Abstract
RsaE is a regulatory RNA highly conserved amongst Firmicutes that lowers the amount of mRNAs associated with the TCA cycle and folate metabolism. A search for new RsaE targets in Staphylococcus aureus revealed that in addition to previously described substrates, RsaE down-regulates several genes associated with arginine catabolism. In particular, RsaE targets the arginase rocF mRNA via direct interactions involving G-rich motifs. Two duplicated C-rich motifs of RsaE can independently downregulate rocF expression. The faster growth rate of ΔrsaE compared to its parental strain in media containing amino acids as sole carbon source points to an underlying role for RsaE in amino acid catabolism. Collectively, the data support a model in which RsaE acts as a global regulator of functions associated with metabolic adaptation.
INTRODUCTION
Staphylococcus aureus is a major human opportunistic pathogen that is responsible for diseases ranging from minor skin infections to life-threatening septicemia and toxic shock syndrome. Its virulence is associated with its ability to survive in different ecological niches and to coordinate the expression of virulence factors. Two alternative sigma factors, σB and σS, contribute to adaptation (1,2), with σB modulating the expression of numerous virulence factors and is required for long-term persistence in vivo (3). The expression of virulence genes is affected by energy resources and the use of carbon sources is tightly regulated by global regulators such as CcpA and CodY (4–6). Numerous trans-acting regulators, including regulatory RNAs (sRNAs), contribute to this adaptation (7,8).
sRNAs are highly abundant in bacteria (9,10). Many of them modulate mRNA translation and stability by base-pairing with one or several targeted RNAs. They act on almost all cellular functions, contributing to adaptation and bacterial homeostasis (11). In S. aureus, the functions and targets of most sRNAs are unknown. RNAIII is one exception; this 514-nucleotide (nt) sRNA has been extensively studied and is a paradigm for regulatory RNAs affecting virulence (12). A second 102-nt sRNA, RsaE, drew the attention of researchers because of its high conservation in the Bacillales order (13). In Bacillus subtilis, the RsaE homolog RoxS downregulates transcripts associated with redox reactions, contributing to the cellular NAD+/NADH balance (14,15). In S. aureus, RsaE affects the expression of several genes involved in oligopeptide transport, folate metabolism and the TCA cycle (16,17). In vitro experiments on some targets suggested that RsaE and RoxS bind mRNA Shine-Dalgarno (SD) sequences to prevent the formation of ribosomal initiation complexes (14,16,17). A C-rich motif, repeated in RsaE, and present in other staphylococcal sRNAs, reportedly contributes to an RNA-RNA pairing mechanism that is common to different sRNAs (17). These duplex formations generate RNase III cleavage sites that irreversibly prevent translation (7,14). In contrast to numerous Gram-negative bacteria, in B. subtilis and Staphylococcus aureus, the RNA chaperone Hfq is likely not required for sRNA-dependent regulations (18–20).
rsaE transcription is induced in mid- or late-exponential phase as observed in various strains. Its expression is significantly reduced post-exponentially in strain N315 and clinical isolates (16,17,21). rsaE is regulated by the two-component system SrrAB (14), which is likely activated in response to reduced menaquinone (22).
Here, by combining different approaches and selecting common candidates, we identified new targets of RsaE. We investigated RsaE mechanism of action and demonstrated that it exerts direct post-transcriptional regulation on a gene associated with arginine catabolism. Growth analysis in media containing amino acids as the only carbon source further reveals that the absence of RsaE results in enhanced growth rate, which is consistent with the up-regulation of associated catabolic pathways.
MATERIALS AND METHODS
Bacterial strains, plasmids and growth conditions
This study was performed using HG003, a model strain for S. aureus regulation (23), and constructed derivatives (Supplementary Table S1). For HG003 gene description, the NCTC8325 nomenclature retrieved from Genbank file CP00025.1 was used. Engineered plasmids were constructed in Escherichia coli DH5α, transferred to RN4220 (a strain transformable by exogenous DNA) and subsequently to HG003. Chromosomal gene modifications (deletions and point mutations) were performed using pMAD2 derivatives as described (24). Conditional gene expression was obtained by cloning genes under the xyl/tetO promoter in pRMC2 (25). Translational regulations were studied by gene fusions with untranslated transcriptional regions (UTRs) + the first codons cloned downstream the rpoB promoter as reported (26) using pTCV-PrpoB-lac (Supplementary Table S2), a pTCV-lac derivative (27). Plasmids (Supplementary Table S2) were engineered mainly by the Gibson assembly method (28) with the indicated appropriate primers (Supplementary Table S3).
Staphylococcus aureus strains were routinely grown aerobically in Brain Heart Infusion (BHI) broth at 37°C. DH5α was grown aerobically in Luria-Bertani (LB) broth at 37°C. Antibiotics were added to media as needed: ampicillin 100 μg/mL, chloramphenicol 20 μg/ml for E. coli; chloramphenicol 10 μg/ml and erythromycin 5 μg/ml for RN4220; chloramphenicol 5 μg/ml and erythromycin 0.5 μg/ml for HG003. Expression of rsaE and mutated rsaE under the control of promoter Pxyl/tetO (pRMC2RsaE and mutated derivatives) was induced by the addition of anhydrotetracycline (aTc, 1 μM) to culture medium.
Growth in complete defined medium (CDM) and metabolite analyses
S. aureus HG003 and HG003 ΔrsaE were grown aerobically (250 rpm and 10:1 flask-to- volume ratio) in CDM lacking glucose as previously described (29,30). The CDM contains 18 amino acids excluding glutamine and asparagine. For amino acid analysis, 1 ml of bacterial culture was collected during growth and centrifuged at 14 000 g to collect supernatant. Supernatant was subsequently filtered through an Amicon Ultra Centrifugal filter (Amicon; 3000 molecular weight cut off). Amino acid concentration was determined using a Hitachi L-8800 amino acid analyzer at the University of Nebraska Medical Center Protein Structure Core Facility. Statistical analyses were performed using PC SAS version 9.4. The statistical level of significance was set to 0.05 for all analyses. For each amino acid, a general linear model was fit that included a term for group (ΔrsaE versus wild-type) and the continuous covariate OD. Residuals from the model were tested for normality using the Wilks-Shapiro test.
Total RNA extraction
Overnight cultures were diluted 1000 times and incubated in BHI at 37°C. For HG003 and HG003 ΔrsaE, bacteria were harvested at OD600 of 6. For HG003 ΔrsaE pRMC2RsaE, at OD600 of 0.5, the culture was split in two halves and aTc (1 μM) was added to one of them for rsaE induction. Induced and non-induced cultures were harvested 5 min after aTc addition. Total RNAs were extracted as described (19). Samples were prepared in triplicates and treated with TURBO DNase (Ambion) according to manufacturer's instructions. Ten microgram of RNA were processed using MICROBExpress (Ambion) according to manufacturer's instructions to remove ribosomal RNA. RNA-seq libraries were generated with the TruSeq SBS Kit v3 (Illumina) and sequenced using a HiSeq to generate paired-end 100-nt reads.
Hybrid-trap-seq
The procedure is summarized in Supplementary Figure S1. Synthetic RNAs were generated with the T7 MEGAshortscript kit (Ambion) according to manufacturer's instructions, using sRNA genes that were PCR-amplified with specific oligonucleotides (Supplementary Table S3). Synthetic RNAs were 3′-end biotinylated as described (31) using biotinamidocaproyl hydrazide (Sigma-Aldrich, B3770). After the biotinylation reaction, full length RNAs were purified on 5% urea PAGE as recommended in the T7 MEGAshortscript kit procedure. Running Hybrid-trap-seq requires a pool of total RNA extracts to be used as prey. Total RNA samples were extracted in 16 different biological conditions (referred to as 16-condition RNA pool): (i) eight samples grown in BHI at OD600 of 0.6, 1.8, 3.3, 4.5, 7.2, 9.8 and 12.8, and late stationary phase (24 h), (ii) seven samples grown under stress conditions (cold shock, heat shock, oxygen limitation, alkaline stress, peroxide stress, disulfide stress, iron-depletion) and (iii) one sample from colonies on BHI-agar plates. The complete description of the 16-condition RNA pool is available in the GEO database (GEO accession number: GSE104971) (10). Twenty μg of each of the 16 total RNA extracts were pooled to obtain the combined RNA extract sample used for the procedure. Hybrid-trap-seq experiments were then carried out as follows: MasterBeads pre-coated with streptavidin (Ademtech, Pessac-France) were equilibrated in binding buffer (20 mM Tris–HCl, 0.5 M NaCl pH 8) and incubated 10 min at 20°C with 100 pmol of biotinylated sRNA. Unbound sRNAs were removed by magnetic separation, and then sRNA-bound streptavidin beads were washed twice with binding buffer. Fifty μg of 16-condition RNA pool were mixed with the sRNA-bound beads. After 15 min at 45°C, followed by 15 min at room temperature, unbound RNAs were removed by magnetic separation. The RNA-bound beads were washed twice with wash buffer (7 mM Tris–HCl pH 8, NaCl 0.17 M). RNAs were then eluted in RNase-free water. RNA samples were analyzed by Illumina high throughput sequencing. Five oriented libraries were generated from the four Hybrid-trap eluted RNA samples and the 16-condition RNA pool depleted of rRNA used as prey, as described (32). Libraries were sequenced using Illumina Genome Analyzer IIx to generate single-end 40-nt reads, which were then analyzed.
Read mapping and differential expression analysis
Quality control of all transcriptome samples using FastQC (v0.10.1) confirmed that the reads could be directly used for the mapping step. Reads were aligned to the S. aureus NCTC8325 chromosome sequence (CP000253.1) using bowtie2 mapper (v2.1.0) and default values (33). Log-read coverage profiles were computed using in-house shell scripts as described (32) and visualized in the Artemis viewer (34). Uniquely mapped read counts per gene were calculated for each dataset with feature Counts (1.5.0-p1, with options specifying paired-end and stranded library) (35) using the list of CDS retrieved from Refseq CP000253.1. The list of CDSs differentially expressed between two conditions (i.e. ΔrsaE mutant versus wild-type strain; induced versus non-induced RsaE expression) was supplied by SARTools v1.3.0 (36) using DESeq2 (37), with a false discovery rate (FDR) set at 0.05. Genes with a fold change >1.33 were considered as differentially expressed. Due to an outlier library identified by PCA and P-value analysis in the wild-type versus ΔrsaE mutant comparison, differential analysis of these conditions was performed using only replicates 1 and 3 and corrected for batch effect between pairs of replicates. The differential expression analysis between with and without induction of the RsaE expression was done with the three replicates. To identify RNAs specifically enriched with RsaE as sRNA-bait, Hybrid-trap-seq samples were analyzed as follow. Mapping was performed on the list of CDS retrieved from Refseq CP000253.1 and the 44 HG003 bona fide sRNA list recently established (10) using bowtie with parameters -p 12 -S -m 1 –q. Read counts per gene were then calculated with HTSeq-count (http://htseq.readthedocs.io/). Raw read counts of the four datasets (namely, RsaE, RsaA, RsaH and RNAIII used as bait) were normalized using DESeq and fold changes were computed from normalized read counts of RsaE-bait dataset to normalized read counts of control datasets (i.e. the three other sRNA datasets). Genes considered as putative sRNA targets displayed fold changes ≥10.
Computational analysis of RNA-RNA hybrids
Target RNA-sRNA interactions were predicted using the IntaRNA package (38); scoring is based on hybridization free energy and accessibility of the interaction sites in both RNA molecules (IntaRNA V 2.1.0 through the web interface at http://rna.informatik.uni-freiburg.de with default parameters). For Figure 2, complete sRNA sequences were used as input, and each putative target comprised the region from the transcriptional start site (TSS) to 150 nucleotides past the start codon. When possible, TSSs were defined from sequencing of the 16-condition RNA pool as described (32). Coordinates of relevant transcripts are provided (Supplementary Table S4). We searched for consensus motifs in the twelve putative target sequences and rsaE (in antisense) using the MEME suite V.4.9.0 (39) run locally with options -dna -minw 4 -maxw 10. Input sequence fragments were from TSS to ATG+150. The standard SD motif was identified by MEME using as input the 1114 S. aureus sequences for which a TSS could be identified by the above protocol.
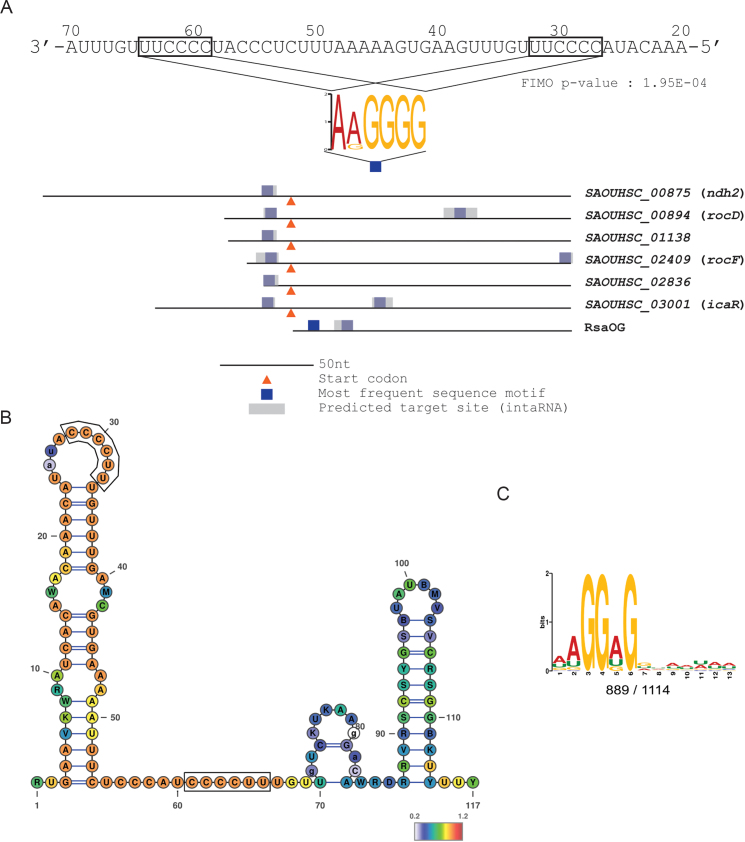
Figure 2.
Search of putative RsaE binding motifs. A) MEME (39) sequence motif found in RsaE Hybrid-trap seq targets (Table 2) shown as a colored logo. The motif is connected to its location in target RNAs (colored boxes) and to its complementary site in RsaE. Blue and grey areas in targets correspond to regions of RsaE interaction predicted by MEME (39) and IntaRNA (38), respectively. Orange arrows indicate the position of translation initiation codons. Coordinates of the consensus motif positions on the putative sRNA targets are presented in Supplementary Table S4. B) RsaE secondary structure predicted from a structural alignment of homologous sequences, as provided by the RFAM database (60). Color scale indicates evolutionary conservation at each position (red: most conserved). Boxes around sequences indicate predicted seed matching regions. Structures are drawn using Varna (61). RsaE RFAM entries: RF01820. An experimental structure was reported (17). C) Model for the standard S. aureus SD motif, identified by MEME (39) using all HG003 available 5′ UTR regions (see Materials and Method).
Quantitative reverse transcriptase PCR and northern blots
qRT–PCR experiments were performed on a subset of putative targets selected among the most enriched mRNAs of RsaE Hybrid-trap-seq set. Experiments were performed on biological triplicates and data were analyzed as described; the geometric mean of 4 genes (recA, gyrA, glyA and ftsZ) was used to normalize the samples (40). All Northern blots were performed as described in duplicates (41–43). Samples were separated by either PAGE or agarose gels and probed with 32P-labeled PCR probes using the Megaprime DNA labeling system (GE Healthcare) (for primer used, see Supplementary Table S3).
β-Galactosidase assay
The effect of RsaE on translation was tested by means of gene fusions with lacZ (pTCV-PrpoB-1138-lac and pTCV-PrpoB-rocD-lac) by β-galactosidase assays with two experimental settings: by comparing i) the ΔrsaE and its parental strains and ii) the ΔrsaE strain carrying pRMC2RsaE with and without RsaE induction. Overnight cultures grown in BHI supplemented with chloramphenicol and kanamycin (when necessary) were diluted 1000-fold in the same medium at 37°C. ΔrsaE and its parental strain were sampled at OD600 ∼7. For ΔrsaE carrying pRMC2RsaE, aTc (1 μM) was added to the medium at OD600 ∼0.5; cultures were sampled after a 15 min induction and frozen. β-galactosidase activity was measured with MUG (4-methylumbelliferyl beta-d-galactopyranoside, Sigma-Aldrich) as described (44). Briefly, cell pellets were resuspended in 500 μl ABT buffer (100 mM NaCl, 60 mM K2HPO4, 40 mM KH2PO4, 0.1% triton X100) and 10 mg/ml MUG. After 1 h incubation at 25°C, the reaction was stopped by addition of 500 μl Na2CO3 0.4 M. OD600 and fluorescence (excitation: 365 nm; emission: 455 nm) were measured with CLARIOStar (Monochromator Microplate Reader). The fluorescence of each sample was normalized by its OD600. Translation efficiency was estimated as a percentage of normalized fluorescent units of bacterial cells relative to the wild type strain or to no aTc addition depending on the experiment. Each condition culture was done in triplicate.
RESULTS
Modulation of RsaE expression uncovers putative RsaE targets
Most characterized sRNAs modulate the stability of targeted mRNAs, directly, or indirectly via translation inhibition. To find new RsaE targets, we analyzed the transcriptomes of (i) S. aureus HG003 compared to its isogenic ΔrsaE mutant, and (ii) ΔrsaE containing rsaE controlled by an inducible promoter (PtetO-rsaE) grown under non-induced and induced conditions (Supplementary Table S1). For the first couple (wild-type and ΔrsaE mutant), both cultures were in steady state and adapted to the presence or absence of RsaE. In contrast, the short induction time (5 min) used with the PtetO-rsaE system enabled us to identify primary effects of RsaE accumulation, i.e. putative direct targets, with minimized detection of secondary targets. Transcriptome alterations were determined through differential expression analysis of RNA-seq data.
Twenty-five mRNAs organized in 20 transcription units were both up-regulated in the mutant strain and down-regulated upon RsaE expression and are thus considered as possible targets (Table 1). Among them, expression of SAOUHSC_00951, SAOUHSC_01138, folD, fhs, gcvT-gcvPA-gcvPB, rocF, citB and sucC-sucD mRNAs was previously reported to be modulated by RsaE in RN6390, a σB deficient strain, and direct regulation was demonstrated in vitro for SAOUHSC_00951 and sucD mRNAs (16,17). The present results confirm that in HG003, a σB repaired strain, the amounts of transcripts encoding enzymes from the TCA cycle and folate pathway are also down-regulated when RsaE is expressed. Interestingly, among the newly identified mRNAs (Table 1), sucA-sucB, fumC and mqo1 mRNAs encode other enzymes of the TCA cycle, namely dihydrolipoamide succinyltransferase and 2-oxoglutarate dehydrogenase, fumarase and malate quinone oxidoreductase, thus extending the regulatory role of RsaE on this pathway. In addition to these, ald2 mRNA, which encodes an alanine dehydrogenase converting L-alanine to pyruvate, was found to be regulated by RsaE. Also, rocF, encoding the arginase that converts arginine to ornithine, was among the mRNAs that were most affected by the absence or accumulation of RsaE (Table 1). These observations suggest that RsaE is also involved in amino acid catabolism.
Table 1.
RNAs down-regulated by RsaE
| Locus tag* | Gene | Annotation | − RsaE** | + RsaE*** |
|---|---|---|---|---|
| TCA cycle | ||||
| SAOUHSC_01216 | sucC | Succinyl-CoA synthetase, beta subunit | 2.69 | 0.64 |
| SAOUHSC_01218 | sucD | Succinyl-CoA synthetase, alpha subunit | 2.40 | 0.63 |
| SAOUHSC_01347 | citB | Aconitate hydratase 1 | 1.82 | 0.38 |
| SAOUHSC_01416 | sucB | 2-oxoglutarate dehydrogenase, E2 component | 1.35 | 0.53 |
| SAOUHSC_01418 | sucA | 2-oxoglutarate dehydrogenase, E1 component | 1.36 | 0.48 |
| SAOUHSC_01983 | fumC | Fumarate hydratase | 1.49 | 0.42 |
| Malate metabolism | ||||
| SAOUHSC_00698 | Putative malate transporter | 2.31 | 0.59 | |
| SAOUHSC_02647 | mqo1 | Malate:quinone-oxidoreductase | 1.38 | 0.56 |
| Glycine cleavage system | ||||
| SAOUHSC_01632 | gcvPB | Glycine cleavage system P-protein subunit ll | 1.37 | 0.35 |
| SAOUHSC_01633 | gcvPA | Glycine cleavage system P-protein subunit I | 1.40 | 0.28 |
| SAOUHSC_01634 | gcvT | Glycine cleavage system T protein | 1.42 | 0.27 |
| Tetrahydrofolate metabolism | ||||
| SAOUHSC_01007 | folD | Tetrahydrofolate dehydrogenase | 1.38 | 0.59 |
| SAOUHSC_01845 | fhs | Formate-tetrahydrofolate ligase | 2.32 | 0.10 |
| Amino-acid metabolism | ||||
| SAOUHSC_01818 | ald2 | Alanine dehydrogenase | 3.09 | 0.48 |
| SAOUHSC_02409 | rocF | Arginase | 2.28 | 0.31 |
| Others | ||||
| SAOUHSC_00094 | sasD | Cell-wall-anchored protein | 1.48 | 0.34 |
| SAOUHSC_00204 | hmp | Globin domain containing protein | 1.39 | 0.60 |
| SAOUHSC_00690 | Conserved hypothetical protein | 1.38 | 0.63 | |
| SAOUHSC_00699 | phrB | Deoxyribodipyrimidine photolyase | 1.82 | 0.74 |
| SAOUHSC_00838 | Truncated conserved hypothetical protein | 1.38 | 0.60 | |
| SAOUHSC_00951 | Putative RNA ligase or phosphoesterase | 2.67 | 0.56 | |
| SAOUHSC_01137 | yjjG | Putative HAD-hydrolase | 1.42 | 0.50 |
| SAOUHSC_01138 | Putative N-acetyltransferase | 1.78 | 0.40 | |
| SAOUHSC_01964 | traP | Signal transduction protein TRAP | 1.45 | 0.43 |
| SAOUHSC_02754 | ABC transporter, ATP-binding protein | 1.44 | 0.74 | |
* List of genes differentially expressed (FDR < 0.05, fold change > 1.33 or < 0.75) in both transcriptome comparisons.
** Fold change expression of HG003 ΔrsaE relative to wild-type.
*** Fold change expression of HG003 ΔrsaE pRMC2RsaE induced relative to not induced.
Fold changes and adjusted P-values are available in the GEO database (GEO accession number GSE106457: GSE106456_Supplemental_table_1.txt for the ‘ΔrsaE relative to wild-type’ comparison; GSE106456_Supplemental_table_2.txt for the ‘ΔrsaE pRMC2RsaE induced relative to not induced’ comparison).
In vitro trapping of putative RsaE-targets
We used a genome-wide approach to identify substrates that interact directly with RsaE, by extending an in vitro method we previously developed to trap sRNA targets using sRNAs as bait (42). Briefly, a synthetic sRNA was produced, biotinylated and fixed to streptavidin-associated magnetic beads. The resulting ‘bait’ was incubated with total RNA extracts, washed, and RNAs bound to sRNAs were eluted. Recovered RNAs were converted to cDNAs and identified on DNA chips. The method is successful if sRNA targets are well expressed; otherwise, sRNA-targets may be masked by RNA background noise. To overcome this difficulty, (i) RNA-seq rather than DNA-chips was used for target identification, which improved detection (threshold and linearity) and (ii) we hypothesized that nonspecific RNA signals recovered by this protocol would be the same with different sRNAs used as baits; therefore, background noise could be subtracted by a differential analysis of data obtained with several different baits. The modified protocol is called Hybrid-trap-seq (Supplementary Figure S1). Total HG003 RNAs were extracted from 16 growth conditions and pooled together. Hybrid-trap-seq experiments were run in parallel with four baits: RsaE, and three other sRNAs, RNAIII, RsaA and RsaH. The background noise due to nonspecific RNA binding was filtered out from the RsaE Hybrid-trap-seq dataset by a differential analysis using the three other sRNA datasets. This procedure identified eleven mRNAs and one sRNA that accumulated more reads (>10-fold difference) with the RsaE trap than with the three other sRNAs used as baits (Figure 1 and Supplementary Table S2).
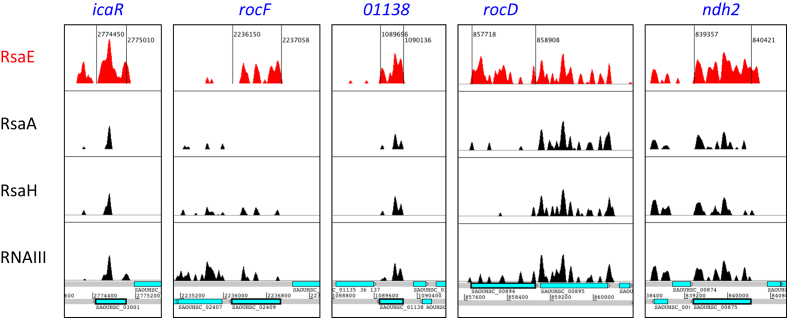
Figure 1.
Examples of regions with RsaE-dependent read enrichment. Artemis genome viewer windows showing read density profiles of RsaE-trapped RNAs obtained with Hybrid-trap-seq (Table 1) and found significantly modulated in at least one of the two transcriptomic studies (GSE106457). The red-filled coverage corresponds to RNAs trapped by RsaE. Black-filled coverages correspond to RNAs also retained using unrelated sRNAs (RsaA, RsaH and RNAIII) used to define the level of background noise resulting from nonspecific association. Bottom panel corresponds to genome annotation with blue boxes indicating open reading frames.
RsaE targets SD-like motifs
The twelve RNAs selectively trapped by RsaE are putative primary targets. The regions potentially interacting with RsaE were therefore investigated in silico. A predominant motif akin to SD sequences (AAGGGG) was present in seven out of twelve targets, with at least one motif being located at the SD site for the mRNAs. The AAGGGG motif has two exact complementary sequences in RsaE (CCCCTT). Interestingly, the putative RsaE binding motif is present twice in rocD, rocF, icaR and rsaOG, suggesting a possible double seed contact with these putative targets (Figure 2A). The anti-SD-like motifs in RsaE are single-stranded (17) hence accessible to base-pairing, and are located in evolutionary conserved regions (Figure 2B).
Altogether, four independent lines of evidence (motif enrichment, complementarity with sRNA, accessibility and conservation) support the idea that RsaE operates through a seed binding mechanism targeting SD-like regions. A question raised by the use of an SD-like seed region is how specific recognition can be achieved. The RsaE motifs in putative targets (AAGGGG) differ from the canonical S. aureus SD motif (AAGGAG, Figure 2C). These differences are matched by specific complementary bases and likely expand from the seed region to allow substrate discrimination as suggested by IntaRNA predicted target sites (Figure 2A and Supplementary Table S4).
Combining experimental approaches to confidently select new RsaE targets
Hybrid-trap-seq experiments are equivalent to genome-wide RNA-RNA retardation assays, with the same caveat: do putative targets uncovered in vitro correspond to real in vivo targets? Among the eleven putative RsaE mRNA targets identified by Hybrid-trap-seq (Table 2), levels of SAOUHSC_1138 and rocF mRNAs were significantly increased in the absence of RsaE and decreased when RsaE expression was induced (Table 1). rocD, ndh2 and icaR mRNAs trapped by RsaE showed significantly decreased levels when RsaE expression was induced (0.36, 0.44 and 0.56, FDR < 0.05, respectively). RNA duplexes are predicted in silico between RsaE and each of these mRNAs. Altogether, these results suggest a direct interaction of RsaE with SAOUHSC_1138, rocF, rocD, ndh2 and icaR mRNAs (Figure 3).
Table 2.
RsaE-trapped RNAs
| Locus tag | Gene | Annotation | Fold change |
|---|---|---|---|
| SAOUHSC_03001 | icaR | ica operon transcriptional regulator | 46.6 |
| rsaOG | CcpA regulated sRNA | 31.1 | |
| SAOUHSC_01546 | Conserved hypothetical phage protein | 25.5 | |
| SAOUHSC_01016 | purN | Phosphoribosylglycinamide formyltransferase | 20.1 |
| SAOUHSC_02409 | rocF | Arginase | 19.4 |
| SAOUHSC_02836 | Acetyltransferase (GNAT) family protein | 18.9 | |
| SAOUHSC_01543 | Phage phi related protein | 16.1 | |
| SAOUHSC_01138 | Putative N-acetyltransferase | 11.9 | |
| SAOUHSC_00875 | ndh2 | Pyridine nucleotide-disulfide oxidoreductase | 11.9 |
| SAOUHSC_01017 | purH | Phosphoribosylaminoimidazolecarboxamide formyltransferase / IMP cyclohydrolase | 10.9 |
| SAOUHSC_00894 | rocD | Ornithine aminotransferase | 10.7 |
| SAOUHSC_00819 | cspC | Cold shock domain protein | 10.4 |
Full results are available under GEO accession number GSE106327.
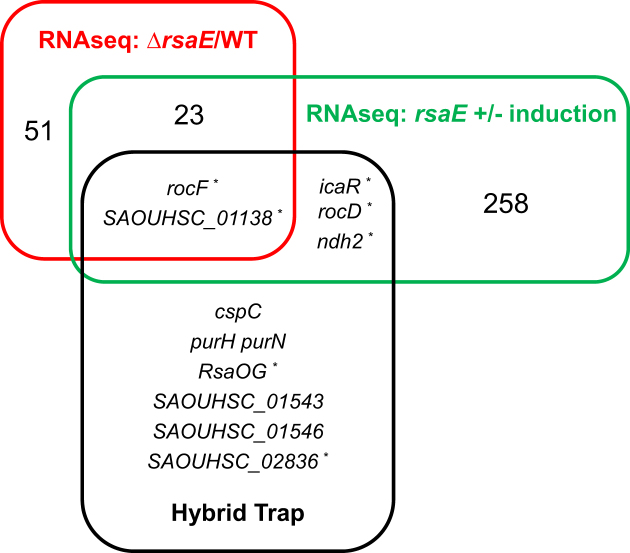
Figure 3.
Combining experiments to uncover RsaE-targets. Venn diagram showing the overlap between different methods for prediction of putative RsaE targets: transcriptomic (ΔrsaE versus HG003, red; ΔrsaE pRsaE with versus without induction, green; threshold 1.33, P< 0.05) and Hybrid-trap-seq experiments (black, min. 10-fold variation). *, RNAs with G-rich motifs.
The level of six other mRNAs identified by Hybrid-trap-seq was not significantly modulated by the RsaE status. In these cases, a direct RsaE interaction with these transcripts may modulate their translation without affecting their stability. However, a complementary sequence with RsaE was only found for RsaOG and SAOUHSC_02836, which encodes a putative acetyl transferase active on phosphinothricin, a glutamate analogue (45).
rocF and SAOUHSC_1138 mRNAs, found by three approaches, are most likely RsaE direct targets (Figure 3). rocD mRNA encodes the ornithine-oxo-acid transaminase and RocD acts together with RocF and RocA to interconvert arginine to glutamate (30). rocF and rocD are genetically unlinked but are both down-regulated by RsaE and their mRNAs were also trapped in vitro by RsaE; this functional convergence reinforces our hypothesis of a direct involvement of RsaE in the control of arginine metabolism.
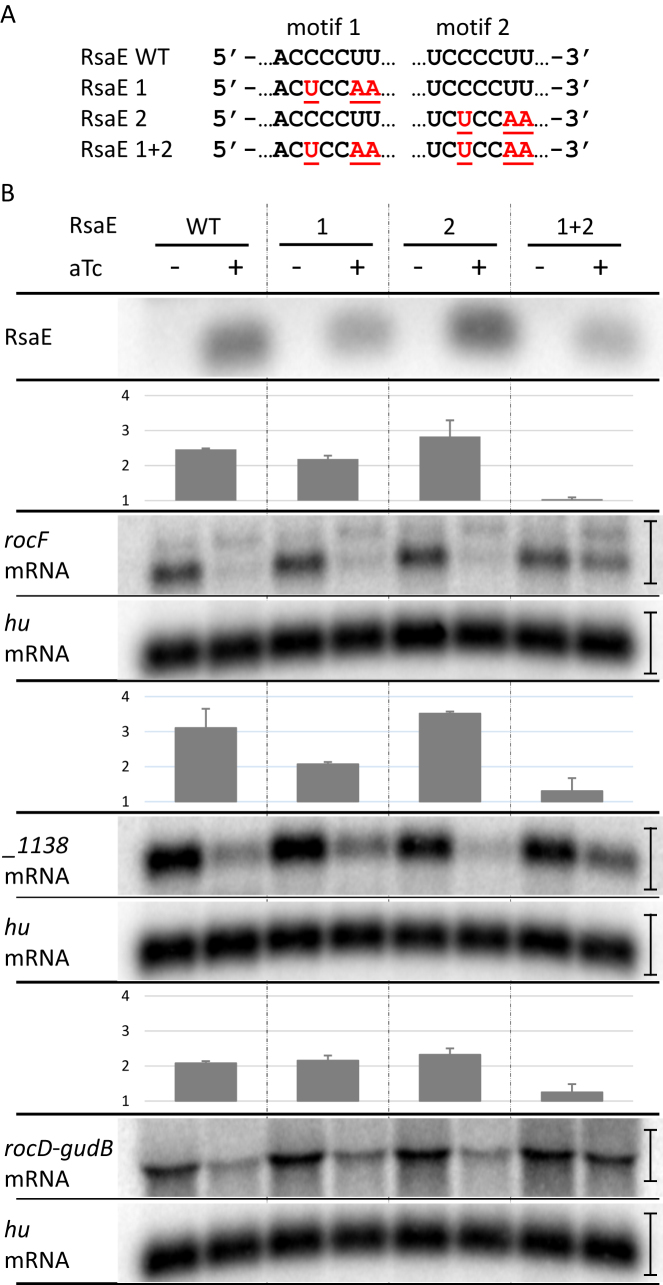
Two motifs of RsaE contribute independently to the down-regulation of its substrates
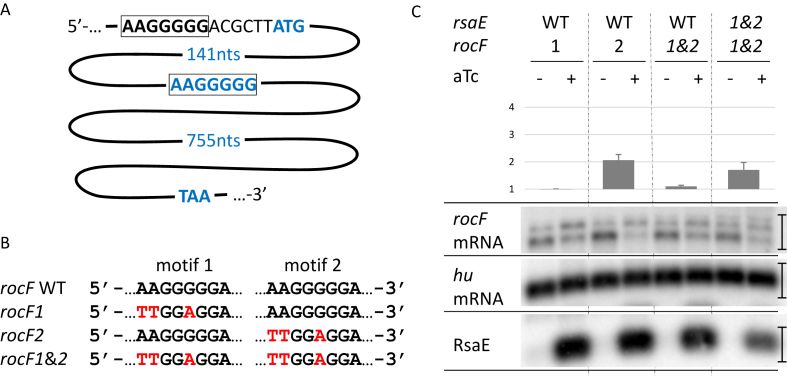
The repetition of identical binding motifs is an unusual feature for sRNAs. We therefore tested the contribution of each CCCCUU sequence of RsaE on target degradation. For this study, we chose to monitor rocF and SAOUHSC_01138 mRNAs as the most likely primary RsaE targets (Figure 3) as well as rocD mRNA which is involved in the same metabolic pathway as rocF mRNA. We confirmed their down-regulation upon RsaE induction by Northern blots (Figure 4B) and qRT-PCR (Supplementary Table S5). Two bands were detected when rocF mRNA was probed. They may come from either alternative promoters or processing events. The lower band is predominant and the higher is moderately affected by RsaE; however, both were summed for quantification. The first CCCCTT motif of rsaE was changed to CTCCAA leading to the rsaE1 allele (Figure 4A). Induction of rsaE1 still resulted in the decrease of rocF, SAOUHSC_01138 and rocD-gudB mRNAs, showing that the first motif was not essential for RsaE regulatory activity, although the rsaE1 allele was expressed to a lesser amount (Figure 4B). Mutations in the first motif likely affect RsaE stability, judging by expression levels. The second CCCCTT rsaE motif was also changed to CTCCAA to create the rsaE2 allele (Figure 4A). Induction of rsaE2 also resulted in the disappearance of rocF, SAOUHSC_01138 and rocD-gudB mRNAs showing that the second motif was not essential for RsaE activity against these mRNAs (Figure 4B). However, induction of a rsaE allele mutated for both motifs (rsaE1&2) did not affect mRNA quantities of the three targets (Figure 4B). We concluded that the two RsaE motifs can act independently against rocF and rocD-gudB mRNAs. However, on SAOUHSC_1138 mRNA, RsaE motif 1 was more active than RsaE motif 2 (Figure 4B) indicating that both motifs may act differently according to substrates.
Figure 4.
Two RsaE repeated motifs contribute independently to its activity. (A) RsaE mutations introduced in motifs 1 and 2. Wild-type sequence is in black, mutations are in red. (B) Effect of RsaE and mutated RsaE induction on rocF, rocD and SAOUHSC_1138 mRNA levels. Total RNAs were extracted from strains expressing conditionally RsaE upon the addition of aTc (1 μM) to the medium. (–), without aTc addition; (+), with 5 min aTc induction. RsaE, rocF mRNA, rocD mRNA and SAOUHSC_1138 RNAs were visualized by Northern blot experiments as indicated. Hu mRNA was used as a loading control. Histograms indicate target repression folds expressed as a ratio of target intensity upon non-induction relative to induction of rsaE and mutated rsaE. Vertical bars on the right indicate the width of regions around Northern bands selected to calculate 32P signal intensity with ImageJ software. rocF mRNA signals are located between markers 1000 and 1500 nt. Error bars represent standard deviations of the repression folds calculated using biological duplicates.
RsaE targets mRNA SD sequence and translation
Some mRNAs targeted by RsaE have two cognate putative RsaE binding sites (Figure 2A and Supplementary Figure S2). We tested whether changes in these sites on the rocF mRNA alter RsaE activity. The first putative binding motif is within a non-canonical SD sequence AAGGGGG (Figure 5A). The motif was changed to TTGGAGG, leading to rocF1 allele (Figure 5B). This modification created a canonical SD sequence to maintain rocF translation. The second putative motif was 141 nucleotides downstream from the translational start codon. It was changed from AAGGGGG to TTGGAGG leading to rocF2 allele and consequently altering the RocF amino acid composition (K49L and G50E). The combination of both alleles, rocF1&2, was also constructed (Figure 5B). All these mutations were introduced by allelic replacement at the rocF locus in HG003 ΔrsaE. These strains were then transformed with pRMC2RsaE and the amount of rocF mRNA was assayed by northern blot (Figure 5C). As for wild-type rocF mRNA (Figure 4B), two bands were detected when rocF mRNA was probed, but mutations affecting rocF mRNA resulted in an increase of the higher band. Mutations may affect rocF mRNA structure and translation, which in turn affect the ratio between each band. As indicated above, both rocF mRNA bands were summed for quantification. rocF downregulation by RsaE was effective for rocF2 but less for rocF1 and rocF1&2 carrying strains, suggesting that RsaE preferentially pairs to the rocF mRNA SD sequence. One prediction from this pairing is that mutations in rsaE restoring base-pair complementarity with rocF1&2 would restore RsaE activity. To test this, HG003 ΔrsaE rocF1&2 was transformed with pRMC2RsaE1&2. Induction of RsaE1&2 led indeed to a decreased quantity of rocF1&2 mRNA (Figure 5C). The effect with the mutated alleles is less pronounced than with wild-type alleles, possibly due to structural constraints that affect pairing, and/or susceptibility of the complex to ribonucleases. Altogether, these experiments demonstrate an in vivo direct pairing between RsaE and the rocF mRNA SD sequence suggesting that RsaE acts at the level of translation and indirectly affects stability.
Figure 5.
RsaE targets the SD sequence of rocF mRNA. (A) Schematic representation of the two duplicated motifs complementary of RsaE in rocF mRNA. The first one is within the SD sequence (black and squared), the second one within the coding sequence (blue and squared). (nts), nucleotides. (B) rocF mutations introduced in motifs 1 and 2. The wild-type sequence is in black, mutations are in red. Mutated rocF alleles were introduced in HG003 chromosome by allelic exchange at the rocF locus. (C) Effect of RsaE and RsaE1&2 induction on rocF and mutated rocF mRNAs. For figure explanation and experimental conditions see Figure 4 legend.
The SD region was also the predicted RsaE pairing site for other mRNA targets (Figure 2A and Supplementary Figure S2) suggesting that they are also regulated by RsaE at the translation level. To determine if this is indeed the case, translational fusions between SAOUHSC_1138 and rocD mRNA 5′-end region and lacZ were constructed and introduced in (i) ΔrsaE and its parental strain and (ii) in ΔrsaE pRMC2RsaE. Absence of RsaE lead to an increase of β-galactosidase activity, while a 15-min induction of rsaE resulted in a significant decrease of β-galactosidase activity, confirming that RsaE acts negatively on rocD and SAOUHSC_01138 mRNA translation (Supplementary Figure S3).
RsaE contributes to amino acid catabolism regulation
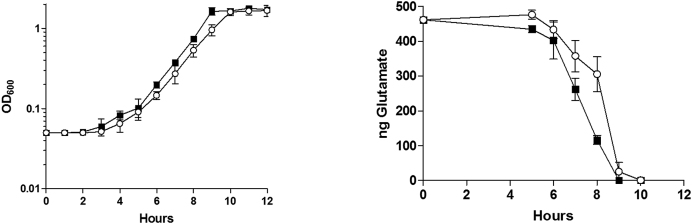
Despite being highly conserved within the Firmicutes phylum and having putative and demonstrated targets, no physiological phenotype was thus far associated with an rsaE deletion. Indeed, no competitive growth difference between HG003 and its ΔrsaE derivative was observed in rich media. However, if RsaE functions to repress amino acid catabolism, we would predict that ΔrsaE would have an increased growth rate in media containing only amino acids as a carbon source. Indeed, the ΔrsaE strain had a faster growth rate than HG003 in CDM, which contains amino acids as the sole carbon source (Figure 6). Of those amino acids that are known to be rapidly catabolized in CDM (30), glutamate (Figure 6) as well as threonine, serine, alanine, glycine, proline and aspartate were catabolized more rapidly during exponential phase in the absence of RsaE (Supplementary Figure S4). When percent amino acid consumed were corrected for growth, the rates of consumption were not significantly different between ΔrsaE and WT HG003 documenting that the amount of amino acid consumed between the two strains was proportional to the biomass (Supplementary Figure S5). Collectively, these data suggest that the absence of RsaE leads to the upregulation of enzymes contributing to amino acid catabolism, which in turn stimulates growth rate.
Figure 6.
Growth and glutamate consumption of HG003 and HG003 ΔrsaE. HG003 (white dot) and HG003 ΔrsaE (black square). Left panel: growth in CDM. HG003 and HG003 ΔrsaE doubling time are 69 min (±1.1) and 58 min (±1.8), respectively. Right panel: Glutamate consumption following aerobic growth in CDM. Glutamate concentrations (ng/μl) were measured in the supernatant at the indicated time of growth. Experiments were performed three times and the error bars represent the standard deviation of the means.
DISCUSSION
Finding sRNA targets and function remain challenging. Here, by combining transcriptomic analyses, in vitro trapping and bioinformatics, we bring to light new RsaE features.
RsaE is a paradigm for regulatory RNAs in Firmicutes
RsaE is remarkably conserved in the Staphylococcaceae and Bacillaceae families (Supplementary Table S6) and has been studied in two representative species S. aureus and B. subtilis, respectively. In S. aureus, RsaE modulates transcripts of enzymes involved in the TCA cycle, the glycine cleavage system, and tetrahydrofolate metabolism (16,17). In B. subtilis, the RsaE homolog RoxS downregulates transcripts associated with oxido-reduction reactions and is proposed to partly control NAD+/NADH balance (14,15).
Our transcriptomic analyses provide a confirmation of previously proposed RsaE targets. They also reveal additional RsaE-regulated steps of the TCA cycle (i.e. sucAB, fumC and mqo1). All TCA cycle steps except succinate-fumarate conversion are subject to RsaE downregulation. In addition, importantly, we provide new evidence that RsaE regulates arginine catabolism.
RsaE targets the arginine catabolism by a direct interaction with the rocF mRNA
RsaE affects numerous transcripts, but to date, formal demonstrations of physiological sRNA/mRNA interactions were limited by difficulties in performing staphylococcal genetic studies. In vitro evidence based on gel retardation assays and toe-printing experiments, associated with transcriptomic and proteomic studies, indicated that RsaE acts by a direct pairing with sucCD, SA0873 (SAOUHSC_00951), opp3B (17) and opp3A mRNAs (16). Here, the in vivo effect of RsaE on rocF mRNA was demonstrated by transcriptomic, northern blot and qRT-PCR experiments. Further evidence for a direct interaction was given in vitro by a RsaE-dependent trapping of rocF mRNA, and in vivo by a targeted mutagenesis on predicted intermolecular RsaE/rocF mRNA pairings.
The RsaE duplicated motifs can have common and specific targets
Some sRNAs exert their regulatory activity by pairing to targets with two complementary regions [e.g. (46)]. However, RsaE harbors two identical motifs that may recognize identical target sequences. In B. subtilis, pairing of RoxS with its targets was addressed via a mutational study of C-rich regions (CRR1 to 4) (14,15). RoxS and RsaE sequences are almost identical (except for their terminators), and two repeat sequences of 10 nucleotides, referred to here as RsaE motifs 1 and 2, correspond to CRR1 and CRR3 in B. subtilis, respectively. Motif 1 is within a loop of a stem–loop structure while motif 2 is an unpaired sequence between two stems (Figure 3B). Both sequences are predicted to pair to complementary G-rich sequences and indeed we observed that both motifs 1 and 2 can exert independent regulatory functions. However, motif 1 had a stronger effect on one target (SAOUHSC_01138 mRNA), suggesting that presentation of the motif by a stem may promote the sRNA/mRNA pairing. On the contrary, RoxS requires the integrity of CRR3 (equivalent to motif 2) but not CRR1 to inhibit the formation of a translation initiation complex on the ppnKB target mRNA (14). Also, RoxS CRR3 but not CRR1 was shown to be involved in yflS RNA recognition (15). The presence of two conserved motifs in RsaE and its orthologs is probably not fortuitous, as they can target the same substrate (e.g. motifs 1 and 2 on rocF mRNA) or dedicated substrates (e.g. CCR3 on ppnKB mRNA). Sequences surrounding each motif might extend RsaE/mRNA pairing and be essential for recognition of certain substrates (Supplementary Figure S2 and Supplementary Table S4). Note that IntaRNA rank first RsaE motif 2 for interactions with rocF and rocD mRNAs (Supplementary Figure S2) while both motifs seem equivalent for down-regulation. Numerous factors not considered by target prediction software such as RNA chaperones, RNA helicases, RNA sponges and nucleases strongly affect the sRNA/mRNA interactions.
How RsaE binding leads to substrate regulation remains to be elucidated. In B. subtilis, RNase Y contributes to RoxS processing and RoxS-target degradation (14). However, in a S. aureus strain lacking RNase Y, RsaE quantity was not increased and no significant RsaE-target enrichment was observed (47). In contrast, RsaE and putative substrates where found associated with RNase III, suggesting a role for this double strand specific RNase in RsaE-dependent regulations (48).
RsaE and amino acid metabolism
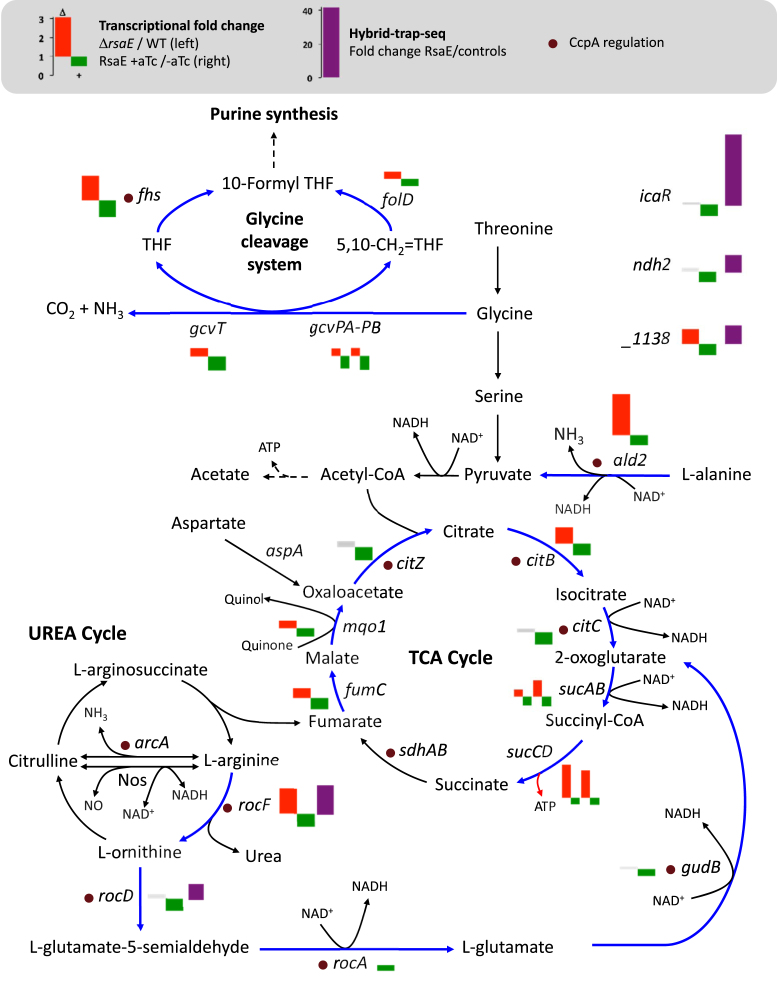
The gcvT-gcvPA-gcvPB operon encodes glycine decarboxylase enzymes providing methylene groups for the one-carbon metabolism and ald2 encodes an alanine dehydrogenase, a deaminating enzyme converting alanine to pyruvate. The corresponding mRNAs of both pathways are downregulated by RsaE. This observation is also true for the arginase rocF mRNA. In agreement with these results, in the absence of RsaE, HG003 grows at a faster rate in media containing amino acids as the sole carbon sources. It is known that pathways essential to growth on secondary carbon sources, such as catabolic pathways generating pyruvate from alanine and glycine, and the TCA cycle are regulated by RsaE. These results, in addition to the demonstrated rocF regulation, suggest that RsaE is a general regulator of amino acid catabolism. Besides repressing several enzymes using NAD as cofactor, RsaE reduces amino acid catabolism and may consequently limit the feeding of the TCA cycle (Figure 7). If so, RsaE would contribute to control the NAD+/NADH ratio, a role proposed for RoxS in B. subtilis (15) and therefore conserved among distantly related Firmicutes.
Figure 7.
Metabolic pathways associated with RsaE regulations. RsaE downregulates mRNAs (blue arrows) associated with the glycine cleavage system, the TCA cycle, the urea cycle and amino acid metabolisms. Histograms next to relevant gene names represent transcriptional change values from Table 1 (red boxes correspond to genes up-regulated in ΔrsaE/WT experiments, green boxes to genes down-regulated upon rsaE induction, and grey boxes to genes not significantly regulated). Some RsaE direct interactions on mRNAs are supported by in vitro evidences; histograms with purple boxes correspond to values from Table 2 (Hybrid-trap-seq data). Purple dots indicate CcpA-regulated genes.
Evidence for overlapping RsaE and SrrAB regulons
RsaE expression requires the two-component system SrrAB (14). Low oxygen concentrations or the presence of nitric oxide (NO) is sensed by the membrane protein SrrB, which activates SrrA via a phosphorelay, consequently stimulating rsaE transcription. In B. subtilis, RoxS expression is enhanced by ResDE (SrrAB orthologs), but also repressed by Rex, a redox sensing regulator activated by high NAD+/ NADH ratio (14,15). The roxS Rex binding site is strictly conserved in rsaE, suggesting that in S. aureus, Rex similarly represses rsaE expression (15). If so, full expression of RsaE would require SrrAB activation (low O2 or NO) and a low NAD+/NADH ratio. High levels of NAD+, which is a cofactor for many enzymes downregulated by RsaE (Figure 7) would induce Rex-mediated repression of RsaE in low oxygen conditions. Consequently, these NAD+ consuming metabolic pathways would not be shut down by RsaE.
Disruption of the srrAB operon (also named srhSR) leads to altered growth in anaerobic conditions (49). A proteomic study of S. aureus strain WCUH29 revealed a number of proteins that significantly increase in the absence of SrrAB: in aerobic conditions, SucD, RocD, RocF and SAOUHSC_01138 (alias YlbP) and in anaerobic conditions, SucC, SucD, RocD, RocF, SAOUHSC_01138, LctE, Ndh, Ald2, CitB, FumC (49). With the exception of LctE, all upregulated proteins in ΔsrrAB are also down-regulated by RsaE. Cross-comparison of Throup et al. with the current work suggests that RsaE is the main negative effector of the SrrAB pathway. Recent studies comparing the srrAB and wild-type transcriptomes show a limited overlap with Throup et al. (50,51), but conditions and strains are different. As RsaE binds to SDs and its inhibitory activity is first on translation, proteomics provides an efficient readout of RsaE activation.
RsaE controls CcpA-regulated genes
CcpA is a key regulator of carbon metabolism adaptation in Firmicutes. It represses genes associated with secondary carbon source acquisition or utilization, including TCA cycle and amino acid related genes (30,52–54). Many of these genes are also RsaE-regulated (Figure 7). RsaE, by acting post-transcriptionally, may synergistically shut off gene expression or ensure a negative control during oxygen deprivation on a subset of CcpA-regulated genes. The sRNA RsaOG (43,55) [also known as RsaI (17)] is CcpA-regulated (56, BioRxiv: https://doi.org/10.1101/278127) and trapped by RsaE (Table 2). Using an MS2-affinity purification approach, P. Romby, I. Caldelari and coworkers recently showed that RsaOG binds to several sRNAs including RsaE to possibly regulate them (BioRxiv: https://doi.org/10.1101/278127). Consequently, RsaOG could connect RsaE activity to CcpA regulation.
RsaE function conservation across species
A computer search for putative RsaE targets in Staphylococcus epidermidis (RP62A) and B. subtilis using RNApredator (58) revealed a remarkable target conservation, an unusual feature for sRNAs.
In S. epidermidis, sucD, ylbP (SERP0741), traP (SERP1369), rocD, citB (SERP0921), yjcG (SERP0604) and icaR mRNAs are among the best matches of SD regions targeted by RsaE (Supplementary Table S7).
Although B. subtilis is distantly related to S. aureus, some common targets of RoxS and RsaE, such as sucCD mRNA, were confirmed in both species. Moreover, sucCD mRNA is likely an RsaE target also in S. epidermidis. (Supplementary Table S7). This example in itself supports the idea of a conserved RsaE function across distant species.
In B. subtilis, the arginine catabolic genes have a different genetic organization compared to S. aureus: the transcription of rocABC and rocDEF operons is σL-dependent and under the control of RocR and AhrC (59). Interestingly, the B. subtilis arginine catabolic pathway is possibly controlled by RoxS. rocA and rocG (alias gudB in S. aureus) mRNAs are downregulated by RoxS (14). In addition, rocD SD is a predicted RoxS target (Supplementary Table S7). RoxS activity on rocD mRNA likely affect rocF (named argI in B. subtilis) expression as both genes are in the same operon. These observations suggest that the RsaE regulation of the arginine catabolic pathway may also be conserved between Staphylococcaceae and Bacillaceae.
CONCLUSION
RsaE is a global and highly conserved regulator of central metabolic functions, including the TCA cycle, the glycine cleavage pathway, tetrahydrofolate metabolism and the catabolism of several amino acids including arginine. It affects numerous CcpA regulated genes. RsaE would be a negative effector of the SrrAB regulon, and expectedly be more expressed when oxygen and NAD+ levels are low (14,15). RsaE operates via two seed motifs that may act differentially according to targets. It is likely that RsaE regulation is conserved and acts on the same pathways in other Firmicutes.
DATA AVAILABILITY
Sequencing data from transcriptomes and Hybrid-trap-seq have been deposited in GEO under the accession number GSE106457.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Tim Foster and Friedrich Götz for providing plasmids and strains. We thank Sandy Gruss, Steven Carson and Vinai Thomas for critical reading of the manuscript. We thank Sandy Gruss, David Halpern, Elise Borezée-Durant and Hoega Arden for helpful discussions and warm support. Lastly, we thank Valerie Shostrom for statistical analyses. This work has benefited from the facilities and expertise of the high throughput-sequencing platform of IMAGIF (http://www.i2bc.paris-saclay.fr) and of the INRA MIGALE bioinformatics platform (http://migale.jouy.inra.fr) for computational resources.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Agence Nationale pour la Recherche [ANR-2012-BLAN-1602 (Duplex-Omics); ANR-15-CE12-0003 (sRNA-Fit)]; Fondation pour la Recherche Médicale [DBF20160635724]; National Institutes of Health [NIH/NIAID P01AI083211]. Funding for open access charge: Agence Nationale pour la Recherche [ANR-15-CE12-0003].
Conflict of interest statement. None declared.
REFERENCES
- 1. Bischoff M., Dunman P., Kormanec J., Macapagal D., Murphy E., Mounts W., Berger-Bachi B., Projan S.. Microarray-based analysis of the Staphylococcus aureus sigmaB regulon. J. Bacteriol. 2004; 186:4085–4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shaw L.N., Lindholm C., Prajsnar T.K., Miller H.K., Brown M.C., Golonka E., Stewart G.C., Tarkowski A., Potempa J.. Identification and characterization of sigma, a novel component of the Staphylococcus aureus stress and virulence responses. PLoS One. 2008; 3:e3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tuchscherr L., Loffler B.. Staphylococcus aureus dynamically adapts global regulators and virulence factor expression in the course from acute to chronic infection. Curr. Genet. 2016; 62:15–17. [DOI] [PubMed] [Google Scholar]
- 4. Seidl K., Stucki M., Ruegg M., Goerke C., Wolz C., Harris L., Berger-Bachi B., Bischoff M.. Staphylococcus aureus CcpA affects virulence determinant production and antibiotic resistance. Antimicrob. Agents Chemother. 2006; 50:1183–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Majerczyk C.D., Dunman P.M., Luong T.T., Lee C.Y., Sadykov M.R., Somerville G.A., Bodi K., Sonenshein A.L.. Direct targets of CodY in Staphylococcus aureus. J. Bacteriol. 2010; 192:2861–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pohl K., Francois P., Stenz L., Schlink F., Geiger T., Herbert S., Goerke C., Schrenzel J., Wolz C.. CodY in Staphylococcus aureus: a regulatory link between metabolism and virulence gene expression. J. Bacteriol. 2009; 191:2953–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Felden B., Vandenesch F., Bouloc P., Romby P.. The Staphylococcus aureus RNome and its commitment to virulence. PLoS Pathog. 2011; 7:e1002006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Durica-Mitic S., Gopel Y., Gorke B.. Carbohydrate Utilization in Bacteria: Making the Most Out of Sugars with the Help of Small Regulatory RNAs. Microbiol. Spectrum. 2018; 6:RWR-0013–2017. [DOI] [PubMed] [Google Scholar]
- 9. Barquist L., Vogel J.. Accelerating discovery and functional analysis of small RNAs with new technologies. Annu. Rev. Genet. 2015; 49:367–394. [DOI] [PubMed] [Google Scholar]
- 10. Liu W., Rochat T., Toffano-Nioche C., Le Lam T.N., Bouloc P., Morvan C.. Assessment of Bona Fide sRNAs in Staphylococcus aureus. Front Microbiol. 2018; 9:228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Storz G., Vogel J., Wassarman K.M.. Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell. 2011; 43:880–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Novick R.P., Geisinger E.. Quorum sensing in staphylococci. Annu. Rev. Genet. 2008; 42:541–564. [DOI] [PubMed] [Google Scholar]
- 13. Gautheret D. Handbook of RNA Biochemistry. 2014; Wiley-VCH Verlag GmbH & Co. KGaA; 595–618. [Google Scholar]
- 14. Durand S., Braun F., Lioliou E., Romilly C., Helfer A.C., Kuhn L., Quittot N., Nicolas P., Romby P., Condon C.. A nitric oxide regulated small RNA controls expression of genes involved in redox homeostasis in Bacillus subtilis. PLos Genet. 2015; 11:e1004957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Durand S., Braun F., Helfer A.C., Romby P., Condon C.. sRNA-mediated activation of gene expression by inhibition of 5′-3′ exonucleolytic mRNA degradation. eLife. 2017; 6:e23602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bohn C., Rigoulay C., Chabelskaya S., Sharma C.M., Marchais A., Skorski P., Borezee-Durant E., Barbet R., Jacquet E., Jacq A. et al. . Experimental discovery of small RNAs in Staphylococcus aureus reveals a riboregulator of central metabolism. Nucleic. Acids. Res. 2010; 38:6620–6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Geissmann T., Chevalier C., Cros M.J., Boisset S., Fechter P., Noirot C., Schrenzel J., Francois P., Vandenesch F., Gaspin C. et al. . A search for small noncoding RNAs in Staphylococcus aureus reveals a conserved sequence motif for regulation. Nucleic. Acids. Res. 2009; 37:7239–7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bouloc P., Repoila F.. Fresh layers of RNA-mediated regulation in Gram-positive bacteria. Curr. Opin. Microbiol. 2016; 30:30–35. [DOI] [PubMed] [Google Scholar]
- 19. Bohn C., Rigoulay C., Bouloc P.. No detectable effect of RNA-binding protein Hfq absence in Staphylococcus aureus. BMC Microbiol. 2007; 7:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rochat T., Delumeau O., Figueroa-Bossi N., Noirot P., Bossi L., Dervyn E., Bouloc P.. Tracking the Elusive Function of Bacillus subtilis Hfq. PLoS One. 2015; 10:e0124977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Song J., Lays C., Vandenesch F., Benito Y., Bes M., Chu Y., Lina G., Romby P., Geissmann T., Boisset S.. The expression of small regulatory RNAs in clinical samples reflects the different life styles of Staphylococcus aureus in colonization vs. infection. PLoS One. 2012; 7:e37294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mashruwala A.A., Guchte A.V., Boyd J.M.. Impaired respiration elicits SrrAB-dependent programmed cell lysis and biofilm formation in Staphylococcus aureus. eLife. 2017; 6:e23845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Herbert S., Ziebandt A.K., Ohlsen K., Schafer T., Hecker M., Albrecht D., Novick R., Gotz F.. Repair of global regulators in Staphylococcus aureus 8325 and comparative analysis with other clinical isolates. Infect. Immun. 2010; 78:2877–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Le Lam T.N., Morvan C., Liu W., Bohn C., Jaszczyszyn Y., Bouloc P.. Finding sRNA-associated phenotypes by competition assays: an example with Staphylococcus aureus. Methods. 2017; 117:21–27. [DOI] [PubMed] [Google Scholar]
- 25. Corrigan R.M., Foster T.J.. An improved tetracycline-inducible expression vector for Staphylococcus aureus. Plasmid. 2009; 61:126–129. [DOI] [PubMed] [Google Scholar]
- 26. Huntzinger E., Boisset S., Saveanu C., Benito Y., Geissmann T., Namane A., Lina G., Etienne J., Ehresmann B., Ehresmann C. et al. . Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J. 2005; 24:824–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Poyart C., Trieu-Cuot P.. A broad-host-range mobilizable shuttle vector for the construction of transcriptional fusions to beta-galactosidase in gram-positive bacteria. FEMS Microbiol. Lett. 1997; 156:193–198. [DOI] [PubMed] [Google Scholar]
- 28. Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A. 3rd, Smith H.O.. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009; 6:343–345. [DOI] [PubMed] [Google Scholar]
- 29. Hussain M., Hastings J.G., White P.J.. A chemically defined medium for slime production by coagulase-negative staphylococci. J. Med. Microbiol. 1991; 34:143–147. [DOI] [PubMed] [Google Scholar]
- 30. Halsey C.R., Lei S., Wax J.K., Lehman M.K., Nuxoll A.S., Steinke L., Sadykov M., Powers R., Fey P.D.. Amino acid catabolism in Staphylococcus aureus and the function of carbon catabolite repression. mBio. 2017; 8:e01434-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jestin J.L., Deme E., Jacquier A.. Identification of structural elements critical for inter-domain interactions in a group II self-splicing intron. EMBO J. 1997; 16:2945–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Toffano-Nioche C., Nguyen A.N., Kuchly C., Ott A., Gautheret D., Bouloc P., Jacq A.. Transcriptomic profiling of the oyster pathogen Vibrio splendidus opens a window on the evolutionary dynamics of the small RNA repertoire in the Vibrio genus. RNA. 2012; 18:2201–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rutherford K., Parkhill J., Crook J., Horsnell T., Rice P., Rajandream M.A., Barrell B.. Artemis: sequence visualization and annotation. Bioinformatics. 2000; 16:944–945. [DOI] [PubMed] [Google Scholar]
- 35. Liao Y., Smyth G.K., Shi W.. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014; 30:923–930. [DOI] [PubMed] [Google Scholar]
- 36. Varet H., Brillet-Gueguen L., Coppee J.Y., Dillies M.A.. SARTools: A DESeq2- and EdgeR-Based R Pipeline for Comprehensive Differential Analysis of RNA-Seq Data. PLoS One. 2016; 11:e0157022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Busch A., Richter A.S., Backofen R.. IntaRNA: efficient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions. Bioinformatics. 2008; 24:2849–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S.. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009; 37:W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bury-Mone S., Nomane Y., Reymond N., Barbet R., Jacquet E., Imbeaud S., Jacq A., Bouloc P.. Global analysis of extracytoplasmic stress signaling in Escherichia coli. PLos Genet. 2009; 5:e1000651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sambrook J., Russell D.. Molecular Cloning: a laboratory manual. 2001; 3th ednNY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 42. Douchin V., Bohn C., Bouloc P.. Down-regulation of porins by a small RNA bypasses the essentiality of the regulated intramembrane proteolysis protease RseP in Escherichia coli. J. Biol. Chem. 2006; 281:12253–12259. [DOI] [PubMed] [Google Scholar]
- 43. Marchais A., Naville M., Bohn C., Bouloc P., Gautheret D.. Single-pass classification of all noncoding sequences in a bacterial genome using phylogenetic profiles. Genome Res. 2009; 19:1084–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Horsburgh M.J., Clements M.O., Crossley H., Ingham E., Foster S.J.. PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect. Immun. 2001; 69:3744–3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. VanDrisse C.M., Hentchel K.L., Escalante-Semerena J.C.. Phosphinothricin acetyltransferases identified using in vivo, in itro, and bioinformatic analyses. Appl. Environ. Microbiol. 2016; 82:7041–7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Geisinger E., Adhikari R.P., Jin R., Ross H.F., Novick R.P.. Inhibition of rot translation by RNAIII, a key feature of agr function. Mol. Microbiol. 2006; 61:1038–1048. [DOI] [PubMed] [Google Scholar]
- 47. Marincola G., Schafer T., Behler J., Bernhardt J., Ohlsen K., Goerke C., Wolz C.. RNase Y of Staphylococcus aureus and its role in the activation of virulence genes. Mol. Microbiol. 2012; 85:817–832. [DOI] [PubMed] [Google Scholar]
- 48. Lioliou E., Sharma C.M., Caldelari I., Helfer A.C., Fechter P., Vandenesch F., Vogel J., Romby P.. Global regulatory functions of the Staphylococcus aureus endoribonuclease III in gene expression. PLos Genet. 2012; 8:e1002782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Throup J.P., Zappacosta F., Lunsford R.D., Annan R.S., Carr S.A., Lonsdale J.T., Bryant A.P., McDevitt D., Rosenberg M., Burnham M.K.. The srhSR gene pair from Staphylococcus aureus: genomic and proteomic approaches to the identification and characterization of gene function. Biochemistry. 2001; 40:10392–10401. [DOI] [PubMed] [Google Scholar]
- 50. Kinkel T.L., Roux C.M., Dunman P.M., Fang F.C.. The Staphylococcus aureus SrrAB two-component system promotes resistance to nitrosative stress and hypoxia. mBio. 2013; 4:e00696-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wilde A.D., Snyder D.J., Putnam N.E., Valentino M.D., Hammer N.D., Lonergan Z.R., Hinger S.A., Aysanoa E.E., Blanchard C., Dunman P.M. et al. . Bacterial hypoxic responses revealed as critical determinants of the host-Staphylococcus aureuspathogen outcome by TnSeq analysis of invasive infection. PLoS Pathog. 2015; 11:e1005341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nuxoll A.S., Halouska S.M., Sadykov M.R., Hanke M.L., Bayles K.W., Kielian T., Powers R., Fey P.D.. CcpA regulates arginine biosynthesis in Staphylococcus aureus through repression of proline catabolism. PLoS Pathog. 2012; 8:e1003033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li C., Sun F., Cho H., Yelavarthi V., Sohn C., He C., Schneewind O., Bae T.. CcpA mediates proline auxotrophy and is required for Staphylococcus aureus pathogenesis. J. Bacteriol. 2010; 192:3883–3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gorke B., Stulke J.. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 2008; 6:613–624. [DOI] [PubMed] [Google Scholar]
- 55. Marchais A., Bohn C., Bouloc P., Gautheret D.. RsaOG, a new staphylococcal family of highly transcribed non-coding RNA. RNA Biol. 2010; 7:116–119. [DOI] [PubMed] [Google Scholar]
- 56. Mader U., Nicolas P., Depke M., Pane-Farre J., Debarbouille M., van der Kooi-Pol M.M., Guerin C., Derozier S., Hiron A., Jarmer H. et al. . Staphylococcus aureus Transcriptome Architecture: From Laboratory to Infection-Mimicking Conditions. PLos Genet. 2016; 12:e1005962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Figueroa-Bossi N., Valentini M., Malleret L., Fiorini F., Bossi L.. Caught at its own game: regulatory small RNA inactivated by an inducible transcript mimicking its target. Genes Dev. 2009; 23:2004–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Eggenhofer F., Tafer H., Stadler P.F., Hofacker I.L.. RNApredator: fast accessibility-based prediction of sRNA targets. Nucleic Acids Res. 2011; 39:W149–W154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gardan R., Rapoport G., Debarbouille M.. Role of the transcriptional activator RocR in the arginine-degradation pathway of Bacillus subtilis. Mol. Microbiol. 1997; 24:825–837. [DOI] [PubMed] [Google Scholar]
- 60. Burge S.W., Daub J., Eberhardt R., Tate J., Barquist L., Nawrocki E.P., Eddy S.R., Gardner P.P., Bateman A.. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res. 2013; 41:D226–D232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Darty K., Denise A., Ponty Y.. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics. 2009; 25:1974–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data from transcriptomes and Hybrid-trap-seq have been deposited in GEO under the accession number GSE106457.