Abstract
S-adenosyl-L-methionine-dependent 2′-O-methylati-on of the 3′-terminal nucleotide plays important roles in biogenesis of eukaryotic small non-coding RNAs, such as siRNAs, miRNAs and Piwi-interacting RNAs (piRNAs). Here we demonstrate that, in contrast to Mg2+/Mn2+-dependent plant and bacterial homologues, the Drosophila DmHen1 and human HsHEN1 piRNA methyltransferases require cobalt cations for their enzymatic activity in vitro. We also show for the first time the capacity of the animal Hen1 to catalyse the transfer of a variety of extended chemical groups from synthetic analogues of the AdoMet cofactor onto a wide range (22–80 nt) of single-stranded RNAs permitting their 3′-terminal functionalization and labelling. Moreover, we provide evidence that deletion of a small C-terminal region of the DmHen1 protein further increases its modification efficiency and abolishes a modest 3′-terminal nucleotide bias observed for the full-length protein. Finally, we show that fluorophore-tagged ssRNA molecules are successfully detected in fluorescence resonance energy transfer assays both individually and in a total RNA mixture. The presented DmHen1-assisted RNA labelling provides a solid basis for developing novel chemo-enzymatic approaches for in vitro studies and in vivo monitoring of single-stranded RNA pools.
INTRODUCTION
RNA molecules play a central role in mediating gene regulation and converting the genetic information from DNA into amino acid sequence of peptides or proteins (1). Basic biological and medical studies seek to analyse RNA functions and subcellular localization, which demands efficient post-transcriptional labelling methods for manipulating and profiling RNAs. While non-covalent approaches for single-stranded RNA labelling are hybridization-based or utilize RNA-binding proteins or aptamers, covalent chemo-enzymatic labelling typically exploits modification enzymes such as terminal transferases, polymerases, ligases and kinases (2). In the past few years methyltransferase-assisted strategies showed promise for covalent targeted RNA tagging (2, 3). According to the most used approach, named mTAG (4), an unnatural side-chain possessing a functional group or a reporter is transferred onto RNA target from synthetic analogue of cofactor S-adenosyl-L-methionine (AdoMet). The current toolbox of RNA methyltransferases has been adjusted for functionalization of the 5′-cap of mRNA (5, 6), an invariable nucleotide in a particular tRNA (7), a defined internal position of RNA using programmable C/D box RNP complex (8) and for addressable targeting of microRNAs (miRNAs) and small interfering RNAs (siRNAs) (9, 10). However, to date no comparable approach has been appropriated for the derivatization of 3′-termini of single-stranded RNAs.
AdoMet-dependent methylation of the 2′-hydroxyl of the 3′-terminal nucleotide is a part of biogenesis of eukaryotic small non-coding RNAs (ncRNAs), such as siRNAs, miRNAs and Piwi-interacting RNAs (piRNAs) (11). Aberrant modification of 3′-ends has detrimental consequences for the abundance and functionality of endogenous small ncRNAs (12–15). The HEN1 plant 2′-O-methyltransferase appends methyl groups to short 21–24 nt double-stranded miRNAs/miRNAs* and siRNAs/siRNAs* (16–18). In contrast, its animal counterparts exhibit an apparent specificity towards single-stranded piRNAs and siRNAs (Figure 1A) (19–21). Different subfamilies of 2′-O-methyltransferases share a highly conservative catalytic domain essential for the methylation activity but have adopted diverse domain assemblies to facilitate the recognition of specific substrate types. Hence, the plant methyltransferases carry a large (∼700 amino acid residues) N-terminal region, which serves to bind properly-sized double-stranded RNA (dsRNA) substrates and interact with other cellular proteins (22, 23). The animal Hen1 homologues lack the N-terminal dsRNA binding domains but contain an ∼100 residue-long C-terminal region lacking bioinformatically predicted functional motifs. This region appears to mediate interactions of Hen1 with PIWI proteins during piRNA biogenesis (19, 21) and anchoring to the meiotic germline-specific organelle, called nuage (12). The recombinant animal Hen1 promiscuously methylates 20–40 nt long single-stranded RNAs in vitro suggesting no intrinsic structural-functional constraints for the length of the modified substrates (20). It is likely that, other host factors sustain the specific recognition of piRNAs and particular siRNAs by Hen1 in the cell.
Figure 1.
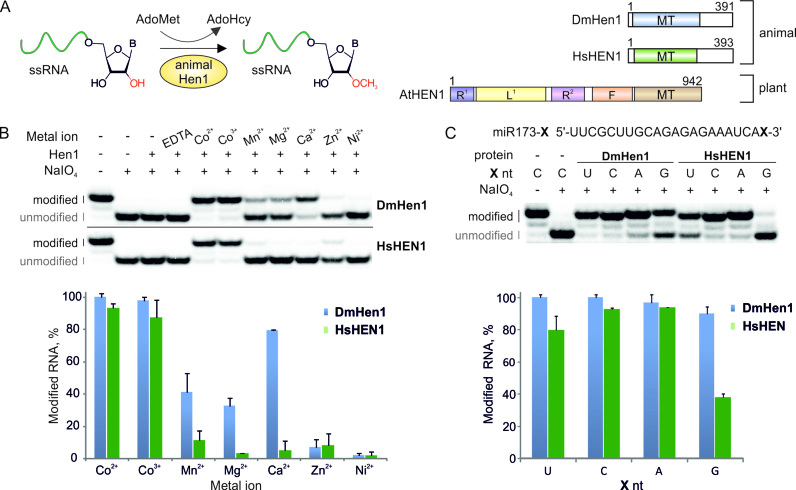
DmHen1 and HsHEN1 methyltransferases require cobalt cations for efficient modification of terminal nucleosides of single-stranded RNA. (A) Schematic representation of animal Hen1 methylation reaction together with structures of Drosophila melanogaster DmHen1, Homo sapiens HsHEN1 and Arabidopsis thaliana AtHEN1 methyltransferases. MT: methyltransferase domain; R1 and R2: dsRNA binding domains; L: La-motif-containing domain; F: FK506 binding protein-like domain. (B) Only the presence of Co2+ or Co3+ confers full methylation of RNA substrate. Reactions were performed using 0.2 μM of 5′-32P-labelled 22-nt miR173, 0.1 mM of AdoMet 1, 10 mM of metal ion in the form of chloride salt (CoCl2 or [Co(NH2)6]Cl3 in the case of Co2+ or Co3+, respectively), except for nickel sulphate and 1 μM of Hen1 proteins. After 30 min of incubation at 37°C, the extent of modification was determined by NaIO4-mediated oxidation/β-elimination as described in (24). This treatment resulted in shortening of unmodified RNA by 1 nt and increasing its mobility relatively to the modified one. Modified and unmodified RNA strands are marked in black and grey, accordingly. Histogram of RNA methylation efficiency with different metal cofactors was calculated from duplicate experiments. (C) Co2+ reduces 3′ terminal nucleoside bias in DmHen1 methylation reactions. miR173 with different 3′ terminal nucleosides, namely U, C, A and G, was used as a substrate for DmHen1 or HsHEN1. Results presented in a histogram are average of two experiments.
In this report, we studied enzymatic parameters important for the target RNA modification by the fly DmHen1 and human HsHEN1 piRNA methyltransferases. Our experiments revealed that efficient modification of single-stranded RNAs up to 80 nt is dependent on cobalt cations in vitro. We have developed novel approaches for DmHen1-directed single-step or two-step mTAG labelling resulting in 3′-terminal attachment of fluorophore or biotin reporters to selected single-stranded RNAs using a series of synthetic AdoMet analogues and demonstrated applicability of this approach for fluorescence resonance energy transfer (FRET) analysis of ssRNA molecules.
MATERIALS AND METHODS
Construction of plasmids and purification of recombinant proteins
Detailed description of construction of plasmids carrying recombinant proteins, their expression and purification (24) is given in Supplementary Methods.
Preparation of long RNA substrates and cofactor AdoMet analogues
43 nt and longer RNA substrates were in vitro transcribed using TranscriptAid T7 High Yield Transcription Kit (Thermo Scientific) according to manufacturer’s recommendations. The templates for in vitro transcription were prepared as detailed in Supplementary Methods. Cofactor AdoMet 1 analogues Ado-6-amine 2, Ado-6-azide 3 and Ado-13-biotin 4 were prepared as outlined elsewhere (10, 25). The synthesis of Ado-14-Cy3 5 is described in Supplementary Methods. Chemical structures of cofactors used are shown in Supplementary Figure S1. High-performance liquid chromatography (HPLC) and mass spectrometry (MS) data of synthetic cofactors are given in Supplementary Figure S2.
RNA modification and analysis
RNA modification reactions, analysis of their products on denaturing polyacrylamide gel and HPLC-MS analysis of modified nucleosides are described in detail in Supplementary Methods.
Kinetic analysis of modification reaction
Modification reactions under single turnover conditions were performed with 0.2 μM 32P-RNA, 2 μM Hen1, 0.1 mM AdoMet 1/Ado-6-amine 2/Ado-6-azide 3 or 0.05 mM Ado-13-biotin 4 in the Reaction buffer containing 10 mM Tris–HCl (pH 7.4), 50 mM NaCl, 5% glycerol, 10 mM Co2+ in the form of CoCl2 salt, 0.2 mM DTT, 0.1 mg/ml bovine serum albumin (BSA) and 0.04 u/μl RiboLock RNase inhibitor (Fisher Scientific). After incubation at 37°C for 0–60 min reactions were quenched at certain time points and analysed on denaturing polyacrylamide gel. kchem values were obtained by using first order kinetics and fitting single-exponential equation to single-turnover experimental data from at least two replicates with the GraFit5 software (Erithacus Software).
Cofactor preference analysis
Analysis of DmHen1 preference for AdoMet 1 or its synthetic analogue Ado-6-azide 3 was carried out with 0.2 μM 32P-siR23 (or siR173 according to Plotnikova et al., (18)), 2 μM protein and 0.1 mM cofactor, which gradually changed from 0.1 mM AdoMet 1 to 0.1 mM Ado-6-azide 3 in 10 μM steps. Samples were processed as described above.
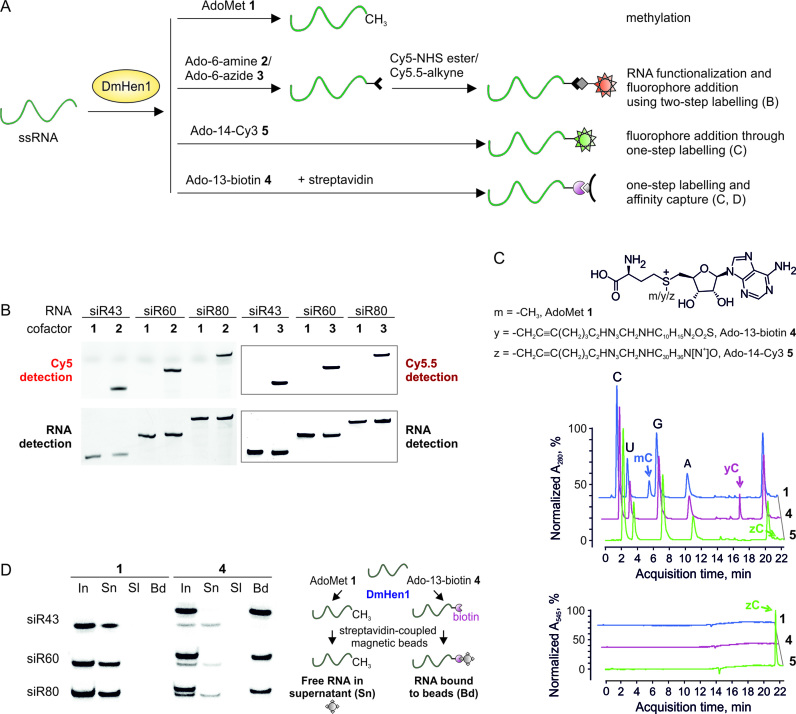
RNA functionalization and fluorophore addition using two-step Hen1-directed mTAG labelling
0.2 μM ssRNAs were incubated with 1 μM DmHen1 and 0.1 mM Ado-6-amine 2/Ado-6-azide 3 or AdoMet 1 (as a control) in the Reaction buffer for 30 min at 37°C. Reactions were quenched with Proteinase K (Thermo Scientific), extracted with phenol-chloroform and precipitated. Amine modified RNA was suspended in 60 mM Borate buffer (pH 8.6) up to 4 μM with 0.05 mg/ml Cy5-649/670-NHS ester (GE Healthcare) and incubated for 1 h at room temperature. A total of 4 μM of azide modified RNA was subsequently used in a click reaction with 0.5 mg/ml Cy5.5-673/707-alkyne (Lumiprobe) in the presence of freshly prepared 3.3 mM CuBr-TBTA and 60% DMSO for 1 h at 40°C. After reactions RNA in all samples was precipitated, dissolved in water, mixed with equal volume of 95% formamide and 10 mM ethylenediaminetetraacetic acid (EDTA) (pH 8.2), heated for 5 min at 85°C and analysed in 10% denaturing PAA gel with 7 M urea. Cy5 and Cy5.5 fluorescence was detected by FLA-5100 Image Reader (Fujifilm) using 635 and 670 nm lasers accordingly, bulk RNA was visualized with 473 nm laser after staining with ethidium bromide.
One-step mTAG RNA labelling and affinity capture
0.05 mM of Ado-13-biotin 4 was used to label 0.01 μM of 32P-RNA substrate in the presence of 1 μM DmHen1 in the Reaction buffer for 30 min at 37°C, were ssRNAs were modified with 0.1 mM AdoMet 1 as a control. After phenol–chloroform extraction and precipitation, RNA was dissolved in water up to 0.2 μM. 10 μl (0.1 mg) of Dynabeads MyOne Streptavidin C1 (Invitrogen) were used per reaction and washed according to manufacturer’s recommendations, then resuspended in 10 μl 2 × binding and wash buffer (10 mM Tris–HCl (pH 7.5), 2 M NaCl, 1 mM EDTA, supplemented with 0.2% Tween-20) and complemented with 5 μl of 0.4 μg/μl tRNA and incubated for 15 min at room temperature at constant rotation. After addition of 5 μl of 0.2 μM biotinylated RNA incubation was continued for additional 15 min. For 23 and 43 nt-long RNA, RNA bound beads were washed according to manufacturer’s recommendations, whilst longer RNA species were washed twice with binding and wash buffer, once with 4 × SSC-formamide (0.6 M NaCl, 0.06 M Na citrate (pH 7.0), 50% formamide) and once with 2 × saline-sodium citrate (SSC). Biotinylated RNA was released from streptavidin beads after 10 min of incubation at 70°C in 95% formamide and 10 mM EDTA (pH 8.2). Samples from each step of experiment, including initial mixture of biotinylated RNA, supernatant collected after RNA incubation with beads and after bead washing and bead bound RNAs were analysed on denaturing 10–13% PAA gel with 7 M urea.
Fluorescence resonance energy transfer (FRET) in solution
1 μM of DmHen1ΔC was used to label 1 μM of siR23 with 3 μM of Ado-14-Cy3 5 or AdoMet 1 as a control in Cy3 reaction buffer composed of 0.1 M Tris–HCl (pH 7.0), 0.2 mM DTT, 0.1 mg/ml BSA, 10 mM Co2+ in the form of CoCl2 salt and 50 μM tryptophan for 1 h at 37°C. Cy3 labelled or methylated RNA was annealed to a complementary DNA with or without Cy5 modification at second or fifth position (DNA-2Cy5 5′-G(Cy5-dT)GATTCTCTCTGCAAGCGTTAA-3′ or DNA-5Cy5 5′-GTGA(Cy5-dT)TCTCTCTGCAAGCGTTAA-3′) in equimolar amount in annealing buffer (7.5 mM HEPES-KOH, 25 mM KCl, 0.5 mM MgCl2, pH 7.4) to a final concentration of 0.2 μM. After diluting RNA to 15 nM in TE buffer (pH 7.0) the fluorescence was analysed with FluoroMax-3 (Jobin Yvon Horiba) spectrofluorimeter with excitation set at 500 or 590 nm and registration of emission from 520 or 610 to 800 nm, respectively, with 1 nm increment, 0.2 s integration time and excitation and emission split of 5 nm. The background fluorescence of RNA-CH3/DNA was subtracted from RNA-CH3/DNA-Cy5, RNA-Cy3/DNA and RNA-Cy3/DNA-Cy5 spectra. Data were analysed in GraphPad Prism software.
For siR23 and let-7a2 labelling and detection through FRET in total RNA the amount of Ado-14-Cy3 5 was increased to 10.3 μM. In addition, the modification reaction was supplemented with 0.5 μg/μl total RNA from HCT116 human colon carcinoma cell line. Total RNA was extracted with RNAzol RT according to manufacturer’s recommendations (Molecular Research Center, Inc.).
RESULTS
Cobalt cations activate animal Hen1 methyltransferases in vitro
Previous studies of various subfamilies of HEN1 defined essential roles of bivalent metal cations for the methylation reaction (11). The enzymatic activities of plant and bacterial HEN1 are strictly dependent on magnesium and manganese ions, correspondingly (22,26). Results presented in Figure 1B indicate that both DmHen1 and HsHEN1 can completely methylate 22-nt single-stranded miR173 identical to Arabidopsis thaliana miRNA in the presence of Co2+ or Co3+ added in the form of CoCl2 or [Co(NH2)6]Cl3 salts, respectively. At the same time, only DmHen1 but not HsHEN1 reaches about 80% methylation efficiency with Ca2+ and is slightly stimulated by Mn2+ and Mg2+. Addition of EDTA to a final concentration of 2 mM inhibits methylation further supporting the assumption that indicated metal ions catalyse the reaction of these enzymes. Hence, we conclude that animal Hen1 are cobalt-dependant methyltransferases in vitro.
Previous studies showed a strong 3′-nt preference (A>C>U>G) of the mouse mHEN1 in the presence of Mg2+ ions (20). Our metal-screening experiments showed that substitution of magnesium for cobalt enhances the catalytic activity of DmHen1 and HsHEN1 up to 40–100% for all RNA substrates and considerably reduces the apparent 3′-end bias, especially in case of the fruit fly methyltransferase (Figure 1C). Thorough kinetic analysis performed under single turnover conditions confirmed that the methyl transfer rates with DmHen1 are only slightly susceptible (3-fold variation of kchem rates) to the nature of the 3′-terminal nucleoside (Table 1). The human methyltransferase also exhibited enhanced activity although with significantly reduced methylation rates at uridine and guanosine as compared to adenosine and cytidine.
Table 1.
Alkylation rate kchem of different 3′-terminal nucleosides in the presence of AdoMet 1 or synthetic cofactor analogues Ado-6-amine 2 and Ado-6-azide 3
| HsHEN1 | DmHen1 | |||
|---|---|---|---|---|
| X nt of miR173-X | k chem, (min−1) | k chem, (min−1) | k chem, (min−1) | k chem, (min−1) |
| (AdoMet 1) | (AdoMet 1) | (Ado-6-amine 2) | (Ado-6-azide 3) | |
| U | 0.16 ± 0.01 | 0.80 ± 0.02 | 1.08 ± 0.09 | 1.17 ± 0.12 |
| C | 0.33 ± 0.03 | 0.61 ± 0.04 | 0.91 ± 0.09 | 1.03 ± 0.05 |
| A | 0.68 ± 0.06 | 0.46 ± 0.02 | 0.54 ± 0.05 | 0.82 ± 0.05 |
| G | 0.08 ± 0.01 | 0.28 ± 0.02 | 0.29 ± 0.02 | 0.47 ± 0.02 |
Models fitted to experimental data are shown in Supplementary Figure S4.
Electrophoretic mobility shift assays revealed that DmHen1 does not form stable complexes with the substrate RNAs irrespective of the presence of cobalt cations and cofactor product S-adenosylhomocysteine (AdoHcy) in the reaction mixture (Supplementary Figure S3). Thus, the sole methyltransferase is incapable of tight RNA binding suggesting that other cellular RNA-binding factor/s are involved in stabilizing its interactions with RNA substrates in vivo.
Transfer of extended side-chains from synthetic AdoMet analogues by animal Hen1 methyltransferases
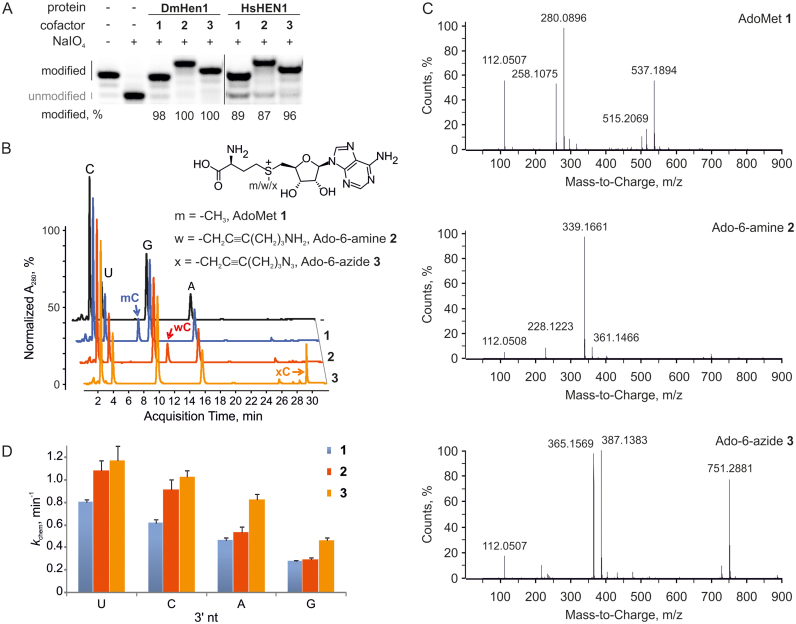
Next, to expand the practical utility of the methyltransferases, we examined their activity under conditions typical for mTAG reactions. For this, the modification of the 3′-terminal nucleoside in single-stranded RNA was monitored in the presence of the natural cofactor AdoMet 1 or its synthetic analogues Ado-6-amine 2 or Ado-6-azide 3 carrying primary amine or azide groups, respectively, in their transferable side chains. We found that both methyltransferases conferred efficient modification of the miR173 substrate (≥87%) regardless of the cofactor used (Figure 2A). Since DmHen1 exhibited a somewhat higher activity in these experiments, it was selected for further studies. Reverse-phase liquid chromatography/mass spectrometry analysis of RNA samples modified with the three cofactors showed the formation of modified nucleosides with molecular mass values matching corresponding alkylated cytidines (Figure 2B and C; Supplementary Table S1), confirming the transfer of intact side-chains onto the RNA substrate. Moreover, a lower electrophoretic mobility of ssRNA modified using Ado-6-amine 2 compared to Ado-6-azide 3 correlates well with the higher positive charge of transferred moiety (Figure 2A).
Figure 2.
Animal RNA 2′-O-methyltransferases can be utilized for efficient transfer of various functional groups from synthetic cofactor analogues to single-stranded RNAs. (A) 0.2 μM miR173 was modified using 100 μM AdoMet 1 or its synthetic analogues Ado-6-amine 2 or Ado-6-azide 3 in the presence of 2 μM DmHen1 or HsHEN1. (B) Reverse-phase HPLC analysis of nucleosides derived from alkylated miR173 shows efficient transfer of intact cofactor side chain to 3′ termini of RNA substrate by DmHen1. 1 μM methyltransferase was used to modify 2 μM miR173 in the presence of 100 μM AdoMet 1 or its analogues. After 1.5 h of incubation at 37°C, RNA was degraded to nucleotides, dephosphorylated and analysed by RP-HPLC. (C) ESI-MS analysis of 2′O-alkylated cytidine. Mass-to-charge ratios of modified 3′ terminal cytidine derivatives are indicated in the spectrum and assigned to particular compounds in Supplementary Table S1. (D) DmHen1 modifies RNA substrates with different 3′ terminal nucleosides with synthetic cofactor analogues at the same or even higher reaction rates kchem as compared to AdoMet 1 under single-turnover conditions.
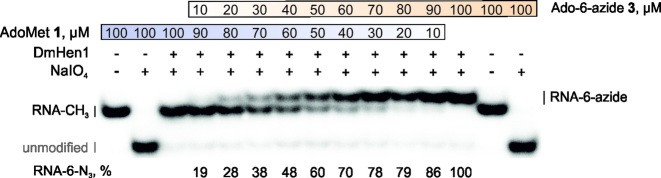
Notably, DmHen1-directed alkylation with synthetic cofactor analogues was nearly as efficient as the methylation reaction (Supplementary Figure S4). Figure 2D and Table 1 show that irrespective of the nature of 3′-terminal nucleotide of the modified substrate, the rates of alkyl-group transfer under single-turnover conditions were even slightly higher than those observed with AdoMet. Cofactor selectivity analysis using a direct competition assay at different AdoMet 1/Ado-6-azide 3 ratios showed no preference for the natural cofactor (Figure 3). This observation indicates that, at sufficiently high concentrations of exogenous cofactor analogue, efficient ssRNAs labelling can be achieved even in complex environments containing endogenous AdoMet such as in situ samples or live cells. Overall, our data demonstrate a high tolerance of DmHen1 towards the side chain size of bound cofactor and its capacity to covalently append a variety of functional groups to ssRNAs.
Figure 3.
DmHen1 shows no preference for natural cofactor or its synthetic analogue. 2 μM of DmHen1 was used to modify 0.2 μM of siR23 in the presence of 100 μM of cofactor in total, were AdoMet 1 and Ado-6-azide 3 were mixed in 10:0, 9:1, 8:2, 7:3, 6:4, 5:5, 4:6, 3:7, 2:8, 1:9 and 0:10 ratios, respectively.
2′-O-methyltransferase DmHen1 alkylates single-stranded RNA regardless of substrate length
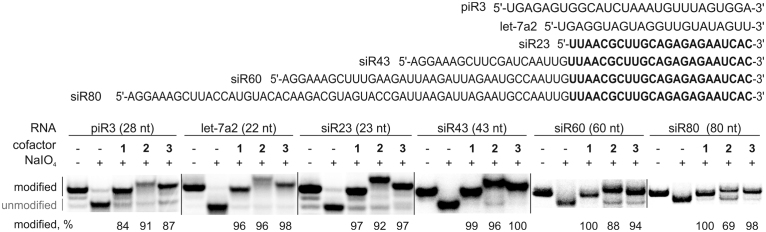
Although animal Hen1 methyltransferases are known to specifically target 22–30 nt piRNAs in vivo (27), the recombinant proteins have been shown to methylate substrates up to 40 nt in length (20,21). To better understand the substrate length preference of the Drosophila methyltransferase in the mTAG reactions, we tested a series of RNA substrates ranging from 23 to 80 nt (Figure 4). Our studies not only confirmed efficient methylation of small ncRNAs (miRNA, siRNA and piRNA) consisting of 22–28 nt but also revealed methylation of longer ssRNA substrates spanning 43–80 nt. The modification efficiency of DmHen1 using cofactors with the extended side-chains was not significantly compromised; the alkylation levels with Ado-6-amine 2 and Ado-6-azide 3 varied within the range of 69–96% and 87–100%, respectively. Additionally, we carried out kinetic studies performed under single-turnover conditions to precisely compare the rates of methyl- and 6-azide-group transfer onto ssRNAs of varied length (Supplementary Figure S5). The kchem values of 23 nt siR23 as well as previously investigated 22 nt miR173-C (Table 1) correspond well to those obtained with longer 43 nt siR43 using either AdoMet 1 or Ado-6-azide 3 (Table 2). Thus, DmHen1 does not only exhibit high tolerance to artificial cofactors with bulky moieties but is also indiscriminative to the length of ssRNA substrates.
Figure 4.
DmHen1 alkylates single-stranded RNA substrates of diverse sequences and wide length range. Experiment was conducted as described in Figure 1, except that 2 μM of DmHen1 was used. siR43, siR60 and siR80 are derivatives of siR23 containing its full sequence at their 3′ ends.
Table 2.
DmHen1 modification rate of different length RNAs in the presence of AdoMet 1 or its synthetic analogues
| Cofactor | k chem, (min−1) (siR23, 23 nt) | k chem, (min−1) (siR43, 43 nt) |
|---|---|---|
| AdoMet1 | 0.78 ± 0.04 | 0.85 ± 0.05 |
| Ado-6-azide3 | 0.99 ± 0.05 | 0.66 ± 0.12 |
| Ado-13-biotin4 | 0.27 ± 0.03 | 0.20 ± 0.03 |
The rate constants kchem were obtained from plots shown in Supplementary Figure S5.
3′-terminal functionalization of ssRNAs using one-step and two-step mTAG labelling
To achieve mTAG labelling of ssRNA substrates, one-step and two-step labelling techniques were developed as schematically explained in Figure 5A. In the first approach, a moiety with a defined reporter group, such as a fluorophore or biotin, is directly transferred onto the substrate. The single-step labelling approach requires fewer sample manipulations, but may be less efficient and costlier due to the complexity of the transferable side chain. Alternatively, two-step mTAG labelling offers a wide selection of reporter groups and commercial reagents for chemoselective conjugation to the 3′-terminus of ssRNA covalently derivatized with a smaller functional entity (28).
Figure 5.
DmHen1-directed functionalization of various length single-stranded RNAs. (A) Principal scheme of two- and one-step ssRNA labelling in comparison to natural reaction of DmHen1. (B) Visualization of ssRNAs labelled by two-step approach on a denaturing PAA gel. 0.2 μM of ssRNAs incubated with 1 μM of DmHen1 and 0.1 mM of Ado-6-amine 2 or Ado-6-azide 3, were treated with Cy5-649/670-NHS ester or Cy5.5-673/707-alkyne, respectively. RNAs methylated using AdoMet 1 served as a control for specific labelling. Cy5 and Cy5.5 fluorescence was detected using 635 and 670 nm lasers accordingly (top panel), bulk RNA was visualized after staining with ethidium bromide (bottom panel). The detailed scheme of current experiment can be found in Supplementary Figure S6. (C) RP-HPLC analysis of nucleosides derived after one-step labelling of miR173 by DmHen1. The top chromatogram shows the absorption at 280 nm, whereas the bottom one highlights the absorption of C attached Cy3 fluorophore at 545 nm. 1 μM of methyltransferase was used to modify 2 μM of miR173 in the presence of 100 μM of AdoMet 1, 50 μM of Ado-13-biotin 4 or 6 μM of Ado-14-Cy3 5. After 1.5 h of incubation at 37°C RNA was degraded to nucleotides, dephosphorylated and analysed by RP-HPLC. (D) Affinity capture of ssRNAs following one-step labelling. 50 μM of Ado-13-biotin 4 was used to label 0.01 μM of RNA substrate in the presence of 1 μM DmHen1. The initial mixture of biotin labelled RNAs (In) was loaded on streptavidin beads and supernatant (Sn) was collected. Following a buffer wash (Sl), streptavidin beads were resuspended in 95% formamide, 10 mM EDTA and heated for 10 min at 70°C to release bound RNA (Bd). As a control ssRNAs were modifies with 100 μM AdoMet 1. The right panel represents an experimental outline.
First, we tested RNA substrates of different length (from 43 to 80 nt) in two-step labelling assays with Cy5 and Cy5.5 fluorophores (Supplementary Figure S6). After methyltransferase-directed RNA modification with Ado-6-amine 2 or Ado-6-azide 3, the fluorophores were conjugated by using suitable N-hydroxysuccinimide ester and alkyne reagents, respectively. Fluorescence signals were only detectable in samples containing cofactors 2 and 3 but not AdoMet 1. The observed cofactor-dependent fluorophore incorporation (Figure 5B) demonstrates that the two-step strategy can be successfully used for chemo-selective ssRNA labelling and visualization.
For single-step labelling, we chemically synthesized AdoMet derivatives Ado-13-biotin 4 and Ado-14-Cy3 5 containing biotin or a Cy3 fluorophore, respectively (Figure 5C). Both cofactors were effectively utilized in the DmHen1-mediated modification reaction. More than 90% of total RNA regardless of its length was biotinylated in the presence of cofactor 4 (Supplementary Figures S7A and 8D). The yield of RNA labelled using cofactor 5 was slightly lower, but well sufficient for further optical detection and studies of RNA (Supplementary Figures S7B and 8D). Reverse-phase HPLC analysis of nucleotides of the mTAG-modified miR173 identified clear peaks (Figure 5C) with m/z values corresponding to cytidine-13-biotin or cytidine-14-Cy3 (Supplementary Figure S8B and C and Table S2). In the latter case, a single peak was detected using 545 nm wavelength (Figure 5C), which corresponds to an absorption maximum characteristic of cofactor 5 (data not shown). Moreover, individual Cy3 labelled RNAs were clearly detectable by fluorescence imaging using 532 nm laser (Supplementary Figure S7B). Finally, to test the functionality of biotin-group transferred onto the ssRNAs, we performed affinity purification experiments. Figure 5D shows that all biotinylated RNAs were selectively retained on streptavidin-coated magnetic beads (Bd fraction), while the methylated RNAs from the control reactions remained entirely in solution (Sn fraction).
Altogether, our experiments demonstrated a novel chemo-enzymatic approach to specifically label 3′-termini of single-stranded RNA through incorporation of desired chemical functionalities by either single-step or two-step protocols.
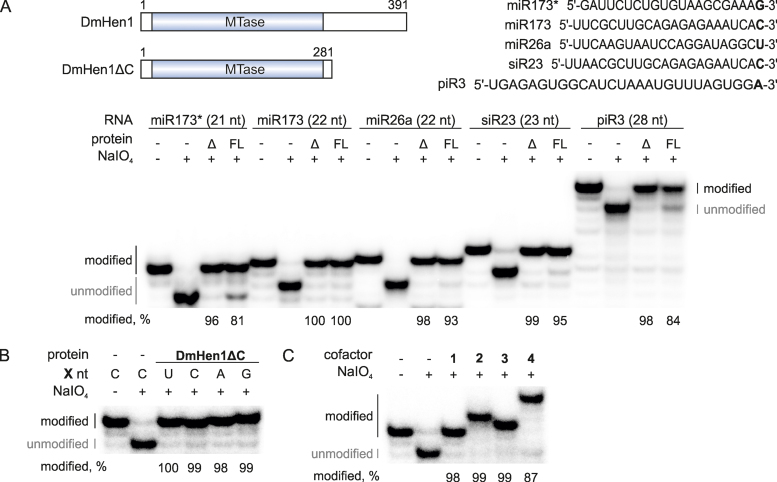
The C-terminal region of DmHen1 is dispensable for the catalytic efficiency in vitro
Sequence alignments of the animal Hen1 indicate that the methyltransferase domain (MT), conserved among plant, fungi, protozoa and bacterial HEN1 proteins, is followed by a C-terminal region of yet unknown function (11). We generated and purified a truncated variant of the protein (residues 1–281), DmHen1ΔC, lacking the C-terminal region and tested its 2′-O-methylation activity with a series of ssRNA substrates ( Figure 6A). Surprisingly, we found that the deletion of the downstream sequence had a stimulating effect on the methylation efficiency in vitro, as compared to the full-length DmHen1. Moreover, DmHen1ΔC exhibited an improved methylation activity (complete modification of ssRNA under standard reaction conditions) with all tested substrates regardless of the nature of 3′-terminal nucleotide (Figure 6B). Furthermore, an increasing bulk of the transferable moiety did not impair the effectiveness of the reaction as complete modification of miR173-C was detected with Ado-6-amine 2, Ado-6-azide 3 or Ado-13-biotin 4 (Figure 6C). The biggest improvement in the alkylation efficiency was observed in the reaction with Ado-14-Cy3 5, which displayed 83–100% modification yields in comparison to 55–81%, identified for wild-type protein (Supplementary Figure 7B and C). Thus, our results demonstrate that the C-terminal region is not required for catalysis, and the DmHen1ΔC protein is better suited for versatile chemo-enzymatic modifications of ssRNA than the full-length methyltransferase.
Figure 6.
C-terminal domain of DmHen1 is dispensable for efficient ssRNA modification. (A) DmHen1ΔC methylates RNA substrates of different length and sequence. On the top, schematic representation of DmHen1 and shorter variant lacking C terminal domain DmHen1ΔC, as well as sequences of ssRNA substrates with different 3′-terminal nucleosides (shown in bold). Bottom, methylated RNA separated on denaturing PAA gel with names and lengths of ssRNA indicated above it. Methylation reactions were performed with 2 μM of full length DmHen1 (FL) or truncated DmHen1ΔC (Δ), 0.2 μM of 32P-labelled RNA and 100 μM of AdoMet 1 at 37°C for 30 min. (B) Removal of C terminal domain abolishes DmHen1 bias towards guanosine resulting in full methylation of miR173 with different terminal nucleosides. (C) DmHen1 methyltransferase domain efficiently transfers bulky side chains from synthetic cofactor analogues onto miR173 substrate in the presence of 100 μM of AdoMet 1, Ado-6-amine 2, Ado-6-azide 3 or 50 μM of Ado-13-biotin 4.
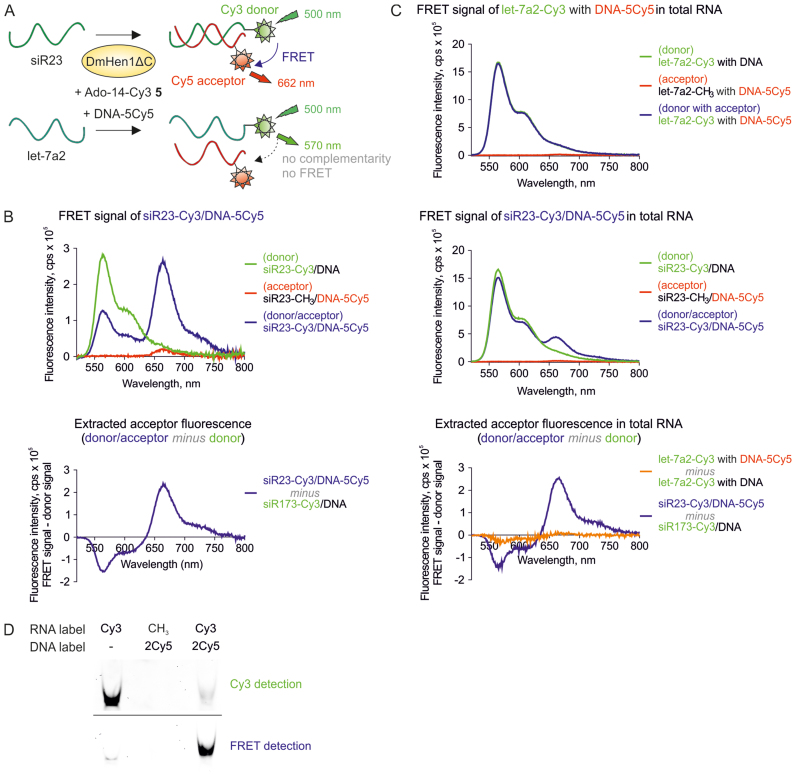
Figure 7.
Selective detection of DmHen1-labelled ssRNA using FRET assay. (A) A schematic illustration of the experiment. ssRNA was 3′-end labelled in one step with Cy3 fluorophore transferred from Ado-14-Cy3 5 cofactor by DmHen1ΔC and later annealed to the siR23-complementary DNA bearing Cy5 at 5th position. The FRET signal at 662 nm appears only if Cy3-RNA donor and DNA-born Cy5 acceptor are close enough for an energy transfer. (B) Detection of FRET signal of Cy3/Cy5 pair in solution. siR23 Cy3-labelled by 1 μM of DmHen1ΔC in the presence of 3 μM of Ado-14-Cy3 was annealed to a complementary DNA in equimolar concentration. The top graph shows the emission spectra of siR23-Cy3/DNA-5Cy5 FRET pair excited at 500 nm and two control samples: donor siR23-Cy3/DNA containing RNA-born Cy3 and intact DNA and acceptor siR23-CH3/DNA-5Cy5-methylated RNA and DNA-born Cy5. The graph at the bottom highlights the extracted acceptor fluorescence acquired by subtracting the donor fluorescence from two-coloured spectra. (C) Specific detection of siR23 in total RNA sample through FRET signal registration. Top: FRET emission at 662 nm did not appear in Cy3-labelled total RNA with let-7a2, which was not complementary to DNA-5Cy5. Middle: the presence of siR23 in the sample is proved by emerging FRET signal. Bottom: extracted acceptor fluorescence at 662 nm is only visible upon addition of siR23. Samples were prepared as described earlier with the exception that 0.5 μg/μl of total RNA from HCT116 human colon carcinoma cell line was added in the labelling reaction and 10 μM of cofactor was used. (D) Visualization of siR23-Cy3/DNA-2Cy5 FRET pair in polyacrylamide gel. Cy3 and FRET signals were detected by exciting the fluorophores with green laser at 532 nm and registering their emission using LPG (575 nm) and LPR (665 nm) filters, respectively.
Application of fluorescence resonance energy transfer (FRET) approach for ssRNA analysis
Single-fluorophore labelling is widely used for detection of biomolecules, however, background signal rising from an excess of unreacted fluorophore could complicate analysis of rare and less abundant species. To overcome the limitations of signal masking, two fluorophore-based FRET approaches are employed for analysis of more complicated biological systems. We performed a proof-of-principle experiment using DmHen1 in FRET assays with Cy5-containing DNA hybridized to a complementary target siR23 covalently labelled with a Cy3 fluorophore (Figure 7A). Following one-step DmHen1ΔC-directed addition of the Cy3 dye to a target ssRNA (Supplementary Figure S9A) and hybridization of a complementary DNA probe, the fluorescence of three differently labelled RNA/DNA duplexes were measured in solution at 500 nm excitation. As shown in Figure 7B and Supplementary Figure S9B, spectra of dually-tagged siR23-Cy3/DNA-5Cy5 relative to donor-tagged siR23-Cy3/DNA had a weaker Cy3-emission signal at 570 nm caused by a strong increment of Cy5-fluorescence at 662 nm. Further, to examine the selectivity of the FRET approach, we spiked total RNA extracted from human colon cancer cells HCT116 with siR23 or let-7a2. DmHen1ΔC effectively deposited the Cy3-label on the both spiked-in single-stranded RNAs (Supplementary Figure S9A). Accordingly, the donor and acceptor fluorescence showed a similar pattern for both siR23 and let-7a2 upon addition of complementary Cy5-DNA probes (Figure 7C). In addition, we found a clearly detectable FRET signal of siR23-Cy3/DNA-5Cy5, despite the presence of a high excess of the unreacted Cy3 dye in the reaction mixture. Moreover, extracted acceptor fluorescence intensity was similar to siR23-Cy3/DNA-5Cy5 alone and in mixture with total RNA but was not registered in samples with let7a2, which is non-complementary to DNA-5Cy5 (bottom panels of Figure 7B and C). Because the energy transfer signal appeared only after target RNA hybridization to a complementary DNA strand, the proposed method is well suited for highly selective detection of desired RNAs.
Finally, we validated the results obtained from DmHen1-labelled FRET samples in solution by a conventional native polyacrylamide gel electrophoresis. Indeed, only a dual-colour sample with both donor and acceptor fluorophores present in the heteroduplex was visualized using a green 532 nm laser with a 665 nm cut-off filter (Figure 7D). Altogether, we demonstrate that the DmHen1-directed labelling can be instrumental for reliable FRET detection of particular ssRNA both in solution and in fixed samples.
DISCUSSION
Cobalt as metal cofactor of the animal Hen1 methyltransferases
Based on the domain arrangement, the HEN1 RNA 2′-O-methyltransferases can be divided into four subfamilies that have different preferences for bivalent metal cofactors (11,29). Magnesium ions are required for the catalytic activity of plant small ncRNA methyltransferases (22), while manganese is essential for a bacterial Hen1 (26). Unexpectedly, the Drosophila and human piRNA methyltransferases appeared to utilize cobalt ions, joining a small group of metal cobalt-dependent enzymes (Figure 1B) (30). Due to chemical interconversion of Co2+ and Co3+ in aqueous buffers containing dithiothreitol (0.2 mM), both ions were present in significant quantities in reaction buffers used in vitro. Given a high conservation of the bivalent metal coordinating amino acids in the HEN1 homologues (29) and an expected effect of the reducing cytosol environment (31) on the highly oxidized cobalt form, we presume that the bona fide reactive species is Co2+. On the other hand, direct participation of Co(III) in catalysis cannot be excluded since the ion was supplied in the form of [Co(NH2)6]3+ complex in which electron donating ligands reduce the positive charge of the hexa-coordinated centre making it electronically similar to Co2+.
The preference of the animal MTases for Co2+ and Co3+ is not exclusive, inasmuch as DmHen1 can, albeit less efficiently, methylate small RNAs using Ca2+ ions. It is possible that distinct metal-dependencies of different subfamilies of the HEN1 methyltransferases derive from their adjustment to a particular cellular environment. Accordingly, a well-established importance of calcium for the spermatogenesis and sperm function could extend also to the regulation of piRNA methylation in male germ cells (32). At this point, it is difficult to verify a clear link between the intracellular cobalt and Hen1 activity, since changes in its physiological concentration have not been established in the germ cells. On the other hand, it is interesting to note, that it is not unusual for enzymes that demonstrate highest activity with Co2+ ions to function with other metal cofactors. This could be exemplified by the rat DL-methylmalonyl-CoA racemase (activity row Co2+> Co3+> Fe2+ > Mn2+) (33), human histone deacetylase 8 (Co2+> Fe2+ > Zn2+ > Ni2+) (34) or Trypanosoma brucei methionine aminopeptidase 1 (Co2+ > Ni2+ > Mn2+> Fe2+> Zn2+) and to various extent with similar activation pattern described for other methionine aminopeptidases where the most potent metal ion depend on the particular enzyme (35).
Substrate promiscuity of the animal Hen1 2′-O-methyltransferases
Our results reveal a broad substrate specificity of the recombinant protein in vitro spanning from 20 to at least 80 nt (Figure 4, Supplementary Figure S5 and Table 2). Moreover, when total RNA is added into the reaction mixture, the alkylation of all its components of all lengths is observed (data not shown). This observation is in agreement with the lack of additional N-terminal domains responsible for substrate size gauging present in the A. thaliana HEN1, AtHEN1 (22). In vivo targets of the animal Hen1 methyltransferases are 22–30 nt long (27). Therefore, in vivo the methylation of specific RNA substrates must be tightly regulated and determined by additional factors, most likely PIWI proteins.
Previous studies have identified significant sensitivity of methylation efficiency to the nature of the 3′-terminal nucleoside for the mouse mHEN1 (20) and Clostridium thermocellum CtHen1 (36). In agreement to this, we also observed a prominent 3′-terminal nucleoside preference of HsHEN1, while DmHen1 showed only a modest bias against guanosine (Figures 1C and 2D, Supplementary Figure S4 and Table 1). Importantly, this bias was eliminated and overall modification activity was increased upon removal of the C-terminal region from the DmHen1 protein, indicating that certain C-terminal residues alter interactions of the substrate with the catalytic domain (Figure 6 and Supplementary Figure S7). Although the C-terminal region appears important for biological function in vivo as protein's localization and potentially interactions with other factors of sRNA biogenesis depends on it (12,37), it turned out to be completely dispensable for the efficient alkyltransferase activity in vitro.
DmHen1 as a tool for mTAG labelling and analysis of ssRNA
In addition to highly efficient and unbiased modification of ssRNA substrates of different composition and length by an engineered Hen1 methyltransferase, DmHen1ΔC, its remarkable promiscuity for AdoMet analogues (Figure 6) demonstrates the robustness of this enabling tool for targeted derivatization and labelling of RNA. To emphasize the practical potential of these reactions, it is important to note that their rate was highly similar to that observed with AdoMet (Tables 1 and 2; Supplementary Figures S4 and 5) and that DmHen1 showed no preference for the natural cofactor in the presence of a synthetic analogue (Figure 3). Using two-step mTAG labelling, we showed efficient covalent derivatization of ssRNA with primary amine or azide functional groups followed by conjugation of desired fluorophores through N-hydroxysuccinimide ester or bio-orthogonal click chemistry, respectively (Figure 5B). While two-step labelling permits a broad choice of reporters to be used after the derivatization step, one-step mTAG reactions offer streamlined processing of inherently fragile RNA under mild conditions. This is well illustrated by FRET analysis of Cy3 labelled RNA in a total RNA pool in the presence of a vast excess of the precursor dye (Figure 7 and Supplementary Figure S9).
Currently post-synthetic RNA detection, visualization and analysis can be achieved using hybridization techniques, aptamers, RNA binding proteins and covalent labelling strategies (2). Covalent mTAG derivatization and labelling has been demonstrated for (i) a specific internal locus in a unique tRNA (7); (ii) programmable sequence-specific internal position in any RNA molecule (8); (iii) 5′-terminal cap structure of mRNA (Supplementary Figure S10) (38,39). For small RNA detection and analysis our group has recently proposed addressable AtHEN1 directed mTAG labelling of miRNA and siRNA in RNA/RNA or RNA/DNA duplexes (9,10). In this context, the current method stands up as the only chemo-enzymatic approach for 3′-terminal single-stranded RNA labelling, which is not restricted to a specific nucleotide, structural element or length of the RNA strand. Since the reaction is only possible with RNA but not DNA (due to the lack of the 2′-hydroxyl group), we envision its further applications in such fields as RNA sequencing after RNA functionalization with a DNA primer (data not shown).
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Mikiko Siomi for a gift of the pGEX-DmHen1 plasmid. The authors thank Audronė Rukšėnaitė for valuable help with MS analysis and Vilius Kurauskas with cloning of HsHEN1.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Research Council of Lithuania [MIP-059/2015 to G.V]. Funding for open access charge: Research Council of Lithuania.
Conflict of interest statement. None declared.
REFERENCES
- 1. Mattick J.S. Challenging the dogma: the hidden layer of non-protein-coding RNAs in complex organisms. Bioessays. 2003; 25:930–939. [DOI] [PubMed] [Google Scholar]
- 2. Anhäuser L., Rentmeister A.. Enzyme-mediated tagging of RNA. Curr. Opin. Biotechnol. 2017; 48:69–76. [DOI] [PubMed] [Google Scholar]
- 3. Deen J., Vranken C., Leen V., Neely R.K., Janssen K.P.F., Hofkens J.. Methyltransferase-Directed labeling of biomolecules and its applications. Angew. Chem. Int. Ed. Engl. 2017; 56:5182–5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lukinavicius G., Lapiene V., Stasevskij Z., Dalhoff C., Weinhold E., Klimasauskas S.. Targeted labeling of DNA by methyltransferase-directed transfer of activated groups (mTAG). J. Am. Chem. Soc. 2007; 129:2758–2759. [DOI] [PubMed] [Google Scholar]
- 5. Muttach F., Rentmeister A.. A biocatalytic cascade for versatile One-Pot modification of mRNA starting from methionine analogues. Angew. Chem. Int. Ed. Engl. 2016; 55:1917–1920. [DOI] [PubMed] [Google Scholar]
- 6. Muttach F., Mäsing F., Studer A., Rentmeister A.. New AdoMet Analogues as tools for enzymatic transfer of Photo-Cross-Linkers and capturing RNA–Protein interactions. Chem. Eur. J. 2017; 23:5988–5993. [DOI] [PubMed] [Google Scholar]
- 7. Motorin Y., Burhenne J., Teimer R., Koynov K., Willnow S., Weinhold E., Helm M.. Expanding the chemical scope of RNA:methyltransferases to site-specific alkynylation of RNA for click labeling. Nucleic Acids Res. 2011; 39:1943–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tomkuviene M., Clouet-d’Orval B., Cerniauskas I., Weinhold E., Klimasauskas S.. Programmable sequence-specific click-labeling of RNA using archaeal box C/D RNP methyltransferases. Nucleic Acids Res. 2012; 40:6765–6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Plotnikova A., Osipenko A., Masevičius V., Vilkaitis G., Klimašauskas S.. Selective covalent labeling of miRNA and siRNA duplexes using HEN1 methyltransferase. J. Am. Chem. Soc. 2014; 136:13550–13553. [DOI] [PubMed] [Google Scholar]
- 10. Osipenko A., Plotnikova A., Nainytė M., Masevičius V., Klimašauskas S., Vilkaitis G.. Oligonucleotide-addressed covalent 3′-terminal derivatization of small RNA strands for enrichment and visualization. Angew. Chem. Int. Ed. Engl. 2017; 56:6507–6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang R.H. Unique 2′-O-methylation by Hen1 in eukaryotic RNA interference and bacterial RNA repair. Biochemistry. 2012; 51:4087–4095. [DOI] [PubMed] [Google Scholar]
- 12. Kamminga L.M., Luteijn M.J., den Broeder M.J., Redl S., Kaaij L.J.T., Roovers E.F., Ladurner P., Berezikov E., Ketting R.F.. Hen1 is required for oocyte development and piRNA stability in zebrafish. EMBO J. 2010; 29:3688–3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhai J., Zhao Y., Simon S.A., Huang S., Petsch K., Arikit S., Pillay M., Ji L., Xie M., Cao X. et al. . Plant microRNAs display differential 3′ truncation and tailing modifications that are ARGONAUTE1 dependent and conserved across species. Plant Cell. 2013; 25:2417–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ren G., Xie M., Zhang S., Vinovskis C., Chen X., Yu B.. Methylation protects microRNAs from an AGO1-associated activity that uridylates 5′ RNA fragments generated by AGO1 cleavage. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:6365–6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lim S.L., Qu Z.P., Kortschak R.D., Lawrence D.M., Geoghegan J., Hempfling A.-L., Bergmann M., Goodnow C.C., Ormandy C.J., Wong L. et al. . HENMT1 and piRNA stability are required for adult male germ cell transposon repression and to define the spermatogenic program in the mouse. PLoS Genet. 2015; 11:e1005620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yu B., Yang Z., Li J., Minakhina S., Yang M., Padgett R.W., Steward R., Chen X.. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005; 307:932–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vilkaitis G., Plotnikova A., Klimasauskas S.. Kinetic and functional analysis of the small RNA methyltransferase HEN1: the catalytic domain is essential for preferential modification of duplex RNA. RNA. 2010; 16:1935–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Plotnikova A., Baranauskė S., Osipenko A., Klimašauskas S., Vilkaitis G.. Mechanistic insights into small RNA recognition and modification by the HEN1 methyltransferase. Biochem. J. 2013; 453:281–290. [DOI] [PubMed] [Google Scholar]
- 19. Horwich M.D., Li C., Matranga C., Vagin V., Farley G., Wang P., Zamore P.D.. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr. Biol. 2007; 17:1265–1272. [DOI] [PubMed] [Google Scholar]
- 20. Kirino Y., Mourelatos Z.. The mouse homolog of HEN1 is a potential methylase for Piwi-interacting RNAs. RNA. 2007; 13:1397–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Saito K., Sakaguchi Y., Suzuki T., Suzuki T., Siomi H., Siomi M.C.. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi- interacting RNAs at their 3′ ends. Genes Dev. 2007; 21:1603–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang Y., Ji L., Huang Q., Vassylyev D.G., Chen X., Ma J.-B.. Structural insights into mechanisms of the small RNA methyltransferase HEN1. Nature. 2009; 461:823–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Baranauskė S., Mickutė M., Plotnikova A., Finke A., Venclovas Č., Klimašauskas S., Vilkaitis G.. Functional mapping of the plant small RNA methyltransferase: HEN1 physically interacts with HYL1 and DICER-LIKE 1 proteins. Nucleic Acids Res. 2015; 43:2802–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang Z., Vilkaitis G., Yu B., Klimasauskas S., Chen X.. Approaches for studying microRNA and small interfering RNA methylation in vitro and in vivo. Methods Enzymol. 2007; 427:139–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Masevičius V., Nainytė M., Klimašauskas S.. Synthesis of S-adenosyl-L-methionine analogs with extended transferable groups for methyltransferase-directed labeling of DNA and RNA. Curr. Protoc. Nucleic Acid Chem. 2016; 64:1.36.1–1.36.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jain R., Shuman S.. Bacterial Hen1 is a 3′ terminal RNA ribose 2′-O-methyltransferase component of a bacterial RNA repair cassette. RNA. 2010; 16:316–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang H., Ma Z., Niu K., Xiao Y., Wu X., Pan C., Zhao Y., Wang K., Zhang Y., Liu N.. Antagonistic roles of Nibbler and Hen1 in modulating piRNA 3′ ends in Drosophila. Dev. Camb. Engl. 2016; 143:530–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tiefenbrunn T.K., Dawson P.E.. Chemoselective ligation techniques: Modern applications of time-honored chemistry. Pept. Sci. 2010; 94:95–106. [DOI] [PubMed] [Google Scholar]
- 29. Mui Chan C., Zhou C., Brunzelle J.C., Huang R.H.. Structural and biochemical insights into 2′-O-methylation at the 3′-terminal nucleotide of RNA by Hen1. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:17699–17704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kobayashi M., Shimizu S.. Cobalt proteins. Eur. J. Biochem. 1999; 261:1–9. [DOI] [PubMed] [Google Scholar]
- 31. López-Mirabal H.R., Winther J.R.. Redox characteristics of the eukaryotic cytosol. Biochim. Biophys. Acta. 2008; 1783:629–640. [DOI] [PubMed] [Google Scholar]
- 32. Correia J., Michelangeli F., Publicover S.. Regulation and roles of Ca2+ stores in human sperm. Reprod. Camb. Engl. 2015; 150:R65–R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stabler S.P., Marcell P.D., Allen R.H.. Isolation and characterization of DL-methylmalonyl-coenzyme A racemase from rat liver. Arch. Biochem. Biophys. 1985; 241:252–264. [DOI] [PubMed] [Google Scholar]
- 34. Gantt S.L., Gattis S.G., Fierke C.A.. Catalytic activity and inhibition of human histone deacetylase 8 is dependent on the identity of the active site metal ion. Biochemistry. 2006; 45:6170–6178. [DOI] [PubMed] [Google Scholar]
- 35. Marschner A., Klein C.D.. Metal promiscuity and metal-dependent substrate preferences of Trypanosoma brucei methionine aminopeptidase 1. Biochimie. 2015; 115:35–43. [DOI] [PubMed] [Google Scholar]
- 36. Jain R., Shuman S.. Active site mapping and substrate specificity of bacterial Hen1, a manganese-dependent 3′ terminal RNA ribose 2′O-methyltransferase. RNA. 2011; 17:429–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kurth H.M., Mochizuki K.. 2′-O-methylation stabilizes Piwi-associated small RNAs and ensures DNA elimination in Tetrahymena. RNA. 2009; 15:675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schulz D., Holstein J.M., Rentmeister A.. A chemo-enzymatic approach for site-specific modification of the RNA cap. Angew. Chem. Int. Ed. Engl. 2013; 52:7874–7878. [DOI] [PubMed] [Google Scholar]
- 39. Holstein J.M., Anhäuser L., Rentmeister A.. Modifying the 5′-Cap for click reactions of eukaryotic mRNA and to tune translation efficiency in living cells. Angew. Chem. Int. Ed. Engl. 2016; 55:10899–10903. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.