Abstract
We have optimized point mutation knock-ins into zebrafish genomic sites using clustered regularly interspaced palindromic repeats (CRISPR)/Cas9 reagents and single-stranded oligodeoxynucleotides. The efficiency of knock-ins was assessed by a novel application of allele-specific polymerase chain reaction and confirmed by high-throughput sequencing. Anti-sense asymmetric oligo design was found to be the most successful optimization strategy. However, cut site proximity to the mutation and phosphorothioate oligo modifications also greatly improved knock-in efficiency. A previously unrecognized risk of off-target trans knock-ins was identified that we obviated through the development of a workflow for correct knock-in detection. Together these strategies greatly facilitate the study of human genetic diseases in zebrafish, with additional applicability to enhance CRISPR-based approaches in other animal model systems.
INTRODUCTION
Strategies employing CRISPR/Cas9 are now used in a large variety of species with one of the greatest advantages being the ability to introduce specific genomic modifications. However, introducing defined point mutations has been a significant challenge in some species because it requires some form of homology-directed repair (HDR) or recombination. Zebrafish (Danio rerio) has been one such species in which these challenges have been recognized, despite a wide adoption of CRISPR/Cas9 technology overall and significant advances in many areas (1–4). The zebrafish is frequently used for both basic developmental biology research and human disease modeling because of its transparency, fecundity, and the availability of well-developed genetic and cell biological tools. For disease modeling, the application of CRISPR/Cas9 to generate missense point mutants of residues conserved between humans and zebrafish can be of particular value, as these types of studies in zebrafish can be significantly more cost-effective and scalable than in other vertebrate model animals, such as mice. However, this potential can only be realized if the current efficiencies of point mutation knock-in strategies can be substantially improved.
The first demonstrations of small mutation knock-ins in zebrafish (5,6) were proof-of-concept that such modifications using single-stranded oligodeoxynucleotides (ssODNs) were possible, but did not show germline transmission of the introduced mutations. Importantly, the feasibility of introducing small modifications using ssODNs was actually established much earlier by previous ground-breaking work employing transcription activator-like effector nucleases (TALENs) (7,8). However, TALEN-based methods were not widely adopted by the field for knock-in generation, likely due to the greater difficulty of using TALENs than producing single guide RNAs (sgRNA) once CRISPR/Cas9 became available. Over the last few years, ssODN-based knock-ins have advanced in several respects. The introduction of inserts encoding protein epitope tags such as HA was accomplished successfully, although the proportion of correctly modified alleles was low (9,10). This problem of low-fidelity insertions occurred in all early studies on ssODN-based knock-ins and no systematic attempt to address this problem has yet been published. The first report of point mutation insertion and germline transmission in zebrafish was published, describing the modification of tardp and fus genes involved in amyotrophic lateral sclerosis (11). These authors were able to introduce a mutation into the fus gene in 1 of 47 founders using a 33-nucleotide (nt) oligo and another mutation into the tardp gene in 3 out of 77 founders using a 100-nt oligo containing sgRNA site mutations. None of the successful knock-ins contained indels but whether this is a representative sample from a real distribution of knock-in alleles is unclear. This paper by Armstrong et al. (11) is important; however, in that it shows that zebrafish CRISPR-based knock-ins are certainly doable and germline transmissible but its methodology is heavily reliant on sequencing of many polymerase chain reaction (PCR) products without an easier pre-screening method.
Several important optimizations have emerged from recent publications on CRISPR/Cas9-mediated knock-ins that have yet to be adopted in vivo. The findings in two recent papers (12,13) suggest the strong inverse relationship between knock-in efficiency and the distance of the modification to the cut site. The most efficient positions for a modification should be located <15 nt and ideally <10 nt away from the cut site, as at a distance of 20 nt away from the cut site, the efficiency drops to 20–30% of the maximum, as observed in murine cells (13). Another important trend has been the focus on the structure of oligos. Asymmetric anti-sense oligos with homology arms of 36 and 90 nt were demonstrated by Richardson et al. (14) to be superior to all other designs of the same size. The oligos in this case are anti-sense to the PAM-containing (non-target) strand, a portion of which was proposed to separate from the Cas9-sgRNA ribonucleoprotein and become available to bind the 36-nt homology arm of the oligo. This event can then facilitate HDR, as evidenced by the highly efficient repair of a mutated EGFP gene (14). Chemical modifications have also been applied to ssODNs. Two phosphorothioate (phosphate (PO) where an oxygen is replaced with a sulphur atom) (PS) linkages at the ends of ssODNs promote generation of knock-ins in human cell lines compared to the oligos with traditional PO-oxygen bonds. Knock-in stimulation was most likely due to blocking the activity of exonucleases, independent of the size of the oligos (15). This result was confirmed in other studies of knock-ins in cell lines (12,16,17). PS-modification of oligos has also been tried in zebrafish (15), but these authors found mainly imprecise knock-ins and did not seek direct evidence of knock-in stimulation. Thus, it is still unclear if PS modifications can be applied in zebrafish knock-in experiments.
In this manuscript, we apply these modifications to improve the ease and efficiency of performing point mutation knock-ins in zebrafish. For detection, we assessed the relative performance of restriction site-based measurement using sites introduced by knock-ins with synonymous mutation and allele-specific PCR (AS-PCR) assays most commonly used for nucleotide polymorphism detection (18). We found that AS-PCR has a dramatically greater sensitivity than the restriction-based method. For achieving improved rates of knock-in generation, we took advantage of the proximity of the cut site to the site of modification in the case of cdh5 G767S mutation. For tp53 mutation knock-ins, we compared symmetric sense and asymmetric anti-sense oligos and found the latter to be substantially more efficient (3- to 10-fold) as measured by AS-PCR and next-generation sequencing (NGS). This is the first demonstration of the efficacy of the anti-sense oligo approach in zebrafish. Upon isolation of zebrafish with correct knock-ins, we confirmed them at the genomic DNA and cDNA levels. This new method of knock-in genotyping also led us to identify an off-target phenomenon of trans knock-ins, when oligos used for knock-ins were inserted into other loci but could still generate false-positive AS-PCR hits. To facilitate the process of distinguishing true knock-ins from trans knock-ins, we used a combination of AS-PCR followed by restriction digestion of PCR amplicons centered on the knock-in sites at the restriction sites introduced as synonymous mutations. Lastly, for the lamin A/C (lmna) knock-in strategy, we demonstrated that PS oligos produce a significant improvement in knock-in efficiency and consistency, as measured by AS-PCR. In sum, by applying these strategies, we have optimized CRISPR/Cas9-based knock-ins in the zebrafish, enhancing the genome editing toolbox for zebrafish researchers to more efficiently model human genetic disorders.
MATERIALS AND METHODS
Animal care and husbandry
Use of zebrafish in this study was approved by the Dalhousie University Committee on Laboratory Animals (Protocols 15–125, 15–134). All zebrafish embryos were maintained in E3 embryo medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4) in 10 cm Petri dishes at 28°C. The tp53 knock-in mutant fish were generated in the casper strain (19) and the cdh5 G767S knock-in line also contained fli1a:EGFP transgene (20).
Design of sgRNAs and mutation donor ssODNs
The process of sgRNA design for the purpose of replacing particular nucleotides in the endogenous genes (point mutation knock-in or knock-in for short thereafter) involved several bioinformatic analyses. We initially performed protein sequence alignments using NCBI BLAST (21) of zebrafish proteins to the corresponding human proteins and identified which residues in zebrafish proteins correspond to amino acids mutated in human proteins. Exons containing the amino acid codons to be modified were located in the genomic and cDNA sequences. The sgRNAs were identified using SSC (22) (http://cistrome.org/SSC/) for efficiency prediction and CC-Top (23) (https://crispr.cos.uni-heidelberg.de/) for off-target prediction. sgRNA sites were also mapped to both genomic and cDNA sequences of the genes and placed into the context of amino acid codons. We then introduced the desired codon mutations and inactivating PAM site or spacer site mutations in the sgRNA sites into the gene and cDNA sequences in silico for the genes to be modified using standard DNA editors such as Vector NTI. Importantly, sgRNA site mutations were synonymous. The tp53 ssODNs also contain artificial silent mutations to introduce restriction sites BanI and MspI using WatCut software (http://watcut.uwaterloo.ca/template.php?act=silent_new), but for other knock-ins we did not pursue this strategy. For producing the sense symmetric ssODNs we copied 123–136 nt from the in silico modified genomic sequence centered on the desired mutation and ensured that the shorter homology arm from the cut site was 60 nt. The anti-sense asymmetric oligos were generated by copying 36 nt to the 5′-end from the cut site of the DNA strand non-complementary to the sgRNA and 90 nucleotides from the same strand in the other direction. This sequence was then reverse-complemented and ordered from Integrated DNA Technologies as 4 nanomole Ultramer oligos. The lmna R471W oligos were 90-nucleotide sense oligos which contained the codon mutation and sgRNA site mutations with homology arms of 60 and 30 nt. The PS-modified oligo had the two POs on each end of the oligo replaced with a PS.
Synthesis of sgRNAs and Cas9 mRNA
The corresponding sgRNAs were generated by performing an overlap-extension PCR of the sense sgRNA oligos (Supplementary Table S1) each combined with Rev-sgRNA-scaffold oligo. sgRNA template synthesis reactions were set up using Taq DNA polymerase (ABM, G009) by combining 10 μl of 10× buffer, 6 μl of 25 mM MgSO4, 2 μl 10 mM dNTP, 5 μl of each oligo at 25 μM, 71 μl water and 1.5 μl of Taq. The PCRs were run with a short program: 94°C for 5 min; 5 cycles: 94°C for 30 s, 55°C for 30 s, 72°C for 30 s. The resulting PCR products were purified using QIAGEN Gel Extraction kit (QIAGEN, 28704) and used for in vitro transcription using MEGAshortscript T7 kit (Thermo Fisher Scientific, AM1354). The sgRNA was purified according to the kit instructions. Cas9 mRNA was made from pT3TS-nCas9n plasmid (24) (Addgene, 46757) after its linearization with XbaI using mMessage mMachine T3 kit (Thermo Fisher Scientific, AM1348) and purified with LiCl precipitation according to the kit instructions.
Knock-in microinjections
All knock-in injections were performed with Cas9 mRNA at 300 ng/μl, sgRNA at 150 ng/μl and single-stranded oligos at 1 μM into 1-cell zebrafish embryos. Assessment of sgRNA efficiencies was performed using either T7 Endonuclease I (NEB, M0302S) digestion according to the manufacturer's protocol or using heteroduplex mobility assay (HMA) (25). All of the oligos mentioned in this section are listed in Supplementary Table S1.
Preparation of embryonic and adult samples for genotyping
Samples for genotyping of embryos (2–3 dpf) were prepared by treating embryos with 0.02% Tricaine in fish medium (E3 medium: 5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4), transferring them into PCR tubes and replacing the fish medium with 40 μl of 50 mM NaOH, heating at 95°C for 10 minl, vortexing, cooling on ice for 2 min and neutralizing with 4 μl of 1 M Tris–HCl pH 8.0. The same procedure was used for genotyping adults except that samples taken were fin clips. Extracts from embryos pools were prepared by combining 50 embryos into a single sample and adding 1 ml of 50 mM NaOH, heating at 95°C for 10 min with manual tube inversion several times, cooling on ice for 5–10 min and neutralization with 110 μl of 1 M Tris–HCl pH 8.0.
PCR assays for genotyping and allele-specific PCR assays
The allele-specific PCR assays that we developed for discriminating between wild-type (WT) and point mutation knock-ins are based on several principles. First, the WT and knock-in detection primers differ by two or more nucleotides (either codon replacement or codon mutation and a PAM site mutation or another silent mutation), one of the mismatches being located at the 3′-most position of the allele-specific primers. Second, the annealing temperature (Tanneal) for the PCR was typically calculated using NEB Tm Calculator online tool (http://tmcalculator.neb.com/#!/) or alternatively, once a sample positive for knock-in was available, the optimal temperature was determined empirically using gradient PCR. This was done in the case of the tp53 R217H knock-in AS-PCR because it initially had high background. Third, we used the touch-down PCR method for all AS-PCRs described in this study, which works as follows: 94°C for 3 min; 10 cycles: 94°C for 30 s, Tanneal + 10 (with 1°C decrease every cycle), 72°C for 30 s, 25 cycles: 94°C for 30 s, Tanneal, 72°C for 30 s. At last, all AS-PCRs were done with Taq polymerase because the error rates are not a concern in this method and, more importantly, due to the fact that proof-reading polymerases may remove mismatches between the knock-in primers and WT genomic DNA leading to false-positive amplification. Tanneal for tp53 R143H, tp53 R217H, cdh5 G767S and lmna R471W was 55, 58, 51 and 51°C, respectively.
Illumina-based sequencing of amplicons from knock-in embryos to quantify mutation rates
PCR products for HiSeq Illumina sequencing were prepared using primers containing all of the relevant priming adapters and indexes according to the relevant experimental design (single or triplicate samples) (Supplementary Table S1). Q5 High-Fidelity 2× Mastermix (NEB, M0492) was used for amplifying PCR products for these high-throughput sequencing analyses. PCR products from individual biological samples were amplified using different indexed primers and then pooled into sequencing samples. The sequencing and initial data processing were done by the Next Generation Sequencing Facility of The Centre for Applied Genomics in Toronto, ON, Canada. FASTQ files containing paired sequencing reads were assembled by FLASH (26) (https://ccb.jhu.edu/software/FLASH/), mapped to the reference amplicons using bowtie2 (27) software (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml) and SAM files were generated. The SAM files were then processed using custom Python scripts (https://github.com/SergeyPry/knock-in_analysis) to categorize the editing events. The counts of different event categories were processed and plotted using R scripts (https://github.com/SergeyPry/knock-in_analysis).
cDNA cloning and sequencing
The heterozygous fish carrying knock-in mutations were outcrossed and the embryos were grown to 30 hpf. RNA was extracted from 50 embryos using RNeasy Mini kit (QIAGEN, 74104). cDNA was produced by mixing 10 μl of total RNA with 4 μl of 2.5 mM dNTP and 2 μl of 100 μM oligodT(18), heating at 70°C for 10 min and cooling on ice. We then added 2 μl of M-MuLV buffer (NEB, M0253S), 0.25 μl of Protector RNAse Inhibitor (Roche, 03335399001), 0.25 μl of M-MuLV reverse transcriptase (NEB, M0253S) and 1.6 μl of water. The synthesis reaction was incubated at 42°C for an hour and for 10 min at 90°C. We used Q5 Hot Start High-Fidelity 2× Master Mix (NEB, M0494S) for amplifying cDNA fragments for tp53 using p53cDNA_for and p53cDNA_rev primers and for cdh5 gene using cdh5_lastExon_for and cdh5_lastExon_rev (Supplementary Table S1). The PCR protocol used was 98°C for 30 s, 35 cycles of: 98°C for 10 s, 64°C for 30 s, 72°C for 30 s and the final extension at 72°C for 2 min. The whole PCR reaction was gel-extracted using QIAGEN Gel Extraction kit (QIAGEN, 28704). The purified PCR was cloned into pME-TA using a previously published protocol (28). The clones were screened by Taq-based colony PCR using universal M13 primers and then sequenced. Each colony was resuspended in 100 μl of sterile water and 10 μl of bacterial suspension were heated at 95°C for 5 min followed by addition of 10 μl of Taq master mix and the standard Taq program was run for 36 cycles with the annealing temperature of 55°C.
RESULTS
Allele-specific PCR efficiently detects point mutation knock-ins in zebrafish introduced with ssODNs and CRISPR/Cas9
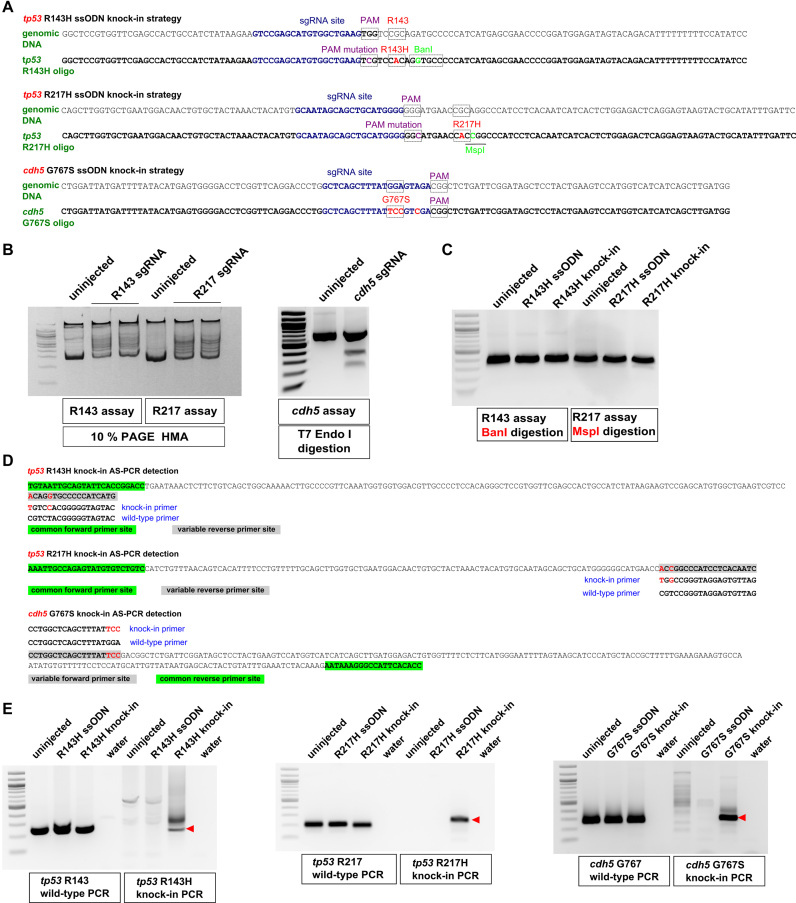
Genetic diseases in humans are frequently caused by point mutations, but until recently these mutations were modeled in laboratory animals using null mutants of the affected genes, which may result in too extreme a phenotype and possibly do not recapitulate the phenotypes seen in human patients. Given the tremendous progress in genome editing, the zebrafish model is poised for the development of effective methods for precise point mutant generation. We aimed at creating defined point mutations, R143H and R217H, in the zebrafish tp53 gene at the positions equivalent to those most frequently mutated in patients with the Li-Fraumeni cancer predisposition syndrome. In another project, we decided to introduce a specific G767S mutation into cdh5, a gene involved in blood vessel development (29). For point mutation knock-ins, we first identified effective sgRNAs close to target codons (Figure 1A). sgRNA activities were demonstrated using the HMA for tp53 sgRNAs and by T7 Endonuclease I digestion for cdh5 sgRNA (Figure 1B). Next, 123 to 136-nt sense symmetric ssODNs were generated that contained the target point mutation, silent PAM site mutations to prevent re-cutting by Cas9, and silent mutations to introduce BanI and MspI restriction sites for tp53 R143H and R217H knock-ins (Figure 1A). By contrast, the cdh5 G767S ssODN contained only one additional silent mutation because the intended mutation would likely disrupt sgRNA binding. To verify whether BanI and MspI can be used for genotyping, we injected either ssODNs alone or knock-in mixes for both tp53 knock-in strategies and then digested relevant PCR products with BanI and MspI, however no cleavage was observed (Figure 1C). This result can be explained by the low knock-in efficiencies. To identify rare targeted alleles, we turned to allele-specific PCR (AS-PCR), a technique used for genotyping single-nucleotide polymorphisms (18). AS-PCR requires a common primer and two detection primers matching either the WT or variant alleles at their 3′-most nucleotides (Figure 1D). For all three knock-in AS-PCR strategies, the WT primer sets produced expected amplicons in all samples, whereas the knock-in primer sets only amplified correct products in the knock-in samples (Figure 1E) confirming the validity of our approach. The stronger signal for cdh5 G767S compared to those observed for tp53 knock-ins is likely due to greater mutation knock-in efficiency closer to the cut sites (12,13). Indeed, the cdh5 knock-in was located at the cut site, while the tp53 knock-ins were 11 and 13 nucleotides away from their cut sites, respectively. This observation supports the contention that knock-ins farther than 10 nt from the cut site are much less efficient (12,13).
Figure 1.
Design of knock-in strategies for tp53 and cdh5 point mutations and the AS-PCR assays for detection. (A) Genomic sequences and donor oligos are shown for sites of tp53 R143H, R217H and cdh5 G767S knock-ins. sgRNA sites are shown in dark blue, PAM sites and target codons are boxed and their mutations are highlighted in magenta and red, respectively. Mutations leading to introduction of BanI (boxed) and MspI (underlined) restriction sites are highlighted in light green. (B) Mutations induced by tp53 sgRNAs were detected by HMA using 10% PAGE analysis. Detection of indel mutations induced by cdh5 sgRNA was performed using the T7 Endonuclease I assay. Comparison of PCR product samples from uninjected zebrafish embryos with those injected with respective sgRNAs shows the degree of sgRNA effectiveness. (C) Restriction analysis of PCR products from uninjected, oligo-injected and tp53 knock-in injected embryos with enzymes introduced into donor oligos. (D) Allele-specific PCR assays for detecting introduced knock-ins are based on the idea that when two or more nucleotides are different between the WT and knock-in alleles, it is possible to design primers that can distinguish the two. Primers common to both WT and knock-in assays are highlighted in green and the site for variable primers is highlighted in gray. The WT and knock-in primers are indicated below or above the site and the nucleotides corresponding to the knock-in allele are in red both in the amplicon and the knock-in primer. (E) Example AS-PCRs are shown that were run on the extracts of pooled embryos from uninjected, oligo-injected, knock-in-injected samples and water. Both WT and knock-in AS-PCRs are shown, with the WT PCRs typically being very strong and indicative of the expected size for the correct knock-in AS-PCR products (indicated with red arrowheads).
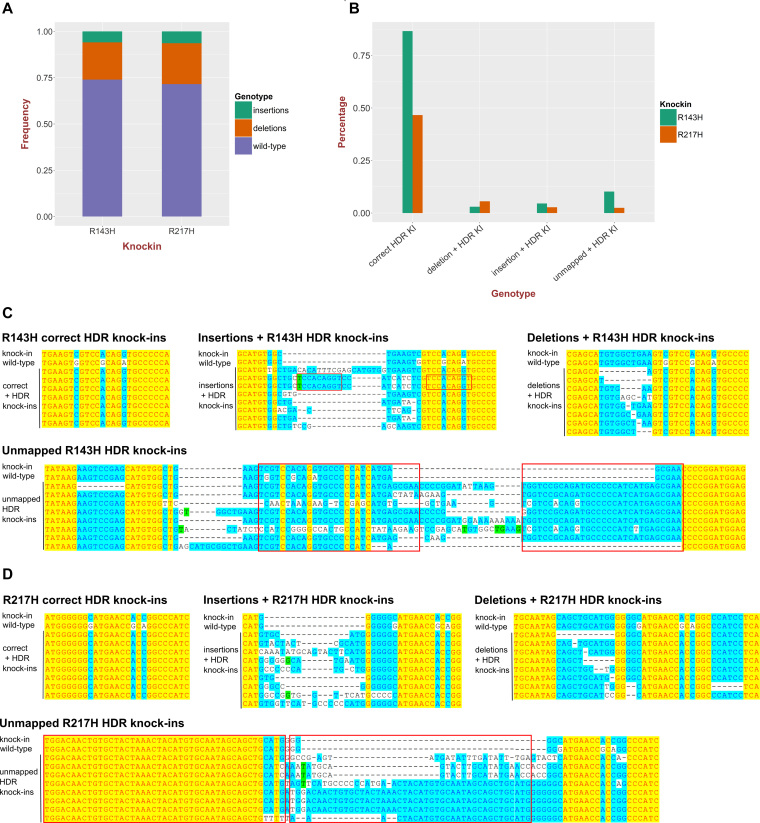
Quantification of knock-in efficiency by next-generation sequencing
The lower efficiency of the tp53 knock-ins relative to cdh5 knock-in prompted us to perform Illumina sequencing of the amplicons around the knock-in sites derived from embryo pools injected with either tp53 R143H or R217H knock-in mixes. We obtained >1*106 reads from each of the tp53 knock-in samples and quantified their indel percentage as 26.1% (R143H) and 28.4% (R217H) (Figure 2A). The limitations of current point mutation knock-in approaches are the low percentage of knock-ins and the presence of additional undesirable mutations. We therefore divided all of the knock-in events into four classes: correct knock-ins, knock-ins with deletions, knock-ins with insertions and knock-ins in unmapped sequence reads (Figure 2B–D). The unmapped knock-in events likely represent an inappropriate insertion of donor oligos into the target loci without recombination. The total percentages for R143H and R217H knock-ins were 1.04 and 0.57%, with the correct knock-ins constituting 83% of total knock-ins for R143H and 81% for R217H (Figure 2B). The high relative percentage of correct knock-in reads suggests that more than 80% of recovered alleles with positive genotyping should contain the correctly modified sequences. However, the alleles with indels and especially unmapped reads (Figure 2C and D) are very variable in sequence as well as size, thus requiring that potential knock-in animals are both genotyped and sequenced at the knock-in site to verify the correctness of modifications.
Figure 2.
High-throughput sequencing analysis of point mutation knock-ins into the tp53 gene. (A) Quantification of the total fractions of small insertions and deletions in tp53 R143H and R217H knock-in injected embryo samples. The proportion of the reads with deletions or insertions represents a measure of sgRNA activity. (B) Measurements of R143H and R217H correct HDR knock-ins, knock-ins with additional insertions and deletions as well as knock-in reads with more aberrant complex events (unmapped). (C) Examples of different classes of R143H knock-ins: correct HDR knock-ins, knock-ins with deletions or insertions and unmapped knock-ins aligned to WT and expected knock-in sequences. (D) Examples of different classes of R217H knock-ins: correct HDR knock-ins, knock-ins with deletions or insertions and unmapped knock-ins aligned to WT and expected knock-in sequences.
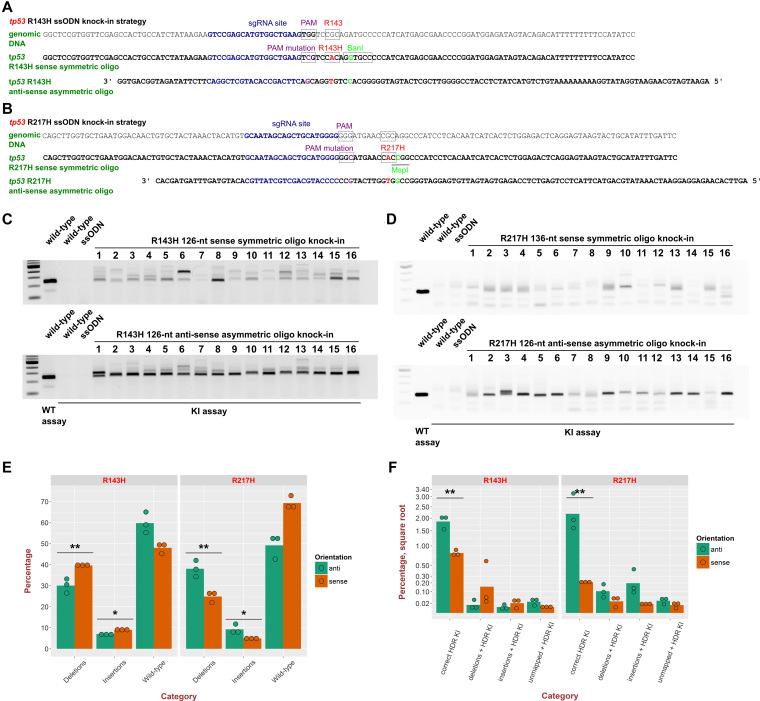
Knock-in efficiencies into tp53 gene are greatly improved using asymmetric anti-sense oligos
Since we initially failed to establish tp53 knock-in zebrafish lines using sense ssODN despite finding one R143H founder that died (Table 1), we sought to improve knock-in efficiency. Richardson et al. (14) found that after Cas9 cuts its genomic DNA site, the DNA strand opposite the sgRNA-binding strand becomes exposed and can interact with the 36-nt homology arm of anti-sense asymmetric oligos, which also have a 90-nt homology arm on the other side. This approach has not been previously employed in zebrafish. We applied this strategy to improve tp53 R143H and R217H knock-ins and compared knock-in efficiencies of the original sense symmetric oligos to those of the anti-sense asymmetric oligos. After knock-in injections with either sense symmetric or anti-sense asymmetric oligos for both R143H (Figure 3A) and R217H knock-ins (Figure 3B), we selected 16 embryos for each type of sample. The semi-quantitative AS-PCR assays show that in cases of both tp53 R143H (Figure 3C) and R217H (Figure 3D), anti-sense asymmetric oligo knock-ins were dramatically more efficient than the sense symmetric ones, as reflected in much higher band intensities and fewer bands of larger sizes that very likely correspond to the unmapped type of knock-in reads. For the tp53 R143H knock-in we also performed knock-ins with anti-sense symmetric and sense asymmetric (reverse complement of the anti-sense asymmetric oligo) to determine the contributions of orientation and symmetry to the knock-in efficiency improvement. Quantification of relative efficiencies of all the four oligo types showed that the anti-sense orientation and asymmetry have additive contributions to improving efficiency (Supplementary Figure S1).
Table 1.
Results of screening point mutation knock-in founders for germline transmission
| Knock-in name | Oligo used | Total number | True positives | False positives |
|---|---|---|---|---|
| tp53 R143H | 126-nt sense symmetric | 30 | 1 | 2 |
| tp53 R143H | 126-nt anti-sense asymmetric | 41 | 2 | 7 |
| tp53 R217H | 136-nt sense symmetric | 38 | 0 | 0 |
| tp53 R217H | 126-nt anti-sense asymmetric | 22 | 2 | 2 |
| cdh5 G767S | 123-nt sense symmetric | 14 | 3 | 1 |
Figure 3.
Enhancement of knock-in efficiency by anti-sense asymmetric oligos. Genomic sequences and donor oligos (sense symmetric and anti-sense asymmetric) are shown for sites of tp53 R143H (A) and R217H (B) knock-ins. All gel image panels show WT embryo genomic DNA extracts run with the tp53 R143H (C) or R217H (D) WT assays to serve as controls for PCR product sizes. As negative controls, knock-in (KI) assays for R143H (C) and R217H (D) were run on WT embryos or embryos injeсted with the respective oligos only. Knock-ins into tp53 to insert the R143H mutation were performed with either 126-nt sense symmetric oligo or with the 126-nt anti-sense asymmetric oligo (36 nt overlapping sgRNA site and 90 nt on the other side) (A and C). In the case of tp53 R217H, the sense symmetric oligo was 136-nt due to a larger distance from the cut site to the mutation and the anti-sense asymmetric one was 126-nt and had the same structure as the asymmetric R143H oligo (B and D). Samples of 16 individual embryos were taken from each knock-in injection and the knock-in assays performed on all of them under the same conditions and at the same time. The gel images shown are representative of at least three independent injections and corresponding comparisons of sense symmetric and anti-sense asymmetric oligo knock-in approaches. (E) Plot of total percentages of insertions, deletions and WT sequence reads at knock-in sites of tp53 R143H and R217H knock-in samples performed either with anti-sense asymmetric (anti) or sense symmetric (sense) ssODNs. (F) Quantification of percentages of different types of knock-ins at knock-in sites of tp53 R143H and R217H knock-in samples performed either with anti or sense ssODNs. Square root was applied to the percentage axis for the knock-in plot to better distinguish different values. Bars indicate the mean level for 3 replicates, whose values are plotted with green or orange circles for anti and sense oligos. The significance of differences in knock-in rates was determined using t-test and the P-value level is indicated above the respective items (**P-value < 0.01, *P-value < 0.05).
To verify improvement in knock-in efficiency using anti-sense asymmetric oligos, we performed high-throughput sequencing on the R143H and R217H knock-in amplicons with three biological replicates for each type of sample (sense or anti-sense). For the tp53 R143H knock-in, overall levels of deletions and insertions were modestly but significantly higher (30.5 versus 39.2% for deletions and 7.5 versus 8.5% for insertions) for the sense knock-in strategy (Figure 3E). However, in the case of tp53 R217H knock-ins the situation was the opposite with both deletions (42 versus 25.9%) and insertions (11.8 versus 5%) being higher in anti-sense knock-in injections (Figure 3E). Despite this variation in indel frequency, the anti-sense asymmetric oligos were significantly more efficient at introducing the correct knock-in modifications, resulting in 2.72-fold stimulation of the tp53 R143H knock-ins (2 versus 0.74%, P < 0.01 t-test) and 9.5-fold stimulation for the tp53 R217H knock-in (1.92 versus 0.2% P < 0.01 t-test) (Figure 3F). These results highlight the application of anti-sense asymmetric oligo design to enhance point mutation knock-in efficiency in zebrafish.
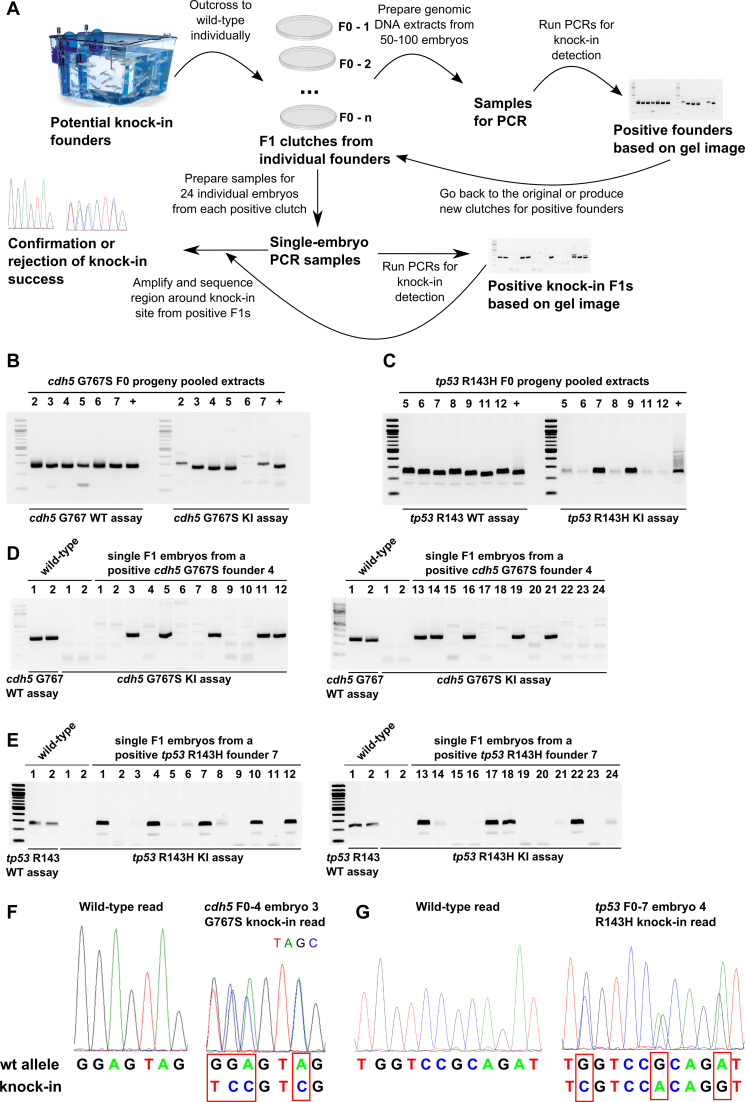
A workflow incorporating AS-PCR for rapid and effective isolation of F1 zebrafish carrying correct knock-ins
In the process of screening more than 100 adult potential founder fish we developed an efficient workflow for isolating correct knock-ins (Figure 4). The first step of the process is to obtain clutches of F1 embryos from randomly selected potential knock-in founders. Genomic DNA extracts are generated from groups of 50 fish at 24–48 hpf stage from each clutch and WT and knock-in AS-PCR assays are applied to all pooled embryo extracts (Figure 4A) as has been done for potential cdh5 G767S (Figure 4B) and tp53 R143H (Figure 4C) knock-in founders. Upon obtaining a positive result for a pooled embryo extract derived from a specific founder, it is recommended to either go back to the clutch derived from this founder or breed the founder again if embryos are not available. One should then prepare 24 individual embryo extracts from a positive founder clutch and at least two WT embryo extracts. The WT embryo extracts are typically run using the WT AS-PCR assay to control for the size of PCR products from the knock-in assay. The lack of knock-in assay amplification on WT samples ensures that the knock-in assay is specific to knock-in events (Figure 4A, D and E). Example applications of the previous step for cdh5 G767S and tp53 R143H positive founders (Figure 4D and E) show the results that can be expected at this stage of the workflow. To complete the workflow, it is also necessary to amplify the genomic region around the knock-in site (‘site assay’) from F1 embryos determined as positive by the knock-in assay. This final step is essential to distinguish between false-positive and true-positive knock-in embryos and founders. This can be accomplished by assessment of sequencing chromatograms, which show double peaks at the expected positions in the case of true knock-ins and WT peaks in WT samples or false-positive knock-in samples (Figure 4F and G).
Figure 4.
AS-PCR-based strategy enables rapid screening and confirmation of potential founders and knock-in F1 embryos. (A) In the first step of this workflow, fish injected with the verified knock-in mix (Cas9, sgRNA and oligo) are grown to adulthood and then outcrossed to WT fish. The clutches derived from the breedings are used to prepare pooled genomic DNA samples from 50–100 embryos. WT and knock-in PCR assays are then run on these samples to identify the founders with detectable levels of germ-line transmission and to provide the size marker for knock-in AS-PCR products, which should be the same size as WT assay products. The information from the first screening round is then used to determine which founders should be bred or which available clutches should be chosen for subsequent screening of 24 individual embryos from each clutch. Knock-in assays are then performed on single-embryo samples and upon obtaining the results, positive embryo extracts are used to amplify a region of DNA surrounding the knock-in site and the resulting PCR products are sent for sequencing to determine if correct knock-in has actually occurred. (B andC) WT and knock-in AS-PCR are shown for cdh5 G767S (B) and tp53 R143H (C) knock-in screening of extracts from embryo clutches produced by potential founders. (D andE) Screening examples of 24 individual embryos from cdh5 G767S and tp53 R143H knock-in founders by AS-PCR. WT and knock-in AS-PCR on two WT embryo samples are shown as controls for PCR product size and as negative controls, respectively. (F) Sequencing chromatograms from WT and heterozygous cdh5 G767S F1 embryos and alignment of the corresponding sequencing reads confirm successful knock-in at the genomic level. (G) Sequencing chromatograms from WT and heterozygous tp53 R143H F1 embryos and alignment of the corresponding sequencing reads confirm successful knock-in at the genomic level.
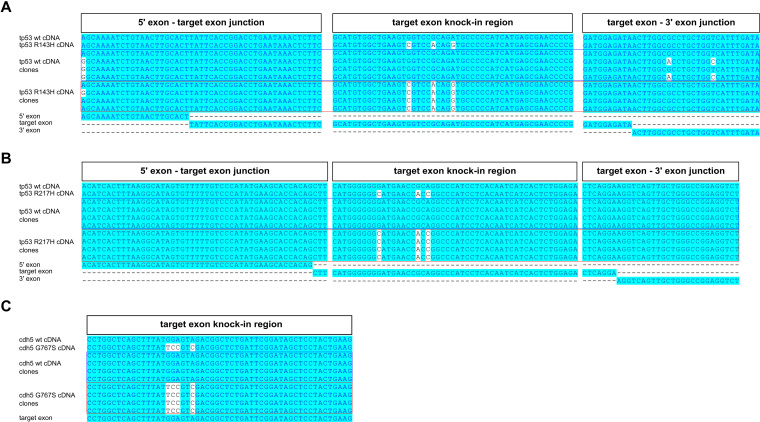
Although sequencing of PCRs from modified genomic regions is highly suggestive that the correct modification was introduced, it does not constitute proof, since it is conceivable that the mutations may interfere with splicing or other process involved in mRNA biogenesis. To prove that the knock-ins we generated correctly modify corresponding mRNAs, we cloned and sequenced cDNA fragments from all three knock-in zebrafish lines. We confirmed that all types of knock-in cDNA clones were present at the expected frequencies (data not shown), sequenced 4 WT and 4 knock-in clones for each of the knock-ins, and aligned the sequences to the theoretical WT and knock-in cDNAs, as well as to the target, and also to flanking exons, in the case of tp53 knock-ins (Figure 5). The resulting alignments for tp53 R143H (Figure 5A), tp53 R217H (Figure 5B) and cdh5 G767S (Figure 5C) confirm that the mutations were faithfully transmitted to mRNAs.
Figure 5.
Knock-in analysis of cDNAs isolated from F1 carrier zebrafish. Analysis of cDNA cloned from F1 knock-in zebrafish with introduced tp53 R143H (A), tp53 R217H (B) or cdh5 G767S (C) mutations was performed by cloning cDNA fragments, identifying bacterial clones carrying either WT cDNA plasmids or those with knock-in mutations introduced, sequencing them and aligning the reads to the expected WT and knock-in cDNA sequences. Each of the alignments was performed with WT and mutant cDNA reference sequences, four WT cDNA clones, four knock-in cDNA clones and relevant exons. For the tp53 R143H and R217H knock-ins (A and B) we show the junction of the 5′ exon and target exon, knock-in region in the target exon and the junction between the target exon and 3′ exon. For the cdh5 G767S knock-in, we only show the target exon alignment because the mutation is at the end of the last exon and therefore unlikely to affect splicing (C).
Identification of false-positive knock-ins and their possible causes
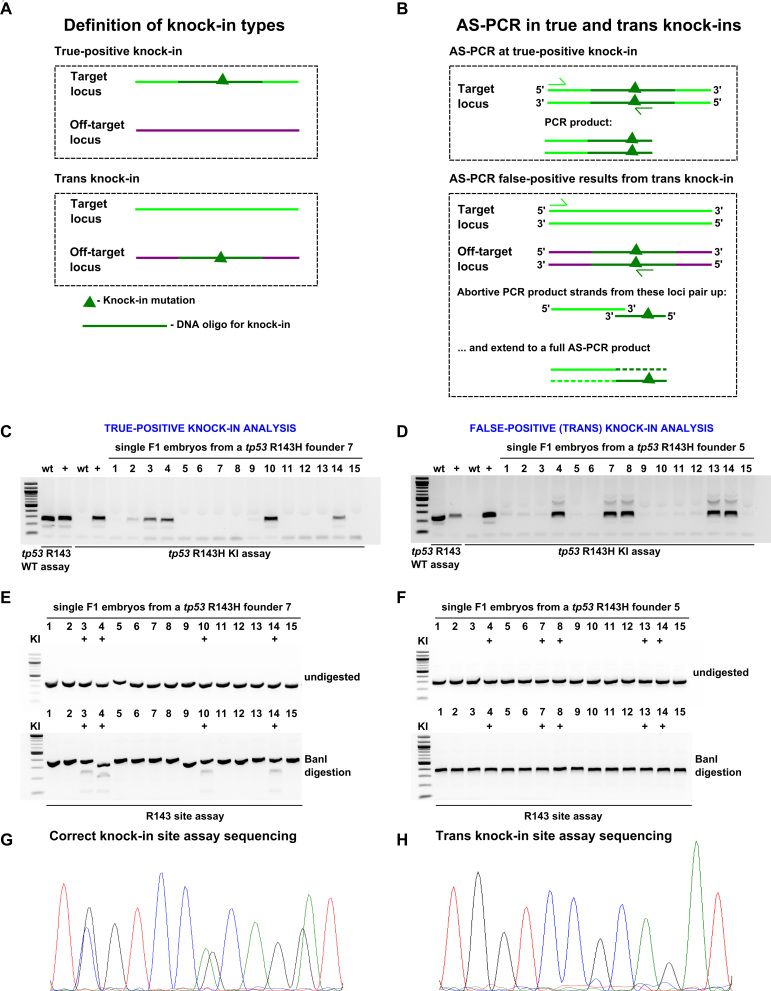
False-positives not related to sample contamination are likely generated due to a genetic modification that is different from the intended one. We sought to efficiently screen out false-positive founders. To explain the false-positive or trans knock-ins, we propose that integration of ssODNs independent HDR occurs at off-target sgRNA sites (Figure 6A). During AS-PCR, either the regular PCR occurs at the true knock-in site or the knock-in detection primer binding at the trans knock-in site can produce single-strand fragments, which may bind single-stranded DNA (ssDNA) molecules derived from the primer binding at the endogenous site and then be extended to a full AS-PCR product (Figure 6B). These models, albeit speculative, emphasize the necessity to verify initial AS-PCR hits. BanI restriction site embedded into the tp53 R143H knock-in provided an independent means of screening F1 heterozygous embryos from a true (#7) and a trans (#5) knock-in founder (Figure 6C–H). Although multiple true and trans knock-in F1 embryos were positive by knock-in AS-PCR (Figure 6C and D), BanI only digested ‘site assay’ amplicons of true knock-in embryos but not those of trans knock-in embryos (Figure 6E and F). This result was further confirmed by sequencing (Figure 6G and H).
Figure 6.
Analysis of true and off-target (trans) knock-ins at tp53 R143H site. (A) Definitions of knock-in types. In true-positive knock-ins, the targeting oligo modifies the intended locus without off-target insertions, whereas in the trans knock-ins, insertion of the oligo occurs at an off-target locus. (B) A model of how AS-PCR can produce PCR products in both true-positive and trans knock-in situations. In the case of a true-positive knock-in case, standard PCR successfully amplifies the expected PCR product. A possible mechanism in the trans knock-in case is presented here involving an abortive PCR product strands from the WT intended knock-in locus and the trans knock-in off-target locus. Since the oligo and WT locus share significant homology, it is conceivable that very short abortive PCR products from the two loci can pair up and become amplified to the full PCR product in the next cycle thus initiating the exponential cycle of amplification leading to large amounts of PCR product visible as an apparent knock-in band. Screening and sequencing verification of true-positive (C, E and G) and trans (D, F and H) knock-ins. (C and D) A set of 15 F1 embryos from positive founders were analysed using tp53 R143H knock-in AS-PCR. WT and positive control samples were run with both the WT and knock-in PCRs as controls for the size of the PCR product and specificity of the assay. (E and F) Knock-in site assay PCRs were run on samples from both founders and then either run undigested (upper panel) or digested with BanI enzyme (lower panel) to detect the BanI site expected to be introduced by correct tp53 R143H mutation knock-in. The samples previously identified as positive for knock-in are marked with ‘+’. (G and H) R143H site assay sequencing for true-positive and trans knock-in samples. Chromatograms show that in the true-positive knock-in base positions, there are double peaks (G), which are absent from the comparable trans knock-in read (H).
Founder genotyping results in Table 1 provide evidence that the anti-sense asymmetric oligo design improved combined tp53 knock-in founder numbers (1 of 68 for the sense symmetric strategy and 4 of 63 for the anti-sense asymmetric design). These data also show that the proportion of trans knock-in founders was much lower in all of our sense knock-in strategies (3 of 82) than in all of the anti-sense knock-ins (9 of 63). This result can be possibly explained by complementarity of anti-sense asymmetric oligos to ssDNA regions generated by off-target sgRNAs. Thus, while the anti-sense oligo approach may enhance point mutation knock-in efficiency, it is with the caveat that a greater frequency of trans knock-ins may also result, necessitating careful evaluation to exclude false positives.
Phosphorothioate linkages at oligo ends stimulate knock-in efficiency and consistency of lmna R471W knock-in
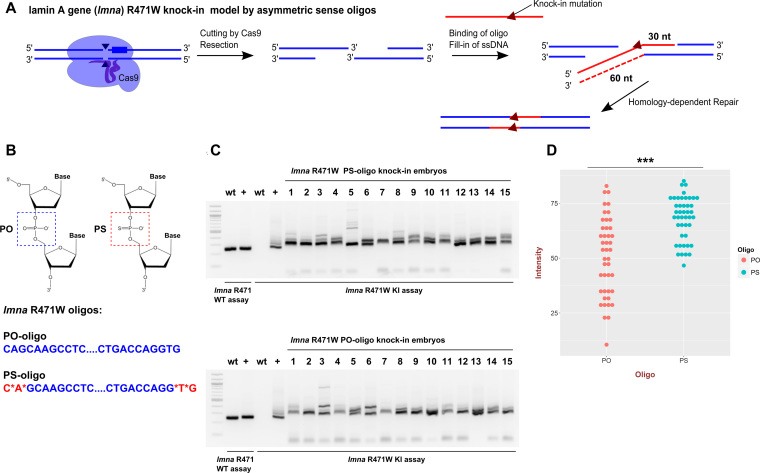
Another attractive, simple and inexpensive way to improve knock-in efficiency is to introduce PS linkages at the oligo ends to block the effect of exonuclease activity. Based on the observations in cell culture that sense asymmetric and anti-sense asymmetric oligos of 97-nt length were equally effective (12), for the lmna R471W knock-in, we designed 90-nt sense asymmetric oligo versions with or without PS modifications that according to the model suggested by this study would stimulate HDR after a CRISPR-induced double-strand break (DSB) (Figure 7A and B). Another reason for choosing a sense asymmetric oligo was to uncouple the PS-mediated effects from the stimulation and potential off-target binding of anti-sense asymmetric oligos since the sense oligos do not have complementarity regions for off-target sgRNA sites. We performed a total of 5 lmna R471W knock-in experiments with regular (PO) and PS oligos and tested 15 injected embryos each time for both types of injection using knock-in AS-PCR. In all of these experiments, we observed stimulation of knock-ins and a decrease in variation in band intensities after the knock-in AS-PCR assay (Figure 7C). The statistical analysis of measured intensities in three of these experiments (44 embryos for each knock-in) showed that there was an ∼1.4-fold significant increase in average intensity (P-value = 3.9*10−7) and a striking shift of measured values in the PS-oligo sample toward the top of the distribution suggesting that in many embryos, stimulation was significantly stronger (Figure 7D). We also followed up this initial result with quantifying germline transmission from PO and PS oligo knock-in founders. The positive founders were defined as those showing a positive signal in the initial extract from 50 embryos and in at least 1 out of 16 single embryos screened. By this measure, PO and PS oligos performed similarly with seven and eight positive founders, out of which two and three were true-positive, respectively (Supplementary Table S1). However, positive founders from the PS-modified oligo knock-in had more positive embryos than the PO founders in total (46/128 versus 25/112; Fisher Exact Test P-value = 0.0236) as well as more correct knock-in embryos (17/128 versus 5/112; Fisher Exact Test P-value = 0.0237). These results validate PS modifications as another approach to stimulate knock-in efficiency in zebrafish and the utility of AS-PCR to measure the extent of improvement.
Figure 7.
PS-modified oligos improve knock-in consistency and efficiency. (A) Targeting scheme for introducing R471W knock-in into lamin A/C gene (lmna) in zebrafish using a sense asymmetric oligo. Red-colored lines indicate the donor oligo or DNA-derived from it and blue lines indicate genomic DNA. (B) Chemical structures of the PO and the PS groups show that one of the oxygens in PO is substituted with a sulphur atom in PS. The PS groups were added in the last two chemical bonds on either end of the PS-oligo for lmna R471W knock-in and the PO-oligo was synthesized in a standard way. (C) An example of gel data for AS-PCR analysis of lmna R471W knock-ins using PS and PO oligos. WT assay serves as a control for size of the products and the knock-in assay detects the modification. (D) Graph of measured intensities of AS-PCR signals for 44 embryos for each of the PO- and PS-oligo knock-in injected groups derived from three independent experiments. The data are aggregated because there was little variation between experiments. The type of oligo is indicated by color and with x-axis label. The ‘***’ indicate the P-value in t-test of < 0.001 (3.9e-07).
DISCUSSION
The generation of point mutants using single-stranded DNA oligos and CRISPR/Cas9 genome editing reagents is an emerging technology in zebrafish and other animal model systems. The use of this technique enables single-nucleotide precision of genome editing experiments and will allow generation of specific disease models and precise mutational analysis of biological processes. Despite their obvious promise, point mutation knock-ins remain inefficient in zebrafish and the methods for testing their efficiency remain laborious or not easily accessible to many labs, such as sequencing individual plasmid clones by Sanger sequencing and NGS of PCR amplicons (11,30). In an early TALEN-based knock-in study in zebrafish, restriction sites were introduced into specific genomic sites and shown to be digested by the corresponding enzyme (7,8), but this approach has not yet been shown to work for CRISPR-based knock-ins in zebrafish. The restriction site introduction as a means of genotyping knock-ins is potentially attractive but these will have to be silent mutations in protein coding genes rather than insertions of complete sites. Moreover, PCR products with silent mutations may behave differently than those with added sequences. In the knock-in studies we describe here, we introduced missense mutations into tp53, cdh5 and lmna genes as well as synonymous mutations in PAM or sgRNA sites to prevent Cas9-mediated cutting or to introduce restriction sites for tp53 knock-ins. Restriction enzymes initially failed to genotype knock-in injected embryos, but were successful at genotyping of F1 heterozygous knock-in embryos. This discrepancy can be explained by the fact that in late PCR cycles, the strands of different PCR products can be randomly shuffled, from which follows that the fraction of PCR products having both strands containing knock-in mutations has a quadratic dependence on the knock-in allele frequency. Thus, at low (1 and 3%) knock-in rates (x), only a very small fraction (x2) (0.01 and 0.09%) of total amplicon products will contain fully complementary strands and become digested. By contrast, in knock-in heterozygotes (50% allele frequency), 25% of PCR product can be digested, which was fully consistent with our results. We therefore switched to allele-specific PCR strategy to detect point mutations in all of our knock-ins and have shown that it is very sensitive to knock-in presence at allele frequencies <0.5%. Similar detection strategies were previously used for epitope-tagging knock-ins (9,10), where one of the primers was specific to the tag inserts and the other was outside of the donor oligo region. Epitope tagging detection PCRs and point mutation AS-PCR assays are conceptually similar. However, the relatively small number of nucleotide differences between the WT and knock-in alleles can make it hard to avoid background amplification of a knock-in assay PCR in WT genomic DNA. We employed a touchdown PCR (31) protocol to make our AS-PCR strategies more specific, which was essential to the success of some AS-PCR assays and improved others. AS-PCR was also recently used in a mouse study of CRISPR/Cas9-based point mutation knock-in approaches (32) supporting the universal utility of this approach.
Overall, our studies of point mutation knock-ins revealed three main methods of improving efficiency. The first method was to reduce the distance between the mutation and Cas9 cut site (13). The first application of AS-PCR also indicated that this strategy may be useful in zebrafish since tp53 knock-ins where the distances were 10 and 13 nt were much less efficient than the cdh5 knock-in where the mutation was located exactly at the cut site. This experiment, although suggestive, could be improved by systematic variation of the mutation position relative to the cut site. The second approach, namely the usage of asymmetric oligos emerged as the key knock-in optimization in zebrafish. Knock-in AS-PCRs showed a very strong stimulation using this strategy in the case of tp53 knock-ins and NGS confirmed the significance of this result and allowed us to measure the extent of stimulation (ca. 3- and 10-fold for R143H and R217H knock-ins, respectively). Another explanation for why asymmetric oligos may function better comes from a well-established model proposing that the protruding single-stranded 3′ regions result from resection of DSBs (33). The team who explored this model performed knock-ins with multiple 97-nt oligos with different homology arms and they found that the most efficient oligos were 97 nt in length and were designed with shorter homology arms (30 nt) complementary to the resected single-stranded DNA ends produced after DSBs (12). These 30–67 asymmetric oligos introduced knock-ins equally well on either side of the DSB, thus supporting the resection model much more than the original model proposed by Richardson et al. (14). Future work will be necessary to establish if stimulation by asymmetric oligos on either strand can be equally effective, but our study provides evidence and an example of how this can be accomplished. However, the stimulation of knock-in efficiency by anti-sense asymmetric ssODNs may not be universal. For example, Moreno-Mateos et al. did not find any difference in knock-in efficiency between sense or anti-sense asymmetric oligos corresponding to the same stretch of genomic DNA when using SpCas9, but they measured that the anti-sense oligos were typically more efficient than the sense ones when DNA was cut by Cpf1 regardless of homology arm lengths ratios (34). Thus, both the genomic site and the nature of the CRISPR-related nuclease may play a role in determining the editing efficiency. In the third optimization, we tested PS modification of oligo ends while performing an lmna R471W knock-in. To uncouple potential effects of this modification from those of anti-sense asymmetric oligos and to avoid possible binding of oligos to ssDNA regions at off-target sgRNA sites, we chose sense asymmetric oligos of 90 nt with or without two PS bonds at either end of the oligo. Indeed, the PS-modified oligo was significantly more efficient and consistent at introducing knock-ins than the standard DNA oligo. Previously, the group that developed PS-mediated knock-in stimulation could only identify some imprecise knock-in events in zebrafish and did not test for knock-in improvement (15). We believe that all of these new optimization methods have utility and may even have multiplicative effects when deployed simultaneously. Therefore, at this stage of genome editing technology development, it is advisable to test several versions of oligos incorporating desired optimizations as well as a non-optimized control oligo in order to determine if the optimized versions behave in the expected way.
In the process of genotyping knock-in founders we developed a general workflow to identify true knock-in founders. The unexpected result that emerged from sequencing single F1 embryos was that there were many false-positive or trans knock-in founders (25–78% of total founder number). These could be screened out by sequencing or restriction digests, but they also revealed a weakness of AS-PCR strategies, which can falsely produce a positive signal likely due to hybridization of single DNA strands from abortive PCR products from target and trans loci. A possible mechanism of trans knock-in origin most likely has to do with off-target sgRNA sites, into which the oligo can ligate. Interestingly, the proportion of trans knock-in founders was much lower in all of our sense knock-in strategies than in all of anti-sense knock-ins. Anti-sense asymmetric oligos may have some complementarity to ssDNA regions generated at off-target sites, but this possibility needs further investigation.
In conclusion, we have provided and validated strategies to optimize and enhance point mutation knock-in efficiency in zebrafish. Proximity of the knock-in mutations to the Cas9 cut sites and anti-sense asymmetric oligos were identified as the most effective optimizations. PS modifications also enhanced knock-in efficiency and improved consistency among different embryos. These optimizations were enabled by AS-PCR assays and NGS. Restriction sites introduced by silent mutation as part of the knock-in process were also very useful for point mutant genotyping but only when they were present at high frequency (e.g. at 50%). We also identified the phenomenon of trans knock-ins, which can be filtered out using digestions of restriction sites introduced with silent mutations. We envision that this work will make point mutation knock-in generation a straightforward procedure accessible to all zebrafish researchers and other model systems researchers given the universal applicability of the methodology for point mutation introduction and optimization across a variety of animal models.
DATA AVAILABILITY
All of the amplicon sequencing datasets have been deposited into the Sequence Read Archive under the accession number SRP126996 and are available at the following web address: https://www.ncbi.nlm.nih.gov/sra/SRP126996.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Gretchen Wagner, Emma Cummings and David Maley for excellent fish care. Sergio Pereira performed Illumina Sequencing of PCR products from knock-in sites. We would like to thank Dr David Langenau and Dr Shawn Burgess for expert peer review of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Terry Fox Research Institute Program Project Grant (to D.M., A.S., J.N.B); Atlantic Opportunities Agency of Canada, Atlantic Innovation Fund Grant (to C.R.M., J.M.R, J.N.B.). Funding for open access charge: Terry Fox Research Institute Program Project Grant (to D.M., A.S., J.N.B); Atlantic Opportunities Agency of Canada, Atlantic Innovation Fund Grant (to C.R.M., J.M.R, J.N.B.).
Conflict of interest statement. None declared.
REFERENCES
- 1. Li M., Zhao L., Page-McCaw P.S., Chen W.. Zebrafish genome engineering using the CRISPR–Cas9 System. Trends Genet. 2016; 32:815–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ceasar S.A., Rajan V., Prykhozhij S. V., Berman J.N., Ignacimuthu S.. Insert, remove or replace: A highly advanced genome editing system using CRISPR/Cas9. Biochim. Biophys. Acta. 2016; 1863:2334–2344. [DOI] [PubMed] [Google Scholar]
- 3. Varshney G.K., Sood R., Burgess S.M.. Understanding and editing the zebrafish genome. Methods Mol. Biol. 2015; 92:1–52. [DOI] [PubMed] [Google Scholar]
- 4. Prykhozhij S. V., Rajan V., Berman J.N.. A guide to computational tools and design strategies for genome editing experiments in zebrafish using CRISPR/Cas9. Zebrafish. 2015; 13:70–73. [DOI] [PubMed] [Google Scholar]
- 5. Gagnon J.A., Valen E., Thyme S.B., Huang P., Ahkmetova L., Pauli A., Montague T.G., Zimmerman S., Richter C., Schier A.F.. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One. 2014; 9:e98186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hwang W.Y., Fu Y., Reyon D., Maeder M.L., Kaini P., Sander J.D., Joung J.K., Peterson R.T., Yeh J.R.J.. Heritable and precise zebrafish genome editing using a CRISPR-Cas system. PLoS One. 2013; 8:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bedell V.M., Ekker S.C.. Using engineered endonucleases to create knockout and knockin zebrafish models. Methods Mol. Biol. 2015; 1239:291–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bedell V.M., Wang Y., Campbell J.M., Poshusta T.L., Starker C.G., Krug R.G. II, Tan W., Penheiter S.G., Ma A.C., Leung A.Y.H. et al. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012; 490:114–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hruscha A., Krawitz P., Rechenberg A., Heinrich V., Hecht J., Haass C., Schmid B.. Efficient CRISPR/Cas9 genome editing with low off-target effects in zebrafish. Development. 2013; 140:4982–4987. [DOI] [PubMed] [Google Scholar]
- 10. Burg L., Zhang K., Bonawitz T., Grajevskaja V., Bellipanni G., Waring R., Balciunas D.. Internal epitope tagging informed by relative lack of sequence conservation. Sci. Rep. 2016; 6:36986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Armstrong G.A.B., Liao M., You Z., Lissouba A., Chen B.E., Drapeau P.. Homology directed knockin of point mutations in the zebrafish tardbp and fus genes in ALS using the CRISPR/Cas9 system. PLoS One. 2016; 11:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liang X., Potter J., Kumar S., Ravinder N., Chesnut J.D.. Enhanced CRISPR/Cas9-mediated precise genome editing by improved design and delivery of gRNA, Cas9 nuclease, and donor DNA. J. Biotechnol. 2016; 241:136–146. [DOI] [PubMed] [Google Scholar]
- 13. Paquet D., Kwart D., Chen A., Sproul A., Jacob S., Teo S., Olsen K.M., Gregg A., Noggle S., Tessier-Lavigne M.. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature. 2016; 533:1–18. [DOI] [PubMed] [Google Scholar]
- 14. Richardson C.D., Ray G.J., DeWitt M. a, Curie G.L., Corn J.E.. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol. 2016; 34:339–344. [DOI] [PubMed] [Google Scholar]
- 15. Renaud J.B., Boix C., Charpentier M., De Cian A., Cochennec J., Duvernois-Berthet E., Perrouault L., Tesson L., Edouard J., Thinard R. et al. Improved genome editing efficiency and flexibility using modified oligonucleotides with TALEN and CRISPR-Cas9 nucleases. Cell Rep. 2016; 14:2263–2272. [DOI] [PubMed] [Google Scholar]
- 16. Bialk P., Rivera-Torres N., Strouse B., Kmiec E.B.. Regulation of gene editing activity directed by single-stranded oligonucleotides and CRISPR/Cas9 systems. PLoS One. 2015; 10:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bialk P., Sansbury B., Rivera-Torres N., Bloh K., Man D., Kmiec E.B.. Analyses of point mutation repair and allelic heterogeneity generated by CRISPR/Cas9 and single-stranded DNA oligonucleotides. Sci. Rep. 2016; 6:32681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gaudet M., Fara A.-G., Beritognolo I., Sabatti M.. Allele-Specific PCR in SNP Genotyping. Methods Mol. Biol. 2009; 578:415–424. [DOI] [PubMed] [Google Scholar]
- 19. White R.M., Sessa A., Burke C., Bowman T., Ceol C., Bourque C., Dovey M., Goessling W., Burns E., Zon L.I.. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell. 2008; 2:183–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lawson N.D., Weinstein B.M.. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev. Biol. 2002; 248:307–318. [DOI] [PubMed] [Google Scholar]
- 21. Johnson M., Zaretskaya I., Raytselis Y., Merezhuk Y., McGinnis S., Madden T.L.. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008; 36:5–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xu H., Xiao T., Chen C., Li W., Meyer C.A., Wu Q., Wu D., Cong L., Zhang F., Liu J.S. et al. Sequence determinants of improved CRISPR sgRNA design. Genome Res. 2015; 25:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stemmer M., Thumberger T., del Sol Keyer M., Wittbrodt J., Mateo J.L.. CCTop: An intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS One. 2015; 10:e0124633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jao L.-E., Wente S.R., Chen W.. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:13904–13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen J., Zhang X., Wang T., Li Z., Guan G., Hong Y.. Efficient detection, quantification and enrichment of subtle allelic alterations. DNA Res. 2012; 19:423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Magoč T., Salzberg S.L.. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011; 27:2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Miles L.B., Verkade H.. TA-cloning vectors for rapid and cheap cloning of zebrafish transgenesis constructs. Zebrafish. 2014; 11:281–282. [DOI] [PubMed] [Google Scholar]
- 29. Larson J.D., Wadman S.A., Chen E., Kerley L., Clark K.J., Eide M., Lippert S., Nasevicius A., Ekker S.C., Hackeff P.B. et al. Expression of VE-cadherin in zebrafish embryos: A new tool to evaluate vascular development. Dev. Dyn. 2004; 231:204–213. [DOI] [PubMed] [Google Scholar]
- 30. Boel A., Steyaert W., De Rocker N., Menten B., Callewaert B., De Paepe A., Coucke P., Willaert A.. BATCH-GE: Batch analysis of Next-Generation sequencing data for genome editing assessment. Sci. Rep. 2016; 6:30330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Korbie D.J., Mattick J.S.. Touchdown PCR for increased specificity and sensitivity in PCR amplification. Nat. Protoc. 2008; 3:13–15. [DOI] [PubMed] [Google Scholar]
- 32. Ma X., Chen C., Veevers J., Zhou X., Ross R.S., Feng W., Chen J.. CRISPR/Cas9-mediated gene manipulation to create single-amino-acid-substituted and floxed mice with a cloning-free method. Sci. Rep. 2017; 7:42244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Symington L.S. Mechanism and regulation of DNA end resection in eukaryotes. Crit. Rev. Biochem. Mol. Biol. 2016; 9238:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Moreno-Mateos M.A., Fernandez J.P., Rouet R., Vejnar C.E., Lane M.A., Mis E., Khokha M.K., Doudna J.A., Giraldez A.J.. CRISPR-Cpf1 mediates efficient homology-directed repair and temperature-controlled genome editing. Nat Commun. 2017; 8:2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the amplicon sequencing datasets have been deposited into the Sequence Read Archive under the accession number SRP126996 and are available at the following web address: https://www.ncbi.nlm.nih.gov/sra/SRP126996.