Abstract
Neural stem cells (NSCs) are a prominent cell source for understanding neural pathogenesis and for developing therapeutic applications to treat neurodegenerative disease because of their regenerative capacity and multipotency. Recently, a variety of cellular reprogramming technologies have been developed to facilitate in vitro generation of NSCs, called induced NSCs (iNSCs). However, the genetic safety aspects of established virus-based reprogramming methods have been considered, and non-integrating reprogramming methods have been developed. Reprogramming with in vitro transcribed (IVT) mRNA is one of the genetically safe reprogramming methods because exogenous mRNA temporally exists in the cell and is not integrated into the chromosome. Here, we successfully generated expandable iNSCs from human umbilical cord blood-derived mesenchymal stem cells (UCB-MSCs) via transfection with IVT mRNA encoding SOX2 (SOX2 mRNA) with properly optimized conditions. We confirmed that generated human UCB-MSC-derived iNSCs (UM-iNSCs) possess characteristics of NSCs, including multipotency and self-renewal capacity. Additionally, we transfected human dermal fibroblasts (HDFs) with SOX2 mRNA. Compared with human embryonic stem cell-derived NSCs, HDFs transfected with SOX2 mRNA exhibited neural reprogramming with similar morphologies and NSC-enriched mRNA levels, but they showed limited proliferation ability. Our results demonstrated that human UCB-MSCs can be used for direct reprogramming into NSCs through transfection with IVT mRNA encoding a single factor, which provides an integration-free reprogramming tool for future therapeutic application.
Keywords: cellular reprogramming, direct conversion, neural stem cell, synthetic mRNA, umbilical cord blood
Introduction
Although induced pluripotent stem cells (iPSCs) can be differentiated into neural stem/progenitor cells, they can generate teratomas in host tissue because of their heterogeneous population, including undifferentiated cells1. Because direct reprogramming bypasses the pluripotent state, it can prevent the risk of teratoma formation2–4. To date, the most widespread approach to reprogramming somatic cells into neural stem/progenitor cell types is based on overexpression of a combination of pluripotency-associated factors, including octamer-binding transcription factor 4 (OCT4), brain-specific homeobox 4 (BRN4), Kruppel-like factor (KLF4), proto-oncogene c-Myc (c-MYC), and sex determining region Y-box 2 (SOX2), with synergistic effects for driving cell fate conversion. In several reports, it has been demonstrated that combination of SOX2 with other transcription factors can directly reprogram mouse or human somatic cells into neural stem/progenitor cell types5–9. Moreover, previous reports have shown that it is possible to directly reprogram mouse or human somatic cells by transducing cells with the single-factor SOX2 using viral methods8,10. They demonstrated that overexpression of a single-factor, SOX2, via a viral method is sufficient to convert human somatic cells into self-renewing and multipotent neural stem cells (NSCs). However, virus-mediated reprogramming entails a high risk of genetic insertion leading to tumor formation in vivo.
To replace the virus-mediated method, a number of transgene-free reprogramming technologies – including non-integrating adenoviral vectors, DNA plasmid-based vectors, and recombinant proteins incorporating cell-penetrating peptides (CPPs) – for transduction have recently been developed. While adenoviral vectors are non-integrating vectors, they can trigger multiple components of the immune response, such as cytotoxic T lymphocyte activation11,12. Although transfection using DNA plasmid-based vectors is safer than using viral vectors, there are concerns about insertional mutagenesis, and it is difficult to completely eliminate the risk of genomic insertion13. It is also difficult to directly introduce reprogramming factors as proteins and peptides into cells because penetrating the lipid bilayer of the cell membrane to enter the intracellular space while maintaining protein tertiary structure is difficult, and direct introduction of proteins causes instability in the extracellular space14. Importantly, these DNA- or protein-based methods depend on repeated administration of transient vectors and thus have shown very low reprogramming efficiency15–19.
In addition to integration-free gene delivery systems, it has been shown that direct transfection of in vitro transcribed (IVT) mRNA-encoding transcription factors can reprogram human somatic cells into pluripotent stem cells, which could be redifferentiated into myogenic cells20 and a retinal lineage21. Importantly, it is reported that human fibroblasts can be directly reprogrammed into hepatocyte-like cells by IVT mRNAs22. Moreover, IVT mRNA-encoding transcription factors can efficiently overexpress the target gene without risk of insertional mutagenesis. Because exogenously transfected mRNA is translated in the cells and only temporally expressed, it is a genetically safe method compared to the other approaches15,23. Moreover, the mRNA-based method does not leave a genetic footprint or have a risk of genome integration, suggesting the potential safety advance of the mRNA-mediated method15,23,24. Therefore, thus far, mRNA-based methodologies are the most suitable for cell therapy and clinical approaches due to the safety aspects13,15.
However, it has a low reprogramming success rate because the influx of exogenous mRNA exists only temporarily. Therefore, previous reports have suggested that daily transfection of mRNA is needed to retain gene expression for cellular reprogramming13,20,25. Nevertheless, such repetitive transfections of exogenous IVT mRNA can activate innate antiviral defense systems in mammalian cells through type I interferons and NF-κB pathways, which activates the dsRNA-dependent protein kinase (PKR), 2′-5′-oligoadenylate synthetase (OAS) and interferon-induced protein with tetratricopeptide (IFIT). By interacting with pattern-recognition receptors such as RIG-I receptor family, these proteins inhibit translation initiation and global protein expression from both endogenous and exogenous mRNA, and lead to pro-inflammatory cytokine responses25–27. To conduct an effective reprogramming process, optimal conditions are needed to maintain gene expression and to minimize the innate immune response.
Non-integrative direct reprogramming into induced NSCs (iNSCs) and induced neurons is promising for neurodegenerative disease therapy. Unlike terminally differentiated induced neurons, iNSCs are more potent for transplantation therapies and investigation of pathology for neurodegenerative disease because of their self-renewal ability and multipotency9,28–32. In our previous research, we successfully generated iNSCs from human dermal fibroblasts (HDFs) and CD34+ cord blood cells via transduction with SOX2-incorporated retrovirus10. As a further study of our previous reports, we used the transcription factor SOX2 as a master direct neural reprogramming factor via a non-integrative gene delivery system.
In this study, we hypothesized that a SOX2 mRNA-mediated method facilitates overexpression of the SOX2 protein in nuclei, and it is sufficient to reprogram the human umbilical cord blood-derived mesenchymal stem cells (UCB-MSCs) into iNSCs available for various clinical approaches without concerns about uncontrolled genetic integrations. First, we optimized the duration and concentration of mRNA to reduce the risk for degradation of exogenous IVT mRNA, and then we quantitatively and temporally controlled the transfection of exogenous IVT mRNA. This facilitated effective expression of exogenous SOX2 protein in human UCB-MSCs. Finally, we successfully obtained expandable iNSCs from human UCB-MSCs that have neuronal characteristics. This mRNA-based neural reprogramming method using IVT mRNA might be applied as an attractive alternative to viral vector-mediated reprogramming methods for generation of therapeutically usable iNSCs.
Materials and Methods
Isolation and Culture of Human UCB-MSCs
All of the human UCB-MSC experiments were performed with approval of the Boramae Hospital Institutional Review Board (IRB) and the Seoul National University IRB (IRB No. 1608/001-021). Human UCB-MSCs were isolated as previously described33. Briefly, to remove red blood cells in human cord blood samples, HetaSep solution (Stem Cell Technologies, Vancouver, British Columbia, Canada) was incubated with the samples at a ratio of 5:1 at room temperature. The supernatant was collected, and mononuclear cells were harvested using Ficoll (Sigma Aldrich, St. Louis, MO, USA) density-gradient centrifugation at 2500 rpm for 20 min. The cells were washed twice in phosphate-buffered saline (PBS). Cells were seeded in KSB-3 Complete medium (Kangstem Biotech, Seoul, Republic of Korea) containing 10% fetal bovine serum (FBS; Gibco BRL, Grand Island, NY, USA) and 1% penicillin/streptomycin (Gibco BRL) and stabilized for 3 days in 5% CO2.
Cell Culture of HDFs and NSCs
HDFs were maintained in fibroblast growth medium-2 with supplied supplements (LONZA, Basel, BS, Switzerland) containing 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (Gibco BRL). H9-NSCs (NA800-100) were purchased from Gibco BRL and cultured in NSC maintenance medium that is a 1:1 mix of Knockout DMEM/F12 basal medium containing StemPro Neural Supplement, recombinant fibroblast growth factor (FGF) and epidermal growth factor (EGF) proteins (Gibco BRL), 1% glutamine (Gibco BRL), 1% penicillin/streptomycin (Gibco BRL), and ReNcell NSC maintenance medium (Millipore, Billerica, MA, USA) with 20 ng/ml FGF (Peprotech, Rocky Hill, NJ, USA), 20 ng/ml EGF (Peprotech), and 1% penicillin/streptomycin (Gibco BRL). These cells were cultured on a dish coated with poly-L-ornithine (PLO; Sigma Aldrich)/fibronectin (FN; BD Biosciences, San Diego, CA, USA) with the 1:1 mixed medium described above.
Generation of Poly-(A) Tailed DNA Fragments and In Vitro Transcribed mRNA
We purchased pcDNA3.3-SOX2 and pcDNA3.3 enhanced green fluorescent protein (EGFP) from ADDGENE (Cambridge, MA, USA), and mRNA-SOX2 was synthesized as previously described34. In brief, plasmid DNAs were used as the template for poly-(A) tail polymerase chain reaction (PCR). The forward and reverse primers 5′-TTG GAC CCT CGT ACA GAA GCT AAT ACG-3′ and 5′-T (120)-CTT CCT ACT CAG GCT TTA TTC AAA GAC CA-3′ were used. After generation of poly-(A) tailed DNA fragments, tail PCR products were purified using a PureLink PCR purification kit (Invitrogen, Carlsbad, CA, USA). RNA was synthesized using a MEGAscript T7 kit (Ambion, Carlsbad, CA, USA) with purified tail PCR product. We also used a Cap/NTP mixture with an m7G(5′)ppp(5′)G ARCA cap analog (New England Biolabs, Manchester, CT, USA), 3′-methylcytidine triphosphate and pseudo-uridine triphosphate (TriLink Biotechnologies, San Diego, CA, USA) following the protocol for generation of modified mRNA. To generate unmodified mRNA, we made a Cap/NTP mixture using ATP, CTP, UTP, and GTP components. Reactions were incubated for 3–6 h at 37°C, and DNase and Antarctic phosphatase (New England Biolabs) were also added. Synthesized mRNAs were purified using MEGAclear spin columns (Ambion) according to the manufacturer’s protocol and quantitated with a NanoDrop spectrophotometer.
Transfection with mRNA and Isolation and Subculture of iNSC Colonies
Cells were seeded at 50,000–100,000 cells in PLO/FN-coated six-well plates with 2 ml of fibroblast growth medium-2 or endothelial growth medium-2 (LONZA) with supplied supplements, except GA-1000 containing 10% FBS without antibiotics. The cells were incubated at 37°C in a CO2 incubator for 24 h. The following day, the medium was exchanged with the same fresh medium. mRNA transfection was performed using a TransIT®–mRNA Transfection Kit (MirusBio, Madison, WI, USA) or polyethylemine (PEI)-conjugated carriers (provided by Prof. Rohidas Arote). Two types of PEI-conjugated carriers, pullulan-PEI and poly-lactitol-PEI, were kindly provided by Rohidas Arote. When neural stem cell-like colonies were generated, we picked and transferred cells into PLO/FN-coated culture dishes containing NSC maintenance medium with 20 ng/ml FGF, 20 ng/ml EGF, and 1% penicillin/streptomycin. After a few days, the cells were dissociated with Accutase (Gibco BRL) and subcultured on non-coated dishes for suspension culture. To produce a homogeneous population, the cells were passed into non-coated dishes and coated dishes by turn.
Cumulative Population-Doubling Level Analysis
In brief, 100,000 cells of each NSC line were seeded in six-well plates in triplicate. The cells were passaged every 3 days (three or four passages), and the same population of cells were seeded as before and counted using trypan blue to detect live cells. The cumulative population-doubling level (CPDL) was calculated based on the formula CPDL = ln (Nf/Ni) ln2, where Ni is the initial number of cells seeded, Nf is the final number of harvested cells, and ln is the natural log. The population-doubling level was calculated by adding to the previous passages.
Colony Formation Assay
The colony-forming assay procedure has been previously described35. In brief, we seeded cells on non-coated 24-well dishes at 1000 cells/well to form primary neurospheres. After 3 days, the primary neurospheres were dissociated and re-plated at the same density for formation of secondary neurospheres.
Neural Differentiation
For neural differentiation, approximately 3000 cells at passages 13–16 were seeded onto PLO/FN-coated coverslips containing NSC maintenance medium with basic fibroblast growth factor (bFGF) and EGF. After 24 h, the medium was changed to specific medium to induce differentiation into three lineages (neurons, astrocytes, and oligodendrocytes). Neurobasal-A medium (Gibco BRL) with N2 and B27 without other growth factors was used for random differentiation. The differentiation medium was prepared as previously described10. Briefly, the neuronal subtype induction medium includes 200 μM ascorbic acid (AA; Sigma Aldrich), 20 ng/ml BDNF (Peprotech), 20 ng/ml GDNF (Peprotech), 20 ng/ml NT3 (Peprotech), 10 ng/ml IGF-1 (Peprotech), and 2 μM 3′,5′-cyclic adenosine monophosphate (cAMP; Sigma). We added FGF8 (Peprotech) and 1 μM Purmorphamin (Sigma) in the neuronal subtype induction medium for dopaminergic neuronal differentiation. The astrocyte induction medium contained 1% FBS. The oligodendrocyte induction medium included 5 μM retinoic acid (RA; Sigma Aldrich), Purmorphamin (Sigma Aldrich), 20 ng/ml PDGF-BB (Peprotech), and 200 ng/ml bFGF (Peprotech). After 3–5 days 60 ng/ml T3 (Sigma Aldrich) and 2 μM cAMP (Sigma Aldrich) were added for maturation, We refreshed half of the medium every other day and analyzed cells 14–21 days after differentiation6,8,10.
RNA Extraction and Quantitative Real-Time PCR
Total RNA was extracted using a PureLink RNA Mini Kit (Ambion) according to the manufacturer’s instructions. Reverse transcription was performed with Accupower RT-PCR premix (Bioneer, Sung Nam, Republic of Korea) according to the manufacturer’s protocol. cDNA, primers, and diethyl pyrocabonate (DEPC) were combined with a PCR premix (Bioneer) for PCR analysis, and PCR products were loaded on 1.5% agarose gels with gel red (Koma, Seoul, Republic of Korea) and detected using a Bio-Rad Gel Doc XR system (Bio-Rad, Hercules, CA, USA). Quantitative real-time SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA) was used for real-time PCR. The relative expression of all individual genes was calculated using the 2-ΔΔCt method and normalized to the endogenous expression level of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Applied Biosystems, Foster City, CA, USA). For detection of SOX2 transcripts, we used a Total-SOX2 primer binding to the Coding DNA Sequence (CDS) and an Endo-SOX2 primer binding to the SOX2 3′-untranslated region (UTR), which does not exist in the SOX2 mRNA. All primers are listed in Supplementary Table 1.
Immunocytochemistry
Cells were washed three times, fixed for 10 min with 4% paraformaldehyde (PFA) in 0.1 M PBS (pH 7.4) at room temperature, and then incubated with 0.25% Triton X-100 (Sigma) for 10 min to permeabilize cells. Then, the cells were blocked with 5% normal goat serum (NGS; Zymed, San Francisco, CA, USA) in PBS for 1 h at room temperature. The cells were incubated overnight at 4°C with one of the following primary antibodies diluted in 5% NGS according to the manufacturer’s recommendations: rabbit-anti-SOX2 (Abcam 1:200, Cambridge, UK), rabbit-anti-PAX6 (Biolegend 1:200), rabbit-anti-Ki67 (Abcam 1:500), mouse-anti-NESTIN (Abcam 1:200), mouse-anti-NF (Cell Signaling Technology 1:400, Danvers, MA, USA), mouse-anti-GFAP (Abcam 1:200), mouse-anti-TH (R&D Systems 1:200, Minneapolis, MN, USA), rabbit-anti-Doublecortin (Abcam 1:500), mouse-anti-Tuj1 (Abcam 1:500), rabbit-anti-MAP2 (Abcam 1:200), mouse-anti-NeuN (Millipore 1:100), and rabbit-anti-MBP (Millipore 1:200). Next day, the cells were incubated with secondary Alexa 488- or Alexa 555-conjugated antibodies (1:1000; Invitrogen) for 1 h at room temperature diluted in PBS. The nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI, Sigma Aldrich) for 10 min. We performed three washes with PBS in the intervals. The images were captured using a confocal microscope (Nikon, Eclipse TE200, Minato, Tokyo, Japan).
Flow Cytometry
Cells were washed in PBS and harvested in a tube. After centrifugation the cells were resuspended in 300 µl of PBS, and the live cells were immediately analyzed using a MACSQuant Analyzer (Miltenyi Biotec, Bergisch Gladbach, Germany).
Microarray
Each cRNA was labeled with cyanine, and target cRNA was fragmented by adding 10X blocking agent and 25X fragmentation buffer and incubating at 60°C for 30 min. The fragmented cRNA was suspended in 2X hybridization buffer and transferred to an assembled Agilent Mouse (V2) Gene Expression 4X 44 K Microarray. The hybridization images were analyzed with an Agilent DNA microarray scanner (Agilent Technologies, Santa clara, CA, USA) and quantitated using Agilent Feature Extraction software 10.7. Fold-changes in genes in all samples were normalized and selected using GeneSpringGX 7.3.1 (Agilent Technologies).
Statistical Analyses
All experiments were conducted more than three times (n ≥ 3), and all data are presented as the mean ± standard deviation (SD) of independent experiments. Statistical analyses were conducted using analysis of variance (ANOVA) in SPSS22 software (International Business Machines Corporation, Armonk, NY, USA), followed by Duncan’s multiple range tests or Student’s t-test. A value of p < 0.05 was considered a significant result (*p < 0.05; **p < 0.01, ***p < 0.001).
Results
Effective Induction of SOX2 Intra-Nuclear Expression Using IVT mRNA
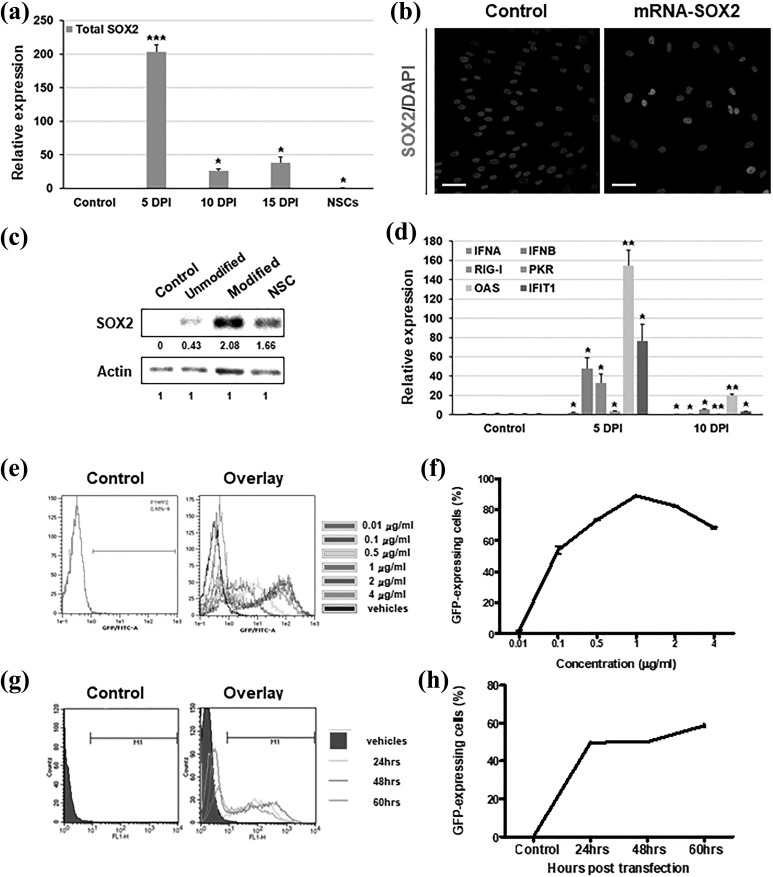
Based on our previous studies, we generated SOX2 mRNA including innate SOX2 CDS, and we then transfected this construct into cells. During transfections of SOX2 mRNA, the gene expression of total SOX2, including both endogenous and exogenous mRNA, was increased approximately 200-fold at 5 days post-induction (DPI). After transfections of SOX2 mRNA, the gene expression level was dramatically decreased and showed 26-fold in comparison to the non-transfected control cells at 10 DPI (Figure 1(a)). Remarkably, the endogenous expression of SOX2 was detected at 15 DPI (Supplementary Figure S1(a)), and the expression level of total SOX2 was also increased again at 15 DPI (Figure 1(a)). Then, we observed that the transfected SOX2 mRNA was not only translated into protein but was also localized in the nucleus (Figure 1(b)). However, the percentage of SOX2-expressing cells after transfection with IVT mRNA was only 25%, and the efficiency needed improvement.
Figure 1.
Optimization of transfection conditions for effective induction of exogenously transfected mRNA. (a) Human UCB-MSCs were transfected with mRNA-encoding SOX2 at 1, 3, and 5 days post-induction (DPI). Quantitative real-time PCR data demonstrated the expression level of total exogenous and endogenous SOX2 at 5, 10, and 15 DPI. (b) Immunocytochemistry data showed a nuclear localization of SOX2 proteins (red) 48 h post-transfection. Nuclei were counterstained with DAPI. Scale bar = 200 μm. (c) Western blot analysis indicated protein expression of SOX2 48 h post-transfection of unmodified and modified mRNA. Relative expression levels were calculated using the Image J system. (d) Relative gene expression levels of innate immune-related genes (INFA, IFNB, RIG-I, PKR, OAS, and IFIT1) were analyzed at 5 and 10 DPI using quantitative real-time PCR. (e,f) A concentration-dependent transfection test was performed with 0.01, 0.1, 0.5, 1, 2, and 4 µg/ml EGFP mRNA. The GFP-positive cells were counted at 48 h after transfection using flow cytometry. (g,h) A time-dependent transfection test was performed at 24, 48, and 60 hours after EGFP mRNA transfection at a dose of 1 µg/ml, and the GFP-positive cells were counted using flow cytometry. For the control, the cells treated with only transfection reagents (control) and the ESC-derived NSCs were used. Error bars represent the standard deviation of reactions repeated more than three times. ***P < 0.001; **P < 0.01; *P < 0.05.
DAPI, 4′,6-diamidino-2-phenylindole; EGFP, enhanced green fluorescent protein; ESC, embryonic stem cell; GFP, green fluorescent protein; NSC, neural stem cell; PCR, polymerase chain reaction; UCB-MSC, umbilical cord blood-derived mesenchymal stem cell.
In prominent studies about modification of IVT mRNA transcripts, modified nucleotides with 5-methylcytidine substituted for cytidine and pseudouridine for uridine showed significant improvement in protein expression, accompanied by reduced activation of the antiviral innate immune system. Therefore, we investigated whether the expression levels of SOX2 and antiviral signaling-related proteins changed depending on the modification of nucleotides. We replaced nucleoside bases with 5-methylcytidine for cytidine and pseudouridine for uridine. After transfection with SOX2 mRNA transcripts containing modified nucleotides, we analyzed the protein expression level using Western blotting and an Image J system. Our results showed that the modified mRNA transcript led to an approximately five-fold increase in expression in comparison to the unmodified transcript (Figure 1(c)). Furthermore, the expression of interferon response genes, including IFNA, IFNB, RIG-I, PKR, and OAS, were increased after five days with two transfections, but diminished further to the expression levels observed in untransfected cells (Figure 1(d)). Interestingly, the expression levels of interferon response genes at two weeks post-transfection were increased as much as those in embryonic stem cell (ESC) derived NSCs (Supplementary Figure S1(b)).
To further optimize the transfection conditions, including concentrations, intervals, and transfection reagents, we transfected cells with IVT mRNA-encoding EGFP (EGFP mRNA) and analyzed the percentage of GFP-expressing live cells using flow cytometry. First, we transfected cells with EGFP mRNA transcripts at various concentrations (0.01, 0.1, 0.5, 1, 2, and 4 μg/ml), and GFP-expressing cells were counted 48 hours post-transfection. In these dose-dependent experiments, the treatments using 1 and 2 μg/ml showed effective fluorescence intensities in the histogram data (Figure 1(e) and Supplementary Figure S1(c)). Of these, the highest number of GFP-expressing cells was observed after treatment with 1 μg/ml, with 88.98% GFP-positive cells (Figure 1(f)). Second, we measured the number of GFP-expressing cells over time. After transfection with GFP mRNA transcripts, measurements at 24, 48, and 60 h post-transfection showed that the percentages of GFP-expressing cells were 49.60%, 50.72%, and 56.06%, respectively (Figure 1(e,f) and Supplementary Figure S1(d)). The highest number of GFP-expressing cells was observed at 60 h, but the overlay data indicated the greatest effective fluorescence intensity at 48 h (Figure 1(e)). It seemed that the translated GFP was divided into daughter cells 48 h post-transfection. Third, we tested the dependence of the transfection efficiency on the transfection technique by using two widely used transfection systems: commercial cationic polymer/lipid formulated reagents and cationic polymer-based gene carriers. Both transfection systems are known to facilitate high transfection efficiency with low cellular toxicity. Among commercial cationic polymer/lipid based reagents, TransIT®–mRNA showed more effective intracellular delivery of exogenous mRNA than Lipofectamine3000 at a concentration of 1 μg/ml EGFP mRNA (data not shown). After transfection with EGFP mRNA transcripts using TransIT®–mRNA, the percentage of GFP-expressing cells was approximately 80%. Then, we also tested two types of PEI-conjugated carriers, which consist of cationic polymers and form complexes with mRNA. Dose-dependent mixtures of pullulan-PEI (PPI) showed the highest GFP expression level at a 1:15 ratio, whereas the GFP-expressing cell percentage was under 20% in all mixtures of poly-lactitol-PEI (PLT) (Supplementary Figure S1(e)). Taken together, we optimized the mRNA transfection protocol with a prominent intracellular delivery system that reduced cytotoxicity and activation of interferon response genes.
Generation of Proliferative NSCs from Human UCB-MSCs Using SOX2-Encoding mRNA
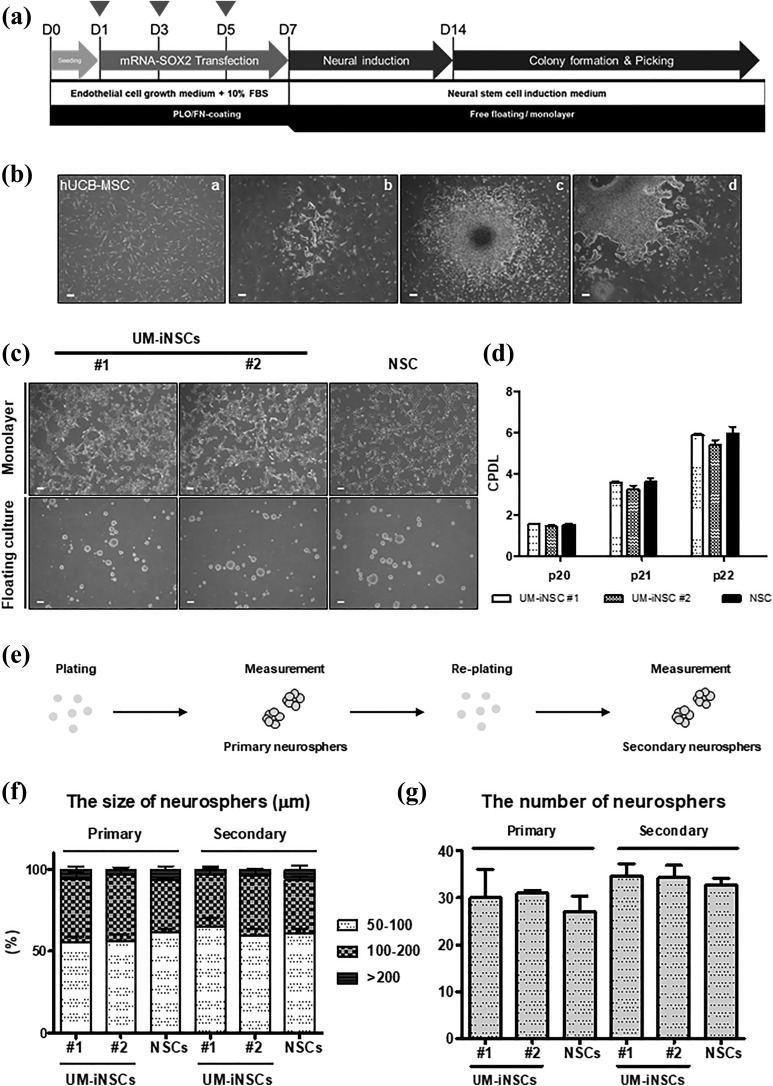
Based on the data above and our previous studies, we developed a direct conversion protocol for NSCs, as illustrated in Figure 2(a). Here, we used human UCB-MSCs as a source for NSC generation (Figure 2(b)). These cells were transfected with SOX2 mRNA three times at intervals of every other day. After 14 days, we observed NSC-like colonies with 0.015% efficiency (Figure 2(b)). Then, we transferred these colonies to coated cell culture dishes that promoted attachment and next passaged these cells to non-coated sphere culture dishes to obtain a homogeneous population of human UCB-MSC-derived iNSCs (UM-iNSCs). On both types of culture dishes, the cells could be maintained as a monolayer or as neurosphere, and their morphologies were similar to those of human ESC-derived NSCs (Figure 2(c) and Supplementary Figure S2(a)).
Figure 2.
Generation of UM-iNSCs from human UCB-MSCs induced by treatment with SOX2 mRNA. (a) A schematic diagram illustrating the procedure for generation of iNSCs from human UCB-MSCs was shown. Arrowheads: SOX2 mRNA transfection. (b) Morphological changes during the reprogramming procedure from UCB-MSCs (i) though NSC-like colonies at 14 DPI (ii) to sub-culture of picked colonies (iii–iv) were observed. (c) The typical morphologies of two UM-iNSC lines (#1 and #2) on an adhesion culture and a floating culture were observed at passage 20. (d) The cumulative CPDL analysis was performed with two UM-iNSC lines to characterize the self-renewal ability. (e) An illustrated schema showed the sphere formation assay procedure. (f,g) the sphere formation assay revealed that there were no significant differences of sizes and numbers between two UM-iNSC lines and ESC-derived NSCs in primary and secondary neurospheres. Error bars represent the standard deviation of reactions repeated more than three times. Scale bar = 200 μm.
CPDL, cumulative population-doubling level; DPI, days post-induction; ESC, embryonic stem cell; NSC, neural stem cell; UCB-MSCs, umbilical cord blood-derived mesenchymal stem cell; UM-iNSC, UCB-MSC-derived induced neural stem cell.
To test the ability of these cells to self-renew and proliferate, which are two of the key characteristics of NSCs, we performed a CPDL experiment in cultures of both UM-iNSCs and ESC-derived NSCs. As expected, there were no significant differences between UM-iNSCs and ESC-derived NSCs at passage numbers 20 to 22 (Figure 2(d)). Moreover, UM-iNSC lines were expandable for more than 50 passages without changes in morphology or growth rate (data not shown). Next, we evaluated secondary neurosphere formation by comparing the size and number of secondary neurospheres between UM-iNSCs and ESC-derived NSCs. To measure these, cells formed primary neurospheres through transfer from coated to non-coated culture dishes, and they were again dissociated and re-plated on non-coated culture dishes to form secondary neurospheres (Figure 2(e)). As a result, the size and number of neurospheres were not significantly different between UM-iNSC lines and ESC-derived NSCs (Figure 2(f,g)). These results demonstrated that overexpression of SOX2 via IVT mRNA transfection is sufficient to reprogram human UCB-MSCs directly into iNSCs. The successfully generated iNSCs from human UCB-MSCs showed stable expansion and neurosphere formation abilities similar to those of human ESC-derived NSCs.
Immunocytochemical Characterizations and Genome-Wide Transcriptional Profiling of UM-iNSCs
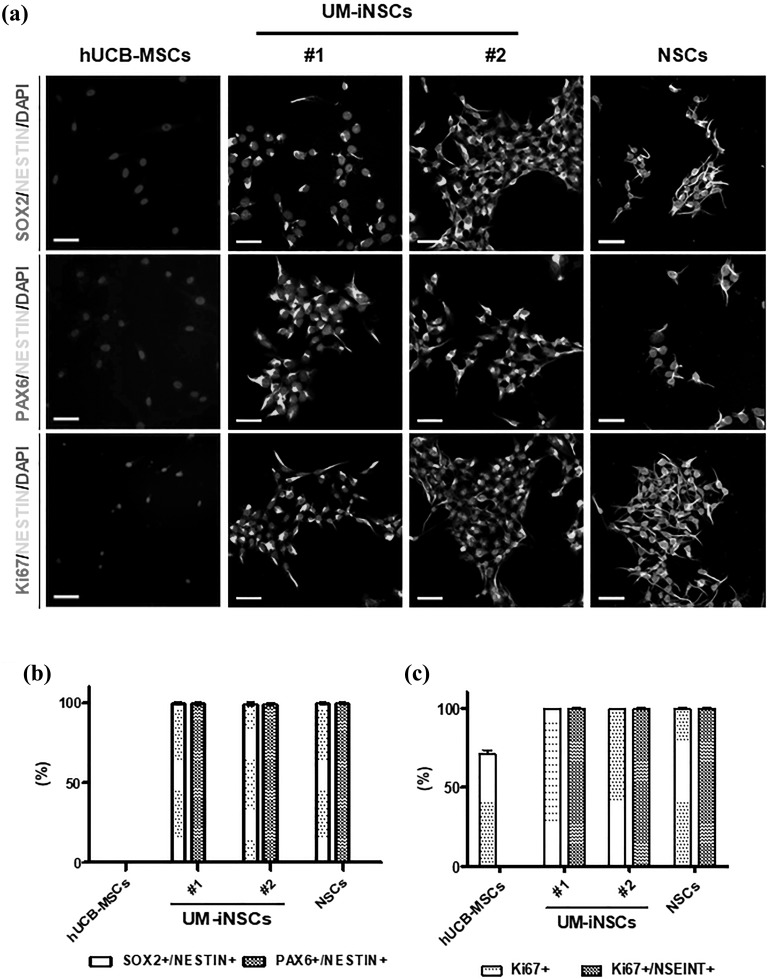
To identify the NSC properties of UM-iNSCs, we investigated the level of protein marker expression in human UCB-MSCs, six cell lines of UM-iNSCs, and ESC-derived NSCs using immunocytochemistry. First, cells were stained against NSC-specific markers, including SOX2, PAX6, and NESTIN, and showed close to 100% SOX2, PAX6, and NESTIN expression in UM-iNSCs and ESC-derived NSCs, whereas their expression was not detected in UCB-MSCs. UM-iNSCs appear to consist of a homogeneous population in which cells express NSC-specific markers (Figure 3(a,b) and Supplementary Figure S2(b)). Second, cells were stained for the proliferation marker Ki-67 to further characterize the self-renewal property of UM-iNSCs. Similar to the NSC-specific markers, UM-iNSCs and ESC-derived NSCs revealed nearly 100% SOX2, PAX6, and NESTIN expression. In particular, the NESTIN-positive cells co-expressed Ki-67 protein in both UM-iNSCs and ESC-derived NSCs (Figure 3(a) and Supplementary Figure S2(B)B). In UCB-MSCs, which are well-known to show a high level of proliferation, Ki-67 expression was detected in 71.03 ± 4.18% of cells. However, the Ki-67/NESTIN-double positive cells were not detected in UCB-MSCs, unless they were directly reprogrammed into UM-iNSCs (Figure 3(c)).
Figure 3.
Characterization of the UM-iNSCs by immunocytochemistry. (a) The UM-iNSCs were stained using antibodies for the NSC-enriched markers (SOX2, PAX6, NESTIN) and a cellular proliferation marker (Ki67). Scale bar = 50 μm. (b,c) Immunocytochemistry data were analyzed using the Image J system and showed percentages of SOX2/PAX6 and NESTIN-double-positive cells, Ki67-positive cells, and Ki67 and NESTIN-double-positive cells in UCB-MSCs, UM-iNSCs, and ESC-derived NSCs, respectively.
ESC, embryonic stem cell; NSC, neural stem cell; UCB-MSC, UCB-MSCs, umbilical cord blood-derived mesenchymal stem cell; UM-iNSC, UCB-MSC-derived induced neural stem cell.
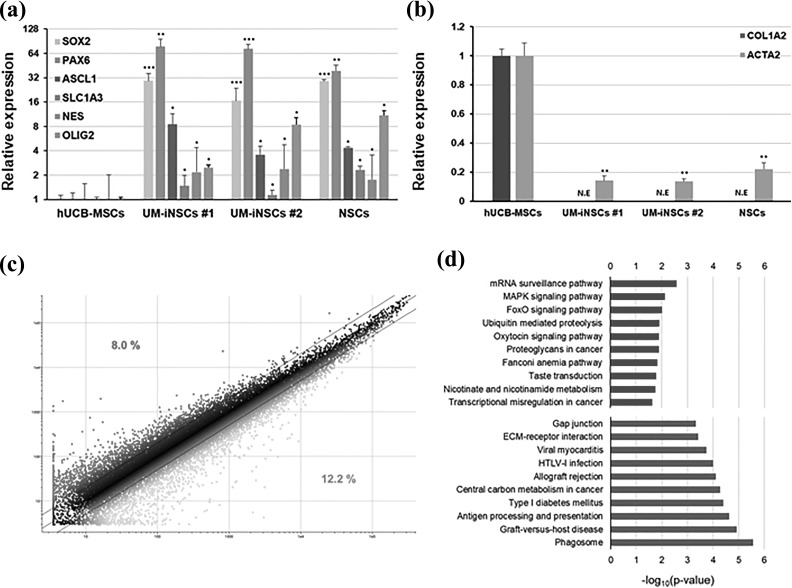
To identify the molecular properties of UM-iNSCs, the gene expression level of NSC-specific genes (SOX2, PAX6, ASCL1, SLC1A3, NES, and OLIG2) and fibroblast-enriched genes (COL1A2 and ACTA2) were compared between UCB-MSCs and UM-iNSCs by using quantitative real-time PCR. After neural reprogramming, compared to human UCB-MSCs, UM-iNSCs showed significant increases in the expression level of each of the NSC-specific genes (Figure 4(a)). Importantly, the mesenchymal cell/fibroblast-enriched genes were significantly decreased in UM-iNSCs (Figure 4(b)). Remarkably, these gene expression patterns of UM-iNSCs were similar to those of ESC-derived NSCs, strongly indicating that gene expression levels were reprogrammed as much as in ESC-derived NSCs. To further identify changes in gene expression patterns after the neural reprogramming, we performed global gene expression profiling between UM-iNSCs and ESC-derived NSCs using a microarray analysis with 34,127 probes in total. Of those without the flag “Absent,” 27,250 probes indicated changes in gene expression in UM-iNSCs compared to ESC-derived NSCs, as illustrated in a scatter plot (Figure 4(c)). Here, 8.0% of the total were identified as being expressed at a more than two-fold higher level, and 12.2% of the total were identified as being expressed at a more than two-fold lower level. These differentially expressed genes were categorized using gene ontology (GO) function enrichment analysis (Figure 4(d)). The relative overexpressed genes in UM-iNSCs were related to mRNA surveillance pathway, MAPK signaling pathway, and FoxO signaling pathway in order of enrichment, whereas the relative downregulated genes were related to phagosome, graft-versus-host disease, and antigen processing and presentation. It seemed that mRNA- and differentiation-related genes were relatively overexpressed and the UCB-MSC-related genes were relatively downregulated in UM-iNSCs compared to ESC-derived NSCs. These data demonstrated that the UCB-MSCs were entirely converted to an NSC fate during the reprogramming process induced by transfection with SOX2 mRNA.
Figure 4.
Genome-wide transcriptional profiling of UM-iNSCs. (a,b) Relative gene expression levels of NSC-specific genes (endogenous SOX2, PAX6, ASCL1, SLC1A3, NES, OLIG2) and mesenchymal cell- or fibroblast-enriched genes (COL1A2, ACTA2) in UM-iNSCs and ESC-derived NSCs were compared to human UCB-MSCs using quantitative real-time PCR. (c) A pair-wise scatter plot indicated differences of genome-wide transcriptional gene expression in UM-iNSCs and ESC-derived NSCs profiled by microarray analysis. Two-fold change difference boundaries are displayed as black lines. (d) GO enrichment analysis in biological processes are shown. Selected GO categories of two-fold increased (red) and two-fold decreased (blue) genes in UM-iNSCs compared to ESC-derived NSCs are listed. Error bars represent the standard deviation of reactions repeated more than three times. ***P < 0.001; **P < 0.01; *P < 0.05.
ESC, embryonic stem cell; GO, gene ontology; NE, no expression; NSC, neural stem cell; PCR, polymerase chain reaction; UCB-MSC, umbilical cord blood-derived mesenchymal stem cell; UM-iNSC, UCB-MSC-derived induced neural stem cell.
Differentiation of UM-iNSCs into Three Major Neural Lineages: Neurons, Astrocytes, and Oligodendrocytes
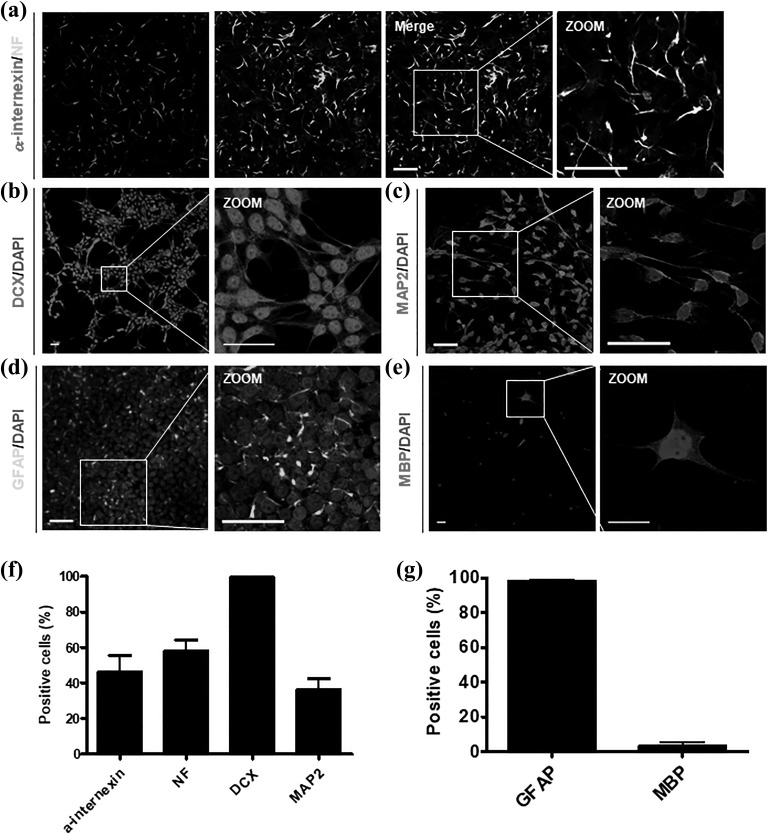
To verify the multipotency of UM-iNSCs, we differentiated the cells into neurons, astrocytes, and oligodendrocytes with specific proper conditions. After 7 days of neuronal differentiation, a-internexin and neuro-filament (NF), which are known as neuronal intermediate filament proteins, and doublecortin (DCX), which is known as a microtubule-associated neuronal migrating protein marker, were positively stained (Figure 5(a,b)). To further investigate whether it is possible to differentiate UM-iNSCs into mature neuronal subtypes, we stained the cells with neuronal maturation markers – neuronal nuclei (NeuN), microtubule-associated protein 2 (MAP2) and choline acetyltransferase (ChAT) – after 14 days of neuronal differentiation. The differentiated cells were positively stained at cell bodies and the nucleus (Figure 5(c) and Supplementary Figure S3(a,b)). Further transcription factors, 1 μM purmorphamine and 100 ng/ml FGF8, facilitate the cells to give rise to dopaminergic neurons that express tyrosine hydroxylase (TH) (Supplementary Figure S3(c)). We counted cells stained by the typical neuronal immature and mature markers α-internexin, NF, DCX and MAP2, and their percentages were 46.21 ± 16.07%, 58.2 ± 10.2%, 99.56 ± 0.51%, and 36.17 ± 10.51%, respectively (Figure 5(f)). Using well-established differentiation protocols, we successfully differentiated cells into glial fibrillary acidic protein (GFAP)-positive astrocytes and oligodendrocyte transcription factor (OLIG2)- and myelin basic protein (MBP)-positive oligodendrocytes (Figure 5(d,e) and Supplementary Figure S3(d)). By counting, GFAP-positive cells were 98.5 ± 0.69% and MBP-positive cells were 3.54 ± 2.32% (Figure 5(g)). Altogether, these data showed that UM-iNSCs can be differentiated into neurons, astrocytes, and oligodendrocytes, demonstrating their multipotency.
Figure 5.
Differentiation capacity of UM-iNSCs into neurons, astrocytes, and oligodendrocytes. Immunocytochemistry results revealed neurons (stained against α-internexin, NF, DCX, MAP2; (a–c)), astrocytes (stained against GFAP; (d)), and oligodendrocytes (stained against MBP; (e)). (f,g) Percentages of NF-, DCX-, and MAP2-positive neuronal cells and GFAP- and MBP-positive neural cells were measured using the Image J system. Error bars represent standard deviation of triplicate reactions. Scale bar = 50 µm.
DCX, doublecortin; GFAP, glial fibrillary acidic protein; MAP2, microtubule-associated protein 2; MBP, myelin basic protein; NF, neuro-filament; UM-iNSC, umbilical cord blood-derived mesenchymal stem cell -derived induced neural stem cell.
Challenges to Reprogramming of HDFs using SOX2-Encoding mRNA Transfections
Using the optimized transfection protocol for IVT SOX2-encoding mRNA and the reprogramming protocol of iNSCs, we investigated direct reprogramming of terminally differentiated human somatic cells – HDFs. Similar to UM-iNSCs, we observed neural stem cell-like colonies at day 7 after mRNA transfections and floating spheres at day 14 (Supplementary Figure S4(a)). Based on gene expression analysis, NSC-specific genes (SOX2, PAX6, ASCL1, SLC1A3, NESTIN, and OLIG2) were significantly increased in HDFs transfected with SOX2-mRNA, and fibroblast-enriched genes (COL1A2 and ACTA2) were significantly decreased, as much as in ESC-derived NSCs (Supplementary Figure S4(b,c)). Although morphology and gene expression patterns were dramatically changed and were similar to those of ESC-derived NSCs, the SOX2-mRNA-transfected HDFs were not expandable and not sufficient for long-term culture. It seems that HDFs were not completely reprogrammed by the neural reprogramming method for human UCB-MSCs using IVT SOX2-mRNA constructs.
Discussion
Previous studies suggested that SOX2 is the master regulatory gene for the preservation of properties of NSCs, including proliferation, self-renewal and neurogenesis, and therefore, SOX2 could play a crucial role in direct reprogramming of somatic cells into neural lineages11,16,36–42. Furthermore, several studies have shown that neural reprogramming using single-factor SOX2 is possible using the viral method8,10. In the meantime, cellular reprogramming techniques are advancing, and mRNA-based technologies for reprogramming are also actively being studied because mRNA-based gene regulation is not concerned with chromosomal-integration. Here, we have explored an optimal concentration and an appropriate interval for transfections of IVT SOX2 mRNA to human UCB-MSCs, and successfully converted into iNSCs. This study is notable for using IVT mRNA, which is an extremely valuable tool for therapeutic applications and for various clinical approaches. In particular, it is, as far as we know, the first approach for direct neural reprogramming using IVT mRNA-encoding SOX2 as a single reprogramming factor.
In this study, we suggested the best conditions for the highest transfection and translation efficiency by screening of the optimal concentration and interval of transfected IVT mRNA. We optimized the concentration that led to maximum GFP-expressing cells without cell death and the most effective interval which reduced the immune response and sustained SOX2 protein expression. However, the low transfection and translation efficiencies of mRNA still remain a challenge. Currently, IVT mRNAs are complexed with structural elements, such as nanoparticles, polymers, or cationic lipids, to further improve intracellular stability and translational efficiency23,43,44.
Not only limitations of transfection and translation efficiency but also stochastic efficiency might have led to the failure of reprogramming45. Recently, various methods have been attempted to increase stochastic efficiency during the reprogramming process. More recently, chemical-mediated methods have been reported that make the cells lose original cellular features and gain multipotent stem cell-specific features by acting as epigenetic modifiers, cell death-related pathway regulators, and metabolic modifiers. Chemicals that play various roles in a cellular pathway have been suggested as options to increase the stochastic efficiency. For example, it is known that a transforming growth factor-beta (TGF-β) inhibitor, including SB431542 and A-83-01, enhances the neural reprogramming efficiency more than seven-fold46. In addition, histone methyltransferase (HMT), including BIX-01294, increases levels of OCT4 and KLF4 and results in efficient reprogramming47. Such chemicals that might enhance the cellular reprogramming efficiency have been investigated in other studies.
As a cell source of direct neural reprogramming, we used UCB-MSCs. In general, MSCs can be derived from umbilical cord blood (UCB), bone marrow (BM), and adipose tissue (AD), and reveal multipotent differentiations into chondrocytes, adipocytes, and osteocytes36. Especially, it is known that MSCs from human UCB show high proliferation and differentiation potency compared to other MSC sources, and there is no ethical problem because collecting MSCs from UCB is a noninvasive method. Moreover, whenever the patients need them, we can clinically use the cells because they can be cryopreserved in a cell bank. Many researchers suggest UCB-MSCs are valuable and need to be studied for their clinical utility and regenerative medicine48.
However, while SOX2 mRNA-transfected HDFs exhibited morphological changes and sustained notable NSC-specific gene expression at the transcriptional level, for direct reprogramming of HDFs it was shown that transfection with the SOX2 mRNA constructs was limited in fully converting HDFs into expandable iNSCs. SOX2 overexpression influences cell proliferation by regulating oncogenic pathways, including Wnt/β-catenin PI3K/mTOR, JAK/STAT3, and EGFR signaling49. However, transfection of HDFs with exogenous SOX2 mRNA was not able to sustain the proliferative state. An introduction of appropriate genes is compulsory to kick-start the reprogramming process, and they have to force the cells to overcome the reprogramming barrier50. In HDFs, forced expression of SOX2 using IVT mRNA is likely insufficient to explosively accelerate the molecular conversion needed for NSC refinement.
Interestingly, we verified that UCB-MSCs have slight expression of NSC-related genes in the RNA. It is known that human UCB-MSCs are not terminally differentiated36. Moreover, some researchers reported that very slight expression of markers of pluripotency in the UCB-MSCs is higher than in BM-derived MSCs but lower than in ESCs36,51. Although we have not tried the direct reprogramming of other cell sources than UCB-MSCs and HDFs, we assume that UCB-MSCs have lower barriers in the process of cellular reprogramming than the others. To clarify these limitations of reprogramming, such as cell type variations, whole-genome expression analyses of initial and intermediate states in the reprogramming processes are needed for understanding reprogramming efficiency.
Most neurodegenerative diseases caused by neuronal dysfunction involve loss of the neuronal population. Due to a lack of molecular studies and therapeutic treatments for diseases, cell therapy and disease modeling by direct reprogramming of disease-specific cells are actively being studied to develop an efficacious alternative. Reprogramming of human UCB-MSCs into iNSCs using a genetically safe mRNA delivery system is a worthwhile technique. Although differentiated neurons from iNSCs have not been functionally proved in the present study, iNSCs were differentiated into mature neurons. As long as it is verified that differentiated neurons are functional, it has remarkable potential for use as a clinical approach, especially for patients with neurodegenerative diseases. Thus, it will be valuable to study mRNA-based methods and application of synthetic mRNA for cellular reprogramming, and continuous study should be done to improve efficiency.
Supplemental Material
Supplemental Material, Revision_final_supple_minor_revision for Single-Factor SOX2 Mediates Direct Neural Reprogramming of Human Mesenchymal Stem Cells via Transfection of In Vitro Transcribed mRNA by Bo-Eun Kim, Soon Won Choi, Ji-Hee Shin, Jae-Jun Kim, Insung Kang, Byung-Chul Lee, Jin Young Lee, Myoung Geun Kook, and Kyung-Sun Kang in Cell Transplantation
Footnotes
Authors Contribution: Bo-Eun Kim, Soon Won Choi, and Ji-Hee Shin contributed equally to this work.
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was carried out with the support of the Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ01100201), Rural Development Administration, Republic of Korea, and partially supported by the Research Institute for Veterinary Science, Seoul National University (SNU, Republic of Korea).
Ethical Approval: All of the human UCB-MSC experiments were performed with approval of the Boramae Hospital Institutional Review Board (IRB) and the Seoul National University IRB (IRB No. 1608/001-021).
Statement of Human and Animal Rights: This article does not contain any studies with human or animal subjects.
Statement of Informed Consent: There are no human subjects in this article and informed consent is not applicable.
Supplemental Material: Supplementary material for this article is available online.
References
- 1. Wolber W, Ahmad R, Choi SW, Eckardt S, McLaughlin KJ, Schmitt J, Geis C, Heckmann M, Siren AL, Muller AM. Phenotype and stability of neural differentiation of androgenetic murine es cell-derived neural progenitor cells. Cell Med. 2013;5:29–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fong CY, Gauthaman K, Bongso A. Teratomas from pluripotent stem cells: A clinical hurdle. J Cell Biochem. 2010;111:769–781. [DOI] [PubMed] [Google Scholar]
- 3. Hou S, Lu P. Direct reprogramming of somatic cells into neural stem cells or neurons for neurological disorders. Neural Regen Res. 2016;11:28–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Miura K, Okada Y, Aoi T, Okada A, Takahashi K, Okita K, Nakagawa M, Koyanagi M, Tanabe K, Ohnuki M, Ogawa D, Ikeda E, Okano H, Yamanaka S. Variation in the safety of induced pluripotent stem cell lines. Nat Biotechnol. 2009;27:743–745. [DOI] [PubMed] [Google Scholar]
- 5. Giorgetti A, Marchetto MC, Li M, Yu D, Fazzina R, Mu Y, Adamo A, Paramonov I, Cardoso JC, Monasterio MB, Bardy C, Cassiani-Ingoni R, Liu GH, Gage FH, Izpisua Belmonte JC. Cord blood-derived neuronal cells by ectopic expression of Sox2 and c-Myc. Proc Natl Acad Sci USA. 2012;109:12556–12561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kim J, Efe JA, Zhu S, Talantova M, Yuan X, Wang S, Lipton SA, Zhang K, Ding S. Direct reprogramming of mouse fibroblasts to neural progenitors. Proc Natl Acad Sci USA. 2011;108:7838–7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lujan E, Chanda S, Ahlenius H, Sudhof TC, Wernig M. Direct conversion of mouse fibroblasts to self-renewing, tripotent neural precursor cells. Proc Natl Acad Sci USA. 2012;109:2527–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ring KL, Tong LM, Balestra ME, Javier R, Andrews-Zwilling Y, Li G, Walker D, Zhang WR, Kreitzer AC, Huang Y. Direct reprogramming of mouse and human fibroblasts into multipotent neural stem cells with a single factor. Cell Stem Cell. 2012;11:100–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Thier M, Worsdorfer P, Lakes YB, Gorris R, Herms S, Opitz T, Seiferling D, Quandel T, Hoffmann P, Nothen MM, Brüstle O, Edenhofer F. Direct conversion of fibroblasts into stably expandable neural stem cells. Cell Stem Cell. 2012;10:473–479. [DOI] [PubMed] [Google Scholar]
- 10. Yu KR, Shin JH, Kim JJ, Koog MG, Lee JY, Choi SW, Kim HS, Seo Y, Lee S, Shin TH, Jee MK, Kim DW, Jung SJ, Shin S, Han DW, Kang KS. rapid and efficient direct conversion of human adult somatic cells into neural stem cells by HMGA2/let-7b. Cell Rep. 2015;10:441–452. [DOI] [PubMed] [Google Scholar]
- 11. Thomas CE, Schiedner G, Kochanek S, Castro MG, Lowenstein PR. Peripheral infection with adenovirus causes unexpected long-term brain inflammation in animals injected intracranially with first-generation, but not with high-capacity, adenovirus vectors: toward realistic long-term neurological gene therapy for chronic diseases. Proc Natl Acad Sci USA. 2000;97:7482–7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Thomas CE, Schiedner G, Kochanek S, Castro MG, Lowenstein PR. Preexisting antiadenoviral immunity is not a barrier to efficient and stable transduction of the brain, mediated by novel high-capacity adenovirus vectors. Hum Gene Ther. 2001;12:839–846. [DOI] [PubMed] [Google Scholar]
- 13. Schlaeger TM, Daheron L, Brickler TR, Entwisle S, Chan K, Cianci A, DeVine A, Ettenger A, Fitzgerald K, Godfrey M, Gupta D, McPherson J, Malwadkar P, Gupta M, Bell B, Doi A, Jung N, Li X, Lynes MS, Brookes E, Cherry AB, Demirbas D, et al. A comparison of non-integrating reprogramming methods. Nat Biotechnol. 2015;33:58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Matsui A, Uchida S, Ishii T, Itaka K, Kataoka K. Messenger RNA-based therapeutics for the treatment of apoptosis-associated diseases. Sci Rep. 2015;5:15810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bernal JA. RNA-based tools for nuclear reprogramming and lineage-conversion: towards clinical applications. J Cardiovasc Transl Res. 2013;6:956–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Favaro R, Valotta M, Ferri AL, Latorre E, Mariani J, Giachino C, Lancini C, Tosetti V, Ottolenghi S, Taylor V, Nicolis SK. Hippocampal development and neural stem cell maintenance require Sox2-dependent regulation of Shh. Nat Neurosci. 2009;12:1248–1256. [DOI] [PubMed] [Google Scholar]
- 17. Jia F, Wilson KD, Sun N, Gupta DM, Huang M, Li Z, Panetta NJ, Chen ZY, Robbins RC, Kay MA, Longaker MT, Wu JC. A nonviral minicircle vector for deriving human iPS cells. Nat Methods. 2010;7:197–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, Thomson JA. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhou H, Wu S, Joo JY, Zhu S, Han DW, Lin T, Trauger S, Bien G, Yao S, Zhu Y, Siuzdak G, Schöler HR, Duan L, Ding S. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4:381–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, Daley GQ, Brack AS, Collins JJ, Cowan C, Schlaeger TM, Rossi DJ. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sridhar A, Ohlemacher SK, Langer KB, Meyer JS. Robust differentiation of mRNA-reprogrammed human induced pluripotent stem cells toward a retinal lineage. Stem Cells Transl Med. 2016;5:417–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Simeonov KP, Uppal H. Direct reprogramming of human fibroblasts to hepatocyte-like cells by synthetic modified mRNAs. PLoS One. 2014;9:e100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guan S, Rosenecker J. Nanotechnologies in delivery of mRNA therapeutics using nonviral vector-based delivery systems. Gene Ther. 2017;24:133–143. [DOI] [PubMed] [Google Scholar]
- 24. Chien KR, Zangi L, Lui KO. Synthetic chemically modified mRNA (modRNA): toward a new technology platform for cardiovascular biology and medicine. Cold Spring Harb Perspect Med. 2014;5:a014035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Steinle H, Behring A, Schlensak C, Wendel HP, Avci-Adali M. Concise review: application of in vitro transcribed messenger RNA for cellular engineering and reprogramming: progress and challenges. Stem Cells. 2017;35:68–79. [DOI] [PubMed] [Google Scholar]
- 26. Loomis KH, Kirschman JL, Bhosle S, Bellamkonda RV, Santangelo PJ. Strategies for modulating innate immune activation and protein production of in vitro transcribed mRNAs. J Mater Chem. B 2016;4:1619–1632. [DOI] [PubMed] [Google Scholar]
- 27. Kumar P, Sweeney TR, Skabkin MA, Skabkina OV, Hellen CU, Pestova TV. Inhibition of translation by IFIT family members is determined by their ability to interact selectively with the 5′-terminal regions of cap0-, cap1- and 5′ppp- mRNAs. Nucleic Acids Res. 2014;42:3228–3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Colasante G, Lignani G, Rubio A, Medrihan L, Yekhlef L, Sessa A, Massimino L, Giannelli SG, Sacchetti S, Caiazzo M, Leo D, Alexopoulou D, Dell’Anno MT, Ciabatti E, Orlando M, Studer M, Dahl A, Gainetdinov RR, Taverna S, Benfenati F, Broccoli V. Rapid conversion of fibroblasts into functional forebrain GABAergic interneurons by direct genetic reprogramming. Cell Stem Cell. 2015;17:719–734. [DOI] [PubMed] [Google Scholar]
- 29. Han DW, Tapia N, Hermann A, Hemmer K, Hoing S, Arauzo-Bravo MJ, Zaehres H, Wu G, Frank S, Moritz S, Greber B, Yang JH, Lee HT, Schwamborn JC, Storch A, Schöler HR. Direct reprogramming of fibroblasts into neural stem cells by defined factors. Cell Stem Cell. 2012;10:465–472. [DOI] [PubMed] [Google Scholar]
- 30. Hou P, Li Y, Zhang X, Liu C, Guan J, Li H, Zhao T, Ye J, Yang W, Liu K, Ge J, Xu J, Zhang Q, Zhao Y, Deng H. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science. 2013;341:651–654. [DOI] [PubMed] [Google Scholar]
- 31. Yoo J, Lee E, Kim HY, Youn DH, Jung J, Kim H, Chang Y, Lee W, Shin J, Baek S, Jang W, Jun W, Kim S, Hong J, Park HJ, Lengner CJ, Moh SH, Kwon Y, Kim J. Electromagnetized gold nanoparticles mediate direct lineage reprogramming into induced dopamine neurons in vivo for Parkinson’s disease therapy. Nat Nanotechnol. 2017;12:1006–1014. [DOI] [PubMed] [Google Scholar]
- 32. Choi DH, Kim JH, Kim SM, Kang K, Han DW, Lee J. Therapeutic potential of induced neural stem cells for Parkinson’s Disease. Int J Mol Sci. 2017;18:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim HS, Shin TH, Lee BC, Yu KR, Seo Y, Lee S, Seo MS, Hong IS, Choi SW, Seo KW, Núñez G, Park JH, Kang KS. Human umbilical cord blood mesenchymal stem cells reduce colitis in mice by activating NOD2 signaling to COX2. Gastroenterology. 2013;145:1392–1403. [DOI] [PubMed] [Google Scholar]
- 34. Mandal PK, Rossi DJ. Reprogramming human fibroblasts to pluripotency using modified mRNA. Nat Protoc. 2013;8:568–582. [DOI] [PubMed] [Google Scholar]
- 35. Sung EA Yu KR, Shin JH, Seo Y, Kim HS, Koog MG, Kang I, Kim JJ, Lee BC, Shin TH, Lee JY, Lee S, Kang TW, Choi SW, Kang KS. Generation of patient specific human neural stem cells from Niemann–Pick disease type C patient-derived fibroblasts. Oncotarget. 2017;8:85428–85441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Arutyunyan I, Elchaninov A, Makarov A, Fatkhudinov T. Umbilical cord as prospective source for mesenchymal stem cell-based therapy. Stem Cells Int. 2016;2016:6901286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bylund M, Andersson E, Novitch BG, Muhr J. Vertebrate neurogenesis is counteracted by Sox1-3 activity. Nat Neurosci. 2003;6:1162–1168. [DOI] [PubMed] [Google Scholar]
- 39. Episkopou V. SOX2 functions in adult neural stem cells. Trends Neurosci. 2005;28:219–221. [DOI] [PubMed] [Google Scholar]
- 40. Graham V, Khudyakov J, Ellis P, Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39:749–765. [DOI] [PubMed] [Google Scholar]
- 41. Maucksch C, Jones KS, Connor B. Concise review: the involvement of SOX2 in direct reprogramming of induced neural stem/precursor cells. Stem Cells Transl Med. 2013;2:579–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rizzino A. Concise review: the Sox2–Oct4 connection. Critical players in a much larger interdependent network integrated at multiple levels. Stem Cells. 2013;31:1033–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Baek S, Oh J, Song J, Choi H, Yoo J, Park GY, Han J, Chang Y, Park H, Kim H, Cho SG, Kim BS, Kim J. Generation of integration-free induced neurons using graphene oxide-polyethylenimine. Small. 2017;13:1601993. [DOI] [PubMed] [Google Scholar]
- 44. Midoux P, Pichon C. Lipid-based mRNA vaccine delivery systems. Expert Rev Vaccines 2015;14:221–234. [DOI] [PubMed] [Google Scholar]
- 45. Yamanaka S. Elite and stochastic models for induced pluripotent stem cell generation. Nature. 2009;460:49–52. [DOI] [PubMed] [Google Scholar]
- 46. Zhu S, Li W, Zhou H, Wei W, Ambasudhan R, Lin T, Kim J, Zhang K, Ding S. Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell. 2010;7:651–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. [DOI] [PubMed] [Google Scholar]
- 48. Nagamura-Inoue T, He H. Umbilical cord-derived mesenchymal stem cells: Their advantages and potential clinical utility. World J Stem Cells. 2014;6:195–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Weina K, Utikal J. SOX2 and cancer: current research and its implications in the clinic. Clin Transl Med. 2014;3:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ebrahimi B. Reprogramming barriers and enhancers: strategies to enhance the efficiency and kinetics of induced pluripotency. Cell Regen (Lond). 2015;4:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fong CY, Chak LL, Biswas A, Tan JH, Gauthaman K, Chan WK, Bongso A. Human Wharton’s jelly stem cells have unique transcriptome profiles compared to human embryonic stem cells and other mesenchymal stem cells. Stem Cell Rev. 2011;7:1–16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, Revision_final_supple_minor_revision for Single-Factor SOX2 Mediates Direct Neural Reprogramming of Human Mesenchymal Stem Cells via Transfection of In Vitro Transcribed mRNA by Bo-Eun Kim, Soon Won Choi, Ji-Hee Shin, Jae-Jun Kim, Insung Kang, Byung-Chul Lee, Jin Young Lee, Myoung Geun Kook, and Kyung-Sun Kang in Cell Transplantation