Abstract
Genomic sequencing of hepatocellular carcinoma (HCC) uncovers a paucity of actionable mutations, underscoring the necessity to exploit epigenetic vulnerabilities for therapeutics. In HCC, EZH2-mediated H3K27me3 represents a major oncogenic chromatin modification, but how it modulates the therapeutic vulnerability of signaling pathways remains unknown. Here, we show EZH2 acts antagonistically to AKT signaling in maintaining H3K27 methylome through epigenetic silencing of IGFBP4. ChIP-seq revealed enrichment of Ezh2/H3K27me3 at silenced loci in HBx-transgenic mouse-derived HCCs, including Igfbp4 whose down-regulation significantly correlated with EZH2 overexpression and poor survivals of HCC patients. Functional characterizations demonstrated potent growth- and invasion-suppressive functions of IGFBP4, which was associated with transcriptomic alterations leading to deregulation of multiple signaling pathways. Mechanistically, IGFBP4 stimulated AKT/EZH2 phosphorylation to abrogate H3K27me3-mediated silencing, forming a reciprocal feedback loop that suppressed core transcription factor networks (FOXA1/HNF1A/HNF4A/KLF9/NR1H4) for normal liver homeostasis. Consequently, the in vivo tumorigenicity of IGFBP4-silenced HCC cells was vulnerable to pharmacological inhibition of EZH2, but not AKT. Our study unveils chromatin regulation of a novel liver tumor suppressor IGFBP4, which constitutes an AKT-EZH2 reciprocal loop in driving H3K27me3-mediated epigenetic reprogramming. Defining the aberrant chromatin landscape of HCC sheds light into the mechanistic basis of effective EZH2-targeted inhibition.
INTRODUCTION
Hepatocellular carcinoma (HCC) is notorious for dismal prognosis and high mortality rate. Despite the escalating trend of incidence in the past decades, treatment options for advanced HCCs remain extremely limited. Such inadequacy of actionable drug targets is attributed to the genomic heterogeneity of HCC, and the complex roles of oncoproteins in signaling networks that possess complementary and redundant functions (1). To circumvent these issues, comprehensive sequencing analyses have been performed on different HCC subtypes to identify the respective genomic abnormalities and explore for new therapeutic vulnerabilities. For instance, a recent large-scale multi-platform study on 363 patients revealed frequent mutations in 26 classical cancer genes (e.g. TP53, RB1, CTNNB1, etc.) and eight potential HCC drivers (e.g. LZTR1, EEF1A1, AZIN1, etc.) (2); a study on 49 cases identified the specific association of ADGRB1 mutation with aflatoxin-related HCCs (3); and a study on 343 samples uncovered the linkages of gene mutations (e.g. RPS6KA3, AXIN1, TSC1/TSC2, etc.) and HCC histological subtypes (4). While these omics studies undoubtedly increase our understanding of HCC pathogenesis and infer novel druggable targets, they corroborate the low recurrence of genetic mutations in large HCC cohorts (mostly <10%), therefore targeting of these genetically altered oncoproteins might only benefit a limited subset of HCC patients.
An alternative yet equally important molecular aberration in HCCs is epigenetic deregulation. Histone modifications and chromatin remodelling are among the important epigenetic mechanisms to safeguard the genome and dynamically adjust gene expression in response to external insults. In HCCs, the master chromatin regulator EZH2, which is the catalytic subunit of the PRC2 complex for histone H3 lysine 27 tri-methylation (H3K27me3), is frequently overexpressed and associates with malignant characteristics. Of clinical significance, EZH2 up-regulation from dysplastic nodule to early HCC is associated with the subsequent development of advanced HCC (5), and EZH2 may be used as a diagnostic biomarker for HCC detection in liver biopsy tissues (6). This evidence suggests that EZH2 may be a potential target for therapeutic treatment of HCCs. Nonetheless, the EZH2-modulated targets in HCC development have yet been fully elucidated, and it remains obscure whether and how the EZH2-instigated effects interrupt or stimulate other cancer signaling cascades (7).
The insulin-like growth factors (IGFs) are proteins with high sequence similarity to insulin. However, unlike insulin, the IGFs in serum and other biological fluids are bound to specific IGF-binding proteins (IGFBPs) which were initially discovered as carrier proteins in serum (8). IGFBP4 is the smallest and a unique member among the human IGFBPs, that it has two extra inter-linked cysteine residues in the variable L-domain (9). Under physiological conditions, IGFBP4 binds to IGF-1 and IGF-II to fine tune their biological effects, but IGFBP4 can also act independently of IGFs to modulate cellular activities such as cell proliferation, survival and migration. The constitutive expression of IGFBP4 in tissues is important, as deregulation of this gene can contribute to cardiogenic defects (10), angiogenesis (11) and other disorders. In terms of malignancies, IGFBP4 has been associated with cancers in lung, breast, stomach, colon, prostate, bone, endocrine systems and nervous tissues, wherein IGFBP4 exerts opposite roles in a context-dependent manner (12). Yet the role of IGFBP4 in HCC remained unknown.
Based on the therapeutic potential and restorability of the EZH2-mediated epigenetic defects in HCC, it is crucial to delineate the EZH2/H3K27me3-modulated genes and signaling pathways in order to evaluate the effectiveness and safety of EZH2-targeted drugs. Previously, we showed that EZH2 functions to suppress Wnt antagonists, thereby activating the oncogenic Wnt/β-catenin signaling to drive HCC cell proliferation (13). Moreover, we demonstrated that EZH2 silences certain tumor-suppressive microRNAs and up-regulates the pro-tumorigenic CCRK in HCCs (14,15). In this study, we aimed to profile the genome-wide EZH2/H3K27me3 targets and the associated signaling networks in a hepatitis B virus X protein (HBx)-transgenic (TG) mouse model, which resembles the male predominant human HCCs (16). Using a top-down approach, we identified IGFBP4 as one of the most consistently ablated genes by EZH2/H3K27me3 in both mouse and human HCCs. Surprisingly, IGFBP4 not only possesses strong tumor-suppressive functions, but also constitutes a double-negative feedback circuitry comprising of AKT and EZH2 signaling. This IGFBP4/AKT/EZH2 regulatory hub epigenetically silences a core set of transcription factors crucial for normal liver homeostasis, thereby providing mechanistic basis for effective EZH2-targeted therapy.
MATERIALS AND METHODS
Patients
HCCs and the matched non-tumor liver tissues were collected from patients during the curative surgeries at the Prince of Wales Hospital in Hong Kong. HCC tissues were taken from the most viable areas of tumors immediately after surgical resection, excluding the necrotic and hemorrhagic areas of tissues. The matched non-tumor tissues were selected at a clear distance from the tumor edge (>5 cm) with no evidence of tumor invasion. Tissues were snap-frozen immediately after resection and stored at −80°C until use. The study protocol complied with the Declaration of Helsinki, and was approved by the Joint Chinese University of Hong Kong-New Territories East Cluster Clinical Research Ethics Committee.
Mice
The experimental uses of all mice were approved by the Animal Experimentation Ethics Committee of the Chinese University of Hong Kong. All mice received humane care according to the criteria outlined in the Guide for the Care and Use of Laboratory Animals (NIH). The strain of HBx-TG mice was fixed to C57BL/6 by backcrossing with the C57BL/6 strain for >20 generations. For xenograft assay one month-old male nude mice were used. 5 × 106 cells were subcutaneously injected into the left and right dorsal flanks of nude mice respectively to induce tumor formation. The growths of tumors were recorded from day 4 after injection. Volume of tumor V (mm3) was calculated by L/2 × W2, where L (mm) is the length of tumor and W (mm) is the width of tumor. Experiment was terminated on day 28 and the mice were sacrificed by chloroform treatment and tumors were isolated for analyses.
Cell culture
Human liver cell lines were maintained in high-glucose Dulbecco's modified Eagle's medium (DMEM; Gibco) supplemented with 10% fetal bovine serum (FBS; Hyclone), 1% Penicillin–Streptomycin (Gibco) and 1% Non-Essential Amino Acids (Gibco). All cells were incubated at 37°C in a humidified chamber containing 5% CO2. The cultures were passaged at pre-confluent densities using a solution of 0.25% trypsin and 1 mM EDTA-Na (Invitrogen).
Chromatin immunoprecipitation (ChIP) assays
Cells grown at confluence of 80% in 150-mm dishes were rinsed twice with cold PBS and cross-linked with 15 ml of 1% formaldehyde for 10 min at room temperature, and then quenched by the addition of 0.75 ml of 2.5 M glycine for 3 min. Cells were rinsed with cold PBS twice and harvested by scraping. Cells were centrifuged at 700 × g for 5 min at 4°C, and lysed in 10 ml cold lysis buffer I (50 mM HEPES–KOH, pH 8.0, 1 mM EDTA, 140 mM NaCl, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100, supplemented with protease inhibitors) and then rotated at 4°C for 10 min. Cell debris was collected by centrifugation at 1350 × g for 5 min at 4°C, and resuspended in 10 ml of cold lysis buffer II (10 mM Tris–HCl, pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, supplemented with protease inhibitors) and then rotated for 10 min at 4°C. Nuclei were collected by centrifugation at 1350 × g for 5 min at 4°C and resuspended in 3.6 ml of lysis buffer III (10 mM Tris–HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% Na-Deoxycholate, 0.5% N-Lauroylsarcosine, supplemented with protease inhibitors). Nine cycles of sonication were performed to fragment DNA using a Bioruptor ultrasonicator (Diagenode). Fifty microliters of cell lysate was saved for input DNA extraction. Five microgram of antibodies attached to 100 μl of Dynabeads Protein G (Invitrogen) was used for protein–DNA complexes immunoprecipitation overnight. Dynabeads were washed five times at 4°C with RIPA buffer (50 mM HEPES–KOH, pH 8.0, 500 mM LiCl, 1 mM EDTA, 1.0% NP-40, 0.7% Na-Deoxycholate), once with 10 mM TE buffer (1 mM EDTA and 50 mM NaCl), collected by centrifugation (960 × g for 3 min at 4°C) and then incubated in elution buffer (50 mM Tris–HCl, pH 8.0, 10 mM EDTA, 1.0% SDS) for 30 min at 65°C. Eluted fragments in the supernatant were collected by centrifugation (16 000 × g for 1 min at room temperature). Eight microliter of 5 M NaCl was added and reversed cross-links at 65°C overnight. An aliquot input (2%) was also subjected to reverse cross-linking in parallel. After that, eluted materials were treated with RNase A solution (TE, pH 8.0, 0.7% SDS, 200 μg/ml RNase A) and protease K solution (TE, pH 8.0, 0.7% SDS, 200 μg/ml protease K) to degrade RNA and protein, respectively, and DNA fragments were purified using Phase Lock Gel (Fisher).
Library preparation for ChIP-sequencing
Library preparation was conducted using The NEBNext® Ultra™ DNA Library Prep Kit (NEB, UK) according to the manufacturer's instructions. In brief, ChIP-DNA was end-polished with end-polishing enzymes: T4 DNA polymerase, Klenow DNA polymerase and T4 polynucleotide kinase to create blunt ends. The end-repaired DNA fragments were A-tailed for the Illumina pipeline and ligated with adapters. The ligated fragments were amplified using adapter-specific primers and the number of cycles were low to reduce duplicates. The amplified PCR products were size selected by AMPure XP beads (Beckman Coulter, USA) and purified. The purified DNA fragment sizes were checked on a BioAnalyzer (Agilent, USA) and used for next-generation sequencing. The ChIP-seq data are accessible at NCBI GEO database: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE113326.
Library preparation for RNA-seq
Library preparation for RNA sequencing was conducted according to the manufacturer's instructions. Briefly, the mRNA and non-coding RNAs were enriched by removing rRNA from total RNA. Then mRNA and non-coding RNAs were fragmented into short fragments (∼200–700 bp). The first-strand cDNA was synthesized by random hexamer-primer. The second strand of cDNA was synthesized with DNA polymerase I. cDNA was purified and ligated with sequencing adapters. The second strand was degraded by UNG (Uracil-N-Glyosylase). The fragments were purified and enriched by PCR amplification. Every 6 RNA-seq libraries were multiplexed and sequenced on 1 lane of Hiseq4000 to obtain 100-base paired-end reads.
RNA-seq data processing
The clean reads from each sample were aligned to the human genome (hg19) or mouse genome (mm10) using TopHat v2.0.11 with default parameters and turning off novel junction search function (17). Random subsampling for each samples were performed with picard tools v1.1.98 (http://broadinstitute.github.io/picard). Different conditions were compared with the Cufflinks v2.2.1 tool cuffdiff with default parameters and geometric normalization (18).
Gene set enrichment analysis (GSEA)
We acknowledge our use of the gene set enrichment analysis, GSEA software, and Molecular Signature Database (MSigDB) (http://www.broad.mit.edu/gsea) (19). GSEA is a robust computational method that was used to determine whether a defined set of genes showed statistically significant, concordant correlation with the IGFBP4-related signaling axis. In the analysis setting, the number of permutations was set to 1000, and the Molecular Signature Database (MSigDB) version was 6.0. In the results, only gene sets that met the false discovery rate (FDR) of 25% and P < 0.05 criteria were considered.
Protein interactome and subcellular localization analyses
The protein interactions were analyzed using esyN (http://www.esyn.org) (20), BioGRID (https://thebiogrid.org) (21), and HumanMine (http://www.humanmine.org) (22). The subcellular localizations of protein candidates were analysed using databases (knowledge channel, experiments channel, text mining channel) from COMPARTMENTS (http://compartments.jensenlab.org) (23).
Xenograft assay
Female nude mice of 6–8 weeks old were subcutaneously injected with 5 × 106 of Huh7 cells. Tumors were allowed to grow until an average volume of about 0.1 cm3, and the mice were then randomly divided into control and treatment groups (n = 4–8 mice/group). The following treatments were administered as follows: vehicle only, MK-2206 (50 mg/kg, oral gavage, once per day), EPZ-6438 (125 mg/kg, oral gavage, twice per day), and the combination group (EPZ-6438 + MK-2206). Tumor sizes were measured twice a week with calipers. Tumor volumes were calculated using the formula (L × W2)/2. The mice were sacrificed after 32 days of treatments.
Statistical method
The Power and Sample Size Program was used to determine the sample size for in vitro experiments. For animal study, AEEC Animal Experimentation Sample Size Calculator was used to estimate the sample size which represents an optimum number to attain statistical significance of P < 0.05 with a 90% probability. The GraphPad Outlier Calculator software was used to exclude outlier. To avoid biased results, treatments were randomized that each experimental unit has a known, often equal, probability of receiving a given treatment. Statistical analysis was performed by GraphPad Prism (GraphPad Software). Mann–Whitney U test was used to analyze the gene expression of clinical samples between two selected groups. Student's t test was used to analyze colony formation assay, cell proliferation, cell cycle analysis and apoptosis. Log-rank (Mantel-Cox) Test was used in survival curve clinical tissues. P value < 0.05 was considered as statistically significant. Graphs were generated GraphPad Prism (GraphPad Software).
Additional materials and methods can be found in the online supplementary data.
RESULTS
Identification of novel EZH2-epigenetically regulated tumor suppressor genes in HBx-TG mouse model
To interrogate the EZH2-driven epigenomic alterations in hepatocarcinogenesis, we employed chromatin profiling and transcriptome analyses using an HBx-TG mouse male model that is prone to HCC formation (16). Wild-type (WT) and HBx-TG mice aged below 10 months were all free of tumors, whereas the 15- to 18 month-old HBx-TG mice developed HCCs. Western blot analysis showed that the normal livers from WT mice expressed low levels of Ezh2 and H3K27me3. By contrast, the liver tissues from HBx-TG mice exhibited consistent expression of HBx, and progressively increasing the expression of Ezh2 and H3K27me3 that peaked in the tumor tissues of 18-month-old TG mice (Figure 1A). The unparalleled expression patterns of HBx and Ezh2 in male HBx-TG mice are concordant with our previous studies, demonstrating that HBx can activate an androgen receptor (AR)-dependent transcriptional signaling pathway, which in turn up-regulates EZH2 to form a self-reinforcing loop, thus resulting in continuous EZH2 induction (14,16). These data demonstrated that the HBx-TG mouse could serve as clinically-relevant HCC model for the identification of novel EZH2-epigenetically regulated genes.
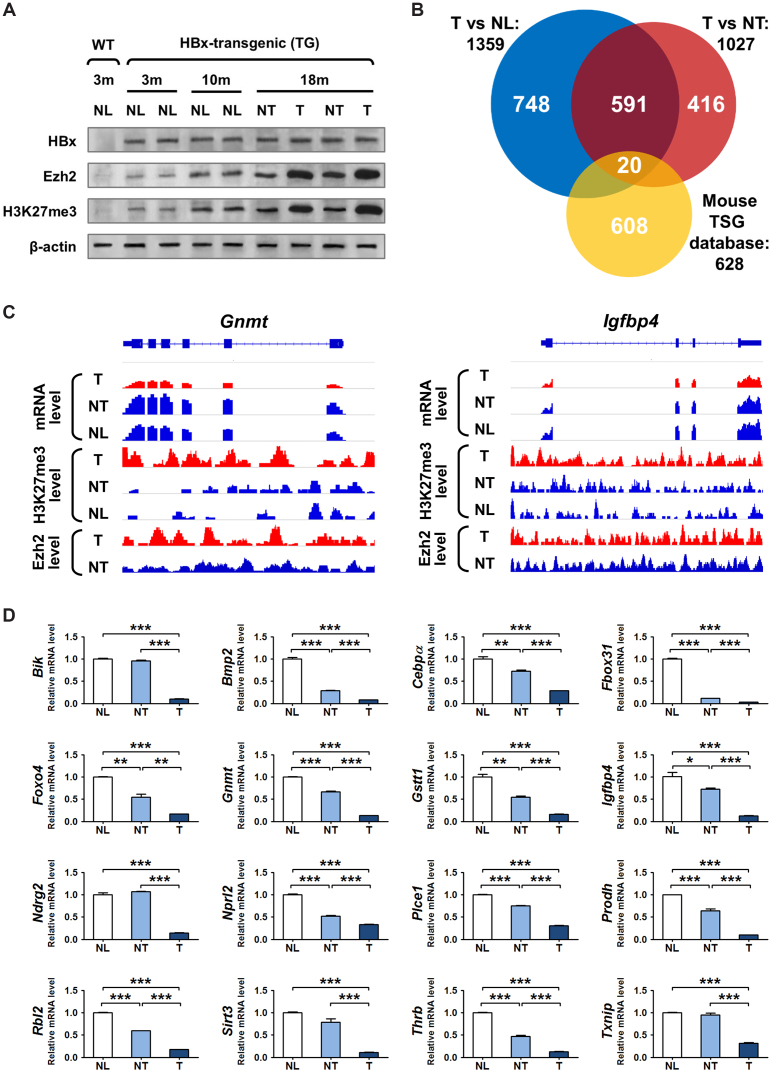
Figure 1.
Deregulation of H3K27 tri-methylation and gene repression in the HBx-TG mouse model. (A) Western blot analysis of the liver tissues collected from WT and HBx-TG mice at different time-points (m = month). Normal (NL), adjacent non-tumor (NT), tumor (T). (B) Venn diagram showing the integration of ChIP-seq, RNA-seq and mouse TSG data: (i) 1359 H3K27me3-occupied and repressed genes in tumor (TG-18m-T) versus normal liver (WT-3m-NL); (ii) 1027 H3K27me3-occupied and repressed genes in tumor (TG-18m-T) versus adjacent non-tumor (TG-18m-NT); and iii) 628 mouse tumor suppressors from published database. (C) Representative diagram showing the RNA-seq and ChIP-seq signals mapped to two candidate genes in the liver tissues of WT-3m-NL, TG-18m-NT and TG-18m-T mice, respectively. (D) In qRT-PCR analysis, 16 novel epigenetically-regulated TSGs displayed significant down-regulation in murine tumors (T) compared with non-tumor (NT) or normal (NL) tissues. *P < 0.05; **P < 0.01; ***P < 0.001.
To map the genome-wide H3K27me3 binding sites, we used the normal liver tissue from WT mouse and a pair of HCC and adjacent non-tumor tissues from HBx-TG mouse for chromatin immunoprecipitation sequencing (ChIP-seq) analysis. We identified 2833 H3K27me3-bound genes in tumor compared with normal liver, and 3525 genes in tumor compared with adjacent non-tumor across the genome. To screen for genes with concurrent H3K27me3 binding and repressed expression, we integrated our ChIP-seq and RNA-seq data from the same tissues, and uncovered 1359 and 1027 H3K27me3-occupied and repressed genes (at least 1.5-fold reduction) in tumor versus normal liver and adjacent non-tumor tissues, respectively (Figure 1B). Among the 611 targets common in both gene lists, 20 have been previously reported to function as tumor suppressor genes (TSGs) (24) (Figure 1B). Consistent with the strong H3K27me3 occupancy, some repressed TSG transcripts e.g. Gnmt and Igfbp4 were also enriched with Ezh2 in tumor (Figure 1C). To validate the accuracy of the RNA-seq data, the expression of these 20 candidate TSGs was examined by qRT-PCR in the original murine tissues. The results showed that 16 potential TSGs were significantly down-regulated in tumors compared with control tissues (Figure 1D; P < 0.05), whereas four candidates did not show significant changes (Supplementary Figure S1A). For further validation, an independent set of liver tissues from 4-month-old WT mice (n = 3) and HBx-TG mice (n = 5) were subjected to quantitative ChIP-PCR and qRT-PCR analyses. We successfully verified at least four TSGs (Gstt1, Igfbp4, Ndrg2 and Sirt3) in our screening cohort that showed concordant gene silencing by H3K27me3 in the livers of HBx-TG mice compared to the WT counterparts (Supplementary Figure S1B and C, P < 0.05). These results demonstrate the accuracy of the ChIP-seq data and suggest that H3K27me3-mediated TSG silencing is an early carcinogenic event.
IGFPB4 is remarkably silenced by EZH2 and inversely correlates with poor prognosis in HCC patients
To confirm whether the H3K27me3-bound and repressed genes (Figure 1D) were subject to EZH2 action, 4 human HCC cell lines (Hep3B, HepG2, Huh7 and PLC5) were treated with or without GSK126, a potent EZH2 inhibitor (25), followed by qRT-PCR analysis. The effect of GSK126 on global H3K27me3 reduction was confirmed in all 4 cell lines by Western blot (Figure 2A). Nine out of the 15 candidate genes (Gstt1 was omitted as it has no human orthologue) were significantly reactivated by GSK126 in at least two cell lines (Figure 2B; P < 0.05), while the remaining genes were generally non-responsive (Supplementary Figure S2A). Next, we examined the transcript levels of these 9 genes in a cohort of human HBV-associated HCC samples, and found that six genes (BIK, FOXO4, IGFBP4, NDRG2, NPRL2, PRODH) were significantly down-regulated in tumors compared with the matched non-tumor tissues (Figure 2C; P < 0.05), while 3 genes showed insignificant changes (Supplementary Figure S2B). The overall screening strategies are shown in Supplementary Table S1. All screened genes except IGFBP4 have been reported to exhibit tumor suppressor functions in the liver (Supplementary Table S2), supporting the critical role of EZH2-mediated silencing in hepatocarcinogenesis.
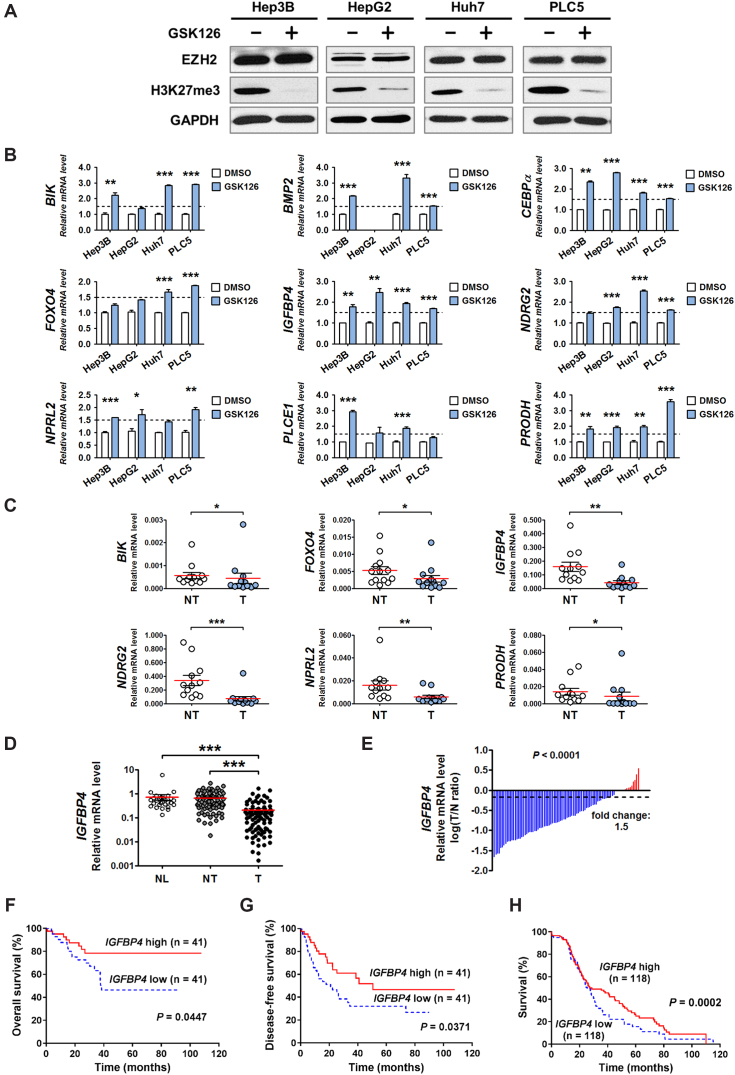
Figure 2.
Identification of IGFBP4 as a novel epigenetically-regulated and clinically significant TSG in human HBV-related HCCs. (A) The suppression effect of GSK126 on EZH2, as evident by H3K27me3 down-regulation, was examined in four HCC cell lines by western blot. (B) In qRT-PCR analysis, nine TSGs exhibited significant up-regulation (>1.5-fold-change) following GSK126 treatment in at least two HCC cell lines. In addition, (C) Six TSGs were significantly down-regulated in HCC tissues compared with paired adjacent non-tumor controls. (D) The IGFBP4 mRNA levels were assessed by qRT-PCR in 29 normal liver tissues (NL) and 82 pairs of adjacent non-tumor (NT) and tumor (T) liver tissues. (E) Comparison of IGFBP4 mRNA expression in paired HCC tissues. The bars (shown in log scale) illustrate the relative IGFBP4 mRNA levels in individual tissue pairs, of which negative and positive values respectively indicate down- and up-regulation of IGFBP4 in HCCs. (F, G) Kaplan–Meier analyses of the (F) overall survival and (G) disease-free survival of 82 HCC patients. (H) Kaplan–Meier analysis of patient survival using TCGA dataset. *P < 0.05; **P < 0.01; ***P < 0.001.
We then focused on elucidating the hitherto unknown function of IGFBP4 in human HCC. qRT-PCR analysis of 29 normal livers and 82 pairs of HBV-associated HCC and matched non-tumor tissues confirmed significant tumoral down-regulation of IGFBP4 (on average 3.43 and 3.13 times lower than normal liver and non-tumor tissues, respectively; Figure 2D; P < 0.001). Indeed, IGFBP4 down-regulation was prevalent in human HCCs (>90%; Figure 2E). More importantly, Kaplan–Meier analysis revealed that HCC patients with low IGFBP4 expression had significantly shorter overall survival (Figure 2F; P < 0.05) and disease-free survival (Figure 2G; P < 0.05), which was further confirmed using The Cancer Genome Atlas (TCGA) dataset (Figure 2H; P < 0.001). Correlation analyses further showed that decreased IGFBP4 mRNA levels were significantly associated with serum AFP level (P < 0.01) and tumor recurrence (P < 0.05) in HCC patients (Supplementary Table S3). These data clearly demonstrate the clinical significance of IGFBP4 silencing in HBV-associated HCCs.
To further investigate whether the EZH2-mediated silencing of IGFBP4 is solely associated with HBV, we analysed the expression patterns of EZH2 and IGFBP4 in HCCs with other etiological factors using TCGA dataset. The results showed that aberrant EZH2 up-regulation (∼6-fold) and IGFBP4 down-regulation (∼2-fold) were remarkably consistent among different HCCs associated with HBV, HCV, alcohol or NAFLD (Supplementary Figure S2C). Moreover, there was a significant negative correlation between EZH2 and IGFBP4 expressions in all HCCs (Supplementary Figure S2D; P < 0.001). This collective information suggests that HBV and other HCC risk factors can commonly trigger EZH2 overexpression that results in IGFBP4 ablation.
IGFBP4 exerts potent tumor suppressor activities in HCC cells
To determine the functional effects of IGFBP4, we transiently transfected HepG2 and Huh7 HCC cells with empty vector or IGFBP4-expression plasmid. IGFBP4 re-expression was confirmed by Western blot (Figure 3A), which evidently inhibited cell growth in both lines as measured by MTS assay (Figure 3B; P < 0.01) and colony formation assay (Figure 3C; P < 0.01). Consistently, matrigel invasion assay demonstrated that both IGFBP4-abundant HepG2 and Huh7 cells were less capable of invading extracellular matrix (ECM) in vitro (Figure 3D; P < 0.01).
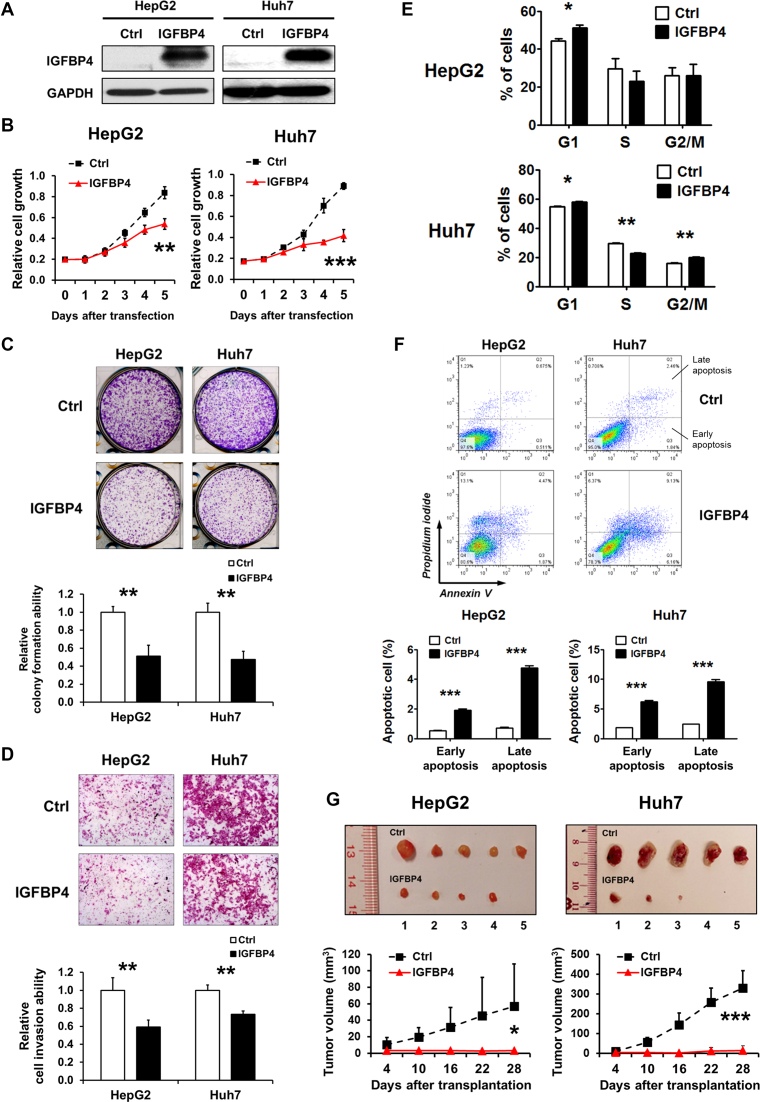
Figure 3.
IGFBP4 is a potent tumor suppressor in HCC. (A) Western blot analysis of HepG2 and Huh7 cells transiently transfected with empty vector or IGFBP4-expression plasmid. (B) Cell growth curves of differently transfected HepG2 and Huh7 cells. The growth rates were analysed by MTS assay. The data represent the results of three independent experiments. (C) The colony formation abilities of differently transfected HepG2 and Huh7 cells. The cell foci were visualized by crystal violet staining. All experiments were performed in triplicate. Quantification of cell foci is presented in the bar chart, wherein colony counts are presented as ±S.D. (D) The cell invasion abilities of differently transfected HepG2 and Huh7 cells were assessed by Matrigel transwell assay. Quantification of the cells migrating through the Matrigel-coated filter is presented in the bar chart. (E) The differently transfected HepG2 and Huh7 cells were stained with propidium iodide (PI) and the DNA contents were analyzed by flow cytometry. The cell numbers in individual cell cycles were calculated using ModFit software. The results shown were from three independent experiments. (F) Apoptotic cells were assessed by flow cytometry using PI and Annexin V staining. Cells in the lower left quadrant (PI low, Annexin V low) are viable, those in the lower right quadrant (PI low, Annexin V high) are early apoptotic, those in the upper left quadrant (PI high, Annexin V low) are necrotic, and those in the upper right quadrant (PI high, Annexin V high) are late apoptotic. Data obtained from three separate experiments are expressed as mean ± S.D. (G) Nude mice were subcutaneously injected with 0.5 × 106 HepG2 or Huh7 cells with or without IGFBP4 re-expression. After implantation for 4 days, tumor volumes were measured every 6 days for a period of 28 days (n = 5). Representative images of dissected tumors and tumor volume quantification are shown. *P < 0.05; **P < 0.01; ***P < 0.001.
To assess whether such IGFBP4-dependent growth suppression was due to altered cell division, we evaluated the cell cycle profiles by FACS. The results showed that re-expression of IGFBP4 caused G1-cell cycle arrest in both HepG2 and Huh7 cells (Figure 3E; P < 0.05). In addition, we performed Annexin V/Propidium iodide (PI) assay to examine the proportions of apoptotic cells, and found that IGFBP4 re-expression significantly promoted early and late apoptosis in both lines (Figure 3F; P < 0.001). We next subcutaneously injected HepG2 and Huh7 cells with or without stable transfection of IGFBP4 into the dorsal flanks of nude mice and measured the growth of xenografts over 4 weeks. We found that the IGFBP4-abundant tumors developed from both lines displayed remarkably slower growth, as evident by >16-fold smaller tumor volumes, when compared with controls (Figure 3G; P < 0.05). These in vitro and in vivo data concordantly demonstrate that IGFBP4 is a potent tumor suppressor in the liver.
IGFBP4 deregulates multiple oncogenic signaling pathways in HCC cells
To delineate the molecular mechanisms underlying the strong tumor-suppressive functions of IGFBP4, we performed RNA-seq on the HepG2 and Huh7 cells with or without IGFBP4 re-expression (Supplementary Table S4). We found that up-regulation of IGFBP4 caused significant transcriptional changes (at least 1.5-fold) of ∼7000 genes in HepG2 cells and Huh7 cells (Figure 4A and B). Gene Set Enrichment Analysis (GSEA) was then used to classify these perturbed genes according to the predefined gene sets in the Molecular Signatures Database (MSigDB). The gene signatures of different chemically and genetically perturbed liver cancers were enriched in both IGFBP4-expressed HepG2 and Huh7 cells (Figure 4C). Consistently, 20 common cancer modules correlated with liver cancer in both lines (Supplementary Figure S3A), which were associated with deregulation of numerous cellular pathways and transcription factors (Supplementary Figure S3B-F). These enrichments paralleled the identification of oncogenic signatures, namely AKT, EGFR, KRAS, p53, PTEN and TGF-β (Figure 4D), supporting the regulatory role of IGFBP4 on major HCC-related pathways.
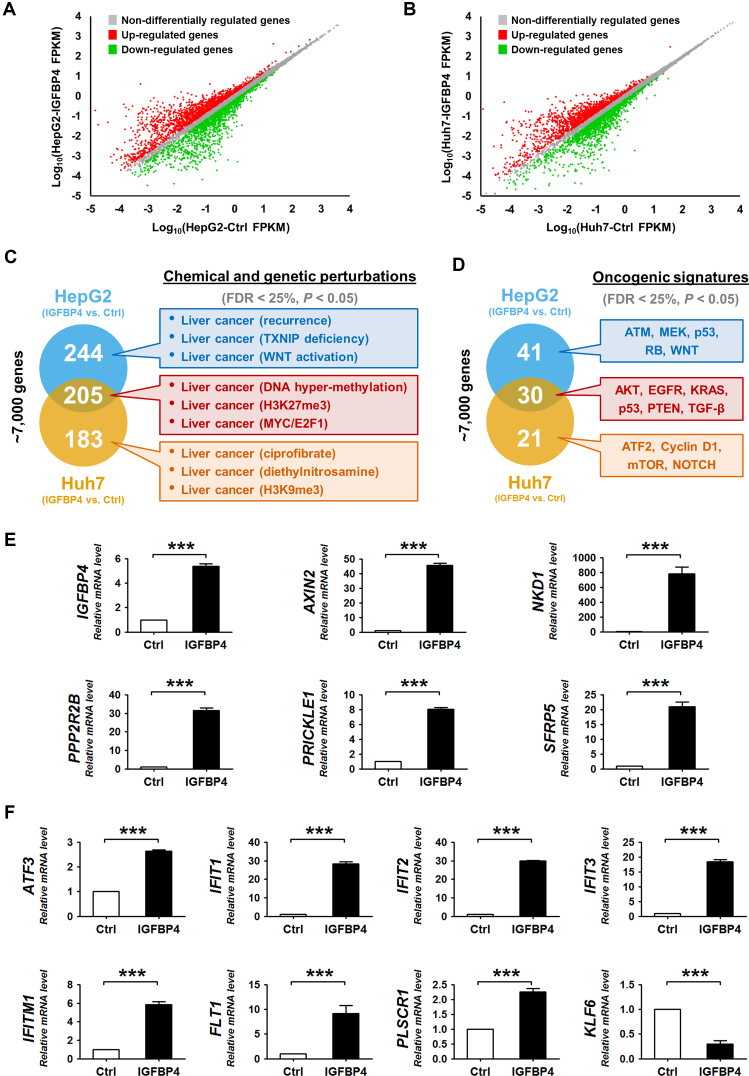
Figure 4.
IGFBP4 modulates oncogenic signaling pathways. (A and B) Scatter plots showing the global gene expression in (A) HepG2 and (B) Huh7 cells with or without IGFBP4 re-expression. The differentially regulated genes (with >1.5-fold change) are marked in green and red, respectively. FPKM = fragments per kilobase of transcript per million mapped reads. (C and D) GSEA analyses of the ∼7000 differentially regulated genes in HepG2 and Huh7 cells with or without IGFBP4 re-expression. These genes were classified according to the predefined gene sets in the MSigDB. Selected enrichment items are highlighted in the colored boxes. (E and F) qRT-PCR analyses of Huh7 cell-derived xenografts with or without IGFBP4 re-expression, in which the relative expression of (E) IGFBP4 and Wnt antagonists, and (F) AKT downstream molecules was examined. ***P < 0.001.
Interestingly, two significantly enriched molecular signatures in IGFBP4-expressing HCC cells, namely H3K27me3 (Figure 4C) and AKT (Figure 4D) point to a potential feedback loop (26). First, IGFBP4 may alleviate H3K27me3-mediated gene silencing. As we previously demonstrated the concordant repression of Wnt antagonists by H3K27me3 in HCC cells (13), we postulated that IGFBP4 re-expression may lead to up-regulation of Wnt antagonists. Indeed, qRT-PCR analysis of the Huh7-derived xenograft tumors showed significant elevation of the transcript levels of multiple Wnt antagonists including AXIN2, NKD1, PPP2R2B, PRICKLE1 and SFRP5 in the IGFBP4-expressing tumors when compared to the controls (Figure 4E; P < 0.001). Second, IGFBP4 may increase AKT signaling to suppress EZH2/H3K27me3 activity (26). In support of this hypothesis, we found that IGFBP4 significantly up-regulated the AKT downstream molecules such as ATF3, IFIT1-3, IFITM1, FLT1 and PLSCR1 (27), but suppressed KLF6 which is negatively-regulated by AKT signaling (28) (Figure 4F; P < 0.001).
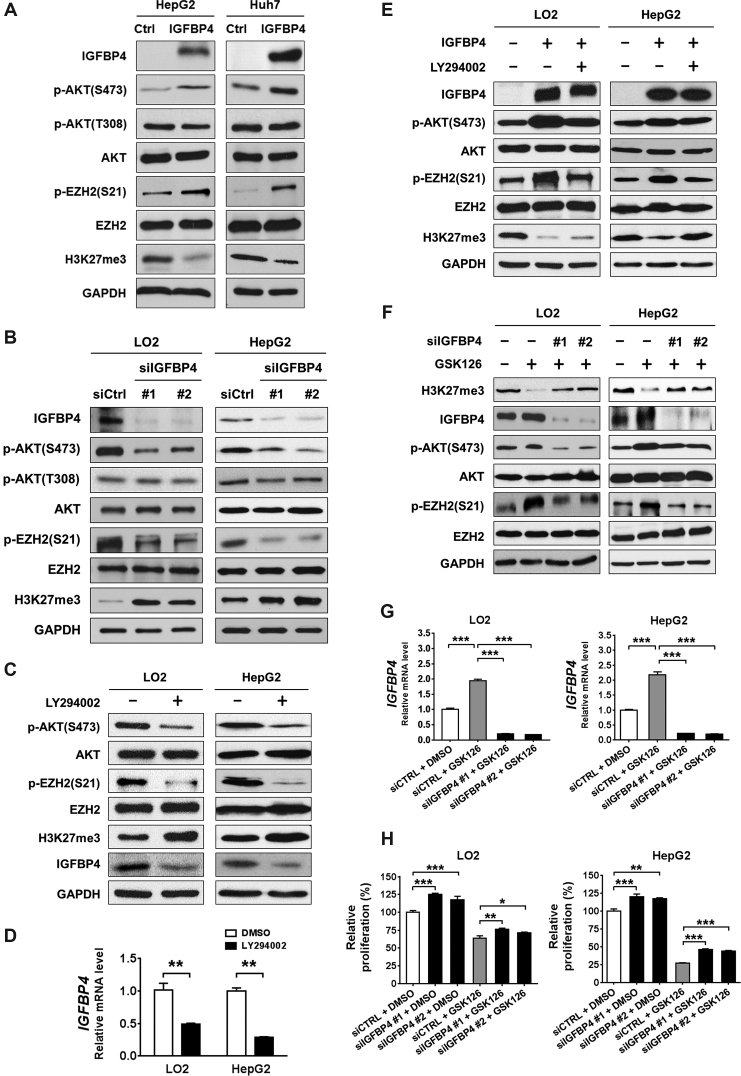
IGFBP4-induced AKT/EZH2 phosphorylation cascade abrogates EZH2/H3K27me3-mediated silencing
We next determined whether IGFBP4 modulates AKT and EZH2/H3K27me3 signaling by a series of ectopic expression/knockdown experiments. IGFBP4 re-expression in HepG2 and Huh7 cells induced phosphorylation of AKT (p-AKT) at Ser473 but not Thr308, phosphorylation of EZH2 (p-EZH2) at Ser21, and reduced global H3K27me3 level, while total AKT and EZH2 levels remained unchanged (Figure 5A). Conversely, knockdown of IGFBP4 by two independent siRNAs decreased the levels of p-AKT and p-EZH2, and up-regulated H3K27me3 levels in LO2 liver cells and HepG2 HCC cells (Figure 5B). To explore the underlying mechanisms, we examined the potential kinase/regulators that might mediate the IGFBP4-induced p-AKT. We found that IGFBP4 re-expression induced PI3K-mTORC2 (Rictor and p-mTOR), but not PTEN, and the downstream signals of p-AKT including mTORC1-RPS6 cascade (Raptor and p-RPS6) (Supplementary Figure S4A). Consistently, knockdown of IGFBP4 caused the opposite changes (Supplementary Figure S4B), suggesting that IGFBP4 activates AKT phosphorylation potentially through PI3K-mTORC2 signaling.
Figure 5.
IGFBP4/AKT/EZH2 form a double-negative feedback loop. (A) Representative Western blots showing the protein expression patterns of HepG2 and Huh7 cells with or without IGFBP4 re-expression, respectively. (B) siIGFBP4 #1 and #2 represent two different siRNAs for IGFBP4 knockdown in LO2 and HepG2 cells. Total cell lysates were analysed by western blot. (C and D) LO2 and HepG2 cells were treated with or without the AKT inhibitor LY294002. (C) Total cell lysates were analysed by Western blot, and (D) the expression changes of IGFBP4 were further confirmed by qRT-PCR in the same cells. (E) LO2 and HepG2 cells were transfected with empty vector or IGFBP4-expression plasmid, together with DMSO or LY294002 treatment. Total cell lysates were analysed by Western blot. (F–H) LO2 and HepG2 cells were transfected with DMSO or the EZH2 inhibitor GSK126, together with or without IGFBP4 knockdown, of which (F) total cell lysates were analysed by western blot, (G) the IGFBP4 transcriptional levels were examined by qRT-PCR and (H) cell proliferation rates were measured by MTS assay. **P < 0.01; ***P < 0.001.
To further verify the feedback loop, we inhibited AKT signaling by LY294002, which not only reduced p-EZH2 level, but also induced H3K27me3, leading to IGFBP4 down-regulation (Figure 5C and D; Supplementary Figure S5A and B; P < 0.01). Notably, AKT inhibition by LY294002 alleviated the suppression of H3K27me3 by IGFBP4 re-expression (Figure 5E; Supplementary Figure S5C). In a complementary experiment, cells were treated with single or combined inhibition of EZH2 and IGFBP4 by GSK126 and siRNA, respectively. EZH2 inhibition by GSK126 not only suppressed the H3K27me3 levels, but also induced IGFBP4 transcription and AKT/EZH2 phosphorylation (Figure 5F and G; Supplementary Figure S5D and E; P < 0.001). More importantly, knockdown of IGFBP4 by two independent siRNAs abrogated GSK126-induced IGFPB4/AKT/EZH2 phosphorylation cascade to partially rescue H3K27me3 (Figure 5F and G; Supplementary Figure S5D and E; P < 0.001). Overall, these data suggest that IGFBP4 feedback inactivates H3K27me3 silencing via the AKT/EZH2 phosphorylation.
Notably, while knockdown of IGFBP4 significantly increased basal cell proliferation (Figure 5H; P < 0.05), IGFBP4 down-regulation also restored GSK126-suppressed cell proliferation to a similar extent (Figure 5H; P < 0.05), demonstrating the functional significance of IGFBP4 in EZH2/H3K27me3 silencing. We further examined the IGFBP4/AKT signaling in a cohort of 21 paired human HCC tumor and non-tumor tissues by Western blot (Supplementary Figure S6A). Despite the relatively small sample size, we identified different subsets of HCCs based on the levels of IGFBP4 expression and AKT phosphorylation (Supplementary Figure S6B). We found that ∼33% of HCCs exhibited higher levels of p-AKT(Ser473) in tumors. Moreover, AKT activation in these tumors was associated with the loss of PTEN (data not shown). On the other hand, ∼48% of HCCs in our cohort exhibited concurrent IGFBP4 silencing and suppressed AKT activity (Supplementary Figure S6C and D; P < 0.05). In corroboration with the inverse relationship between Ezh2/H3K27me3 and Igfbp4/p-Akt/p-Ezh2 in HBx-TG mouse model (Supplementary Figure S6E), our data confirm a double-negative feedback loop between EZH2 and IGFBP4/AKT in maintaining H3K27me3-mediated silencing in HCC.
Aberrant IGFBP4/AKT/EZH2 epigenetic circuitry suppresses transcription factor networks for liver homeostasis
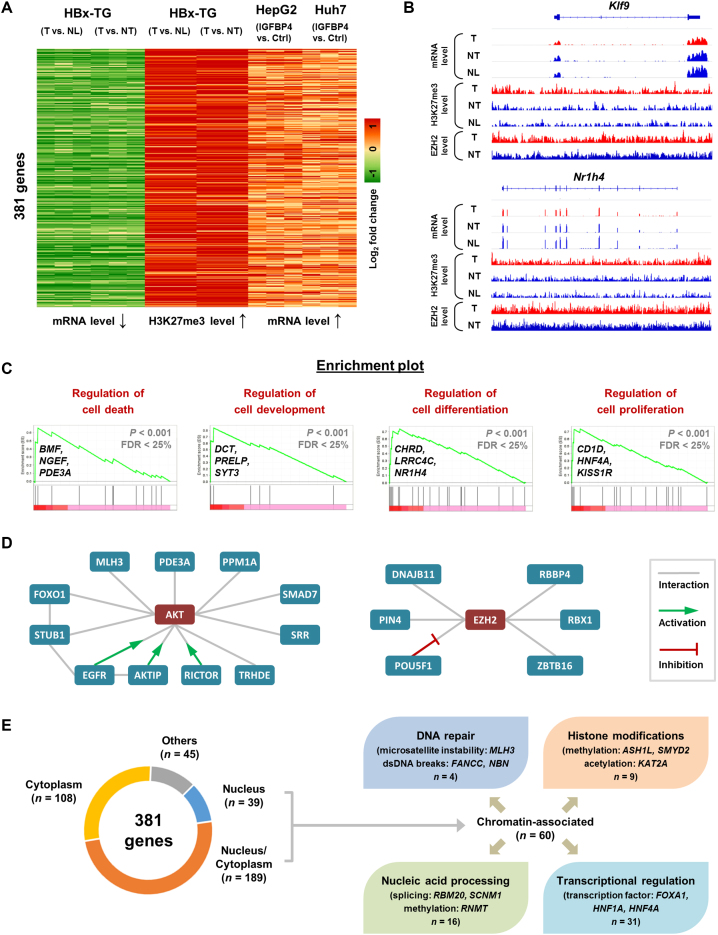
To evaluate the functional significance of the IGFBP4/AKT/EZH2 feedback circuitry, we elucidated the core transcriptional network controlled by this epigenetic circuitry. To this end, we integrated our ChIP-seq and RNA-seq datasets from the HBx-TG mouse (Figure 1) and human HCC cell models (Figure 4), and identified 381 genes that were simultaneously 1) occupied and down-regulated by H3K27me3 in the tumor tissues of HBx-TG mice, and 2) up-regulated in the IGFBP4 re-expressing HepG2 and Huh7 cells (Figure 6A). For examples, the IGFBP4-up-regulated Klf9 and Nr1h4, key transcription factors for normal liver functions (29,30), were markedly down-regulated and bound by EZH2 and H3K27me3 in the HBx-TG HCC model (Figure 6B). We next submitted the 381 target genes to GSEA which revealed significant enrichments in the regulations of cell death (BMF, NGEF, PDE3A), cell development (DCT, PRELP, SYT3), cell differentiation (CHRD, LRRC4C, NR1H4) and cell proliferation (CD1D, HNF4A, KISS1R) (Figure 6C; Supplementary Figure S7A), as well as liver cancer phenotypes and oncogenic signatures (Supplementary Figure S7B and D).
Figure 6.
IGFBP4/AKT/EZH2 signaling drives epigenetic reprogramming in HCC. (A) The RNA-seq and ChIP-seq data of HBx-TG mice (NL, NT and T) and the RNA-seq data of HepG2 and Huh7 cells (with or without IGFBP4 re-expression) were integrated. The 381 genes were selected based on concordant mRNA down-regulation and H3K27me3 enrichment in tumor tissues, and mRNA up-regulation in IGFBP4-abundant cells. (B) Representative diagrams showing the RNA-seq and ChIP-seq signals mapped to two candidate genes in the murine tissues. (C) GSEA analyses of the 381 differentially regulated genes in HepG2 and Huh7 cells (with or without IGFBP4) showed common enrichments in cell growth features, of which four enrichment plots are shown with genes highlighted. (D) The respective interactomes of AKT and EZH2 with candidate proteins encoded by those within 381 target genes. (E) Classification of candidate genes based on the subcellular localizations and chromatin-related functions of their encoded proteins.
In silico protein interaction data further indicate that 98 encoded proteins from the 381 genes form specific interactomes, in which EGFR, STUB1 and POU5F1 act as hubs (interacting with >5 proteins) for functional coordination (Supplementary Figure S8). Intriguingly, these encoded proteins can also interact with the upstream core circuitry. For instance, 11 proteins can bind to AKT, wherein EGFR, AKTIP and RICTOR function as activators (31–33); and six proteins can interact with EZH2, among which POU5F1 acts as negative regulator (34) (Figure 6D). This information supports the notion that additional feedback controls may reinforce the IGFBP4/AKT/EZH2 circuitry. In addition, subcellular classification (Figure 6E) showed that 60/381 gene products have documented roles for genome integrity in the chromatin compartment (Supplementary Table S5), e.g. FANCC for DNA repair, KAT2A for histone modification, RBM20 for nucleic acid processing, and FOXA1 for transcriptional regulation. Overall, our integrative genome-wide analysis implicates that the IGFBP4/AKT/EZH2 epigenetic circuitry suppresses core transcription factor networks that normally regulate liver homeostasis.
EZH2 inhibition suppresses the growth of IGFBP4-silenced HCC tumors
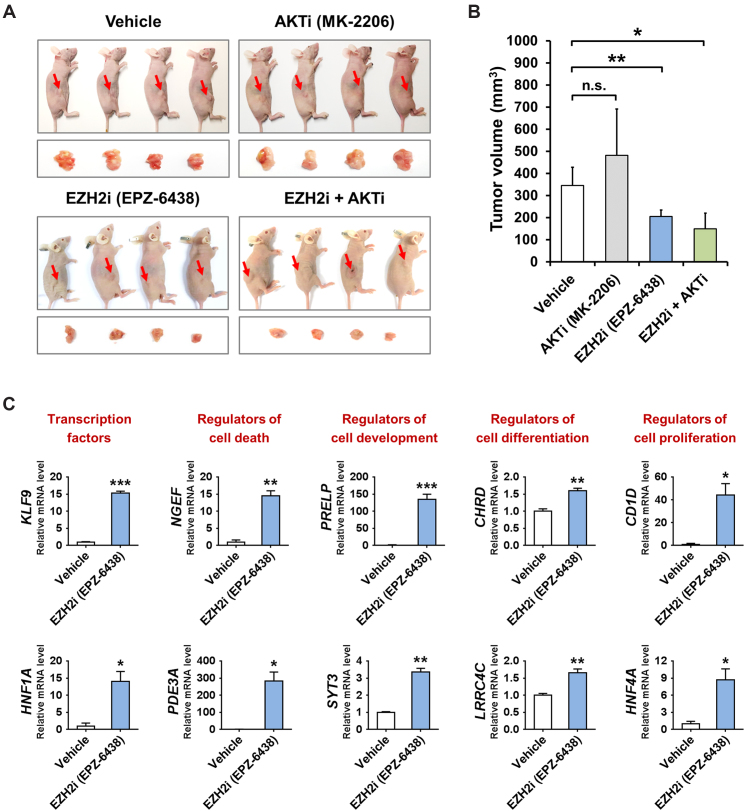
Finally, we investigated the translational relevance of the IGFBP4/AKT/EZH2 feedback circuitry. We examined the in vivo effects of both AKT inhibitor (MK-2206) and EZH2 inhibitor (EPZ-6438) in the Huh7-derived xenografts that exhibited epigenetic silencing of IGFBP4 (Figure 5A). The results showed that MK-2206 exerted no significant effect on xenograft growth (Figure 7A and B), despite the suppression of p-AKT in the dissected tumors (Supplementary Figure S9A-B). In contrast, EPZ-6438, either alone or in combination with MK-2206, was potent in suppressing the tumor growth (Figure 7A and B; ∼40% reduction in tumor volume, P < 0.05). Importantly, EPZ-6438 could significantly reactivate a number of transcription factors and cellular regulators (Figure 7C; P < 0.05) that are aberrantly suppressed in HCCs (Figure 6), demonstrating its potential in rectifying the IGFBP4/AKT/EZH2 epigenetic circuitry for HCC inhibition. These results clearly support that pharmacological targeting of EZH2 represents an effective therapeutic approach for HCC.
Figure 7.
EZH2 inhibitor suppresses HCC growth through epigenetic rectification. (A) The Huh7 cell-derived xenograft tumors were dissected from nude mice treated with or without selective inhibitor of AKT (AKTi) and/or EZH2 (EZH2i), of which volumes were quantified (n = 4) and shown in (B). (C) The expression of transcription factors and cellular regulators in the vehicle-treated and EZH2i-treated tumors was measured by qRT-PCR analysis. *P < 0.05; **P < 0.01; ***P < 0.001.
DISCUSSION
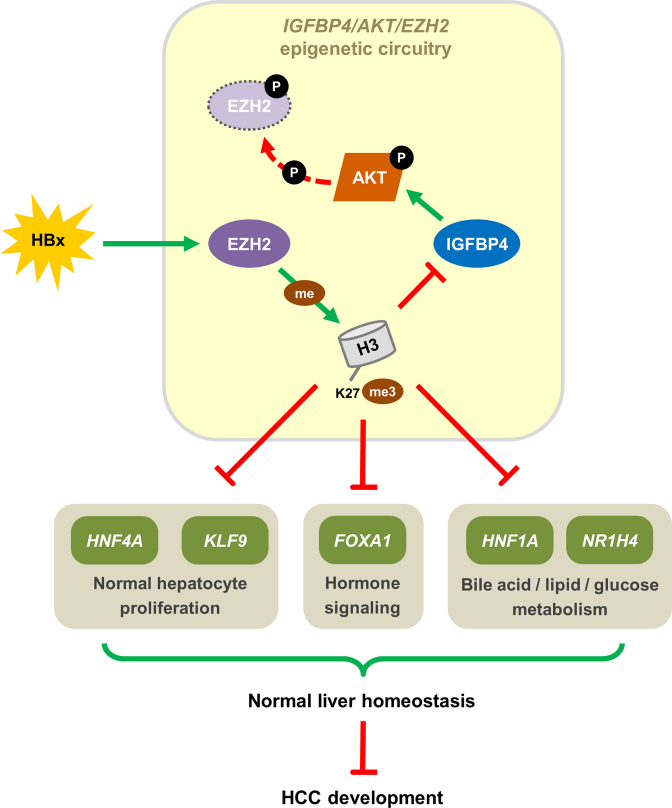
By epigenomic and transcriptomic profiling using genetic mouse model and complementary cell models, we have delineated a novel epigenetic circuitry in HBV-associated hepatocarcinogenesis (Figure 8). Based on our integrated data, IGFBP4 is normally expressed in healthy liver, leading to stimulation of AKT signaling that inactivates EZH2 methyltransferase function, thereby allowing the expression of transcription factors and other proteins that regulate liver homeostasis. However, activation of EZH2/H3K27me3 by HBx, and potentially other HCC etiological factors, silences IGFBP4 and alleviates the AKT brake on EZH2, leading to aberrant epigenetic reprogramming and cancer development. This double-negative feedback loop between AKT and EZH2 represents a major HBx-driven pathogenic mechanism, as demonstrated by the massive changes in the transcriptional and signaling networks following IGFBP4 deregulation. IGFPB4 is not only a potent liver tumor suppressor, but also a prognostic biomarker for HCC. More importantly, targeting EZH2 by specific inhibitor is a promising approach in restoring the IGFBP4-dependent protective signaling to counteract hepatocarcinogenesis.
Figure 8.
The mechanistic pathway of IGFBP4/AKT/EZH2 epigenetic circuitry. Schematic diagram showing the IGFBP4/AKT/EZH2 epigenetic circuitry with molecular and functional consequences.
Similar to other family members, IGFBP4 can play pleiotropic roles in different cancers. In ovarian cancer, IGFBP4 transcript and serum protein levels are significantly elevated across all stages of tumor samples (35). In primary renal cancer, IGFBP4 can promote cell growth by inducing M-CAM and activating β-catenin/TCF signaling together with its downstream effector MMP14 (36). By contrast, IGFBP4 mediates the tumor-suppressive function of SOX9 in colon cancer (37). In the context of bone cancer, rescue of IGFBP4 can suppress the clonogenicity of stromal cells (38). Concordantly, IGFBP4 can promote apoptosis and inhibit tumorigenesis in prostate cancer (39). Paradoxically, IGFBP4 was reported to inhibit angiogenesis and tumorigenesis in glioblastoma (40), but it was also shown to increase cell proliferation, migration and invasion in the same malignancy (41), further highlighting the intricate and context-dependent roles of IGFBP4 in different tissues. Herein we provide evidence that IGFBP4 is a novel and potent tumor suppressor in HCC, effective in causing G1-cell cycle arrest and inducing apoptosis, thereby suppressing cell proliferation, colony formation and cell invasion, and also diminishing xenograft tumor growth in mice. At molecular level, we show for the first time that IGFBP4 is a critical mediator interconnecting the EZH2 and AKT pathways that are important for liver homeostasis, providing mechanistic insights into the strong tumor suppression effects of IGFBP4. These results are consistent with the fact that IGFBP4 is negatively regulated by AR (42) which roots the male-predominance hallmark of HCC (43).
The serine/threonine kinase AKT is well known for its oncogenic effects in many human cancers (44). Upstream factors including PI3K, PDK1 and mTORC2 are responsible for the activation of AKT by binding to and phosphorylating its PH, kinase and C-terminal regulatory domains, respectively. Once activated, AKT can phosphorylate and inactivate a large number of substrate proteins such as caspase-9, FOXO, GSK3β and p27, resulting in elevated cell growth and reduced apoptosis (45). Based on these findings, a number of inhibitors targeting the AKT pathway have been developed as potential cancer drugs (44). Yet the tumorigenic effects of AKT are not universal across all cancer types. Indeed, AKT-dependent inhibition of FOXO and, the concurrently stimulated JNK/c-JUN signaling, markedly decreased leukemic cell growth and leukemia-initiating cell function in acute myeloid leukemia (46). Moreover, a recent study presented strong evidence that knockout of AKT predisposes the liver to cancer formation (47). As an important mediator of the IGFBP4/EZH2/H3K27me3 epigenetic circuitry (Figure 8), AKT may not be suitable for therapeutic targeting as shown by the ineffectiveness of MK-2206 in tumor suppression (Figure 7A–B). In line with this, there was an early termination of a Phase II clinical trial of MK-2206 in treating patients with advanced HCCs (NCT01239355), wherein the median progression-free survival was merely 1.7 (1.6–3.0) months. In another similar trial of MK-2206 (NCT01425879) for patients with severe HCCs/gallbladder carcinomas/bile duct carcinomas, the results were equally disappointing with median progression-free survival of only 1.7 (0.5–5.6) months.
This study addresses an important crosstalk of AKT and EZH2 signaling via chromatin regulation of IGFBP4, and delineates their co-regulated targets in the liver epigenome that possess critical homeostatic roles, thus revealing the mechanistic basis of EZH2-targeted inhibition for HCC therapy. Among the 381 genes directly silenced by EZH2/H3K27me3, at least a subset is known to play critical roles in liver homeostasis. Prime examples include NR1H4 and HNF1A which are central regulators of bile acid/lipid/glucose metabolism (48,49); HNF4A and KLF9 which regulate hepatocyte proliferation and differentiation (29,50); and FOXA1 which controls the hormone signaling and is responsible for the sexual dimorphism of liver cancer (51). Importantly, these transcription factors also act in hierarchical and combinatorial manners at a multitude of gene promoters and enhancers, thereby co-regulate liver-specific gene expression and hence hepatocellular maintenance and housekeeping functions (30). These broad ranges of downstream effects exemplify and underscore the importance of EZH2 in the liver. The upstream role of EZH2 and, the reversibility of H3K27me3-mediated silencing, provide a strong rationale to target this epigenetic regulator for HCC treatment. To date a number of EZH2-targeted inhibitors have been developed and evaluated by in vitro and in vivo assays (52–54). Of note, the EZH2-specific drug EPZ-6438 is currently undergoing a Phase I trial in patients with advanced solid tumors or with relapsed/refractory B-cell lymphoma (55). Given the promising effects of epigenetic drugs in multiple disease models (56), the EZH2-targeted drugs are potentially an effective option to treat human advanced HCCs. Future characterizations of the effects of EZH2-targeted inhibitors on the IGFBP4/AKT/EZH2 signaling and the related functional partners will help determine and optimize their therapeutic values for HCC.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
University Grants Committee through the Collaborative Research Fund [C4017-14G, C4014-14G]; General Research Fund [14102914, 14120816]; Focused Innovations Scheme-Scheme B [1907309] from the Chinese University of Hong Kong (CUHK); National Natural Science Foundation of China [81403272, 81774240]; Shanghai Chenguang Program [15CG48]; Shanghai Rising-Star Program [17QA1403900]; Shanghai training plan of outstanding young medical talents [2017YQ021]; Young Researcher Award, CUHK (to A.C.). Funding for open access charge: Hong Kong.
Conflict of interest statement. None declared.
REFERENCES
- 1. Yap T.A., Omlin A., de Bono J.S.. Development of therapeutic combinations targeting major cancer signaling pathways. J. Clin. Oncol. 2013; 31:1592–1605. [DOI] [PubMed] [Google Scholar]
- 2. Cancer Genome Atlas Research Network Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. 2017; 169:1327–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhang W., He H., Zang M., Wu Q., Zhao H., Lu L.L., Ma P., Zheng H., Wang N., Zhang Y. et al. . Genetic features of Aflatoxin-associated hepatocellular carcinoma. Gastroenterology. 2017; 153:249–262. [DOI] [PubMed] [Google Scholar]
- 4. Calderaro J., Couchy G., Imbeaud S., Amaddeo G., Letouze E., Blanc J.F., Laurent C., Hajji Y., Azoulay D., Bioulac-Sage P. et al. . Histological subtypes of hepatocellular carcinoma are related to gene mutations and molecular tumour classification. J. Hepatol. 2017; 67:727–738. [DOI] [PubMed] [Google Scholar]
- 5. Sciarra A., Di Tommaso L., Nakano M., Destro A., Torzilli G., Donadon M., Maggioni M., Bosari S., Bulfamante G., Matsuda M. et al. . Morphophenotypic changes in human multistep hepatocarcinogenesis with translational implications. J. Hepatol. 2016; 64:87–93. [DOI] [PubMed] [Google Scholar]
- 6. Cai M.Y., Tong Z.T., Zheng F., Liao Y.J., Wang Y., Rao H.L., Chen Y.C., Wu Q.L., Liu Y.H., Guan X.Y. et al. . EZH2 protein: a promising immunomarker for the detection of hepatocellular carcinomas in liver needle biopsies. Gut. 2011; 60:967–976. [DOI] [PubMed] [Google Scholar]
- 7. Tsang D.P., Cheng A.S.. Epigenetic regulation of signaling pathways in cancer: role of the histone methyltransferase EZH2. J. Gastroenterol. Hepatol. 2011; 26:19–27. [DOI] [PubMed] [Google Scholar]
- 8. Zapf J., Waldvogel M., Froesch E.R.. Binding of nonsuppressible insulinlike activity to human serum. Evidence for a carrier protein. Arch. Biochem. Biophys. 1975; 168:638–645. [DOI] [PubMed] [Google Scholar]
- 9. Chelius D., Baldwin M.A., Lu X., Spencer E.M.. Expression, purification and characterization of the structure and disulfide linkages of insulin-like growth factor binding protein-4. J. Endocrinol. 2001; 168:283–296. [DOI] [PubMed] [Google Scholar]
- 10. Zhu W., Shiojima I., Ito Y., Li Z., Ikeda H., Yoshida M., Naito A.T., Nishi J., Ueno H., Umezawa A. et al. . IGFBP-4 is an inhibitor of canonical Wnt signalling required for cardiogenesis. Nature. 2008; 454:345–349. [DOI] [PubMed] [Google Scholar]
- 11. Contois L.W., Nugent D.P., Caron J.M., Cretu A., Tweedie E., Akalu A., Liebes L., Friesel R., Rosen C., Vary C. et al. . Insulin-like growth factor binding protein-4 differentially inhibits growth factor-induced angiogenesis. J. Biol. Chem. 2012; 287:1779–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Durai R., Davies M., Yang W., Yang S.Y., Seifalian A., Goldspink G., Winslet M.. Biology of insulin-like growth factor binding protein-4 and its role in cancer (review). Int. J. Oncol. 2006; 28:1317–1325. [PubMed] [Google Scholar]
- 13. Cheng A.S., Lau S.S., Chen Y., Kondo Y., Li M.S., Feng H., Ching A.K., Cheung K.F., Wong H.K., Tong J.H. et al. . EZH2-mediated concordant repression of Wnt antagonists promotes beta-catenin-dependent hepatocarcinogenesis. Cancer Res. 2011; 71:4028–4039. [DOI] [PubMed] [Google Scholar]
- 14. Feng H., Yu Z., Tian Y., Lee Y.Y., Li M.S., Go M.Y., Cheung Y.S., Lai P.B., Chan A.M., To K.F. et al. . A CCRK-EZH2 epigenetic circuitry drives hepatocarcinogenesis and associates with tumor recurrence and poor survival of patients. J. Hepatol. 2015; 62:1100–1111. [DOI] [PubMed] [Google Scholar]
- 15. Tsang D.P., Wu W.K., Kang W., Lee Y.Y., Wu F., Yu Z., Xiong L., Chan A.W., Tong J.H., Yang W. et al. . Yin Yang 1-mediated epigenetic silencing of tumour-suppressive microRNAs activates nuclear factor-kappaB in hepatocellular carcinoma. J. Pathol. 2016; 238:651–664. [DOI] [PubMed] [Google Scholar]
- 16. Yu Z., Gao Y.Q., Feng H., Lee Y.Y., Li M.S., Tian Y., Go M.Y., Yu D.Y., Cheung Y.S., Lai P.B. et al. . Cell cycle-related kinase mediates viral-host signalling to promote hepatitis B virus-associated hepatocarcinogenesis. Gut. 2014; 63:1793–1804. [DOI] [PubMed] [Google Scholar]
- 17. Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L.. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013; 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L.. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010; 28:511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S. et al. . Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bean D.M., Heimbach J., Ficorella L., Micklem G., Oliver S.G., Favrin G.. esyN: network building, sharing and publishing. PLoS One. 2014; 9:e106035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stark C., Breitkreutz B.J., Reguly T., Boucher L., Breitkreutz A., Tyers M.. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006; 34:D535–D539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Smith R.N., Aleksic J., Butano D., Carr A., Contrino S., Hu F., Lyne M., Lyne R., Kalderimis A., Rutherford K. et al. . InterMine: a flexible data warehouse system for the integration and analysis of heterogeneous biological data. Bioinformatics. 2012; 28:3163–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Binder J.X., Pletscher-Frankild S., Tsafou K., Stolte C., O’Donoghue S.I., Schneider R., Jensen L.J.. COMPARTMENTS: unification and visualization of protein subcellular localization evidence. Database. 2014; 2014:bau012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhao M., Sun J., Zhao Z.. TSGene: a web resource for tumor suppressor genes. Nucleic Acids Res. 2013; 41:D970–D976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McCabe M.T., Ott H.M., Ganji G., Korenchuk S., Thompson C., Van Aller G.S., Liu Y., Graves A.P., Della Pietra A. 3rd, Diaz E. et al. . EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012; 492:108–112. [DOI] [PubMed] [Google Scholar]
- 26. Cha T.L., Zhou B.P., Xia W., Wu Y., Yang C.C., Chen C.T., Ping B., Otte A.P., Hung M.C.. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science. 2005; 310:306–310. [DOI] [PubMed] [Google Scholar]
- 27. Majumder P.K., Febbo P.G., Bikoff R., Berger R., Xue Q., McMahon L.M., Manola J., Brugarolas J., McDonnell T.J., Golub T.R. et al. . mTOR inhibition reverses Akt-dependent prostate intraepithelial neoplasia through regulation of apoptotic and HIF-1-dependent pathways. Nat. Med. 2004; 10:594–601. [DOI] [PubMed] [Google Scholar]
- 28. Sangodkar J., Dhawan N.S., Melville H., Singh V.J., Yuan E., Rana H., Izadmehr S., Farrington C., Mazhar S., Katz S. et al. . Targeting the FOXO1/KLF6 axis regulates EGFR signaling and treatment response. J. Clin. Invest. 2012; 122:2637–2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cvoro A., Devito L., Milton F.A., Noli L., Zhang A., Filippi C., Sakai K., Suh J.H., D H.S., Dhawan A. et al. . A thyroid hormone receptor/KLF9 axis in human hepatocytes and pluripotent stem cells. Stem Cells. 2015; 33:416–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dubois-Chevalier J., Dubois V., Dehondt H., Mazrooei P., Mazuy C., Serandour A.A., Gheeraert C., Guillaume P., Bauge E., Derudas B. et al. . The logic of transcriptional regulator recruitment architecture at cis-regulatory modules controlling liver functions. Genome Res. 2017; 27:985–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Garay C., Judge G., Lucarelli S., Bautista S., Pandey R., Singh T., Antonescu C.N.. Epidermal growth factor-stimulated Akt phosphorylation requires clathrin or ErbB2 but not receptor endocytosis. Mol. Biol. Cell. 2015; 26:3504–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Remy I., Michnick S.W.. Regulation of apoptosis by the Ft1 protein, a new modulator of protein kinase B/Akt. Mol. Cell. Biol. 2004; 24:1493–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sarbassov D.D., Guertin D.A., Ali S.M., Sabatini D.M.. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005; 307:1098–1101. [DOI] [PubMed] [Google Scholar]
- 34. Wu F.R., Zhang Y., Ding B., Lei X.H., Huang J.C., Wang C.H., Liu Y., Wang R., Li W.Y.. H3K27me3 may be associated with Oct4 and Sox2 in mouse preimplantation embryos. Genet. Mol. Res. 2014; 13:10121–10129. [DOI] [PubMed] [Google Scholar]
- 35. Mosig R.A., Lobl M., Senturk E., Shah H., Cohen S., Chudin E., Fruscio R., Marchini S., D’Incalci M., Sachidanandam R. et al. . IGFBP-4 tumor and serum levels are increased across all stages of epithelial ovarian cancer. J. Ovarian Res. 2012; 5:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ueno K., Hirata H., Majid S., Tabatabai Z.L., Hinoda Y., Dahiya R.. IGFBP-4 activates the Wnt/beta-catenin signaling pathway and induces M-CAM expression in human renal cell carcinoma. Int. J. Cancer. 2011; 129:2360–2369. [DOI] [PubMed] [Google Scholar]
- 37. Shi Z., Chiang C.I., Mistretta T.A., Major A., Mori-Akiyama Y.. SOX9 directly regulates IGFBP-4 in the intestinal epithelium. Am. J. Physiol. Gastrointest. Liver Physiol. 2013; 305:G74–G83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fellenberg J., Sahr H., Liu L., Schonsiegel F., Depeweg D., Lehner B., Herr I.. Rescue of silenced UCHL1 and IGFBP4 expression suppresses clonogenicity of giant cell tumor-derived stromal cells. Cancer Lett. 2013; 336:61–67. [DOI] [PubMed] [Google Scholar]
- 39. Damon S.E., Maddison L., Ware J.L., Plymate S.R.. Overexpression of an inhibitory insulin-like growth factor binding protein (IGFBP), IGFBP-4, delays onset of prostate tumor formation. Endocrinology. 1998; 139:3456–3464. [DOI] [PubMed] [Google Scholar]
- 40. Moreno M.J., Ball M., Andrade M.F., McDermid A., Stanimirovic D.B.. Insulin-like growth factor binding protein-4 (IGFBP-4) is a novel anti-angiogenic and anti-tumorigenic mediator secreted by dibutyryl cyclic AMP (dB-cAMP)-differentiated glioblastoma cells. Glia. 2006; 53:845–857. [DOI] [PubMed] [Google Scholar]
- 41. Praveen Kumar V.R., Sehgal P., Thota B., Patil S., Santosh V., Kondaiah P.. Insulin like growth factor binding protein 4 promotes GBM progression and regulates key factors involved in EMT and invasion. J. Neurooncol. 2014; 116:455–464. [DOI] [PubMed] [Google Scholar]
- 42. Gori F., Hofbauer L.C., Conover C.A., Khosla S.. Effects of androgens on the insulin-like growth factor system in an androgen-responsive human osteoblastic cell line. Endocrinology. 1999; 140:5579–5586. [DOI] [PubMed] [Google Scholar]
- 43. Feng H., Cheng A.S., Tsang D.P., Li M.S., Go M.Y., Cheung Y.S., Zhao G.J., Ng S.S., Lin M.C., Yu J. et al. . Cell cycle-related kinase is a direct androgen receptor-regulated gene that drives beta-catenin/T cell factor-dependent hepatocarcinogenesis. J. Clin. Invest. 2011; 121:3159–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cheng J.Q., Lindsley C.W., Cheng G.Z., Yang H., Nicosia S.V.. The Akt/PKB pathway: molecular target for cancer drug discovery. Oncogene. 2005; 24:7482–7492. [DOI] [PubMed] [Google Scholar]
- 45. Faes S., Dormond O.. PI3K and AKT: unfaithful partners in cancer. Int. J. Mol. Sci. 2015; 16:21138–21152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sykes S.M., Lane S.W., Bullinger L., Kalaitzidis D., Yusuf R., Saez B., Ferraro F., Mercier F., Singh H., Brumme K.M. et al. . AKT/FOXO signaling enforces reversible differentiation blockade in myeloid leukemias. Cell. 2011; 146:697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang Q., Yu W.N., Chen X., Peng X.D., Jeon S.M., Birnbaum M.J., Guzman G., Hay N.. Spontaneous hepatocellular carcinoma after the combined deletion of Akt isoforms. Cancer Cell. 2016; 29:523–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ma K., Saha P.K., Chan L., Moore D.D.. Farnesoid X receptor is essential for normal glucose homeostasis. J. Clin. Invest. 2006; 116:1102–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shih D.Q., Bussen M., Sehayek E., Ananthanarayanan M., Shneider B.L., Suchy F.J., Shefer S., Bollileni J.S., Gonzalez F.J., Breslow J.L. et al. . Hepatocyte nuclear factor-1alpha is an essential regulator of bile acid and plasma cholesterol metabolism. Nat. Genet. 2001; 27:375–382. [DOI] [PubMed] [Google Scholar]
- 50. Bonzo J.A., Ferry C.H., Matsubara T., Kim J.H., Gonzalez F.J.. Suppression of hepatocyte proliferation by hepatocyte nuclear factor 4alpha in adult mice. J. Biol. Chem. 2012; 287:7345–7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li Z., Tuteja G., Schug J., Kaestner K.H.. Foxa1 and Foxa2 are essential for sexual dimorphism in liver cancer. Cell. 2012; 148:72–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Knutson S.K., Wigle T.J., Warholic N.M., Sneeringer C.J., Allain C.J., Klaus C.R., Sacks J.D., Raimondi A., Majer C.R., Song J. et al. . A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat. Chem. Biol. 2012; 8:890–896. [DOI] [PubMed] [Google Scholar]
- 53. Konze K.D., Ma A., Li F., Barsyte-Lovejoy D., Parton T., Macnevin C.J., Liu F., Gao C., Huang X.P., Kuznetsova E. et al. . An orally bioavailable chemical probe of the Lysine Methyltransferases EZH2 and EZH1. ACS Chem. Biol. 2013; 8:1324–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Qi W., Chan H., Teng L., Li L., Chuai S., Zhang R., Zeng J., Li M., Fan H., Lin Y. et al. . Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:21360–21365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Knutson S.K., Kawano S., Minoshima Y., Warholic N.M., Huang K.C., Xiao Y., Kadowaki T., Uesugi M., Kuznetsov G., Kumar N. et al. . Selective inhibition of EZH2 by EPZ-6438 leads to potent antitumor activity in EZH2-mutant non-Hodgkin lymphoma. Mol. Cancer Ther. 2014; 13:842–854. [DOI] [PubMed] [Google Scholar]
- 56. Yan W., Herman J.G., Guo M.. Epigenome-based personalized medicine in human cancer. Epigenomics. 2016; 8:119–133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.