Abstract
Significant advances have been made in our understanding of the regulation of cold hardiness. The existence of numerous biophysical and biochemical adaptive mechanisms in perennial woody plants and the complexity their regulation has made the development of methods for managing and improving cold hardiness in perennial woody plants has been very difficult. This may be partially attributed to viewing cold hardiness as a single dimensional response, rather than as a complex phenomenon, involving different mechanisms (avoidance and tolerance), different stages (mid-winter vs. late winter), and having an intimate overlap with the genetic regulation of dormancy. In particular separating the molecular regulation of cold hardiness from growth processes has been challenging. ICE and C-repeat binding factor (CBF), transcription factors (Inducer of CBF expression and CRT-binding factor) have been shown to be an important aspect in the regulation of cold-induced gene expression. Evidence has emerged, however, that they are also intimately involved in the regulation of growth, flowering, dormancy, and stomatal development. This evidence includes the presence of CBF binding motifs in genes regulating these processes, or through cross-talk between the pathways that regulate them. Recent changes in climate that have resulted in erratic episodes of unseasonal warming followed by more seasonal patterns of low temperatures has also highlighted the need to better understand the genetic and molecular regulation of deacclimation, a topic of research that is only more recently being addressed. Environmentally-induced epigenetic regulation of stress responses and seasonal processes such as cold acclimation, deacclimation, and dormancy have been documented but are still poorly understood. Advances in the ability to efficiently generate large DNA and RNA datasets and genetic transformation technologies have greatly increased our ability to explore the regulation of gene expression and explore genetic diversity. Greater knowledge of the interplay between epigenetic and genetic regulation of cold hardiness, along with the application of advanced genetic analyses, such as genome-wide-association-studies (GWAS), are needed to develop strategies for addressing the complex processes associated with cold hardiness in woody plants. A cautionary note is also indicated regarding the time-scale needed to examine and interpret plant response to freezing temperatures if progress is to be made in developing effective approaches for manipulating and improving cold hardiness.
Keywords: freezing tolerance, ice nucleation, cold acclimation, deacclimation, dormancy, C-repeat binding factor (CBF), DAM genes, antifreeze protein (AFP)
Introduction
Ever since the first microscopic observations of the freezing response of cells were made in the latter part of the 19th century and early 20th century (Molisch, 1897; Wiegand, 1906), and it was discovered that plant cells undergo cytorrhysis rather than plasmolysis in response to freezing, an elusive search has been conducted to develop a complete and integrated understanding of cold hardiness and freezing tolerance in plants (Wisniewski et al., 2003; Gusta and Wisniewski, 2013; Arora, 2018). Despite thousands of reports and countless reviews, reliable approaches to improving freezing tolerance, without affecting other aspects of plant development, have yet to be developed, either at the molecular/genetic level or the physiological level. New technologies have allowed us to understand plant response to low temperatures in greater and greater detail, but the picture has greatly increased in complexity.
The lack of progress may be partially attributable to two factors. One factor is interpreting cold hardiness as a singular on/off response rather than a combination of many diverse mechanisms that involve significant structural, biochemical, and genetic adjustments, as well as the complexity of manipulating cold hardiness without having a negative impact on other plant developmental processes. The characteristics of these components are species-specific (often genotype-specific), potentially under separate genetic control. Therefore, it is essential when investigating plant cold hardiness to be cognizant of what aspect of the process is being studied and its potential impact on the aspect of cold hardiness that is deemed to be critical for survival. The second factor, is related to the difficulty of studying the biology of organisms at low temperatures, where the kinetics of reactions, and the time required for processes to reach an equilibrium can be problematic when conducting experiments. As noted in Gusta and Wisniewski (2013), the admonition made by Felix Franks in his book on the biophysics of water at low temperature (Franks, 1985), is very relevant. “Too frequently experimental observations on highly complex systems are based on measurements performed under non-equilibrium conditions and rationalized in terms of elementary textbook science. The degree of undercooling (mostly presented using the incorrect terminology, supercooling), the mechanism of ice nucleation, the growth and type of crystals, their size and distribution, the flow properties of the unfrozen matrix, and long-term effects of aging, all need to be taken into account.” The book published by Franks, 1985 still serves as an invaluable primer on the low temperature biology.
Cold hardiness adaptations in plants have been divided into two general categories, tolerance and avoidance. The former involves transcriptomic reprogramming and a host of subsequent biochemical changes that allow plants to tolerate freezing temperatures and the presence of ice in their tissues, while the latter involves mechanisms that allows pockets of water to remain undercooled (deep supercooling) to very low, sub-zero temperatures (-20 to -40°C), so that the supercooled cells are not exposed to the dehydrative effects associated with a freeze tolerance response (often referred to as extracellular freezing). Deep supercooling is characteristic of the dormant buds of many woody perennials and the xylem parenchyma cells of many temperate tree species. The terms freeze tolerance and avoidance, however, are somewhat inaccurate, though widely used, as in both cases cells are avoiding freezing. In the case of freezing tolerance, this is accomplished by the loss of cellular water to extracellular ice, which then decreases the freezing point of the cytoplasm. In the second case, water is not relocated to sites of extracellular ice, even though extracellular ice is present, but instead remains in a metastable condition, and prone to “flash” intracellular freezing (Fujikawa et al., 2009; Wisniewski et al., 2014b). Processes relevant to these strategies are ice nucleation and propagation (Wisniewski et al., 2009, 2014b), the ability to specifically determine where ice crystals are initiated in plant tissues and what shape they form as they grow (McCully et al., 2004), and the formation of cryoprotective and antifreeze compounds (Duman and Wisniewski, 2014).
Despite the complexity of plant cold hardiness, considerable progress has been made in understanding the various components that comprise cold hardiness (Gusta and Wisniewski, 2013). This mini-review highlights one area where considerable progress has been made in understanding the genetic regulation of cold acclimation, and another topic, deacclimation, that is deserving of considerable more focus due to the erratic patterns of warming and cooling temperatures that have developed in the context of climate change. These highly variable weather patterns have had a major impact on dormancy, cold acclimation, and chilling requirements.
The Molecular Regulation of Plant Cold Hardiness
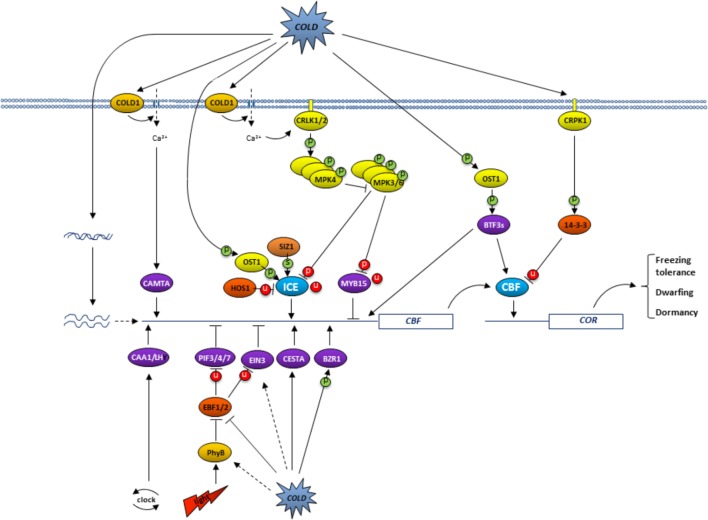
Plants cannot move but rather must adapt to a stressful environment. Genes encoding transcription factors in the model plant Arabidopsis constitute 6–10% of their genome, compared to 5% in humans, and it is therefore not surprising that adaptation to stresses in plants includes a dramatic change in transcriptional cascades (Pireyre and Burow, 2015). The C-repeat binding factor (CBF), transcription factor pathway has been demonstrated to play an exceptionally important role in plant cold acclimation, a process in which low temperatures lead to biochemical and physiological changes that confer freezing tolerance. These changes are largely associated with the expression of so-called Cold Responsive (COR) genes. In Arabidopsis, two or three CBFs co-regulate, often with other transcription factors, more than two-thirds of COR genes (Shi et al., 2017). The increase in frost tolerance under ambient conditions that has been demonstrated to occur in many plants as a result of CBF overexpression, and the decrease in frost tolerance in CBF triple mutants (Jia et al., 2016; Zhao et al., 2016), further underscores the importance of CBF genes. CBF overexpression in herbaceous and tree species, however, can also reduce growth and induce dormancy (Wisniewski et al., 2011, 2014a), thus it is not surprising that CBF activity is tightly regulated and exhibits only short periods of elevated presence in an active form. This regulation occurs at various levels, including transcriptional (transcript quantity and variant), translational (protein quantity), and post-translational (protein activity), and can have an immediate effect, because many changes are made to pre-existing molecules. Detailed insights into the regulation of the CBF pathway in Arabidopsis has only recently been emerging (Figure 1), while limited information for other plant species suggests they have similar but unique regulatory processes of their own. An overview of the information on Arabidopsis, and woody plants when available, is presented in the current mini-review.
FIGURE 1.
Overview of the regulation of the CBF pathway in Arabidopsis. Low temperatures trigger plasma membrane rigidification which leads, presumably via COLD1-like protein, to the opening of Ca2+ channels. The resulting higher calcium levels activate CRLK1/2 (calcium/calmodulin-regulated receptor-like kinase; Yang et al., 2010a,b). In turn, this CRKL1/2 triggers the MEKK1-MMK2-MPK4 cascade to ultimately increase ICE activity, because it inhibits the phosphorylation of ICE1 by MPK3/6 and subsequent ICE1 ubiquitination and degradation (Li et al., 2017a; Zhao et al., 2017). ICE activity is further regulated by low temperatures via OST1 (open stomata 1), induced phosphorylation (Ding et al., 2015), and SIZ1-induced sumoylation (Miura et al., 2007), both of which interfere with HOS1 (high expression of osmotically responsive protein 1), directed ubiquitination and subsequent degradation of ICE (Dong et al., 2006). The resulting active ICE directs CBF expression. Low temperature activated phosphorylation of 14-3-3 proteins by CRPK1 cause the degradation of CBF proteins (Liu et al., 2017). In contrast, cold-induced OST1-directed phosphorylation of BTF3s promotes its binding to CBFs and thereby prevents CBF degradation (Ding et al., 2018). Photoperiod regulates CBF expression via red light perception by PhyB and subsequent degradation of PIF3 (phytochrome-interacting factor 3), thereby relieving its inhibition of CBF expression (Jiang et al., 2017), whereas the circadian clock regulates CCA1 and LHY activity (Dong et al., 2011). Interestingly, PIF3 stability is increased by low temperatures, presumably at a later time to downregulate CBF expression (Jiang et al., 2017). PIF4/7 and EIN3 (Ethylene insensitive 3) downregulate whereas BZR and CESTA upregulate CBF expression, but how this is triggered is not yet known (Shi et al., 2018). Phosphorylation, sumoylation and ubiquitination events are indicated by P, S and U, respectively, with activating modifications in green and inhibiting modifications in red.
Low temperature-induced chromatin modification provides physical access to certain genes and allows their transcription. This apparently includes access to and activation of COR genes by CBFs (Park et al., 2018). CBF gene expression itself is regulated by a large number of transcription factors (Shi et al., 2018). Transcriptional activators include ICE1 and 2 (Inducer of CBF expression 1 and 2), CAMTA 3 and 5 (Calmodulin binding transcription activator 3 and 5), CESTA, BZR1 (Brassinazole-resistant 1), and CCA1/LHY1 (circadian clock-associated 1/late elongated hypocotyl). In contrast, CBF transcription repressors include MYB15, EIN3 (Ethylene insensitive 3), PIF3 (phytochrome-interacting factor 3), and PIF4/7. The activity of these transcription factors is modulated by low temperature, light, and/or a circadian clock in such a way that it results in CBF expression only at specific times during constant low-temperature conditions (Figure 1). Each transduction most likely also involves one or more hormones (Eremina et al., 2016; Barrero-Gill and Salinas, 2017; Li et al., 2017b; Zhou et al., 2017), whereby CBFs also affect hormone levels (Li et al., 2017c), but details are currently relatively sparse. The fact that a gradual or rapid decrease in temperature has slightly different effects make the CBF pathway even more complicated (Kidokoro et al., 2017).
In addition to transcript levels, the type of transcripts can also be altered in response to a cold period. Approximately 60% of intron-containing genes in Arabidopsis were reported to undergo alternative splicing (Marquez et al., 2012), especially under stress conditions. Recent RNAseq analysis for Arabidopsis identified the often rapid cold induction of alternative splicing (AS) of over 2,400 genes, with over 1,600 regulated only at the AS level and therefore not detected in most previous analyses (Calixto et al., 2018). CBF genes do not have introns, thus AS does not directly affect them, however, alternatively spliced transcripts have been detected for PIF7, PHYB, and CAMTA3 (Calixto et al., 2018), which may alter CBF expression.
Once produced, the stability of CBF1/3 proteins is downregulated by their interaction with cold-induced CRPK1-phosphorylated 14-3-3 protein (Liu et al., 2017), and upregulated by their interaction with cold-induced OST1-phosphorylated basic transcription factor 3/BTF3-like protein (BTF3/BTF3L; Ding et al., 2018). While some reports have suggested that Arabidopsis CBF1-3 are equally important, others suggest that Arabidopsis CBF2 and CBF3 play a more important role in directing the cold response (Jia et al., 2016; Zhao et al., 2016; Shi et al., 2017), and adaptation to low temperature in natural populations (Gehan et al., 2015). This apparently occurs through the employment of different regulons. Whereas the main genes upregulated in CBF2 overexpressing plants were related to lipid localization, starch metabolic process, light stimulus response, and regulation of transcription, the genes regulated by CBF3 were mainly related to oxidative stress response (Li et al., 2017c).
The presence of a similar pathway in perennial woody plants, such as poplar, apple, grape, Prunus sp., and eucalyptus, is suggested by the identification of usually a larger number of ICE- and CBF-like genes, increasing the possible further delineation of functions (Wisniewski et al., 2014a). Investigations into their regulation found that ICE RNA levels are not much affected by treatments, suggesting an emphasis on regulation of ICE by post-translational modifications (Wisniewski et al., 2014a). CBF expression is often induced by low temperatures and/or drought or high salt (Wisniewski et al., 2014a), can be affected by the circadian clock (Artlip et al., 2013), and, for some CBFs, is induced by a continuous cold treatment later and/or for a longer time period than reported for the Arabidopsis CBFs (Xiao et al., 2006, 2008; Artlip et al., 2013; Leyva-Pérez et al., 2015; Li et al., 2017d). AS was determined to be a prevalent occurence in the transcription of many genes in apple, orange, and grape, with genes in grape plants showing the most AS events (Sablok et al., 2017). Not much is currently known about AS events, however, in ICE, or COR genes in low temperature conditions, except for ICE transcripts in grape (Rahman et al., 2014). AS, however, has been suggested to regulate responses to environmental stresses in many plants, including Western poplar (Populus trichocarpa) (Filichkin et al., 2018). Amino acid motifs thought to be involved in post-translational modifications have been identified in predicted sequences for ICE and CBF proteins, and functional studies suggest that they are important (Feng et al., 2012; Nguyen et al., 2016; Carlow et al., 2017), but much more studies are needed to determine when and how they regulate ICE and CBF activity. Together the collective studies suggest that woody perennial plants have a CBF-like pathway similar to Arabidopsis (Benedict et al., 2006), even including a trade-off between growth and cold stress tolerance (Ibanez et al., 2010; Tillett et al., 2011; Nguyen et al., 2016) but details of CBF regulation in woody plants is still very limited.
Prospects for the Generation of Plants with Enhanced Freezing Tolerance
The tight regulation of CBF activity is largely lost in transgenic plants using a CBF construct driven by a constitutive 35S promoter, thus the use of natural promoters is preferred. Recent reports suggest that plants with constitutive brassinosteroid (BR) response display higher CBF expression but no signs of growth retardation (Li et al., 2017b). Therefore, it may be possible to avoid the reduced growth associated with CBF overexpression (Artlip et al., 2014), if the “correct” CBF or CBF regulators are utilized. Recently, Dong et al. (2017) conducted a meta-analysis of the effect of CBF overexpression on temperature stress tolerance and related responses. In that study data from 75 published articles were analyzed to determine the impact of a host of factors such as origin of the CBF gene, promoter used to drive expression, the method of stress evaluation, etc., on temperature response and associated indicators, such as electrolyte leakage, growth, chlorophyll fluorescence, sugar and proline levels, etc., Results indicated that 7 of 8 measured variables were significantly modulated in CBF (DREB)-transgenic plants, while two of the eight parameters were only modulated in non-stressed plants. The measured parameters were modulated by 32% or more by various experimental variables. The modulating variables included, acclimated vs. non-acclimated, type of promoter, duration of stress and its severity, source of the donor gene, and whether the donor and recipient were the same genus. CBF overexpression had a consistent negative impact on plant height, a reduction in electrolyte leakage, and positive impact on survival. The impact was evident in both acclimated and non-acclimated plants, although the greatest impact was observed in acclimated plants. Such analyses may provide a more comprehensive understanding of how to best utilize CBF genes with modified promoters to improve freezing stress tolerance.
An alternative, enterprising approach is the modification of endogenous CBF genes into variants that lead to a higher frost tolerance using a clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein 9 nuclease (Cas9)-like system which has recently been optimized for use in vegetatively propagated perennial plants (Chen et al., 2018). For example, existing sequences could be modified based on variants present in more frost tolerant cultivars or species (Carlow et al., 2017; Li et al., 2017d), and thereby change their regulation. Because of the precise change, the resulting plants may not be considered genetically modified by government regulatory agencies and may be more acceptable to the general public.
Deacclimation (DA) Response, a Critical Factor for Winter-Survival
The ability to increase freeze-tolerance in temperate- and boreal-zone woody perennials (fruit and forest tree species) via autumnal cold acclimation is undoubtedly the first line of defense against harsh and long winters. Seasonally induced freeze-tolerance is lost under relatively warmer conditions in a process called ‘deacclimation’, a process that typically occurs in response to spring-warming. The maintenance of a sufficient level of cold-induced freeze-tolerance until the danger of killing frosts is passed, however, is an imperative to avoid frost-damage. For example, erratic temperature fluctuations, i.e., sudden winter-warming or premature spring-like conditions followed by more “normal” freezing temperatures, could render partially or fully deacclimated tissues vulnerable to freeze-damage. Indeed, the frequency of such fluctuations has been increasing (Jentsch et al., 2007; IPCC, 2014), and some of the most devastating killer-frosts across North America have been attributed to such events, e.g., Easter freeze of 2007 (Gu et al., 2008), Mother’s Day freeze of 2010, killer frost of 2012, and the polar vortex of 2014. Field simulations of winter-warming events have also confirmed their damaging effects on overwintering perennials (Taulavuori et al., 1997; Bokhorst et al., 2009, 2010).
Dormancy Status and Spring Phenology (Budbreak) in Relation to DA
Temperate trees have evolved the ability to tolerate harsh winters by undergoing a period of endodormancy (rest), during which cold-acclimated meristems are less prone to DA when trees are exposed to unseasonal episodes of warming (Kalberer et al., 2006, 2007a,b). Buds of native temperate and boreal trees must satisfy a genetically defined chilling requirement to exit endodormancy (Richardson et al., 1974). Post-endodormancy, buds enter an ecodormant state where they must be exposed to a genetically defined threshold of warming (‘heat units’) (Charrier et al., 2011), for the resumption of meristematic activity and growth to occur (budbreak or spring phenology). Ecodormant buds are substantially more sensitive to warmer temperatures and DA than endodormant buds (Kalberer et al., 2007a). This sensitivity increases progressively as the period of ecodormancy increases, finally culminating in complete DA and spring budbreak (Kalberer et al., 2006; Arora and Taulavuori, 2016). Any shift in this annual cycle of spring phenology could potentially increase the risk of trees encountering frost injury (Vitasse et al., 2014 and references therein). Whether an increased risk occurs depends on a variety of internal factors, including species, chilling requirement, the genetic ability to resist deacclimation in response to transient, unseasonal episodes of warm temperatures, and the capacity to reacclimate. External (environmental) mitigating factors include, temperature fluctuations (intensity and timing), and the region/site (latitude and altitude) where the trees are located (Pagter and Arora, 2013; Arora and Taulavuori, 2016; Vitasse et al., 2018b). The sensitivity of ecodormant buds to deacclimating temperatures has also been reported to increase with increasing photoperiod in spring in species such as European beech (Fagus sylvatica) (Vitasse and Basler, 2013). There is ample evidence that warming trends in recent decades have advanced spring budbreak and leaf development in many plant species growing in cold regions (Penuelas and Filella, 2001; Menzel et al., 2006, and references therein). Some studies have indicated, however, that the degree of advancement in spring phenology appears to be declining in the recent years (Yu et al., 2010; Fu et al., 2015).
Advances in spring phenology due to climate change can occur under two scenarios. In the first scenario, a faster than normal accumulation of heat-units by ecodormant buds occurs due to earlier and warmer spring-like temperatures (Cleland et al., 2007). This could render prematurely deacclimated buds vulnerable to subsequent spring frosts. The second, somewhat ignored and paradoxical scenario, involves a more rapid fulfillment of chilling requirement due to warmer winter temperatures than has occurred in more typical, historical winters in certain regions. For example, tree species in northern latitudes or high elevations could experience a greater level of ‘dormancy-breaking chill units’ since the warmer winter temperatures could expose trees to temperatures >0°C that are more effective in breaking endodormancy and reduce exposure to sub-freezing temperatures that do not contribute to chill unit accumulation (Hanninen, 2006). Based on this premise, spring phenology could be expected to advance more rapidly in historically colder areas under warming climate conditions. This would result in premature deacclimation and a greater risk of spring freeze-damage. Indeed, Vitasse et al. (2018a) reported that spring phenology in fruit (apple, cherry), and forest (Norway spruce and European beech) trees has advanced at a faster rate during 1975–2016 at fifty high elevation locations in Switzerland than in other temperate locations. The authors argued that even if the frequency and severity of late spring frosts remains unchanged in the future or changes less than the spring phenology of plants, deacclimated organs may be more exposed to a greater number of freeze-damage events (Vitasse et al., 2018a).
On the other hand, proponents of a possible decline in the advancement or delay in spring phenology by warming climate make their case as follows. They suggest that elevated winter temperature may result in a chilling-deficit, i.e., reduced duration and/or sum of cold. And since heat unit requirement for spring phenology is believed to be inversely correlated with the chill accumulation during dormancy (Harrington et al., 2010; Laube et al., 2014), any reduction in accumulated chilling would result in higher heat-unit requirement, thus slowing down the advance in, or delaying, the spring leaf-unfolding (Yu et al., 2010; Fu et al., 2015). One of these studies (Fu et al., 2015) noted that while spring phenology for seven deciduous forest tree species had advanced by ∼4 d during 1980–1994 in Europe, this response has decreased by ∼40% (to 2.3 d) during 1999–2013 (Pan European Phenology Network). A caveat that must be added for such an observation to be practically and widely applicable, however, is that the temperatures during warmer winters have to be high enough to cause real chilling deficit, i.e., either negate accumulated chilling or be ineffective to meet the chilling requirement.
Although the focus of this mini-review is not on bud dormancy, it is relevant to note that several studies have associated the genetic regulation of chilling requirement and dormancy with Dormancy Associated MADs-box (DAM) genes in peach (Bielenberg et al., 2008; Li et al., 2009) apple (Wu et al., 2017a), pear (Saito et al., 2013), apricot (Sasaki et al., 2011), leafy spurge (Euphorbia esula) (Horvath et al., 2010), kiwifruit (Wu et al., 2011, 2017b), and tea plant (Camellia sinensis) (Hao et al., 2017). Bud-dormancy associated candidate genes have also been identified in blackcurrant (Ribes nigrum) by Hedley et al. (2010) and Yordanov et al. (2014) identified an early budbreak (EBB) gene in poplar that was an APETALA2/Ethylene responsive transcription factor responsible for early bud flush.
C-repeat binding factor-binding motifs have been identified in promoters of several DAM genes and an EBB homolog in apple (Wisniewski et al., 2015a), as well as other plant species. A comprehensive analysis of DAM genes in the ornamental woody plant, P. mume (Chinese plum, Japanese apricot) demonstrated an interaction between CBFs and DAM genes, especially PMCBF1 - PMDAM1 (Zhao et al., 2018) and CBF expression lowered whereas MADS-box gene (1 and 3) expression increased in almond flower buds after bud break (Barros et al., 2012). Wisniewski et al. (2015a) noted that overexpression of a peach CBF gene (PpCBF1) in apple altered the expression of DAM, EBB, and RGL (DELLA) genes and that some members of each of these gene families contained C-repeat regions in their promoter regions that are the target sites for CBF. They provided a model linking CBF expression with the regulation of dormancy, bud-break, freezing tolerance, and growth. Interestingly, a subsequent study indicated that the impact of the apple transgenic rootstock overexpressing CBF was not graft-transmissible and thus did not affect the cold hardiness of dormancy of the scion cultivar grafted to the transgenic ‘M.26’ rootstock, although growth and flowering were significantly impacted (Artlip et al., 2016).
Current research has highlighted the impact of dormancy status and spring phenology on the propensity of trees to deacclimate. Spring-phenology is an outcome influenced by both the chilling and heat unit requirements of overwintering tree species. The fact that chill- and heat-units can be satisfied by the same temperatures for certain species (Cooke et al., 2012 and references therein) makes their combined effect on spring phenology even more complex. It is therefore critical to include dormancy-status and the interactions between chilling and heat requirements as key parameters in models designed to predict the relationship between deacclimation response and freezing-tolerance.
Future Directions
The past 50 years of research has provided a wealth of information on the genetic and molecular regulation of plant cold hardiness, as well as the regulation of dormancy. These advances have been fueled by new technologies associated with high-throughput sequencing, genetic mapping, and transformation technologies. In particular the regulatory role of CBF genes in freezing tolerance, and of DAM genes in the regulation of chilling requirement stand out as major advances. The discovery that CBF activity is regulated continually and at various levels, helps explain, at least in part, why slightly different treatment of plants with respect to light, duration and rate of low temperature treatment for example, lead to different outcomes with respect to their frost tolerance. Notable advances have also been made with the use of high-resolution infrared thermography in our understanding of ice nucleation and propagation (Wisniewski et al., 2014b, 2015b), and the properties of antifreeze proteins (Duman and Wisniewski, 2014). Despite these advances, significant improvements in plant cold hardiness have been elusive and problematic due to the complexity of this trait and its intimate connection to other plant developmental processes, especially growth and flowering. In addition, the relatively new field of epigenetics has demonstrated the key role that the environment can play on imprinting plant response to abiotic stress (Kumar, 2018).
Future studies will need to better understand the cross-talk that occurs between different plant developmental processes and how it can be manipulated in a prescribed manner. A key question will be whether processes that determine cold hardiness can be separated from processes that restrict growth. Can CBF genes be regulated in a manner that removes their negative impact on growth and reproductive output? What are the genetic mechanisms that can be used to tease these processes apart? Which CBF gene or gene variant present in more frost tolerant species is best targeted for manipulation and how can epigenetic modifications affecting their activity best be harnessed?
Although, not as glamorous, a comprehensive understanding of the underlying biophysical mechanisms responsible for freeze avoidance, especially in woody plants, is still lacking. Deep supercooling of xylem parenchyma (Wisniewski, 1995; Fujikawa et al., 2009) and floral buds (Kuprian et al., 2016, 2017) is an integral aspect of the cold hardiness of many temperate tree species, especially fruit trees, however, few advances have been made on this topic over the past 30 years. What new technologies that can be applied to better understand how, when, and where ice is initiated in plants, how it is propagated, and how the size and shape of ice crystals are regulated. Genetic studies of the inheritance of avoidance traits, such as supercooling, have yet to be conducted, but would provide very useful information. An integrated approach that takes into account the complexity of traits that contribute to plant cold hardiness will be needed to achieve advances that can be translated into practical solutions that address the challenges of a rapidly changing climate.
Author Contributions
MW was responsible for the general overview of the opinions stated in the manuscript and any faults or shortcoming in logic directly fall on him. AN contributed the overview of CBF regulation, and RA the information on the importance of deacclimation in a changing climate. All authors reviewed and agreed with the final version of the submitted manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors apologize to all colleagues whose relevant work is not quoted due to space limitations.
Footnotes
Funding. Research on CBF in fruit trees in the Wisniewski lab was supported by the United States Department of Agriculture – Agricultural Research Service (USDA-ARS).
References
- Arora R., Taulavuori K. (2016). Increased risk of freeze damage in woody perennials vis-à-vis climate change: importance of deacclimation and dormancy response. Front. Environ. Sci. 4:44 10.3389/fenvs.2016.00044 [DOI] [Google Scholar]
- Arora R. (2018). Mechanism of freeze-thaw injury and recovery: a cool retrospective and warming up to new ideas. Plant Sci. 270 301–313. 10.1016/j.plantsci.2018.03.002 [DOI] [PubMed] [Google Scholar]
- Artlip T. A., Wisniewski M., Bassett C. L., Norelli J. L. (2013). CBF gene expression in peach leaf and bark tissues is gated by a circadian clock. Tree Physiol. 33 866–877. 10.1093/treephys/tpt056 [DOI] [PubMed] [Google Scholar]
- Artlip T., Wisniewski M., Arora R., Norelli J. (2016). An apple rootstock overexpressing a peach CBF gene alters growth and flowering in the scion but does not impact cold hardiness or dormancy. Hort. Res. 3:16006. 10.1038/hortres.2016.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artlip T., Wisniewski M., Norelli J. (2014). Field evaluations of apple overexpressing a peach CBF gene confirms its effect on cold hardiness, dormancy, and growth. Environ. Exp. Bot. 106 79–86. 10.1038/hortres.2016.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero-Gill J., Salinas J. (2017). CBFs at the crossroads of plant hormone signaling in cold stress response. Mol. Plant. 10 542–544. 10.1016/j.molp.2017.03.004 [DOI] [PubMed] [Google Scholar]
- Barros P. M., Gonçalves N., Saibo N. J. M., Oliveira M. M. (2012). Cold acclimation and floral development in almond bud break: insights into the regulatory pathways. J. Exp. Biol. 63 4585–4596. 10.1093/jxb/ers144 [DOI] [PubMed] [Google Scholar]
- Benedict C., Skinner J. S., Meng R., Chang Y., Bhalerao R., Huner N. P. A., et al. (2006). The CBF1-dependent low temperature signaling pathway, regulon and increase in freeze tolerance are conserved in Populus spp. Plant Cell Environ. 29 1259–1272. 10.1111/j.1365-3040.2006.01505 [DOI] [PubMed] [Google Scholar]
- Bielenberg D. G., Wang Y., Li Z., Zhebentyayeva T., Fan S., Reighard G., et al. (2008). Sequencing and annotation of the evergrowing locus in peach (Prunus persica L. Batsch) reveals a cluster of six MADS-box transcription factors as candidate genes for regulation of terminal bud formation. Tree Genet. Genomes 4 495–507. 10.1007/s11295-007-0126-9 [DOI] [Google Scholar]
- Bokhorst S., Bjerke J. W., Davey M. P., Taulavuori K., Taulavuori E., Laine K., et al. (2010). Impacts of extreme winter warming events on plant physiology in a sub-Arctic heath community. Physiol. Plant. 140 128–140. 10.1111/j.1399-3054.2010.01386.x [DOI] [PubMed] [Google Scholar]
- Bokhorst S., Bjerke J. W., Tommervik H., Callaghan T. V., Phoenix G. K. (2009). Winter warming events damage sub-Arctic vegetation: consistent evidence from an experimental manipulation and natural event. J. Ecol. 97 1408–1415. 10.1111/j.1365-2745.2009.01554.x [DOI] [Google Scholar]
- Calixto C. P. G., Guo W., James A. B., Tzioutziou N. A., Entizne J. C., Panter P. E., et al. (2018). Rapid and dynamic alternative splicing impacts the Arabidopsis cold response transcriptome. Plant Cell 30 1424–1444. 10.1105/tpc.18.00177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlow C. E., Faultless J. T., Lee C., Siddiqua M., Edge A., Nassuth A. (2017). Nuclear localization and transactivation by Vitis CBF transcription factors are regulated by combinations of conserved amino acid domains. Plant Phys. Biochem. 118 306–319. 10.1016/j.plaphy.2017.06.027 [DOI] [PubMed] [Google Scholar]
- Charrier G., Bonhomme M., Lacointe A., Ameglio T. (2011). Are budburst dates, dormancy and cold acclimation in walnut trees (Juglans regia L.) under mainly genotypic or environmental control? Int. J. Biometeorol. 55 763–774. 10.1007/s00484-011-0470-1 [DOI] [PubMed] [Google Scholar]
- Chen L., Li W., Katin-Grazzini L., Ding J., Gu X., Li Y., et al. (2018). A method for the production and expedient screening of CRISPR/Cas9-mediated non-transgenic mutant plants. Hort. Res. 5:13. 10.1038/s41438-018-0023-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleland E. E., Chuine I., Menzel A., Mooney H. A., Schwartz M. D. (2007). Shifting plant phenology in response to global change. Trends Ecol. Evol. 22 357–365. 10.1016/j.tree.2007.04.003 [DOI] [PubMed] [Google Scholar]
- Cooke J. E., Eriksson M. E., Juntilla O. (2012). The dynamic nature of bud dormancy in trees: environmental control and molecular mechanisms. Plant Cell Environ. 35 1707–1728. 10.1111/j.1365-3040.2012.02552.x [DOI] [PubMed] [Google Scholar]
- Ding Y., Jia Y., Shi Y., Zhang X., Song C., Gong Z., et al. (2018). OST1-mediated BTF3L phosphorylation positively regulates CBFs during plant cold responses. EMBO J. 37:e982288. 10.15252/embj.201798228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Li H., Zhang X., Xie Q., Gong Z., Yang S. (2015). OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis. Dev. Cell 32 278–289. 10.1016/j.devcel.2014.12.023 [DOI] [PubMed] [Google Scholar]
- Dong C., Ma Y., Wisniewski M., Cheng Z.-M. (2017). Meta-analysis of the effect of overexpression of CBF/DREB family genes on drought stress response. Environ. Exp. Bot. 142 1–14. 10.1016/j.envexpbot.2017.07.014 [DOI] [Google Scholar]
- Dong C. H., Agarwal M., Zhang Y., Xie Q., Zhu J. K. (2006). The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. U.S.A. 103 8281–8286. 10.1073/pnas.0602874103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong M. A., Farre E. M., Thomashow M. F. (2011). Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the C-repeat binding factor (CBF) pathway in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108 7241–7246. 10.1073/pnas.1103741108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duman J. G., Wisniewski M. (2014). The use of antifreeze proteins for frost protection in sensitive crop plants. Environ. Exp. Bot. 106 60–69. 10.1016/j.envexpbot.2014.01.001 [DOI] [Google Scholar]
- Eremina M., Rozhon W., Poppenberger B. (2016). Hormonal control of cold stress responses in plants. Cell. Mol. Life Sci. 73 797–810. 10.1007/s00018-015-2089-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X.-M., Zhao Q., Zhao L.-L., Qiao Y., Xie X.-B., Li H.-F., et al. (2012). The cold-induced basic helix-loop-helix transcription factor gene MdCIbHLH1encodes an ICE-like protein in apple. BMC Plant Biol. 12:22. 10.1186/1471-2229-12-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filichkin S. A., Hamilton M., Dharmawardhana P. D., Singh S. K., Sullivan C., Ben-Hur A., et al. (2018). Abiotic stresses modulate landscape of poplar transcriptome via alternative splicing, differential intron retention, and isoform ratio switching. Front. Plant Sci. 9:5. 10.3389/fpls.2018.00005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks F. (1985). Biophysics and Biochemistry at Low Temperatures. Cambridge: Cambridge University Press. [Google Scholar]
- Fu Y. H., Zhao H., Piao S., Peaucelle M., Peng S., Zhou G., et al. (2015). Declining global warming effects on the phenology of spring leaf unfolding. Nature 526 104–107. 10.1038/nature15402 [DOI] [PubMed] [Google Scholar]
- Fujikawa S., Kasuga J., Takata N., Arakawa K. (2009). “Factors related to changes of deep supercooling capability in xylem parenchyma cells of trees,” in Plant Cold Hardiness: From the Laboratory to the Field, eds Gusta L. V., Wisniewski M. E., Tanino K. (Oxfordshire: CABI; ), 29–42. [Google Scholar]
- Gehan M. A., Park S., Gilmour S. J., An C., Lee C.-M., Thomashow M. F. (2015). Natural variation in the C-repeat binding factor cold response pathway correlates with local adaptation of Arabidopsis ecotypes. Plant J. 84 682–693. 10.1111/tpj.13027 [DOI] [PubMed] [Google Scholar]
- Gu I., Hanson P. J., Post W. M., Kaiser D. P., Yang B., Nemani R., et al. (2008). The 2007 eastern US spring freeze: increased cold damage in a warming world? Bioscience 58 253–262. 10.1641/B580311 [DOI] [Google Scholar]
- Gusta L. V., Wisniewski M. (2013). Understanding plant cold hardiness: an opinion. Physiol. Plant. 147 4–14. 10.1111/j.1399-3054.2012.01611.x [DOI] [PubMed] [Google Scholar]
- Hanninen H. (2006). Climate warming and the risk of frost damage to boreal forest trees: identification of critical ecophysiological traits. Tree Physiol. 26 889–898. 10.1093/treephys/26.7.889 [DOI] [PubMed] [Google Scholar]
- Hao X., Yang Y., Yue C., Wang L., Horvath D. P., Wang X. (2017). Comprehensive transcriptome analyses reveal differential gene expression profiles of Camellia sinensis axillary buds at para-, endo-, ecodormancy, and bud flush stages. Front. Plant Sci. 8:553. 10.3389/fpls.2017.00553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington C. A., Gould P. J., St. Clair J. B. (2010). Modeling the effects of winter environment on dormancy release of Douglas-fir. For. Ecol. Manag. 259 798–808. 10.1016/j.foreco.2009.06.018 [DOI] [Google Scholar]
- Hedley P. E., Russell J. R., Jorgensen L., Gordon S., Morris J. A., Hackett C. A., et al. (2010). Candidate genes associated with bud dormancy release in blackcurrant (Ribes nigurum L.). BMC Plant Biol. 10:202. 10.1186/1471-2229-10-202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath D. P., Sung S., Kim D., Chao W., Anderson J. (2010). Characterization, expression and function of DORMANCY ASSOCIATED MADS-BOX genes from leafy spurge. Plant Mol. Biol. 73 169–179. 10.1007/s11103-009-9596-5 [DOI] [PubMed] [Google Scholar]
- Ibanez C., Kozarewa I., Johansson M., Ogren E., Rohde A., Eriksson M. E. (2010). Circadian clock components regulate entry and affect exit of seasonal dormancy as well as winter hardiness in populus trees. Plant Physiol. 153 1823–1833. 10.1104/pp.110.158220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- IPCC (2014). Impacts, Adaptation, and Vulnerability. Part A: Global and Sectoral Aspects. Contribution of Working Group II of the Fifth Assessment Report of the Intergovernmental Panel on Climate Change (IPCC), eds Field C. B.et al. (Cambridge: Cambridge University Press; ). [Google Scholar]
- Jentsch A., Kreyling J., Beierkuhnlein C. (2007). A new generation of climate-change experiments: events, not trends. Front. Ecol. Environ. 5:7 10.1890/1540 [DOI] [Google Scholar]
- Jia Y., Ding Y., Shi Y., Zhang X., Gong Z., Yang S. (2016). The cbfs triple mutants reveal the essential functions of CBFs in cold acclimation and allow the definition of CBF regulons in Arabidopsis. New Phytol. 212 345–353. 10.1111/nph.14088 [DOI] [PubMed] [Google Scholar]
- Jiang B., Shi Y., Zhang X., Xin X., Qi L., Guo H., et al. (2017). PIF3 is a negative regulator of the CBF pathway and freezing tolerance in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 114 E6695–E6702. 10.1073/pnas.1706226114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalberer S. R., Leyva-Estrada N., Krebs S. L., Arora R. (2007a). Cold hardiness of floral buds of deciduous azaleas: dehardening, rehardening, and endodormancy in late winter. J. Amer. Soc. Hort. Sci. 132 73–79. [Google Scholar]
- Kalberer S. R., Leyva-Estrada N., Krebs S. L., Arora R. (2007b). Frost dehardening and rehardening of floral buds of deciduous azaleas depend on genotypic biogeography. Environ. Exp. Bot. 59 264–275. 10.1016/j.envexpbot.2006.02.001 [DOI] [Google Scholar]
- Kalberer S. R., Wisniewski M., Arora R. (2006). Deacclimation and reacclimation of cold-hardy plants: current understanding and emerging concepts. Plant Sci. 171 3–16. 10.1016/j.plantsci.2006.02.013 [DOI] [Google Scholar]
- Kidokoro S., Yoneda K., Takasaki H., Takahashi F., Shinozaki K., Yamaguchi-Shinozaki K. (2017). Different cold-signaling pathways function in the responses to rapid and gradual decreases in temperature. Plant Cell 29 760–774. 10.1105/tpc.16.00669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S. (2018). Epigenetic memory of stress responses in plants. J. Phytochem. Biochem. 2:1. [Google Scholar]
- Kuprian E., Koch S., Munkler S., Resnyak A., Buchner O., Oberhammer M., et al. (2017). Does winter desiccation account for seasonal increases in supercooling capacity of Norway spruce bud primordia. Tree Physiol. 38 591–601. 10.1093treephys/tpx142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuprian E., Tuong T. D., Pfaller K., Wagner J., Livingston D. P., III, Neuner G. (2016). Persistent supercooling of reproductive shoots in enabled by structural ice barriers being activie despite and intact xylem connection. PLoS One 11:e0163160. 10.1371/journal.pone.0163160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laube J., Sparks T. H., Estrella N., Hofler J. F., Ankerst D. P., Menzel A. (2014). Chilling outweighs photoperiod in preventing precocious spring development. Glob. Change Biol. 20 170–182. 10.1111/gcb.12360 [DOI] [PubMed] [Google Scholar]
- Leyva-Pérez M. L., Valverde-Corredor A., Valderrama R., Jiménez-Ruiz J., Muñoz-Merida A., Trelles O., et al. (2015). Early and delayed long-term transcriptional changes and short-term transient responses during cold acclimation in olive leaves. DNA Res. 22 1–11. 10.1093/dnares/dsu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A., Zhou M., Wei D., Chen H., You C., Lin J. (2017c). Transcriptome profiling reveals the negative regulation of multiple plant hormone signaling pathways elicited by overexpression of C-repeat binding factors. Front. Plant Sci. 8:1647. 10.3389/fpls.2017.01647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Ding Y., Shi Y., Zhang X., Zhang S., Gong Z., et al. (2017a). MPK3-and MPK6-mediated ICE1 phosphorylation negatively regulates ICE1 stability and freezing tolerance in Arabidopsis. Dev. Cell 43 630–642. 10.17632/jyxknn3gmr.1 [DOI] [PubMed] [Google Scholar]
- Li H., Ye K., Shi Y., Cheng J., Zhang X., Yang S. (2017b). BZR1 positively regulates freezing tolerance via CBF-dependent and CBF independent pathways in Arabidopsis. Mol. Plant. 10 545–559. 10.1016/j.molp.2017.01.004 [DOI] [PubMed] [Google Scholar]
- Li Y., Song Y., Xu B., Xie J., Zhang D. (2017d). Poplar CBF1 functions specifically in an integrated cold regulatory network. Tree Physiol. 37 98–115. 10.1093/treephys/tpw079 [DOI] [PubMed] [Google Scholar]
- Li Z., Reighard G., Abbott A., Bielenberg D. (2009). Dormancy-associated MADS genes from the EVG locus of peach (Prunus perica L. Batsch.) have distinct seasonal and photoperiodic expression patterns. J. Exp. Bot. 60 3521–3530. 10.1093/jxb/erp195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Jia Y., Ding Y., Shi Y., Li Z., Guo Y., et al. (2017). Plasma membrane CRPK1-mediated phosphorylation of 14-3-3 proteins induces their nuclear import to fine-tune CBF signaling during cold response. Mol. Cell 66 117.e5–128.e5. 10.1016/j.molcel.2017.02.016 [DOI] [PubMed] [Google Scholar]
- Marquez Y., Brown J. W., Simpson C., Barta A., Kalyna M. (2012). Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 22 1184–1195. 10.1101/gr.134106.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCully M. E., Canny M. J., Huang C. X. (2004). The management of extracellular ice by petioles of frost-resistant herbaceous plants. Ann. Bot. 94 665–674. 10.1093/aob/mch191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel A., Sparks T. H., Estrella N., Koch E., Aasa A., Ahas R., et al. (2006). European phenological response to climate change matches the warming pattern. Glob. Change Biol. 12 1969–1976. 10.1111/J.1365-2486.2006.01193.X [DOI] [Google Scholar]
- Miura K., Jin J. B., Lee J., Yoo C. Y., Stirm V., Miura T., et al. (2007). SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19 1403–1414. 10.1105/tpc.106.048397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molisch H. (1897). Untersuchungen über das erfrieren der Pflanzen (reprinted in English in 1982). Cryo Lett. 3 332–390. [Google Scholar]
- Nguyen H. C., Cao P. B., San Clemente H., Ployet R., Mounet F., Ladouce N., et al. (2016). Special trends in CBF and DREB2 groups in Eucalyptus gunnii vs Eucalyptus grandis suggest that CBF are master players in the trade-off between growth and stress resistance. Physiol. Plant. 4 445–467. 10.1111/ppl.12529 [DOI] [PubMed] [Google Scholar]
- Pagter M., Arora R. (2013). Winter survival and deacclimation of perennials under warming climate: physiological perspectives. Physiol. Plant. 147 75–87. 10.1111/j.1399-3054.2012.01650.x [DOI] [PubMed] [Google Scholar]
- Park J., Lim C. J., Shen M., Park H. J., Cha J.-Y., Iniesto E., et al. (2018). Epigenetic switch from repressive to permissive chromatin in response to cold stress. Proc. Natl. Acad. Sci. U.S.A. 115 E5400–E5409. 10.1073/pnas.1721241115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penuelas J., Filella I. (2001). Responses to a warming world. Science 294 793–795. 10.1126/science.1066860 [DOI] [PubMed] [Google Scholar]
- Pireyre M., Burow M. (2015). Regulation of MYB and bHLH transcription Factors: a glance at the protein level. Mol. Plant. 8 378–388. 10.1016/j.molp.2014.11.022 [DOI] [PubMed] [Google Scholar]
- Rahman M. A., Moody M. A., Nassuth A. (2014). Grape contains 4 ICE genes whose expression includes alternative polyadenylation, leading to transcripts encoding at least 7 different ICE proteins. Environ. Exp. Bot. 106 70–78. 10.1016/j.envexpbot.2014.01.003 [DOI] [Google Scholar]
- Richardson E. A., Seeley S. D., Walker D. R. (1974). A model for estimating the completion of rest for ‘Redhaven’ and ‘Elberta’ peach trees. Hortscience 9 331–332. 26849277 [Google Scholar]
- Sablok G., Powell B., Braessler J., Yu F., Min X. J. (2017). Comparative landscape of alternative splicing in fruit plants. Curr. Plant Biol. 9-10 29–36. 10.1016/j.cpb.2017.06.001 [DOI] [Google Scholar]
- Saito T., Bai S., Ito A., Sakamoto D., Saito T., EwaUbi B., et al. (2013). Expression and genomic structure of the dormancy-associated MADS box genes MADS13 in Japanese pears (Pyrus pyrifolia Nakia) that differ in their chilling requirement for endodormancy release. Tree Physiol. 33 654–667. 10.1093/treephys/tpt037 [DOI] [PubMed] [Google Scholar]
- Sasaki R., Yamane H., Ooka T., Jotatsu H., Kitamura Y., Akagi T., et al. (2011). Functional and expressional analyses of PmDAM genes associated with endodormancy in Japanese apricot. Plant Physiol. 157 485–497. 10.1104/pp.111.181982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Ding Y., Yang D. (2018). Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci. 23 623–637. 10.1016/j.tplants.2018.04.002 [DOI] [PubMed] [Google Scholar]
- Shi Y., Huang J., Sun T., Wang X., Zhu C., Ai Y., et al. (2017). The precise regulation of different COR genes by individual CBF transcription factors in Arabidopsis thaliana. J. Integr. Plant Biol. 59 118–133. 10.1111/jipb.12515 [DOI] [PubMed] [Google Scholar]
- Taulavuori K., Laine K., Taulavuori E., Pakonen T., Saari E. (1997). Accelerated dehardening in the bilberry (Vaccinium myrtillus L.) induced by a small elevation in air temperature. Environ. Pollut. 98 91–95. 10.1016/s0269-7491(97)00115-2 [DOI] [PubMed] [Google Scholar]
- Tillett R. L., Wheatley M. D., Tattersall E. A., Schlauch K. A., Cramer G. R., Cushman J. C. (2011). The Vitis vinifera C-repeat binding protein 4 (VvCBF4) transcriptional factor enhances freezing tolerance in wine grape. Plant Biotechnol. J. 10 105–124. 10.1111/j.1467-7652.2011.00648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitasse Y., Basler D. (2013). What role for photoperiod in the bud burst phenology of European beech. J. For. Res. 132 1–8. 10.1007/s10342-012-0661-2 [DOI] [Google Scholar]
- Vitasse Y., Lenz A., Korner C. (2014). The interaction between freezing tolerance and phenology in temperate deciduous trees. Front. Plant Sci. 5:541. 10.3389/fpls.2014.00541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitasse Y., Schneider L., Rixen C., Christen D., Rebetez M. (2018a). Increase in the risk of exposure of forest and fruit trees to spring frosts at higher elevations in Switzerland over the last four decades. Agric. For. Meteorol. 248 60–69. 10.1016/j.agrformet.2017.09.005 [DOI] [Google Scholar]
- Vitasse Y., Signarbieux C., Fu Y. H. (2018b). Global warming leads to more uniform spring phenology across elevations. Proc. Natl. Acad. Sci. U.S.A. 115 1004–1008. 10.1073/pnas.1717342115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand K. M. (1906). The occurrence of ice in plant tissue. Plant World 9 25–39. [Google Scholar]
- Wisniewski M. (1995). “Deep supercooling in woody plants and the role of cell wall structure,” in Biological Ice Nucleation and Its Applications, eds Lee R. E., Warren G. J., Gusta L. V. (Minneapolis, MN: APS Press; ),163–181. [Google Scholar]
- Wisniewski M., Basset C. L., Gusta L. V. (2003). An overview of cold hardiness in woody plants: seeing the forest through the trees. Hortscience 38 952–959. [Google Scholar]
- Wisniewski M., Gusta L. V., Fuller M. P., Karlson D. (2009). “Ice nucleation, propagation, and deep supercooling: the lost tribes of freezing studies,” in Plant Cold Hardiness: From the Laboratory to the Field, eds Gusta L. V., Wisniewski M. E., Tanino K. (Cambridge: CAB International; ), 1–11. [Google Scholar]
- Wisniewski M., Gusta L. V., Neuner G. (2014a). Adaptive mechanisms of freeze avoidance in plants: a brief update. Environ. Exp. Bot. 99 133–140. 10.1016/j.envexpbot.2013.11.011 [DOI] [Google Scholar]
- Wisniewski M., Nassuth A., Teulières C., Marque C., Rowland J., Cao P. B., et al. (2014b). Genomics of cold hardiness in woody plants. Crit. Rev. Plant Sci. 33 92–124. 10.1080/07352689.2014.870408 [DOI] [Google Scholar]
- Wisniewski M., Neuner G., Gusta L. V. (2015a). The use of high-resolution infrared thermography (HRIT) for the study of ice nucleation and propagation in plants. J. Vis. Exp. 99:52703. 10.3791/52703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski M., Norelli J., Artlip T. (2015b). Overexpression of a peach CBF gene in apple: a model for understanding the integration of growth, dormancy, and cold hardiness in woody plants. Front. Plant Sci. 6:85. 10.3389/fpls.2015.00085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski M., Norelli J., Bassett C., Artlip T., Macarisn D. (2011). Ectopic expression of a novel peach (Prunus persica) CBF transcription actor in apple (Malus x domestica) results in short-day induced dormancy and increased cold hardiness. Planta 233 971–983. 10.1007/s00425-011-1358-3 [DOI] [PubMed] [Google Scholar]
- Wu R., Tomes S., Karunairetnam S., Tustin S. D., Hellens R., Allen A., et al. (2017a). SVP-like MADS box genes control dormancy and budbreak in apple. Front. Plant Sci. 8:477. 10.3389/fpls.2017.00477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R., Want T., Warren B., Allan A., Macknight R. C., Varkonyi-Gasic E. (2017b). Kiwifruit SVP2 gene prevents premature budbreak during dormancy. J. Exp. Bot. 68 1071–1082. 10.1093/jxb/erx014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R.-M., Walton E., Richardsons A. G., Wood M., Hellens R. P., Varkonyi-Gasic E. (2011). Conservation and divergence of four kiwifruit SVP-like MADS-box genes suggest distinct roles in kiwifruit bud dormancy and flowering. J. Exp. Bot. 63 797–807. 10.1093/jxb/err304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H., Siddiqua M., Braybrook S., Nassuth A. (2006). Three grape CBF/DREB1 genes are regulated by low temperature, drought and abscisic acid. Plant Cell Environ. 29 1410–1421. 10.1111/j.1365-3040.2006.01524 [DOI] [PubMed] [Google Scholar]
- Xiao H., Tattersall E., Siddiqua M., Cramer G. R., Nassuth A. (2008). CBF4 is a unique member of the CBF transcription factor family of Vitis vinifera and Vitis riparia. Plant Cell Environ. 31 1–10. 10.1111/j.1365-3040.2007.01741 [DOI] [PubMed] [Google Scholar]
- Yang T., Chaudhuri S., Yang L., Du L., Poovaiah B. W. (2010a). A calcium/ calmodulin-regulated member of the receptor-like kinase family confers cold tolerance in plants. J. Biol. Chem. 285 7119–7126. 10.1074/jbc.M109.035659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T., Shad Ali G., Yang L., Du L., Reddy A. S., Poovaiah B. W. (2010b). Calcium/calmodulin-regulated receptor-like kinase CRLK1 interacts with MEKK1 in plants. Plant Signal. Behav. 5 991–994. 10.4161/psb.5.8.12225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yordanov Y. S., Ma C., Strauss S. H., Buscov V. B. (2014). EARLY BUD-BREAK 1 (EBB1) is a regulator of release from seasonal dormancy in poplar trees. Proc. Natl. Acad. Sci. U.S.A. 111 10001–10006. 10.1073/pnas.1405621111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Luedeling E., Xu J. (2010). Winter and spring warming result in delayed spring phenology on the Tibetan Plateau. Proc. Natl. Acad. Sci. U.S.A. 107 22151–22156. 10.1073/pnas.1012490107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Wang P., Si T., Hsu C.-C., Wang L., Zayed O., et al. (2017). MAP kinase cascades regulate the cold response by modulating ICE1 protein stability. Dev. Cell 43 618–629. 10.1016/j.devcel.2017.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Zhang Z., Xie S., Si T., Li Y., Zhu J. K. (2016). Mutational evidence for the critical role of CBF transcription factors in cold acclimation in Arabidopsis. Plant Physiol. 171 2744–2759. 10.1104/pp.16.00533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K., Zhou Y., Li Y., Zhuo X., Ahmad S., Han Y., et al. (2018). Crosstalk of PmCBFs and PmDAMs based on the changes of phytohormones under seasonal cold stress in the stem of Prunus mume. Int. J. Mol. Sci. 19:15. 10.3390/ijms19020015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Chen H., Wei D., Ma H., Lin J. (2017). Arabidopsis CBF3 and DELLAs positively regulate each other in response to low temperature. Sci. Rep. 7:39819. 10.1038/srep39819 [DOI] [PMC free article] [PubMed] [Google Scholar]