Abstract
SOX9 is known as a crucial transcription factor for various developmental processes and for tissue homeostasis. We examined here its potential role in alternative splicing by analyzing global splicing changes, using RNA-seq of colon tumor cells. We show that SOX9 knockdown alters the splicing of hundreds of genes without affecting their expression levels, revealing that SOX9 controls distinct splicing and transcriptional programs. SOX9 does not affect splicing patterns through the control of splicing factors expression. We identify mutants that uncouple SOX9 splicing function from its transcriptional activity. We demonstrate that SOX9 binds to RNA and associates with several RNA-binding proteins, including the core exon junction complex component Y14. Half of SOX9 splicing targets are also modulated by Y14 and are no longer regulated by SOX9 upon Y14 depletion. Altogether, our work reveals that SOX9 is a moonlighting protein which modulates either transcription or splicing of distinct sets of targets.
INTRODUCTION
SOX9 is a member of the SOX proteins family for SRY-related HMG (high-mobility group) proteins (1). Since its discovery 30 years ago, SOX9 has been described as a key player during embryogenesis, especially in the maintenance of the progenitor pool and in cell differentiation (2), chondrogenesis (3), male sex determination (4), neural development (5, 6) and biliary morphogenesis (7). SOX9 is crucial, not only during development but also in mature organs, particularly in stem cells. Indeed, SOX9 has important roles in homeostasis and maintenance of the pool of progenitors in various tissues (2). In the intestinal epithelium, SOX9 is mostly expressed in progenitor cells at the bottom of the crypts, as well as in differentiated Paneth cells where it controls their differentiation (8,9). Consistent with SOX9 pleiotropic roles during development and in adulthood, deregulation of SOX9 expression has physiopathological consequences. SOX9 heterozygous mutations cause campomelic dysplasia (1), a lethal disorder that involves severe skeletal malformations and sex reversal. In contrast, SOX9 overexpression leads to fibrosis in the liver and SOX9 is overexpressed in various types of cancer, including colorectal cancer (2). SOX9 has been shown to have oncogenic properties. It drives breast cancer dissemination and endocrine resistance (10), regulates lung cancer cell plasticity (11) and promotes metastasis in colon carcinoma (12). However, the exact role of SOX9 in tumorigenesis remains debated, particularly its effect on cell proliferation. For instance, SOX9 overexpression promotes (13,14) or suppresses (8,15) cell proliferation depending on the tumor type, the cell line or the basal level of SOX9 expression.
SOX transcription factors bend DNA through the interaction of their HMG domains with the minor groove of the DNA helix at the consensus-binding motif (A/T)(A/T)CAA(A/T)G (16). SOX proteins are pioneer factors as they are able to bind compact silent chromatin and recruit non-pioneer transcription factors to drive cell fate decisions (17). Recent ChIP-seq analyses in a developmental context (14,18) and in a colorectal cancer cell line (19) have reported that SOX9 binds to different sites and modulates expression of distinct genes, depending on which partners it associates with. Therefore, the SOX9 regulatory networks are more complex than expected and likely depend on cellular context.
Fifteen years ago, the Sassone–Corsi group demonstrated a direct role for SRY, SOX6 and SOX9 in splicing using in vitro splicing assay (20). Later, SOX9 was shown to cooperate with the RNA-binding protein p54nrb/NONO to modulate the splicing of the SOX9 transcriptional target Col2a1 (21). More recently, a global analysis has shown that SOX9 depletion leads to splicing changes in Sertoli cells (18). However, none of these studies addressed how SOX9 regulates alternative splicing and, most importantly, whether this function of SOX9 is coupled to its transcriptional activity. Here, we demonstrate that SOX9 affects alternative splicing of hundreds of genes independently of its transcriptional activity. We also show that SOX9 modifies splicing patterns through its association with bona fide splicing factors, including the exon junction complex (EJC) component Y14.
MATERIALS AND METHODS
Antibodies and plasmids
For proximity ligation assay (PLA), we used mouse monoclonal anti-SOX9 (Sigma-Aldrich), anti-p54nrb (BD Transduction Laboratories™), anti-PSF (Sigma-Aldrich) and anti-Y14 (Abcam) antibodies, as well as polyclonal rabbit anti-SAM68 (Santa Cruz Biotechnology, INC), anti-PSP1 (22) and anti-SOX9 (Merck) antibodies. A rabbit anti-FLAG (Sigma-Aldrich) antibody was used for RNA immunoprecipitation assays. For western blots, we used a rabbit anti-SOX9 antibody (Merck) to detect the endogenous SOX9 protein, monoclonal anti-FLAG M2 (Sigma-Aldrich) to detect overexpressed FLAG-SOX9 mutants, as well as rabbit polyclonal anti-GFP (Torrey pines Biolabs Inc.), anti-PSF (Atlas Antibodies), anti-GAPDH (Cell Signaling) and mouse anti gamma-tubulin (Sigma) antibodies.
N-terminally FLAG-tagged wild-type (wt) SOX9 was cloned into pcDNA3 vector (23) and used to generate SOX9 mutants using the QuickChange® II XL site-directed mutagenesis kit (Agilent Technologies). Point mutations were made to generate the indicated amino acid changes. Deletion mutants were obtained by inserting stop codons. SOX9 W143R and MiniSOX9 constructs were previously described (24). The ZDHHC16 minigene, containing exon 7, its flanking introns and exons 6 and 8, as well as the SOX9 mutants DelDIM, K68E and R94H were generated by gene synthesis and cloned into pcDNA3.1 vector (GenScript). The EIF4A3, MAGOH and Y14 open reading frames were cloned downstream of the GFP coding region into the peGFP-C3 plasmid.
Cell culture and transfections
DLD-1 and HEK293T cells were maintained in DMEM Glutamax (ThermoFisher Scientific) and supplemented with penicillin/streptomycin (Thermo Fisher Scientific) and 10% FCS (EUROBIO) at 37°C in a 5% CO2 atmosphere.
For siRNA knockdown, DLD-1 or HEK293T cells were plated in 6-well plates 24 h before transfection and transfected with 50 nM of siRNA using INTERFERin® (Polyplus Transfections) according to the manufacturer’s protocol. Cells were then harvested 72 h after transfection for RNA isolation and protein analysis. HEK293T cells were transfected with SOX9 plasmids 48 h later.
For plasmid transfection, pcDNA3-SOX9 wt or SOX9 mutants and pcDNA3-ZDHHC16-Minigene constructs were transfected into HEK293T cells in 6-well plates. 400 000 cells were plated 24 h prior to transfection with a mix of 1 μg plasmid DNA and 6 μl of jetPEI® (Polyplus Transfections). Cells were harvested 24 h later for RNA isolation or protein analysis.
Luciferase assay
HEK293T cells were co-transfected as described above with 0.5 μg wt or mutant SOX9 constructs and 0.5 μg SOX reporter (SOX) or control vector (SAC). A total of 0.025 μg pRLSV-Renilla was used as an internal control. Luciferase assays were performed with the Dual-Luciferase® Reporter Assay System (Promega) according to the manufacturer’s instructions. Luciferase activities in cell lysates were normalized relative to the Renilla luciferase activity, and the indicated activities represent the SOX/SAC ratio, indicative of the SOX9 binding site-specific activity. The ‘SOX-luciferase’ reporter construct consists of seven copies of the AACAAAG SOX-binding sequence, inserted upstream of a minimal herpes simplex thymidine kinase promoter. The ‘SAC-luciferase’ control construct consists of seven copies of the CCGCGGT sequence.
Proximity Ligation assay (PLA)
The association between SOX9 and its potential partners was tested by PLA using the Duolink® green kit (Sigma-Aldrich) according to manufacturer’s instructions. Green fluorescence indicates the association of the two proteins of interest if they are no further than 40 nm apart. As a control for non-specific fluorescent background, each primary antibody was used alone. Briefly, DLD-1 cells were plated on glass coverslips at various densities and fixed 24 h later with 4% paraformaldehyde (PFA) for 10 min, then permeabilized in phosphate-buffered saline (PBS)/Triton 1% for 5 min and then blocked for 45 min in PBS/bovine serum albumin (BSA) 5% at room temperature. Cells were then incubated with primary antibodies against SOX9 and its potential partners (for antibodies references see above). Antibodies were diluted in PBS/BSA 10% buffer and incubated for 1 h at room temperature. After incubation with the mouse and rabbit secondary antibodies conjugated to PLA probes, ligation and amplification, cells were mounted in ProLong® Gold antifade medium supplemented with Hoechst (Sigma-Aldrich) at a concentration of 1 μg/ml. Fluorescence was analyzed using a ZEISS Axioimager Z2 wide-field microscope.
RNA extraction and reverse transcription
Total RNA extraction was performed using an RNeasy Plus Mini Kit (Qiagen) according to the manufacturer’s instructions after homogenization of each sample with a QIAshredder column (Qiagen). A DNase step was additionally performed during RNA isolation as described by the manufacturer. After RNA denaturation during 5 min at 65°C, 1 to 2 μg of total RNA was converted to cDNA using SuperScript™ III (Thermo Fisher Scientific) for 1 h at 50°C and the enzyme was then inactivated for 5 min at 85°C.
PCR and quantitative RT-PCR
cDNAs were diluted 5-fold and 2 μl of diluted cDNA were then used for each polymerase chain reaction (PCR) or quamtitative PCR reaction. All primers were designed using Primer3 with default parameters (see a list in Supplementary Table S1).
PCR reactions were performed using the Taq CORE kit (MP Biomedicals) according to the manufacturer protocol. A first cycle of 15 min at 95°C was followed by 35 cycles of 30 s at 94°C, 30 s at 55°C and 30s at 72°C. The reaction was ended with an extension step for 3 min at 72°C. Visualization and quantification of amplified products was performed with a LabChip HT DNA assay on an automated microfluidic station (25) (Caliper, Hopkinton, MA, USA). For all experiments, quantifications of PCR triplicates are shown as a mean of their percent spliced in (PSI) or percentage spliced exclusion (PSE). The ΔPSI value was obtained by subtracting the PSI value of SOX9 KD from that of the corresponding control.
Quantitative RT-PCR reactions were performed using LightCycler® 480 SYBR Green Master Mix and amplified on the LightCycler® 480 instrument (Roche) with primers listed in Supplemental Table S1. Relative levels of gene expression were analyzed using the 2ΔΔCt method and compared to the expression of the human housekeeping gene MRPL19. All analyses were performed according to the MIQE guidelines (26).
RNA-seq and bioinformatics
High-throughput sequencing of RNA libraries was performed following the standard protocol from Fasteris (www.fasteris.com). All sequencing runs were performed on an Illumina HiSeq 2500 with a minimum percentage of bases above a quality score of 30 (Q30) of about 90%. Sequencing length was 100 bases for siSOX9 RNAseq and 125 bases for siY14.
Raw sequences were evaluated with FastQC for the quality of the sequencing and for sequencing adaptor contamination. Low quality sequencing reads were trimmed and sequencing adaptor sequences were removed with fastq-mcf (www.expressionanalysis.github.io/ea-utils/) (Parameters: -D 100 -l 50 for siSOX9-RNA-seq experiments and -l 125 for siY14 RNA-seq experiments).
High quality sequencing reads were then aligned with Tophat2 (Bowtie1) (https://ccb.jhu.edu/software/tophat/index.shtml) and Hisat2 (https://ccb.jhu.edu/software/hisat2/index.shtml) for siSOX9 reads and siY14 reads, respectively. The BAM files produced by the aligners were sorted with samtools. Differential alternative splicing events were quantified with rMATS (http://rnaseq-mats.sourceforge.net/) (parameters: -t single -len 100 for siSOX9 or -len 125 for siY14).
Aligned reads (BAM files) were converted into SAM format with samtools view (http://samtools.sourceforge.net/), and read counting of transcripts was performed with htseq-count (http://chipster.csc.fi/manual/htseq-count.html). Differential expression analyses were then performed with DESeq on three replicates per condition (http://bioconductor.org/packages/release/bioc/html/DESeq.html).
ChIP-seq data from Shi Z. et al (19) were retrieved through SRA (sequence repository archive) under the reference numbers SRR1663101 and SRR1663102. SRA files were converted into the original fastq files with the Fastq-dump utility. Fastq-mcf was used to remove duplicate reads from the input and SOX9-ChIP sequencing files (parameter: -D 49). Reads were then mapped to the hg19 human reference genome with Bowtie (more than 96% aligned reads). Enriched ChIP-seq peaks were analyzed with Macs14 (http://liulab.dfci.harvard.edu/MACS/00README.html) (Parameters: -t ChIP-SOX9_uniq.sam -c Input-SOX9_uniq.sam -n ChIP-SOX9_uniq_peaks_pval_1e-7 -f SAM -g hs -s 50 -w -S -p 1e-7). ChIP-seq first peak distance from the transcription start sites (TSS) of SOX9 splicing targets or the center of SOX9-dependent alternatively spliced exons were computed with the Bedtools closest utility (http://bedtools.readthedocs.io/en/latest/content/tools/closest.html?highlight=closest) (Parameter: -d). Closest peak distances from spliced exons or TSS were compared to closest peak distances from randomly generated bed sequences with the Bedtools shuffle utility (Parameters: -i SE_MATS.bed -g hg19.chrom). Twenty runs of shuffled sequences were used to compute the expected random peak distances.
RNA-IP
HEK293T cells were transfected with wt SOX9 or its mutants together with the ZDHHC16 minigene reporter constructs. Twenty-four hours later, cells were harvested and cross-linked for 10 min in 1% formaldehyde and the reaction was blocked with 10 mM final Glycine. Nuclear extracts were then prepared using, successively, a lysis buffer (Tris 50 mM pH8, NaCl 100 mM, MgCl2 5 mM, 0.5% NP-40, Protease inhibitor (Roche) and RNAsin 50U/ml (Promega)) and FA lysis buffer (Tris 50 mM pH8, NaCl 140 mM, ethylenediaminetetraacetic acid (EDTA) 1 mM, 1% Triton X-100, 0.1% Na-deoxycholate, Protease inhibitor (Roche) and RNAsin 50U/ml (Promega)). Extracts were then sonicated for 8 min (30s on/30s off) in a Bioruptor® twin sonicator (Diagenode). The nuclear extracts were then incubated with 10 μg of rabbit anti-FLAG antibody overnight and 2 h with Dynabeads™ coupled with Protein A (Thermo Fisher Scientific) on a rotating wheel at 4°C. Beads were then washed in RIPA buffer (FA lysis buffer with 0.1% sodium dodecyl sulphate (SDS)), RIPA high salt buffer (RIPA buffer with NaCl 500mM) and Tris/EDTA 10:1 and eluted in NaHCO3 100 mM with SDS 1%. The reverse crosslink reaction was performed for 2 h at 65°C with proteinase K (Thermo Fisher Scientific) and RNA was extracted using TRIzol according to the manufacturer instructions (Thermo Fisher Scientific). Any contaminating DNA was then removed using DNA-free™ DNA Removal Kit and RNA was reverse-transcribed using a SuperScript® VILO™ cDNA synthesis kit as recommended (Thermo Fisher Scientific). Quantitative PCR was then performed as described above and results are expressed as a percentage of the input.
Co-immunoprecipitation
HEK293T cells were transfected with wt SOX9 and pEGFP-C3 plasmids expressing GFP alone or in fusion with EIF4A3, MAGOH or Y14. Twenty-four hours later, cells were harvested with a lysis buffer containing Tris 50 mM pH8, NaCl 100 mM, MgCl2 5 mM, 0.5% NP-40 and protease inhibitors (Roche). After centrifugation, protein extracts were incubated with GFP-Trap® agarose beads (Chromotek) and beads were washed four times in Tris 50 mM buffer.
Statistics
Differences between groups in Figure 3 and Supplementary Figure S4 were analyzed relatively to wt SOX9 or control conditions using one-way ANOVA test with a Dunnett test for post-hoc comparisons. In Figure 6, differences between groups were tested using one-way ANOVA with a Bonferroni post-hoc multiple comparison test. ns = non significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
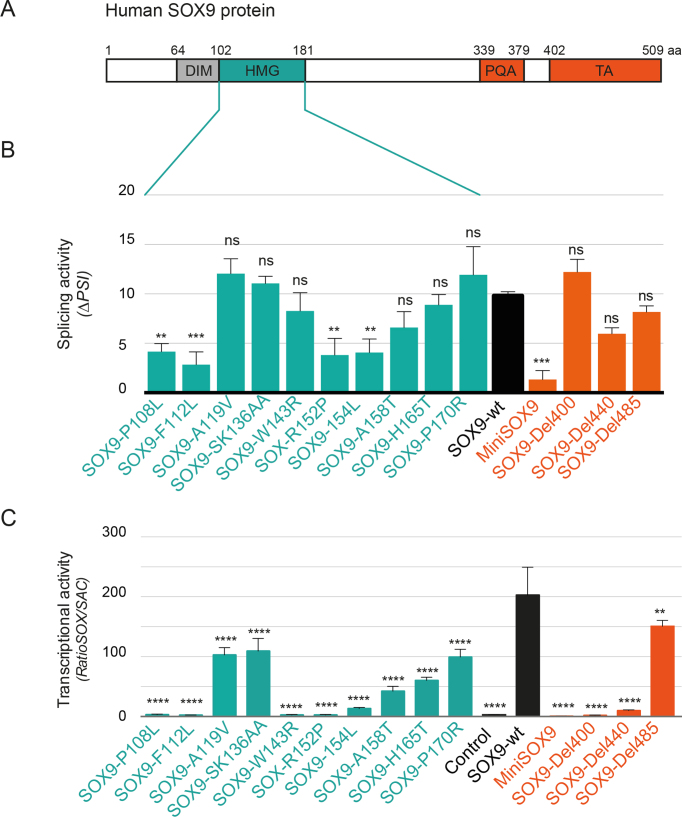
Figure 3.
The HMG and C-terminal domains of SOX9 are required for its splicing activity. (A) Domain organization of the human SOX9 protein, including the dimerization domain (DIM) in gray, the HMG domain in green and the proline-, glutamine- and alanine-rich domain (PQA) and the TA in orange. Amino acids positions are indicated. (B) Mutational analysis of SOX9 splicing activity. HEK293T cells were transfected with wt SOX9 or SOX9 mutants (point mutations in the HMG domain (green) and C-terminal deletions (orange)). The levels of the PTBP2 mRNA variants were then evaluated by RT-PCR and quantified as PSI. ΔPSI represented here were normalized to their corresponding control conditions. Data are shown as means ± SEM (n ≥ 8). ***P < 0.001 and **P < 0.01 compared with SOX9-wt (one-way ANOVA test with a Dunnett post-test). (C) Mutational analysis of SOX9 transcriptional activity. HEK293T cells were co-transfected with wt or SOX9 mutants and either with SOX-luciferase or SAC-luciferase reporter constructs. The ratio of SOX/SAC luciferase activity is shown. Each experiment was performed in triplicates and repeated at least three times, and representative examples are shown. Data are presented as means ± SEM. ****P < 0.0001 and **P < 0.01 compared to SOX9-wt (one-way ANOVA test with a Dunnett post-test).
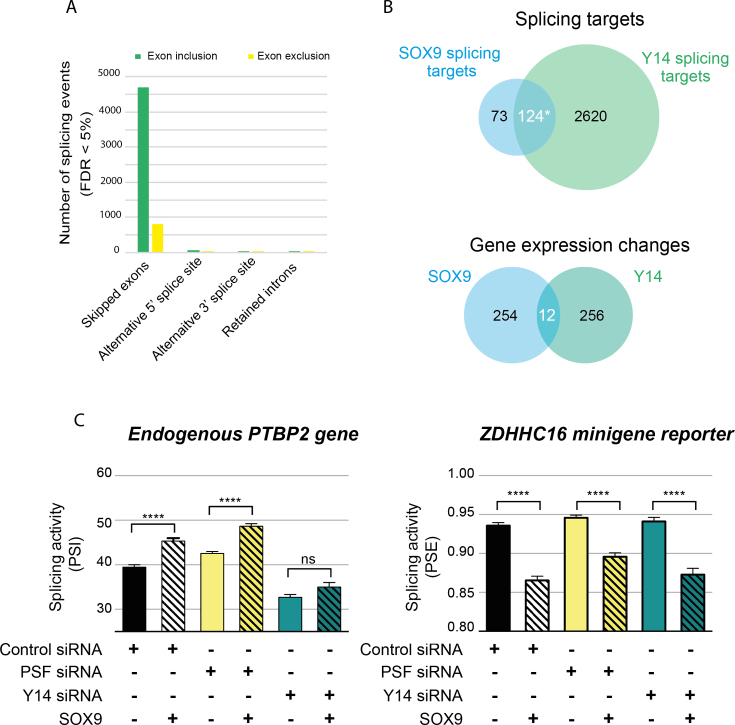
Figure 6.
SOX9 and Y14 regulate the splicing of a common set of genes. (A) MATS analysis of RNA-seq data of DLD-1 cells upon Y14 knockdown. The inhibition of Y14 expression induces thousands of splicing changes, which are mostly in exon cassettes. In green, splicing events with a positive ΔPSI in MATS analysis represent the induction of exon inclusion by Y14. In contrast, exon exclusions are represented in yellow. (B) Venn diagrams showing the overlap of SOX9 and Y14 splicing targets and SOX9 and Y14 gene expression changes. Y14 shares 60% of SOX9 splicing targets but <5% of its transcriptional targets. *The probability of incorrectly concluding that SOX9 and Y14 splicing targets overlap was determined using a two-sided t-test against the bootstrapped distribution of common targets between SOX9 and 2744 randomly sampled genes and is P = 1.3e–142. (C) Functional interaction between SOX9 and Y14. HEK293T cells were co-transfected with control, anti-PSF or anti-Y14 siRNAs, together with either a control plasmid or wt SOX9. The levels of mRNA variants of the endogenous PTBP2 and ZDHHC16 reporter minigene were quantified by RT-PCR. PSI and PSE were then calculated for PTBP2 and ZDHHC16, respectively. Data are presented as means ± SEM (n ≥ 5). ****P < 0.0001 with a one-way ANOVA test with Bonferroni’s multiple comparison test.
We tested the hypothesis that SOX9 and Y14 could interact with the same targets by chance with the following procedure (Figure 6). We repeatedly selected 2744 random targets from all human genes. One hundred of these random samples were compared with the set of SOX9 interacting targets to count the number of common targets. The normal distribution of these counts was used in a two-sided t-test with the number of common interacting targets of SOX9 and Y14 (124). The P-value is 1.3e-142 and considered significant.
RESULTS
SOX9 knockdown induces alternative splicing changes
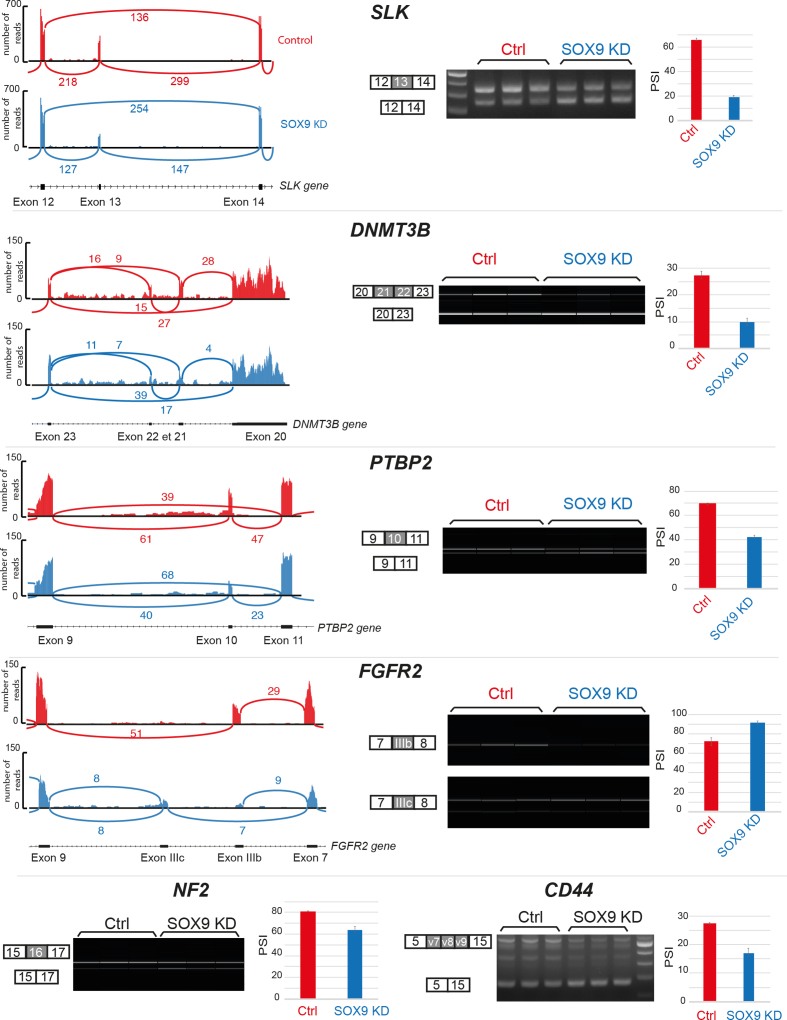
To evaluate the effect of SOX9 on alternative splicing at a transcriptome scale, we performed RNA-seq experiments in DLD-1 cells in which SOX9 is knockdown. DLD-1 colon tumor cells, which express SOX9 strongly, were transfected with a SOX9-targeting siRNA, leading to a reduction of SOX9 mRNA levels of about 70% and of SOX9 protein levels of about 90% (Supplementary Figure S1A and B). We generated over 150 million reads for control and SOX9 knockdown (SOX9 KD) samples each. We analyzed alternative exon usage by MATS analysis (27) and found that the inhibition of SOX9 induced hundreds of splicing changes (Supplementary Figure S1C and Table S2). Changes were mostly skipped exons (243 events) and for a minor part, alternative 5′ and 3′ splice site and retained introns (28 events). SOX9 affected exon cassette inclusion and exclusion equally, with 138 and 105 events, respectively (Supplementary Figure S1C). We confirmed the effect of SOX9 on splicing of a restricted number of specific targets. To this aim, we used RT-PCR to test 21 coding exons that are longer than 10 bases, detected at P < 0.05, a false discovery rate (FDR) < 10% and a differential inclusion level > 30%. These criteria defined a signature of the major splicing targets of SOX9 (Figure 1). This signature comprises six alternatively spliced genes: SLK exon 13, DNMT3B exons 21 and 22, PTBP2 exon 10, FGFR2 exons IIIb and IIIc, NF2 exon 16 and CD44 exons v7, v8, v9. RT-PCR validation of these six genes is presented in Figure 1, including the corresponding sashimi plots that illustrate the RNA-seq data. As an example, the sashimi plot of SLK shows that the amount of transcripts excluding exon 13 increased 2-fold in SOX9 KD cells, while the amount of transcripts including exon 13 decreased to the same extent, leading to a differential level of exon inclusion of 34%, as calculated by the MATS algorithm. We indeed observed a similar difference in the PSI (ΔPSI) score after quantification by RT-PCR (45%, Figure 1). Interestingly SOX9 KD did not significantly affect the overall levels of SLK transcripts (Supplementary Figure S1D). We validated this signature using a second siRNA against SOX9, in two additional colorectal cell lines (HCT116, SW480) and a mammary tumor cell line(MCF7) (Supplementary Figure S1E and F).
Figure 1.
SOX9 knockdown modifies splicing patterns in colon tumor cells. Sashimi plots, obtained from RNA-seq datasets, show the major splicing targets of SOX9 as examples and include SLK, DNMT3B, PTBP2 and FGFR2. Histograms represent the number of reads per exons and the reported values indicate the number of sequencing reads per splice junctions. RT-PCR validations are shown using standard electrophoresis (SLK and CD44) and virtual migration using a Caliper (DNMT3B, PTBP2, FGFR2 and NF2). Histograms show quantifications of each RT-PCR measurements, computed as a mean of PSI from three biological replicates ± SD.
Overall, these results suggest that SOX9 modulates the splicing pattern of specific transcripts and defines a consistent and specific splicing signature.
SOX9 affects splicing independently of its transcription factor activity
SOX9 is a well-characterized transcription factor (3,17). Therefore, we first suspected that the observed splicing changes are a consequence of an altered expression of splicing factors and/or regulators, which would in turn modulate alternative splicing. We thus performed a RNA-seq differential expression analysis to determine the set of transcripts which expression depends on SOX9 (Figure 2 and Supplementary Table S2). Genes whose expression changes at least 2-fold, with a P-value adjusted <0.05 (Figure 2A) were considered significantly deregulated. Remarkably, whereas SOX9 knockdown affects the expression of hundreds of genes, we found no significant expression changes of the 309 known splicing regulators expressed in DLD-1 cells (green dots in Figure 2A). Distribution of their mRNA levels followed an x = y linear fit (slope = 0.9997) between control and SOX9 KD conditions (Supplementary Figure S2). This suggests that SOX9 does not control the expression of splicing regulators and that it may have a more direct role in alternative splicing. Interestingly, SOX9 regulates the splicing of some splicing factors (Supplementary Table S2; i.e. PTBP1, PTBP2, MBNL1, ESRP1, SR proteins and hnRNPs). Some splicing changes observed upon SOX9 KD might thus result from the alternative splicing of specific splicing factors.
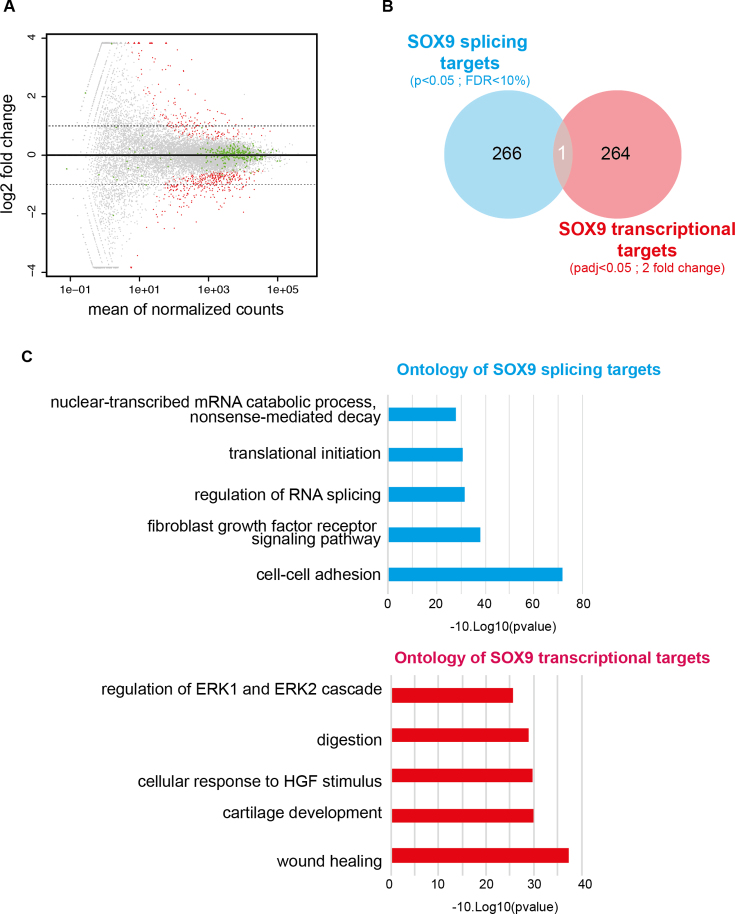
Figure 2.
SOX9 regulates splicing independently of its transcriptional activity. (A) Mean Average (MA) plot of the transcriptomic analysis of DLD-1 cells upon SOX9 depletion. Each gene (gray dots) is plotted according to its level of expression (x-axis) and to the log2 of its fold change upon SOX9 inhibition (y-axis). Red dots show gene expression changes with a padj < 0.05. The 2-fold thresholds are depicted using gray dotted lines. Green dots highlight expression changes of the 309 splicing factors expressed in DLD-1 cells. (B) Venn diagram showing the overlap between SOX9 splicing targets and SOX9 transcriptional targets. SOX9 controls both the expression and the splicing of only one gene. (C) Ontology analysis (DAVID) of SOX9 splicing and transcriptional targets. SOX9 transcriptional targets are enriched in signaling and developmental response genes whereas its splicing targets are associated with mRNA production, translation and cell–cell adhesion gene categories.
It has been previously described that SOX9 regulates both the splicing and the transcription of Col2a1 in chondrocytes (21). To determine whether SOX9 regulates the splicing of its transcriptional targets globally, we compared its transcriptional targets (padj < 0.05 and 2-fold threshold) to its splicing targets (P < 0.05 and False Discovery Rate (FDR) < 10%). Clearly, we observed that the SOX9-dependent splicing and transcriptional programs are distinct. CCDC50 was the only gene for which both transcription and alternative splicing are affected (Figure 2B and Supplementary Table S2). We conclude from this analysis that SOX9 controls distinct transcriptional and splicing programs. Supporting this idea, splicing and transcriptional SOX9 targets distribute in different ontology classes. SOX9 transcriptional targets are enriched in genes implicated in signaling and developmental responses, whereas SOX9 splicing targets are associated with mRNA metabolism, including splicing, surveillance, decay, and translation, and cell–cell adhesion.
Altogether, our results suggest that SOX9 regulates splicing independently of its transcriptional activity and that these two functions of SOX9 likely impacts distinct cellular processes.
SOX9 HMG and C-terminal domains are involved in its splicing activity
We next sought to identify which domains of SOX9 are involved in splicing. We selected 10 mutants with substitutions in the HMG domain that are known to affect DNA binding, four serial deletions in the C-terminal transactivation domain (TA), two substitutions in the dimerization domain or its complete deletion (Figure 3A). Most of these mutations cause campomelic dysplasia (28,29) and/or have been reported in colorectal cancer patients (30). Of note, the MiniSOX9 isoform corresponds to a C-terminal deletion from amino acid 229 and is also a splice variant produced by the SOX9 locus (24). Importantly, all SOX9 mutants were expressed at least as much as wt SOX9 (Supplementary Figure S3).
We measured the splicing activity of SOX9 mutants using either endogenous PTBP2 (Figure 3B) or the ZDHHC16 minigene reporter construct (Supplementary Figure S4A). The latter consists of ZDHHC16 alternative exon 8 with its flanking exons and introns under the control of a constitutive promoter. All 14 HMG and C-terminal SOX9 mutants tested showed reduced splicing activity on the minigene reporter (Supplementary Figure S4A). Only five of them showed significantly reduced activity on PTBP2 splicing (SOX9-P108L, -F112L, -R152P, -F154L and MiniSOX9; Figure 3B). Substitutions or deletion of the dimerization domain did not affect SOX9 splicing activity on the minigene reporter and on the endogenous PTBP2 gene (Supplementary Figure S4D). These results indicate that both the HMG and the C-terminal domains are involved in SOX9 splicing activity. In contrast, SOX9 dimerization domain might not contribute to this function.
We next confirmed that, as expected from previous work, the HMG and C-terminal domain mutants show transcriptional defects, using the SOX luciferase reporter assay (SOX/SAC ratio) (24). Half of the HMG mutations and three out of four C-terminal deletions abolished SOX9 transcriptional activity while the others had only partial effects (Figure 3C). Most importantly, the three mutants SOX9-W143R, -Del400 and –Del440 showed almost no transcriptional activity whereas their splicing activities are similar to that of wt SOX9 (Figure 3). In conclusion, we uncoupled the transcriptional and splicing activities of SOX9 with a single amino acid substitution in the HMG domain (SOX9-W143R). This observation strengthens our conclusion that SOX9 regulates alternative splicing and transcription independently.
SOX9 binds the RNA of its regulated exons and the DNA of the corresponding promoters
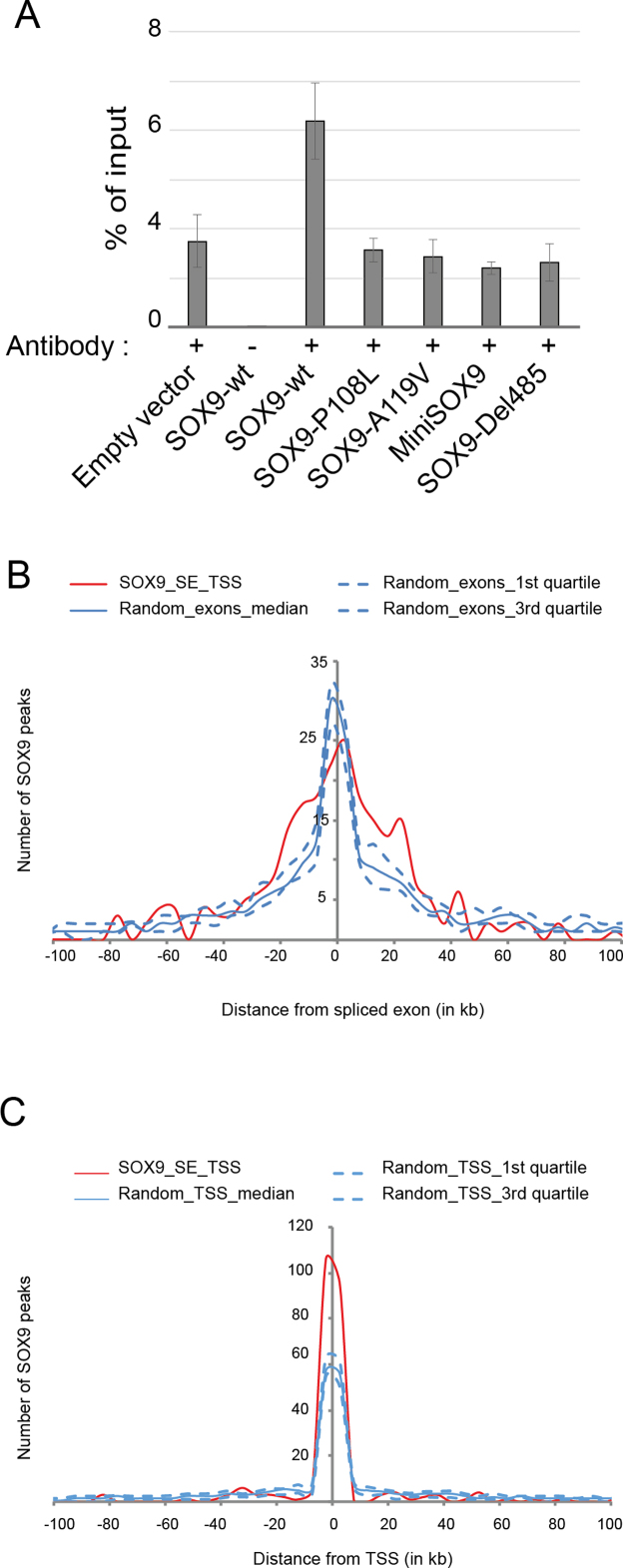
It has been previously speculated that the SOX family of proteins bind RNA (18,20,31). Because we found that SOX9 regulates alternative splicing, we used RNA immunoprecipitation to determine if SOX9 binds RNA. We transfected HEK293T cells with the ZDHHC16 minigene and FLAG-SOX9 constructs and immunoprecipitated SOX9-bound RNAs using a polyclonal FLAG antibody (Figure 4A). We observed that wt SOX9 binds the ZDHHC16 minigene RNA, as compared to control samples, including cells expressing empty FLAG vector or an IP without FLAG antibody. Interestingly, two HMG point mutations (P108L and A119V) and two C-terminal deletions (MiniSOX9 and Del485) reduced RNA binding of SOX9, suggesting that both the HMG and C-terminal domains are necessary for binding to RNA. However, we noted that SOX9 interaction with RNA is dispensable for its splicing activity on PTBP2, as shown in the SOX9-A119V and SOX9-Del485 mutants, which do not bind RNA but show normal splicing activity. This indicates that the splicing activity does not strictly rely on SOX9 capacity to bind RNA.
Figure 4.
Analysis of SOX9 RNA- and DNA-binding. (A) SOX9 RNA-immunoprecipitation. wt and mutants (P108L, A119V, MiniSOX9, Del485) FLAG-tagged SOX9 were co-transfected with the ZDHHC16 minigene in HEK293T cells. FLAG-SOX9-wt immunoprecipitates RNA whereas FLAG-SOX9-mutants are comparable to the empty vector control condition. Enrichment of SOX9 binding sites in HT-29 cells at SOX9-dependent exon cassettes (B) and at TSS (C) of SOX9-splicing targets. A list of SOX9 enrichment peaks was obtained from the analysis of previously published ChIP-seq data from Shi Z. et al., (19). Graphs represents the specific distribution of SOX9 peaks around SOX9-dependent (red) or random (blue) exon-cassettes in C and around TSS of SOX9 splicing targets (red) or random TSS (blue) in B.
SOX9 does not contain a bona fide RNA-binding domain, suggesting that its binding to RNA is not direct. We hypothesized that SOX9 binds the DNA of its splicing target genes to recruit splicing regulators, which then bind to RNA and modulate splicing patterns. To test this possibility, we took advantage of available SOX9 ChIP-seq data from colon tumor HT-29 cells (19) and searched for an enrichment of SOX9 peaks either close to SOX9-dependent exon cassettes or close to TSS. SOX9 peaks were enriched in the vicinity of the TSS of SOX9 splicing targets (Figure 4C), but not of exon cassettes (Figure 4B). As shown in Figure 2B, the enrichment on TSS is not associated with transcriptional changes of the corresponding genes. Altogether these results indicate that SOX9 binds the promoters of its splicing targets, where it regulates splicing of the transcribed RNAs, rather than transcription itself.
SOX9 associates with RNA-binding proteins
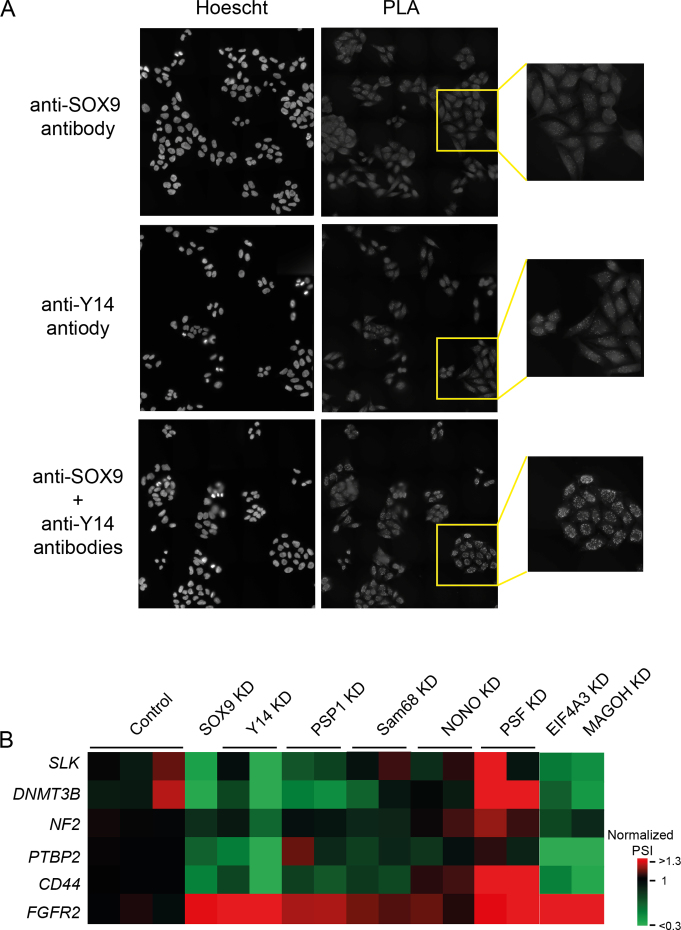
We next determined with which RNA-binding proteins SOX9 interacts to regulate splicing. We took advantage of data from the literature (21) and a preliminary proteomic analysis performed in our laboratory. Interaction between endogenous proteins was tested directly in DLD-1 cells by PLA, in which a fluorescent signal is obtained only if the two proteins are in close proximity (<40 nm). Using this technique, we confirmed the interaction of SOX9 and p54nrb/NONO in the nucleus of DLD-1 cells (Supplementary Figure S5). Indeed, the combination of anti-SOX9 and anti-NONO antibodies produced fluorescent foci as compared to each antibody alone (Supplementary Figure S5). We also found that SOX9 associates with PSP1/PSPC1 and PSF/SFPQ, two RNA-binding proteins reported to co-localize with p54nrb/NONO (32). SOX9 also associated with the Sam68/KHDRBS3 and Y14/RBM8A RNA-binding proteins (Figure 5A and Supplementary Figure S5).
Figure 5.
RNA-binding proteins associate with SOX9 and modulate SOX9 splicing signature. (A) Interaction between endogenous SOX9 and Y14 was tested in vivo using PLA, in which a fluorescent signal is obtained only if the two proteins are in close proximity (<40 nm). PLA using only one of each primary antibody shows the unspecific background signal. Nuclei were counterstained with Hoechst. (B) Heat map representation of the effect of the knockdown of SOX9 partners on the SOX9 splicing signature. Each column represents one individual siRNA except the MAGOH and EIF4A3 columns for which a pool of siRNAs was used. Each line represents an alternative splicing event. Results are expressed in PSI normalized to the mean of the three control experiments.
We next examined the impact of the depletion of these RNA-binding proteins on the SOX9 splicing signature that we defined (Figure 1). We used 2 different siRNAs to knockdown Y14, PSP1, Sam68, NONO and PSF, inducing a maximum mRNA depletion of about 82, 67, 88, 90, 82%, respectively (Supplementary Figure S6). Western blot confirmed that siRNA treatments depleted each protein accordingly, but that SOX9 protein levels remained largely unaffected (Supplementary Figure S7). Several depletions clearly impacted the alternative splicing of SOX9 signature targets (Figure 5B). In particular, Y14 depletion mimicked SOX9 depletion. PSP1 and Sam68 depletions showed similar effects as SOX9 depletion, although to a lesser extent. PSF depletion induced opposite effects, as compared to SOX9 depletion, indicating that PSF may antagonize SOX9 splicing activity. Of the five SOX9 partners, we found that the depletion of only NONO had no effect on SOX9 splicing signature. Overall, these results suggest that SOX9 modulates splicing by recruiting a regulatory complex comprising Y14, PSP1 and Sam68 to specific targets.
Y14 is part of the core EJC that is deposited at the exon/exon junctions of mRNAs following splicing (33). EJC is a tetrameric complex, composed of Y14, EIF4A3, MAGOH and MLN51, and has established roles in non-sense mRNA decay, transport, translation and more recently, in the regulation of alternative splicing (34–38). We thus tested other proteins of the EJC complex and found that knockdown of EIF4A3 and MAGOH elicited changes in the SOX9 splicing signature similar to those elicited by Y14 or SOX9 depletion. We also confirmed by co-immunoprecipitation assay that SOX9 interacted not only with Y14 but also with EIF4A3 and MAGOH (Supplementary Figure S8). Therefore, our findings support a model by which SOX9 associates with the EJC complex at specific targets to regulate alternative splicing independently of transcription.
SOX9 and Y14 regulate the alternative splicing of a common set of genes
To establish that Y14 and SOX9 proteins control alternative splicing of the same target genes in DLD-1 cells, we used RNA-seq to compare changes in splicing observed upon depletion of either Y14 or SOX9. In total, we generated 154, 164 and 158 million reads for control, Y14 knockdown with siRNA 1 and with siRNA 2 samples, respectively. We analyzed alternative exon usage using MATS analysis as described above (27). Y14 depletion modulated thousands of splicing events, which mostly corresponded to exon cassettes (Figure 6A and Supplementary Table S2). We found many more splicing changes than the previous global analysis of Y14 knockdown in HeLa cells (38). For example, we found 5510 exon cassette changes in 2744 genes in DLD-1 cells compared to 367 exons in 342 genes that were found in the aforementioned HeLa cell study. Importantly, we confirmed half of the target genes identified in the HeLa study (38).
Y14 knockdown affected the alternative splicing patterns of over 60% of SOX9 splicing targets (Figure 6B). This overlap is significantly more than one would expect to find by chance. Indeed, the probability to observe such an overlap using multiple random drawings of 2744 genes from the human genome is 1.3e−142. Specifically, Y14 knockdown affected the splicing pattern of 40% (97 out of 243 events) of the exon cassettes that are regulated by SOX9. Among those, SOX9 and Y14 modulated about 80% of exclusion or inclusion events in the same direction. This overlap is restricted to splicing because the expression of <5% of SOX9 transcriptional targets are also affected upon Y14 knockdown (Figure 6B).
These observations suggest that a significant proportion of SOX9-dependent splicing events may depend on Y14. To test this, we determined the effect of Y14 on the SOX9-dependent splicing of the endogenous PTBP2 and ZDHHC16 minigene reporter genes in HEK293T cells. We observed that SOX9-dependent splicing of PTBP2 is abolished in the absence of Y14 (Figure 6C). In contrast, PSF knockdown showed no detectable effect on SOX9-dependent splicing of PTBP2. Importantly, knockdown of either PSF or Y14 had no effect on SOX9-dependent splicing of the ZDHHC16 minigene reporter. These results are consistent with our observation that this PTBP2 exon is a common splicing target of both SOX9 and Y14, whereas the ZDHHC16 exon is targeted by SOX9, but not Y14. Altogether, these findings indicate that the splicing activity of SOX9 requires Y14 at some exons, though not at all its targets.
DISCUSSION
In this study, we show that SOX9 modulates the splicing of a large subset of genes in mammalian cells. Three prior studies suggested that SOX9 is implicated in alternative splicing (18,20,21). Our analysis identified only 7 out of the 70 SOX9-dependent exon cassettes that were reporter in a previous study (18). This discrepancy likely results from the use of different cell lines, as well as possible differences in RNA-seq depth and choice of splicing analysis algorithms. Nevertheless, our work extends these reports and better describes the role of SOX9 in splicing. Unexpectedly, genome-wide data mining and mutational analyses provide strong evidence that the splicing activity of SOX9 is uncoupled from its function as a transcription factor. First, SOX9 modulates splicing patterns without affecting the level of expression of splicing factors. Second, SOX9 regulates the expression levels and the alternative splicing patterns of two distinct sets of genes, revealing its ability to control separate splicing and transcriptional programs. Third, we identified three SOX9 mutants that are transcriptionally inactive, but show normal splicing activity. Notably, a single amino acid substitution in the HMG domain allowed us to genetically uncouple SOX9 splicing function from its transcriptional activity. Conversely, SOX9 dimerization domain is dispensable for its splicing activity but not for its function as a transcription factor (39). Altogether, these results demonstrate that SOX9 has two independent functions, splicing and transcriptional regulation. Therefore, our study expands the repertoire of cellular processes that may be controlled by SOX9. The HMG domain mutations that discriminate SOX9 splicing from its transcription activities will be of great interest for future studies aimed at deciphering the cellular and physiological relevance of each function of SOX9.
Mechanistic analyses of the role of SOX9 in alternative splicing suggest a model in which SOX9 is recruited to promoters of specific target genes to either activate transcription or modulate alternative splicing of target mRNAs. Mining of published ChIP-seq data (19) revealed an enrichment of SOX9 peaks close to promoters of its splicing targets, rather than in gene bodies, close to alternatively spliced exons. We propose that SOX9 forms an mRNP complex, which includes the RNA-binding protein Y14, to promote the splicing of nascent transcripts in close proximity to promoters. The significance of promoter-proximal alternative splicing opens up interesting questions for future investigations. For example, it is possible that SOX9 brings promoter specificity to alternative splicing events, such that splicing is restricted to specific tissues or responds to signaling cues.
Y14 is a core member of the EJC complex that is deposited after splicing on exon/exon junction and regulates the decay, the export and the translation of produced mRNAs (33). Interestingly, the core EJC components have been recently implicated in intron retention in Drosophila (34,35) and in pre-mRNA splicing in Xenopus (40) and mammalian cells (37,38). As previously described (37,38), the three core EJC proteins appear to act together on splicing and indeed we find that like Y14, EIF4A3 and MAGOH affect SOX9 splicing signature. The exact mechanism by which EJC components influence alternative splicing remains to be elucidated. Two models have been proposed. The first model is based on the coupling between RNA Polymerase II transcription and mRNA processing (33,41–44). EJC knockdown has been shown to increase RNA Pol II transcription speed at some genes, which in turn modifies their splicing patterns (38). According to the kinetic model of co-transcriptional splicing, EJC proteins slow down transcription elongation, which, in turn, offers a longer time frame to include alternative exons. Our analysis reveals that 85% of SOX9 target cassette exons are less included upon Y14 knockdown. However, unlike Y14, SOX9 knockdown induces as much cassette exon inclusion as exclusion. In addition, 70% of SOX9-dependent splicing events are absent from a list of alternatively spliced exons that are sensitive to pol II transcription speed (45). Thus, modulation of RNA Pol II elongation rate is unlikely to account for the general role of SOX9 in alternative splicing. The second model postulates that EJC proteins acts as bona fide splicing cofactors by recruiting other splicing factors and/or by stabilizing large mRNP complexes onto pre-mRNAs and influence splicing decisions (41,46). SOX9 may therefore act as part of such mRNP complexes, together with Y14. Eighty percent of all the exons of the genome are associated with EJC proteins (47). SOX9 may therefore confer gene specificity to the alternative splicing activity of EJC and possibly other splicing factors.
Overall, our work uncovered a previously unidentified role for SOX9 in the regulation of alternative splicing. Surprisingly, this function of SOX9 is independent from its transcriptional activity. Rather, SOX9 interacts with and requires specific RNA-binding proteins such as Y14, to modulate alternative splicing. It is conceivable that SOX9 regulates splicing through several, non-mutually exclusive mechanisms. A thorough analysis of specific splicing events will definitely help to shed light on the exact role of SOX9. Additionally, we found that the HMG domain of SOX9 is involved in the regulation of alternative splicing. This new function might thus be conserved in other members of the SOX family, which share the HMG domain and provide novel insights into the essential role of the SOX transcription factors in development and disease.
DATA AVAILABILITY
Processed and raw data of RNA-seq experiments have been deposited on GEO under accession number: GSE104103.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dr D. Lleres (IGMM, Montpellier, France) for providing anti-PSP1 antibody and to Dr V. Gire and F. Tomas for their help in western blot analyses. We thank Dr D. Helmlinger for discussion and critical reading of the manuscript. We also thank V. Georget and S. De Rossi from Montpellier RIO Imaging microscopy facility and S. Fromont and F. Lionneton from Montpellier Genomic Collection facility for the quantification with the Caliper LabCHIP.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Fondation ARC pour la Recherche sur le cancer [Projet ARC N°SFI20111203667]; la Ligue contre le Cancer (appel à projet Languedoc-Roussillon). Funding for open access charges: Institut National du Cancer [INCa N°2017-69] .
Conflict of interest statement. None declared.
REFERENCES
- 1. Wagner T., Wirth J., Meyer J., Zabel B., Held M., Zimmer J., Pasantes J., Bricarelli F.D., Keutel J., Hustert E. et al. Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell. 1994; 79:1111–1120. [DOI] [PubMed] [Google Scholar]
- 2. Jo A., Denduluri S., Zhang B., Wang Z., Yin L., Yan Z., Kang R., Shi L.L., Mok J., Lee M.J. et al. The versatile functions of Sox9 in development, stem cells, and human diseases. Genes Dis. 2014; 1:149–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Akiyama H., Chaboissier M.C., Martin J.F., Schedl A., de Crombrugghe B.. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 2002; 16:2813–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chaboissier M.C., Kobayashi A., Vidal V.I., Lutzkendorf S., van de Kant H.J., Wegner M., de Rooij D.G., Behringer R.R., Schedl A.. Functional analysis of Sox8 and Sox9 during sex determination in the mouse. Development. 2004; 131:1891–901. [DOI] [PubMed] [Google Scholar]
- 5. Cheung M., Chaboissier M.C., Mynett A., Hirst E., Schedl A., Briscoe J.. The transcriptional control of trunk neural crest induction, survival, and delamination. Dev. Cell. 2005; 8:179–192. [DOI] [PubMed] [Google Scholar]
- 6. Weider M., Wegner M.. SoxE factors: transcriptional regulators of neural differentiation and nervous system development. Semin. Cell Dev. Biol. 2017; 63:35–42. [DOI] [PubMed] [Google Scholar]
- 7. Antoniou A., Raynaud P., Cordi S., Zong Y., Tronche F., Stanger B.Z., Jacquemin P., Pierreux C.E., Clotman F., Lemaigre F.P.. Intrahepatic bile ducts develop according to a new mode of tubulogenesis regulated by the transcription factor SOX9. Gastroenterology. 2009; 136:2325–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bastide P., Darido C., Pannequin J., Kist R., Robine S., Marty-Double C., Bibeau F., Scherer G., Joubert D., Hollande F. et al. Sox9 regulates cell proliferation and is required for Paneth cell differentiation in the intestinal epithelium. J. Cell Biol. 2007; 178:635–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mori-Akiyama Y., van den Born M., van Es J.H., Hamilton S.R., Adams H.P., Zhang J., Clevers H., de Crombrugghe B.. SOX9 is required for the differentiation of paneth cells in the intestinal epithelium. Gastroenterology. 2007; 133:539–546. [DOI] [PubMed] [Google Scholar]
- 10. Jeselsohn R., Cornwell M., Pun M., Buchwalter G., Nguyen M., Bango C., Huang Y., Kuang Y., Paweletz C., Fu X. et al. Embryonic transcription factor SOX9 drives breast cancer endocrine resistance. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E4482–E4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lin S.-C., Chou Y.-T., Jiang S.S., Chang J.-L., Chung C.-H., Kao Y.-R., Chang I.-S., Wu C.-W.. Epigenetic switch between SOX2 and SOX9 regulates cancer cell plasticity. Cancer Res. 2016; 76:7036–7048. [DOI] [PubMed] [Google Scholar]
- 12. Shen Z., Deng H., Fang Y., Zhu X., Ye G.-T., Yan L., Liu H., Li G.. Identification of the interplay between SOX9 and S100P in the metastasis and invasion of colon carcinoma. Oncotarget. 2015; 6:20672–20684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Matheu A., Collado M., Wise C., Manterola L., Cekaite L., Tye A.J., Canamero M., Bujanda L., Schedl A., Cheah K.S. et al. Oncogenicity of the developmental transcription factor Sox9. Cancer Res. 2012; 72:1301–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Larsimont J.-C., Youssef K.K., Sánchez-Danés A., Sukumaran V., Defrance M., Delatte B., Liagre M., Baatsen P., Marine J.-C., Lippens S. et al. Sox9 controls Self-Renewal of oncogene targeted cells and links tumor initiation and invasion. Cell Stem Cell. 2015; 17:60–73. [DOI] [PubMed] [Google Scholar]
- 15. Shi Z., Chiang C.I., Mistretta T.A., Major A., Mori-Akiyama Y.. SOX9 directly regulates IGFBP-4 in the intestinal epithelium. Am. J. Physiol. Gastrointest Liver Physiol. 2013; 305:G74–G83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mertin S., McDowall S.G., Harley V.R.. The DNA-binding specificity of SOX9 and other SOX proteins. Nucleic Acids Res. 1999; 27:1359–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hou L., Srivastava Y., Jauch R.. Molecular basis for the genome engagement by Sox proteins. Semin. Cell Dev. Biol. 2017; 63:2–12. [DOI] [PubMed] [Google Scholar]
- 18. Rahmoun M., Lavery R., Laurent-Chaballier S., Bellora N., Philip G.K., Rossitto M., Symon A., Pailhoux E., Cammas F., Chung J. et al. In mammalian foetal testes, SOX9 regulates expression of its target genes by binding to genomic regions with conserved signatures. Nucleic Acids Res. 2017; 45:7191–7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shi Z., Chiang C.-I., Labhart P., Zhao Y., Yang J., Mistretta T.-A., Henning S.J., Maity S.N., Mori-Akiyama Y.. Context-specific role of SOX9 in NF-Y mediated gene regulation in colorectal cancer cells. Nucleic Acids Res. 2015; 43:6257–6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ohe K., Lalli E., Sassone-Corsi P.. A direct role of SRY and SOX proteins in pre-mRNA splicing. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:1146–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hata K., Nishimura R., Muramatsu S., Matsuda A., Matsubara T., Amano K., Ikeda F., Harley V.R., Yoneda T.. Paraspeckle protein p54nrb links Sox9-mediated transcription with RNA processing during chondrogenesis in mice. J. Clin. Invest. 2008; 118:3098–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fox A.H., Lam Y.W., Leung A.K.L., Lyon C.E., Andersen J., Mann M., Lamond A.I.. Paraspeckles: a novel nuclear domain. Curr. Biol. 2002; 12:13–25. [DOI] [PubMed] [Google Scholar]
- 23. Südbeck P., Schmitz M.L., Baeuerle P.A., Scherer G.. Sex reversal by loss of the C-terminal transactivation domain of human SOX9. Nat. Genet. 1996; 13:230–232. [DOI] [PubMed] [Google Scholar]
- 24. Abdel-Samad R., Zalzali H., Rammah C., Giraud J., Naudin C., Dupasquier S., Poulat F., Boizet-Bonhoure B., Lumbroso S., Mouzat K. et al. MiniSOX9, a dominant-negative variant in colon cancer cells. Oncogene. 2011; 30:2493–2503. [DOI] [PubMed] [Google Scholar]
- 25. Klinck R., Bramard A., Inkel L., Dufresne-Martin G., Gervais-Bird J., Madden R., Paquet E.R., Koh C., Venables J.P., Prinos P. et al. Multiple alternative splicing markers for ovarian cancer. Cancer Res. 2008; 68:657–663. [DOI] [PubMed] [Google Scholar]
- 26. Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L. et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009; 55:611–622. [DOI] [PubMed] [Google Scholar]
- 27. Shen S., Park J.W., Huang J., Dittmar K.A., Lu Z., Zhou Q., Carstens R.P., Xing Y.. MATS: a Bayesian framework for flexible detection of differential alternative splicing from RNA-Seq data. Nucleic Acids Res. 2012; 40:e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McDowall S., Argentaro A., Ranganathan S., Weller P., Mertin S., Mansour S., Tolmie J., Harley V.. Functional and structural studies of wild type SOX9 and mutations causing campomelic dysplasia. J. Biol. Chem. 1999; 274:24023–24030. [DOI] [PubMed] [Google Scholar]
- 29. Meyer J., Südbeck P., Held M., Wagner T., Schmitz M.L., Bricarelli F.D., Eggermont E., Friedrich U., Haas O.A., Kobelt A. et al. Mutational analysis of the SOX9 gene in campomelic dysplasia and autosomal sex reversal: lack of genotype/phenotype correlations. Hum. Mol. Genet. 1997; 6:91–98. [DOI] [PubMed] [Google Scholar]
- 30. Prévostel C., Rammah-Bouazza C., Trauchessec H., Canterel-Thouennon L., Busson M., Ychou M., Blache P.. SOX9 is an atypical intestinal tumor suppressor controlling the oncogenic Wnt/ß-catenin signaling. Oncotarget. 2016; 7:82228–82243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ohe K., Tamai K.T., Parvinen M., Sassone-Corsi P.. DAX-1 and SOX6 molecular interplay results in an antagonistic effect in pre-mRNA splicing. Dev. Dyn. 2009; 238:1595–1604. [DOI] [PubMed] [Google Scholar]
- 32. Fox A.H., Lamond A.I.. Paraspeckles. Cold Spring Harb. Perspect. Biol. 2010; 2:a000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hir H.L., Saulière J., Wang Z.. The exon junction complex as a node of post-transcriptional networks. Nat. Rev. Mol. Cell Biol. 2016; 17:41–54. [DOI] [PubMed] [Google Scholar]
- 34. Ashton-Beaucage D., Udell C.M., Lavoie H., Baril C., Lefrançois M., Chagnon P., Gendron P., Caron-Lizotte O., Bonneil É., Thibault P. et al. The exon junction complex controls the splicing of mapk and other long Intron-Containing transcripts in drosophila. Cell. 2010; 143:251–262. [DOI] [PubMed] [Google Scholar]
- 35. Roignant J.-Y., Treisman J.E.. Exon junction complex subunits are required to splice drosophila MAP kinase, a large heterochromatic gene. Cell. 2010; 143:238–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hayashi R., Handler D., Ish-Horowicz D., Brennecke J.. The exon junction complex is required for definition and excision of neighboring introns in Drosophila. Genes Dev. 2014; 28:1772–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Michelle L., Cloutier A., Toutant J., Shkreta L., Thibault P., Durand M., Garneau D., Gendron D., Lapointe E., Couture S. et al. Proteins associated with the exon junction complex also control the alternative splicing of apoptotic regulators. Mol. Cell. Biol. 2012; 32:954–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang Z., Murigneux V., Hir H.L.. Transcriptome-wide modulation of splicing by the exon junction complex. Genome Biol. 2014; 15:551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Coustry F., Oh C., Hattori T., Maity S.N., de Crombrugghe B., Yasuda H.. The dimerization domain of SOX9 is required for transcription activation of a chondrocyte-specific chromatin DNA template. Nucleic Acids Res. 2010; 38:6018–6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Haremaki T., Weinstein D.C.. Eif4a3 is required for accurate splicing of the Xenopus laevis ryanodine receptor pre-mRNA. Dev. Biol. 2012; 372:103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Woodward L.A., Mabin J.W., Gangras P., Singh G.. The exon junction complex: a lifelong guardian of mRNA fate. Wiley Interdiscip. Rev. RNA. 2017; 8:doi:10.1002/wrna.1411. [DOI] [PubMed] [Google Scholar]
- 42. Carrillo Oesterreich F., Bieberstein N., Neugebauer K.M.. Pause locally, splice globally. Trends Cell Biol. 2011; 21:328–335. [DOI] [PubMed] [Google Scholar]
- 43. Moore M.J., Proudfoot N.J.. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009; 136:688–700. [DOI] [PubMed] [Google Scholar]
- 44. Bentley D.L. Coupling mRNA processing with transcription in time and space. Nat. Rev. Genet. 2014; 15:163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fong N., Kim H., Zhou Y., Ji X., Qiu J., Saldi T., Diener K., Jones K., Fu X.-D., Bentley D.L.. Pre-mRNA splicing is facilitated by an optimal RNA polymerase II elongation rate. Genes Dev. 2014; 28:2663–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Singh G., Kucukural A., Cenik C., Leszyk J.D., Shaffer S.A., Weng Z., Moore M.J.. The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell. 2012; 151:750–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Saulière J., Murigneux V., Wang Z., Marquenet E., Barbosa I., Le Tonquèze O., Audic Y., Paillard L., Roest Crollius H., Le Hir H.. CLIP-seq of eIF4AIII reveals transcriptome-wide mapping of the human exon junction complex. Nat. Struct. Mol. Biol. 2012; 19:1124–1131. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Processed and raw data of RNA-seq experiments have been deposited on GEO under accession number: GSE104103.