Abstract
As adapter molecules to convert the nucleic acid information into the amino acid sequence, tRNAs play a central role in protein synthesis. To fulfill this function in a reliable way, tRNAs exhibit highly conserved structural features common in all organisms and in all cellular compartments active in translation. However, in mitochondria of metazoans, certain dramatic deviations from the consensus tRNA structure are described, where some tRNAs lack the D- or T-arm without losing their function. In Enoplea, this miniaturization comes to an extreme, and functional mitochondrial tRNAs can lack both arms, leading to a considerable size reduction. Here, we investigate the secondary and tertiary structure of two such armless tRNAs from Romanomermis culicivorax. Despite their high AU content, the transcripts fold into a single and surprisingly stable hairpin structure, deviating from standard tRNAs. The three-dimensional form is boomerang-like and diverges from the standard L-shape. These results indicate that such unconventional miniaturized tRNAs can still fold into a tRNA-like shape, although their length and secondary structure are very unusual. They highlight the remarkable flexibility of the protein synthesis apparatus and suggest that the translational machinery of Enoplea mitochondria may show compensatory adaptations to accommodate these armless tRNAs for efficient translation.
INTRODUCTION
The essential role of tRNAs in protein synthesis is clearly mirrored in their conserved structural features found in all three kingdoms of life. Even in the translational machinery of chloroplasts and mitochondria of most eukaryotes, the tRNA molecules exhibit the typical cloverleaf structure and the three-dimensional (3D) L-shape. Due to the many interaction partners of tRNAs, their conserved structure is regarded as a prerequisite for efficient and correct binding to maturation enzymes, elongation factors and ribosomes (1–4). Accordingly, mutations in mitochondrial tRNA genes frequently lead to serious diseases like myopathies and neurological disorders in humans (5–8). Yet, deviations from canonical tRNA structures are known (Figure 1), especially in metazoan mitochondria, where the lack of the D-arm is described for tRNASer(AGY) in mammals (9–11). Furthermore, in nematode mitochondria, many tRNAs were identified that lack either the D- or the T-arm (12–14). Even further reductions were observed in mitochondrial tRNAs of Acariformes (mites) and Enoplea (a class of Nematoda) (15–18). Here, the loss of both the D- and the T-arm leads to the shortest tRNA molecules identified so far, with a minimal size of 45 nt, including the CCA terminus (18). While the actual length and sequence of most of these tRNAs are currently only computer-based predictions, at least in the roundworm Romanomermis culicivorax, several tRNAs were identified as processed mitochondrial transcripts that carry a non-encoded post-transcriptionally added CCA end—a strong indication that these armless miniaturized tRNAs are indeed functional and not massively edited into a more standard-like tRNA form (18).
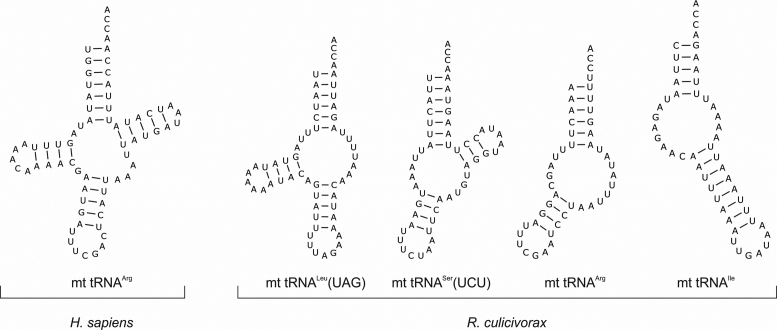
Figure 1.
Secondary structure predictions of bizarre armless tRNAs. While almost all human mt tRNAs fold into the standard cloverleaf structure as exemplarily indicated for mt tRNAArg, the bizarre miniaturized tRNAs from Romanomermis culicivorax lack either the T- (11 tRNAs; example: mt tRNALeu(UAG)), the D- (2 tRNAs; example: mt tRNASer(UCU)) or both arms (9 tRNAs; examples: mt tRNAArg and mt tRNAIle). Hence, all mitochondrial tRNAs of this organism show strong deviations from the classical cloverleaf structure.
Hence, mitochondrial tRNAs can show massive structural deviations from the standard shape, where D- and T-arms are lost and replaced by short single-stranded regions (Figure 1), and it is currently not clear how the corresponding translational apparatus co-evolved in order to accept such bizarre armless tRNAs for synthesizing mitochondrially encoded proteins. While an in vitro translation system is not available for a detailed analysis of these transcripts, information about secondary structure and 3D shape of the tRNA transcripts can give a first idea about their functionality as translational adapter molecules. Here, we show that the computationally predicted hairpin-like secondary structures of two of these tRNAs are indeed formed and exhibit a rather stable conformation. In addition, the 3D shape in solution is boomerang-like but deviates from the canonical L-form described for standard tRNAs. Yet, the anticodon loop and the 3′-end of the acceptor stem exhibit a distance similar to that found in conventional tRNAs. These results indicate that the required adaptation of the translational machinery in Enoplea mitochondria to the unusual features of these tRNAs might not be as dramatic as initially suspected.
MATERIALS AND METHODS
tRNA 2D structure predictions
RNA secondary structures were calculated using Mfold and RNAfold (19,20). Structure presentations were done using VARNA (21).
Preparation of tRNAs
Romanomermis culicivorax mt tRNAArg and mt tRNAIle sequences as identified by Wende et al. (18) were used to generate transcription templates by overlap extension polymerase chain reaction (PCR) and were cloned into a pCR®2.1-TOPO plasmid by QuikChange site-directed mutagenesis (Agilent, Germany), where mutations are introduced by primers in a DNA amplification step with subsequent template removal by DpnI digestion. In vitro transcripts, including that of Saccharomyces cerevisiae tRNAPhe lacking its CCA tail, were prepared as described (22,23). Reaction products were separated on a denaturing (8 M urea) polyacrylamide gel, isolated and radioactively labeled at the 5′-end as described (24). Native baker’s yeast cytosolic tRNAPhe was purchased from Sigma (cat. number R4018) and was solubilized in water at a concentration of 7.5 mg/ml.
Structural analysis by in-line probing
A total of 40 pmol 5′-labeled tRNA were incubated in a 10 μl volume containing 50 mM Tris/HCl (pH 8.5) and 20 mM MgCl2. 2 μl samples were taken after 15, 24 and 48 h. Alkaline hydrolysis was carried out at 90°C for 2 min in a 10 μl reaction volume containing 100 mM Na2CO3/NaHCO3 (pH 9.8) and 40 pmol 5′-labeled RNA. Probing reactions were stopped by the addition of 5 μl RNA loading dye (10 mM Tris/HCl, pH 7.6, 80% formamide, 0.25% bromophenol blue, 0.25% xylene cyanol), reaction products were separated on 15% denaturing PAA gels.
NMR measurements
A total of 1.05 mg of tRNAArg and 1.17 mg of tRNAIle were dissolved in 500 μl buffer containing 25 mM potassium phosphate and 50 mM KCl at pH 6.2. The final concentration of each tRNA was 100 μM. Where indicated, MgCl2 was added to a final concentration of 2 mM. Nuclear magnetic resonance (NMR) spectra were recorded on a Bruker Avance III 950 MHz NMR spectrometer equipped with a cryogenic triple resonance probe head. 1D-1H proton NMR spectra with Jump-Return water suppression were recorded at temperatures between 5°C and 25°C (25).
Small angle X-ray scattering
tRNAArg and tRNAIle samples were concentrated to a final concentration of 6 mg/ml (tRNAArg) and 3.5 mg/ml (tRNAIle) in 200 μl buffer containing 50 mM HEPES-NaOH, pH 7.4 and 10 mM MgCl2 using centrifugation in an Amicon Ultra-4 (cut off 3 kDa) at 5000 rpm. SAXS measurements were performed at the SWING beamline (SOLEIL synchrotron, Saint-Aubin, France) with a beam wavelength of λ = 1.033 Å and a 17 × 17 cm2 low-noise Aviex CCD detector. A total of 70 and 50 μl of solutions containing tRNAArg and tRNAIle, respectively, were loaded onto an SEC HPLC column (BioSEC3–150, Agilent) upstream the SAXS cell in order to separate monomers from potential oligomers and aggregates. Miniaturized tRNAs were compared to the native form (Sigma) and to a transcript of cytosolic yeast tRNAPhe analyzed with the same setup (injection of 30 μl at 7.5 mg/ml and 70 μl at 1.7 mg/ml, respectively).
SEC elution was performed using an Agilent HPLC system at 0.2 ml/min in 50 mM HEPES-NaOH, pH 7.4, and 10 mM MgCl2 (low salt conditions), or in a same buffer supplemented with 150 mM KCl (high salt conditions). Samples were exposed to X-rays directly downstream to the SEC column to record the corresponding scattering signal (240 images of 1 sec exposure at 0.5 sec interval). The scattering signal from the buffer was collected at the beginning of the run (90 images) to be subtracted from the RNA signal. SAXS images were processed with FOXTROT to generate individual curves (subtracted from the buffer signal) and plots corresponding to I(0) and gyration radius Rg as a function of frames (26). US-SOMO was used to separate the scattering signal corresponding of successive molecular species in overlapping regions of the chromatogram by Gaussian decomposition as described (27) and curves from consecutive images showing similar Rg were averaged.
Resulting SAXS profiles were analyzed with the package ATSAS 2.8.2 (28), including various tools to evaluate structural parameters such as radius of gyration Rg from Guinier plots (PRIMUS; (29)), as well as the particle distance distribution function p(r) (GNOM; (30)) and associated maximum distance Dmax and Porod volume. Ab initio reconstructions of 3D pseudo-atomic models were performed with DAMMIF (31) using data up to q = 0.25 Å−1. All-atom models of armless tRNAs were built using RNAcomposer (32). Based on a normal mode analysis (NMA) carried out with ElNeMo (33), tRNA models were perturbed to generate conformations that better fitted experimental SAXS profiles owing to the goodness-of-fit (Chi2) determined with CRYSOL (34). Selected conformers were handled in PyMol 1.8.6 (35) using the SASpy plugin (36) and were minimized in Phenix (37). Ab initio pseudo-atomic and all-atom models were compared and represented with PyMOL 1.8.6 (35). Experimental SAXS data of miniaturized tRNAArg and tRNAIle and derived models have been deposited in the SASBDB (38).
RESULTS
Structure analysis by in-line probing
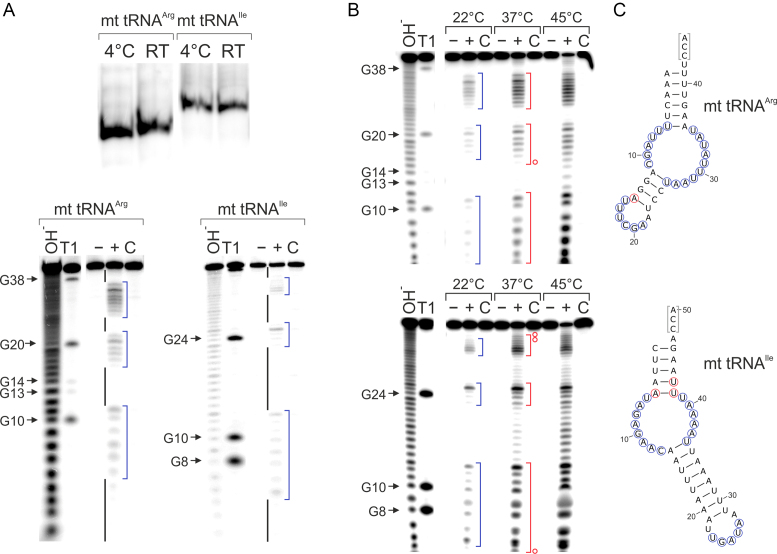
For the structural investigation of armless mitochondrial (mt) tRNAs of R. culicivorax, we focused on tRNAArg and tRNAIle, where the complete in vivo transcript sequence was identified by 5′- and 3′-RACE analysis (18). To avoid conformational heterogeneity and a corresponding increase of noise in the data used to model the tRNA structures, transcripts without dangling CCA ends were generated, excluding artificial interaction signatures in NMR spectra and ensuring compactness of the molecules for SAXS analysis. Further, we intended to study transcripts as close as possible to the miniaturized tRNA molecules released from precursor transcripts, before the introduction of modifications and CCA ends. In vitro transcripts of mt tRNAArg and mt tRNAIle with homogeneous ends lacking the CCA terminus were prepared according to Mörl et al. (23), resulting in transcript lengths of 42 nt (tRNAArg) and 47 nt (tRNAIle). For internally labeling of the tRNAs, transcription was carried out in the presence of α-32P-ATP. The isolated tRNAs were separated on a native polyacrylamide gel at 4°C and at room temperature. Autoradiography shows that tRNAArg as well as tRNAIle run at both temperatures as one defined single band, indicating the formation of a single conformation in both cases (Figure 2A). Hence, the tRNAs fold into one preferred structure, and the AU richness of both transcripts (tRNAArg: 76%; tRNAIle: 87%) does not result in a mixture of conformations.
Figure 2.
Structural analysis of in vitro transcripts of mt tRNAIle and mt tRNAArg. (A) Top, radiolabeled in vitro transcribed mt tRNAs for isoleucine and arginine migrate as single bands in native polyacrylamide gel electrophoresis, representing the formation a single stable structural conformer at 4°C as well as at room temperature. Bottom, in-line probing of both in vitro transcripts. Black bars indicate base-paired regions, blue brackets unpaired elements. The cleavage pattern is in very good agreement with the calculated secondary structures shown in (C). (B) In-line probing at different temperatures. Blue brackets indicate single-stranded regions at 22°C, red brackets at 37°C. At the increased temperatures, a slight fraying in the anticodon stem of tRNAArg and in the acceptor stem of tRNAIle can be observed, indicated by the red circles in (C). (C) Proposed secondary structures. Blue circles indicate cleavage at 22°C (corresponding to blue brackets in the gels), red circles at 37°C (equivalent to red brackets). The CCA ends that are not present in the in vitro transcripts are indicated in gray brackets. −, tRNA preparation without incubation; +, tRNA incubated under in-line probing conditions; C, incubation control in water; OH−: alkaline hydrolysis; T1: G-specific cleavage by RNase T1.
In order to determine the secondary structure of these tRNA species, 5′-end-labeled in vitro transcripts were prepared and analyzed by in-line probing (39). To obtain information about the stability of the individual secondary structures, in-line probing was performed for 24 h at 22, 37 and 45°C. Cleavage products were separated on a denaturing polyacrylamide gel and visualized by autoradiography (Figure 2A and B). Due to the massive size reduction, we did not use the tRNA standard numbering (40,41). Instead, the numbering of the individual nucleotides corresponds to the actual positions in the transcripts. Interestingly, both tRNA transcripts show secondary structures almost identical to the predicted ones (18). At 22°C, tRNAArg has an acceptor stem consisting of 6 bp (A1-U41 to U6-A36; U6 is somewhat scissile, probably due to some fraying of the stem) and an anticodon stem of 4 bp (A12-U26 to A15-U23). The connector elements located between acceptor and anticodon stems consist of five (U7 to C11) and nine (A27 to U35) unpaired residues, respectively. At 37°C, these base paired elements are stable, and only position A15 becomes scissile, indicating a slight fraying of the corresponding base pairs. At a further increased temperature of 45°C, this secondary structure starts to unfold. The secondary structure of tRNAIle at 22°C has only 5 bp in the acceptor stem (C1-G46 to A5-U42) and an unusually elongated anticodon stem with 9 bp (A14-U36 to U22-A28). This results in a contraction of the anticodon loop from the standard seven unpaired bases to a more condensed form with five unpaired residues, as no cleavage occurred at positions U22 and A28. A reason for this loop collapse is probably the missing modifications in the in vitro transcript, as it was shown for canonical tRNAs that modification of a highly conserved purine at the standard position 37 (corresponding to position 27 in tRNAIle) keeps the loop in an open conformation (42–44). The connectors are eight (U6 to C13) and five (A37 to U41) nucleotides long and structurally flexible/unpaired, as indicated by the cleavage pattern. Similar to tRNAArg, tRNAIle has an almost unchanged secondary structure at 37°C. Only in the acceptor stem, positions A5, U42 and U43 are scissile, indicating a similar helix fraying as found for tRNAArg. However, at 45°C, tRNAIle starts to unfold, as it is cleaved at many additional positions.
NMR analysis
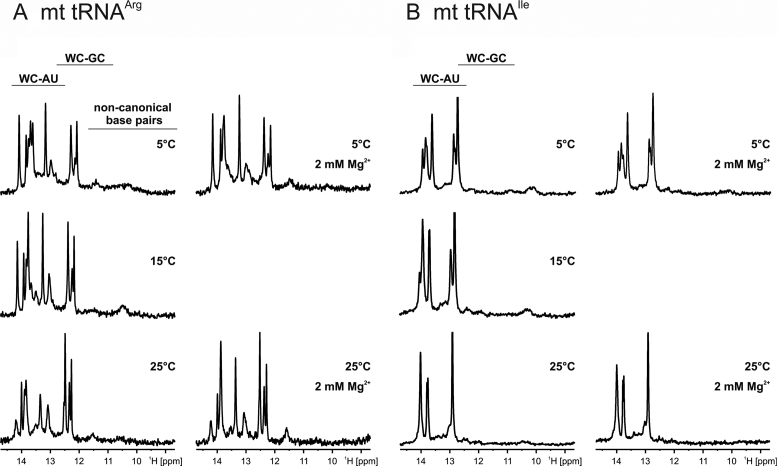
The imino proton region of 1D-1H spectra of tRNAArg in H2O at different temperatures is shown in Figure 3A. The spectra are characterized by several imino group hydrogen resonances appearing between 12.2 and 14.2 ppm. Imino proton signals of uridines in typical Watson–Crick A-U base pairs are normally found downfield of ∼12.5 ppm, while the imino protons of guanines in Watson–Crick G-C base pairs show signals downfield of ∼ 11.8 ppm (25). Hence, all imino proton signals visible in these spectra stem from nucleotides that participate in canonical Watson–Crick base pair interactions. Based on the known chemical shift differences between uridine and guanine imino protons in Watson–Crick base pairs, the spectra reveal the presence of three Watson–Crick G-C base pairs and seven to eight Watson–Crick A-U base pairs in the secondary structure of tRNAArg, in agreement with the results of the in-line probing experiments. No evidence is seen for the presence of stable non-Watson–Crick base pairing in this RNA, since imino proton signals with chemical shifts upfield of 12 ppm are absent from the spectra. This suggests that the connector elements are not involved in additional tertiary interactions, again in agreement with the in-line probing data. At higher temperatures (20°C and 25°C), some of the imino proton signals start to broaden as expected due to increased fraying at the ends of helices. It has been shown that millimolar Mg2+ concentrations have the ability to stabilize RNA tertiary structures (45,46). However, the addition of Mg2+ to tRNAArg led only to slight chemical shift changes for some of the imino proton signals. Importantly, no new imino proton signals appeared in the spectra, indicating that the presence of 2 mM Mg2+ did not induce the formation of additional base pairings or tertiary interactions.
Figure 3.
NMR analysis of in vitro transcribed mt tRNAArg and mt tRNAIle. Imino proton regions of 1D-1H NMR-spectra at different temperatures and in the absence (left) and the presence (right) of 2 mM Mg2+ for (A) mt tRNAArg and (B) mt tRNAIle. The typical chemical shift regions for U imino protons in Watson–Crick A-U and G imino protons in Watson–Crick G-C as well as for imino protons from non-canonical base pairs are indicated on top.
The imino proton spectra of mt tRNAIle (Figure 3B) show only imino proton signals of uridines in Watson–Crick A-U base pairs at all temperatures measured. Due to putative signal overlap caused by similarities in the chemical environment for the central A-U base pairs in the two helical elements of the predicted secondary structure, the number of signals cannot be quantified accurately. The imino proton signal of G46 in the C1-G46 base pair is most likely absent due to increased fraying of terminal base pairings as often observed in NMR studies of RNA (25). The addition of 2 mM Mg2+ has no significant effect on the imino proton spectra and therefore on the structure and stability of mt tRNAIle.
Small angle X-ray scattering analysis (SAXS)
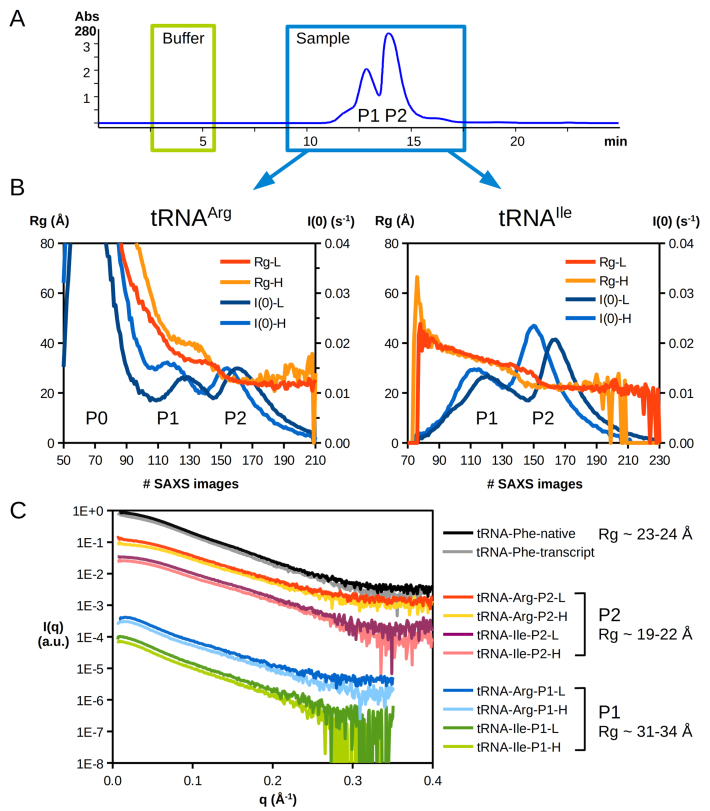
Both armless tRNA transcripts were further studied by small-angle X-ray scattering (SAXS) and were compared to the (gold-standard) S. cerevisiae cytosolic tRNAPhe in its native form and as a transcript lacking the CCA end (like the armless species). Size exclusion chromatography (SEC) was used upstream of the SAXS cell to separate individual populations and to analyze monodisperse samples. A typical HPLC-SEC chromatogram is shown in Figure 4A. Because we observed some aggregation in the initial buffer, especially for tRNAArg, both armless tRNAs were also analyzed in the same buffer supplemented with 150 mM KCl to increase the ionic strength. However, the presence of salt did not significantly change the behavior of the tRNAs, except for a slight shift in the elution times of the main populations (Figure 4B). The X-ray scattering signal recorded along the SEC elution indicates the presence of two to three distinct populations.
Figure 4.
SEC-SAXS analysis of armless tRNAs. (A) Upstream to SAXS analysis, different molecular entities present in the samples were separated by HPLC-SEC. A typical chromatogram corresponding to tRNAArg in low salt conditions is displayed with the absorbance signal at 280 nm (other chromatograms are shown in Supplementary Figure S1). Green and blue boxes indicate time windows of SAXS data collection for the buffer (90 images) and the sample signals (240 images), respectively. All samples presented two elution peaks (P1 and P2). (B) The evolution of scattering intensity I(0) and the gyration radius Rg is plotted along the dataset. Two main populations are observed (P1 and P2), as in the SEC-UV signal, both in low (L) and high (H) salt concentrations (see ‘Materials and Methods’ section), as well as aggregates (P0) in the case of tRNAArg. (C) SAXS profiles of tRNAs after solvent subtraction and averaging. For clarity, intensity curves represented in arbitrary units (a.u.) are offset and grouped by similar types of population (P1, P2). Values of Rg derived from the Guinier plot analysis and Porod volumes (see Table 1) indicate that monomeric tRNAs elute in the second peak (P2), whereas the first peak (P1) likely corresponds to tRNA dimers.
SAXS profiles collected in peak 2 are typical of rod-like objects with a radius of gyration (Rg, molecular size obtained from the slope of Guinier representation ln[I(q)] versus q2 at very small q angles) of 19–22 Å and are similar to that obtained with the canonical tRNAPhe (native or transcript, Rg ≈ 23–24 Å), while those measured in peak 1 exhibit a different shape, and their Guinier analysis indicates a larger Rg (Rg ≈ 31–34 Å). The analysis of the Porod invariant (estimation of particle volume derived from the Porod representation I(q)*q4 versus q) strongly suggest that peak 1 contains a dimeric form with a Porod volume twice as large as that of tRNA monomers present in peak 2 (Table 1). Large aggregates observed with tRNAArg display an Rg of about 130–150 Å (the limited number of experimental points in the Guinier region did not allow a precise estimation for these molecular entities). Noticeably, increasing the KCl concentration hardly affects the SAXS profiles in general nor the tendency of tRNAArg to form large aggregates (peak 0).
Table 1.
Structural parameters of tRNAs analyzed by SAXS
| Sample | Peak | Rg Guinier(Å) | Rg p(r)(Å) | Dmax(Å) | Porod volume(Å3) | Oligomer |
|---|---|---|---|---|---|---|
| tRNAArg + KCl | #1 | 31.5 ± 0.2 | 33.6 ± 0.1 | 125 | 38.2 103 | dimer |
| tRNAArg | #1 | 31.6 ± 0.2 | 33.7 ± 0.1 | 125 | 37.7 103 | dimer |
| tRNAIle+ KCl | #1 | 33.7 ± 0.2 | 35.1 ± 0.1 | 123 | 47.4 103 | dimer |
| tRNAIle | #1 | 32.7 ± 0.2 | 34.9 ± 0.1 | 122 | 47.2 103 | dimer |
| tRNAArg + KCl | #2 | 18.9 ± 0.1 | 19.5 ± 0.1 | 64 | 18.6 103 | monomer |
| tRNAArg | #2 | 20.2 ± 0.1 | 20.8 ± 0.1 | 67 | 20.2 103 | monomer |
| tRNAIle+ KCl | #2 | 20.6 ± 0.1 | 21.4 ± 0.1 | 69 | 18.6 103 | monomer |
| tRNAIle | #2 | 20.2 ± 0.1 | 20.9 ± 0.1 | 67 | 19.7 103 | monomer |
| tRNAPhe transcript | 22.6 ± 0.1 | 22.8 ± 0.1 | 74 | 28.6 103 | monomer | |
| tRNAPhe native | 23.3 ± 0.1 | 24.0 ± 0.1 | 81 | 29.6 103 | monomer |
The shape of SAXS profiles is related to the shape of a molecule. The SAXS analysis for peak 2 thus confirms that armless tRNAs adopt a shape close to that of a canonical tRNA, since their SAXS profiles are similar to that of tRNAPhe (Figure 4C). The reduction of Rg (Table 1) is in agreement with compact RNAs with shorter sequences (42–47 nt instead of 72 or 75, for the tRNAPhe transcript or native form, respectively) and that of the maximum distance (Dmax) determined in the pair-distribution function p(r) (Supplementary Figure S2), is in line with the absence of the CCA tail in the prepared transcripts.
Ensembles of ab initio pseudo-atomic models reproducing experimental SAXS curves were generated for the three transcripts. Supplementary Figure S3 shows representative 3D models obtained for tRNAArg, tRNAIle and tRNAPhe. These models look quite similar in size and shape to the crystal structure of tRNAPhe (PDB ID: 1EHZ) (47).
Atomic models of armless tRNAs were built based on the secondary structure proposed by Wende et al. (18) that we verified here by in-line probing and NMR. These initial models were then disturbed along their normal modes using the elastic network approach (33) to select tRNA conformers that better fitted the experimental data (Supplementary Figure S4). The result of the ab initio and all-atom modeling is summarized in Figure 5, showing the superposition of two models for each tRNA transcript. Overall, armless tRNAs present a slightly more compact and straight shape, but a size comparable to that of canonical tRNAs.
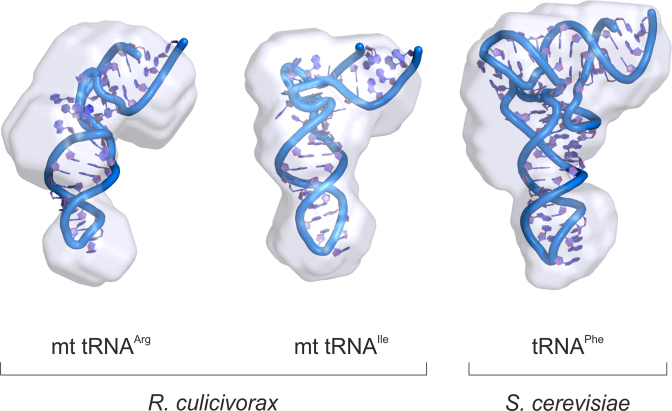
Figure 5.
Shape of armless tRNAs. All-atom models (see Supplementary Figure S4) of Romanomermis culicivorax mitochondrial tRNAArg and tRNAIle are compared with that of Saccharomyces cerevisiae cytosolic tRNAPhe. These models are fitted into the volume of corresponding pseudo-atomic models (see Supplementary Figure S3). Note that all models, alike analyzed molecules, lack their 3′ CCA tail. They are displayed at the same scale, illustrating the shape similarity of armless tRNAs with canonical tRNAs.
DISCUSSION
In many metazoans, mitochondrially encoded tRNAs show considerable deviations in length and secondary structures. In particular, mammalian tRNASer(AGY) was the first identified bizarre tRNA, where the D-arm is replaced by a single-stranded connector region (9,10). Similarly, truncated mitochondrial tRNAs were discovered in nematodes, where either the T arm and the variable loop or the D-arm is missing (12,13). With a length of 54 nt, mt tRNASer(AGY) from Ascaris suum was believed to represent the shortest functional tRNA possible (48). Yet, sequence analyses of additional mitochondrial genomes in other nematode (like R. culicivorax) and arthropod species (like Dermatophagoides farina and Steganacarus magnus) predicted even shorter tRNAs, with the most truncated tRNAArg (42 nt) in R. culicivorax (15,16,49). At least in the latter species, several of the mt tRNAs were identified at the tRNA level, indicating that these transcripts represent indeed functional, yet highly abnormal, tRNA molecules (18).
The presented structural analysis indicates that the in vitro transcripts of tRNAArg and tRNAIle, two of the identified transcripts in R. culicivorax, fold into a single conformation that is nearly identical to the predicted hairpin-like structure (Figure 2) (18). A consequence of the high AU content of the transcripts is that stabilizing GC base pairs are quite rare. tRNAArg carries a single C4-G38 in the acceptor stem, and two G13-C25 and G14-C24 in the anticodon domain. In tRNAIle, the only CG base pair is found at position 1–46 in the acceptor stem (Figure 2). Furthermore, the predicted free energy for the secondary structures of these tRNAs is substantially lower (tRNAArg: −4.40 kcal/mol; tRNAIle: −1.9 kcal/mol) compared to the (more canonical) human mitochondrial tRNAArg (−6.0 kcal/mol; calculations were done using RNAeval from the Vienna RNA package (20)). Yet, both bizarre tRNA molecules fold into a single and rather stable structure that shows remarkable robustness against cleavage over a prolonged incubation time (up to 48 h), supporting the observation of stable single conformers (Figure 2A). Surprisingly, the tRNA structures were not only stable at low temperatures (22°C) corresponding to the optimal growth temperature of R. culicivorax (50), but remained unchanged up to 37°C. Only at 45°C, a denaturation of both transcripts was observed, correlating with the upper growth limit of Romanomermis. The considerable stability of these mitochondrial armless tRNAs is a very particular characteristic compared to other in vitro transcribed tRNAs (51–53).
Furthermore, the structural probing as well as the NMR analysis of both mitochondrial tRNAs identified only canonical Watson–Crick base pairings, while tertiary interactions were not recognized. Hence, it is highly likely that these transcripts indeed fold into a shape that exclusively consists of the described base-paired and single-stranded elements. In general, the 3D structure of classical tRNAs is stabilized by long-range base pair interactions and base stacking, especially between D- and T-loop regions (54). However, based on our results, this seems not to be the case in the characterized tRNAs. The replacements for the D- and the T-arm obviously represent structurally flexible and unpaired connector modules as they were described for D- or T-arm replacements in metazoan mt tRNAs (48,55,56). While both transcripts share a stem-bulge-stem architecture, single- as well as double-stranded regions show considerable differences in size. In both tRNAs, the acceptor stems are rather similar with 6 (tRNAArg) and 5 bp (tRNAIle), respectively. The anticodon stems, however, show a dramatic size difference with four and nine base pairs. A ninth base pair U22-A28 in tRNAIle, as it is mapped in the in-line probing experiment (Figure 2), results from the frequently observed anticodon loop collapse of in vitro transcribed tRNA, where base modifications downstream of the anticodon are missing that usually eliminate such additional base pairs (43,57). In tRNAArg, the connector replacing the D-arm consists of five unpaired nucleotides, while in tRNAIle, it contains eight residues. The T-arm replacing connector, however, has a complementary size, with 9 nt in tRNAArg and 5 nt in tRNAIle. Whether this size complementarity (D connector 5 nt/T connector 9 nt in tRNAArg and D connector 8 nt/T connector 5 nt in tRNAIle) is essential and a prerequisite for a similar 3D shape remains to be clarified. Surprisingly, the Romanomermis tRNAs show significant differences to other mitochondrial tRNAs lacking D- or T-arms. According to Steinberg et al., tRNAs with D-arm replacements show a conserved length of both strands in the anticodon arm, with nine residues between anticodon loop and T-stem and ten residues between anticodon loop and acceptor stem, and these distances seem to be essential for a proper folding of the bizarre tRNA (48). In the transcripts described here, however, these distances are not found, and our in-line probing and NMR data do not provide any evidence for the existence of stable tertiary interactions in the connector regions. This is remarkable since such interactions usually stabilize 3D structures of RNAs, and in particular tRNAs (1,58). Yet, the SAXS data indicate that both tRNAArg and tRNAIle fold into a 3D architecture that looks very similar to the shapes described for tRNAs that lack only the D- or T-arm (48,55,56), and the characteristic Rg of armless tRNA molecules (19–21 Å) is only slightly lower compared to a canonical tRNA (23 Å), indicating a more compact shape. This is in line with the observed sequence reduction in the central part of the tRNA body.
Another interesting parameter is the Dmax of the tRNA molecules derived from the pair–distribution function. This maximum distance represents the distance between residues 35 at the apex of the anticodon loop and the 3′ extremity. Dmax is estimated at 81 Å for the native tRNAPhe, as observed by Rambo and Tainer (59), which is close to the distance (75 Å) between the ends of the two arms in the crystal structure. While the latter adopts an L-shape with an angle of 90° (PDB ID: 1EHZ) (47), it has also been demonstrated that the tRNAPhe core can exhibit higher intrinsic flexibility in solution (60). We also observe that the model that best describes the data is more open than the crystal structure (Supplementary Figure S4). This Dmax value likely reflects the opening of the angle of the tRNA molecule, which appears slightly more extended in solution than in a constraint context of a crystal packing. This intrinsic flexibility is probably even more pronounced in armless tRNAs, with their internal bulge region, high A/U content and absence of tertiary interactions. With a Dmax of 64–69 Å, they are still close to a tRNAPhe transcript (Dmax 74 Å) which is also deprived of CCA tail. If we consider an increased intrinsic flexibility for armless tRNAs with a CCA tail representing an extension of ∼10–12 Å, the size of these molecules in solution would be comparable to that of classical tRNAs.
Interestingly, these tRNAs with single arm replacements (D- or T-arm) are found in mitochondrial systems where the residual tRNAs are rather similar to the canonical tRNA cloverleaf structure and L-shaped 3D form. Hence, it is believed that the single connector elements in these transcripts have an increased flexibility in order to allow the formation of an extended 3D tRNA shape, bringing anticodon loop and 3′-terminus into a conserved distance that is required for acceptance by the ribosome and, consequently, participation in translation. This shape deviates from the classical L form and is more boomerang-like (48,55,56). However, this seems to be a general characteristic of R. culicivorax and other Enoplea, since their mitochondrial genome does not encode any canonical tRNA, and all 22 mt tRNAs being either D-, T- or completely armless (16).
In the mitochondrial D- or T-arm- lacking tRNAs from Bos taurus and A. suum, several post-transcriptional nucleoside modifications are described. In the anticodon loop, modifications like 5-carboxymethylaminomethyluridine (cmnm5U) at position 34 are involved in wobble interaction with mRNA codons (61), while at position 37, predominantly 1-methylguanine (m1G), N6-threonylcarbamoyladenosie (t6A) are found, keeping the anticodon loop in an open conformation, including a U-turn that is required for efficient translation (43,44,62,63). Modifications that have an impact on the overall structure are 1-methyladenosine (m1A9) in tRNAs lacking the T-loop, and several pseudouridine (Ψ) positions in acceptor- and anticodon stems (63). While m1A9 dos not contribute to the overall stability of the tRNAs, it is well documented that Ψ exerts such an effect (44,64–66). However, in the case of the mitochondrial tRNAs from R. culicivorax, no modifications were identified yet, and it is possible that similarly modified positions contribute to the structural stability and functionality of these transcripts.
While the translational machinery in several mitochondrial systems is quite well characterized, it is completely uncharted in Enoplea. In arthropods and nematodes like Caenorhabditis elegans, a specialized EF-Tu variant was identified that carries a C-terminal extension to specifically interact with T-armless tRNAs, while the standard mitochondrial counterpart binds only canonical tRNAs with both D- and T-arms (67,68). It is highly likely that a similar, maybe even more pronounced, adaptation took place in R. culicivorax. However, as an annotated genome of this organism is not yet available, the corresponding EF-Tu genes are not accessible for such a comparison. A further indication that loss of RNA regions in the translational machinery is replaced and compensated by an increase in protein composition is found in mitochondrial ribosomes. Indeed, additional ribosomal proteins without recognizable homologs in prokaryotic or eukaryotic cytoplasmic proteomes have evolved to replace rRNA elements (69). Again, in nematode mitochondria, the ribosomes are even more striking in their rRNA loss and are much more protein-enriched, and it seems that this increased protein part compensates for the size reduction in the mitochondrial ribosomal RNA (70–72). Whether such compensations are also important for the functionality of miniaturized armless tRNAs in translation remains to be clarified. Clearly, mitochondria of nematodes, and especially Enoplea, bear many unresolved and interesting conundrums in their highly specialized protein synthesis, rendering these organisms worthwhile to be studied in detail. As they indicate that the translational machinery is highly tolerant in terms of size and substrate (tRNA) specificity, such systems will shed light on the evolutionary adaptability of protein synthesis.
DATA AVAILABILITY
SAXS data of armless tRNAs and derived models were deposited in the SASBDB with the accession codes SASDDF7 and SASDDG7.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge P. Roblin and the team of the SWING beamline (SOLEIL synchrotron, Saint-Aubin, France) for synchrotron beam time provision, assistance during data collection and processing, as well as R. de Wijn (IBMC, Strasbourg) for performing the SAXS analysis of the tRNAPhe transcript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft (DFG) [Mo 634/10-1]; French Centre National de la Recherche Scientifique; University of Strasbourg; LabEx consortia ‘MitoCross’ [ANR-11-LABX-0057_MITOCROSS]; ‘NetRNA’ [ANR-10-LABX-0036_NETRNA]; Excellence initiative (IdEx) of the University of Strasbourg in the frame of the French National Program ‘Investissements d’Avenir’; Franco-German University (DFH-UFA PhD track program UdS-Bio and Co-tutelle de Thèse CT-06-16); PROCOPE Hubert Curien cooperation program (French Ministry of Foreign Affair and DAAD); Center of Biomolecular Magnetic Resonance (BMRZ); Goethe-University Frankfurt; State of Hessen. We acknowledge support from the Deutsche Forschungsgemeinschaft (DFG) and Leipzig University within the program of Open Access Publishing.
Conflict of interest statement. None declared.
REFERENCES
- 1. Giegé R., Jühling F., Pütz J., Stadler P., Sauter C., Florentz C.. Structure of transfer RNAs. Similarity and variability. Wiley Interdiscip. Rev. RNA. 2012; 3:37–61. [DOI] [PubMed] [Google Scholar]
- 2. Watanabe Y.-I., Suematsu T., Ohtsuki T.. Losing the stem-loop structure from metazoan mitochondrial tRNAs and co-evolution of interacting factors. Front. Genet. 2014; 5:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barciszewska M.Z., Perrigue P.M., Barciszewski J.. tRNA–the golden standard in molecular biology. Mol. Biosyst. 2016; 12:12–17. [DOI] [PubMed] [Google Scholar]
- 4. Fernández-Millán P., Schelcher C., Chihade J., Masquida B., Giegé P., Sauter C.. Transfer RNA. From pioneering crystallographic studies to contemporary tRNA biology. Arch. Biochem. Biophys. 2016; 602:95–105. [DOI] [PubMed] [Google Scholar]
- 5. Florentz C., Sohm B., Tryoen-Tóth P., Pütz J., Sissler M.. Human mitochondrial tRNAs in health and disease. Cell. Mol. Life Sci. 2003; 60:1356–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Suzuki T., Nagao A., Suzuki T.. Human mitochondrial tRNAs: biogenesis, function, structural aspects, and diseases. Annu. Rev. Genet. 2011; 45:299–329. [DOI] [PubMed] [Google Scholar]
- 7. Abbott J.A., Francklyn C.S., Robey-Bond S.M.. Transfer RNA and human disease. Front. Genet. 2014; 5:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Russell O., Turnbull D.. Mitochondrial DNA disease-molecular insights and potential routes to a cure. Exp. Cell Res. 2014; 325:38–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Arcari P., Brownlee G.G.. The nucleotide sequence of a small (3S) seryl-tRNA (anticodon GCU) from beef heart mitochondria. Nucleic Acids Res. 1980; 8:5207–5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. de Bruijn M.H., Schreier P.H., Eperon I.C., Barrell B.G., Chen E.Y., Armstrong P.W., Wong J.F., Roe B.A.. A mammalian mitochondrial serine transfer RNA lacking the “dihydrouridine” loop and stem. Nucleic Acids Res. 1980; 8:5213–5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ueda T., Ohta T., Watanabe K.. Large scale isolation and some properties of AGY-specific serine tRNA from bovine heart mitochondria. J. Biochem. 1985; 98:1275–1284. [DOI] [PubMed] [Google Scholar]
- 12. Wolstenholme D.R., Macfarlane J.L., Okimoto R., Clary D.O., Wahleithner J.A.. Bizarre tRNAs inferred from DNA sequences of mitochondrial genomes of nematode worms. Proc. Natl. Acad. Sci. U.S.A. 1987; 84:1324–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Okimoto R., Wolstenholme D.R.. A set of tRNAs that lack either the T psi C arm or the dihydrouridine arm. Towards a minimal tRNA adaptor. EMBO J. 1990; 9:3405–3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Watanabe Y., Tsurui H., Ueda T., Furushima R., Takamiya S., Kita K., Nishikawa K., Watanabe K.. Primary and higher order structures of nematode (Ascaris suum) mitochondrial tRNAs lacking either the T or D stem. J. Biol. Chem. 1994; 269:22902–22906. [PubMed] [Google Scholar]
- 15. Klimov P.B., Oconnor B.M.. Improved tRNA prediction in the American house dust mite reveals widespread occurrence of extremely short minimal tRNAs in acariform mites. BMC Genomics. 2009; 10:598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jühling F., Pütz J., Florentz C., Stadler P.F.. Armless mitochondrial tRNAs in Enoplea (Nematoda). RNA Biol. 2012; 9:1161–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Palopoli M.F., Minot S., Pei D., Satterly A., Endrizzi J.. Complete mitochondrial genomes of the human follicle mites Demodex brevis and D. folliculorum. Novel gene arrangement, truncated tRNA genes, and ancient divergence between species. BMC Genomics. 2014; 15:1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wende S., Platzer E.G., Jühling F., Pütz J., Florentz C., Stadler P.F., Mörl M.. Biological evidence for the world's smallest tRNAs. Biochimie. 2014; 100:151–158. [DOI] [PubMed] [Google Scholar]
- 19. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003; 31:3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lorenz R., Bernhart S.H., Höner zu, Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L.. ViennaRNA Package 2.0. Algorithm. Mol. Biol. 2011; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Darty K., Denise A., Ponty Y.. VARNA. Interactive drawing and editing of the RNA secondary structure. Bioinformatics. 2009; 25:1974–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schürer H., Lang K., Schuster J., Mörl M.. A universal method to produce in vitro transcripts with homogeneous 3′ ends. Nucleic Acids Res. 2002; 30:e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mörl M., Lizano E., Willkomm D.K., Hartmann R.K.. Hartmann RK, Bindereif A, Schön A, Westhof E. Production of RNAs with Homogeneous 5′- and 3′-Ends. Handbook of RNA Biochemistry. 2012; Weinheim; Chichester: Wiley-VCH; 22–35. [Google Scholar]
- 24. Domin G., Findeiß S., Wachsmuth M., Will S., Stadler P.F., Mörl M.. Applicability of a computational design approach for synthetic riboswitches. Nucleic Acids Res. 2017; 45:4108–4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fürtig B., Richter C., Wöhnert J., Schwalbe H.. NMR spectroscopy of RNA. Chembiochem. 2003; 4:936–962. [DOI] [PubMed] [Google Scholar]
- 26. David G., Pérez J.. Combined sampler robot and high-performance liquid chromatography. A fully automated system for biological small-angle X-ray scattering experiments at the Synchrotron SOLEIL SWING beamline. J. Appl. Crystallogr. 2009; 42:892–900. [Google Scholar]
- 27. Brookes E., Vachette P., Rocco M., Pérez J.. US-SOMO HPLC-SAXS module. Dealing with capillary fouling and extraction of pure component patterns from poorly resolved SEC-SAXS data. J. Appl. Crystallogr. 2016; 49:1827–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Franke D., Petoukhov M.V., Konarev P.V., Panjkovich A., Tuukkanen A., Mertens H.D.T., Kikhney A.G., Hajizadeh N.R., Franklin J.M., Jeffries C.M et al. . ATSAS 2.8. A comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J. Appl. Crystallogr. 2017; 50:1212–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Konarev P.V., Volkov V.V., Sokolova A.V., Koch M.H.J., Svergun D.I.. PRIMUS. A Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003; 36:1277–1282. [Google Scholar]
- 30. Svergun D.I. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992; 25:495–503. [Google Scholar]
- 31. Franke D., Svergun D.I.. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2009; 42:342–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Popenda M., Szachniuk M., Antczak M., Purzycka K.J., Lukasiak P., Bartol N., Blazewicz J., Adamiak R.W.. Automated 3D structure composition for large RNAs. Nucleic Acids Res. 2012; 40:e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Suhre K., Sanejouand Y.-H.. ElNemo. A normal mode web server for protein movement analysis and the generation of templates for molecular replacement. Nucleic Acids Res. 2004; 32:W610–W614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Svergun D., Barberato C., Koch M.H.J.. CRYSOL – a Program to Evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 1995; 28:768–773. [Google Scholar]
- 35. Schrödinger L.L.C. The PyMOL molecular graphics system. 2015; Version 1.8.
- 36. Panjkovich A., Svergun D.I.. SASpy. A PyMOL plugin for manipulation and refinement of hybrid models against small angle X-ray scattering data. Bioinformatics. 2016; 32:2062–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.-W., Kapral G.J., Grosse-Kunstleve R.W et al. . PHENIX. A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Valentini E., Kikhney A.G., Previtali G., Jeffries C.M., Svergun D.I.. SASBDB, a repository for biological small-angle scattering data. Nucleic Acids Res. 2015; 43:D357–D363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Regulski E.E., Breaker R.R.. In-line probing analysis of riboswitches. Methods Mol. Biol. 2008; 419:53–67. [DOI] [PubMed] [Google Scholar]
- 40. Sprinzl M., Horn C., Brown M., Ioudovitch A., Steinberg S.. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1998; 26:148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jühling F., Mörl M., Hartmann R.K., Sprinzl M., Stadler P.F., Pütz J.. tRNAdb 2009. Compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009; 37:D159–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cabello-Villegas J., Winkler M.E., Nikonowicz E.P.. Solution conformations of unmodified and A37N6-dimethylallyl modified anticodon stem-loops of Escherichia coli tRNAPhe. J. Mol. Biol. 2002; 319:1015–1034. [DOI] [PubMed] [Google Scholar]
- 43. Agris P.F. Bringing order to translation: the contributions of transfer RNA anticodon-domain modifications. EMBO Rep. 2008; 9:629–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lorenz C., Lünse C.E., Mörl M.. tRNA Modifications. Impact on structure and thermal adaptation. Biomolecules. 2017; 7:E35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Römer R., Hach R.. tRNA conformation and magnesium binding. A study of a yeast phenylalanine-specific tRNA by a fluorescent indicator and differential melting curves. Eur. J. Biochem. 1975; 55:271–284. [DOI] [PubMed] [Google Scholar]
- 46. Stein A., Crothers D.M.. Conformational changes of transfer RNA. The role of magnesium(II). Biochemistry. 1976; 15:160–168. [DOI] [PubMed] [Google Scholar]
- 47. Shi H., Moore P.B.. The crystal structure of yeast phenylalanine tRNA at 1.93 A resolution: a classic structure revisited. RNA. 2000; 6:1091–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Steinberg S., Gautheret D., Cedergren R.. Fitting the structurally diverse animal mitochondrial tRNAs(Ser) to common three-dimensional constraints. J. Mol. Biol. 1994; 236:982–989. [DOI] [PubMed] [Google Scholar]
- 49. Domes K., Maraun M., Scheu S., Cameron S.L.. The complete mitochondrial genome of the sexual oribatid mite Steganacarus magnus. Genome rearrangements and loss of tRNAs. BMC Genomics. 2008; 9:532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hughes D.S., Platzer E.G.. Temperature effects on the parasitic phase of Romanomermis culicivorax in Culex pipiens. J. Nematol. 1977; 9:173–175. [PMC free article] [PubMed] [Google Scholar]
- 51. Bhaskaran H., Rodriguez-Hernandez A., Perona J.J.. Kinetics of tRNA folding monitored by aminoacylation. RNA. 2012; 18:569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Derrick W.B., Horowitz J.. Probing structural differences between native and in vitro transcribed Escherichia coli valine transfer RNA. Evidence for stable base modification-dependent conformers. Nucleic Acids Res. 1993; 21:4948–4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sampson J.R., Uhlenbeck O.C.. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl. Acad. Sci. U.S.A. 1988; 85:1033–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Giegé R. Toward a more complete view of tRNA biology. Nat. Struct. Mol. Biol. 2008; 15:1007–1014. [DOI] [PubMed] [Google Scholar]
- 55. Wolstenholme D.R., Okimoto R., Macfarlane J.L.. Nucleotide correlations that suggest tertiary interactions in the TV-replacement loop-containing mitochondrial tRNAs of the nematodes, Caenorhabditis elegans and Ascaris suum. Nucleic Acids Res. 1994; 22:4300–4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ohtsuki T., Kawai G., Watanabe K.. The minimal tRNA. Unique structure of Ascaris suum mitochondrial tRNA(Ser) (UCU) having a short T arm and lacking the entire D arm. FEBS Lett. 2002; 514:37–43. [DOI] [PubMed] [Google Scholar]
- 57. Cabello-Villegas J., Tworowska I., Nikonowicz E.P.. Metal ion stabilization of the U-turn of the A37 N6-dimethylallyl-modified anticodon stem-loop of Escherichia coli tRNAPhe. Biochemistry. 2004; 43:55–66. [DOI] [PubMed] [Google Scholar]
- 58. Westhof E., Auffinger P.. Transfer RNA structure. Encyclopedia of Life Sciences. 2012; Chichester: John Wiley & Sons. [Google Scholar]
- 59. Rambo R.P., Tainer J.A.. Improving small-angle X-ray scattering data for structural analyses of the RNA world. RNA. 2010; 16:638–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Friederich M.W., Vacano E., Hagerman P.J.. Global flexibility of tertiary structure in RNA. Yeast tRNAPhe as a model system. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:3572–3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sakurai M., Ohtsuki T., Suzuki T., Watanabe K.. Unusual usage of wobble modifications in mitochondrial tRNAs of the nematode Ascaris suum. FEBS Lett. 2005; 579:2767–2772. [DOI] [PubMed] [Google Scholar]
- 62. Dao V., Guenther R., Malkiewicz A., Nawrot B., Sochacka E., Kraszewski A., Jankowska J., Everett K., Agris P.F.. Ribosome binding of DNA analogs of tRNA requires base modifications and supports the “extended anticodon”. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:2125–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sakurai M., Ohtsuki T., Watanabe K.. Modification at position 9 with 1-methyladenosine is crucial for structure and function of nematode mitochondrial tRNAs lacking the entire T-arm. Nucleic Acids Res. 2005; 33:1653–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Arnez J.G., Steitz T.A.. Crystal structure of unmodified tRNA(Gln) complexed with glutaminyl-tRNA synthetase and ATP suggests a possible role for pseudo-uridines in stabilization of RNA structure. Biochemistry. 1994; 33:7560–7567. [DOI] [PubMed] [Google Scholar]
- 65. Davis D.R. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res. 1995; 23:5020–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kierzek E., Malgowska M., Lisowiec J., Turner D.H., Gdaniec Z., Kierzek R.. The contribution of pseudouridine to stabilities and structure of RNAs. Nucleic Acids Res. 2014; 42:3492–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ohtsuki T., Watanabe Y., Takemoto C., Kawai G., Ueda T., Kita K., Kojima S., Kaziro Y., Nyborg J., Watanabe K.. An “elongated” translation elongation factor Tu for truncated tRNAs in nematode mitochondria. J. Biol. Chem. 2001; 276:21571–21577. [DOI] [PubMed] [Google Scholar]
- 68. Sakurai M., Watanabe Y.-I., Watanabe K., Ohtsuki T.. A protein extension to shorten RNA. Elongated elongation factor-Tu recognizes the D-arm of T-armless tRNAs in nematode mitochondria. Biochem. J. 2006; 399:249–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. O’Brien T.W. Evolution of a protein-rich mitochondrial ribosome. Implications for human genetic disease. Gene. 2002; 286:73–79. [DOI] [PubMed] [Google Scholar]
- 70. Suzuki T., Terasaki M., Takemoto-Hori C., Hanada T., Ueda T., Wada A., Watanabe K.. Structural compensation for the deficit of rRNA with proteins in the mammalian mitochondrial ribosome. Systematic analysis of protein components of the large ribosomal subunit from mammalian mitochondria. J. Biol. Chem. 2001; 276:21724–21736. [DOI] [PubMed] [Google Scholar]
- 71. Zhao F., Ohtsuki T., Yamada K., Yoshinari S., Kita K., Watanabe Y.-I., Watanabe K.. Isolation and physiochemical properties of protein-rich nematode mitochondrial ribosomes. Biochemistry. 2005; 44:9232–9237. [DOI] [PubMed] [Google Scholar]
- 72. Watanabe K. Unique features of animal mitochondrial translation systems. The non-universal genetic code, unusual features of the translational apparatus and their relevance to human mitochondrial diseases. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2010; 86:11–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
SAXS data of armless tRNAs and derived models were deposited in the SASBDB with the accession codes SASDDF7 and SASDDG7.