Abstract
Polycomb group (PcG) and Trithorax group (TrxG) proteins are essential for maintaining epigenetic memory in both embryonic stem cells and differentiated cells. To date, how they are localized to hundreds of specific target genes within a vertebrate genome had remained elusive. Here, by focusing on short cis-acting DNA elements of single functions, we discovered three classes of response elements in human genome: Polycomb response elements (PREs), Trithorax response elements (TREs) and Polycomb/Trithorax response elements (P/TREs). In particular, the four PREs (PRE14, 29, 39 and 48) are the first set of, to our knowledge, bona fide vertebrate PREs ever discovered, while many previously reported Drosophila or vertebrate PREs are likely P/TREs. We further demonstrated that YY1 and CpG islands are specifically enriched in the four TREs (PRE30, 41, 44 and 55), but not in the PREs. The three classes of response elements as unraveled in this study should guide further global investigation and open new doors for a deeper understanding of PcG and TrxG mechanisms in vertebrates.
INTRODUCTION
Polycomb group (PcG) and Trithorax group (TrxG) proteins were originally discovered in Drosophila where their mutations resulted in improper body plans (1–5). Their crucial importance in development is exemplified by early lethality in mouse embryos upon deleting some of these proteins (6). PcG proteins are present as two major multi-protein complexes, polycomb repressive complex (PRC) 1 and 2. In mammals, PRC1 is further subdivided into canonical (cPRC1) and non-canonical (ncPRC1) complexes (7). PRC2 catalyzes the hallmark repressive mark of histone 3 Lys27 trimethylation (H3K27me3). Human PRC2 core complex contains the catalytic subunit Enhancer of Zeste Homolog 1 or 2 (EZH1 or EZH2), Suppressor of Zeste 12 homolog (SUZ12), Embryonic Ectoderm Development (EED) and Retinoblastoma-Binding Protein 4 or 7 (RBBP4 or RBBP7), while PRC2.1 and PRC2.2 contain additional but distinct accessory proteins (6,8–17). Other PcG complexes include Pleiohomeotic (Pho)-repressive complex (PhoRC), SCM-related gene containing four MBT domains (dSfmbt) and Polycomb repressive deubiquitinase complex (PR-DUB) (9). TrxG proteins are also present as several multi-subunit COMPASS (complex of proteins associated with Set1) -like complexes. Drosophila Trithorax (Trx) within the COMPASS-like complex is responsible for the placement of the hallmark activation marks of H3K4me3 on target genes (18). Mixed-lineage leukemia (MLL) 1/2 are the human homologues of Drosophila Trx (9,14,17,19).
From Drosophila to humans, PcG and TrxG complexes regulate the expression of hundreds of developmentally important, evolutionarily conserved target genes in each genome in a highly sequence-specific fashion. Principles for genomic targeting of these complexes initially emerged from earlier studies on Polycomb and Trithorax response elements (PRE/TREs) in Drosophila HOX clusters (9,11,12,18,20). Drosophila PcG and TrxG complexes were frequently found to co-occupy these PRE/TREs (21–25), and switch from repressed to activated states was observed after a brief transcriptional activity (26–29). Therefore, these elements are also termed ‘cellular memory modules’ (CMMs). A CMM of 219 base pair (bp) was identified in Drosophila Fab-7 region that regulates the Abdominal-B gene (30). Drosophila PRE/TREs were found to contain complex combinations of recognition motifs for transcription factors such as Pho, Engrailed (En), GAGA factor (GAF) and Zeste (Z) (18).
Despite the high conservation of PcG and TrxG complexes as well as their target genes from Drosophila to humans, the PRE/TREs that are signatures for their target genes are not evolutionarily conserved, which had severely halted the study of PcG and TrxG-mediated epigenetic regulation in vertebrates (as comprehensively reviewed in (12)). The first two reported vertebrate PRE/TREs were Kr3kb in mouse genome that was found to recruit PcG in Drosophila and mouse cells and modulate reporter gene expression in a PcG and TrxG-dependent manner (31), and a 1.8-kilobase pair (kb) D11.12 (between human HOXD11 and HOXD12) that endorsed PcG-dependent gene silencing (32).
Despite these and other pioneering studies (reviewed in (12,33)), the mechanisms of PcG and TrxG recruitment in vertebrates had remained elusive and many controversies existed. For instance, some studies suggested a role of CpG islands in recruiting PRC2 complex to vertebrate PREs (34–39). Indeed, insertion of GC-rich and CpG-rich sequences (1000 bp) at ectopic sites was shown to be sufficient to lead to both H3K4me3 and H3K27me3 marks (40). However, many other studies strongly argued against such a role for CpG islands (31,41,42). In addition, although Pho was believed to participate in recruiting PcG proteins in Drosophila, the roles of its mammalian homologue Yin Yang 1 (YY1) in this capacity have been under heated debate, in particular its binding motif was found to be depleted in mammalian polycomb domains (34,43).
We reasoned that the longstanding difficulties and controversies in the field are partially due to the large sizes of cis-acting DNA segments used in previous studies. We further postulated that within each of these large-size cis-acting DNA segments, there exist short, modular building blocks of cis-acting DNA elements. Each such cis-acting DNA element performs a single function and harbors a specific signature for recognition by PcG and/or TrxG recruitment factors. Discovery and detailed characterization of these modular units will provide the foundations towards a unified mechanistic understanding of PcG and TrxG recruitment and regulation that had been so far unattainable.
In this study, by taking a ‘reductionist’ approach, we have obtained multiple lines of solid evidence that unraveled, for the first time, three classes of PRE/TREs in human genome: PREs, TREs and P/TREs. In this context, PRE/TREs are used as a collective term for all response elements recognized by either PRC2 or MLL1/2-TrxG or both and contain three classes: PREs are short cis-acting DNA elements that are occupied by PRC2 proteins and contain H3K27me3 marks, TREs are occupied by MLL1/2-TrxG proteins and contain H3K4me3 marks, and P/TREs attract both PRC2 and MLL1/2-TrxG complexes and harbor H3K27me3 and H3K4me3 marks. Furthermore, this ‘reductionist’ approach allowed unequivocal demonstrations that YY1 and CpG islands co-localize to TREs, but not PREs, that are characterized in this study, suggesting the possible roles of YY1 and CpG islands in recruiting human MLL1/2–TrxG complex.
MATERIALS AND METHODS
Materials
Human HeLa cells and 293T cells were purchased from American Type Culture Collection (Catalog # ATCC CCL-2 and CRL-3216, respectively). Human Embryonic Stem Cell H9 total RNA was purchased from ScienCell Research Laboratories (Catalog # 5825). pGL4.74[hRLuc/TK] (Catalog # E6921) and pGL4.28[luc2CP/minP/Hygro] (Catalog # E8461) were purchased from Promega Corp. pCAGGS-flpE-puro (coding for wild-type flippase (FLPe) recombinase) and AAVS1 hPGK-puroR-pA donor were gifts from Dr. Rudolf Jaenisch (Addgene plasmid # 20733 and 22072, respectively) (44). pSpCas9(BB)-2A-Puro (PX459) was a gift from Dr. Feng Zhang (Addgene plasmid # 48139) (45). YY1pLuc vector (32) was kindly provided by Dr Robert E. Kingston at Massachusetts General Hospital. Human genomic DNA (Catalog # 11691112001) was purchased from Roche Diagnostics. Human SUZ12 shRNA (Catalog # V2LHS_74301) was purchased from Open Biosystems, Inc.
The following antibodies were used for western blotting: Anti-EZH2 from BD Biosciences (Catalog # 612666); anti-SUZ12 from Santa Cruz (Catalog # SC-67105); anti-EED from Millipore (Catalog # 05-1320); anti-WDR5 from Abcam (Catalog # ab56919); anti-MLL1 from Santa Cruz (Catalog # SC-20153); anti-Tubulin from Sigma (Catalog # T8328), anti-TATA Binding Protein from Abcam (Catalog # ab818), anti-Mouse-IgG-HRP from Sigma (Catalog # A2554), and anti-Rabbit-IgG-HRP from Sigma (Catalog # A0545).
The following antibodies were used for chromatin immunoprecipitation (ChIP): normal mouse IgG from Millipore (Catalog # 17-662), normal rabbit IgG from Santa Cruz (Catalog # sc-2027), anti-EZH2 from Millipore (Catalog # 17-10044), anti-EED from Millipore (Catalog # 05-1320), anti-WDR5 from Bethyl Lab (Catalog # 429A), anti-MLL1 from Santa Cruz (Catalog # sc-20153), anti-H3 from Active Motif (Catalog # 61277), anti-H3K4me3 from Active Motif (Catalog # 39915), anti-H3K4me3 from Active Motif (Catalog # 61017), and anti-YY1 from Santa Cruz (Catalog # sc-7341 X).
We also used the following kits and reagents in this study: Zyppy Plasmid Miniprep Kit (Zymo Research, Catalog # D4037); HiFi HotStart ReadyMix PCR Kit (KAPA Biosystems, Catalog # KK2602); Lipofectamine 3000 (Invitrogen, Catalog # L3000015); QuickExtract DNA Extraction Solution (Epicentre, Catalog # QE0905T); RNeasy Kit (Qiagen, Catalog # 74104); SuperScript III First Strand Synthesis Kit (Invitrogen, Catalog # 18080-051); protease inhibitor cocktails (Sigma-Aldrich, Catalog # 4693159001); Dual-Luciferase Reporter Assay System (Promega, Catalog # E1960); DMEM medium (Lonza, Catalog # 12-604Q); Fetal bovine serum (FBS) (HyClone, Catalog # SH30910.03); and protein A/G magnetic beads (Thermo Scientific, Catalog # 88803).
Dual luciferase reporter gene assay
A pair of FLPe recognition target (FRT) sites (46) were assembled from DNA oligos (47) synthesized by Integrated DNA Technologies and inserted immediately upstream of the YY1 enhancer and mini promoter of YY1pLuc, giving rise to 2FRT-YY1pLuc. A FLAG-tag was inserted into the C-terminus of FLPe in pCAGGS-flpE-puro to facilitate quantification of the expression level. We also made an inactive FLPe mutant (FLP(m)) with a deletion at residues 277–372 by QuikChange polymerase chain reaction (PCR).
Each of the top 61 predictions was amplified from human genomic DNA (Roche Diagnostics) and subcloned into the 2FRT-YY1pLuc vector flanked by two FRT sites, yielding a recombinant plasmid 2FRT-PRE/TRE-YY1pLuc. In addition, a second plasmid pGL4.74[hRluc/TK] (Promega) containing a Renilla luciferase gene under the control of an upstream thymidine kinase (TK) promoter was co-transfected. This allowed the signal of firefly luciferase to be normalized against that of Renilla luciferase from the same sample to account for any variations in transfection and expression.
Before transfection, HeLa cells were seeded in a 96-well plate at 20 000 cells/well with 100 μl/well DMEM medium (Lonza) and 10% FBS (HyClone), and incubated at 37°C with 5% CO2. Three plasmids, 2FRT-PRE/TRE-YY1pLuc, pGL4.74[hRluc/TK] and pCAGGS-flpE-puro coding for wild-type FLPe (named as FLP(+)) or a C-terminally truncated FLP(m), were co-transfected at a ratio of 200:1:40 (total DNA of 100 ng/well) using Lipofectamine 3000 (Invitrogen). In FLP(–) samples, only two plasmids, 2FRT-PRE/TRE-YY1pLuc and pGL4.74[hRluc/TK], were used for transfection. At 40 h post transfection, firefly and Renilla luciferases were measured with the Dual-Luciferase Reporter Assay System (Promega) using FLUOstar Omega (BMG LABTECH). To account for variability among experiments, firefly luciferase signals were divided by the Renilla luciferase signals to yield the relative luciferase signals (RLU). The RLU signal of each FLP(+) or FLP(m) sample was further normalized against that of FLP(-) sample for the same 2FRT-YY1pLuc or 2FRT-PRE/TRE-YY1pLuc construct to yield the Normalized RLU shown in some dual luciferase assay figures. As PREs or TREs were supposed to repress or activate gene expression, respectively, the partial excision of inserted DNA fragments by FLPe in FLP(+) samples was expected to increase the firefly luciferase signals for PREs, and decrease the firefly luciferase signals for TREs.
Lentiviral shRNA knockdown
Human SUZ12 shRNA (V2LHS_74301) in pGIPZ lentiviral vector was purchased from Open Biosystems, Inc. The shRNA target sequences for WDR5 were synthesized and annealed in vitro, and then inserted into the XhoI/HpaI sites of the pGIPZ vector. The targeting sequences are: SUZ12: 5′-CATGCATGACTTTAATCTT-3′; WDR5: 5′-GTGGAAGAGTGACTGCTAA-3′. To produce lentivirus, the shRNA-pGIPZ plasmid was co-transfected with psPAX2 and pMD2.G (at a ratio of 4 μg:3.6 μg:0.4 μg) using Lipofectamine 3000 into 293T cells in a six-well plate. Supernatants containing virus were filtered and added to HeLa cells every 24 h for 3 days. Infected cells were selected by 2 μg/ml puromycin for at least 14 days.
ChIP quantitative real-time PCR (ChIP-qPCR)
The ChIP protocol provided by Agilent (https://www.agilent.com/cs/library/usermanuals/Public/G4481-90010_ChIP-on-chip_11.3.pdf) was followed with minor modifications. Briefly, HeLa cells were cross-linked with 1% formaldehyde for 15 min and then lysed with Lysis Buffer (10 mM Tris•HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% sodium deoxycholate, 0.5% N-Lauroylsarcosine sodium). The chromatin DNA was sheared by sonication (Misonix 4000, Amplitude 30, power on 30 sec, 40 cycles) to get genomic DNA fragments in the range of 100–600 bp in size. The sheared chromatin fragments were incubated with specific antibody or control IgG-conjugated protein A/G magnetic beads (Thermo Scientific) for overnight at 4°C. The immunoprecipitated chromatin was washed and eluted, crosslinking was reversed, and RNA and protein were degraded by RNase A and Protease K, respectively. The final DNA fragments were purified by phenol:choloroform (24:1) extraction and ethanol precipitation. DNA purified from 1% of starting chromatin in the same way except the immunoprecipitation step was used as internal control in the following qPCR.
qPCR was performed with primers designed by Primer3 (http://bioinfo.ut.ee/primer3-0.4.0/primer3/) and UCSC In-Silico PCR (https://genome.ucsc.edu/cgi-bin/hgPcr). According to the manufacturers’ suggestions, MYT1 or HOXA9 was used as a positive control for ChIP-qPCR.
The qPCR reaction was performed in three or more biological replicates for each sample on the Eco Real-Time PCR System (Illumina) with FastStart Universal SYBR Green Master (Roche Diagnostics) according to the manufacturers’ instructions. For each reaction, the following components were included: 1 μl template, 1 μl primer pair mix (5 μM for each primer), 3 μl H2O and 5 μl SYBR Green Mix (2×). The 10 μl reaction mixture was added to one well of the 48-well qPCR plates (Illumina). The thermo-cycling parameters were as follows: 50°C for 2 min, 95°C for 10 min, followed by 45 cycles of 95°C for 15 s, 60°C for 1 min, melt curve analysis performed between 55 and 95°C for 15 min at a ramp-rate of 1.6°C/s. The EcoStudy Software v5.0 (Illumina) was used to calculate the threshold cycle (Ct) value, which is defined as the number of cycles at which the fluorescence signal is significantly above the threshold. The percent (%) input was calculated from ChIP-qPCR results for each genomic locus from specific antibodies or control rabbit or mouse IgG according to https://www.thermofisher.com/us/en/home/life-science/epigenetics-noncoding-rna-research/chromatin-remodeling/chromatin-immunoprecipitation-chip/chip-analysis.html. Since control rabbit or mouse IgG antibody gave different basal nonspecific binding at different genomic loci, always at very small values, the % input of immunoprecipitated chromatin in each sample after subtracting that of the corresponding rabbit or mouse IgG control was shown. The % input of immunoprecipitated chromatin by anti-H3 antibody at each locus was also shown along with that of H3 marks (H3K27me3 or H3K4me3). Significant enrichment of a given H3 mark at candidate PRE/TREs was concluded by comparing the relative enrichment of H3 marks over H3 to that of Gene Desert.
Expression analysis by reverse transcription (RT)-qPCR
Total RNA was prepared according to the manufacturer's instructions using RNeasy Kit (Qiagen). The extracted RNA concentration and purity were assessed with UV spectroscopy (absorbance at 260 and 280 nm). The RNA was reverse-transcribed using SuperScript III First Strand Synthesis Kit (Invitrogen) per manufacturer's instructions. Briefly, 2 μg of total RNA was reverse-transcribed into first-stand cDNA with 50 ng random hexamers in a 20 μl reaction system. The cDNA was serially diluted and used as template in qPCR. The qPCR was performed as in the ChIP-qPCR section. The endogenous housekeeping gene GAPDH was used as the reference.
The Pfaffl method, an improved comparative Ct method that accounts for variations in primer efficiency, was employed to process the relative gene expression data (48). Briefly, the Ct values of the gene transcript of the sample were subtracted from those of the control to obtain ΔCt. The correct primer efficiency (E) was calculated using a standard curve which was obtained from qPCR reactions using 1:10 serially diluted cDNA as template. The target gene expression ratio was then calculated with the Pfaffl equation: ratio = (Etarget)ΔCttarget/(EGAPDH)ΔCtGAPDH.
Integration of 2FRT-PRE/TRE-YY1pLuc cassettes into the AAVS1 locus of human chromosome
The Clustered, Regularly Interspaced, Short Palindromic Repeats and Cas9 endonuclease (CRISPR-Cas9) system was used to integrate 2FRT-PRE/TRE-YY1pLuc cassette into the adeno-associated virus integration site 1 (AAVS1) locus of human chromosome. The AAVS1 guide oligo (5′-GGGGCCACTAGGGACAGGAT-3′) was annealed and ligated via the Bbs I site into pSpCas9(BB)-2A-Puro (PX459) bearing both Cas9 and the remainder of the gRNA (Addgene), yielding the Cas9/gRNA plasmid pSpCas9-Puro/AAVS1_gRNA. To construct the donor plasmid, the 2FRT-PRE/TRE-YY1pLuc fragment was first transferred from the 2FRT-YY1pLuc vector (Kpn I/Fse I) to pGL4.28[luc2CP/minP/Hygro] vector (Promega) to get 2FRT-PRE/TRE-YY1pLuc-pGL4.28 which included a hygromycin selection marker. The AAVS1 homologous arms were amplified from AAVS1 hPGK-puroR-pA donor (Addgene) and used as mega-primers in the QuikChange PCR to be inserted into 2FRT-PRE/TRE-YY1pLuc-pGL4.28 vector, giving rise to the donor vector 2FRT-PRE/TRE-YY1pLuc-pGL4.28-AAVS1-Arms.
HeLa cells were seeded in six-well plates (5 × 105 cells/well) one day before transfection. The plasmids pSpCas9-Puro/AAVS1_gRNA (2 μg) and 2FRT-PRE/TRE-YY1pLuc-pGL4.28-AAVS1-Arms donor (2 μg) were co-transfected into the cells with Lipofectamine 3000 (Invitrogen). At 24 h post transfection, the medium was changed to DMEM with 10% FBS and 2 μg/ml puromycin. After selection with puromycin for two days, the cells were resuspended, serial diluted and plated on 96-well plates at a final concentration of 0.5 cell/well. After 2 weeks of further selection with 200 μg/ml hygromycin, the survived single colonies were subjected to DNA isolation and genotyping.
For genotyping, genomic DNA from isolated clonal cell lines was extracted by QuickExtract DNA Extraction Solution (Epicentre) following the manufacturer's instruction. Homology Directed Repair (HDR) was detected via PCR amplification of junction region, followed by Sanger sequencing. The confirmed clonal cell lines were maintained in DMEM medium with 10% FBS plus 100 μg/mL hygromycin.
Knockout of PREs and P/TRE from human genome in HeLa cells
To knock out the PRE or P/TRE fragment from the genome of HeLa cells, two gRNAs flanked by the desired region were designed for each element with online CRISPR Design Tools: http://tools.genome-engineering.org (45). gRNA-1 was constructed into pSpCas9(BB)-2A-Puro (PX459) and gRNA-2 was inserted into pSpCas9p-2A-Blast. The pSpCas9p-2A-Blast vector was constructed from pSpCas9(BB)-2A-Puro (PX459) by replacing the puromycin-resistance gene with the blasticidin-resistance gene by QuikChange mutagenesis. The blasticidin-resistance gene was assembled from DNA oligos (47) synthesized by Integrated DNA Technologies.
Two microgram pSpCas9(BB)-2A-Puro-gRNA-1 and 2 μg pSpCas9p-2A-Blast-gRNA-2 were co-transfected into the HeLa cells with Lipofectamine 3000 (Invitrogen). At 24 h post transfection, the medium was changed to DMEM with 10% FBS plus 2 μg/ml puromycin and 5 μg/ml blasticidin. After two days of selection, the cells were resuspended, serial diluted and plated into 96-well plates at a final concentration of 0.5 cell/well. The cells were allowed to expand in DMEM with 10% FBS for 2–3 weeks before genotyping.
Homozygous PRE or P/TRE knockout HeLa clones were detected via PCR amplification of the deletion region, followed by Sanger sequencing of the modified region. The validated clonal cell lines were maintained in DMEM medium with 10% FBS.
Genome-wide analysis of PRE/TREs in K562 cells
We chose human K562 cell line as it was the only one with data for ChIP-seq experiments for all the following: H3K27me3, H3K4me3, YY1 and EZH2 through the UCSC Genome browser (hg19). Additionally, CpG island data for K562 was also available from UCSC Genome Browser (hg19). The ChIP-seq data for KMT2B (MLL2) were taken from the ENCODE project experiment ENCSR735JCD in the ENCFF693DIG peak set. From these data, we first retrieved regions that harbor individual marks by imposing a size limit of 416 bp for EZH2, H3K27me3, MLL2 and H3K4me3, and 500 bp for YY1 and CpG islands. Overlapping regions (with >25% overlap) were then defined using the UCSC table browser intersection feature for H3K4me3 and MLL2 (denoted as MK4(25)), or H3K27me3 and EZH2 (denoted as EK27(25)). If overlap was found, the coordinates from the protein, either EZH2 or MLL2, were passed on to MK4(25) or EK27(25), respectively. Then the P/TRE group was obtained by imposing a full 100% overlap between the groups of MK4(25) and EK27(25). The group of DNA fragments in MK4(25) after subtracting any EZH2 or H3K27me3 peaks was termed as TREs, while the group of EK27(25) after subtracting any MLL2 or H3K4me3 peaks was termed as PREs. In total, we retrieved 15 173 PREs (416 bp or smaller), 10 693 TREs (416 bp or smaller), 107 P/TREs (416 bp or smaller), 1309 YY1 peaks and 12 852 CpG islands (500 bp or smaller). The overlaps of YY1 or CpG islands with each of the PREs, TREs, or P/TREs were then defined.
RESULTS
Prediction of candidate human PRE/TREs by EpiPredictor
We first used EpiPredictor, a bioinformatics tool that we developed earlier (49), to predict human candidate PRE/TREs. Due to the lack of well-characterized human transcription factors known to recruit PRC2 or MLL1/2-TrxG complexes, we started from Drosophila transcription factors with these functions. Our previous study found that the combination of GAF, Pho, En and Zeste recognition motifs in EpiPredictor performed the best in identifying Drosophila PRE candidates (49). Therefore, we used the same sets of DNA recognition motifs (Supplementary Table S1) to scan human genome. The 61 top-ranked DNA fragments (Supplementary Table S2) were selected for experimental investigations.
Characterization of candidate PRE/TREs by dual luciferase reporter assay
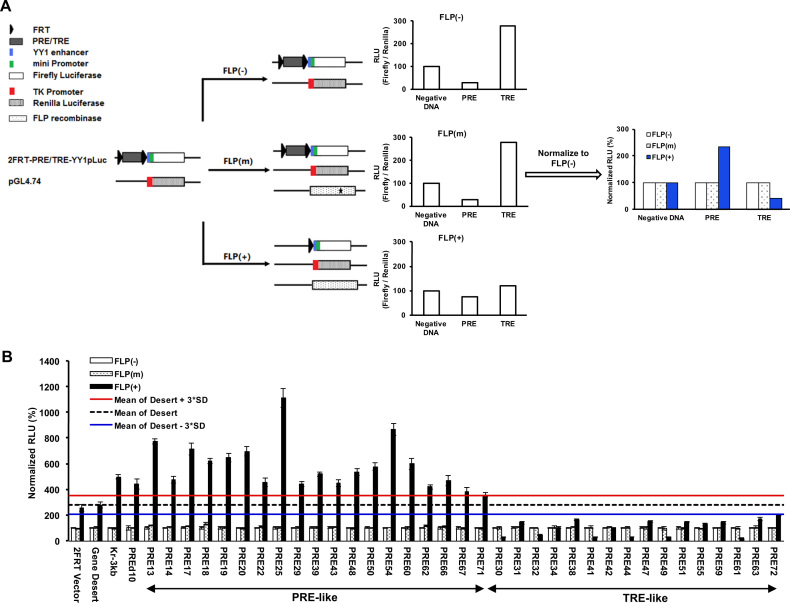
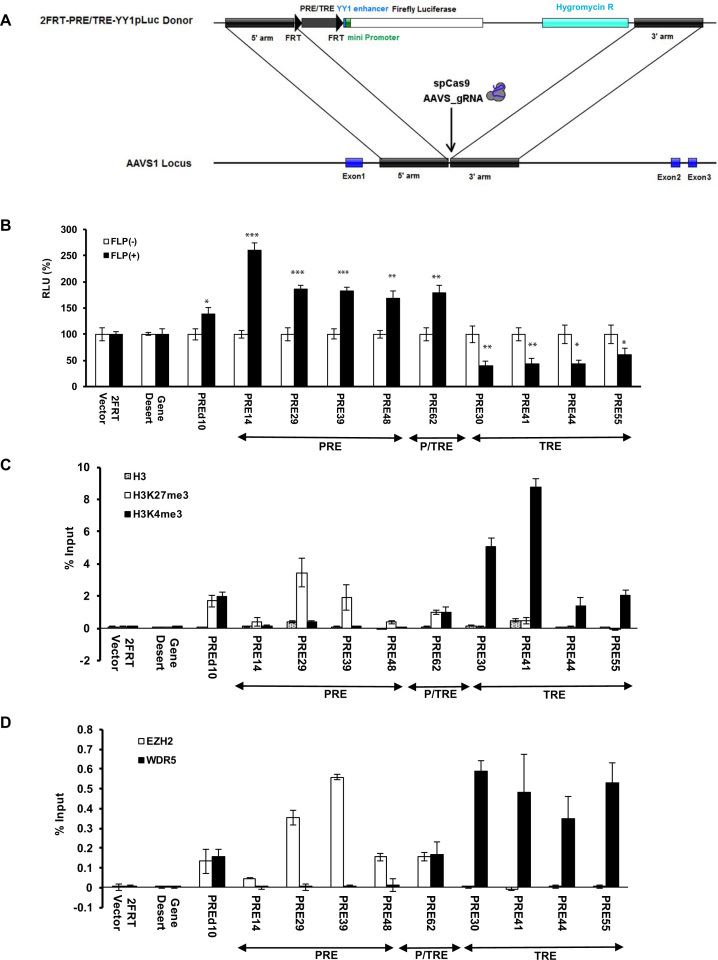
To determine whether these top-ranked predictions by EpiPredictor are indeed PRE/TREs that regulate gene expression, we developed a transient dual luciferase assay in cultured human HeLa cells with the help of Dr Robert Kingston (32) (Figure 1A). The use of HeLa cells instead of embryonic stem cells was to avoid the predominance of the bivalent H3K4me3/H3K27me3 domains on the genome (34,50). In the assay, each of the top 61 predictions flanked by two FRT sites was inserted immediately upstream of the YY1 enhancer and mini promoter that control the expression of firefly luciferase. For negative controls, we used the empty 2FRT-YY1pLuc vector (2FRT Vector) or the vector inserted with a 200-bp fragment from chr1:81132056–81132255 (Gene Desert) that was not associated with any epigenetic marks or proteins in ChIP-seq tracks on Genome Brower. Kr3kb from mouse and PREd10 from human that have been reported as PREs (31,42) were used as positive controls. FLPe recombinase in FLP(+) samples partially excised out the DNA insert between the two FRT sites (Supplementary Figure S1a), leading to changes in firefly luciferase activity (Supplementary Figure S1b). The significantly increased firefly luciferase activity of Kr3kb and PREd10 in FLP(+) samples confirmed their PRE-like behaviors and validated our transient dual luciferase assay in cultured human HeLa cells (Figure 1B).
Figure 1.
Screening of candidate human PRE/TREs by dual luciferase reporter assay. (A) Overview of the screening strategy. Two plasmids were common for all samples: 2FRT-YY1pLuc and pGL4.74. Each of the candidate human PRE/TREs was inserted between the two FRT sequences in 2FRT-YY1pLuc to yield 2FRT-PRE/TRE-YY1pLuc. pGL4.74 contains a Renilla luciferase gene under the control of an upstream TK promoter. This allowed the signal of firefly luciferase to be normalized against that of Renilla luciferase from the same sample, yielding relative luciferase activity (RLU), to account for any variations in transfection and expression. In FLP(+) samples, an additional plasmid harboring the wild-type FLPe recombinase gene was co-transfected with the two luciferase vectors to allow for partial excision of candidate PRE/TRE at the two FRT sites. In FLP(m), a plasmid containing a C-terminally truncated, inactive FLPe (shown with an asterisk in the FLPe gene) was used in the transfection to serve as a negative control. The RLU values of the FLP(m) and FLP(+) samples were normalized against that of the FLP(-) for the same DNA insert to give rise to Normalized RLU. As PREs or TREs were supposed to repress or activate gene expression, respectively, the partial excision of inserted DNA fragments by FLPe in FLP(+) samples should increase the firefly luciferase signals for PREs, and decrease the firefly luciferase signals for TREs. (B) Results of dual luciferase assay in HeLa cells. The data were represented as mean ± SD (standard deviations) from at least three biological replicates that were performed on different samples on different days. The mean signal of the Gene Desert FLP(+) sample and its plus and minus 3*SD were shown as black dashed line or red or blue solid line, respectively. Candidate PRE/TREs were selected as PRE-like or TRE-like if the Normalized RLU values of their FLP(+) samples were above the red line or below the blue line, respectively.
One key design of the dual luciferase assay in conjunction with FLPe recombinase is to detect any changes in firefly luciferase activity upon excision of a DNA fragment upstream of the YY1 enhancer and mini promoter that control firefly luciferase transcription. Although the excision of DNA fragment located between the two FRT sites were not 100%, only the plasmids that were excised by FLPe contributed to the observed changes of luciferase activity. In both the 2FRT-vector and the Gene Desert constructs, we noticed an increased level of luciferase activity for FLP(+) samples than their corresponding FLP(-) and FLP(m) samples (Figure 1B). The higher luciferase activity of the FLP(+) samples in these cases is likely the result of FLPe-mediated recombination that potentiated the mini promoter. However, the fact that the FLP(+) samples of both the 2FRT-vector and the Gene Desert fragment had a similar level of luciferase activity confirmed that this is the baseline luciferase signals for all FLP(+) samples of PRE/TRE-containing firefly vectors, because they share the same recombination end product, the firefly vector with just one FRT. Consequently, we used the FLP(+) sample of the Gene Desert fragment plus or minus 3 × standard deviations (3*SD) values from multiple independent measurements to define PRE-like or TRE-like DNA fragments, respectively.
By comparing the mean ± 3*SD values of the FLP(+) sample of the Gene Desert fragment, we found that 19 (31.1%) DNA inserts behaved like PREs where their partial removal by FLPe significantly enhanced the firefly luciferase reporter gene expression, and 16 (26.2%) DNA inserts that behaved like TREs where their partial excision by FLPe significantly reduced the expression of firefly luciferase in HeLa cells (Figure 1b, Supplementary Figure S1B). These together accounted for about 57% of the 61 top-ranked predictions.
Recruitment of PRC2 and MLL1/2-TrxG complexes to endogenous genomic loci
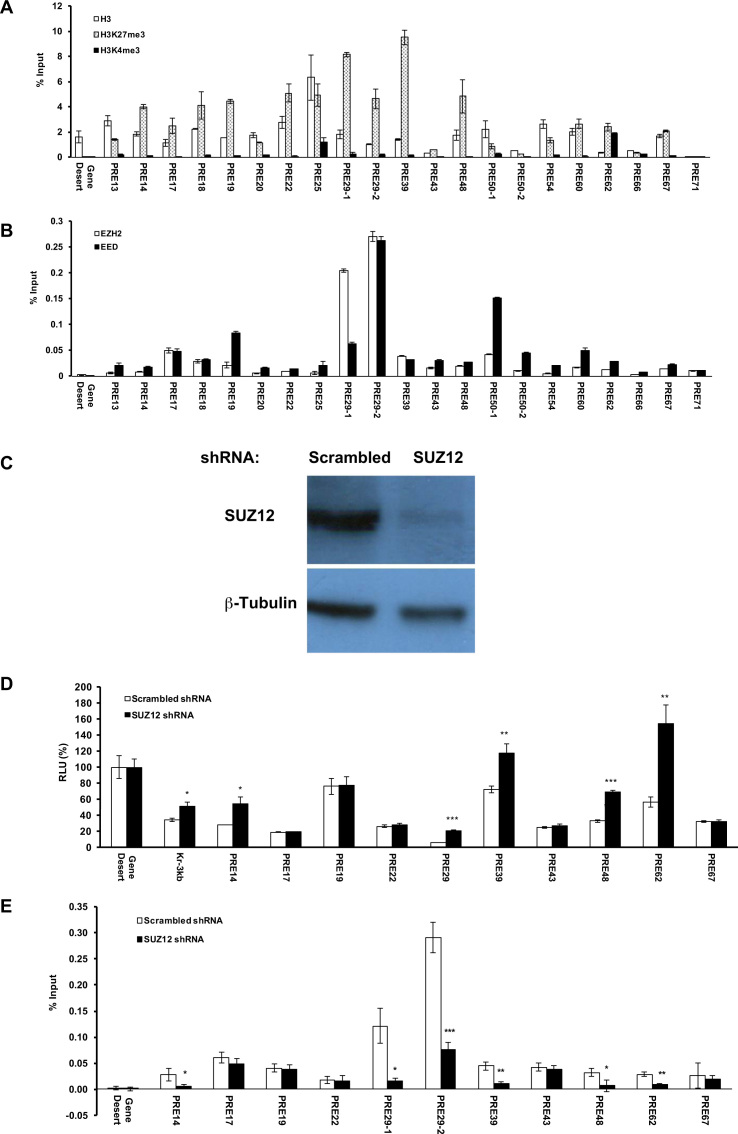
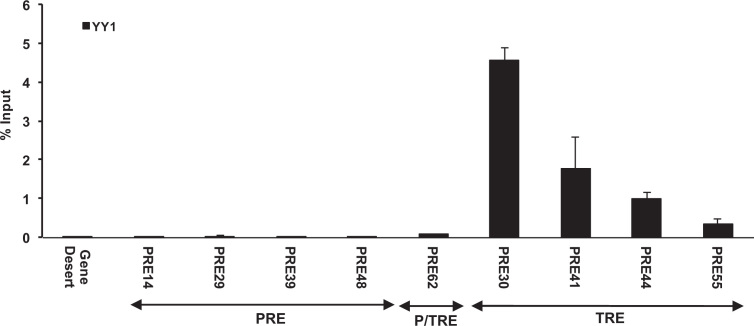
Woo et al. clearly demonstrated that the repression conferred by D11.12 on transfected plasmid DNA was mediated by PcG proteins (32). Since our primary focus was on the behaviors of candidate PRE/TREs in their native environment, next we examined whether the DNA fragments with PRE- or TRE-like behaviors in the dual luciferase assay carry corresponding H3K27me3 or H3K4me3 marks at the endogenous loci and are enriched with key components of PRC2 or MLL1/2-TrxG complexes that catalyzes these marks (primers are listed in Supplementary Table S3). Comparing to the ratio of % input of immunoprecipitated chromatin fragments by anti-H3K27me3 versus anti-H3 antibody for Gene Desert, all the 19 DNA fragments with PRE-like behavior in the dual luciferase assay had significant enrichment for H3K27me3 over H3 (Figure 2A). In addition, all except PRE62 had very low enrichment for H3K4me3 marks, confirming their PRE-like behaviors (Figure 2A). Furthermore, except for PRE13, 20, 25, 54 and 66, all PRE-like DNA fragments with strong H3K27me3 signals had strong ChIP signals for both EZH2 and EED (Figure 2B). In HeLa cells in which the expression of SUZ12 was knocked down using shRNA lentiviruses (Figure 2C), five PRE-like DNA fragments (PRE14, 29, 39, 48 and 62) as well as mouse Kr3kb exhibited statistically significantly higher luciferase activity than those in cells with scrambled shRNA (Figure 2D). In addition, the endogenous loci of PRE14, 29, 39, 48 and 62 all showed statistically significant decrease of EED signals upon SUZ12 knockdown (Figure 2E).
Figure 2.
Enrichment of H3 marks and PRC2 at the endogenous loci of PRE-like DNA fragments. (A) ChIP-qPCR of H3 (open bars), H3K27me3 (dotted open bars) and H3K4me3 (filled solid bars) at the endogenous loci of PRE-like DNA fragments in HeLa cells shown as % input. (B) ChIP-qPCR of EZH2 (open bars) and EED (filled solid bars) shown as % input. (C) The expression of SUZ12 in SUZ12 shRNA knockdown cell line was significantly reduced as compared to that in control cell line treated with scrambled shRNA. (D) The luciferase activity of PRE-like DNA fragments in SUZ12 knockdown cells (filled solid bars) compared to control cell line treated with scrambled shRNA (open bars). The RLU at Gene Desert was considered as 100%. (E) ChIP-qPCR of EED at the endogenous loci of PRE-like DNA fragments in SUZ12 knockdown cell line shown as % input. In a, b, d, e, data were represented as mean ± SD from at least three biological replicates that were performed on different samples on different days. The −1, −2 labels in a, b, e reflect the use of multiple primers in qPCR as listed in Supplementary Table S3. The P values of Student's t-test comparing SUZ12 shRNA and scrambled shRNA (D, E) were showed as *P < 0.05, **P < 0.005 and ***P < 0.001.
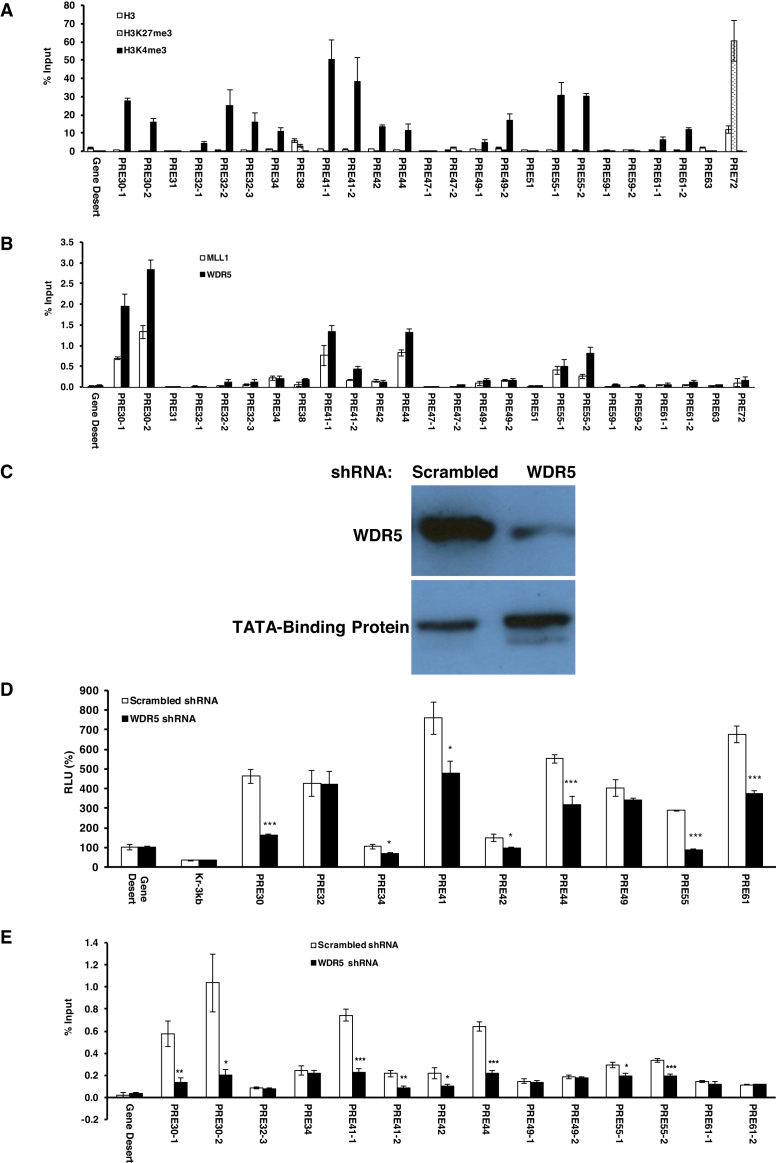
On the other hand, of the 16 DNA fragments with TRE-like behavior in the dual luciferase assay, nine of them (PRE30, 32, 34, 41, 42, 44, 49, 55 and 61) had significant enrichments for H3K4me3 over H3 than that of Gene Desert (Figure 3A). Among them, four DNA fragments (PRE30, 41, 44 and 55) had significant enrichment in ChIP signals for both MLL1 and WD repeat-containing protein 5 (WDR5), which are key components of human COMPASS complexes (Figure 3B). Additionally, in WDR5-knockdown HeLa cells (Figure 3C), all of PRE30, 41, 44 and 55 exhibited significant decrease in terms of luciferase activity (Figure 3D) and enrichment of MLL1 at their endogenous loci than in cells with scrambled shRNA (Figure 3E).
Figure 3.
Enrichment of H3 marks and MLL1/2-TrxG complex at the endogenous loci of TRE-like DNA fragments. (A) ChIP-qPCR of H3 (open bars), H3K27me3 (dotted open bars) and H3K4me3 (filled solid bars) at the endogenous loci of TRE-like DNA fragments in HeLa cells shown as % input. (B) ChIP-qPCR of MLL1 (open bars) and WDR5 (filled solid bars) shown as % input. (C) The expression of WDR5 in WDR5 shRNA knockdown cell nuclear extract was significantly reduced as compared to that in control cell line treated with scrambled shRNA. (D) The luciferase activity of TRE-like DNA fragments in WDR5 knockdown cells (filled solid bars) compared to control cell line treated with scrambled shRNA (open bars). The RLU at Gene Desert was considered as 100%. (E) ChIP-qPCR of MLL1 at the endogenous loci of TRE-like DNA fragments in WDR5 knockdown cell line shown as % input. In a, b, d, e, data were represented as mean ± SD from at least three biological replicates that were performed on different samples on different days. The −1, −2 labels in a, b, e reflect the use of multiple primers in qPCR as listed in Supplementary Table S3. The P values of Student's t-test comparing WDR5 shRNA and scrambled shRNA (D, E) were showed as *P < 0.05, **P < 0.005 and ***P < 0.001.
Collectively, we have identified five putative PREs (PRE14, 29, 39, 48 and 62) that were enriched for PRC2 complex, had a high H3K27me3 level and repressed luciferase expression, and four putative TREs (PRE30, 41, 44 and 55) that were enriched for MLL1/2-TrxG complex, had a high H3K4me3 level and activated luciferase expression. However, PRE62 was different from all other putative PREs: it had a high level of enrichment for both H3K4me3 and H3K27me3 marks (Figure 2A) at the endogenous locus (see more later).
Identification of core sequences of putative PREs and TREs
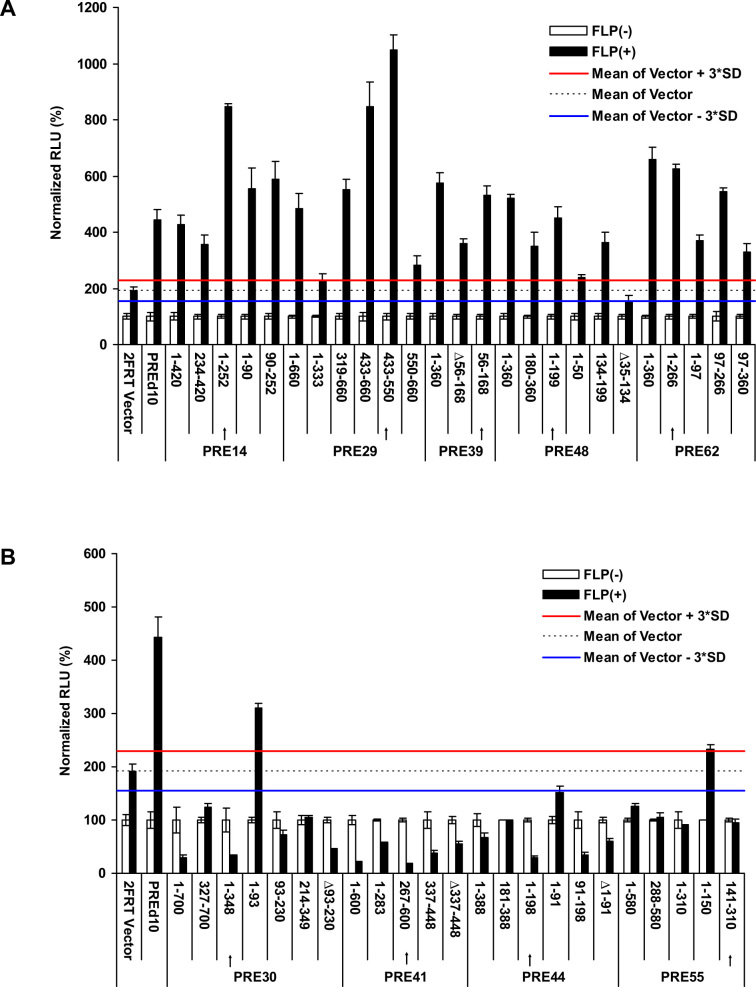
The five putative PREs (PRE14, 29, 39, 48 and 62) and four putative TREs (PRE30, 41, 44, 55) were of the size of 360–700 bp long. Previous studies have shown that mouse DNA sequences as short as 220 bp can recruit H3K27me3 marks (38). We asked whether these DNA fragments can be further truncated while maintaining the observed PRE or TRE properties. We tested the luciferase activity of multiple truncations and identified the functional core for each individual DNA fragment (Figure 4A, B). The core DNA sequence was defined as the shortest fragment with the highest activity in repression (for putative PREs, Figure 4A) or activation (for putative TREs, Figure 4B) among all tested truncations for a given DNA fragment. Sometimes, the core DNA sequences even had higher activity than the full-length DNA fragment, examples including PRE14 (Figure 4A) and PRE44 (Figure 4B). These core DNA sequences were in the range of 113–266 bp for putative PREs and 170–348 bp for putative TREs. Therefore, the repression or activation activities of these putative PREs or TREs, respectively, were localized in short DNA fragments.
Figure 4.
Define the core sequences of putative PREs and TREs. (A) Normalized RLU of various truncations of putative PREs. (B) Normalized RLU of various truncations of putative TREs. In a and b, the arrows indicated the regions corresponding to the core sequences. For each sample, the signals of FLP(-) was deemed as 100% against which the signal of the FLP(+) sample was normalized. The data were represented as mean ± SD from at least three replicates. The mean signal of the 2FRT Vector FLP(+) sample and its plus and minus 3*SD were shown as black dashed line or red or blue solid line, respectively. The firefly luciferase signals were considered as significantly changed if the Normalized RLU values were above the red line (for PREs, a) or below the blue line (for TREs, b).
Validation of putative PREs and TREs in an identical genomic environment
PREs are sensitive to genomic location (51,52). To further validate these putative PREs and TREs in a well-controlled genomic environment, we integrated each core sequence with luciferase reporter cassette into the AAVS1 site in human chromosome 2 by the CRISPR-Cas9 system (Figure 5A, Supplementary Figure S2). AAVS1 has an open chromatin structure, is transcription-competent and represents a well-validated genomic location for testing cellular functions of a DNA fragment (53). Most importantly, there is no known adverse effect on the cells from the DNA fragment inserted at AAVS1. Following Cas9 cleavage and HDR, the colonies with desired inserts were selected by flanking PCR and verified by Sanger sequencing (Supplementary Figure S2). Dual luciferase activities were measured for a representative clone of each core sequence (Figure 5B). Clearly, comparing to their respective FLP(-) samples, the FLP(+) samples of cells stably carrying PRE14, 29, 39, 48, 62 core sequences at the AAVS1 site exhibited significantly higher expression of firefly luciferase, while those of cells integrated with PRE30, 41, 44, 55 core sequences showed significantly reduced luciferase expression (Figure 5B). Furthermore, except for PRE62, all putative PREs (PRE14, 29, 39 and 48) had significant enrichment for EZH2 and H3K27me3 and no enrichment for WDR5 or H3K4me3 (Figure 5C, D). On the contrary, all putative TREs (PRE30, 41, 44 and 55) had no enrichment for EZH2 or H3K27me3 but significant enrichment for WDR5 and H3K4me3 (Figure 5C, D). Strikingly, PRE62 again exhibited a unique behavior for being enriched for all of EZH2, H3K27me3, WDR5 and H3K4me3. In addition, we also included PREd10 as a positive control for its shorter sequence (1.4 kb) than Kr3kb. Clearly, PREd10 behaved like PRE62 with significant enrichment for all of EZH2, H3K27me3, WDR5 and H3K4me3, while exhibiting repressed luciferase activity (Figure 5B–D).
Figure 5.
Effects of putative PREs and TREs on transcription in an identical genomic environment. A). Schematic illustration of the strategy that integrated the core sequences of the putative PREs and TREs (2FRT-PRE/TRE-YY1pLuc donor) into AAVS1 locus by CRISPR-Cas9. B). Dual luciferases assay in HeLa cells carrying stably integrated core sequences comparing FLP(-) samples (open bars) and FLP(+) samples (filled solid bars). Data are represented as mean ± SD from at least three replicates. The P value of Student's t-test comparing FLP(+) and FLP(-) for each integrated core sequence was shown as * for P<0.05, ** for P<0.005 and *** for P<0.001. C). ChIP-qPCR of H3 (dotted open bars), H3K27me3 (open bars) and H3K4me3 (filled solid bars) at the promoter region downstream of the integrated core sequences shown as % input. D). ChIP-qPCR results of EZH2 (open bars) and WDR5 (filled solid bars) at the promoter region downstream of the integrated core sequences shown as % input.
All these data together provided multiple lines of evidence for the existence of three distinct classes of PRE/TREs in human HeLa cells: PREs such as PRE14, 29, 39 and 48, TREs such as PRE30, 41, 44 and 55, and P/TREs such as PRE62 and PREd10. The enrichment data of all but one (PRE14) of these PRE/TREs in human HeLa cells are consistent with those of HeLa S3 available in Genome Browser (Supplementary Figure S3). The discrepancy observed for PRE14 between HeLa cells in our hands and HeLa S3 cells from Genome Browser may reflect some of the differences between these two cell lines. Future studies are needed to investigate whether the bivalent phenotype of PREd10 is localized to a shorter DNA region. Interestingly, we found that PRE30, 44 and 55 are within the boundary of super enhancers while PRE14 and 41 are located within enhancers found in HeLa cells (54).
TREs are enriched with YY1 motifs
DNA sequence-specific transcription factors are expected to be at least partially responsible for recruiting PRC2 and/or MLL1/2-TrxG proteins to their genomic targets (55). However, we knew very little of the transcription factors that perform these functions in mammalian cells. The DNA recognition motifs used in the initial search for candidate human PRE/TREs (Supplementary Table S1) were mapped on the core sequences of the nine PREs, P/TRE and TREs (Supplementary Table S4). Interestingly, we noticed a high and constant enrichment of the DNA motifs for Pho and Zeste among the TRE class, however a constant enrichment of DNA motifs was not found for the PRE class (Supplementary Table S4). We selected YY1, which is the human homologue of Drosophila Pho and exhibits a good level of expression in HeLa cells, for experimental verification. The ChIP-qPCR results clearly demonstrated the predominant preference of YY1 at all four TREs, to a much lesser degree at the P/TRE (PRE62), but not at any of the four PREs (Figure 6). These data confirmed previous studies that YY1 is unlikely a recruiter of mammalian PRC2 complexes (36,56,57), at least for the four PREs identified in this study.
Figure 6.
Enrichments of transcription factor YY1 at the endogenous loci of the nine human PRE/TREs shown as % input.
Interestingly, among the ∼40% of the elements that did not have activity in the luciferase assay (Supplementary Figure S1b), some elements such as PRE40, 65 or 73 also contained a similar level of enrichment for Pho and Zeste binding motifs as the four TREs (Supplementary Table S2). Therefore, the enrichment of Pho and Zeste motifs may be necessary, but not sufficient, for the activation activity mediated by MLL1/2-TrxG complex. Instead, additional unknown DNA binding factors with distinct binding motifs may be involved as well. Identifying the unknown binding factors involved in PRC2 and/or MLL1/2-TrxG recruitment will be the subjects of future studies.
TREs are enriched with CpG islands
More detailed analysis of the identified core sequences of the nine PRE/TRE fragments revealed that their GC contents were significantly distinct from each other. In particular, the four PREs (PRE14, 29, 39 and 48) had an average GC content of 55.0±3.8%, the P/TRE (PRE62) had a GC content of 47%, while the four TREs (PRE30, 41, 44 and 55) had an average GC content of 66.0 ± 4.2% (with a P value of 0.008 from the four PREs in two-tailed Student's t-test) (Supplementary Table S5). Despite the much longer length, the overall GC content of PREd10, at 42.9%, closely resembled that of PRE62.
In order to investigate whether the GC content is a determining factor for the repression or activation behaviors of the DNA fragments, we compared the GC contents of all 61 candidate PRE/TREs in Supplementary Table S6. We found that the GC content by itself is not directly related to the repression or activation activity of the DNA fragments. For instance, in terms of GC contents, PRE23, 24, 26, 27, 28, 33, 37, 45, 56, 57, 58, 65, 70, 73 and 74 are all close to the averaged GC contents for the four PREs, PRE16, 36, 40, 46, 53, 64, 69 are close to the GC content of 47% for the P/TRE, while PRE21, 52, 68, particularly their smaller fragments such as PRE21(1–156), PRE52(222–377), and PRE68(216–371), are close to the GC contents for the four TREs (Supplementary Table S6). However, all these DNA fragments did not exhibit significant luciferase activity upon FLPe excision in FLP(+) samples (Supplementary Table S6, Supplementary Figure S1b).
Interestingly, we found all TREs (PRE30, 41, 44 and 55) are located within or partially overlap with CpG islands (Supplementary Table S2). In sharp contrast, the four PREs, PRE14, 29, 39 and 48, do not overlap with any CpG islands, and are at a distance of 7.4, 35.9, 408.0 and 4.5 kb away from the nearest CpG islands, respectively. On the other hand, the P/TREs PRE62 and PREd10 are about 1.3 and 3.0 kb away from the closest CpG islands, respectively. However, CpG islands also co-localize with many other fragments (Supplementary Table S2), which, nevertheless, failed the tests for TREs. Examples included PRE34 and PRE68. One noticeable difference of PRE34 and PRE68 with the four final TREs is the absence of Pho motifs in the former (Supplementary Table S2). These data collectively suggest that CpG islands may be necessary, but not sufficient, for MLL1/2-TrxG-mediated activation.
Identification of endogenous target genes
We examined the expression level of genes within ±500 kb of each of the nine PRE/TREs by using HeLa cells harboring SUZ12 or WDR5 shRNA to discover their endogenous targets. Statistically significant increase of transcription upon SUZ12 knockdown allowed us to define the following target genes: AK023819 and LINC01819 (PRE14), carbohydrate sulfotransferase 2 (CHST2) and AK097380 (PRE29), LOC101928441 (PRE39), potassium calcium-activated channel subfamily M alpha 1 (KCNMA1) (PRE48), AK093368, AK127336 and BC127870 (PRE62) (Figure 7A, Supplementary Tables S7 and S8). Except for CHST2 and KCNMA1, all other targets are currently annotated as long non-coding RNAs. CHST2 was previously found to be among SUZ12-silenced regions in a cell-type-specific manner (58), while KCNMA1 was also previously shown to be a PRC2 target in lymphoma cells (59). In sharp contrast, the expression level of all genes proximal to the TRE class (PRE30, 41, 44 and 55) was unaffected by SUZ12 knockdown. On the other hand, WDR5 knockdown led to no significant expression changes for the targets of the PRE class, but significantly decreased transcription levels for the following target genes of the P/TRE and TRE classes: AK093368 and BC127780 (PRE62), AKT1 Substrate 1 (AKT1S1) and TBC1 domain family member 17 (TBC1D17) (PRE30), the nuclear transcription cofactor host cell factor 1 (HCF1) and transmembrane protein 187 (TMEM187) (PRE41), receptor of activated protein C kinase 1 (GNB2L1) (PRE44), heterogeneous nuclear ribonucleoproteins A2/B1 (HNRNPA2B1) and chromobox protein homolog 3 (CBX3) (PRE55) (Figure 7B, Supplementary Tables S7, S8). All these targets of the TRE class (PRE30, 41, 44 and 55) are housekeeping genes. Among them, the transcription pattern of TBC1D17 was previously found to be associated with H3K4me3 in liver (60). Furthermore, a genome locus encompassing PRE41 was previously found to regulate HCF1 (61).
Figure 7.
Identification of endogenous targets regulated by the nine human PRE/TREs.A). RT-qPCR of neighboring genes in SUZ12 knockdown HeLa cells. The genes with significantly increased transcription than in control cell line treated with scrambled shRNA were deemed as PRE or P/TRE targets. B). RT-qPCR of neighboring genes in WDR5 knockdown HeLa cells. The genes with significantly decreased transcription than in control cell line treated with scrambled shRNA were deemed as P/TRE or TRE targets. C). Schematic illustration of the strategy used to knockout PRE and P/TRE fragments from the genome loci in HeLa cells by CRISPR-Cas9. D). RT-qPCR of neighboring genes in PRE or P/TRE-knockout HeLa cells. In all panels, the expression data were represented relative to that of housekeeping GAPDH deemed as 1.0 (Relative Expression), and the 0.5 and 2.0 expression levels are shown as blue and red dashed lines, respectively. The P values of Student's t-test comparing the expression levels of target genes in knockdown (A, B) or knockout (D) HeLa cells and control cells were showed as * for P<0.05, ** for P<0.005 and *** for P<0.001.
From Supplementary Table S8, we noticed a distinct feature in terms of the distance of these three classes of PRE/TREs to the transcription start site (TSS) of the targets. While the TREs (PRE30, 41, 44 and 55) partially overlap with or are very close to the TSS of target genes, this distance is in the range of 6–142 kb for PREs (PRE14, 29, 39 and 48) and 3–19 kb for P/TRE (PRE62). The longer distance of PREs and P/TRE to TSSs allowed us to knock out the core sequences using the CRISPR-Cas9 system (Figure 7C). Relative to the wild-type cells, the knockout resulted in an increased transcription level (Figure 7D) of all the targets of the PRE and P/TRE classes identified in Figure 7A. These results thus provided additional evidence for the endogenous gene targets of the PRE and P/TRE classes.
Genome-wide identification and characterization of PRE/TREs in human K562 cells
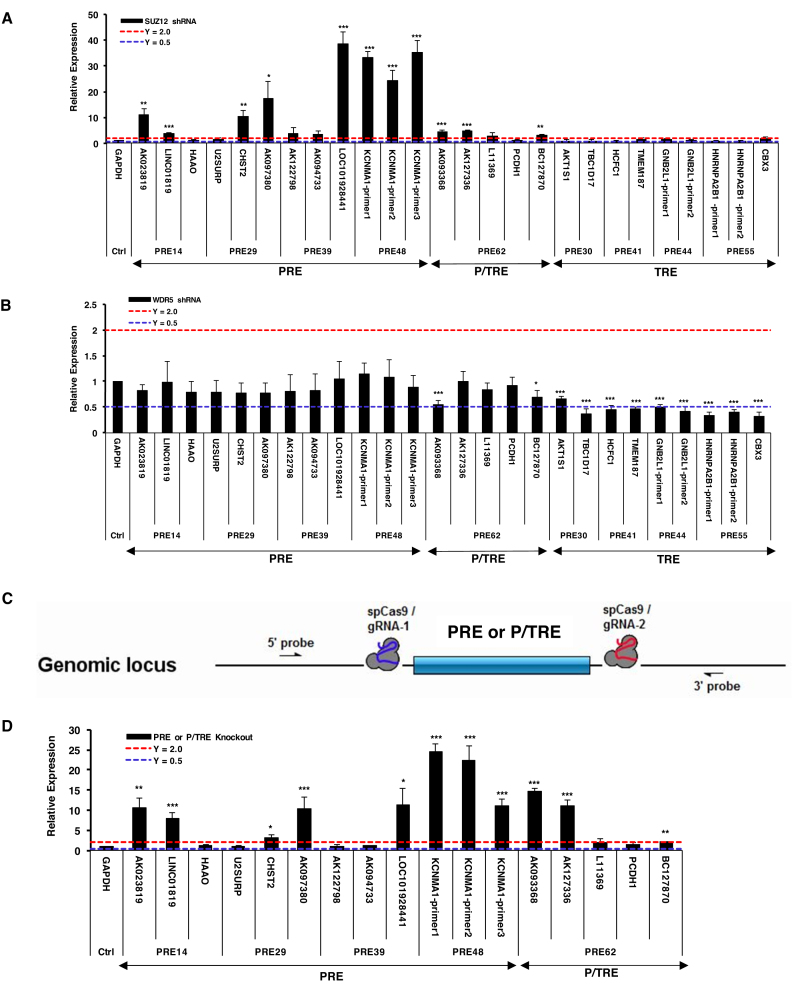
The nine cis-regulatory DNA sequences characterized in this study have two outstanding features: (a) they are of short sizes (113–348 bp); (b) each is enriched for histone marks (H3K27me3 for PREs, H3K4me3 for TREs, and both H3K27me3/ H3K4me3 for P/TREs) and the corresponding histone-modifying protein complexes. We next employed these two features to identify PRE/TREs on a genome-wide scale in human K562 cells using publically available data. By applying stringent criteria, we retrieved 15,173 putative PREs (416 bp or smaller) that were uniquely enriched for both EZH2 and H3K27me3, 10 693 putative TREs (416 bp or smaller) that were specifically enriched for MLL2 and H3K4me3, and 107 putative P/TREs that contained fully overlapping EZH2/H3K27me3 and MLL2/H3K4me3 marks (Supplementary Table S9). Interestingly, out of the nine PRE/TREs identified in this study, the region of PRE30 was among the putative TREs in K562 cells. The requirement of completely overlapping EZH2/H3K27me3 and MLL2/H3K4me3 marks in the 107 P/TREs was to ensure a genuine bivalent state, which obviously led to a substantial underestimate of the P/TRE class in the genome.
These genome-wide response elements were then used to overlap with 1309 YY1 peaks (Figure 8A–C). Strikingly, in marked contrast to the high percentage of TREs that had overlapping YY1 signals, at 3.1%, only 0.02% PREs had overlapping YY1 signals (Figure 8A and B). In addition, one of the 107 P/TREs (at 0.9%) also had overlapping YY1 signals (Figure 8C). Although the tendency of YY1 enrichment in TREs was clear, the rather small number of YY1 peaks in the original ChIP-seq data nevertheless prevented a solid conclusion. We also investigated the overlap of these three classes of genome-wide PRE/TREs with 12 852 CpG islands (500 bp or smaller). In sharp contrast to the large percentage of TREs and P/TREs, at 6.6% and 4.7%, respectively, that overlapped with CpG islands, there was only 0.3% PREs overlapping with CpG islands (Figure 8D–F). These data, together with our observation on the nine PRE/TREs, suggest that CpG islands are unlikely the key features responsible for recruiting PRC2 proteins to human PREs.
Figure 8.
Genome-wide analysis of K562 cell line. (A–C) Venn diagram showing the overlaps of YY1 with genome-wide putative PREs (A), TREs (B) and P/TREs (C). (D–F). Venn diagram showing the overlaps of CpG islands with genome-wide putative PREs (D), TREs (E) and P/TREs (F).
DISCUSSION
Three classes of PRE/TREs in human genome
The three classes discovered in this study are fundamentally distinct from each other in many aspects: (i) The distance to TSS. All members of the PRE class are located at 6–142 kb away from the TSS of the endogenous targets. The P/TRE (PRE62) is 3–19 kb away from the TSS of its targets, while the members of the TRE class are very close to, and often overlap with, the TSS site of their targets. (ii) The GC contents. The GC contents of the four PREs (PRE14, 29, 39 and 48) were ∼11% lower than those of the four TREs (PRE30, 41, 44 and 55). Similarly, we randomly selected 105 PREs and 105 TREs retrieved from genome-wide analysis of human K562 cell line and found that the GC contents of the PREs were ∼15% lower than those of TREs. (iii) Co-localization with CpG islands. All TREs (PRE30, 41, 44 and 55) are located within or partially overlap with CpG islands, while all PREs are 4.5 to 408 kb away from the nearest CpG islands. Similarly, in the genome-wide analysis of human K562 cell line, CpG islands were found to be specifically enriched in TREs, slightly less enriched in P/TREs, but at a very low level in PREs (Figure 8D–F). (iv) Their endogenous targets. The target genes of the four PREs (PRE14, 29, 39 and 48) are predominantly long non-coding RNAs. The target genes of the P/TRE (PRE62) are also non-coding RNAs. In marked contrast, the target genes of the four TREs (PRE30, 41, 44 and 55) are exclusively housekeeping genes. 5). Recognition by transcription factors. YY1 was found to be specifically enriched in all TREs (PRE30, 41, 44 and 55), but not in the four PREs (PRE14, 29, 39 and 48). Furthermore, in genome-wide analysis, YY1 was found to co-localize with TREs, but at a very low level with PREs. All these intrinsic differences among the three classes of PRE/TREs suggest that they indeed carry out distinct functions and likely represent the basic building blocks for PRC2 and MLL1/2-TrxG functions in the cell.
Whether the same three classes of PRE/TREs exist in the genome of Drosophila and other species awaits to be investigated. The previously identified Drosophila PRE/TREs in HOX clusters, also known as CMMs that are co-occupied by PcG and TrxG proteins and can switch from repressed states to activated states, likely behave as the P/TRE class in this three-class system of human response elements. The bivalent domains found in mammalian pluripotent or multipotent cells (34,50,62–68) probably harbor many modular P/TREs. However, the discovery of the P/TRE class in HeLa cells, together with the dual functionality of cis-acting elements as reported by Maini et al. and Erceg et al. (69–72), suggests that the bivalent marks of H3K27me3 and H3K4me3 have a broader role in regulating gene expression in embryonic stem cells, throughout the development and in differentiated cells.
The four PREs (PRE14, 29, 39 and 48) as described here are the first set of bona fide vertebrate PREs that recruit PRC2 proteins, are enriched for H3K27me3 marks and exhibit repressed gene expression. In marked contrast, many previously reported vertebrate PREs (reviewed in (12)) were only probed for the enrichment of PcG proteins and H3K27me3 marks. These PREs may need to be re-visited to re-classify them using the three-class system as revealed here. For instance, the previously identified PREd10 (42) in fact belongs to the P/TRE class when the involvement of MLL1/2-TrxG proteins was considered (Figure 5C, D). In addition, more members of the TRE class of human response elements are likely to be found at the TSSs of other housekeeping genes. Thus, the discovery of the three classes of human response elements not only points to much broader cellular functions that are regulated by PRC2 and MLL1/2-TrxG proteins than previously appreciated, but also suggests a new standard for characterizing vertebrate response elements.
The role of YY1 in recruiting MLL1/2-TrxG complexes
An important unresolved issue in the PcG and TrxG field is how these protein complexes are recruited to their specific genomic targets. One central controversy was around YY1. Although its Drosophila homologue, Pho, has been implicated in PcG recruitment (73), YY1’s binding motif was puzzlingly depleted in mammalian polycomb domains (34,43). Our results from both detailed characterization of the nine PRE/TREs and genome-wide analysis of human K562 cell line suggested that YY1 is likely specifically enriched at TREs, but not at PREs. These new findings, together with previous observations that YY1 was found to bind at active promoters in human genome but not PcG target genes (57), point to possibly different roles of human YY1 and Drosophila Pho in PRC2 and TrxG-mediated epigenetic regulation. Further in-depth studies are needed for mechanistic insights into these differences.
The role of CpG islands in recruiting MLL1/2-TrxG complexes
Another pivotal controversy in the field was the role of CpG islands in recruiting PRC2 complexes. Previous studies have suggested a role of CpG islands in recruiting PRC2 (34–38), while other studies argued against such a role (31,41,42). However, all these studies examined relatively large chromatin regions. Our studies on much smaller DNA fragments (113–348 bp) revealed that all four PREs (PRE14, 29, 39 and 48) are 4.5 kb to 408 kb away from the closest CpG islands. In marked contrast, all TREs (PRE30, 41, 44 and 55) are located within or partially overlap with CpG islands. Furthermore, genome-wide analysis of K562 cells found that the putative TREs and P/TREs harbor more than 20-fold enrichment of CpG islands than the putative PREs. These data collectively suggested that CpG islands are probably involved in recruiting MLL1/2-TrxG complexes to human TRE and P/TRE sites instead. This preliminary conclusion agrees very well with the overwhelming genome-wide overlaps between CpG islands and H3K4me3 peaks, but minimal overlaps between CpG and H3K27me3 (74,75). Previous studies suggested that insertion of GC-rich and CpG-rich sequences (1,000 bp) at ectopic sites was sufficient to recruit both H3K4me3 and H3K27me3 marks (40) and short DNA sequences (500–1,000 bp) capable of recruiting H3K27me3 marks also had enrichment for H3K4me3 (38). One possible explanation for these observations is that the CpG islands recruit MLL1/2-TrxG complexes to P/TREs, which somehow leads to the recruitment of PRC2 complex and the addition of H3K27me3 marks. However, the detailed mechanisms underlying this process await future elucidation.
The ‘reductionist’ approach for dissecting complex gene regulations
All the information dictating the development of a human zygote into a well-developed body is stored in the same genome that is shared by all types of cells in the body. Therefore, in order to comprehensively and precisely instruct specific gene expression in different cells, human genes carry long cis-regulatory DNA regions including promoters and enhancers with highly complex organization of binding sites for transcription factors and other regulatory factors. Consequently, the use of long cis-regulatory DNA regions mingled with multiple functions was partially responsible for the difficulties and confusions in the field of PRC2 and MLL1/2-TrxG-mediated regulation. By focusing on shorter DNA fragments of single functions in this study, the ‘reductionist’ approach is powerful in distinguishing classes of fundamentally different PRE/TREs in human genome, and in unraveling the underlying DNA signatures and transcription factors required for recruitment. Future investigations into these basic building blocks of gene regulations will provide the much-needed foundations for reconciling many controversies in the PcG and TrxG field and to elucidate how various combinations of such building blocks collectively endorse robust responses in the cell.
Supplementary Material
ACKNOWLEDGEMENTS
Authors Contributions: J.M. and Q.W. conceived and supervised the project. J.Z. and B.D.K. performed the prediction using EpiPredictor. J.D. performed all experimental characterizations and wrote the draft. B.D.K. performed genome-wide analysis of K562 cell line. J.D., B.D.K., J.M. and Q.W. analyzed the results and wrote the paper. All authors agreed on the final manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R01-GM067801, R01-GM116280, R01-GM127628] (J.M.); Welch Foundation [Q-1512] (J.M.); National Institutes of Health [R01-AI067839, R01-GM116280, R01-GM127628] (Q.W.); Welch Foundation [Q-1826] (Q.W.); postdoctoral training fellowships from the Keck Center Computational Cancer Biology Training Program of the Gulf Coast Consortia funded by CPRIT [RP101489] (B.D.K. and J.Z.). Funding for open access charge: The publication charges will be paid from the corresponding author's start-up fund.
Conflict of interest statement. None declared.
REFERENCES
- 1. Lewis P. Pc: Polycomb. Drosoph. Inf. Ser. 1949; 21:69. [Google Scholar]
- 2. Duncan I. Polycomblike: A gene that appears to be required for the normal expression of the bithorax and Antennapedia gene complexes of Drosophila melunoguster. Genetics. 1982; 102:49–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ingham P., Whittle J.R.S.. Trithorax: A new homeotic mutation of Drosophila melanogaster causing transformations of abdominal and thoracic imaginal segments. Mol. Gen. Genet. 1980; 179:607–614. [Google Scholar]
- 4. Kuzin B., Tillib S., Sedkov Y., Mizrokhi L., Mazo A.. The Drosophila trithorax gene encodes a chromosomal protein and directly regulates the region-specific homeotic gene fork head. Gene Dev. 1994; 8:2478–2490. [DOI] [PubMed] [Google Scholar]
- 5. Jürgens G. A group of genes controlling the spatial expression of the bithorax complex in Drosophila. Nature. 1985; 316:153–155. [Google Scholar]
- 6. Margueron R., Reinberg D.. The Polycomb complex PRC2 and its mark in life. Nature. 2011; 469:343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blackledge N.P., Farcas A.M., Kondo T., King H.W., McGouran J.F., Hanssen L.L., Ito S., Cooper S., Kondo K., Koseki Y. et al. . Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell. 2014; 157:1445–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kassis J.A., Brown J.L.. Polycomb group response elements in Drosophila and vertebrates. Adv. Genet. 2013; 81:83–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schuettengruber B., Bourbon H.M., Di Croce L., Cavalli G.. Genome regulation by Polycomb and Trithorax: 70 years and counting. Cell. 2017; 171:34–57. [DOI] [PubMed] [Google Scholar]
- 10. Aranda S., Mas G., Di Croce L.. Regulation of gene transcription by Polycomb proteins. Sci. Adv. 2015; 1:e1500737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Simon J.A., Kingston R.E.. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell. 2013; 49:808–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bauer M., Trupke J., Ringrose L.. The quest for mammalian Polycomb response elements: are we there yet. Chromosoma. 2016; 125:471–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Whitcomb S.J., Basu A., Allis C.D., Bernstein E.. Polycomb Group proteins: an evolutionary perspective. Trends Genet. 2007; 23:494–502. [DOI] [PubMed] [Google Scholar]
- 14. Geisler S.J., Paro R.. Trithorax and Polycomb group-dependent regulation: a tale of opposing activities. Development. 2015; 142:2876–2887. [DOI] [PubMed] [Google Scholar]
- 15. Chittock E.C., Latwiel S., Miller T.C., Muller C.W.. Molecular architecture of polycomb repressive complexes. Biochem. Soc. Trans. 2017; 45:193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Comet I., Riising E.M., Leblanc B., Helin K.. Maintaining cell identity: PRC2-mediated regulation of transcription and cancer. Nat. Rev. Cancer. 2016; 16:803–810. [DOI] [PubMed] [Google Scholar]
- 17. Piunti A., Shilatifard A.. Epigenetic balance of gene expression by Polycomb and COMPASS families. Science. 2016; 352:aad9780. [DOI] [PubMed] [Google Scholar]
- 18. Ringrose L., Paro R.. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development. 2007; 134:223–232. [DOI] [PubMed] [Google Scholar]
- 19. Kingston R.E., Tamkun J.W.. Transcriptional regulation by trithorax-group proteins. Cold Spring Harb. Perspect. Biol. 2014; 6:a019349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Entrevan M., Schuettengruber B., Cavalli G.. Regulation of genome architecture and function by Polycomb proteins. Trends Cell Biol. 2016; 26:511–525. [DOI] [PubMed] [Google Scholar]
- 21. Chang Y.L., King B.O., O’Connor M., Mazo A., Huang D.H.. Functional reconstruction of trans regulation of the Ultrabithorax promoter by the products of two antagonistic genes, trithorax and Polycomb. Mol. Cell. Biol. 1995; 15:6601–6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Orlando V., Jane E.P., Chinwalla V., Harte P.J., Paro R.. Binding of trithorax and Polycomb proteins to the bithorax complex: dynamic changes during early Drosophila embryogenesis. EMBO J. 1998; 17:5141–5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gindhart J.G. Jr., Kaufman T.C.. Identification of Polycomb and trithorax group responsive elements in the regulatory region of the Drosophila homeotic gene Sex combs reduced. Genetics. 1995; 139:797–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chinwalla V., Jane E.P., Harte P.J.. The Drosophila trithorax protein binds to specific chromosomal sites and is co-localized with Polycomb at many sites. EMBO J. 1995; 14:2056–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Strutt H., Cavalli G., Paro R.. Co-localization of Polycomb protein and GAGA factor on regulatory elements responsible for the maintenance of homeotic gene expression. EMBO J. 1997; 16:3621–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rank G., Prestel M., Paro R.. Transcription through intergenic chromosomal memory elements of the Drosophila bithorax complex correlates with an epigenetic switch. Mol. Cell. Biol. 2002; 22:8026–8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cavalli G., Paro R.. The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell. 1998; 93:505–518. [DOI] [PubMed] [Google Scholar]
- 28. Cavalli G., Paro R.. Epigenetic inheritance of active chromatin after removal of the main transactivator. Science. 1999; 286:955–958. [DOI] [PubMed] [Google Scholar]
- 29. Maurange C., Paro R.. A cellular memory module conveys epigenetic inheritance of hedgehog expression during Drosophila wing imaginal disc development. Genes Dev. 2002; 16:2672–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dejardin J., Cavalli G.. Chromatin inheritance upon Zeste-mediated Brahma recruitment at a minimal cellular memory module. EMBO J. 2004; 23:857–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sing A., Pannell D., Karaiskakis A., Sturgeon K., Djabali M., Ellis J., Lipshitz H.D., Cordes S.P.. A vertebrate Polycomb response element governs segmentation of the posterior hindbrain. Cell. 2009; 138:885–897. [DOI] [PubMed] [Google Scholar]
- 32. Woo C.J., Kharchenko P.V., Daheron L., Park P.J., Kingston R.E.. A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell. 2010; 140:99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. van Kruijsbergen I., Hontelez S., Veenstra G.J.. Recruiting polycomb to chromatin. Int. J. Biochem. Cell Biol. 2015; 67:177–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ku M., Koche R.P., Rheinbay E., Mendenhall E.M., Endoh M., Mikkelsen T.S., Presser A., Nusbaum C., Xie X., Chi A.S. et al. . Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLos Genet. 2008; 4:e1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lynch M.D., Smith A.J., De Gobbi M., Flenley M., Hughes J.R., Vernimmen D., Ayyub H., Sharpe J.A., Sloane-Stanley J.A., Sutherland L. et al. . An interspecies analysis reveals a key role for unmethylated CpG dinucleotides in vertebrate Polycomb complex recruitment. EMBO J. 2012; 31:317–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mendenhall E.M., Koche R.P., Truong T., Zhou V.W., Issac B., Chi A.S., Ku M., Bernstein B.E.. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLos Genet. 2010; 6:e1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tanay A., O’Donnell A.H., Damelin M., Bestor T.H.. Hyperconserved CpG domains underlie Polycomb-binding sites. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:5521–5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jermann P., Hoerner L., Burger L., Schubeler D.. Short sequences can efficiently recruit histone H3 lysine 27 trimethylation in the absence of enhancer activity and DNA methylation. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E3415–E3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Basu A., Dasari V., Mishra R.K., Khosla S.. The CpG island encompassing the promoter and first exon of human DNMT3L gene is a PcG/TrX response element (PRE). PLoS One. 2014; 9:e93561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wachter E., Quante T., Merusi C., Arczewska A., Stewart F., Webb S., Bird A.. Synthetic CpG islands reveal DNA sequence determinants of chromatin structure. Elife. 2014; 3:e03397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. van Heeringen S.J., Akkers R.C., van Kruijsbergen I., Arif M.A., Hanssen L.L., Sharifi N., Veenstra G.J.. Principles of nucleation of H3K27 methylation during embryonic development. Genome Res. 2014; 24:401–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schorderet P., Lonfat N., Darbellay F., Tschopp P., Gitto S., Soshnikova N., Duboule D.. A genetic approach to the recruitment of PRC2 at the HoxD locus. PLos Genet. 2013; 9:e1003951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu Y., Shao Z., Yuan G.C.. Prediction of Polycomb target genes in mouse embryonic stem cells. Genomics. 2010; 96:17–26. [DOI] [PubMed] [Google Scholar]
- 44. Beard C., Hochedlinger K., Plath K., Wutz A., Jaenisch R.. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006; 44:23–28. [DOI] [PubMed] [Google Scholar]
- 45. Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F.. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013; 8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lacroix C., Giovannini D., Combe A., Bargieri D.Y., Spath S., Panchal D., Tawk L., Thiberge S., Carvalho T.G., Barale J.C. et al. . FLP/FRT-mediated conditional mutagenesis in pre-erythrocytic stages of Plasmodium berghei. Nat. Protoc. 2011; 6:1412–1428. [DOI] [PubMed] [Google Scholar]
- 47. Stemmer W.P., Crameri A., Ha K.D., Brennan T.M., Heyneker H.L.. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene. 1995; 164:49–53. [DOI] [PubMed] [Google Scholar]
- 48. Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001; 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zeng J., Kirk B.D., Gou Y., Wang Q., Ma J.. Genome-wide polycomb target gene prediction in Drosophila melanogaster. Nucleic Acids Res. 2012; 40:5848–5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K. et al. . A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006; 125:315–326. [DOI] [PubMed] [Google Scholar]
- 51. Okulski H., Druck B., Bhalerao S., Ringrose L.. Quantitative analysis of polycomb response elements (PREs) at identical genomic locations distinguishes contributions of PRE sequence and genomic environment. Epigenet. Chromatin. 2011; 4:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Steffen P.A., Ringrose L.. What are memories made of? How Polycomb and Trithorax proteins mediate epigenetic memory. Nat. Rev. Mol. Cell Biol. 2014; 15:340–356. [DOI] [PubMed] [Google Scholar]
- 53. Sadelain M., Papapetrou E.P., Bushman F.D.. Safe harbours for the integration of new DNA in the human genome. Nat. Rev. Cancer. 2011; 12:51–58. [DOI] [PubMed] [Google Scholar]
- 54. Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-Andre V., Sigova A.A., Hoke H.A., Young R.A.. Super-enhancers in the control of cell identity and disease. Cell. 2013; 155:934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Klose R.J., Cooper S., Farcas A.M., Blackledge N.P., Brockdorff N.. Chromatin sampling–an emerging perspective on targeting polycomb repressor proteins. PLos Genet. 2013; 9:e1003717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Vella P., Barozzi I., Cuomo A., Bonaldi T., Pasini D.. Yin Yang 1 extends the Myc-related transcription factors network in embryonic stem cells. Nucleic Acids Res. 2012; 40:3403–3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kahn T.G., Stenberg P., Pirrotta V., Schwartz Y.B.. Combinatorial interactions are required for the efficient recruitment of pho repressive complex (PhoRC) to polycomb response elements. PLos Genet. 2014; 10:e1004495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Squazzo S.L., O’Geen H., Komashko V.M., Krig S.R., Jin V.X., Jang S.W., Margueron R., Reinberg D., Green R., Farnham P.J.. Suz12 binds to silenced regions of the genome in a cell-type-specific manner. Genome Res. 2006; 16:890–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Knutson S.K., Wigle T.J., Warholic N.M., Sneeringer C.J., Allain C.J., Klaus C.R., Sacks J.D., Raimondi A., Majer C.R., Song J. et al. . A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat. Chem. Biol. 2012; 8:890–896. [DOI] [PubMed] [Google Scholar]
- 60. Valekunja U.K., Edgar R.S., Oklejewicz M., van der Horst G.T., O’Neill J.S., Tamanini F., Turner D.J., Reddy A.B.. Histone methyltransferase MLL3 contributes to genome-scale circadian transcription. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:1554–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Huang L., Jolly L.A., Willis-Owen S., Gardner A., Kumar R., Douglas E., Shoubridge C., Wieczorek D., Tzschach A., Cohen M. et al. . A noncoding, regulatory mutation implicates HCFC1 in nonsyndromic intellectual disability. Am. J. Hum. Genet. 2012; 91:694–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Azuara V., Perry P., Sauer S., Spivakov M., Jorgensen H.F., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M. et al. . Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 2006; 8:532–538. [DOI] [PubMed] [Google Scholar]
- 63. Sanz L.A., Chamberlain S., Sabourin J.C., Henckel A., Magnuson T., Hugnot J.P., Feil R., Arnaud P.. A mono-allelic bivalent chromatin domain controls tissue-specific imprinting at Grb10. EMBO J. 2008; 27:2523–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Vastenhouw N.L., Schier A.F.. Bivalent histone modifications in early embryogenesis. Curr. Opin. Cell Biol. 2012; 24:374–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Roh T.Y., Cuddapah S., Cui K., Zhao K.. The genomic landscape of histone modifications in human T cells. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:15782–15787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pan G., Tian S., Nie J., Yang C., Ruotti V., Wei H., Jonsdottir G.A., Stewart R., Thomson J.A.. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007; 1:299–312. [DOI] [PubMed] [Google Scholar]
- 67. Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P. et al. . Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007; 448:553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhao X.D., Han X., Chew J.L., Liu J., Chiu K.P., Choo A., Orlov Y.L., Sung W.K., Shahab A., Kuznetsov V.A. et al. . Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell. 2007; 1:286–298. [DOI] [PubMed] [Google Scholar]
- 69. Bengani H., Mendiratta S., Maini J., Vasanthi D., Sultana H., Ghasemi M., Ahluwalia J., Ramachandran S., Mishra R.K., Brahmachari V.. Identification and validation of a putative polycomb responsive element in the human genome. PLoS One. 2013; 8:e67217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Maini J., Ghasemi M., Yandhuri D., Thakur S.S., Brahmachari V.. Human PRE-PIK3C2B, an intronic cis-element with dual function of activation and repression. Biochim. Biophys. Acta. 2017; 1860:196–204. [DOI] [PubMed] [Google Scholar]
- 71. Erceg J., Pakozdi T., Marco-Ferreres R., Ghavi-Helm Y., Girardot C., Bracken A.P., Furlong E.E.. Dual functionality of cis-regulatory elements as developmental enhancers and Polycomb response elements. Genes Dev. 2017; 31:590–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Jaensch E.S., Kundu S., Kingston R.E.. Multitasking by Polycomb response elements. Genes Dev. 2017; 31:1069–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Fritsch C., Brown J.L., Kassis J.A., Muller J.. The DNA-binding polycomb group protein pleiohomeotic mediates silencing of a Drosophila homeotic gene. Development. 1999; 126:3905–3913. [DOI] [PubMed] [Google Scholar]
- 74. Thomson J.P., Skene P.J., Selfridge J., Clouaire T., Guy J., Webb S., Kerr A.R., Deaton A., Andrews R., James K.D. et al. . CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature. 2010; 464:1082–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Illingworth R.S., Gruenewald-Schneider U., Webb S., Kerr A.R., James K.D., Turner D.J., Smith C., Harrison D.J., Andrews R., Bird A.P. et al. . Orphan CpG islands identify numerous conserved promoters in the mammalian genome. PLoS Genet. 2010; 6:e1001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.