Abstract
Derivatives of 5-hydroxyuridine (ho5U), such as 5-methoxyuridine (mo5U) and 5-oxyacetyluridine (cmo5U), are ubiquitous modifications of the wobble position of bacterial tRNA that are believed to enhance translational fidelity by the ribosome. In gram-negative bacteria, the last step in the biosynthesis of cmo5U from ho5U involves the unique metabolite carboxy S-adenosylmethionine (Cx-SAM) and the carboxymethyl transferase CmoB. However, the equivalent position in the tRNA of Gram-positive bacteria is instead mo5U, where the methyl group is derived from SAM and installed by an unknown methyltransferase. By utilizing a cmoB-deficient strain of Escherichia coli as a host and assaying for the formation of mo5U in total RNA isolates with methyltransferases of unknown function from Bacillus subtilis, we found that this modification is installed by the enzyme TrmR (formerly known as YrrM). Furthermore, X-ray crystal structures of TrmR with and without the anticodon stemloop of tRNAAla have been determined, which provide insight into both sequence and structure specificity in the interactions of TrmR with tRNA.
INTRODUCTION
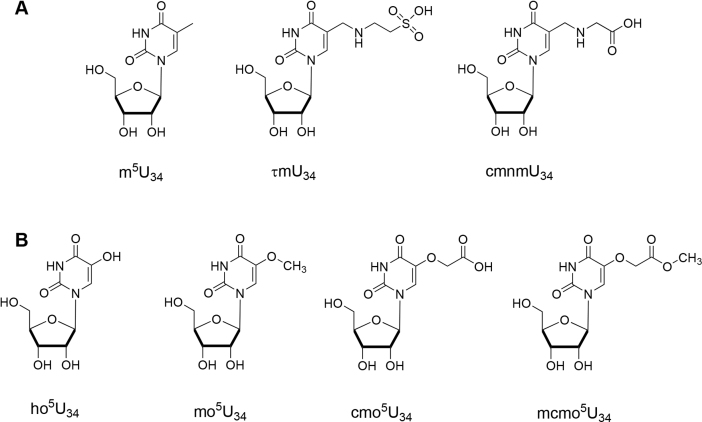
Degeneracy of the genetic codon occurs through non-canonical Watson–Crick pairings between the 3′ nucleotide of a codon triplet in mRNA and the ‘wobble’ nucleoside at the 5′-end of an anticodon in tRNA; the first two nucleosides of the codon are conserved, but divergence occurs at the third codon (1). Modifications of tRNA within anticodon stemloop (ASL) promote non-Watson–Crick pairings of the wobble nucleoside with the cognate codon (2). The C5 position of uridine nucleobases in the wobble position of tRNA are frequently targeted for modification (3,4). These modifications range from methylation (m), to more structurally complicated like taurinomethylation (τm) and carboxymethylaminomethylation (cmnm), which require multiple enzymatic steps for their formation (Figure 1A). The uridine nucleobase in the wobble position (U34) of several tRNAs is hydroxylated at the C5 position by an unknown enzyme, which is then further elaborated with methyl, carboxymethyl and carboxymethylmethoxy modifications (Figure 1B). These oxyalkylated uridine modifications are widely conserved among bacterial tRNA, where the biological significance has been associated with expanded base-pairing specificity during translation. Uridines at the wobble position pair with the third codon A and G in the wobble hypothesis (1). However, modified tRNAs with wobble xo5U (x = methyl, carboxymethyl or carboxymethylmethoxy) usually recognize three codons ending with U, A, G and, in some instances, C (5). Furthermore, it has been observed that xo5U modified tRNASer can read the native UCU and UCG Ser codons more efficiently than the unmodified form (6). Thus, 5-oxyalkylation enables broader binding capacities of wobble uridines to efficiently decode -A and -C and -G ending codons (7,8).
Figure 1.
Examples of the wobble uridine modification at C5. Chemical structures of modified uridine at the wobble position (U34) is shown with (A) alkylation or (B) O-alkylation at C5 of pyrimidine. 5-methyluridine (m5U), 5-taurinomethyluridine (τm5U), 5-carboxymethylaminomethyluridine (cmnm5U), 5-hydroxyuridine (ho5U), 5-methoxyuridine (mo5U), 5-carboxymethyluridine (cmo5U), 5-carboxymethoxyuridine (mcmo5U).
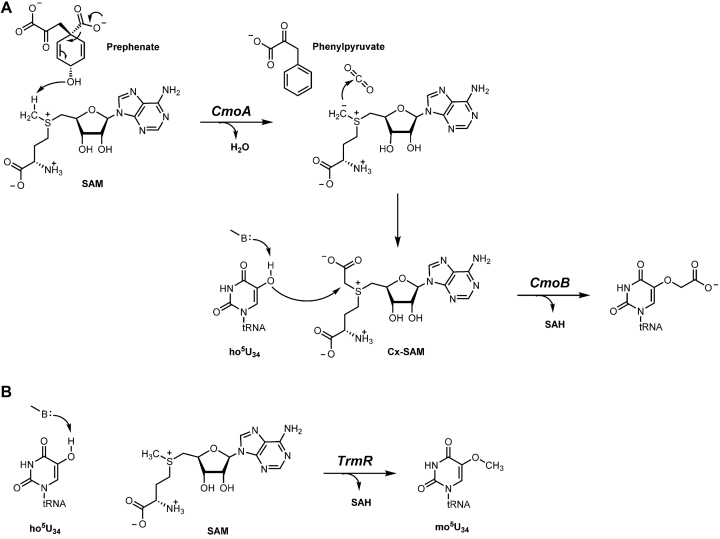
5-oxyacetyluridine (cmo5U) is one of the most abundant modified tRNA bases in Escherichia coli (9) and has been identified at the wobble position of Ala-, Pro-, Ser-, Thr- and Val-specific tRNA isoacceptors (10). Biosynthesis of cmo5U involves the enzymes CmoA and CmoB, which were predicted, based on sequence homology, to be S-adenosyl-l-methionine (SAM) dependent methyltransferases (5). However, we have recently shown that CmoA produces carboxy-SAM (Cx-SAM)—a naturally occurring SAM analog—from prephenate and SAM (11) Subsequently, CmoB transfers the carboxymethyl group from Cx-SAM to 5-hydroxyuridine (ho5U) (Figure 2A). Although CmoB is capable of utilizing SAM instead of Cx-SAM, the reaction is extremely slow and likely not physiological (12). Indeed, 5-methoxyuridine is not observed in E. coli, unless the biosynthesis of Cx-SAM has been compromised by disrupting cmoA gene. The wobble uridines of equivalent isoacceptors in Bacillus subtilis are modified to 5-methoxyuridine (mo5U) (13–16) Notably, biosynthetic pathways leading to mo5U and cmo5U have been shown to share a common intermediate, ho5U (17) A BLAST search of the B. subtilis genome with either E. coli cmoA or cmoB does not yield homologues sequences, indicating that B. subtilis has a unique methyltransferase for installing the methyl group on ho5U.
Figure 2.
Proposed biosynthetic pathways for the synthesis of cmo5U and mo5U. (A) In Gram-negative bacteria, Cx-SAM is synthesized from SAM and prephenate by CmoA. Subsequently, CmoB directs carboxymethyltransferation at the 5-hydroxyl group of ho5U34, which is generated by an unknown pathway. (B) TrmR transfers the methyl group from SAM onto ho5U34 of equivalent isoacceptors in Gram-positives. B denotes a general base.
By screening a set of previously uncharacterized SAM-dependent methyltransferases, we report that TrmR (previously annotated as YrrM, Uniprot ID: O32036) catalyzes the methylation of ho5U containing tRNA in B. subtilis (Figure 2B). Furthermore, we have determined crystal structures of TrmR complexed with SAM and the ASL of tRNAAla, as well as TrmR complexed with S-adenosyl-l-homocysteine (SAH) at 1.70 and 2.27 Å resolution, respectively. These structures provide a molecular basis for the distinct mode of tRNA recognition to promote the efficient methyltransfer by TrmR. To our knowledge, this is the first structure of a wobble-nucleotide-specific methyltransferase complexed with the cognate tRNA substrate.
MATERIALS AND METHODS
Cloning and protein purification
The trmR gene was amplified from genomic DNA of B. subtilis 168 strain using the forward primer (AGAAGGAGATATAACTGTGACTGACCGGTATGAACAAATA) and the reverse primer (GTGGTGGTGATGGTGATGGCCTCACCTCTTCTTTTTACTA) and Phusion DNA polymerase (Thermo Scientific) in a standard polymerase chain reaction (PCR) reaction. The amplified PCR fragment was purified by gel extraction (Qiagen) and cloned into LIC-pLATE31 (Thermo Scientific) and verified by the DNA sequence analysis (Macrogen, Korea). Escherichia coli BL21 (DE3) cells were transformed with vectors harboring the trmR gene, grown in LB containing 100 mg/ml ampicillin at 37°C and induced with 0.5 mM isopropyl -D-1-thiogalactopyranoside (IPTG) at an OD600 of ∼1. Cells were incubated overnight at 25°C and subsequently harvested by centrifugation. The cell pellet was resuspended in lysate buffer (25 mM HEPES, pH 7.5 and 150 mM NaCl, 2 mM dithiothreitol, 10% glycerol) and lysed by sonication for 15 min. The lysate was cleared by centrifugation (39 000 g for 15 min), and the supernatants were applied to HisTrap HP columns (GE Healthcare) charged with Ni2+. The column was washed with buffer A, binding buffer (25 mM HEPES, pH 7.5 and 150 mM NaCl, 10 mM imidazole) and eluted with a linear gradient (0.01–0.5 M) of imidazole. The recombinant protein was eluted with ∼30% buffer B and further purified by size exclusion chromatography on a HiLoad Superdex 75pg column (GE Healthcare) equilibrated with buffer C (25 mM HEPES, pH 7.5 and 150 mM NaCl). Final purity of the eluted fraction was over 98% as verified by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis. An extinction coefficient of ϵ280 = 23.4 cm−1mM−1 was used to calculate the yield of the TrmR as calculated from the amino acid sequence.
Crystallization and structure determination
For tRNA-free TrmR sample, a mixture of purified TrmR was with 5 mM MgCl2, and 2 mM SAH (Sigma-Aldrich) was crystallized by sitting drop vapor diffusion at room temperature by mixing 0.8 μl of the protein sample with 0.8 μl of reservoir solution (0.2 M Lithium sulfate monohydrate, 0.1 M TRIS hydrochloride pH 7.5, 30% w/v PEG 4000) and equilibrating over 60 μl of reservoir solution. Crystals were cryoprotected by addition of 25% glycerol, mounted in a nylon loop and flash-cooled in liquid nitrogen. X-ray data were collected on an ADSC QUANTUM 270 detector at the Pohang Accelerator Laboratory (PAL) beam line 7A, and processed with HKL2000 (18). Single wavelength diffraction data were collected at a wavelength λ = 0.9793 Å, which were scaled in a space group P21212 (Table 1). Molecular replacement was performed using the structure of O-methyltransferase from Bacillus Halodurans TrmR (PDB ID: 2GPY) as the search model with MOLREP (19). Subsequent model building and refinement were performed with Coot (20) and REFMAC5 (21). The final model was refined to 2.27 Å with Rwork = 0.213 and Rfree = 0.266. All residues are in allowed regions, and no outliers were found in a Ramachandran plot.
Table 1.
Crystallographic statistics
| TrmR:tRNAAla | RNA-free TrmR | |
|---|---|---|
| Data collection | ||
| Wavelength (Å) | 0.9793 | 0.9793 |
| Space group | P 32 2 1 | P 21 21 2 |
| Cell dimensions | ||
| a, b, c (Å) | 64.98 64.98 126.68 | 76.87 116.74 43.44 |
| α, β, γ (°) | 90 90 120 | 90 90 90 |
| Resolution (Å) | 50.0–1.70 (1.73–1.70) | 50.0–2.27 (2.31–2.27) |
| R merge | 0.090 (>1) | 0.091 (0.41) |
| I/σI | 33.8 (1.5) | 20.7 (4.0) |
| CC1/2* | 0.449 | 0.976 |
| Completeness (%) | 94.1 (81.4) | 99.2 (97.6) |
| Redundancy | 17.5 (9.5) | 6.1 (5.1) |
| Refinement | ||
| Resolution (Å) | 50.0–1.70 | 30.0–2.27 |
| R work/Rfree | 0.193/0.222 | 0.216/0.266 |
| No. atoms | ||
| Protein | 1792 | 3204 |
| RNA | 317 | - |
| Ligand/ion | 32 | 54 |
| Water | 162 | 88 |
| B-factors (Å2) | ||
| Protein | 36.47 | 48.20 |
| RNA | 113.9 | - |
| Water | 45.99 | 43.11 |
| R.m.s deviations | ||
| Bond lengths (Å) | 0.009 | 0.009 |
| Bond angles (°) | 0.955 | 1.188 |
| Ramanchandran Analysis | ||
| Favored (%) | 97.66 | 95.66 |
| Allowed (%) | 1.87 | 4.34 |
| Outliers (%) | 0.47 | 0 |
Crystallization of tRNA–TrmR complex was attempted by mixing 1:1.5 molar ratio of TrmR and alanine-specific 17-mer ASL tRNA (5′-rCrCrUrGrCrUrUrUrGrCrArCrGrCrArGrG-3′), which was purchased from Integrated DNA Technology (Coralville, USA). The protein–RNA complex solution also containing 2 mM SAM (Sigma-Aldrich) was crystallized by sitting drop vapor diffusion method as described for tRNA-free TrmR sample. The crystals of tRNA–TrmR complex appeared in a condition containing 0.4 M Ammonium phosphate monobasic (Hampton Research, USA) in the reservoir solution. Crystals were cryoprotected by addition of 25% glycerol and stored in liquid nitrogen. X-ray diffraction data, consistent with a space group P3221, were collected at the beam line 7A of PAL using a wavelength λ = 0.9793 Å on an ADSC QUANTUM 270 detector. Phases were obtained by molecular replacement using Molrep (19) using the tRNA-free TrmR structure as an initial model. Subsequent model building and refinement were performed with Coot (20) and REFMAC5 (21). The final model was refined to 1.70 Å with Rwork = 0.193 and Rfree = 0.222.
Determination of molecular weight of TrmR by size exclusion chromatography
The molecular weight of recombinant TrmR was determined by gel filtration using Superdex 75 pg column (GE Healthcare, Life Sciences) on NGC FPLC system (Biorad). The Gel Filtration Standard kit (Biorad) was used for column calibration and molecular weight determination. In each experiment, 1 mg of purified recombinant TrmR (calculated molecular weight = 26 kD) was loaded on the column. The mobile phase (25 mM HEPES, pH 7.5, containing 150 mM sodium chloride) for column equilibration and protein elution was used at a flow rate of 1 ml/min.
In vitro assay of TrmR
The O-methyltransfer activity of TrmR was examined with a solution containing 100 mM NaCl, 10 mM Sodium Phosphate (pH 7.5), 2 mM MgCl2, 0.1 mM SAM, 10 μM TrmR and total RNA 100 μg extracted from trmR-mutant B. subtilis cells (National BioResource Project Japan (NBRP), Japan, strain number MGNA-A853). A total volume of the assay was 50 μl, which was incubated at room temperature overnight. Total RNA was extracted, digested and analysed as described previously (11). In brief, cellular RNA was purified with Trizol (Ambion) according to the manufacturer’s instruction. One unit of P1 nuclease (Sigma-Aldrich) was added to the assay solution and incubated at 65°C for 30 min to convert polynucleotides into 5′-nucleotide monophosphates. Then, one unit of Antarctic Phosphatase (New England Biolab) was added to the assay solution and incubated at 37°C for 1 h. The P1 nuclease and Antarctic Phosphatase-treated sample was injected into a 6545XT AdvancedBio Agilent LC-QTOF-MS for MS/MS analysis in the positive mode to identify the modified nucleotides. To identify mo5U, the mass of these nucleotides (calculated m/z = 275.0879) were included in the parent ion inclusion list for MS/MS. An aliquot of 20 μl sample was injected onto the HPLC column (Agilent C18 Column, 100 Å, 3.5 μm, 4.6 mm × 150 mm) coupled to Agilent 1200 HPLC (Agilent) to generate a gradient with a 0.3 ml·min−1 flow rate. Solvent A was DW, 0.1% formic acid (FA); solvent B consisted of 100% acetonitrile, 0.1% FA. After desalting for 3 min with 5% B, the nucleotides were eluted at 0.3 ml·min−1 with a 5–25% gradient for 15 min, 25–100% for 5 min.
Genetic complementation of trmR
For the complementation assay, expression plasmids with an inducible promotor (T5) for E. coli were constructed by inserting trmR gene between BamHI and SmaI sites on pQE30a. The pQE30a-trmR was used to transform ΔcmoB cell; empty pQE30a plasmids were used to transform ΔcmoB cell as a control. Transformed cells were typically grown in 50 ml LB media at 37°C, which were harvested by centrifugation at 3214 g for 20 min. Total RNA was extracted by treating with one unit of P1 nuclease at 65°C for 30 min and one unit of Antarctic Phosphatase at 37°C for 1 h. The nucleoside samples were analysed using LC-MS (Agilent C18 column, 100 Å, 3.5 μm, 4.6 mm × 150 mm coupled to a 6545XT AdvancedBio Agilent LC-QTOF-MS) in positive mode. A linear gradient of 5 to 25% acetonitrile and 0.1% FA was used over 15 min.
RESULTS
Genetic screening for the ho5U methyltransferse
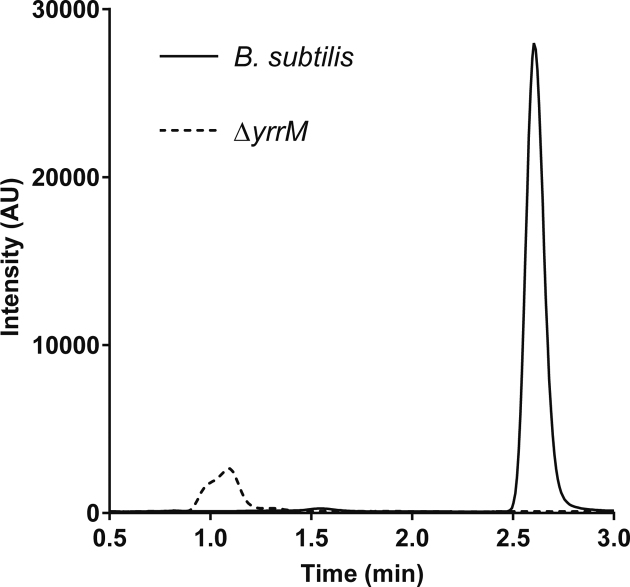
Using the sequence of CmoB from E. coli in a BLAST search of the B. subtilis genome fails to identify a 5-hydroxyuridine methyltransferase. Therefore, we set out to examine the in vivo production of mo5U from a group of eight single gene knock-outs of B. subtilis methyltransferases (yrrM, yodH, ydaC, yibH, yrhH, ycgJ, yacO and yrrT). These eight were chosen because they lacked full functional annotation in NCBI database. Total RNAs were extracted from each mutant, digested with Nuclease P1, dephosphorylated with Antartic phosphatase, and then analyzed by LC-MS looking for the presence of mo5U. All mutant strains, except for ΔyrrM, contained mo5U (Figure 3 and Supplementary Figure S1), suggesting yrrM is likely to account for the in vivo methylation of ho5U in B. subtilis.
Figure 3.
Biosynthesis of mo5U by yrrM (or trmR). Presence of mo5U in total RNA extracted from Bacillus subtilis and the single knockout mutants was examined by triple-quadruple (QqQ) mass spectrometry in positive ion mode. Multiple reaction monitoring of mo5U is quantified by tracing the fragmentation of 5-methoxyuracil (mass-to-charge ratio 143.046), and an exemplary chromatogram of the parent strain is displayed (left), whereas no equivalent transition was observed in the sample prepared from ΔyrrM, or ΔtrmR.
Because previous in vitro experiments showed that the crude lysate from B. subtilis can utilize E. coli tRNA to produce mo5U (17), we tested whether mo5U was synthesized in cmoB-deficient E. coli by yrrM complementation. Indeed, analysis of RNAs from ΔcmoB cells transformed with the B. subtilis YrrM-expressing plasmid showed the presence of mo5U (Supplementary Figure S2A). The genetic data, which have been further verified by in vitro assays and X-ray crystallography as shown below, supports that yrrM is the ho5U methyltransferase in tRNA modification and we reassign the nomenclature of yrrM to trmR hereafter.
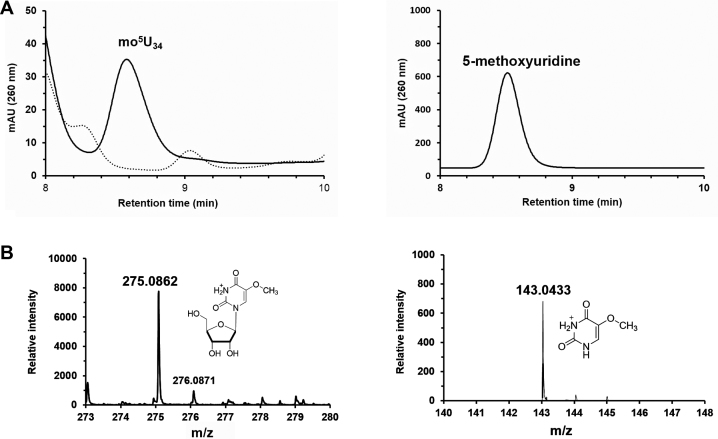
TrmR is able to methylate ho5U-containing tRNA in vitro
The mo5U formation by TrmR has been tested by measuring the in vitro activity of the recombinant protein with SAM and ho5U-containing tRNAs, which had been prepared from ΔtrmR cells. After overnight incubation of reaction mixture at room temperature, the tRNAs were digested and analyzed. Incorporation of the methyl group into the substrate tRNA was observed as mo5U by HPLC, and further confirmed by mass spectrometry (Figure 4). In order to confirm the in vivo specificity of TrmR on the corresponding tRNA isoacceptors of E. coli, the in vitro activity of recombinant TrmR was examined with ho5U-containing tRNA prepared from cmoB- knockout E. coli cells. The formation of mo5U within E. coli tRNA was indeed TrmR dependent, consistent with the gene complementation assay and the previous observation (17) (Supplementary Figure S2B).
Figure 4.
In vitro mo5U forming activity of TrmR. (A) On the left, HPLC analysis of TrmR directed the formation of mo5U (solid line) versus control which contains no TrmR (dashed line). The peak corresponding mo5U eluted from the C-18 column at 8.6 min which has been confirmed by spiking with an authentic compound (right panel). (B) The assay sample was analyzed by LC-MS/MS in positive mode, where the peak corresponding to mo5U ([C10H15N2O7]+, calculated m/z = 275.088) was selected (left) and the subsequent fragmentation by ESI was confirmed as detection of 5-methoxyuracil ([C5H7N2O3]+, calculated m/z = 143.046) in MS/MS (right).
Crystallization of TrmR with the substrates
Our initial efforts to crystalize recombinant TrmR with the hexahistidine tag located in the N-terminus was not successful in generating high-quality crystals for further structure determination, typically yielding highly mosaic diffraction to 7–8 Å. Interestingly, when recombinant TrmR fused with the C-terminal hexahistidine tag was used, well-diffracting crystals were found in multiple conditions from the initial crystallization screens. Crystals of RNA-free TrmR was obtained in presence of both MgCl2 and SAH. Magnesium chloride was included in our crystallization trials since several structural homologs of TrmR have been shown to contain a divalent metal ion in the active site (22–24). The crystals of TrmR: ASL of tRNAAla: SAM ternary complex, hereafter denoted as TrmR:tRNAASL, were obtained using chemically synthesized 17mer ASL of an alanine-specific tRNA, and SAM. Of note, unlike RNA-free TrmR crystals, the average half-life of TrmR:tRNAASL crystals was much shorter (<48 h) and exposure to air during cryo protection and freezing step promoted even faster decay within minutes. It is speculated that the observed instability of the crystals resulted from the fragile nature of the bound tRNA stemloop. Crystallographic statistics for both structures are summarized in Table 1.
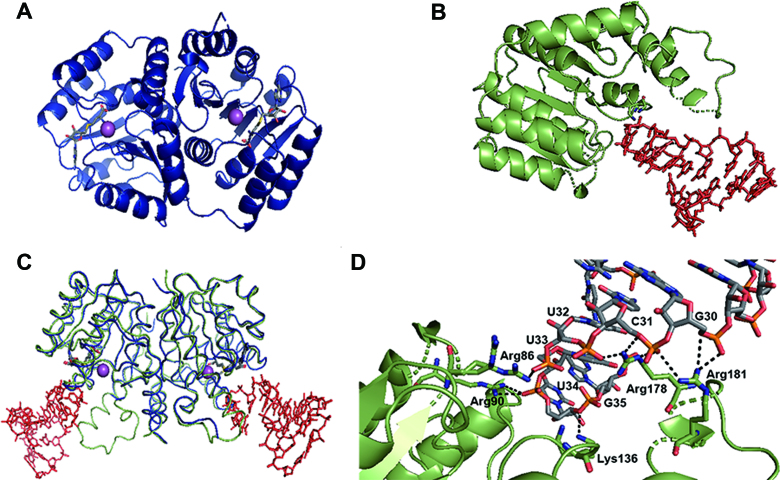
The overall structure of TrmR
The protein adopts a characteristic Rossmann-fold, with seven β-strands flanked by two layers of α-helices (Figure 5A and B). There is no significant difference in the overall conformation of TrmR between TrmR:tRNAASL and the RNA-free TrmR structures, with RMSD of 0.61 Å of Cα (Figure 5C). Using the coordinates of RNA-free TrmR structure as a probe, a search for structural homologs on DALI server (25) showed that the top 30 hits belong to nine proteins, with RMSD and Z-value ranging from 1.5 to 2.5 Å, and from 30.1 to 25.6, respectively. The protein with the highest structural similarity is putative caffeoyl-CoA O-methyltransferase from Staphylococcus aureus (PDB ID: 3NTV), sharing 38% sequence identity. Eight other proteins are a member of either O-methyltransferase or caffeoyl-CoA O-methyltransferase family.
Figure 5.
Overall structures of TrmR. The ribbon presentation of the crystal structure of (A) RNA-free TrmR (navy) with SAH (gray stick) and magnesium ion (sphere, magenta), and (B) TrmR (green) bound to tRNAASL (red stick) with SAM (gray stick). (C) Overlaid structures of RNA-free TrmR (blue) and TrmR:tRNAASL (green and red). (D) Ionic/hydrogen bond interactions between ASL tRNA and a set of basic residues of TrmR in the crystal structure of TrmR–ASLAla complex. The ASL is shown with their carbon atoms colored gray, nitrogen atoms colored blue, oxygen atoms colored red and phosphorous atoms colored orange. Hydrogen bond is indicated by dashed lines.
Whereas a homodimer is presented as the asymmetric subunit in RNA-free TrmR structure, it is a monomer in TrmR:tRNAASL structure. However, essentially identical dimeric interactions between symmetry-related pairs are observed in TrmR:tRNAASL crystalline as in RNA-free structure. The oligomerization state of the protein in solution was examined by size exclusion chromatography, and the purified protein eluted at the retention volume corresponding to the molecular weight of ∼44 kD, consistent with a homodimer (Supplementary Figure S3). The buried surface area (26) of the reconstituted dimer is 1982 Å2 in TrmR:tRNAASL structure, whereas that of the RNA-free structure is 1652 Å2, substantially smaller because of disordered amino acid residues around the dimeric interface in the absence of tRNA. These residues participate in tRNA recognition which will be discussed in more detail later.
In TrmR:tRNAASL structure, 50 amino acid residues from each monomer are contributing to homodimeric interactions, where twenty of those contact with the other subunit through hydrogen bonding within 3.2 Å. Electrostatic potential mapped on the surface of TrmR:tRNAASL structure illustrates that the protein dimerization is mostly driven by hydrophobic interactions (Supplementary Figure S4A). Ionic interactions are minor at the protein-protein dimer interface, where a single pair of residues, Asp-3 and Arg-4, participates in forming a salt bridge between subunits (Supplementary Figure S4B).
Recognition of tRNA binding
An initial Fo – Fc differential Fourier density map shows electron density for most of the backbone phosphate atoms as well as the purine or pyrimidine side chains of the tRNAASL near protein interface (Supplementary Figure S5); however, the overall electron densities of tRNAASL are weaker compared to those of the protein in TrmR:tRNAASL structure, presumably because of a high degree of disorder or low binding affinity of tRNA, as reflected in the elevated average B-factor for tRNA compared to that for the protein (113.9 and 36.47 Å2, respectively). Out of 17 nucleotides of the ASL, only seven (U29 through G35, numbering based on B. subtilis tRNAala sequence) are in contact with 12 amino acid residues of the protein, conferring buried surface area of 379.9 Å2. Nucleotides C27, C28, G42 and G43 are located at the lattice interface, making contacts with the symmetry-related pair of TrmR. However, C36 through A41 do not interact with the protein nor contribute to crystal packing. While all amino acids except for the initial methionine are accountable in TrmR:tRNAASL structure, electron densities corresponding to amino acid residues 174–182 and 164–185 were not visible in chain A and chain B of RNA-free structure, respectively (Figure 5C). These disordered residues in RNA-free structure are well defined in TrmR:tRNAASL structure upon binding with tRNA. Recognition of tRNA by TrmR mainly occurs through a series of ionic/hydrogen bonding interactions between the side chains of key basic residues and phosphate backbone of polynucleotides; e.g. G30-R181, C31-R181, U32-R178, U33-R86, U34-R90 and G35-K136 (Figure 5D). Of note, there is no significant salt bridge or hydrogen bond between the protein and any base/ribose moiety of tRNA.
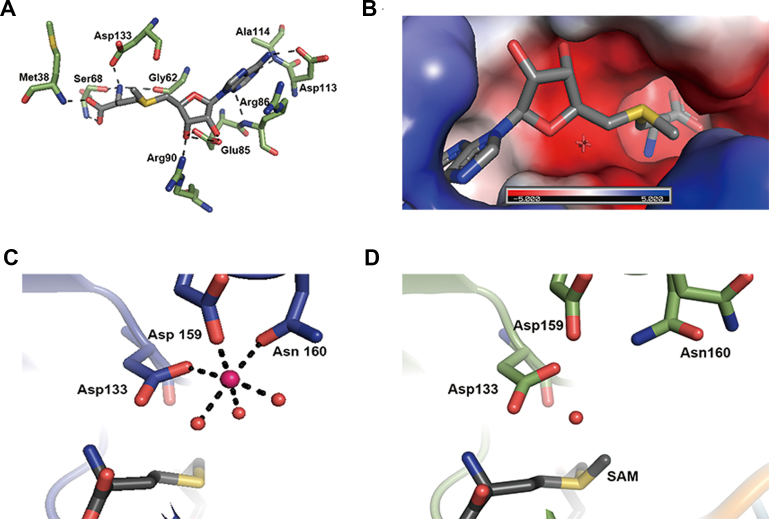
SAM/SAH binding pocket
The SAM binding site is located within a canonical nucleotide binding site of Rossmann motif, where nine amino acid residues (Met-38, Gly-62, Ser-68, Glu-85, Arg-86, Arg-90, Asp-113, Ala-114 and Asp-133) make contacts with SAM through ionic and/or hydrogen bonding interactions (Figure 6A). SAH binding to TrrM is nearly indistinguishable from that of SAM, which involves an identical set of residues. Apparently, the binding mode of SAM/SAH is not affected by the tRNA substrate, where SAM and tRNA are apart at least 4.6 Å. The distance between CH3 of the S-methyl group in SAM and C5 of unmodified U34 is 8.6 Å, which appears to be somewhat distant for 5-hydroxyl group of the wobble uridine to attack on SAM. Electrostatic potential around the SAM binding pocket is mostly negative, facilitating the docking of the positively charged methyl donor and metal ion as described below (Figure 6B).
Figure 6.
Ligand/metal binding by TrmR. (A) Close-up view of SAM-binding site in the crystal structure of Bacillus subtilis. SAM is shown in dark gray. TrmR amino acid residues that interact with SAM are shown in green. Hydrogen bonds are shown as dashed lines. (B) TrmR model colored according to the distribution of electrostatic potential, from red (−5 kT) to blue (5 kT). The SAM-binding pocket is largely negatively charged. (C) Coordination of magnesium ion in RNA-free TrmR structure, where the metal ion (magenta) is coordinated by six ligands; i.e. Asp-133, Asp-159 and Asn-160 residues and three ordered water molecules. (D) An absence of magnesium ion in TrmR:tRNAASL structure. Note Asn160 is in two rotameric states.
Mg2+ coordination in RNA-free structure
A magnesium ion was found near the binding site of SAH in each subunit of RNA-free structure, ∼5.5 Å apart from the sulfur atom of SAH. In one subunit, the metal center adopts the characteristic octahedral geometry, where side chains of three amino acid residues, Asp-133, Asp-159 and Asn-160 and three water molecules coordinate with magnesium ion (Figure 6C). The arrangement of the metal center in the other subunit is similar, except Asn-160 interacts with the metal ion in a bidentate fashion, presenting overall hepta-coordination of the metal ion. Conversely, no equivalent protein-metal interaction was observed in TrmR:tRNAASL structure even though MgCl2 was added during crystallization step. The side chain of Asn-160 assumes dual rotamer configuration, exhibiting the structural plasticity in Mg2+-free TrmR:tRNAASL structure (Figure 6D), which is the only notable conformational difference around the metal coordination site between two structures.
DISCUSSION
We have identified TrmR (formerly known as YrrM) in B. subtilis as the protein responsible for methylation of the hypomodified nucleotide at the wobble position of tRNA from ho5U to mo5U. This hypermodified nucleotide is present in tRNAs capable of decoding all of the codons for Ala, Pro, Thr, Val or Ser. At the equivalent location, it is substituted by cmo5U in Gram-negative bacteria. It is noteworthy that in B. subtilis genome, there are multiple copies of genes encoding tRNA which would contain mo5U; out of 86 tRNA genes, 18 transcribe mo5U containing tRNA (Supplementary Figure S6A) (27). For example, three genes transcribe the sole proline-specific tRNA, the anticodon of which is mo5UGG. Therefore, B. subtilis exclusively relies on this tRNA to decode all four codons for proline, and in general exhibits major preference toward mo5U-containing anticodon among other isoacceptors for decoding alanine, threonine, and valine. 5-methoxyuridine is indeed the most abundant wobble modification in this organism, which is present in ∼25% of total tRNA (28).
Primary sequences and secondary structures of five isoacceptors of ASL tRNA are shown in Supplementary Figure S6B, all containing 5-methoxyuridine at the wobble position in B. subtilis. Of interest, a common sequence feature shared among these ASLs can be defined by a stretch of four nucleotides, Py32-U33-mo5U34-Pu35, where Py and Pu stand for pyrimidine or purine, respectively. This short motif indeed appears to be critical in dictating the recognizable conformation of the anticodon loop in the structure of TrmR:tRNAASL. The 2′-hydroxyl group of U33 interacts via hydrogen bonding with the N7 of G35, which is also likely to occur if substituted with another purine base, A35. As described in the ‘Results’ section, a panel of basic residues of TrmR interacts with the phosphate backbone of tRNA, mainly encompassing G30–G35. Therefore, the enzyme appears to recognize the substrate tRNA by sensing the overall conformation of the ASL, rather than by directly interacting with specific bases within. This short sequence feature is also common to cmo5U containing tRNA in E. coli (3). We have shown that TrmR is able to methylate hypomodified E. coli tRNA isoacceptors in vivo and in vitro, supporting our structure-guided specificity.
Certain structural homologs of TrmR contain a divalent metal ion in the active site (e.g. Mg2+, Ca2+, Mn2+ and Fe2+) (22–24,29–31). In some cases, the chelation of a metal ion has been associated with a specific function of the enzyme, such as increasing the nucleophilicity of an oxygen atom during methylation. Indeed, we located magnesium ions in the TrrM RNA-free structure. It is tempting to hypothesize that the 5-hydroxyl group of ho5U becomes activated prior to the nucleophilic attack on SAM through a similar mechanism. However, the in vitro formation of mo5U by TrmR was shown to be unaffected by the presence or absence of magnesium, implying a divalent ion is dispensable for mo5U forming activity by TrmR. This observation is further supported by the lack of magnesium ion coordination in TrmR:tRNAASL structure even though MgCl2 was present. Activation of the nucleophile through metal coordination may not be the stringent requirement for TrmR, where deprotonation of the 5-hydroxyl group is supposed to be quite feasible in physiological conditions, where the pKa is estimated around 7.6 (32). However, another possibility is that the lack of ho5U in our ASL tRNA mimic affects the binding of magnesium ion. Studies are currently underway to address this issue.
In the TrmR:tRNAASL structure, U34 is not in optimal conformation to efficiently serve as a nucleophile. The wobble uracil base is stacked on the base of G35, and makes non-polar interactions with the backbone atoms of a short peptide loop (composed of His-34, Val-35 and Pro-36), collectively promoting the inactive conformation (Supplementary Figure S7A). Base-flipping within tRNA has been reported in several examples, where the target nucleotide is required to be accessible for enzyme-dependent modification (33–36). A competent conformation of the wobble 5-hydroxyuridine in solution should be readily achieved to prime TrmR-dependent methyltransfer, as illustrated in a base-flipped model as shown in Supplementary Figure S7B. Our rigid-body model appears to well accommodate the flipped-out wobble uridine, and the 5-hydroxyl group would have clear access toward the S-methyl group of SAM for the subsequent nucleophilic attack. Because the U34 of tRNAASL structure does not have the 5-hydroxyl group, it is possible that the tRNA is bound in an inactive state. Hydroxylation at C5 may provide additional energy to overcome the transition barrier between the inactive pose to the active pose.
Homology search of B. subtilis TrmR protein (E-value cut off ∼1.0E-41, sequence identity >∼40%) demonstrates that sequence neighbors are found exclusively among Firmicutes, most of which belong to Bacilli (84.7%), followed by clostridiales (11.7%) and erysipelotrichaceae (2.1%). Analysis of the sequence similarity network (37) among members of methyltransferase families PF01596 (Methyltrans_3, containing TrmR) and PF08003 (Methyltrans_9, containing CmoB) demonstrates that there exist TrmR related orthologs in a subgroup of PF01596 (Supplementary Figure S8), separate from the cluster containing CmoB. Among the resides identified to be critical in binding to tRNA in the crystal structure of TrmR, Arg-86, Arg-90 and Arg-178 are highly conserved specifically for TrmR orthologues. However, these arginine residues are missing in other members of PF01596, for example, caffeoyl-CoA 3-O-methyltransferase (PDB ID: 1SUI, 1SUS), which is involved in the biosynthesis of feruloylated polysaccharides in plants (38), or MdmC (UniProt ID: Q00719), which methylates the 4-hydroxyl group of the lactone ring of midecamycin and other macrolide antibiotics (39). Our analysis estimates TrmR orthologues constitute approximately 3% of PF01596, a family of O-methyltransferases.
Intriguingly, wobble mo5U has been reported in threonine-specific isoacceptor from Mycobacterium bovis BCG, although the authors claimed that the modification is directed by BCG_2975c, a homolog of E. coli CmoB (40). In this report, M. bovis is able to modulate the modification of tRNAthr in response to the availability of oxygen, where BCG_2975c converts the wobble ho5U to mo5U or cmo5U in normoxic or hypoxic condition, respectively. SAM-dependent methyltransfer activity of CmoB is quite limited, however, because of electrostatic repulsion between positively charged SAM and the binding surface (12). The M. bovis homolog is not evolutionarily close to E. coli CmoB (51% sequence coverage, E-value = 2E-5), therefore may possess distinct specificity toward SAM and Cx-SAM. Alternatively, it is possible that B. bovis BCG genome encodes a standalone ho5U-methyltransferase, analogous to B. subtilis TrmR. Further biochemical and structural validation will be necessary to gain full insights into how these remotely related proteins contribute to wobble uridine modification in Mycobacteria. Recently, UHPLC-MS/MS-based detection of mo5U in human neural stem cell RNA has been reported, despite the concentration was estimated to be nearly four-orders of magnitude lower than the most abundant nucleoside, G (41). Moreover, whether this modified nucleoside has originated from tRNA is unclear, much less the exact position in the sequence context of RNA that contains this modification. How widely mo5U is conserved in nature and what other cellular function is possibly embedded beyond being a part of translational machinery remains to be elucidated.
DATA AVAILABILITY
Atomic coordinates and structure factors for the RNA-free and tRNA stem loop-bound crystal structures of TrmR are deposited in the Protein Data Bank under the accession codes 5ZW3 and 5ZW4, respectively.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Pohang Accelerator Laboratory and the beamline 7A scientists for support with the X-ray diffraction data collection.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Research Foundation of Korea [NRF-2016R1D1A1B03930716 to J.K., and H.R.]; GIST Research Institute (GRI) Grant (to J.K., H.R); National Institutes of Health [R21 AI133329 to T.L.G., S.C.A., P01 GM118303–01 to S.C.A., U54 GM093342 to S.C.A., U54 GM094662 to S.C.A.]; Price Family Foundation (to S.C.A.). Funding for open access charge: GIST Research Institute (GRI) Grant.
Conflict of interest statement. None declared.
REFERENCES
- 1. Crick F.H.C. Codon—anticodon pairing: the wobble hypothesis. J. Mol. Biol. 1966; 19:548–555. [DOI] [PubMed] [Google Scholar]
- 2. Agris P.F. Wobble position modified nucleosides evolved to select transfer RNA codon recognition: A modified-wobble hypothesis. Biochimie. 1991; 73:1345–1349. [DOI] [PubMed] [Google Scholar]
- 3. Machnicka M.A., Milanowska K., Oglou O.O., Purta E., Kurkowska M., Olchowik A., Januszewski W., Kalinowski S., Dunin-Horkawicz S., Rother K.M. et al. . MODOMICS: A database of RNA modification pathways—2013 update. Nucleic Acids Res. 2013; 41:262–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Takai K., Yokoyama S.. Roles of 5-substituents of tRNA wobble uridines in the recognition of purine-ending codons. Nucleic Acids Res. 2003; 31:6383–6391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nasvall S.J., Chen P., Bjork G.R.. The modified wobble nucleoside uridine-5-oxyacetic acid in tRNAPro(cmo5UGG) promotes reading of all four proline codons in vivo. RNA. 2004; 10:1662–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Takai K., Okumura S., Hosono K., Yokoyama S., Takaku H.. A single uridine modification at the wobble position of an artificial tRNA enhances wobbling in an Escherichia coli cell-free translation system. FEBS Lett. 1999; 447:1–4. [DOI] [PubMed] [Google Scholar]
- 7. Armengod M.E., Meseguer S., Villarroya M., Prado S., Ruiz-partida R.. Modification of the wobble uridine in bacterial and mitochondrial tRNAs reading NNA/NNG triplets of 2-codon boxes. RNA Biol. 2014; 11:1495–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Weixlbaumer A., Iv F.V.M., Dziergowska A., Malkiewicz A., Vendeix F.A.P., Agris P.F., Ramakrishnan V.. Mechanism for expanding the decoding capacity of transfer RNAs by modification of uridines. 2007; 14:498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dong H., Nilsson L., Kurland C.G.. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J. Mol. Biol. 1996; 260:649–663. [DOI] [PubMed] [Google Scholar]
- 10. UteKothe M.V.R. Codon Reading by tRNAAla with modified Uridine in the wobble position. Mol. Cell. 2007; 25:167–174. [DOI] [PubMed] [Google Scholar]
- 11. Kim J., Xiao H., Bonanno J.B., Kalyanaraman C., Brown S., Tang X., Al-Obaidi N.F., Patskovsky Y., Babbitt P.C., Jacobson M.P. et al. . Structure-guided discovery of the metabolite carboxy-SAM that modulates tRNA function. Nature. 2013; 498:123–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim J., Xiao H., Koh J., Wang Y., Bonanno J.B., Thomas K., Babbitt P.C., Brown S., Lee Y.S., Almo S.C.. Determinants of the CmoB carboxymethyl transferase utilized for selective tRNA wobble modification. Nucleic Acids Res. 2015; 43:4602–4613.25855808 [Google Scholar]
- 13. Murao K., Hasegawa T., Ishikura H.. 5-Methoxyuridine: A New minor constituent located in the first position of the anticodon of tRNAAla, tRNAThr, and tRNAVal from Bacillus Subtilis. Nucleic Acids Res. 1976; 3:2851–2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hasegawa T., Murao K., Ishikura H.. The nucleotide sequence of proline tRNAmo5UGG from Bacillus subtilis. Biochem. Int. 1985; 10:663–671. [PubMed] [Google Scholar]
- 15. Matsugi J., Jia H.T., Murao K., Ishikura H.. Nucleotide sequences of serine tRNAs from Bacillus subtilis. Biochim. Biophys. Acta. 1992; 1130:333–335. [DOI] [PubMed] [Google Scholar]
- 16. Murao K., Hasegawa T., Ishikura H.. Nucleotide sequence of valine tRNA mo5UAC from Bacillus subtilis. Nucleic Acids Res. 1982; 10:715–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Murao K., Ishikura H., Albani M., Kersten H.. On the biosynthesis of 5-methoxyuridine and uridine-5-oxyacetic acid in specific procaryotic transfer RNAs. Nucleic Acids Res. 1978; 5:1273–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Otwinowski Z., Minor W.. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 19. Vagin A., Teplyakov A.. Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:22–25. [DOI] [PubMed] [Google Scholar]
- 20. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Cryst. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 21. Murshudov G.N., Vagin A.A., Dodson E.J.. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997; 53:240–255. [DOI] [PubMed] [Google Scholar]
- 22. Kopycki J.G., Rauh D., Chumanevich A.A., Neumann P., Vogt T., Stubbs M.T.. Biochemical and structural analysis of substrate promiscuity in plant Mg2 + -dependent O -methyltransferases. J. Mol. Biol. 2008; 378:154–164. [DOI] [PubMed] [Google Scholar]
- 23. Kopycki J.G., Stubbs M.T., Brandt W., Hagemann M., Porzel A.. Functional and structural characterization of a cation-dependent O-methyltransferase from the cyanobacterium synechocystis sp. strain PCC 6803. J. Biol. Chem. 2008; 283:20888–20896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Walker A.M., Sattler S.A., Regner M., Jones J.P., Ralph J., Vermerris W., Sattler S.E., Kang C.. The structure and catalytic mechanism of Sorghum bicolor Caffeoyl-CoA O-methyltransferase. Plant Physiol. 2016; 172:78–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Holm L., Rosenstrom P.. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010; 38:W545–W549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Krissinel E., Henrick K.. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007; 372:774–797. [DOI] [PubMed] [Google Scholar]
- 27. Chan P.P., Lowe T.M.. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 2016; 44:D184–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kanaya S., Yamada Y., Kudo Y., Ikemura T.. Studies of codon usage and tRNA genes of 18 unicellular organisms and quantification of Bacillus subtilis tRNAs: gene expression level and species-specific diversity of codon usage based on multivariate analysis. Gene. 1999; 238:143–155. [DOI] [PubMed] [Google Scholar]
- 29. Ferrer J., Zubieta C., Dixon R.A., Noel J.P.. Crystal structures of alfalfa caffeoyl coenzyme A 3-O-methyltransferase. Plant Physiol. 2005; 137:1009–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chávez A.S.O., Fairman J.W., Felsheim R.F., Nelson C.M., Herron M.J., Higgins L., Burkhardt N.Y., Oliver J.D., Markowski W., Kurtti T.J. et al. . An O-methyltransferase is required for infection of tick cells by Anaplasma phagocytophilum. PLos Pathog. 2015; 11:e1005248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chatterjee D., Kudlinzki D., Linhard V., Saxena K., Schieborr U., Gande S.L., Philip J., Wo J., Abele R., Rogov V. V et al. . Structure and biophysical characterization of the S-adenosylmethionine-dependent O-methyltransferase PaMTH1, a putative enzyme accumulating during senescence of Podospora anserina. J. Biol. Chem. 2015; 290:16415–16430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Francois C.J. La, Jang Y.H., Cagin T., Goddard W.A., Sowers L.C.. Conformation and proton configuration of pyrimidine deoxynucleoside oxidation damage products in water. Chem. Res. Toxicol. 2000; 13:462–470. [DOI] [PubMed] [Google Scholar]
- 33. Losey H.C., Ruthenburg A.J., Verdine G.L.. Crystal structure of Staphylococcus aureus tRNA adenosine deaminase TadA in complex with RNA. Nat. Struct. Mol. Biol. 2006; 13:153–159. [DOI] [PubMed] [Google Scholar]
- 34. Hoang C., Ferré-D’Amaré A.R.. Cocrystal structure of a tRNA Ψ55 pseudouridine synthase. Cell. 2017; 107:929–939. [DOI] [PubMed] [Google Scholar]
- 35. Hoang C., Chen J., Vizthum C.A., Kandel J.M., Hamilton C.S., Mueller E.G., Ferré-D’Amaré A.R.. Crystal structure of pseudouridine synthase RluA: Indirect sequence readout through protein-induced RNA structure. Mol. Cell. 2017; 24:535–545. [DOI] [PubMed] [Google Scholar]
- 36. Schwalm E.L., Grove T.L., Booker S.J., Boal A.K.. Crystallographic capture of a radical S-adenosylmethionine enzyme in the act of modifying tRNA. Science. 2016; 352:309–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shannon P., Markiel A., Ozier O, Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T.. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003; 13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ferrer J.-L.J., Zubieta C., Dixon R.R.A., Noel J.J.P.. Crystal structures of alfalfa caffeoyl coenzyme A 3-O-methyltransferase. Plant Physiol. 2005; 137:1009–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hara O., Hutchinson C.R.. A macrolide 3-O-acyltransferase gene from the midecamycin-producing species Streptomyces mycarofaciens. J. Bacteriol. 1992; 174:5141–5144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chionh Y.H., McBee M., Babu I.R., Hia F., Lin W., Zhao W., Cao J., Dziergowska A., Malkiewicz A., Begley T.J. et al. . tRNA-mediated codon-biased translation in mycobacterial hypoxic persistence. Nat. Commun. 2016; 7:13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Basanta-Sanchez M., Temple S., Ansari S.A., D’Amico A., Agris P.F.. Attomole quantification and global profile of RNA modifications: epitranscriptome of human neural stem cells. Nucleic Acids Res. 2015; 44:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for the RNA-free and tRNA stem loop-bound crystal structures of TrmR are deposited in the Protein Data Bank under the accession codes 5ZW3 and 5ZW4, respectively.