Abstract
The small molecule cyclic di-GMP (c-di-GMP) is known to affect bacterial gene expression in myriad ways. In Vibrio cholerae in vivo, the presence of c-di-GMP together with the response regulator VpsR results in transcription from PvpsL, a promoter of biofilm biosynthesis genes. VpsR shares homology with enhancer binding proteins that activate σ54-RNA polymerase (RNAP), but it lacks conserved residues needed to bind to σ54-RNAP and to hydrolyze adenosine triphosphate, and PvpsL transcription does not require σ54 in vivo. Consequently, the mechanism of this activation has not been clear. Using an in vitro transcription system, we demonstrate activation of PvspL in the presence of VpsR, c-di-GMP and σ70-RNAP. c-di-GMP does not significantly change the affinity of VpsR for PvpsL DNA or the DNase I footprint of VpsR on the DNA, and it is not required for VpsR to dimerize. However, DNase I and KMnO4 footprints reveal that the σ70-RNAP/VpsR/c-di-GMP complex on PvpsL adopts a different conformation from that formed by σ70-RNAP alone, with c-di-GMP or with VpsR. Our results suggest that c-di-GMP is required for VpsR to generate the specific protein–DNA architecture needed for activated transcription, a previously unrecognized role for c-di-GMP in gene expression.
INTRODUCTION
Biofilm formation and its persistence on catheters, pacemakers, sutures and other indwelling medical devices account for the vast majority of the two million healthcare-associated annual infections and approximately 100 000 deaths per year in the USA (1). These biofilm-based infections impose an estimated annual $94 billion in excess medical costs (2). Forming on both biotic and abiotic surfaces, biofilms are aggregates of microbial communities encased by a matrix of extracellular polymeric substances (3). Biofilms, which are formed by almost all bacteria, play a significant role in environmental persistence, dissemination and transmission as well as protection from environmental stressors such as nutrient limitation, predation and bacteriophages (4–8). However, most concerning of all, biofilms dramatically decrease susceptibility to antimicrobial agents, posing a serious threat to public health.
Because biofilms are recalcitrant to conventional antibiotic therapies and represent a major clinical obstacle, it is essential to understand the molecular mechanisms responsible for biofilm gene expression. A central regulator of biofilm formation is the second messenger cyclic dimeric guanosine monophosphate (c-di-GMP). Present in about 85% of bacteria, c-di-GMP is synthesized by diguanylate cyclases (DGCs), which typically contain a conserved GGDEF motif, and is degraded by phosphodiesterases, which contain a conserved EAL or HD-GYP motif (9). Generally, high levels of c-di-GMP increase biofilm formation and decrease motility, while low levels of c-di-GMP exert the opposite effect (10). Along with biofilm formation and motility, c-di-GMP also regulates a diverse array of phenotypes including quorum sensing, virulence, cell-cycle control, secretion, bacterial predation and stress responses (10). Although c-di-GMP has been extensively studied since its discovery in 1987 (11) and many groups have studied the mechanisms by which c-di-GMP interacts with effectors (12–19), mechanism(s) by which c-di-GMP might be needed to directly modulate RNA polymerase (RNAP) in transcription have not been elucidated.
Catalyzing transcription is the multi-subunit enzyme RNAP. Bacterial RNAP is an ∼500 kDa enzyme comprised of two large subunits (beta and beta’), two alpha subunits, one omega subunit and a promoter specificity factor, σ (20). Although the primary σ, such as σ70 in Escherichia coli, is used for the expression of most genes during exponential growth, alternate σ factors, which are either related to σ70 or belong to the σ54 family, are used under other growth conditions or times of stress (20). The first step in transcription is initiation, a multi-step process that can be controlled by various regulators. During transcription initiation with σ70-RNAP, polymerase first binds to double-stranded DNA elements in the −10 and −35 regions, forming closed complex that is typically unstable (21–23). Isomerization to the open complex proceeds rapidly and requires unwinding and bending of the DNA, major conformational changes within RNAP, and formation of the transcription bubble from −11 to ∼+3 (21). Upon addition of ribonucleoside triphosphates (rNTPs), the complex transitions to the initiating complex where small abortive RNAs are synthesized and released prior to promoter clearance (21). While RNAP catalyzes transcription efficiently at promoters with optimal −35 and −10 consensus sequences, activators are typically needed to regulate promoters with suboptimal sequences. Some activators additionally use second messenger molecules such as cyclic adenosine monophosphate, guanosine pentaphosphate or c-di-GMP to modulate gene expression (24).
In Vibrio cholerae, an important pathogen that causes the acute diarrhea disease cholera and uses biofilms to aid in environmental transmission, survival and pathogenesis, VpsR is the master regulator that activates biofilm gene transcription in vivo in the presence of high levels of c-di-GMP and also binds c-di-GMP with a Kd(app) of 1.6 μM in vitro (25–31). VpsR is known to activate promoters for vpsL and vpsT, genes within the biofilm biosynthesis operons. Furthermore, VpsR also directly activates expression of other phenotypes in response to c-di-GMP such as acetoin biosynthesis, the transcription factor tfoY and the eps operon encoding the type II secretion system (28,32,33), suggesting that this transcription factor is the hub for a central network of c-di-GMP transcriptional control in V. cholerae. However, despite the abundance of evidence showing the positive regulatory role of VpsR and c-di-GMP in activating gene expression in vivo, previous work has not recapitulated this result in vitro.
Based on amino acid sequence homology, VpsR has been classified as an atypical enhancer binding protein (EBP) (28,31). Classic EBPs utilize σ54 to activate transcription and are comprised of three conserved domains: an N-terminal receiver (REC) domain, a central AAA+ domain (Adenosine triphosphatase (ATPase) associated with diverse cellular activities) involved in ATP hydrolysis and binding to σ54 and a C-terminal helix-turn-helix DNA-binding domain (34). Although VpsR has overall homology to EBPs, several residues known to be required for specific EBP functions are not conserved. Not only does VpsR lack the GAFTGA motif involved in binding to σ54, but the highly conserved aspartate (D) and glutamate (E) residues in the Walker B domain involved in ATP hydrolysis are asparagine (N) and aspartate (D) residues in VpsR (Supplementary Figure S1). Furthermore, microarray analyses demonstrate that transcription from promoters known to be regulated by VpsR does not change in a σ54 (rpoN-) mutant (30), and sequence analyses indicate that the VpsR-activated promoters do not contain the well-conserved −24 GG and −12 GC consensus sequences utilized by σ54-RNAP. Instead, some of these promoters have reasonable matches to the consensus −10 element of promoters dependent on a primary sigma factor, such as σ70.
Here we have developed an in vitro transcription system demonstrating activated transcription from the VpsR-activated promoter for the vpsL gene (PvpsL) in the presence of VpsR, c-di-GMP and σ70-RNAP. We have used DNase I and KMnO4 footprinting to characterize the protein–DNA complex made by σ70-RNAP alone with PvpsL versus complexes made by σ70-RNAP with VpsR and/or c-di-GMP. Surprisingly, we find that c-di-GMP together with VpsR is needed to generate the correct protein–DNA interactions required for an active transcription complex with σ70-RNAP. Our results provide a new paradigm in c-di-GMP-dependent transcription activation.
MATERIALS AND METHODS
DNA
pMLH06 (vpsL short promoter) and pMLH07 (vpsL long promoter) contain the vpsL promoter from −97 to +213 and from −393 to +213, respectively, cloned into the EcoRI and HindIII restriction enzyme sites of pRLG770 (35). pMLH09 (vpsL-lux long promoter) and pMLH10 (vpsL-lux short promoter) contain the vpsL promoter from −393 to +213 and from −97 to +213, respectively, cloned into the SpeI and BamHI restriction sites of pBBRlux (36). pMLH17 was generated by cloning the wildtype vpsR gene into the EcoRI and HindIII restriction sites of pHERD20T (37). Inserts were obtained as polymerase chain reaction (PCR) products, which had been amplified with primers from V. cholerae genomic DNA (BH1514) using Pfu Turbo polymerase (Stratagene). Inserts and vectors were digested with the appropriate restriction enzymes and cloning was performed using standard techniques. Primer sequences are available upon request.
pMLH11 is a pET28b(+) derivative (Novagen) that contains vpsR cloned between the NdeI and XhoI restriction sites. PCR was used to amplify the pET28b(+) vector for restrictionless cloning. Gibson Assembly Master Mix (New England Biolabs) was used to assemble the PCR products and vectors according to manufacturer’s instructions.
pCMW75 contains an active Vibrio harveyi DGC, qrgB, and was used for expression of high levels of c-di-GMP in gene reporter assays (29). pCMW98 contains an inactive V. harveyi DGC, qrgB, and was used for expression of low levels of c-di-GMP (29).
Fragments containing PvpsL used for electrophoretic mobility shift assays (EMSAs) and DNase I footprinting were obtained as PCR products using Pfu Turbo polymerase (Stratagene) and upstream and downstream PCR primers, which anneal from positions −97 to +113 relative to the transcription start site (TSS). To radiolabel the DNA, non-template or template primer was treated with T4 polynucleotide kinase (Affymetrix) in the presence of [γ-32P] ATP prior to PCR. The radiolabeled PCR products were purified as described (38).
Strains and growth conditions
E. coli ElectroMAX DH10B (Invitrogen) or E. coli DH5α (New England Biolabs) were used for cloning, and BL21(DE3) or Rosetta2 (DE3) pLysS (New England Biolabs) were used for protein production. The V. cholerae strains, ΔvpsL and ΔvpsLΔvpsR, used in this study were derived from the El Tor biotype strain C6707str2 (39). For lux-fusion assays, strains containing a mutation in vpsL were used to prevent cellular aggregation, allowing us to obtain accurate readings of reporter gene expression at high levels of c-di-GMP by preventing cellular aggregation (29). High c-di-GMP was synthesized by production of the DGC QrgB from the plasmid pCMW75 after addition of 0.5 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) (29). Strains were grown at 37°C in Luria-Bertani broth (LB) (1% tryptone, 0.5% yeast extract, 1% NaCl at pH 7.5). LB agar medium contained 1.5% (wt/vol) granulated agar (Acumedia). Antibiotics were added at the following concentrations: ampicillin at 100 μg/ml, chloramphenicol at 100 μg/ml and kanamycin at 100 μg/ml. For lux fusion assays shown in Supplementary Figure S2, E. coli S17-λpir (40) or V. cholerae strains containing indicated plasmids were grown overnight in LB medium supplemented with appropriate antibiotics. Cells were then diluted 1:200 in fresh LB supplemented with appropriate antibiotics and grown to an OD600 of ∼0.5. A final concentration of 0.2% arabinose and/or 1 mM IPTG was added to the medium to induce VpsR and/or c-di-GMP synthesis, respectively. Luminescence was measured using an Envision multilabel counter (PerkinElmer) and lux expression was reported in relative luminescent units (RLU; counts min−1 ml−1/OD600 unit). Assays were repeated with at least two biological replicates and three technical replicates.
Proteins
E. coli RNAP core was purchased from Epicenter Technologies. E. coli σ70 was purified as previously described (41). VpsR protein was isolated from Rosetta2 (DE3)/pLysS (Novagen) containing pMLH11, which was grown at 37°C with shaking at 220 rpm in 200 ml of LB containing 50 μg/ml kanamycin and 25 μg/ml chloramphenicol to an OD600 of ∼0.5. Cultures were placed on ice, IPTG (final concentration of 0.5 mM) was added, and the cells were then incubated at 16°C with shaking at 150 rpm for 16 h. After centrifugation at 13 000 × g, cells were harvested and then sonicated in 30 ml of sonication buffer [20 mM sodium phosphate (pH 7.8), 400 mM NaCl and 7 mM β-mercaptoethanol]. Centrifugation at 17 500 × g removed insoluble materials, and the soluble fraction was subjected to chromatography on a 1 ml Ni-NTA column (Qiagen). The column was washed with wash buffer [20 mM sodium phosphate (pH 6.0), 400 mM NaCl and 7 mM β-mercaptoethanol] containing increasing amounts of imidazole: 0 mM (10 ml), 5 mM (10 ml), 50 mM (5 ml), 100 mM (5 ml), 150 mM (5 ml), 200 mM (5ml) and 250 mM (5 ml). Purified VpsR eluted with fractions containing 150–200 mM imidazole. These fractions were pooled and dialyzed in VpsR buffer [20 mM sodium phosphate (pH 7.8), 150 mM NaCl, 7 mM β-mercaptoethanol and 20% glycerol] prior to storage at −80°C. Protein concentrations were determined by comparison with known amounts of RNAP core after sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and gel staining with Colloidal Blue (Invitrogen). To determine if VpsR co-purified with c-di-GMP, 1 μM VpsR (100 μl) was heated at 95°C for 5 min and pelleted by centrifugation. The resulting supernatant was examined using ultrahigh-pressure liquid chromatography (UPLC)–tandem mass spectrometry (MS-MS) as previously described (42).
Thin-layer chromatography (TLC) ATPase assay
Reactions (2.5 μl) containing 3 pmol of VpsR, 20 μM [γ-32P] ATP at 2 × 105 dpm/pmol, 50 μM c-di-GMP (when indicated) and 1 × transcription buffer [40 mM Tris-acetate (pH 7.9), 150 mM potassium glutamate, 4 mM magnesium acetate, 0.1 mM ethylenediaminetetraacetic acid (EDTA) (pH 8.0), 0.01 mM DTT, and 100 μg/ml bovine serum albumin (BSA)] were incubated for 10 min at 37°C and aliquots (2 μl) were spotted on polyetherimide (PEI) membranes (Sigma) and allowed to dry. Calf intestinal phosphatase was used as a positive control while buffer alone or BSA were used as negative controls. PEI plates were developed in 0.85M KH2PO4 (pH 3.4), autoradiographed, and the images scanned using a Powerlook 2100XL densitometer.
BS3 crosslinking
A solution of VpsR buffer containing 1.5 μM VpsR, 5 mM BS3 crosslinker (Thermo Scientific), and as indicated, 50 μM c-di-GMP was incubated at room temperature for 30 min. Reactions were quenched with the addition of Tris-Cl (pH 7.5) to a final concentration of 0.1 M. Proteins were separated by SDS-PAGE on a 10–20% (wt/vol) Tris-tricine gel (Invitrogen) and stained with Colloidal Blue (Invitrogen). To mimic transcription conditions, 5 mM BS3 was added to a solution containing 0.6 μM VpsR and 12.5 μM c-di-GMP in transcription buffer. After 30 min of incubation at room temperature, reactions were quenched as described above. Proteins were separated on 10–20% (wt/vol) Tricine gels (Invitrogen) and stained with SilverXpress Silver Stain (Invitrogen) according to manufacturer's instructions.
Electrophoretic mobility shift assays
Protein-DNA complexes were formed by incubating 5 nM 32P-labeled DNA, and as indicated, VpsR (final concentration from 0.2 μM to 2 μM), 0.16 μM reconstituted RNAP (σ:core ratio of 2.5:1), and unless indicated otherwise, 50 μM c-di-GMP (final volume of 10 to 20 μl) at 37°C for 10 min in transcription buffer. A 1 μl solution of 1 mg/ml poly(dI-dC) or 500 μg/ml heparin was added to VpsR-DNA complexes or transcription complexes, respectively. Reactions containing VpsR-DNA complexes were loaded onto 5% (wt/vol) non-denaturing polyacrylamide gels already running at 100 V in 1 X Tris/borate/EDTA (TBE) buffer. Samples were electrophoresed for 1.5 h. Transcription complexes were loaded onto 4% (wt/vol) non-denaturing, polyacrylamide gels already running at 100 V in 1 × TBE buffer. After loading, voltage was increased from 100 to 380 V, and samples were electrophoresed for 3 h. After autoradiography, films were scanned on a Powerlook 2100XL densitometer and analyzed with Quantity One software (Bio-Rad). Kd(app)s were calculated as the concentration of VpsR needed to shift 50% of the free DNA.
In vitro transcriptions
Multiple and single round in vitro transcriptions were performed in 5 μl reactions containing 0.02 pmol supercoiled template, 0–3.0 pmol VpsR, 0–50 μM c-di-GMP, reconstituted RNAP (0.2 pmol σ70 plus 0.05 pmol core) and transcription buffer. Unless otherwise indicated, samples were incubated at 37°C for 10 min prior to the addition of a solution (1 μl) containing rNTPs (2.86 mM ATP, GTP, CTP and 71 μM [α-32P] UTP at 5 × 104 dpm/pmol) with and without 500 ng heparin. After incubation for 10 min at 37°C, reactions were collected on dry ice, formamide load solution (15 μl) was added and aliquots were electrophoresed on 4% (wt/vol) polyacrylamide, 7 M urea denaturing gels for 2500 V-h in 0.5 × TBE buffer. After electrophoresis, gels were exposed to X-ray films, films were scanned and radioactivity was quantified as described above.
Primer extensions
Primer extension analyses of RNA generated in vitro were performed according to manufacturer’s instructions (Promega) by using AMV Reverse Transcriptase. A sample (5 μl) of the in vitro transcription reaction was added to 6 μl of primer mixture containing 2 × AMV Primer Extension buffer and 2 pmol 32P-labeled primer, which annealed +103 bp downstream of the +1 transcriptional start site. Aliquots were electrophoresed on 8% (wt/vol) polyacrylamide, 7 M urea denaturing gels for 4000 V-h in  × TBE. Densitometry and quantification were performed as described above.
× TBE. Densitometry and quantification were performed as described above.
In vivo RNA was obtained from WN310 containing pMLH17 and pCMW75 or pCMW98 grown in LB with 100 μg/ml ampicillin and 100 μg/ml kanamycin at 37°C with shaking at 220 rpm to an OD600 of ∼0.5. Cells were harvested by centrifugation and RNA was extracted using the RNeasy kit (Qiagen), and an on-column DNase I digestion (Qiagen) was performed according to manufacturer's instructions. After elution, 5 μg total RNA in 5 μl was added to the primer mixture and subsequent steps for primer extension reactions were performed as described above.
DNase I footprinting
Solutions were assembled as described for EMSAs using 0.04 μM DNA, and as indicated, 1.4 μM VpsR, 50 μM c-di-GMP and/or 0.16 μM reconstituted RNAP (σ:core ratio of 2.5:1). After incubation with poly(dI-dC) (complexes lacking RNAP) or heparin (complexes with RNAP) for 15 s, 0.3 U of DNase I in 1.5 μl was added. Solutions were incubated for an additional 30 s at 37°C and then immediately loaded onto 4% (wt/vol) non-denaturing, polyacrylamide gels already running at 100 V in 1 × TBE buffer. Upon loading samples, voltage was increased from 100 to 380 V, and samples were electrophoresed for 3 h. After autoradiography, the protein/DNA complexes were excised and extracted DNA was electrophoresed on denaturing gels as described (43).
Potassium permanganate footprinting
For potassium permanganate (KMnO4) footprinting, solutions were assembled as described for DNase I footprinting. After addition of 500 ng of heparin, KMnO4 was added to a final concentration of 2.5 mM, solutions were incubated at 37°C for 2.5 min, quenched with 5 μl of 14 M 2-mercaptoethanol and further processed as described (43).
RESULTS
In the presence of c-di-GMP, VpsR activates σ70/RNAP at the vpsL promoter (PvpsL) in vitro
It has previously been demonstrated in V. cholerae that deletion of vpsR eliminates expression of a PvpsL-driven lux (31,44) and that high levels of c-di-GMP yield greater levels of PvpsL-lux transcription (29). Thus, we sought to analyze the effects of both VpsR and c-di-GMP at PvpsL using gene reporter fusion assays. We find that the presence of VpsR and c-di-GMP activates PvpsL-lux expression in either V. cholerae (Supplementary Figure S2A) (28,29,44) or E. coli (Supplementary Figure S2B). Thus, no specific Vibrio factors other than VpsR are required for activation. Consequently, we established an in vitro transcription system and performed various footprinting assays using E. coli RNAP. These analyses are detailed below and summarized in Figure 1. It should be noted that the TSS, which we determined by primer extension analyses of both in vivo RNA isolated from V. cholerae and in vitro RNA described below, differs from previously reported locations (28,31).
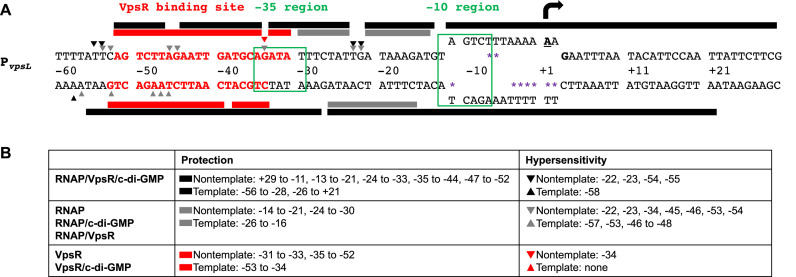
Figure 1.
Summary of in vitro primer extension and DNase I and KMnO4 footprinting. (A) Sequence of PvpsL from −60 to +30. Bold and underlined A with black arrow at +1 and bold G (+3) represent the TSS determined by primer extensions; the −10 element and the −35 region are labeled and boxed in green; sequences in bold and red denote the VpsR binding site. Protection sites from DNase I footprinting and hypersensitive sites are depicted as rectangular boxes and triangles, respectively, either above (non-template) or below (template) the sequences: gray, RNAP with or without c-di-GMP or VpsR; black, RNAP with VpsR and c-di-GMP; red, VpsR with or without c-di-GMP. The open transcription bubble detected using KMnO4 footprinting is shown as separated ssDNA from position −11 to +2 with sites of KMnO4 cleavage indicated as purple asterisks. (B) Summary of positions of protection and hypersensitive sites on non-template and template strand DNA.
As a transcription template, we constructed pMLH06, which contains V. cholerae DNA from −97 to +213 relative to the vpsL TSS inserted upstream of the rrnBT1 terminator. Previous EMSAs and DNase I footprinting analyses indicated that VpsR binds to PvpsL at both a promoter distal site (−297 to −336) and a promoter proximal site (−31 to −52). However, promoter-fusion expression studies have demonstrated that the downstream site is sufficient for activation (28,31).
Using E. coli core RNAP reconstituted with σ70, we found that the presence of VpsR and c-di-GMP activates transcription at PvpsL by ∼7-fold (Figure 2). Both VpsR and c-di-GMP are required for this activation. Addition of varying amounts of either c-di-GMP alone or VpsR alone to RNAP does not alter the basal level observed with RNAP alone, while addition of both c-di-GMP and VpsR to RNAP results in a dose-dependent activation (Figure 2 and Supplementary Figure S3). We also determined that only the downstream VpsR binding site is required for this activation in vitro (Supplementary Figure S4A), consistent with the results obtained with lux-fusion assays (Supplementary Figure S4B) as well as other studies (28,31).
Figure 2.
VpsR and c-di-GMP activate transcription at PvpsL by ∼7-fold in vitro. (A) Representative gel showing PvpsL RNA obtained after multiple round in vitro transcription reactions using plasmid template PvpsL with RNAP alone (lane 1), RNAP and c-di-GMP (lane 2), RNAP and VpsR (lane 3), and RNAP, VpsR, and c-di-GMP (lane 4). (B) Graph showing the level of PvpsL transcription relative to that with RNAP alone (basal) obtained from at least three independent experiments (one-way ANOVA with Tukey’s HSD (honest significant difference) posthoc analysis, *P < 0.05).
Primer extension analyses and KMnO4 footprinting identifies the TSS of PvpsL
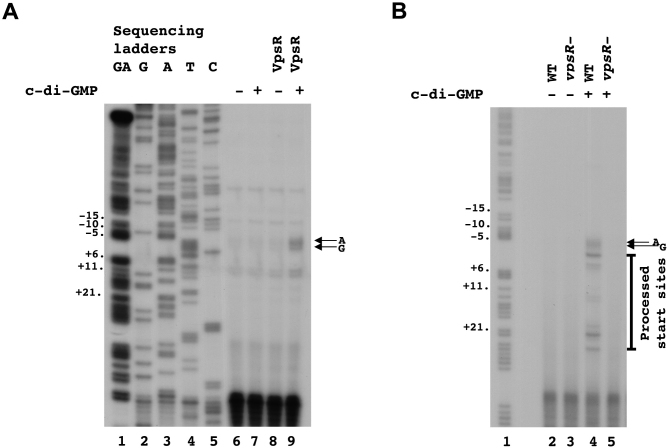
Previous primer extension analyses of V. cholerae RNA isolated from cells have identified multiple 5′ ends for the RNA occurring upstream of the vpsL coding sequence. These included an A, a T (most abundant), a G and an A nucleotide, located 37, 39, 57 and 59 bases upstream of the assigned GUG translation start site, respectively (31,45). However, these positions were determined using exponentially growing V. cholerae, which should have low levels of c-di-GMP. To determine the TSS in vitro, we performed primer extensions using RNA synthesized from our in vitro transcription reactions. This analysis identified two 5′ ends whose presence is stimulated when reactions contain RNAP together with both VpsR and c-di-GMP (Figure 3A): the ‘A’ located 59 bases upstream of the vpsL GUG, which was one of the ends observed previously and is indicated as the +1 in Figure 1, and the ‘G’ located 57 bases upstream of the GUG. To assign the TSS in the presence of high levels of c-di-GMP in vivo, we isolated RNA from V. cholerae grown with high intracellular concentrations of c-di-GMP. We again observed these two sites as well as other downstream 5′-ends (Figure 3B). Given that the farthest upstream end(s) seen in vivo align with start sites seen in vitro and the TSS determined by KMnO4 footprinting (below), we propose that the farthest upstream sites are in fact the start of the vpsL RNA and the other ends observed in vivo arise from RNA processing or degradation.
Figure 3.
Identification of +1 TSS using in vitro and in vivo primer extensions. RNA was isolated from in vitro transcriptions (A) or V. cholerae (B). Two major primer extension products, which are observed only in the presence of both VpsR and c-di-GMP are indicated with arrows.
In a transcription open complex, the single-stranded (ss) transcription bubble typically occurs from −11 to ∼+3 (22,23). KMnO4 footprinting, which selectively oxidizes thymines in ssDNA, is considered the ‘gold’ standard for observing the position of this transcription bubble and by extension the position of the +1 TSS (46). In this analysis, the ‘T’ at position −11 on the template strand marks the start of the ss region of the DNA within the open complex and is thus the farthest upstream reactive T in the analysis. Reactive thymines can extend to the end of the bubble (position ∼+3). In our assay, we challenged complexes with the addition of heparin, which typically destabilizes closed complexes but does not impact open complexes. Thus, we conclude that any oxidized thymines we are observing arise from the stable open complex.
When we incubated PvpsLwith RNAP/VpsR/c-di-GMP, we observed reactive thymines on the template strand at positions −11 and −4 to +2 (Figure 4A, lane 2) and reactive thymines on the non-template strand at positions −6 and −7 (Figure 4B, lane 2) relative to the ‘A’ that is 59 bases upstream of the GUG. These reactive bases identify the open complex and are consistent with our identification of the TSS at position −59 relative to start of the gene. Furthermore, this analysis defines the σ70 −10 recognition element of PvpsL as −12TAGTCT−7.
Figure 4.
KMnO4 footprinting assigns the +1 TSS at the A located 59 bp upstream of the vpsL translation start site. (A) Reactive thymines within the transcription bubble are observed at positions −11, −4, −3, -2, −1, +1 and +2 on template DNA. (B) Reactive thymines within the transcription bubble are also observed at positions −6 and −7 on non-template DNA. GA corresponds to G+A ladder.
We also used KMnO4 footprinting to investigate open complex formation when only some of the components are present. As expected from the basal transcription that we observed with RNAP alone (Figure 2), we observed a low level of reactive thymines at the same positions in the complexes made by RNAP in the absence of c-di-GMP and VpsR (Supplementary Figure S5, lane 2). Addition of either VpsR or c-di-GMP to RNAP did not stimulate this basal level of transcription (Figure 2) or the amount of reactive thymines (Supplementary Figure S5, lanes 4 and 6). Thus, KMnO4 analyses indicate that RNAP together with both VpsR and c-di-GMP is needed to form the maximum level of open complex a PvpsL, consistent with our in vitro transcription results (Figure 2). Interestingly, these analyses also indicated the presence of reactive thymines at other positions within the basal complexes (Supplementary Figure S5, lanes 2, 4 and 6), which were relatively much less abundant in the activated complex (Supplementary Figure S5, lane 8). It is possible then that in the absence of both VpsR and c-di-GMP, RNAP is promiscuous in promoter choice. Nevertheless, it is important to note that RNAP, VpsR and c-di-GMP form an open complex in the absence of ATP. This is unlike classic EBPs which use ATPase activity to drive open complex formation. To investigate whether VpsR has ATPase activity, we assayed ATP hydrolysis in the presence and absence of c-di-GMP (Supplementary Figure S6). No ATPase activity was detected.
In the presence of VpsR and c-di-GMP, RNAP rapidly forms a heparin-resistant, stable complex at PvpsL
To determine the rate of PvpsL open complex formation in the presence of RNAP, VpsR and c-di-GMP, we incubated PvpsL DNA with these components for various times before adding heparin together with rNTPs. As the addition of heparin will destabilize any unstable complexes, the subsequent level of single round transcription reflects the level of stable complex present at that time point. As seen from our KMnO4 analyses (Figure 4), this represents the open complex. As seen in Supplementary Figure S7A, no additional activation was observed after the first time point of 1 min, indicating that the open complex forms rapidly in the presence of VpsR and c-di-GMP.
To determine the stability of the open complex, we incubated proteins and DNA for 10 min and then added heparin for varying time periods prior to the addition of rNTPs. Either a one min or 15 min heparin incubation yielded similar levels of transcription (Supplementary Figure S7B), indicating that the open complex is stable for at least several min. The stability of open complex to heparin challenge was not dependent upon the addition of c-di-GMP or VpsR as basal expression exhibited equivalent stability to heparin addition at one versus 15 min although total transcript levels were reduced. Thus, we conclude that RNAP rapidly forms a stable open complex at the vpsL promoter and the amount of open complex formation is stimulated by VpsR and c-di-GMP.
Dimerization and DNA binding by VpsR with and without c-di-GMP are similar
Multiple studies have found that many transcription factors require c-di-GMP binding to facilitate dimerization or the formation of higher order structures (12–15). Consequently, we used BS3 crosslinking, which generates non-specific amine to amine covalent bonds, to investigate whether VpsR forms oligomers and if so, whether this formation is affected by c-di-GMP. As seen in Figure 5A, VpsR dimers are observed in the presence of BS3 crosslinker, and the amount of this crosslinked species is similar in the presence or absence of c-di-GMP. To make sure that our transcription conditions did not affect these results, we also tested the BS3 crosslinking using the same protein concentration and buffer conditions that were used for transcription. In this case, the presence of Tris buffer, which quenches the crosslinking reaction, reduces the overall amount of crosslinking, but again there is no significant difference in the presence or absence of c-di-GMP (Figure 5B). On these silver stained gels, three crosslinked bands are observed with and without c-di-GMP. We infer that these bands represent different crosslinked VpsR dimer conformations using different amines since BS3 is a non-specific amine to amine crosslinker. These results indicate that unlike other characterized c-di-GMP binding proteins (12–15), VpsR does not require c-di-GMP to dimerize. However, it is possible that the presence of c-di-GMP could change the conformation of the formed dimers.
Figure 5.
VpsR forms dimers in vitro with or without c-di-GMP. (A) Samples containing 1.5 μM VpsR with and without 50 μM c-di-GMP were treated with the chemical crosslinker BS3 as indicated and separated on a 10–20% (wt/vol) Tricine gel that was stained with Colloidal Blue. (B) Samples containing 0.6 μM VpsR with and without 12.5 μM c-di-GMP in transcription buffer were treated with the chemical crosslinker BS3 as indicated and separated on a 10–20% (wt/vol) Tricine gel that was silver stained. Far left lane of each panel contains marker proteins, whose molecular weights are indicated. Black arrows indicate position of VpsR monomer (∼50 kDa) and gray arrows indicate position of VpsR dimer (∼100 kDa). Each sample was repeated independently three times, and a representative gel image is shown.
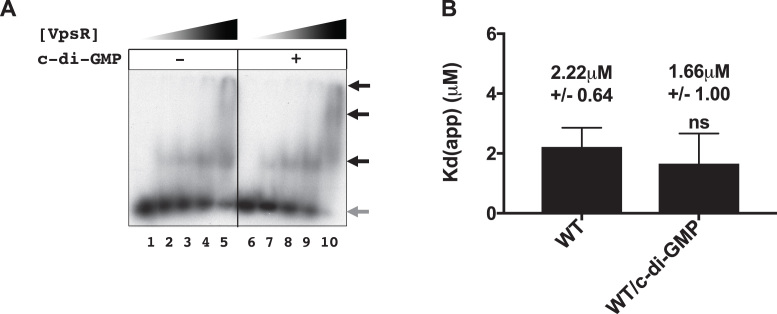
Along with oligomerization, c-di-GMP also plays an important role in helping transcription factors bind the DNA (12–19). Thus, we asked whether the ability of c-di-GMP to stimulate open complex formation could arise by promoting the interaction of VpsR with the DNA. We tested this possibility by determining the apparent dissociation constant (Kd(app)) for VpsR binding to PvpsL in the presence or absence of c-di-GMP. Kd(app)s were calculated by determining the concentration of VpsR needed to shift 50% of the free DNA. We found that the presence of c-di-GMP did not enhance VpsR binding to the DNA [2.22 μM (±0.64 μM) without c-di-GMP, 1.66 μM (±1.00 μM) with c-di-GMP (Figure 6)], indicating that c-di-GMP does not alter the affinity of VpsR for the DNA.Our results are consistent with a previous study that did not observe differences in VpsR binding to vpsL in the presence or absence of c-di-GMP using EMSAs, though these experiments were only done at one concentration of VpsR and dissociation constants were not measured (31).
Figure 6.
VpsR binds PvpsL DNA with similar affinity with or without c-di-GMP. Representative gels showing the retardation of 32P-labeled DNA harboring -97 to +103 of PvpsL with increasing VpsR concentrations from 0 to 2 μM either in the absence (lanes 1–5) or presence (lanes 6–10) of 50 μM c-di-GMP. Black arrows indicate retarded complexes while gray arrow indicates free DNA. (B) Quantitation of EMSAs. Apparent DNA-binding dissociation constants (Kd(app)) were calculated as the concentration of VpsR needed to retard 50% of the free DNA. Values from at least three EMSAs were analyzed using one-way ANOVA with Tukey’s HSD posthoc analysis (ns, not significant).
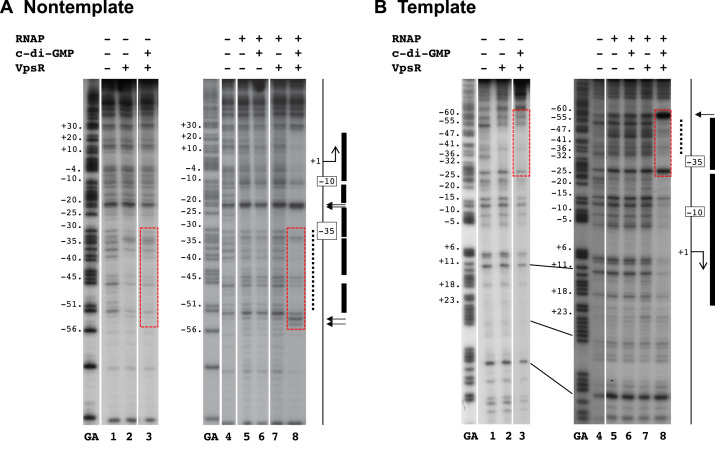
We also performed DNase I footprinting to determine whether there are different VpsR–DNA contacts in the presence or absence of c-di-GMP. To make sure that we were only observing the footprint of the stable protein complex of interest, we challenged the complexes with poly(dI-dC), treated them with DNase I and then isolated the complexes from EMSA gels before isolating the DNA. Similar to a previous study, which identified the proximal VpsR binding site from −31 to −52 using non-template PvpsL in the absence of c-di-GMP (31), we found that VpsR with or without c-di-GMP protected the DNA from −31 to −52 on non-template PvpsL and from −34 to −53 on template PvpsL (Figure 7A and B, lanes 2 and 3). Thus, we did not observe any significant differences between the VpsR–DNA contacts whether c-di-GMP was present or absent.
Figure 7.
DNase I footprinting of PvpsL complexes on (A) nontemplate DNA and (B) template DNA. GA corresponds to G+A ladder. VpsR, c-di-GMP and/or RNAP are present as indicated. To the right of each gel image, a schematic indicates the −10 and −35 regions and the +1. The VpsR binding site is indicated as a dashed black line. DNase I protection regions and hypersensitive sites seen with the activated complex of RNAP, VpsR, c-di-GMP and DNA are depicted as black rectangles and horizontal arrows, respectively. The dashed red boxes indicate the regions of DNA where the protection/enhancement within and immediately adjacent to the VpsR binding site changes when comparing complexes containing RNAP with or without VpsR or c-di-GMP to the activated complex. (See text for details.)
It is important to note that in the previous study, DNase I footprinting was performed in the absence of any competitor (31), while here we used poly(dI-dC) and isolated footprinting complexes from EMSA gels. We observed no protection when complexes made with VpsR ± c-di-GMP were challenged with heparin (data not shown), even though the VpsR/RNAP/c-di-GMP/PvpsL transcription complex is stable to heparin challenge (Supplementary Figure S7). We conclude that the presence of RNAP with c-di-GMP stabilizes VpsR binding to the DNA, forming a heparin-resistant complex.
DNase I footprinting analyses suggest that c-di-GMP is needed to form the active transcriptional protein/DNA architecture at PvpsL
While RNAP alone, RNAP/c-di-GMP and RNAP/VpsR all yield basal transcription from PvpsL, activated transcription requires RNAP, VpsR and c-di-GMP. To investigate whether the protein–DNA interactions differed between the basal and activated transcription complexes, we performed DNase I footprinting. Again, we challenged the complexes with heparin and extracted the stable complexes from EMSA gels before isolating the DNA (Supplementary Figure S8) to ensure that we were observing contacts made within the stable open complex. The observed protection patterns and hypersensitive sites are summarized in Figure 1.
DNase I footprints of basal complexes formed with RNAP alone, RNAP/c-di-GMP or RNAP/VpsR at PvpsL were similar. On the non-template strand, protection was observed from −14 to −21 and −24 to −30 with hypersensitive sites at −22, −23, −34, −45, −46, −53, −54 (Figure 7A, lane 5–7). On the template strand, protection was present from −26 to −16 with hypersensitive sites at −57, −53 and −46 to −48 (Figure 7B, lane 5–7). Because the KMnO4 footprinting (detailed above) indicated the presence of an open bubble in these heparin resistant complexes, we conclude that these are the contacts present within open complexes for basal transcription at PvpsL. The presence of neither VpsR nor c-di-GMP alone to RNAP significantly affects these contacts.
In contrast, the activated complex of RNAP/VpsR/c-di-GMP at PvpsL generated distinct footprints. On the non-template strand, strong protection was observed from +29 to −11, −13 to −21, −24 to −33, −35 to −44 and −47 to -52 with hypersensitive sites at −22, −23, −54 and −55 (Figure 7A, lane 8). On the template strand, strong protection was seen from −56 to −28 and −26 to +21 with a hypersensitive site at −58 (Figure 7B, lane 8). This pattern is consistent with the formation of a typical stable open complex of RNAP and an activator or in our case, RNAP and VpsR/c-di-GMP at PvpsL.
Interestingly, a comparison of the DNase I footprints obtained with VpsR/RNAP/c-di-GMP versus VpsR/c-di-GMP reveals differences in the protection/cleavage patterns within the VpsR/c-di-GMP binding site of −31 to −53 as well as within the immediate upstream and downstream regions (compare patterns in Figure 1 and regions within the red dashed boxes in Figure 7). For example, on the non-template strand, the addition of RNAP to VpsR/c-di-GMP yielded enhanced protection downstream and within the downstream portion of the binding site (−25 to −32 and −35 to −40) and enhanced cleavage in the upstream portion (−45, −54 and −55). On the template strand, addition of RNAP to VpsR/c-di-GMP yielded enhanced cleavage (−46 to −48) within the VpsR/c-di-GMP-binding site and more protection (−53 to −55) and a hypersensitive site (−58) upstream of the binding site. Because footprints between VpsR alone versus VpsR/c-di-GMP are identical, both RNAP and c-di-GMP are required to facilitate these protein–DNA contact changes within the VpsR-binding site in the activated complex. Thus, these results suggest that the binding of VpsR to its DNA site is altered by the presence of both RNAP and c-di-GMP and/or that contacts between RNAP and the DNA are altered by the presence of both VpsR/c-di-GMP.
Taken together, the footprints suggest that the transcription complex formed by VpsR, c-di-GMP and RNAP at PvpsL is competent because it achieves a different architecture. The presence of RNAP alone or with either c-di-GMP or VpsR does not generate this particular protein–DNA conformation.
DISCUSSION
Biofilm formation by bacteria imposes an enormous medical cost, both in suffering and in the price of treatment. Consequently, understanding the regulation of biofilm formation is crucial to the prevention and treatment of bacterial disease. A central player in biofilm formation is the second messenger c-di-GMP, which has previously been shown to be required for the activity of several transcriptional activators including VpsR, the master regulator of biofilm formation in V. cholerae. By developing the first in vitro transcription assay with c-di-GMP, we have demonstrated that c-di-GMP works with VpsR in a novel way to stimulate transcription by RNAP at PvpsL, a promoter for biofilm biogenesis genes. Surprisingly, unlike other characterized regulators that use c-di-GMP, such as Klebsiella pneumoniae MrkH, Mycobacterium smegmatis LtmA, Streptomyces coelicolor BldD, V. cholerae VpsT and Pseudomonas aeruginosa FleQ and BrlR (12–19), VpsR does not require c-di-GMP to oligomerize or bind to the DNA. VpsR dimers form with or without c-di-GMP, and the presence of the second messenger does not substantially affect the affinity of VpsR for the DNA or the protein–DNA contacts made by VpsR alone at PvpsL. Instead, c-di-GMP is needed to observe distinct protein–DNA contacts within the activated transcription complex of σ70-RNAP/VpsR/c-di-GMP. How the presence of c-di-GMP results in these contacts is not clear. However, it could be needed to generate a particular VpsR conformation that is active for transcription and/or by promoting needed contacts between VpsR and σ70-RNAP. In fact, the position of the VpsR binding site immediately upstream of the −35 region suggests that VpsR should function as a Class II activator that can interact with σ70 region 4 and/or alpha CTDs.
Besides the novelty of activation, VpsR is also unusual as an atypical EBP. Classic EBPs interact with σ54-RNAP at a promoter, utilizing ATPase to form homomeric hexamers to generate the energy needed to form a stable open complex (47,48). However, VpsR, like other atypical EBPs, lacks the GAFTGA motif needed for interaction with σ54-RNAP and has non-conserved amino acids in the Walker B motif involved in ATP hydrolysis. Atypical EBPs that utilize σ70 rather than σ54 may represent an evolutionary link between these two very different σ class families. To date, five atypical EBPs have been characterized: E. coli TyrR, Rhodobacter capsulatus HupR, Myxococcus xanthus HsfA, Pseudomonas putida PhhR and Brucella abortus NtrX (49–54). While all five atypical EBPs contain variations in the GAFTGA motif responsible for binding to σ54, some contain non-consensus Walker A or Walker B motifs involved in ATP binding and hydrolysis (55). Recently, the crystal structure of B. abortus NtrX was solved, representing the first full-length crystal structure of a NtrC-like response regulator as well as the first full-length crystal structure of an atypical EBP. However, unlike VpsR, NtrX functions as a repressor at the pYX promoter and does not bind c-di-GMP (54). Thus, it appears that atypical EBPs may function by varied mechanisms. Nevertheless, the remaining four atypical EBPs work with σ70-RNAP in the absence of c-di-GMP to activate transcription. How the activity of these non-canonical EBPs is regulated remains to be determined for most of these transcription factors, but we show here that VpsR represents the first EBP and first atypical EBP that is dependent on a second messenger to directly activate transcription with RNAP.
In addition to VpsR, two other EBPs, FlrA in V. cholerae and FleQ from P. aeruginosa, are also directly controlled by c-di-GMP. However, unlike VpsR, these regulators are typical EBPs and contain conserved elements needed for σ54-dependent transcription. While binding of c-di-GMP to FlrA inhibits its ability to bind to the flrBC promoter to promote transcription activation (56), binding of c-di-GMP to FleQ has more complex effects. FleQ can regulate transcription at promoters containing σ54 or σ70 elements in P. aeruginosa, but it is unclear whether FleQ directly activates transcription with these sigma factors in vitro. Like FlrA, binding of c-di-GMP to FleQ represses flagellar genes in P. aeruginosa, but also derepresses and activates the pel biofilm extracellular polysaccharide gene cluster in vivo (14,57,58). Thus, VpsR, FlrA and FleQ appear to function on a continuum with each transcription factor having different dependencies on σ54 or σ70 as well as different responses to c-di-GMP. While FlrA and FleQ bind c-di-GMP via conserved arginine residues that flank a central cavity between the N-terminal receiver domain and central AAA+ domain (14), VpsR lacks these arginines, instead having a methionine and glutamate at those positions. The mechanism by which VpsR binds to c-di-GMP is therefore unknown.
Along with the proximal VpsR binding site from −31 to −53 at PvpsL, interestingly, a second VpsR binding site lies far upstream of PvpsL at −297 to −336. These binding sites differ in both sequence, length and protection intensities. Using DNase I footprinting with VpsR alone in the absence of c-di-GMP, the protection pattern was stronger at the distal site versus the proximal site (31). While VpsR protected the sequence TTTCTCAAAAATAATTCAATGTAAATCCAAAATGTAATAC at the distal site, VpsR protected the sequence AGTCTTAGAATTGATGCAGATA at the proximal site (31). Although this distal site has no effect in our in vitro transcription assays with purified proteins as well as no effect in transcriptional fusion studies when truncated, it appears that VpsR binding here is needed to relieve H-NS repression in vivo (31). The downstream portion of the distal VpsR binding site overlaps the first distal H-NS binding site (45). Thus, we speculate that at PvpsL, VpsR acts as an anti-H-NS repressor, blocking H-NS binding at the distal promoter site. In between the proximal and distal VpsR binding sites, a VpsT binding site is present from −238 to −192. Previous work demonstrates that VpsT acts solely as an antirepressor of H-NS at PvpsL and in vitro transcription studies in our laboratory show that VpsT does not directly activate transcription at PvpsL (data not shown). This allows for additional H-NS regulation in which both VpsR-binding to the distal promoter site and VpsT-binding downstream together help prevent H-NS from first binding the site overlapping the distal VpsR binding site. Upon freeing the DNA from H-NS binding, VpsR/c-di-GMP may then bind to the proximal binding site to directly activate transcription with RNAP. Other VpsR sites appear at various locations relative to the TSS of various genes. VpsR binds and regulates vpsT with a site at −149 to −119, aphA with a site at −88 to −70, and epsC with a site from −50 to −33 (28,32,59), and in silico analyses have identified conserved VpsR boxes present at other locations, including promoters for rbmA, rbmB, rbmC, rbmE, vpsU, vpsR, cdgC and bap1 (31). H-NS sites have also been identified at some of these promoters (vpsL, vpsT, rbmA, rbmB and rbmC (45,60,61)). Thus, we speculate that in general, promoter distal VpsR binding sites may correlate with a role in relieving H-NS repression, while promoter proximal sites may correlate with VpsR/c-di-GMP activation with RNAP. Such a mechanism may be similar to that used by Salmonella typhimurium SsrB. During biofilm formation, SsrB binds the DNA and displaces H-NS to relieve H-NS silencing and enable transcription activation of csgD, the master regulator of biofilms (62). However, at promoters of Salmonella Pathogenicity Island-2 SPI-2 genes, SsrB interacts with RNAP to activate transcription (63). The role of VpsR’s diverse and numerous binding sites remain unclear and future studies in determining the differing roles of VpsR in transcriptional activation versus relieving H-NS repression are in progress.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Hongwei Yu for plasmid pHERD20T, Geoffrey Severin for quantification of c-di-GMP using mass spectrometry, Michael Cashel for supplies and help on TLC assays and Kyung Moon for advice on protocols/protein purification. We also thank members of the Hinton and Waters laboratory for helpful discussions and comments on the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [F30GM123632 to M.L.H., R01GM109259 to C.M.W., in part]; National Science Foundation [MCB1253684 to C.M.W.]; Michigan State University DO/PhD Program (to M.L.H.); Intramural Research Program of the NIH, National Institute of Diabetes and Digestive and Kidney Diseases (to M.L.H., D.M.H.). Funding for open access charge: R01GM109259.
Conflict of interest statement. None declared.
REFERENCES
- 1. Bryers J.D. Medical biofilms. Biotechnol. Bioeng. 2008; 100:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wolcott R.D., Rhoads D.D., Bennett M.E., Wolcott B.M., Gogokhia L., Costerton J.W., Dowd S.E.. Chronic wounds and the medical biofilm paradigm. J. Wound Care. 2010; 19:45–46. [DOI] [PubMed] [Google Scholar]
- 3. Teschler J.K., Zamorano-Sanchez D., Utada A.S., Warner C.J., Wong G.C., Linington R.G., Yildiz F.H.. Living in the matrix: assembly and control of Vibrio cholerae biofilms. Nat. Rev. Microbiol. 2015; 13:255–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Alam M., Islam A., Bhuiyan N.A., Rahim N., Hossain A., Khan G.Y., Ahmed D., Watanabe H., Izumiya H., Faruque A.S. et al. . Clonal transmission, dual peak, and off-season cholera in Bangladesh. Infect. Ecol. Epidemiol. 2011; 1:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Alam M., Sultana M., Nair G.B., Siddique A.K., Hasan N.A., Sack R.B., Sack D.A., Ahmed K.U., Sadique A., Watanabe H. et al. . Viable but nonculturable Vibrio cholerae O1 in biofilms in the aquatic environment and their role in cholera transmission. PNAS. 2007; 104:17801–17806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Colwell R.R., Huq A., Islam M.S., Aziz K.M., Yunus M., Khan N.H., Mahmud A., Sack R.B., Nair G.B., Chakraborty J. et al. . Reduction of cholera in Bangladeshi villages by simple filtration. PNAS. 2003; 100:1051–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huq A., Small E.B., West P.A., Huq M.I., Rahman R., Colwell R.R.. Ecological relationships between Vibrio cholerae and planktonic crustacean copepods. Appl. Environ. Microbiol. 1983; 45:275–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Islam M.S., Jahid M.I., Rahman M.M., Rahman M.Z., Islam M.S., Kabir M.S., Sack D.A., Schoolnik G.K.. Biofilm acts as a microenvironment for plankton-associated Vibrio cholerae in the aquatic environment of Bangladesh. Microbiol. Immunol. 2007; 51:369–379. [DOI] [PubMed] [Google Scholar]
- 9. Ryjenkov D.A., Tarutina M., Moskvin O.V., Gomelsky M.. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 2005; 187:1792–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Romling U., Galperin M.Y., Gomelsky M.. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 2013; 77:1–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ross P., Weinhouse H., Aloni Y., Michaeli D., Weinberger-Ohana P., Mayer R., Braun S., de Vroom E., van der Marel G.A., van Boom J.H. et al. . Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature. 1987; 325:279–281. [DOI] [PubMed] [Google Scholar]
- 12. Krasteva P.V., Fong J.C., Shikuma N.J., Beyhan S., Navarro M.V., Yildiz F.H., Sondermann H.. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science. 2010; 327:866–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tschowri N., Schumacher M.A., Schlimpert S., Chinnam N.B., Findlay K.C., Brennan R.G., Buttner M.J.. Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development. Cell. 2014; 158:1136–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Matsuyama B.Y., Krasteva P.V., Baraquet C., Harwood C.S., Sondermann H., Navarro M.V.. Mechanistic insights into c-di-GMP-dependent control of the biofilm regulator FleQ from Pseudomonas aeruginosa. PNAS. 2016; 113:E209–E218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schumacher M.A., Zeng W.. Structures of the activator of K. pneumonia biofilm formation, MrkH, indicates PilZ domains involved in c-di-GMP and DNA binding. PNAS. 2016; 113:10067–10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wilksch J.J., Yang J., Clements A., Gabbe J.L., Short K.R., Cao H., Cavaliere R., James C.E., Whitchurch C.B., Schembri M.A. et al. . MrkH, a novel c-di-GMP-dependent transcriptional activator, controls Klebsiella pneumoniae biofilm formation by regulating type 3 fimbriae expression. PLoS Pathog. 2011; 7:e1002204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li W., He Z.G.. LtmA, a novel cyclic di-GMP-responsive activator, broadly regulates the expression of lipid transport and metabolism genes in Mycobacterium smegmatis. Nucleic Acids Res. 2012; 40:11292–11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chambers J.R., Liao J., Schurr M.J., Sauer K.. BrlR from Pseudomonas aeruginosa is a c-di-GMP-responsive transcription factor. Mol. Microbiol. 2014; 92:471–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schumacher M.A., Zeng W., Findlay K.C., Buttner M.J., Brennan R.G., Tschowri N.. The Streptomyces master regulator BldD binds c-di-GMP sequentially to create a functional BldD2-(c-di-GMP)4 complex. Nucleic Acids Res. 2017; 45:6923–6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee D.J., Minchin S.D., Busby S.J.. Activating transcription in bacteria. Annu. Rev. Microbiol. 2012; 66:125–152. [DOI] [PubMed] [Google Scholar]
- 21. Decker K.B., Hinton D.M.. Transcription regulation at the core: similarities among bacterial, archaeal, and eukaryotic RNA polymerases. Annu. Rev. Microbiol. 2013; 67:113–139. [DOI] [PubMed] [Google Scholar]
- 22. Feklistov A., Darst S.A.. Structural basis for promoter-10 element recognition by the bacterial RNA polymerase sigma subunit. Cell. 2011; 147:1257–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang Y., Feng Y., Chatterjee S., Tuske S., Ho M.X., Arnold E., Ebright R.H.. Structural basis of transcription initiation. Science. 2012; 338:1076–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kalia D., Merey G., Nakayama S., Zheng Y., Zhou J., Luo Y., Guo M., Roembke B.T., Sintim H.O.. Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP, (p)ppGpp signaling in bacteria and implications in pathogenesis. Chem. Soc. Rev. 2013; 42:305–341. [DOI] [PubMed] [Google Scholar]
- 25. Beyhan S., Bilecen K., Salama S.R., Casper-Lindley C., Yildiz F.H.. Regulation of rugosity and biofilm formation in Vibrio cholerae: comparison of VpsT and VpsR regulons and epistasis analysis of vpsT, vpsR, and hapR. J. Bacteriol. 2007; 189:388–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lauriano C.M., Ghosh C., Correa N.E., Klose K.E.. The sodium-driven flagellar motor controls exopolysaccharide expression in Vibrio cholerae. J. Bacteriol. 2004; 186:4864–4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rashid M.H., Rajanna C., Zhang D., Pasquale V., Magder L.S., Ali A., Dumontet S., Karaolis D.K.. Role of exopolysaccharide, the rugose phenotype and VpsR in the pathogenesis of epidemic Vibrio cholerae. FEMS Microbiol. Lett. 2004; 230:105–113. [DOI] [PubMed] [Google Scholar]
- 28. Srivastava D., Harris R.C., Waters C.M.. Integration of cyclic di-GMP and quorum sensing in the control of vpsT and aphA in Vibrio cholerae. J. Bacteriol. 2011; 193:6331–6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Waters C.M., Lu W., Rabinowitz J.D., Bassler B.L.. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J. Bacteriol. 2008; 190:2527–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yildiz F.H., Dolganov N.A., Schoolnik G.K.. VpsR, a member of the response regulators of the Two-Component regulatory systems, is required for expression of vps biosynthesis genes and EPS(ETr)-Associated phenotypes in vibrio cholerae O1 El Tor. J. Bacteriol. 2001; 183:1716–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zamorano-Sanchez D., Fong J.C., Kilic S., Erill I., Yildiz F.H.. Identification and characterization of VpsR and VpsT binding sites in Vibrio cholerae. J. Bacteriol. 2015; 197:1221–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sloup R.E., Konal A.E., Severin G.B., Korir M.L., Bagdasarian M.M., Bagdasarian M., Waters C.M.. Cyclic Di-GMP and VpsR induce the expression of type II secretion in vibrio cholerae. J. Bacteriol. 2017; 199:e00106-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pursley B.R., Maiden M.M., Hsieh M.L., Fernandez N.L., Severin G.B., Waters C.M.. Cyclic di-GMP regulates TfoY in vibrio cholerae to control motility by both transcriptional and posttranscriptional mechanisms. J. Bacteriol. 2018; 200:578–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rappas M., Bose D., Zhang X.. Bacterial enhancer-binding proteins: unlocking sigma54-dependent gene transcription. Curr. Opin. Struct. Biol. 2007; 17:110–116. [DOI] [PubMed] [Google Scholar]
- 35. Ross W., Thompson J.F., Newlands J.T., Gourse R.L.. E. coli Fis protein activates ribosomal RNA transcription in vitro and in vivo. EMBO J. 1990; 9:3733–3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hammer B.K., Bassler B.L.. Regulatory small RNAs circumvent the conventional quorum sensing pathway in pandemic Vibrio cholerae. PNAS. 2007; 104:11145–11149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Qiu D., Damron F.H., Mima T., Schweizer H.P., Yu H.D.. PBAD-based shuttle vectors for functional analysis of toxic and highly regulated genes in Pseudomonas and Burkholderia spp. and other bacteria. Appl. Environ. Microbiol. 2008; 74:7422–7426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. March-Amegadzie R., Hinton D.M.. The bacteriophage T4 middle promoter PuvsX: analysis of regions important for binding of the T4 transcriptional activator MotA and for activation of transcription. Mol. Microbiol. 1995; 15:649–660. [DOI] [PubMed] [Google Scholar]
- 39. Thelin K.H., Taylor R.K.. Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains. Infect. Immun. 1996; 64:2853–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. de Lorenzo V., Timmis K.N.. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 1994; 235:386–405. [DOI] [PubMed] [Google Scholar]
- 41. Hsieh M.L., James T.D., Knipling L., Waddell M.B., White S., Hinton D.M.. Architecture of the bacteriophage T4 activator MotA/promoter DNA interaction during sigma appropriation. J. Biol. Chem. 2013; 288:27607–27618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Massie J.P., Reynolds E.L., Koestler B.J., Cong J.P., Agostoni M., Waters C.M.. Quantification of high-specificity cyclic diguanylate signaling. PNAS. 2012; 109:12746–12751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Boulanger A., Moon K., Decker K.B., Chen Q., Knipling L., Stibitz S., Hinton D.M.. Bordetella pertussis fim3 gene regulation by BvgA: phosphorylation controls the formation of inactive vs. active transcription complexes. PNAS. 2015; 112:E526–E535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Teschler J.K., Cheng A.T., Yildiz F.H.. The two-component signal transduction system VxrAB positively regulates vibrio cholerae biofilm formation. J. Bacteriol. 2017; 199:e00139-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ayala J.C., Wang H., Silva A.J., Benitez J.A.. Repression by H-NS of genes required for the biosynthesis of the Vibrio cholerae biofilm matrix is modulated by the second messenger cyclic diguanylic acid. Mol. Microbiol. 2015; 97:630–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hook-Barnard I.G., Hinton D.M.. Transcription initiation by mix and match Elements: Flexibility for polymerase binding to bacterial promoters. Gene Regul. Syst. Biol. 2007; 1:275–293. [PMC free article] [PubMed] [Google Scholar]
- 47. Bush M., Dixon R.. The role of bacterial enhancer binding proteins as specialized activators of sigma54-dependent transcription. Microbiol. Mol. Biol. Rev. 2012; 76:497–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Joly N., Zhang N., Buck M., Zhang X.. Coupling AAA protein function to regulated gene expression. Biochim. Biophys. Acta. 2012; 1823:108–116. [DOI] [PubMed] [Google Scholar]
- 49. Pittard J., Camakaris H., Yang J.. The TyrR regulon. Mol. Microbiol. 2005; 55:16–26. [DOI] [PubMed] [Google Scholar]
- 50. Studholme D.J., Dixon R.. Domain architectures of sigma54-dependent transcriptional activators. J. Bacteriol. 2003; 185:1757–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Herrera M.C., Ramos J.L.. Catabolism of phenylalanine by Pseudomonas putida: the NtrC-family PhhR regulator binds to two sites upstream from the phhA gene and stimulates transcription with sigma70. J. Mol. Biol. 2007; 366:1374–1386. [DOI] [PubMed] [Google Scholar]
- 52. Dischert W., Vignais P.M., Colbeau A.. The synthesis of Rhodobacter capsulatus HupSL hydrogenase is regulated by the two-component HupT/HupR system. Mol. Microbiol. 1999; 34:995–1006. [DOI] [PubMed] [Google Scholar]
- 53. Ueki T., Inouye S.. Transcriptional activation of a heat-shock gene, lonD, of Myxococcus xanthus by a two component histidine-aspartate phosphorelay system. J. Biol. Chem. 2002; 277:6170–6177. [DOI] [PubMed] [Google Scholar]
- 54. Fernandez I., Cornaciu I., Carrica M.D., Uchikawa E., Hoffmann G., Sieira R., Marquez J.A., Goldbaum F.A.. Three-Dimensional structure of Full-Length NtrX, an unusual member of the NtrC family of response regulators. J. Mol. Biol. 2017; 429:1192–1212. [DOI] [PubMed] [Google Scholar]
- 55. Ghosh T., Bose D., Zhang X.. Mechanisms for activating bacterial RNA polymerase. FEMS Microbiol. Rev. 2010; 34:611–627. [DOI] [PubMed] [Google Scholar]
- 56. Srivastava D., Hsieh M.L., Khataokar A., Neiditch M.B., Waters C.M.. Cyclic di-GMP inhibits Vibrio cholerae motility by repressing induction of transcription and inducing extracellular polysaccharide production. Mol. Microbiol. 2013; 90:1262–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Baraquet C., Harwood C.S.. FleQ DNA binding consensus sequence revealed by studies of FleQ-Dependent regulation of biofilm gene expression in pseudomonas aeruginosa. J. Bacteriol. 2015; 198:178–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Baraquet C., Harwood C.S.. Cyclic diguanosine monophosphate represses bacterial flagella synthesis by interacting with the Walker A motif of the enhancer-binding protein FleQ. PNAS. 2013; 110:18478–18483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lin W., Kovacikova G., Skorupski K.. The quorum sensing regulator HapR downregulates the expression of the virulence gene transcription factor AphA in Vibrio cholerae by antagonizing Lrp- and VpsR-mediated activation. Mol. Microbiol. 2007; 64:953–967. [DOI] [PubMed] [Google Scholar]
- 60. Ayala J.C., Wang H., Benitez J.A., Silva A.J.. RNA-Seq analysis and whole genome DNA-binding profile of the Vibrio cholerae histone-like nucleoid structuring protein (H-NS). Genomics Data. 2015; 5:147–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wang H., Ayala J.C., Silva A.J., Benitez J.A.. The histone-like nucleoid structuring protein (H-NS) is a repressor of Vibrio cholerae exopolysaccharide biosynthesis (vps) genes. Appl. Environ. Microbiol. 2012; 78:2482–2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Desai S.K., Winardhi R.S., Periasamy S., Dykas M.M., Jie Y., Kenney L.J.. The horizontally-acquired response regulator SsrB drives a Salmonella lifestyle switch by relieving biofilm silencing. Elife. 2016; 5:e10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Walthers D., Li Y., Liu Y., Anand G., Yan J., Kenney L.J.. Salmonella enterica response regulator SsrB relieves H-NS silencing by displacing H-NS bound in polymerization mode and directly activates transcription. J. Biol. Chem. 2011; 286:1895–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.