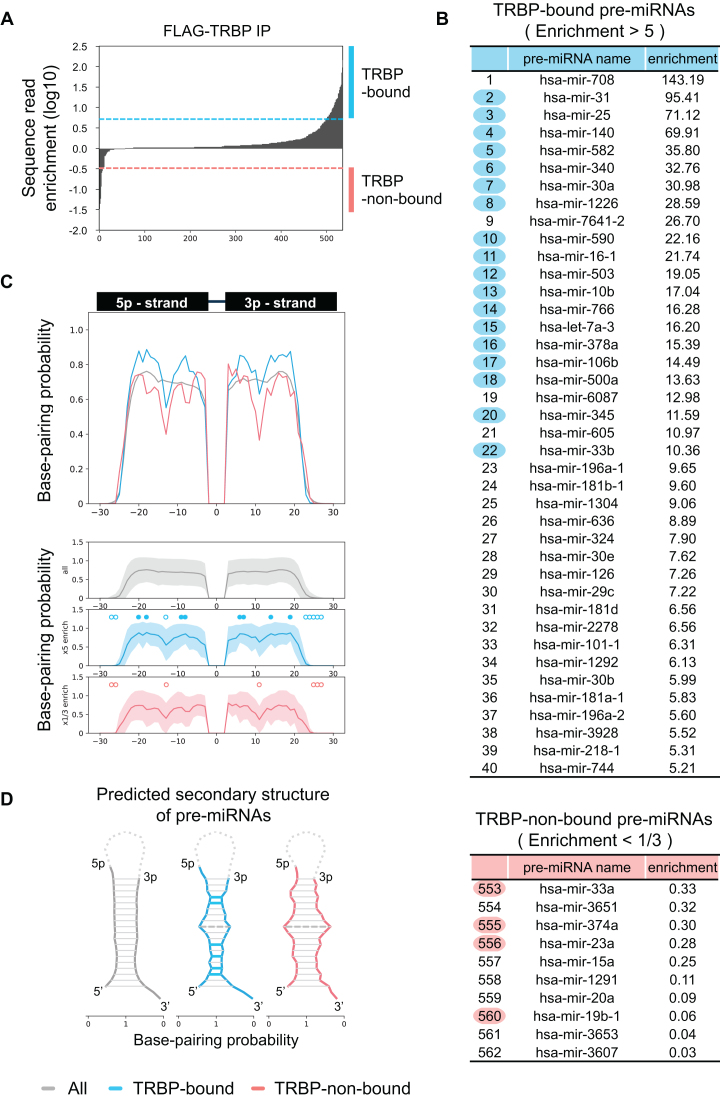
TRBP has a binding preference for specific types of pre-miRNAs with tight base pairing
TRBP is a dsRNA-binding protein that has no specific sequence preference for siRNA with perfect complementarity in base pairing; however, it is not able to bind single-stranded RNA (31). Such binding properties suggests that TRBP may have a binding preference to specific types of endogenous miRNAs, because endogenous miRNA duplexes have a wide variety of secondary structures other than perfect base-pairing, including bulges and mismatches that are composed of single-stranded RNA. To investigate if TRBP binds to particular types of pre-miRNAs, we performed RNA sequencing analysis of pre-miRNAs bound to TRBP. We established doxycycline-inducible FLAG-tagged TRBP expressing stable cell lines (Flp-In 293 TRBP cells) and performed immunoprecipitation with anti-FLAG antibody to collect TRBP-bound pre-miRNA (Supplementary Figure S4). The binding preference was analyzed using TRBP-bound and -non-bound pre-miRNA, classified by the enrichment of the number of sequence reads compared with input RNA before immunoprecipitation. In this study, pre-miRNA that preferably bound to TRBP were defined as pre-miRNA with sequence read enrichment >5, and those not bound to TRBP were defined as pre-miRNA with sequence read enrichment <1/3 (Figure 5A). Using these values, we identified 40 TRBP-bound and 10 TRBP-non-bound pre-miRNAs (Figure 5B). To identify the specific structural characteristics of TRBP-bound and -non-bound pre-miRNAs, we calculated the base-pairing probability (BPP), which provides the probability for each base-pair with respect to an ensemble of RNA secondary structures for predicting accurate RNA secondary structure. The mean BPP values at stem regions of all pre-miRNAs, TRBP-bound and non-bound pre-miRNAs were calculated using the max values of fluctuating BPPs at each nucleotide, calculated by CentroidFold (39) (Figure 5C). The BPP values in the stem regions of both 5p- and 3p-strands of TRBP-bound pre-miRNAs were high compared to all pre-miRNAs, except for the central regions, but those of TRBP-non-bound pre-miRNAs had a tendency to be low. These results are reflected in the structures of pre-miRNAs (Figure 5D). As shown in Figure 5D, the nucleotides with high BPP form strong base-pairing, but those with low BPP form weak base-pairing. These results clearly distinguished between pre-miRNA that preferably bound to TRBP and pre-miRNA not bound to TRBP in the context of miRNA secondary structures. The stem region of TRBP-bound pre-miRNA had tight base-pairing, while that of TRBP-non-bound pre-miRNA had weak base-pairing.
Figure 5.
TRBP has a binding preference for specific miRNAs with stems showing tight base-pairing. (A) Enrichment analysis of TRBP-bound pre-miRNAs. Immunoprecipitation was performed with anti-FLAG antibody using Flp-In 293 FLAG-TRBP stable cells, and TRBP-bound RNAs were purified from the immunoprecipitates. RNA sequencing was performed using the small RNA fraction (15–110 nucleotides, nts). Sequence read enrichment by immunoprecipitation was calculated and compared with input RNA not subjected to immunoprecipitation. A total of 562 pre-miRNAs were immunoprecipitated with TRBP. (B) The top 40 TRBP-bound pre-miRNAs and bottom 10 TRBP-non-bound pre-miRNAs. Blue or pink circles indicate pre-miRNAs used for expression analysis of target transcripts on Figure 6C. (C) Base-pairing probability (BPP) values of all pre-miRNAs (gray), the top 40 TRBP-bound pre-miRNAs (blue, sequence read enrichment by immunoprecipitation > 5) and bottom 10 TRBP-non-bound miRNAs (pink, sequence read enrichment by immunoprecipitation < 1/3) were calculated using the max value of base-pairing calculated by CentroidFold. Filled or open circles indicate the nt region that is significantly high and low, respectively, by Student's t-test (*P < 0.05). The nt positions −20, −18, −9, −8, +6, +7, +14 and +19 in TRBP-bound pre-miRNAs showed significantly high BPP values, and positions −27, −26, −13, +23, +24, +25, +26, and +27 showed significantly low BPP values compared to all pre-miRNAs. The nt positions −27, −26, −13, +11, +25, +26 and +27 in TRBP-non-bound pre-miRNAs showed significantly low BPP values compared to all pre-miRNAs. The 3′-terminal nt of 5p miRNA is nt position ‘−3′ and the 5′-terminal nt of 3p miRNA is nt position ‘3’. In the lower graphs, the light gray, blue, and pink areas above and below the solid lines show the standard deviations. (D) Predicted secondary structures of all pre-miRNA (gray), TRBP-bound pre-miRNA (blue), and TRBP-non-bound pre-miRNA (pink) based on BPP values. The horizontal lines between 5p and 3p miRNAs represent base-pairing, and solid or dotted colored lines indicate the positions that show significantly high and low BPP values, respectively, by Student's t-test (*P < 0.05).
LGP2 regulates miRNA-mediated gene expression of endogenous genes in human cells
To evaluate if LGP2 regulates miRNA-mediated gene expression of endogenous genes, we analyzed the conversion of gene expression profiles in WT and LGP2−/− HeLa cells by microarray analysis. Almost no remarkable changes were observed in the global gene expression profiles between WT and LGP2−/− cells (Figure 6A). Next, we analyzed the expression profiles of target transcripts predicted to be repressed by mature miRNAs derived from TRBP-bound pre-miRNAs, which are shown to be expressed in HeLa cells (45) and their mature sequences are registered in miRBase Release 21 (1). The target genes of each miRNA were predicted using TargetScan (46) and the top 100 high-score genes were used as the targets. MA plots, showing the mean log2 of signal intensities in LGP2−/− cells relative to those in WT cells (M value) and the averaged log10 signal intensities in WT and LGP2−/− cells (A value), and a cumulative distribution of the predicted target transcripts of one of the TRBP-bound miRNAs, miR-106b, were shown (Figure 6B). They revealed that the predicted targets were obviously downregulated in LGP2−/− cells compared with WT cells, while the predicted targets of miR-19b-1, a TRBP-non-bound miRNA, were upregulated (Figure 6B). The differences in the integral values of cumulative distribution between all transcripts and predicted targets were calculated on all of the predicted targets of high-ranked (sequence read enrichment by immunoprecipitation > 10) TRBP-bound pre-miRNAs, including miR-106b, and low-ranked (sequence read enrichment by immunoprecipitation < 1/3) TRBP-non-bound pre-miRNAs including miR-19b-1 (Figure 6C). The results showed that the predicted targets of TRBP-bound miRNAs had a strong tendency to be downregulated in LGP2−/− cells compared with WT cells, indicating that the RNA silencing activities of TRBP-bound miRNAs are enhanced in the absence of LGP2. Among TRBP-bound miRNAs, miR-106b had the strongest effect on downregulating the predicted targets, but an additional eight miRNAs also had significant effects, suggesting that TRBP binding to pre-miRNA is indispensable for maturation and silencing activity. A TRBP-non-bound miRNA, miR-19b-1, upregulated the predicted target genes, although other TRBP-non-bound miRNAs did not show similar effects. Thus, upregulation of the predicted targets of miR-19b-1 was not due to specific regulation. Northern blot analysis showed that the amount of mature miR-106b was certainly decreased by LGP2 overexpression (Figure 6D) and increased in LGP2−/− cells (Figure 6E), indicating that LGP2 repressed the maturation of miR-106b. In contrast, the amount of mature miR-19b-1 was not changed regardless of LGP2 expression. The miR-106b maturation was repressed by LGP2 in dose-dependent manner in LGP2−/− cells transfected with expression plasmid encoding FLAG-LGP2 with different concentrations (Supplementary Figure S5A), and the expression of ATPase/Helicase domain (ΔCTD) is sufficient to repress the miR-106b maturation (Supplementary Figure S5B). In LGP2−/− cells, the expression levels of the predicted target genes of miR-106b, DERL2 and ZNFX1, were significantly reduced (Figure 6F). In contrast, the expression levels of predicted target genes of miR-19b-1, QKI and RAB18, were not significantly changed or slightly increased, consistent with microarray results (Figure 6G). To investigate whether the repression of these genes was responsible for the upregulation of miR-106b in the absence of LGP2, a miR-106b inhibitor (TuD-miR-106b), which functions as a decoy of miR-106b (34), was introduced into LGP2−/− cells. TuD-miR-106b rescued the downregulation of DERL2 and ZNFX1 expression in LGP2−/− cells (Figure 6F, left two panels), and the overexpression of LGP2 also rescued the downregulation (Figure 6F, right panel), indicating that LGP2 represses RNA silencing activity by suppressing the maturation of miR-106b.
Figure 6.
LGP2 regulates miRNA-mediated gene expression of endogenous genes in human cells. (A) XY plot of microarray signals of all transcripts expressed in WT and LGP2−/− cells. (B) MA plots (above) and cumulative distributions (below) of all transcripts in LGP2−/− cells normalized by their expression levels in WT cells. The predicted targets of miR-106b (left) and miR-19b-1 (right) are colored blue and pink, respectively. ZNFX1 and DERL2 are the predicted targets of miR-106b, but not miR-19b-1. (C) The differences of integral values of cumulative distribution between all transcripts and predicted targets were calculated on all of the predicted targets of each TRBP-bound pre-miRNA (sequence read enrichment by immunoprecipitation > 10) and TRBP-non-bound pre-miRNA (sequence read enrichment by immunoprecipitation < 1/3), and represented as fold change (log2). P-values were determined by Wilcoxon test (*P < 0.05). (D) Northern blot of endogenous mature miR-106b and miR-19b-1 in HeLa cells transfected with empty vector or an expression plasmid encoding FLAG-LGP2. The bar graph shows the quantified signal intensities of miR-106b, miR-19b-1 or U6 as control. (E) Northern blot of endogenous mature miR-106b and miR-19b in WT and LGP2−/− cells. The bar graph shows the quantified signal intensities of miR-106b, miR-19b-1 or U6 as control. (F) Relative mRNA levels of miR-106b targets, DERL2 and ZNFX1, quantified by qRT-PCR in WT and LGP2−/− cells. WT and LGP2−/− cells were transfected with either control plasmid (TuD-shuttle), a plasmid encoding miR-106b inhibitor (TuD-miR-106b), or FLAG-LGP2 expression plasmid. (G) Relative mRNA levels of miR-19b target gene, QKI and RAB18, quantified by qRT-PCR in WT and LGP2−/− cells. The experiments were performed in triplicate and P-values were determined by Student's t-test (*P < 0.05).
DISCUSSION
To the best of our knowledge, this is the first report to show that the RLR, LGP2, is a modulator of the RNA silencing activity directed by endogenous miRNA (Figure 7). We demonstrated that LGP2 binds to the dsRNA-binding sites of the RNA silencing enhancer, TRBP (Figure 3C). This competitive binding leads to the release of TRBP-bound pre-miRNA and results in the inhibition of pre-miRNA recruitment by TRBP (Figure 4B), miRNA maturation by Dicer/TRBP (Figure 4C), and subsequent RNA silencing activities (Figure 4D). Furthermore, TRBP is known to be a dsRNA-binding protein but it was shown here to have no sequence preference for binding; however, it was revealed that TRBP discriminates between miRNAs based on their secondary structures. TRBP had a strong binding preference for pre-miRNAs, whose stem region had tight base-pairing except for the center region (Figure 5). TRBP consists of three dsRBDs, and the N-terminal domains dsRBD1 and dsRBD2 contain typical α-β-β-β-α folds, which are necessary to bind dsRNAs (47). Structural analysis revealed that the canonical dsRBD recognizes a total of about 16 bp of RNA duplex that conserves a similar electrostatic potential charge distribution. The positive electrostatic potential charge in the central region recognizes the major groove in the RNA duplex, and the neutral and negative electrostatic potential surfaces in both terminals recognize two successive minor grooves in both sides (47). The difference in electrostatic potential charges on the surface may inform the preferred binding site in the RNA duplex. Strictly tight base-pairing may be necessary for interacting with the negatively charged TRBP surface, but weak base-pairing is acceptable or preferable for interaction with positively charged surfaces.
Figure 7.
Regulatory mechanism of RNA silencing by LGP2. The left panel shows the RNA silencing pathway in the absence of LGP2. TRBP recruits TRBP-bound pre-miRNA to Dicer to promote miRNA maturation. Mature miRNA is loaded onto AGO protein to form active RISC, resulting in the repression of target genes. The right panel shows the inhibitory mechanism of the RNA silencing pathway in the presence of LGP2. LGP2 interacts with the dsRNA-binding site of TRBP to prevent pre-miRNA recruitment, resulting in the repression of TRBP-bound pre-miRNA activity, which leads to target gene upregulation.
Our results revealed that LGP2 repressed specific TRBP-bound miRNA activities by competitively interacting with TRBP (Figure 6), resulting in selective regulation of gene expression. miR-106b had greatly repressed function in the presence of LGP2 (Figure 6). Previous reports showed that miR-106b promotes cell proliferation and inhibits apoptosis in lung cancer cells, vascular endothelial cells, and glioma cells (48–50), and the deletion of a miRNA cluster including miR-106b was lethal in mice (51). In addition, it was revealed that another TRBP-bound miRNA, let-7a, could regulate the cell cycle, proliferation, and apoptosis in multiple cancer cell lines (52). Furthermore, miR-16 was shown to target B cell lymphoma 2 (BCL2), which is an anti-apoptotic gene, in cells from patients diagnosed with chronic lymphocytic leukemia (53). Thus, LGP2 may regulate miRNA-mediated gene expression networks with certain functions, such as apoptosis or cell growth, to protect cells from pathogens.
van der Veen et al. reported that LGP2 had no impact on miRNA processing by in vitro assay (54). They showed that the processing of let-7a and miR-16, which are revealed to be TRBP-bound miRNAs in our study, are not inhibited by LGP2 even in the presence of Dicer. Their results strongly support our conclusion that LGP2 does not interact with Dicer in the absence of TRBP (Figure 1C), and LGP2-TRBP interaction is essential for inhibition of processing of TRBP-bound miRNAs. The dsRNA-binding affinity of TRBP is greatly higher than that of Dicer (55), suggesting that the substrate recruitment is strongly affected by TRBP in vivo. However, they also showed that LGP2 interacts with Dicer via its C-terminal domain, which is an essential domain for RNA binding, and inhibits Dicer-dependent processing of long dsRNA. The interaction between LGP2 and Dicer is weakened but not abolished by RNase treatment. Then, they deduced that long dsRNA-bound CTD domain of LGP2 undergoes structural rearrangements that allow it to associate with Dicer to inhibit its activity, then further strengthened in a cooperative manner by the presence of the helicase domain (54). Thus, the processing of miRNAs and long dsRNAs might be differently regulated by LGP2.
During viral infection, RIG-I and MDA5 recognize different types of RNA viruses to produce antiviral cytokines, including type I IFN, and the molecular mechanism of the signaling pathway has been identified. However, the role of LGP2 was controversial. Previous reports showed positive or negative effects of LGP2 on RLR signaling. Satoh et al. reported that LGP2 positively regulated RIG-I- and MDA5-mediated antiviral responses (56), whereas Rothenfusser et al., Saito et al. and Komuro et al. reported that LGP2 negatively regulated RIG-I-mediated dsRNA recognition (57–59). Venkataraman et al. reported that type I IFN production was promoted in Lgp2−/- mice in response to polyI:C stimulation and vesicular stomatitis virus (VSV) infection, whereas its response to encephalomyocarditis virus (EMCV) was suppressed (60). Our results revealed the molecular mechanism of a primary function of LGP2 function, and this suggests that endogenous gene expression might be regulated by miRNAs that are well disciplined by LGP2-TRBP interaction during viral infection. Komuro et al. also reported that TRBP interacted with LGP2 but not with related RIG-I-like receptors, RIG-I or MDA5 (61). Furthermore, overexpressed TRBP increased Cardiovirus-triggered interferon promoter activity only when LGP2 and MDA5 are co-expressed but not MDA5 alone. Thus, their results indicated that TRBP is important for Cardiovirus-triggered IFN responses mediated by the LGP2/MDA5 RNA recognition system, but is not involved in Sendai virus-triggered IFN response mediated by RIG-I. This may indicate that infection by specific viruses represses RNA silencing activity. Further studies are needed to identify the function of target genes regulated by LGP2.
TRBP functions not only as an RNA silencing enhancer, but also as a repressor of PKR. Some types of viral infections induce the formation of cytoplasmic granular aggregates, termed antiviral stress granules (avSG), which may function as a platform for viral RNA sensing and downstream signaling activation (35). The activation of PKR is required for avSG formation, suggesting that LGP2-TRBP interaction may lead to avSG formation and the activation of downstream signaling pathways.
In summary, we found that a novel function of LGP2, a cytoplasmic viral sensor protein, is the modulation of RNA silencing. Our results suggest that there is a crosstalk between RNA silencing and RLR signaling, and this crosstalk machinery may function as the antiviral defense system in mammalian cells.
DATA AVAILABILITY
Microarray data is registered with GEO, accession number GSE113028. RNA sequence data is now accessible via DDBJ Sequence Read Archive of National Institute of Genetics, Japan, accession number DRA006762.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr T. Fujita for kindly providing LGP2 expression plasmid, Dr H. Iba and Dr T. Haraguchi for kindly providing microRNA inhibitor (TuD-decoy), and Dr K. Saigo for helpful comments and discussion. T.T. and K.U.-T. designed the study at first, and T.T., Y.N., K.O, M.Y. and K.U.-T. discussed the methods, results, and the experimental plans. T.T., Y.N., K.O., F.M. and C.K. performed the experiments, Y.S. performed RNA sequence analysis, and T.T. and Y.N. analyzed microarray and RNA sequence data. The manuscript was drafted by T.T. and K.U.-T., and T.T., M.Y., and K.U.-T. were involved in reviewing the manuscript. All authors read and approved the final manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Education, Culture, Sports, Science and Technology of Japan [21310123, 21115004, 15H04319, 16H14640, 221S0002, 16H06279 to K.U.-T., 15K19124, 18K15178 to T.T.]; Ichiro Kanehara Foundation, the Inamori Foundation, the Uehara Memorial Foundation, Japan Science and Technology Agency (to T.T.); Suzuken Memorial Foundation and Japan Health & Research Institute (to K.U.-T.); Joint Research Center for Promotion of Basic and Applied Medical Sciences (to T.T., K.O., M.Y. and K.U.-T.). Funding for open access charge: Ministry of Education, Culture, Sports, Science and Technology.
Conflict of interest statement. None declared.
REFERENCES
- 1. Kozomara A., Griffiths-Jones S.. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014; 42:D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yi R., Qin Y., Macara I.G., Cullen B.R.. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003; 17:3011–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lund E., Güttinger S., Calado A., Dahlberg J.E., Kutay U.. Nuclear export of microRNA precursors. Science. 2004; 303:95–98. [DOI] [PubMed] [Google Scholar]
- 4. Doi N., Zenno S., Ueda R., Ohki-Hamazaki H., Ui-Tei K., Saigo K.. Short-interfering-RNA-mediated gene silencing in mammalian cells requires Dicer and eIF2C translation initiation factors. Curr. Biol. 2003; 13:41–46. [DOI] [PubMed] [Google Scholar]
- 5. Liu J., Carmell M.A., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J.. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004; 305:1437–1441. [DOI] [PubMed] [Google Scholar]
- 6. Rivas F.V., Tolia N.H., Song J.J., Aragon J.P., Liu J., Hannon G.J., Joshua-Tor L.. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 2005; 12:340–349. [DOI] [PubMed] [Google Scholar]
- 7. Chendrimada T.P., Gregory R.I., Kumaraswamy E., Norman J., Cooch N., Nishikura K., Shiekhatter R.. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005; 436:740–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Haase A.D., Jaskiewicz L., Zhang H., Lainé S., Sack R., Gatignol A., Filipowicz W.. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005; 6:961–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gatignol A., Buckler-White A., Berkhout B., Jeang K.T.. Characterization of a human TAR RNA-binding protein that activates the HIV-1 LTR. Science. 1991; 251:1597–1600. [DOI] [PubMed] [Google Scholar]
- 10. Park H., Davies M.V., Langland J.O., Chang H.W., Nam Y.S., Tartaglia J., Paoletti E., Jacobs B.L., Kaufman R.J., Venkatesan S.. TAR RNA-binding protein is an inhibitor of the interferon-induced protein kinase PKR. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:4713–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cosentino G.P., Venkatesan S., Serluca F.C., Green S.R., Mathews M.B., Sonenberg N.. Double-stranded-RNA-dependent protein kinase and TAR RNA-binding protein form homo- and heterodimers in vivo. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:9445–9449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yoneyama M., Fujita T.. Recognition of viral nucleic acids in innate immunity. Rev. Med. Virol. 2010; 20:4–22. [DOI] [PubMed] [Google Scholar]
- 13. Yoneyama M., Onomoto K., Jogi M., Akaboshi T., Fujita T.. Viral RNA detection by RIG-I-like receptors. Curr. Opin. Immunol. 2015; 32:48–53. [DOI] [PubMed] [Google Scholar]
- 14. Alexopoulou L., Holt A.C., Medzhitov R., Flavell R.A.. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001; 413:732–738. [DOI] [PubMed] [Google Scholar]
- 15. Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M., Taira K., Akira S., Fujita T.. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004; 5:730–737. [DOI] [PubMed] [Google Scholar]
- 16. Yoneyama M., Kikuchi M., Matsumoto K., Imaizumi T., Miyagishi M., Taira K., Foy E., Loo Y.M., Gale M. Jr, Akira S. et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005; 175:2851–2858. [DOI] [PubMed] [Google Scholar]
- 17. Hornung V., Ellegast J., Kim S., Brzózka K., Jung A., Kato H., Poeck H., Akira S., Conzelmann K.K., Schlee M. et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006; 314:994–997. [DOI] [PubMed] [Google Scholar]
- 18. Pichlmair A., Schulz O., Tan C.P., Näslund T.I., Liljeström P., Weber F., Reis e Sousa C.. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006; 314:997–1001. [DOI] [PubMed] [Google Scholar]
- 19. Takahasi K., Yoneyama M., Nishihori T., Hirai R., Kumeta H., Narita R., Gale M. Jr, Inagaki F., Fujita T.. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol. Cell. 2008; 29:428–440. [DOI] [PubMed] [Google Scholar]
- 20. Wang Y., Ludwig J., Schuberth C., Goldeck M., Schlee M., Li H., Juranek S., Sheng G., Micura R., Tuschl T. et al. Structural and functional insights into 5′-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat. Struct. Mol. Biol. 2010; 17:781–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goubau D., Schlee M., Deddouche S., Pruijssers A.J., Zillinger T., Goldeck M., Schuberth C., Van der Veen A.G., Fujimura T., Rehwinkel J. et al. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5′-diphosphates. Nature. 2014; 514:372–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kato H., Takeuchi O., Mikamo-Satoh E., Hirai R., Kawai T., Matsushita K., Hiiragi A., Dermody T.S., Fujita T., Akira S.. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 2008; 205:1601–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yamamoto M., Sato S., Hemmi H., Hoshino K., Kaisho T., Sanjo H., Takeuchi O., Sugiyama M., Okabe M., Takeda K. et al. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003; 301:640–643. [DOI] [PubMed] [Google Scholar]
- 24. Kawai T., Takahashi K., Sato S., Coban C., Kumar H., Kato H., Ishiim K.J., Takeuchi O., Akira S.. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 2005; 6:981–988. [DOI] [PubMed] [Google Scholar]
- 25. Seth R.B., Sun L., Ea C.K., Chen Z.J.. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005; 122:669–682. [DOI] [PubMed] [Google Scholar]
- 26. Vazquez C., Horner S.M.. MAVS coordination of antiviral innate immunity. J. Virol. 2015; 89:6974–6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maillard P.V., Ciaudo C., Marchais A., Li Y., Jay F., Ding S.W., Voinnet O.. Antiviral RNA interference in mammalian cells. Science. 2013; 342:235–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li Y., Lu J., Han Y., Fan X., Ding S.W.. RNA interference functions as an antiviral immunity mechanism in mammals. Science. 2013; 342:231–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Seo G.J., Kincaid R.P., Phanaksri T., Burke J.M., Pare J.M., Cox J.E., Hsiang T.Y., Krug R.M., Sullivan C.S.. Reciprocal inhibition between intracellular antiviral signaling and the RNAi machinery in mammalian cells. Cell Host Microbe. 2013; 14:435–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pare J.M., Sullivan C.S.. Distinct antiviral responses in pluripotent versus differentiated cells. PLoS Pathog. 2014; 10:e1003865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Takahashi T., Miyakawa T., Zenno S., Nishi K., Tanokura M., Ui-Tei K.. Distinguishable in vitro binding mode of monomeric TRBP and dimeric PACT with siRNA. PLoS One. 2013; 8:e63434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ui-Tei K., Naito Y., Takahashi F., Haraguchi T., Ohki-Hamazaki H., Juni A., Ueda R., Saigo K.. Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res. 2004; 32:936–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Narita R., Takahasi K., Murakami E., Hirano E., Yamamoto S.P., Yoneyama M., Kato H., Fujita T.. A novel function of human Pumilio proteins in cytoplasmic sensing of viral infection. PLoS Pathog. 2014; 10:e1004417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Haraguchi T., Ozaki Y., Iba H.. Vectors expressing efficient RNA decoys achieve the long-term suppression of specific microRNA activity in mammalian cells. Nucleic Acids Res. 2009; 37:e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Onomoto K., Jogi M., Yoo J.S., Narita R., Morimoto S., Takemura A., Sambhara S., Kawaguchi A., Osari S., Nagata K. et al. Critical role of an antiviral stress granule containing RIG-I and PKR in viral detection and innate immunity. PLoS One. 2012; 7:e43031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Naito Y., Hino K., Bono H., Ui-Tei K.. CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics. 2015; 31:1120–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ui-Tei K., Maruyama S., Nakano Y.. Enhancement of single guide RNA transcription for efficient CRISPR/Cas-based genomic engineering. Genome. 2017; 60:537–545. [DOI] [PubMed] [Google Scholar]
- 38. Anders S., Pyl P.T., Huber W.. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015; 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sato K., Hamada M., Asai K., Mituyama T.. CENTROIDFOLD: a web server for RNA secondary structure prediction. Nucleic Acids Res. 2009; 37:W277–W280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Daniels S.M., Melendez-Peña C.E., Scarborough R.J., Daher A., Christensen H.S., El Far M., Lainé S., Gatignol A.. Characterization of the TRBP domain required for dicer interaction and function in RNA interference. BMC Mol. Biol. 2009; 10:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E.. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012; 337:816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Takahashi T., Zenno S., Ishibashi O., Takizawa T., Saigo K., Ui-Tei K.. Interactions between the non-seed region of siRNA and RNA-binding RLC/RISC proteins, Ago and TRBP, in mammalian cells. Nucleic Acids Res. 2014; 42:5256–5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Daviet L., Erard M., Dorin D., Duarte M., Vaquero C., Gatignol A.. Analysis of a binding difference between the two dsRNA-binding domains in TRBP reveals the modular function of a KR-helix motif. Eur. J. Biochem. 2000; 267:2419–2431. [DOI] [PubMed] [Google Scholar]
- 44. Machitani M., Sakurai F., Wakabayashi K., Takayama K., Tachibana M., Mizuguchi H.. Type I interferon impede short hairpin RNA-Mediated RNAi via inhibition of dicer-mediated processing to small interfering RNA. Mol. Ther. Nucleic Acids. 2017; 6:173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Panwar B., Omenn G.S., Guan Y.. miRmine: a database of human miRNA expression profiles. Bioinformatics. 2017; 33:1554–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Agarwal V., Bell G.W., Nam J.W., Bartel D.P.. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015; 4:e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yang S.W., Chen H.Y., Yang J., Machida S., Chua N.H., Yuan Y.A.. Structure of Arabidopsis HYPONASTIC LEAVES1 and its molecular implications for miRNA processing. Structure. 2010; 18:594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wei K., Pan C., Yao G., Liu B., Ma T., Xia Y., Jiang W., Chen L., Chen Y.. MiR-106b-5p promotes proliferation and inhibits apoptosis by regulating BTG3 in non-small cell lung cancer. Cell Physiol. Biochem. 2017; 44:1545–1558. [DOI] [PubMed] [Google Scholar]
- 49. Zhang J., Li S.F., Chen H., Song J.X.. MiR-106b-5p inhibits tumor necrosis factor-α-induced apoptosis by targeting phosphatase and tensin homolog deleted on chromosome 10 in vascular endothelial cells. Chin. Med. J. (Engl.). 2016; 129:1406–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liu F., Gong J., Huang W., Wang Z., Wang M., Yang J., Wu C., Wu Z., Han B.. MicroRNA-106b-5p boosts glioma tumorigensis by targeting multiple tumor suppressor genes. Oncogene. 2014; 33:4813–4822. [DOI] [PubMed] [Google Scholar]
- 51. Ventura A., Young A.G., Winslow M.M., Lintault L., Meissner A., Erkeland S.J., Newman J., Bronson R.T., Crowley D., Stone J.R. et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008; 132:875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Johnson S.M., Grosshans H., Shingara J., Byrom M., Jarvis R., Cheng A., Labourier E., Reinert K.L., Brown D., Slack F.J.. RAS is regulated by the let-7 microRNA family. Cell. 2005; 120:635–647. [DOI] [PubMed] [Google Scholar]
- 53. Cimmino A., Calin G.A., Fabbri M., Iorio M.V., Ferracin M., Shimizu M., Wojcik S.E., Aqeilan R.I., Zupo S., Dono M. et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:13944–13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. van der Veen A.G., Maillard P.V., Schmidt J.M., Lee S.A., Deddouche-Grass S., Borg A., Kjær S., Snijders A.P., Reis E Sousa C.. The RIG-I-like receptor LGP2 inhibits Dicer-dependent processing of long double-stranded RNA and blocks RNA interference in mammalian cells. EMBO J. 2018; 37:e97479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lee H.Y., Zhou K., Smith A.M., Noland C.L., Doudna J.A.. Differential roles of human Dicer-binding proteins TRBP and PACT in small RNA processing. Nucleic Acids Res. 2013; 41:6568–6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Satoh T., Kato H., Kumagai Y., Yoneyama M., Sato S., Matsushita K., Tsujimura T., Fujita T., Akira S., Takeuchi O.. LGP2 is a positive regulator of RIG-I and MDA5-mediated antiviral responses. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:1512–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rothenfusser S., Goutagny N., DiPerna G., Gong M., Monks B.G., Schoenemeyer A., Yamamoto M., Akira S., Fitzgerald K.A.. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J. Immunol. 2005; 175:5260–5268. [DOI] [PubMed] [Google Scholar]
- 58. Saito T., Hirai R., Loo Y.M., Owen D., Johnson C.L., Sinha S.C., Akira S., Fujita T., Gale M. Jr.. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:582–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Komuro A., Horvath C.M.. RNA- and virus-independent inhibition of antiviral signaling by RNA helicase LGP2. J. Virol. 2006; 80:12332–12342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Venkataraman T., Valdes M., Elsby R., Kakuta S., Caceres G., Saijo S., Iwakura Y., Barber G.N.. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J. Immunol. 2007; 178:6444–6455. [DOI] [PubMed] [Google Scholar]
- 61. Komuro A., Homma Y., Negoro T., Barber G.N., Horvath C.M.. The TAR-RNA binding protein is required for immunoresponses triggered by Cardiovirus infection. Biochem. Biophys. Res. Commun. 2016; 480:187–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microarray data is registered with GEO, accession number GSE113028. RNA sequence data is now accessible via DDBJ Sequence Read Archive of National Institute of Genetics, Japan, accession number DRA006762.