Abstract
Despite the prime importance of telomeres in chromosome stability, significant mysteries surround the architecture of telomeric chromatin. Through micrococcal nuclease mapping, we show that fission yeast chromosome ends are assembled into distinct protected structures (‘telosomes’) encompassing the telomeric DNA repeats and over half a kilobase of subtelomeric DNA. Telosome formation depends on the conserved telomeric proteins Taz1 and Rap1, and surprisingly, RNA. Although yeast telomeres have long been thought to be free of histones, we show that this is not the case; telomere repeats contain histones. While telomeric histone H3 bears the heterochromatic lys9-methyl mark, we show that this mark is dispensable for telosome formation. Therefore, telomeric chromatin is organized at an architectural level, in which telomere-binding proteins and RNAs impose a unique nucleosome arrangement, and a second level, in which histone modifications are superimposed upon the higher order architecture.
INTRODUCTION
Telomere composition is uniquely tailored to execute the diverse functions ascribed to chromosome ends, including chromosome end-replication and telomerase recruitment, meiotic bouquet formation, and the prevention of chromosome end-fusion (1). Tracts of tandemly repeated double-stranded (ds) G-rich telomeric DNA repeats (TTAGGG in vertebrates and related repeats in other eukaryotes), terminating in a single-strand (ss) G-rich overhang, form the foundation for recruitment of six proteins, known collectively as shelterin. Mammalian shelterin comprises the ds telomere-binding proteins TRF1 and TRF2, the ss telomere-binding proteins POT1 and TPP1, the TIN2 ss-ds bridging factor and the TRF2-interacting protein RAP1 (2,3). The analogous complex in fission yeast comprises the ds telomere-binding protein Taz1 (ortholog of TRF1/2), the ss telomere-binding proteins Pot1 and Tpz1, the Pot1-interacting factor Ccq1, the ss-ds bridging factor Poz1 and the Taz1-interacting protein Rap1 (4). Transcripts of the telomere and subtelomere, known as TERRA, also associate with chromosome ends. While the function of this long noncoding RNA (lncRNA) is not fully understood, a growing body of evidence suggests important modulatory roles in telomere maintenance and protection (5,6).
Current pressing issues include how telomere components function within the context of a higher order chromatin structure and how this structure adapts to accommodate the plethora of telomere functions. The prevailing model of mammalian telomere architecture implicates a structure termed the t-loop, in which the telomeric region folds back and the 3′ ss overhang invades the subterminal ds telomeric DNA repeats (7). This chromosome end arrangement has been evinced by electron microscopy and superresolution microscopy of psoralen-crosslinked telomeric DNA, and a role in chromosome end protection has been suggested by the observation that the presence of t-loops requires TRF2 (8). However, even if this model is applicable to Schizosaccharomyces pombe, an organism with shorter telomeres and thus fewer homologous repeats for the ss terminus to invade, questions about the distribution of proteins at telomeres remain.
How histones fit into this picture and in particular, how sequence-specific binding proteins access telomeric DNA in the context of nucleosomes, have not been resolved. In vitro studies have demonstrated that telomeric nucleosomes are much less stable than bulk nucleosomes due to the sequence and periodicity of telomere repeats (9). The low affinity of telomeric DNA for nucleosome wrapping may be overcome, or perhaps capitalized upon, by telomere-specific binding proteins that package the telomere into a functional chromosomal domain. Recent in vitro studies have demonstrated that TRF2 binds with much lower affinity than TRF1 to nucleosomal DNA, and that these affinities are differentially altered by histone modifications; accordingly, TRF1 and TRF2 binding appears to remodel nucleosome spacing or mobility on model DNA templates (10). Moreover, TRF1 has been suggested to alter the structure of the nucleosome itself (11). Nonetheless, a typical ‘beads-on-a-string’ nucleosome array has been observed by EM of telomere-enriched chromatin from chicken erythrocytes and mouse splenocytes, suggesting that nucleosomal wrapping of telomeric DNA can be achieved in vivo (12).
Telomere architecture has also been investigated using classical chromatin-mapping techniques, in which the pattern of micrococcal nuclease (MNase) digestion of intact chromatin preparations is analysed. Mammalian telomeres present a particular challenge to the MNase approach, as the telomeric DNA sequence comprises five to dozens of kilobases of 6 bp tandem repeats. Hence, sequence-based analysis of chromatin structure, which represents an average of the entire telomeric region as well as interstitial telomere regions, does not distinguish distal versus centromere-proximal telomeric regions. The bulk of telomere repeats in mammals are incorporated into nucleosomes that have a shorter repeat length than the genome average but exhibit periodic, well-defined spacing (13,14). However, an alternative structure has been observed in some mammalian cells that harbor shorter telomeres, suggesting that when the terminus represents a significant fraction of the total telomere repeats, a distinct structure can be seen at the ‘business end’ of the telomere (15). Further support for an alternative telomere arrangement came from examination of telomere ultrastructure in organisms with fewer telomere repeats, where MNase mapping revealed a ‘non-nucleosomal’ structure termed the ‘telosome’. In S. cerevisiae, the telosome comprises a variable region encompassing the terminal 245–400 bp of the chromosome (16). The termini of Oxytricha and Tetrahymena chromosomes are also protected from MNase digestion in structures distinct from canonical nucleosomal arrays (17,18). The nature of these telosomal structures is not entirely understood, though it is thought that binding by telomere-specific proteins could exclude nucleosomes or block access to nucleases. The centromere-proximal 3–10% of Tetrahymena telomere repeats appear in nucleosomal arrays, leading to the suggestion that differences between the nucleosomal nature of mammalian versus ciliate telomeres are quantitative rather than qualitative (19). Intriguingly, the presence of histones within such telosomes has neither been explicitly demonstrated nor fully refuted.
Fission yeast telomeres are highly similar to human telomeres not only in composition but also in terms of functional principles (Figure 1A). Like the TRFs, Taz1 is required for protection from end-fusions, semi-conservative replication of telomeric DNA, telomerase regulation and meiotic bouquet formation (20–23). The loss of Taz1 leads as well to an increase in G-rich and C-rich telomeric and subtelomeric RNA, indicating a role in telomere transcriptional regulation (24). Moreover, like budding yeast and mammalian telomeres, fission yeast telomeres are heterochromatic as initially revealed via telomere position effect (TPE), in which genes placed adjacent to telomeres are silenced in a Taz1-dependent manner (20). Accordingly, canonical heterochromatic histone marks, such as histone H3K9Me2, are seen in subtelomeric chromatin (25,26).
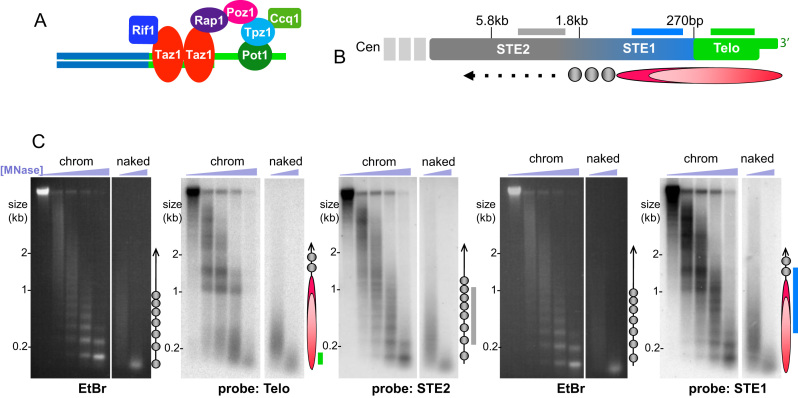
Figure 1.
Unique chromatin structure at telomeres. (A) Schematic of fission yeast telomere components. S. pombe shelterin, comprising Taz1, Rap1, Poz1, Tpz1, Ccq1 and Pot1, is highly similar to mammalian shelterin. (B) Schematic of subtelomeric (STE1 and STE2) and telomeric (Telo) regions, with distance from chromosome end indicated above. Probes used are depicted as lines below each region (not to scale). Below the schematic is a ball-and-stick diagram of the dimensions of the telosome mapped in this study. Circles indicate canonical nucleosomes; ovals depict the telosome. (C) Gel and Southern blot analysis of subtelomeric and telomeric regions following treatment of intact chromatin with increasing concentrations of MNase (‘chromatin’). Naked DNA was also treated with MNase to control for sequence specificity. Duplicate samples were electrophoresed on the same gel and EtBr stained. The EtBr images in the left and 4th panel show the pattern of bulk genomic chromatin, attesting to the intactness of the chromatin preparation. Each gel was subjected to Southern blot analysis using the probes indicated below each panel. The STE2 probe was applied after stripping the Telo blot. The ball-and-stick schematics to the right of each blot depict the chromatin structure, to scale with the blots; circles and ovals as in (B) with probe positions indicated by lines whose colors correspond to the designations in (B). Note that the ball-and-stick diagrams reflect the different probe positions for each blot; for instance, the STE1 probe hybridizes to more centromere-proximal genomic regions that start to contain canonical nucleosomes, hence the appearance of mononucleosome bands on the STE1-probed blot.
We sought to exploit the relatively tractable telomeres of fission yeast to define telomeric chromatin structure and its determinants through MNase mapping. Our findings reveal the presence of a distinct protected structure at the chromosome end whose size scales with telomere length and whose presence relies on conserved telomere proteins. Interestingly, we uncover a role for RNA in telosome structure. Moreover, we find that despite the packaging of telomeres in a manner distinct from canonical nucleosomal arrays, the telosome contains histones, adding a further layer of complexity to the telomere ultrastructure.
MATERIALS AND METHODS
Fission yeast strains
Strains are listed in Supplementary Table S3. Standard media and growth conditions were used.
Nuclear isolation, chromatin treatment and Southern blotting
Nuclei were isolated largely according to Simpson et al. (27) and Experiment 18 (28). Briefly, 1 L of freshly cultured cells at 0.6 × 107 cells/ml were resuspended in S buffer (1.4 M sorbitol, 40 mM HEPES, 0.5 mM MgCl2 pH 6.5), incubated in S buffer with 10 mM β-Me and treated with zymolyase for 20 min. After washing with S buffer, cells were resusupended in 20 ml F buffer (18% Ficoll 400, 20 mM PIPES, 0.5 mM MgCl2 pH 6.5) and spun over 20 ml GF buffer (7% Ficoll 400, 20% glycerol, 20 mM PIPES, 0.5 mM MgCl2) at 20 000g for 30 min. Pellets were then resuspended in F buffer, spun at 3000g for 15 min to remove debris, then centrifuged at 20 000g for 25 min. Nuclear pellets were then resuspended in D buffer (10 mM HEPES pH 7.4, 0.5 mM MgCl2 and 0.05 mM CaCl2). Nuclei were split into separate reactions and treated with various concentrations of micrococcal nuclease (Thermo Scientific) ranging from 0 to 500 units/ml for 10 min at 37°C. Reactions were stopped by adding 10 mM EDTA and placing tubes on ice. DNA was then isolated using standard procedures. In RNA removal experiments, nuclei were resuspended in D buffer and incubated with 0.03 μg/μl RNase A (Sigma) for 10 or 30 min at 37°C. RNase was removed by washing nuclei in D buffer twice and adding 400u RNasin (Promega). MNase digestion was then performed as described above. Southern blot analyses were performed as described previously using telomere probes that contain only telomere repeat sequence (24). STE1 probe was isolated from pNSU70 after digestion with Apa1 and EcoRI, and STE2 was isolated from pNSU70 after digestion with NsiI. Naked DNA was prepared using a standard genomic DNA extraction as for Southern blot analysis. Genomic DNA was treated with MNase (7.5 and 100 units/ml) under the same conditions as above for nuclei, phenol–chloroform extracted, ethanol precipitated and included in subsequent Southern blot analysis.
ChIP and ChIP-seq
Anti-histone H3-K9Me2 antibody (Abcam) was used to perform ChIP for dot blots and Southerns as described previously (29).
For telomere enrichment analysis by ChIP-seq, 1 L of cells at 0.6 × 107 cells/ml were crosslinked with 1% formaldehyde for 15 min at 32°C, and ChIP carried out with the anti-histone H3-K9Me2. DNA was then prepared for sequencing using the TruSeq ChIP library preparation kit. Sequencing was performed on the Illumina HiSeq 2500 platform and typically generated ∼46 million 101-bp paired-end reads per sample. Raw reads from each sample were adapter-trimmed with cutadapt (version 1.9.1) (30) and subsequently filtered for the telomeric repeat motif ‘GTTACAGG’ (31). De novo assembly was performed independently for each sample on the motif filtered paired-end reads with both SPAdes (version 3.9.0; kmer = 97) (32) and ABySS (version 1.9.0; kmer = 95) (33). Upon manual inspection, we were able to identify 10 candidate telomeric contigs (five for each replicate) across the replicate groups that were assembled independently within both the input and IP samples and/or by both SPAdes and ABySS.
The S. pombe genome was reconstructed to include the assembled contigs, three nuclear chromosomes, the mitochondrial chromosome, mating type region, gap filling sequences between SPBPB21E7.09 and SPBPB10D8.01 in chromosome 2 (contig AB325691) (34), and a set of small insert clones generated from a telomere plasmid library. Sequence files were downloaded from Pombase (35). BWA (version 0.5.9-r16) (36) with default parameters was used to perform genome-wide mapping of the adapter-trimmed reads. Discordant, multi-mapped and soft-clipped read pairs were removed, leaving only those that mapped to the same contig in the correct orientation, had zero mismatches in both reads, and a maximum insert size of 1 kb.
RESULTS
Non-canonical chromatin structure of the S. pombe telomere
As MNase preferentially cleaves linker regions between nucleosomes, limited MNase digestion of intact chromatin followed by purification of the underlying DNA yields a ladder of digestion products representing a ‘beads-on-a-string’ protection pattern. Ethidium bromide (EtBr) staining of electrophoresed fission yeast MNase digestion products (Figure 1) yields bands consistent with the reported average nucleosomal repeat length of ∼156 bp (37). Subjecting this gel to Southern blot analysis using a probe for telomere repeats (Figure 1C) reveals the telomeric chromatin arrangement. In marked contrast to the ladders representing mono-, di-, tri- and higher order nucleosome arrays seen for the bulk genome, two predominant diffuse bands of ∼1.0 and 1.4 kb are detected with the telomere probe. We hereafter refer to this alternative structure as the telosome (16). The difference between this MNase digestion pattern and that of naked telomeric DNA reveals that the digestion pattern is not due to sequence (Figure 1c).
As telomere repeats constitute only 300 ± 50 bp of the chromosome end, the MNase-resistant fragments of ∼1 and 1.4 kb contain ∼700–1100 bp of subtelomeric sequences, which comprise semi-repetitive elements that extend ∼20 kb toward the centromere. Indeed, hybridization of the blot with a probe to the most telomere-proximal subtelomeric region (STE1, Figure 1B and C) reveals the same protected fragments at 1.0 and 1.4 kb, confirming that they incorporate subtelomeric sequences along with telomere repeats. In addition, a faint nucleosomal ladder becomes detectable with the STE1 probe, likely reflecting hybridization not only to the most distal part of the subtelomere, but also to more centromere-proximal regions of the subtelomere that are not included in the telosome. Using the STE2 probe (Figure 1B and C), which hybridizes to more centromere-proximal subtelomeric elements, the telosome is no longer detectable, confirming that the telosome comprises only the most distal region of the chromosome.
In some experiments (e.g. Figure 1C), we observe additional telomeric hybridization centered at ∼0.2 kb. As these species are variable and do not appear in other experiments (e.g. Figure 2A below and (38)), they likely represent replicating or dynamic telomeres that are quasi-resistant to MNase, or variable levels of protease activity in our chromatin preparations. Nonetheless, we cannot discount the possibility that these ∼0.2 kb species comprise substructures of the 1.0/1.4 kb telosome.
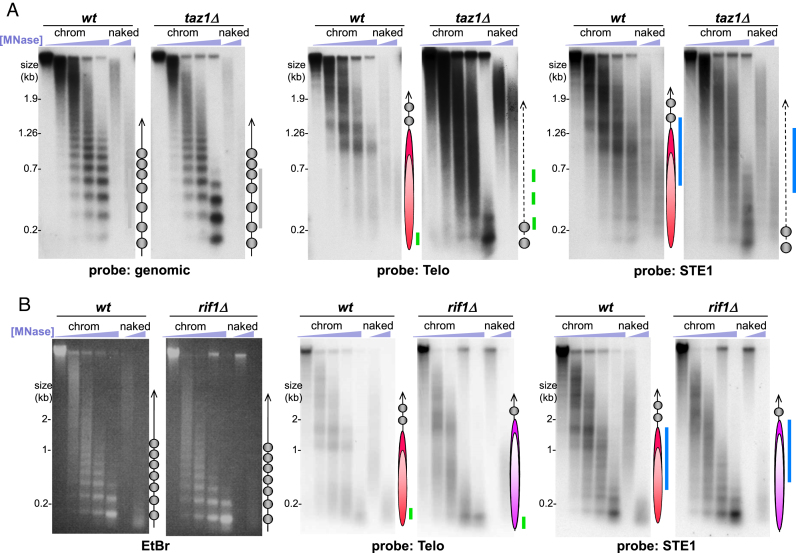
Figure 2.
Telosomes require Taz1 and scale in size with telomere length. (A) Southern blot analysis of MNase-digested chromatin. Probes are designated below each pair of blots. Bulk chromatin is represented by a probe for the gene cut7+. The ball-and-stick diagrams are as in Figure 1C. The disorganized structure at taz1Δ telomeres is represented by a dotted line. (B) Analysis of chromatin from wt and rif1Δ cells. Designations as in Figure 1; the longer ovals in violet represent the larger telosomes in rif1Δ cells.
The inclusion of 700–1100 bp of subtelomeric DNA (the subtelomeric elements or STE, upstream of the telomere repeat region) in a protected structure prompted us to ask whether the subtelomere itself defines the internal edge of the telosome, and indeed whether the subtelomere is necessary for telosome formation. For this, we utilized a strain in which only Chromosome III (Chr III) contains telomeres, as Chr I and II are circularized (see Supplementary Figure S1). STE are absent from Chr III, on which the telomeres at either end are directly abutted by rDNA repeats. A MNase digestion pattern similar to that of the wt telosome is detectable in this strain (Supplementary Figure S1), implying that chromosomes lacking STE can form telosomes. The altered size of the telosome bands in this strain could reflect a difference in telomere length stemming from reintroduction of telomerase following survival of telomere loss (see Supplementary Figure S1), or a subtle difference conferred by the abutting rDNA; the decreased intensity of telosome bands presumably reflects the relative deficit of telomeres in this strain harbouring only one linear chromosome. Nevertheless, the presence of telosomes lacking STE indicates that STE sequences are dispensable for telosome formation.
Taz1 and Rap1 are required for telosome structure
What components of the telomere confer the unique structure of the telosome? To define the telosomal determinants, we examined the MNase cleavage profiles of cells lacking Taz1. Strikingly, while bulk chromatin is unaffected by Taz1 loss, neither a telosomal pattern nor an extensive nucleosomal array is detectable at taz1Δ telomeres (Figure 2A, Supplementary Figure S2). Instead, we observe a diffuse smear of MNase digestion products along with mono- and di-nucleosomes; note that the intensity of hybridization with the telomere probe is enhanced in taz1Δ relative to wt, due to the 5- to 10-fold increase in telomere length in this setting. Importantly, loss of the telosome pattern is also apparent when hybridizing the same blot with a STE1 probe whose cognate sequence is not amplified in a taz1Δ setting (Figure 2A, Supplementary Figure S2). Hence, the diffuse pattern is not simply a result of the pronounced telomere lengthening seen in the absence of Taz1. The lack of a detectable telosome in the absence of Taz1 indicates that Taz1 is required for telosome formation or integrity. Our observation that extensive nucleosomal arrays fail to replace the telosome at taz1Δ telomeres suggests that nucleosome formation is obstructed by an array of ill-defined particles containing non-telomere-specific proteins whose residual affinities for telomere sequences rival the low affinity of histones for these sequences. Alternatively, taz1Δ telomeres may be largely unprotected from MNase. A paucity of nucleosomes could stem from the excessive levels of G-rich ssDNA at taz1Δ telomeres (39), which may exclude nucleosomal arrays or associate with histones in a non-periodic manner.
To address the possibility that single-strandedness is responsible for the diffuse pattern of taz1Δ telomeric MNase digests, we deleted rad50+ in a taz1Δ strain, as Rad50 is necessary for excessive ssDNA accumulation in cells lacking Taz1 (39). Deletion of rad50+ yields no detectable alteration of the telosome pattern in wt or taz1Δ cells, indicating that the chromatin pattern at taz1Δ telomeres is not a consequence of excessive ssDNA accumulation (Supplementary Figure S3).
A subset of Taz1’s functions is mediated by its interacting partner Rap1. Indeed, the telosome pattern is disrupted in rap1Δ cells, indicating that Taz1 and Rap1 act together in organizing the telosome (Supplementary Figure S4). This result allows us to parse telomere functions that correlate or fail to correlate with telosome formation. For example, progression of replication forks through telomere repeats requires Taz1 but not Rap1, while telomere length regulation and prevention of telomeric non-homologous end-joining (NHEJ) require both (21,23,40). Hence, our observation that rap1Δ telomeres lack detectable telosomes suggests that telosomes are not required for telomeric fork progression. Conversely, the telomerase-dependent hyper-elongation and loss of protection from chromosome end-fusion in taz1Δ and rap1Δ cells accompanies loss of the telosome, hinting at a role for the telosome in protecting the chromosome end from inappropriate telomerase-mediated elongation and NHEJ.
Telosome size scales with telomere length
To further investigate telosomal determinants and the interdependency of telosome formation and telomere function, we examined rif1Δ cells, which harbor telomeres ∼2-fold longer than wt but retain Taz1, Rap1 and protection from inappropriate DNA-damage repair (DDR) reactions at telomeres (40,41). Intriguingly, protection patterns reminiscent of wt are seen for rif1Δ telomeres but fragment sizes are shifted upwards, from 1.0 and 1.4 kb in wt to 1.7 and 2.1 kb in the rif1Δ setting (Figure 2B). Therefore, the telosome can accommodate larger amounts of telomeric DNA, and this is reflected in a corresponding increase in telosome size; this observation is consistent with the larger telosome fragments seen in strains harbouring only the longer telomeres found on Chr III (Supplementary Figure S1). We infer that telosome formation does not prevent the moderate telomere elongation conferred by rif1 deletion, nor does it enforce late replication of telomeres (which requires Rif1) (42).
Chromatin structure of an internally placed stretch of telomere repeats
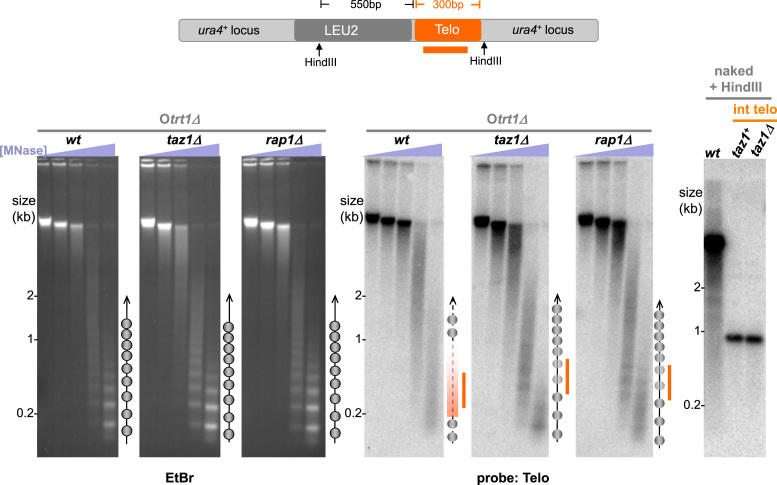
To address the role of ‘endedness’ in telosome formation as well as in nucleosome array exclusion in the absence of the telosome, we investigated the MNase protection pattern induced by a 300 bp stretch of telomeric repeats inserted at an internal chromosome site, the ura4+ locus on Chr III (Figure 3). Previous work has shown that Taz1 binds these internal telomere repeat stretches and confers a local position effect, silencing nearby genes (43). To avoid confusion between the internal telomere stretch and natural telomeres, we utilized a telomerase-minus strain harbouring circular chromosomes that lack endogenous telomeres. Curiously, we observe neither a discrete telosomal pattern nor a ladder of bands representing nucleosomal arrays at the internal telomere; rather, a diffuse pattern is seen (Figure 3). Interestingly, deletion of either taz1+ or rap1+ in this strain leads to a more distinct nucleosome array pattern, indicating that the noncanonical MNase pattern of the internal telomere is dictated by the telomeric dsDNA binding proteins. The residual indistinctness of the pattern in taz1Δ or rap1Δ backgrounds could be due to the poor affinity of nucleosomes for telomere repeats or to some level of redundancy between Taz1 and Rap1 for conferring the exclusion of canonical nucleosome patterns. Hence, the presence of Taz1/Rap1 either prevents the formation of nucleosomal arrays or alters the accessibility of linker regions. However, the discrete telosome structure found at bona fide chromosome ends appears to require some feature, like the telomeric 3′ overhang, that is unique to the chromosome terminus.
Figure 3.
Alternative chromatin structure at internal telomeric DNA stretches. Analysis of MNase-digested chromatin in telomerase-minus strains harboring circular chromosomes (Otrt1Δ) with an internally inserted stretch of telomere repeats at the ura4+ locus on Chr III. EtBr-stained gels (left) were subjected to Southern blot analysis with a telomere repeat probe (right). Ball-and-stick schematics are as in Figure 1C. The internal telomeric DNA lacks a discrete structure in an otherwise wt setting, while deletion of taz1+ or rap1+ leads to a more distinct nucleosomal pattern, although more diffuse than the bulk genomic nucleosomal pattern. Importantly, the internal telomeric DNA runs as a homogenous, stable band on Southern blots of HindIII-digested DNA both before and after taz1 deletion (diagram, and right-hand blot) despite the modest degree of replication fork stalling seen at the internal telomere in the absence of Taz1 (21).
The Taz1/Rap1-dependent indistinctness conferred by the inserted telomere repeat stretch resembles the MNase digestion pattern seen at the central core of the centromere, where the histone H3 variant Cnp1 forms unique nucleosomal complexes that confer centromere function (38,44–48). Despite the packaging of centromeric DNA into nucleosomes, their alternative structural composition renders this region ‘indecipherable’ by MNase digestion. Thus, centromeric chromatin structure may lend clues about possible alternative arrangements of telomeric chromatin; however, in order to interpret these clues, we were compelled to ascertain definitively whether fission yeast telosomes harbor histone proteins.
Fission yeast telomeres contain histones
To determine whether telomere repeats associate with histones, we performed ChIP with an antibody against histone H3-lys9-dimethyl (H3K9Me2), reasoning that if histones exist in the telomere, they likely carry this modification, which characterizes all fission yeast heterochromatin. Dot-blot analysis revealed that telomere sequences are enriched in H3K9Me2 immunoprecipitates when input chromatin fragments are sonicated to a small size, between 100 and 600 bp (Figure 4A), suggesting that telomeres are indeed incorporated into H3K9Me2-containing nucleosomes. However, it remained possible that the sonicated fragments hybridizing with the telomere probe also contain subtelomeric sequences.
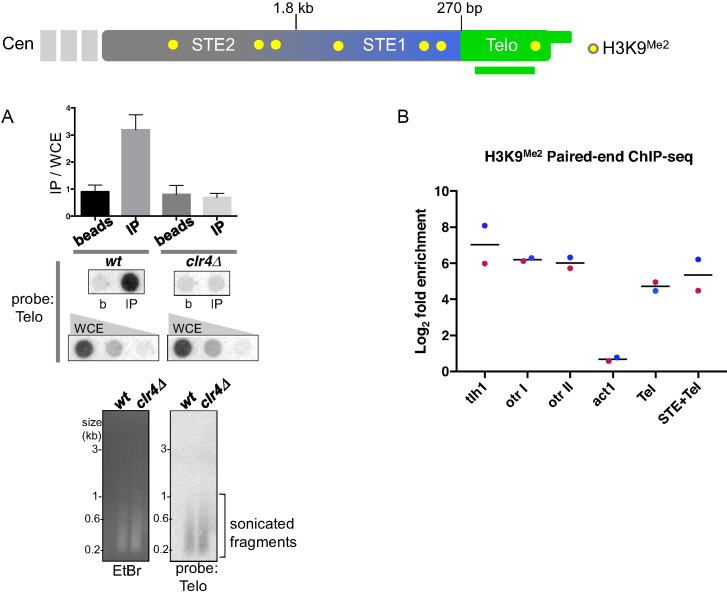
Figure 4.
Fission yeast telomeres contain histones. (A) Schematic of the chromosome end. Histone H3K9Me2 is present in the STE1 and STE2 regions (25) but its presence in the telomeric DNA repeat region has not been previously defined. ChIP was performed using an antibody against histone H3K9Me2 followed by dot blot analysis using a telomere repeat probe. WCE, whole cell extract; b, beads-alone; IP, H3K9Me2 immunoprecipitate. Quantitation of the dot blot is shown in the graph above. The gel and Southern blot below show sonicated fragments of the bulk genome; most fragments are 100–400 bp in size. (B) Paired-end ChIP-seq definitively shows the presence of histones in the telomere-repeat region. Fold enrichment is reported as the ratio of the number of perfectly mapped paired-end reads in the IP versus input, normalized by sequencing depth and region length. For the region labeled ‘Tel’, paired-end reads that mapped wholly within the telomeric portion of the assembled contigs were counted and each of the contig enrichment values were averaged; the mean fold-enrichment for all contigs is represented by a black bar. Enrichment of reads overlapping the boundary of the subtelomere and telomere within the assembled contigs is shown in ‘STE+Tel’. Non-telomeric heterochromatic regions of the genome, including the centromere-proximal subtelomere (tlh1) and outer centromeric regions (otr I: SPNCRNA.231, otr II: SPNCRNA.373), and a euchromatic region (act1) are shown for comparison; blue (replicate 1) and red (replicate 2) dots represent the fold enrichment for each replicate.
To determine whether immunoprecipitates of H3K9Me2 harbor fragments that contain telomere repeats but lack subtelomeric sequences, we performed paired-end ChIP-Seq of immunoprecipitated fragments in duplicate (replicates R1 and R2) (Figure 4B, Supplementary Table S2). Due to the heterogeneity of telomere sequences in a population, we needed to first generate a reference for mapping telomere sequences. We filtered raw reads from input and IP samples for a common variant (GTTACAGG) of the degenerate telomere repeat sequence, then performed de novo assembly for each sample and selected 5 telomere contigs for each experiment (Supplementary Table S1) (see Materials and Methods for details). We found that the level of enrichment of telomere-only sequence (Tel) reads not only far exceeds that seen at a euchromatic locus (act1) but also is comparable to that of previously well-characterized heterochromatic sequences such as the centromeric outer repeats (otr I and II) and the subtelomeric helicase-encoding sequences tlh1 (Figure 4B) (25,26,49,50). Interestingly, the telomeric enrichments in TelR1 and TelR2 were unique to their respective contigs; R1 telomere sequences were not enriched in R2 and vice versa (Supplementary Table S2). This supports previous observations that specific telomere sequences vary between different fission yeast cultures (51), due mainly to variation introduced by telomerase, which adds the degenerate sequence (TTAC(A)GG(G1–4) (52,53). By enforcing strict alignment criteria, any small sequence difference in the reads will prevent alignment. Therefore, while the telomere sequences in R1 and R2 are indeed highly similar, reads that do not align perfectly will fail to map to non-corresponding reference contigs. We also examined reads that included telomere and distal subtelomere (STE+Tel, Figure 4B) as our MNase mapping shows that such regions are included in the telosome; for such telomere/distal STE reads, we observe enrichment similar to that seen for the telomere-only reads. Hence, the telomeric DNA repeats themselves, as well as the distal subtelomere, are incorporated into chromatin harboring H3K9Me2.
RNA, but not heterochromatin, is a determinant of telosome structure
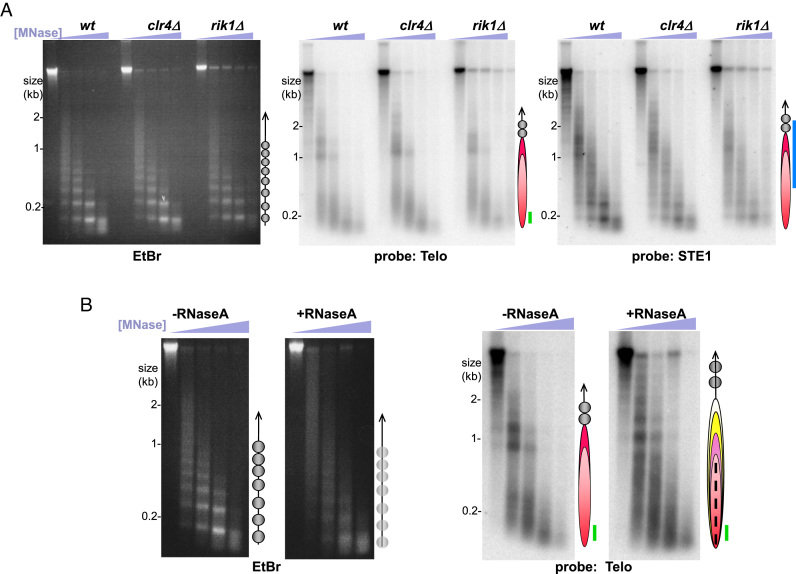
Our ChIP-seq results indicate not only that telomeres contain histones, but also that these histones are H3K9 methylated. This observation prompted us to investigate whether methylation of histone H3, an essential step in heterochromatin formation, is required for telosome formation. We queried the telomere protection pattern in cells lacking the sole fission yeast histone H3K9 methyltransferase, Clr4 (Figure 5A). Telosomal bands are unchanged in the absence of Clr4, demonstrating that histone H3K9 methylation is dispensable for telosome formation. Similarly, the telosome remains intact in cells lacking Rik1, another component essential for heterochromatin formation (Figure 5A). Hence, the histone methylation we observe within the telomere does not contribute to the telosome structure. The converse scenario, in which the telosome contributes to telomeric heterochromatin formation, remains possible. Taz1 is necessary for heterochromatin formation at the distal end of the telomere while the RNAi machinery governs more centromere-proximal subtelomeric heterochromatin formation (25). The biophysical properties of the telosome (considered below) may promote the activity of the heterochromatin assembly machinery at the distal chromosome end.
Figure 5.
RNA, but not H3K9 methylation, is a critical component of telosome structure. (A) Analysis of chromatin in strains deficient for heterochromatin assembly, with designations as in Figure 1. (B) Analysis of chromatin untreated or treated with RNase A prior to MNase digestion. Bulk RNase-treated chromatin exhibits a more diffuse pattern, hence the less-defined circles in the schematic. RNase A treatment alters the telosome pattern, as reflected by the more numerous telosome particles in the schematic; the dotted line within the red elipse represents the diffuse digestion products, suggesting smaller, more mobile or more frequently dismantled particles.
In addition to histone methylation and other marks of heterochromatin, chromosome ends are associated with telomeric RNAs including TERRA (54,55). To determine whether such transcripts contribute to the higher order structure of the telomere, we subjected isolated nuclei to mock-treatment or treatment with RNase A prior to MNase digestion. Consistent with previous work demonstrating that RNA is an integral component of chromatin (56–58), RNase A treatment affected the bulk genomic MNase digestion pattern (Figure 5B). While a nucleosomal ladder is still detectable following RNase A treatment, the bands are more diffuse, suggesting that global nucleosome periodicity is less well-defined in the absence of RNA. In contrast, RNase A treatment results in a conspicuous, discrete alteration in telomeric MNase digestion products, easily visualized on the blot (Figure 5B). While the two predominant bands at 1.4 and 1.0 kb appear in the absence of RNase A, treatment with the RNase exposes new sites of MNase accessibility and shifts band positions such that the predominant bands are slightly larger than those seen for untreated telosomes. In addition, a diffuse pattern of MNase products appears below 0.9 kb upon RNase A treatment.
As removal of RNAs by RNase A exposes some sites to MNase digestion while camouflaging others, we considered the possibility that unbridled association with RNA confers the disordered MNase digestion pattern of taz1Δ telomeres. This is particularly relevant as loss of Taz1 results in a high abundance of telomeric and subtelomeric transcripts (24). However, RNase treatment fails to confer a significant change in the MNase pattern of taz1Δ chromatin, suggesting that nucleosomal array exclusion is not a result of excess RNA (Supplementary Figure S5). Hence, the altered pattern conferred by RNase A suggests that subtelomeric and/or telomeric RNAs associate directly with telosomes, perhaps altering the path of DNA around nucleosomes or regulating the positions of telomere-binding proteins. Treatment of intact chromatin with RNase H does not lead to detectable alterations in telosomal (nor bulk) MNase digestion patterns (data not shown), suggesting that RNA/DNA double helices are not key determinants of telosome structure.
DISCUSSION
What features of the telosome could explain the MNase pattern we observe? The presence of histones in the telomere repeat region suggests that rather than excluding nucleosomes, Taz1 and Rap1 enforce a folded chromatin fiber in which the linker regions are occluded either by telomere proteins themselves or by an alternative wrapping pattern that disfavors MNase access. In budding yeast and flies, centromeric nucleosomes have been shown to induce positive supercoils in centromeric DNA, rather than the negative supercoils induced by nucleosomes throughout the majority of the genome (59,60). The above-mentioned similarity between the digestion pattern of the internal telomere repeat stretch and the centromere suggests the intriguing possibility that telomeres share an alternative torsional state with centromeres, a possibility that we are exploring. Conceivably, such altered supercoiling could propagate into nearby sequences, explaining both the extension of the telosome past the telomere repeat region and the transitional nature of the subtelomeric chromatin as it acquires a canonical nucleosome array pattern more centromere-proximally.
Intriguingly, TRF2 has been shown to wrap naked telomeric DNA around itself in vitro in a conformation that generates positive supercoiling, propagating a propensity for single-strandedness along adjacent DNA segments and in turn, promoting strand invasion by ssDNA (forming t-loops) (61,62). While the extent to which this wrapping activity affects nucleosomal DNA is unclear, mutations that abolish the activity compromise TRF2’s ability to form t-loops and to protect telomeres from ATM activation. TRF2’s wrapping ability has also been shown to promote telomeric invasion by telomeric RNA (63). The residues that confer TRF2 wrapping appear to be absent in Taz1; nonetheless, it is conceivable that structural homology exists and could contribute to Taz1-mediated regulation of telomeric torsional state.
An alternative, and not mutually exclusive, explanation for the nature of the telosome could invoke phase separation behavior at telomeres. Chromatin regions that associate with human or Drosophila HP1α show such behavior, forming liquid droplet domains that exclude some proteins while allowing free diffusion of others (64,65). Intriguingly, association with RNA is known to change the parameters of liquid droplet formation, often promoting phase separation (66,67). The marked change in MNase accessibility of telomeric chromatin following RNase treatment (Figure 5B) could suggest a model in which the boundaries of the telosome are constrained by RNA, perhaps TERRA; the removal of RNA may not only expose specific sites within the telosome to MNase, but may also allow a telosomal ‘phase’ to expand.
Whether occlusion, torsion or phase separation underlies telosome structure, the altered architecture at the chromosome end is likely to impact telomere function. Indeed, the dependence of telosome structure on Taz1 harkens to the TRF2-dependent t-loops in organisms with longer telomeres, and several potential parallels can be drawn. In the absence of TRF2, t-loops are compromised, correlating with exposure of telomeres to inappropriate DDR reactions analogous to those encountered by taz1Δ cells. POT1, the telomeric ssDNA binding protein, has been suggested to stabilize t-loops by binding the DNA strand extruded by 3′ end invasion into subterminal dsDNA. In fission yeast, deletion or inactivation of pot1+ leads to rampant resection and telomere loss, thus preventing us from studying its impact on telosome formation (68,69); however, the discreteness of the chromatin pattern at natural telomeres relative to internal telomere stretches hints at a role for endedness (and by extension, ssDNA in complex with binding proteins) in ‘sealing’ the telosome-specific architecture.
The uniqueness of the chromosome end is highlighted by the alternative higher order telomere structure reported herein. The dependence of telosome formation on Taz1 reinforces the idea that telomeric DNA sequences specify the telosome, while the known roles of Taz1 in safeguarding chromosome ends underline the importance of defining the biophysical basis for the telosomal pattern. Our results provide a framework for such definition, in which telomere-binding proteins enforce a non-canonical histone-containing chromatin fold whose precise dimensions are dictated by RNA transcripts and the presence of the chromosome end. Our previous observations that centromeres and telomeres have some interchangeable functions (70) and that nontelomeric heterochromatic repeats can fulfil chromosome end-protection roles previously thought specific to telomere repeats (71) further underscore the need to define the features that unify and distinguish these prominent chromosomal landmarks.
DATA AVAILABILITY
Data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO series accession number GSE10807.
Supplementary Material
ACKNOWLEDGEMENTS
We thank our lab members, both from the early 1990s when this work was initiated in the Cech lab and more recent years, for discussions and support. Special mention goes to Creighton Tuzon and Luis Valente, whose experiments anticipated the effects (or lack thereof) of Rik1 and Rap1 on telosomes. We thank Cancer Research UK, the European Research Council, and the NIH for funding. T.R.C. is an investigator of the Howard Hughes Medical Institute.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Howard Hughes Medical Institute; Cancer Research UK intramural funding; European Research Council [250326]; National Cancer Institute intramural program. Funding for open access charge: National Cancer Institute. This work was also funded in part by the Francis Crick Institute which receives its core funding from Cancer Research UK [FC01121]; the UK Medical Research Council [FC01121]; and the Wellcome Trust [FC01121].
Conflict of interest statement. None declared.
REFERENCES
- 1. Jain D., Cooper J.P.. Telomeric strategies: means to an end. Annu. Rev. Genet. 2010; 44:243–269. [DOI] [PubMed] [Google Scholar]
- 2. de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005; 19:2100–2110. [DOI] [PubMed] [Google Scholar]
- 3. Lim C.Z., Zaug A.J., Kim H.J., Cech T.R.. Reconstitution of human shelterin complexes reveals unexpected stoichiometry and dual pathways to enhance telomerase processivity. Nat. Commun. 2017; 8:1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Miyoshi T., Kanoh J., Saito M., Ishikawa F.. Fission yeast Pot1-Tpp1 protects telomeres and regulates telomere length. Science. 2008; 320:1341–1344. [DOI] [PubMed] [Google Scholar]
- 5. Azzalin C.M., Lingner J.. Telomere functions grounding on TERRA firma. Trends Cell Biol. 2015; 25:29–36. [DOI] [PubMed] [Google Scholar]
- 6. Rippe K., Luke B.. TERRA and the state of the telomere. Nat. Struct. Mol. Biol. 2015; 22:853–858. [DOI] [PubMed] [Google Scholar]
- 7. Griffith J.D., Comeau L., Rosenfield S., Stansel R.M., Bianchi A., Moss H., de Lange T.. Mammalian telomeres end in a large duplex loop. Cell. 1999; 97:503–514. [DOI] [PubMed] [Google Scholar]
- 8. Doksani Y., Wu J.Y., de Lange T., Zhuang X.. Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell. 2013; 155:345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cacchione S., Cerone M.A., Savino M.. In vitro low propensity to form nucleosomes of four telomeric sequences. FEBS Lett. 1997; 400:37–41. [DOI] [PubMed] [Google Scholar]
- 10. Galati A., Micheli E., Alicata C., Ingegnere T., Cicconi A., Pusch M.C., Giraud-Panis M.J., Gilson E., Cacchione S.. TRF1 and TRF2 binding to telomeres is modulated by nucleosomal organization. Nucleic Acids Res. 2015; 43:5824–5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Galati A., Rossetti L., Pisano S., Chapman L., Rhodes D., Savino M., Cacchione S.. The human telomeric protein TRF1 specifically recognizes nucleosomal binding sites and alters nucleosome structure. J. Mol. Biol. 2006; 360:377–385. [DOI] [PubMed] [Google Scholar]
- 12. Nikitina T., Woodcock C.L.. Closed chromatin loops at the ends of chromosomes. J. Cell Biol. 2004; 166:161–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Makarov V.L., Lejnine S., Bedoyan J., Langmore J.P.. Nucleosomal organization of telomere-specific chromatin in rat. Cell. 1993; 73:775–787. [DOI] [PubMed] [Google Scholar]
- 14. Wu P., de Lange T.. No overt nucleosome eviction at deprotected telomeres. Mol. Cell. Biol. 2008; 28:5724–5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tommerup H., Dousmanis A., de Lange T.. Unusual chromatin in human telomeres. Mol. Cell. Biol. 1994; 14:5777–5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wright J.H., Gottschling D.E., Zakian V.A.. Saccharomyces telomeres assume a non-nucleosomal chromatin structure. Genes Dev. 1992; 6:197–210. [DOI] [PubMed] [Google Scholar]
- 17. Budarf M.L., Blackburn E.H.. Chromatin structure of the telomeric region and 3′-nontranscribed spacer of Tetrahymena ribosomal RNA genes. J. Biol. Chem. 1986; 261:363–369. [PubMed] [Google Scholar]
- 18. Gottschling D.E., Cech T.R.. Chromatin structure of the molecular ends of Oxytricha macronuclear DNA: phased nucleosomes and a telomeric complex. Cell. 1984; 38:501–510. [DOI] [PubMed] [Google Scholar]
- 19. Cohen P., Blackburn E.H.. Two types of telomeric chromatin in Tetrahymena thermophila. J. Mol. Biol. 1998; 280:327–344. [DOI] [PubMed] [Google Scholar]
- 20. Cooper J.P., Nimmo E.R., Allshire R.C., Cech T.R.. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature. 1997; 385:744–747. [DOI] [PubMed] [Google Scholar]
- 21. Miller K.M., Rog O., Cooper J.P.. Semi-conservative DNA replication through telomeres requires Taz1. Nature. 2006; 440:824–828. [DOI] [PubMed] [Google Scholar]
- 22. Tomita K., Cooper J.P.. The telomere bouquet controls the meiotic spindle. Cell. 2007; 130:113–126. [DOI] [PubMed] [Google Scholar]
- 23. Ferreira M.G., Cooper J.P.. The fission yeast Taz1 protein protects chromosomes from Ku-dependent end-to-end fusions. Mol. Cell. 2001; 7:55–63. [DOI] [PubMed] [Google Scholar]
- 24. Greenwood J., Cooper J.P.. Non-coding telomeric and subtelomeric transcripts are differentially regulated by telomeric and heterochromatin assembly factors in fission yeast. Nucleic Acids Res. 2012; 40:2956–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kanoh J., Sadaie M., Urano T., Ishikawa F.. Telomere binding protein Taz1 establishes Swi6 heterochromatin independently of RNAi at telomeres. Curr. Biol.: CB. 2005; 15:1808–1819. [DOI] [PubMed] [Google Scholar]
- 26. Cam H.P., Sugiyama T., Chen E.S., Chen X., FitzGerald P.C., Grewal S.I.. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat. Genet. 2005; 37:809–819. [DOI] [PubMed] [Google Scholar]
- 27. Shimizu M., Roth S.Y., Szent-Gyorgyi C., Simpson R.T.. Nucleosomes are positioned with base pair precision adjacent to the alpha 2 operator in Saccharomyces cerevisiae. EMBO J. 1991; 10:3033–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Alfa C., Fantes P., Hyams J., McLeod M., Warbrick E.. 1993; Cold Spring Harbor Laboratory Press. [Google Scholar]
- 29. Dehe P.M., Rog O., Ferreira M.G., Greenwood J., Cooper J.P.. Taz1 enforces cell-cycle regulation of telomere synthesis. Mol. Cell. 2012; 46:797–808. [DOI] [PubMed] [Google Scholar]
- 30. M M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011; 17:10–12. [Google Scholar]
- 31. Duffy M., Chambers A.. DNA-protein interactions at the telomeric repeats of Schizosaccharomyces pombe. Nucleic Acids Res. 1996; 24:1412–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D. et al. . SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012; 19:455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Simpson J.T., Wong K., Jackman S.D., Schein J.E., Jones S.J., Birol I.. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009; 19:1117–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sasaki M., Idiris A., Tada A., Kumagai H., Giga-Hama Y., Tohda H.. The gap-filling sequence on the left arm of chromosome 2 in fission yeast Schizosaccharomyces pombe. Yeast. 2008; 25:673–679. [DOI] [PubMed] [Google Scholar]
- 35. Wood V., Harris M.A., McDowall M.D., Rutherford K., Vaughan B.W., Staines D.M., Aslett M., Lock A., Bahler J., Kersey P.J. et al. . PomBase: a comprehensive online resource for fission yeast. Nucleic Acids Res. 2012; 40:D695–D699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li H., Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics. 2009; 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Godde J.S., Widom J.. Chromatin structure of Schizosaccharomyces pombe. A nucleosome repeat length that is shorter than the chromatosomal DNA length. J. Mol. Biol. 1992; 226:1009–1025. [DOI] [PubMed] [Google Scholar]
- 38. Chikashige Y., Kinoshita N., Nakaseko Y., Matsumoto T., Murakami S., Niwa O., Yanagida M.. Composite motifs and repeat symmetry in S. pombe centromeres: direct analysis by integration of NotI restriction sites. Cell. 1989; 57:739–751. [DOI] [PubMed] [Google Scholar]
- 39. Tomita K., Matsuura A., Caspari T., Carr A.M., Akamatsu Y., Iwasaki H., Mizuno K., Ohta K., Uritani M., Ushimaru T. et al. . Competition between the Rad50 complex and the Ku heterodimer reveals a role for Exo1 in processing double-strand breaks but not telomeres. Mol. Cell. Biol. 2003; 23:5186–5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Miller K.M., Ferreira M.G., Cooper J.P.. Taz1, Rap1 and Rif1 act both interdependently and independently to maintain telomeres. EMBO J. 2005; 24:3128–3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kanoh J., Ishikawa F.. spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr. Biol.: CB. 2001; 11:1624–1630. [DOI] [PubMed] [Google Scholar]
- 42. Hayano M., Kanoh Y., Matsumoto S., Renard-Guillet C., Shirahige K., Masai H.. Rif1 is a global regulator of timing of replication origin firing in fission yeast. Genes Dev. 2012; 26:137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Miyoshi T., Sadaie M., Kanoh J., Ishikawa F.. Telomeric DNA ends are essential for the localization of Ku at telomeres in fission yeast. J. Biol. Chem. 2003; 278:1924–1931. [DOI] [PubMed] [Google Scholar]
- 44. Clarke L., Baum M., Marschall L.G., Ngan V.K., Steiner N.C.. Structure and function of Schizosaccharomyces pombe centromeres. Cold Spring Harb. Symp. Quant. Biol. 1993; 58:687–695. [DOI] [PubMed] [Google Scholar]
- 45. Goshima G., Saitoh S., Yanagida M.. Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes Dev. 1999; 13:1664–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Marschall L.G., Clarke L.. A novel cis-acting centromeric DNA element affects S. pombe centromeric chromatin structure at a distance. J. Cell Biol. 1995; 128:445–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Polizzi C., Clarke L.. The chromatin structure of centromeres from fission yeast: differentiation of the central core that correlates with function. J. Cell Biol. 1991; 112:191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Takahashi K., Murakami S., Chikashige Y., Funabiki H., Niwa O., Yanagida M.. A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol. Biol. Cell. 1992; 3:819–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nakayama J., Rice J.C., Strahl B.D., Allis C.D., Grewal S.I.. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001; 292:110–113. [DOI] [PubMed] [Google Scholar]
- 50. Bannister A.J., Zegerman P., Partridge J.F., Miska E.A., Thomas J.O., Allshire R.C., Kouzarides T.. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001; 410:120–124. [DOI] [PubMed] [Google Scholar]
- 51. Bennett H.W., Liu N., Hu Y., King M.C.. TeloPCR-seq: a high-throughput sequencing approach for telomeres. FEBS Lett. 2016; 590:4159–4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Leonardi J., Box J.A., Bunch J.T., Baumann P.. TER1, the RNA subunit of fission yeast telomerase. Nat. Struct. Mol. Biol. 2008; 15:26–33. [DOI] [PubMed] [Google Scholar]
- 53. Webb C.Z., Zakian V.A.. Identification and characterization of the Schizosaccharomyces pombe TER1 telomerase RNA. Nat. Struct. Mol. Biol. 2008; 15:34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Azzalin C.M., Reichenbach P., Khoriauli L., Giulotto E., Lingner J.. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007; 318:798–801. [DOI] [PubMed] [Google Scholar]
- 55. Bah A., Azzalin C.M.. The telomeric transcriptome: from fission yeast to mammals. Int. J. Biochem. Cell Biol. 2012; 44:1055–1059. [DOI] [PubMed] [Google Scholar]
- 56. Mondal T., Rasmussen M., Pandey G.K., Isaksson A., Kanduri C.. Characterization of the RNA content of chromatin. Genome Res. 2010; 20:899–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nickerson J.A., Krochmalnic G., Wan K.M., Penman S.. Chromatin architecture and nuclear RNA. PNAS. 1989; 86:177–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rodriguez-Campos A., Azorin F.. RNA is an integral component of chromatin that contributes to its structural organization. PLoS One. 2007; 2:e1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Furuyama T., Henikoff S.. Centromeric nucleosomes induce positive DNA supercoils. Cell. 2009; 138:104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Henikoff S., Furuyama T.. The unconventional structure of centromeric nucleosomes. Chromosoma. 2012; 121:341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Amiard S., Doudeau M., Pinte S., Poulet A., Lenain C., Faivre-Moskalenko C., Angelov D., Hug N., Vindigni A., Bouvet P. et al. . A topological mechanism for TRF2-enhanced strand invasion. Nat. Struct. Mol. Biol. 2007; 14:147–154. [DOI] [PubMed] [Google Scholar]
- 62. Benarroch-Popivker D., Pisano S., Mendez-Bermudez A., Lototska L., Kaur P., Bauwens S., Djerbi N., Latrick C.M., Fraisier V., Pei B. et al. . TRF2-Mediated control of telomere DNA topology as a mechanism for Chromosome-End protection. Mol. Cell. 2016; 61:274–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lee Y.W., Arora R., Wischnewski H., Azzalin C.M.. TRF1 participates in chromosome end protection by averting TRF2-dependent telomeric R loops. Nat. Struct. Mol. Biol. 2018; 25:147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Larson A.G., Elnatan D., Keenen M.M., Trnka M.J., Johnston J.B., Burlingame A.L., Agard D.A., Redding S., Narlikar G.J.. Liquid droplet formation by HP1alpha suggests a role for phase separation in heterochromatin. Nature. 2017; 547:236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Strom A.R., Emelyanov A.V., Mir M., Fyodorov D.V., Darzacq X., Karpen G.H.. Phase separation drives heterochromatin domain formation. Nature. 2017; 547:241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Shin Y., Brangwynne C.P.. Liquid phase condensation in cell physiology and disease. Science. 2017; 357:eaaf4382. [DOI] [PubMed] [Google Scholar]
- 67. Van Treeck B., Protter D.S.W., Matheny T., Khong A., Link C.D., Parker R.. RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. PNAS. 2018; 115:2734–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Baumann P., Cech T.R.. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001; 292:1171–1175. [DOI] [PubMed] [Google Scholar]
- 69. Pitt C.W., Cooper J.P.. Pot1 inactivation leads to rampant telomere resection and loss in one cell cycle. Nucleic Acids Res. 2010; 38:6968–6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Fennell A., Fernandez-Alvarez A., Tomita K., Cooper J.P.. Telomeres and centromeres have interchangeable roles in promoting meiotic spindle formation. J. Cell Biol. 2015; 208:415–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Jain D., Hebden A.K., Nakamura T.M., Miller K.M., Cooper J.P.. HAATI survivors replace canonical telomeres with blocks of generic heterochromatin. Nature. 2010; 467:223–227. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO series accession number GSE10807.