Abstract
During cell division, maintenance of chromatin features from the parental genome requires their proper establishment on its newly synthetized copy. The loss of epigenetic marks within heterochromatin, typically enriched in repetitive elements, endangers genome stability and permits chromosomal rearrangements via recombination. However, how histone modifications associated with heterochromatin are maintained across mitosis remains poorly understood. KAP1 is known to act as a scaffold for a repressor complex that mediates local heterochromatin formation, and was previously demonstrated to play an important role during DNA repair. Accordingly, we investigated a putative role for this protein in the replication of heterochromatic regions. We first found that KAP1 associates with several DNA replication factors including PCNA, MCM3 and MCM6. We then observed that these interactions are promoted by KAP1 phosphorylation on serine 473 during S phase. Finally, we could demonstrate that KAP1 forms a complex with PCNA and the histone-lysine methyltransferase Suv39h1 to reinstate heterochromatin after DNA replication.
INTRODUCTION
Heterochromatin is paramount to the stability of eukaryotic genomes, which abound in repetitive units such as satellite repeats and transposable elements (1,2). Loss of control over these regions can lead to transcriptional perturbations, abnormal splicing and DNA recombination, events all at the root of oncogenic transformation (3–8). Heterochromatin is largely transcriptionally inactive and typically described as either constitutive or facultative. Facultative heterochromatin is made up of locus- and cell type-specific domains, the epigenetic features of which may be regulated, whereas its constitutive counterpart is traditionally more extensive, ubiquitous and permanent, such as in pericentromeric regions of chromosomes (9). In mammalian genomes, constitutive heterochromatic regions are typically marked with post-translational modifications (PTM) of histones such as tri-methylation of histone H3 on lysine 9 (10) and 64 (11) (H3K9me3 and H3K64me3 respectively), and of histone H4 on lysine 20 (H4K20me3) (12–14). Furthermore, they are enriched in non-histone proteins such as Heterochromatin Protein 1 (HP1) (15) and KAP1 (KRAB-associated protein 1, also known as TRIM28 or Tif1β) (16), and the underlying DNA is generally methylated (17). H3K9 tri-methylation plays a pivotal role in the organization of this heterochromatin, as it promotes the binding of HP1 (15) and of the histone-lysine methyltransferases (KMT) Suv39h1 (Suppressor of Variegation 3–9 Homolog 1) (18) and Suv420h1 (Suppressor of Variegation 420 Homolog 1) (12). Moreover, heterochromatin-associated non-coding RNAs (ncRNAs) play an important role in the regulation and formation of constitutive heterochromatin by stabilizing Suv39h1, which can instate H3K9me3 (19–21), and KAP1 itself can associate with all five KMTs so far identified in mammals, namely, SETDB1 (SET Domain Bifurcated 1), GLP, and G9a in addition to Suv39h1/h2. These enzymes act differentially on their substrate, with GLP/G9a and SETDB1 capable of mediating mono, di and tri-methylation of H3K9, whereas Suv39h1/h2 can only deposit a third methyl group on previously mono- and di-methylated H3K9 (22–27). Heterochromatic regions are mostly replicated during late S phase and chromatin marks present on the parental DNA must be recapitulated in the daughter cell at the outset of this process. KAP1 was previously found to participate in these events by ensuring the maintenance of H3K9me3 as part of a complex also comprising CAF1 (chromatin-associated factor 1) and HP1, which recruits the KMT SETDB1 responsible for mono-methylating histone H3 before its association with the newly synthesized DNA. Moreover, SMARCAD1 (a SWI/SNF-like protein) and CHD can be associated with KAP1 in order to maintain H3K9 methylation and heterochromatin integrity during cell division at pericentromeric and subtelomeric heterochromatin (27–29). However, how these heterochromatin marks are maintained during cell division is not known. Here, we reveal that KAP1 becomes phosphorylated on serine 473 (to generate pS473KAP1) during late S phase, triggering the formation of a PCNA-KAP1-Suv39h1 complex that plays an essential role in the maintenance of heterochromatin-associated histone modifications across cell division.
MATERIALS AND METHODS
Cell culture, antibodies and reagents
Cells were maintained at 37°C in a 7% CO2 incubator in medium containing 100U/ml penicillin-streptomycin, for NIH3T3, MEF, HeLa and 293T cells Dulbecco's modified Eagle's medium (DMEM, Gibco BRL), for K562 cells RPMI-1640, all with 10% fetal bovine serum (FBS), and for H1 ESCs (WA01, WiCell) mTesRI (StemCell Technologies) on hESC-qualified Matrigel (BD Biosciences). WT and Suv39h1/h2 KO MEFs were kindly provided by Pr Thomas Jenuwein. Anti-pS473KAP1 antibody was produced in rabbits using a peptide coupled to KLH with the following sequence: krsrSgegevsgl (Eurogentech) and used for WB (tested in the Supplementary Figure S1F) and poly6446 was used for IF. Other antibodies were as follows: Anti-KAP1 (ab10483 for ChIP and IF and MAB3662 for WB and Co-IP), anti-H3K9me3 (ab8898 for ChIP and WB and active motif 39765 for IF), anti-H4K20me3 (ab9053 for ChIP, IF and WB), anti-H3K64me3 (C15410211 for ChIP), anti-H3K9me1 (ab9045 for WB), anti-H3K9me2 (ab1220 for WB), anti-H3pS10pT11 (ab32107 for WB), anti-panH4ac (06-866 for WB), anti-panH3ac (06-599 for WB), anti-H3 (ab1791 for ChIP and WB), anti-HA (3F10 for IF and HA.11 for WB), anti-FLAG (M2 for WB), anti-PCNA (PC10 for IF and IP and ab-1 for WB), anti-MCM3 (ab4460 for WB), anti-MCM5 (sc-165995 for WB), anti-MCM6 (ab184147 for WB), anti-RCF4 (sc-20996), anti-HP1α (1H5 for ChIP and IF and 2G9 for WB), anti-HP1β (ab40828 for ChIP and 1G9 for WB), anti-HP1γ (42s2 for ChIP and 1G6 for WB), anti-Suv39h1 (07-550 for ChIP validated in (30) and D11B6 for WB), and anti-Suv420h1 (61415 for ChIP). Cell synchronization was achieved by treatment with 0.4μg/ml Nocodazole (M1404) during 10 hrs or 5mM Hydroxyurea (H8627). Serum starvation was performed by reducing FBS concentration to 0.1% for 72 h.
Chromatin immunoprecipitation (ChIP)
Cells were harvested, washed with PBS, fixed in 2 ml fixation buffer (10 min in 1% formaldehyde, PBS), quenched with 10 ml 250 mM Tris–HCl pH 8, washed three times with PBS, pelleted, lysed by resuspension on ice in 1 ml sonication buffer (10 mM Tris pH 8, 1 mM EDTA, 0.2% SDS, protease inhibitors), transferred to TC 12 × 12 tubes (Covaris) and sonicated (Covaris settings: 20 min, 5% duty cycle, 140 W, 200 cycles). Sonication was assessed by reverse cross-linking (37°C RNAse A at 1 μg/μl 1 h, then 65°C, Proteinase K at 400 ng/μl, overnight), followed by DNA extraction. Fragment size (between 200 and 400 bp) was checked on a Bioanalyzer (Agilent 2100). Immunoprecipitations were performed in IP buffer (10 mM Tris at pH 8, 1 mM EDTA, 0.1% SDS, 150 mM NaCl, 10% Triton X-100 and protease inhibitors) overnight. Chromatin was reverse cross-linked (65°C, Proteinase K at 400 ng/μl, overnight) and DNA was further extracted for analysis. ChIP samples were used for SYBER Green qPCR (Applied Biosystems) or library preparation for sequencing as previously described (31).
RNA extraction and RT-qPCR
RNA was extracted with High Pure RNA Isolation Kit from Roche. All RT-qPCR reactions were performed with independent biological duplicates using random hexamers, and each cDNA was tested in triplicate with SYBR Green mix (Applied Biosystems) and primers listed in Supplementary Table S2. Negative controls without reverse transcriptase were processed in parallel.
Protein and histone extraction
Whole-cell lysates were extracted in 8 M ureas lysis buffer. Histone extraction was achieved according Abcam's histone extraction protocols (http://www.abcam.com/protocols/histone-extraction-protocol-for-western-blot). Harvest cells were resuspended and lysed in Triton Extraction Buffer (0.5% Triton X100; 2 mM PMSF; 0.02% NaN3). Histone extraction was achieved with 0.2N HCL over night at 4°C. Protein estimation was determined using a ND-1000 spectrophotometer (Nanodrop, Thermo Fisher Scientific).
Lentiviral vector production and transduction
Production and titration lentiviral vectors were achieved as previously described (32). Transduction was performed as previously described (33) (see http://tronolab.epfl.ch/lentivectors for details).
Cell fractionation
KAP1 KO MEFs were lysed in soluble buffer (0.1% NP40, 10 mM Tris–HCl pH 8, 5 mM EDTA, 0.5 mM EGTA, 1 mM PMSF, protease inhibitor) during 3 min at 4°C, then centrifuged 30 s at 6000 rpm at 4°C (supernatant = soluble fraction). The pellet was re-incubated with nucleus buffer (250 mM NaCl, 50 mM Tris–HCl pH 8, 5 mM EDTA, 0.1% NP40, 1 mM PMSF, protease inhibitor) during 30 min at 4°C, then centrifuged 15 min at 14 000 rpm and 4°C (supernatant = nucleus). The pellet was re-suspended in chromatin buffer (250 mM NaCl, 50 mM Tris–HCl pH 8, 5 mM EDTA, 0.1% NP40, 1 mM PMSF, protease inhibitor), sonicated and incubated (125 mM NaCl, 50 mM Tris–HCl pH 8, 5 mM EDTA, 0.1% NP40, 1 mM PMSF, protease inhibitor), 1 h at 37°C with 10 U micrococcal nuclease (88216), then centrifuged 15 min at 14 000 rpm at 4°C (supernatant = chromatin).
RESULTS
KAP1 interacts with proteins involved in DNA replication
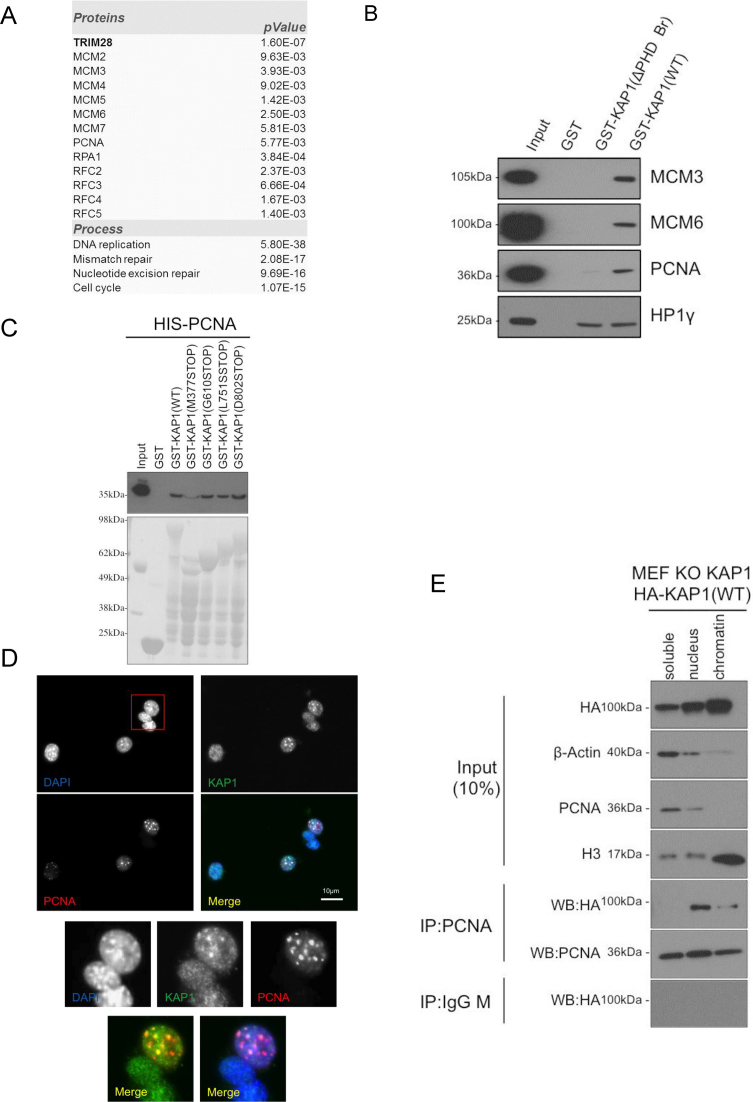
In order to determine the comprehensive set of proteins interacting with KAP1, we performed affinity-purification of endogenous KAP1 coupled to mass spectrometry, using human K562 and embryonic stem cells (hESC) as substrates. We identified several polypeptides significantly enriched in the precipitated fraction, some of which were previously identified as KAP1 interactors, such as HP1γ (in hESC) or KRAB-containing zinc finger proteins (KZFPs) (34) (Supplementary Table S1). We also detected the presence of proteins critical for DNA replication, such as PCNA (Proliferating Cell Nuclear Antigen), RPA1 (Replication Protein A1), and several subunits of the MCM (MiniChromosome Maintenance) and RFC (Replication Factor complex) complexes (Figure 1A). We validated these results by performing GST pull-down assays with recombinant GST-KAP1 proteins, and then probed the importance of different KAP1 domains for these interactions. Wild type GST-KAP1 (WT) bound PCNA, MCM3 and MCM6 proteins, confirming the mass spectrometry data. These interactions were compromised when using a C-terminally deleted form of KAP1 (GST-KAP1(ΔPHD Br)), which still could bind HP1γ (Figure 1B). Similar results were obtained when using HEK293T (293T) protein extracts as substrates for the GST pull-down, and use of HP1 binding-defective GST-KAP1 mutant indicated that HP1 proteins were not involved in these interactions (Supplementary Figure S1A). We next asked which domain of KAP1 is required to interact with PCNA by using recombinant HIS-PCNA and GST-KAP1 mutants (Supplementary Figure S1B). Among GST-KAP1 mutants tested, only GST-KAP1(M377STOP) was not able to interact with HIS-PCNA suggesting that the RBCC domain is not mediating PCNA association (Figure 1C). Often PCNA partners contain a PIP (PCNA Interacting Protein) motif (Q-x-x-H-x-x-A-A, where H represents any hydrophobic amino acids, notably methionine, leucine, isoleucine or valine, and A an aromatic amino acids such as phenylalanine or tyrosine) and we found a putative PIP motif between amino acids 502–509 of KAP1 (QPPVFKVF). However, GST pull-down assays performed with GST-KAP1 point or deletion mutants targeting this putative PIP motif indicated that this region is not required for recruiting PCNA (Supplementary Figure S1C). Thus, an undefined motif located between amino acids 495 and 610 of KAP1 can interact with PCNA (Figure 1B and C). Moreover, immunofluorescence (IF) coupled with confocal microscopy demonstrated that KAP1 and PCNA co-localized only when PCNA was found within DAPI dense regions at mid/late S phase, but not in early S phase (Figure 1D and Supplementary Figure S1D). Moreover, this interaction was found by cell fractionation to occur mainly within the nuclear fraction and weakly on chromatin fraction (Figure 1E), and not to be mediated by DNA since it resisted DNase treatment (Supplementary Figure S1E).
Figure 1.
KAP1 interacts with replication factors. (A) Main KAP1-interacting proteins detected by mass spectrometry in hESC and K562 cells, with indication of some over-represented biological processes. Data were generated with a Scaffold software with option quantitative values calculating P-value with unpaired, two tailed t-test. (B and C) Recombinant GST, GST-KAP1 and mutants were incubated for pull-down assays with (B) NIH3T3 cell extracts or (C) recombinant HIS-PCNA. Western blotting (WB) was performed with indicated antibodies. (D) Immunofluorescence microscopy was performed with indicated antibodies in NIH3T3 cells. Representative of 159 cells examined, 53.5% of which were in S-phase (27.7% early, 15.7% mid, 10% late). (E) KAP1 KO MEFs complemented with HA-KAP1(WT) were subjected to fractionation to yield soluble, nucleus and chromatin fractions, before immunoprecipitation (IP) and WB with indicated antibodies.
S phase-specific KAP1 phosphorylation on serine 473 promotes interaction with DNA replication factors
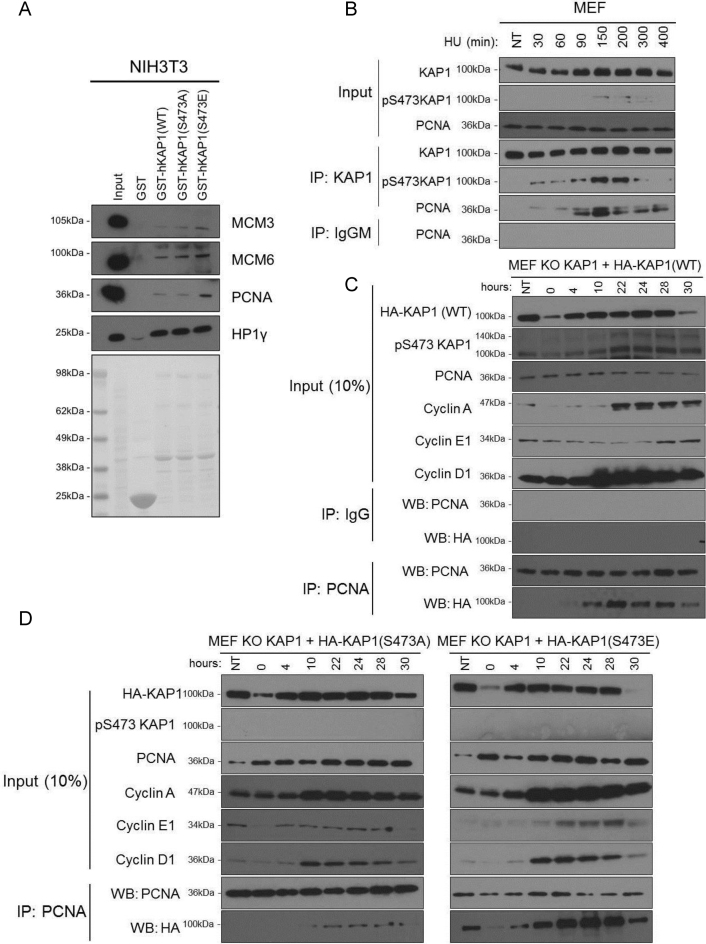
KAP1 can be subjected to several post-translational modifications (PTM) including sumoylation, ubiquitination and phosphorylation. Among these, phosphorylation on serine 473 (S473) was observed in the setting of DNA damage (35), metabolic stress (36), T cell receptor activation (37,38) and KSHV (Kaposi's sarcoma-associated herpervirus) infection (39), and to be mediated by PKCδ (Protein Kinase C δ) during S phase (40). We thus examined the influence of the cell cycle on this KAP1 PTM by performing IF with a pS473KAP1-specific antibody on NIH3T3 cells synchronized by 3 days of serum starvation followed by serum re-addition. This revealed that KAP1 became phosphorylated on S473 two hours after serum addition and stayed so for about 5 h, yielding an IF pattern that overlapped with that of PCNA (Supplementary Figure S2A). This prompted us to ask if KAP1 association with PCNA and other components of the DNA replication machinery was influenced by this PTM. To this aim, we used phospho-null (S473A) and phospho-mimetic (S473E) GST-KAP1 mutants. GST-KAP1(S473E) interacted more strongly than the other two derivatives with PCNA and, to a lesser extent, MCM proteins, whether human or murine cellular extracts were used as capture substrates, whereas all three variants associated equally well with HP1γ, which served as a positive control (Figure 2A and Supplementary Figure S2B). The S473A mutation did not abrogate PCNA recruitment in this system, but this might have been due to oligomerization of the mutant protein with its wild type counterpart, with secondary recruitment of PCNA. We next performed co-immunoprecipitation (co-IP) studies with both endogenous PCNA and KAP1 in mouse embryonic fibroblasts (MEFs) synchronized/arrested in S phase following treatment with hydroxyurea (HU). We observed that KAP1 increasingly associated with PCNA between 90 and 150 min of HU treatment, then decreased. Interestingly, this interaction coincided with KAP1 phosphorylation on serine 473 (Figure 2B). We then performed co-IP studies on synchronized Kap1 KO MEFs complemented with either HA-KAP1(WT), HA-KAP1(S473A) or HA-KAP1(S473E). After serum addition, PCNA increasingly associated with HA-KAP1(WT), peaking at 22 h, again remarkably coinciding with KAP1 S473 phosphorylation in these cells, which have a significantly longer cycle compared with NIH3T3 fibroblasts (Figure 2C). Similar results were obtained when using extracts from NIH3T3 cells overexpressing HA-KAP1(WT) as substrates for the co-IP (Supplementary Figure S2C). In contrast, the association of the phospho-null HA-KAP1(S473A) with PCNA was weak and restricted to 22–28 h after serum addition (Figure 2D, left panel), whereas the phospho-mimetic HA-KAP1(S473E) constitutively interacted with PCNA (Figure 2D, right panel). These results were confirmed by determining the fraction of total input HA-KAP1 that was co-immunoprecipitated with PCNA (Supplementary Figure S2D). Of note, levels of all KAP1 forms (WT, S473A and S473E) were significantly decreased at the end of the serum starvation period and 30 h after serum re-addition, suggesting their regulation by the cell cycle (Figure 2C and D). The temporal pattern of KAP1-PCNA co-immunoprecipitation remarkably coincided with that of KAP1 S473 phosphorylation and this association preferentially occurred during S phase as demonstrated by FACS analysis (Supplementary Figure S2E). Confirming this result, PCNA interacted with transfected HA-KAP1 (WT) and (S473E) but much less with the phospho-null S473A mutant in wild type MEFs synchronized/arrested in S phase by HU treatment (Supplementary Figure S2F). Confirming the stimulation of PCNA-KAP1 association by phosphorylation, levels of HA-KAP1 co-immunoprecipitated with PCNA were considerably decreased by phosphatase treatment (Supplementary Figure S2G). Taken together, these results demonstrate that KAP1 phosphorylation on S473, an S phase-specific modification, governs its interactions with components of the DNA replication complex.
Figure 2.
Phosphorylation-stimulated interaction of KAP1 with DNA replication factors. (A) Recombinant GST, GST-KAP1 and mutants were incubated with NIH3T3 cell extracts for pull down assays, and WB performed with indicated antibodies. Results representative of 3 independent experiments. (B) MEFs were treated with HU for indicated numbers of minutes and cell extracts subjected to co-IP/WB with antibodies against indicated proteins. (C and D) Kap1 KO MEFs were complemented with HA-tagged KAP1 WT, S473A or S473E and synchronized by serum starvation for 72 hrs, before performing IP/WB at indicated number of hrs post-serum re-addition. (B, C and D) are each representative of two independent experiments.
KAP1 participates in the maintenance of heterochromatin across cell division
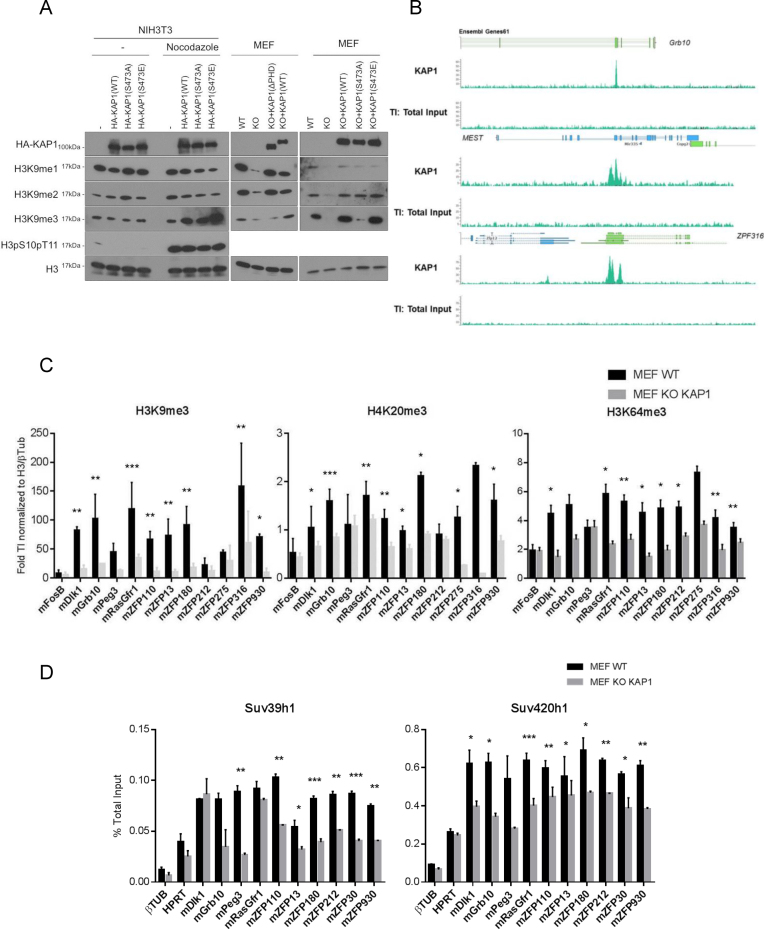
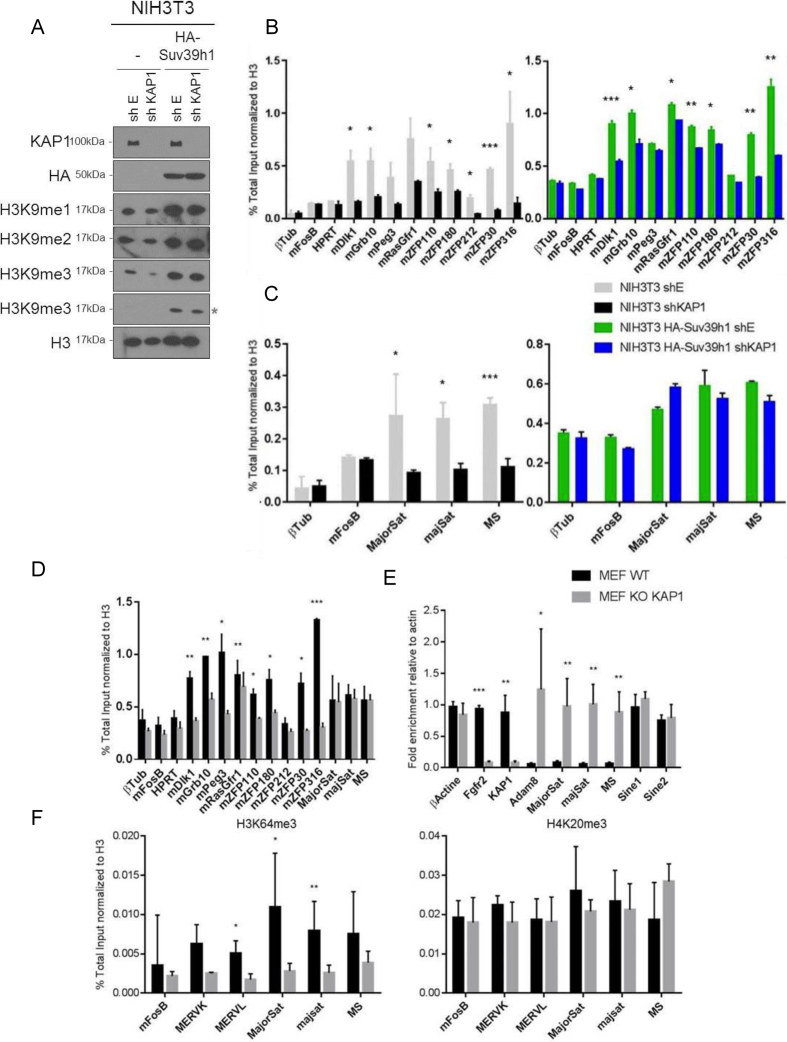
Given the roles previously proposed for KAP1 in the maintenance of H3K9 tri-methylation (27–29), we next explored the impact of mutations affecting its interaction with PCNA on the maintenance of this heterochromatic mark. For this, we carried out histone extraction in NIH3T3 cells overexpressing wild type or mutant versions of HA-KAP1. We first verified that steady-state levels of H3K9me3 were not modified in the presence of these proteins in asynchronous cell populations. Then, following addition of nocodazole, a compound that blocks the cell cycle in G2/M, we found that overexpression of wild type, S473A and S473E KAP1 increased levels of H3K9me3 (Figure 3A, left panel), although with a stronger effect for the phospho-mimetic mutant. Similar observations were made in 293T cells (Supplementary Figure S3A). These results suggest that the gain of H3K9me3 induced by KAP1 overexpression might occur after S phase or in G2. To explore further the influence of KAP1 on H3K9 mono- and di-methylation, we used WT and Kap1 KO MEFs. A decrease of all three methylated forms of H3K9 was observed in the knockout cells. Complementation with KAP1(WT) and KAP1(S473E) restored wild type levels of di- and tri-methylation, although effects on H3K9me1 levels were only modest. In contrast, the phospho-null KAP1(S473A) and plant homeodomain deletion (ΔPHD) mutants rescued H3K9 mono- and di-methylation but only very weakly affected levels of tri-methylation (Figure 3A, middle and right panel). We confirmed H3K9me3 reduction in NIH3T3 cells treated with a short hairpin RNA targeting KAP1 (shKAP1) (Supplementary Figure S3B). Moreover, in these cells as well, complementation with the phospho-null KAP1 mutant only weakly increased H3K9me3 (Figure 3A, right panel), consistent with previous data demonstrating that KAP1 is required to provide H3K9me1/2 as substrate for Suv39h1 (27). We next asked whether this reduction of H3K9me3 was genome wide or a local effect restricted to the surroundings of KAP1 binding sites. For this, we first performed KAP1 chromatin immunoprecipitation followed by deep sequencing (ChIP-Seq) in WT MEFs. As expected (41,42), KAP1 was detected at the 3′ end of zinc finger protein genes (e.g. 3′ZFP316) and over imprinting control regions (e.g. Grb10 and MEST) (Figure 3B and Supplementary Figure S3C). We then compared by ChIP-qPCR the distribution of several heterochromatin marks at these loci in WT and Kap1 KO MEFs. H3K9me3, H4K20me3 and H3K64me3, a modification within the globular domain of H3 found at pericentromeric heterochromatin (11,43), were all enriched over KAP1-binding regions in wild type cells and markedly decreased at these sites in their Kap1 KO counterparts (Figure 3C). The KMTs Suv39h1 and Suv420h1, involved in tri-methylation of respectively H3K9 and H4K20, were also less enriched at these loci in the absence of KAP1 (Figure 3D). HP1 proteins recognize H3K9me3 via their chromodomain (15). In Kap1 KD NIH3T3 cells, HP1α was also delocalized from pericentromeric heterochromatin domains (Supplementary Figure S3D), consistent with the observed decrease of H3K9me3 (Supplementary Figure S3B). Importantly, total levels of a large series of heterochromatin regulators were not significantly altered in Kap1 KO MEFs (Supplementary Figure S3E). This indicated that the decreased DNA association of Suv39h1, Suv420h, H3K9me3, H4K20me3 and HP1α resulted primarily from a defect in the genomic recruitment of these proteins. Therefore, KAP1 contributes to the maintenance of all forms of H3K9 methylation (mono-, di-, and tri-), with pS473KAP1 specifically required to preserve H3K9me3. Moreover, at genomic loci normally bound by KAP1, absence of this regulator results in loss of both writers (KMT) and readers (HP1) of these heterochromatin histone marks.
Figure 3.
KAP1 maintains heterochromatin-associated histone modifications and factors. (A) Left, NIH3T3 cells expressing indicated KAP1 variants were treated or not with nocodazole for 10 h before performing WB on histone extracts with indicated antibodies. Right, same experiment on WT and Kap1 KO MEFs without nocodazole treatment. (B) Screenshot of selected KAP1 ChIP-Seq peaks in WT MEFs. (C and D) ChIP-qPCR with indicated antibodies on pre-mapped KAP1-binding regions. Data are means ± s.e.m. from three independent experiments. Significance of the differences was estimated using Student's t‐test. ***P< 0.01, 0.01 < **P< 0.05 and 0.05 < *P< 0.1.
Differential KAP1 requirements for H3K9me3 and H4K20me3 maintenance
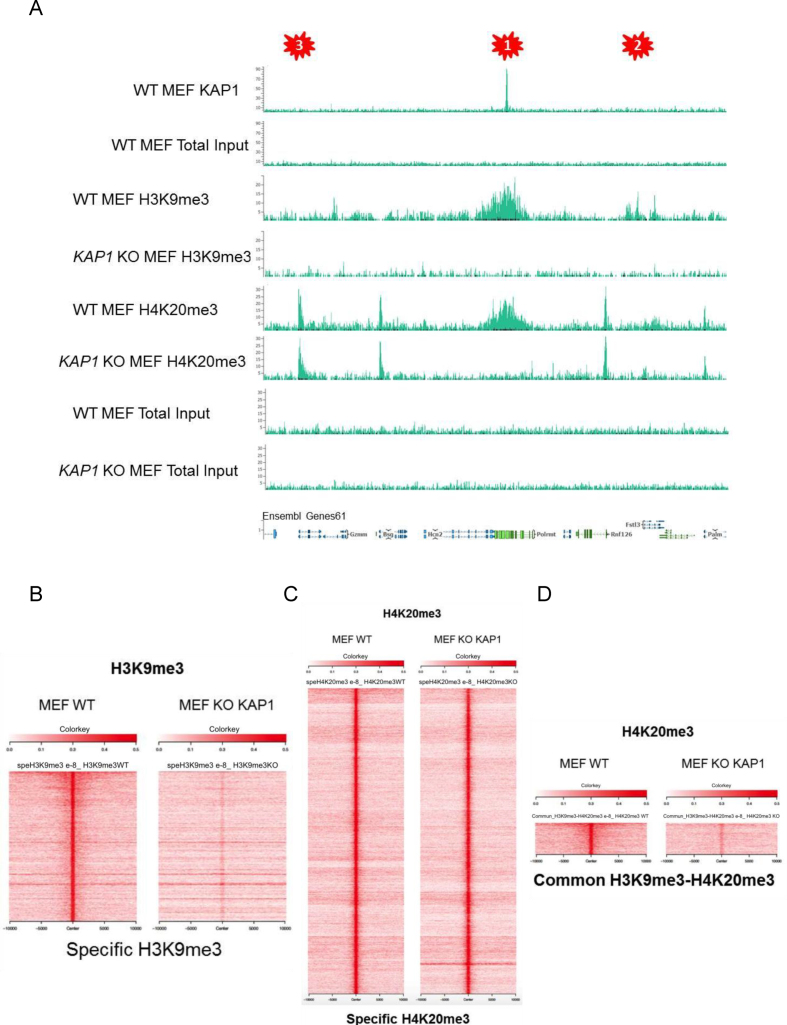
We found KAP1 to be required for maintaining H3K9me3 and H4K20me3 at its genomic targets (Figure 3C). However, histone extraction and IF studies detected a global decrease in levels of the former but not the latter histone mark following Kap1 KD in NIH3T3 cells (Supplementary Figure S4A and B). This suggested that the KAP1-dependent accumulation of H3K9me3, but not H4K20me3, might extend beyond KAP1 binding sites. We thus performed H3K9me3 and H4K20me3-specific ChIP-Seq in WT and Kap1 KO MEFs and could identify and characterize three patterns (Figure 4A): (i) KAP1 binding regions with loss of H3K9me3 and H4K20me3 upon KAP1 depletion; (ii) H3K9me3 and H4K20me3 enriched sites without detectable KAP1 at baseline, but losing theses marks in the absence of the scaffold protein and (iii) domains with only H4K20me3, for which KAP1 depletion had no effect. Thus, the accumulation of H3K9me3 seemed to be globally KAP1-dependent, whereas that of H4K20me3 appeared to require this protein only at some sites, where it was coupled with H3K9me3. To explore this point further, we mapped all H3K9me3- and H4K20me3-enriched regions in WT and Kap1 KO MEFs. This confirmed an overall reduction of H3K9me3 in the absence of KAP1, not limited to genomic loci normally bound by KAP1 (Figure 4A and B), which contrasted with H4K20me3 (Figure 4A and C). We then defined 723 regions enriched both with H3K9me3 and H4K20me3 (Supplementary Figure S4C), and could determine that H4K20me3 was lost only at these loci upon KAP1 KO (Figure 4A and D). H4K20me3 is a mark previously described as enriched within pericentromeric heterochromatin and generally associated with H3K9me3. However, we found this modification also present downstream of transcription start sites (TSS), where KAP1 depletion had no effect (Supplementary Figure S4D). The TSS-association of H4K20me3 was previously reported, and appears to regulate pol II release (44). Taken together, our results indicate that KAP1 plays a critical role for the overall maintenance of H3K9me3, and indirectly that of H4K20me3 at H3K9me3-positive loci.
Figure 4.
Differential KAP1-mediated maintenance of H3K9me3 and H4K20me3. (A) KAP1, H3K9me3 and H4K20me3 ChIP-Seq peaks in WT and Kap1 KO MEFs, pointing on top to regions (1, 2 and 3 asterisk) with differential patterns. (B–D) Heatmaps representing the raw data –/+10 kb around the middle of histone mark peaks in WT and Kap1 KO MEFs. H3K9me3/H4K20me3-positive regions were defined with a stringent P-value (10e–8), and specific loci as positive according to this criterion in one dataset and negative even with low stringency P-value in the other data set (<10e–8 versus >10e–3).
KAP1 partners up with Suv39h1 to maintain heterochromatin during DNA replication
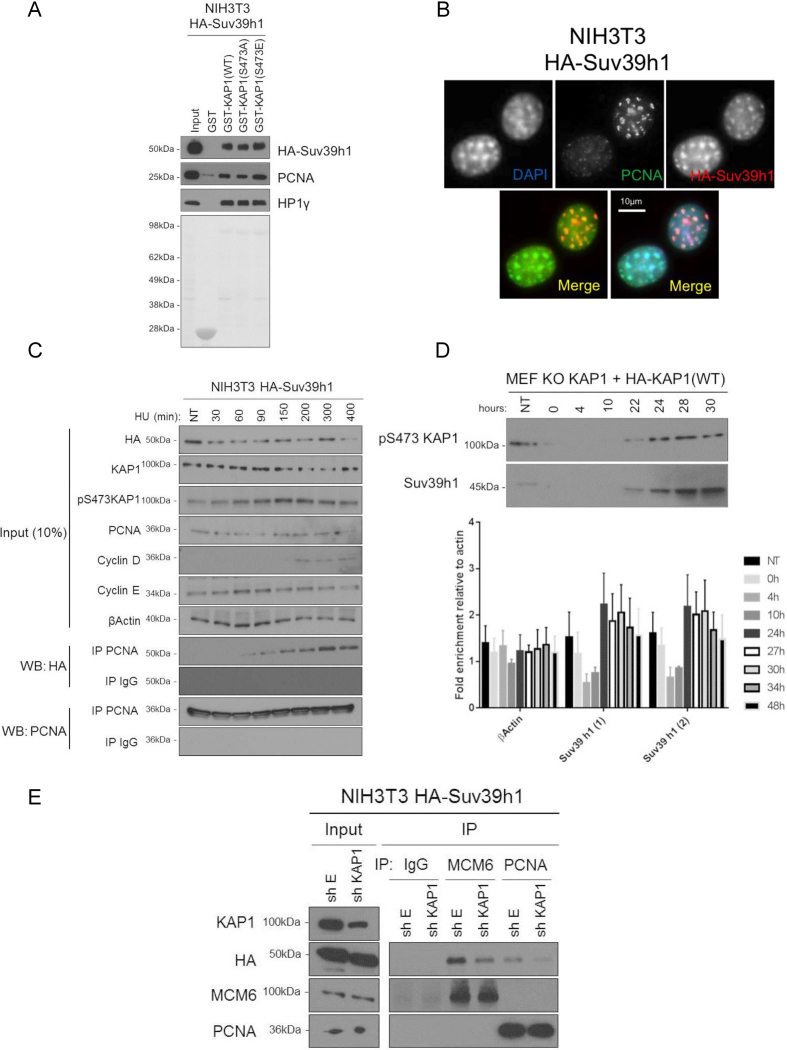
We next asked which histone KMT is recruited by KAP1 to maintain H3K9me3 during DNA replication. In mammalian cells, five such enzymes have been identified: SETDB1, GLP, G9a and Suv39h1/h2, all of which have been detected in complexes with KAP1 by mass spectrometry (30). G9a was unlikely to be the KMT involved in the phenomenon described here, because we previously observed that KAP1 phosphorylation on S473 prevents its recruitment, at least within MyoD transcription complexes (45). Still, overexpressing each one of these methyltransferases resulted in increased levels of H3K9 methylation in 293T cells, although Suv39h1 had the strongest effect (Supplementary Figure S5A). Moreover, pull-down assays with WT, S473A or S473E GST-KAP1 on NIH3T3 or HeLa protein extracts expressing HA-tagged Suv39h1 revealed that this enzyme interacted most strongly with the phospho-mimetic KAP1 mutant (S473E), and most weakly with its phospho-null S473A counterpart. (Figure 5A and Supplementary Figure S5B). Recently, both Suv39h1 and KAP1 were described as potential components of nascent chromatin (46,47). This prompted us to ask whether Suv39h1 might also interact with DNA replication factors. Co-localization of Suv39h1 and PCNA at DAPI-intense loci was first documented by immunofluorescence microscopy (Figure 5B). We next performed co-IP studies on NIH3T3 overexpressing HA-Suv39h1 synchronized by HU treatment. Expression of Cyclin E and Cyclin D ascertained that these cells were well synchronized and in S phase. PCNA and HA-Suv39h1 were found to interact increasingly after HU addition, with a peak at 300 min (Figure 5C). Similar results were obtained in HA-Suv39h1-overspressing NIH3T3, and HeLa cells synchronized by serum starvation/re-addition (Supplementary Figure S5C and D). We next examined the expression profile of Suv39h1 during the cell cycle in KAP1 KO MEFs complemented with HA-KAP1(WT). This revealed that Suv39h1 levels peaked during S phase (24 hrs and 22 hrs after serum addition for mRNA and protein, respectively), when KAP1 and PCNA interact (Figure 5D). These results suggest that Suv39h1, when expressed in S phase, might associate with PCNA. Furthermore, knocking down Kap1 in NIH3T3 cells overexpressing HA-Suv39h1 resulted in reducing, albeit not abrogating, the Suv39h1-PCNA interaction (Figure 5E). In contrast, GST pull-down assays performed with Suv39h1/2 KO MEFs demonstrated that the interaction of PCNA or MCM6 with KAP1 is independent of these KMTs (Supplementary Figure S5E). These data suggest that p473KAP1 serves as a bridge between Suv39h1 and PCNA. We next investigated if KAP1 and Suv39h1 were recruited on newly synthesized DNA. For this, we performed ChIP-qPCR on control and Kap1 KD NIH3T3 cells synchronized by serum starvation, using antibodies against KAP1, Suv39h1 and H3K9me3, focusing on strong KAP1 targets (e.g. Dlk1, Supplementary Figure S5F). In control cells, KAP1 genomic recruitment was increased shortly (2 h) after serum addition, Suv39h1 enrichment was sustained for several hours and H3K9me3 levels remained robust. In contrast, in KAP1-depleted cells, Suv39h1 recruitment was only transient, dropping 4–5 h after serum addition, and H3K9me3 levels were lower than in controls cells throughout the several hour-long experiment. Together, these data are consistent with a model whereby phosphorylation of serine 473 of KAP1 during S phase fosters the recruitment of Suv39h1 by PCNA.
Figure 5.
pS473KAP1 dependent Suv39h1-replication factors complex formation. (A) Recombinant GST, GST-KAP1 and mutants were incubated with NIH3T3 cell extracts overexpressing HA-Suv39h1. WB was performed with indicated antibodies. (B) Immunofluorescence with DAPI or indicated antibodies in NIH3T3 cells overexpressing HA-Suv39h1. (C) NIH3T3 cells overexpressing HA-Suv39h1 were synchronized/arrested by HU treatment. Co-IP experiments and WB were performed with indicated antibodies at indicated minutes post-HU addition. (D) Nuclear protein extraction and RNA extractions were performed in KAP1 KO MEFs complemented with HA-KAP1 (WT), arrested by serum starvation for 72 h and released to cell cycle with serum addition then WB were performed with indicated antibodies. Levels of mRNA for the indicated genes were measured by RT-qPCR at indicated times post-serum addition. Two different primers for Suv39h1 were used in this experiment. (E) Co-IP experiments and WB were performed in NIH3T3 cells overexpressing HA-Suv39h1 and transduced with shE or shKAP1 with indicated antibodies.
Differential effects of KAP1 and Suv39h1 at major satellite repeats
Even though MEFs stably deleted for Kap1 could be generated, acute KAP1 depletion was highly toxic in several cell lines including NIH3T3, where it induced cell death within less than 10 days (Supplementary Figure S6A). We reasoned that if KAP1 and Suv39h1 were functionally linked, elevating levels of Suv39h1 might attenuate this effect. We thus induced HA-Suv39h1 overexpression in Kap1 KD NIH3T3 cells, and remarkably observed a rescue of their toxic phenotype (Supplementary Figure S6A), with cells further displaying wild type levels of H3K9me3 (Figure 6A and Supplementary Figure S6B). To determine whether this effect was maintained throughout the cell cycle, we cultured HA-Suv39h1 overexpressing NIH3T3 cells depleted for KAP1 during 3 weeks and monitored H3K9me3 levels every three days. H3K9me3 levels were maintained up to 15 days after Kap1 KD, but gradually decreased thereafter (Supplementary Figure S6C). These results indicate that Suv39h1 overexpression can compensate for KAP1 depletion in global maintenance of heterochromatin, although this effect is progressively lost as cell divisions accumulate. We next examined the relative KAP1-dependence and Suv39h1-mediated rescue of H3K9 tri-methylation at different subsets of genomic loci. For this, we compared WT and Kap1 KD NIH3T3 cells overexpressing or not Suv39h1. We found a marked drop in H3K9me3 at KAP1 binding sites upon Kap1 KD, which was only partly rescued by Suv39h1 overexpression, at least at the time point examined (15 days post-KD) (Figure 6B). The recent analysis of MEFs stably deleted for Suv39h1/h2 served as a basis for implicating these histone methyltransferases in the establishment and maintenance H3K9me3 at constitutive heterochromatin, including major satellite repeats (48). When we analyzed H3K9me3 ChIP-Seq data in WT and Suv39h1/h2 KO MEFs (H3K9me3 ChIP-Seq from (48)), we observed that depletion of Suv39h1/h2 did not lead to an overall reduction in H3K9me3, contrasting with what was found in Kap1 KO MEFs (Supplementary Figure S6D and Figure 4B). Furthermore, while KAP1 depletion reduced H3K9me3 levels at major satellite repeats, Suv39h1 overexpression completely abrogated this phenomenon (Figure 6C), consistent with a major role for this KMT in the maintenance of constitutive heterochromatin (48). When we repeated ChIP-qPCR experiments in stable Kap1 KO MEFs, there was a decrease in H3K9me3 at sites normally bound by the regulator (as illustrated in Figure 3C), but not at major satellite repeats (Figure 6D). Correspondingly, the presence HP1α and HP1β was dramatically decreased at the former, but not at the latter type of loci (Supplementary Figure S6E). Together, these data indicate that cell death and loss of heterochromatin integrity induced by KAP1 depletion can be rescued by Suv39h1 overexpression. Noteworthy, in spite of their persistent association with H3K9me3, the expression of major satellite transcripts increased in Kap1 KO MEFs (Figure 6E). It suggests that this chromatin mark does not primarily influence transcription from these loci. We then examined by ChIP-qPCR the enrichment in H3K64me3 and H4K20me3, two other marks of constitutive heterochromatin. While H4K20me3, as H3K9me3, was lost at KAP1 binding sites but preserved at major satellite repeats in Kap1 KO MEFs, H3K64me3 was reduced at both places (Figures 3C and 6F). Thus, the H3K64me3 histone mark appears to play an important role in repressing transcription at major satellite repeats.
Figure 6.
Suv39h1 overexpression overcomes cell death and restores H3K9me3 depletion induced by KAP1 depletion at major satellite regions. (A) NIH3T3 cells overexpressing or not HA-Suv39h1 were transduced with lentiviral vectors expressing (shKAP1) or not (shE) Kap1-targeting small hairpin RNAs. Histone were extracted and tested by WB with indicated antibodies. (*) Corresponds to lower exposure time. (B–D) H3K9me3-specific ChIP-qPCR in indicated cell lines. Data are means ± s.e.m. from two independent experiments. Student's t‐test, ***P< 0.01, 0.01 < **P< 0.05 and 0.05 < *P< 0.1 (three different primers used for major satellite). (E) RT-qPCR measurement of indicated transcripts in WT and Kap1 KO MEFs. (F) H3K64me3 (left) and H4K20me3 (right) -specific ChIP-qPCR at indicated loci. Data are means ± s.e.m. from three independent experiments. Student's t‐test, ***P< 0.01, 0.01 < **P< 0.05 and 0.05 < *P< 0.1.
DISCUSSION
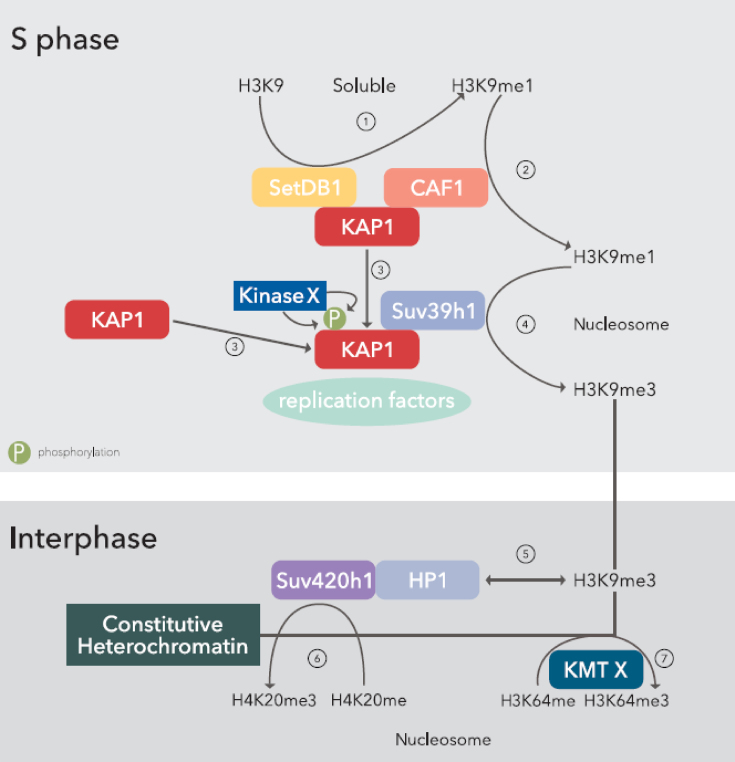
This work demonstrates a central role for KAP1 in the maintenance of heterochromatin during DNA replication. In contrast to previous work focused on the role of KAP1 in the maintenance of H3K9me3 during cell division at pericentromeric and subtelomeric heterochromatin (27–29), we demonstrate here that KAP1 phosphorylation on serine 473 is required to maintain H3K9me3 globally and that this modification also promotes H4K20me3 and H3K64me3. Furthermore, we reveal that KAP1 is not required for maintenance of only H3K9me3 but of all three methylated forms of H3K9. Based on our result and previous report, we propose a model for heterochromatin maintenance across cell division whereby, during the late phase of DNA synthesis, KAP1 is phosphorylated on serine 473, which triggers its association with PCNA and other components of the DNA replication machinery within heterochromatic regions. The Suv39h1 KMT is in turn recruited, enabling the tri-methylation on lysine 9 of newly incorporated histone H3 molecules, after they have been mono-methylated by the CAF1–KAP1–HP1–SETDB1 complex (27). The H3K9me3 mark is then recognized by HP1, and Suv420h1 is recruited, which establishes H4K20me3 (12). Finally, H3K64 tri-methylation ensues, through the action of an enzyme that remains to be identified, which results in transcriptional silencing notably at satellite repeats (Figure 7). We also observed that Suv39h1 overexpression could rescue cell death induced by KAP1 depletion by preserving heterochromatin integrity.
Figure 7.
A model for heterochromatin maintenance across cell division. During S phase, the complex SETDB1-KAP1-CAF1 induces the mono-methylation of lysine 9 on histone H3 (1), which is incorporated into nucleosomes via CAF1 (2). During late S phase (3), an unknown kinase phosphorylates KAP1 on serine 473, which promotes interaction with PCNA and other replication factors. At the replication fork, KAP1 recruits Suv39h1, which mediates the formation of H3K9me3 (4). During Interphase, nucleosomal H3K9me3 is recognized by HP1 (5), which in turn tethers Suv420h1, responsible for the tri-methylation of H4K20 (6), whereas an unknown KMT mediates the formation of H3K64me3 in an H3K9me3-dependant manner (7).
Previously, Fritsch et al. found KAP1 associated with H3K9 KMT such as GLP/G9a, SETDB1 and Suv39h1/h2 (30). However, we found only EHMT1 (G9a) in our mass spectrometry data. The difference could stem from our immune-precipitating endogenous KAP1 for our affinity purification / mass spectrometry, whereas they used an overexpressed and FLAG-tagged form of the KMT to recover KAP1.
How KAP1 is specifically targeted to heterochromatic regions during DNA synthesis is unclear since PCNA is found within both euchromatin and heterochromatin. In our IF experiment, we never observed PCNA-KAP1 co-localization at early S phase, when euchromatin is known to be replicating. It could be that during heterochromatin replication the kinase responsible for phosphorylating KAP1 on S473 is activated, since this modification enhances KAP1 association with the DNA replication complex. KAP1 phosphorylation on S473 and S824 prevents its interaction with the KMTs G9a and SETDB1, respectively (49,50). During DNA damage, KAP1 is first phosphorylated on S824, and then on S473 by ATM-Chk2 or ATR-Chk1 (35,51,52). It could be that a phospho-switch mechanism is at play following double strand break (DSB), with phosphorylation first on S824 to relax chromatin and facilitate the recruitment of DNA repair factors (53,54), and secondly on S473 to re-establish heterochromatin marks via the PCNA-pS473KAP1-Suv39h1 complex (Figure 7).
Our demonstration that pS473KAP1 acts as bridge between Suv39h1 and PCNA fits with the detection of both KAP1 and Suv39h1 at the replication fork (46,47). Furthermore, our finding that H3K9 tri- but not mono- or di-methylation requires KAP1 phosphorylation on S473, a modification needed to recruit Suv39h1, is consistent with results indicating that CAF1-HP1-KAP1-SETDB1 can induce mono-methylation of newly synthesized histone H3 on lysine 9 (27) and implicates KAP1 at several steps of the H3K9 methylation process through two and perhaps three distinct KMT partners, G9a being apparently involved at some loci (28).
KAP1 depletion was previously noted to be toxic in a number of cells (31,55–57), and here Kap1 KD caused the death of NIH3T3 cells. This effect could be avoided at least temporarily by overexpressing Suv39h1, which further prevented loss of H3K9me3 at major satellite repeats, consistent with a major role for this KMT in the maintenance of H3K9me3 at constitutive heterochromatin (48). More surprisingly, stable Kap1 knockout MEFs cell lines could be developed, which displayed normal levels of H3K9me3, H4K20me3 and HP1 at major satellite repeats, indicating possible compensatory mechanism. Nevertheless, transcription from these regions was aberrantly activated in these cells, coinciding with a decrease in H3K64me3. Acetylation at this histone residue (H3K64ac) was recently reported to facilitate transcription (58) and to provide the underlying DNA with enhancer activity (59). The modulation of major satellite transcription likely plays an important role in the establishment, formation and spreading of heterochromatin. RNAs emanating from heterochromatin can guide HP1 proteins to these regions (60–62). Moreover, in fission yeast, expression of long noncoding RNA (LncRNA) transcribed from constitutive heterochromatin is required for the establishment and spreading of repressive marks via the RNAi pathway (63). On the contrary, Borderline, a LncRNA transcribed at the border between heterochromatin and euchromatin, inhibits the spreading of the former into the latter in Schizosaccharomyces pombe (64). Supporting our model, it was very recently proposed that major satellite repeat transcripts, by forming RNA:DNA hybrids, can stabilize the chromatin retention of Suv39h (20,21). Together, these results warrant future experiments exploring the relationship between KAP1, H3K64me3 and major satellite repeats expression.
KAP1 contributes to genome stability through a variety of effects, whether by silencing the endovirome (3–6,56,57), participating in DNA repair (35,51–54) or ensuring the maintenance of heterochromatin across cell division (the present work and (27–29)), which likely minimizes recombination between repetitive sequences (65). As such, KAP1 truly represents a key factor in the preservation of genome integrity.
DATA AVAILABILITY
All next-generation sequencing data have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) database under the accession number GEO: GSE 86391.
Supplementary Material
ACKNOWLEDGEMENTS
We thank S. Ait-Si-Ali for providing all KMT expression plasmids, C. Green for the His-PCNA, C. Rachez for the FLAG-KAP1 and F. Rauscher for the GST-KAP1(RVEE) vectors, T. Jenuwein for WT and Suv39h1/h2 KO MEFs, M. Friedli, E. Allemand, O. Mauger, A. De Iaco for discussion, C. Raclot and B. Quentin for technical assistance, the University of Lausanne Genomics Core Facility for sequencing and Yuri Kim for drawing Figure 7.
Authors Contributions: S.M.J. conceived the study, designed and performed experiments, analyzed and interpreted data, and wrote the manuscript. A.K., J-P.Q. and B.R. designed, performed and analyzed experiments. J.P. and J.D. performed bioinformatics studies. A.C and P.T. made intellectual contributions. G.A. designed experiments, analyzed and interpreted data. D.T. conceived the study, designed experiments, interpreted data and wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swiss National Foundation and the European Research Council [ERC-2010-AdG-268721 “KRABnKAP” and ERC-2015-AdG-694658 “TRANSPOS-X” to D.T.]; la Ligue National contre le Cancer (Equipe labellisée Ligue) [ANR-11-LABX-0044 DEEP and ANR-10-IDEX-0001-02 PSL, ANR-12-BSV5-0022-02 ’CHAPINHIB’, ANR-14-CE16-0009 ‘Epicure’, ANR-14-CE10-0013 ‘CELLECTCHIP’, EU project 678563 ‘EPOCH28’, ERC-2015-AdG-694694 ‘ChromADICT’, ANR-16-CE15-0018 ‘CHRODYT’, ANR-16-CE-12-0024 ‘CHIFT’ and ANR-16-CE11-0028 ‘REPLICAF’ to G.A.]. Funding for open access charge: Swiss National Foundation and the European Research Council.
Conflict of interest statement. None declared.
REFERENCES
- 1. Saksouk N., Simboeck E., Dejardin J.. Constitutive heterochromatin formation and transcription in mammals. Epigenet. Chromatin. 2015; 8:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lippman Z., Gendrel A.V., Black M., Vaughn M.W., Dedhia N., McCombie W.R., Lavine K., Mittal V., May B., Kasschau K.D. et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004; 430:471–476. [DOI] [PubMed] [Google Scholar]
- 3. Goering W., Ribarska T., Schulz W.A.. Selective changes of retroelement expression in human prostate cancer. Carcinogenesis. 2011; 32:1484–1492. [DOI] [PubMed] [Google Scholar]
- 4. Schulz W.A. L1 retrotransposons in human cancers. J. Biomed. Biotechnol. 2006; 2006:83672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang Y., Jiang J., Li Q., Ma H., Xu Z., Gao Y.. KAP1 is overexpressed in hepatocellular carcinoma and its clinical significance. Int. J. Clin. Oncol. 2016; 21:927–933. [DOI] [PubMed] [Google Scholar]
- 6. Ayarpadikannan S., Kim H.S.. The impact of transposable elements in genome evolution and genetic instability and their implications in various diseases. Genomics Inform. 2014; 12:98–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Solyom S., Kazazian H.H. Jr. Mobile elements in the human genome: implications for disease. Genome Med. 2012; 4:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Klement K., Goodarzi A.A.. DNA double strand break responses and chromatin alterations within the aging cell. Exp. Cell Res. 2014; 329:42–52. [DOI] [PubMed] [Google Scholar]
- 9. Allshire R.C., Madhani H.D.. Ten principles of heterochromatin formation and function. Nat. Rev. Mol. Cell Biol. 2018; 19:229–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Martens J.H., O'Sullivan R.J., Braunschweig U., Opravil S., Radolf M., Steinlein P., Jenuwein T.. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 2005; 24:800–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Daujat S., Weiss T., Mohn F., Lange U.C., Ziegler-Birling C., Zeissler U., Lappe M., Schubeler D., Torres-Padilla M.E., Schneider R.. H3K64 trimethylation marks heterochromatin and is dynamically remodeled during developmental reprogramming. Nat. Struct. Mol. Biol. 2009; 16:777–781. [DOI] [PubMed] [Google Scholar]
- 12. Schotta G., Lachner M., Sarma K., Ebert A., Sengupta R., Reuter G., Reinberg D., Jenuwein T.. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004; 18:1251–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P. et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007; 448:553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Trojer P., Reinberg D.. Facultative heterochromatin: is there a distinctive molecular signature. Mol. Cell. 2007; 28:1–13. [DOI] [PubMed] [Google Scholar]
- 15. Lachner M., O’Carroll D., Rea S., Mechtler K., Jenuwein T.. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001; 410:116–120. [DOI] [PubMed] [Google Scholar]
- 16. Cheng C.T., Kuo C.Y., Ann D.K.. KAPtain in charge of multiple missions: emerging roles of KAP1. World J. Biol. Chem. 2014; 5:308–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bartova E., Krejci J., Harnicarova A., Galiova G., Kozubek S.. Histone modifications and nuclear architecture: a review. J. Histochem. Cytochem. 2008; 56:711–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Melcher M., Schmid M., Aagaard L., Selenko P., Laible G., Jenuwein T.. Structure-function analysis of SUV39H1 reveals a dominant role in heterochromatin organization, chromosome segregation, and mitotic progression. Mol. Cell. Biol. 2000; 20:3728–3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Johnson W.L., Straight A.F.. RNA-mediated regulation of heterochromatin. Curr. Opin. Cell Biol. 2017; 46:102–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Johnson W.L., Yewdell W.T., Bell J.C., McNulty S.M., Duda Z., O’Neill R.J., Sullivan B.A., Straight A.F.. RNA-dependent stabilization of SUV39H1 at constitutive heterochromatin. Elife. 2017; 6:e25299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Velazquez Camacho O., Galan C., Swist-Rosowska K., Ching R., Gamalinda M., Karabiber F., De La Rosa-Velazquez I., Engist B., Koschorz B., Shukeir N. et al. Major satellite repeat RNA stabilize heterochromatin retention of Suv39h enzymes by RNA-nucleosome association and RNA:DNA hybrid formation. Elife. 2017; 6:e25293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rivera C., Saavedra F., Alvarez F., Diaz-Celis C., Ugalde V., Li J., Forne I., Gurard-Levin Z.A., Almouzni G., Imhof A. et al. Methylation of histone H3 lysine 9 occurs during translation. Nucleic Acids Res. 2015; 43:9097–9106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schultz D.C., Ayyanathan K., Negorev D., Maul G.G., Rauscher F.J. 3rd. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002; 16:919–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tachibana M., Sugimoto K., Fukushima T., Shinkai Y.. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem. 2001; 276:25309–25317. [DOI] [PubMed] [Google Scholar]
- 25. Tachibana M., Ueda J., Fukuda M., Takeda N., Ohta T., Iwanari H., Sakihama T., Kodama T., Hamakubo T., Shinkai Y.. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 2005; 19:815–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rea S., Eisenhaber F., O’Carroll D., Strahl B.D., Sun Z.W., Schmid M., Opravil S., Mechtler K., Ponting C.P., Allis C.D. et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000; 406:593–599. [DOI] [PubMed] [Google Scholar]
- 27. Loyola A., Tagami H., Bonaldi T., Roche D., Quivy J.P., Imhof A., Nakatani Y., Dent S.Y., Almouzni G.. The HP1alpha-CAF1-SetDB1-containing complex provides H3K9me1 for Suv39-mediated K9me3 in pericentric heterochromatin. EMBO Rep. 2009; 10:769–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rowbotham S.P., Barki L., Neves-Costa A., Santos F., Dean W., Hawkes N., Choudhary P., Will W.R., Webster J., Oxley D. et al. Maintenance of silent chromatin through replication requires SWI/SNF-like chromatin remodeler SMARCAD1. Mol. Cell. 2011; 42:285–296. [DOI] [PubMed] [Google Scholar]
- 29. Klement K., Luijsterburg M.S., Pinder J.B., Cena C.S., Del Nero V., Wintersinger C.M., Dellaire G., van Attikum H., Goodarzi A.A.. Opposing ISWI- and CHD-class chromatin remodeling activities orchestrate heterochromatic DNA repair. J. Cell Biol. 2014; 207:717–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fritsch L., Robin P., Mathieu J.R., Souidi M., Hinaux H., Rougeulle C., Harel-Bellan A., Ameyar-Zazoua M., Ait-Si-Ali S.. A subset of the histone H3 lysine 9 methyltransferases Suv39h1, G9a, GLP, and SETDB1 participate in a multimeric complex. Mol. Cell. 2010; 37:46–56. [DOI] [PubMed] [Google Scholar]
- 31. Turelli P., Castro-Diaz N., Marzetta F., Kapopoulou A., Raclot C., Duc J., Tieng V., Quenneville S., Trono D.. Interplay of TRIM28 and DNA methylation in controlling human endogenous retroelements. Genome Res. 2014; 24:1260–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Barde I., Verp S., Offner S., Trono D.. Lentiviral vector mediated transgenesis. Curr. Protoc. Mouse Biol. 2011; 1:169–184. [DOI] [PubMed] [Google Scholar]
- 33. Barde I., Rauwel B., Marin-Florez R.M., Corsinotti A., Laurenti E., Verp S., Offner S., Marquis J., Kapopoulou A., Vanicek J. et al. A KRAB/KAP1-miRNA cascade regulates erythropoiesis through stage-specific control of mitophagy. Science. 2013; 340:350–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ryan R.F., Schultz D.C., Ayyanathan K., Singh P.B., Friedman J.R., Fredericks W.J., Rauscher F.J. 3rd. KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Kruppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol. Cell. Biol. 1999; 19:4366–4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. White D., Rafalska-Metcalf I.U., Ivanov A.V., Corsinotti A., Peng H., Lee S.C., Trono D., Janicki S.M., Rauscher F.J. 3rd. The ATM substrate KAP1 controls DNA repair in heterochromatin: regulation by HP1 proteins and serine 473/824 phosphorylation. Mol. Cancer Res. 2012; 10:401–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cheng C.T., Kuo C.Y., Ouyang C., Li C.F., Chung Y., Chan D.C., Kung H.J., Ann D.K.. Metabolic stress-induced phosphorylation of KAP1 Ser473 blocks mitochondrial fusion in breast cancer cells. Cancer Res. 2016; 76:5006–5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhou X.F., Yu J., Chang M., Zhang M., Zhou D., Cammas F., Sun S.C.. TRIM28 mediates chromatin modifications at the TCRalpha enhancer and regulates the development of T and natural killer T cells. PNAS. 2012; 109:20083–20088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chikuma S., Suita N., Okazaki I.M., Shibayama S., Honjo T.. TRIM28 prevents autoinflammatory T cell development in vivo. Nat. Immunol. 2012; 13:596–603. [DOI] [PubMed] [Google Scholar]
- 39. King C.A. Kaposi's sarcoma-associated herpesvirus kaposin B induces unique monophosphorylation of STAT3 at serine 727 and MK2-mediated inactivation of the STAT3 transcriptional repressor TRIM28. J. Virol. 2013; 87:8779–8791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chang C.W., Chou H.Y., Lin Y.S., Huang K.H., Chang C.J., Hsu T.C., Lee S.C.. Phosphorylation at Ser473 regulates heterochromatin protein 1 binding and corepressor function of TIF1beta/KAP1. BMC Mol. Biol. 2008; 9:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Quenneville S., Verde G., Corsinotti A., Kapopoulou A., Jakobsson J., Offner S., Baglivo I., Pedone P.V., Grimaldi G., Riccio A. et al. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol. Cell. 2011; 44:361–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. O’Geen H., Squazzo S.L., Iyengar S., Blahnik K., Rinn J.L., Chang H.Y., Green R., Farnham P.J.. Genome-wide analysis of KAP1 binding suggests autoregulation of KRAB-ZNFs. PLos Genet. 2007; 3:e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lange U.C., Siebert S., Wossidlo M., Weiss T., Ziegler-Birling C., Walter J., Torres-Padilla M.E., Daujat S., Schneider R.. Dissecting the role of H3K64me3 in mouse pericentromeric heterochromatin. Nat. Commun. 2013; 4:2233. [DOI] [PubMed] [Google Scholar]
- 44. Kapoor-Vazirani P., Kagey J.D., Vertino P.M.. SUV420H2-mediated H4K20 trimethylation enforces RNA polymerase II promoter-proximal pausing by blocking hMOF-dependent H4K16 acetylation. Mol. Cell. Biol. 2011; 31:1594–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Singh K., Cassano M., Planet E., Sebastian S., Jang S.M., Sohi G., Faralli H., Choi J., Youn H.D., Dilworth F.J. et al. A KAP1 phosphorylation switch controls MyoD function during skeletal muscle differentiation. Genes Dev. 2015; 29:513–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Alabert C., Bukowski-Wills J.C., Lee S.B., Kustatscher G., Nakamura K., de Lima Alves F., Menard P., Mejlvang J., Rappsilber J., Groth A.. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat. Cell Biol. 2014; 16:281–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Alabert C., Barth T.K., Reveron-Gomez N., Sidoli S., Schmidt A., Jensen O.N., Imhof A., Groth A.. Two distinct modes for propagation of histone PTMs across the cell cycle. Genes Dev. 2015; 29:585–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bulut-Karslioglu A., De La Rosa-Velazquez I.A., Ramirez F., Barenboim M., Onishi-Seebacher M., Arand J., Galan C., Winter G.E., Engist B., Gerle B. et al. Suv39h-dependent H3K9me3 marks intact retrotransposons and silences LINE elements in mouse embryonic stem cells. Mol. Cell. 2014; 55:277–290. [DOI] [PubMed] [Google Scholar]
- 49. Ivanov A.V., Peng H., Yurchenko V., Yap K.L., Negorev D.G., Schultz D.C., Psulkowski E., Fredericks W.J., White D.E., Maul G.G. et al. PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol. Cell. 2007; 28:823–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zeng L., Yap K.L., Ivanov A.V., Wang X., Mujtaba S., Plotnikova O., Rauscher F.J. 3rd, Zhou M.M.. Structural insights into human KAP1 PHD finger-bromodomain and its role in gene silencing. Nat. Struct. Mol. Biol. 2008; 15:626–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hu C., Zhang S., Gao X., Gao X., Xu X., Lv Y., Zhang Y., Zhu Z., Zhang C., Li Q. et al. Roles of Kruppel-associated Box (KRAB)-associated co-repressor KAP1 Ser-473 phosphorylation in DNA damage response. J. Biol. Chem. 2012; 287:18937–18952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bolderson E., Savage K.I., Mahen R., Pisupati V., Graham M.E., Richard D.J., Robinson P.J., Venkitaraman A.R., Khanna K.K.. Kruppel-associated Box (KRAB)-associated co-repressor (KAP-1) Ser-473 phosphorylation regulates heterochromatin protein 1beta (HP1-beta) mobilization and DNA repair in heterochromatin. J. Biol. Chem. 2012; 287:28122–28131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Goodarzi A.A., Noon A.T., Deckbar D., Ziv Y., Shiloh Y., Lobrich M., Jeggo P.A.. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol. Cell. 2008; 31:167–177. [DOI] [PubMed] [Google Scholar]
- 54. Ziv Y., Bielopolski D., Galanty Y., Lukas C., Taya Y., Schultz D.C., Lukas J., Bekker-Jensen S., Bartek J., Shiloh Y.. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat. Cell Biol. 2006; 8:870–876. [DOI] [PubMed] [Google Scholar]
- 55. Iyengar S., Ivanov A.V., Jin V.X., Rauscher F.J. 3rd, Farnham P.J.. Functional analysis of KAP1 genomic recruitment. Mol. Cell. Biol. 2011; 31:1833–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rowe H.M., Jakobsson J., Mesnard D., Rougemont J., Reynard S., Aktas T., Maillard P.V., Layard-Liesching H., Verp S., Marquis J. et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010; 463:237–240. [DOI] [PubMed] [Google Scholar]
- 57. Ecco G., Cassano M., Kauzlaric A., Duc J., Coluccio A., Offner S., Imbeault M., Rowe H.M., Turelli P., Trono D.. Transposable elements and their KRAB-ZFP controllers regulate gene expression in adult tissues. Dev. Cell. 2016; 36:611–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Di Cerbo V., Mohn F., Ryan D.P., Montellier E., Kacem S., Tropberger P., Kallis E., Holzner M., Hoerner L., Feldmann A. et al. Acetylation of histone H3 at lysine 64 regulates nucleosome dynamics and facilitates transcription. Elife. 2014; 3:e01632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pradeepa M.M., Grimes G.R., Kumar Y., Olley G., Taylor G.C., Schneider R., Bickmore W.A.. Histone H3 globular domain acetylation identifies a new class of enhancers. Nat. Genet. 2016; 48:681–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Maison C., Bailly D., Peters A.H., Quivy J.P., Roche D., Taddei A., Lachner M., Jenuwein T., Almouzni G.. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 2002; 30:329–334. [DOI] [PubMed] [Google Scholar]
- 61. Maison C., Bailly D., Roche D., Montes de Oca R., Probst A.V., Vassias I., Dingli F., Lombard B., Loew D., Quivy J.P. et al. SUMOylation promotes de novo targeting of HP1alpha to pericentric heterochromatin. Nat. Genet. 2011; 43:220–227. [DOI] [PubMed] [Google Scholar]
- 62. Muchardt C., Guilleme M., Seeler J.S., Trouche D., Dejean A., Yaniv M.. Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1alpha. EMBO Rep. 2002; 3:975–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Matzke M.A., Birchler J.A.. RNAi-mediated pathways in the nucleus. Nat. Rev. Genet. 2005; 6:24–35. [DOI] [PubMed] [Google Scholar]
- 64. Keller C., Kulasegaran-Shylini R., Shimada Y., Hotz H.R., Buhler M.. Noncoding RNAs prevent spreading of a repressive histone mark. Nat. Struct. Mol. Biol. 2013; 20:994–1000. [DOI] [PubMed] [Google Scholar]
- 65. Vogel M.J., Guelen L., de Wit E., Peric-Hupkes D., Loden M., Talhout W., Feenstra M., Abbas B., Classen A.K., van Steensel B.. Human heterochromatin proteins form large domains containing KRAB-ZNF genes. Genome Res. 2006; 16:1493–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All next-generation sequencing data have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) database under the accession number GEO: GSE 86391.